Abstract
The need for international collaboration in rodent pathology has evolved since the 1970s and was initially driven by the new field of toxicologic pathology. First initiated by the World Health Organization’s International Agency for Research on Cancer for rodents, it has evolved to include pathology of the major species (rats, mice, guinea pigs, nonhuman primates, pigs, dogs, fish, rabbits) used in medical research, safety assessment, and mouse pathology. The collaborative effort today is driven by the needs of the regulatory agencies in multiple countries, and by needs of research involving genetically engineered animals, for “basic” research and for more translational preclinical models of human disease. These efforts led to the establishment of an international rodent pathology nomenclature program. Since that time, multiple collaborations for standardization of laboratory animal pathology nomenclature and diagnostic criteria have been developed, and just a few are described herein. Recently, approaches to a nomenclature that is amenable to sophisticated computation have been made available and implemented for large-scale programs in functional genomics and aging. Most terminologies continue to evolve as the science of human and veterinary pathology continues to develop, but standardization and successful implementation remain critical for scientific communication now as ever in the history of veterinary nosology.
Keywords: International Agency for Research on Cancer, International Harmonization of Nomenclature and Diagnostic Criteria, International Mouse Phenotyping Consortium, mouse pathology ontology, National Cancer Institute Mouse Models of Human Cancer Consortium, National Toxicology Program Nonneoplastic Lesion Atlas, nomenclature, standard for exchange of nonclinical data
History of International Laboratory Animal Pathology Nomenclature
The use of any standardized nomenclature for rodent pathology perhaps began in the 1970s with the publication of a series of tumor pathology books (on mice, rats, and hamsters) by the International Agency for Research in Cancer (IARC), World Health Organization (WHO), Lyon, France. Dr. Vladimir Turusov, a medical pathologist, was the initial editor of a book series and chapter authors were invited from Europe, Japan, and the USA.1,2,3,4,5 In the 1990s, Ulrich Mohr was editor for the second and third IARC series on rats and mice.6,7 In these series, international committees of pathologists prepared monographs on tumors of each organ system for rats and one book for mice. In the early 1990s, committees for nomenclature of tumors and nonproliferative lesions for each organ system of rats and mice were established as Guides for Toxicologic Pathology, a system of standardized nomenclature and diagnostic criteria, by a collaboration of the Armed Forces Institute of Pathology, American Registry of Pathology, and the Society of Toxicologic Pathology (STP). The first published series, on rat pathology, was published by the Armed Forces Institute of Pathology and are presently online (https://toxpath.org/ssndc.asp). From 1983 to1996, The International Life Sciences Institute sponsored a series of 13 monographs on pathology of laboratory animals led by T. C. Jones, U. Mohr, and R. D. Hunt;8https://link.springer.com/bookseries/780). During the same period, the National Toxicology Program (NTP) staff pathologists, contractors, and collaborators published two books on rat and mouse pathology.9,10 These efforts led to the establishment of an international rodent pathology nomenclature program (International Harmonization of Nomenclature and Diagnostic Criteria [INHAND]) involving several of the national societies of toxicologic pathology.
INHAND for Use in Toxicology Safety Assessment
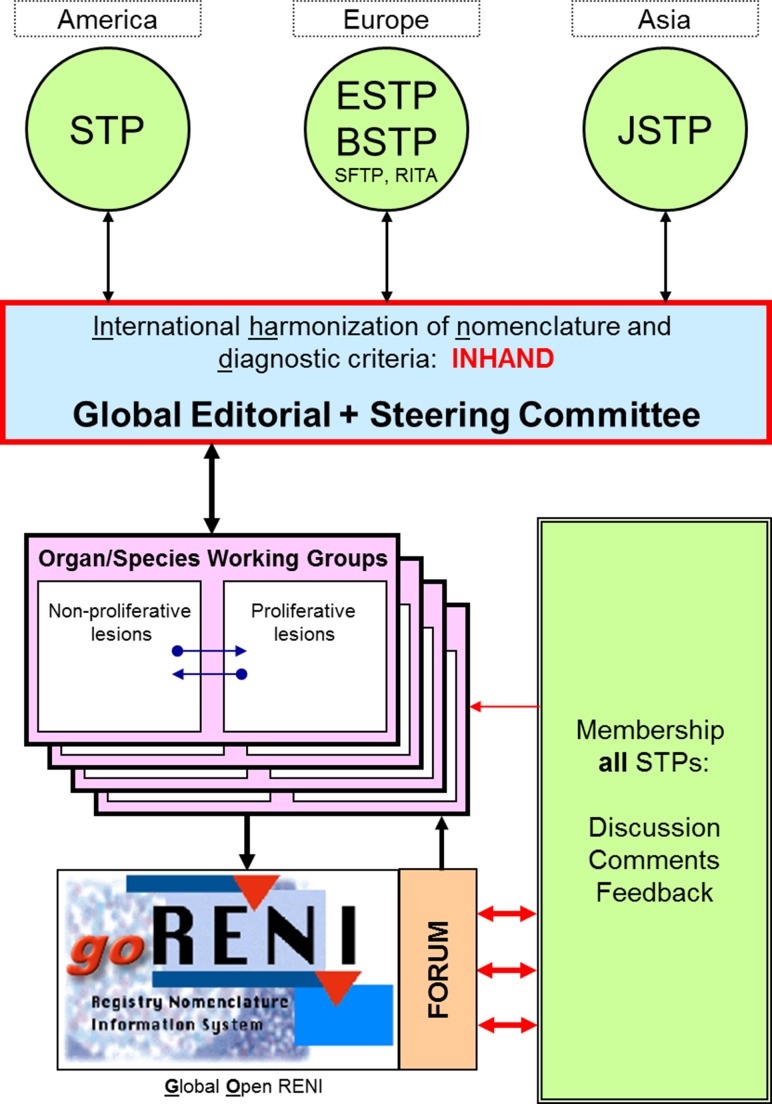
In 2005, the Strategic and Regulatory Policy Committee of the STP determined that there was need for a revision of the earlier Standardized System of Nomenclature and Diagnostic Criteria guides. The European Society of Toxicologic Pathology (ESTP), in conjunction with the Registry of Industrial Toxicology Animal-data (RITA), endorsed the proposal in late 2005. In 2006, the Japanese Society of Toxicologic Pathology (JSTP) and the British Society of Toxicologic Pathology (BSTP) joined the initiative, providing a truly global participation. Members of these major Societies of Toxicologic Pathology (JSTP, BSTP, ESTP, and STP) and RITA are now engaged in an international collaborative effort (INHAND) to codify and publish uniform nomenclature for both proliferative and nonproliferative lesions in laboratory rodents. Several features unique to this effort include: (1) a truly international scope, (2) implementation of an open comment period allowing a wide group of toxicologic pathologists the opportunity to provide input, (3) inclusion of neoplastic and nonneoplastic terminology, and (4) availability in a web-based format along with publication in society journals. Project oversight is provided by the Global Editorial and Steering Committee (GESC), which consists of members from each of the major Societies of Toxicologic Pathology (Figure 1). Each rodent organ system or nonrodent species organ working group consists of a chairperson and members from each of the major Societies of Toxicologic Pathology, drawing upon a diversity of experience and background with individuals from industry, academia, and government.
