Significance
A flavodoxin from the domain Archaea has been characterized. It is the first of any flavodoxin shown to stabilize an anionic semiquinone, providing a platform for understanding how the protein environment modulates the reduction potentials of flavins. The unusual flavodoxin plays an electron transport role in the pathway of acetate conversion to methane in Methanosarcina acetivorans, a model methanogen for investigating the process by which two-thirds of the 1 billion metric tons of methane are produced annually in Earth’s anaerobic biospheres with a substantial contribution to global warming effecting climate change. Homologs of the gene encoding the flavodoxin are uniformly distributed in diverse acetotrophic methanogens consistent with a wider range of electron transport functions awaiting discovery.
Keywords: electron transport, methanogenesis, anaerobic, greenhouse gas, global warming
Abstract
Flavodoxins, electron transfer proteins essential for diverse metabolisms in microbes from the domain Bacteria, are extensively characterized. Remarkably, although genomic annotations of flavodoxins are widespread in microbes from the domain Archaea, none have been isolated and characterized. Herein is described the structural, biochemical, and physiological characterization of an unusual flavodoxin (FldA) from Methanosarcina acetivorans, an acetate-utilizing methane-producing microbe of the domain Archaea. In contrast to all flavodoxins, FldA is homodimeric, markedly less acidic, and stabilizes an anionic semiquinone. The crystal structure reveals an flavin mononucleotide (FMN) binding site unique from all other flavodoxins that provides a rationale for stabilization of the anionic semiquinone and a remarkably low reduction potentials for both the oxidized/semiquinone (−301 mV) and semiquinone/hydroquinone couples (−464 mV). FldA is up-regulated in acetate-grown versus methanol-grown cells and shown here to substitute for ferredoxin in mediating the transfer of low potential electrons from the carbonyl of acetate to the membrane-bound electron transport chain that generates ion gradients driving ATP synthesis. FldA offers potential advantages over ferredoxin by (i) sparing iron for abundant iron-sulfur proteins essential for acetotrophic growth and (ii) resilience to oxidative damage.
Flavodoxins (Fld) are electron-transfer proteins essential for diverse metabolisms in microbes from the domain Bacteria, whereas organisms from the domain Eukarya contain multidomain flavoproteins evolved from ancestral flavodoxin genes (1, 2). Flds were discovered over 50 y ago in Cyanobacteria and Clostridia growing in low-iron conditions where they serve as electron carriers in enzyme systems operating at potentials near that of the hydrogen electrode (3, 4). Flds contain 1 molecule of redox active flavin mononucleotide (FMN) that is noncovalently bound. All Flds characterized are highly acidic proteins containing between 140 and 180 residues that are divided into short-chain and long-chain types differing by a 20-residue loop of yet unknown function (2). The protein environment of FMN stabilizes the neutral form of the semiquinone (sq), producing dramatic shifts in the reduction potentials for each of 2 1-electron reductions of the flavin. The FMN of Flds cycle between the sq and hydroquinone (hq) for which the reduction potentials are below −101 mV for free flavin (5, 6).
Genome annotations for Flds are widespread among microbes in the domain Archaea. Although characterizations of Flds from the domain Bacteria are abundant, none from Archaea are reported. Metabolically diverse species of the domain Archaea are present in a variety of environments where they play major roles in the biogeochemical cycling of carbon, nitrogen, and sulfur. Methane-producing species (methanogens) are the largest group in the domain for which annotations of genomes include an abundance of Flds (https://www.ncbi.nlm.nih.gov/pubmed) not yet investigated. Methanogens are terminal organisms of microbial food chains that decompose plant biomass to methane in Earth’s diverse anaerobic environments. The process produces an annual 109 metric tons of methane of which two-thirds derives from the methyl group of acetate by acetotrophic species. As atmospheric methane accounts for about 30% of net anthropogenic radiative forcing, acetotrophic methanogens are responsible for a substantial share of global warming and adverse climate change (7). The evolution of efficient acetotrophic methanogens, represented by Methanosarcina acetivorans, is proposed to have produced a methanogenic burst in the end-Permian carbon cycle, contributing to a dramatic increase in global warming and Earth’s greatest mass extinction (8).
M. acetivorans has emerged as a model for understanding the biochemistry of acetotrophic Methanosarcina species. In the acetoclastic pathway, the C-C bond of acetate is cleaved by the CO dehydrogenase/acetyl-CoA decarbonylase (CODH/ACD), yielding a methyl group that is reduced to methane with electrons derived from oxidation of the carbonyl group to CO2 that is also catalyzed by the CODH/ACD (9). Electrons are channeled through a membrane-bound electron transport chain that begins with the Rnf complex and generates sodium and proton gradients driving ATP synthesis (10–13). Initially guided by findings that ferredoxin (Fdx) is an electron donor to Rnf homologs in the domain Bacteria, a Fdx isolated from M. acetivorans was shown to mediate electron transfer between CODH/ACD and Rnf (13). However, the genome is annotated with several Flds of which transcriptomic analyses show a 3-fold increased expression in acetate-grown versus methanol-grown cells of the Fld encoded by MA1799 (FldA) signaling an electron transport role in the acetoclastic pathway (14). Here, we report the structural, biochemical, and physiological characterization of FldA. The results show that: (i) FldA mediates electron transfer between CODH/ACD and the membrane-bound electron transport chain, (ii) homologs are widely distributed among acetotrophic methanogens, and (iii) the properties of FldA are distinct from all characterized Flds, which includes the unprecedented stabilization of an anionic sq.
Results
Expression and Purification.
His6-tagged FldA was produced in Escherichia coli and aerobically purified to homogeneity. SDS/PAGE (SI Appendix, Fig. S1) indicated a monomer molecular mass of 16 kDa consistent with the calculated value of 16,363. Size-exclusion chromatography estimated a native molecular mass of 35.8 kDa, which indicated a dimer in solution.
Spectral Analysis and Cofactor Determination.
