Abstract
Background
Analysis of circulating tumor nucleic acids in plasma of Non-Small Cell Lung Cancer (NSCLC) patients is the most widespread and documented form of "liquid biopsy" and provides real-time information on the molecular profile of the tumor without an invasive tissue biopsy.
Methods
Liquid biopsy analysis was requested by the referral physician in 121 NSCLC patients at diagnosis and was performed using a sensitive Next Generation Sequencing assay. Additionally, a comparative analysis of NSCLC patients at relapse following EGFR Tyrosine Kinase Inhibitor (TKIs) treatment was performed in 50 patients by both the cobas and NGS platforms.
Results
At least one mutation was identified in almost 49% of the cases by the NGS approach in NSCLC patients analyzed at diagnosis. In 36 cases with paired tissue available a high concordance of 86.11% was observed for clinically relevant mutations, with a Positive Predictive Value (PPV) of 88.89%. Furthermore, a concordance rate of 82% between cobas and the NGS approach for the EGFR sensitizing mutations (in exons 18, 19, 21) was observed in patients with acquired resistance to EGFR TKIs, while this concordance was 94% for the p.T790M mutation, with NGS being able to detect this mutation in three 3 additional patients.
Conclusions
This study indicates the feasibility of circulating tumor nucleic acids (ctNA) analysis as a tumor biopsy surrogate in clinical practice for NSCLC personalized treatment decision making. The use of new sensitive NGS techniques can reliably detect tumor-derived mutations in liquid biopsy and provide clinically relevant information both before and after targeted treatment in patients with NSCLC. Thus, it could aid physicians in treatment decision making in clinical practice.
Introduction
Non-Small Cell Lung Cancer (NSCLC) is one of the most common and aggressive tumor types, nevertheless, it is also the tumor type with the majority of approved targeted agents available. [1, 2]. Analysis of tumor molecular profile can provide predictive information to guide treatment in these patients. Due to frequent low availability of tissue, simultaneous analysis of targetable mutations using multigene tests seems to be the most appropriate approach, saving both time and tissue availability. Next-generation sequencing (NGS) analysis is a robust technology that has been widely used for the detection of aberrations in genes that can be used as biomarkers of response to treatment, such as EGFR, ALK, ROS1, BRAF, NTRK1,2 & 3, ERBB2, RET and MET [3, 4].
For decades the only available material for molecular profiling was considered the patient’s Formalin Fixed Paraffin Embedded (FFPE) tumor tissue. The FFPE material has several advantages since it is a widely available material, easy to use and store [5, 6]. In addition, it provides the possibility of selecting the most appropriate cancer tissue either by microdissection or by macrodissection, increasing the sensitivity of somatic mutation detection assays [7, 8]. However, FFPE material also has several disadvantages, such as its unavailability in cases of inoperable tumors and its inability in some cases to capture the tumor’s heterogeneity [9]. Moreover, the genetic material obtained due to paraffin processing of tissue, is sometimes of poor quality and not adequate for molecular analysis [5, 10]. Most importantly, the tumor’s molecular profile is altered, mainly following targeted therapy and those alterations cannot be detected by analyzing the primary tumor material [11–13] but require invasive tissue re-biopsies.
The presence of neoplastic characteristics in the plasma DNA of cancer patients was first reported back in 1989 [14]. In the following years, several studies have shown that the analysis of cell-free tumor-derived nucleic acids in cancer patient’s body fluids (plasma, serum, Bronchoalveolar lavage, urine, stool, etc.) can be used to detect tumor specific alterations [15]. The term Liquid Biopsy has emerged indicating the use of these non- or minimally invasive materials for tumor characterization. The mutation status detected in a liquid biopsy reflects the status present in the patient’s tumor. Additionally, circulating tumor nucleic acids (ctNA) analysis could eventually detect more somatic alterations compared to the analysis of a specific area in a FFPE tissue, since it originates from the whole tumor’s area and/or metastasis present in the patient’s body, thus being more representative of intra and inter-tumor heterogeneity [16–18].
The use of plasma samples for ctNA analysis has nowadays become feasible due to the development of sensitive molecular techniques that can detect with high accuracy minimal amounts of ctNAs that are present in this material. For this purpose a variety of methods have been used, including digital PCR, Real-time PCR, Arms PCR, BEAMing technology and Next Generation Sequencing (NGS) [19–21].
Currently, the only FDA (Food and Drug Administration) approved test for EGFR mutation detection in plasma is the cobas® EGFR Mutation Test v2. Even though this approach meets the prerequisites for reliable detection of tumor-specific mutations in plasma, it has the disadvantage of being a single gene test analyzing specific mutations [20, 22–24]. On the other hand, NGS is the only methodology that can be used for the simultaneous analysis of multiple genes or even the entire cancer genome. This is of great importance in the era of individualized treatment, where the number of genes and gene alterations, targeted by drugs is increasing continuously, especially in tumors with many targetable gene alterations present such as NSCLC. Thus, NGS can be used to provide a broad molecular profile either in tissue or in plasma. Since it is the only method that provides a comprehensive analysis of many biomarkers simultaneously, it is becoming a tool of great clinical utility. The importance of such an approach is more prominent for tumors such as NSCLC, with limited tissue available but with an abundance of biomarkers that could guide treatment decisions. The simultaneous analysis of all tumor variations can be cost and time saving without the waste of valuable tissue material. Thus, liquid biopsy analysis can be used for tumor monitoring and for early detection of molecular relapse. Furthermore, the presence of resistance mutations due to treatment can be identified and eventually lead to the modification of the treatment plan in these patients [3, 25].
The most prevalent resistance mutation in this setting is the T790M in exon 20 of the EGFR gene, which can be targeted by third-generation TKIs. However, several other gene alterations are also implicated in Tyrosine Kinase Inhibitors (TKIs) resistance and NGS is the most appropriate method for the analysis of these resistance mechanisms. Moreover, resistance mutations that arise following treatment with third-generation TKIs (such as the C797S mutation in the EGFR gene) can also be detected by this methodology [11, 26]. Plasma ctNA NGS analysis has been shown to be a reliable alternative to tissue biopsy analysis and its use in clinical practice is constantly increasing [27, 28].
The aim of this study was to investigate the feasibility of ctNA analysis in everyday practice, using a sensitive NGS approach in patients with NSCLC. The plasma mutation distribution in newly diagnosed patients was calculated. Furthermore, a comparison among the results obtained by cobas and NGS was carried out in patients at progression on treatment with EGFR TKIs.
Material and methods
Patient selection
In the current study, plasma liquid biopsy analysis was conducted using the NGS and/or cobas methods in 171 NSCLC patients who were referred to our laboratory between January 2017 and June 2019. Of those 121 were newly diagnosed without a previous treatment assigned. In the majority of the cases (85), a liquid biopsy analysis was requested due to the unavailability of a tissue sample, while in 36 patients both tissue and liquid biopsy were available. Additionally, 50 patients that were positive for an EGFR sensitizing mutation at diagnosis and had received targeted treatment were tested by both the cobas EGFR mutation test and NGS, at relapse (Fig 1).
Fig 1. Study design.
The study was approved by the ethical committee of “Agii Anargiri” Cancer Hospital. All patients gave informed consent for molecular analysis in blood and tissue, in accordance with the Declaration of Helsinki.