Figure 1.
Organizational map of INHAND.
The Rodent Organ Working Groups have the responsibility to prepare the nomenclature guidelines for both proliferative and nonproliferative lesions of rats and mice for their assigned organ system—15 total systems. The Non-rodent Species Working Groups (NRWGs) cover terminology specific to a species as well as noting diagnostic criteria that may be different from rodents for common lesions. The NRWGs include nonhuman primate, dog, minipig, rabbit, and fish. In addition to lesions that occur spontaneously, the groups are asked to determine if there are common, xenobiotic-induced lesions for which standardized nomenclature might be needed. The working groups draw heavily from existing nomenclature documents, websites, and publications including prior work of the RITA and the Standardized System of Nomenclature and Diagnostic Criteria. For each diagnostic entity, the working groups select a preferred diagnosis and acceptable alternative diagnoses, provide diagnostic criteria and differential diagnosis, prepare representative photomicrographs, and also provide a comment section with key references. In general, working groups develop nomenclature that is primarily descriptive in nature and denote findings that can be documented from the review of routine histologic specimens. Incorporating specific diagnostic entities such as an infectious disease or that imply a process that cannot be ascertained from routine histologic specimens (e.g., phospholipidosis) is generally not recommended.
Finalized nomenclature is available to toxicologic pathologists and the broader scientific community in both electronic and print forms. The print-based publications are available in the toxicologic pathology journals: Toxicologic Pathology, the official journal of STP, BSTP, and ESTP (http://journals.sagepub.com/home/tpx) and the Journal of Toxicologic Pathology, which is the official journal of JSTP (https://www.jstage.jst.go.jp/browse/tox). Electronic access is via the global open Registry Nomenclature Information System (goRENI) website (https://www.goreni.org) or the journal websites (https://www.toxpath.org/inhand.asp#pubg or https://www.jstage.jst.go.jp/browse/tox).
Substantial progress has been made to date; 12 of the 15 rodent organ systems have been published: Respiratory System,11 Hepatobiliary System,12 Urinary System,13 Nervous System,14 Mammary, Zymbal’s, Preputial and Clitoral Glands,15 Male Reproductive System,16 Soft Tissue,17 Integument,18 Female Reproductive System,19 Digestive System,20 Cardiovascular System,21 and Skeletal System and Tooth.22 To address consistent terminology for cell death, Recommendations from an Apoptosis/Necrosis Working Group was published.23 The Endocrine and Special Senses Systems and the Hematopoietic and Lymphoid System are scheduled for publication in 2018/2019.
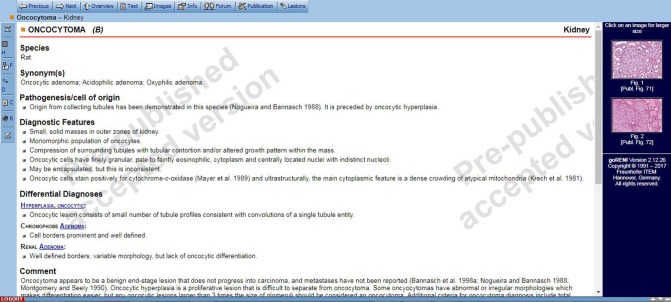
An important aspect of the INHAND project is utilization of goRENI. ESTP offered access to an open version of goRENI to serve as a platform. Access to goRENI is restricted to members of the participating STPs.24,25 Once access is granted, pathologists can navigate by organ systems and select a diagnosis they would like to view. Within the goRENI system, each diagnostic entity is referred to as a manuscript. An example is provided in (Figure 2) of the written information and photographic illustrations provided for a kidney oncocytoma.
Figure 2.
Example of a goRENI manuscript.
Although the published INHAND nomenclature for each organ system is expected to be very comprehensive, it is recognized that additional lesions may need to be included, inaccuracies corrected as they become apparent, or changes to terminology made based on new scientific information. To address this, a formal change control process was implemented in 2013 and is available on www.goreni.org and each pathology society website. Society members are encouraged to submit recommendations for changes to the nomenclature systems and provide justifications for such changes through this mechanism. Updates will be posted on goRENI, and this will be the source for the most current information.
The GESC and STP, BSTP, ESTP, and JSTP leadership recognize the significant efforts of all of those serving on the rodent Organ Working Groups and NRWGs and look forward to working with the global toxicologic pathology community as additional systems are drafted, reviewed, and completed. The international scope and review of the INHAND documents will provide a strong framework for use by pathologists and regulatory agencies that are engaged in the safety assessment of drugs, biologics, and chemicals.
