ABSTRACT
Some antibodies exhibit elevated viscosity at high concentrations, making them poorly suited for therapeutic applications requiring administration by injection such as subcutaneous or ocular delivery. Here we studied an anti-IL-13/IL-17 bispecific IgG4 antibody, which has anomalously high viscosity compared to its parent monospecific antibodies. The viscosity of the bispecific IgG4 in solution was decreased by only ~30% in the presence of NaCl, suggesting electrostatic interactions are insufficient to fully explain the drivers of viscosity. Intriguingly, addition of arginine-HCl reduced the viscosity of the bispecific IgG4 by ~50% to its parent IgG level. These data suggest that beyond electrostatics, additional types of interactions such as cation-π and/or π-π may contribute to high viscosity more significantly than previously understood. Molecular dynamics simulations of antibody fragments in the mixed solution of free arginine and explicit water were conducted to identify hotspots involved in self-interactions. Exposed surface aromatic amino acids displayed an increased number of contacts with arginine. Mutagenesis of the majority of aromatic residues pinpointed by molecular dynamics simulations effectively decreased the solution’s viscosity when tested experimentally. This mutational method to reduce the viscosity of a bispecific antibody was extended to a monospecific anti-GCGR IgG1 antibody with elevated viscosity. In all cases, point mutants were readily identified that both reduced viscosity and retained antigen-binding affinity. These studies demonstrate a new approach to mitigate high viscosity of some antibodies by mutagenesis of surface-exposed aromatic residues on complementarity-determining regions that may facilitate some clinical applications.
KEYWORDS: Bispecific IgG, IgG, viscosity
Introduction
Antibodies are the largest class of biologic drugs, with over 80 antibodies approved1 for the treatment of a broad range of human diseases.2 Most approved antibody drugs are delivered by either intravenous (IV) infusion or subcutaneous (SC) injection.2 SC delivery is increasingly used because it can potentially decrease administration time from hours to minutes, increase patient convenience by enabling at-home injection and reduce associated health-care costs.3,4 A notable challenge with SC administration is injection volumes, which are typically small (up to 1.5 mL) and necessitate the formulation of antibodies at high concentration (>150 mg/mL) to deliver the required dose. Non-covalent interactions of antibodies at high concentrations can lead to high viscosity, making injection difficult and painful.5 Furthermore, elevated viscosity of antibody solutions can lead to problems in manufacturing, including ultrafiltration and diafiltration steps, as well as vialing.5–7
Research involving alteration of pH or ionic strength, and the addition of excipients with different physical properties have improved our understanding and helped managed the high viscosity of some monospecific antibodies.8–18 Additionally, some studies focused on the effect of net charge, charged patches and molecular dipole of antibodies on solution viscosity.19–21 Recently, efforts have been applied to protein engineering and in silico-centered approaches during the early development of antibodies to optimize lead drug candidates with improved high-concentration solution behaviors.22–28 While the viscosity of monospecific IgG has been extensively studied, novel antibody formats are increasingly used to enable novel or enhanced clinical potential.2 For example, bispecific antibodies (BsAb) can support novel mechanisms of action that are not available to monospecific antibodies and are being widely pursued as next-generation antibody drugs.29–31 In contrast to monospecific antibodies, much less is known about the viscosity properties of BsAb.32 This growing clinical importance of both BsAb and SC delivery motivated our study of the viscosity of a BsAb.
We investigated the viscosity of a bispecific IgG (BsIgG), as this represents one of the most commonly used of many different possible BsAb formats.29,30 An anti-IL-13/IL-17 BsIgG4 was previously found to have twice the viscosity of its parental monospecific anti-IL-13 IgG4 and anti-IL-17 IgG1 antibodies for reasons that are not well understood.32 To characterize viscosity, we used arginine-HCl, an extensively used formulation excipient that effectively reduced viscosity.14,33 The effect of arginine led us to study possible interactions of arginine with surface-exposed aromatic residues in variable regions by molecular dynamics (MD) simulations. Systematic mutational analysis was performed of aromatic amino acid residues predicted by MD simulation, thereby reducing the high viscosity of antibody solutions. Lastly, our findings were extended to an anti-GCGR monospecific IgG1 antibody with elevated viscosity at high concentrations.
Results
Viscosity of antibody fragments
We assembled a collection of full-length monospecific and bispecific antibodies and fragments of these molecules to investigate the determinants of viscosity (Figure 1(a)). Rheometry confirmed a previous report32 indicating that both anti-IL-13/IL-17 BsIgG4 and anti-IL-13 IgG4 and IL-17 IgG1 mixtures have higher viscosities than individual anti-IL-13 IgG4 or IL-17 IgG1 antibodies (Figure 1(b)). The viscosities of various antibody fragments were assessed to dissect the regions of antibodies contributing to high viscosity. Specifically, antigen-binding fragments (Fab and F(ab’)2) were generated by enzymatic proteolysis and purified to eliminate crystallizable fragments (Fcs). The viscosity of the F(ab’)2 was comparable to the corresponding IgG antibody for anti-IL-13, anti-IL-17, anti-IL-13 plus anti-IL-17 mixture and the anti-IL-13/IL-17 bispecific (Figure 1(b)). These observations suggest that the Fc region makes at most only minor contributions to the observed elevated viscosity. In contrast to the bivalent F(ab’)2, monovalent Fab displayed significantly reduced viscosity compared to the corresponding individual IgG or the IgG mixture (Figure 1(b)). These findings are consistent with the notion that bivalency is needed to form networks of self-interacting molecules that can lead to high viscosity.39
Figure 1.
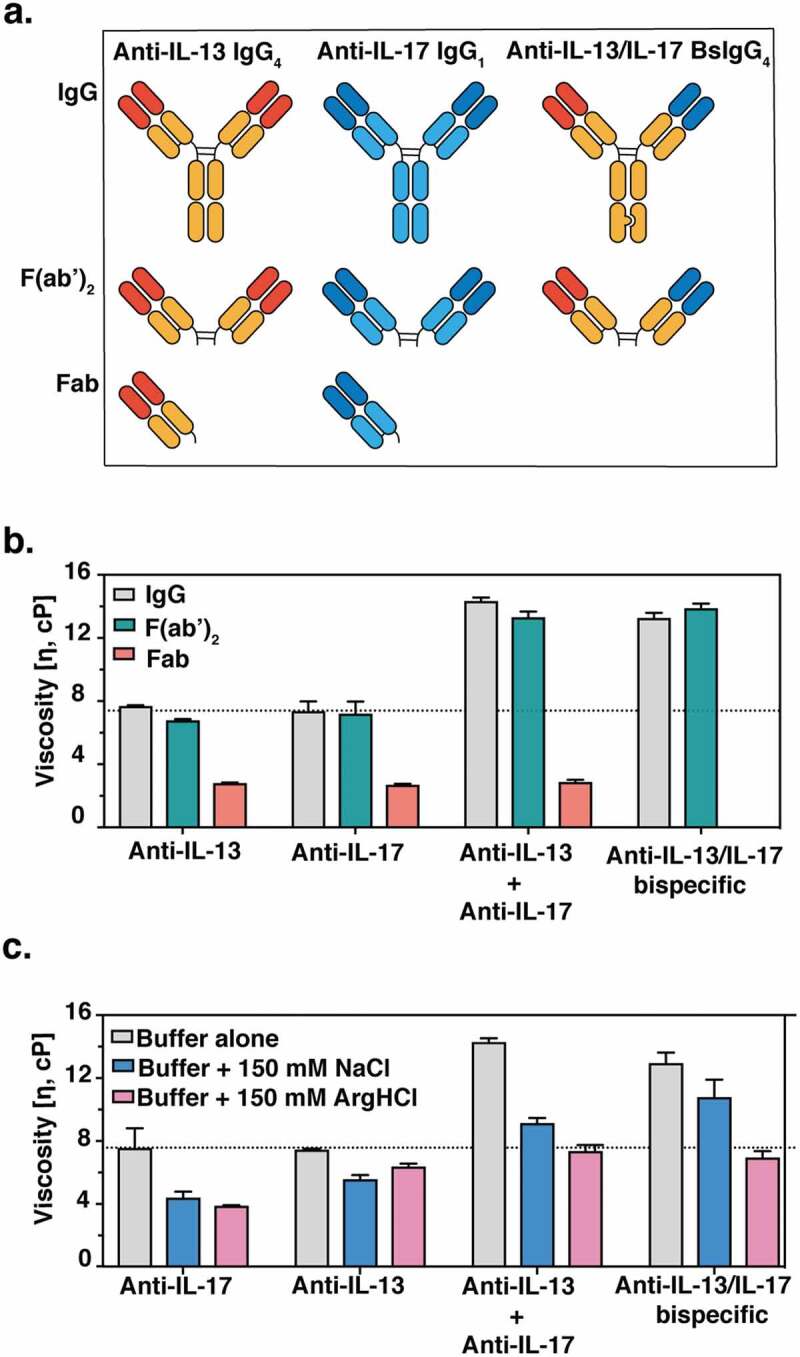
(a) Cartoon representation of monospecific and bispecific antibody formats investigated. The isotype for the anti-IL-17 antibody is IgG1, whereas the anti-IL-13 and BsIgG antibodies are both IgG4. All antibodies have κ light chains. Half-antibodies containing knobs-into-holes mutations34 (‘knob’ mutation (T366W) in the anti-IL-13 arm or ‘hole’ mutations (T366S:L368A:Y407V) in the anti-IL-17arm) were expressed and purified separately before assembly in vitro as described previously.35,36 A hinge stabilizing mutation (S228P) was introduced into the IgG4 to attenuate Fab arm exchange.37,38 (b) Viscosity of fragments (F(ab’)2 and Fab) or full-length anti-IL-13/IL-17 bispecific or its parent antibodies are shown. (c) The effect of 150 mM NaCl or 150 mM Arg-HCl on solution viscosity is shown. The dotted line represents the viscosity of parent IgG antibody level as a reference. Data shown are the mean values (n = 3, ± SD) measured by rheometer at 23ºC and 150 mg/mL total protein concentration in 20 mM His-HCl pH 6.0 (buffer) in the absence or presence of excipients.
