Abstract
High mobility group box 1 (HMGB1) is an abundant non-histone nuclear protein that functions as a structural protein of chromatin, regulating genome replication and recombination, mRNA transcription and DNA repair. HMGB1 has been implicated in the tumorigenesis of various cancer types, and the upregulation of HMGB1 has been demonstrated in glioma cells. However, the association between HMGB1 and the mitotic chromosomes in glioma remains uncharacterized. In the present study, the sub-cellular localization of HMGB1 in glioma tissues and cells was investigated. In addition, enhanced green fluorescent protein (EGFP)-tagging of the human HMGB1 protein and chromosome spreading were used to investigate the combination of HMGB1 with mitotic chromosomes. The results of the current study indicated that HMGB1 was localized to the nucleus and the cytoplasm, and it was determined to combine with the condensed chromosomes of proliferating cells in paraformaldehyde (PFA)-fixed glioma tissues. However, HMGB1 was also associated with interphase (but not mitotic chromosomes) when fixed with chilled methanol and 5% (v/v) acetic acid or PFA in vitro. Data from live cell imaging and chromosome spreading indicated the association of HMGB1 with mitotic chromosomes in glioma cells. The present results suggest that HMGB1 combines with mitotic chromosomes in glioma cells, and that the use of fixatives may result in the dissociation of the HMGB1-DNA interaction. Therefore, in live specimens and chromosome spreads, EGFP fusion proteins may represent an accurate indicator for the determination of the correct localization and interaction of HMGB1 in glioma cells.
Keywords: high mobility group box 1, paraformaldehyde, mitotic chromosome, glioma, enhanced green fluorescent protein
Introduction
High mobility group box 1 (HMGB1) is a class of non-histone chromatin protein that is constitutively expressed in the nuclei of both cancerous and normal cells. It serves a regulatory role in numerous cellular processes including DNA repair, gene transcription, the construction and stability of nucleosome structure, cell differentiation, proliferation, migration and apoptosis, signal transduction, and tumour occurrence, growth, metastasis and invasion (1,2). HMGB1 possesses two homologous DNA-binding motifs known as the HMG boxes (A and B) and an acidic C-terminal tail. These motifs are able to recognize and preferentially bind distorted DNA structures, such as four-way junctions (4H) (3) cisplatin-modified DNA (4), hemi-catenated DNA loops (5) and supercoiled DNA (6). The DNA-binding capability of HMGB1 also suggests its involvement in DNA-dependent nuclear processes, including transcription, DNA repair and recombination.
HMGB1 has been indicated to be upregulated in various solid tumour types, and high HMGB1 expression levels may lead to the abnormal expression of certain genes associated with tumorigenesis (7,8). Additionally, high HMGB1 expression inhibits apoptotic signalling, which may promote tumour development (9,10). Furthermore, extracellular HMGB1 promotes cancer cell proliferation, energy metabolism and angiogenesis, as well as the synthesis of inflammatory factors, whilst inhibiting the host anticancer immune response, and contributing to cancer progression (11). Gliomas are the most common malignant brain tumours and are associated with a high mortality rate (12). Several studies have investigated the association between HMGB1 and glioma, and determined that HMGB1 mRNA and protein levels are significantly higher in glioma tissues and glioma cell lines, compared with adjacent normal tissues and immortalized human astrocytes (13,14). Moreover, HMGB1 expression in glioblastoma multiforme (GBM) cell lines can activate the AKT and ERK signalling pathways, promoting GBM-cell invasion via an autocrine pathway (15). HMGB1 has also been reported to function as a structural protein in nuclear chromatin, regulating genome replication and recombination, mRNA transcription and DNA repair (16). Previously, it was revealed that proliferating glioma cells in GBM tissues exhibited an upregulation in HMGB1 expression (17), suggesting that HMGB1 expression may correlate with mitosis in glioma cells.
Paraformaldehyde (PFA) is a chemical fixative widely used in immunohistochemistry (IHC) and immunocytochemistry (ICC), with the ability to preserve cell morphology via the cross-linking of different biomolecules. Nevertheless, PFA fixation has been suggested to affect the interaction between HMGB1 and mitotic chromosomes in both animal and plant cells (18,19). In the present study, the effect of PFA fixation on this interaction was analysed in human glioma cells; the localization of HMGB1 in mitotic cells was investigated using enhanced green fluorescence protein (EGFP)-tagged proteins and chromosome spreading. Chilled methanol, supplemented with 5% (v/v) acetic acid (20), was used as an alternative fixative to further determine the association between HMGB1 and mitotic chromosomes in glioma cells.
