The initiation of mitochondrial protein synthesis fine-tunes the assembly of respiratory complexes and energy production.
Abstract
Mammalian mitochondrial ribosomes are unique molecular machines that translate 11 leaderless mRNAs; however, it is not clear how mitoribosomes initiate translation, since mitochondrial mRNAs lack untranslated regions. Mitochondrial translation initiation shares similarities with prokaryotes, such as the formation of a ternary complex of fMet-tRNAMet, mRNA and the 28S subunit, but differs in the requirements for initiation factors. Mitochondria have two initiation factors: MTIF2, which closes the decoding center and stabilizes the binding of the fMet-tRNAMet to the leaderless mRNAs, and MTIF3, whose role is not clear. We show that MTIF3 is essential for survival and that heart- and skeletal muscle–specific loss of MTIF3 causes cardiomyopathy. We identify increased but uncoordinated mitochondrial protein synthesis in mice lacking MTIF3, resulting in loss of specific respiratory complexes. Ribosome profiling shows that MTIF3 is required for recognition and regulation of translation initiation of mitochondrial mRNAs and for coordinated assembly of OXPHOS complexes in vivo.
INTRODUCTION
Mitochondria are the product of the endosymbiosis between an ancestral alpha-proteobacterium and a proto-eukaryotic cell, and the circular mitochondrial genome is a relic of this union. The mitochondrial DNA (mtDNA) encodes a small subset of proteins that are components of the oxidative phosphorylation (OXPHOS) system, vital for energy production. Consequently, mitochondria have to import all the proteins that they require for gene expression and protein synthesis, including mitochondrial ribosomal proteins and translation factors that are essential for the production of the mtDNA-encoded proteins. Therefore, nuclear and mitochondrial gene expression require tight coordination to enable correct assembly and function of the electron transport chain.
In animals, the gene organization is compact within the mtDNA and lacks introns; the genome encodes 2 ribosomal RNAs (rRNAs) required for the assembly of the mitochondrial ribosomes and 22 transfer RNAs (tRNAs) necessary for the synthesis of the 13 OXPHOS polypeptides that are encoded on 11 leaderless mRNAs (1). Mitochondrial protein synthesis in animals is unique in that the ribosomes have acquired mitochondria-specific proteins (2–4) and translate mRNAs that lack conventional 5′ and 3′ untranslated regions (UTRs) and Shine-Dalgarno sequences found in bacterial mRNAs (5). Because mitochondrial mRNAs (mt-mRNAs) lack UTRs, it is still not clear how mitoribosomes dock and recognize their start codons, suggesting alternative mechanisms to translate leaderless mRNAs exist. It is likely that mS39 (PTCD3) via its pentatricopeptide repeats interacts with the incoming mRNAs and facilitates their recruitment to the small subunit (6, 7). Nevertheless, protein synthesis in mitochondria is overall similar to bacteria, beginning with an initiation phase, followed by polypeptide elongation, termination, and ribosome recycling.
Translation initiation is the rate-limiting and most highly regulated phase in protein synthesis. In bacteria, translation initiation begins with the disassociation of the ribosome into its small (SSU) and large (LSU) subunits, and then in the presence of mRNA, formylated tRNAMet (fMet-tRNAMet), and three initiation factors IF1, IF2, and IF3, the initiation complex can form to initiate protein synthesis (8). Mitochondria use only two initiation factors, MTIF2 and MTIF3, where a 37–amino acid insertion in MTIF2 serves the role of IF1, allowing it to substitute its function (9), as it can interact with the same ribosomal site (6, 10).
Ribosome dissociation marks the beginning of translation initiation, and in bacteria, this is catalyzed by elongation factor EF-G and ribosomal recycling factor RRF (11). To prevent the reassembly of the two subunits, bacterial IF3 passively binds to the SSU and thereby begins the process of initiation complex formation (12). The ability of IF3 to enhance subunit dissociation is attributed to the specific sites that this factor occupies on the 16S rRNA, which are essential binding sites for the 50S subunit (13). In contrast, little is known about the function of MTIF3. In vitro systems using purified MTIF3 have proposed that initiation of translation in mitochondria could begin with MTIF3 binding to the 28S SSU of the mitoribosome, facilitating the active disassociation of the subunits (14). Mammalian MTIF3 has diverged considerably from its eubacterial counterpart (15), and the active role of MTIF3 in subunit dissociation is attributed to features of its N- and C-terminal extensions (16). One of the essential roles of bacterial IF3 is its ability to proofread the codon-anticodon interaction between the mRNA and initiator tRNA at the P site, and it subsequently inhibits initiation on noncanonical start codons (17). Currently, it is not known if MTIF3 plays a similar role in proofreading during mitochondrial translation initiation or if MTIF3 can destabilize the initiation complex in the absence of mRNA binding.
To understand the physiological function of mammalian MTIF3 in vivo, we generated conditional Mtif3 knockout mice, because we identified that constitutive knockout of Mtif3 is embryonic lethal. We show that MTIF3 is essential for coordinated translation initiation and its loss reduces specific respiratory complexes, compromising OXPHOS function, which causes dilated cardiomyopathy.
RESULTS
Loss of MTIF3 leads to dilated cardiomyopathy
A conditional knockout allele of the mouse Mtif3 gene was generated by flanking exon 3 with loxP sites in embryonic stem cells (fig. S1A). Mice with germline transmission of this allele (Mtif3+/loxP-neo) were crossed with transgenic mice expressing the Flp recombinase to excise the neomycin cassette (fig. S1A). Mice expressing Cre recombinase under the control of the β-actin promoter were crossed to Mtif3+/loxP mice to generate heterozygous Mtif3 knockout mice (Mtif3+/−). We intercrossed the Mtif3+/− mice for 6 months and only observed Mtif3+/− and Mtif3+/+ mice in Mendelian proportions, but not homozygous knockout mice (Mtif3−/−), suggesting that these mice were not viable. Embryonic lethality has been identified previously for many other proteins involved in regulating mitochondrial gene expression (18–22). We analyzed the embryos from intercrossed Mtif3+/− mice and found that the loss of MTIF3 in mice caused embryonic lethality at E8.5 (Fig. 1B), indicating that MTIF3 is required for survival.
Fig. 1. Heart and skeletal muscle conditional knockout of mouse Mtif3 causes cardiomyopathy.
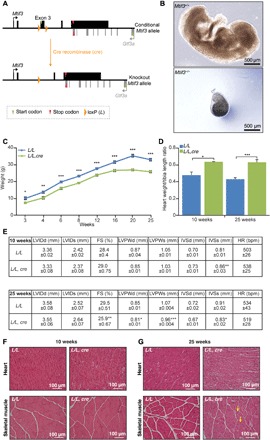
(A) Schematic showing the homologous recombination at the Mtif3 locus to generate conditional knockout mice. LoxP sites were introduced to allow the deletion of exon 3 by Cre recombinase. (B) Development of embryos of Mtif3+/+ and constitutive Mtif3−/− mice at embryonic day 8.5 (E8.5). (C) Weight differences between control (L/L) and knockout (L/L, cre) mice from 3 to 25 weeks of age. (D) Heart weight–to–tibia length ratio in control (L/L) and knockout (L/L, cre) mice at 10 and 25 weeks. (E) Echocardiographic parameters for control (L/L), and knockout (L/L, cre), 10- and 25-week-old mice. LVIDd, left ventricular end diastolic diameter; LVIDs, left ventricular end systolic diameter; FS, fractional shortening; LVPWd, left ventricular posterior wall in diastole; LVPWs, left ventricular posterior wall in systole; IVSd, intraventricular septum in diastole; IVSs, intraventricular septum in systole; HR, heart rate. (F) Heart and skeletal muscle sections cut to 5-μm thickness from 10-week-old and (G) 25-week-old L/L and L/L, cre mice were stained with H&E; yellow arrows show centralized nuclei in the skeletal muscle. Scale bars, 100 μm. All values are means ± SEM of n = 5. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
To produce heart- and skeletal muscle–specific Mtif3 knockout mice (Mtif3loxP/loxP, cre+), we crossed Mtif3+/loxP mice with transgenic mice expressing Cre recombinase under the control of the muscle creatinine kinase promoter (Ckmm-cre) (Fig. 1A and fig. S1A). The heart- and skeletal muscle–specific MTIF3 knockout mice (L/L, cre) develop a heart defect by 25 weeks compared to the control (L/L) mice and lack MTIF3 (fig. S1B). The knockout mice are significantly lighter compared to the control mice from 3 weeks of age to 25 weeks when they have to be sacrificed for ethical reasons because they develop severe cardiomyopathy (Fig. 1C), determined by increased heart weight/tibial length (Fig. 1D) and echocardiography (Fig. 1E). Echocardiography of the 25-week-old knockout mice showed a significant decrease in their fractional shortening and a significant decrease in left ventricular posterior wall (LVPW) and intraventricular septum (IVS) (thinning of the heart walls) compared to control mice (Fig. 1E). Hematoxylin and eosin (H&E) staining of the 10- and 25-week-old knockout mice showed cellular disarray and necrotic foci, further confirming the development of dilated cardiomyopathy in the absence of MTIF3 (Fig. 1, F and G). In addition, we identified decreased size of specific fibers and centralized nuclei in the skeletal muscle of 25-week-old knockout mice compared to controls by H&E staining (Fig. 1G).