Cell-free Total Nucleic Acids (cfTNA) isolation
10 ml of blood samples were collected in Cell-Free DNA BCT tubes (Streck, La Vista, NE), that are used to stabilize Cell free Total Nucleic Acids (cfTNA). Plasma was separated from the cellular fraction by centrifugation twice at 1800 rcf for 10 min at 4 oC. cfTNA was isolated from 2 ml of plasma using QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s instructions. cfTNAs’ concentration was measured with the use of the Qubit™ 2.0 Fluorometer in combination with the Qubit™ ssDNA Assay Kit (Thermo Fisher Scientific).
Tissue selection and DNA extraction
Hematoxylin and eosin stained sections of formalin-fixed and paraffin-embedded (FFPE) tumor biopsies from all samples were reviewed to ensure a tumor cell content of >75%, where possible and the tumor area was marked by a pathologist. Genomic DNA was extracted from unstained 10 μm thick FFPE sections using the QIAmp FFPE tissue kit (Qiagen). The DNA concentration of all samples was determined spectrophotometrically (NanoDrop2000, Thermo Fisher Scientific). FFPE extracted DNA concentration was measured using the Qubit™ 2.0 Fluorometer in combination with the Qubit™ dsDNA HS assay kit (Thermo Fischer Scientific).
Next generation sequencing libraries preparation
The NGS for FFPE DNA analysis was conducted using a custom Ion AmpliSeq panel which was based on the Ion Ampliseq Colon and Lung Cancer Research Panel v2 with an additional amplicon in the MET gene (to include the exon 14 skipping mutations) and two amplicons of exons 2 and 3 of the HRAS gene [29]. Fusion RNA transcript analysis was performed using the Ion AmpliSeq™ RNA Fusion Lung Cancer Research Panel (Thermo Fisher Scientific). Details of the target regions of the 23-gene panel are available upon request. The genes analyzed include AKT1, ALK, BRAF, CTNNB1, DDR2, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, KRAS, MAP2K1, MET, NOTCH, NRAS, PIK3CA, PTEN, SMAD4, STK11, TP53, HRAS. An amplicon library was generated from 10ng of total FFPE extracted DNA or 6 μl of ctDNA, using the Ion AmpliSeq Library Kit 2.0 (Thermo Fischer Scientific) according to the manufacturer’s instructions. Briefly, amplicon amplification was performed using Ion Ampliseq™ HiFi Master Mix (Thermo Fischer Scientific). The amplicons were then digested with FUPA reagent and barcoded with the IonCode™ Barcode Adapters 1–384 Kit (Thermo Fischer Scientific). Subsequently, the amplified products were purified from the other reaction components using Agencourt AMPure XP PCR purification system (Beckman Coulter).
The NGS for cfTNA analysis was conducted using Oncomine Lung Cell-Free Total Nucleic Acid Research Assay (Thermo Fisher Scientific). The assay uses random molecular tags that are used as unique molecular identifiers (UMI) to uniquely label each molecule prior to library amplification. In this way, randomly incorporated errors can be distinguished (and removed) from true variants, increasing the method’s accuracy and minimizing false positives. The assay includes the analysis of Hotspot genes (Single Nucleotide Variations and short insertions/deletions): ALK, BRAF, EGFR, ERBB2 (HER2), KRAS, MAP2K1, MET, NRAS, PIK3CA, ROS1, and TP53. Furthermore, the assay includes the analysis of RNA fusions that involve 3 fusion driver genes: ALK, RET, ROS1. Copy number variations are also calculated for the following genes: MET and ERBB2. cfTNA concentration was measured with the Qubit™ 2.0 Fluorometer using the Qubit™ ssDNA Assay Kit (Thermo Fischer Scientific). Library preparation was performed according to the manufacturer’s provided protocol. Briefly, Reverse Transcription of the cfTNA was carried out using SuperScript™ VILO™ Master Mix. The Cell-free total nucleic acid reverse transcription products were amplified with a 2 cycle PCR to copy each strand of an original DNA fragment into a fragment with random sequences (Tags) and A/P1 adaptors attached to 5’/3’ ends. Following that, a bead purification step was performed using Agencourt AMPure XP PCR purification system (Beckman Coulter, Inc., Brea, CA, USA). Subsequently, an 18 cycle amplification of the tagged amplicons was performed with a unique barcode for each sample, using the Tag Sequencing Barcode Set (Thermo Fischer Scientific). Purification and size selection of the barcoded library was achieved using Agencourt AMPure XP PCR purification system. Finally, the concentration of each library was determined by real-time PCR, using the Ion Library TaqMan® Quantitation Kit (Thermo Fischer Scientific)
Successively, 100pmoles of the DNA or ctTNA libraries were separately combined and clonally amplified on Ion sphere™ particles (ISP) by emulsion PCR performed on the Ion One Touch™ 2 instrument with the Ion 540™ Kit-OT2 (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Quality control was performed using the Ion Sphere™ Quality Control kit (Thermo Fisher Scientific) to ensure that 10–30% of template-positive ISP were generated in the emulsion PCR. Finally, the template-positive Ion Sphere™ particles were enriched using the Ion OneTouch™ ES instrument, loaded on an Ion 540™ Chip and sequenced on an Ion GeneStudio™ S5 Prime System Sequencer according to the manufacturer’s instructions. All samples were processed in duplicate. In case of a discordant result a new duplicate NGS experiment was performed. A variant was considered as true when it was detected in at least 3 out of the 4 NGS runs.
NGS data analysis
NGS data analysis was performed with the Ion Reporter™ 5.10.1.0 software directly from within Torrent Suite™ 5.10.1 software (Thermo Fisher Scientific), followed by manual inspection, along with the commercial analysis software Sequence Pilot version 4.3.0 (JSI medical systems, Ettenheim, Germany). The coverage analysis was performed using the coverage analysis plug-in v5.0.4.0. The statistics generated from this plugin were used to evaluate the quality of each library in the sequencing run. For FFPE DNA libraries, the copy number variation (CNV) analysis was performed using the Ion Reporter™ Software. CNVs were reported based on their copy number relative to the control sample used. The software reports all possible CNVs assigning a score, with scores >10 indicating high-confidence CNVs. This value was used as a threshold for identifying a copy number amplification. cfTNA mutations detection, RNA fusion and CNV analysis were performed with the Ion Reporter software using a manufacturer’s provided workflow. (Oncomine TagSeq Lung Liquid Biopsy w2.1—Single Sample, Thermo Fisher Scientific).
EGFR mutation analysis by cobas
EGFR mutation analysis was conducted using the cobas® cfDNA Sample Preparation Kit followed by the cobas® EGFR Mutation Test v2 (Roche Molecular Diagnostics), according to the manufacturer’s instructions. The amplification and detection of the mutations was performed in the cobas® 4800 System (Roche Molecular Diagnostics).
Statistical analysis
For the comparison of mutations detected in paired plasma and tissue biopsy analysis Sensitivity, Specificity, Positive Predictive Value (PPV), and Concordance were calculated. True positives (TP) and true negatives (TN) were defined using the results of the tissue biopsy analysis as a gold standard. False positives (FP) and false negatives (FN) were calculated as the number of mutations detected and not detected respectively by the plasma analysis. Statistics were performed with SPSS (version 20. IBM SPSS STATISTICS). The p-values were based on Fisher’s Exact Test. A p-value <0.05 was considered to be statistically significant.
