Abstract
Background
Efficient conversion of plant biomass to commodity chemicals is an important challenge that needs to be solved to enable a sustainable bioeconomy. Deconstruction of biomass to sugars and lignin yields a wide variety of low molecular weight carbon substrates that need to be funneled to product. Pseudomonas putida KT2440 has emerged as a potential platform for bioconversion of lignin and the other components of plant biomass. However, P. putida is unable to natively utilize several of the common sugars in hydrolysate streams, including galactose.
Results
In this work, we integrated a De Ley–Doudoroff catabolic pathway for galactose catabolism into the chromosome of P. putida KT2440, using genes from several different organisms. We found that the galactonate catabolic pathway alone (DgoKAD) supported slow growth of P. putida on galactose. Further integration of genes to convert galactose to galactonate and to optimize the transporter expression level resulted in a growth rate of 0.371 h−1. Additionally, the best-performing strain was demonstrated to co-utilize galactose with glucose.
Conclusions
We have engineered P. putida to catabolize galactose, which will allow future engineered strains to convert more plant biomass carbon to products of interest. Further, by demonstrating co-utilization of glucose and galactose, continuous bioconversion processes for mixed sugar streams are now possible.
Keywords: Galactose, Co-utilization, De Ley–Doudoroff, Pseudomonas putida KT2440
Background
The vast majority of global fuels and platform chemicals are produced from petroleum. However, petroleum is a finite resource, so synthesizing platform chemicals from renewable feedstocks is needed for a sustainable future. Biological valorization of sugars and lignin from plant-based biomass to commodity chemicals is a potential route to renewable and sustainable alternatives. Although different feedstocks and pretreatment processes yield different available substrates for microbial conversion, several sugars are regularly detected in hydrolysate streams [1, 2]. While glucose is typically the most abundant, xylose, galactose, mannose, and arabinose are all present as well at different concentrations. In order to effectively convert these sugars to product, an ideal organism would at a minimum require high tolerance to inhibitors and rapid sugar catabolism.
Pseudomonas putida KT2440 is emerging as a new favorite synthetic biology chassis for biocatalysis of deconstructed biomass [3]. P. putida KT2440 has a wide range of genetic tools available [4–7], a well-characterized metabolism well suited for redox-intensive transformations [8–11], demonstrated ability to host a variety of heterologous pathways in vivo for a vastly enlarged biochemical work space [12, 13], and established scale-up capabilities. For example, P. putida KT2440 has been engineered to grow anoxically [14], to catabolize novel substrates [15–17], and to synthesize a diverse array of chemicals [3, 12]. Moreover, P. putida KT2440 has been successfully engineered to utilize both of the common hemicellulosic pentoses: xylose and arabinose [18–20]. However, P. putida KT2440 has not been engineered to catabolize galactose, the next most abundant sugar in many hemicelluloses, which can be up to 3% of total sugars in plant biomass [21]. It will be important to capture this carbon for an efficient bioconversion process.