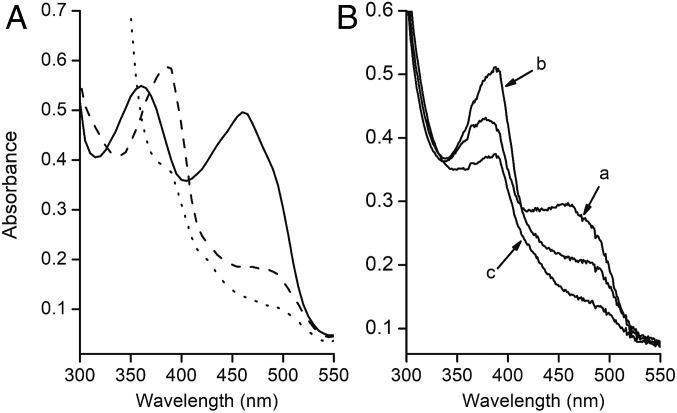
The UV-visible absorption spectrum (Fig. 1A) of as-purified FldA showed maxima at 360 and 460 nm with a shoulder at 490 nm typical of oxidized Flds. The ε460 was found to be 12.8 ± 0.5 mM−1·cm−1, which is characteristic of Flds with a full complement of flavin (15). Partial reduction with sodium dithionite yielded a spectrum showing a peak and pronounced shoulder with maxima at 387 and 480 nm, indicative of flavin anionic sq unprecedented for Flds. Further reduction produced a spectrum of the hq form. The spectrum was identical to the as-purified protein when fully reduced FldA was oxidized with potassium ferricyanide.
Fig. 1.
Spectral changes accompanying reduction of FldA. (A) Reduction with dithionite. As-purified FldA (50 µM) was contained in 50 mM Mops (pH 7.0). Solid line, as-purified aerobically; dashed line, partial reduction with sodium dithionite; dotted line, complete reduction with excess sodium dithionite. (B) Reduction with CODH. The 700-μL reaction mixture contained 60 μg of CODH where indicated, 23 μM as-purified oxidized FldA, and 50 mM NaCl contained in 100 mM potassium phosphate buffer (pH 7.0) under 1 atm CO in a sealed cuvette. The reaction was initiated with CODH and spectra recorded with a Cary Model 50 BIO UV-visible spectrophotometer. Spectra: oxidized as-purified (a), semiquinone 0.7 min after addition CODH (b), hydroquinone 3.5 min after addition of CODH (c).
Reduction Potentials.
Potentiometric titrations at pH 7.0 revealed Em values of −301 ± 5.06 mV for the ox/sq couple and −464 ± 2.61 mV for the sq/hq couple (SI Appendix, Fig. S2).
Crystal Structure.
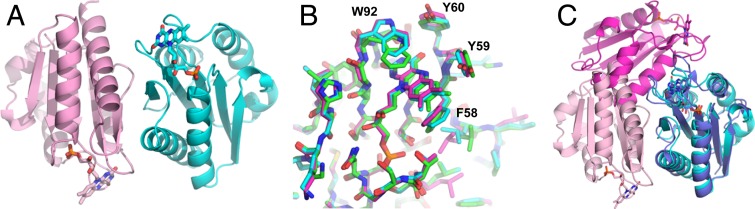
The crystal structure of oxidized FldA, solved to 1.68-Å resolution (SI Appendix, Table S1), contained 3 FldA monomers in the asymmetric unit (Fig. 2 and SI Appendix, Fig. S3). All 3 monomers contained an FMN molecule. The overall structure and FMN binding pocket of the monomer structures are nearly identical (0.161∼0.231 Å r.m.s.d.) without any particularly local conformation differences. Among the 3 FldA molecules in the asymmetric unit, protomers A and B share a large molecular surface (891.2 Å2). Molecule C also forms a dimer with its symmetry related protomer C molecule joined by a molecular interface of 884 Å2. The dimeric structure is in agreement with the native molecular mass in solution. The monomer is folded into a sandwich configuration with 5 β-sheets in the middle flanked by either 2 or 3 α-helices similar to generic short-chain Flds (16–19) (Fig. 2). Each monomer contains 1 FMN located near the surface arranged head-to-tail where the heads contain the FMN molecules separated by 33 Å (Fig. 2). An overlay of the 3 FldA protomers at the FMN binding site reveals identical structures except for displacement of the W92 side chain in protomer C relative to W92 in protomers A and B (Fig. 2B) since W92 of protomer C contacts with a symmetry related molecule.
Fig. 2.
The overall structure of FldA from M. acetivorans. (A) Structure of the FldA dimer. The FMN molecules are shown in stick representation. (B) Overlay of the FMN binding sites of 3 protomers found in the asymmetric unit. (C) Overlay of dimeric Flds from D. gigas and M. acetivorans. The D. gigas structure was determined with PDB ID code 4HEQ (17). The FMN molecules are shown in stick representation. The D. gigas protomers are shown in orange and blue. The M. acetivorans protomers are shown in teal and pink.
The FldA structure joins the Fld from Desulfovibrio gigas as the only other WT dimeric Fld structure (17). The overall and secondary structures of monomers are similar with an r.m.s.d. between FldA and the D. gigas Fld of 1.562 Å for the Cα of 143 residues (Fig. 2C). A notable difference is the length of loops surrounding FMN. The D. gigas Fld has longer loops compared with FldA from M. acetivorans. Importantly, the environment of FMN in the D. gigas Fld is more negative than for FldA. An overlay of the dimers reveals further differences in configuration (Fig. 2C). Unlike FldA, the D. gigas Fld monomers are oriented head-to-head bringing the FMN molecules to within 17 Å.
