Significance
Honey bees and other social bees harbor specialized gut microbiota dominated by 5 coevolved bacterial clusters. Bees eat pollen, which contains diverse polysaccharides, energy-rich substrates potentially digested by gut bacteria. Polysaccharide degradation genes were identified in genome sequences of cultured bacteria and in metagenomic data, revealing that Bifidobacterium and Gilliamella digest polysaccharides in the honey bee gut. In both, individual strains vary in these abilities. Polysaccharide-degrading genes are clustered within Bifidobacterium genomes and are expressed in response to specific substrates. Other bee gut bacterial species cannot degrade polysaccharides, and some species rely on others for amino acids. This work provides insight into how bacterial species diverge into different ecological niches within the gut of their hosts.
Keywords: honey bee, gut microbiota, polysaccharide, amino acid, symbiosis
Abstract
Bees acquire carbohydrates from nectar and lipids; and amino acids from pollen, which also contains polysaccharides including cellulose, hemicellulose, and pectin. These potential energy sources could be degraded and fermented through microbial enzymatic activity, resulting in short chain fatty acids available to hosts. However, the contributions of individual microbiota members to polysaccharide digestion have remained unclear. Through analysis of bacterial isolate genomes and a metagenome of the honey bee gut microbiota, we identify that Bifidobacterium and Gilliamella are the principal degraders of hemicellulose and pectin. Both Bifidobacterium and Gilliamella show extensive strain-level diversity in gene repertoires linked to polysaccharide digestion. Strains from honey bees possess more such genes than strains from bumble bees. In Bifidobacterium, genes encoding carbohydrate-active enzymes are colocated within loci devoted to polysaccharide utilization, as in Bacteroides from the human gut. Carbohydrate-active enzyme-encoding gene expressions are up-regulated in response to particular hemicelluloses both in vitro and in vivo. Metabolomic analyses document that bees experimentally colonized by different strains generate distinctive gut metabolomic profiles, with enrichment for specific monosaccharides, corresponding to predictions from genomic data. The other 3 core gut species clusters (Snodgrassella and 2 Lactobacillus clusters) possess few or no genes for polysaccharide digestion. Together, these findings indicate that strain composition within individual hosts determines the metabolic capabilities and potentially affects host nutrition. Furthermore, the niche specialization revealed by our study may promote overall community stability in the gut microbiomes of bees.
Plant polysaccharides are abundant in many animal diets, but animals generally lack enzymes to digest these substrates (1). Instead, many rely on gut bacteria to break diverse polysaccharide bonds (2) and to release sugars or short chain fatty acids that can be absorbed by hosts (3). In humans, dietary polysaccharides shape gut microbial ecology and, in turn, host physiology and health (4). As in humans, honey bees (Apis mellifera) host a conserved bacterial community in the distal gut that can digest polysaccharides (5). Hindgut compartments are densely colonized by 5 bacterial clades (6). Snodgrassella alvi, a microaerophilic species of Betaproteobacteria, adheres to the ileum wall and consumes acetate and oxygen, generating an anoxic lumen (5). Snodgrassella is topped with a layer of Gilliamella, saccharolytic fermenters of the Gammaproteobacteria (7). Two clades of Lactobacillus, called Firm-4 and Firm-5, are present in the lumen of the hindgut, along with Bifidobacterium species. These 5 bacterial groups are the dominant members in gut communities of a large clade of social bees, including honey bees (genus Apis), bumble bees (genus Bombus), and stingless bees (tribe Meliponini) (8).
Bees feed solely on floral nectar, which provides sugars, and pollen, which provides amino acids, lipids, and vitamins and which is required for normal weight gain in newly emerged adult honey bees (9). The pollen cytoplasm is surrounded by several layers: the pollenkit; the exine layer, consisting of sporopollenin; and the intine, composed of polysaccharides, i.e., cellulose, hemicellulose, and pectin (10). These polysaccharides contain diverse chemical bonds, and their structures are highly variable depending on plant species (11). Pollen cell walls mainly contain type I rhamnogalacturonan (RG-I) and homogalacturonan, which are the major components of pectin. They also have arabinogalactan as well as monosaccharides, including mannose, xylose, galactose, and arabinose (12, 13). Pectins are classified into 3 major groups: homogalacturonan, RG-I, and rhamnogalacturonan II (14). Homogalacturonan is the most abundant pectic polymer and consists of linear chains of α-1,4-linked d-galacturonate residues, which can be methyl esterified or acetylated. The enzymes degrading the main chain of pectin can be divided into hydrolases and lyases. Polygalacturonases (GH28) act by hydrolysis, whereas pectin/pectate lyases cleave pectin using a β-elimination mechanism (15). Pectate lyases are assigned to 5 PL families: PL1, PL2, PL3, PL9, and PL10. In addition, PL22 cleaves only short oligogalacturonates, and PL4 specifically targets the RG-I backbone (16). As a whole, the bee gut microbiota possesses a diverse repertoire of carbohydrate-active enzymes (CAZymes) (17, 18). Specifically, some strains of G. apicola harbor pectate lyase genes and can break down pectin in vitro (18), and other bacterial lineages also may participate in polysaccharide digestion (17, 19). Studies to date have been based largely on metatranscriptomic and metagenomic datasets, preventing elucidation of gene repertoires of particular strains as needed for a clear picture of how community members divide up the many enzymatic steps of polysaccharide degradation.
Here, we investigate the potential for digestion of pollen-derived polysaccharides of all core members of the bee gut microbiota. We analyze genomes of bacterial isolates from Apis and Bombus guts and identify genes encoding glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterase (CE), glycosyl transferase (GT), and carbohydrate-binding module (CBM), as defined in the CAZy database (20). Bifidobacterium and Gilliamella are implicated as the primary degraders of hemicellulose and pectin, while Snodgrassella and Lactobacillus play little or no role in polysaccharide digestion. These findings are further supported by analyses of a metagenomic dataset for the A. mellifera gut microbiota and by experimental trials conducted in vitro and in vivo.
Results
Gut Bacteria of Bees Have Distinct Repertoires of Carbohydrate-Active Enzymes.
