Abstract
In the past decade, in vitro evolution techniques have been used to improve the performance or alter the activity of a number of different enzymes and have generated enzymes de novo. In this review, we provide an overview of the available in vitro methods, their application and some general considerations for enzyme engineering in vitro. We discuss the advantages of in vitro over in vivo approaches and focus on ribosome display, mRNA display, DNA display technologies and in vitro compartmentalization methods. This review aims to help researchers determine which approach is best suited for their own experimental needs and to highlight that in vitro methods offer a promising route for enzyme engineering.
Keywords: enzyme, directed evolution, in vitro selection, ribosome display, mRNA display, DNA display, in vitro compartmentalization
Introduction
In vitro enzyme evolution offers a means to engineer enzymes by exploring enormous libraries of protein variants that exceed the capabilities of in vivo methods. The development of cell-free protein production systems made it possible to evolve enzymes outside of cells, in a test tube. In vitro evolution techniques have been used to improve existing enzymes and, in addition, have enabled the generation of biocatalysts de novo from a non-catalytic protein library.
All methods used for enzyme evolution require that each protein in a pool of mutants can be traced back to its encoding gene for identification, and potentially for the purpose of amplification, expression and further evolution [1]. A stable genotype-phenotype linkage allows for many enzyme variants to be mixed in a single reservoir while maintaining the ability to amplify genes of individual desired variants. Those variants are isolated from the reservoir using suitable screening or selection approaches. In the case of in vivo evolution methods, the genotype and phenotype are linked as the protein and its gene are contained in the same cell. With partial in vitro methods, proteins are translated by the host’s cellular machinery and then displayed in an extracellular fashion, for example on the surface of a phage in the phage display approach. In contrast, the methods described in this review are carried out entirely in vitro and do not require any step to be performed inside a host cell. The crucial genotype-phenotype link is maintained through either a direct physical link or through artificial compartmentalization. This review will discuss in vitro enzyme evolution technologies such as ribosome display, mRNA display, in vitro compartmentalization (IVC), and different versions of DNA display (Fig. 1). We will focus on protein enzymes only and therefore exclude the work on in vitro evolution of ribozymes and deoxyribozymes [2,3]. We will first describe general features common to all in vitro protein evolution strategies and then review the individual methods in more detail, highlighting examples of enzymes that have been evolved. We will also include several in vitro evolution strategies that have so far only been used for proof of principle model selections. We believe that this overview and the number of general considerations presented here will provide the reader with sufficient information to decide which of these broadly applicable in vitro techniques may be best suited for their own research. We hope to encourage scientists to harness the advantages of in vitro methods to evolve their specific enzyme of interest.
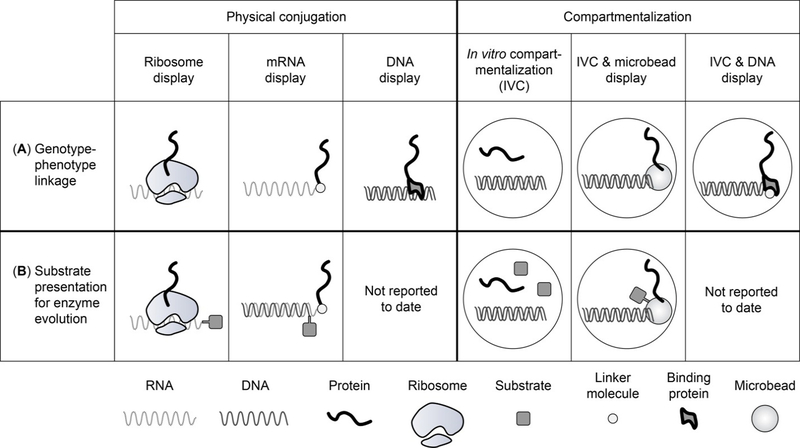
Fig. 1.
Overview of methods for the in vitro selection or screening of proteins discussed in this review. (A) The top row shows different strategies to establish the crucial linkage between gene and protein. (B) The bottom row illustrates the introduction of substrate into the selection scheme to enable the evolution of enzymes.
Benefits of in vitro evolution
In vitro methodologies have several advantages over in vivo and partial in vitro methods because they are not limited by cell survival, growth, or function. The three main advantages are: (1) the ability to work with larger libraries of variants, (2) the tolerance to conditions that would be deleterious to cell survival, and (3) the ability to directly manipulate the DNA after each round of evolution.
As in vitro evolution is not dependent on library transformation into a host, the number of unique sequences that can be evaluated in a single experiment exceeds in vivo approaches. The largest reported in vitro libraries contain 1014 DNA sequences [4]. By comparison, phage display libraries produce up to 1010 unique variants in a single transformation [5]. Library sizes up to 1012 variants were reported for phage display by the pooling of dozens of separate transformations, but such scale-up may not be feasible for most laboratories [6]. Most typical library sizes for in vivo selections are between 106 and 108 variants. Because in vitro evolution can search a larger sequence space, it is particularly well suited for isolating beneficial enzyme mutations that may be very rare.
The evolution of enzymes in vitro greatly expands the range of substrates and environmental conditions that can be investigated. The presentation of substrate to the enzymes is simplified as no cell walls have to be crossed, which are impermeable to many potential substrates. Most importantly, substrates and enzymes can be used that would be toxic to a cell [7]. Furthermore, enzymes can be engineered with in vitro methods for increased stability under extreme conditions of pH, temperature, ion concentration, or in the presence of denaturants or organic solvents. In vitro evolution also allows for a more accurate representation of enzyme performance. Cellular evolution, in contrast, can generate complex phenotypes that falsely suggest increased activity through increased enzyme accumulation, rather than improved catalysis [8].
Finally, in vitro evolution allows for direct manipulation of the DNA library between each round of evolution. Unlike in vivo methods that require time-consuming purification of the target gene, DNA from in vitro evolution is amplified directly through PCR. This facilitates the introduction of diversity through methods like error-prone PCR or in vitro recombination. In comparison, in vivo methods may introduce genetic diversity by using microbial strains deficient in DNA repair pathways to eliminate the need for DNA purification. However, these mutations may occur anywhere in the genome, necessitating a low mutation rate for continued survival [9]. Thus, by combining a selection or screen with methods to add genetic diversity, full Darwinian evolution can be carried out more conveniently in vitro.
While in vitro evolution greatly expands the tools available for the creation and engineering of new enzymes, in vivo approaches have certain advantages, too. As in vitro methods require purification of the genotype/phenotype components, in vivo evolution may involve fewer discrete steps. Furthermore, some enzymes are being developed for in vivo use, such as enzymes that need to function within a metabolic pathway. Those enzymes could initially be evolved in vitro, but ultimately need to be evolved in their native environment to optimize their intracellular compatibility. Thus, the two approaches can complement each other.