Materials and methods
Glioma tissues and cell lines
The GBM tissue microarray (TMA) was obtained from Wuhan Aiwei Biotechnology Co. Ltd., and comprised 60 GBMs in each section. A total of 41 paraffin-embedded glioma tissue sections (5-µm thick; 25 from males and 16 from females; median age: 47.7 years) were obtained between January 2017 and December 2018 from the Department of Histology of the No. 988 Hospital of Joint Logistic Support Force (Zhengzhou, China). All glioma samples were fixed with 4% paraformaldehyde (PFA) for ~24 h at 4°C and evaluated by two experienced pathologists according to the World Health Organisation classification (2016) (21). The Research Ethics Committees of the General Hospital of Chinese People's Liberation Army (Beijing, China) and the No. 988 Hospital of Joint Logistic Support Force and Zhengzhou University (Zhengzhou, China) reviewed and approved the study according to the principles expressed in the Declaration of Helsinki. Informed written consent was obtained from each patient, prior to surgery.
The HA1800 astrocyte cell line was purchased from Shanghai Ji Ning Industrial Co., Ltd., and three human glioma cell lines [U251-MG, U87-MG (glioblastoma of unknown origin) and U118-MG (glioblastoma of unknown origin)] were acquired from China Infrastructure of Cell Line Resources (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences). All four cell lines were tested for mycoplasma contamination and the three glioma cell lines were authenticated by STR profiling. The cells were subsequently cultured in Dulbecco's modified Eagle's medium (DMEM; cat. no. 04-052-1ACS) supplemented with 10% foetal bovine serum (FBS; cat. no. 04-001-1ACS, both Biological Industries) and 1% penicillin-streptomycin, and maintained at 37°C (5% CO2) in a humidified incubator.
IHC
The paraffin-embedded glioma tissue sections were deparaffnized with xylene (100%, twice for 5 min) and rehydrated by gradient ethanol (100%, twice for 5 min; 95%, twice for 5 min; 90% for 5 min; and 80% for 5 min) at room temperature (RT). Subsequently, antigen retrieval was performed using citrate solution (pH 6.0) at 95°C for 15 min. The slides were then treated with methanol containing 3% H2O2 to quench endogenous peroxidase activity, before being washed three times with phosphate buffered saline (PBS) for 5 min each. The sections were incubated in a blocking solution [5% bovine serum albumin (BSA; Beijing Solarbio Science and Technology co. Ltd., cat. no. A8020) plus 0.3% Triton X-100 in PBS] for 1 h at RT, and then incubated with a rabbit anti-human HMGB1 primary antibody (1:1,000; cat. no. ab18256; Abcam) overnight at 4°C. The sections were then rinsed with PBS and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG polyclonal antibody (1:1,000; OriGene Technologies, Inc.; cat. no. ZB-23) for 2 h at RT. Subsequently, 3,3′-diaminobenzidine (DAB) staining was performed; DAB was dropped onto the slides, which were then incubated at 37°C for 3 min. The sections were then counterstained using haematoxylin, washed with distilled water, differentiated using 1% hydrochloric acid (in 70% ethanol) and mounted with neutral resin. For the fluorescence-labelled reaction, the Alexa Fluor 488 donkey anti-rabbit IgG secondary antibody (1:1,000; Abcam; cat. no. ab150073) was used. The nuclei were stained with DAPI (1:5,000; Beijing Solarbio Science & Technology Co. Ltd.; cat. no. C0060) and mounted using 50% (v/v) glycerol with PBS. Images were captured and analysed using a confocal microscope (magnifications, ×40 and ×100; cat. no. U-TBI90; Olympus Corporation).
ICC
The glioma and astrocyte cell lines were cultured over sterile glass coverslips and then fixed for 20 min using: i) 4% PFA at RT; or ii) methanol with 5% (v/v) acetic acid at −20°C. The cells were then rehydrated and permeabilized using 0.5% Triton X-100 (in PBS) for 5 min at RT, prior to being blocked with 3% BSA for 1 h at RT. The cells were then incubated with a HMGB1 primary antibody (1:1,000 diluted in 1% BSA; Abcam; cat. no. ab18256) overnight at 4°C, followed by subsequent incubation with Alexa Fluor 488 donkey anti-rabbit IgG (1:1,000; Abcam; cat. no. ab150073). The cells were then counterstained using DAPI. Coverslips were mounted in 50% (v/v) glycerol with PBS, and observed under an Olympus confocal microscope (magnification, ×100; cat. no. U-TBI90; Olympus Corporation).
Cell transfection
Glioma and astrocyte cells were cultured to 80% confluence in six-well culture plates, and then transfected with 2 µg pEGFP-C1 or pEGFP-HMGB1-C1 using Simple-Fect Transfection Reagent (cat. no. profect-01; Zhengzhou Kebang Biological Technology Co., Ltd.), according to the manufacturer's protocol.