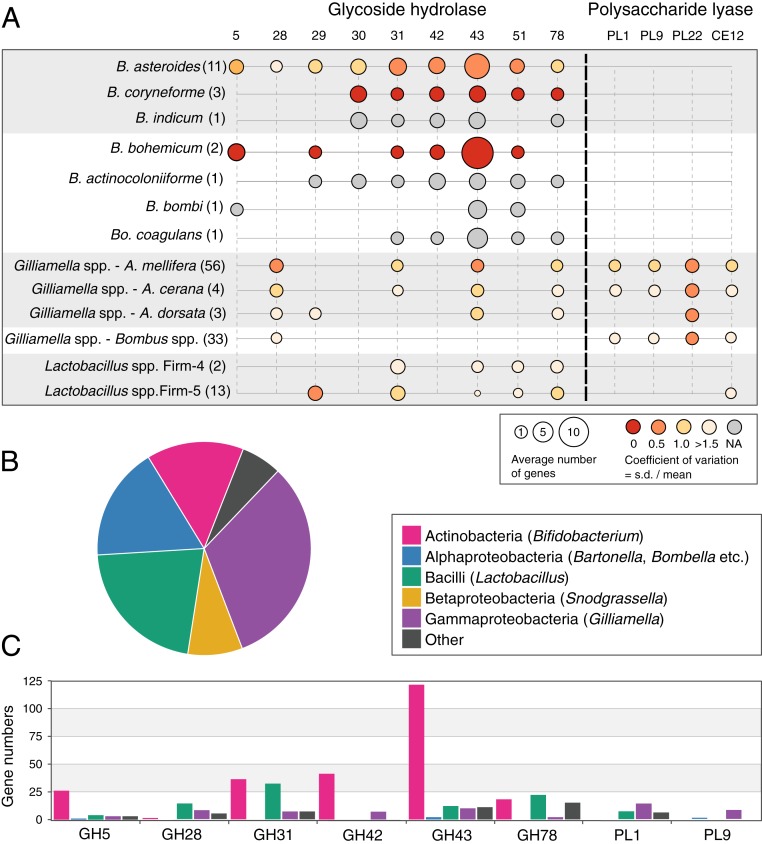
To investigate the carbohydrate-digestive capacities of the bacterial species in bee guts, we identified genes underlying CAZymes in 231 genomes of isolates originating from both Apis and Bombus hosts (Dataset S1). We found divergent CAZyme profiles for bee gut bacterial species. Bifidobacterium species possess many more GH genes than species of any other genus, while PL genes occur exclusively in a subset of Gilliamella strains (Fig. 1A).
Fig. 1.
(A) The average relative abundance of GH and PL families in the genomes of Bifidobacterium, Gilliamella, and Lactobacillus from bee guts. Gilliamella genomes from A. mellifera, Apis cerana, and Bombus species are shown separately. A full list of the distribution of all GH/PL families in all analyzed genomes is given in Dataset S2. The numbers of genomes analyzed for each species are indicated in parentheses. The circle size indicates the average gene numbers of the GH/PL family per genome. The circle color represents the relative variability in number of GH/PL across genomes, calculated as the coefficient of variation (ratio of SD to the mean). (B) Relative abundances of bacterial groups in the gut microbiota of A. mellifera based on the best BLASTP hit distribution of 614,276 CDSs from the metagenomic data. Names in parentheses indicate likely bee gut taxa from which the CDSs derive. (C) Distributions of select GHs across different bacterial genera based on metagenomic data.
In the analyzed Gilliamella genomes, we identified GH28, PL1, PL9, PL22, and an acetylesterase, CE12. No other bee gut bacteria possess PLs, although 2 strains of the Lactobacillus Firm-5 group have a single CE12 gene (Fig. 1A). Although the CBM32 family is known to play a role in pectin degradation (15), we only identified CBM13, CBM50, and CBM67 families in the bee gut species (Dataset S2).
GHs are specifically enriched in bifidobacterial species of the bee gut. GH31 (exo-α-glycosidases with activity for α-glucosides, α-xylosides, and α-galactosides), GH42 (β-galactosidase), GH43 (α-l-arabinofuranosidase, β-d-xylosidase, α-l-arabinanase, β-d-galactosidase), GH51 (cleaving terminal, nonreducing α-l-arabinofuranose residues from arabinose-containing compounds), and GH5 subfamily 4 (xyloglucanase) and 18 (β-mannosidase) were identified in most of the Bifidobacterium genomes. Interestingly, we also identified GH28 and GH78 (rhamnogalacturonases) in the genomes of Bifidobacterium, which suggests that they also contribute to pectin degradation.
To confirm the distinct glycan niches of different bee gut bacteria, we sequenced the metagenome of the gut microbiota of bees from a colony of A. mellifera, resulting in an assembly with total length of 250 Mb (SI Appendix, Table S1). The bacterial community was dominated by the known core members (Fig. 1B), as expected based on previous studies (e.g., refs. 8 and 18). We identified 2,656 CAZyme-coding genes in this assembly. Analysis of taxon affiliations (Materials and Methods) revealed that most GH genes belonged to Bifidobacterium and most PLs to Gilliamella (Fig. 1C). These assignments fit with the genomic results from isolates and confirm that GHs and PLs are specifically enriched in Bifidobacterium and Gilliamella, respectively.
Bee Gut Bifidobacterium Exhibit Strain-Level Variation in Their GH Repertoires.