Results
Mutation distribution in plasma of NSCLC patients
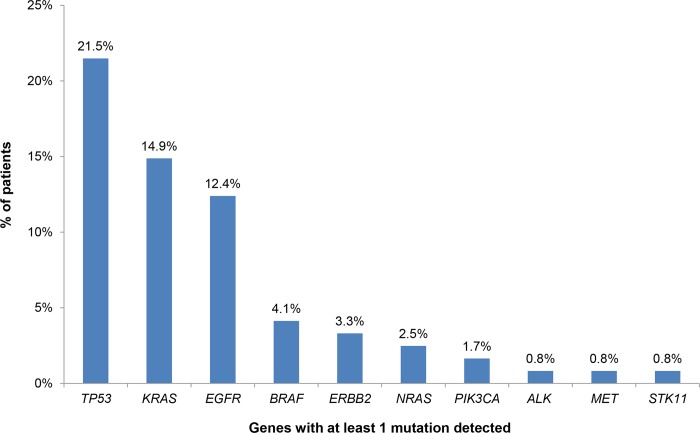
Among the 121 patients with newly diagnosed metastatic NSCLC who underwent liquid biopsy analysis, 82 (67.8%) of them were male and 39 (32.2%) were female. The mean age of diagnosis was 67 years. At least one mutation was detected in the plasma of 59 patients (48.76%), of which 74.58% presented only one mutation and 25.42% presented two or more mutations (S1 Table). The percentage of patients with a plasma mutation was similar between males and females (48.78% and 48.72% respectively). TP53 mutations were the most common alterations (detected in 21.49% of the patients), followed by KRAS and EGFR gene mutations detected in 14.88% and 12.39% of the cases respectively (Fig 2). The EGFR mutation frequency did not vary significantly between males and females (10.76% and 15.38% among males and females respectively).
Fig 2. Mutation distribution in plasma samples of 121 NSCLC patients at diagnosis.
EGFR mutation distribution for exons 18, 19 and 21 was 20,00%, 46.67% and 33.33% respectively. In 2 samples a second mutation in exon 20 (p.S768I) was also present in addition to the sensitizing mutation (exons 18 and 21). In vitro and in vivo studies have shown that the double mutant receptor is sensitive to treatment with EGFR TKIs [30].
The findings obtained by this approach could be used for the assignment of the appropriate treatment based on patients’ molecular profile. The gene alterations analyzed by the NGS methodology used were selected based on their increased prevalence in NSCLC and their possible use as biomarkers of response to targeted treatment. Thus, we have categorized them based on their actionability in 4 categories: those related to on-label treatments, those considered as emerging biomarkers with increasing evidence of predictive value for targeted treatments, those related to clinical trials and those with unknown actionability. Hence, each patient can be assigned to one of these categories on the basis of his mutation profile. In the case of multiple mutations present in the same patient, the categorization is based on the most clinically significant mutation. Using this categorization we observed that 14.88% of the individuals with at least one mutation detected, carried a mutation related to an approved treatment for NSCLC. These include sensitizing mutations in the EGFR gene, the p.V600E mutation in the BRAF gene and ALK fusions (in 12.39%, 1.65% and 0.83% of the patients respectively). In 3.31% of the patients tested, a mutation in the ERBB2 gene was detected. There is increasing evidence for the utility of the alterations in this gene as a biomarker of response to targeted treatment and thus it is reported as an emerging biomarker for NSCLC in the NCCN guidelines (www.nccn.org). Furthermore, in 37 patients (30.58%) the mutations detected could be related to off-label treatment or a clinical trial. More precisely, these mutations were located in the TP53, KRAS, NRAS, BRAF (non-V600E), STK11 and PIK3CA genes.
Concordance in mutation distribution between tissue and plasma
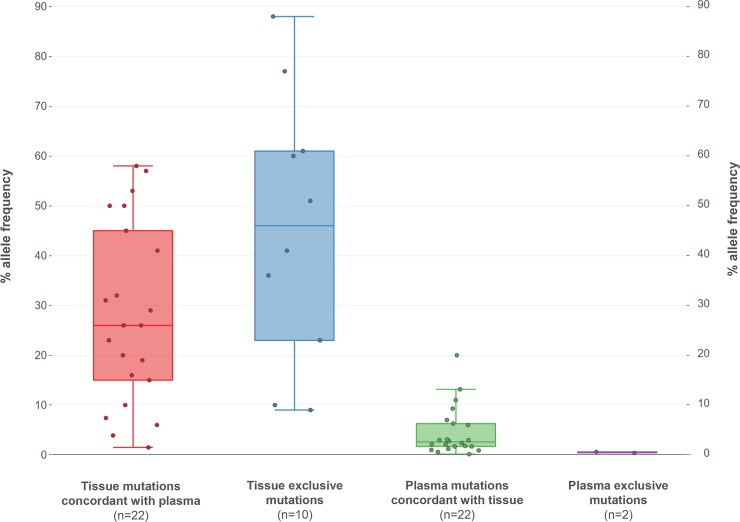
In the 36 patients with concomitant tissue and liquid biopsy available, a total of 34 mutations were detected. Of them, 22 mutations were detected in both materials, 10 mutations were detected in tissue only and 2 mutations were detected in plasma only (Fig 3). The mean mutated allele fraction was 33.62% for somatic mutations detected in tumor biopsy, while this value, as expected, was significantly lower in plasma samples (4.22%). However, the detection of a mutation in plasma did not correlate with its percentage in tissue as there was not a statistically significant difference in the % allele frequency between tissue exclusive mutations and concordant with plasma tissue mutations (p = 0.2695) (Fig 3).
Fig 3. Median allele frequency of mutations detected in tissue and plasma grouped based on their concordance among the two materials.
The concordance between these 2 materials varied depending on the gene analyzed. The lowest concordance rate in the mutational status between tissue and plasma was observed for the TP53 gene (67.44%), where 7 out of the 13 mutations detected in tissue were not present in the plasma (sensitivity 46.15%, specificity 100%) (Table 1, Fig 4). On the contrary, the concordance rate between tissue-plasma for the KRAS and NRAS status was 94.74%, with an 85.71% sensitivity of RAS mutation detection in plasma in case of a positive tissue result and 100% specificity. Notably, all 5 EGFR mutations detected in the tissue were also present in plasma (sensitivity 100%); however, in 2 cases, an EGFR mutation was detected in plasma while it was not present in the tissue sample. Considering the tissue as the gold standard material, we could assign a specificity rate for these mutations of 93.94%.
Table 1. Sensitivity, specificity, PPV (Positive Predictive Value) and concordance for mutations detected in paired plasma and tissue biopsy analysis.
| Gene(s) | Sensitivity | Specificity | PPV | Concordance |
|---|---|---|---|---|
| KRAS/NRAS | 85.71% | 100% | 100% | 94.74% |
| EGFR | 100% | 93.94% | 60% | 94.44% |
| TP53 | 46.15% | 100% | 46.15% | 67.44% |
| Clinically Significant Genes | 84.21% | 88.24% | 88.89% | 86.11% |
| All Genes | 68.75% | 83.33% | 91.67 | 72.73% |
Fig 4. Comparison of the mutation distribution in plasma and tissue of 36 patients with newly diagnosed NSCLC.