Excipient effect on viscosity
To test whether electrostatic interactions are involved in high viscosity, 150 mM NaCl was used to screen for solvent-exposed charged amino acids. Interestingly, the viscosity of the anti-IL-13/IL-17 BsIgG4 was decreased by only ~30% (Figure 1(c)), suggesting the involvement of other types of intermolecular interactions in addition to electrostatic ones. This finding is in agreement with a previous light scattering study, where Rayleigh scattering profiles for the anti-IL-13/IL-17 BsIgG4 were not significantly affected by the change in ionic strength, suggesting that short-ranged non-electrostatic attractive interactions play a significant role in the net protein–protein interactions of BsIgG.32 To further characterize viscosity, arginine-HCl was used. Despite having the same charge as Na+ ions, arginine successfully reduced the viscosity of anti-IL-13/IL-17 BsIgG4 by 50% to the level of its constituent parent monospecific antibodies (Figure 1(c)). We also investigated possible synergistic effects of NaCl and arginine but observed no additional reduction in viscosity relative to arginine alone (Figure S2(b)). A similar effect was observed when guanidine-HCl was used instead of arginine-HCl (Figure S2(a)). Since our results showed that NaCl was not as effective as arginine or guanidine in disrupting intermolecular interactions and mitigating high viscosity, we hypothesized that cation-π and/or π-π interactions may contribute significantly to the elevated viscosity.
X-ray structure determination and MD simulations
Computational methods were used to better understand the mechanism by which arginine reduced antibody viscosity. Specifically, MD simulations were used to investigate potential interactions of arginine in solution with solvent-exposed aromatic residues on the antibody variable domains. The anti-IL-13 and anti-IL-17 Fab structures, minus their corresponding antigens, were used here for MD simulations. The X-ray crystallographic structure of a Fab of lebrikizumab, a humanized anti-IL-13 antibody, bound to IL-13 was previously determined at 1.9 Å resolution (Figure 2(a), PDB 4I77).40 The crystallographic structure of a Fab of the human anti-IL-17 antibody MCAF5352A41 bound to IL-17FF was solved here at 2.75 Å resolution (Figure 2(b), PDB 6PGG, Table S1 and Figure S1). Fab rather than IgG or BsIgG were used to model the interactions between the variable domain residues and arginine, as these excipient-complementary-determining regions (CDR) residue interactions are highly localized.
Figure 2.
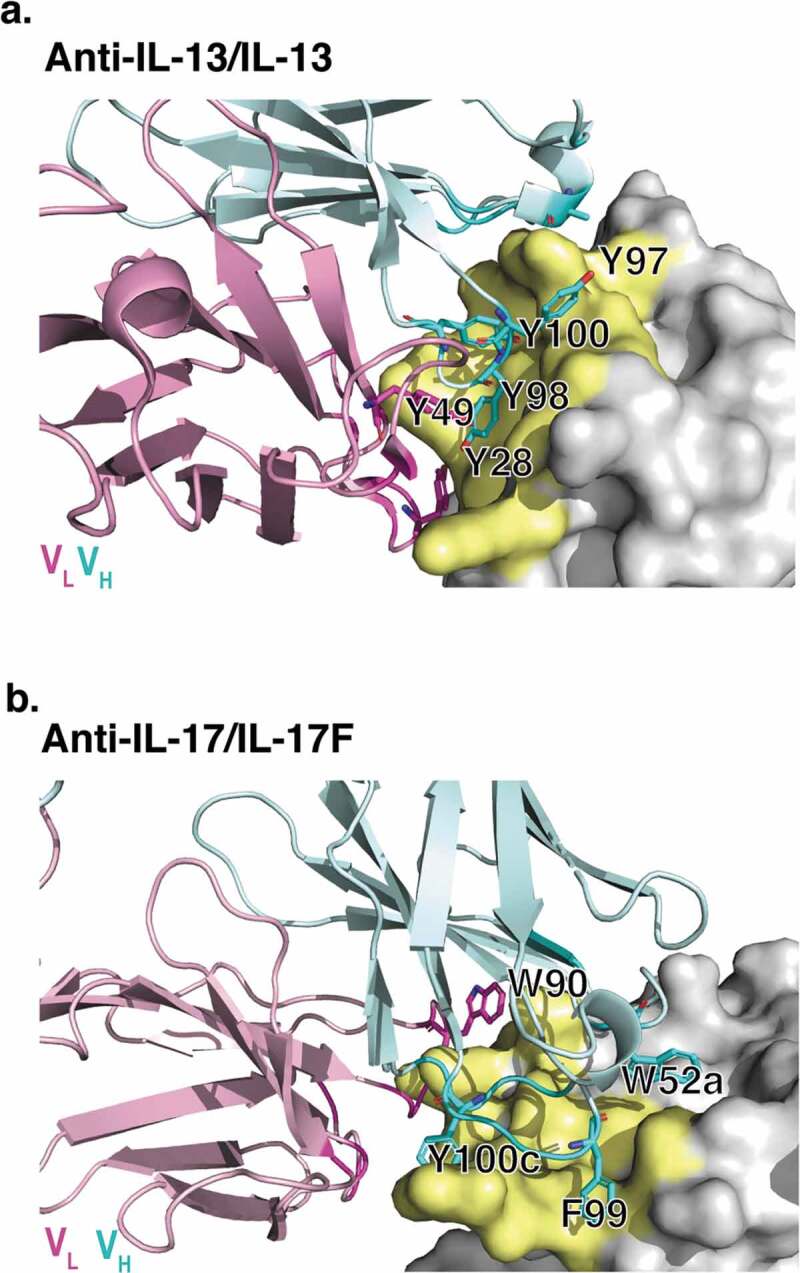
X-ray crystallographic structures of anti-IL-13 (lebrikizumab)40 and anti-IL-17 (MCAF5352A41) antibody Fabs complexed with their respective ligands displaying aromatic residues chosen for mutational studies. (a) View of anti-IL-13/IL-13 interface depicting important viscosity reducing residues (PDB 4I77).40 The anti-IL13 VH is cyan and VL is light pink, with dark cyan and magenta side chains within 4 Å of IL-13. IL-13 is gray with yellow epitope within 4 Å of anti-IL-13. (b) View of anti-IL-17/IL-17F interface depicting important residues (PDB 6PPG). Anti-IL-17F VH is cyan and VL is light pink, with dark cyan and magenta side chains within 4 Å of IL-17. IL-17F is gray with yellow epitope within 4 Å of anti-IL-17F. See the Supplementary Materials for the sequences for the variable domains for the anti-IL-13 (Figure S6) and anti-IL17 (Figure S7) antibodies.
