Abstract
Objective
MicroRNAs (miRNAs) play an integral role in maintaining beta cell function and identity. Deciphering their targets and precise role, however, remains challenging. In this study, we aimed to identify miRNAs and their downstream targets involved in the regeneration of islet beta cells following partial pancreatectomy in mice.
Methods
RNA from laser capture microdissected (LCM) islets of partially pancreatectomized and sham-operated mice were profiled with microarrays to identify putative miRNAs implicated in beta cell regeneration. Altered expression of the selected miRNAs, including miR-132, was verified by RT-PCR. Potential targets of miR-132 were selected through bioinformatic data mining. Predicted miR-132 targets were validated for their changed RNA, protein expression levels, and signaling upon miR-132 knockdown and/or overexpression in mouse MIN6 and human EndoC-βH1 insulinoma cells. The ability of miR-132 to foster beta cell proliferation in vivo was further assessed in pancreatectomized miR-132−/− and control mice.
Results
Partial pancreatectomy significantly increased the number of BrdU+/insulin+ islet cells. Microarray profiling revealed that 14 miRNAs, including miR-132 and -141, were significantly upregulated in the LCM islets of the partially pancreatectomized mice compared to the LCM islets of the control mice. In the same comparison, miR-760 was the only downregulated miRNA. The changed expression of these miRNAs in the islets of the partially pancreatectomized mice was confirmed by RT-PCR only in the case of miR-132 and -141. Based on previous knowledge of its function, we focused our attention on miR-132. Downregulation of miR-132 reduced the proliferation of MIN6 cells while enhancing the levels of pro-apoptotic cleaved caspase-9. The opposite was observed in miR-132 overexpressing MIN6 cells. Microarray profiling, RT-PCR, and immunoblotting of the latter cells demonstrated their downregulated expression of Pten with concomitant increased levels of pro-proliferative factors phospho-Akt and phospho-Creb and inactivation of pro-apoptotic Foxo3a via its phosphorylation. Downregulation of Pten was further confirmed in the LCM islets of pancreatectomized mice compared to the sham-operated mice. Moreover, overexpression of miR-132 correlated with increased proliferation of EndoC-βH1 cells. The regeneration of beta cells following partial pancreatectomy was lower in the miR-132/212−/− mice than the control littermates.
Conclusions
This study provides compelling evidence about the critical role of miR-132 for the regeneration of mouse islet beta cells through the downregulation of its target Pten. Hence, the miR-132/Pten/Akt/Foxo3 signaling pathway may represent a suitable target to enhance beta cell mass.
Keywords: miR-132, β cell regeneration, Apoptosis, Pten/Akt/Foxo3a, Pancreatectomy
Abbreviations: miRNA, microRNA; miR-132, microRNA 132; Pten, phosphatase and tensin homolog; pHH3, phosphohistone H3; Akt, protein kinase B; Foxo3a, Forkhead box O3a; RT-PCR, reverse transcription polymerase chain reaction; T2D, type 2 diabetes; BrdU, bromodeoxyuridine; LCM, laser capture microscopy; IPA, Ingenuity Pathway Analysis; Pi3k, phosphatidylinositol-4,5-bisphosphate 3-kinase; Ras, rat sarcoma; Raf, rapidly accelerated fibrosarcoma; Mapk1, mitogen-activated protein kinase 1; Sos1, Son of Sevenless homolog 1; Gnb1, G protein subunit beta 1; Nras, neuroblastoma RAS viral oncogene homolog; Pik3r1, phosphoinositide-3 kinase regulatory subunit 1; Gnb, G protein subunit beta; Fgfr3, fibroblast growth factor receptor 3; Creb, cAMP response element-binding protein; GSIS, glucose-stimulated insulin secretion
Highlights
-
•
miR-132 is induced in mouse islets upon partial pancreatectomy.
-
•
miR-132 promotes regeneration of β-cells in vivo following partial pancreatectomy.
-
•
miR-132 fosters in vitro proliferation/survival through Pten/Akt/Foxo3 signaling.
-
•
Downstream targets of miR-132 were identified in pancreatic β-cells.
-
•
miR-132−/− mice have impaired β-cell proliferation.
1. Introduction
MiRNAs belong to the class of short non-coding RNAs that regulate gene expression by annealing to 3′-untranslated region sequences in target mRNAs and inducing their post-transcriptional repression [1]. The functional importance of miRNAs has been extensively investigated in recent years and their altered expression has been implicated in a wide range of diseases, including cancer [2], cardiovascular disease [3,4], and diabetes [5,6]. Increasing evidence indicates that miRNA-dependent post-transcriptional gene silencing is implicated in regulating beta cell function and can potentially be linked to the development of diabetes [7]. In pancreatic beta cells, the changed expression of miRNAs correlates with profound impairment of insulin secretion [8]. RNA sequencing of human islets detected 346 miRNAs, including 40 that were enriched compared to other tissues [9]. miR-375 is the most highly expressed miRNA in human and mouse pancreatic islets. Its downregulation inhibits pancreatic islet development in Xenopus laevis [10], while its global inactivation in mice leads to decreased beta cell mass and ultimately diabetes [11,12]. MiR-132 also plays a key role in beta cell function. Its expression is altered in different mouse models of type 2 diabetes (T2D) [[13], [14], [15]], and its overexpression is correlated with improved glucose-stimulated insulin release from dissociated rat islet cells [15] and enhanced beta cell proliferation and survival [[14], [15], [16]]. In PC12 cells, another endocrine cell model, miR-132 controls cell survival via direct regulation of Pten, Foxo3a, and p300 signaling [17]. However, in pancreatic islets, the functional relevance of miR-132 in vivo and its downstream targets remain unknown and its involvement in beta cell regeneration in vivo has not been investigated. To identify the major miRNAs and their downstream targets involved in beta cell proliferation, we analyzed the profile of miRNAs differentially expressed in laser capture microdissected (LCM) islets of partially pancreatectomized mice compared to LCM islets of sham-operated mice.
2. Methods
2.1. Mice
The miR-132/212−/− mice were a gift from K. Chowdhury and was generated by homologous recombination of the genomic region encoding pre-miR132 and pre-miR-212 [18]. The miR-132/212−/− mice used in this study had been backcrossed into the C57Bl/6N background for at least seven generations. All animal protocols were approved by the institutional animal care and use committee at the Faculty of Medicine of TU Dresden and all experiments were conducted in accordance with relevant guidelines and regulations.
2.2. Mouse partial pancreatectomy
Thirteen to 19 week-old male C57Bl/6N mice with body weights of 28–34 g were subjected to a 75% partial pancreatectomy (3 mice) or sham operated (4 mice) as described [19], except for anesthesia, which was administered using a small rodents' anesthesia unit (Harvard Apparatus Ltd., Holliston, MA, USA) for mask inhalation of isoflurane (Baxter Deutschland GmbH, Unterschleiβheim, Germany) at a concentration of 4.5–5% for induction and 2–2.5% for maintenance of anesthesia with an airflow rate of 200 ml/min. For perioperative analgesia, buprenorphine (0.05 mg/kg bodyweight) was administered subcutaneously. At the end of surgery, Alzet 1007D mini-osmotic pumps (Alza, Cupertino, CA, USA) were implanted intraperitoneally to deliver 50 μg μl−1 BrdU (Sigma–Aldrich, St. Louis, MO, USA) in 50% DMSO at a rate of 0.5 μl h−1 for 7 days. Blood glucose levels were measured daily with a Glucotrend glucometer (Roche Diagnostics, Basel, Switzerland). A second set of sham or partial pancreatectomies was performed on 4 wild-type mice for RNA extraction from LCM islets isolation and validation of miRNA expression by RT-PCR. Furthermore, 12 wild-type and 12 miR-132/212−/− mice were equally divided in four groups, each consisting of 6 animals, for sham or partial pancreatectomy as previously described. Pancreatic tissue from these mice was collected 7 days after surgery and processed for BrdU labeling as follows.
2.3. BrdU staining of pancreatic tissue sections
The pancreatectomized and sham-operated mice were anesthetized 7 days post-surgery as described. After fixation by intracardial perfusion with 4% PFA, the pancreas was removed, fixed overnight in 10% neutral formalin, embedded in paraffin, and cut into 10-μm thin serial sections. Immunostaining was performed as described in [19].
2.4. Intraperitoneal glucose tolerance test (IpGTT)
IpGTTs were conducted two days before surgery and six days after surgery to assess differences between the wild-type and mutant mice and between the pancreatectomized and sham-operated animals. After an overnight fast of 10 h, the mice were injected with 2 g/kg body weight of 20% glucose solution. Glucose levels were measured in blood samples collected from the tail veins at 0, 15, 30, 60, 120, and 180 min after glucose injection.
2.5. Laser capture microdissection of islets after partial pancreatectomy
The mice were sacrificed seven days post-surgery. The remnant pancreas was excised for serial sectioning (40 sections/mouse) and staining with cresyl violet to locate the islets. Adjacent unstained sections were then used to the count BrdU+ cells prior to islet core excision by laser capture microdissection with a PALM MicroBeam Laser Capture System (Zeiss, Feldbach, Switzerland).
2.6. MicroRNA profiling of LCM islets with microarrays and RT-PCR
RNA was isolated from the LCM islets and 100 ng was used for the microarray analysis of differentially expressed microRNAs. The hybridization was performed using Agilent Mouse miRNA Microarray 8 × 15 K 019,119, which covered 3p of 589 distinct microRNAs targets. Expression of the selected microRNAs was further tested by RT-PCR using RNA extracted from an independent set of LCM islets from another group of C57Bl/6N pancreatectomized and sham-operated mice. cDNA obtained from the reverse transcription of LCM-isolated RNA was used as template for real-time PCR analysis using miScript primer assays in combination with a miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany).
