Significance
The diversity and ecology of RNA viruses is severely understudied in complex environments. Here we investigate the diversity and community patterns of soil RNA viruses by analyzing assembled metatranscriptomes. We tracked RNA viral and host communities for 22 d in 2 soil environments central to carbon cycling, the rhizosphere and detritosphere. This work is an important step toward understanding the factors that drive RNA viral communities. The main hosts in our system may be Fungi and Proteobacteria; this is in contrast to the ocean, where diatoms and other single cellular eukaryotes are primary hosts for RNA viruses. These results greatly expand the known diversity of viruses and contribute to understanding microbial interactions in soil with major implications for carbon cycling.
Keywords: virus, phage, soil, rhizosphere, metatranscriptome
Abstract
Viruses impact nearly all organisms on Earth, with ripples of influence in agriculture, health, and biogeochemical processes. However, very little is known about RNA viruses in an environmental context, and even less is known about their diversity and ecology in soil, 1 of the most complex microbial systems. Here, we assembled 48 individual metatranscriptomes from 4 habitats within a planted soil sampled over a 22-d time series: Rhizosphere alone, detritosphere alone, rhizosphere with added root detritus, and unamended soil (4 time points and 3 biological replicates). We resolved the RNA viral community, uncovering a high diversity of viral sequences. We also investigated possible host organisms by analyzing metatranscriptome marker genes. Based on viral phylogeny, much of the diversity was Narnaviridae that may parasitize fungi or Leviviridae, which may infect Proteobacteria. Both host and viral communities appear to be highly dynamic, and rapidly diverged depending on experimental conditions. The viral and host communities were structured based on the presence of root litter. Clear temporal dynamics by Leviviridae and their hosts indicated that viruses were replicating. With this time-resolved analysis, we show that RNA viruses are diverse, abundant, and active in soil. When viral infection causes host cell death, it may mobilize cell carbon in a process that may represent an overlooked component of soil carbon cycling.
We have much to learn about soil, the world’s most diverse and enigmatic microbial habitat. With the advent of metaomics techniques, the diversity, ecology, and impact of bacteria and archaea in soil is being rapidly revealed (1, 2). However, we have only begun to study viruses in soil despite community interest and their probable importance to soil functioning and carbon storage (3). Recent work on double-stranded DNA bacteriophage (phage) in soil has begun to explore their diversity and host interactions (4–6). However, other viral members of the soil community, such as RNA-based phage and eukaryotic RNA viruses, are largely unknown. RNA viruses, those with genomes encoded on RNA, are less studied in environmental contexts because they are not captured in the more common DNA sequencing studies. In some systems, the impact of RNA viruses on community and ecosystem processes has been proposed to rival or exceed the impact of DNA viruses (7, 8). The majority of soil RNA viral work has been single-host focused for agriculturally relevant crops (9) or crop pathogens (10). Environmental RNA virus studies have focused on less complex or more tractable systems, such as marine environments and the human and animal gut (11–14). To our knowledge, no sequencing-based RNA viral community analyses have investigated soil.
Viruses may be major players in biogeochemical cycling. However, much of what is known about viral impacts on elemental cycling comes from aquatic systems. Marine phages can lyse up to one-third of bacteria in ocean waters per day, releasing a huge amount of carbon (15–17). The released components include dissolved organic carbon that is readily metabolized by heterotrophic bacteria, but largely inaccessible to eukaryotic grazers and higher trophic levels (18). This phenomenon, termed “the viral shunt” (18, 19), is thought to sustain up to 55% of heterotrophic bacterial production in marine systems (20). However, some organic particles released through viral lysis aggregate and sink to the deep ocean, where they are sequestered from the atmosphere (21). Most studies investigating viral impacts on carbon cycling have focused on DNA phages, while the extent and contribution of RNA viruses on carbon cycling remains unclear in most ecosystems. Several studies have indicated RNA viruses may outnumber DNA viruses in some instances, a possible hint to their influence in select ecosystems (7, 8, 22). We suspect that, by analogy, lysis of organisms by RNA viruses may represent a large contribution to carbon flow in soils. The process of cell lysis and biomass carbon liberation likely stimulates heterotrophic consumption, with a substantial portion of the carbon lost to the atmosphere as CO2. The liberated cellular debris may be protected from microbial access, through interaction with mineral surfaces or occlusion within soil aggregates (generation of mineral–organic associations) (23, 24). These processes could conceivably result in carbon stabilization and ultimately long-term soil carbon persistence, although in truth we have much to learn about the processes influencing soil carbon stabilization.
Understanding the diversity and ecology of soil viruses may contribute to advances in biotechnology. Viral genomes have been mined for biopesticides and self-assembling nanomaterials (25, 26). Viruses have been proposed, and used, as biocontrol agents for culling invasive organisms, including fire ants and moths (27, 28). Viruses are also being investigated as biocontrol agents for devastating plant pathogens, such as Fusarium sp., Botrytis cinereal, and Rosellinia necatrix (10, 29–34). Novel, environmentally derived viruses may be a source for new biotechnology tools and biocontrol agents.
Here, we used assembled metatranscriptomic data from a California annual grassland soil to reconstruct RNA viral genome sequences and constrain their possible hosts. We searched the assembled sequences for the RNA-dependent RNA polymerase (RdRp), a conserved gene found in all RNA viruses that lack a DNA stage (generally referred to as Baltimore type III–V RNA viruses). Although recent work tracing the deep evolutionary history of RNA viruses has merged viral families into supergroups (35–37), our focus was not on resolving deep phylogenetic viral branches. Rather, our objectives were to investigate fine-scale diversity of RNA viruses, their putative hosts, and their possible functional importance in soil. Thus, we relied heavily on Shi et al. (36), the most comprehensive metatranscriptomic study of RNA virus diversity so far, for taxonomic classification and to delineate viruses from previously undocumented clades.
The diversity of soil eukaryotes that may serve as hosts for RNA viruses remains largely understudied. Generally, RNA viruses are known to infect fungi, plants, animals and the many clades of single-celled eukaryotes, in addition to some Proteobacteria. Soil eukaryotic studies have relied heavily on primer-based sequencing or visual classification, methods that can impart biases and miss novel organisms. Using the genomic information contained in our assembled metatranscriptomes, we identified many clades of eukaryotes without reliance on primers or microscopy. In contrast to many other environmental viral studies, which sequence extracted viral particles, we sequenced whole samples, which included viral genomes and possible host transcripts. This method allowed us to explore both extracellular viruses that would be sequenced in the standard method and viruses within host cells, such as during the infection process or latent infections. We tracked both viral and eukaryotic communities in key soil habitats: The rhizosphere (soil influenced by the root), detritosphere (soil influenced by decaying particulate organic matter; bulk+litter), a combination of the 2 (rhizosphere+litter), and unamended soil (bulk) over time. Using a relatively short sampling timescale (3, 6, 12, and 22 d) allowed us to investigate viral and host community dynamics. Our research places direct constraints on the timescales for virus dynamics and provides a genomic view of RNA viruses in soil.
Results
Experimental Design.
This analysis utilized data generated by a previous study on microbial niche differentiation in soil habitats (38). Briefly, wild oat (Avena fatua), an annual grass common in Mediterranean climates, was grown in microcosms with a sidecar, allowing us to track root age and sample directly from the rhizosphere (39). The experimental set-up used microcosms, half of which included soil mixed with dried ground A. fatua root litter and the other half contained soil without litter amendment. All microcosms contained bulk soil bags, which excluded roots, providing control soil uninfluenced by the root. Once A. fatua was mature, roots were allowed to enter the sidecar and the growth of individual roots was tracked. We destructively harvested rhizosphere soil (and paired bulk soil) that had been in contact with the root for 3, 6, 12, and 22 d. In total, we sampled paired rhizosphere and bulk samples from 4 time points with 2 treatments (with and without litter), with 3 biological replicates, for a total of 48 samples for metatranscriptome sequencing. Using rRNA-depleted RNA, we sequenced a total of 408 Gbp (average of 8.7 Gbp per sample).