Loss of MTIF3 leads to dysregulated mitochondrial RNA metabolism
We investigated mitochondrial RNA (mtRNA) metabolism using northern blotting and show that loss of MTIF3 caused specific changes in mtRNAs in both the heart (fig. S2A) and skeletal muscle (fig. S3A) compared to controls in both 10- and 25-week-old mice. Specific mtRNAs such as mt-Nd3, mt-Co2, and mt-Co3 were reduced, whereas 12S rRNA and Atp8/6 were increased and the remaining transcripts were unaffected (fig. S2A). We corroborated these findings by measuring the abundance of mature mRNAs and rRNAs in the hearts and skeletal muscle of control and knockout mice by quantitative reverse transcription polymerase chain reaction (qRT-PCR) (figs. S2B and S3B).
Next, we carried out RNA sequencing (RNA-Seq) to analyze the effects of MTIF3 loss on the entire mitochondrial transcriptome, as we have done previously (18, 19, 23). RNA-Seq analyses revealed that the 5′ regions of mitochondrial transcripts were substantially increased (Fig. 2A). We identified that the stability of mt-mRNAs progressively decreased in a 5′-3′ orientation such that the read coverage at 5′ mRNA ends was increased and the 3′ ends were decreased in the knockout mice (Fig. 2B), suggesting that MTIF3 might be required for efficient translation, and therefore protection, of mt-mRNAs.
Fig. 2. Transcriptome analysis of mitochondrial transcripts by RNA-Seq.
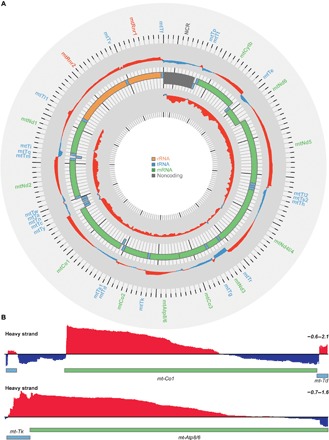
(A) Complete map of changes in mitochondrial transcript abundance determined by RNA-Seq coverage from control (L/L) and knockout (L/L, cre) mice on heavy (outer track) and light (inner track) strands. Increases are shown in red, and decreases are shown in blue (log2[RPMKO/RPMWT]; scale, −1.0 to 2.0). The mitochondrial genome is displayed in the central track, with the nucleotide position in base pairs displayed across the exterior; rRNAs are displayed in orange, mRNAs are in green, tRNAs are in blue, and the noncoding region (NCR) is in gray. (B) Genome browser view of the mean RNA-Seq coverage (log2[RPMKO/RPMWT] of Co1 and Atp8/6 mRNAs (scale, −0.6 to 2.1).
MTIF3 is required for coordinated translation of mt-mRNAs and biogenes of the OXPHOS system
We measured de novo mitochondrial protein synthesis of the 13 mtDNA-encoded respiratory complex subunits in 10- and 25-week-old knockout and control mice and identified a marked increase in translation levels in the absence of MTIF3 (Fig. 3, A and B, and fig. S3C). Upon closer inspection, we identified that translation of specific mitochondrial proteins such as Cyt b, COXI, and ND1 was significantly increased, whereas the levels of others such as ND3 were reduced. This is consistent with the decreased levels of mt-Nd3 (fig. S2), indicating that, in the absence of MTIF3, this mRNA is not efficiently translated and therefore its stability is reduced, possibly because of its noncanonical start codon or short length. In addition, there were specific lower–molecular weight proteins that were decreased in the knockout mice but not in control mice, suggesting that their stability was reduced. Despite the increased de novo protein synthesis, chase labeling revealed reduced stability of the newly synthesized mitochondrial proteins in the 25-week-old knockout mice (Fig. 3B). We pretreated mitochondria with puromycin to measure the rate of translation initiation and found that the rate of de novo protein synthesis initiation from 5 to 60 min was increased in the absence of MTIF3 (fig. S4A). These findings suggest that MTIF3 is required for coordinating the rate of translation of mt-mRNAs.
Fig. 3. Loss of MTIF3 results in uncoordinated mitochondrial protein synthesis.
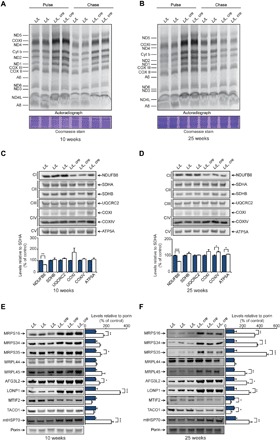
(A) Levels of de novo protein synthesis were measured in heart mitochondria from control (L/L) and knockout (L/L, cre) 10-week-old mice by pulse and chase incorporation of 35S-labeled cysteine and methionine. Mitochondrial protein was separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE), stained with Coomassie, and visualized by autoradiography. Representative gels of five independent biological experiments are shown. (B) Levels of de novo protein synthesis in heart mitochondria from 25-week-old control (L/L) and knockout (L/L, cre) mice, determined as in (A). Mitochondrial proteins from isolated heart mitochondria of control (L/L) and knockout (L/L, cre) 10-week-old (C) and 25-week-old (D) mice were resolved on 4 to 20% SDS-PAGE gels and immunoblotted using antibodies to investigate the steady-state levels of OXPHOS proteins. SDHA was used as a loading control. Levels of mitoribosomal proteins, proteases, and MTIF2 proteins from isolated heart mitochondria of control (L/L) and knockout (L/L, cre) 10-week-old (E) and 25-week-old (F) mice. Porin was used as a loading control. All values are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test.
Immunoblotting of mitochondria and nuclear-encoded OXPHOS subunits showed specific decrease of NDUFB8, the nuclear-encoded subunit of complex I, in both 10- and 25-week-old knockout mice (Fig. 3, C and D). The level of the mitochondrially encoded COXI subunit was not different between the control and knockout mice (Fig. 3, C and D). Similarly, the levels of the complex II subunits SDHA and SDHB were not different between control and knockout mice (Fig. 3, C and D). These findings were corroborated in skeletal muscle (fig. S3D). Immunoblotting of nuclear-encoded proteins that are required for mitochondrial gene expression showed that loss of MTIF3 increases the levels of mitoribosomal proteins (Fig. 3, E and F), likely in an attempt to cope with impaired mitochondrial translation initiation. Increased levels of mitochondrial ribosomal proteins likely contribute to the increased levels of mitochondrial proteases and the chaperone mtHSP70 also identified in the knockout mice (Fig. 3, E and F). However, MTIF2 was significantly reduced in the absence of MTIF3 (Fig. 3, E and F), indicating that these factors depend on each other for their stability. Mass spectrometry analyses of the mitochondrial proteomes in control and knockout mice revealed that complex I subunits were specifically reduced in both 10- and 25-week-old knockout mice (Fig. 4A and fig. S5). In addition, we confirmed that mitoribosomal proteins and translation factors were increased in the 10- and 25-week-old knockout mice (Fig. 4A and fig. S5).
Fig. 4. Loss of MTIF3 affects the stability and assembly of the mitochondrial OXPHOS complexes.