2.7. Gene expression analysis of LCM islets using microarrays
RNA was isolated from the LCM islets of three C57Bl/6N pancreatectomized and two sham-operated mice using an RNeasy Mini Kit from Qiagen as previously described. The RNA quality was assessed using an RNA 6000 Pico Kit from Agilent. A total of 100 ng RNA per condition were used for hybridization using the Affymetrix GeneChip Mouse Genome 430 2.0 Array according to the manufacturer's instructions. The gene expression signals were normalized for the β-actin levels and analyzed with ANOVA for statistically significant differences.
2.8. Cell culture
Mouse MIN6 cells, a gift from Dr. Jun-ichi Miyazaki (Osaka University, Osaka, Japan), were cultured as described [20] in 25 mmol/L glucose Dulbecco's modified Eagle's medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and supplemented with 15% fetal bovine serum (Gibco), 100 U/mL penicillin, 100 U/mL streptomycin (Sigma–Aldrich), and 70 μM β-mercaptoethanol (Sigma–Aldrich) at 37 °C in a humidified atmosphere with 5% CO2. EndoC-βH1 cells were cultured as described in [21]. HEK293T cells were cultured in 25 mmol/L glucose Dulbecco's modified Eagle's medium (Gibco) and supplemented with 10% fetal bovine serum (Gibco), 100 U/mL penicillin, 100 U/mL streptomycin (Sigma–Aldrich), and 0.1 mM non-essential amino acids (Gibco) at 37 °C in a humidified atmosphere with 5% CO2.
2.9. Cloning of miR-132 in pacAd5 shuttle vector
An adenovirus vector for miR-132 overexpression was generated using the RAPAd miRNA Adenoviral Expression System (Cell Biolabs, San Diego, CA, USA). Following the kit's instructions, the mmu-miR-132 precursor sequence, obtained from https://www.miRBase.org (GGTAACAGTCTACAGCCATGGTCGCCC), was PCR amplified from mouse genomic DNA including a ∼100 bp flanking region on each side (forward: 5′-TCGAGGATCCTCCCTGTGGGTTGCGGTGGG-3′; reverse: 5′-TCGAGCTAGCACATCGAATGTTGCGTCGCCGC-3′) and then cloned into the human β-globin Intron of the kit's pacAd5-miR-GFP-Puro vector via BamHI/NheI digestion. This human β-globin intron containing mmu-miR-132 precursor was then subcloned into the kit's pacAd5-CMV-eGFP vector via PCR amplification (forward: 5′-TGCAACCGGTGCCAGAACACAGGTACACATAT-3′; reverse:5′-TGCAACCGGTCGTGCTTTGCCAAAGTGATG-3′) and AgeI digestion to obtain a, miR-132 overexpressing shuttle vector with a CMV promoter. The empty pacAd5-CMV-eGFP vector was used to produce a control virus.
2.10. Adenovirus production in HEK293T cells
The adenoviral backbone vector pacAd5-9.2-100 from the RAPAd miRNA Adenoviral Expression System (Cell Biolabs) and both shuttle vectors pacAd5-CMV-eGFP and pacAd5-CMV-mir132-eGFP were PacI linearized and co-transfected into HEK293T cells using X-tremeGENE 9 DNA transfection reagent (Roche). Adenoviral particles collected from the medium were aliquoted and stored at −80 °C. Virus titers were determined with an Adeno-X Rapid Titer Kit (Clontech, Mountain View, CA, USA). All steps were carried out according to the manufacturer's instructions.
2.11. BrdU labeling of miR-132 silenced or overexpressing culture cells
For the silencing of the miR-132 in MIN6 cells, the following siRNA oligo was used: 5′-CGACCAUGGCUGUAGACUGUUACC-3 (Riboxx, Radebeul, Germany). A scrambled siRNA oligo was used as a control. The MIN6 cells were transfected in a 6-well plate with 100 nM miR-132 or the scrambled siRNA oligos using Dharmafect4 (Thermo Fisher Scientific) as a transfection reagent on day 1 after seeding. On day 4, after siRNA oligo removal, the cells were either harvested for Western blotting or further incubated with BrdU (Roche, Basel, Switzerland) for 8 h and then fixed with 4% paraformaldehyde (PFA). For miR-132 overexpression, the MIN6 and EndoC-βH1 cells were transduced for 16 or 4 h, respectively, with CMV-eGFP or CMV-miR132-eGFP (MOI: 6.4 × 103). On day 4, miR-132 overexpressing MIN6 cells were also incubated with BrdU for 8 h and then fixed with 4% PFA. After transduction, the EndoC-βH1 cells were incubated for 48 h in medium with DMSO (1/10,000) plus or minus 10 μM harmine (Sigma–Aldrich) and then for additional 72 h with BrdU before fixation with 4% PFA. The cells were then stained with BrdU according to the manufacturer's instructions. This protocol was also used to immunostain the EndoC-βH1 cells for phosphohistone H3 (Cell Signaling, Danvers, MA, USA).
2.12. Insulin secretion
The MIN6 cells were transfected on day 1 with Mock or pEZX-miR-132 plasmid expressing the mature form of miR-132 from a commercial vector (Gene Copoeia, Rockville, MD, USA). Two days post-transfection, the cells were incubated in growth serum-free media with or without 0.5 mM palmitate/0.5% BSA for 24 h. On day 3 post-transfection, the cells were pre-incubated for 1 h at 37 °C in buffer containing 120 mmol/l NaCl, 5 mmol/l KCl, 2 mmol/l CaCl2, 1 mmol/l MgCl2, 24 mmol/l NaHCO3, 0.1% (wt./vol.) BSA, 15 mmol/l HEPES, and pH 7.4 with 0 mM glucose (resting media). After pre-incubation, the cells were re-incubated either in resting or stimulating media (25 mM glucose) for a 1.5 h. The media were collected and the cells were harvested and lysed in acidified ethanol (75% [vol./vol.] ethanol and 0.55% [vol./vol.] HCl). The amount of intracellular and released insulin was measured using a radioimmunoassay (Merck Millipore, Billerica, MA, USA).
2.13. Caspase-3/7 assay
The MIN6 cells transduced either with CMV-eGFP or CMV-miR132-eGFP were cultured in a 12-well plate. On day 3, the cells were incubated in fresh media with or without 1 μM staurosporine for 12 h. On day 4, the cells were harvested and lysed using reagents from a Caspase-Glo 3/7 assay (Promega, Madison, WI, USA). The cleavage of non-fluorescent rhodamine 110 bis-(N-CBZ-l-aspartyl-l-glutamyl-l-valyl-aspartic acid amide) (Z-DEVD-R110) substrate and its conversion to fluorescent rhodamine 110, which was proportional to the caspase-3/7 activity, was quantified according to the manufacturer's instructions.
2.14. Cell extraction and immunoblotting
The MIN6 cells were harvested at 4 °C in RIPA buffer (50 mM Tris·HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor mixture; Sigma–Aldrich) for total protein extraction. Insoluble material was removed by centrifugation. Aliquots of 20 μg were separated by SDS-PAGE as previously described [18]. The source, species, and dilutions of antibodies used for immunoblotting are listed in ESM Table 1.
2.15. RNA isolation from MIN6 and EndoC-BH1 cells for microRNA measurement
The cells were harvested after washing them once in Dulbecco's PBS (Sigma–Aldrich) by scraping and pelleting them via centrifugation at 3,000×g at 4 °C for 5 min. The cell pellets were immediately stored at −80 °C. MiRNA isolation was performed with a mirVana miRNA Isolation Kit (Ambion, Foster City, CA, USA) according to the manufacturer's instructions. Both the total RNA and microRNA-enriched fractions were isolated. RNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer (PEQLAB/VWR, Darmstadt, Germany). The samples were stored at −80 °C.
2.16. TaqMan microRNA assay
Reverse transcription of 15 ng miRNA/reaction of the miRNA-enriched fraction into cDNA was performed with a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) using the miR-132-specific or the U6 snRNA-specific stem-looped RT primer of the TaqMan MicroRNA Assay hsa-miR-132 (Applied Biosystems, Cat.# 4427975, Assay# 000457; Thermo Fisher Scientific, Waltham, MA, USA) or the TaqMan Micro RNA Control Assay U6 snRNA (Applied Biosystems, Cat # 4427975, Assay# 001973; Thermo Fisher Scientific, Waltham, MA, USA). Both transcriptions were conducted for each sample. Real-time PCR was then performed with TaqMan Universal PCR Master Mix (Applied Biosystems/Thermo Fisher Scientific, Waltham, MA, USA) on an Aria MX Real-Time PCR System (Agilent Technologies) using the small RNA-specific primer mixes of the previously described TaqMan assays. All of the steps were carried out according to the manufacturer's instructions.
2.17. Real-time PCR
The cDNA samples were obtained by reverse transcription of 1 μg total RNA using M-MLV Reverse Transcriptase (Promega). Quantitative real-time PCR was then performed with GoTaq qPCR Master Mix (Promega) according to the manufacturer's instructions using the oligonucleotides listed in ESM Table 2.