Twenty independent 500 ns MD trajectories for each anti-IL-13 or anti-IL-17 Fab solvated in free arginine and explicit water were generated at pH 6.0, which was the same as the experimental pH for viscosity measurement and also close to the formulation pH (see Supplementary Material for details of the MD method). The average contact number of arginine with residues was calculated for each heavy (VH) and light (VL) variable domain residue for both antibodies (Figure 3). Residues showing greater than two standard deviations (>2σ) of the average contact number with arginine are considered potential residues involved in intermolecular interactions. As expected, mostly aromatic residues in the CDRs experienced larger average contact numbers with free arginine.
Figure 3.
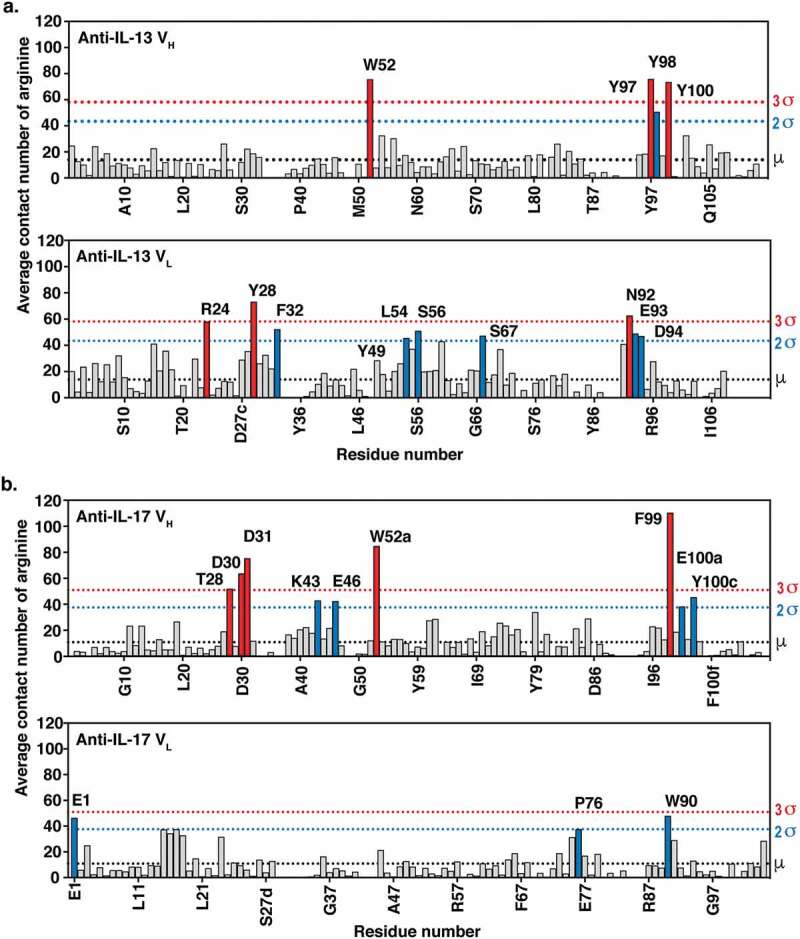
MD simulations of (a) anti-IL-13 (lebrikizumab40) and (b) anti-IL-17 (MCAF5352A41) variable heavy (VH) and variable light (VL) domains in the presence of arginine. Data shown are the mean number of arginine contacts from 20 independent simulations. Residues that show >2σ or >3σ deviations from the mean (μ) number of arginine contact number of all residues in chains are highlighted. Kabat numbering is used.42.
Viscosity and antigen-binding affinity assessment of anti-IL-13/IL-17 antibody variants
Mutagenesis studies were performed using full-length IgG antibodies. Antibody variants were designed based upon MD simulations and molecular modeling. The aim was to alter the intermolecular electrostatic, cation-π and/or π-π interactions without affecting overall IgG structure. Designed substitutions were those predicted in BioLuminate to be the most energetically stable, namely: R/K/D/E → Q and Y/W → L/Q. Aromatic CDR residues selected for mutation in the anti-IL-13 and anti-IL-17 antibodies are highlighted in Figure 2(a,b), respectively. All the surface-exposed aromatic residues in the CDRs were mutated, independent of MD assessment of interaction with free arginine. All variants that could be expressed in sufficient quantities were studied further. MD simulations of selected anti-IL-13 variants, namely Y97L, Y98L, and Y100Q, confirmed a significant decrease in number of arginine contacts compared to the parent antibody (Figure S4).
Monospecific anti-IL-13 IgG4 or anti-IL-17 IgG1 variants were mixed with anti-IL-17 IgG1 or anti-IL-13 parent IgG4, respectively, at a 1:1 ratio and a final total concentration of 150 mg/mL. The viscosity was measured by rheometer and compared to that of a mixture of the anti-IL-13 and anti-IL-17 parent IgG (Figure 4). Remarkably, replacement of some single aromatic CDR residues (Figure 2) in either antibody reduced the viscosity to the level of the corresponding parent IgG (Figure 4(c)). In contrast, replacement of acidic or basic amino acid residues led to at most small reductions in viscosity (Figure 4(a,b)). In addition, mutation of equivalent aromatic residues in the BsIgG showed a significant reduction in the solution viscosity (Figure 4(d)) as they did in the parent monospecific IgG mixture (Figure 4(c)).
Figure 4.
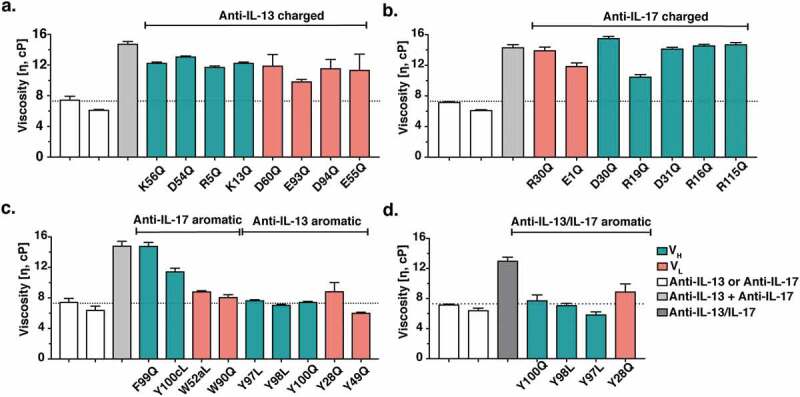
Viscosity of antibody variants. Monospecific anti-IL-13 IgG4 and anti-IL-17 IgG1 antibody variants were mixed with the parent anti-IL-17 IgG1 and anti-IL-13 IgG4 antibody counterparts, respectively, at a 1:1 ratio and a final total concentration of 150 mg/mL. The viscosity of the solutions was then measured by rheometer at 23ºC. (a) Viscosity of mixtures of anti-IL-13 charged variants and the anti-IL-17 parent antibody. (b) Viscosity of mixtures of anti-IL-17 charged variants and the anti-IL-13 parent antibody. (c) Viscosity of mixtures of anti-IL-13 and anti-IL-17 aromatic variants with their parent IgG antibody counterpart. (d) Viscosity of anti-IL-13/IL-17 BsIgG4 variants at 150 mg/mL final concentration. Data shown are the mean values (n = 2, ± SD) measured in 20 mM His-HCl buffer at pH 6.0. The dotted line shows the parent antibody viscosity level as a reference.