Eukaryotic RNA Viruses.
We used profile hidden Markov models (HMM) to search our assembled metatranscriptomes and found a total of 3,884 unique viral RdRp sequences (dereplicated at 99% amino acid sequence identity; AAI). This includes 1,350 predicted RNA bacteriophage (phage) viruses that infect bacteria, and 2,534 predicted viruses that infect eukaryotes.
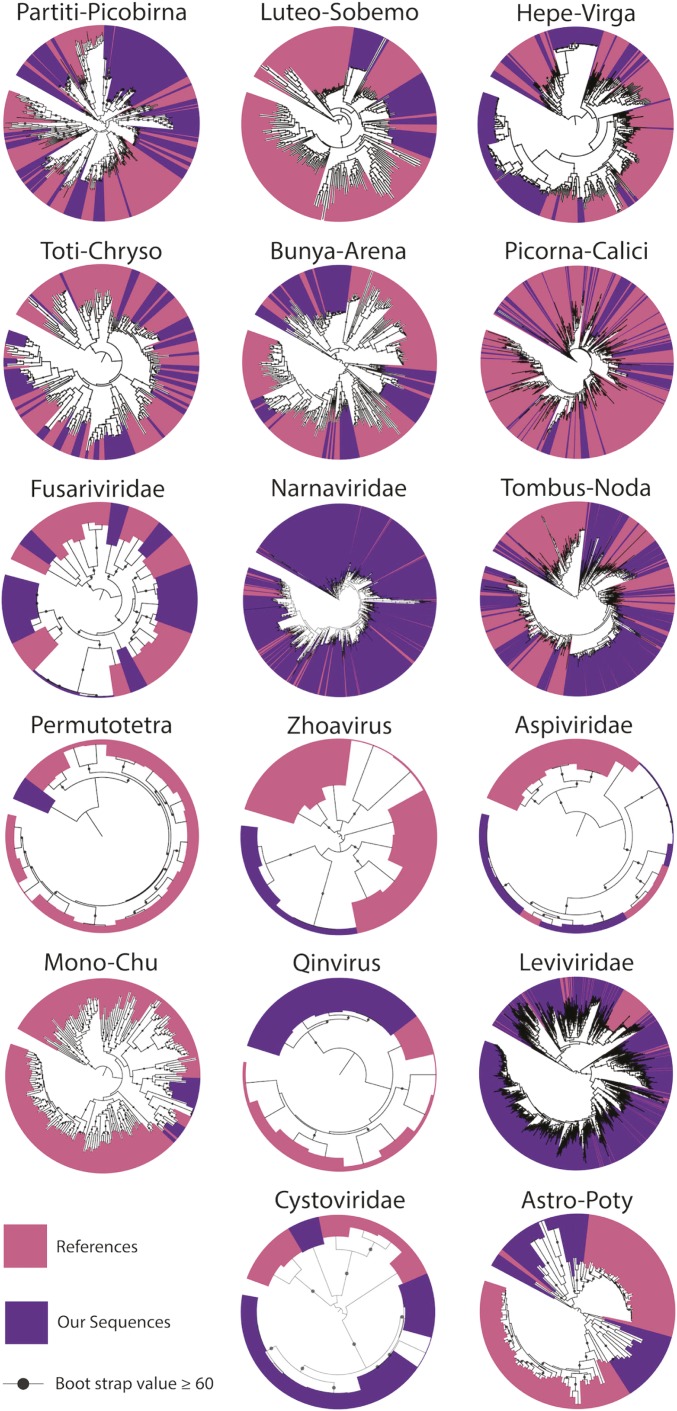
Our eukaryotic viruses group into 15 major clades that span the majority of known viral diversity (Fig. 1). Many were included into the supergroup-like clades defined by Shi et al. (36). For the remainder, we constructed phylogenetic trees to define additional viral families.
Fig. 1.
Phylogenetic trees representing clades of RNA viruses (based on RdRp) identified in our California annual grassland experimental soil. Within each tree, the RdRp sequences we identified are colored purple and previously described sequences are in pink. Trees are all midpoint rooted. Trees with predicted fungal infecting clades are presented in SI Appendix, Fig. S1.
Overall, in trees that include both existing and newly generated sequences, we noted a strong grouping of our RNA viral sequences into “fans of diversity,” much like the seed head of a dandelion. Many of these fans included a single reference sequence previously used to propose a new viral family. For example, in the Hepe-Virga, sequences grouped with the Agaricus bisporus virus 16 [proposed family Ambsetviridae (40)], in the Astro-Poty clade sequences group with Bufivirus UC1 from wastewater (37) and in the Partiti-Picobirna, sequences cluster with Purpureocillium lilacinum nonsegmented virus 1 (41). We substantially expanded the Barnaviridae (Lueto-Sobemo superfamily) and Mymonaviridae (Tombus-noda superfamily) families (42, 43), which replicate in fungi, and the newly proposed Zhaovirus and Qinvirus families, which are found in invertebrates (36).
We predict that fungi are the most common hosts for many of our reported RNA viruses (mycoviruses) (SI Appendix, Fig. S1). We are most confident when they fall into the Barnaviridae, Megabirnaviridae, Quadriviridae, and mitoviruses, groups currently thought to only infect fungi (44, 45). The most frequently encountered virus in our dataset, accounting for over 50% of the eukaryotic viral strains identified, came from the mitovirus genus in the Narnaviridae family. Mitoviruses are linear single-stranded RNA viruses that replicate within fungal mitochondria and spread vertically through spores and horizontally through hyphal fusion (33, 46). We suspect that most of the mitoviruses we detect are infecting fungi because, although some mitoviral sequences have been found integrated into the genomes of plants, they are frequently truncated and not transcribed (46, 47). Mitoviruses were also the most abundant viral clade in every sample (SI Appendix, Fig. S2).
Until recently, the Narnaviridae group, which includes mitoviruses, were thought to only encode an RdRp, but a recent discovery suggests some narnaviruses (only distantly related to mitoviruses) encode additional proteins, including capsids and helicases (36). We identified some mitovirus genomes with possible additional genes, which could play a part in infection efficiency or infection of new hosts. However, the majority of mitoviruses we identified contained only a single RdRp gene. We predicted several additional proteins on some near-complete mitoviral genomes (SI Appendix, Fig. S3). These putative genes are small (average 79 amino acids), and functions could not be predicted for them. Sometimes the additional genes are predicted to be transcribed in the same direction as RdRp and in other cases they are predicted to be transcribed in the opposite direction (SI Appendix, Fig. S3).
We reconstructed many sequences for viruses that likely infect eukaryotes other than fungi. For the Picorna–Calici, hosts are likely vertebrates, insects, algae, and plants, based on viral phylogenetic placements (48). In the Tombus-Noda tree, many of the RdRps group with umbraviruses, well-recognized plant viruses. Other RdRp sequences group with sequences from complex environmental samples where the hosts are unknown.
RNA viruses from some previously defined superfamilies were conspicuously absent. We did not identify any soil RNA viruses belonging in the Nidovirales-like, Reoviridae-like or Orthomyxoviridae-like superfamilies. The absence of Reoviridae is interesting, as these RNA viruses can infect fungi (49). We also did not confidently identify any Ortervirales, retroviruses that replicate with a DNA intermediate. We identified many reverse transcriptases, but none of the corresponding scaffolds encoded capsid proteins, so the sequences may be retrotransposons or fragments of retroviruses.