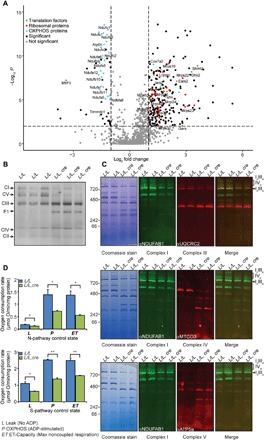
(A) Quantitative proteomic analysis of mitochondrial proteins from control (L/L) and knockout (L/L, cre) 25-week-old mice. (B) Isolated heart mitochondria from 25-week-old mice were treated with 1% n-dodecyl-β-d-maltoside, resolved on 4 to 16% native bis-tris gels, and immunoblotted with the blue native OXPHOS cocktail antibody. (C) Isolated heart mitochondria from 25-week-old mice were treated with 1% digitonin, resolved on 4 to 16% native bis-tris gels, and immunoblotted to show complexes I and III (upper panels), complexes I and IV (middle panels), and complexes I and V (lower panels). (D) Oxygen consumption through the N-pathway and S-pathway using either pyruvate, glutamate, malate, or succinate as substrates in the absence or presence of mitochondrial inhibitors, and 2 μM carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) was measured for leak (L), OXPHOS capacity (P), and ET capacity (ET) states in heart mitochondria from control (L/L) and knockout (L/L, cre) 25-week-old mice using an Oroboros oxygen electrode. All values are means ± SEM. *P < 0.05, **P < 0.01, Student’s t test.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) followed by immunoblotting further indicated that loss of MTIF3 affected the assembly and stability of the proton-translocating OXPHOS complexes, and increased the levels of complex II (Fig. 4B). There was disassembly of complex V and the release of the ATPase (adenosine triphosphatase) inhibitory factor 1 (ATPIF1) (Fig. 4B), which binds to β-F1-ATPase in response to cellular and pH changes and inhibits the hydrolysis of adenosine triphosphate (ATP) to conserve energy use and is typically found in other models of severe OXPHOS dysfunction (24). Because we observed increased levels of complex III in the knockout mice using DDM to solubilize the individual OXPHOS complexes, we investigated the effects of MTIF3 loss on supercomplex assembly and the association of complex III within the respirasome. We used 1% digitonin to solubilize mitochondria and show that the supercomplexes as well as subcomplexes containing complexes I, III, and IV are significantly reduced in the knockout mice compared to controls (Fig. 4C). Furthermore, we showed that complex V is significantly reduced (Fig. 4C). De novo 35S labeling of OXPHOS complexes resolved by BN-PAGE and visualized by autoradiography showed that, in the absence of MTIF3, the rate of mitochondrial translation is not coordinated with the assembly of the OXPHOS complexes, resulting in reduced stability of de novo assembled OXPHOS complexes (fig. S4B). We resolved mitochondrial lysates on sucrose gradients to detect the distribution of the respiratory complexes in the control and knockout mice. We identify that only smaller subcomplexes of complexes I, V, and IV are found in the earlier, less sucrose-dense fractions of the gradient in the knockout mice compared to the control mice (fig. S4C). These findings indicate that the assembly of these complexes is impaired in the absence of MTIF3, indicating that the rate of mitochondrial protein synthesis is regulated by MTIF3, and this regulation is required for the coordinated assembly of nuclear and mitochondrial OXPHOS polypeptides into the OXPHOS complexes.
Consistent with impaired assembly of the OXPHOS system, mitochondrial oxygen consumption for both the N-pathway that delivers electrons from NADH (reduced form of nicotinamide adenine dinucleotide) to complex I and the S-pathway that delivers electrons from succinate to complexes II and III was significantly decreased in the leak (L), OXPHOS capacity (P), and electron transport (ET) capacity (ET) states in the knockout mice compared to control mice (Fig. 4D). These findings show that MTIF3 regulates the rate of mitochondrial protein synthesis, and this is necessary for the coordinated assembly of nuclear and mitochondrial OXPHOS polypeptides and consequently OXPHOS function.
Mitochondrial ribosome assembly is not affected by loss of MTIF3, but the engagement with mt-mRNAs is reduced
To investigate the effects of MTIF3 loss on mitochondrial ribosome assembly, we resolved the ribosomal subunits and translating ribosomes on 10 to 30% sucrose gradients and immunoblotted against specific mitoribosomal proteins that make up the small and large ribosomal subunits. Loss of MTIF3 did not impair the assembly of mitochondrial ribosomes or their subunits (Fig. 5A). Instead, we identified reduced association of mature mRNAs with translating ribosomes in the knockout mice when we measured the distribution of the mRNAs in the gradients by qRT-PCR (Fig. 5B). We also identified a shift in the small ribosomal subunit from fraction 7 to fraction 6 in the knockout mice (Fig. 5A), possibly resulting from the loss of MTIF3 and reduction in the levels of MTIF2 that associate with this subunit (6). We confirmed this shift when we measured the distribution of the rRNAs and mRNAs within the gradients by qRT-PCR, where the levels of the 12S rRNA were also found in fraction 6 consistent with the immunoblot results (Fig. 5B).
Fig. 5. MTIF3 does not affect mitochondrial ribosome assembly but alters the interactions between mitoribosomes and mt-mRNAs.
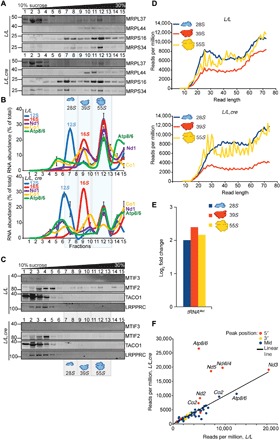
(A) Mitochondrial ribosome subunits and translating ribosomes were resolved on 10 to 30% sucrose gradients in control (L/L) and knockout (L/L, cre) 25-week-old mice. MTIF2 and the mitochondrial ribosomal protein markers of the small (MRPS34 and MRPS16) and large (MRPL44 and MRPL37) ribosomal subunits were detected by immunoblotting. The data are representative of results from four independent biological experiments. (B) The distributions of the 12S and 16S rRNAs and mRNAs in sucrose gradients were analyzed by qRT-PCR. The data are expressed as a percentage total of RNA abundance and show results from three independent biological replicates. (C) Mitochondrial ribosome subunits and translating ribosomes were resolved on 10 to 30% sucrose gradients in control (L/L) and knockout (L/L, cre) 25-week-old mice. MTIF3, MTIF2, TACO1, and LRPPRC were detected by immunoblotting. The data are representative of results from four independent biological experiments. (D) The normalized read length profile (reads per million) of the mitoribosome and its subunits on mRNAs is altered in the absence of MTIF3. (E) The association of tRNA-Met with the mitoribosome and its subunits is increased in the absence of MTIF3. (F) Enriched peaks at the 5′ end of mt-Atp8/6, mt-Nd4l/4, and mt-Nd5 mRNAs associating with the translating ribosome indicate ribosomal stalling. Peaks were considered 5′ or 3′ if they partially or fully overlapped a 100-nt window centered on the transcript 5′ or 3′ ends.
MTIF3 has been found closely associated with MTIF2 and the mitoribosome (6, 25); therefore, we investigated the association of these two factors with mitoribosomes and how the loss of MTIF3 would affect the association of MTIF2 with the mitoribosome in the sucrose gradients. Although MTIF3 was absent in the gradient from the Mtif3 knockout mice, we found that it localized to the upper fractions but not mitoribosomal fractions of the sucrose gradient in the control mice, indicating that its interactions with the SSU are transient in vivo (Fig. 5C). MTIF2 migrated with the mitoribosomal fractions as well as some earlier fractions; however, the association with the translating ribosomes was markedly lost in the Mtif3 knockout mice (Fig. 5C). We also identified that the levels of the translational activator of cytochrome oxidase 1 (TACO1) were reduced (Figs. 3F and 5C) in the knockout mice and that the low levels associated with mitoribosomes were lost, indicating that uncoordinated translation affects the interactions of the mitoribosome with its protein synthesis factors. The levels and distribution of the leucine-rich pentatricopeptide repeat-containing protein (LRPPRC) were not changed between the knockout and control mice, likely because LRPPRC is required to relax the secondary structures of mRNAs (26) and to facilitate poly(A) tail maintenance (21).