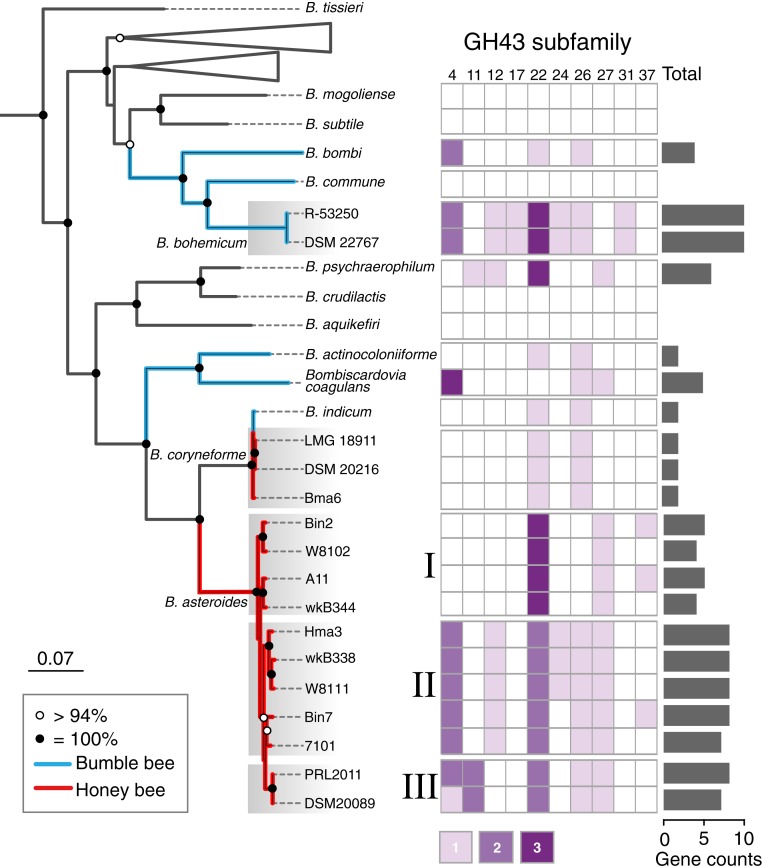
The Bifidobacterium genomes show obvious intraspecific and interspecific differences in repertoires of genes involved in carbohydrate metabolism (Fig. 1A). To further investigate the GH profiles of Bifidobacterium strains, we performed phylogenomic analysis on the Bifidobacterium species isolated from both Apis and Bombus together with representative genomes of described species from the genus (21). A phylogeny based on shared ribosomal protein genes showed that Bifidobacterium species from the bee gut form separate lineages (Fig. 2 and SI Appendix, Fig. S1A), corroborating previous findings (22). Further, strains from Apis hosts generally cluster separately from those from Bombus hosts, as found for other bee gut bacteria (23, 24). Bifidobacterium from Bombus are derived from 2 distinct phylogenetic lineages. B. actinocoloniiforme, B. coagulans, and B. indicum are more closely related to strains from Apis, while B. bombi, B. commune, and B. bohemicum clustered together with other members of the genus. The strains within subclades have high (>93%) average nucleotide identities (ANI), whereas ANI between genomes from different subclades are lower (90–92%) (SI Appendix, Fig. S1B), suggesting that these subclades represent separate species (25).
Fig. 2.
Phylogenetic tree of Bifidobacterium strains using the maximum-likelihood algorithm based on the concatenation of 101 core protein sequences (29,909 amino acid positions). Bootstrap values are indicated on nodes, and strains from Apis or Bombus are indicated by branch color. Strains of Bifidobacterium coryneforme and 3 clusters of Bifidobacterium asteroides are indicated by gray shading. A full tree is shown in SI Appendix, Fig. S1. The heatmap shows the numbers of genes belonging to GH43 subfamilies in each strain. The total numbers of GH43 genes are shown by bar plots.
GH43 members are assumed to be involved in degradation of hemicellulosic backbones or debranching hemicellulose and pectin polymers (26). Notably, GH43 is the most abundant GH family in bee gut bifidobacterial species (Fig. 1A). We identified 10 subfamilies of GH43 in these genomes (26) (SI Appendix, Fig. S2). The GH43 repertoires varied among species, and closely related strains always shared similar profiles. Strains R-53250 and DSM 22767 of B. bohemicum have the most GH43 genes (10 per genome); other strains from Bombus have 0–5 GH43 genes per genome (Fig. 2). According to the GH43 profiles, the 9 strains of B. asteroides from Apis formed 3 subclades, consistent with the phylogenomic analysis. Although some bifidobacterial species harbor fewer GHs, almost all possess genes for metabolizing monosaccharides that are released through polysaccharide breakdown (SI Appendix, Fig. S3). The distribution and frequency of GH43 subfamily genes across the Bifidobacterium genomes implies a history of gene duplications and deletions or of frequent horizontal gene transfer. To investigate the latter possibility, we reconstructed the phylogenetic relationships of genes encoding GH43 subfamily 34, a single-copy gene present in all Bifidobacterium strains except Bin2 (SI Appendix, Fig. S4). The phylogenetic histories of GH43-34 genes are similar to the phylogeny for the genome overall, suggesting that these genes have been solely or primarily vertically transmitted within Bifidobacterium and have not been exchanged or acquired multiple times.
GH Genes of Bifidobacterium Are Organized into Polysaccharide Utilization Loci and Expressed in Response to Different Hemicelluloses.
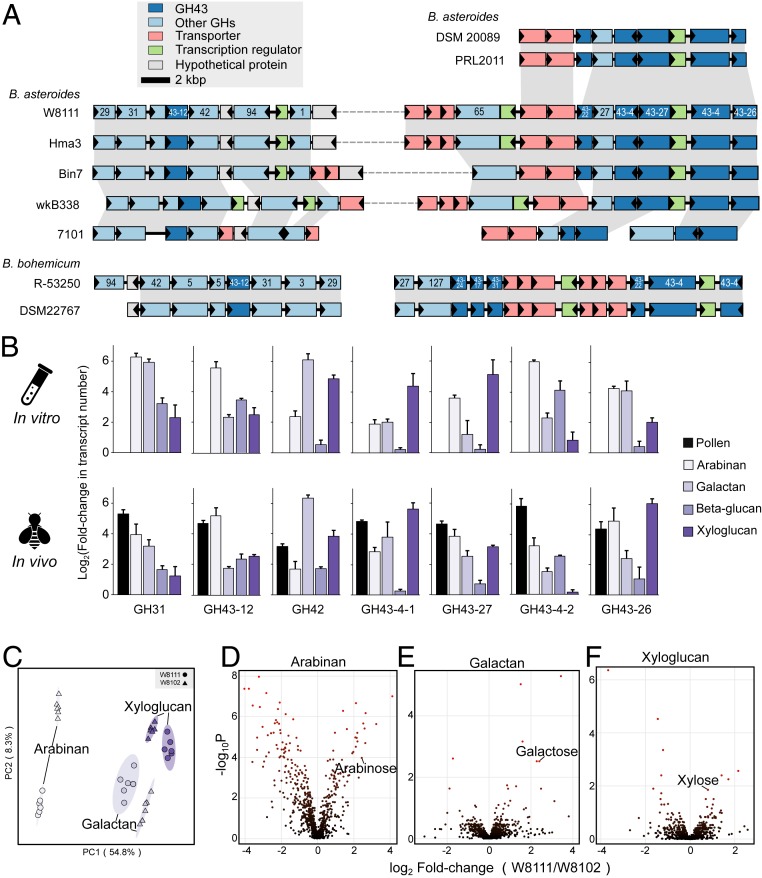
In Bacteroidetes from the human gut, CAZyme-encoding genes are colocalized and coregulated together with genes encoding regulators and transporters, forming polysaccharide utilization loci (PULs), which enable the saccharification of complex carbohydrates (27). We explored the arrangements of GHs in genomes of Bifidobacterium species from bees. B. asteroides clades II and III from Apis and B. bohemicum from Bombus contain PUL-like structures in their genomes (Fig. 3A). Interestingly, B. asteroides clade II strains have the largest gene cluster, made of 12–13 GH genes together with genes encoding transporters and transcriptional regulators, while B. asteroides clade III strains possess only a subset of these genes. In B. bohemicum, the cluster is divided into 2 loci, in which GH43 is the most abundant family. Genes within these regions were syntenic among strains from Apis and Bombus. PUL-like loci are absent from other strains, which have fewer GH43 genes.
