Abstract
Background
Jatropha curcas is an oil-bearing plant, and has seeds with high oil content (~ 40%). Several advantages, such as easy genetic transformation and short generation duration, have led to the emergence of J. curcas as a model for woody energy plants. With the development of high-throughput sequencing, the genome of Jatropha curcas has been sequenced by different groups and a mass of transcriptome data was released. How to integrate and analyze these omics data is crucial for functional genomics research on J. curcas.
Results
By establishing pipelines for processing novel gene identification, gene function annotation, and gene network construction, we systematically integrated and analyzed a series of J. curcas transcriptome data. Based on these data, we constructed a J. curcas database (JCDB), which not only includes general gene information, gene functional annotation, gene interaction networks, and gene expression matrices but also provides tools for browsing, searching, and downloading data, as well as online BLAST, the JBrowse genome browser, ID conversion, heatmaps, and gene network analysis tools.
Conclusions
JCDB is the most comprehensive and well annotated knowledge base for J. curcas. We believe it will make a valuable contribution to the functional genomics study of J. curcas. The database is accessible at http://jcdb.xtbg.ac.cn.
Keywords: Jatropha curcas, Woody energy plant, Functional genomics, Database
Background
Jatropha curcas is a perennial shrub belonging to the Euphorbiaceae family. It is a tropical species that is native to Mexico and Central America and now thrives in Latin America, Africa, India, and South East Asia [1–5]. As a multi-functional plant, it has been used in traditional medicine and for hedges, animal feed, and firewood [6–9]. With the gradual depletion and cost escalation of fossil energy resources, J. curcas is now attracting much attention for its potential use for biofuel production, because of its high seed oil content (the seeds of J. curcas contain ~ 40% oil) [10], easy propagation, rapid growth, and ability to grow in a wide range of conditions, including degraded, sodic, alkaline, and contaminated soils [7, 11].
J. curcas has a relatively small genome, which is organized in 22 chromosomes (2n) [12]. The J. curcas genome has been sequenced by four groups worldwide [13–17]. For the RefSeq representative version from the Wu laboratory, the assembled genome is 320.5 Mb [15]. J. curcas also has several advantages, including easy genetic transformation and short generation duration, which make it an attractive wood energy model plant for function genome analysis, particular among the Euphorbiaceae [18–20]. J. curcas is also a potential model for studies of flower sex determination in monoecious trees, as most J. curcas germplasms are monoecious, bearing male and female flowers on the same inflorescence [21, 22].
In recent years, there have been significant advances in the application of transcriptome analysis to J. curcas [22–31]. Using bioinformatics tools and a comprehensive knowledge database to integrate all these genome and transcriptome data is crucial for further functional genomics research on J. curcas. Advances in J. curcas research have led to the creation of several J. curcas genetic information resources. For instance, the Jatropha Genome Database (JAT_r4.5) focuses on the J. curcas genome sequence and annotation [13], and KaPPA-View4 is a KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway viewer for J. curcas [32]. Although each of these resources provides valuable information, there is a lack of database unification and integration of the J. curcas genome and transcriptome with a broad set of multi-omics analysis results, such as gene functional annotation, gene expression matrices, and gene interaction networks.
In this study, we constructed a J. curcas database (JCDB) that is dedicated to providing a comprehensive platform for J. curcas functional genomics research. By establishing pipelines for processing novel gene identification, gene function annotation, gene expression level quantification, and gene network construction, we systematically integrated and analyzed a series of J. curcas transcriptome data, which were used to generate JCDB. The database includes general gene information (including genomic coordinates and sequences), gene functional annotation (including gene ontology (GO), KEGG, Pfam, and InterPro), gene interaction networks (gene co-expression and protein-protein interaction (PPI) networks), and gene expression matrices. We also provide tools for browsing, searching, and downloading all data, as well as user-friendly web services such as BLAST, the JBrowse genome browser, ID conversion, heatmaps, and gene network analysis tools. In the case studies presented here, we demonstrate the possibility of using JCDB to mine genes related to flowering and lipid synthesis pathways in J. curcas. We believe that JCDB represents a valuable and unique resource for further functional genomics studies of J. curcas.