Standard for Exchange of Nonclinical Data (SEND)
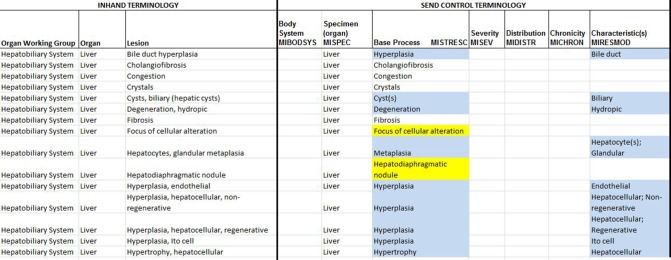
SEND is a standardized procedure for submitting data from nonclinical studies to the Food and Drug Administration (FDA) electronically and in a standardized format.26 During 2011, INHAND GESC representatives attended meetings with representatives of the FDA Center for Drug Evaluation and Research, Clinical Data Interchange Standards Consortium, and the National Cancer Institute (NCI) Enterprise Vocabulary Services to initiate integration of INHAND terminology as the preferred terminology for SEND. INHAND GESC representatives work with the SEND Controlled Terminology committee to provide definitions for base processes and modifiers associated with the INHAND published terminology. Any issues or questions are presented to the full GESC and/or appropriate INHAND Working Group for resolution. The initial list for the SEND codelist of nonneoplastic (NONNEO) microscopic pathology contains terms from published INHAND organ systems. The list will continue to grow as INHAND publishes additional organ systems. Some terms on the NONNEO codelist may look different from how they have been presented in the INHAND publications. Terms on the NONNEO codelist are mostly generic and can be used across tissues, where appropriate. INHAND published terms have been modified to fit the SEND standard in some cases by being broken into base process and modifiers. For example, the INHAND term necrosis, zonal would be separated into NECROSIS for population in MISTRESC (Microscopic Standardized Result) and ZONAL in MIDISTR (Microscopic Distribution). Tissue-specific terms from INHAND are included on the NONNEO codelist when it is important to use the exact term representing a spectrum of tissue changes (e.g., focus of cellular alteration). In the process of mapping terms from INHAND to SEND, some inconsistencies have been noted for the same term across several organ systems (e.g., thrombus vs. thrombosis). These will be harmonized using the new change control process, and the most current terminology will be available on the goRENI website. An example of a nomenclature map with INHAND terminology is shown in (Figure 3). The most current SEND controlled terminology can be found at the National Cancer Institute (NCI) Enterprise Vocabulary Services site: https://evs.nci.nih.gov/ftp1/CDISC/SEND/.
Figure 3.
Example of nomenclature map with INHAND terminology.
NTP Nonneoplastic Lesion Atlas
Assessing the carcinogenicity of agents of concern in its rodent models has been at the core of the NTP’s testing program; however, in recent years, the NTP has increased its focus on nonneoplastic lesions, many of which have been linked to occupational or environmental exposures. Diagnosing nonneoplastic lesions in toxicity studies presents a challenge in that there can be variation in terminology and diagnostic strategy. With nonneoplastic lesions, there are often several related lesions present concurrently, such as inflammation, necrosis or degeneration, fibrosis, and regeneration. Some pathologists record each lesion individually, while others record the predominant lesion, or the primary process, and describe other lesions or features in the pathology narrative. Additionally, it can be difficult to determine which is the primary lesion or which is the primary process. Also, the terminology used by different pathologists can vary based on training, experience, and personal opinion. In an effort to standardize the nomenclature and diagnostic strategy for NTP studies, the NTP created the Nonneoplastic Lesion Atlas (NNLA). The goal of the NNLA is to create a more consistent database of nonneoplastic lesions, which would allow comparison across studies, facilitate data mining, and allow for the generation of historical control data for some nonneoplastic lesions.
The NNLA is an online guide for the diagnosis and recording of nonneoplastic lesions in studies conducted by the NTP. It is organized by organ system and subdivided by tissue. Each page discusses a single lesion and provides recommendations for terminology and diagnostic strategy. The NNLA also provides references, links to related lesions, other useful information about the lesions, and thousands of zoomable photos of the lesions. The NTP has made every effort to be consistent with the terminology presented in the INHAND in Rats and Mice. The NNLA can be a valuable supplement to the INHAND documents.
Since the NNLA is an online document, it can (and will) be updated as the field of toxicologic pathology changes. It is searchable, downloadable, and available at https://ntp.niehs.nih.gov/nnl/. Though its main purpose is as a guide for toxicologic pathologists reading NTP studies, it is available free for public use around the world. It can be used by any toxicologic or pathology laboratory wanting to standardize their own database, by other scientists evaluating tissues, and by students as a training aid.
NCI Mouse Models of Human Cancer Consortium (MMHCC Nomenclature)
The advent of genetic engineering opened up a new era in animal research. Suddenly, the cell and molecular biologist could determine the effect of genetic mutations and selected engineered mutations in living mammalian organisms. Testing your mutation in a mouse fulfilled the “modern Koch’s postulates.” Rodents were once valued in cancer research because they spontaneously developed neoplasia in specific organs. In fact, they led the way to the understanding of oncogenic viruses. However, after investigators became equipped with an endless list of genes, they wished to know whether their gene(s) caused cancer in their favorite organ and whether or not their tumors resembled the comparable human tumor.
The NCI started exploring these questions by organizing and convening a Breast Cancer Consensus Meeting in Annapolis, Maryland in 1999. The meeting included oncologists, modelers, breast pathologists, and comparative or veterinary pathologists. The pathologists were tasked with developing a taxonomy and vocabulary that could be used to compare and contrast breast cancers in human and genetically engineered mice. The pathologists responded with a recommended taxonomy and a landmark paper.27
With NCI’s organization of the MMHCC groups, under the NCI Division of Cancer Biology’s extramural grant program, each of nine organ systems was tasked with developing comparable (human and mouse) consensus pathology meetings. In preparation, the MMHCC met with the NCI vocabulary informatics experts to discuss the classification and nomenclature for each organ system. This exercise was accompanied by consensus meetings for each organ composed of committees of medical and veterinary pathologists and PhD researchers from various medical and veterinary colleges, government, and private research institutes. In some cases, the meetings were repeated with follow-up meetings. In some instances, specific subsets of issues, such as preneoplasia, were addressed.28
The strength of the resulting nomenclatures was that the similarities and differences in the anatomy, physiology, and pathology of diseases were compared and contrasted by experts and the information became generally available. While instances of similarities in tumor histopathology between the two species have been recorded, the classifications generally lacked the granularity needed to satisfy the investigators. The MMHCC classification was merged with the NCI vocabularies and no longer exists as an independent taxonomy. The results of each pathology committee group were published in refereed journals.29,30,31,32,33,34,35,36,37,38 These publications report the comparative pathology for organ-specific carcinogenesis for the purpose of developing mouse models of cancer.
Computable Terminology: Development and Implementation of the Mouse Pathology Ontology (MPATH) for Use in Mouse Research
MPATH is an online structured vocabulary of mutant and transgenic mouse pathological lesions and processes (http://bioportal.bioontology.org/ontologies/MPATH). The historical motivation for the generation of MPATH was a response to the initiation of a database project, funded by the European Commission, to produce a definitive image resource for rodent pathology called Pathbase (http://www.pathbase.net.39 The community tasked to develop this resource was drawn from experts in the fields of rodent toxicopathology and human anatomic pathology. This grouping expanded considerably in number and specialty as the ontology grew.40 When MPATH was developed (1999–2004), ontologies were being created and implemented for the Mouse Genome Informatics and related databases covering the areas of gene function, phenotype,41 and anatomy.42 It became clear in these and other areas that standard terminologies, computable using biosemantic techniques,43,44,45 were highly desirable. The ontology that emerged from these efforts, MPATH, was manually created by pathologists over a period of a decade from 1999 to 2010 with the help of colleagues from across Europe, North America, and Japan.