Fig. 3.
Genomic organization, expression, and function of genes involved in digestion of polysaccharides. (A) Syntenic loci of GHs, transporter genes, and transcriptional regulators in B. asteroides and Bifidobacterium bohemicum strains. Homologous genes are connected by gray bars, and the GH family number is shown for respective genes. (B) Gene expression profiles of GHs in response to different hemicellulose substrates. Error bars represent SDs of 3 biological replicates. (C) Results of partial least squares discriminant analysis based on 687 metabolites detected from guts of W8111-colonized or W8103-colonized bees fed on sucrose supplemented with arabinan, galactan, or xyloglucan. Six biological replicates for each treatment. (D–F) Volcano plots of differentially abundant metabolites identified in guts of monoassociated bees fed on different polysaccharides.
To define if the PUL-like regions are active in polysaccharide degradation, we first tested ability of strain W8111 from B. asteroides clade II to use various hemicelluloses by growing it on chemically defined medium supplemented with arabinan, galactan, β-glucan, xyloglucan, or xylan. Cells grew on all hemicelluloses tested, although they achieved lower cell densities than on glucose (SI Appendix, Fig. S5A). In contrast, B. asteroides W8102 with a lower GH index could not grow on any of the hemicelluloses (SI Appendix, Fig. S6). Thus, the PUL-like locus appears to enable polysaccharide digestion.
We subsequently quantified expressions of W8111 PUL-like genes both in vitro and in the guts of monoassociated bees fed sucrose solution supplemented with pollen, or with one of the hemicelluloses. Microbiota-free bees were inoculated with pure cultures of W8111 and W8102, resulting in the establishment of ∼107 bacterial cells per gut 3 d postinoculation (SI Appendix, Fig. S5B). All 7 GH family genes assayed were up-regulated in response to at least 1 hemicellulose source in vitro (Fig. 3D). Furthermore, expression of all 7 genes increased in vivo when bees were fed pollen, which is rich in plant cell wall glycans. Transcriptional responses of specific GH genes depended on which hemicellulose was present. GH42 expression increased 90-fold in the presence of galactan in vivo (Fig. 3D). This family has been characterized in probiotic bifidobacteria as a β-galactosidase, catalyzing the release of galactose from the nonreducing end of different β-d-galactosides such as galactan (28). We also tested 5 GH43 genes belonging to different subfamilies. Genes from subfamilies GH43-12 and GH43-27 were up-regulated by all hemicellulose substrates, while GH43-26 did not respond to β-glucan. Strain W8111 harbors 2 genes of subfamily GH43-4; interestingly, they responded differently to the hemicelluloses. GH43-4-1 increased expression 38-fold in response to xyloglucan, while GH43-4-2 responded more to arabinan than to xyloglucan in vivo. Together, these findings provide evidence that GH genes can be broadly involved in degradation of multiple major classes of hemicelluloses, or highly specific to certain classes of hemicelluloses. The elevated gene expression in vivo in response to pollen suggests that these glycans are present in ingested pollen and are degraded by Bifidobacterium in the bee rectum.
To confirm that the encoded enzymes actually perform the biochemical functions, we performed untargeted metabolomic analyses of gut contents from W8111- and W8102-colonized bees fed on different polysaccharides. The metabolomic profiles are different between bees that were fed on same diets but were experimentally monocolonized with different Bifidobacterium strains (Fig. 3C). Notably, we found differences in arabinose, galactose, and xylose, depending on the Bifidobacterium strain colonizing the gut. These monosaccharides are significantly higher in W8111-colonized bees fed on hemicellulose (Fig. 3 D–F). Elevated levels of these metabolites confirm that Bifidobacterium with high GH index are able to digest hemicellulose.
Gilliamella Is the Primary Degrader of Pectin in the Bee Gut.
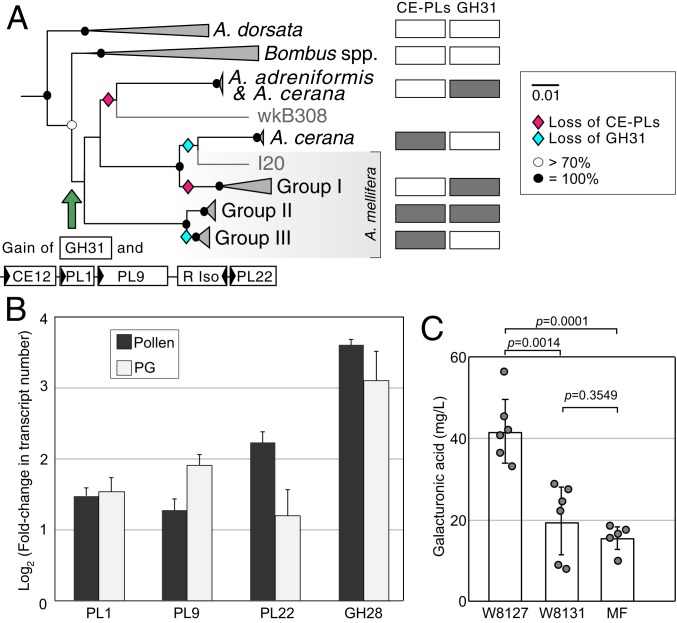
Pectinases can be classified as esterases, eliminative depolymerases (PLs), and hydrolytic depolymerases (GHs). The CAZy-based annotation of Gilliamella identified that the pectin (PL1) and pectate (PL9) lyases, the oligogalacturonate lyase (PL22), and one acetylesterase (CE12) were colocalized in 1 genomic region together with a ribose isomerase gene, forming a modular structure (Fig. 4A). Based on comparative genomic analysis, strains from Apis and Bombus fall into distinct monophyletic groups (SI Appendix, Fig. S7) (23). Almost all Gilliamella possess the PL22 gene, but PL1, PL9, and CE12 genes were mainly absent from Gilliamella from Bombus hosts and were completely absent from the cluster corresponding to Gilliamella apis strains (group I) as well as Gilliamella from Apis dorsata (Fig. 4A and SI Appendix, Fig. S8). The distributions of other GHs were the same across Gilliamella strains, except that GH31 (SI Appendix, Fig. S8), which has transglycosylation activity and might be involved in pectin degradation (29), was identified only in groups II and III and 2 strains from A. cerana and Apis andreniformis. These results suggest that the PLs and GHs related to pectin degradation were acquired by one sublineage of Gilliamella and were lost several times subsequently (Fig. 4A). However, CE8 is absent from Gilliamella genomes, and it is not clear how PL9 and polygalacturonase cleave the bonds attached to methylesterified residues (30). Thus, bee gut Bifidobacterium with abundant GHs might hydrolyze short residual pectin with extensive glycosyl side chains, which could have sterically protected the backbone from degradation by Gilliamella.