Live cell microscopy
Live cell microscopy was performed 48 h after transfection. Briefly, the transfected cells were incubated for 10 min at 37°C in culture medium containing 0.2 µg/ml Hoechst 33342. They were then washed before the sub-nuclear compartmentalization and mitosis were evaluated under an Olympus confocal microscope (magnification, ×100).
Chromosome spreading
The preparation of chromosome spreads was performed according to pre-described methods (20). U87-MG, U118-MG and HA1800 cells were incubated for 4 h at 37°C separately, in medium (DMEM; cat. no. 04-052-1ACS] supplemented with 10% FBS (cat. no. 04-001-1ACS), both from Biological Industries, and 1% penicillin-streptomycin containing 0.1 µg/ml nocodazole. They were subsequently dislodged by swirling the media, centrifuged at 600 × g for 5 min at 4°C, resuspended in hypotonic buffer [0.25% (w/v) sodium citrate and 37.5 mM KCl] and incubated for 20 min at 37°C. The cells were then pelleted at 900 × g for 10 min. Following resuspension in 0.5 ml hypotonic buffer, 5 ml ME buffer (methanol and glacial acetic acid, 4:1) was added and gently mixed. The cells were then re-pelleted and resuspended in fresh ME buffer (methanol and glacial acetic acid, 4:1). After 1 h incubation at RT, the cells were centrifuged at 900 × g for 10 min, spread onto the pre-cooled (−20°C) and dry coverslips (cat. no. EF.188105; Jiangsu Shitai Experimental Equipment Co., Ltd.) and then air-dried. The chromosome spreads were then incubated in blocking buffer and subjected to immunodetection using an anti-HMGB1 antibody, according to the methodology described above.
Western blotting
Cells were lysed using radioimmunoprecipitation assay buffer and the protein concentration was determined using the BCA Protein Assay kit (Beijing Solarbio Science & Technology Co., Ltd., cat. no. PC0020). A total of 20 µg from each sample was separated via SDS-PAGE on a 10% gel. The separated proteins were then transferred onto PVDF membranes (EMD Millipore; cat. no. IPVH00010), which were blocked for 1 h at RT in 5% non-fat dry milk [in Tris-buffered saline wash buffer and Tween 20 (TBST)]. The membranes were then incubated at 4°C overnight with the following primary antibodies: anti-HMGB1 (Abcam; cat. no. ab18256; 1:2,000) and anti-β-actin (OriGene Technologies, Inc.; cat. no. TA-09; 1:2,000). Following primary incubation, the membranes were incubated at RT for 2 h with horseradish peroxidase-conjugated goat anti-rabbit/mouse IgG polyclonal secondary antibodies (ZSGB-Bio; cat. no. ZB-23; 1:5,000). Enhanced chemiluminescent substrate (Dalian Meilun Biology Technology Co., Ltd.; cat no. MA0186-Sep-25D) was used to conduct autoradiography and visualise the protein bands. The signal intensity of each band was determined using Image J software (National Institutes of Health; version 1.4.3.67).
Statistical analysis
Data are presented as the mean ± standard deviation (SD). Statistical analyses were performed using one-way ANOVA and Tukey's multiple comparison test with GraphPad Prism 5 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
HMGB1 combines with condensed chromosomes in the proliferating cells of PFA-fixed glioma tissues
The cellular localization of HMGB1 in PFA-fixed glioma tissues was examined, and HMGB1 was determined to localize to both the nuclei (Fig. 1A-a) and cytoplasm (Fig. 1A-b) of glioma tissue cells. Within the nuclei in interphase (asterisks, Fig. 1B), HMGB1 was primarily aggregated in close proximity to the chromatin blocks, with additional, diffuse expression throughout these blocks. The HMGB1 expression pattern in the TMA sections was then examined, indicating diffuse HMGB1 expression in the cytoplasm of some mitotic cells. However, DAB staining could not determine whether HMGB1 was combined with the condensed chromosomes (Fig. 1C). To confirm localization, a co-staining IHC assay was performed for HMGB1 and the proliferation marker Ki67. Consequently, HMGB1 was discovered to combine with condensed chromosomes in Ki67-positive proliferating glioma cells (Fig. 1D). Collectively, these results indicated that HMGB1 combined with the chromosomes of proliferating cells of PFA-fixed glioma tissues.
Figure 1.