In summary in both tissue and plasma, the detection of clinically significant mutations was possible. Considering only genes with approved or emerging targeted treatments available, such as EGFR, BRAF and ERBB2 and genes with prognostic relevance such as KRAS and NRAS genes, we can observe a concordance rate among tissue and plasma for their mutational status of 86.11% with a Positive Predictive Value (PPV) of 88.89%.
Concordance of plasma mutation distribution between cobas and NGS approaches
Another point of investigation of this study was the feasibility of liquid biopsy analysis in patients that harbored an EGFR mutation at diagnosis and have relapsed following treatment with EGFR TKIs. In these cases, a very common clinical practice is the use of the CE-IVD cobas® EGFR Mutation Test, in order to detect the T790M resistance mutation in the EGFR gene. Thus, we compared the results obtained by cobas and by our NGS approach in patients that were referred to our laboratory, following treatment with EGFR TKIs, due to suspected insensitivity to the given treatment. Of note, in the majority of the cases, the initial EGFR mutation analysis was performed in a different laboratory, thus we were not aware of the primary sensitizing EGFR mutation nor in position to verify it.
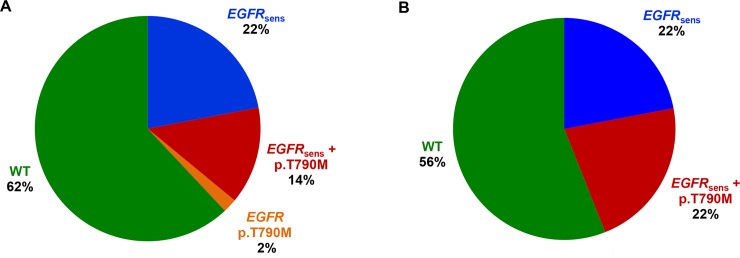
When the EGFR analysis by cobas was applied in this patients’ group, an EGFR mutation was detected in 38% of the cases, while the primary sensitizing EGFR mutation was absent in 62% of the cases. Among patients with an EGFR mutation detected, 11 harbored a sensitizing EGFR mutation, 7 harbored an EGFR sensitizing mutation plus the T790M resistance mutation, and 1 harbored the T790M resistance mutation alone (Table 2, Fig 5).
Table 2. Gene mutation results obtained by cobas and NGS methods in 50 consecutive NSCLC patients referred for T790M resistance mutation analysis due to relapse after TKI treatment.
| PATIENT | EGFR analysis by cobasa | EGFR analysis by NGSb | Other Gene Alterations detected by NGSc |
|---|---|---|---|
| 1 | Ex19Del (12.82) | Ex19Del (p.L747_S752del) [9.62%] | BRAF p.V600K [0.6%] |
| 2 | p.L858R (6.00) | Wild Type | KRAS p.G12V [0.5%] |
| 3 | p.S768l (5.49), p.G719X (2.54) | p.G719S [5.23%] | NONE |
| 4 | p.L858R (6.97) | p.L858R[6.72%] | NONE |
| 5 | p.L861Q (5.21) | p.L861Q [0.38%] | NONE |
| 6 | Ex19Del (19.01) | Ex19Del (p.Glu746_Ala750del) [27.71%] | TP53 p.Y220C [0.9%] |
| 7 | Ex19Del (17.49) | Ex19Del p.Glu746_Ala750del [23.44%] | NONE |
| 8 | Ex19Del (5.99) | Ex19Del p.E746_A750del [9.12%] | TP53 p.R175H [1.1%] |
| 9 | Ex19Del (5.99) | Wild Type | NONE |
| 10 | Ex19Del (10.12) | Ex19Del (p.E746_A750del) [20.57%], p.T790M [0.55%] | NONE |
| 11 | EX19Del (10.96), p.T790M (15.28) | Ex19Del (p.L747_A750delinsP) (24.42%), p.T790M (12.78%) | NONE |
| 12 | EX19Del (6.00), p.T790M (8.33) | Ex19Del(p.L747_T751del) (0.82%), p.T790M (1.24%) | NONE |
| 13 | p.L858R (13.02), p.T790M (16.84) | p.L858R (2.44%), p.T790M (4.55%) | NONE |
| 14 | EX19Del (12.04), p.T790M (17.02) | Ex19Del (p.E746_A750del) (11.55%), p.T790M (10.66%) | NONE |
| 15 | EX19Del (14.56), p.T790M (12.73) | p.L858R (4.45%), p.T790M (3.12%) | NONE |
| 16 | EX19Del, p.T790M (11.2) | Ex19Del (p.L747_T751>Q) (11.52%), p.T790M (2.45%) | NONE |
| 17 | EX19Del (11.36), p.T790M (16.30) | Ex19Del (p.E746_A750del) (2.98%), p.T790M (3.21%) | NONE |
| 18 | EGFR p.T790M (5.66) | Ex19Del (p.E746_A750del) (1.04%), p.T790M (1.56%) | NONE |
| 19 | Wild Type | p.L858R [9.63%], p.T790M [1.24%] | NONE |
| 20 | Wild Type | Wild Type | BRAF p.V600E [0.8%] |
| 21 | Wild Type | Wild Type | NONE |
| 22 | Wild Type | Wild Type | NONE |
| 23 | Wild Type | Wild Type | TP53 p.C176S [1.9%] |
| 24 | Wild Type | Wild Type | NONE |
| 25 | Wild Type | Wild Type | MET amplification [1.3] |
| 26 | Wild Type | Wild Type | NONE |
| 27 | Wild Type | Wild Type | KRAS p.G12D [0.7%] |
| 28 | Wild Type | Wild Type | NONE |
| 29 | Wild Type | Ex19Del (p.E746_A750del) [9.24%], p.T790M [1.65%] | NONE |
| 30 | Wild Type | Wild Type | NONE |
| 31 | Wild Type | Wild Type | NONE |
| 32 | Wild Type | Wild Type | NONE |
| 33 | Wild Type | Wild Type | NONE |
| 34 | Wild Type | Wild Type | MET AMPLIFICATION [1.64] |
| 35 | Wild Type | Wild Type | NONE |
| 36 | Wild Type | Wild Type | NONE |
| 37 | Wild Type | Wild Type | NRAS p.G13S [0.82%] |
| 38 | Wild Type | Ex19Del (p.E746_S752delinsV) [0.72%] | TP53 p.G245D [0.83%], MET p.Y1248H [0.94%], KRAS p.G12C [0.63%], PIK3CA p.E542K [1.02%] |
| 39 | Wild Type | Wild Type | NONE |
| 40 | Wild Type | Wild Type | TP53 p.R249fs [3.82%] |
| 41 | Wild Type | Wild Type | NONE |
| 42 | Wild Type | Wild Type | NONE |
| 43 | Wild Type | Wild Type | TP53 p.R282W [2.54%] |
| 44 | Wild Type | Wild Type | NONE |
| 45 | Wild Type | p.G719A [4.12%] | KRAS p.G12V [2.33%], BRAF p.V600E [1.64%], TP53 p.R175H [1.23%] |
| 46 | Wild Type | Ex19Del (p.E746_S750del) [0.47%] | MAP2K1 p.E203K [0.82%] |
| 47 | Wild Type | Wild Type | NONE |
| 48 | Wild Type | Wild Type | NONE |
| 49 | Wild Type | p.G719A [0.93%] | PIK3CA p.E542G [0.44%], TP53 p.R175H [0.85%], KRAS p.G12C [0.66%] |
| 50 | Wild Type | Wild Type | NONE |
a. The semi-quantitative index of the cobas test is provided in brackets.
b. Allele frequency of EGFR gene detected by the NGS method is given in brackets.
c. Allele frequency of each gene alteration and/or the CNV ratio of each amplification detected is given in brackets.