MPATH was recently adopted for the computational capture of gross and microscopic anatomic pathology from the primary phenotyping program of the European Mouse Disease Clinic project46 and subsequently the International Mouse Phenotyping Consortium’s (IMPC) globally coordinated mutant strain production and phenotyping program,and implemented as the terminology in the Jackson Laboratory’s Nathan Shock Aging Institute large scale mouse lifespan study.47,48,49 For both projects, further development of the ontology was undertaken with changes in the structure of several areas and a large increase in the number of terms included, together with an expansion of the textual class definitions.
MPATH is constructed according to ontological “good practice” rules and is consistent with the Open Biological and Biomedical Ontology (OBO) Foundry principles.50 One criterion adopted from the OBO Foundry is the separation of physical pathological entities and pathological processes. Such a separation also facilitates automated reasoning across the ontology and integration and interoperability with OBO Foundry ontologies. MPATH therefore consists of two major branches, one containing pathological processes and another containing pathological structures. Each branch is itself constructed as a taxonomy of classes linked by logical axioms, or relations.51 The majority of axioms in MPATH are subclass (or “is-a”) relations, asserting that a given class is a subclass of a parental class, for example, choriocarcinoma “is-a” carcinoma. These axioms allow computational work to be done with the data, bridging different groups and expanding how searches are done. The hierarchical organization of MPATH will be familiar to anyone with experience of taxonomies, and the branches within are derived from traditional histopathological classifications of lesions and processes. MPATH does not contain classes representing a “disease,” which often include many lesions and have distinct etiological origins; this type of entity is more usefully captured with a disease ontology such as the Human Disease Ontology (DO) (52,53http://www.diseaseontology.org). There are strong arguments, mainly from experience in toxicologic pathology, that a descriptive (anatomic) coding rather than diagnostic is the most useful way to code and analyze pathology-based observations and in fact MPATH can be used in the computational definitions of higher order disease classes from other ontologies such as the DO.
MPATH currently has 888 classes in a hierarchy nine layers deep that may be obtained from its repository (https://raw.githubusercontent.com/PaulNSchofield/mpath/master/mpath.obo). Currently, over 90% of the classes have textual definitions. The ontology was specifically designed for use by trained histopathologists. However, MPATH has the additional advantage to permit computation and is amenable to machine presentation in data capture software.54
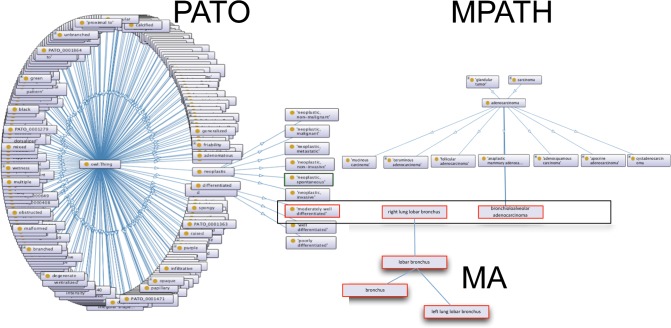
Within a single hierarchy it is not feasible to capture, in a precomposed way, a class for every type of lesion of every subtype and stage for every tissue in which it occurs. This would give rise to ontology “bloat,” and it becomes very difficult to handle for humans and computers alike. To solve this problem, we have included some of the more common precomposed classes in MPATH; for example, the names of many neoplasms contain anatomic information such as brochioloalveolar adenocarcinoma. For cases where such a precomposed term is not available, users are able to use a postcomposition approach. In the postcomposition of ontology classes different elements of the description are taken from different ontologies and used to create a formal computational statement to describe an observation. In the case of MPATH, classes may be combined with those from the mouse adult anatomy ontology (MA; https://bioportal.bioontology.org/ontologies/MA) to describe the location of the lesion; they may then be further qualified with one or more classes that characterize qualities, from the Phenotype and Trait Ontology (PATO; http://agroportal.lirmm.fr/ontologies/PATO).55 PATO is an ontology of qualities that qualify or provide formal attributes to an entity or a process, such as color, texture, or more complex qualities such as malignancy.56 The PATO subset derived for use for histopathology can be found as a “slim” in the main PATO file available on from the code repository (https://raw.githubusercontent.com/pato-ontology/pato/master/pato.obo). An example of a postcomposed term using PATO, MA, and MPATH is shown in (Figure 4).
Figure 4.
Schematic diagram describing post-composition strategy for lesions. Classes are taken from PATO, MPATH, and MA and combined to form a formal statement describing the lesion and its location. A similar process may be used for gross pathology as well; PATO contains appropriate macroscopic qualifiers for this purpose such as color, texture, size, and shape.
The computational advantages of using ontologies for terminological coding are very significant. At the simplest level, the subclass relations provided by the hierarchy allow for query expansion and for coding of a less specific parent term when there is some doubt as to which term is appropriate. The use of standardized terms—the class labels—allows the ontology to be used for text mining57 and greatly facilitates the process of coding. However, the more important advantages lie in the ability to classify, combine, and split lesions for analysis. When a large experiment is coded using MPATH (and other ontologies such as MA), it becomes possible to quantify the occurrence of specific cancers, as well as all cancers, and cancer types automatically without having to recode or manually recalculate the primary coding. Similarly it is possible to compute overrepresentation of particular lesions,58 types of lesions, or anatomical location of lesions in one group of animals versus another. We can use MPATH to precisely calculate the similarity in disease profiles between two animals or groups of animals using semantic similarity measures. Furthermore, it becomes possible to combine and semantically integrate different datasets that use MPATH for coding pathology even if the investigators worked at different levels of granularity or different geographic locations.
Experience of coding a study using MPATH and MA has been very positive. In a large-scale aging study conducted on 28 inbred strains, the type and diversity of spontaneous diseases that aging mice develop were captured using the Mouse Disease Information System system.47,48 In addition to MA and MPATH, the terminologies in Mouse Disease Information System were designed to be extended to take user-defined diagnostic terms, such as pseudoxanthoma elasticans, to allow targeted searches to be done on specific disease entities.59,54,60 A total of 20,885 different diagnoses were made by the same pathologist from reading approximately 50,000 slides from 2000 individual mice, with an average of 12 diagnoses per mouse in the study. These data have already been successfully utilized for a series of genome-wide association and other studies.61,62,63 Work in progress is generating a comprehensive quantitative survey of disease frequency across the lifespan of these strains.