Immunostaining of HMGB1 in glioma tissues. (A) Localization of HMGB1 in the nuclei (a) and cytoplasm (b). Scale bar=25 µm. (B) Localization of HMGB1 in the nuclei. Asterisks signify chromatin blocks. Scale bar=5 µm. (C) HMGB1 localization in the mitotic cells of TMA sections. Scale bar=5 µm. (D) Double-labelling of HMGB1 and Ki67. Scale bar=10 µm. DAPI was used to stain the nuclei. HMGB1, high mobility group box 1; TMA, tissue microarray.
HMGB1 dissociates from mitotic chromosomes in fixative-treated glioma cells
Ki67 is preferentially expressed during the late G1, S, G2 and M phases of the cell cycle (22). Therefore, Ki67-positive cells may include not only mitotic cells, but also those in interphase. To investigate the relationship between HMGB1 and mitotic chromosomes in vitro, the intracellular localization of HMGB1 was determined in glioma cells and HA1800 astrocytes. HMGB1 protein expression levels were elucidated using western blotting, and no significant difference was observed between HMGB1 expression in glioma cells and HA1800 astrocytes (Fig. S1).
The localization of HMGB1 in HA1800 astrocytes and three glioma cell lines (all fixed with PFA) was then investigated. The results revealed that HMGB1 was detected in the nuclei, with diffuse expression in the nucleoli (Fig. 2A). Notably, the majority of HMGB1 was found to be dissociated from the mitotic chromosomes, with certain cells showing only a minor fraction of HMGB1 expression with condensed chromatin in the nuclei, at each phase of mitosis (Fig. 2A). Considering the potential limitations of PFA, which have been highlighted in previous literature (18–20), an alternative fixation procedure (chilled methanol with 5% (v/v) acetic acid) (20) was conducted. However, the staining results were similar to those obtained with PFA fixation, implying that this was also an unsuitable method of fixing glioma cells (Fig. 2B). In summary, the results of the current study demonstrated that HMGB1 was dissociated from mitotic chromosomes in fixative-treated glioma cells and astrocytes.
Figure 2.
Comparative analysis of cell fixation procedures for determining the optimum conditions for immunodetection of mitotic chromatin-associated HMGB1. The astrocyte HA1800 cell line and three glioma cell lines (U251, U118 and U87) were used for the immunodetection of HMGB1 during mitosis. (A) HMGB1 localization in interphase and mitotic nuclei in PFA-fixed astrocytes and glioma cells. Scale bar=10 µm. (B) HMGB1 localization in interphase and mitotic nuclei in astrocytes and glioma cells fixed with chilled methanol with 5% (v/v) acetic acid. Scale bar=10 µm. DAPI was used to stain the nuclei. HMGB1, high mobility group box 1; PFA, paraformaldehyde; Ip, interphase; Pp, prophase; Mp, metaphase; Ap, anaphase; Tp, telophase.
HMGB1 combines with mitotic chromosomes in both live glioma cells and mitotic chromosome spreads
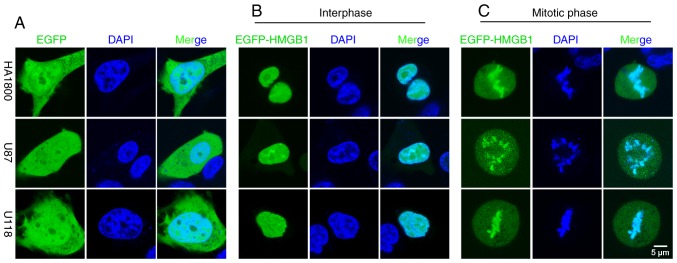
To avoid artefacts that may have been introduced by the fixation procedures, an EGFP-tagged human HMGB1 (EGFP-HMGB1) plasmid was constructed and transfected into the glioma (U87-MG and U118-MG) and astrocyte (HA1800) cell lines; EGFP localization was subsequently determined. In the live cells, EGFP localized to both the cytoplasm and nuclei during interphase (Fig. 3A). In EGFP-HMGB1 transfected cells, there was no significant difference between the localization of HMGB1 between fixed and unfixed cells in interphase. In both cases, HMGB1 localized to the nuclei and diffused into the nucleoli (Figs. 2A and 3B). The present results were consistent with previously reported studies (16). However, there was a discrepancy in the results between fixed and unfixed mitotic cells; diffuse expression was detected in the cytoplasm, but compacted EGFP expression was detected along the metaphase plate (Fig. 3C), indicating an association between HMGB1 and the mitotic chromosomes in glioma cells and astrocytes.
Figure 3.
Combination of EGFP-HMGB1 with mitotic chromosomes without paraformaldehyde fixation. (A) Live interphase cells expressing free EGFP. (B) Live interphase cells expressing EGFP-HMGB1. (C) Live mitotic cells expressing EGFP-HMGB1. Scale bar=5 µm. Hoechst was used to stain the nuclei. HMGB1, high mobility group box 1; EGFP, enhanced green fluorescent protein.