The CDRs of antibodies are commonly enriched in aromatic residues that contribute to antigen-binding affinity.43 Therefore, to compare variant affinities to parent antibodies we determined the dissociation constant (KD) values for antigens binding to antibodies by surface plasmon resonance (SPR). Two anti-IL-13 variants were identified that reduced viscosity to about that of the parent IgG4 (Figure 4(c)), VL Y49Q and VH Y97L, and had no measurable effect on the binding affinity for IL-13 (Table 1). In contrast, the anti-IL-17 variants that reduced viscosity also reduced the binding affinity for IL-17 by at least threefold (Table 2).
Table 1.
Antigen-binding kinetics of anti-IL-13 antibody variants. Binding kinetics of anti-IL-13 variants for IL-13 were measured by SPR at 37°C, pH 7.4 and data were fitted to a 1:1 binding model. *Variants that reduced viscosity and have comparable KD as the parent antibody. Dissociation rate constants (koff values) ≤ 1 × 10−4 s−1 could not be reliably measured by SPR under the experimental conditions used.
| Variant | kon, s−1 M−1 x 106 | koff, s−1 x 10−6 | KD, pM |
|---|---|---|---|
| Parent | 2.16 ± 0.04 | ≤ 1 × 10−4 | <10 |
| VH Y97L* | 2.08 ± 0.11 | ≤ 1 × 10−4 | <10 |
| VL Y49Q* | 2.34 ± 0.06 | ≤ 1 × 10−4 | <10 |
| VH Y98L | 1.16 ± 0.04 | 87.6 ± 2.7 | 75.3 ± 0.2 |
| VL Y28Q | 0.58 ± 0.05 | 159 ± 11 | 275 ± 3 |
| VH Y100Q | 1.00 ± 0.01 | 3,310 ± 99 | 3,320 ± 95 |
Table 2.
Antigen-binding kinetics of anti-IL-17 antibody variants. Binding kinetics of the anti-IL-17 variants for IL-17FF were measured by SPR at 37°C, pH 7.4 and data fitted to a 1:1 binding model.
| Variant | kon, s−1 M−1 x 107 | koff, s−1 x 10−4 | KD, pM |
|---|---|---|---|
| Parent | 5.68 ± 0.22 | 4.42 ± 0.30 | 7.78 ± 0.23 |
| VL W90Q | 3.92 ± 0.62 | 9.20 ± 2.12 | 23.4 ± 3.7 |
| VH Y100cL | 4.04 ± 0.14 | 10.8 ± 1.07 | 26.6 ± 1.8 |
| VH W52aL | 8.10 ± 0.39 | 26.5 ± 2.32 | 32.6 ± 1.9 |
| VH F99Q | 4.74 ± 1.37 | 73.8 ± 12.09 | 161 ± 38 |
Viscosity, antigen-binding affinity assessment and MD simulations of an anti-GCGR antibody
To test the generality of our findings, we studied an IgG1 antibody with high viscosity at high concentration. The anti-GCGR (G protein-coupled glucagon receptor) antibody44 displays an abnormal elevated viscosity at high concentrations (Figure S2(c)). The addition of 150 mM NaCl to a solution of the anti-GCCR IgG1 resulted in only a minor decrease (~10%) in the viscosity (Figure S2(c)). All 16 aromatic residues within the variable regions of the anti-GCGR IgG1 were mutated to alanine to investigate a possible role for aromatic residues in the high viscosity of this antibody. Twelve out of 16 mutants were expressed in sufficient quantities to be used for further studies. Viscosity was measured at 180 mg/mL final concentration in 20 mM histidine-acetate buffer at pH 5.5. Seven different alanine variants were identified that substantially lowered viscosity (~2 to ~8-fold reduction) (Table 3). This result strongly suggests the general involvement of aromatic amino acid residues in the intermolecular interactions underlying the high viscosity displayed by concentrated antibody solutions. Two of these seven variants, namely VL Y55A and Y91A, also retained antigen-binding affinity for GCGR, a desirable attribute for a potential antibody clinical candidate (Table 3).
Table 3.
Antigen-binding kinetics and viscosity of anti-GCGR antibody variants. Binding kinetics of the anti-GCGR antibody variants for GCGR were measured by SPR at 37°C, pH 7.4 and data fitted to a 1:1 binding model. *Variants that reduced viscosity by >2-fold and maintained KD within 2-fold of the parent antibody. The viscosity was measured at 180 mg/mL final concentration in 20 mM histidine-acetate buffer at pH 5.5.
| Variant | kon s−1 M−1 x 105 | koff s−1 x 10−4 | KD nM | Viscosity cP |
|---|---|---|---|---|
| Parent | 5.66 ± 0.47 | 3.43 ± 0.08 | 0.61 ± 0.06 | 60.6 ± 9.9 |
| VH Y59A | 21.0 ± 2.1 | 6.60 ± 0.14 | 0.32 ± 0.03 | 84 ± 11 |
| VL Y55A* | 6.18 ± 0.50 | 4.53 ± 0.09 | 0.74 ± 0.05 | 27.0 ± 0.2 |
| VL Y91A* | 5.69 ± 0.56 | 5.63 ± 0.06 | 0.84 ± 0.08 | 20.0 ± 0.5 |
| VL Y96A | 6.74 ± 0.41 | 5.40 ± 0.08 | 0.93 ± 0.08 | 51.4 ± 0.6 |
| VH F51A | 4.47 ± 0.39 | 6.43 ± 0.08 | 1.45 ± 0.15 | 75.7 ± 2.8 |
| VH Y79A | 2.41 ± 0.54 | 10.40 ± 0.35 | 1.84 ± 0.14 | 42.2 ± 1.1 |
| VH Y100dA | 5.80 ± 0.67 | 29.0 ± 1.1 | 5.03 ± 0.41 | 23.1 ± 0.5 |
| VH W100aA | 2.41 ± 0.09 | 26.1 ± 0.3 | 10.8 ± 0.3 | 7.95 ± 0.1 |
| VH F100eA | 7.78 ± 0.86 | 109 ± 5 | 14.1 ± 0.9 | 65.2 ± 0.4 |
| VH Y54A | 0.57 ± 0.28 | 23.0 ± 3.5 | 46 ± 17 | 14.3 ± 0.3 |
| VH Y33A | 3.68 ± 0.47 | 193.0 ± 1.0 | 53.0 ± 7.0 | 35.3 ± 0.8 |
| VH Y31A | 0.24 ± 0.16 | 43.7 ± 16.1 | 206 ± 71 | 19.2 ± 0.8 |
No crystallographic structure was available for the anti-GCGR antibody, so we built a homology model as an alternative starting point for MD simulations by using Molecular Operating Environment45 (MOE, 2018 release) from the Chemical Computing Group. Ten independent 500 ns MD trajectories for the anti-GCGR Fab homology model solvated in free arginine and explicit water were generated at pH 5.5. The mean contact number of arginine with residues was calculated for each heavy (VH) and light (VL) variable domain residue (Figure S5). Unlike the case of the anti-IL-13/IL-17 BsIgG, only 7 of 12 aromatic CDR residues were predicted to have a high number of contacts with arginine.
Discussion
At high concentrations, some antibodies tend to form non-covalent multimers, presumably reflecting intermolecular interactions, thereby elevating the viscosity.46 Such high viscosity can be a major obstacle for antibody therapeutics because it can hamper their manufacture and therapeutic administration by SC or intravitreal injection. Enabling facile SC delivery is attractive for antibody therapeutics that require frequent administration, particularly when combined with a pre-filled syringe and autoinjector medical device.3
The injection volume for SC delivery of antibodies is commonly limited to a volume of ~1.5 mL, which typically necessitates high antibody concentration to deliver the required dose.3 Higher SC injection volumes (up to ~13 mL) can be achieved by coformulations with recombinant human hyaluronidase, which hydrolyzes hyaluronan and opens up the interstitial space.47 Thus, the use of hyaluronidase permits higher antibody doses to be delivered SC. In June 2017, rituximab coformulated with human hyaluronidase became the first such combination product to be approved by the US Food and Drug Administration (FDA) for SC delivery.48 Growing adoption of this approach is suggested by the FDA approval of trastuzumab coformulated with human hyaluronidase in 2019.49 An alternative and potentially complementary strategy to allow higher SC antibody doses is through lowering viscosity to enable SC injection at higher antibody concentration. Addition of hyaluronidase is not a viable option in some cases, such as for intravitreal injection. Therefore, it is of high importance to understand the nature of reversible intermolecular interactions at the molecular level to efficiently overcome the high viscosity challenge.