Standardized Histopathology Terminology Implemented by the IMPC
The IMPC was established in 2011 as a global consortium of large-scale mouse production and phenotyping centers.64 It consists of 19 research institutions and 5 national funders from 11 countries (http://www.mousephenotype.org/data/documentation/aboutImpc#howdoesimpcwork). The Consortium’s 10-year goal is to generate a “knockout” mutant for every protein coding gene in the mouse genome in an effort to characterize the phenotype(s) that each gene confers. All mutant strains as live mice (if available) or cryopreserved sperm and phenotype data are freely available to the public, including summary data for a cohort compared with multiple wildtype controls. To overcome any potential issue of publication bias, the IMPC’s phenotype data includes all negative results as well as positive findings, and an automated statistical analysis tool65 is used to ensure the validity of the post quality-controlled data made available to the research community. The IMPC’s web portal (http://www.mousephenotype.org) provides a unified single point of access to the production and phenotyping data and enables researchers to formulate hypotheses for biomedical and translational research as well as purpose-driven preclinical studies. In the past 5 years, data for more than 4000 genes have been captured by 10 IMPC centers around the world.
The IMPC’s standardized phenotyping pipeline has been carefully designed, validated, and implemented at each participating center. An International Mouse Phenotyping Resource of Standardised Screens protocol including procedure, data type description, and metadata are available for every test through the IMPC’s portal (http://www.impc.org/impress). Cohorts of at least seven female and seven male adult mutants from each strain enter the pipeline at 4 weeks of age. Then a sequential set of clinical phenotyping tests to assay all major adult organ systems and most areas of major human disease is performed to identify abnormal phenotypes of functional, biological, or disease relevance. The majority of IMPC tests are mandatory in the pipeline; however, several optional tests have been standardized to use by individual centers where specialized equipment and expertise is available. At 16 weeks of age, a standardized panel of terminal tests, including an optional histopathology test, is done to complete the pipeline (66; https://www.mousephenotype.org/impress/protocol/276/7).
Similar to all of the clinical and terminal tests that the IMPC phenotyping pipeline uses, the histopathology test, and the data it generates, must be high-throughput, robust, and standardized to facilitate reliable and reproducible downstream analysis by the global research community.67,68 Histopathology has always played a pivotal role in hypothesis-driven studies, purpose-driven translational investigations, and preclinical assessment of mouse models, providing important insight into the morphological (structural) consequences and mechanisms of gene function or dysfunction and therapeutic effect and safety. In the context of the IMPC’s high-throughput phenotyping pipeline, the histopathology test’s objectives using a panel of tissues (25 required for female mice; 26 required for male mice) collected from 2 female and 2 male mutant mice from each strain are to:
Identify abnormalities (“lesions”) correlated with clinical phenotype (e.g., clinical ataxia, cerebellar histopathology).
Identify significant abnormalities not directly correlated with clinical phenotype, often the result of gene pleiotropy whereby a single mutate gene causes multi-system changes (e.g., clinical ataxia, liver histopathology).
Identify significant abnormalities that are novel findings in strains with no identified clinical phenotype.
Classify any of these findings as “not significant” (interpreted by the histopathologist to be background-related or incidental) or “significant” (interpreted by the histopathologist to not be a background-related finding; e.g., low-incidence retinal dysplasia) or incidental finding (e.g., focal hepatic microgranuloma).
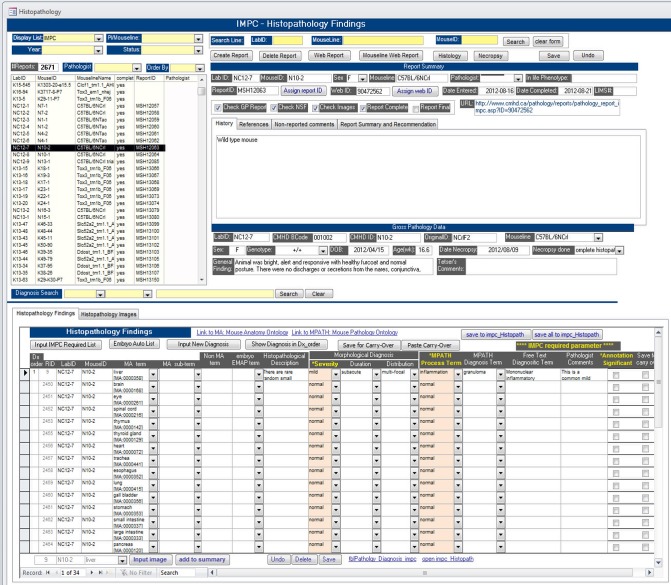
To achieve the same objectives for histopathology data of standardization, quality-control, and semantic standards (i.e., machine-search ability by web portal users) required by the IMPC, the Consortium’s Morphology Working Group has developed a histopathology ontology that is a compilation of three well-established ontologies described in other sections of this paper; MA, PATO, and MPATH. The data capture, annotation, and storage system developed at The Centre for Phenogenomics (TCP; http://www.phenogenomics.ca) in Toronto is provided as an example of integration, implementation, and use of the IMPC histopathology ontology within an IMPC center. Briefly, TCP histopathology data acquisition work flow supported by the system’s user interface (Figure 5) and integration of MA, PATO, and MPATH ontologies within the database typically includes several steps:
Select a mouseID on the worklist for review.
The required tissue list is auto-populated.
Each individual tissue row includes the MA term name and term ID (e.g., liver [MA:00,003581]) and entry fields with dropdown lists to select PATO descriptors (Severity, Duration, Distribution), MPATH Process Terms (e.g., inflammation [MPATH:212]), and MPATH Diagnosis (entity) Terms (e.g., granuloma [MPATH:847]).
Add Free Text Diagnostic Term if necessary, add Pathologist Comments if appropriate, and toggle the Significance Score check-box (i.e., unchecked equals Not Significant, checked equals Significant).
Figure 5.
Screen shot of TCP’s histopathology data entry user interface used to annotate IMPC strains.