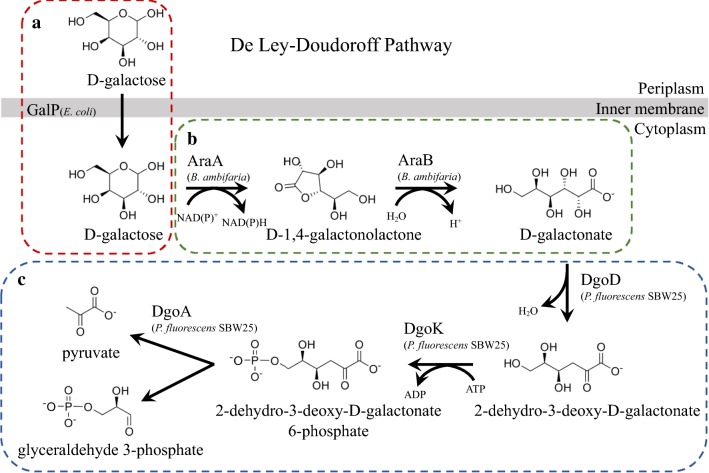
There are two common pathways for galactose catabolism in bacteria: the Leloir (LL) pathway and the De Ley–Doudoroff (DLD) pathway [2]. In the LL pathway, galactose is phosphorylated and then converted to glucose-1-phosphate through a cyclic pair of transferase/epimerase reactions with uridyl monophosphate intermediates [22, 23]. The DLD pathway, on the other hand, mirrors the Entner–Doudoroff (ED) pathway for glucose catabolism used by P. putida KT2440, wherein the sugar is ultimately converted to glyceraldehyde 3-phosphate (G3P) and pyruvate (PYR) [24]. The DLD pathway can be separated into three parts: transport (Fig. 1a), galactose conversion to galactonate (Fig. 1b), and galactonate to G3P and PYR (Fig. 1c). Transport of galactose into the cell is relatively well studied with numerous sugar transporters reported to have activity on galactose as either a primary or secondary substrate; for example, GalP from Escherichia coli is a sugar-proton symporter of both galactose and glucose [25]. The second portion of the DLD pathway, where galactose is converted to galactonate, is less well characterized. Some organisms have been described to have these activities. Although enzymes with dehydrogenase and lactonase activity on galactose and 1,4-galactonolactone have been identified, such as AraAB from Burkholderia ambifaria, no sequence of a specific galactonolactonase has been identified [26–30]. For the last portion of the DLD pathway, three enzymatic steps of dehydration, phosphorylation, and subsequent aldol cleavage are performed by DgoD, DgoK, and DgoA, respectively (Fig. 1c) [31]. Homologs of these proteins are encoded in a wide variety of organisms, including many pseudomonads such as Pseudomonas fluorescens SBW25. Interestingly, these genes are even encoded in some organisms that use the LL pathway like E. coli, where the dgoKAD operon is a separately regulated pathway only for galactonate catabolism. However, to the best of our knowledge, the complete DLD pathway has not been successfully introduced into an organism that does not natively utilize galactose and allowed for growth with galactose as the sole carbon source.
Fig. 1.
Components of the DLD galactose pathway used in this study. a Transport across the inner membrane, outlined in red. b Galactose conversion to galactonate, outlined in green. c Galactonate conversion to pyruvate and glyceraldehyde 3-phosphate, outlined in blue. GalP galactose–proton symporter, DgoK 2-dehydro-3-deoxygalactonokinase, DgoA 2-dehydro-3-deoxy-6-phosphogalactonate aldolase, DgoD d-galactonate dehydratase, AraA l-arabinose 1-dehydrogenase/d-galactose dehydrogenase, AraB, l-arabinolactonase/d-galactonolactonase
To expand the substrate range of P. putida KT2440 to include galactose, in this study, we harness the less commonly used DLD pathway of galactose catabolism. We chose this pathway because it is observed in pseudomonads, and the products of the DLD pathway (G3P and PYR) are the same as the ED pathway natively used by P. putida KT2440 for glucose catabolism. Additionally, the DLD pathway catabolizes galactose via different metabolic intermediates relative to glucose, whereas the Leloir pathway uses the exact same intermediates and may compete for the same flux space. Therefore, the DLD pathway might lead to better sugar co-utilization. Here we built a functional DLD pathway using genes from E. coli, P. fluorescens SBW25, and B. ambifaria. We then demonstrated the ability of this strain to utilize galactose alone and co-utilize galactose and glucose.