We studied an anti-IL-13/IL-17 BsIgG4 and the corresponding monospecific IgG to elucidate the molecular basis of elevated viscosity. First, we dissected the parts of the antibody involved in intermolecular interactions by studying antibody fragments. For the parent anti-IL-13 IgG4 or IL-17 IgG1 antibodies, the viscosity was reduced following proteolysis to monovalent Fabs, but not upon proteolysis to bivalent F(ab’)2 fragments. These data suggest that a network of bivalent molecules interacting via Fab regions (i.e., anti-IL-17 Fab or anti-IL-13 Fab with themselves) may contribute to the viscosity of these monospecific IgG antibodies. The anti-IL-13/IL-17 BsIgG4 had elevated viscosity compared to its parent IgG as previously shown32 and confirmed here as did the corresponding bispecific F(ab’)2 (Figure 1(b)). This observation is consistent with the notion39 that a network of bivalent molecules interacting via different Fab regions (i.e., anti-IL-17 Fab with anti-IL-13 Fab) may contribute to high viscosity. In contrast, bispecific antibodies whose Fabs (monovalent for each specificity) do not interact are predicted not to form networks and have low viscosity, since they behave like Fabs.50
Next, we studied the molecular basis of high viscosity at the amino acid level. To delineate the chemical nature of the intermolecular interactions that drive the high viscosity of some antibodies, the surrounding environment of the antibodies can be changed by tuning the ionic strength of the solvent using various excipients such as salts or amino acids.8,14,33 Alternatively, the antibody molecule can be engineered to tune the surface charges to disrupt the intermolecular interactions. A large variety of intermolecular interactions, such as electrostatic, hydrophobic, dipole-dipole, hydrogen bonding and van der Waals interactions, have been suggested to contribute to the high viscosity characteristic of concentrated antibody solutions.11,14,26,51,52 Even though long-range electrostatic intermolecular interactions are dominant, raising the ionic strength of antibody solutions to screen charges does not consistently reduce the viscosity of antibodies, indicating the importance of other forces for some antibodies. In a previously reported study, the viscosity of an antibody known as mAbC was reduced by ~80% in the presence of 100 mM NaCl.53 However, the viscosity of anti-IL-13/IL-17 BsIgG4 and anti-GCGR IgG1 was reduced dramatically when arginine-HCl was added to the solution, but only modestly by NaCl. No synergistic effect of NaCl and arginine-HCl was seen in the case of anti-IL-13/IL-17 BsIgG4; the addition of 75 mM of each salt reduced viscosity better than NaCl alone, but the effect was similar to that of 150 mM arginine-HCl. Guanidine-HCl was as effective as arginine-HCl in reducing viscosity of the antibody solution.
Replacement of acidic or basic amino acid residues led to at most small reductions in viscosity. This may reflect that there are ~50 arginine or lysine residues in the Fab structures, so changing individual charged residues is compensated by the large number of remaining positively charged residues. An alternative potential explanation is that π-π interactions contribute to elevated viscosity.
Recent years have seen a growing appreciation of a noncovalent interaction known as cation-π or π-π in protein sciences,54,55 with the cation being either a lysine or arginine residue and the π-electrons contributed by the aromatic rings of tryptophan, tyrosine and more rarely phenylalanine. It has been shown computationally that in aqueous environments a cation–π interaction is stronger than a salt-bridge,56 which might explain the ineffectiveness of NaCl (relative to arginine or guanidine) in reducing the viscosity.
Inspired by the observation that antibody CDRs commonly include surface aromatic residues and that arginine can interact with the quadrupole ring of aromatic residues, we studied the possible involvement of aromatic residues in high viscosities. MD simulations demonstrated that indeed arginine makes significant contacts with surface aromatic residues. Further, alteration of most of the aromatic residues by point mutations reduced viscosity to the level similar to parent antibody, or to the level of arginine excipient effect. Specifically, anti-IL-13 VH residues Y97, Y98, Y100, and VL Y28 and anti-IL-17 residues VH W52 and VL W90 all were expected to make significant arginine contacts and their mutation led to low-viscosity solutions. The specific mechanism of single mutant viscosity reduction is unknown but is likely due to the removal of weak intermolecular interactions between Fabs. However, the observation that the point mutant viscosity reduction effect is similar to arginine excipient suggests the involvement of both cationic and aromatic residues. Furthermore, the fact that all of the aromatic to leucine mutations reduced viscosity, despite the hydrophobic nature of leucine, suggests that π-electrons in aromatic rings of tyrosine and tryptophan residues were fundamental to high viscosity and presumably intermolecular interactions.
We observed some inconsistencies between predictions from MD simulations and experimental measurements. For example, anti-IL-13 residue VL Y49 was predicted to not make extensive arginine contacts, but a Y49Q mutant did reduce viscosity, whereas mutating highly arginine-contacting residues anti-IL-17 VH F99 did not lead to a significant reduction in viscosity. This may be attributable to several possibilities. For the F99 residue, it may simply be that the model over-predicts the impact of the arginine contacts in the interaction. Indeed, previous research has shown that phenylalanine residues are rarely found in cation-π sites, and the electrostatics for engaging in this type of bonding are less favorable for the phenyl ring in phenylalanine relative to the indole and phenol rings found in tryptophan and tyrosine residues, respectively.57 As for the unexpected behavior of the Y49Q mutant, this may be due to simulation failure or changes to the structure upon mutation.
The anti-GCGR antibody study strengthens the idea of involvement of aromatic amino acid residues in the intermolecular interactions underlying the elevated viscosity displayed by antibody solutions. Nine of 12 mutants reduced viscosity. Three of these had minimal impact on antigen binding and could be considered leads in antibody development. Antigen-binding affinity maturation methodologies could be used to potentially improve the binding of the other mutants.58,59
In this work, we have shown that point mutations of select aromatic residues may dramatically reduce viscosity. Advancing these engineered antibodies into clinical development would require assessment and mitigation of immunogenicity risk60,61and any manufacturing liabilities,62–64 which are beyond the scope of this study. Indeed, assessment of whether these viscosity-lowering mutants also have improved behavior with respect to other properties affected by protein–protein interactions (e.g., aggregation, opalescence, turbidity) could further streamline the removal of antibody liabilities and provide insight into the interactions involved in these phenomena.
Our approach for risk mitigation of elevated viscosity at high protein concentration may be beneficial to early stages of antibody drug development, such as for clinical candidate selection. However, there are some challenges to overcome for such early-stage applications. First, traditional viscosity measurements require substantial amounts of protein, typically tens of milligrams. Several technologies require less protein, and models have been developed to address this protein supply constraint, including microrheology,32,65 SAXS66, and correlation to diffusion interaction parameter.67 Second, crystal structures of antibody candidates are not always available for MD analysis. However, homology models of the Fabs are readily generated68 and can be used as the starting point for the analysis, as exemplified by the anti-GCGR antibody here. While the MD simulation may be less reliable with homology models than for high-resolution crystallographic structures, several variants with reduced viscosity were identified here for the anti-GCGR antibody.
In summary, data presented here strongly suggest that intermolecular interactions mostly mediated by surface accessible aromatic amino acids are major contributors to the high viscosity observed for some antibody at high concentrations. Aromatic amino acid residues may engage in a complex intermolecular interaction that requires a precise geometric arrangement for forming networks. Our experimental and MD simulation results set the basis for a targeted strategy to alter solvent-exposed aromatic residues of antibodies to decrease high viscosity in cases where the viscosity is not affected by high ionic strength. This process could easily be incorporated at either humanization or affinity maturation steps of clinical antibody candidate generation.
Materials and methods
Plasmid construction and mutagenesis
Antibodies were previously cloned into mammalian or bacterial expression vectors as described previously.69–71 The BsIgG was engineered with IgG4 isotype using knobs-into-holes mutations on heavy chains (‘knob’ mutation (T366W)34 in the anti-IL-13 arm or ‘hole’ mutations (T366S:L368A:Y407V)34 in the anti-IL-17arm). Residue scanning for aromatic and charged residue positions was computed with all the amino acids excluding aromatic and charged ones by using the Residue Scanning functionality in BioLuminate (version 3.3.012, Schrödinger, LLC, New York, NY, 2012).72 The chosen amino acid substitutions were generated by PCR-based mutagenesis using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA) and the desired modifications were confirmed by DNA sequencing.