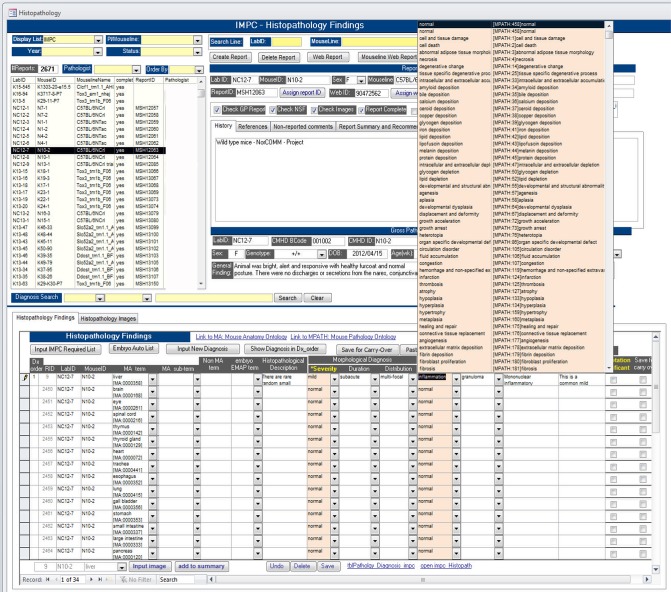
Note that certain Term fields are auto-populated for efficient workflow (if no findings to annotate, no entry effort required) and to comply with the minimum required dataset for successful upload to the IMPC Data Center. Additional functionality includes parsed dropdown lists (e.g., liver row’s MPATH Process Terms and MPATH Diagnostic Terms only provide term options applicable to liver pathology) (Figure 6).
Figure 6.
Screen shot of TCP’s histopathology data entry user interface. Note in the example for annotating a liver section that the dropdown selection list for MPATH Process Terms provides only terms applicable to liver histopathology.
The IMPC histopathology ontology described here is fully integrated in the IMPC’s central database. Therefore, any center in the consortium that is doing histopathology at International Mouse Phenotyping Resource of Standardised Screens standards can upload data compliant with the standardization, quality-control, and machine-readable requirements of the IMPC. Using MA, PATO, and MPATH, each well-established, publically available, and actively curated extant ontology was, and is, essential to this process. Data display at the portal is in active development.
Acknowledgments
For INHAND, in addition to the leaders of supporting societies, we appreciate the contributions from Drs. Wolfgang Kaufmann, Ian Pyrah, Julia Baker, Peter Mann, Alys Bradley, Matthew Jacobsen, Susanne Rittinghausen, Thomas Nolte, Christine Ruehl-Fehler, Rupert Kellner, John L. Vahle, Dawn G. Goodman, Emily Meseck, Ronald Herbert, Shim-mo Hayashi, Takanori Harada, and Katsuhiko Yoshizawa.
C.M. acknowledges the contributions from all members of the IMPC’s Morphology Work Group, appreciates and recognizes the effort and feedback from the entire IMPC, and especially thanks the Pathology Core Team and Informatics Team at The Centre for Phenogenomics for their database development and support. This work was supported by Government of Canada through Genome Canada and Ontario Genomics (OGI-051) (C.M.). P.N.S. acknowledges the support of the Commission of the European Community for Pathbase and MPATH QLRI-1999-CT-0320 and the continued contributions over its technical development from Michael Gruenberger and all the pathologists who have generously given their expertise and time over the years. R.H. acknowledges the support of King Abdullah University of Science and Technology (KAUST), and J.P.S. acknowledges support from the Ellison Medical Foundation, and the National Institutes of Health (CA34196, CA089713, and AG038070-05). For S.E. and M.C., this research was supported (in part) by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS).
References
- 1. Turusov VS, ed. Pathology of Tumours in Laboratory Animals. I. Part I—Tumours of the Rat. Lyon: IARC; 1973. [Google Scholar]
- 2. Turusov VS, ed. Pathology of Tumours in Laboratory Animals. II. Tumours of the Mouse. Lyon: IARC; 1980; p 682. [Google Scholar]
- 3. Turusov VS, ed. Pathology of Tumours in Laboratory Animals. III. The Hamster. Lyon: IARC; 1983; p 472. [Google Scholar]
- 4. Turusov VS, Mohr U, eds. Pathology of Tumours in Laboratory Animals. I. Tumours of the Rat, 2nd ed Lyon: IARC; 1990; p 766. [Google Scholar]
- 5. Turusov VS, Mohr U, eds. Pathology of Tumours in Laboratory Animals. I. Tumours of the Rat, 2nd ed Lyon: IARC; 1994; p 776. [Google Scholar]
- 6. Mohr U, ed. International Classification of Rodent Tumours. Part I. The Rat. IARC Scientific Publications; 1997; No. 122. [PubMed] [Google Scholar]
- 7. Mohr U, ed. International Classification of Rodent Tumours: The Mouse. Heidelberg: Springer-Verlag; 2001; p 474. [Google Scholar]
- 8. Jones TC, Mohr U, Hunt RD. Monographs on Pathology of Laboratory Animals. Berlin: Springer-Verlag; (volumes by organ system or species). 1983. –1996. [Google Scholar]
- 9. Boorman GA, Eustis SL, Elwell MR, Montgomery CA, MacKenzie WF. San Diego, CA: Academic Press, Inc; 1990; p 1–580. [Google Scholar]
- 10. Maronpot RR, Boorman GA, Baul BW. Pathology of the Mouse. Vienna, IL: Cache River Press; 1999; p 1–699. [Google Scholar]
- 11. Renne R, Brix A, Harkema J, Herbert R, Kittel B, Lewis D, March T, Nagano K, Pino M, Rittinghausen S et al. Proliferative and nonproliferative lesions of the rat and mouse respiratory tract. Toxicol Pathol 2009;37(7 Suppl):5S–73S. [DOI] [PubMed] [Google Scholar]
- 12. Thoolen B, Maronpot RR, Harada T, Nyska A, Rousseaux C, Nolte T, Malarkey DE, Kaufmann W, Küttler K, Deschl U et al. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol 2010;38(7 Suppl):5S–81S. [DOI] [PubMed] [Google Scholar]
- 13. Frazier KS, Seely JC, Hard GC, Betton G, Burnett R, Nakatsuji S, Nishikawa A, Durchfeld-Meyer B, Bube A. Proliferative and nonproliferative lesions of the rat and mouse urinary system. Toxicol Pathol 2012;40(4 Suppl):14S–86S. [DOI] [PubMed] [Google Scholar]
- 14. Kaufmann W, Bolon B, Bradley A, Butt M, Czasch S, Garman RH, George C, Gröters S, Krinke G, Little P et al. Proliferative and nonproliferative lesions of the rat and mouse central and peripheral nervous systems. Toxicol Pathol 2012;40(4 Suppl):87S–157S. [DOI] [PubMed] [Google Scholar]
- 15. Rudmann D, Cardiff R, Chouinard L, Goodman D, Küttler K, Marxfeld H, Molinolo A, Treumann S, Yoshizawa K, INHAND Mammary Zs, Preputial, and Clitoral Gland Organ Working Group . Proliferative and nonproliferative lesions of the rat and mouse mammary, Zymbal’s, preputial, and clitoral glands. Toxicol Pathol 2012;40(6 Suppl):7S–39S. [DOI] [PubMed] [Google Scholar]
- 16. Creasy D, Bube A, de Rijk E, et al. Proliferative and nonproliferative lesions of the rat and mouse male reproductive system. Toxicol Pathol 2012;40(6 Suppl):40S–121S. [DOI] [PubMed] [Google Scholar]