General workflow for in vitro methods
All in vitro directed evolution methods follow a similar scheme. The initial DNA library encoding the protein variants is transcribed and translated, either sequentially or in a one-pot reaction. Next, the genotype-phenotype link and then genotype-phenotype-substrate link is established. This may be accomplished through a physical connection (ribosome display, mRNA display, and DNA display), or through compartmentalization (IVC, IVC-based DNA and microbead displays) (Fig. 1). Active enzyme variants that convert substrate to product result in co-localization of genotype-phenotype with product and are then isolated by screening or selection. Finally, the genotype is recovered and either analyzed directly by sequencing or subjected to additional diversification for subsequent rounds of evolution. In the following sections, we will briefly discuss aspects of the construction of DNA libraries, present the different in vitro methods individually and discuss their application for enzyme engineering.
Library construction
In any directed evolution procedure, the size and quality of the starting DNA library are of great importance as they affect the probability of finding the desired mutant. Numerous reviews are available to guide researchers through library design and construction [27–31]. Here, we will highlight only a few general considerations and point out aspects specific to in vitro evolution methods.
Although in vitro selection methods can sift through comparably large libraries of trillions of mutants, the sheer size of the protein sequence space prevents us from sampling more than an exceedingly small fraction of all possibilities. For example, the largest protein libraries used to date contain about 1013 variants (Table 1). This vast number of mutants will just be enough to include one molecule of all possible combinations for a sequence of ten amino acid positions. In comparison, most natural proteins are more than 100 amino acids in length. Therefore, libraries of mutants should be designed wisely to increase the chances of success in a directed evolution experiment. Accordingly, one should consider randomizing specific amino acid positions by using degenerate codons. Instead of randomizing positions with NNN codons (N=A,C,G,T), NNK codons (K=G,T), NNS codons (S=C,G) or even a reduced alphabet of NDT codons (D=A,G,T) can be used to reduce oversampling caused by codon degeneracy [32]. The use of degenerate codons can also reduce the likelihood of introducing unintended stop codons. For example, the NNN codon includes three stop codons whereas the NNK or NNS codons include only one. Alternatively, a given library can be assembled from fragments that have been pre-selected to decrease the occurrence of premature stop codons [4]. More recently, DNA synthesis via phosphoramidite trinucleotides has become commercially available [33]. Codon by codon synthesis using trinucleotides offers full control of the library composition by defining the set of desired amino acid mutations at any position while avoiding stop codons (Note 1). This method is still costly but will likely become affordable in the near future [34].
Table 1.
Comparison of in vitro technologies discussed in this review.
| Method | Genotype-phenotype link | Reported variants in single experiment | Results |
|---|---|---|---|
| Ribosome display | Non-covalent complex of mRNA-ribosome-protein | ~1013 | Proof of concept selections for sialyltransferase [10], beta-lactamase [11], dihydrofolate reductase [12], DNA ligase [13] and sortase [14] |
| mRNA display | Covalent fusions of mRNA-protein via puromycin | ~1013 | Selection for de novo RNA ligase [15,16] |
| DNA display | Covalent or non-covalent complex of DNA-protein | ~1012 | Proof of concept selection of binders, but no enzymes [17,18] |
| In vitro compartmentalization (IVC) | Spatial confinement | ~109 (selection) ~106–108 (screening by FACS/microfluidics) |
Selection for methyltransferase [19] and restriction nuclease [20]; Proof of concept screening for β–galactosidase [21,22] |
| IVC & microbead display | Non-covalent complex of DNA-microbead-protein | ~109 (selection) ~106–108 (screening by FACS/microfluidics) |
Screening for phosphotriesterase [23]; Proof of concept screening for hydrogenase [24]; Proof of concept selection of biotin ligase [25] |
| IVC & DNA display | Covalent or non-covalent complex of DNA-protein | ~108–109 |
Selection of antibodies as heterodimers, but no enzymes [26] |
In order to use a DNA library for a specific in vitro evolution technique, the sequences at both termini of the DNA have to be made compatible to the method of choice. The 5’-end includes promoter and enhancer sequences necessary to facilitate transcription and translation, respectively (Note 2). The nature of these sequences depends on the type of transcription and translation system used (Note 3). Other sequence elements might be included such as a terminator, stabilizing hairpins, affinity purification tags or sequences that are specific to the particular in vitro evolution method [35] (Note 4).
Methods for in vitro enzyme evolution
Ribosome Display.
The ribosome display technology creates the genotype-phenotype linkage through a ternary complex of a stalled ribosome, the translated protein and its encoding mRNA (Fig. 1). The complex is stabilized by high magnesium concentrations and low temperatures. Ribosome display was initially described for the purification of specific mRNA sequences based on immunoprecipitation of the encoded protein [36]. Subsequently, this method was developed further to select and evolve peptides and proteins [37,38]. Although ribosome display has mostly been used for selection of binders, several model selections for enzymatic activity have been reported and will be reviewed here in more detail.
Several criteria must be met in order to generate ribosome-displayed proteins. Most importantly, the terminal stop codon of the gene of interest must be removed. This will prevent the ribosome from dissociating and releasing the nascent protein and will instead promote stalling of the ribosome and therefore maintain the ternary complex. Stem-loop structures are often added to flank the gene on both termini to increase RNA stability during translation and subsequent manipulations. Since the protein is not released from the ribosome, a C-terminal protein spacer (>100 amino acids) is added to ensure that the displayed protein has exited the protein-conducting channel of the ribosome and can fold properly. Typically, ribosome-displayed proteins are generated through sequential transcription and translation, as coupled transcription/translation systems can result in 100-fold reduced protein yield [38,39]. The translation is stopped by decreasing the temperature and increasing the Mg2+ concentration to stabilize the ternary complex. To maintain the genotype-phenotype linkage, the subsequent selection process also has to be performed at low temperatures and in presence of elevated Mg2+ concentrations. The ribosome-displayed proteins are mostly used in selections without any additional purification. The RNA is recovered after the selection by dissociating the ternary complex through chelation of Mg2+ with EDTA. A detailed protocol has been published elsewhere [40] (Note 5).
