CRISPR-Cas (CRISPR-associated) systems confer adaptive immunity in prokaryotes to infectious agents. CRISPR-Cas activity is mediated by RNA-guided effector proteins capable of surveilling and silencing foreign nucleic acid, such as by cleaving the genome of an infecting phage.1 Due to their programmable nature and robust endonuclease activity, Class 2 single-effector Cas proteins, such as Cas9, are now also revolutionizing our ability to edit genomes and, more broadly, localize any desired activity to the genome of a cell.2 Realization of the broad utility of Cas proteins has thus set off a search to find Cas variants with enhanced functions – such as higher activity, potential for therapeutic delivery, nucleic acid detection, etc. – and to understand how these highly functionalized effectors systems arose from the evolutionary arms race between prokaryotic host and pathogen.
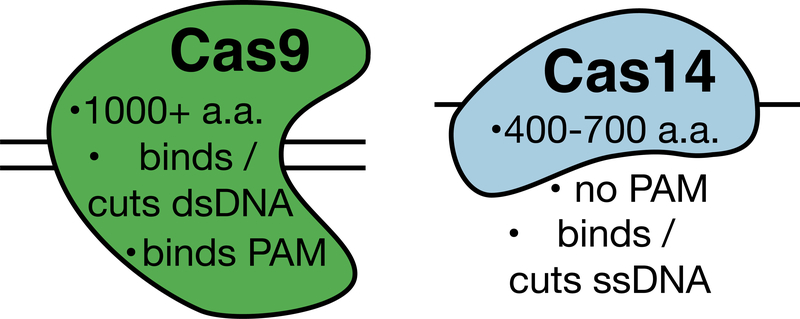
Phylogenetic analysis suggests DNA-binding Class 2 Cas proteins evolved from an ancestral gene that possessed the signature of a RuvC-like endonuclease domain3. In a recent paper, Doudna, Banfield and colleagues exploit this logic to identify a simpler, smaller Cas protein, that represents a snapshot of the Cas evolutionary process and, surprisingly, offers several biotechnological advantages relative to Cas proteins characterized so far. 4 Briefly, by sifting through terabase scale sequencing datasets for genes possessing a RuvC-like domain and nearby to a CRISPR locus, the researchers identified a set of highly novel, putative single effector Cas proteins. Termed Cas14s, these variants are reminiscent of other Type V Cas proteins, including the well-characterized Cas12a protein and recently discovered CasX variant, but are notable for their extremely small size, being roughly ~400–700, rather than the typical 1000+ amino acids of Class 2 variants (Figure).
Figure.
Schematic of properties of newly discovered Cas14 relative to the well-characterized Cas9.
Characterization of Cas14 also revealed several unique biochemical activities. Class 2 Cas effectors possess a strict so-called protospacer adjacent motif (PAM) DNA binding requirement. Unlike RNA-guided binding to complementarity target DNA, PAM binding is hardwired into the Cas protein. Binding to a DNA locus thus requires both complementarity between guide RNA and DNA target and the proper PAM sequence adjacent to the target DNA site at a location that it can interact with the PAM-binding elements of the Cas protein. PAM binding therefore provides a mechanism for ensuring a genomically-encoded CRISPR-Cas system does not target itself. The guide RNA gene locus necessarily does not possess a PAM sequence and cannot be recognized by an active effector. Surprisingly – given that PAM binding is an assumed activity of Cas effectors - the researchers could detect no PAM-binding requirement for Cas14.
This result raised a mechanistic question – if there is no PAM binding requirement, what is the true substrate for Cas14? Here, too, the researchers found a surprising result when activity assays revealed that Cas14 cleaved only ssDNA, and not dsDNA or ssRNA. Cas proteins can often tolerate mismatches in target binding but PAM binding is an absolute requirement, so this result raised another mechanistic question. Is Cas14 sensitive to mismatches in a location dependent fashion? The researchers tiled two nucleotide mismatches across various ssDNA substrates to find that, yes, Cas14 is sensitive and in a unique way. Unlike other Cas proteins - and for reasons that remain unknown - Cas14 is highly susceptible to mismatches in a roughly six basepair stretch in the middle of its target region.
These properties make Cas14 particularly unique for nucleic acid detection. Recent work has shown that many Cas proteins are capable of cleaving an oligonucleotide in trans after binding a complementary target nucleic acid. Addition of a fluorophore and quencher pair to the oligonucleotide can act as a fluorescent reporter of target binding and therefore as a detector of nucleic acid sequence without the need for amplification. Doudna and colleagues found that Cas14 also possesses trans cleavage activity. Moreover, since Cas14 doesn’t require a PAM, yet is still highly sensitive to mismatches in the middle of its target region, Cas14 is uniquely suited for sequence detection. Specifically, the researchers showed that Cas14 is robustly capable of distinguishing single nucleotide polymorphisms, such as variants of human HERC2 responsible for eye color.
Thus, the molecular mechanisms inherent to Cas14 are useful for biotechnological applications and raise a number of important questions for further inquiry. The discovery of Cas14 lends empirical credence to the hypothesis that Class 2 effectors arose through a RuvC-like ancestor. Moreover, as supported by a recent discovery of other Type V effectors, an evolutionary view comprising RuvC may be a productive route to the discovery of even more variants.5 It does not, though, tell us how these effectors gain their other functionalities. Indeed, the remaining activities of guide RNA binding, target recognition, and PAM binding occur from domains that are not conserved. These have likely evolved through the accretion and harmonization of a variety of domains in a molecule-specific fashion. The presence of such diverse extant sequences that share similar functions (e.g. Cas9 and Cas12a), suggests this is a problem that can be answered through a variety of convergent means. Looking towards the future, it also implies that the evolutionary insights provided by Cas14 and related proteins, along with the proper molecular biological tools, will enable biochemists to tap into the inherently modularity of Cas proteins in order to refine and maximize their biotechnological utility.
References
- (1).Bhaya D, Davison M, and Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. [DOI] [PubMed] [Google Scholar]
- (2).Doudna JA, and Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- (3).Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, and Koonin EV (2017) Diversity and evolution of class 2 CRISPR–Cas systems Nature Publishing Group; 15, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, Cofsky JC, Kyrpides NC, Banfield JF, and Doudna JA (2018) Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, Garrity AJ, Chong S, Makarova KS, Koonin EV, Cheng DR, and Scott DA (2019) Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]