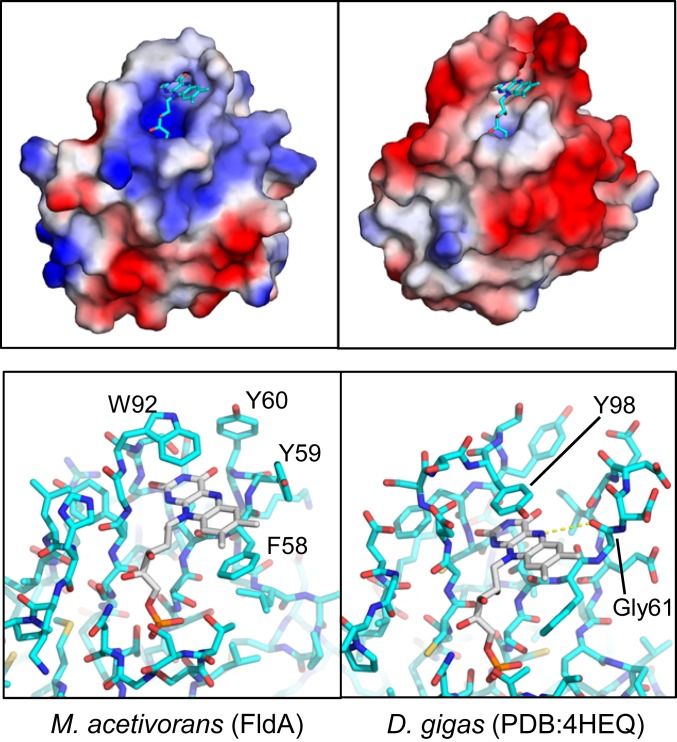
FldA has properties distinct from all generic short-chain monomeric Flds of the domain Bacteria with important implications for the biochemistry and physiology. FldA is more positively charged in comparison to generic Flds having negatively charged FMN-binding pockets (Fig. 3). FldA has a calculated pI of 5.75 in stark contrast to an analysis of 38 generic Fld sequences, which reveals a median pI value of 4.5 ± 0.6 (20). The difference is more pronounced in the FMN binding region exemplified in the electrostatic energy landscape of FldA compared to the generic Fld from D. gigas of the domain Bacteria (Fig. 3). SI Appendix, Fig. S4 shows residues interacting with the FMN of FldA from M. acetivorans. Notably, no acidic side chains interact with the FMN in contrast to generic Flds from the domain Bacteria (17, 21). Importantly, the FMN environment of FldA from M. acetivorans is distinct from generic Flds in that a glycine is absent in the position adjacent to the N5 atom of the isoalloxazine ring (Fig. 3). The si and re faces of the FldA ring are flanked by W92 and F58 forming π−π stacking interactions (Fig. 3 and SI Appendix, Fig. S4), similar to generic Flds where the isoalloxazine rings are flanked by aromatic residues (22).
Fig. 3.
Comparison of FldA from M. acetivorans (Left) and the generic Fld from D. gigas of the domain Bacteria (Right). Prepared using PyMOL. (Upper) Comparison of the electrostatic potentials. FMN molecules are shown in stick representation. Blue, basic; red, acidic; white, neutral. FldA has an overall net charge of −3 (pI = 5.75) compared to the net charge of −17 (pI = 4.08) for D. gigas Fld. (Lower) Comparison of FMN binding sites.
Physiology.
Electron transport in the acetoclastic pathway of methanogenesis for M. acetivorans originates with the 5-subunit (αβγδε) CODH/ACD complex (SI Appendix, Fig. S5). The ACD component (βγδ) cleaves acetyl-CoA, and the CODH component (αε) oxidizes the carbonyl group to CO2 (23). CODH also oxidizes CO, a surrogate for the carbonyl group. The electrons are transferred to the Rnf complex that initiates a membrane-bound electron transport chain culminating in reduction of the heterodisulfide (CoMS-SCoB) of coenzyme M (HSCoM) and coenzyme B (HSCoB) catalyzed by the heterodisulfide reductase HdrDE.
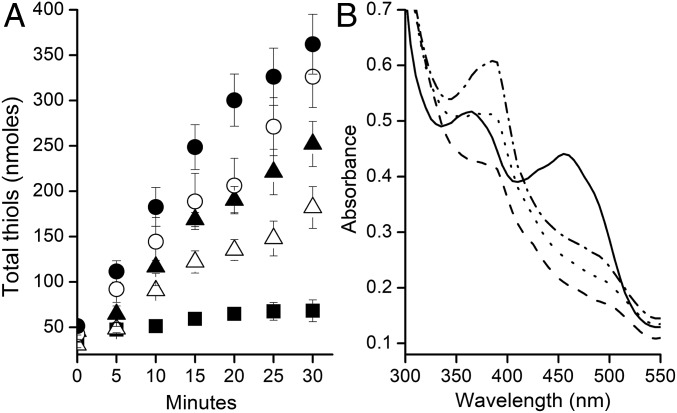
The CODH component of CODH/ACD oxidized CO and reduced the FMN of FldA to the hq form with intermediate formation of the anionic sq (Fig. 1B), predicting a role in electron transfer to the membrane culminating in reduction of CoMS-SCoB. Fig. 4A shows a time course for reduction of CoMS-SCoB by membranes dependent on CO, CODH, and either Fdx or FldA. The initial rates of FldA-dependent reduction were 74 and 82% of the Fdx-dependent rates with the electron carriers at 3.5 and 7.0 μM. Fig. 4B shows emergence of the sq form of FMN on completion of the reaction with excess CoMS-SCoB, a result indicating 1-electron transfer from the hq form. The results establish that FldA is competent in mediating electron transfer between CODH/ACD and the membrane-bound electron transport chain supporting acetotrophic growth. Furthermore, FldA replaced Fdx as an electron acceptor to the coenzyme F420H2-dependent electron bifurcating heterodisulfide reductase HdrA2B2C2 (SI Appendix, Fig. S6) proposed to function in Fe(III)-dependent respiratory methanotrophic and acetotrophic growth of M. acetivorans (24, 25).
Fig. 4.