Ribosome display has been utilized in a number of model selections for enzymatic activity. Most selections were performed by selecting for binding to an immobilized substrate, substrate analog, or inhibitor. These model selections demonstrated enrichment of the desired enzyme (10 to 100-fold per round of selection) compared to an inactive control (Table 1) [10–12,14]. While enzyme selection strategies based on binding can be successful in isolating enzymes with known properties (e.g. searching through metagenomic libraries for a desired activity), they are not well suited for changing substrate specificity or substantially improving activity [41,42]. In one example of a truly product-driven model selection, ribosome display has been employed for isolation of a T4 DNA ligase [13]. Active enzymes able to ligate a DNA adaptor to the 3’-end of their encoding mRNA were selectively amplified via an adaptor-specific primer and were enriched 40-fold over known inactive mutants. Similar to this selection approach, the 3’-end of the mRNA could be used for the attachment of alternative substrates which would allow for a selection of other catalysts by ribosome display.
mRNA display.
mRNA-displayed proteins are covalently attached to their encoding mRNA via the small linker molecule puromycin (Fig. 1) [43,44]. This stable covalent link allows for the selection of proteins under a wide range of conditions. mRNA display has been used to select for a novel enzymatic activity from a non-catalytic library of randomized proteins [15,16]. This is the first example of a de novo enzyme generated by directed evolution from a naïve library. In addition, and similar to ribosome display, mRNA display has been widely employed for isolation of binders [45].
Central to the mRNA display method is the modification of the stop codon-free 3’-end of the messenger RNA with a puromycin-containing DNA linker prior to translation [46,47]. During the subsequent in vitro translation, the ribosome synthesizes the polypeptide until it reaches the DNA-puromycin-modified 3’-end of the mRNA where it stalls. Puromycin, which is an antibiotic that mimics the aminoacyl end of tRNA, enters the ribosome and becomes covalently attached to the C-terminus of the nascent polypeptide. The resulting mRNA-displayed proteins are typically purified from unfused proteins and mRNA using purification tags. The mRNA-displayed proteins are reverse transcribed to produce the cDNA. Reverse transcription also minimizes potential RNA secondary structure and increases RNA stability. Detailed protocols on mRNA display have been published recently [48,16,49]. Through slight modifications of the mRNA display protocol, covalent fusions of protein and encoding cDNA can be generated (cDNA display) [50,51].
mRNA display is the first directed evolution method that has produced an entirely artificial enzyme without a predecessor in nature. Starting from a non-catalytic protein scaffold containing two zinc fingers with each loop randomized [52], the authors isolated an RNA ligase enzyme that catalyzes the splinted ligation of a 5’-triphosphorylated RNA strand to the 3’-hydroxyl end of a second RNA [15,16]. This particular catalytic activity has not been reported in any natural enzyme. For this selection, product formation was the only selection criterion. The authors attached one of the substrates to the mRNA-displayed protein during the reverse transcription step forming a protein-mRNA-cDNA-substrate complex. The incubation with the second substrate, which was labeled with biotin, allowed any active enzymes to ligate the biotin moiety to their own cDNA enabling the selective immobilization on streptavidin beads. The isolated enzyme accelerates the reaction more than 106 fold. The ligase shows multiple turnover, although the selection scheme only requires a single catalytic event. While this example has been the only reported application of mRNA display for the isolation of enzymes to date, the general selection scheme is applicable for a wide range of bond-forming reactions. Furthermore, variations of this scheme have been proposed to apply mRNA display to the evolution of enzymes for bond-breaking and other modification reactions [53].
In vitro compartmentalization (IVC).
Directed evolution by in vitro compartmentalization mimics in vivo evolution inside a cell by using water-in-oil emulsions to enclose proteins and their encoding DNA within the same droplet compartment thereby creating the genotype-phenotype link through spatial confinement [54]. IVC has been employed not only in several model enzymes selections, but also to improve the performance of existing enzymes through screening and selection methods.
Compartmentalization by droplet formation is achieved by stirring an aqueous solution of genes and a coupled transcription/translation (TS/TL) system into a mixture of mineral oil and surfactants [55]. The DNA concentration is chosen such that the average droplet contains no more than a single gene. The low volume of the droplets (5–10 femtoliters) corresponds to a low nanomolar concentration of the single DNA molecule, which is efficiently transcribed and translated inside the droplet [54,56,22]. Although droplet composition is similar across different IVC experiments, in some cases the oil/surfactant mixtures need to be optimized for compatibility with the specific TS/TL solution used and the enzymatic activity that is being evolved [54,57]. It has been shown that the droplets are stable up to 100°C for many days and do not exchange DNA or protein between each other [54,58]. Detailed protocols for the IVC method have been published [55].
IVC-based selections have been used to evolve enzymes that process nucleic acid substrates. Here, the encoding DNA is also the substrate for the enzyme and the selection is dependent on successful DNA modification. In one approach, the activity of the methyltransferase (M.HaeIII) was improved toward a non-native, although already recognized, DNA sequence [19]. A library of variants of M.HaeIII was made by mutating the DNA contacting residues. The 3’-end of the DNA library was modified with a biotin moiety and connected to the remaining gene via the target methylation site that can be cleaved by endonuclease NheI unless the site is has been methylated by M.HaeIII. Therefore, only methylated genes were not cleaved by NheI and were captured on streptavidin beads. A similar approach was used for the model selection of a restriction endonuclease activity from a randomized library of the restriction enzyme FokI. Three specific residues were randomized in the catalytic domain, and cleavage sites for FokI were introduced in the 3’-UTR [20]. Only the genes coding for an active FokI variant were cleaved and captured on beads after incorporation of biotinylated deoxyuracil triphosphate at the cohesive ends generated by the restriction enzyme.
The IVC methodology has also been used in combination with screening approaches. This allows for the evolution of enzymes for non-nucleic acid related reactions, but also reduces the number of mutants that can be interrogated compared to selection strategies. In the screening approach, either fluorescence activated cell sorting (FACS) or microfluidics-based droplet sorting are used to separate active and inactive enzymes based on the conversion of non-fluorescent substrate into fluorescent product. For FACS mediated screening, water-in-oil-in-water emulsions (double emulsions) are generated since FACS instrumentation is incompatible with oil as the main medium [59]. Exploiting this principle, the very low β-galactosidase activity of the Ebg enzyme from E. coli was increased at least 300-fold by in vitro evolution using a commercially available fluorogenic substrate [22]. Recently, the same researchers reported a model enrichment of β-galactosidase using a home-made microfluidic system [21]. Although the throughput in the microfluidic system is about 10-fold less than in FACS-based screening, this loss is offset by other advantages. First, the microfluidic system generates highly monodisperse droplets, enabling quantitative kinetic analysis [60,21]. Second, the authors utilized microfluidic components that allowed them to fuse droplets together and introduce new content into droplets. This conferred multiple benefits as the authors were able to perform emulsion PCR in droplets and then merge them with droplets containing the TS/TL mix. By generating about 30,000 gene copies per droplet prior to TS/TL, low enzymatic activity is more likely to be detected due to the elevated enzyme concentration [21]. Furthermore, reagents can be readily added to the droplets after translation, in case the translation conditions are not compatible with enzymatic assay [61]. The use of microfluidics is a promising route for IVC-based enzyme engineering due to the modularity and potential for customization of individual components. However, in contrast to commercially available FACS instruments, assembly of microfluidics devices still requires substantial expertise.