2.18. Dual luciferase reporter assay
Analysis of the TargetScan Mouse 7.2 database (http://www.targetscan.org) to predict the miR-132 targets revealed the presence in Mapk1 and Pten of 4 and 6 highly conserved regions for pairing with miR-132/212, respectively. Of these regions, 2 in Mapk1 and 3 in Pten were specific for miR-132 only. Oligonucleotides encompassing the sequences containing the predicted miR-132 binding sites in Mapk1 (5′-CCTTCAACGGCCTTCAGCATGACTGTTACAAGGAGAAGGGCGAGGTGTTCAGCTTTCAACGGAAGCTTGGCAATCC-3′), Pten (5′-CCTTCACATTAACTGTTAGACGGCCTTCAGTTGCACTG TTAAACGGCCTTCATTTAAATACTGTTAAACGGAAGCTTGGCAATCC-3′) or a control scrambled sequence (5′-CCTTCAACGGCCTTCATGGCTTGGGCACAAGGAGAAGGGCGAG GTATGGCTTGGACAACGGAAGCTTGGCAATCC-3′) were subcloned into the XhoI/NotI sites at the 3′ prime end of Renilla luciferase in the bicistronic vector psiCHECK2 (Promega). The dual luciferase activity assay was performed according to the manufacturer's instructions.
2.19. Statistical analyses
The statistical analyses were performed using unpaired Student's t-test unless otherwise stated. The results are presented as mean SE unless otherwise stated. A value of p ≤ 0.05 was considered significant. Error bars show the standard deviations from at least three independent experiments unless otherwise stated. Histograms were prepared with Microsoft Excel (Microsoft, Redmond, WA, USA) or GraphPad Prism.
3. Results
3.1. MiR-132 is upregulated in proliferating islet cells of pancreatectomized mice
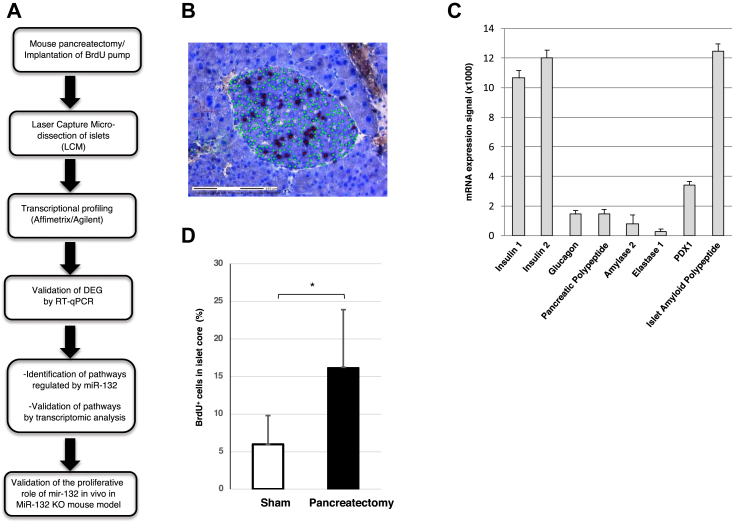
To identify the key miRNAs involved in beta cell proliferation, we used partially pancreatectomized mice (n = 3) as a model for beta cell regeneration (Figure 1A). The removal of 70–80% of the pancreas is a well-established procedure for inducing the replication of beta cells in the remaining pancreas [19,22]. Proliferating cells were stained with 25 μg/h BrdU continuously delivered by an osmotic mini-pump implanted in the abdomen at the time of partial pancreatectomy. This approach ensures the labeling of every dividing cell [19]. As controls, 4 mice were similarly implanted with an osmotic mini-pump for BrdU delivery but only underwent total splenectomy. Seven days post-surgery, all of the mice were sacrificed. Their remnant pancreas were excised for serial sectioning (40 sections/mouse) and staining with cresyl violet to locate the islets. Adjacent unstained sections were then used to count BrdU+ cells in the islet cores, which in rodents consist mainly of beta cells [23] (Figure 1B) prior to islet core excision by laser capture microdissection (LCM). Accordingly, total RNA extracted from the LCM islets was enriched for beta cell-specific transcripts Ins1, Ins2, Pdx1, and IAPP (Figure 1C). As expected, in the partially pancreatectomized mice, the fraction of islet core BrdU+ cells/total islet core cells determined by nuclear counting was significantly higher than in the sham-operated mice (Figure 1D). The RNA extracted from the LCM islet cores was then profiled using microarrays. Fourteen miRNAs were differently expressed (cut-off values: p ≤ 0.05; FC ≥ 1.5) in the islet cores of the partially pancreatectomized mice compared to the sham-operated mice (Table 1). All of the miRNAs were upregulated, except miR-760, which was reduced by 2.28-fold. The expression levels of all 14 differentially expressed miRNAs were further quantified by real-time PCR (RT-PCR) and the significantly changed expression of miR-132 and miR-141 was validated (ESM Table 3). Given the role of miR-132 in the replication in vitro of primary islet cells [15] and other cell types, including glioma cells [24] and epidermal keratinocytes [25], we focused on its potential involvement and mode of action in the regulation of beta cell proliferation.
Figure 1.
Quantification of BrdU+cells in islets of partially pancreatectomized mice. (A) Overview of the experimental design. (B) A tissue section of the remnant pancreas excised 1 week after partial pancreatectomy stained with cresyl violet to reveal the islet core. The tissue was labeled with DAPI (green) and anti-BrdU antibodies (black) to detect the cell nuclei and replicating cells, respectively. (C) Enrichment of beta cell-specific markers shown by Q-PCR of mRNA extracted from laser capture microdissected islets. (D) Percentage of BrdU+ cells in the islet cores. The analysis included 7 mice, of which 4 were sham-operated and 3 were partially pancreatectomized. 40 slices/group were counted for BrdU+ cells. *p < 0.05.
Table 1.
miRNAs differentially expressed in islet cores of partially pancreatectomized mice.
| Systematic name | p-value | FC | Px1 | Px1 | Px3 | S1 | S2 | S3 | S4 |
|---|---|---|---|---|---|---|---|---|---|
| mmu-mir-205* | 0.009 | 4.91 | 6.5011 | 12.7521 | 9.4035 | 1 | 5.34,742 | 2.0243 | 1 |
| mmu-mir-132* | 0.005 | 4.63 | 85.442 | 90.0032 | 71.439 | 26.371 | 16.9079 | 22.282 | 8.6324 |
| mmu-mir-186* | 0.016 | 2.5 | 4.0622 | 5.34,086 | 3.1434 | 1.7227 | 1.1133 | 1.9388 | 1.686 |
| mmu-mir-129-3p* | 0.022 | 1.99 | 213.55 | 272.406 | 121.26 | 118.28 | 100.958 | 92.346 | 68.376 |
| mmu-mir-690* | 0.03 | 1.79 | 49.413 | 48.5373 | 30.88 | 23.957 | 39.5096 | 16.126 | 17.206 |
| mmu-mir-130b* | 0.014 | 1.77 | 42.937 | 40.5664 | 29.545 | 22.405 | 28.2766 | 18.247 | 14.679 |
| mmu-mir-300* | 0.045 | 1.77 | 2.9997 | 4.06448 | 2.006 | 1.8972 | 1.78,003 | 1.8814 | 1 |
| mmu-mir-598* | 0.049 | 1.75 | 18.357 | 22.9878 | 14.596 | 12.314 | 10.6534 | 12.703 | 6.3097 |
| mmu-mir-431* | 0.013 | 1.65 | 23.393 | 27.3673 | 17.432 | 14.414 | 15.546 | 13.637 | 9.7251 |
| mmu-mir-329* | 0.035 | 1.64 | 25.756 | 29.392 | 19.037 | 17.004 | 16.0322 | 15.797 | 9.9605 |
| mmu-mir-154* | 0.029 | 1.63 | 19.404 | 19.675 | 15.306 | 10.545 | 11.4188 | 13.067 | 8.2187 |
| mmu-mir-141* | 0.04 | 1.59 | 878.04 | 970.708 | 655.42 | 566.11 | 672.314 | 515.55 | 318.15 |
| mmu-mir-200c* | 0.039 | 1.58 | 976.04 | 997.919 | 672.69 | 608.97 | 681.16 | 530.91 | 361.34 |
| mmu-mir-375 | 0.107 | 1.48 | 6291.99 | 7241.707 | 4643.13 | 4778.29 | 4398.79 | 4440.34 | 2461.28 |
| mmu-mir-760* | 0.041 | - 2.28 | 12.088 | 13.0156 | 4.6472 | 12.305 | 37.794 | 19.394 | 17.248 |
FC: Fold change.
Px (1–3): 3 Pancreatectomized mice S; (1–4): 4 control sham-operated mice.
* p<0.05.