Dependence on FldA or Fdx to mediate electron transfer from CO dehydrogenase to the membrane-bound electron transport chain reducing CoMS-SCoB. (A) Time course of the reaction monitoring the products HSCoM and HSCoB. The anaerobic assay mixtures (0.2 mL in a 2-mL sealed serum vial) contained 42 µg of CO dehydrogenase, 4.0 mM CoMS-SCoB, 50 mM NaCl, and the indicated concentrations of either Fdx or FldA contained in 100 mM potassium phosphate buffer (pH 7.0). The atmosphere was 100% CO. Reactions were initiated by adding membranes (200 µg of protein). Activity was determined by measuring appearance of the free thiols, HSCoM, and HSCoB. Symbols: (Δ) 3.5 μM FldA, (▲) 7 μM FldA, (○) 3.5 μM Fdx, (●) 7 μM Fdx, (■) minus FldA or Fdx. (B) Spectral changes in FldA. Solid line, as-purified oxidized FldA; dashed line, hydroquinone after reduction with CO and CODH; dotted line, immediately after addition of membranes; dash-dot line, completion of the reaction.
Bioinformatics.
A BLASTp search was conducted with M. acetivorans FldA as the query while constraining searches to Flds represented in the National Center for Biotechnology Information nonredundant database. The results retrieved FldA homologs only from metabolically diverse methanogens (SI Appendix, Fig. S7). The results underscore the uniqueness of FldA compared to generic Flds. A distance tree of alignments of FldA and generic Flds from the domain Bacteria (SI Appendix, Fig. S7) revealed 2 clades (I and II) of FldA homologs in which clade I was comprised of acetotrophic Methanosarcina species consistent with a role for FldA as shown here for M. acetivorans. The only exception in clade I is the FldA homolog from Methanoperedens nitroreducens, a species capable of oxidizing methane and reducing nitrate, suggesting a potential role for this FldA homolog in the anaerobic oxidation of methane (26). A distant FldA homolog (clade III) was retrieved from Methanosaeta concilii. Methanosaeta is the only other genera capable of metabolizing acetate consistent with a role for FldA homologs in acetotrophic methanogens (27). All FldA homologs in clade II were from nonacetotrophic species that utilize either H2/CO2 or methylotrophic substrates (methanol, methylamines, and dimethyl sulfide) for growth and methanogenesis. These results suggest potential roles for FldA homologs in diverse metabolisms of methanogens. Sequence alignments show that FldA homologs in clade I and clade II, and the homolog from the acetotroph M. concilii, share the S56TFYY60 FMN binding motif of FldA from M. acetivorans but are missing the cognate glycine residue that stabilizes the neutral sq in generic Flds (SI Appendix, Fig. S8). This result is consistent with the FldA homologs stabilizing an anionic sq. The chain lengths of FldA (149 residues) and homologs range from 144 to 156 residues, similar to generic short-chain Flds. However, the tree shows that generic short-chain Flds from diverse species (clade IV) are only distantly related to FldA with sequence identities less than 35%, which further underscores the unique position of FldA homologs in the Fld family. Clade IV Flds include a generic short-chain Fld (147 residues) from M. acetivorans for which expression of the gene (MA2699) is up-regulated severalfold in methanol-grown versus acetate-grown cells, suggesting a role other than in acetotrophic growth (14).
Discussion
The FMN Environment of FldA Is Unprecedented for Flavodoxins.
The biochemical, structural, and physiological characterization of FldA from M. acetivorans is an Fld characterized from the domain Archaea. Although the overall crystal structure establishes FldA as a member of the Fld family, several unique properties distinguish it from generic Flds of the domain Bacteria.
The protein environment of generic short-chain Flds is responsible for shifting the Esq/hq for free FMN to more negative values between −372 and −440 mV (28), thereby endowing Flds with the ability to substitute for the low-potential 1-electron carrier Fdx (24). The negative Esq/hq for generic Flds is accomplished by stabilization of a neutral sq and destabilization of the anionic hq dependent on residues proximal to the flavin. Several acidic residues clustered around the FMN binding site provide an unfavorable electrostatic environment, which is a major factor destabilizing the anionic hq (29). The absence of positively charged residues adjacent to the phosphate dianion of FMN also contributes to the negatively charged environment (30). The isoalloxazine ring system is in an electron-rich hydrophobic environment where at least 1 aromatic residue forms a π–π stacking interaction, further contributing to destabilization of the anionic hq (31). Finally, the more negative value for Esq/hq of FMN in generic Flds is further dependent on a neutral sq stabilized by a hydrogen bond between the protonated N5 atom of FMN and a polypeptide carbonyl group facilitated by a glycine residue adjacent to the N5 atom (28, 32–36).
Similar to generic Flds, the isoalloxazine ring of FMN in FldA is sandwiched by aromatic residues and the negative charge of the phosphate moiety is uncompensated by positively charged residues. However, in contrast to generic Flds, the FMN environment of FldA is less negatively charged, favoring stabilization of the anionic sq. As in generic Flds, unfavorable π–π stacking interactions between the isoalloxazine ring system of FMN and aromatic residues W92 and F58 are anticipated to destabilize the FMN anionic hq in FldA. Also similar to generic Flds, the uncompensated negative charge of the phosphate group of FldA is expected to contribute to the destabilization, albeit to a lesser extent than π–π stacking interactions. The anionic sq is a common intermediate for both Eox/sq and Esq/hq; thus, the inability of FldA to stabilize a blue neutral sq discourages the first 1-electron reduction while encouraging the second reductive step as shown for G57 variants of Clostridium beijerinckii Fld (37). This explains the lowering of both the Eox/sq and Esq/hq potentials of FMN in FldA below that of free FMN that is unusual for generic Flds of the domain Bacteria that stabilize a blue neutral sq.