IVC has also been used in conjunction with in vivo enzyme evolution by generating compartments that contain cells. To keep the focus of this review we are not discussing this in vivo application.
DNA display.
Strategies that either directly or indirectly establish a physical link between the DNA and the encoded protein are referred to as DNA display (Table 2). Although several different DNA display methods have been developed, only the IVC-mediated microbead display has been used to evolve enzymes. This method generates the genotype-phenotype link through the capture of DNA and its translated protein onto the same streptavidin-coated microbeads inside a droplet (Fig. 1) [23,24]. This approach requires multiple biotinylated reagents such as primers, antibody and reaction substrate in order to capture the template DNA, the protein modified with an epitope tag and the substrate onto the microbead, respectively.
Table 2.
DNA display methods. Only the microbead display has been used to evolve enzymes.
| Method | Principle of attachment | DNA - point of attachment | Protein fusion partner |
|---|---|---|---|
| Microbead display [23,25,24] | Non-covalent binding of DNA to streptavidin microbead and of HA-tagged protein via anti-HA antibody to same bead, IVC is needed | Biotinylated | HA-tag |
| STABLE [62,26] |
Non-covalent attachment of protein to DNA, IVC is needed |
Biotinylated | Streptavidin |
| CIS-display [17] |
Non-covalent attachment of protein to DNA |
RepA gene | DNA replication initiator (RepA) |
| Covalent DNA display [63,64] | Covalent attachment of enzyme to suicide inhibitor that is linked to DNA, IVC is needed | Modified with 5-fluoro-deoxycytidine |
HaeIII methyltransferase |
| Covalent antibody display [18] | Covalent attachment of enzyme to DNA |
P2A gene | Endonuclease P2A |
| SNAP display [65,66] | Covalent attachment of enzyme to suicide inhibitor that is linked to DNA, IVC is needed | Modified with benzyl guanine | SNAP-tag |
Using microbead display, Tawfik and Griffiths improved the catalytic performance of an already very efficient phosphotriesterase enzyme 63-fold (kcat > 105 s−1) through FACS-based screening [23]. This work demonstrated the ability to generate, break and regenerate the IVC droplets and purify the genotype-phenotype-product attached to the microbeads. Furthermore, a substrate was used that carried a photo-caged biotin. Therefore, the substrate stays in solution until the biotin is uncaged, which causes the immobilization of substrate and resulting product on the beads. Incubation with a fluorescent product-specific antibody enabled the specific labeling and isolation by FACS of only those microbeads to which functional enzymes and their coding DNA were attached [23].
In a different proof of concept experiment, a modified microbead display protocol was performed as a selection instead of a screen, thereby potentially harnessing larger library sizes [25]. In this experiment, an active biotin ligase was enriched from a mixture of inactive genes. Following product formation and immobilization, the purified microbeads were incubated with product-specific antibodies that were conjugated to a cleavable, gene-specific PCR primer instead of a fluorophore. Re-emulsification and droplet PCR with a solution lacking this primer resulted in a 20-fold enrichment of the desired genes.
Another microbead display model screen employing FACS used an indirect readout for activity to isolate [FeFe] hydrogenases [24]. Because the hydrogenase activity (H2 breakdown) is difficult to measure directly, the authors employed a redox-sensitive dye that can generate a fluorescent signal. Purified microbeads carrying the immobilized DNA and enzymes were re-compartmentalized in the presence of the redox dye. This dye was modified with a C12-alkyl chain and therefore interacts non-specifically with the hydrophobic polystyrene beads. Hydrogenase activity resulted in fluorescence of the dye and enabled flow cytometric sorting of the microbeads to recover the DNA of active enzymes, yielding a 20-fold enrichment over inactive genes. This proof of concept study used microfluidics to generate mono-disperse droplets and microbeads with a larger diameter (5.6 μm rather than 1 μm) to increase the bead surface allowing more fluorescent substrate to bind, thereby improving the signal to noise ratio. The indirect readout as described here could be applied to other screening strategies if environmentally sensitive fluorophores are available (pH, redox potential).
Presently, only microbead display has been employed to evolve enzymes. Yet other DNA display methods could potentially be used for this purpose. In contrast to microbead display, all other DNA display methods directly attach the protein to its encoding gene via a fusion protein which binds to a specific DNA sequence within the parent gene or to a small molecule attached to the parent gene (Table 2). The IVC method is often used in conjunction with DNA display as the physical genotype-phenotype linkage allows for the microcompartments to be broken up and generated again in order to introduce new components into the system (e.g. substrates). However, two proof-of-concept studies conducted without IVC demonstrated the production of DNA-displayed proteins solely by incubating templates with the E. coli cell extract [17,18].
General principles and comparison of different methods
In the previous section, we highlighted individual examples for the use of in vitro enzyme evolution. In this section, we will compare the different methods and discuss aspects that several methods have in common.