Results and discussion
Heterologous expression of DgoKAD allows growth on galactose
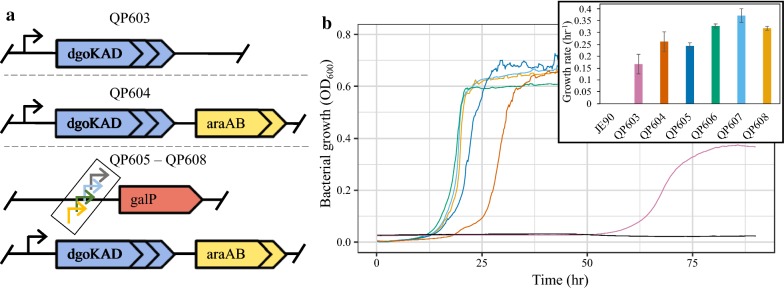
Wild-type P. putida KT2440 does not grow on galactose or galactonate, so we first explored what portions of the DLD pathway were required for galactose catabolism [32]. Glucose dehydrogenases in pseudomonads can have a wide substrate range, and there are many uncharacterized and promiscuous sugar transporters in P. putida KT2440 [18]. We therefore hypothesized that side activity of native enzymes may be sufficient for galactose transport and conversion to galactonate. Therefore, we introduced a galactonate conversion pathway into the chromosome of P. putida by expressing the dgoKAD operon from P. fluorescens SBW25 under its native promoter, creating strain QP603 (Fig. 2a, Table 1). We inoculated strain QP603 into minimal medium with galactose as the sole carbon and energy source and observed slow growth on galactose after a 52-h lag phase (Fig. 2b, Additional file 1: Table S1). No accumulation of galactonate or any other molecule was seen in the supernatant by HPLC. This demonstrates that expression of dgoKAD is sufficient for galactose catabolism and that native systems must be capable of galactose transport and oxidation to galactonate at a low level.
Fig. 2.
Schematic of strains constructed and their growth on galactose. a Pictorial representation of genotypes for strains QP603–QP608 (see “Materials and methods” for additional details). b Growth of each strain on MME media with 10 mM galactose as the sole carbon source. Strains JE90 (parent strain), black; QP603 (JE90::dgoKAD), pink; QP604 (QP603::araAB), dark orange; QP605 (QP604::P1548-galP), dark blue; QP606 (QP604::P3079-galP), green; QP607 (QP604::Plac-galP), light blue; QP608 (QP604::Ptac-galP), yellow. The inset shows the measured growth rate for each strain. Strains were grown at 30 °C aerobically in a 48-well microtiter plate in biological triplicate with measurements every 10 min
Table 1.
Strains and plasmids used in this study
| Strains | Genotype/plasmid | Source |
|---|---|---|
| JE90 | P. putida KT2440 ∆PP_4740::PtacBXB1int-attBBxB1 | [17] |
| QP603 | JE90 ∆PP_0545::dgoKAD | This work |
| QP604 | JE90 ∆PP_0545::dgoKAD:araAB | This work |
| QP605 | QP604 attBBxB1::pQP344 | This work |
| QP606 | QP604 attBBxB1::pQP345 | This work |
| QP607 | QP604 attBBxB1::pQP346 | This work |
| QP608 | QP604 attBBxB1::pQP347 | This work |
| E. coli F’IQ | E. coli F’IQ | NEB |
| Plasmids | ||
| pK18mobsacB | pUC origin, KanR, origin of transfer, sacB counter selectable marker | [18] |
| pJE1045 | “Cargo” plasmid for chromosomal integration, BxB1 attP, pUC origin, KanR, and Ptac:mNeongreen | [17] |
| pJE1553 | pk18mobsacB based, for ∆PP_0545::dgoKAD mutation | This work |
| pQP348 | pk18mobsacB based, for ∆PP_0545::dgoKAD:araAB mutation | This work |
| pQP344 | pJE1045 with PPP_1548:galPppopt | This work |
| pQP345 | pJE1045 with PPP_3079:galPppopt | This work |
| pQP346 | pJE1045 with Plac:galPppopt | This work |
| pQP347 | pJE1045 with Ptac:galPppopt | This work |
Growth rate improved by pathway expansion
We next examined improving the growth rate of strain QP603. While the side activity of native enzymes could perform the transport, dehydrogenase, and lactonase activities of the DLD pathway, they could be rate limiting. We therefore introduced enzymes to convert intracellular galactose to galactonate. Because no galactose-specific dehydrogenase and lactonase are yet to be identified, we assembled our pathway using the best-characterized galactonolactonase and its associated galactose dehydrogenase that are currently known—an arabinose dehydrogenase, AraA, and arabinonolactonase, AraB, from the oxidative arabinose pathway of B. ambifaria [26, 27, 30]. We introduced codon-optimized versions of araA and araB into strain QP603 at the end of the dgoKAD operon, resulting in a longer operon of five genes dgoKAD:araAB and a new strain QP604 (Fig. 2a). This strain grew 60% faster than strain QP603 and had a substantially reduced lag phase (Fig. 2b, Additional file 1: Table S1). Unfortunately, strain QP604 still experienced a significant lag of approximately 26 h, and the growth rate was still slower than that of glucose catabolism despite having a similar energetic yield and producing the same central metabolites, G3P and PYR.