Residues interacting with FMN explains the stabilization of an anionic sq in FldA. Unlike generic Flds, a carboxyl group adjacent to the N5 atom FMN is absent in FldA that would otherwise hydrogen bond to the N5-bound proton and stabilize a neutral sq. Furthermore, no positively charged residues are near the N1-C2-O2 atoms of FMN that would stabilize the anionic sq as reported for several flavoproteins other than Flds (38–40). Finally, no electron density attributed to a metal cation was detected in the crystal structure of FldA that might also assist in stabilizing the anionic sq as proposed for the flavodoxin-like NrdI (20). Thus, the less overall negative charge of FldA and absence of a glycine residue adjacent to the N5 atom of FMN contributes to stabilization of the anionic sq, which is unmatched for generic Flds from the domain Bacteria. It appears that the archaea evolved a mechanism for modulating the reduction potential of flavin in Flds to values more negative than free flavin, which is distinct from that of Flds in the domain Bacteria. Finally, the unconventional protein environment of FMN in FldA provides an experimental platform for determination of how flavoproteins modulate the reduction potentials of free FMN.
Physiological Relevance of FldA and Homologs.
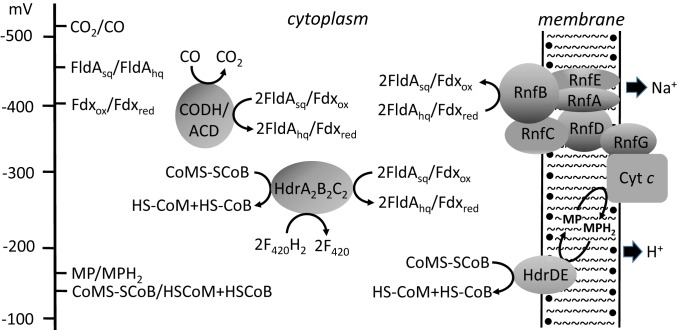
The results presented here show that FldA can replace Fdx in mediating electron transport from CODH/ACD to the membrane-bound electron transport chain essential for acetotrophic growth (Fig. 5) (12, 13). The Esq/hq of FldA (−464 mV) approximates the CO2/CO couple (Em = −540 mV) of CODH/ACD comparable to Fdx (Em = −414 mV) consistent with the results. The findings (Fig. 4B and SI Appendix, Fig. S6) that support 1-electron transfer roles for FldAhq (Fig. 5) are analogous to the generic Fld from Acidaminococcus fermentans with roles for the Fldhq in electron bifurcation and 1-electron reduction of RnfB (41).
Fig. 5.
Proposed roles of FldA in electron transport of acetate-grown M. acetivorans. CODH/ACD, CO dehydrogenase/acetyl-CoA decarbonylase; CoMS-SCoB, heterodisulfide of HS-CoM and HS-CoB; Cyt c; multiheme cytochrome c; Fdxox, oxidized ferredoxin; Fdxred, reduced ferredoxin; FldAhq, 2-electron reduced; FldAsq, one-electron reduced; HdrA2B2C2, F420H2-dependent heterodisulfide reductase; HdrDE, heterodisulfide reductase; HS-CoB, coenzyme B; HS-CoM, coenzyme M; MP, methanophenazine; RnfA-G, subunits of the Rnf complex.
A role for FldA in acetotrophic growth of M. acetivorans is supported by a 3-fold up-regulation in acetate-grown versus methanol-grown cells producing methane. FldA is also up-regulated in M. acetivorans cultured with methanol as the electron donor and anthraquione-2,6-disulfonate as electron acceptor for nonmethanogenic respiratory growth dependent on Rnf and cytochrome c, which further supports an electron transport role (42). FldA may provide an advantage in the marine environment where M. acetivorans was isolated (43). Iron levels in seawater are extremely low (0.02–1 nM), predicting an iron-sparing role for FldA to augment the supply for abundant iron-sulfur enzymes essential for growth (44, 45). Finally, Flds are more resistant to inactivation during oxidative stress than are Fdxs (46–48), consistent with a role for FldA in the exceptional oxidative stress tolerance of M. acetivorans (49).
FldA homologs may also play a role in alternative electron transport pathways of acetotrophic methanogens such as Methanosarcina barkeri that utilizes CODH/ACD in the acetoclastic pathway. However, unlike M. acetivorans and most acetotrophic species in clade I that contain the Rnf complex (SI Appendix, Fig. S8), M. barkeri instead utilizes a proton-translocating electron transport pathway involving H2-producing and H2-consuming hydrogenases that may interact with FldA homologs (9).
Conclusions.
An Fld from the domain Archaea (FldA) has been biochemically, structurally, and physiologically characterized. The stabilization of an anionic sq that modulates the reduction potential of FMN is unique to the domain Archaea. The unique environment of FMN in FldA offers a platform on which to interrogate the role of active site residues in modulating the reduction potential of free flavin in flavoproteins. The findings show that FldA is capable of replacing Fdx and has a potential advantage by sparing iron for abundant iron-sulfur proteins essential for growth and by a greater resilience to oxidative damage. Finally, bioinformatics analyses support a role for FldA homologs in metabolically diverse methanogens suggesting diverse roles.
Materials and Methods
Materials.
Fdx, membranes, and the CO dehydrogenase component of CODH/ACD from M. acetivorans were purified as previously described (13, 50). CoM-S-S-CoB was a gift of Jan Keltjens, Radboud University, The Netherlands. All other chemicals were purchased from Sigma-Aldrich. Primers were from Integrated DNA Technologies. Resins and columns required for chromatographic separations were from GE Healthcare.
General Methods.
Anaerobic procedures were performed in a Coy anaerobic chamber at 25 °C as described previously (13, 50). Free thiols were determined with DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] as described elsewhere (51). Iron and acid-labile sulfur were quantitated as described elsewhere (52, 53). The flavin content of FldA was determined by UV-visible and fluorescence spectrometry as described elsewhere (54). Protein concentrations were determined using the Bradford and BCA methods (55). Protein parameters were determined with the ExPASy ProtParam tool (https://web.expasy.org).