Fig 5. EGFR mutation distribution in ctDNA of 50 patients with an EGFR mutation at diagnosis that have relapsed following TKI treatment.
A. Analysis by cobas B. Analysis by NGS.
EGFR analysis by NGS in the same cohort revealed the presence of an EGFR mutation in 44% (22/50) of the cases. In the mutation positive group, 50% of the patients carried a sensitizing mutation while a sensitizing plus a resistance mutation was present in the rest of the cases.
The concordance rate between the two methods in the EGFR mutation analysis was 82% for the EGFR sensitizing mutations (in exons 18, 19, 21). In some cases the discordances concerned low frequency variants with an allele frequency <1% (as measured by NGS) or with a cobas semi-quantitative index < 6. In 2 cases an EGFR sensitizing mutation was not detected by NGS while it was detected by cobas. Furthermore, in 6 cases the sensitizing mutation was only detectable by the NGS methodology.
For the p.T790M mutation, a higher concordance between the two approaches was observed (94%), with the NGS analysis being able to detect the p.T790M mutation in 3 additional patients.
In contrast to the cobas assay, NGS analysis is a multigene assay that is also providing information for other important and possibly clinically relevant cancer genes. Such approach allows the detection of additional mutations besides the T790M mutation that could contribute to resistance to EGFR TKI treatment. This is of significant importance in cases without the T790M mutation detected. In our cohort, in the 39 patients without the T790M mutation (10 with only an EGFR sensitizing mutation present and 32 without any mutation detected), mutations in the KRAS and BRAF, PIK3CA, MAP2K1 genes as well as a MET amplification (Table 2) where identified. These alterations could explain the unresponsiveness to treatment in these patients, thus clarifying the resistance mechanism. Additionally, some of these alterations such as BRAF and MET amplification are targetable and could be used for the enrollment of these patients to clinical trials (Fig 6).
Fig 6. NGS-cobas comparison in 50 EGFR mutant NSCLC patients in relapse following EGFR TKIs treatment.
EGFR sens.: EGFR sensitizing mutations in exons 18, 19, 21.
Discussion
Several previous studies have shown the clinical utility of liquid biopsy analysis using multigene approaches [31–33]. However, its predictive value for genes other than EGFR is still not widely recognized [34, 35]. In the current study, we report a single center’s experience of using this type of analysis for the detection of clinically significant alterations in NSCLC patients. Hence, plasma analysis of 171 NSCLC patients was performed using a sensitive NGS assay that analyses hotspot regions of 12 genes frequently altered in NSCLC and fusions in ALK, ROS1 and RET genes.
The NGS method applied in this study employs random molecular tags that are used as unique molecular identifiers (UMI) to uniquely label each molecule prior to library amplification. In this way, randomly incorporated errors can be distinguished (and removed) from true variants, increasing the method’s accuracy and minimizing false positives [34, 36, 37]. The variant detection limit of the assay is 0.1%, with 90% sensitivity and >98% specificity for Single nucleotide variation hotspots and small insertions/deletions [27, 38]. Nevertheless, cautious selection of the appropriate NGS method is indispensable for obtaining accurate molecular analysis results using plasma as a starting material. Not all methods have the same sensitivity and specificity and traditional amplicon based NGS techniques without the addition of unique molecular identifiers seem to be less appropriate for use in liquid biopsy analysis [39]
Multi-gene analysis can be achieved in less than a week, allowing physicians to make quick treatment decisions. Additionally, the clinical information provided is superior compared to that obtained from the analysis of only one gene. The price of such analysis depends mainly on the number of genes analyzed and the methodology used. In any case, the cost per gene is much lower than the single-gene analysis. Therefore, we believe that plasma NGS methods analyzing a small group of targetable genes specific for each tumor type are cost-effective. Thus, their application in clinical practice is feasible.
The simultaneous analysis of many therapeutic targets by this methodology in NSCLC, a tumor type with various treatment options available, is of great clinical significance. NGS technology rendered this analysis possible even in a material such as plasma with minute concentrations of genetic material available. However, physicians often require the examination only of traditional biomarkers such as EGFR and ALK. Nevertheless, if the analysis was limited to these genes, a mutation would have been detected in only 13.2% of the cases (Fig 7). Furthermore, a recently approved biomarker for NSCLC is the p.V600E mutation in the BRAF gene. The analysis of this variant increases the percentage of patients positive for an approved biomarker to 14.89% (Fig 7). ERBB2 mutations in NSCLC, mainly consist of exon 20 in frame insertions and are present in 1–3% of the patients [40]. Additionally MET exon14 skipping mutations and amplification are also present in 1–4% of the cases [41–43]. Both ERBB2 and MET alterations are considered emerging biomarkers of responsiveness to treatment and are included in the NCCN guidelines (www.nccn.org). In our cohort ERBB2 and MET alterations were present in 3.31% and 0.83% of the cases respectively (Fig 7).
Fig 7. Apportionment of the 59 NSCLC patients with at least one variant identified in liquid biopsy analysis, using 4 different biomarker categories: traditional targeted treatment biomarkers (EGFR, ALK); all approved treatment associated biomarkers (EGFR, ALK, BRAF); approved and emerging treatment biomarkers (EGFR, ALK, BRAF, HER2); approved, emerging treatment & clinical trials associated biomarkers (EGFR, ALK, BRAF, HER2, KRAS, NRAS, MET, PIK3CA, STK11,TP53).
In case a patient harbors more than one mutation the categorization is based on the more clinically significant variant.
Further biomarker analysis could also contribute to the clarification of the patient’s genetic profile and give additional clinical important information. For example the analysis of KRAS mutations, which is present in 15–25% of the patients, is suspected to be a prognostic biomarker of worse treatment outcome in NSCLC adenocarcinoma [44]. Additionally, there are studies showing that patients harboring this mutation are unlike to respond to targeted treatment with EGFR TKIs [45, 46]. Clinical trials also exist for other biomarkers in our cohort (www.clinicaltrials.gov). The BFAST phase II/III clinical trial is an example of such an effort, aiming to investigate the clinical utility of actionable alterations’ identification using plasma NGS analysis without the influence of tissue-based testing (NCT03178552). NSCLC patients with an actionable alteration detected in plasma are assigned to the matched targeted treatment or immunotherapy and treatment’s benefit is calculated. Recently, the first results announced from this study in the ALK-positive cohort indicated that liquid biopsy analysis can efficiently detect such alteration and the plasma positivity in ALK rearrangements is highly predictive of alectinib response.