We classified 2 clades within the Bunya-Arena viral superfamily as novel, given that they exhibit sequence divergence comparable to that which separates known RNA viral families (SI Appendix). In both cases, the scaffolds only encode the polymerase with a Bunyavirus RNA-dependent RNA polymerase domain. This genome structure is shared by Hubei myriapoda virus 6 (36) and Ixodes scapularis-associated virus 3 (50).
Eukaryotic Hosts.
As viruses replicate within their host, changes in virus abundance levels implicate introduction of new vectors or hosts, shifts in infected and uninfected host abundance levels, changes in infected host physiology, or changes in host susceptibility to infection. To better understand the diversity and ecology of RNA viral hosts and carriers in soil, we used the mitochondrially encoded cytochrome C oxidase subunit 1 (Cox1) gene as a marker to define the eukaryotic populations present (51–56). However, mitochondrial transcription is determined by metabolic activity of a cell (57) and can be impacted by the switch to a symbiotic state [e.g., in arbuscular mycorrhizal fungi (58)] or nutrient sensing and hormonal signals [e.g., in animals (59)]. Thus, the number of reads mapped to the Cox1 genes is determined by the number of organisms present and the organism’s activity.
We identified 726 eukaryotic Cox1 sequences and clustered them at 98% AAI to approximate a species-level view. The eukaryotes we discovered span the diversity of known soil organisms. Not surprisingly, sequences from A. fatua were the most abundant Cox1 transcripts in many samples. In some samples, the most abundant Cox1 were from an Enchytraeidae-related worm or an Amoebozoa sp. Other samples were dominated by a mixture of fungi, Amoebozoa, Viridiplantae, and unknown eukaryotes.
Amoebozoa were the most diverse eukaryotic clade in this dataset, >25% of the identified eukaryotes, followed by species of fungi and unknown eukaryotes. The many unknown eukaryotes reflect the lack of environmental Cox1 sequences from micro- and mesoeukaryotes in public databases. For this reason, we reconstructed 18S rRNA gene sequences for classification, as they are better represented than Cox1 in public databases. However, the transcripts we analyzed had been depleted in bacterial and plant ribosomal RNA (via Ribo-Zero rRNA Removal kits), which undoubtedly influenced the composition and relative amount of other rRNA sequences. As such, the identified 18S rRNA sequences could not be used for abundance measurement or as a quantitative measure of diversity, only for identification of eukaryotes not identified using COX1. In total we identify 521 distinct species based on 18S rRNA sequences (after clustering at 98% nucleic acid identity to approximate species groups) (Table 1). The results revealed the presence of Centroheliozoa, Malawimonadidae, and Jakobida not identified on the Cox1 analyses and many more species of Heterolobosea, Euglenozoa, and Rhizaria.
Table 1.
Number of eukaryotes identified based on marker genes and significant correlations between eukaryotes and RNA viruses
| Significant cooccurrences | ||||||||||
| Clade | Cox1 | 18S rRNA* | Narnaviridae | Bunya-Arena | Tombus-Noda | Luteo-Sobemo | Toti-Chryso | Picorna-Calici | Leviviridae | Parti-Picobirna |
| Amoebozoa | 193 | 295 | 14 | 16 | 3 | 2 | 3 | 1 | ||
| Fungi | 174 | 33 | 15 | 11 | 2 | 1 | 2 | 2 | ||
| Unknown eukaryotes | 108 | 14 | 1 | 1 | ||||||
| Metazoa | 85 | 28 | ||||||||
| Stramenopiles | 55 | 17 | 11 | 6 | 3 | 2 | 1 | |||
| Alveolata | 54 | 14 | ||||||||
| Heterolobosea | 11 | 41 | ||||||||
| Nucleariidae | 11 | 0 | 11 | 2 | 5 | 1 | ||||
| Cryptophyta | 9 | 1 | ||||||||
| Euglenozoa | 7 | 28 | ||||||||
| Viridiplantae | 6 | 2 | ||||||||
| Choanoflagellida | 5 | 8 | 1 | 3 | 1 | |||||
| Rhizaria | 3 | 21 | ||||||||
| Apusozoa | 2 | 1 | ||||||||
| Alveidia | 2 | 0 | ||||||||
| Ichthyosporea | 1 | 0 | ||||||||
| Centroheliozoa | 0 | 11 | ||||||||
| Malawimonadidae | 0 | 6 | ||||||||
| Jakobida | 0 | 1 | ||||||||
The number of statistically significant correlations is the number of positive cooccurrence edges between a eukaryote and virus derived from the network analysis (SI Appendix, Fig. S11).
Note that the absolute values may be skewed by rRNA depletion.
Eukaryotic Virus and Host Ecology.
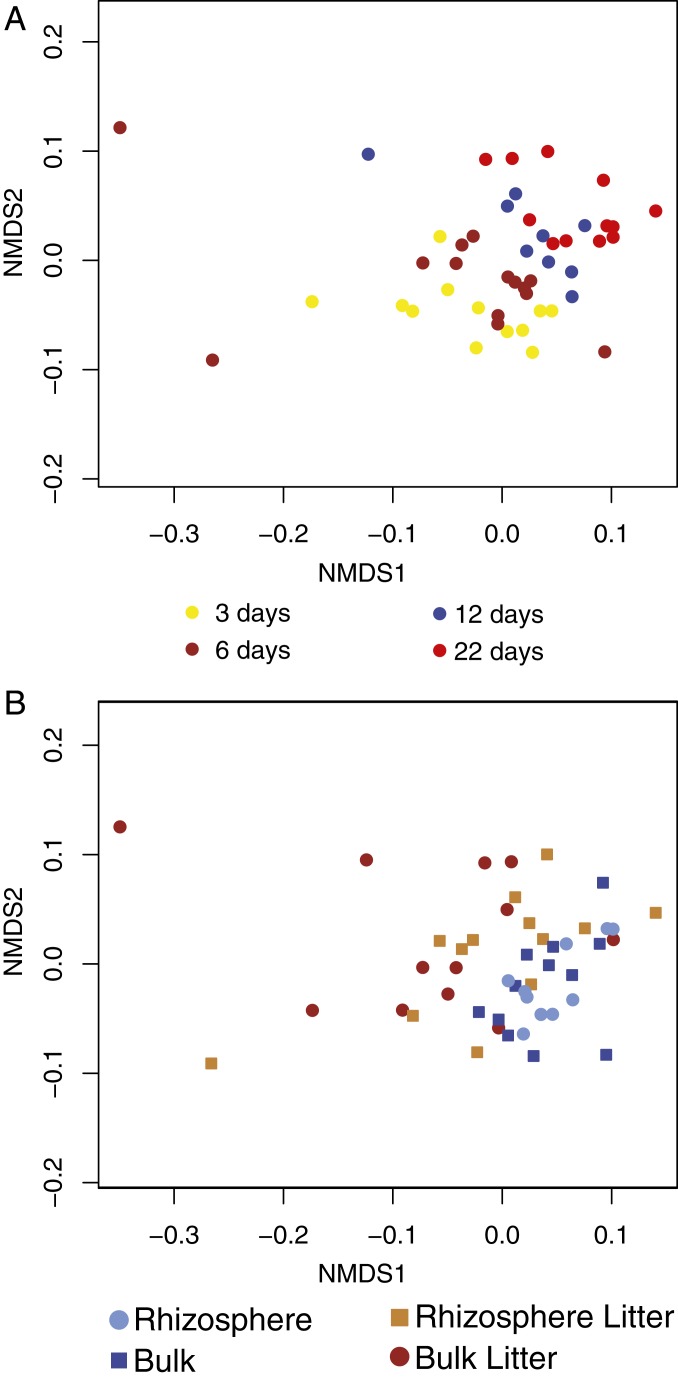
The presence of root litter shaped the soil’s eukaryotic community structure, as measured by Cox1 gene transcript abundance with A. fatua Cox1 sequences removed (Fig. 2A) (PERMANOVA R2 0.17, P < 9.999e-05). Separation of the communities was evident by the time of the first sampling (3 d) and persisted to the end of the 22-d long experiment (SI Appendix, Fig. S4).