Cloning Expression and Purification of FldA.
The full-length sequence encoding FldA (MA1799, MA_RS09355) was amplified from M. acetivorans genomic DNA by PCR using Phusion Taq polymerase with sense (5′TTGTTGCATATGTGGAGAAAACAATTATTG) and antisense (5′GATGATCTCGAGCACACTTACTGCTTGTG) primers containing NdeI and XhoI restriction sites (underlined sequences), respectively. The amplified fragments were cloned into the pET 22b(+) (Novagen) expression vector yielding pET22B-FldA for use in generating the C-terminal His6-tagged protein FldA-His6. The recombinant FldA was expressed in E. coli Rosetta pLacI cells harboring pET22B-FldA. E. coli was cultured at 37 °C in Luria-Bertani broth containing 100 mg of ampicillin per liter. When the optical density at 600 nm reached 0.6, expression was induced by adding 1 mM (final concentration) isopropyl-d-thiogalactoside to the culture, and the cells further cultured at 16 °C for 16 h. The induced cells were collected by centrifugation and stored at −20 °C.
Purification of recombinant FldA was performed aerobically. Approximately 15 g of thawed cells were resuspended in 50 mM Tris-Cl (pH 8.0) containing 500 mM NaCl and 20 mM imidazole and lysed by sonication with a QSonica sonicator, by applying 7 cycles of 45-W pulses for 25 s each with interval of 10 s. Cell debris was removed by centrifugation at 10,000 × g for 30 min at 4 °C. The supernatant solution was loaded onto a Ni-Sepharose high-performance column (1.6 × 13 cm) equilibrated with 50 mM Tris-Cl (pH 8.0), containing 500 mM NaCl and 20 mM imidazole. The column was then washed with 3 column volumes of 50 mM Tris-Cl (pH 8.0) containing 500 mM NaCl and 40 mM imidazole. FldA was batch eluted with 50 mM Tris-Cl (pH 8.0) containing 500 mM NaCl and 250 mM imidazole. FldA-containing fractions were identified by their characteristic yellow coloration and their UV-visible absorbance spectra. These fractions were concentrated using an Amicon 3 kDa MWCO filter and stored at −20 °C. The protein was purified to electrophoretic homogeneity by loading onto a Superdex 75 HR 10/30 column connected to an Akta FPLC system (GE Healthcare). The column was preequilibrated with 50 mM Tris⋅HCl (pH 8) containing 150 mM NaCl and 10% glycerol. FldA eluted with the same buffer as a single isolated peak. Fractions containing FldA were collected and concentrated by using an Amicon 3 kDa MWCO filter and stored at −80 °C.
Redox Titrations.
Please see SI Appendix.
Structure Determination.
FldA was crystallized by vapor diffusion in hanging drops with microseeding at 22 °C against a reservoir containing 0.1 M Bis-Tris (pH 6.5), 0.2 M ammonium acetate, and 20–26% (wt/vol) 2-methyl-2,4-pentanediol (MPD). Crystals reached their full size within 2 d and were yellow in color because of the presence of FMN. For cryocrystallography, the crystals were harvested from a drop, soaked 5 min in 0.1 M Bis-Tris (pH 6.5), 0.2 M ammonium acetate, and 32% (wt/vol) MPD, and then flash-cooled by immersion in liquid nitrogen. Complete 2.34-Å resolution diffraction data were collected by synchrotron radiation (F1 line) at the MacCHESS facility (Cornell University) and processed with HKL2000 (56). Primitive hexagonal space group P6122 crystals (a = b = 112.77 Å, c = 173.72 Ǻ) contained 3 17-kDa FldA molecules per asymmetric unit. The structure was determined by molecular replacement. The search model was derived from the A-type flavoprotein (FprA) from Moorella thermoacetica (PDB ID code 1YCF) via removal of its N-terminal domain (search model contains residues 252–399 and 1 FMN) (57). A molecular replacement solution includes 3 FldA molecules in an asymmetric unit. The electron density map was calculated using phases from the molecular replacement, and it was further improved using the density modification program Resolve (58) which includes 3-fold noncrystallographic symmetry (NCS) restraints. The resulting electron density map has several deviations from the molecular replacement solution, indicating that model bias was effectively removed by density modification. A near complete model was built automatically by using Resolve and a complete model was built by Coot (59). The positional refinement with NCS restraints was performed with refmac5 and Phenix (60, 61). Finally, positional refinement without NCS restraint was carried out, and FMN and water molecules were added to the model. The final model contains 3 FldA molecules (A, residues 1–144; B, residues 1–143; C, residues 1–144) Each molecule has 1 FMN, 1 MPD, 2 acetate, and 474 water molecules (Rwork = 15.8%; Rfree = 17.3%).
Enzyme Assays.
Assays were conducted anaerobically with degassed buffers in an N2 atmosphere. Reduction of FldA or Fdx was monitored at 460 nm or 395 nm, respectively. Initial rates were determined with molar extinction coefficients of 12,800 determined for FldA and 30,600 for Fdx based on clostridal Fdx (62).
Data Availability.
The X-ray crystal structure coordinates and structure factors have been deposited in the Protein Data Bank (ID code 5WID).
Supplementary Material
Acknowledgments
We thank the staff at the Cornell High Energy Synchrotron Source (CHESS)/ Macromolecular X-ray science at the Cornell High Energy Synchrotron Source (MacCHESS) for support of crystallographic data collection. CHESS is supported by the National Science Foundation (NSF) and NIH/National Institute of General Medical Sciences (NIGMS) via NSF Award D-\MR-1332208, and the MacCHESS resource is supported by NIGMS Award GM-103485. This work was supported by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grants DE-FG02-95ER20198 MOD16 (to J.G.F.) and DE-SC0010575 (to J.H.G.), and NIH/NIGMS Grant R01-GM087350 (to K.S.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The X-ray crystal structure coordinates and structure factors have been deposited in the Protein Data Bank (ID code 5WID).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908578116/-/DCSupplemental.