Chromosomal details can be resolved more accurately using chromosome spreading, compared with a whole-cell visualisation techniques; thus, chromosome spreads were prepared from U87-MG and U118-MG glioma cells treated with nocodazole to initiate cell cycle arrest. HMGB1 was found to be significantly associated with mitotic chromosomes, and distributed non-uniformly along the condensed chromatin (Fig. 4). However, multiple HMGB1-positive areas were detected in close proximity to the mitotic chromosome. This may be attributable to the use of methanol/glacial acetic acid during fixation (for the preparation of the chromosome spreads). Although the reason for the uneven distribution of HMGB1 in the mitotic chromosomes requires further investigation, the results using live cells and mitotic chromosome spreading techniques revealed that HMGB1 combines with mitotic chromatin in all of the cell lines analysed.
Figure 4.

Distribution of immunodetected HMGB1 on chromosome spreads. HMGB1 is unevenly co-localized with the mitotic chromosomes in both U87 and U118 glioma cells. Scale bar=10 µm. DAPI was used to stain the nuclei. HMGB1, high mobility group box 1.
Discussion
HMGB1 is a non-histone chromatin-binding protein, which has been proven to regulate the tumorigenesis and progression of multiple cancer types, including mesothelioma, gastric and colorectal cancer and hepatocellular carcinoma (23,24). The results of the present study, and those of a study conducted by Wang et al (25), indicated that HMGB1 expression was upregulated in glioma tissues. HMGB1 is typically expressed in the nucleus of normal cells. However, in tumour cells it may be localized to the nucleus, cytoplasm or extracellular space, regulating gene transcription and the autophagic and inflammatory pathways associated with tumour cell proliferation (25,26). Consequently, the detection of both nuclear and cytoplasmic HMGB1 in the glioma tissues used in the present study was unsurprising.
In interphase nuclei, HMGB1 exhibits a differential distribution pattern between cells from glioma tissues and cultured glioma cells; HMGB1 accumulated in the vicinity of, or distributed diffusely on the chromatin blocks in cells from glioma tissues. Whereas in cultured glioma cells, the distribution of HMGB1 almost entirely overlapped with DAPI or Hoechst staining, confirming that the protein is distributed throughout the entire nucleus in glioma cells, in vitro. It is speculated that this discrepancy is due to the different distribution patterns of chromatin in glioma tissues, compared with those in cultured glioma cells. For example, in glioma tissues, chromatin is unevenly distributed and packaged in the interphase nuclei. Conversely, in the nuclei of glioma cells in vitro, it is more evenly distributed. The diffuse HMGB1 expression observed in the chromatin blocks may represent an artefact of PFA fixation, in accordance with the results of the present study, which indicated that treatment with PFA may lead to the dissociation of HMGB1 from compacted chromatin. The dissociation of HMGB1 from the compacted chromosomes suggests a weak affinity of HMGB1 for chromatin, such that the transition from the bound to unbound state is rapid and transient. This status-change feature of HMGB1 may result in the ability of glioma cells to adapt rapidly to various environmental stressors. It has been demonstrated that the acetylation of HMGB1 weakens its binding affinity to DNA, promoting its migration to various tissues and resulting in the secretion of proinflammatory cytokines (27).
In the present study, although the majority of HMGB1-positive nuclei were at the interphase stage in glioma tissues, a proportion of proliferating cells also expressed HMGB1. Moreover, HMGB1 was localized in the condensed chromosomes of certain Ki67-positive proliferating cells in PFA-fixed glioma tissue sections. However, this result was not consistent in PFA-fixed glioma cells in vitro.
PFA is a frequently-used fixative in microscopic studies. However, it has been discovered to create artefacts due to the disruption of cellular structures and lowered protein antigenicity (28). In fact, certain studies have suggested that PFA fixation may affect the interaction between mammalian HMGB proteins and mitotic chromosomes (18,19). Kumar et al (20) proposed that chilled methanol (−20°C) with 5% (v/v) acetic acid was a suitable alternative fixative for mitotic chromatin. Therefore, this fixative was applied to re-investigate the binding of HMGB1 to the mitotic chromosomes in glioma cells. Counterintuitively, HMGB1 failed to bind the mitotic chromosomes. This may be because this fixation method was also unsuitable for the observation of glioma cells; it was thus hypothesized that that live-cell imaging of fluorescently-tagged proteins may represent an improved method for the observation of HMGB1-chromatin interactions, as it would bypass any potential artefacts caused by the fixation process (18,29). Therefore, EGFP-tagged hHMGB1 plasmids were transfected into live astrocyte and glioma cells, and binding of HMGB1 to the mitotic chromosomes was observed. Moreover, a chromosomal spread assay confirmed the binding of HMGB1 to the mitotic chromosomes. Thus, the results of the present study suggest that HMGB1 is a component of the mitotic chromosome, and that the use of fixatives may disrupt its affinity for mitotic chromosomes in glioma cells.