3.2. MiR-132 promotes the proliferation and survival of mouse insulinoma MIN6 cells
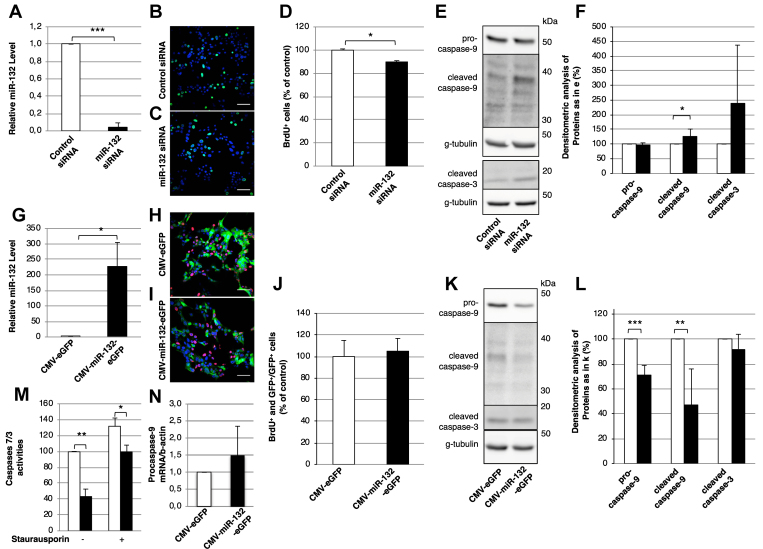
We first downregulated the miR-132 expression in the mouse insulinoma MIN6 cells using an anti-microRNA approach. Reduced expression of miR-132 by 90% (n = 3, Figure 2A), as assessed by RT-PCR, correlated with a modest yet significant reduction (89% ± 6.47%) in the percentage of the BrdU+ cells relative to the control cells (100% ± 8.16%, n = 6, p-value = 0.039) that had been transfected with control siRNA (Figure 2B–D). MiR-132 depletion correlated also enabled increased detection of cleaved caspase-9 (n = 6; Figure 2E,F), while the levels of cleaved caspase-3 did not significantly change. Conversely, overexpression of miR-132 with a bicistronic adenovirus vector also encoding for eGFP (n = 3. Figure 2G) was not associated with a further increase in the proliferation of the MIN6 cells, presumably due to their neoplastic state (n = 3, Figure 2H–J). Notably, overexpression of miR-132 also correlated with reduced levels of pro- and cleaved caspase-9 (n = 6, Figure 2K, L), consistent with the anti-apoptotic role of the latter. The anti-apoptotic role of miR-132 was corroborated by reduced staurosporine-induced apoptosis in cells overexpressing miR-132 (Figure 2M), consistent with the decreased levels of cleaved caspase-9 [26] observed in these cells (Figure 2K). The miR-132-induced decrease in pro- and cleaved caspase-9 expression was likely post-transcriptional, since in the miR-132 overexpressing cells, the levels of procaspase-9 mRNA, as measured by RT-PCR, did not significantly change relative to the control cells (Figure 2N).
Figure 2.
MiR-132 regulates the proliferation and caspase levels in MIN6 cells. (A) Levels of miR-132 in MIN6 cells treated with control (white bar) or miR-132 (black bar) siRNA as determined by RT-PCR. (B-C) Representative confocal images of BrdU+ cells (green) in MIN6 cells treated with control (B) or miR-132 (C) siRNA. (D) Number of BrdU+ in control vs miR-132 depleted MIN6 cells counted from images as in (B-C). (E) Representative immunoblots for pro- and cleaved caspase-9, cleaved caspase-3, and γ-tubulin as loading controls in extracts of MIN6 cells treated with control or miR-132 siRNA. (F) Densitometric quantification of immunoblots as in (E). White bars: control siRNA-treated cells; black bars: miR-132 siRNA-treated cells. (G) Expression levels of miR-132 in MIN6 cells transduced with adenovirus vectors for the co-expression of miR-132 and eGFP (black bar) or eGFP alone (white bar) as detected by RT-PCR. (H-I) Representative confocal images of BrdU+ (red) and eGFP+ (green) MIN6 cells transduced with adenovirus vectors for the co-expression of miR-132 and eGFP (i) or eGFP alone. Nuclei were detected with DAPI (blue). (J) Number of BrdU+ and GFP+ cells normalized to GFP+ cells in MIN6 cells transduced with eGFP (white bars) or miR-132 and eGFP (black bars) counted from images as in (H-I). (K) Representative immunoblots for pro- and cleaved caspase-9, cleaved caspase-3, and γ-tubulin as loading controls in extracts of MIN6 cells transduced with adenovirus vectors for the co-expression of miR-132 and eGFP or eGFP alone. (L) Densitometric quantification of immunoblots as in (K). (M) caspase-3/7 enzymatic activity was assayed by measuring the cleavage of Z-DEVD-R110 substrate via spectrophotometry as described in the materials and methods section. (N) Expression of procaspase-9 mRNA in MIN6 cells overexpressing miR-132 (white bar) and eGFP (black bar) as measured by RT-PCR. n = 3.
We further found that in the MIN6 cells, overexpression of miR-132 neither affected glucose-stimulated insulin secretion nor prevented its 35% reduction upon treatment of the cells with 0.5 mM palmitate for 24 h (ESM Fig. 1).
3.3. MiR-132 promotes the proliferation of human EndoC-βH1 cells
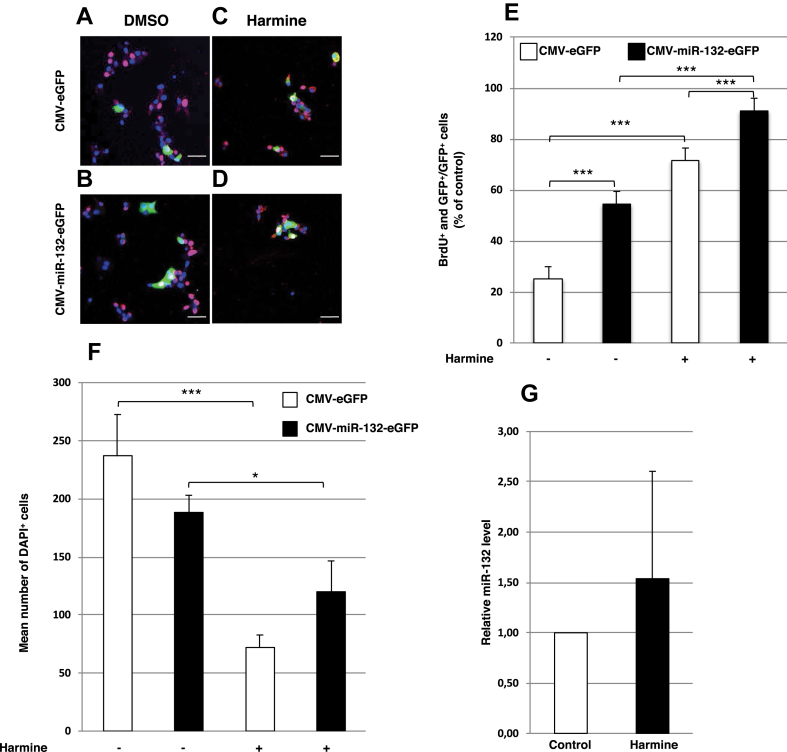
To further assess its role in beta cell proliferation, we overexpressed miR-132 together with eGFP in human EndoC-βH1 cells, which proliferate less than MIN6 cells (Figure 3). The miR-132 overexpression in these cells correlated with a ∼2-fold (118%, p < 0.001) increase in the number of BrdU+ and GFP+ cells (Figure 3A,B, and E) without a reduction in the number of cells left on the coverslip 5 days after transfection compared to GFP+ cells treated with 1.4 mM DMSO alone for 48 h prior to BrdU labeling (p = 0.262; Figure 3F). In comparison, the known inducer of β cell proliferation harmine [27] increased the number of BrdU+, GFP+ cells by ∼3-fold (187%, p < 0.001; Figure 3A,C, and E) without increasing the miR-132 levels as measured by RT-PCR (Figure 3G). Treatment with 10 μM harmine in 1.4 mM DMSO for 48 h, however, also correlated with a pronounced reduction (−69%, p < 0.001) in the total number of remaining cells on day 5 after treatment as measured with DAPI staining of the nuclei (Figure 3F). Combination of harmine and miR-132 overexpression further increased the number of BrdU+ and GFP+ cells by 27% (p < 0.001) compared to harmine alone (Figure 3C,D, and E). In this case, the number of cells remaining on day 5 was still reduced relative to the control (−49%, p < 0.001) and miR-132 overexpressing (−36%, p = 0.031) cells (Figure 3F). Notably, 91% of the cells treated with harmine and transduced with miR-132 were proliferating compared to 71% of the cells treated with harmine only, consistent with miR-132's role in promoting cell proliferation. Intriguingly, however, among the cells treated with harmine and overexpressing miR-132, as assessed by being GFP+, the number of cells immunostained for phosphohistone H3 (pHH3), a marker of the late G2 and M phases, was reduced 3-fold compared to the DMSO-treated cells (ESM Fig. 2).
Figure 3.
Effect of miR-132 on beta cell proliferation in EndoC-βH1 cells. (A–D) Representative confocal images of BrdU+ (red) and eGFP+ (green) in EndoC-βH1 cells transduced with adenovirus vectors for control eGFP (A and C) or miR-132-eGFP (B and D) expression either in 1.4 mM DMSO (A and B) or 10 μM harmine (C and D). Nuclei were detected with DAPI (blue). (E) BrdU+ and GFP+ EndoC-βH1 cells normalized to GFP+ cells in control or miR-132 overexpressing cells treated as described in (A-D). (F) Mean number of DAPI+ cells of EndoC-βH1 cells/field transduced as described in (A–D). White bars: cells transduced with CMV-eGFP adenovirus vector; black bars: cells transduced with CMV-miR-132/eGFP adenovirus vector. (G) Relative miR-132 level in EndoC-βH1 cells treated with 1.4 mM DMSO or 10 μM harmine. Data shown in (A–D) were collected from 15 to 20 images with 1,000–4,000 cells for each condition. Scale bars in (A–D) = 50 μm *p < 0.05, **p < 0.01, **p < 0.001.