- 17. Greaves P, Chouinard L, Ernst H, Mecklenburg L, Pruimboom-Brees IM, Rinke M, Rittinghausen S, Thibault S, Von Erichsen J, Yoshida T. Proliferative and non-proliferative lesions of the rat and mouse soft tissue, skeletal muscle and mesothelium. J Toxicol Pathol 2013;26(3 Suppl):1S–26S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mecklenburg L, Kusewitt D, Kolly C, Treumann S, Adams ET, Diegel K, Yamate J, Kaufmann W, Müller S, Danilenko D et al. Proliferative and non-proliferative lesions of the rat and mouse integument. J Toxicol Pathol 2013;26(3 Suppl):27S–57S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dixon D, Alison R, Bach U, et al. Nonproliferative and proliferative lesions of the rat and mouse female reproductive system. J Toxicol Pathol 2014;27(3–4 Suppl):1S–107S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nolte T, Brander-Weber P, Dangler C, Deschl U, Elwell MR, Greaves P, Hailey R, Leach MW, Pandiri AR, Rogers A et al. Nonproliferative and proliferative lesions of the gastrointestinal tract, pancreas and salivary glands of the rat and mouse. J Toxicol Pathol 2016;29(1 Suppl):1S–125S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berridge BR, Mowat V, Nagai H, et al. Non-proliferative and proliferative lesions of the cardiovascular system of the rat and mouse. J Toxicol Pathol 2016;29(3 Suppl):1S–47S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fossey S, Vahle J, Long P, Schelling S, Ernst H, Boyce RW, Jolette J, Bolon B, Bendele A, Rinke M et al. Nonproliferative and proliferative lesions of the rat and mouse skeletal tissues (bones, joints, and teeth). J Toxicol Pathol 2016;29(3 Suppl):49S–103S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elmore SA, Dixon D, Hailey JR, et al. Recommendations from the INHAND Apoptosis/Necrosis Working Group. Toxicol Pathol 2016;44(2):173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mann PC, Vahle J, Keenan CM, Baker JF, Bradley AE, Goodman DG, Harada T, Herbert R, Kaufmann W, Kellner R et al. International harmonization of toxicologic pathology nomenclature: An overview and review of basic principles. Toxicol Pathol 2012;40(4 Suppl):7S–13S. [DOI] [PubMed] [Google Scholar]
- 25. Vahle J, Bradley A, Harada T, Herbert R, Kaufmann W, Kellner R, Mann P, Pyrah I, Rittinghausen S, Tanaka T. The international nomenclature project: An update. Toxicol Pathol 2009;37(5):694–7. [DOI] [PubMed] [Google Scholar]
- 26. Keenan CM, Goodman DG. Regulatory Forum commentary: Through the looking glass—SENDing the pathology data we have INHAND. Toxicol Pathol 2014;42(5):807–10. [DOI] [PubMed] [Google Scholar]
- 27. Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, et al. The mammary pathology of genetically engineered mice: The consensus report and recommendations from the Annapolis meeting. Oncogene 2000;19(8):968–88. [DOI] [PubMed] [Google Scholar]
- 28. Cardiff RD, Anver MR, Boivin GP, Bosenberg MW, Maronpot RR, Molinolo AA, et al. Precancer in mice: Animal models used to understand, prevent, and treat human precancers. Toxicol Pathol 2006;34(6):699–707. [DOI] [PubMed] [Google Scholar]
- 29. Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, et al. Pathology of mouse models of intestinal cancer: Consensus report and recommendations. Gastroenterology 2003;124(3):762–77. [DOI] [PubMed] [Google Scholar]
- 30. Hruban RH, Adsay NV, Albores-Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: Consensus report and recommendations. Cancer Res 2006;66(1):95–106. [DOI] [PubMed] [Google Scholar]
- 31. Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: The consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res 2013;73(9):2718–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood 2002;100(1):238–45. [DOI] [PubMed] [Google Scholar]
- 33. Morse HC 3rd, Anver MR, Fredrickson TN, Haines DC, Harris AW, Harris NL, et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 2002;100(1):246–58. [DOI] [PubMed] [Google Scholar]
- 34. Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, et al. Classification of proliferative pulmonary lesions of the mouse: Recommendations of the mouse models of human cancers consortium. Cancer Res 2004;64(7):2307–16. [DOI] [PubMed] [Google Scholar]
- 35. Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, et al. Prostate pathology of genetically engineered mice: Definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res 2004;64(6):2270–305. [DOI] [PubMed] [Google Scholar]
- 36. Stemmer-Rachamimov AO, Louis DN, Nielsen GP, Antonescu CR, Borowsky AD, Bronson RT, et al. Comparative pathology of nerve sheath tumors in mouse models and humans. Cancer Res 2004;64(10):3718–24. [DOI] [PubMed] [Google Scholar]
- 37. Washington MK, Powell AE, Sullivan R, Sundberg JP, Wright N, Coffey RJ, Dove WF. Pathology of rodent models of intestinal cancer: Progress report and recommendations. Gastroenterology 2013;144(4):705–17. 10.1053/j.gastro.2013.01.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weiss WA, Israel M, Cobbs C, Holland E, James CD, Louis DN, et al. Neuropathology of genetically engineered mice: Consensus report and recommendations from an international forum. Oncogene 2002;21(49):7453–63. [DOI] [PubMed] [Google Scholar]
- 39. Schofield PN, Bard JB, Booth C, Boniver J, Covelli V, Delvenne P, Ellender M, Engstrom W, Goessner W, Gruenberger M et al. Pathbase: A database of mutant mouse pathology. Nucleic Acids Res 2004;32(Database issue):D512–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schofield PN, Gruenberger M, Sundberg JP. Pathbase and the MPATH ontology. Community resources for mouse histopathology. Vet Pathol 2010;47(6):1016–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith CL, Goldsmith CA, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol 2005;6(1):R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ringwald M, Eppig JT, Begley DA, Corradi JP, McCright IJ, Hayamizu TF, Hill DP, Kadin JA, Richardson JE. The Mouse Gene Expression Database (GXD). Nucleic Acids Res 2001;29(1):98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bodenreider O, Smith B, Burgun A. The ontology-epistemology divide: A case study in medical terminology. Form Ontol Inf Syst 2004;2004:185–195. [PMC free article] [PubMed] [Google Scholar]