In the present study, it was observed that HMGB1 was bound to the condensed chromosomes of proliferating glioma cells in vivo fixed with PFA, and it is hypothesized that this result was due to the possible manipulation of cells by fixation. HMGB1 protein in cultured cells may be more accessible to manipulation by fixatives, compared with those in vivo. Moreover, Ki67 is present during all active phases of the cell cycle (G1, S, G2 and M) (22); therefore, the HMGB1 and Ki67 double-positively stained cells may not have been in the mitotic stage of the cell cycle. Both PFA and chilled methanol with 5% acetic acid can create artefacts during fixative cross-linking, which may explain the dissociation of HMGB1 from condensed mitotic chromosomes in glioma cells. Thus, novel fixation procedures are required to accurately determine the association between HMGB1 and mitotic chromosomes. Applying high pressure freezing or embedding cells in resin may represent improved methods for preserving cellular structure (30,31). Moreover, chromosome spreading is a more accurate method of resolving chromosomal details (20). Thus, this method was used in the present study to observe the uneven distribution of HMGB1 on mitotic chromosomes. However, only the combination of HMGB1 with mitotic chromosomes in cultured glioma cells was elucidated. The histological effect on mitotic chromosomes in glioma tissues should be investigated in future studies.
Chromosomal spreading is an alternative technique used to assess protein binding to mitotic chromosomes in the tissues. However, it is first necessary to sort glioma cells from glioma tissues via flow cytometry, after which the subsequent procedures are similar to those used for cultured cells. Notably, the treatment of chromosomes with fixatives (methanol/glacial acetic acid) is a routine step during chromosomal spread assays. This treatment may lead to the dissociation of protein from the mitotic chromosomes, as demonstrated in the present study. Hence, the identification of novel fixation procedures would be beneficial in the preparation of chromosomal spread specimens.
During mitosis, transcription is halted, chromatin condenses and the majority of basal transcription factors (TFs) are reportedly excluded from the chromosomes. However, a class of TFs was discovered that are continuously bound to mitotic chromosomes, suggesting a potential mechanism for the maintenance of certain transcriptional programmes throughout the cell cycle, termed mitotic bookmarking (32). Accordingly, HMGB1 (a nuclear protein involved in the transcription-level regulation of various genes) may serve as a mitotic bookmarker for daughter glioma cells, enabling the re-establishment of the original transcription programme in glioma cells. Nevertheless, the combination of HMGB1 with mitotic chromosomes in astrocytes was also detected. Therefore, the discrepancy of mitotic chromosomal binding to HMGB1 between normal glial and glioma cells should be investigated in future studies, using techniques such as chromatin immunoprecipitation. In addition, determination of the role of HMGB1 binding to enable glioma cells to ‘remember’ their identity would increase our understanding of glioma development and progression.
In the present study, HMGB1 was typically expressed in the nuclei during interphase, whilst it dissociated from the condensed chromosomes during the mitotic phase of all four cell lines, following PFA treatment. It was observed that, although the majority of HMGB1 had dispersed into the cytoplasm, a minor fraction of HMGB1 remained in the nucleus, where it combined with the condensed chromosomes of all four cell types (Fig. 2A). It is possible that PFA did not comprehensively disrupt the interaction of HMGB1 with all of the condensed chromosomes. However, the difference in the binding affinity of HMGB1 to mitotic chromosomes, between normal astrocytes and glioma cell lines, is yet to be elucidated. Moreover, no significant difference was observed between the total expression level of HMGB1 in the four cell lines (Fig. S1). This was inconsistent with the result in glioma tissues, in which HMGB1 expression was upregulated (17). The presence of alternative stimulating factors in vivo may provide a possible explanation for this difference.
The present study revealed that HMGB1 was constitutively expressed in the nuclei of four cell lines under non-stimulating conditions, which differed from the diffuse expression (in the nuclei, cytoplasm and extracellular space) observed in glioma tissues (17). It has been revealed that glioma cells secrete numerous chemokines, cytokines and growth factors that promote the infiltration of non-neoplastic cells, creating a specific tumor microenvironment that influences the biological properties of glioma cells (33). As a highly conserved nuclear protein, HMGB1 is a chromatin-binding factor that is able to alter DNA structure and promote access to transcriptional protein assemblies on specific DNA targets (1,34,35). Therefore, the difference in HMGB1 function between the nuclei of normal astrocytes and glioma cells should be investigated in future studies.