3.4. miR-132 regulates Pten signaling in MIN6 cells and in islets from pancreatectomized mice
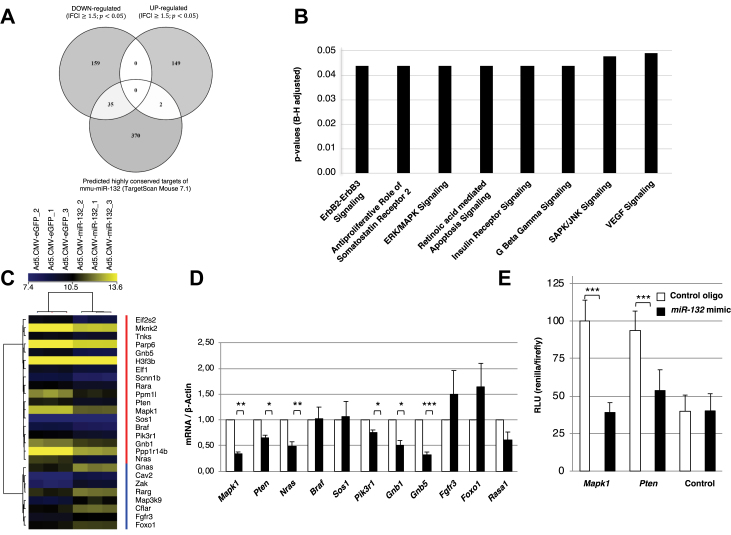
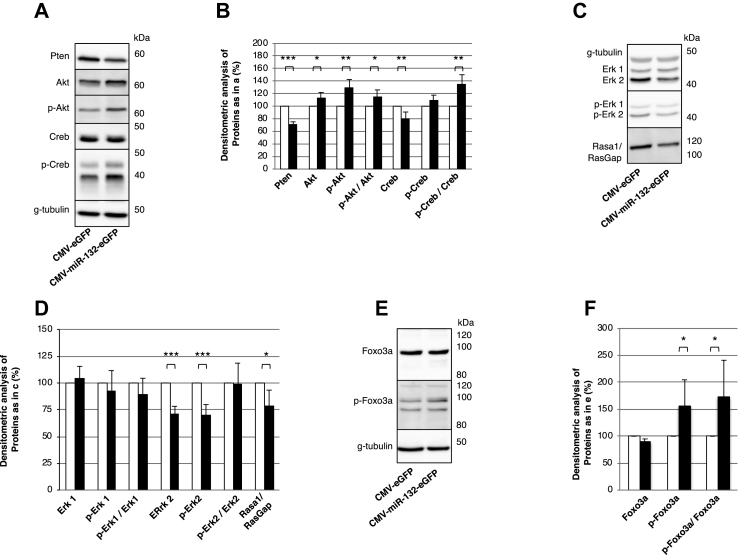
Next, we assessed the downstream targets of miR-132 in MIN6 cells. Microarray gene expression analysis of miR-132 overexpressing MIN6 cells identified 345 unique differentially expressed genes (cut-off values: p < 0.05, FC ≥ 1.5), with 194 (56.2%) as being downregulated and 151 (43.8%) upregulated (Figure 4A and ESM Table 4). Using the TargetScan Mouse 7.2 database, 31 of the down- and 2 of the upregulated genes were predicted to contain highly conserved binding sites for miR-132 (Figure 4A and ESM Table 5). Further evaluation of the regulated genes with Ingenuity Pathway Analysis revealed 8 regulated pathways (Figure 4B and ESM Table 6), which included 26 of the 345 differentially expressed genes, mostly related to cell proliferation and survival (Figure 4C and ESM Table 7). Among these 26 genes, the top 10 most represented genes in the 8 signaling pathways were further selected for validation of their mRNA levels by RT-PCR. As a control, we also assessed the mRNA expression levels of Rasa1, an established target of miR-132 [28]. As shown in Figure 4, Figure 6 out of 10 of the selected genes, namely Mapk1/Erk2, Pten, Nras, Pik3r1, Gnb1, and Gnb5, were confirmed to be downregulated upon miR-132 overexpression. Among those genes, Mapk1 and Pten were also among the 31 predicted targets for miR-132 binding (ESM Table 5). Notably, Mapk1, also known as Erk2, is a serine-threonine kinase located downstream of the tumor-suppressor phosphatase Pten and both genes play a critical role in the control of cell proliferation and survival [29,30]. We then used dual luciferase assays to test whether miR-132-mediated downregulation of Mapk1 and Pten occurs through a direct binding of miR-132 to their mRNA 3′-UTR. As shown in Figure 4E, these analyses indicated that miR-132 overexpression inhibited the activity of Renilla luciferase constructs bearing the seed regions of either Mapk1 or Pten by 61% and 46%, respectively. We further verified whether the islet expression of the miR-132 target Pten was altered upon pancreatectomy. In the LCM islets of the pancreatectomized mice, the levels of Pten mRNA were reduced (p = 0.012) compared to the LCM islets of the sham-operated mice (ESM Fig. 3A). Similar to the Min6 cells, the expression of Nras and Pik3r1 was also reduced in the LCM islets of the pancreatectomized mice. Moreover, this analysis revealed that the upregulation in the LCM islets of the pancreatectomized mice of genes implicated in the regulation of cell proliferation and the cell cycle such as cyclins d1, d2, and stathmin1 (Stmn1) (ESM Fig. 3b).
Figure 4.
Identification and validation of miR-132 target genes in MIN6 cells. (A) Venn diagram showing the number of down- and upregulated genes in MIN6 cells transduced with CMV-miR-132/eGFP adenovirus vector compared to MIN6 cells transduced with CMV-eGFP adenovirus vector and their overlap with predicted highly conserved murine mouse targets of miR-132. (B) Ingenuity Pathway Analysis of significantly altered pathways in MIN6 cells overexpressing miR-132 and eGFP compared to MIN6 cells overexpressing eGFP alone. (C) Heat map of the 26 (red vertical bar: cluster of downregulated genes; blue vertical bar: cluster of upregulated genes) among the 345 differentially expressed genes included in the 8 regulated pathways indicated in (B) and related to cell proliferation and survival in MIN6 cells overexpressing miR-132 and eGFP relative to MIN6 cells overexpressing eGFP alone. The brightest shades of yellow and blue reflect the higher and lower expression levels of a given gene as indicated in the top horizontal bar. The results are from 3 independent microarray analyses. (D) Validation by RT-PCR of the expression levels of the top 10 most represented genes in the 8 signaling pathways shown in (B) and among the 26 regulated genes shown in (C). White bars: expression in MIN6 cells overexpressing eGFP alone; black bars: expression in MIN6 cells overexpressing miR-132 and eGFP. (E) Min6 cells were co-transfected with 1.5 ug of bicistronic vector psiCHECK-2 in which Pten-3′-UTR or Mapk1-3′-UTR were inserted downstream of the Renilla translational stop codon along with 100 nM of either control or miR-132 mimic oligonucleotide. Co-expressed firefly luciferase was used for the normalization of Renilla luciferase expression, and the luciferase activity of the control was set as 100% The data represent mean ± standard error from 3 separate experiments. *p < 0.05, **p < 0.01, **p < 0.001. (B) The significance was assessed using the Benjamini–Hochberg test.
Figure 6.
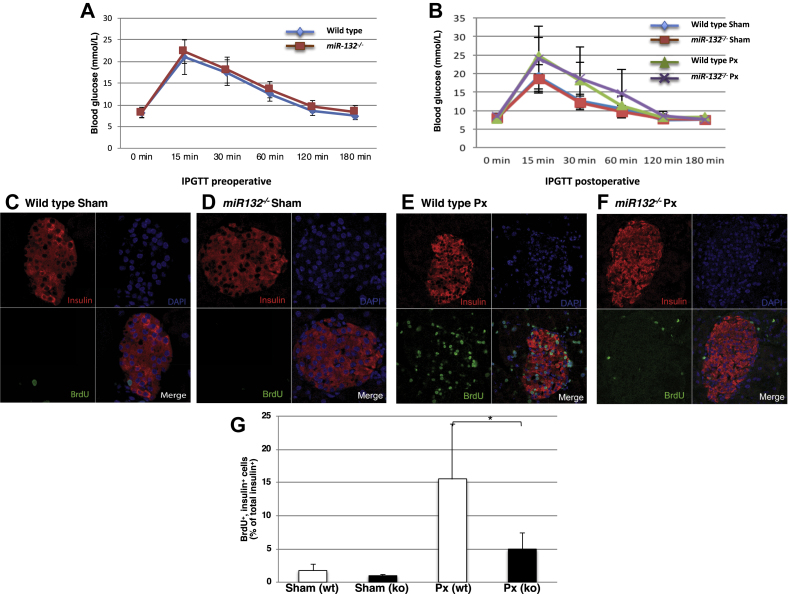
Impaired pancreatic beta cell regeneration in miR-132−/−mice. (A) Expression of blood glucose levels in wild-type (blue line) and miR-132−/− (red line) mice during an intraperitoneal glucose tolerance test conducted two days before surgery. (B) Expression of blood glucose levels in wild-type and miR-132−/− partially pancreatectomized or sham-operated mice during an intraperitoneal glucose tolerance test performed six days after surgery. Sham-operated wild-type mice: blue line; partially pancreatectomized wild-type mice: green line; sham-operated miR-132−/− mice: red line; partially pancreatectomized miR-132−/− mice: purple line. (C–F) Representative confocal images of paraffin pancreatic sections from wild-type sham-operated (C) or partially pancreatectomized mice (E) or from miR-132−/− sham-operated (D) or partially pancreatectomized (F) mice. Sections were immunolabeled for insulin (red) and BrdU (green), while nuclei were detected with DAPI (blue). (G) Percentage of BrdU+ and insulin+ cells. White bars: wild-type mice; black bars: miR-132−/− mice. Data are from 3 independent series with two mice in each group. Up to 50 islets were counted for each mouse for a total of 250–300 islets per group.