Fig. 4.
(A) Phylogenetic tree of Gilliamella isolates using the maximum-likelihood algorithm based on the concatenation of 106 core protein sequences (32,259 amino acid positions). A full phylogenetic tree is shown in SI Appendix, Fig. S6. The presence/absence of the loci containing genes for PLs and CE12 and the gene for GH31 gene are indicated by open/closed boxes. (B) In vivo gene expression profiles of genes of G. apicola W8127 in response to pollen and polygalacturonic acid (PG) fed to the monoinoculated bees relative to controls (sucrose syrup). Error bars represent SDs of 3 biological replicates. (C) Gut concentrations of galacturonic acid of monoassociated and microbiota-free (MF) bees fed on polygalacturonic acid (n = 5 to 6). Tested by Mann–Whitney u test.
To determine whether genes underlying pectin degradation are expressed in response to pectin in the diet, we measured transcriptional responses of PL1, PL9, PL22, and GH28 genes in vivo. Supplementation with pollen or polygalacturonic acid resulted in up-regulation of all of these genes in G. apicola W8127 in monoassociated bees (Fig. 4B), supporting a role of Gilliamella in degradation of dietary pectin. Since the increase in gene expression was not dramatic, we confirmed the capacity of strain W8127 for pectin degradation by determining the gut concentration of d-galacturonic acid of gnotobiotic bees. Concentrations of d-galacturonic acid in gut contents of W8127-colonized bees were higher than concentrations in those associated with W8131 and microbiota-free bees (Fig. 4C and SI Appendix, Fig. S6). This result indicates that Gilliamella with PL-CE gene clusters are responsible for breakdown of pectin in the gut.
Certain Bee Gut Symbionts Do Not Synthesize Their Own Amino Acids.
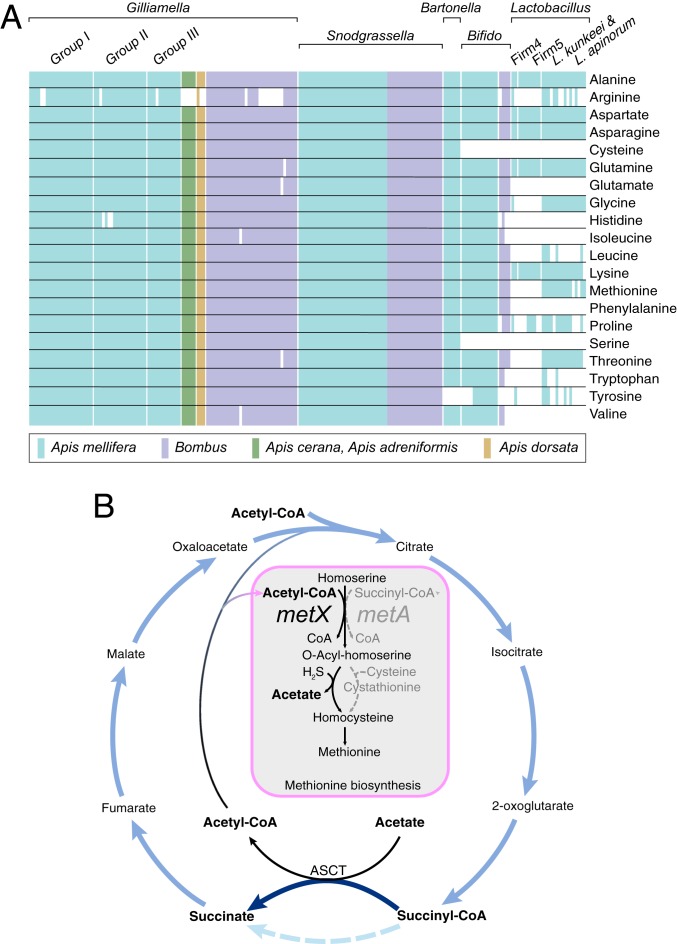
The provisioning of amino acids, particularly protein amino acids that animals cannot synthesize, is a prominent function of many insect symbionts (31). We investigated genes related to synthesis of amino acids in bee gut bacterial genomes (Dataset S3). Almost all Gilliamella and Snodgrassella strains possess all genes required for synthesis of the 20 protein amino acids, except that argG (needed for arginine biosynthesis) and hisB (needed for histidine biosynthesis) were not detected in some Gilliamella strains. These pathways are otherwise intact in those strains, suggesting that failure to detect these genes reflects incompleteness or fragmentation of the genome sequences, or the presence of alternative, uncharacterized enzymes that perform these steps. In contrast to Snodgrassella and Gilliamella strains, many amino acid biosynthetic pathways were absent from Lactobacillus isolates (Fig. 5A). For Bifidobacterium, most strains from Apis spp. are predicted to be able to synthesize most amino acids, whereas strains from Bombus lack many of the required genes.
Fig. 5.
(A) Presence and absence of genes underlying amino acid synthesis in the genomes of 231 bacterial isolates from bee guts. Colored boxes indicate the presence of all genes required for a pathway, and white boxes indicate the absence of genes for a pathway. (B) The TCA cycle with the ASCT pathway and the methionine synthesis pathway found in Snodgrassella strains and Bartonella strains, which incorporates acetyl-CoA and acetate rather than succinyl-CoA.