Several studies have confirmed the clinical utility of ctNA analysis for the detection of tumor originated mutations in NSCLC as well as in other tumor types [3, 19, 47–50]. In accordance with these results, in the 36 patients with concurrent tissue and plasma available we observed a 72.73% concordance rate among the two materials in the mutations detected. This concordance is even higher when considering the clinically relevant mutations with an approved or investigational targeted treatment available or with prognostic value (EGFR, BRAF, ERBB2, KRAS, NRAS). The higher concordance rate could be attributed to the fact that these are driver mutations that occur early in cancer development [51]. A sensitivity of 84.2% was achieved for these mutations with a concordance rate of 88.9%. This is of particular importance since these mutations will presumably be used for treatment decision making. Even though, the tissue is considered as the gold standard material for specificity calculation in tissue versus liquid biopsy concordance studies, this assumption does not take into account tumor heterogeneity, that could result in omission of mutations when using tissue biopsies [52]. This is even more prominent for NSCLC, which presents several tissue sampling issues since the majority of the cases are inoperable and the analysis is often performed using limited size biopsies with less than 100–150 tumor cells present [53]. This was also the case of our two negative EGFR tissue results with discordant plasma outcomes. Unfortunately, the physicians relied on the tissue result, since it was considered more trustworthy for treatment decision, rather on the liquid biopsy result. In the same way, no targeted treatment was administrated in the 3 patients with ERBB2 mutations detected (2 in liquid biopsy only and one in both tissue and plasma), even though these variations are considered emerging biomarkers of response to ado trastuzumab emtansine [54] (www.nccn.org/). This was attributed by the physicians to the absence of approval of this regiment for NSCLC and to the costly and/or lengthy procedures required for administrating the drug off-label or for the enrollment in a clinical trial.
In a recent study though, it has been shown that plasma driven targeted treatment administration in NSCLC patients showed effectiveness in 36 of the 42 patients with evaluable results (85.7%), including stable disease partial and complete response in 16, 19 and 1 case respectively. Furthermore, the depth of RECIST response was not correlated to the mutation allele frequency in plasma 49]. Similarly, Reckamp and colleagues showed that patients’ response to third generation EGFR TKI is achieved, irrespectively of whether the T790M mutation is detected in tissue, plasma or urine samples [55]. Hopefully, the issue of liquid biopsy results’ credibility will be resolved soon, since currently there are more than 130 ongoing clinical trials that are exploring the role of such analysis in different tumor types and probably upon completion will highlight its importance, especially in cases with no tissue available (www.clinicaltrial.gov). Recently, results from the SOLAR-1 phase III randomized controlled trial (NCT02437318) indicated a significant improvement in Progression Free Survival (PFS) from the addition of alpelisib to the fulvestrant treatment in HER2 negative, Hormonal Receptors (HR) positive Breast Cancers patients who harbored a PIK3CA mutation accessed either using tissue or plasma. Thus, PIK3CA became the second targetable gene (after EGFR) with an approved drug available using liquid biopsy analysis for the detection of the alteration targeted [56]. The accumulation of such data enhances the position of the non-invasive tumor molecular profiling approach and will eventually lead to more drug approvals based on liquid biopsy biomarkers.
The most recognized application of ctDNA analysis is in the detection of mutations that arise due to the EGFR TKIs targeted treatment, with the p.T790M mutation being the best studied resistant mechanism, since it provides sensitivity to third-generation EGFR TKIs [57]. Osimertinib, was the first third-generation EGFR TKI, to be approved for patients that harbor the p.T790M mutation and have progressed following EGFR TKIs treatment. Recently it has also been approved as first-line therapy in EGFR mutated NSCLC patients, irrespectively of the T790M mutation status [58, 59]. Nonetheless, several issues concerning this TKI remain unclear such as the resistance mechanism of this treatment when received in first line and the consequences of its prolonged administration. The ongoing MELROSE: Phase 2 clinical trial aims to clarify the resistance mechanisms of these medications, analyzing both tissue and plasma ctDNA (NCT03865511). The first and only FDA approved kit for EGFR mutation analysis is the cobas® EGFR Mutation Test v2 from Roche Diagnostics, which is a real-time PCR assay designed to detect 42 EGFR mutations in exons 18, 19, 20 and 21, including the resistant T790M mutation. This assay has shown good performance in EGFR mutation analysis across different studies [20, 22–24]. However, it should be noted that the sensitivity of this assay does not exceed 77% for the EGFR exon 19 and 21 mutations, with the sensitivity being much lower, around 61% for the pT790M resistance mutation (20). As a result, if a p.T790M mutation is present in plasma there is almost 40% of probability to miss it, by the cobas analysis. An additional cause of missing tissue mutations in plasma is the phenomenon of Non-Shedding tumors, that release only limited amount of ctDNA in circulation [60]. Thus, current guidelines recommend a re-biopsy in case of negative plasma result when accessing resistance in NSCLC patients at relapse upon EGFR TKI treatment (www.nccn.org) [61]. In order to reduce the incidence of invasive re-biopsies in these cases the use of more sensitive techniques should be applied with NGS providing an excellent alternative to the cobas method. In the recent years several modified NGS approaches with increased sensitivity and specificity have been applied for ctDNA analysis [27, 28, 62], showing a better accuracy on the plasma for the T790M mutation identification [63]. In addition to the increased sensitivity, the NGS methodology offers the advantage of being a multigene assay, providing more clinically relevant information compared to a single gene test. It is known that p.T790M mutation is only one of the existing resistant mechanisms developed by the tumor following to EGFR TKI treatment. In 45–50% of the cases, the resistance arises through different mechanisms and can be due to EGFR-independent mechanisms, including alterations in genes involved in alternative pathways (such as PIK3CA, BRAF, KRAS, ERBB2, and MET) and histological or phenotypic transformation [11, 64].
In order to evaluate the concordance rate among these two platforms, 50 NSCLC patients that were positive for an EGFR sensitizing mutation at diagnosis and had stopped responding to EGFR TKI treatment were analyzed by both cobas and NGS platforms. An 82% concordance was observed in EGFR sensitizing mutations identification (exons 18, 19, 21) between the two assays, in plasma. The concordance for the p.T790M mutation was higher reaching a percentage of 94%. In 6 cases the sensitizing mutation was detected only by NGS analysis, whereas only 2 sensitizing EGFR mutations were missed by this approach. Most importantly, 3 additional p.T790M mutations were identifiable only by the NGS methodology. In these cases a third-generation EGFR TKI could be used to overcome resistance to TKI treatment, revealing the importance of using more sensitive approaches for plasma analysis. In one case, an eight-month response to osimertinib was achieved, and in a subsequent liquid biopsy analysis recommended by the physician due to loss of responsiveness, the C797S mutation was detected, which is responsible for resistance to third-generation EGFR TKIs. This mutation would have been missed if the analysis was carried out by cobas since it is not included in the mutations analyzed by this assay.
Moreover, NGS analysis of the 42 patients without the p.T790M resistance mutation, revealed mutations in other genes. Those alterations were located in TP53, KRAS, BRAF, MET, PIK3CA, NRAS, and MAP2K1 genes, in 19.05%, 11.90%, 7.14%, 7.14%, 4.76%, 2.38% and 2.38% of the cases respectively. The majority of these alterations have been reported previously as resistance mechanisms to EGFR treatment [11, 64]. Therefore, the resistance mechanism could be elucidated in more patients by the use of a multigene assay. Additionally, among the alterations detected in this cohort, the BRAF p.V600 mutations and the MET amplifications are candidate predictive markers of response to BRAF and MET inhibitors respectively in this patients’ setting [41, 65, 66]. Thus, the addition of such inhibitors could be considered to overcome EGFR TKI resistance in these patients [67, 68].