3.5. MiR-132 regulates Pten/Akt/Foxo3 signaling in MIN6 cells
Next, we tested whether the overexpression of miR-132 affected the protein levels of Pten and Mapk1/Erk2. Immunoblotting of the MIN6 cells transduced with a miR-132/eGFP viral vector confirmed the downregulation of Pten in parallel with the upregulation of its targets Akt and phospho-Akt (S473) (Figure 5A,B), while the Akt mRNA levels were unchanged (ESM Fig. 4). Furthermore, the levels of the Akt substrate Creb and phospho-Creb (S133) were unchanged, but the phospho-Creb/Creb ratio increased. Likewise, the overexpression of miR-132 correlated with reduced expression of Mapk1/Erk2, phospho-Mapk1/Erk2, and Rasa1/RasGAP, but not Erk1 and phospho-Erk1 (Figure 5C,D). As the downregulation of miR-132 correlated with elevated cleaved caspase-9 levels, we further tested whether its overexpression affected the expression of Foxo3a, a key mediator of apoptosis. Immunoblotting of miR-132 overexpressing cells for Foxo3a showed that its inhibitory phosphorylation increased (Figure 5E–F), while its mRNA (ESM Fig. 4) and total protein (Figure 5E–F) levels did not change.
Figure 5.
Validation of gene profiling data in MIN6 cells overexpressing miR132. (A, C, and E) Representative immunoblots for the indicated proteins in cell extracts of MIN6 cells overexpressing miR-132 and eGFP or eGFP alone. (B, D, and F) Densitometric quantification of the indicated proteins as detected by immunblots in (A), (C), and (E), respectively. Data are from 6 independent experiments. *p < 0.05, **p < 0.01, **p < 0.001.
3.6. Deletion of miR-132/212 impairs islet beta cell proliferation in pancreatectomized mice
As MIN6 and EndoC-βH1 are tumor cell lines, their increased proliferation upon miR-132 overexpression does not necessary imply that miR-132 fosters the proliferation of primary beta cells in vivo. In addition, the cell cycle machinery of these insulinoma cells might be affected by adenoviral transduction. To verify whether miR-132 positively affects beta cell regeneration in vivo, we measured the beta cell proliferation in the partially pancreatectomized or sham-operated miR-132/212−/− mice and the control littermates (6 mice/group). Intraperitoneal glucose tolerance tests prior and 6 days after surgery showed no difference between the control and miR-132/212−/− mice (Figure 6A,B). Daily blood glucose measurements, in particular, demonstrated a comparable modest decrease in glycemia in the partially pancreatectomized wild-type and miR-132/212−/− mice relative to the sham-operated mice on the first day post-surgery, followed by a complete normalization of glycemia by the end of the 1-week-long protocol (ESM Fig. 5). Seven days after surgery, the mice were sacrificed, the remnant pancreas excised, and the BrdU+/insulin+ beta cells were counted (Figure 6C–F and ESM Table 8). As assessed by immunostaining for insulin, the average number of beta cells/islet of the wild-type (31.9 beta cells/islet) and miR-132/212−/− (31.7 beta cells/islet) partially pancreatectomized mice was increased relative to their sham-operated counterparts (wild-type: 23.8 beta cells/islet; miR-132/212−/−: 26.8 beta cells/islet). Likewise, the number BrdU+ insulin+ beta cells increased in both groups of partially pancreatectomized mice compared to the sham-operated mice (Figure 6G, ESM Table 8). However, in the partially pancreatectomized miR-132/212−/− mice, there were fewer BrdU+/insulin+ cells than in the partially pancreatectomized wild-type mice (Figure 6G and ESM Table 8). These data provide conclusive evidence that the miR-132/212 cluster, and conceivably miRNA-132 alone (see discussion), exerts a positive role on the regeneration of mouse beta cells in vivo (Figure 7).
Figure 7.
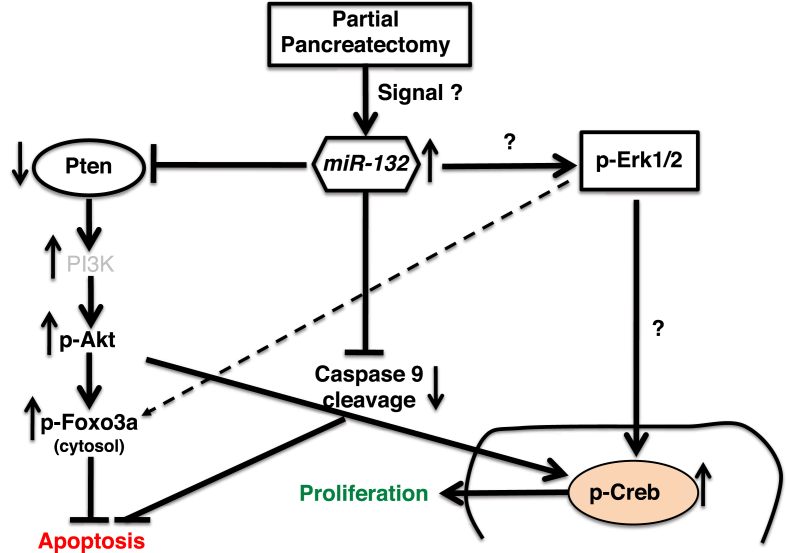
Working model illustrating the miR-132 mediated control of cell survival and proliferation of pancreatic beta cells. Islet beta cell proliferation following partial pancreatectomy correlates with the upregulation of miR-132. MiR-132 reduces the expression of Pten, which in turn enhances Foxo3a phosphorylation. Phosphorylated Foxo3a is sequestered in the cytosol and its transcriptional activity of pro-apoptotic genes such as caspases is reduced. Repression of Pten activity also increases the levels of P-Akt and P-Creb and thus enhances cell proliferation. However, the involvement of Mapk/Erk signaling in miR-132-mediated beta cell proliferation is uncertain. Experimental data presented in this manuscript and literature-based knowledge are displayed as solid and dashed lines, respectively.
4. Discussion
MiR-132 is known to control many cellular processes in various tissues, including neuronal morphogenesis and the regulation of circadian rhythm. MiR-132 altered expression correlates with several neurological disorders, such as Alzheimer's and Huntington's diseases [31]. Thus, most of our knowledge regarding miR-132 regulation and biological functions emerged from studies on neural cells, while less is known about the downstream targets of miR-132 in pancreatic beta cells. MiR-132 and miR-212 have identical seed sequences but very few targets in common [32]. In this study, we identified miR-132 as one of the mostly upregulated miRNAs with a 5-fold expression change in RNA extracted from LCM islets of partially pancreatectomized mice, a condition that induces beta cell proliferation. Moreover, analysis of mRNAs for well-established pancreatic islet or exocrine cell-specific markers showed a clear enrichment in our islet extracts of beta cell specific genes. The microarray analysis did not reveal changes in miR-212 expression although it was arrayed on the chip. Whether this was due to its lower expression than miR-132, as shown previously in rat insulinoma INS-1 cells [33], cannot be excluded. Notably, the levels of miRNAs from the same cluster can differ due to post-transcriptional mechanisms [34]. Moreover, although miR-132 and miR-212 share the same seed sequence, they differ in their 3p sequence. This non-seed region plays an important role in the selection of target mRNAs and on the regulatory features of each miRNA [32]. Furthermore, in hippocampal neuronal cells, the overexpression of miR-132 but not that of miR-212 correlated with the induction of genes related to cell proliferation. As microarray analyses may yield false positives, we further verified the increased expression of miR-132 in the islets of the partially pancreatectomized mice by RT-PCR. This finding is in agreement with previous data showing the elevated expression of miR-132 in different models of type 2 diabetes, including db/db, high-fat diet-fed [15], and ob/ob mice [12,35]. Among our list of differentially expressed miRNAs in the islets of the partially pancreatectomized mice, miR-205 showed the greatest change (5-fold). Interestingly, miR-205 has also been reported to be the miRNA with the highest expression change in the hepatocytes of mice with obesity-induced type 2 diabetes [35]. However, we did not detect significant changes in the expression of miR-375, which is abundant in pancreatic islets and regulates insulin secretion and beta cell proliferation [12]. Previous research indicated that miR-132 is highly expressed in neurons and may regulate their differentiation [36,37]. More recent work in primary neurons and PC12 cells suggested that miR-132 controls cell survival by direct regulation of Pten, Foxo3a, and p300 [16]. In our study, we confirmed that the increased levels of miR-132 did not correlate with changes in pro-caspase-9 expression, suggesting that miR-132 controls apoptotic genes post-transcriptionally.
To ascertain the downstream targets of miR-132 in beta cells, we first investigated whether miR-132 has a proliferative role in insulinoma MIN6 cells. MiR-132 downregulation correlated with a modest but significant inhibition of their proliferation. Inhibition of miR-132 expression correlated with an increase in cleaved caspase-9, while the latter was reduced upon upregulation of miR-132. Increased expression of miR-132 was also associated with enhanced replication of human EndoC-βH1 cells. At variance with harmine, at least in our in vitro experimental settings, its positive effect on cell proliferation was not associated with a detrimental impact on cell viability. Intriguingly, miR-132/212 was also the only miRNA cluster among 250 miRNAs to be selectively upregulated in INS-1 cells and isolated rodent and human islets in response to GLP-1, which promotes β cell proliferation and neogenesis in animal models of diabetes [38]. Our data showed a dual role of miR-132 in cell proliferation and apoptosis. Whether the main function of miR-132 is to regulate cell proliferation or survival remains a challenging question. However, evidence that 91% of miR-132 EndoC-βH1 cells treated with harmine were BrdU+ compared to 71% of EndoC-βH1 cells treated with harmine alone suggested that miR-132 increases cell proliferation. Moreover, the fact that miR-132 induced inhibition of apoptosis is less pronounced 12 h post-staurosporine treatment than at time 0 confirmed the anti-apoptotic role of miR-132 but also suggested that the main role of miR-132 is to promote cell proliferation.