S. alvi has an alternative TCA cycle, which utilizes acetate:succinate CoA-transferase (ASCT) for the synthesis of succinate (32). We found that both S. alvi and Bartonella apis from the bee gut possess genes encoding ASCT; interestingly, they both have the homoserine O-acetyltransferase gene (metX) but not the O-succinyltransferase gene (metA) (Fig. 5B and Dataset S3). Thus, they appeared to use acetyl-CoA, rather than succinyl-CoA, as the acyl substrate to activate the γ-hydroxyl group of homoserine. For the incorporation of sulfur, they appear to synthesize homocysteine directly from acyl-homoserine and hydrogen sulfide through sulfhydrylation, which does not rely on cysteine as the sulfur donor. These pathways are characteristic of Corynebacterium glutamicum, which has been widely used for the fermentative production of amino acids (33).
Discussion
We document striking niche segregation among the species and strains of gut-dwelling bacteria in social bees. Our analysis of 231 isolates, combined with transcriptomic and metabolomic experiments, has allowed us to assign capabilities to specific members of the bee gut microbiota. We discovered that strains have very different capabilities to digest pollen, and that this variation occurs both within and between host bee species.
Simple sugars such as glucose and fructose, the main nutrients in nectar, are absorbed in the host midgut (34). Consequently, the main energy and carbon sources available to hindgut bacteria are recalcitrant substrates such as the pollen intine, which consists of cellulose microfibrils embedded in a matrix of pectin and hemicellulose. We found that genes for hemicellulose and pectin breakdown are common in genomes of honey bee hindgut isolates, whereas GH families known to degrade cellulose are scarce. Our findings corroborate an early histochemical study, showing that hemicellulose and pectic acids were digested, but cellulose and sporopollenin were not lysed (35). However, some insects including honey bees possess endogenous GH9 cellulase, which might be involved in degradation of the inner cellulosic wall of the pollen (36, 37). Metabolomic analyses of gut homogenates also provided evidence that honey bee gut bacteria can utilize some outer pollen wall components (38). Altogether, these results indicate that the honey bee gut microbiota digests complex carbohydrates, thereby acquiring energy. Some of the released short chain fatty acids are absorbed by the host and appear to contribute to host health (5).
Five core bacterial members of the microbiome are ubiquitous and stable across both honey bee and bumble bee species and have evolved with them as they have diversified during the last 80 million years (7, 8). We hypothesize that these core bacteria occupy distinct metabolic niches, enabling them to coexist in a resource-limited environment. Supporting this hypothesis within A. mellifera, some G. apicola, chiefly found in the ileum, can digest pectin, while some Bifidobacterium strains, mainly located downstream in the rectum, possess GHs for digesting hemicellulose and are predicted to hydrolyze glycosyl side chains of pectin (39, 40). This substrate partitioning and physical separation of gut bacteria may facilitate bacterial energy acquisition and may support host nutrition, as has been proposed for the human gut (41). GHs involved in pectin hydrolysis were also identified in other genera of bee gut bacteria, in particular, some Lactobacillus and Bifidobacterium species possess GH5, GH78, and GH28 genes (Fig. 1 and Dataset S2), although the occurrence of GH genes is limited and highly variable among Lactobacillus strains (24). Thus, degradation of pectin including extensive glycosyl side chains requires a diverse array of enzymes (40); collaborative actions of both PLs (PL1, PL9, PL22) and GHs (GH43, GH5, GH78, GH28) might be required for degradation of pectin in the bee gut (16).
Gilliamella strains are prominent members of bee gut communities and together form a distinct clade that has evolved with social bee hosts (9). Gilliamella strains have diverged in carbohydrate-utilization capabilities, which can differ strikingly both within and between host species. Gilliamella strains were previously found to vary in abilities to metabolize monosaccharides (23). We found that this variation extends to pectin digestion, due to histories of gain and loss of PL and GH31 genes. Notably, presence of these genes differs between the closely related G. apicola and G. apis, which coexist in many or all A. mellifera workers, implying niche separation within individual hosts. Also striking is the contrast between Apis and Bombus in pectin degradation abilities of their Gilliamella (Fig. 1). This finding that certain Apis-derived Gilliamella strains have greater ability than Bombus-derived strains to degrade pectin parallels previous results showing more extensive abilities for use of associated monosaccharides in Apis-derived strains (23).
Bifidobacteria are some of the dominant gut commensals of humans and all mammals analyzed to date (42, 43). Their genomes reflect their dependence on glycan substrates (44, 45). Within bee guts, Bifidobacterium species possess a large number of GH genes associated with hemicellulose and possibly pectin digestion. In general, gut-associated Bifidobacterium are enriched in GH43 (46, 47). While Bifidobacterium from bees possess fewer GH genes than strains from other hosts (47), all hemicelluloses tested can be used by strains, such as W8111, which had the highest GH43 index. Bifidobacterial GH43 is predicted to be secreted and to exhibit extracellular activity resulting in release of monosaccharides that might be accessed by other gut members (48), resulting in glycan cross-feeding, as reported for Bifidobacterium under in vitro conditions (49). Such cooperative metabolism may increase the efficiency of substrate utilization by the overall community (50).
Complex carbohydrates are composed of diverse monosaccharide subunits and glycosidic linkages (51). A distinctive feature of genomes of human gut-dwelling Bacteroidetes is the presence of PULs, clusters of colocalized, coregulated genes, which are responsible for the detection, sequestration, digestion, and transport of polysaccharides (27, 52). In bee gut Bifidobacteria strains, the GHs cluster with genes encoding transporters and transcriptional regulators. They form CAZyme gene clusters, which have been defined as a general term of PULs and have been found in more bacteria in addition to Bacteroidetes (53). These colocalized genes are not expressed together as a single locus but, rather, are expressed independently in response to specific monosaccharides and glycosidic linkages. Similar observations have been made for expression of PULs from several marine bacterial species (54).
Gut bacteria often provide amino acids to hosts. In humans, this provisioning may have implications for disorders, such as insulin resistance (55). In insects, dietary amino acids are absorbed in the host midgut (56), resulting in their depletion in the hindgut. This depletion was evident in a mutagenesis study showing that S. alvi genes underlying amino acid biosynthesis are required for colonization of the bee ileum (57). However, nitrogen is abundant in the form of wastes (uric acid and ammonia) that enter the gut via the Malpighian tubules, at the midgut-ileum junction. These nitrogenous waste products appear to be recycled within the ileum by Gilliamella and Snodgrassella, which possess pathways for synthesis of all protein amino acids. Other core bee gut species lack pathways for synthesis of many or most protein amino acids; these species may rely on production by coresident bacteria. Potentially, this nitrogen recycling also contributes to bee nutrition; however, whether amino acids synthesized in the hindgut can be absorbed by hosts is unknown.