MTIF3 is required for correct preinitiation complex formation revealed by mitoribosome footprinting
To identify the molecular role of MTIF3 in mitochondrial protein synthesis, we carried out mitoribosomal profiling followed by limited ribonuclease (RNase) T1 digestion of native complexes and subsequent RNA-Seq of preserved RNA footprints (27, 28). We identified a broad range of mRNA fragment sizes associated with mitoribosomal subunits and translating mitoribosomes whose distributions shifted in response to loss of MTIF3 (Fig. 5D). Most notably, there was a shift in the size of mRNA footprints that associated with the 28S subunit from 25 to 40 nucleotides (nt) to 35 to 55 nt in the Mtif3 knockout mice (Fig. 5D and fig. S6, A to C), suggesting that MTIF3 is required for efficient scanning of mt-mRNAs and 55S formation. In addition, we identified that MTIF3 is required for the removal of the tRNAMet that associates with the 28S subunit in the absence of mRNAs, as we found persistent accumulation of this tRNA in the ribosomal subunits of the Mtif3 knockout mice (Fig. 5E) compared to other tRNAs (fig. S6D). This suggests that MTIF3 surveys the preinitiation translation complex formation to ensure that mRNAs are bound before the binding of fMet-tRNAMet. We identified enriched footprints from actively translating ribosomes at the 5′ ends, predominantly of mt-Atp8/6, mt-Nd4l/4, and mt-Nd5, indicating ribosomal stalling at these sites in the absence of MTIF3 (Fig. 5F).
Next, we compared the differential enrichment of footprints from the small and large ribosomal subunits to the actively translating mitoribosome between the control and knockout mice and identified the differences between them (Fig. 6A). There was increased mitoribosome stalling at specific mRNAs, most notably mt-Atp8/6, mt-Nd4l/4, and mt-Nd5. This increase was also notable in the SSU fractions, suggesting that there is increased association of the 28S subunit with these mRNAs in the absence of MTIF3. On the basis of our findings, we suggest that MTIF3 regulates the order of the preinitiation complex formation (Fig. 6B). MTIF3 is required for the removal of the fMet-tRNAMet from the SSU if an mRNA has not been bound by this subunit. Once the 28S subunit has associated with an mRNA, MTIF3 accommodates the mRNA, with transcript-specific affinities, at which point MTIF2 and fMet-tRNAMet are delivered to the subunit and MTIF3 is displaced from the preinitiation complex to stimulate association with the 39S subunit and initiation of protein synthesis. In the absence of MTIF3, there is increased association of the SSU with both fMet-tRNAMet and specific mRNAs and reduced association of mRNAs with the large subunit (Fig. 6B, lower panel). We propose that MTIF3 is required for molecular proofreading, and without it, translation proceeds at an accelerated rate that outcompetes the rate of OXPHOS assembly. The increased rate of protein synthesis is at the expense of fidelity of translation initiation that can result installing at the 5′ ends of mRNAs, exposing the remaining portions of the mRNA, leaving them prone to degradation (Fig. 6C).
Fig. 6. The mechanism of MTIF3 and its role in mRNA recognition and translation initiation.
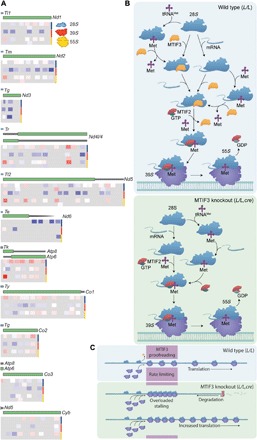
(A) Mitochondrial ribosome profiling shows the distribution of footprints on mt-mRNAs in the SSU (first two tracks), large subunit (middle two tracks), and mitoribosome (bottom two tracks) that are enriched (red) or depleted (blue) in the knockout compared to control mice. Double tracks per ribosomal fraction were used to distinguish partially overlapping footprints on the mRNAs. (B) Schematic showing the proposed roles of MTIF3 in mitochondrial translation (wild type, L/L). MTIF3 prevents the translation initiation complex formation if it is bound by a tRNA in the absence of mRNA. Only small ribosomal subunits that have bound mRNA before the recruitment of tRNA and MTIF2 are able to proceed from translation initiation to elongation. In the absence of MTIF3 (MTIF3 knockout, L/L,cre) preinitiation complexes cannot remove tRNAMet and positioning of the mRNA and its start codon cannot be monitored, so these complexes are able to participate in mature initiation complex formation. GDP, guanosine diphosphate; GTP, guanosine triphosphate. (C) Without the molecular proofreading steps performed by MTIF3, translation initiation proceeds at an accelerated rate but at the expense of fidelity. When fidelity of initiation is compromised, initiation complexes can stall at the 5′ ends of mRNAs, leaving the remainder of the mRNA prone to degradation by 3′-5′ exoribonucleases.
DISCUSSION
Here, we show that MTIF3 is an essential protein for embryo development and survival. Mice with heart- and skeletal muscle–specific knockout of the Mtif3 gene develop dilated cardiomyopathy. Loss of mtRNA-binding proteins in vivo most commonly causes death by 16 weeks of age; however, we have noticed that loss of proteins involved in mitochondrial translation (22, 29, 30), such as MTIF3, causes death beyond 20 weeks of age. This is possibly because mitochondrial ribosomes are particularly stable in vivo and defects in protein synthesis only become apparent once they are turned over. As protein synthesis becomes compromised in the absence of MTIF3, the levels of a subset of mt-mRNAs were increased, potentially as a compensatory measure to maintain mitochondrial biogenesis. This has been observed before in models where mitochondrial translation is impaired (22, 23, 31). In contrast to these examples, in the Mtif3 knockout mice, the increased mRNAs were those with increased levels of bound ribosomal subunits and up-regulated protein synthesis, indicating that translation and mRNA stability are linked in mammalian mitochondria. However, the increase in production of these proteins was not able to overcome the loss of MTIF3 in the longer term, as the OXPHOS complexes were not assembled and resulted in compromised respiration and decreased energy production. Our proteomic data indicate that the levels of OXPHOS polypeptides differ markedly in the knockout mice compared to controls, indicating that OXPHOS proteins need to be produced in correct stoichiometry to assemble the respiratory complexes. The stalled ribosomes that we observed at 5′ ends of mRNAs may also sequester factors that are rate limiting for OXPHOS complex assembly. Furthermore, the increased levels of the mitochondrial proteases, LONP1 and AFG3L2, and the chaperone mtHSP70 in the Mtif3 knockout mice indicate an imbalance in the OXPHOS polypeptides and are a consequence of the impaired assembly of the OXPHOS system. This was further evident from the de novo translation where some of the mitochondrially encoded polypeptides were enriched in the absence of MTIF3, but their stability was reduced, likely because they were not able to be used for OXPHOS complex assembly.
In vitro studies have shown that MTIF3 associates with the small ribosomal subunit in the presence of a cross-linker (32). Unlike MTIF2 that comigrated with the mitochondrial ribosomal subunits and the translating ribosome, MTIF3 does not comigrate with the same, suggesting that the interaction of this factor with the ribosome may be particularly transient during translation initiation. The abundance and the stability of both MTIF3 and MTIF2 are dependent on each other, because we found significantly reduced levels of MTIF2 in the absence of MTIF3. The loss of MTIF3 likely contributes to uncoordinated translation in mitochondria, as the levels of MTIF2 associated with the translating ribosomes are diminished. Nevertheless, it is remarkable that mitochondrial translation can be maintained in the absence of MTIF3 and low levels of MTIF2.