Because we had introduced all the catabolic parts of the DLD pathway, we hypothesized that growth on galactose may now be limited by substrate uptake. We selected the E. coli GalP to study the impact of transport on growth rate in strain QP604. A codon-optimized galP was introduced into the BxB1 attB site of strain QP604 using site-specific recombination with four different promoters to generate strains QP605–QP608 [6]. Promoters of increasing strength were used to express galP, including the upstream regions of PP_1548 and PP_3079 and the E. coli lac and tac promoters (Additional file 1: Table S2), resulting in strains QP605 to QP608, respectively. All four strains showed a significantly reduced duration of lag phase (P value < 0.001) and the growth improved in strains QP606–QP608 (P value < 0.01) when compared to the parent QP604 for growth on galactose (Fig. 2b, Additional file 1: Table S1). Expression of galP with the lac promoter in strain QP607 had the greatest increase in growth rate relative to QP604, 41%, with an overall growth rate of 0.371 ± 0.03 h−1. This expression optimization for the galactose transporter suggests that growth rate improves with higher galP expression up to the strength of the lac promoter. However, growth rate decreased when using the very highly expressed tac promoter, suggesting that overexpression of this transporter can become toxic to our engineered cells. Comparatively, the WT grown under identical conditions but with glucose as the carbon source grew at 0.87 ± 0.09 h−1, about 2.5-fold faster. Overall, while the introduction of dgoKAD was sufficient to supply growth on galactose, the complete DLD pathway including a transporter was required for rapid catabolism of galactose as the sole carbon source.
Galactose is co-utilized with glucose
Co-utilization allows for faster and potentially continuous approaches to bioprocessing, making it important for future commercialization. We therefore sought to determine whether galactose could be co-utilized with glucose. To test whether co-utilization indeed occurs, we measured sugar utilization of strain QP607 in shake flasks with equimolar amounts of glucose and galactose. Both glucose and galactose were simultaneously utilized, no additional peaks such as for galactonate accumulation were observed in the supernatant quantification, and the growth of strain QP607 did not have a diauxic shape (Fig. 3). Together, this evidence demonstrates co-utilization of glucose and galactose in strain QP607. It is not surprising strain QP607 was able to co-utilize galactose and glucose simultaneously. P. putida does not natively utilize galactose, so the heterologous DLD pathway should be unregulated in P. putida. Furthermore, the pathway produces the same products as the natively utilized ED pathway for glucose catabolism, so it was expected the DLD pathway would be able to seamlessly integrate into central metabolism when the strain is growing on glucose. However, we did observe that glucose was utilized more rapidly than galactose in QP607. Fortunately, because galactose is a less abundant sugar than glucose in lignocellulose (approximately 1:30 ratio in corn stover hydrolysate [2]), the current slower utilization rate of galactose should still be sufficient for most real-world settings. Based on the similarity to the ED pathway and the lack of detected products, the galactose was presumably completely oxidized to CO2 via the TCA cycle in these strains.
Fig. 3.