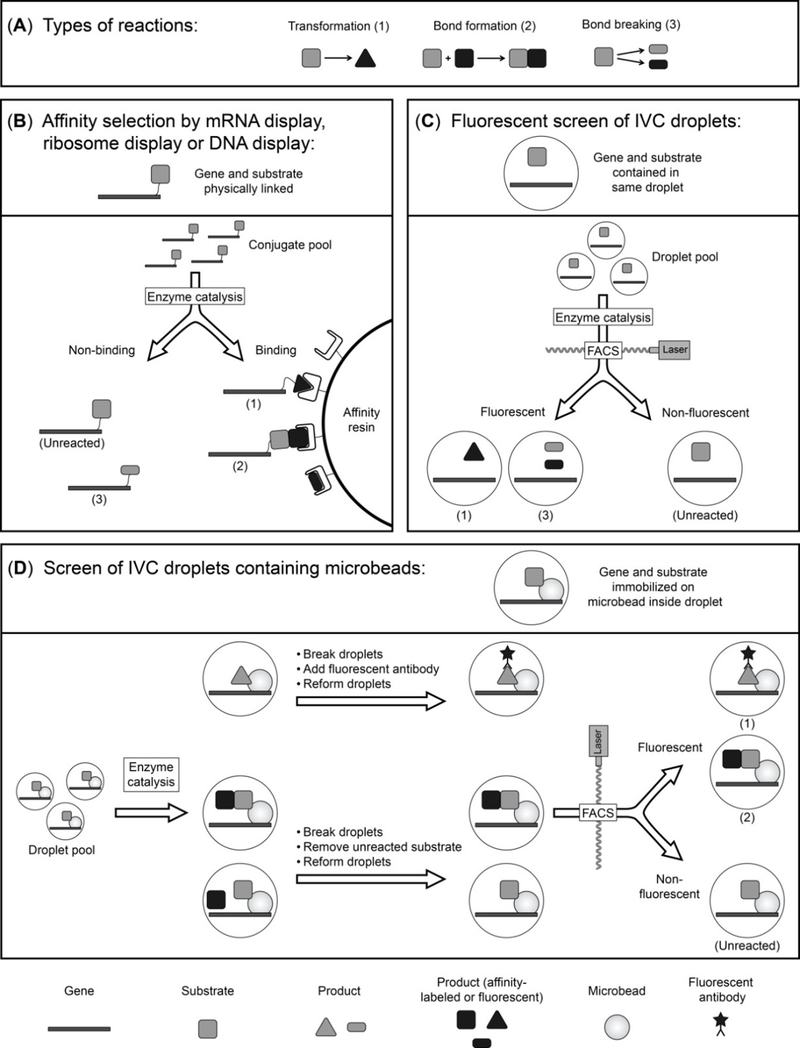
The types of reactions catalyzed by enzymes can be divided into transformation reactions, bond-forming reactions and bond-breaking reactions (Fig. 2A). Depending on the reaction type, the strategy by which enzymes can be selected varies slightly. In general, affinity selections are used to isolate enzymes by methods that create a physical link between phenotype and genotype such as ribosome display, mRNA display and DNA display (Fig. 1 and Fig. 2B). To enable an enzyme affinity selection, the substrate has to be linked to the gene-enzyme complex. Enzymes for a transformation reaction can then be isolated if a product-specific affinity reagent, such as an antibody, is available (reaction type 1). Via the antibody, the ternary complex of product, active enzyme and gene is separated from inactive variants through immobilization (Note 6). In the case of an affinity selection for bond-forming enzymes (reaction type 2), the second substrate carries a selectable moiety. Only proteins that catalyze the bond formation between two substrates will attach this moiety to the gene-protein-substrate complex and can therefore be isolated. For bond-breaking reactions (reaction type 3), the whole complex of gene, protein and substrate is immobilized via the substrate and only variants that cleave the bond will be released and selected (Note 7). In contrast to affinity selections, the IVC methodology mostly employs fluorescent screening to isolate evolved enzyme variants either by FACS or microfluidics (Fig. 2C and D). This can be achieved if the product of the reaction becomes fluorescent or a fluorescent product-specific antibody is available. Alternatively, the second substrate, which will be attached in a bond-forming reaction to the gene-microbead-substrate-complex, is fluorescent.
Fig. 2.
Isolation of enzymatic activities using in vitro technologies. (A) Types of enzymatic activities that can be evolved using in vitro approaches. (B) Affinity selection of physically linked gene-substrate/product conjugates. The enzyme itself is also linked to the gene-substrate complex, but is omitted from the figure for improved clarity. (C) Screen of IVC droplets that become fluorescent as a result of catalysis by the enzyme (not shown) contained in same compartment. Separation is achieved through fluorescence activated cell sorting (FACS) or microfluidics. (D) Screen for enzyme catalysis by FACS of IVC droplets containing microbeads. The enzyme contained in each compartment is not shown to improve clarity. Numbers in brackets refer to the type of activity as shown in (A).
For any enzyme evolution experiment regardless of which methodology is used, the specific selection or screening strategy has to be customized with respect to the underlying reaction. In the case of affinity selections, the need to link the substrate to the gene-complex without substantially changing the nature of the substrate can be challenging especially for small substrates (Notes 8-10). On the other hand, suitable fluorophores that enable the screening of IVC droplets might not be compatible with some types of chemical reactions.
Two important questions have to be considered when deciding on which enzyme evolution strategy to use: Is the desired mutant potentially very rare such as a mutant exhibiting a novel activity? Or, alternatively, is the goal of the evolution experiment to generate a highly proficient enzyme? Selection strategies can search larger libraries and are therefore more likely to discover rare mutants, compared to screening approaches. At the same time, affinity selections only select for a single turnover event and cannot evolve an enzyme for high substrate affinity as the substrate is linked to the enzyme and therefore present at a high local concentration. In contrast, IVC-based screening methods can directly evolve an enzyme for high turnover and substrate affinity, yet, the library size of screening methods is several orders of magnitude smaller than those of selections. Therefore, it might be most beneficial to combine the two strategies and first use an affinity selection method to isolate potentially rare enzyme variants with altered activity or substrate specificity and then switch to an IVC-based screening method to optimize enzymatic proficiency.
Conclusions
Enzyme engineering by in vitro enzyme evolution has made tremendous progress in the past decade. We now have a range of powerful in vitro methods available that can efficiently evolve biocatalysts in a test tube by searching protein libraries orders of magnitude larger than those used by conventional in vivo evolution approaches. In vitro enzyme evolution is uniquely suited to address two of the greatest challenges in biocatalyst design: de novo generation of novel activity, and activity within harsh environments. Applying the repertoire of in vitro evolution methods, exciting new examples of enzyme engineering are expected to emerge, thereby solving problems in biocatalysis that have previously been difficult to address.
Footnotes
NOTES:
Codon usage and compatibility. Depending on the translation system used, the codon usage can vary substantially. During library construction it is important to try to avoid rare codons that would reduce the translation yield. Furthermore, enzymes evolved using eukaryotic systems (e.g. rabbit reticulocyte) might employ codons that cause difficulty with protein expression and evolution in prokaryotes, requiring the use of strains that supplement rare/eukaryotic tRNAs.
Increasing in vitro translation yields. Translation can be controlled or improved by enhancer sequences such as a ribosome binding site for E. coli-based cell free extracts [19], by an AMV enhancer for eukaryotic systems, or a TMV translation enhancer for both eukaryotic systems and E. coli-based systems [67]. Furthermore, the optimization of translation conditions (lysate, salt, and template concentrations) can also increase the protein yield.
Translation systems. Several commercial and homemade options are available for in vitro protein translation such as E. coli, rabbit reticulocyte and wheat germ lysates. E. coli translation systems are attractive because of their low cost and high protein production but suffer from abundant nuclease contamination and simple folding machinery. Rabbit reticulocyte lysates conversely are expensive but robust in promoting proper folding and contain fewer nucleases. Wheat germ lysate provides an intermediate between rabbit reticulocyte and E. coli extracts in that it is both inexpensive and promotes folding but it requires fine optimization of ionic concentrations for each gene. Minimal reconstituted systems of purified individual components (e.g. commercially available PURE kits) have also been employed and have been shown to improve stability and efficiency of mRNA-based methods [68]. Such systems are also more amenable to unnatural amino acid incorporation. Ultimately, the nature of the in vitro method and the enzyme of interest dictate the type of lysate that might be best suited for the desired application. For review articles see [69–71].