Fig. 2.
Nonmetric multidimensional scaling ordination of soil eukaryotic communities in response to 4 soil resource treatments, based on coverage of the Cox1 gene (A. fatua sequences removed) (A), and ordination of eukaryotic RNA viral communities with coverage calculated at the scaffold level (B).
We compared the abundances of eukaryotic Cox1 transcripts in rhizosphere+litter, bulk+litter, and rhizosphere samples to the bulk samples in order to identify species statistically enriched in each case (SI Appendix, Fig. S5). The results showed that the presence of root litter and rhizosphere enriched for many Amoebozoa and fungi. However, enrichment patterns indicate that litter had a greater selective force on more individual species than the presence of a growing root.
The eukaryotic RNA viral community was influenced by some of the experimental treatments. Root litter had a significant effect on the eukaryotic RNA viral community (PERMANOVA P < 9.999e-05); the presence of growing roots had no detectable impact (PERMANOVA P < 0.07) (Fig. 2B). The distinction between added litter vs. no litter samples was evident within 3 d, implying that the viral and eukaryotic communities changed at similar rates. Differences persisted to the end of the 22-d long experiment. We observed the same patterns even when the Narnaviridae were removed from the dataset, indicating that these numerous, unusual host-bound viruses were not causing the separation of the eukaryotic viral community based on the presence of root litter (SI Appendix, Fig. S6).
We identified a smaller number of enriched eukaryotic RNA viruses compared the number of enriched eukaryotes (SI Appendix, Fig. S7). This may be due to the substantial heterogeneity in viral abundance patterns, even within replicates. The RNA viruses displayed a higher level of microheterogeneity than the eukaryotes. In contrast, transcripts for individual eukaryotes were more ubiquitous across samples. The viral strains most frequently enriched relative to bulk soil were from the mitoviruses. This is unsurprising, as the abundance of the obligately intermitochondrial mitoviruses is tied to the abundance and activity of the host and fungi responded strongly to the specific treatments.
To explore possible correlations between the hosts and RNA viruses we created a cooccurrence network. The network was constructed from positive cooccurrences between the RNA viruses and hosts (SI Appendix, Fig. S8). We noted the total number of edges, statistically significant undirected vertices between eukaryotes and RNA virus in the network, between the possible hosts and RNA viruses (Table 1), some eukaryotes and RNA viruses have multiple edges, which explains the high numbers seen in some less diverse clades, such as the Nucleariidae.
RNA Phage, Potential Hosts, and Ecology.
Two families of RNA viruses are known to infect bacteria, the Cystoviridae, which infect Pseudomonas sp. (60, 61), and the Leviviridae, which infect Gammaproteobacteria and Alphaproteobacteria (62–64). We used a marker gene approach to find RdRps for these RNA phage in the assembled transcripts. After dereplication, we identified 12 Cystoviridae RdRp sequences and 1,338 unique Leviviridae RdRp sequences. This is a significant increase in the known diversity of this group, as there are currently just over 200 Leviviridae RdRp sequences in public databases. Some of our sequences group with Allolevivirus and Levivirus, well-studied genera, but importantly, other novel Leviviridae sequences resolved new clades (Fig. 1).
The specific Leviviridae taxa enriched in bulk soil are relatively phylogenetically novel (SI Appendix, Fig. S8) and have unconventional genetic architectures based on metagenomic reconstruction. The most abundant Leviviridae sequence in 46 of 48 samples groups into a predominantly novel branch. The near-complete 4,668-bp genome of this abundant RNA phage encodes 4 nonoverlapping genes, as opposed to the frequently overlapping and fewer genes found in other Leviviridae (SI Appendix, Fig. S9, Top). Like Enterobacteria phage M, it may also encode a +1 frameshift lysis protein inside its RdRp gene (65). The genome of a related Leviviridae is even more complex, with 5 genes and possibly 2 frameshift lysis proteins (SI Appendix, Fig. S9, Bottom). However, additional biochemical work will be needed to resolve the function of these predicted proteins.
We used ribosomal protein S3 (rpS3) phylogeny to identify possible hosts for Leviviridae viruses. All previously documented Leviviridae infect Proteobacteria, so we narrowed our analysis to these bacteria, which represent a possible pool of hosts for the Leviviridae, although other clades cannot be ruled out. In our samples, we identified 355 species of Proteobacteria, which differed in abundance between rhizosphere, bulk, rhizosphere+litter, and bulk+litter treatments (PERMANOVA P < 9.999e-05) (SI Appendix, Fig. S10A) after only 3 d of root growth (SI Appendix, Fig. S10B). The Leviviridae communities also diverged within 3 d and changed over time (PERMANOVA P < 0.02) (Fig. 3A). The communities separated based on the presence or absence of root litter (PERMANOVA P < 9.999e-05), whereas the influence of growing roots was undetectable (PERMANOVA P < 0.09) (Fig. 3B).
Fig. 3.
Nonmetric multidimensional scaling ordination of the Leviviridae community identified in an annual grassland soil exposed to 4 resource treatments, colored by time (A) and treatment (B).
Discussion
To our knowledge, no prior study has genomically investigated the ecology and diversity of RNA viruses in soils. Using assembled metatranscriptomes, we uncovered a vast diversity of RNA viruses (>3,000 sequences) from a single grassland soil and discovered several possible unique families. Some viruses grouped phylogenetically with viral families previously proposed based on only a single member, indicating soils may hold much of the diversity of these understudied groups. The viruses we reconstructed came from nearly all known groups of RNA viruses. We identified hundreds of eukaryotes and bacteria that may represent RNA viral hosts or vectors that can pass infections among plants and other organisms. We know that the transfer of viral agents can have agriculturally relevant effects (66–68) but there are likely many unexplored impacts on the soil community and host-mediated process, such as nutrient cycling.
Many of the RNA viruses we identified group phylogenetically with fungal viruses (mycoviruses), and fungi appear to be one of the main RNA viral hosts in this soil. The apparent dominance of mycoviruses in our dataset provides a striking contrast to aquatic systems, where Picornavirales dominate (14). The most diverse mycoviruses in our samples are mitoviruses (Narnaviridae) which, despite their simplicity, can have significant impacts on fungal fitness and physiology (33, 69, 70).
Previous studies of environmental RNA virus ecology have primarily addressed marine systems. The RNA viruses of oceans appear to be dominated by Picornavirales, thought to infect diatoms and other single-cell eukaryotes (7, 14, 71). While our samples did contain Picornavirales, they did not appear to be dominant in the soil we sampled. In addition to Picornavirales, some aquatic systems contain plant, human, and livestock RNA viruses that may have been introduced through runoff; we identified no viruses known to infect humans (14, 72). Some aquatic studies have noted viruses related to RNA fungal viruses, however not at the abundances or diversity we have in our soil (7). Again, in contrast to the diversity of RNA phage that we found in soil, there is only a single report of an RNA phage from an aquatic ecosystem (71). Compared to marine systems, soil virology, especially investigations of RNA viruses, is in its infancy.