Antibody expression and purification
The parent monospecific and bispecific35 antibodies (anti-IL-13 (IgG4), anti-IL-17 (IgG1) and anti-IL-13/IL-17 (BsIgG4) with κ light chains) were previously produced by Genentech, Inc. Variant monospecific antibodies were transiently expressed in 0.5 L cultures of Expi293F cells and two-step purified by protein A followed by size exclusion chromatography (SEC) as previously described.73 Variant half-antibodies containing knobs-into-holes mutations)34 were expressed in Expi293F cells, purified and then each was assembled with corresponding wildtype half-antibodies into BsIgG in vitro and characterized as described previously.35,36
Antibody fragments
Fabs were generated by digesting 10 mg/mL of IgG antibodies with 0.1 mg/mL papain (Life Technologies, Carlsbad, CA) in 20 mM sodium phosphate, 10 mM EDTA and 20 mM cysteine pH 7.0 buffer solution at 37ºC for 3 h. F(ab’)2 fragments were produced by digesting 10 mg/mL of IgG antibodies with 1 mg/mL IdeS (Genentech) in phosphate-buffered saline pH 7.4 buffer at 37ºC for 3 h. After confirming cleavage by SDS-PAGE, Fab or F(ab’)2 were separated from Fcs by CH1-XL affinity matrix; antibody fragments CH1 region binds the resin. The solution containing antibody fragments of interest was incubated with the resin at room temperature rocking for 3 h and loaded onto a column. Next, the column was washed with 5 times the column volume of a phosphate-buffered saline solution at pH 7.4 and eluted by passing a 100 mM sodium acetate buffer solution at pH 3.0. The isolated fragments were further examined by SEC and time-of-flight liquid chromatography-mass spectrometry (Agilent Technologies, Santa Clara, CA). The final products were ≥97% monomer as judged by SEC chromatography.
Buffer exchange
The buffer solution of interest was prepared as explained previously.32 The IgG or Fab samples were buffer-exchanged several times by using Slide-A-Lyzer Mini dialysis devices (10 kDa molecular weight cutoff, Thermo Fisher Scientific). The dialyzed antibody samples were concentrated to 150 mg/mL using 10 kDa MWCO Amicon-Ultra centrifugal tubes (Millipore) by centrifugation at 3,200 g.
Concentration determination
The concentration of antibody samples concentration was determined by Lunatic UV/V polychromatic spectrophotometer with 0.1–0.7 mm High Lunatic Chip (dynamic range of 0.03–275 OD). The concentration was calculated using an absorptivity determined by quantitative amino acid analysis.
Viscosity measurements
Viscosity measurements were performed on Physica MCR501 rheometer (Anton Paar, Graz, Austria) using CP-25-1 cone-and-plate with a 25 mm diameter and 1.007 angle. 0.07 mL protein sample was pipetted onto the sample plate and cone was lowered to measuring gap to reach uniform contact with the sample. Sample evaporation was prevented by using an evaporation hood and the temperature was controlled by using a thermostat system in H-PTD200 Peltier system. When the temperature reached 23ºC, the samples went through shear-rate sweeps ramping from 10 to 10,000 s−1. The reported value is the average of the 2–3 shear rate sweeps of 1–2 samples at 1,000 s−1. Data analysis was performed with RheoPlus software (Anton Paar, Graz, Austria).
Binding assay using surface plasmon resonance
The antigen-binding kinetics of the antibody variants were measured using SPR on a Biacore T200 instrument (GE Healthcare, Chicago, IL). Antibody variants were captured on protein A sensor chip. Sensorgrams from passage of human IL-13 at concentrations of 0, 3.13, 6.25, 12.50, 25.0, and 50.0 nM, human IL-17 at concentrations of 0, 0.014, 0.041, 0.123, 0.37, 1.11, 3.33, 10 nM, and human GCGR extracellular domain at concentrations of 0, 0.41, 1.23, 3.7, 11.1, 33.3, 100 nM were recorded using an injection time of 3 min with a flow rate of 30 μL/min, at 37°C, and with a running buffer of 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20. After injection, disassociation of the antigens was monitored for 600–900 s in running buffer. The surface was regenerated between binding cycles with a 30-μL injection of 10 mM glycine pH 1.5 buffer. The primary sensorgrams were corrected using a blank cell run and by zero-point run (0-nM antigen buffer). A 1:1 Langmuir binding model was used to analyze corrected sensorgrams with software provided by the manufacturer to calculate the kinetic and binding constants.
X-ray crystallography
The IL-17FF−Fab (20 mg/mL in 25 mM Tris pH 7.5, and 50 mM sodium chloride) was crystallized in hanging drops with equal volumes of well solution (30% polyethylene glycol (PEG) 400, 0.1 M cadmium chloride, and 0.1 M sodium acetate, pH 4.6). Crystallographic data were collected at beamline 9 − 2 at the Stanford Synchrotron Radiation Laboratory (Stanford, CA), processed with HKL2000, solved and refined using the CCP4 suite of programs. For detailed methods refer to Supplementary Material and Table S1.
Abbreviations
- BsAb
bispecific antibodies
- BsIgG
bispecific IgG
- CDR
complementary-determining region
- GCGR
G-protein coupled glucagon receptor
- IV
intravenous
- MD
molecular dynamics
- PDB
Protein Data Bank
- SC
subcutaneous
- SEC
size-exclusion chromatography
- SPR
surface plasmon resonance
- VL
variable light
- VH
variable heavy
Acknowledgments
We thank members of the Research Materials group in the Cell Culture Department at Genentech for mammalian and bacterial antibody expressions. We thank Christoph Spiess and Michael Dillon for supplying antibody expression vectors and David Cain for assistance with the rheometer. We thank Patrick Lupardus for X-ray crystallographic data collection. The anti-IL17 antibody used in this study was provided under license from Novimmune SA. SLAC National Accelerator Laboratory is acknowledged for the use of the Stanford Synchrotron Radiation Lightsource. SLAC National Accelerator Laboratory is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515.
Disclosure of potential conflicts of interest
All authors are current or former employees of Genentech, Inc, which develops and commercializes therapeutics including bispecific antibodies.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.The Antibody Society . List of approved antibodies. [cited 2019. Nov 19]. https://www.antibodysociety.org/resources/approved-antibodies/.