UTR reconstitution between rounds of evolution. The 5’-UTR, and in some cases 3’-UTR [20], are lost during transcription and translation and need to be reconstituted before a subsequent round. This can be done by PCR [15], overlap extension PCR [20] or by a coupled uracil excision–ligation strategy [72].
Ribosome display variations. Several modifications to the ribosome display technology can increase the stability of the ternary complex and were recently highlighted in Methods in Molecular Biology [73]. Ribosome-inactivation display (RID) utilizes a ricin toxin to inactivate and stall the ribosome, resulting in covalent attachment of protein to the ribosome. Although use of the ricin toxin substantially increases the total gene size by about 1kb, the ribosomes no longer need to be kept at low temperature and high salt concentrations to maintain the genotype-phenotype link [74]. Alternatively, the use of translation mixtures assembled from purified components [68] or depleted of transfer-messenger RNA (tmRNA) [38] have also been used to increase yield and stability of the ternary complex by eliminating stalled ribosome rescue mechanisms.
Minimizing non-specific interactions during immobilization. Attachment of large biopolymers to the enzyme of interest (e.g. DNA-, RNA- or ribosome display) may result in non-specific interaction with the resins used during immobilization and purification, lowering the overall enrichment of the desired enzymes. With the exception of pure IVC and mRNA display, all in vitro methods require use of fusion proteins, further increasing size of the genotype-phenotype complex. Non-specific interactions with resins can be counteracted by including an excess of salmon sperm DNA, tRNA or BSA in the buffers. Recently, it was suggested to use of poly-lysine “wrappers” as coatings to mask the negatively charged RNA and minimize its impact on the outcome of selections [75]; however, this strategy has not yet been applied to in vitro evolution of enzymes. Furthermore, non-specific resin interactions can be selected against by first incubating the genotype-phenotype complex with the resin alone and then using only the flow-through for the real selection resin that is modified with the appropriate capture agents.
Counter-selections. Alternating cycles of selection and counter-selection can be used to ensure that the bond-forming or bond-breaking activity occurs at the desired site within the substrate. For example, when isolating nucleases or proteases, counter selections should be employed to improve the specificity of the enzyme towards a desired sequence within the recognition site. Previous work has shown that selections for bond-breaking activity without counter-selection can enrich for catalysts that break bonds outside of the expected region [76,77]. In one case, the selection for a peptidase resulted in the isolation of DNA nucleases instead [76].
For small molecule substrates, the site of modification may be close to the site recognized or acted upon by an enzyme. As with any directed evolution experiment, it is important to confirm that the isolated enzyme also processes the unmodified substrate.
Spacers. Attachment of the genotype, substrate, or additional protein domains to the enzyme of interest requires sufficient spacing to minimize impact of these fusions on the enzyme’s performance. Depending on the location of the protein termini, simple flexible linkers composed of GnSn [4] or rigid linkers using the (EAAAAK)n motif can be used to provide appropriate spacing [78]. Similarly, polyethylene glycol spacers or alkyl chains of varied length can be used as connectors [3].
The decision of whether to modify the substrate or employ product specific antibodies depends on the substrate size and antibody availability. For example, the use of product specific antibodies would be preferred for small molecule substrates where the derivatization may affect the binding to the enzyme. However, if the substrate is a large molecule, it may be simpler to derivatize the substrate (without affecting the enzyme’s performance) than to generate a product-specific antibody.
References
- 1.Cohen N, Abramov S, Dror Y, Freeman A (2001) In vitro enzyme evolution: the screening challenge of isolating the one in a million. Trends Biotechnol 19:507–510. [DOI] [PubMed] [Google Scholar]
- 2.Fiammengo R, Jäschke A (2005) Nucleic acid enzymes. Curr Opin Biotechol 16:614–621. [DOI] [PubMed] [Google Scholar]
- 3.Joyce GF (2004) Directed evolution of nucleic acid enzymes. Annu Rev Biochem 73:791–836. [DOI] [PubMed] [Google Scholar]
- 4.Cho G, Keefe AD, Liu RH, Wilson DS, Szostak JW (2000) Constructing high complexity synthetic libraries of long ORFs using in vitro selection. J Mol Biol 297:309–319. [DOI] [PubMed] [Google Scholar]
- 5.Kehoe JW, Kay BK (2005) Filamentous phage display in the new millennium. Chem Rev 105:4056–4072. [DOI] [PubMed] [Google Scholar]
- 6.Sidhu SS, Lowman HB, Cunningham BC, Wells JA (2000) Phage display for selection of novel binding peptides. Methods Enzymol 328:333–363. [DOI] [PubMed] [Google Scholar]
- 7.Renesto P, Raoult D (2003) From genes to proteins - in vitro expression of rickettsial proteins. Ann NY Acad Sci 990:642–652. [DOI] [PubMed] [Google Scholar]
- 8.Bulter T, Alcalde M, Sieber V, Meinhold P, Schlachtbauer C, Arnold FH (2003) Functional expression of a fungal laccase in Saccharomyces cerevisiae by directed evolution. Appl Environ Microbiol 69:987–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chusacultanachai S, Yuthavong Y (1994) Random mutagenesis strategies for construction of large and diverse clone libraries of mutated DNA fragments. Methods Mol Biol 270:319–333. [DOI] [PubMed] [Google Scholar]
- 10.Bieberich E, Kapitonov D, Tencomnao T, Yu RK (2000) Protein-ribosome-mRNA display: affinity isolation of enzyme-ribosome-mRNA complexes and cDNA cloning in a single-tube reaction. Anal Biochem 287:294–298. [DOI] [PubMed] [Google Scholar]
- 11.Amstutz P, Pelletier JN, Guggisberg A, Jermutus L, Cesaro-Tadic S, Zahnd C, Plückthun A (2002) In vitro selection for catalytic activity with ribosome display. J Am Chem Soc 124:9396–9403. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi F, Ebihara T, Mie M, Yanagida Y, Endo Y, Kobatake E, Aizawa M (2002) Ribosome display for selection of active dihydrofolate reductase mutants using immobilized methotrexate on agarose beads. FEBS Lett 514:106–110. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi F, Funabashi H, Mie M, Endo Y, Sawasaki T, Aizawa M, Kobatake E (2005) Activity-based in vitro selection of T4 DNA ligase. Biochem Biophys Res Commun 336:987–993. [DOI] [PubMed] [Google Scholar]