In our soil, both eukaryotic RNA viral and eukaryotic communities were impacted by the presence of decaying root litter. By the first sampling time point, the eukaryotic and eukaryote-associated RNA viral communities were different in the presence and absence of litter. Saprophytic fungi appeared to respond favorably to the dead litter biomass. In contrast, dissolved organic compounds exuded by roots may be less accessible to nonfungal soil eukaryotes. Accordingly, roots had little impact on the nonplant eukaryotic community composition. Addition of decaying plant biomass promoted the activity of detritivores, which likely immigrated into the litter from surrounding soil, increased transcription levels, grew, and/or germinated from spores. However, the shifts in the eukaryotic RNA viral community we observed are likely due to proliferation following increased activity of their hosts, although they may have also been transported by vectors.
We conducted a network analysis between the RNA viruses and possible hosts to provide new hypotheses about interacting partners. The correlation between virus and host provides possible associations and fodder for future studies. However, overinterpretation of network analysis results, especially in complex systems, should be avoided (73). Within the RNA virus–host cooccurrence network the fungi had the greatest number of cooccurrence links with the Narnaviridae, which provides another piece of evidence that fungi may be hosts for RNA viruses in this soil. Other eukaryotes had cooccurrence links with the Narnaviridae as well, but it is not possible to tell if these viruses are infecting or simply cooccurring with the eukaryote. The Amoebozoa had the largest number of cooccurrence linkages, including 3 with Leviviridae, which are unlikely to infect the Amoebozoa themselves but may be associated with a cooccurring bacteria. We know of only 1 RNA virus that infects Amoebozoa, a Mononegavirales (Mono-Chu) (74). These results suggest a possible link between Amoebozoa and the viral Bunya-Arena clade. The largest module (cluster of linages) contained a phylogenetically diverse array of eukaryotes and viruses including single virus types connected to multiple clades of hosts; this may represent a group of cooccurring entities that respond to similar environmental or biotic conditions (SI Appendix, Fig. S11).
RNA phage, which replicate in bacteria, have received relatively little study of their ecological context. Here, we provide a glimpse into their ecology and diversity in soil. Given that over half of all of the identified RdRp sequences in our dataset were Leviviridae, we encourage additional studies (in other soil types, land uses, tillage practices, and cover vegetations) to assess whether soils generally host a large diversity of Leviviridae. As Leviviridae do not have a known lysogenic life stage, most of their population changes likely reflect replication in their hosts. Since Leviviridae communities and their hosts became distinctly different within the first 3 d of our experiment, we conclude that these RNA viruses infected and replicated within days. Infections over this timescale likely have dramatic effects on host communities, and thus soil ecology.
RNA viruses and RNA phage were heterogeneously distributed across samples, including replicates. In contrast, eukaryotes and bacteria appeared to have more even transcript abundances across samples. In combination, these observations suggest that virus and phage abundances are not solely determined by the presence of hosts, but also factors—such as sporadic blooms or variation in viral resistance levels in the host population —that lead to patchy viral distribution patterns in soil. These patterns may be analogous to those documented in marine virus blooms, although the exact causes of these events are still unknown (13, 75). In addition, viruses have different release rates and persistence times in soil, which may be impacted by a variety of factors, such as soil type, viral strain, host physiology, microsite of release, clay content, and extracellular enzymes. This effect would be most pronounced for viruses with an extracellular life stage and less impactful on obligate intracellular viruses like the Narnaviridae.
In general, the magnitude of viral impacts on the soil carbon cycle is underexplored. Little research has been done on phage-induced bacterial lysis in soil and even less on viral-induced death of fungi and other eukaryotes, which can contain an equal or greater biomass compared to bacteria in some soils (76). In addition to lysis, viruses of fungi can have subtle yet profound effects on fungal biology, pathogenesis, mating success, toxin production, symbiotic relationships, and other physiological effects (77–79). These effects of viruses on fungal biology could have impacts for fungal functioning and thus soil performance as a whole. In our samples, the high diversity and abundance of identified RNA viruses, combined with their dynamic population changes, indicates that there was substantial viral replication. We hypothesize that proliferation of these lytic RNA phage and RNA viruses will have substantial impacts on the form, abundance, and distribution of carbon compounds in soil and on the biology and fitness of soil fungi. For example, lysis of host bacterial cells and viral-induced cell death of eukaryotes will cause release of dissolved low molecular weight carbon compounds. These will likely be quickly consumed by nearby bacteria and much of the carbon respired back to the atmosphere, mostly as CO2. However, as happens when marine snow settles into the deep ocean, a portion of the cellular and viral debris and soluble carbon released may be stabilized in soil. For example, bacterial and eukaryotic lipids and polysaccharides could adhere to mineral surfaces or become occluded within soil aggregates.
Conclusions
By reconstructing soil metatranscriptomes drawn from multiple soil habitats and time points within the context of a controlled experiment we greatly increased the known diversity of RNA viruses. Phylogenetic analyses suggest that fungi are the most common hosts for RNA viruses in the studied grassland soil. While the diversity of hosts for RNA phage remain to be definitively identified, they likely also include Proteobacteria. Shifts in eukaryote, RNA phage, and RNA viral abundances over a few-day period reveal that entire soil communities can rapidly respond to altered resource availability. Our experiments indicate that the form of carbon inputs (root-derived low molecular weight C inputs versus macromolecular carbon compound in litter) may impact eukaryotic and bacterial abundance patterns, and that these in turn may be a major determinant of RNA viral and RNA phage dynamics.
Methods
Experimental Design.
Wild oat (A. fatua) was grown in microcosms with a clear sidecar designed to allow access and visual tracking of the soil and rhizosphere (39, 80). Experimental soil was collected from Hopland Research and Extension Center (Hopland, CA). The soil is a fine-loamy, mixed, active, mesic Typic Haploxeralf, supporting dominant stands of Avena species (81, 82). Microcosms were filled with soil at field bulk density and seedlings were planted and grown for 6 wk in the main chamber before the start of the experiment. Six days before the start of the experiment the divider separating the main chamber and the sidecar was removed and the sidecar was packed with soil. The litter-amended microcosms received 0.4 g of dried A. fatua root litter mixed with 50 g of Hopland soil. Bulk soil was contained in 1-μm mesh bags embedded into the microcosm, which allowed solutes to pass but not roots. Experimental design and soil edaphic characteristics are fully explained in a companion publication, which provides additional details regarding sample collection protocol, RNA extraction and processing, and sequencing (38); what follows is a brief description of the methods.
Sample Collection.
The ages of individual roots were tracked to collect rhizosphere soil that had been influenced by the root for 3, 6, 12, and 22 d. Three replicate microcosms were destructively harvested for paired rhizosphere and bulk soil. Rhizosphere soil was cut out from the rest of the soil along the edge of the root hair zone (<2 mm from the main root). Root sections and adhering soil were placed immediately in ice-cold Lifeguard Soil Preservation Reagent (MoBio) and soil was agitated off the root by vortexing for 2 min on medium speed and pelleted; bulk soil was treated in the same manner. Pelleted soils were frozen on dry ice and stored at −80 °C.
RNA Extraction.
RNA was extracted from 0.5 g of frozen soil using a phenol-chloroform extraction protocol (83). Then Qiagen AllPrep kits to were used to separate DNA from RNA. RNA was treated with TURBO DNase (Thermo Fisher Scientific) following the manufacturer’s protocol to remove residual DNA, and was concentrated using an ethanol precipitation.
Sequencing.
Metatranscriptome libraries were prepared and sequenced at the Joint Genome Institute. Ribosomal RNA was depleted from 1 µg of total RNA using the Ribo-Zero rRNA Removal Kit (Epicentre) for Plants and Bacteria. RNA was sequenced on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v3, and Illumina’s cBot instrument to generate a clustered flowcell for sequencing. Sequencing of the flowcell used a TruSeq SBS sequencing kit, v3, following a 2 × 150 indexed run recipe.