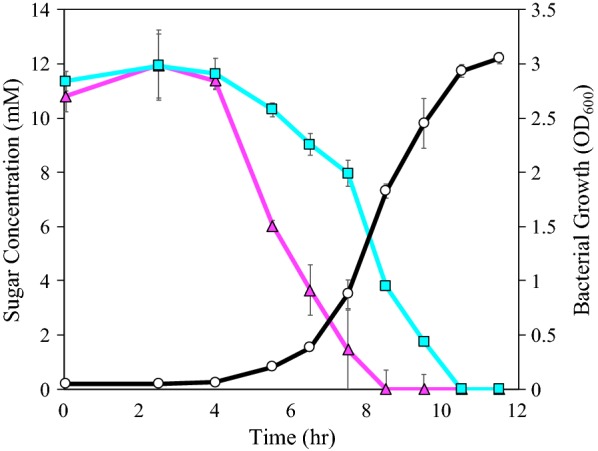
Shake flask characterization of strain QP607 growth on glucose and galactose. HPLC data for glucose and galactose concentrations are shown in magenta triangles, and cyan squares, respectively. Bacterial growth, as measured by a change in optical density, is shown in black circles
While the current level of pathway functionality is likely sufficient for most applications, additional research could lead to faster galactose catabolism. In this work, we primarily focused on tuning expression of the transporter because although substrate uptake is critical, membrane protein overexpression can be toxic. Similar tuning of the rest of the genes in the pathway could further improve the growth rate. Other approaches such as adaptive laboratory evolution would also likely result in more rapid galactose catabolism. Finally, metabolomics studies could help reveal how the newly introduced DLD pathway integrates with the cyclic EDEMP pathway, which may be critical for future metabolic engineering efforts to divert flux away from growth and toward product formation.
Conclusions
We have expanded the potential for total hydrolysate biocatalysis by P. putida KT2440, an emerging model organism for synthetic biology and biomass valorization, by introducing the DLD pathway for galactose catabolism. In doing so, we have determined the enzymes required for rapid catabolism and optimized the expression level of galactose transport. Furthermore, we have shown that this pathway allows co-consumption of galactose with a preferred substrate such as glucose. This work not only further demonstrates the strength of P. putida as a modular biocatalysis chassis, but also benefits the community developing bioprocesses for total hydrolysate conversion by expanding the catabolic sugar profile of P. putida. We plan to expand this work in the future by incorporating the previously demonstrated catabolic pathways for xylose and arabinose into our galactose utilizing strain.
Materials and methods
Strain construction
Pseudomonas putida strain JE90 (P. putida KT2440 ∆hsdR::BxB1int-attB [6]) was the parent for all strains made in this study. For the plasmids to insert dgoKAD and araAB into the chromosome, the genes were cloned into pK18mobsacB [33] flanked by 1 kb sequences identical to the upstream and downstream region of PP_0545, a non-specific aldehyde dehydrogenase. These regions are used for homologous recombination to replace PP_0545 with the pathway genes. Primers were from (Eurofins Genomics, Louisville, KY), and gBlocks for codon-optimized araAB from B. ambifaria ATCC BAA-244 and galP from E. coli were synthesized by Integrated DNA Technologies. Genomic DNA was used to amplify dgoKAD, and its native promoter from P. fluorescens SBW25 and gDNA from P. putida strain JE90 was amplified using Phusion High Fidelity Polymerase (Thermo Fisher Scientific, Waltham, MA) when relevant. Plasmids were constructed via Gibson assembly with the NEBuilder HiFi DNA assembly master mix (New England Biolabs (NEB), Ipswich, MA), DNA was extracted from agarose gels with Zymo gel extraction kit (Zymo Research, Irvine, CA) and transformed into NEB F’IQ E. coli chemically competent cells following the manufacturers guidelines (NEB). Plasmid sequences were confirmed via sequencing by Eurofins Genomics, and annotated plasmid sequences are available in Additional files 2, 3, 4, 5, 6, and 7. Strain genotypes and plasmids used in this study are listed in Table 1. When necessary, all antibiotic selections were performed with 50 µg/mL kanamycin. Plasmids were harvested with the geneJET miniprep kit (Thermo Fisher Scientific). For seamless chromosomal editing to introduce dgoKAD and araAB cassettes into P. putida, pK18mobsacB-derived plasmids were transformed into P. putida and mutants were selected using kanamycin selection and sucrose counter-selection as previously described [34]. The promoter–transporter cassettes were integrated using BxB1 phage integration as previously described [6].