- 14.Quinn DJ, Cunningham S, Walker B, Scott CJ (2008) Activity-based selection of a proteolytic species using ribosome display. Biochem Biophys Res Commun 370:77–81. [DOI] [PubMed] [Google Scholar]
- 15.Seelig B, Szostak JW (2007) Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature 448:828–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seelig B (2011) mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat Protoc 6:540–552. [DOI] [PubMed] [Google Scholar]
- 17.Odegrip R, Coomber D, Eldridge B, Hederer R, Kuhlman PA, Ullman C, FitzGerald K, McGregor D (2004) CIS display: in vitro selection of peptides from libraries of protein-DNA complexes. Proc Natl Acad Sci USA 101:2806–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reiersen H, Lobersli I, Loset GA, Hvattum E, Simonsen B, Stacy JE, McGregor D, FitzGerald K, Welschof M, Brekke OH, Marvik OJ (2005) Covalent antibody display - an in vitro antibody-DNA library selection system. Nucleic Acids Res 33:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen HM, Tawfik DS, Griffiths AD (2004) Altering the sequence specificity of HaeIII methyltransferase by directed evolution using in vitro compartmentalization. Protein Eng Des Sel 17:3–11. [DOI] [PubMed] [Google Scholar]
- 20.Doi N, Kumadaki S, Oishi Y, Matsumura N, Yanagawa H (2004) In vitro selection of restriction endonucleases by in vitro compartmentalization. Nucleic Acids Res 32:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fallah-Araghi A, Baret JC, Ryckelynck M, Griffiths AD (2012) A completely in vitro ultrahigh-throughput droplet-based microfluidic screening system for protein engineering and directed evolution. Lab Chip 12:882–891. [DOI] [PubMed] [Google Scholar]
- 22.Mastrobattista E, Taly V, Chanudet E, Treacy P, Kelly BT, Griffiths AD (2005) High-throughput screening of enzyme libraries: in vitro evolution of a beta-galactosidase by fluorescence-activated sorting of double emulsions. Chem Biol 12:1291–1300. [DOI] [PubMed] [Google Scholar]
- 23.Griffiths AD, Tawfik DS (2003) Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J 22:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stapleton JA, Swartz JR (2010) Development of an in vitro compartmentalization screen for high-throughput directed evolution of [FeFe] hydrogenases. Plos One 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly BT, Griffiths AD (2007) Selective gene amplification. Protein Eng Des Sel 20:577–581. [DOI] [PubMed] [Google Scholar]
- 26.Sumida T, Doi N, Yanagawa H (2009) Bicistronic DNA display for in vitro selection of Fab fragments. Nucleic Acids Res 37:e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold FH, Georgiou G (2003) Directed evolution library creation: methods and protocols, vol 231 Methods in Molecular Biology. Humana Press Inc, Toyota, NJ. [Google Scholar]
- 28.Labrou NE (2010) Random mutagenesis methods for in vitro directed enzyme evolution. Curr Protein Pept Sci 11:91–100. [DOI] [PubMed] [Google Scholar]
- 29.Shivange AV, Marienhagen J, Mundhada H, Schenk A, Schwaneberg U (2009) Advances in generating functional diversity for directed protein evolution. Curr Opin Chem Biol 13:19–25. [DOI] [PubMed] [Google Scholar]
- 30.Denault M, Pelletier JN (2006) Protein library design and screening. Methods Mol Biol 352:127–154. [DOI] [PubMed] [Google Scholar]
- 31.Neylon C (2004) Chemical and biochemical strategies for the randomization of protein encoding DNA sequences: library construction methods for directed evolution. Nucleic Acids Res 32:1448–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reetz MT, Kahakeaw D, Lohmer R (2008) Addressing the numbers problem in directed evolution. Chembiochem 9:1797–1804. [DOI] [PubMed] [Google Scholar]
- 33.Virnekas B, Ge LM, Pluckthun A, Schneider KC, Wellnhofer G, Moroney SE (1994) Trinucleotide phosphoramidites - ideal reagents for the synthesis of mixed oligonucleotides for random mutagenesis. Nucleic Acids Res 22:5600–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janczyk M, Appel B, Springstubbe D, Fritz HJ, Muller S (2012) A new and convenient approach for the preparation of beta-cyanoethyl protected trinucleotide phosphoramidites. Org Biomol Chem 10:1510–1513. [DOI] [PubMed] [Google Scholar]
- 35.Ahn JH, Kang TJ, Kim DM (2008) Tuning the expression level of recombinant proteins by modulating mRNA stability in a cell-free protein synthesis system. Biotechnol Bioeng 101:422–427. [DOI] [PubMed] [Google Scholar]
- 36.Schechter I (1973) Biologically and chemically pure mRNA coding for a mouse immunoglobulin L-chain prepared with the aid of antibodies and immobilized oligothymidine. Proc Natl Acad Sci USA 70:2256–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattheakis LC, Bhatt RR, Dower WJ (1994) An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci USA 91:9022–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanes J, Plückthun A (1997) In vitro selection and evolution of functional proteins by using ribosome display. Proc Natl Acad Sci USA 94:4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lipovsek D, Plückthun A (2004) In vitro protein evolution by ribosome display and mRNA display J Immunol Methods 290:51–67. [DOI] [PubMed] [Google Scholar]
- 40.Zahnd C, Amstutz P, Plückthun A (2007) Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods 4:269–279. [DOI] [PubMed] [Google Scholar]
- 41.Jestin JL, Kaminski PA (2004) Directed enzyme evolution and selections for catalysis based on product formation. J Biotechnol 113:85–103. [DOI] [PubMed] [Google Scholar]
- 42.Turner NJ (2009) Directed evolution drives the next generation of biocatalysts. Nat Chem Biol 5:568–574. [DOI] [PubMed] [Google Scholar]
- 43.Roberts RW, Szostak JW (1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc Natl Acad Sci USA 94:12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H (1997) In vitro virus: bonding of mRNA bearing puromycin at the 3’-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett 414:405–408. [DOI] [PubMed] [Google Scholar]