Construction and content
Transcriptome data retrieving and processing
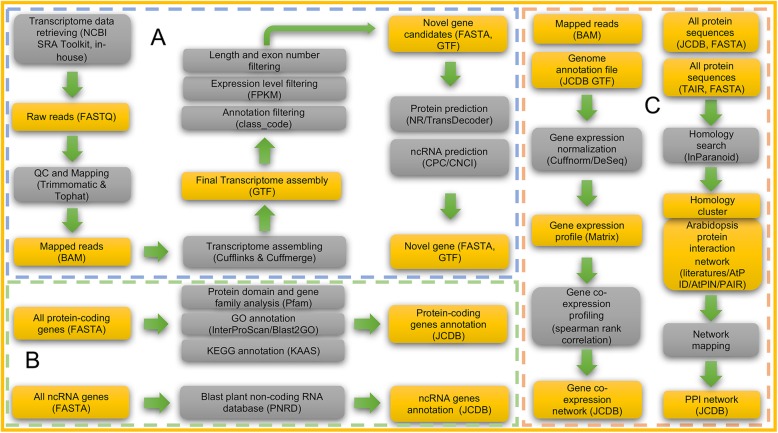
To acquire comprehensive genomic information for J. curcas, we developed a pipeline for transcriptome data collection, integration, and novel gene identification, including non-coding RNAs (Fig. 1a). First, publicly available transcriptome data of J. curcas were downloaded from NCBI’s Sequence Read Archive (SRA) database. Detailed information was collated for each sample, including experimental description, organizational information, and references. (Additional file 1). The SRA data was dumped into the FASTQ format using the fastq-dump utility from the NCBI SRA Toolkit v.2.5.2 [33]. Raw reads were quality trimmed using Trimmomatic (version 0.32) with parameters “LEADING:20 TRAILING:20 MINLEN:36” [34]. Then, all clean reads were mapped onto the J. curcas genome (JatCur_1.0) [15] using TopHat 2 (version 2.1.0), with default parameters except maximum intron length, which was set to 20,000 bp [35]. Next, the mapped reads were assembled using Cufflinks (version 2.2.1) with the RefSeq genome as a guide, and a combined transcriptome assembly was generated using Cuffmerge [36]. Finally, genes that were identified by Cuffcompare as non-overlapping with known genes, having more than one exon, longer than 200 bp, and with FPKM (fragments per kilobase per million) greater than 0.1 were considered as novel gene candidates.
Fig. 1.
JCDB pipelines for data retrieval and processing. a Novel gene discovery pipeline. b Coding and non-coding gene (ncRNA) annotation pipeline. c Gene co-expression and PPI network construction pipeline
Novel protein-coding and non-coding gene identification
As shown in Fig. 1a, novel transcript sequences were first used as query for a BLASTX search against the NCBI non-redundant protein (NR) database with default parameters. Then, open reading frames (ORFs) of these matches were identified using TransDecoder v4.1.0 (https://github.com/TransDecoder/TransDecoder). Matches with a completed ORF were annotated as protein-coding genes. Non-coding genes were further identified using CPC (Version 0.9-r2) [37] and CNCI (Version 2) [38] among the genes not matching the NCBI NR database. The remaining genes were annotated as transcripts of unknown coding potential (TUCPs).
Protein-coding and novel non-coding gene annotation
All the protein-coding and novel non-coding genes in JCDB were annotated using the in-house gene annotation pipeline (Fig. 1b). For the annotation of protein-coding genes, Pfam [39] was used for protein domain and gene family analysis. GO annotations were assigned using InterProScan [40] and Blast2GO [41]. KEGG annotations were assigned using the online service KAAS [42]. For the annotation of novel non-coding genes, we downloaded all small non-coding RNA and long non-coding RNA (lncRNA) sequences from the plant ncRNA database PNRD [43] and annotated the JCDB novel non-coding genes using a BLAST search with default parameters. In total, there were 27 novel non-coding genes with BLAST hits to PNRD, including 22 microRNA (miRNA) host genes, two long intergenic non-coding RNAs (lincRNAs), and three lncRNAs of unknown type.
Co-expression network construction
As shown in Fig. 1c, for conventional RNA-Seq data, gene expression profiles were identified and normalized using Cuffnorm [36]. For digital gene expression data, read count tables were created using htseq-count in the HTSeq toolkit [44] and then normalized using the DESeq method [45]. The two types of expression matrix were merged and normalized again using the upper-quartile method [44]. A gene co-expression network was constructed using the Spearman’s rank correlation coefficients of gene pairs across the samples. Gene pairs with correlation value higher than 0.6 and adjusted P-value less than 0.01 were regarded as showing co-expression.