In conclusion, this study provides evidence of the feasibility of ctNA analysis as a tumor biopsy surrogate in clinical practice for NSCLC personalized treatment decision making. NGS is a technology that can provide a comprehensive molecular characterization of the tumor using both tissue and plasma. The applicability of this approach in clinical practice is shown by the significant percentage of patients with a targetable mutation detected both before and after targeted treatment. Thus, it could aid physicians in treatment decision making in clinical practice.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
GeneKor MSA provided support in the form of salaries for authors EI, NT, KT, VM, PS, AS, EK, GT and GN but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Garinet S, Laurent-Puig P, Blons H, Oudart JB. Current and Future Molecular Testing in NSCLC, What Can We Expect from New Sequencing Technologies? J Clin Med. 2018;7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coco S, Truini A, Vanni I, Dal Bello MG, Alama A, Rijavec E, et al. Next generation sequencing in non-small cell lung cancer: new avenues toward the personalized medicine. Curr Drug Targets. 2015;16(1):47–59. 10.2174/1389450116666141210094640 [DOI] [PubMed] [Google Scholar]
- 3.Thompson JC, Yee SS, Troxel AB, Savitch SL, Fan R, Balli D, et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin Cancer Res. 2016;22(23):5772–82. 10.1158/1078-0432.CCR-16-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruglyak KM, Lin E, Ong FS. Next-Generation Sequencing and Applications to the Diagnosis and Treatment of Lung Cancer. Adv Exp Med Biol. 2016;890:123–36. 10.1007/978-3-319-24932-2_7 [DOI] [PubMed] [Google Scholar]
- 5.Zhang P, Lehmann BD, Shyr Y, Guo Y. The Utilization of Formalin Fixed-Paraffin-Embedded Specimens in High Throughput Genomic Studies. Int J Genomics. 2017;2017:1926304 10.1155/2017/1926304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive—gene expression in paraffin-embedded tissue. J Pathol. 2001;195(1):66–71. [DOI] [PubMed] [Google Scholar]
- 7.Shin HT, Choi YL, Yun JW, Kim NKD, Kim SY, Jeon HJ, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun. 2017;8(1):1377 10.1038/s41467-017-01470-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh K, Wallace WA. Molecular pathology in lung cancer: a guide to the techniques used in clinical practice. Histopathology. 2014;65(6):731–41. 10.1111/his.12505 [DOI] [PubMed] [Google Scholar]
- 9.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501(7467):355–64. 10.1038/nature12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015;61(1):64–71. 10.1373/clinchem.2014.223040 [DOI] [PubMed] [Google Scholar]
- 11.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17(1):38 10.1186/s12943-018-0777-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):827. [DOI] [PubMed] [Google Scholar]
- 13.Giampieri R, Scartozzi M, Del Prete M, Maccaroni E, Bittoni A, Faloppi L, et al. Molecular biomarkers of resistance to anti-EGFR treatment in metastatic colorectal cancer, from classical to innovation. Crit Rev Oncol Hematol. 2013;88(2):272–83. 10.1016/j.critrevonc.2013.05.008 [DOI] [PubMed] [Google Scholar]
- 14.Stroun M, Anker P, Maurice P, Lyautey J, Lederrey C, Beljanski M. Neoplastic characteristics of the DNA found in the plasma of cancer patients. Oncology. 1989;46(5):318–22. 10.1159/000226740 [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Vigil IG, Moreno-Martinez AK, Wang JY, Roehrl MHA, Barrera-Saldana HA. The dawn of the liquid biopsy in the fight against cancer. Oncotarget. 2018;9(2):2912–22. 10.18632/oncotarget.23131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathai RA, Vidya RVS, Reddy BS, Thomas L, Udupa K, Kolesar J, et al. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J Clin Med. 2019;8(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francis G, Stein S. Circulating Cell-Free Tumour DNA in the Management of Cancer. Int J Mol Sci. 2015;16(6):14122–42. 10.3390/ijms160614122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 19.Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet. 2019;20(2):71–88. 10.1038/s41576-018-0071-5 [DOI] [PubMed] [Google Scholar]
- 20.Komatsubara KM, Sacher AG. Circulating Tumor DNA as a Liquid Biopsy: Current Clinical Applications and Future Directions. Oncology (Williston Park). 2017;31(8):618–27. [PubMed] [Google Scholar]
- 21.Cabel L, Proudhon C, Mariani P, Tzanis D, Beinse G, Bieche I, et al. Circulating tumor cells and circulating tumor DNA: What surgical oncologists need to know? Eur J Surg Oncol. 2017;43(5):949–62. 10.1016/j.ejso.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 22.Keppens C, Palma JF, Das PM, Scudder S, Wen W, Normanno N, et al. Detection of EGFR Variants in Plasma: A Multilaboratory Comparison of a Real-Time PCR EGFR Mutation Test in Europe. J Mol Diagn. 2018;20(4):483–94. 10.1016/j.jmoldx.2018.03.006 [DOI] [PubMed] [Google Scholar]
- 23.Xu T, Kang X, You X, Dai L, Tian D, Yan W, et al. Cross-Platform Comparison of Four Leading Technologies for Detecting EGFR Mutations in Circulating Tumor DNA from Non-Small Cell Lung Carcinoma Patient Plasma. Theranostics. 2017;7(6):1437–46. 10.7150/thno.16558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–53. 10.1097/JTO.0000000000000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guibert N, Hu Y, Feeney N, Kuang Y, Plagnol V, Jones G, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol. 2018;29(4):1049–55. 10.1093/annonc/mdy005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart EL, Tan SZ, Liu G, Tsao MS. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res. 2015;4(1):67–81. 10.3978/j.issn.2218-6751.2014.11.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dono M, De Luca G, Lastraioli S, Anselmi G, Dal Bello MG, Coco S, et al. Tag-based next generation sequencing: a feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients. Mol Med. 2019;25(1):15 10.1186/s10020-019-0082-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paweletz CP, Sacher AG, Raymond CK, Alden RS, O'Connell A, Mach SL, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res. 2016;22(4):915–22. 10.1158/1078-0432.CCR-15-1627-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsoulos N, Papadopoulou E, Metaxa-Mariatou V, Tsaousis G, Efstathiadou C, Tounta G, et al. Tumor molecular profiling of NSCLC patients using next generation sequencing. Oncology reports. 2017;38(6):3419–29. 10.3892/or.2017.6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leventakos K, Kipp BR, Rumilla KM, Winters JL, Yi ES, Mansfield AS. S768I Mutation in EGFR in Patients with Lung Cancer. J Thorac Oncol. 2016;11(10):1798–801. 10.1016/j.jtho.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019;5(2):173–80. 10.1001/jamaoncol.2018.4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YC, Zhou Q, Wu YL. The emerging roles of NGS-based liquid biopsy in non-small cell lung cancer. J Hematol Oncol. 2017;10(1):167 10.1186/s13045-017-0536-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malapelle U, Pisapia P, Rocco D, Smeraglio R, di Spirito M, Bellevicine C, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res. 2016;5(5):505–10. 10.21037/tlcr.2016.10.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Volckmar AL, Sultmann H, Riediger A, Fioretos T, Schirmacher P, Endris V, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer. 2018;57(3):123–39. 10.1002/gcc.22517 [DOI] [PubMed] [Google Scholar]