Growth medium
Utilization of galactose and glucose was tested aerobically in the MOPS-buffered minimal medium MME, which consisted of (per liter): 1.6 g K2HPO4·3 H2O, 4.2 g 3-(N-morpholino)propanesulfonic acid (MOPS), 0.25 g NaCl, 0.50 g NH4Cl, 0.10 g MgSO4·7 H20, 0.01 g CaCl2·2 H2O, and 1 mL of a 1000× trace element solution, which consisted of (per liter): 1.00 mL concentrated HCl, 0.50 g Na4EDTA·H2O, 2 g FeCl3, 0.05 g H3BO3, 0.05 g ZnCl2, 0.03 g CuCl2·2H2O, 0.05 g MnCl2·4H2O, 0.05 g (NH4)2MoO4, 0.05 g CoCl2·6H2O, and 0.05 g NiCl2·6H2O. Glucose and galactose were added as growth substrates at the concentrations detailed below.
Plate reader growth assays
Strains were grown from single colonies in LB overnight at 30 °C with shaking at 250 rpm. Cells were washed in substrate-free MME medium and a 1% inoculum was transferred to MME medium supplemented with 10 mM galactose. After the cells had reached stationary phase, 10 μL of each sample were further passaged into 500 μL MME medium supplemented with 10 mM galactose in a 48-well plate (Greiner Bio-One). Edge wells of the plate were filled with 700 μL media and not used for data collection to minimize the impact of evaporation. Data were collected on an Epoch2 plate reader (BioTek, Winooski, VT) with fast continuous double orbital shaking at 30 °C aerobically. A temperature gradient of 1 °C was added to minimize condensation. Measurement of OD600 was performed every 10 min. Growth rates were calculated with CurveFitter software [35] using only linear regions of growth on a log(OD600) vs time plot, and only OD600 values below 25% OD600 max. The lag phase was also calculated with the CurveFitter software. P values were calculated with Student’s t test.
Shake flask growth assay
Strain QP607 was grown to mid-log phase in MME medium supplemented with 12 mM glucose and 12 mM galactose. The cells were transferred to a 125-mL flask containing 25 mL of the glucose–galactose MME medium at 30 °C with 250-rpm shaking. The cells were monitored for growth, and periodically 1 mL was sampled to measure OD600 and for HPLC analysis.
Galactose and glucose HPLC quantification
Samples were filtered with 0.2-μm Corning Costar Spin-X centrifuge tube filters and then acidified with H2SO4 to a final concentration of 5 mM. The samples were run on a Waters HPLC equipped with refractive index detector and a Supelcogel H 6% column with a 0.6 mL/min flow rate of 5 mM H2SO4 in water as the running buffer at 60 °C. Sugar concentrations were determined by comparison to a standard curve.
Supplementary information
Additional file 1. Supplemental Methods and Tables.
Additional file 2. Plasmid map for pJE1553.
Additional file 3. Plasmid map for pQP348.
Additional file 4. Plasmid map for pQP344.
Additional file 5. Plasmid map for pQP345.
Additional file 6. Plasmid map for pQP346.
Additional file 7. Plasmid map for pQP347.
Acknowledgements
Not applicable.
Notice
This manuscript has been authored by UT-Battelle, LLC, under Contract No. DE-AC0500OR22725 with the U.S. Department of Energy. The US Government retains and the publisher, by accepting the article for publication, acknowledges that the US Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for the US Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Abbreviations
- LL pathway
Leloir pathway
- DLD pathway
De Ley–Doudoroff pathway
- ED pathway
Entner–Doudoroff pathway
- G3P
glyceraldehyde-3-phosphate
- PYR
pyruvate
Authors’ contributions
GLPV helped design and perform experiments, analyze the data, and write the manuscript. JRE helped design and perform experiments, analyze the data, and edit the manuscript. JMB helped perform experiments and analyze the data. AMG helped design the study, analyze the data, and write the manuscript. All authors read and approved the final manuscript.
Funding
Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the US DOE under contract DE-AC05-00OR22725. Funding was provided by the U.S. Department of Energy Office of Energy Efficiency and Renewable Energy the Bioenergy Technologies Office via the Agile BioFoundry project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its additional information files.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
GLPV, JRE, and AMG are applying for a patent relating to the content of the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
George L. Peabody V and Joshua R. Elmore contributed equally to this work
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13068-019-1627-0.