Transcriptomic analyses of mitochondrial gene expression identified increased transcription, and the stability of mt-mRNAs was skewed toward the 5′ end but decreased in the 3′ end direction. Although the engagement of mt-mRNAs with the mitoribosome was altered, productive translation can occur; however, the processivity of translation in the absence of MTIF3 may not be efficient enough to protect mRNAs from degradation in the 3′ end direction. Furthermore, the footprints of mitoribosomes at the 5′ ends of mRNAs in the absence of MTIF3 provide evidence that translation initiation complexes can be stalled at the 5′ ends of some mRNAs, which may protect their 5′ ends of mRNAs from cleavage by endogenous RNases. Although mitochondrial protein synthesis was increased, the differences in protein levels of the mitochondrially encoded subunits suggested that translation was uncoordinated without the initiation factors. The RNA chaperone complex, LRPPRC/SLIRP (SRA stem-loop interacting RNA-binding protein) (26), is also required for coordinated protein synthesis of mt-mRNAs (21, 33); however, the loss of this complex decreases the stability of mRNAs (21). Moreover, uncoordinated translation in the absence of LRPPRC did not follow the same trends as those observed when MTIF3 was lost. This highlights the distinct mechanistic roles played by these two proteins in mitochondrial protein synthesis, where the LRPPRC/SLIRP complex is required to relax the secondary structures of mRNAs for their translation, as opposed to MTIF3 that is required for recognition of the distinct start codons of mRNAs.
In Escherichia coli, IF3 is essential for protein synthesis through its several roles in dissociation of the 70S monosome into individual subunits, mediating the codon-anticodon interactions of the fMet-tRNA, formation of the initiation complex, and positioning of the mRNA start codon in the P-site of the SSU (12). We show that the mammalian MTIF3 is also essential for cell health and function, as its loss is not compatible with life. The role of MTIF3, however, is consistent with the differences between the E. coli and mitochondrial ribosomes and their transcriptomes. We do not find any evidence that MTIF3 is required for dissociation of the monosome, as we do not observe any differences in the distribution of the ribosomal subunits and translating ribosomes in our sucrose gradients. There is a small shift of the SSU reflective of the loss of MTIF3 and its association with this subunit. In vitro studies have shown that the C-terminal extension of MTIF3 is required for the dissociation of the fMet-tRNAMet bound to the 28S subunit in the absence of mRNA (34). However, the association of the 28S subunit with mRNAs independently of fMet-tRNAMet or MTIF2 and the dissociation of this complex do not require MTIF3 (34). Our in vivo work shows that, in the absence of MTIF3 protein, synthesis can proceed; however, we observe persistent fMet-tRNAMet association with the ribosomal subunits. This is consistent with our finding that MTIF3 is required for the efficient formation of a productive initiation complex.
The role of MTIF3 was assumed to be similar to that of the bacterial IF3 in subunit disassociation by binding to the SSU and preventing subunit reassembly (35). However, here, we have found that loss of MTIF3 in mammalian mitochondria does not affect association of the ribosomal subunits, or the assembled monosome, suggesting that MTIF3 does not function in ribosome disassembly and other factors have taken on this role within mitochondria. Bacterial ribosome disassembly involves a ribosome recycling factor (RRF) that binds to the 70S ribosome, where guanosine triphosphate (GTP) hydrolysis, catalyzed by elongation factor G (EF-G), leads to the separation of the ribosomal subunits (11). It may be that, in mammals, the mitochondrial RRF (mtRRF) may be important in mammalian mitoribosomal disassembly along with one of the EF-G proteins, EF-G2mt (also known as mtRRF2), which mediates ribosome recycling (36, 37). Both of these proteins are significantly elevated in mice lacking MTIF3 (table S1), suggesting that they may deal with mitoribosomal subunit turnover as a way to cope with increased mitochondrial translation.
Mitoribosome profiling revealed that MTIF3 is required for productive translation initiation as well as coordinated translation of mRNAs. The increased stalling of the mitoribosome at the 5′ ends of specific mRNAs suggests that MTIF3 may be required for regulating relative translation rates by affecting the recruitment of mRNAs to the ribosome-binding site (Fig. 6C). The mitoribosome profiling revealed that, depending on mRNA levels and their discrimination by the ribosome, they are more likely to join the 28S subunit before or after the initiation factors, which is consistent with bacterial ribosomes. In bacteria, ribosomes are in excess to mRNAs, where keeping the subunits apart after termination is likely more important. In contrast, because rRNAs and mRNAs are produced from a common transcript in mammalian mitochondria, and consequently not produced in excess, enforcing the dissociation of the subunits may not be critical.
In mitochondria, formation of the initiation complex involves binding of MTIF2, MTIF3, fMet-tRNAMet, and mRNA to the small ribosomal subunit. These factors can bind in distinct orders, providing parallel modes of initiation complex assembly (see Fig. 6B). In E. coli, this phenomenon is also observed, and initiation factors are thought to be important for conformational rearrangements that transition from initial docking to maturation of a productive initiation complex (38, 39). Consistent with this idea are our observations of stalled 28S subunits at the 5′ ends of mRNAs and increased association of tRNAMet with ribosomes in the absence of MTIF3. In E. coli, IF3 plays an important role in the fidelity of translation initiation, enabling the differentiation between canonical and noncanonical initiation codons (38). In the absence of IF3, bacterial ribosomes are able to initiate translation at noncanonical initiation codons at a much faster rate, approaching that of canonical translation initiation (38), while the rate of docking between small and large ribosomal subunits, with initiator tRNA and IF2, increases 60-fold when IF3 is absent (40). This is consistent with our finding that lack of MTIF3 results in an increase in the rate of mitochondrial translation initiation at the expense of proofreading and coordination. Despite increased protein synthesis, there was reduced stability of mitochondrial-encoded proteins, as they cannot be effectively assembled into OXPHOS complexes. This indicates that both nuclear and mitochondrial OXPHOS polypeptides need to be produced in coordinated stoichiometries to promote respiratory complex assembly and MTIF3 is required to coordinate protein synthesis of the mitochondrial OXPHOS subunits. As well as compromising the stoichiometries of mitochondria- and nuclear-encoded OXPHOS polypeptides, because of a significant increase in the rate of mitochondrial translation, the loss of MTIF3 could impair the fidelity of protein synthesis within mitochondria in two distinct ways. First, without MTIF3, initiation might occur more frequently at noncanonical initiation codons, as has been observed in E. coli in the absence of IF3 (38), producing out-of-frame peptides or proteins with alternative N termini that would be rapidly degraded, resulting in decreased protein stability and consequently OXPHOS formation. Second, as we observed degradation of the 3′ ends of mt-mRNAs due to stalling of ribosomes at their 5′ ends, a variety of unstable C-terminally truncated protein products might be produced from these partially degraded mRNAs. In both cases, experimental testing of these possibilities would be very challenging, as there are no current methods that enable the insertion of reporter genes into mammalian mtDNA or a fully functioning reconstituted mammalian mitochondrial translation system.
Here, we provide the first in vivo evidence for the role of MTIF3 in mitochondrial translation initiation. We show that MTIF3 coordinates efficient translation initiation by surveilling the correct preinitiation complex formation and accommodating the mRNAs within the SSU. Cryo–electron microscopy structures of MTIF3 in complex with mRNAs will be critical to reveal the exact molecular interactions between MTIF3 and the remaining components of the preinitiation complex.
MATERIALS AND METHODS
Generation of conditional Mtif3 mice
Mtif3 transgenic mice on a C57BL/6N background were generated by the European Mouse Mutant Archive (Biomodels, Austria). The puromycin cassette was removed by mating Mtif3+/loxP-neo mice with transgenic mice ubiquitously expressing Flp recombinase. The resulting Mtif3+/loxP mice were mated with mice ubiquitously expressing Cre recombinase to generate heterozygous knockout Mtif3+/− mice that were bred with each other to identify that the homozygous loss of Mtif3 was embryonic lethal. Heart- and skeletal muscle–specific knockout mice were generated by crossing Mtif3loxP/loxP mice with transgenic mice expressing Cre under the control of the muscle creatinine kinase promoter (Ckmm-cre). Double heterozygous mice (Mtif3loxP/+, +/Ckmm) were mated with Mtif3loxP/loxP mice to generate heart-specific knockout (Mtif3loxP/loxP, +/Ckmm) and control mice (Mtif3loxP/loxP). Mice were housed in standard cages (45 cm by 29 cm by 12 cm) under a 12-hour light/dark schedule (lights on 7:00 a.m. to 7:00 p.m.) in controlled environmental conditions of 22° ± 2°C and 50 + 10% relative humidity and fed a normal chow diet (Rat & Mouse Chow, Specialty Foods, Glen Forrest, Western Australia), and water was provided ad libitum. The study was approved by the Animal Ethics Committee of the University of Western Australia (UWA) and performed in accordance with Principles of Laboratory Care [National Health and Medical Research Council (NHMRC), Australian Code for the Care and Use of Animals for Scientific Purposes, ed. 8, 2013].