In conclusion, the results of the present study suggest that HMGB1 combines with mitotic chromosomes in glioma cells. However, the use of fixatives leads to the dissociation of HMGB1 from mitotic chromosomes. Additionally, EGFP-tagged HMGB1 proteins in live glioma cells imitated the localization of endogenous HMGB1 protein at different mitotic stages. Chromosome spreading is a technique that may also be applied to investigate the combination of HMGB1 with mitotic chromosomes. A proportion of studies on glioma have used fixatives to treat tissues or cells. Considering the artefacts induced by fixatives, the biological function of HMGB1, especially with regard to its sub-cellular localization, should be carefully reconsidered.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81402455) and the Key Scientific Research Projects of Higher Education Institutions in Henan Province (grant no. 20A310020). The funding sources had no influence on the study design or the collection, analysis and interpretation of data, or manuscript writing.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
Study concept and design was conceived by LJ, YH and AY. Acquisition of data was performed by LJ, HS, YS, WF, GW and XL. LJ, HS and YS analysed and interpreted the data. LJ, HS and YS conducted the statistical analyses. Writing of the manuscript was conducted by AY, LJ and YH. All authors approved the final version of the manuscript.
Ethics approval and consent to participate
The Research Ethics Committees of the General Hospital of Chinese People's Liberation Army (Beijing, China), No. 988 Hospital of Joint Logistic Support Force; and Zhengzhou University (Zhengzhou, Henan Province, China) reviewed and approved the study according to the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from each participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tang D, Kang R, Zeh HJ, 3rd, Lotze MT. High-mobility group box 1 and cancer. Biochim Biophys Acta. 2010;1799:131–140. doi: 10.1016/j.bbagrm.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belgrano FS, de Abreu da Silva IC, Bastos de Oliveira FM, Fantappie MR, Mohana-Borges R. Role of the acidic tail of high mobility group protein B1 (HMGB1) in protein stability and DNA bending. PLoS One. 2013;8:e79572. doi: 10.1371/journal.pone.0079572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi ME, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 4.Park S, Lippard SJ. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry. 2011;50:2567–2574. doi: 10.1021/bi2000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaouen S, de Koning L, Gaillard C, Muselikova-Polanska E, Stros M, Strauss F. Determinants of specific binding of HMGB1 protein to hemicatenated DNA loops. J Mol Biol. 2005;353:822–837. doi: 10.1016/j.jmb.2005.08.073. [DOI] [PubMed] [Google Scholar]
- 6.Ugrinova I, Pashev IG, Pasheva EA. Post-synthetic acetylation of HMGB1 protein modulates its interactions with supercoiled DNA. Mol Biol Rep. 2009;36:1399–1404. doi: 10.1007/s11033-008-9327-z. [DOI] [PubMed] [Google Scholar]
- 7.Völp K, Brezniceanu ML, Bösser S, Brabletz T, Kirchner T, Göttel D, Joos S, Zörnig M. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut. 2006;55:234–242. doi: 10.1136/gut.2004.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi YR, Kim H, Kang HJ, Kim NG, Kim JJ, Park KS, Paik YK, Kim HO, Kim H. Overexpression of high mobility group box 1 in gastrointestinal stromal tumors with KIT mutation. Cancer Res. 2003;63:2188–2193. [PubMed] [Google Scholar]
- 9.Wagner KU, Schmidt JW. The two faces of Janus kinases and their respective STATs in mammary gland development and cancer. J Carcinog. 2011;10:32. doi: 10.4103/1477-3163.90677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guerriero JL, Ditsworth D, Catanzaro JM, Sabino G, Furie MB, Kew RR, Crawford HC, Zong WX. DNA alkylating therapy induces tumor regression through an HMGB1-mediated activation of innate immunity. J Immunol. 2011;186:3517–3526. doi: 10.4049/jimmunol.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang R, Zhang Q, Zeh HJ, 3rd, Lotze MT, Tang D. HMGB1 in cancer: Good, bad, or both? Clin Cancer Res. 2013;19:4046–4057. doi: 10.1158/1078-0432.CCR-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidu RA, Wu M, Su Z, Xu H. Paradoxical role of high mobility group Box 1 in glioma: A suppressor or a promoter? Oncol Rev. 2017;11:325. doi: 10.4081/oncol.2017.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Liu C, Hou R. Knockdown of HMGB1 improves apoptosis and suppresses proliferation and invasion of glioma cells. Chin J Cancer Res. 2014;26:658–668. doi: 10.3978/j.issn.1000-9604.2014.12.