References
- 1.Saini JK, Saini R, Tewari L. Lignocellulosic agriculture wastes as biomass feedstocks for second-generation bioethanol production: concepts and recent developments. 3 Biotech. 2015;5(4):337–353. doi: 10.1007/s13205-014-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaves JE, Presley GN, Michener JKJP. Modular engineering of biomass degradation pathways. Processes. 2019;7(4):230. doi: 10.3390/pr7040230. [DOI] [Google Scholar]
- 3.Nikel PI, de Lorenzo V. Pseudomonas putida as a functional chassis for industrial biocatalysis: from native biochemistry to trans-metabolism. Metab Eng. 2018;50:142–155. doi: 10.1016/j.ymben.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio T, de Lorenzo V, Martinez-Garcia E. CRISPR/Cas9-based counterselection boosts recombineering efficiency in Pseudomonas putida. Biotechnol J. 2018;13(5):e1700161. doi: 10.1002/biot.201700161. [DOI] [PubMed] [Google Scholar]
- 5.Wirth NT, Kozaeva E, Nikel PI. Accelerated genome engineering of Pseudomonas putida by I-SceI-mediated recombination and CRISPR–Cas9 counterselection. Microb Biotechnol. 2019 doi: 10.1111/1751-7915.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elmore JR, et al. Development of a high efficiency integration system and promoter library for rapid modification of Pseudomonas putida KT2440. Metab Eng Commun. 2017;5:1–8. doi: 10.1016/j.meteno.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zobel S, et al. Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth Biol. 2015;4(12):1341–1351. doi: 10.1021/acssynbio.5b00058. [DOI] [PubMed] [Google Scholar]
- 8.Akkaya O, et al. The metabolic redox regime of Pseudomonas putida tunes its evolvability toward novel xenobiotic substrates. MBio. 2018;9(4):e01512-18. doi: 10.1128/mBio.01512-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aslan S, Noor E, Bar-Even A. Holistic bioengineering: rewiring central metabolism for enhanced bioproduction. Biochem J. 2017;474(23):3935–3950. doi: 10.1042/BCJ20170377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavarria M, et al. The Entner–Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ Microbiol. 2013;15(6):1772–1785. doi: 10.1111/1462-2920.12069. [DOI] [PubMed] [Google Scholar]
- 11.Nikel PI, et al. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner–Doudoroff, Embden–Meyerhof–Parnas, and pentose phosphate pathways. J Biol Chem. 2015;290(43):25920–25932. doi: 10.1074/jbc.M115.687749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domrose A, et al. Rapid generation of recombinant Pseudomonas putida secondary metabolite producers using yTREX. Synth Syst Biotechnol. 2017;2(4):310–319. doi: 10.1016/j.synbio.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CW, et al. Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule. 2019;3:1523–1537. doi: 10.1016/j.joule.2019.05.011. [DOI] [Google Scholar]
- 14.Yu S, et al. Improved performance of Pseudomonas putida in a bioelectrochemical system through overexpression of periplasmic glucose dehydrogenase. Biotechnol Bioeng. 2018;115(1):145–155. doi: 10.1002/bit.26433. [DOI] [PubMed] [Google Scholar]
- 15.Linger JG, et al. Conversion of levoglucosan and cellobiosan by Pseudomonas putida KT2440. Metab Eng Commun. 2016;3:24–29. doi: 10.1016/j.meteno.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarnieri MT, et al. Conversion and assimilation of furfural and 5-(hydroxymethyl)furfural by Pseudomonas putida KT2440. Metab Eng Commun. 2017;4:22–28. doi: 10.1016/j.meteno.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franden MA, et al. Engineering Pseudomonas putida KT2440 for efficient ethylene glycol utilization. Metab Eng. 2018;48:197–207. doi: 10.1016/j.ymben.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Dvorak P, de Lorenzo V. Refactoring the upper sugar metabolism of Pseudomonas putida for co-utilization of cellobiose, xylose, and glucose. Metab Eng. 2018;48:94–108. doi: 10.1016/j.ymben.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Le Meur S, et al. Production of medium-chain-length polyhydroxyalkanoates by sequential feeding of xylose and octanoic acid in engineered Pseudomonas putida KT2440. BMC Biotechnol. 2012;12:53. doi: 10.1186/1472-6750-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Growth of engineered Pseudomonas putida KT2440 on glucose, xylose, and arabinose: hemicellulose hydrolysates and their major sugars as sustainable carbon sources. GCB Bioenergy. 2019;11(1):249–259. doi: 10.1111/gcbb.12590. [DOI] [Google Scholar]