Methionine is an essential amino acid and a precursor for cysteine and other metabolic intermediates (58); it affects starvation resistance and lifespan in Drosophila (59, 60). Methionine can be synthesized by most bee gut bacteria, but they employ different pathways. Our results suggest that bee gut species use different acyl substrates for activation of the γ-hydroxyl group of homoserine (Fig. 5B). Moreover, Snodgrassella and Bartonella utilize acetyl-CoA rather than succinyl-CoA, which coincides with their use of ASCT rather than acetate kinase and phosphotransacetylase for the activation of acetate to acetyl-CoA. Desulfobacter postgatei and Desulfuromonas acetoxidans appear to couple the activation of acetate stoichiometrically with the formation of succinate from succinyl-CoA as an intermediate, through which the oxidative process saves the consumption of ATP (61). Recently, this pathway was found to be widespread in bacteria, especially species inhabiting acetate-rich gut environments (32). Based on our current findings and previous results, we hypothesize that these 2 pathways might be related and advantageous in such environments.
Our study reveals parallels between the honey bee hindgut microbiota and that of the human distal gut (62). In both, a community of microorganisms has evolved with hosts and with one another in an environment rich in plant polysaccharides that represent a major source of energy. Access to this resource is highly dependent on bacterial abilities to degrade and ferment these polysaccharides. In both systems, hosts absorb short chain fatty acids resulting from polysaccharide degradation (5). In both, different members contribute distinct functions for both carbohydrate utilization and biosynthetic contributions. The codependence of species performing different tasks can give an impression of evolved cooperation, although these same patterns may instead reflect competition and exploitation among community members. In either case, the division of labor may enable efficient substrate metabolism as well as community stability, thus benefitting hosts.
Materials and Methods
Detailed protocols are available in SI Appendix, SI Materials and Methods. Two strains of B. asteroides and 2 strains of G. apicola were isolated from gut homogenate of A. mellifera collected in Jilin, China, in 2019. The genome sequences of pure isolates and the metagenomic data from honey bee guts collected in New Haven, CT, in 2012 were obtained on the Illumina HiSeq platform. The assembled metagenome was annotated on the Integrated Microbial Genomes with Microbiomes system, and the taxon affiliations of the CAZymes were assigned by searching the National Center for Biotechnology Information (NCBI) nr protein database with BLAST (63). Genomic data for isolates from this study and those retrieved from the NCBI database were all annotated against the dbCAN2 database using an HMM search approach (64). Microbiota-free bees were obtained as described by Powell et al. (65). Monoassociated bees were obtained by feeding microbiota-free bees with pure cultures of B. asteroides or G. apicola strains. Microbiota-free and monoassociated bees were fed sucrose solution for 2 d. Before diet treatments, bees were starved for 4 h, then were fed arabinan, galactan, β-glucan, xyloglucan, or polygalacturonic acid dissolved in sucrose syrup. Following 24 h of these treatments, whole guts were dissected, and RNA was extracted. Expression levels of the bacterial genes encoding GHs and PLs were determined by qPCR from RNA extracted from the gut homogenate. Metabolomic profiles were determined from extracted gut compartments using GC-MS as described previously (5).
Data Availability.
Genome sequences obtained here have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. VMHJ00000000–VMHM00000000) (66–69). All other data are included in the manuscript, SI Appendix, and Datasets S1–S3.
Supplementary Material
Acknowledgments
H.Z. is supported by National Natural Science Foundation of China Project 31870472. N.A.M. is supported by NIH Grant R01GM108477.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Genome sequences obtained here have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. VMHJ00000000–VMHM00000000.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916224116/-/DCSupplemental.
References
- 1.Wybouw N., Pauchet Y., Heckel D. G., Van Leeuwen T., Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 8, 1785–1801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A., Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Ríos-Covián D., et al. , Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makki K., Deehan E. C., Walter J., Bäckhed F., The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23, 705–715 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Zheng H., Powell J. E., Steele M. I., Dietrich C., Moran N. A., Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 4775–4780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinson V. G., Moy J., Moran N. A., Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol. 78, 2830–2840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong W. K., Moran N. A., Gut microbial communities of social bees. Nat. Rev. Microbiol. 14, 374–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwong W. K., et al. , Dynamic microbiome evolution in social bees. Sci. Adv. 3, e1600513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodschneider R., Crailsheim K., Nutrition and health in honey bees. Apidologie 41, 278–294 (2010). [Google Scholar]
- 10.Roulston T. H., Cane J. H., Pollen nutritional content and digestibility for animals. Plant Syst. Evol. 222, 187–209 (2000). [Google Scholar]
- 11.Heinze T., Petzold-Welcke K., van Dam J. E. G., “Polysaccharides: Molecular and supramolecular structures. Terminology” in The European Polysaccharide Network of Excellence (EPNOE), Navard P., Ed. (Springer, 2012), pp. 23–64. [Google Scholar]
- 12.Li X., et al. , Pectic bee pollen polysaccharide from Rosa rugosa alleviates diet-induced hepatic steatosis and insulin resistance via induction of AMPK/mTOR-mediated autophagy. Molecules 22, 699 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F., et al. , Isolation, characterization and antitumor effect on DU145 cells of a main polysaccharide in pollen of Chinese wolfberry. Molecules 23, 2430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridley B. L., O’Neill M. A., Mohnen D., Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57, 929–967 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Abbott D. W., Gilbert H. J., Boraston A. B., The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate β-elimination. J. Biol. Chem. 285, 39029–39038 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azadi P., O’Neill M. A., Bergmann C., Darvill A. G., Albersheim P., The backbone of the pectic polysaccharide rhamnogalacturonan I is cleaved by an endohydrolase and an endolyase. Glycobiology 5, 783–789 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Lee F. J., Rusch D. B., Stewart F. J., Mattila H. R., Newton I. L., Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol. 17, 796–815 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Engel P., Martinson V. G., Moran N. A., Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U.S.A. 109, 11002–11007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellegaard K. M., et al. , Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16, 284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B., The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milani C., et al. , Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80, 6290–6302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lugli G. A., et al. , Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18, 568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng H., et al. , Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. MBio 7, e01326–e01316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellegaard K. M., et al. ; SAGE class 2016-17 , Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol. Ecol. 28, 2224–2237 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Goris J., et al. , DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Mewis K., Lenfant N., Lombard V., Henrissat B., Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 82, 1686–1692 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grondin J. M., Tamura K., Déjean G., Abbott D. W., Brumer H., Polysaccharide utilization loci: Fueling microbial communities. J. Bacteriol. 