Sequence Analysis.
Reads were trimmed using Sickle (https://github.com/najoshi/sickle) and BBtools (https://sourceforge.net/projects/bbmap/) was used to remove Illumina adapters and trace contaminants. The reads from all 48 samples were assembled individually using IDBA-UD with default settings (84). Genes were predicted using Prodigal in the anonymous mode (85).
To find the host marker genes, Cox1 and rpS3, we used an HMM profile from Pfam (86), Cox1 (PF00115), and HMMs for Bacteria and Archaea (https://github.com/AJProbst/rpS3_trckr) and searched using hmmsearch (-E 0.00001) from the HMMER suite (87). The identified proteins were classified using both National Center for Biotechnology Information (NCBI) blast and by making trees. Once classified, we predicted genes again using Prodigal in the single mode with the appropriate translation table. The Cox1 protein sequences were dereplicated and clustered at 98% AAI representing an estimated species-level designation (88, 89). Assembly errors were found and fixed in the scaffolds containing the Cox1 using ra2.py (https://github.com/christophertbrown/fix_assembly_errors) (90). The bacterial and archaeal rps3 genes was clustered at 99% AAI, representing species-level differences (91), using USEARCH (-cluster_fast) (92). The Cox1 and rpS3 trees were generated using references from NCBI, the protein sequences were then aligned using MAFFT v7.402 (93) with the E-INS-i option on Cipres (94). Then, alignments were trimmed for the conserved domain manually on Geneious and automatedly trimmed using trimAl (95, 96). The tree was made using RAxML (97) with the JTT protein substitution model (98). The trees were analyzed and figures were generated using iTOL (99). The 18s genes were found and the alignment was generated using ssu_tree.py (https://github.com/christophertbrown/bioscripts27), which searches for rRNA genes using an HMM method then the sequences were dereplicated and clustered at 98% nucleic acid identity, again representing a possible species level designation (100, 101), and aligned using SSU-ALIGN (102). Assembly errors were found and fixed in the scaffolds containing the 18S using ra2.py (https://github.com/christophertbrown/fix_assembly_errors) (90). The tree was generated using the approach described above.
We identified the RNA viral scaffolds using a combined profile HMM approach, using HMMs from Pfam (86) for different types of RdRps. The Leviviridae were identified using RNA_replicase_B (PF03431) and scaffolding errors were identified with ra2.py. There were no useable HMMs for the Cystoviridae, so we generated our own. We aligned public RdRp sequences from the Cystoviridae using MAFFT v7.402 (E-INS-i) on Cipres and generated an HMM using hmmbuild from the HMMER suite with default settings (87, 93, 94). The Cystoviridae HMM is publicly available on the figshare repository corresponding to this report. We used many publicly available HMMs to find the RdRp of eukaryotic RNA viruses (Mononeg_RNA_pol [PF00946], RdRP_5 [PF07925], Flavi_NS5 [PF00972], Bunya_RdRp [PF04196], Mitovir_RNA_pol [PF05919], RdRP_1 [PF00680], RdRP_2 [PF00978], RdRP_3 [PF00998], RdRP_4 [PF02123], RVT_1 [PF00078], RVT_2 [PF07727], Viral_RdRp_C [PF17501], and Birna_RdRp [PF04197]). The RdRp genes were initially classified using a blast and tree-building method to determine the correct translation table to use for Prodigal in the single mode (85). For mitoviruses we used translation table 4, which to our knowledge is used by all mitoviruses. However, the additional mitoviral genes we predicted may have been incorrectly called if these genomes use a modified genetic code, as occurs in some fungi (103, 104). We examined gene predictions for indications of this (e.g., interruption of the RdRp gene) but as this phenomenon was not identified, no alternative codes were used for gene prediction. RdRp amino acid sequences were dereplicated and clustered at 99% amino acid identity using USEARCH (92). We used previously published alignments for many viral families (36) and added our sequences and key references using the Mafft v7.402 with the seed (93) and E-INS-i options on Cipres (94). For viral clades without published alignments or where the published alignment was inappropriate (Fusariviridae, Narnaviridae, Leviviridae, and Cystoviridae) we generated our own alignments using reference sequences from NCBI and the same alignment and tree-building steps as described above.
To identify lysis proteins in the Leviviridae genomes we used the Geneious ORF prediction to find all possible ORFs (96). The amino acid sequences were run through PSORTb v3.0.2 with the gram-negative setting to find possible lysis proteins (105).
To obtain coverage values for the viruses, we mapped against the entire scaffold containing the RdRp gene. For the presumed hosts we only mapped reads to the ORF, for the rpS3 and Cox1. Raw reads from all samples were mapped using Bowtie2 (–sensitive and–rfg 200,300 options), then reads were filtered for 2 mismatches using calculate_breadth.py (https://github.com/banfieldlab/mattolm-public-scripts/blob/master/calculate_breadth.py) (106). Coverage values were converted to read counts and normalized with DESeq2 (107). Ordinations were generated from DESeq2 normalized count data in R. The data were ordinated using nonmetric multidimensional scaling (R package: vegan) and significantly different clusters were determined using adonis (108). Using DESeq2 we determined the viral and eukaryotic habitat enrichments in the treatments (rhizosphere+litter, bulk+litter, and rhizosphere) relative to bulk soil at each time point. We also conducted the opposite test, identifying enrichments in bulk soil by comparing normalized abundance to each treatment (rhizosphere+litter, bulk+litter, and rhizosphere) at each time point. P values were corrected for multiple comparisons using a Benjamini–Hochberg correction (109).
Network analysis was conducted using the DESeq2 normalized coverage for the eukaryotic viruses and bacterial viruses and the eukaryotic Cox1 and bacterial rpS3. These values were filtered for coverage greater than 1,000. Spearman’s rank correlation coefficient values were computed in R using the Hmisc package; the P values were corrected for multiple testing (109, 110). The data were then stringently filtered to contain only organisms present in 20 or greater samples, a corrected P value of 0.0001 or less, a correlation value of 0.7 or greater, and removed all host–host correlations. These data were then fed into Cytoscape and a network was constructed using default settings (111). Only clusters that contained at least 1 virus as the dependent variable and 1 host as the independent variable were analyzed and presented.
Supplementary Material
Acknowledgments
This work was made possible by the support and expertise of the personnel at the Hopland Research and Extension Center (Hopland, CA). Plants were grown at the University of California, Berkeley Oxford Tract Greenhouse Facility. Sampling efforts and technical expertise were provided by Shengjing Shi, Donald Herman, and Katerina Estera-Molina. Methods and analysis input was provided by Ella Sieradzki and Alexander Probst. Spencer Diamond assisted in network analysis. Helpful comments were provided by Jonathan Schilling and the MycoClub at the University of Minnesota. We thank the reviewers for their constructive comments and input. This research was supported by the US Department of Energy Office of Science, Office of Biological and Environmental Research Genomic Science program under Awards DE-SC0010570 and DOE-SC0016247 (to M.K.F.), DOE-SC10010566 (to J.F.B.), and SCW1589 and SCW1632 (to J.P.-R.). E.P.S. was supported by a National Science Foundation fellowship, Grant CZP EAR-1331940 for the Eel River Critical Zone Observatory, and Award DOE-SC0016247. Work conducted at Lawrence Livermore National Laboratory was contributed under the auspices of the US Department of Energy under Contract DE-AC52-07NA27344. Sequencing was conducted as part of Community Sequencing Awards 1487 (to J.P.-R.) and 1472 (to M.K.F.) by the US Department of Energy Joint Genome Institute, a Department of Energy Office of Science User Facility, supported under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no competing interest.