Protein-protein interaction network construction
Arabidopsis protein interactions were collected from the literature [46–48] and from three databases (AtPID 5.0 [49], AtPIN 9.0 [50], and PAIR 3.0 [51]), giving a total of 18,037 Arabidopsis genes and 241,468 interactions. Arabidopsis protein sequences were downloaded from TAIR10 [52]. The pairwise similarity matching tool InParanoid [53] with default settings was used to find orthologous groups between the J. curcas and Arabidopsis proteomes. The J. curcas PPI network was inferred from the Arabidopsis PPI network [46–51] by homology mapping (Fig. 1c).
System implementation
The JCDB server was built using Apache/2.4.6 (CentOS), PHP (version 5.4.16), and relational database MySQL (version 5.5.48). The entity relationship diagram is provided in Additional file 2. The physical server was a 4 Intel(R) Xeon(R) CPU E5–2640 v3 @ 2.60 GHz with 8 GB RAM. All data and information were stored in MySQL tables to facilitate efficient management, search, and display. A combination of Thinkphp (version 3.2), Bootstrap (version 3.3.7), and JQuery (version 3.3.7) were used to construct the website. The network was visualized using Cytoscape.js (version 3.8).
Utility and discussion
Search JCDB
The ‘Search page of JCDB (Fig. 2a) provides three different types of search services. ‘Keyword Search’ uses keywords including gene types (such as protein_coding and ncRNA), gene symbols (such as bZIP, myb, and bHLH), and gene/transcript/protein IDs (such as JCDBG00001, JCDBR00001, and JCDBP00001) from JCDB or other databases (such as RefSeq, JAT_r4.5, and GenBank). ‘Position Search’ finds genes/transcripts/proteins located in one specific genomic region specified by the users. ‘Network Search’ provides a gene’s direct network neighbors in the PPI or co-expression network.
Fig. 2.
Screenshots of the JCDB online tools. a Keyword search, position search, and network search. b JCDBtools, the web-based toolkit. c JBrowse, the genome browser. d Online BLAST search
JCDBTools
JCDBTools is a web-based toolkit that provides five tools to help molecular biologists use JCDB more efficiently (Fig. 2b). ‘Sequence Retrieving’ can be used to retrieve genome sequences by providing genomic coordinates. ‘ID Conversion’ converts gene/transcript/protein IDs between JCDB and other databases (including RefSeq, JAT_r4.5, and GenBank). ‘Heatmap’ can be used to retrieve the gene expression patterns of a group of genes from different samples. ‘Network Construction’ can be used to extract a sub-network for user-specified genes from the global PPI or co-expression network. ‘Neighbor Gene Extraction’ can be used to extract the nearest neighbors of a sub-network in the global PPI or co-expression network.
JBrowse
JCDB integrates genome browser JBrowse [54] to provide easy-to-use panning and zooming navigation of the J. curcas reference genome (Fig. 2c). JBrowse includes various tracks, such as the J. curcas genome sequence, gene annotation GFF files from JCDB and RefSeq, and transcriptome-aligned BAM files for different samples.
BLAST service
The BLAST server (Fig. 2d) was implemented using ViroBLAST [55], which is a user-friendly tool for interfacing with the command-line NCBI BLAST+ toolkits. For user convenience, JCDB BLAST provides nucleotide databases (RefSeq genome/RNA, JCDB gene/RNA, and GenBank RNA/CDS) and protein databases (JCDB Protein, GenBank Protein, and RefSeq Protein).
Browse JCDB
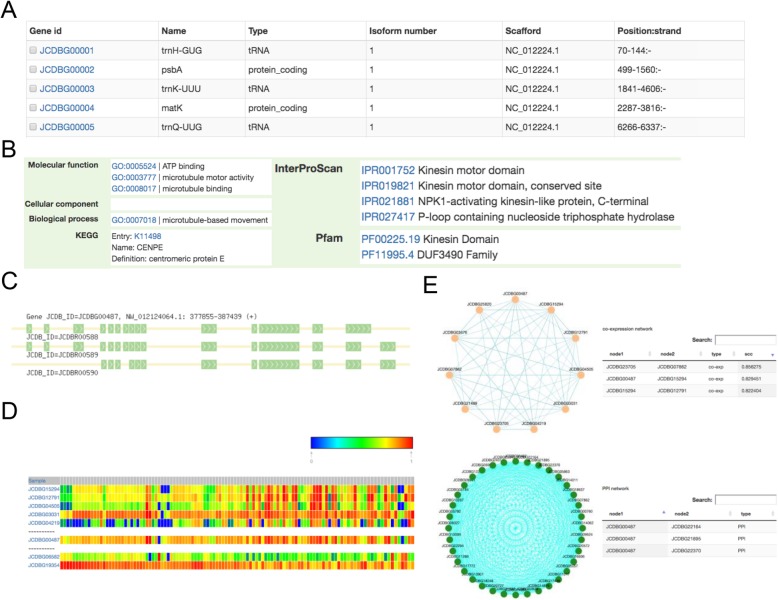
Users can browse all JCDB genes directly on the ‘Browse’ page (Fig. 3a), which provides basic annotations for each gene, such as gene name, gene type, and genomic location. Users can also select and download FASTA files for genes if required. Detailed information page for a specific gene can be accessed by clicking on the gene ID. For each gene, JCDB aims to provide as much comprehensive information as possible, including detailed GO, KEGG, InterPro, and Pfam functional annotations (Fig. 3b); structural information for each gene isoform (Fig. 3c); gene expression heatmaps (Fig. 3d); and co-expression and PPI sub-networks (Fig. 3e). In the gene expression heatmap panel, users can select the number of co-expressed genes that they want to display. In the gene sub-network panel, users can click and drag each gene node to move it, or click each gene ID to redirect to its detail page. The network is also displayed as a table on the right-hand side with a search function. Users can sort the table by column.