- 44. Lord PW, Stevens RD, Brass A, Goble CA. Semantic similarity measures as tools for exploring the gene ontology. Pac Symp Biocomput 2003;601–12. [DOI] [PubMed] [Google Scholar]
- 45. Smith B, Williams J, Schulze-Kremer S. The ontology of the gene ontology. AMIA Annu Symp Proc 2003;2003:609–13. [PMC free article] [PubMed] [Google Scholar]
- 46. Ayadi A, Birling MC, Bottomley J, Bussell J, Fuchs H, Fray M, Gailus-Durner V, Greenaway S, Houghton R, Karp N et al. Mouse large-scale phenotyping initiatives: Overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome 2012;23(9–10):600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sundberg JP, Berndt A, Sundberg BA, Silva K, Kennedy V, Bronson R, Yuan R, Paigen B, Harrison D, Schofield P. The mouse as a model for understanding chronic diseases of aging: The histopathologic basis of aging in inbred mice. Pathobiology of Aging & Age-related Diseases 2011;1:71719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sundberg JP, Berndt A, Sundberg BA, Silva KA, Kennedy V, Smith RS, Cooper TK, Schofield PN. Approaches to investigating complex genetic traits in a large-scale inbred mouse aging study. Vet Pathol 2016;53(2):456–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yuan R, Tsaih S-T, Petkova SB, deEvsikova CM, Xing S, Marion MA, Bogue MA, Mills KD, Peters LL, Bult CJ et al. Aging in inbred strains of mice: Study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell 2009;8(3):277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ et al. The OBO Foundry: Coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 2007;25(11):1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith B, Ceusters W, Klagges B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in biomedical ontologies. Genome Biol 2005;6(5):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kibbe WA, Arze C, Felix V, Mitraka E, Bolton E, Fu G, Mungall CJ, Binder JX, Malone J, Vasant D et al. Disease ontology 2015 update: An expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res 2015;43(Database issue):D1071–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bello SM, Shimoyama M, Mitraka E, Laulederkind SJF, Smith CL, Eppig JT, Schriml LM. Disease ontology: Improving and unifying disease annotations across species. Disease Models and Mechanisms 2018; 1–9. (http://dmm.biologists.org/content/11/3/dmm032839). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sundberg B, Schofield P, Gruenberger M, Sundberg J. A data capture tool for mouse pathology phenotyping. Vet Pathol 2009;46(6) 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gkoutos GV, Schofield PN, Hoehndorf R. The anatomy of phenotype ontologies: Principles, properties and applications. Brief Bioinform. 2018;19(5) 1008–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gkoutos GV, Green EC, Mallon AM, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol 2005;6(1):R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoehndorf R, Schofield PN, Gkoutos GV. Analysis of the human diseasome using phenotype similarity between common, genetic, and infectious diseases. Sci Rep 2015;5:10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hoehndorf R, Gkoutos GV, Schofield PN. Datamining with ontologies. Methods Mol Biol 2016;1415:385–97. [DOI] [PubMed] [Google Scholar]
- 59. Sundberg JP, Sundberg BA, Schofield P. Integrating mouse anatomy and pathology ontologies into a phenotyping database: Tools for data capture and training. Mammalian Genome 2008;19(6):413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sundberg JP, Sundberg BA, Gruenberger M, Schofield PN. A data capture tool for mouse phenotyping: Obtaining a ‘virtual second opinion’ on skin and hair disease. Exp Dermatol 2010;19(6):595. [Google Scholar]
- 61. Berndt A, Sundberg BA, Silva KA, Kennedy VE, Richardson MA, Li Q, Bronson RT, Uitto J, Sundberg JP. Phenotypic characterization of the KK/HlJ inbred mouse strain. Vet Pathol 2014;51(4):846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berndt A, Ackert-Bicknell C, Silva KA, Kennedy VE, Sundberg BA, Cates JM, Schofield PN, Sundberg JP. Genetic determinants of fibro-osseous lesions in aged inbred mice. Exp Mol Pathol 2016;100(1):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li Q, Berndt A, Sundberg BA, Silva KA, Kennedy VE, Cario CL, Richardson MA, Chase TH, Schofield PN, Uitto J et al. Mouse genome-wide association study identifies polymorphisms on chromosomes 4, 11, and 15 for age-related cardiac fibrosis. Mamm Genome 2016;27(5–6):179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: The International Mouse Phenotyping Consortium. Dis Model Mech 2012;5(3):289–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kurbatova N, Mason JC, Morgan H, Meehan TF, Karp NA. PhenStat: A tool kit for standardized analysis of high throughput phenotypic Data. PLoS One 2015;10(7):e0131274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Adissu HA, Estabel J, Sunter D, Tuck E, Hooks Y, Carragher DM, Clarke K, Karp NA, Sanger Mouse Genetics Project, Newbigging S et al. Histopathology reveals correlative and unique phenotypes in a high throughput mouse phenotyping screen. Dis Model Mech 2014;7(5):515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mallon AM, Iyer V, Melvin D, Morgan H, Parkinson H, Brown SD, Flicek P, Skarnes WC. Accessing data from the International Mouse Phenotyping Consortium: State of the art and future plans. Mamm Genome 2012;23(9–10):641–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ring N, Meehan TF, Blake A, Brown J, Chen CK, Conte N, Di Fenza A, Fiegel T, Horner N, Jacobsen JO et al. A mouse informatics platform for phenotypic and translational discovery. Mamm Genome 2015;26(9–10):413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]