Mitochondrial isolation
Mitochondria were isolated from homogenized hearts or skeletal muscle and isolated by differential centrifugation as described previously (18, 41) and detailed in the Supplementary Materials.
Sucrose gradient fractionation
Sucrose gradient fractionation was carried out as described previously (18, 41) and detailed in the Supplementary Materials.
Immunoblotting
Specific proteins were detected using rabbit antibodies against Afg3l2 (14631-1-AP, Proteintech, diluted 1:500), CCDC44 (TACO1) (21147-1-AP, Proteintech, diluted 1:500), LONP1 (15440-1-AP, Proteintech, diluted 1:500), HSPA9 (mtHSP70) (PA5-48035, Thermo Fisher Scientific, diluted 1:1000), LRPPRC (sc66844, Santa Cruz Biotechnology; diluted 1:1000), MRPL45 (15682-1-AP, Proteintech, diluted 1:1000), MRPL44 (16394-1-AP, Proteintech, diluted 1:500), MRPL37 (15190-1-AP, Proteintech, diluted 1:500), MRPS35 (16457-1-AP, Proteintech, diluted 1:500), MRPS34 (HPA042112-100 μl, Sigma, diluted 1:500), MRPS16 (16735-1-AP, Proteintech, diluted 1:1000), MTIF3 (14219-1-AP, Proteintech, diluted 1:500), and MTIF2 (LS-C164664/56970, LSBio, diluted 1:500) and mouse antibodies against ATP5a (ab14748, Abcam, diluted 1:1000), COXI (ab14705, Abcam, diluted 1:1000), COXIV (ab14744, Abcam, diluted 1:500), NDUFA9 (ab14713, Abcam, diluted 1:1000), SDHA (ab14715, Abcam, diluted 1:1000), Total OXPHOS Antibody Cocktail (ab110413, Abcam, diluted 1:5000), Total OXPHOS Blue Native Antibody Cocktail (ab110412, Abcam, diluted 1:5000), UQCRC2 (ab14745, Abcam, diluted 1:1000), and VDAC1/Porin (ab14734, Abcam, diluted 1:1000) in Odyssey Blocking Buffer (Li-Cor). IR Dye 800CW Goat Anti-Rabbit IgG or IRDye 680LT Goat Anti-Mouse IgG (Li-Cor) secondary antibodies were used, and the immunoblots were visualized using the Odyssey Infrared Imaging System (Li-Cor).
RNA isolation, northern blotting, and qRT-PCR
RNA was isolated from total hearts or heart mitochondria, and northern blotting and qRT-PCR were performed as described previously (18, 41). RNA was isolated using the miRNeasy Mini Kit (Qiagen) incorporating an on-column RNase-free deoxyribonuclease (DNase) digestion to remove all DNA. RNA (3 μg) was resolved on 1.2% agarose formaldehyde gels, then transferred to 0.45-μm Hybond-N+ nitrocellulose membrane (GE Lifesciences), and hybridized with biotinylated oligonucleotide probes specific to mouse mt-mRNAs, rRNAs, and tRNAs (42). Hybridizations were carried out overnight at 50°C in 5× SSC, 20 mM Na2HPO4, 7% SDS, and heparin (100 μg ml−1), followed by washing. The signal was detected using streptavidin-linked infrared-labeled antibody [diluted 1:2000 in 3× SSC, 5% SDS, and 25 mM Na2HPO4 (pH 7.5)] using the Odyssey Infrared Imaging System (Li-Cor). Complementary DNA (cDNA) was prepared using the QuantiTect Reverse Transcription Kit (Qiagen) and used as a template in the subsequent PCR that was performed using a Corbett Rotor-Gene 6000 instrument with SensiMix SYBR mix (Bioline) and normalized to 18S rRNA.
RNA sequencing
Strand-specific RNA-Seq was performed on mtRNA from Mtif3 control and knockout mice using the Illumina MiSeq platform, according to the Illumina TruSeq RNA-Seq protocol as we have done previously (18, 19, 23). Raw reads were trimmed of adapter sequences using cutadapt v1.18 (43), filtering out reads with a length of <15 nt. Trimmed reads were mapped against the NuMTs masked mouse genome sequence (mm10) using segemehl v0.3.4 (44). Strand-specific coverage of the primary alignments of properly paired, full-length RNA fragments (excluding those with a length of >500 nt) across the mitochondrial genome was calculated with samtools v1.7 (45) and bedtools genomecov v2.26.0 (46), normalized to library size.
Ribosome profiling
Sucrose gradient fractions corresponding to the small and large subunits as well as the assembled ribosome were isolated and digested with RNase T1 (16.7 U/ml, Ambion) for 30 min at 37°C. RNA was extracted from digested fractions using the miRNeasy Mini Kit (Qiagen) incorporating an on-column RNase-free DNase digestion to remove all DNA. RNA was then concentrated using the Oligo Clean & Concentrator Kit (Zymo). RNA-Seq libraries for ribosome profiling were prepared using the Illumina Small RNA Sample Prep Kit, with a size selection step for 15- to 90-nt RNAs. Ribosome bound footprints were identified using a modification of the peak calling approach of Holmqvist et al. (47). Raw reads were merged, removing adapter sequences, with BBMerge (48), and initially mapped against cytosolic and mitochondrial rRNA and tRNA sequences using segemehl v0.3.4 with default parameters, and unmapped reads were subsequently mapped to the NuMTs masked mouse genome sequence (mm10) similarly. tRNA counts were normalized to the total count of reads mapped to rRNA and tRNA sequences in each fraction. To derive profiling peaks for each sample, appropriately formatted BED files for mitochondrial alignments were used as input for blockbuster (49) (-distance 1 -minBlockHeight 100 -minClusterHeight 500), the resulting cluster regions were resolved into individual peaks according to Holmqvist et al. (47), and the number of reads that map to each peak region in each fraction was counted with featureCounts v1.6.3 (50) (-s 1 --fracOverlap 0.9 --largestOverlap). The two sets of footprints were merged by selecting the knockout footprints with increased abundance and the control footprints with decreased abundance. Read length profiles and footprint interval read counts were normalized to the total count of reads mapped to coding sequences in all fractions per sample.
Translation assay
In organello translation assays were carried out in isolated heart mitochondria as described before (18, 19, 23, 41). Heart or skeletal muscle mitochondria (500 μg) were incubated in 750 μl of translation buffer [100 mM mannitol, 10 mM succinate, 80 mM KCl, 5 mM MgCl2, 1 mM KPi, 25 mM Hepes (pH 7.4), 5 mM ATP, 20 μM GTP, 6 mM creatine phosphate, creatine kinase (60 μg/ml), and all amino acids (60 μg/ml) except methionine and cysteine]. Mitochondria were supplemented with 150 μCi of [35S]methionine/cysteine (PerkinElmer) for 1 hour at 37°C. After labeling, mitochondria were washed in translation buffer and resuspended in radioimmunoprecipitation assay (RIPA) lysis buffer. Protein concentration was measured, and 50 μg of mitochondrial protein was resolved by SDS-PAGE and visualized by autoradiography.
Blue native polyacrylamide gel electrophoresis
BN-PAGE was carried out using isolated mitochondria from hearts and skeletal muscle as described previously (18). BN-PAGE gels were analyzed by transferring to polyvinylidene difluoride and immunoblotting against the respiratory complexes or by drying the gel and exposing to a phosphoimaging cassette.
Respiratory chain function and complex activity
The mitochondrial oxygen consumption flux was measured with an Oxygraph-2k device (Oroboros Instruments), as previously described (18, 24).
Histology
Mouse hearts and skeletal muscle were analyzed as described previously (51). Mouse hearts were fixed with 10% neutral buffered formalin for 24 hours, washed in phosphate-buffered saline, and stored in 70% ethanol. Tissues were embedded in paraffin, sectioned using a microtome to 5-μm thickness, and transferred to positively charged slides. Slides were heated for 2 hours at 60°C and treated with xylene, xylene and ethanol (1:1), and decreasing concentrations of ethanol (100, 95, 80, and 60%) before they were washed in distilled H2O. The H&E staining was performed as described before (51). Coverslips were attached using DPX mounting media (Scharlau), and images were acquired using a Nikon Ti Eclipse inverted microscope using a Nikon 20× objective.