Data deposition The sequences reported in this paper have been deposited in the GenBank database (accession nos. MN032676–MN036333 and MK945893–MK946421). Read files can be accessed through the Joint Genome Institute genome portal using GOLD study ID: Gs0110148. Supplementary datasets 1–6 are available at Figshare: https://figshare.com/projects/Metatranscriptomic_reconstruction_reveals_RNA_viruses_with_the_potential_to_shape_carbon_cycling_in_soil/63722.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908291116/-/DCSupplemental.
References
- 1.Butterfield C. N., et al. , Proteogenomic analyses indicate bacterial methylotrophy and archaeal heterotrophy are prevalent below the grass root zone. PeerJ 4, e2687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diamond S., et al. , Mediterranean grassland soil C-N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms. Nat. Microbiol. 4, 1356–1367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson K. E., Fuhrmann J. J., Wommack K. E., Radosevich M., Viruses in soil ecosystems: An unknown quantity within an unexplored territory. Annu. Rev. Virol. 4, 201–219 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Emerson J. B., et al. , Host-linked soil viral ecology along a permafrost thaw gradient. Nat. Microbiol. 3, 870–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trubl G., et al. , Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3, e00076-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williamson K. E., Radosevich M., Wommack K. E., Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 71, 3119–3125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moniruzzaman M., et al. , Virus-host relationships of marine single-celled eukaryotes resolved from metatranscriptomics. Nat. Commun. 8, 16054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steward G. F., et al. , Are we missing half of the viruses in the ocean? ISME J. 7, 672–679 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andika I. B., Kondo H., Sun L., Interplays between soil-borne plant viruses and RNA silencing-mediated antiviral defense in roots. Front. Microbiol. 7, 1458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang R., et al. , A novel single-stranded RNA virus isolated from a phytopathogenic filamentous fungus, Rosellinia necatrix, with similarity to hypo-like viruses. Front. Microbiol. 5, 360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Culley A. I., et al. , The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. MBio 5, e01210–e01214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day J. M., Ballard L. L., Duke M. V., Scheffler B. E., Zsak L., Metagenomic analysis of the Turkey gut RNA virus community. Virol. J. 7, 313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustavsen J. A., Winget D. M., Tian X., Suttle C. A., High temporal and spatial diversity in marine RNA viruses implies that they have an important role in mortality and structuring plankton communities. Front. Microbiol. 5, 703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeigler Allen L., et al. , The Baltic Sea virome: Diversity and transcriptional activity of DNA and RNA viruses. mSystems 2, e00125-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuhrman J. A., Marine viruses and their biogeochemical and ecological effects. Nature 399, 541–548 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Lang A. S., Rise M. L., Culley A. I., Steward G. F., RNA viruses in the sea. FEMS Microbiol. Rev. 33, 295–323 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Suttle C. A., Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 5, 801–812 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Wilhelm S. W., Suttle C. A., Viruses and nutrient cycles in the sea: Viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788 (1999). [Google Scholar]
- 19.Brum J. R., Sullivan M. B., Rising to the challenge: Accelerated pace of discovery transforms marine virology. Nat. Rev. Microbiol. 13, 147–159 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Winget D. M., et al. , Repeating patterns of virioplankton production within an estuarine ecosystem. Proc. Natl. Acad. Sci. U.S.A. 108, 11506–11511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guidi L., et al. ; Tara Oceans coordinators , Plankton networks driving carbon export in the oligotrophic ocean. Nature 532, 465–470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stough J. M. A., et al. , Diversity of active viral infections within the sphagnum microbiome. Appl. Environ. Microbiol. 84, 1124–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dondini M., Van Groenigen K.-J., Del Galdo I., Jones M. B., Carbon sequestration under miscanthus: A study of 13C distribution in soil aggregates. Glob. Change Biol. Bioenergy 1, 321–330 (2009). [Google Scholar]
- 24.Kawahigashi M., Kaiser K., Rodionov A., Guggenberger G., Sorption of dissolved organic matter by mineral soils of the Siberian forest tundra. Glob. Change Biol. 12, 1868–1877 (2006). [Google Scholar]
- 25.Glare T., et al. , Have biopesticides come of age? Trends Biotechnol. 30, 250–258 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Wen A. M., Steinmetz N. F., Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev. 45, 4074–4126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison R. L., Keena M. A., Rowley D. L., Classification, genetic variation and pathogenicity of Lymantria dispar nucleopolyhedrovirus isolates from Asia, Europe, and North America. J. Invertebr. Pathol. 116, 27–35 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Valles S. M., Porter S. D., Calcaterra L. A., Prospecting for viral natural enemies of the fire ant Solenopsis invicta in Argentina. PLoS One 13, e0192377 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Álvarez P., Vainio E. J., Botella L., Hantula J., Diez J. J., Three mitovirus strains infecting a single isolate of Fusarium circinatum are the first putative members of the family Narnaviridae detected in a fungus of the genus Fusarium. Arch. Virol. 159, 2153–2155 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Mu F., et al. , Virome characterization of a collection of S. sclerotiorum from Australia. Front. Microbiol. 8, 2540 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osaki H., Sasaki A., Nomiyama K., Tomioka K., Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 52, 835–847 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Wang L., Zhang J., Zhang H., Qiu D., Guo L., Two novel relative double-stranded RNA mycoviruses infecting Fusarium poae strain SX63. Int. J. Mol. Sci. 17, E641 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M., Zhang L., Li G., Jiang D., Ghabrial S. A., Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 406, 117–126 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Wu M. D., et al. , Hypovirulence and double-stranded RNA in Botrytis cinerea. Phytopathology 97, 1590–1599 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Koonin E. V., The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72, 2197–2206 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Shi M., et al. , Redefining the invertebrate RNA virosphere. Nature 540, 539–543 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Wolf Y. I., et al. , Origins and evolution of the global RNA virome. MBio 9, e02329-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuccio E. E., et al. , Niche differentiation is spatially and temporally regulated in the rhizosphere. bioRxiv:10.1101/611863 (18 April 2019). [DOI] [PMC free article] [PubMed]
- 39.Jaeger C. H. 3rd, Lindow S. E., Miller W., Clark E., Firestone M. K., Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl. Environ. Microbiol. 65, 2685–2690 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deakin G., et al. , Multiple viral infections in Agaricus bisporus—Characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci. Rep. 7, 2469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herrero N., A novel monopartite dsRNA virus isolated from the entomopathogenic and nematophagous fungus Purpureocillium lilacinum. Arch. Virol. 161, 3375–3384 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Afonso C. L., et al. , Taxonomy of the order Mononegavirales: Update 2016. Arch. Virol. 161, 2351–2360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cañizares M. C., López-Escudero F. J., Pérez-Artés E., García-Pedrajas M. D., Characterization of a novel single-stranded RNA mycovirus related to invertebrate viruses from the plant pathogen Verticillium dahliae. Arch. Virol. 163, 771–776 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Adams M. J., et al. , Changes to taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 162, 2505–2538 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Roossinck M. J., Evolutionary and ecological links between plant and fungal viruses. New Phytol. 221, 86–92 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Silva S. R., et al. , The mitochondrial genome of the terrestrial carnivorous plant Utricularia reniformis (Lentibulariaceae): Structure, comparative analysis and evolutionary landmarks. PLoS One 12, e0180484 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nibert M. L., Vong M., Fugate K. K., Debat H. J., Evidence for contemporary plant mitoviruses. Virology 518, 14–24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Gall O., et al. , Picornavirales, a proposed order of positive-sense single-stranded RNA viruses with a pseudo-T = 3 virion architecture. Arch. Virol. 