The role of miR-132 in beta cell proliferation has already been shown but mostly using oligonucleotides or adenovirus-mediated overexpression of miR-132 on dispersed islets [15] or in mouse models [16], respectively. Our studies confirmed this finding and validated for the first time the ability of miR-132 to regulate the proliferation of β cells in vivo following partial pancreatectomy, a well-established surgical procedure known to induce beta cell regeneration. Such a model system is of special interest for the identification of mechanisms for the regeneration or expansion of β cell masses in adults.
The role of miR-132 in the potentiation of GSIS has been demonstrated in rat-dispersed islets transfected with miR-132 oligonucleotide [15]. In our study, the modulation of miR-132 expression did not show a change in GSIS. Moreover, the overexpression of miR-132 in cells incubated with an elevated concentration of palmitate did not improve GSIS.
Our search for downstream targets of miR-132 indicated that the expression of Pten was significantly downregulated in miR-132 overexpressing cells. Furthermore, analysis of the target genes in the LCM islets of the pancreactectomized mice further validated the downregulation of Pten in proliferating pancreatic beta cells. The downregulation of Pten, which was predicted to be a direct target of miR-132, was further confirmed by dual luciferase assays. Akt, the main survival signaling pathway known to be antagonized by Pten, was also identified and experimentally validated to be a target of miR-132. P-Akt is a major activator of Foxo family proteins, which are members of the Forkhead superfamily of winged helix transcription factors controlling cellular metabolism [39], stress responses, DNA damage repair, and cell death [40]. Phosphorylation of Foxo3a, in turn, promotes its cytoplasmic retention by 14-3-3 proteins, and thereby the inhibition of its transcriptional activity [41]. In agreement with this, we found an inverse correlation between Pten expression and increased phosphorylation of Akt (P-Akt) and Foxo3a (P-Foxo3a), which is consistent with increased cell proliferation and reduced apoptosis. Of significance was also the induced phosphorylation of Creb, a transcription factor involved in various cellular process including beta cell survival [42] and proliferation [43]. Inhibition of Creb activation resulted in cell cycle arrest at the G2/M phase [44], suggesting that P-Creb may foster the progression through the checkpoint G2/M. Interestingly, analysis of genes differentially expressed in the LCM islets of the pancreatectomized mice revealed the upregulation of cell cycle regulators including cyclin d1 and d2. Cyclin d1 induction is required during the G1 phase for the cell to initiate DNA synthesis [45]. Among cell cycle regulators, we also noted the upregulation of Stmn1. Notably, the upregulated expression of Stmn1, which is a microtubule modifier protein, has been shown to shorten the G2 phase [46]. Intriguingly, immunostaining of EndoC-βH1 cells for pHH3, a marker of both the late G2 phase and the M phase of the cell cycle [47] revealed a reduced number of pHH3+ cells upon the overexpression of miR-132 combined with harmine treatment. A plausible explanation is that miR-132 facilitates the entry into the cell cycle with a shortening of its duration.
Many miRNAs have been associated with developmental regulation of pancreatic beta cell proliferation or differentiation (for example, miR-375 [10], miR-7 [48], miR-124a [49], miR-24 [50], let-7a [51], miR-26a [52], miR-184 [53], miR-195, miR-15, miR-16 [54], and miR-132 [15,37]. MiR-132 has also been found to be differentially expressed in T2D models such as obesity-induced diabetes [37] that are associated with increased metabolic demands on beta cells, a condition known to promote beta cell proliferation. Constitutive deletion of miR-132 in mice resulted in deficient endocrine development [18], while the restricted deletion of the miR-13/212 locus in adult hippocampus caused a dramatic decrease in the dendrite length, arborization, and spine density [55]. In this study, we demonstrated that the regeneration of beta cells in pancreatectomized miR-132/212−/− mice is reduced, conceivably through its control of the Pten/Pi3K/Akt signaling pathway. The increased or reduced beta cell proliferation in insulinoma cells overexpressing miR-132 or in miR-132/212−/− mice, respectively, cannot not be attributed to the sole action of miR-132. Indeed, although miR-132 is highly evolutionarily conserved, studies showed only a partial overlap of its downstream targets in mouse and human islet alpha cells, in which miR-132 also favored proliferation [56]. This observation raised the hypothesis that miR-132 acts in concert with other miRNAs that regulate cell proliferation. For instance, altered expression of miR-205, which is upregulated 5-fold in LCM islets of partially pancreatectomized mice, has also been correlated with cell proliferation of various cellular tissues and cell lines and with cancer [57,58]. MiR-205, in particular, has also been shown to enhance Akt signaling by indirectly targeting Pten and Phlpp2 [59]. Thus, the complexity of miRNA networks with interconnected positive and negative regulatory loops makes it difficult to ascribe induced proliferation to a single miRNA. Nonetheless, we have shown that the deletion of the miR-132/212 locus alone is sufficient to hamper the regeneration of mouse beta cells in vivo.
In conclusion, we have identified miR-132 as a critical epigenetic factor for control of beta cell replication after partial pancreatectomy. Targeted therapies for the expansion of beta cell mass in type 1 and type 2 diabetes are actively sought, and in this context, miR-132 appears to be a worthy candidate for consideration. Along these lines, it would be especially interesting to determine which and how extracellular signals promote its expression in beta cells.
Contribution statement
HM, MS, and SK conceived the study and the experimental design. GH profiled the gene expression, conducted the data mining, and validated the target genes with the assistance of KG, JM, KPK, and SW. SH, CK, and JM performed the pancreatectomy, implanted the BrdU pumps, and immunostained the pancreatic islets under the supervision of SK. KC generated and provided the miR-132−/− mice. PR provided advice for cell transduction with viral vectors. HM, MS, and SK wrote the manuscript. HM, MS, and SK are responsible for the study's integrity.
Data availability
Original microarray data are accessible through the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Funding
This study was partially supported with funds from the German Center for Diabetes Research (DZD e.V.) by the German Ministry for Education and Research to MS and SK and by a MeDDrive grant from the Carl Gustav Carus Faculty of Medicine at TU Dresden to SW.
Acknowledgments
We are grateful to Yannis Kalaidzidis for assistance with the statistical analysis of the data; Dr. Jun-ichi Miyazaki (Osaka University, Japan) and Dr. Raphael Scharfmann (INSERM, Paris) for providing the MIN6 and EndoC-βH1 cells, respectively; Julia Jarrells in the Sequencing Facility at the Max Planck Institute for Molecular Cell Biology and Genetics (Dresden, Germany); Anke Sönmez (Dresden, Germany) for help with the cell culture, Katja Pfriem (Dresden, Germany) for administrative assistance, and all of the colleagues in the Departments of General, Thoracic, and Vascular Surgery in the Faculty of Medicine at TU Dresden and the Department of Surgery in Erlangen for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.11.012.
Contributor Information
Michele Solimena, Email: michele.solimena@tu-dresden.de.