- 2.Carter PJ, Lazar GA.. Next generation antibody drugs: pursuit of the ‘high-hanging fruit’. Nat Rev Drug Discov. 2018;17:197–11. [DOI] [PubMed] [Google Scholar]
- 3.Viola M, Sequeira J, Seiça R, Veiga F, Serra J, Santos AC, Ribeiro AJ. Subcutaneous delivery of monoclonal antibodies: how do we get there? J Control Release. 2018;286:301–14. [DOI] [PubMed] [Google Scholar]
- 4.Jin JF, Zhu LL, Chen M, Xu HM, Wang HF, Feng XQ, Zhu XP, Zhou Q. The optimal choice of medication administration route regarding intravenous, intramuscular, and subcutaneous injection. Patient Prefer Adher. 2015;9:923–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Wang D, Mason B, Rossomando T, Li N, Liu D, Cheung JK, Xu W, Raghava S, Katiyar A, et al. Structure, heterogeneity and developability assessment of therapeutic antibodies. MAbs. 2019;11:239–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93:1390–402. [DOI] [PubMed] [Google Scholar]
- 7.Hung JJ, Borwankar AU, Dear BJ, Truskett TM, Johnston KP. High concentration tangential flow ultrafiltration of stable monoclonal antibody solutions with low viscosities. J Membrane Sci. 2016;508:113–26. [Google Scholar]
- 8.Liu J, Nguyen MD, Andya JD, Shire SJ. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J Pharm Sci. 2005;94:1928–40. [DOI] [PubMed] [Google Scholar]
- 9.Yadav S, Liu J, Shire SJ, Kalonia DS. Specific interactions in high concentration antibody solutions resulting in high viscosity. J Pharm Sci. 2010;99:1152–68. [DOI] [PubMed] [Google Scholar]
- 10.Yadav S, Shire SJ, Kalonia DS. Viscosity behavior of high-concentration monoclonal antibody solutions: correlation with interaction parameter and electroviscous effects. J Pharm Sci-Us. 2012;101:998–1011. [DOI] [PubMed] [Google Scholar]
- 11.Du W, Klibanov AM. Hydrophobic salts markedly diminish viscosity of concentrated protein solutions. Biotechnol Bioeng. 2011;108:632–36. [DOI] [PubMed] [Google Scholar]
- 12.He F, Woods CE, Litowski JR, Roschen LA, Gadgil HS, Razinkov VI, Kerwin BA. Effect of sugar molecules on the viscosity of high concentration monoclonal antibody solutions. Pharm Res-Dordr. 2011;28:1552–60. [DOI] [PubMed] [Google Scholar]
- 13.Esfandiary R, Hayes DB, Parupudi A, Casas-Finet J, Bai S, Samra HS, Shah AU, Sathish HA. A systematic multitechnique approach for detection and characterization of reversible self-association during formulation development of therapeutic antibodies. J Pharm Sci. 2013;102:62–72. [DOI] [PubMed] [Google Scholar]
- 14.Esfandiary R, Parupudi A, Casas-Finet J, Gadre D, Sathish H. Mechanism of reversible self-association of a monoclonal antibody: role of electrostatic and hydrophobic interactions. J Pharm Sci. 2015;104:577–86. [DOI] [PubMed] [Google Scholar]
- 15.Mason BD, Zhang L, Remmele RL Jr., Zhang J. Opalescence of an IgG2 monoclonal antibody solution as it relates to liquid-liquid phase separation. J Pharm Sci. 2011;100:4587–96. [DOI] [PubMed] [Google Scholar]
- 16.Melander W, Horvath C. Salt effect on hydrophobic interactions in precipitation and chromatography of proteins: an interpretation of the lyotropic series. Arch Biochem Biophys. 1977;183:200–15. [DOI] [PubMed] [Google Scholar]
- 17.Nishi H, Miyajima M, Nakagami H, Noda M, Uchiyama S, Fukui K. Phase separation of an IgG1 antibody solution under a low ionic strength condition. Pharm Res. 2010;27:1348–60. [DOI] [PubMed] [Google Scholar]
- 18.Pindrus M, Shire SJ, Kelley RF, Demeule B, Wong R, Xu Y, Yadav S. Solubility challenges in high concentration monoclonal antibody formulations: relationship with amino acid sequence and intermolecular interactions. Mol Pharm. 2015;12:3896–907. [DOI] [PubMed] [Google Scholar]
- 19.Yadav S, Sreedhara A, Kanai S, Liu J, Lien S, Lowman H, Kalonia DS, Shire SJ. Establishing a link between amino acid sequences and self-associating and viscoelastic behavior of two closely related monoclonal antibodies. Pharm Res-Dordr. 2011;28:1750–64. [DOI] [PubMed] [Google Scholar]
- 20.Yadav S, Shire SJ, Kalonia DS. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J Pharm Sci. 2010;99:4812–29. [DOI] [PubMed] [Google Scholar]
- 21.Chari R, Jerath K, Badkar AV, Kalonia DS. Long- and short-range electrostatic interactions affect the rheology of highly concentrated antibody solutions. Pharm Res-Dordr. 2009;26:2607–18. [DOI] [PubMed] [Google Scholar]
- 22.Chow CK, Allan BW, Chai Q, Atwell S, Lu J. Therapeutic antibody engineering to improve viscosity and phase separation guided by crystal structure. Mol Pharm. 2016;13:915–23. [DOI] [PubMed] [Google Scholar]
- 23.Shan L, Mody N, Sormani P, Rosenthal KL, Damschroder MM, Esfandiary R. Developability assessment of engineered monoclonal antibody variants with a complex self-association behavior using complementary analytical and in silico tools. Mol Pharm. 2018;15:5697–710. [DOI] [PubMed] [Google Scholar]
- 24.Geng SB, Wittekind M, Vigil A, Tessier PM. Measurements of monoclonal antibody self-association are correlated with complex biophysical properties. Mol Pharm. 2016;13:1636–45. [DOI] [PubMed] [Google Scholar]
- 25.Geoghegan JC, Fleming R, Damschroder M, Bishop SM, Sathish HA, Esfandiary R. Mitigation of reversible self-association and viscosity in a human IgG1 monoclonal antibody by rational, structure-guided Fv engineering. MAbs. 2016;8:941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buck PM, Chaudhri A, Kumar S, Singh SK. Highly viscous antibody solutions are a consequence of network formation caused by domain-domain electrostatic complementarities: insights from coarse-grained simulations. Mol Pharm. 2015;12:127–39. [DOI] [PubMed] [Google Scholar]
- 27.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci U S A. 2009;106:11937–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols P, Li L, Kumar S, Buck PM, Singh SK, Goswami S, Balthazor B, Conley TR, Sek D, Allen MJ. Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions. MAbs. 2015;7:212–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol. 2015;67:95–106. [DOI] [PubMed] [Google Scholar]
- 30.Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs. 2017;9:182–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labrijn AF, Janmaat ML, Reichert JM, Parren P. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov. 2019;18:585–608. [DOI] [PubMed] [Google Scholar]
- 32.Woldeyes MA, Josephson LL, Leiske DL, Galush WJ, Roberts CJ, Furst EM. Viscosities and protein interactions of bispecific antibodies and their monospecific mixtures. Mol Pharm. 2018;15:4745–55. [DOI] [PubMed] [Google Scholar]
- 33.Inoue N, Takai E, Arakawa T, Shiraki K. Specific decrease in solution viscosity of antibodies by arginine for therapeutic formulations. Mol Pharm. 2014;11:1889–96. [DOI] [PubMed] [Google Scholar]
- 34.Atwell S, Ridgway JB, Wells JA, Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol. 1997;270:26–35. [DOI] [PubMed] [Google Scholar]
- 35.Spiess C, Bevers J 3rd, Jackman J, Chiang N, Nakamura G, Dillon M, Liu H, Molina P, Elliott JM, Shatz W, et al. Development of a human IgG4 bispecific antibody for dual targeting of interleukin-4 (IL-4) and interleukin-13 (IL-13) cytokines. J Biol Chem. 2013;288:26583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiess C, Merchant M, Huang A, Zheng Z, Yang NY, Peng J, Ellerman D, Shatz W, Reilly D, Yansura DG, et al. Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat Biotechnol. 2013;31:753–58. [DOI] [PubMed] [Google Scholar]
- 37.Stubenrauch K, Wessels U, Regula JT, Kettenberger H, Schleypen J, Kohnert U. Impact of molecular processing in the hinge region of therapeutic IgG4 antibodies on disposition profiles in cynomolgus monkeys. Drug Metab Dispos. 2010;38:84–91. [DOI] [PubMed] [Google Scholar]
- 38.Labrijn AF, Buijsse AO, van den Bremer ET, Verwilligen AY, Bleeker WK, Thorpe SJ, Killestein J, Polman CH, Aalberse RC, Schuurman J, et al. Therapeutic IgG4 antibodies engage in Fab-arm exchange with endogenous human IgG4 in vivo. Nat Biotechnol. 2009;27:767–71. [DOI] [PubMed] [Google Scholar]
- 39.Kanai S, Liu J, Patapoff TW, Shire SJ. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J Pharm Sci. 2008;97:4219–27. [DOI] [PubMed] [Google Scholar]
- 40.Ultsch M, Bevers J, Nakamura G, Vandlen R, Kelley RF, Wu LC, Eigenbrot C. Structural basis of signaling blockade by anti-IL-13 antibody Lebrikizumab. J Mol Biol. 2013;425:1330–39. [DOI] [PubMed] [Google Scholar]
- 41.Peng K, Wang Y, Siradze K, Erickson R, Fischer SK, Staton TL. Measurement of IL-17AA and IL-17FF as pharmacodynamic biomarkers to demonstrate target engagement in the phase I study of MCAF5352A. Aaps J. 2019;21:9. [DOI] [PubMed] [Google Scholar]
- 42.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Bethesda (MD): NIH; 1991. [Google Scholar]
- 43.Traxlmayr MW, Kiefer JD, Srinivas RR, Lobner E, Tisdale AW, Mehta NK, Yang NJ, Tidor B, Wittrup KD. Strong enrichment of aromatic residues in binding sites from a charge-neutralized hyperthermostable Sso7d scaffold library. J Biol Chem. 2016;291:22496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koth CM, Murray JM, Mukund S, Madjidi A, Minn A, Clarke HJ, Wong T, Chiang V, Luis E, Estevez A, et al. Molecular basis for negative regulation of the glucagon receptor. Proc Natl Acad Sci U S A. 2012;109:14393–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Molecular Operating Environment (MOE) 2013.08 . Chemical Computing Group ULC, 1010 Sherbooke St. West, suite 910, Montreal, QC, Canada, H3A 2R7. 2018., 2018.