Fig. 3.
Screenshots of the browse and detail information pages. a The Browse page. b Detailed gene functional annotations. c Gene structural information. d Gene expression heatmap. e Gene co-expression network and PPI network
Database statistics
Statistics for JCDB are summarized in Table 1. The current database release contains a total of 25,297 genes and 33,785 transcripts, including protein-coding genes (22,446, about 89%), non-coding genes (2391, about 9%), and TUCP genes (460, about 2%). Compared with existing J. curcas databases [13, 15, 32], JCDB includes more non-coding genes and more annotation information, as well as unique gene networks and expression profiles (Table 2). In JCDB, about 58, 40, and 74% of genes have GO, KEGG, and Pfam annotations, respectively; there are also about 90% genes in the co-expression network, 38% genes in the PPI network, and 114 expression profiles for 25,297 genes. Users can freely download all the above annotation files via the Download page.
Table 1.
Gene statistics and data integrated in JCDB
| Category | Number |
|---|---|
| Genes/transcripts | |
| All | 25,297/33,785 |
| Protein-coding | 22,446 (89%) |
| Non-coding | 2391 (9%) |
| TUCP | 460 (2%) |
| Gene annotation | |
| Gene ontology | 14,714 (58%) |
| KEGG pathway | 10,217 (40%) |
| Pfam domain | 18,829 (74%) |
| Genes in network | |
| Co-expression network | 22,749 (90%) |
| PPI network | 9602 (38%) |
| Expression profiles | 114 |
Table 2.
Comparison of gene annotations in JCDB with other Jatropha databases
Case studies
JCDB provides a comprehensive platform for J. curcas functional genomics research by integrating information from various sources, including gene functional annotations and gene interaction networks, and various tools including BLAST search and gene network analysis. Here, we demonstrate the use of the information and tools provided by JCDB to mine some important gene pathways in J. curcas.
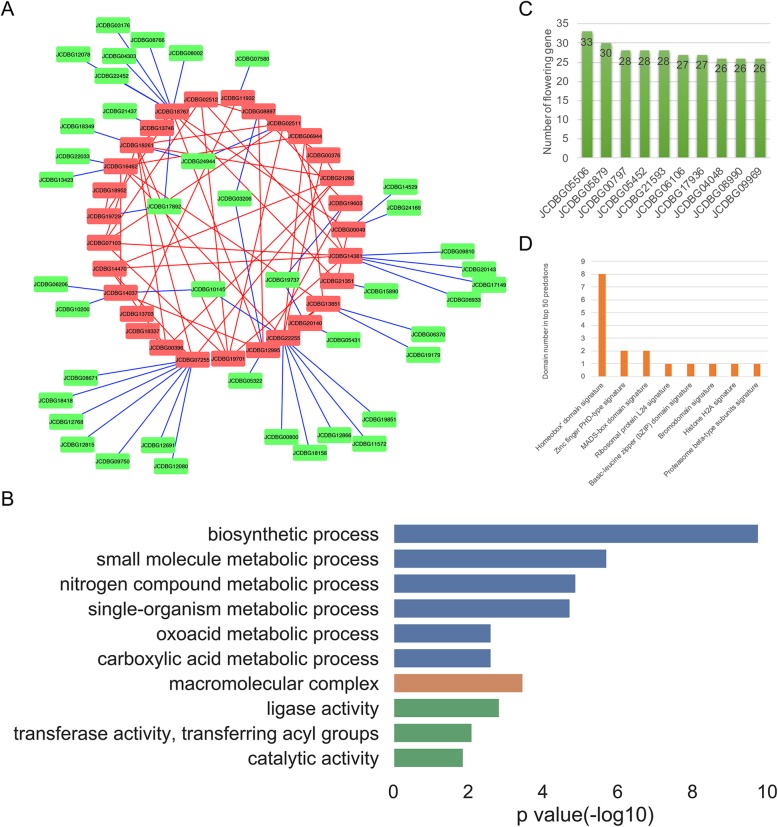
In order to better understand the genetic control of fatty acid and lipid biosynthesis in J. curcas, we collected 132 oil-related genes from Arabidopsis and identified oil-related gene candidates in J. curcas using the JCDB BLAST search. Using the ‘Network Construction’ function in JCDBTools, we obtained a J. curcas oil-related gene sub-network, which showed that these J. curcas oil-related genes were closely connected (Fig. 4a). We also used the ‘Neighbor Gene Extraction’ function in JCDBTools to find J. curcas-specific oil-related genes. We first extracted all the nearest neighbors of the known oil-related genes and then retained those that interacted with known oil-related genes in both the PPI and co-expression networks. We examined the GO annotations of these J. curcas specific oil-related gene candidates using GOATOOLS [56] (Fig. 4b). Consistent with our assumption, these genes appeared to be related to oil synthesis. The top enriched GO terms for biological process (BP) included biosynthetic process, small molecule metabolic process, and oxoacid and carboxylic acid metabolic process; the top cellular component (CC) term was macromolecular complex; and the top molecular function (MF) terms were ligase activity, transferase activity, transferring acyl groups, and catalytic activity.
Fig. 4.