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta P, Ghosh S, Nagarajan A, Mehta VS, Sen E. β-defensin-3 negatively regulates TLR4-HMGB1 axis mediated HLA-G expression in IL-1β treated glioma cells. Cell Signal. 2013;25:682–689. doi: 10.1016/j.cellsig.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Cheng P, Ma Y, Gao Z, Duan L. High mobility group box 1 (HMGB1) predicts invasion and poor prognosis of glioblastoma multiforme via activating AKT signaling in an autocrine pathway. Med Sci Monit. 2018;24:8916–8924. doi: 10.12659/MSM.912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, et al. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell. 2007;27:829–841. doi: 10.1016/j.molcel.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia L, Song Y, Song H, Wang G, Fan W, Li X, Zheng H, Yao A. Overexpression of high mobility group box 1 (HMGB1) has no correlation with the prognosis in glioma. Biomark Med. 2019;13:851–863. doi: 10.2217/bmm-2019-0031. [DOI] [PubMed] [Google Scholar]
- 18.Li MW, Zhou L, Lam HM. Paraformaldehyde fixation may lead to misinterpretation of the subcellular localization of plant high mobility group box proteins. PLoS One. 2015;10:e135033. doi: 10.1371/journal.pone.0135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, Marechal V. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol Biol Cell. 2003;14:3414–3426. doi: 10.1091/mbc.e02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S, Chaturvedi NK, Kumar S, Tyagi RK. Agonist-mediated docking of androgen receptor onto the mitotic chromatin platform discriminates intrinsic mode of action of prostate cancer drugs. Biochim Biophys Acta. 2008;1783:59–73. doi: 10.1016/j.bbamcr.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 22.Bruno S, Darzynkiewicz Z. Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Prolif. 1992;25:31–40. doi: 10.1111/j.1365-2184.1992.tb01435.x. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Jiang Z, Yan J, Ying S. HMGB1 as a potential biomarker and therapeutic target for malignant mesothelioma. Dis Markers. 2019;2019:4183157. doi: 10.1155/2019/4183157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu T, Zhang W, Yang G, Li H, Chen Q, Song R, Zhao L. HMGB1 overexpression as a prognostic factor for survival in cancer: A meta-analysis and systematic review. Oncotarget. 2016;7:50417–50427. doi: 10.18632/oncotarget.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang XJ, Zhou SL, Fu XD, Zhang YY, Liang B, Shou JX, Wang JY, Ma J. Clinical and prognostic significance of high-mobility group box-1 in human gliomas. Exp Ther Med. 2015;9:513–518. doi: 10.3892/etm.2014.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He SJ, Cheng J, Feng X, Yu Y, Tian L, Huang Q. The dual role and therapeutic potential of high-mobility group box 1 in cancer. Oncotarget. 2017;8:64534–64550. doi: 10.18632/oncotarget.17885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo ZS, Liu Z, Bartlett DL, Tang D, Lotze MT. Life after death: Targeting high mobility group box 1 in emergent cancer therapies. Am J Cancer Res. 2013;3:1–20. [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson SM, Bacic A. Preparation of plant cells for transmission electron microscopy to optimize immunogold labeling of carbohydrate and protein epitopes. Nat Protoc. 2012;7:1716–1727. doi: 10.1038/nprot.2012.096. [DOI] [PubMed] [Google Scholar]
- 29.Raccaud M, Friman ET, Alber AB, Agarwal H, Deluz C, Kuhn T, Gebhardt JCM, Suter DM. Mitotic chromosome binding predicts transcription factor properties in interphase. Nat Commun. 2019;10:487. doi: 10.1038/s41467-019-08417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald KL, Auer M. High-pressure freezing, cellular tomography, and structural cell biology. Biotechniques. 2006;41:137–141. doi: 10.2144/000112226. [DOI] [PubMed] [Google Scholar]
- 31.Xiong H, Zhou Z, Zhu M, Lv X, Li A, Li S, Li L, Yang T, Wang S, Yang Z, et al. Chemical reactivation of quenched fluorescent protein molecules enables resin-embedded fluorescence microimaging. Nat Commun. 2014;5:3992. doi: 10.1038/ncomms4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teves SS, An L, Hansen AS, Xie L, Darzacq X, Tjian R. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016;5(pii):e22280. doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97:498–518. doi: 10.1038/labinvest.2017.19. [DOI] [PubMed] [Google Scholar]
- 34.Lotze MT, Tracey KJ. Tracey High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 35.Müller S, Scaffidi P, Degryse B, Bonaldi T, Ronfani L, Agresti A, Beltrame M, Bianchi ME. New EMBO members' review: The double life of HMGB1 chromatin protein: architectural factor and extracellular signal. EMBO J. 2001;20:4337–4340. doi: 10.1093/emboj/20.16.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.