153, 715–727 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Hillman B. I., Supyani S., Kondo H., Suzuki N., A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J. Virol. 78, 892–898 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokarz R., et al. , Identification of novel viruses in Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks. mSphere 3, e00614-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Damon C., et al. , Performance of the COX1 gene as a marker for the study of metabolically active Pezizomycotina and Agaricomycetes fungal communities from the analysis of soil RNA. FEMS Microbiol. Ecol. 74, 693–705 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Hebert P. D. N., Cywinska A., Ball S. L., deWaard J. R., Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Min X. J., Hickey D. A., Assessing the effect of varying sequence length on DNA barcoding of fungi. Mol. Ecol. Notes 7, 365–373 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robba L., Russell S. J., Barker G. L., Brodie J., Assessing the use of the mitochondrial cox1 marker for use in DNA barcoding of red algae (Rhodophyta). Am. J. Bot. 93, 1101–1108 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Robideau G. P., et al. , DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11, 1002–1011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi M., et al. , High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J. Virol. 91, e00680-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amiott E. A., Jaehning J. A., Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol. Cell 22, 329–338 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Tamasloukht M., et al. , Root factors induce mitochondrial-related gene expression and fungal respiration during the developmental switch from asymbiosis to presymbiosis in the arbuscular mycorrhizal fungus Gigaspora rosea. Plant Physiol. 131, 1468–1478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blomain E. S., McMahon S. B., Dynamic regulation of mitochondrial transcription as a mechanism of cellular adaptation. Biochim. Biophys. Acta 1819, 1075–1079 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elbeaino T., Digiaro M., Mielke-Ehret N., Muehlbach H. P., Martelli G. P.; ICTV Report Consortium , ICTV virus taxonomy profile: Fimoviridae. J. Gen. Virol. 99, 1478–1479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mäntynen S., Sundberg L. R., Poranen M. M., Recognition of six additional cystoviruses: Pseudomonas virus phi6 is no longer the sole species of the family Cystoviridae. Arch. Virol. 163, 1117–1124 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Kazaks A., Voronkova T., Rumnieks J., Dishlers A., Tars K., Genome structure of caulobacter phage phiCb5. J. Virol. 85, 4628–4631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klovins J., Overbeek G. P., van den Worm S. H. E., Ackermann H. W., van Duin J., Nucleotide sequence of a ssRNA phage from Acinetobacter: Kinship to coliphages. J. Gen. Virol. 83, 1523–1533 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Krishnamurthy S. R., Janowski A. B., Zhao G., Barouch D., Wang D., Hyperexpansion of RNA bacteriophage diversity. PLoS Biol. 14, e1002409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rumnieks J., Tars K., Diversity of pili-specific bacteriophages: Genome sequence of IncM plasmid-dependent RNA phage M. BMC Microbiol. 12, 277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flores H., Chapman R. A., Population development of Xiphinema americanum in relation to its role as a vector of tobacco ringspot virus. Phytopathology 58, 814–817 (1968). [Google Scholar]
- 67.Ruark C. L., Gardner M., Mitchum M. G., Davis E. L., Sit T. L., Novel RNA viruses within plant parasitic cyst nematodes. PLoS One 13, e0193881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitfield A. E., Falk B. W., Rotenberg D., Insect vector-mediated transmission of plant viruses. Virology 479–480, 278–289 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Ghabrial S. A., Suzuki N., Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384 (2009). [DOI] [PubMed] [Google Scholar]
- 70.Rogers H. J., Buck K. W., Brasier C. M., A mitochondrial target for double-stranded RNA in diseased isolates of the fungus that causes Dutch elm disease. Nature 329, 558–560 (1987). [Google Scholar]
- 71.Culley A., New insight into the RNA aquatic virosphere via viromics. Virus Res. 244, 84–89 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Djikeng A., Kuzmickas R., Anderson N. G., Spiro D. J., Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4, e7264 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carr A., Diener C., Baliga N. S., Gibbons S. M., Use and abuse of correlation analyses in microbial ecology. ISME J. 13, 2647–2655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bird R. G., McCaul T. F., The rhabdoviruses of Entamoeba histolytica and Entamoeba invadens. Ann. Trop. Med. Parasitol. 70, 81–93 (1976). [DOI] [PubMed] [Google Scholar]
- 75.Seymour J. R., Seuront L., Doubell M., Waters R. L., Mitchell J. G., Microscale patchiness of virioplankton. J. Mar. Biol. Assoc. U. K. 86, 551–561 (2006). [Google Scholar]
- 76.Bar-On Y. M., Phillips R., Milo R., The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marquez L. M., Redman R. S., Rodriguez R. J., Roossinck M. J., A virus in a fungus in a plant: Three-way symbiosis required for thermal tolerance. Science 315, 513–515 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Zhang L., Baasiri R. A., Van Alfen N. K., Viral repression of fungal pheromone precursor gene expression. Mol. Cell. Biol. 18, 953–959 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rodríguez-Cousiño N., et al. , A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded rna virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 77, 1822–1832 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeAngelis K. M., et al. , Selective progressive response of soil microbial community to wild oat roots. ISME J. 3, 168–178 (2009). [DOI] [PubMed] [Google Scholar]
- 81.Whitman T., et al. , Microbial community assembly differs across minerals in a rhizosphere microcosm. Environ. Microbiol. 20, 4444–4460 (2018). [DOI] [PubMed] [Google Scholar]
- 82.Placella S. A., Brodie E. L., Firestone M. K., Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. U.S.A. 109, 10931–10936 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barnard R. L., Osborne C. A., Firestone M. K., Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 7, 2229–2241 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Hyatt D., et al. , Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El-Gebali S., et al. , The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Finn R. D., Clements J., Eddy S. R., HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 39 W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Courty P. E., Franc A., Pierrat J. C., Garbaye J., Temporal changes in the ectomycorrhizal community in two soil horizons of a temperate oak forest. Appl. Environ. Microbiol. 74, 5792–5801 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leray M., et al. , A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 10, 34 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown C. T., et al. , Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Sharon I., et al. , Accurate, multi-kb reads resolve complex populations and detect rare microorganisms. Genome Res. 25, 534–543 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edgar R. C., Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 93.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miller M. A., Pfeiffer W., Schwartz T., Creating the CIPRES science gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), (Institute of Electrical & Electronic Engineers, New York, 2010), pp 1–8.
- 95.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kearse M., et al. , Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jones D. T., Taylor W. R., Thornton J. M., The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282 (1992). [DOI] [PubMed] [Google Scholar]
- 99.Letunic I., Bork P., Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hadziavdic K., et al. , Characterization of the 18S rRNA gene for designing universal eukaryote specific primers. PLoS One 9, e87624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu S., Xiong J., Yu Y., Taxonomic resolutions based on 18S rRNA genes: A case study of subclass copepoda. PLoS One 10, e0131498 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nawrocki E. P., “Structural RNA homology search and alignment using covariance models,” PhD thesis: 282, Washington University in St. Louis, MO (2009).
- 103.Forget L., Ustinova J., Wang Z., Huss V. A. R., Lang B. F., Hyaloraphidium curvatum: A linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol. Biol. Evol. 19, 310–319 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Laforest M. J., Roewer I., Lang B. F., Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 25, 626–632 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu N. Y., et al. , PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oksanen J., et al. , Package “vegan” Title Community Ecology Package, version 2.5-4 (2019). Available at: https://cran.r-project.org/web/packages/vegan/vegan.pdf. Accessed 2 April 2019.
- 109.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. Roy. Statist. Soc. 57, 289–300 (1995). [Google Scholar]
- 110.Harrell F. E., Package “Hmisc,” Version 4.2-0 (2019). https://github.com/harrelfe/Hmisc. Accessed 27 June 2019.
- 111.Shannon P., et al. , Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.