References
- 1.Pierella Karlusich J. J., Lodeyro A. F., Carrillo N., The long goodbye: The rise and fall of flavodoxin during plant evolution. J. Exp. Bot. 65, 5161–5178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sancho J., Flavodoxins: Sequence, folding, binding, function and beyond. Cell. Mol. Life Sci. 63, 855–864 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smillie R. M., Isolation of two proteins with chloroplast ferredoxin activity from a blue-green alga. Biochem. Biophys. Res. Commun. 20, 621–629 (1965). [DOI] [PubMed] [Google Scholar]
- 4.Knight E. Jr, D’Eustachio A. J., Hardy R. W., Flavodoxin: A flavoprotein with ferredoxin activity from Clostrium pasteurianum. Biochim. Biophys. Acta 113, 626–628 (1966). [DOI] [PubMed] [Google Scholar]
- 5.Anderson R. F., Energetics of the one-electron reduction steps of riboflavin, FMN and FAD to their fully reduced forms. Biochim. Biophys. Acta 722, 158–162 (1983). [DOI] [PubMed] [Google Scholar]
- 6.Mayhew S. G., The effects of pH and semiquinone formation on the oxidation-reduction potentials of flavin mononucleotide. A reappraisal. Eur. J. Biochem. 265, 698–702 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Conrad R., The global methane cycle: Recent advances in understanding the microbial processes involved. Environ. Microbiol. Rep. 1, 285–292 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Rothman D. H., et al. , Methanogenic burst in the end-Permian carbon cycle. Proc. Natl. Acad. Sci. U.S.A. 111, 5462–5467 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welte C., Deppenmeier U., Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837, 1130–1147 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Schlegel K., Welte C., Deppenmeier U., Muller V., Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Li Q., et al. , Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188, 702–710 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suharti S., Wang M., de Vries S., Ferry J. G., Characterization of the RnfB and RnfG subunits of the Rnf complex from the archaeon Methanosarcina acetivorans. PLoS One 9, e97966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M., Tomb J. F., Ferry J. G., Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L., et al. , Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome. Res. 6, 759–771 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayhew S. G., Tollin G., “General properties of flavodoxins” in Chemistry and Biochemistry of Flavoenzymes, Muller F., Ed. (CRC Press, Boca Raton, Ann Arbor, London, 1992), vol. 3, pp. 389–426. [Google Scholar]
- 16.Romero A., et al. , Crystal structure of flavodoxin from Desulfovibrio desulfuricans ATCC 27774 in two oxidation states. Eur. J. Biochem. 239, 190–196 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Hsieh Y. C., Chia T. S., Fun H. K., Chen C. J., Crystal structure of dimeric flavodoxin from Desulfovibrio gigas suggests a potential binding region for the electron-transferring partner. Int. J. Mol. Sci. 14, 1667–1683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith W. W., et al. , Structure of oxidized flavodoxin from Anacystis nidulans. J. Mol. Biol. 165, 737–753 (1983). [DOI] [PubMed] [Google Scholar]
- 19.Ludwig M. L., Andersen R. D., Mayhew S. G., Massey V., The structure of a clostridial flavodoxin. I. Crystallographic characterization of the oxidized and semiquinone forms. J. Biol. Chem. 244, 6047–6048 (1969). [PubMed] [Google Scholar]
- 20.Johansson R., et al. , High-resolution crystal structures of the flavoprotein NrdI in oxidized and reduced states–An unusual flavodoxin. Structural biology. FEBS J. 277, 4265–4277 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Bradley L. H., Swenson R. P., Role of glutamate-59 hydrogen bonded to N(3)H of the flavin mononucleotide cofactor in the modulation of the redox potentials of the Clostridium beijerinckii flavodoxin. Glutamate-59 is not responsible for the pH dependency but contributes to the stabilization of the flavin semiquinone. Biochemistry 38, 12377–12386 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z., Swenson R. P., The cumulative electrostatic effect of aromatic stacking interactions and the negative electrostatic environment of the flavin mononucleotide binding site is a major determinant of the reduction potential for the flavodoxin from Desulfovibrio vulgaris [Hildenborough]. Biochemistry 35, 15980–15988 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Abbanat D. R., Ferry J. G., Resolution of component proteins in an enzyme complex from Methanosarcina thermophila catalyzing the synthesis or cleavage of acetyl-CoA. Proc. Natl. Acad. Sci. U.S.A. 88, 3272–3276 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prakash D., Chauhan S. S., Ferry J. G., Life on the thermodynamic edge: Respiratory growth of an acetotrophic methanogen. Sci. Adv. 5, w9059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Z., Joshi P., Gorski C. A., Ferry J. G., A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9, 1642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haroon M. F., et al. , Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500, 567–570 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Smith K. S., Ingram-Smith C., Methanosaeta, the forgotten methanogen? Trends Microbiol. 15, 150–155 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Hoover D. M., et al. , Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials. J. Mol. Biol. 294, 725–743 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Zhou Z., Swenson R. P., Electrostatic effects of surface acidic amino acid residues on the oxidation-reduction potentials of the flavodoxin from Desulfovibrio vulgaris (Hildenborough). Biochemistry 34, 3183–3192 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Vervoort J., et al. , Properties of the complexes of riboflavin 3′,5′-bisphosphate and the apoflavodoxins from Megasphaera elsdenii and Desulfovibrio vulgaris. Eur. J. Biochem. 161, 749–756 (1986). [DOI] [PubMed] [Google Scholar]
- 31.Swenson R. P., Krey G. D., Site-directed mutagenesis of tyrosine-98 in the flavodoxin from Desulfovibrio vulgaris (Hildenborough): Regulation of oxidation-reduction properties of the bound FMN cofactor by aromatic, solvent, and electrostatic interactions. Biochemistry 33, 8505–8514 (1994). [DOI] [PubMed] [Google Scholar]