Stephan Kersting, Email: stephan.kersting@uk-erlangen.de.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta M.K., Halley C., Duan Z.H., Lappe J., Viterna J., Jana S. miRNA-548c: a specific signature in circulating PBMCs from dilated cardiomyopathy patients. Journal of Molecular and Cellular Cardiology. 2013;62:131–141. doi: 10.1016/j.yjmcc.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Z.P., Chen J., Seok H.Y., Zhang Z., Kataoka M., Hu X. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circulation Research. 2013;112(9):1234–1243. doi: 10.1161/CIRCRESAHA.112.300682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guay C., Regazzi R. Role of islet microRNAs in diabetes: which model for which question? Diabetologia. 2015;58(3):456–463. doi: 10.1007/s00125-014-3471-x. [DOI] [PubMed] [Google Scholar]
- 6.Plaisance V., Waeber G., Regazzi R., Abderrahmani A. Role of microRNAs in islet beta-cell compensation and failure during diabetes. Journal of Diabetes Research. 2014;2014:618652. doi: 10.1155/2014/618652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roggli E., Gattesco S., Caille D., Briet C., Boitard C., Meda P. Changes in microRNA expression contribute to pancreatic β-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61(7):1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melkman-Zehavi T., Oren R., Kredo-Russo S., Shapira T., Mandelbaum A.D., Rivkin N. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. The EMBO Journal. 2011;30(5):835–845. doi: 10.1038/emboj.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Bunt M., Gaulton K.J., Parts L., Moran I., Johnson O.R., Lindgren C.M. The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0055272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biology. 2007;5(8) doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latreille M., Herrmanns K., Renwick N., Tuschl T., Malecki M.T., McCarthy M.I. miR-375 gene dosage in pancreatic beta-cells: implications for regulation of beta-cell mass and biomarker development. Journal of Molecular Medicine. 2015;93(10):1159–1169. doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esguerra J.L., Bolmeson C., Cilio C.M., Eliasson L. Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One. 2011;6(4) doi: 10.1371/journal.pone.0018613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacovetti C., Abderrahmani A., Parnaud G., Jonas J.C., Peyot M.L., Cornu M. MicroRNAs contribute to compensatory beta cell expansion during pregnancy and obesity. Journal of Clinical Investigation. 2012;122(10):3541–3551. doi: 10.1172/JCI64151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nesca V., Guay C., Jacovetti C., Menoud V., Peyot M.L., Laybutt D.R. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia. 2013;56(10):2203–2212. doi: 10.1007/s00125-013-2993-y. [DOI] [PubMed] [Google Scholar]
- 16.Mulder N.L., Havinga R., Kluiver J.L., Groen A.K., Kruit J.K. AAV8-mediated gene transfer of microRNA-132 improves beta-cell function in mice fed a high fat diet. Journal of Endocrinology. 2019;240(2):123–132. doi: 10.1530/JOE-18-0287. [DOI] [PubMed] [Google Scholar]
- 17.Wong H.K., Veremeyko T., Patel N., Lemere C.A., Walsh D.M., Esau C. De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer's disease. Human Molecular Genetics. 2013;22(15):3077–3092. doi: 10.1093/hmg/ddt164. [DOI] [PubMed] [Google Scholar]
- 18.Ucar A., Vafaizadeh V., Jarry H., Fiedler J., Klemmt P.A., Thum T. miR-212 and miR-132 are required for epithelial stromal interactions necessary for mouse mammary gland development. Nature Genetics. 2010;42(12):1101–1108. doi: 10.1038/ng.709. [DOI] [PubMed] [Google Scholar]
- 19.Mziaut H., Kersting S., Knoch K.P., Fan W.H., Trajkovski M., Erdmann K. ICA512 signaling enhances pancreatic beta-cell proliferation by regulating cyclins D through STATs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(2):674–679. doi: 10.1073/pnas.0710931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 21.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonner-Weir S., Baxter L.A., Schuppin G.T., Smith F.E. A second pathway for regeneration of adult exocrine and endocrine pancreas. A possible recapitulation of embryonic development. Diabetes. 1993;42(12):1715–1720. doi: 10.2337/diab.42.12.1715. [DOI] [PubMed] [Google Scholar]
- 23.Elayat A.A., el-Naggar M.M., Tahir M. An immunocytochemical and morphometric study of the rat pancreatic islets. Journal of Anatomy. 1995;186(3):629–637. [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S., Wang Y., Ni C., Meng G., Sheng X. HLF/miR-132/TTK axis regulates cell proliferation, metastasis and radiosensitivity of glioma cells. Biomedicine & Pharmacotherapy. 2016;83:898–904. doi: 10.1016/j.biopha.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Li D., Wang A., Liu X., Meisgen F., Grünler J., Botusan I.R. MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. Journal of Clinical Investigation. 2015;125(8):3008–3026. doi: 10.1172/JCI79052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X.D., Gillespie S.K., Hersey P. Staurosporine induces apoptosis of melanoma by both caspase-dependent and -independent apoptotic pathways. Molecular Cancer Therapeutics. 2004;3(2):187–197. [PubMed] [Google Scholar]
- 27.Wang P., Alvarez-Perez J.C., Felsenfeld D.P., Liu H., Sivendrn S., Bender A. A high-throughput chemical screen reveals that harmine-mediated inhibition of DYRK1A increases human pancreatic beta cell replication. Nature Medicine. 2015;21(4):383–388. doi: 10.1038/nm.3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anand S., Majeti B.K., Acevedo L.M., Murphy E.A., Mukthavaram R., Scheppke L. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nature Medicine. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deb T.B., Barndt R.J., Zuo A.H., Sengupta S., Coticchia C.M., Johnson M.D. PTEN-mediated ERK1/2 inhibition and paradoxical cellular proliferation following Pnck overexpression. Cell Cycle. 2014;13(6):961–973. doi: 10.4161/cc.27837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Y.C., Yu H.S., Chai C.Y. Roles of oxidative stress and the ERK1/2, PTEN and p70S6K signaling pathways in arsenite-induced autophagy. Toxicology Letters. 2015;239(3):172–181. doi: 10.1016/j.toxlet.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Lee S.T., Chu K., Im W.S., Yoon H.J., Im J.Y., Park J.E. Altered microRNA regulation in Huntington's disease models. Experimental Neurology. 2011;227(1):172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 32.Hansen K.F., Sakamoto K., Aten S., Snider K.H., Loeser J., Hesse A.M. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learning & Memory. 2016;23(2):61–71. doi: 10.1101/lm.039578.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang J., Li J., Keller M.P., Hohmeier H.E., Wang Y., Feng Y. Induction of miR-132 and miR-212 expression by Glucagon-Like Peptide 1 (GLP-1) in rodent and human pancreatic β-cells. Molecular Endocrinology. 2015;29(9):1243–1253. doi: 10.1210/me.2014-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis B.N., Hilyard A.C., Lagna G., Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao E., Keller M.P., Rabaglia M.E., Oler A.T., Stapleton D.S., Schueler K.L. Obesity and genetics regulate microRNAs in islets, liver, and adipose of diabetic mice. Mammalian Genome. 2009;20(8):476–485. doi: 10.1007/s00335-009-9217-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(45):16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wayman G.A., Davare M., Ando H., Fortin D., Varlamova O., Cheng H.Y.M. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(26):9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shang J., Li J., Keller M.P., Hohmeier H.E., Wang Y., Feng Y. Induction of miR-132 and miR-212 expression by Glucagon-Like Peptide 1 (GLP-1) in rodent and human pancreatic β-cells. Molecular Endocrinology. 2015;29(9):1243–1253. doi: 10.1210/me.2014-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haeusler R.A., Hartil K., Vaitheesvaran B., Arrieta-Cruz I., Knight C.M., Cook J.R. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nature Communications. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glauser D.A., Schlegel W. The emerging role of FOXO transcription factors in pancreatic beta cells. Journal of Endocrinology. 2007;193(2):195–207. doi: 10.1677/JOE-06-0191. [DOI] [PubMed] [Google Scholar]
- 41.Brunet A., Bonni A., Zigmond M.J., Lin M.Z., Juo P., Hu L.S. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 42.Durkin J.P., Whitfield J.F. Characterization of G1 transit induced by the mitogenic-oncogenic Ki-ras gene product. Molecular and Cellular Biology. 1986;6(5):1386–1392. doi: 10.1128/mcb.6.5.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobrowolski S., Harter M., Stacey D.W. Cellular ras activity is required for passage through multiple points of the G0/G1 phase in BALB/c 3T3 cells. Molecular and Cellular Biology. 1994;14(8):5441–5449. doi: 10.1128/mcb.14.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Downward J. Routine role for ras. Current Biology. 1997;7(4):R258–R260. doi: 10.1016/s0960-9822(06)00116-3. [DOI] [PubMed] [Google Scholar]
- 45.Yang K., Hitomi M., Dennis W., Stacey D.W. Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Division. 2006;1:32. doi: 10.1186/1747-1028-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carney B.K., Silva V.C., Cassimeris L. The microtubule cytoskeleton is required for a G2 cell cycle delay in cancer cells lacking stathmin and p53. Cytoskeleton. 2012;69(5):278–289. doi: 10.1002/cm.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Hooser A., Goodrich D.W., Allis C.D., Brinkley B.R., Mancini M.A. Histone H3 phosphorylation is required for the initiation, but not maintenance, of mammalian chromosome condensation. Journal of Cell Science. 1998;111(23):3497–3506. doi: 10.1242/jcs.111.23.3497. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y., Liu J., Liu C., Naji A., Stoffers D.A. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic β-cells. Diabetes. 2013;62(3):887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baroukh N., Ravier M.A., Loder M.K., Hill E.V., Bounacer A., Scharfmann R. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. Journal of Biological Chemistry. 2007;282(27):19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- 50.Vijayaraghavan J., Maggi E.C., Crabtree J.S. miR-24 regulates menin in the endocrine pancreas. American Journal of Physiology Endocrinology and Metabolism. 2014;307(1):E84–E92. doi: 10.1152/ajpendo.00542.2013. [DOI] [PubMed] [Google Scholar]
- 51.Gurung B., Muhammad A.B., Hua X. Menin is required for optimal processing of the microRNA let-7a. Journal of Biological Chemistry. 2014;289(14):9902–9908. doi: 10.1074/jbc.M113.520692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu X., Jin L., Wang X., Luo A., Hu J., Zheng X. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(44):17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tattikota S.G., Rathjen T., McAnulty S.J., Wessels H.H., Akerman I., Van De Bunt M. Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metabolism. 2014;19(1):122–134. doi: 10.1016/j.cmet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joglekar M.V., Parekh V.S., Mehta S., Bhonde R.R., Hardikar A.A. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Developmental Biology. 2007;311(2):603–612. doi: 10.1016/j.ydbio.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 55.Magill S.T., Cambronne X.A., Luikart B.W., Lioy D.T., Leighton B.H., Westbrook G.L. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(47):20382–20387. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dusaulcy R., Handgraaf S., Visentin F., Vesin C., Philippe J., Gosmain Yvan. miR-132-3p is a positive regulator of alpha-cell mass and is downregulated in obese hyperglycemic mice. Molecular Metabolism. 2019;22:84–95. doi: 10.1016/j.molmet.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lu J., Lin Y., Li F., Ye H., Zhou R., Jin Y. miR-205 suppresses tumor growth, invasion, and epithelial-mesenchymal transition by targeting SEMA4C in hepatocellular carcinoma. The FASEB Journal. 2018;32(11):6123–6134. doi: 10.1096/fj.201800113R. [DOI] [PubMed] [Google Scholar]
- 58.Lu Z., Xu Y., Yao Y., Jiang S. miR-205-5p contributes to paclitaxel resistance and progression of endometrial cancer by downregulating FOXO1. Oncology Research Featuring Preclinical and Clinical Cancer Therapeutics. 2019 doi: 10.3727/096504018X15452187888839. [DOI] [PubMed] [Google Scholar]
- 59.Cai J., Fang L., Huang Y., Li R., Yuan J., Yang Y. AKT signaling is constitutively activated in various cancers, due in large part to loss-of-function in the PTEN and PHLPP phosphatases that act as tumor suppressor. Cancer Research. 2013;73(17):5402–5415. doi: 10.1158/0008-5472.CAN-13-0297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original microarray data are accessible through the GEO database (https://www.ncbi.nlm.nih.gov/geo/).