- 35.Hofman P, Popper HH. Pathologists and liquid biopsies: to be or not to be? Virchows Arch. 2016;469(6):601–9. 10.1007/s00428-016-2004-z [DOI] [PubMed] [Google Scholar]
- 36.Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–8. 10.1016/j.csbj.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartels S, Persing S, Hasemeier B, Schipper E, Kreipe H, Lehmann U. Molecular Analysis of Circulating Cell-Free DNA from Lung Cancer Patients in Routine Laboratory Practice: A Cross-Platform Comparison of Three Different Molecular Methods for Mutation Detection. J Mol Diagn. 2017;19(5):722–32. 10.1016/j.jmoldx.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 38.Vollbrecht C, Lehmann A, Lenze D, Hummel M. Validation and comparison of two NGS assays for the detection of EGFR T790M resistance mutation in liquid biopsies of NSCLC patients. Oncotarget. 2018;9(26):18529–39. 10.18632/oncotarget.24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salk JJ, Schmitt MW, Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19(5):269–85. 10.1038/nrg.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arcila ME, Chaft JE, Nafa K, Roy-Chowdhuri S, Lau C, Zaidinski M, et al. Prevalence, clinicopathologic associations, and molecular spectrum of ERBB2 (HER2) tyrosine kinase mutations in lung adenocarcinomas. Clin Cancer Res. 2012;18(18):4910–8. 10.1158/1078-0432.CCR-12-0912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Awad MM, Oxnard GR, Jackman DM, Savukoski DO, Hall D, Shivdasani P, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol. 2016;34(7):721–30. 10.1200/JCO.2015.63.4600 [DOI] [PubMed] [Google Scholar]
- 42.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5(8):850–9. 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- 43.Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol. 2009;4(1):5–11. 10.1097/JTO.0b013e3181913e0e [DOI] [PubMed] [Google Scholar]
- 44.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92(1):131–9. 10.1038/sj.bjc.6602258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garzon M, Villatoro S, Teixido C, Mayo C, Martinez A, de Los Llanos Gil M, et al. KRAS mutations in the circulating free DNA (cfDNA) of non-small cell lung cancer (NSCLC) patients. Transl Lung Cancer Res. 2016;5(5):511–6. 10.21037/tlcr.2016.10.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood K, Hensing T, Malik R, Salgia R. Prognostic and Predictive Value in KRAS in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016;2(6):805–12. 10.1001/jamaoncol.2016.0405 [DOI] [PubMed] [Google Scholar]
- 47.Li BT, Janku F, Jung B, Hou C, Madwani K, Alden R, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30(4):597–603. 10.1093/annonc/mdz046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anagnostou V, Forde PM, White JR, Niknafs N, Hruban C, Naidoo J, et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res. 2019;79(6):1214–25. 10.1158/0008-5472.CAN-18-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oellerich M, Schutz E, Beck J, Kanzow P, Plowman PN, Weiss GJ, et al. Using circulating cell-free DNA to monitor personalized cancer therapy. Crit Rev Clin Lab Sci. 2017;54(3):205–18. 10.1080/10408363.2017.1299683 [DOI] [PubMed] [Google Scholar]
- 51.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med. 2015;7(283):283ra54 10.1126/scitranslmed.aaa1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matkovic V, Fontana D, Tominac C, Goel P, Chesnut CH, 3rd. Factors that influence peak bone mass formation: a study of calcium balance and the inheritance of bone mass in adolescent females. Am J Clin Nutr. 1990;52(5):878–88. 10.1093/ajcn/52.5.878 [DOI] [PubMed] [Google Scholar]
- 53.Kim L, Tsao MS. Tumour tissue sampling for lung cancer management in the era of personalised therapy: what is good enough for molecular testing? Eur Respir J. 2014;44(4):1011–22. 10.1183/09031936.00197013 [DOI] [PubMed] [Google Scholar]
- 54.Li BT, Shen R, Buonocore D, Olah ZT, Ni A, Ginsberg MS, et al. Ado-Trastuzumab Emtansine for Patients With HER2-Mutant Lung Cancers: Results From a Phase II Basket Trial. J Clin Oncol. 2018;36(24):2532–7. 10.1200/JCO.2018.77.9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol. 2016;11(10):1690–700. 10.1016/j.jtho.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 56.Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2019;380(20):1929–40. 10.1056/NEJMoa1813904 [DOI] [PubMed] [Google Scholar]
- 57.Ricciuti B. Osimertinib for EGFR-mutant non-small cell lung cancer: place in therapy and future perspectives. J Thorac Dis. 2019;11(Suppl 3):S249–S52. 10.21037/jtd.2019.01.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.FDA. FDA approves osimertinib for first-line treatment of metastatic NSCLC with most common EGFR mutations 2018 [04/19/2018]. Available from: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-osimertinib-first-line-treatment-metastatic-nsclc-most-common-egfr-mutations.
- 59.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378(2):113–25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 60.Lissa D, Robles AI. Comprehensive genomic analysis of circulating tumor DNA for patients with advanced non-small cell lung cancer. Ann Transl Med. 2019;7(5):80 10.21037/atm.2018.12.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stockley T, Souza CA, Cheema PK, Melosky B, Kamel-Reid S, Tsao MS, et al. Evidence-based best practices for EGFR T790M testing in lung cancer in Canada. Curr Oncol. 2018;25(2):163–9. 10.3747/co.25.4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pennell NA, Arcila ME, Gandara DR, West H. Biomarker Testing for Patients With Advanced Non-Small Cell Lung Cancer: Real-World Issues and Tough Choices. Am Soc Clin Oncol Educ Book. 2019;39:531–42. 10.1200/EDBK_237863 [DOI] [PubMed] [Google Scholar]
- 63.Passiglia F, Rizzo S, Di Maio M, Galvano A, Badalamenti G, Listi A, et al. The diagnostic accuracy of circulating tumor DNA for the detection of EGFR-T790M mutation in NSCLC: a systematic review and meta-analysis. Sci Rep. 2018;8(1):13379 10.1038/s41598-018-30780-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Del Re M, Tiseo M, Bordi P, D'Incecco A, Camerini A, Petrini I, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget. 2017;8(8):13611–9. 10.18632/oncotarget.6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Odogwu L, Mathieu L, Blumenthal G, Larkins E, Goldberg KB, Griffin N, et al. FDA Approval Summary: Dabrafenib and Trametinib for the Treatment of Metastatic Non-Small Cell Lung Cancers Harboring BRAF V600E Mutations. Oncologist. 2018;23(6):740–5. 10.1634/theoncologist.2017-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ou SH, Kwak EL, Siwak-Tapp C, Dy J, Bergethon K, Clark JW, et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J Thorac Oncol. 2011;6(5):942–6. 10.1097/JTO.0b013e31821528d3 [DOI] [PubMed] [Google Scholar]
- 67.Nagano T, Tachihara M, Nishimura Y. Mechanism of Resistance to Epidermal Growth Factor Receptor-Tyrosine Kinase Inhibitors and a Potential Treatment Strategy. Cells. 2018;7(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang Z, Yang N, Ou Q, Xiang Y, Jiang T, Wu X, et al. Investigating Novel Resistance Mechanisms to Third-Generation EGFR Tyrosine Kinase Inhibitor Osimertinib in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2018;24(13):3097–107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.