199, e00860–e00816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viborg A. H., et al. , Distinct substrate specificities of three glycoside hydrolase family 42 β-galactosidases from Bifidobacterium longum subsp. infantis ATCC 15697. Glycobiology 24, 208–216 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Larsbrink J., Izumi A., Hemsworth G. R., Davies G. J., Brumer H., Structural enzymology of Cellvibrio japonicus Agd31B protein reveals α-transglucosylase activity in glycoside hydrolase family 31. J. Biol. Chem. 287, 43288–43299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Vries R. P., Visser J., Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol. Mol. Biol. Rev. 65, 497–522 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran N. A., McCutcheon J. P., Nakabachi A., Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Kwong W. K., Zheng H., Moran N. A., Convergent evolution of a modified, acetate-driven TCA cycle in bacteria. Nat. Microbiol. 2, 17067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee H. S., Hwang B. J., Methionine biosynthesis and its regulation in Corynebacterium glutamicum: Parallel pathways of transsulfuration and direct sulfhydrylation. Appl. Microbiol. Biotechnol. 62, 459–467 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Crailsheim K., Intestinal transport of sugars in the honeybee (Apis mellifera L.). J. Insect Physiol. 34, 839–845 (1988). [Google Scholar]
- 35.Klungness L. M., Peng Y. S., A histochemical-study of pollen digestion in the alimentary canal of honeybees (Apis mellifera L.). J. Insect Physiol. 30, 511–521 (1984). [Google Scholar]
- 36.Watanabe H., Tokuda G., Cellulolytic systems in insects. Annu. Rev. Entomol. 55, 609–632 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Kunieda T., et al. , Carbohydrate metabolism genes and pathways in insects: Insights from the honey bee genome. Insect Mol. Biol. 15, 563–576 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kešnerová L., et al. , Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol. 15, e2003467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ndeh D., et al. , Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morra M., et al. , Effects on interfacial properties and cell adhesion of surface modification by pectic hairy regions. Biomacromolecules 5, 2094–2104 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Pereira F. C., Berry D., Microbial nutrient niches in the gut. Environ. Microbiol. 19, 1366–1378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milani C., et al. , Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 11, 2834–2847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turroni F., et al. , Molecular dialogue between the human gut microbiota and the host: A Lactobacillus and Bifidobacterium perspective. Cell. Mol. Life Sci. 71, 183–203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura M., Canchaya C., Fitzgerald G. F., Gupta R. S., van Sinderen D., Genomics as a means to understand bacterial phylogeny and ecological adaptation: The case of bifidobacteria. Antonie van Leeuwenhoek 91, 351–372 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Ventura M., Turroni F., Motherway M. O., MacSharry J., van Sinderen D., Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20, 467–476 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Lugli G. A., et al. , Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl. Environ. Microbiol. 80, 6383–6394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milani C., et al. , Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci. Rep. 5, 15782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turroni F., Milani C., van Sinderen D., Ventura M., Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: An example of possible human-microbe co-evolution. Gut Microb. 2, 183–189 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Turroni F., et al. , Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 6, 1030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turroni F., et al. , Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 10, 1656–1668 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caffall K. H., Mohnen D., The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Martens E. C., Koropatkin N. M., Smith T. J., Gordon J. I., Complex glycan catabolism by the human gut microbiota: The Bacteroidetes sus-like paradigm. J. Biol. Chem. 284, 24673–24677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang L., et al. , dbCAN-seq: A database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46, D516–D521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kabisch A., et al. , Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii’ KT0803. ISME J. 8, 1492–1502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neis E. P., Dejong C. H., Rensen S. S., The role of microbial amino acid metabolism in host metabolism. Nutrients 7, 2930–2946 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfersberger M. G., Amino acid transport in insects. Annu. Rev. Entomol. 45, 111–120 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Powell J. E., Leonard S. P., Kwong W. K., Engel P., Moran N. A., Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. U.S.A. 113, 13887–13892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Willke T., Methionine production–A critical review. Appl. Microbiol. Biotechnol. 98, 9893–9914 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Lee B. C., et al. , Methionine restriction extends lifespan of Drosophila melanogaster under conditions of low amino-acid status. Nat. Commun. 5, 3592 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grandison R. C., Piper M. D., Partridge L., Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature 462, 1061–1064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thauer R. K., Möller-Zinkhan D., Spormann A. M., Biochemistry of acetate catabolism in anaerobic chemotrophic bacteria. Annu. Rev. Microbiol. 43, 43–67 (1989). [DOI] [PubMed] [Google Scholar]
- 62.Zheng H., Steele M. I., Leonard S. P., Motta E. V. S., Moran N. A., Honey bees as models for gut microbiota research. Lab Anim. (NY) 47, 317–325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Markowitz V. M., et al. , IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, D115–D122 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H., et al. , dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powell J. E., Martinson V. G., Urban-Mead K., Moran N. A., Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 80, 7378–7387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng H., et al. , Whole genome shotgun sequencing of Bifidobacterium asteroides strain W8111. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/VMHJ00000000.1. Deposited 28 July 2019.
- 67.Zheng H., et al. , Whole genome shotgun sequencing of Bifidobacterium asteroides strain W8102. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/VMHK00000000.1. Deposited 28 July 2019.
- 68.Zheng H., et al. , Whole genome shotgun sequencing of Gilliamella apicola strain W8131. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/VMHL00000000.1. Deposited 28 July 2019.
- 69.Zheng H., et al. , Whole genome shotgun sequencing of Gilliamella apicola strain W8127. GenBank. https://www.ncbi.nlm.nih.gov/nuccore/VMHM00000000.1. Deposited 28 July 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences obtained here have been deposited in GenBank, https://www.ncbi.nlm.nih.gov/genbank/ (accession nos. VMHJ00000000–VMHM00000000) (66–69). All other data are included in the manuscript, SI Appendix, and Datasets S1–S3.