- 21.Hu Z, et al. Chemical profiles of switchgrass. Bioresour Technol. 2010;101(9):3253–3257. doi: 10.1016/j.biortech.2009.12.033. [DOI] [PubMed] [Google Scholar]
- 22.Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10(4):461–470. doi: 10.1096/fasebj.10.4.8647345. [DOI] [PubMed] [Google Scholar]
- 23.Leloir LF. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch Biochem Biophys. 1951;33(2):186–190. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- 24.De Ley J, Doudoroff M. The metabolism of d-galactose in Pseudomonas saccharophila. J Biol Chem. 1957;227(2):745–757. [PubMed] [Google Scholar]
- 25.Patching SG, et al. Relative substrate affinities of wild-type and mutant forms of the Escherichia coli sugar transporter GalP determined by solid-state NMR. Mol Membr Biol. 2008;25(6–7):474–484. doi: 10.1080/09687680802371963. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe S, Kodaki T, Makino K. Cloning, expression, and characterization of bacterial l-arabinose 1-dehydrogenase involved in an alternative pathway of l-arabinose metabolism. J Biol Chem. 2006;281(5):2612–2623. doi: 10.1074/jbc.M506477200. [DOI] [PubMed] [Google Scholar]
- 27.Watanabe S, et al. Identification and characterization of l-arabonate dehydratase, l-2-keto-3-deoxyarabonate dehydratase, and l-arabinolactonase involved in an alternative pathway of l-arabinose metabolism. Novel evolutionary insight into sugar metabolism. J Biol Chem. 2006;281(44):33521–33536. doi: 10.1074/jbc.M606727200. [DOI] [PubMed] [Google Scholar]
- 28.Blachnitzky EO, Wengenmayer F, Kurz G. D-Galactose dehydrogenase from Pseudomonas fluorescens. Purification, properties and structure. Eur J Biochem. 1974;47(2):235–250. doi: 10.1111/j.1432-1033.1974.tb03687.x. [DOI] [PubMed] [Google Scholar]
- 29.Sperka S, et al. Complete nucleotide sequence of Pseudomonas fluorescensd-galactose dehydrogenase gene. Nucleic Acids Res. 1989;17(13):5402. doi: 10.1093/nar/17.13.5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe S, Kodaki T, Makino K. A novel alpha-ketoglutaric semialdehyde dehydrogenase: evolutionary insight into an alternative pathway of bacterial l-arabinose metabolism. J Biol Chem. 2006;281(39):28876–28888. doi: 10.1074/jbc.M602585200. [DOI] [PubMed] [Google Scholar]
- 31.Deacon J, Cooper RA. D-Galactonate utilisation by enteric bacteria. The catabolic pathway in Escherichia coli. FEBS Lett. 1977;77(2):201–205. doi: 10.1016/0014-5793(77)80234-2. [DOI] [PubMed] [Google Scholar]
- 32.Puchalka J, et al. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol. 2008;4(10):e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marx CJ. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res Notes. 2008;1:1. doi: 10.1186/1756-0500-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson CW, Beckham GT. Aromatic catabolic pathway selection for optimal production of pyruvate and lactate from lignin. Metab Eng. 2015;28:240–247. doi: 10.1016/j.ymben.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Delaney NF, et al. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS ONE. 2013;8(4):e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental Methods and Tables.
Additional file 2. Plasmid map for pJE1553.
Additional file 3. Plasmid map for pQP348.
Additional file 4. Plasmid map for pQP344.
Additional file 5. Plasmid map for pQP345.
Additional file 6. Plasmid map for pQP346.
Additional file 7. Plasmid map for pQP347.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional information files.