- 46.Tomar DS, Kumar S, Singh SK, Goswami S, Li L. Molecular basis of high viscosity in concentrated antibody solutions: strategies for high concentration drug product development. Mabs. 2016;8:216–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Locke KW, Maneval DC, LaBarre MJ. ENHANZE® drug delivery technology: a novel approach to subcutaneous administration using recombinant human hyaluronidase PH20. Drug Deliv. 2019;26:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yelvington BJ. Subcutaneous Rituximab in follicular lymphoma, chronic lymphocytic leukemia, and diffuse large B-cell lymphoma. J Adv Pract Oncol. 2018;9:530–34. [PMC free article] [PubMed] [Google Scholar]
- 49.Dent S, Ammendolea C, Christofides A, Edwards S, Incekol D, Pourmirza B, Kfoury S, Poirier B. A multidisciplinary perspective on the subcutaneous administration of trastuzumab in HER2-positive breast cancer. Curr Oncol. 2019;26:e70–e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kastelic M, Dill KA, Kalyuzhnyi YV, Vlachy V. Controlling the viscosities of antibody solutions through control of their binding sites. J Mol Liq. 2018;270:234–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arora J, Hickey JM, Majumdar R, Esfandiary R, Bishop SM, Samra HS, Middaugh CR, Weis DD, Volkin DB. Hydrogen exchange mass spectrometry reveals protein interfaces and distant dynamic coupling effects during the reversible self-association of an IgG1 monoclonal antibody. MAbs. 2015;7:525–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arora J, Hu Y, Esfandiary R, Sathish HA, Bishop SM, Joshi SB, Middaugh CR, Volkin DB, Weis DD. Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity. MAbs. 2016;8:1561–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sudrik C, Cloutier T, Pham P, Samra HS, Trout BL. Preferential interactions of trehalose, L-arginine.HCl and sodium chloride with therapeutically relevant IgG1 monoclonal antibodies. MAbs. 2017;9:1155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dougherty DA. A computational study of cation-pi interactions vs salt bridges in aqueous media: implications for protein engineering. Peptides for the New Millennium. 2000;550–52.
- 55.McGaughey GB, Gagné M, Rappé AK. pi-stacking interactions alive and well in proteins. J Biol Chem. 1998;273:15458–63. [DOI] [PubMed] [Google Scholar]
- 56.Gallivan JP, Dougherty DA. A computational study of cation-pi interactions vs salt bridges in aqueous media: implications for protein engineering. J Am Chem Soc. 2000;122:870–74. [Google Scholar]
- 57.Mecozzi S, West AP Jr., Dougherty DA. Cation-pi interactions in aromatics of biological and medicinal interest: electrostatic potential surfaces as a useful qualitative guide. Proc Natl Acad Sci U S A. 1996;93:10566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennedy PJ, Oliveira C, Granja PL, Sarmento B. Monoclonal antibodies: technologies for early discovery and engineering. Crit Rev Biotechnol. 2018;38:394–408. [DOI] [PubMed] [Google Scholar]
- 59.Li B, Fouts AE, Stengel K, Luan P, Dillon M, Liang WC, Feierbach B, Kelley RF, Hötzel I. In vitro affinity maturation of a natural human antibody overcomes a barrier to in vivo affinity maturation. MAbs. 2014;6:437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber CA, Mehta PJ, Ardito M, Moise L, Martin B, De Groot AS. T cell epitope: friend or foe? Immunogenicity of biologics in context. Adv Drug Deliv Rev. 2009;61:965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quarmby V, Phung QT, Lill JR. MAPPs for the identification of immunogenic hotspots of biotherapeutics; an overview of the technology and its application to the biopharmaceutical arena. Expert Rev Proteomics. 2018;15:733–48. [DOI] [PubMed] [Google Scholar]
- 62.Haberger M, Bomans K, Diepold K, Hook M, Gassner J, Schlothauer T, Zwick A, Spick C, Kepert JF, Hienz B, et al. Assessment of chemical modifications of sites in the CDRs of recombinant antibodies Susceptibility vs. functionality of critical quality attributes. Mabs. 2014;6:327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DiCara DM, Andersen N, Chan R, Ernst JA, Ayalon G, Lazar GA, Agard NJ, Hilderbrand A, Hotzel I. High-throughput screening of antibody variants for chemical stability: identification of deamidation-resistant mutants. Mabs. 2018;10:1073–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang XY, Xu W, Dukleska S, Benchaar S, Mengisen S, Antochshuk V, Cheung J, Mann L, Babadjanova Z, Rowand J, et al. Developability studies before initiation of process development Improving manufacturability of monoclonal antibodies. Mabs. 2013;5:787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Josephson LL, Galush WJ, Furst EM. Parallel temperature-dependent microrheological measurements in a microfluidic chip. Biomicrofluidics. 2016;10:043503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuda M, Watanabe A, Hayasaka A, Muraoka M, Hori Y, Yamazaki T, Imaeda Y, Koga A. Small-scale screening method for low-viscosity antibody solutions using small-angle X-ray scattering. Eur J Pharm Biopharm. 2017;112:132–37. [DOI] [PubMed] [Google Scholar]
- 67.Tomar DS, Singh SK, Li L, Broulidakis MP, Kumar S. In Silico Prediction of Diffusion Interaction Parameter (kD), a Key Indicator of Antibody Solution Behaviors. Pharm Res. 2018;35:193. [DOI] [PubMed] [Google Scholar]
- 68.Zhu K, Day T, Warshaviak D, Murrett C, Friesner R, Pearlman D. Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction. Proteins. 2014;82:1646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simmons LC, Reilly D, Klimowski L, Raju TS, Meng G, Sims P, Hong K, Shields RL, Damico LA, Rancatore P, et al. Expression of full-length immunoglobulins in Escherichia coli: rapid and efficient production of aglycosylated antibodies. J Immunol Methods. 2002;263:133–47. [DOI] [PubMed] [Google Scholar]
- 70.Eaton DL, Wood WI, Eaton D, Hass PE, Hollingshead P, Wion K, Mather J, Lawn RM, Vehar GA, Gorman C. Construction and characterization of an active factor VIII variant lacking the central one-third of the molecule. Biochem. 1986;25:8343–47. [DOI] [PubMed] [Google Scholar]
- 71.Polson AG, Yu SF, Elkins K, Zheng B, Clark S, Ingle GS, Slaga DS, Giere L, Du C, Tan C, et al. Antibody-drug conjugates targeted to CD79 for the treatment of non-Hodgkin lymphoma. Blood. 2007;110:616–23. [DOI] [PubMed] [Google Scholar]
- 72.Beard H, Cholleti A, Pearlman D, Sherman W, Loving KA. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS One. 2013;8:e82849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bos AB, Luan P, Duque JN, Reilly D, Harms PD, Wong AW. Optimization and automation of an end-to-end high throughput microscale transient protein production process. Biotechnol Bioeng. 2015;112:1832–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- The Antibody Society . List of approved antibodies. [cited 2019. Nov 19]. https://www.antibodysociety.org/resources/approved-antibodies/.