Echocardiography
Echocardiography (ECG) was performed on male mice under light methoxyflurane anesthesia with the use of an i13L probe on a Vivid 7 Dimension system (GE Healthcare), as described before (20).
Peptide digestion and cleanup for label-free mass spectrometry
Mitochondrial proteins (100 μg) were resuspended in lysis buffer [6 M guanidinium chloride, 2.5 mM tris(2-carboxyethyl)phosphine hydrochloride, 10 mM chloroacetamide, and 100 mM tris-HCl]. After lysis, samples were diluted 1:10 in 20 mM tris-HCl (pH 8.0), and 100 μg of protein was mixed with 1 μg of Trypsin Gold (Promega) and incubated overnight at 37°C to achieve complete digestion. Peptides were cleaned with homemade STAGEtips (52) (Empore Octadecyl C18; 3M, Germany) and eluted in 60% acetonitrile/0.1% formic acid buffer. Samples were dried in a SpeedVac apparatus (Eppendorf Concentrator plus 5305) at 45°C, the peptides were suspended with 0.1% formic acid, and 1.5 μg of peptides was analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS).
LC-MS/MS analysis
For mass spectrometric analysis, peptides were separated on a 50-cm-long, 75–μm–internal diameter EASY-spray PepMap C18 column (Thermo Fisher Scientific) using a Dionex UltiMate 3000 Nano-UHPLC system (Thermo Fisher Scientific). The column was maintained at 50°C. Buffers A and B were 0.1% formic acid in water and 0.1% formic acid in acetonitrile, respectively. Peptides were separated on a segmented gradient from 3 to 10% buffer B for 8 min, from 10 to 25% buffer B for 44 min, from 25 to 40% buffer B for 10 min, and from 40 to 95% buffer B for 12 min, at 300 nl/min. Eluting peptides were analyzed on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). The instrument was operated in a data-dependent “Top Speed” mode, with cycle times of 2 s. The “Universal Method” template was used with some modifications. Peptide precursor mass/charge ratio (m/z) measurements (MS1) were carried out at 60,000 resolution in the m/z range of 300 to 1500. The MS1 AGC target was set to 1 × 106, and the maximum injection time was set to 300 ms. Precursor priority was set to “Most intense,” and precursors with charge states 2 to 7 only were selected for higher-energy collisional dissociation fragmentation. Fragmentation was carried out using 27% collision energy. The m/z values of the peptide fragments were measured in the Orbitrap using an AGC target of 5 × 104 and 40-ms maximum injection time. The option “Inject ions for All Available Parallelizable Time” was enabled. This option varies the maximum injection time and ion targets on the fly based on the available parallelizable time.
LC-MS/MS data analysis
The Proteome Discoverer (PD; v.2.2.0388; Thermo Fisher Scientific) proteomics software was used to process raw (Xcalibur) MS/MS data generated using the Orbitrap Fusion mass spectrometer. Label-free quantification analysis was carried out in PD using the standard processing and consensus workflow templates, respectively, without applying a normalization procedure according to the software instructions. The processing workflow template contained the Minora Feature Detector node and Sequest HT engine search nodes, and the consensus workflow contained the Feature Mapper and Precursor Ions Quantifier nodes. Peptide fragmentation spectra were searched against the mouse reference proteome from the UniProt database (UP00000589, release-2018_11) (53). The resulting output file contained protein abundance values that were not normalized, and in this file, we identified housekeeping proteins, excluding mitochondrial related proteins, that were used for our normalization using an in-house script. Next, we adjusted specific parameters in the consensus workflow of PD to carry out the normalization. In the Precursor Ions Quantifier node, the “Normalization and Scaling” tab was changed as follows: (i) the “Normalization Mode” was changed to the “Specific Protein Amount,” (ii) a FASTA file of selected housekeeping genes was selected in the “Proteins For Normalization,” and (iii) the “Scaling Mode” was left to “None.” Additionally, the “Ratio calculation” was set to the “Summed Abundance Based,” and the “Hypothesis Test” was set to the “ANOVA (Individual Proteins)” in the “Quan Rollup Hypothesis Testing” tab. The consensus workflow analyses of the data with these parameters generated a file that contained normalized protein abundance values. The data were visualized using R 3.5.1 (2018-07-02), the EnhancedVolcano (v1.0.1) package (54), by plotting the P values less than 0.01 and log fold change less than −1 and greater than +1 for the significantly changing proteins.
Supplementary Material
Acknowledgments
We thank N. Ban and E. Kummer for helpful discussions and advice. We thank M. Jung for technical assistance. Funding: This project was supported by fellowships and project grants from the NHMRC (to L.A.H., A.F., and O.R.), the ARC (DP170103000 to A.F. and O.R.), and the Cancer Council Western Australia (to O.R. and A.F.). D.L.R., L.A.H., and I.K. are supported by UWA Postgraduate Scholarships. K.L.P. was supported by an NHMRC Scholarship and Mito Foundation top-up scholarship. T.R.R. is a CSIRO Future Fellow, Bright Spark Foundation Honorary Fellow, and Raine Foundation Grant recipient. Author contributions: All authors conducted and analyzed the experiments. L.A.H., D.L.R., O.R., and A.F. wrote the manuscript, and the other authors edited and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The Gene Expression Omnibus (GEO) accession number for the data reported in this paper is GSE131154. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaay2118/DC1
Fig. S1. Genetic deletion of Mtif3 in mice (related to Fig. 1).
Fig. S2. mtRNA abundance is perturbed in the absence of MTIF3 (related to Fig. 3).
Fig. S3. The relative abundance of OXPHOS steady-state proteins levels in control and Mtif3 knockout mouse mitochondria (related to Fig. 3).
Fig. S4. Mitochondrial translation initiation requires MTIF3 for assembly of OXPHOS complexes (related to Figs. 3 and 5).
Fig. S5. Changes in the mitochondrial proteome of 10-week-old Mtif3 knockout mice (related to Fig. 4).
Fig. S6. The effects of MTIF3 loss on mRNA recognition (related to Fig. 5).
Table S1. Mitochondrial proteome of Mtif3 knockout mice compared to control mice.