- 45.Leemhuis H, Stein V, Griffiths AD, Hollfelder F (2005) New genotype-phenotype linkages for directed evolution of functional proteins. Curr Opin Struct Biol 15:472–478. [DOI] [PubMed] [Google Scholar]
- 46.Kurz M, Gu K, Lohse PA (2000) Psoralen photo-crosslinked mRNA–puromycin conjugates: a novel template for the rapid and facile preparation of mRNA–protein fusions Nucleic Acids Res 28:e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu RH, Barrick JE, Szostak JW, Roberts RW (2000) Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol 318:268–293. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi TT, Roberts RW (2009) In vitro selection of protein and peptide libraries using mRNA display. Methods Mol Biol 535:293–314. [DOI] [PubMed] [Google Scholar]
- 49.Cotten SW, Zou JW, Valencia CA, Liu RH (2011) Selection of proteins with desired properties from natural proteome libraries using mRNA display. Nat Protoc 6:1163–1182. [DOI] [PubMed] [Google Scholar]
- 50.Kurz M, Gu K, Al-Gawari A, Lohse PA (2001) cDNA - Protein fusions: covalent protein-gene conjugates for the in vitro selection of peptides and proteins. Chembiochem 2:666–672. [DOI] [PubMed] [Google Scholar]
- 51.Ueno S, Nemoto N (2012) cDNA display: rapid stabilization of mRNA display. Methods Mol Biol 805:113–135. [DOI] [PubMed] [Google Scholar]
- 52.Cho GS, Szostak JW (2006) Directed evolution of ATP binding proteins from a zinc finger domain by using mRNA display. Chem Biol 13:139–147. [DOI] [PubMed] [Google Scholar]
- 53.Golynskiy MV, Seelig B (2010) De novo enzymes: from computational design to mRNA display. Trends Biotechnol 28:340–345. [DOI] [PubMed] [Google Scholar]
- 54.Tawfik DS, Griffiths AD (1998) Man-made cell-like compartments for molecular evolution. Nat Biotechol 16:652–656. [DOI] [PubMed] [Google Scholar]
- 55.Miller OJ, Bernath K, Agresti JJ, Amitai G, Kelly BT, Mastrobattista E, Taly V, Magdassi S, Tawfik DS, Griffiths AD (2006) Directed evolution by in vitro compartmentalization. Nat Methods 3:561–570. [DOI] [PubMed] [Google Scholar]
- 56.Bernath K, Hai MT, Mastrobattista E, Griffiths AD, Magdassi S, Tawfik DS (2004) In vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal Biochem 325:151–157. [DOI] [PubMed] [Google Scholar]
- 57.Ghadessy FJ, Holliger P (2004) A novel emulsion mixture for in vitro compartmentalization of transcription and translation in the rabbit reticulocyte system. Protein Eng Des Sel 17:201–204. [DOI] [PubMed] [Google Scholar]
- 58.Ghadessy FJ, Ong JL, Holliger P (2001) Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci USA 98:4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenstein M (2006) Tiny droplets make a big splash. Nat Methods 3:71. [DOI] [PubMed] [Google Scholar]
- 60.Song H, Ismagilov RF (2003) Millisecond kinetics using nanoliters of reagents. J Am Chem Soc 125:14613–14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazutis L, J.C. B, Treacy P, Skhiri Y, Fallah Araghi A, Ryckelynck M, Taly V, Griffiths AD(2009) Multi-step microfluidic droplet processing: kinetic analysis of an in vitro translated enzyme Lab Chip 9:2902–2908. [DOI] [PubMed] [Google Scholar]
- 62.Doi N, Yanagawa H (1999) STABLE: protein-DNA fusion system for screening of combinatorial protein libraries in vitro. FEBS Lett 457:227–230. [DOI] [PubMed] [Google Scholar]
- 63.Bertschinger J, Neri D (2004) Covalent DNA display as a novel tool for directed evolution of proteins in vitro. Protein Eng Des Sel 17:699–707. [DOI] [PubMed] [Google Scholar]
- 64.Bertschinger J, Grabulovski D, Neri D (2007) Selection of single domain binding proteins by covalent DNA display. Protein Eng Des Sel 20:57–68. [DOI] [PubMed] [Google Scholar]
- 65.Stein V, Sielaff I, Johnsson K, Hollfelder F (2007) A covalent chemical genotype-phenotype linkage for in vitro protein evolution. Chembiochem 8:2191–2194. [DOI] [PubMed] [Google Scholar]
- 66.Kaltenbach M, Stein V, Hollfelder F (2011) SNAP dendrimers: multivalent protein display on dendrimer-like DNA for directed evolution. Chembiochem 12:2208–2216. [DOI] [PubMed] [Google Scholar]
- 67.Gallie DR, Kado CI (1989) A translational enhancer derived from Tobacco Mosaic-Virus is functionally equivalent to a Shine-Dalgarno sequence. Proc Natl Acad Sci USA 86:129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villemagne D, Jackson R, Douthwaite JA (2006) Highly efficient ribosome display selection by use of purified components for in vitro translation. J Immunol Methods 313:140–148. [DOI] [PubMed] [Google Scholar]
- 69.Jagus R, Joshi B, Miyamoto S, Beckler GS (1998) In vitro translation Curr Protoc Cell Biol. John Wiley & Sons, Inc:11.12.11–11.12.25. [DOI] [PubMed] [Google Scholar]
- 70.Hillebrecht JR, Chong SR (2008) A comparative study of protein synthesis in in vitro systems: from the prokaryotic reconstituted to the eukaryotic extract-based. BMC Biotechnol 8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He MY, He YZ, Luo Q, Wang MR (2011) From DNA to protein: no living cells required. Process Biochem 46:615–620. [Google Scholar]
- 72.Stein V, Hollfelder F (2009) An efficient method to assemble linear DNA templates for in vitro screening and selection systems. Nucleic Acids Res 37:e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Douthwaite JA, Jackson RH (2011) Ribosome display and related technologies: methods and protocols, vol 805 Methods in Molecular Biology. Humana Press, New York. [Google Scholar]
- 74.Fujita S, Zhou JM, K T (2007) Ribosome-Inactivation display system. Methods Mol Biol 352:221–236. [DOI] [PubMed] [Google Scholar]
- 75.Lamboy JA, Tam PY, Lee LS, Jackson PJ, Avrantinis SK, Lee HJ, Corn RM, Weiss GA (2008) Chemical and genetic wrappers for improved phage and RNA display. Chembiochem 9:2846–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chandra M, Sachdeva A, Silverman SK (2009) DNA-catalyzed sequence-specific hydrolysis of DNA. Nat Chem Biol 5:718–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang TP, Su YC, Chen Y, Liou YM, Lin KL, Wang EC, Hwang LC, Wang YM, Chen YH (2012) In vitro selection and characterization of a novel Zn(II)-dependent phosphorothiolate thiolesterase ribozyme. Biochem 51:496–510. [DOI] [PubMed] [Google Scholar]
- 78.Yonezawa M, Doi N, Higashinakagawa T, Yanagawa H (2004) DNA display of biologically active proteins for in vitro protein selection. J Biochem 135:285–288. [DOI] [PubMed] [Google Scholar]