- 32.O’Farrell P. A., et al. , Modulation of the redox potentials of FMN in Desulfovibrio vulgaris flavodoxin: Thermodynamic properties and crystal structures of glycine-61 mutants. Biochemistry 37, 8405–8416 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Watt W., Tulinsky A., Swenson R. P., Watenpaugh K. D., Comparison of the crystal structures of a flavodoxin in its three oxidation states at cryogenic temperatures. J. Mol. Biol. 218, 195–208 (1991). [DOI] [PubMed] [Google Scholar]
- 34.Ludwig M. L., et al. , Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: The role of conformation changes. Biochemistry 36, 1259–1280 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Ishikita H., Influence of the protein environment on the redox potentials of flavodoxins from Clostridium beijerinckii. J. Biol. Chem. 282, 25240–25246 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Ishikita H., Redox potential difference between Desulfovibrio vulgaris and Clostridium beijerinckii flavodoxins. Biochemistry 47, 4394–4402 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Chang F. C., Swenson R. P., The midpoint potentials for the oxidized-semiquinone couple for Gly57 mutants of the Clostridium beijerinckii flavodoxin correlate with changes in the hydrogen-bonding interaction with the proton on N(5) of the reduced flavin mononucleotide cofactor as measured by NMR chemical shift temperature dependencies. Biochemistry 38, 7168–7176 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Talfournier F., et al. , Alpha Arg-237 in Methylophilus methylotrophus (sp. W3A1) electron-transferring flavoprotein affords approximately 200-millivolt stabilization of the FAD anionic semiquinone and a kinetic block on full reduction to the dihydroquinone. J. Biol. Chem. 276, 20190–20196 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Lindqvist Y., Brändén C. I., The active site of spinach glycolate oxidase. J. Biol. Chem. 264, 3624–3628 (1989). [PubMed] [Google Scholar]
- 40.Wierenga R. K., Drenth J., Schulz G. E., Comparison of the three-dimensional protein and nucleotide structure of the FAD-binding domain of p-hydroxybenzoate hydroxylase with the FAD- as well as NADPH-binding domains of glutathione reductase. J. Mol. Biol. 167, 725–739 (1983). [DOI] [PubMed] [Google Scholar]
- 41.Chowdhury N. P., Klomann K., Seubert A., Buckel W., Reduction of flavodoxin by electron bifurcation and sodium ion-dependent re-oxidation by NAD+ catalysed by ferredoxin:NAD+ reductase (Rnf). J. Biol. Chem. 291, 11993–12002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes D. E., et al. , A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer. MBio 10, e00789–e00719. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowers K. R., Baron S. F., Ferry J. G., Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47, 971–978 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner D. R., Hunter K. A., de Baar H. J. W., “Introduction” in The Biogeochemistry of Iron in Seawater, Turner D. R., Hunter K. A., Eds. (John Wiley & Sons, Chichester, UK, 2001), pp. 1–7. [Google Scholar]
- 45.Ferry J. G., How to make a living exhaling methane. Annu. Rev. Microbiol. 64, 453–473 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Moyano A. J., et al. , A long-chain flavodoxin protects Pseudomonas aeruginosa from oxidative stress and host bacterial clearance. PLoS Genet. 10, e1004163 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coba de la Peña T., Redondo F. J., Fillat M. F., Lucas M. M., Pueyo J. J., Flavodoxin overexpression confers tolerance to oxidative stress in beneficial soil bacteria and improves survival in the presence of the herbicides paraquat and atrazine. J. Appl. Microbiol. 115, 236–246 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Lodeyro A. F., Ceccoli R. D., Pierella Karlusich J. J., Carrillo N., The importance of flavodoxin for environmental stress tolerance in photosynthetic microorganisms and transgenic plants. Mechanism, evolution and biotechnological potential. FEBS Lett. 586, 2917–2924 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Horne A. J., Lessner D. J., Assessment of the oxidant tolerance of Methanosarcina acetivorans. FEMS Microbiol. Lett. 343, 13–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan Z., Wang M., Ferry J. G., A Ferredoxin- and F420H2-dependent, electron-bifurcating, heterodisulfide reductase with homologs in the domains Bacteria and Archaea. MBio 8, e02285-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ellman G. L., A colorimetric method for determining low concentrations of mercaptans. Arch. Biochem. Biophys. 74, 443–450 (1958). [DOI] [PubMed] [Google Scholar]
- 52.Eskelinen S., Haikonen M., Räisänen S., Ferene-S as the chromogen for serum iron determinations. Scand. J. Clin. Lab. Invest. 43, 453–455 (1983). [PubMed] [Google Scholar]
- 53.Siegel L. M., A direct microdetermination for sulfide. Anal. Biochem. 11, 126–132 (1965). [DOI] [PubMed] [Google Scholar]
- 54.Kozioł J., Fluorometric analyses of riboflavin and its coenzymes. Methods Enzymol. 18, 253–285 (1971). [Google Scholar]
- 55.Bradford M. M., Williams W. L., New, rapid, sensitive method for protein determination. Fed. Proc. 35, 274 (1976). [Google Scholar]
- 56.Otwinowski Z., Minor W., Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Silaghi-Dumitrescu R., Kurtz D. M. Jr, Ljungdahl L. G., Lanzilotta W. N., X-ray crystal structures of Moorella thermoacetica FprA. Novel diiron site structure and mechanistic insights into a scavenging nitric oxide reductase. Biochemistry 44, 6492–6501 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger T. C., Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Murshudov G. N., Vagin A. A., Dodson E. J., Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Afonine P. V., et al. , Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong J.-S., Rabinowitz J. C., Molar extinction coefficient and iron and sulfide content of clostridial ferredoxin. J. Biol. Chem. 245, 4982–4987 (1970). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The X-ray crystal structure coordinates and structure factors have been deposited in the Protein Data Bank (ID code 5WID).