REFERENCES AND NOTES
- 1.Rackham O., Mercer T. R., Filipovska A., The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. WIREs RNA 3, 675–695 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Greber B. J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N., The complete structure of the 55S mammalian mitochondrial ribosome. Science 348, 303–308 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Amunts A., Brown A., Toots J., Scheres S. H. W., Ramakrishnan V., The structure of the human mitochondrial ribosome. Science 348, 95–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T., Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J. Biol. Chem. 276, 33181–33195 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Lee R. G., Rudler D. L., Rackham O., Filipovska A., Is mitochondrial gene expression coordinated or stochastic? Biochem. Soc. Trans. 46, 1239–1246 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kummer E., Leibundgut M., Rackham O., Lee R. G., Boehringer D., Filipovska A., Ban N., Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature 560, 263–267 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Davies S. M. K., Rackham O., Shearwood A. M. J., Hamilton K. L., Narsai R., Whelan J., Filipovska A., Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 583, 1853–1858 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Ramakrishnan V., Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Gaur R., Grasso D., Datta P. P., Krishna P. D. V., Das G., Spencer A., Agrawal R. K., Spremulli L., Varshney U., A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol. Cell 29, 180–190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yassin A. S., Haque M. E., Datta P. P., Elmore K., Banavali N. K., Spremulli L. L., Agrawal R. K., Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc. Natl. Acad. Sci. U.S.A. 108, 3918–3923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zavialov A. V., Hauryliuk V. V., Ehrenberg M., Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol. Cell 18, 675–686 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Simonetti A., Marzi S., Jenner L., Myasnikov A., Romby P., Yusupova G., Klaholz B. P., Yusupov M., A structural view of translation initiation in bacteria. Cell. Mol. Life Sci. 66, 423–436 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moazed D., Samaha R. R., Gualerzi C., Noller H. F., Specific protection of 16S rRNA by translational initiation factors. J. Mol. Biol. 248, 207–210 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Christian B. E., Spremulli L. L., Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry 48, 3269–3278 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ayyub S. A., S. L. A., Dobriyal D., Aluri S., Spremulli L. L., Varshney U., Fidelity of translation in the presence of mammalian mitochondrial initiation factor 3. Mitochondrion 39, 1–8 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Derbikova K., Kuzmenko A., Levitskii S., Klimontova M., Chicherin I., Baleva M., Krasheninnikov I., Kamenski P., Biological and evolutionary significance of terminal extensions of mitochondrial translation initiation factor 3. Int. J. Mol. Sci. 19, 3861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinnel T., Sacerdot C., Graffe M., Blanquet S., Springer M., Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 290, 825–837 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Rackham O., Busch J. D., Matic S., Siira S. J., Kuznetsova I., Atanassov I., Ermer J. A., Shearwood A. M. J., Richman T. R., Stewart J. B., Mourier A., Milenkovic D., Larsson N. G., Filipovska A., Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 16, 1874–1890 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Siira S. J., Rossetti G., Richman T. R., Perks K., Ermer J. A., Kuznetsova I., Hughes L., Shearwood A. J., Viola H. M., Hool L. C., Rackham O., Filipovska A., Concerted regulation of mitochondrial and nuclear non-coding RNAs by a dual-targeted RNase Z. EMBO Rep. 19, e46198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perks K. L., Ferreira N., Richman T. R., Ermer J. A., Kuznetsova I., Shearwood A. M. J., Lee R. G., Viola H. M., Johnstone V. P. A., Matthews V., Hool L. C., Rackham O., Filipovska A., Adult-onset obesity is triggered by impaired mitochondrial gene expression. Sci. Adv. 3, e1700677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruzzenente B., Metodiev M. D., Wredenberg A., Bratic A., Park C. B., Cámara Y., Milenkovic D., Zickermann V., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Brandt U., Stewart J. B., Gustafsson C. M., Larsson N. G., LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 31, 443–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cámara Y. Y., Asin-Cayuela J., Park C. B., Metodiev M. D., Shi Y., Ruzzenente B., Kukat C., Habermann B., Wibom R., Hultenby K., Franz T., Erdjument-Bromage H., Tempst P., Hallberg B. M., Gustafsson C. M., Larsson N. G., MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 13, 527–539 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Perks K. L., Rossetti G., Kuznetsova I., Hughes L. A., Ermer J. A., Ferreira N., Busch J. D., Rudler D. L., Spahr H., Schöndorf T., Shearwood A. M. J., Viola H. M., Siira S. J., Hool L. C., Milenkovic D., Larsson N. G., Rackham O., Filipovska A., PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep. 23, 127–142 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Mourier A., Ruzzenente B., Brandt T., Kühlbrandt W., Larsson N.-G., Loss of LRPPRC causes ATP synthase deficiency. Hum. Mol. Genet. 23, 2580–2592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koc E., Spremulli L., Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 277, 35541–35549 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Siira S. J., Spåhr H., Shearwood A. M. J., Ruzzenente B., Larsson N. G., Rackham O., Filipovska A., LRPPRC-mediated folding of the mitochondrial transcriptome. Nat. Commun. 8, 1532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ingolia N. T., Ghaemmaghami S., Newman J. R. S., Weissman J. S., Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G., Mercer T. R., Shearwood A. M. J., Siira S. J., Hibbs M. E., Mattick J. S., Rackham O., Filipovska A., Mapping of mitochondrial RNA-protein interactions by digital RNase footprinting. Cell Rep. 5, 839–848 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Metodiev M. D., Spåhr H., Loguercio Polosa P., Meharg C., Becker C., Altmueller J., Habermann B., Larsson N.-G., Ruzzenente B., NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLOS Genet. 10, e1004110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metodiev M., Lesko N., Park C. B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., Larsson N. G., Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 9, 386–397 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Park C. B., Asin-Cayuela J., Cámara Y., Shi Y., Pellegrini M., Gaspari M., Wibom R., Hultenby K., Erdjument-Bromage H., Tempst P., Falkenberg M., Gustafsson C. M., Larsson N. G., MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell 130, 273–285 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Haque M. E., Koc H., Cimen H., Koc E. C., Spremulli L. L., Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim. Biophys. Acta 1814, 1779–1784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasarman F., Brunel-Guitton C., Antonicka H., Wai T., Shoubridge E. A.; LSFC Consortium , LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol. Biol. Cell 21, 1315–1323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhargava K., Spremulli L. L., Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nucleic Acids Res. 33, 7011–7018 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koripella R. K., Sharma M. R., Haque M. E., Risteff P., Spremulli L. L., Agrawal R. K., Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit. iScience 12, 76–86 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rorbach J., Richter R., Wessels H. J., Wydro M., Pekalski M., Farhoud M., Kühl I., Gaisne M., Bonnefoy N., Smeitink J. A., Lightowlers R. N., Chrzanowska-Lightowlers Z. M. A., The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 36, 5787–5799 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuboi M., Morita H., Nozaki Y., Akama K., Ueda T., Ito K., Nierhaus K. H., Takeuchi N., EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol. Cell 35, 502–510 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Grigoriadou C., Marzi S., Pan D., Gualerzi C. O., Cooperman B. S., The translational fidelity function of IF3 during transition from the 30 S initiation complex to the 70 S initiation complex. J. Mol. Biol. 373, 551–561 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milón P., Carotti M., Konevega A. L., Wintermeyer W., Rodnina M. V., Gualerzi C. O., The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 11, 312–316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antoun A., Pavlov M. Y., Lovmar M., Ehrenberg M., How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 25, 2539–2550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richman T. R., Spåhr H., Ermer J. A., Davies S. M. K., Viola H. M., Bates K. A., Papadimitriou J., Hool L. C., Rodger J., Larsson N. G., Rackham O., Filipovska A., Loss of the RNA-binding protein TACO1 causes late-onset mitochondrial dysfunction in mice. Nat. Commun. 7, 11884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rackham O., Davies S. M. K., Shearwood A. M. J., Hamilton K. L., Whelan J., Filipovska A., Pentatricopeptide repeat domain protein 1 lowers the levels of mitochondrial leucine tRNAs in cells. Nucleic Acids Res. 37, 5859–5867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 44.Hoffmann S., Otto C., Kurtz S., Sharma C. M., Khaitovich P., Vogel J., Stadler P. F., Hackermüller J., Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLOS Comput. Biol. 5, e1000502 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Nat. Commun. 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmqvist E., Wright P. R., Li L., Bischler T., Barquist L., Reinhardt R., Backofen R., Vogel J., Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bushnell B., Rood J., Singer E., BBMerge—Accurate paired shotgun read merging via overlap. PLOS ONE 12, e0185056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langenberger D., Bermudez-Santana C., Hertel J., Hoffmann S., Khaitovich P., Stadler P. F., Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics 25, 2298–2301 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Richman T. R., Ermer J. A., Davies S. M. K., Perks K. L., Viola H. M., Shearwood A. M. J., Hool L. C., Rackham O., Filipovska A., Mutation in MRPS34 compromises protein synthesis and causes mitochondrial dysfunction. PLOS Genet. 11, e1005089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 53.UniProt Consortium , UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. Allaire, RStudio: Integrated development environment for R (RStudio Inc., 2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/12/eaay2118/DC1
Fig. S1. Genetic deletion of Mtif3 in mice (related to Fig. 1).
Fig. S2. mtRNA abundance is perturbed in the absence of MTIF3 (related to Fig. 3).
Fig. S3. The relative abundance of OXPHOS steady-state proteins levels in control and Mtif3 knockout mouse mitochondria (related to Fig. 3).
Fig. S4. Mitochondrial translation initiation requires MTIF3 for assembly of OXPHOS complexes (related to Figs. 3 and 5).
Fig. S5. Changes in the mitochondrial proteome of 10-week-old Mtif3 knockout mice (related to Fig. 4).
Fig. S6. The effects of MTIF3 loss on mRNA recognition (related to Fig. 5).
Table S1. Mitochondrial proteome of Mtif3 knockout mice compared to control mice.