Case studies: gene function prediction using JCDBTools. a Sub-network of oil-related genes in J. curcas (red: known, green: prediction). b GO enrichment analysis of predicted oil-related genes (blue: BP, orange: CC, green: MF). c Numbers of known flowering-related genes interacting with predicted flowering-related genes (top 10). d Protein domain information for the top 50 predicted flowering-related genes
We also investigated the flowering-related pathway in J. curcas. By manually reviewing the published literature, we identified 303 flowering-related genes of Arabidopsis. Then, using the same method, a total of 187 flowering-related genes in J. curcas were identified through homologous search, and the nearest neighbors and sub-network of these known flowering-related genes were also obtained. In the sub-network, the J. curcas-specific flowering-related gene candidates were closely connected with the known flowering-related genes. All the top 10 candidates had more than 25 interactions, including JCDBG05506 (Fig. 4c). Searching for this gene in JCDB revealed that JCDBG05506 is a MADS-box protein, with annotations including “FLOWERING LOCUS C” and “transcription factor”. Furthermore, we counted the protein domain annotations of the top 50 J. curcas-specific flowering-related gene candidates and found eight genes containing a homeobox domain, as well as two genes containing the zinc finger PHD-type domain and two genes containing the MADS-box domain (Fig. 4d). All of these protein domains are reported to be related to flowering [56–58].
Conclusions
The plant J. curcas has attracted much attention worldwide owing to its potential for biofuel production. However, current databases for J. curcas did not effectively integrate multiple data sources and lacked useful tools for data presentation and analysis, and thus could not meet the needs of functional genomics study. For these reasons, we built JCDB, an integrated knowledge base, which includes not only basic gene information but also gene functional annotations, gene expression profiles, and gene network information. JCDB also provides a user-friendly platform for data presentation and analysis, offering a variety of tools including BLAST, the JBrowse genome browser, and JCDBTools. JCDB is the most comprehensive and well-annotated database available for J. curcas functional genomics research. Future work will include developing new tools to assist users with in-depth exploration of JCDB. We believe JCDB will continue to provide a valuable and unique resource for J. curcas functional genomics studies.
Supplementary information
Additional file 1. The transcriptome data sources of Jatropha curcas.
Additional file 2. The entity relationship diagram of JCDB.
Acknowledgments
Data analysis was supported by the HPC Platform, The Public Technology Service Center of XTBG, CAS, China.
About this supplement
This article has been published as part of BMC Genomics, Volume 20 Supplement 9, 2019: 18th International Conference on Bioinformatics. The full contents of the supplement are available at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-20-supplement-9.
Abbreviations
- BP
Biological process
- CC
Cellular component
- GO
Gene ontology
- JCDB
Jatropha curcas database
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNA
Long non-coding RNA
- MF
Molecular function
- ORFs
Open reading frames
- PPI
Protein-protein interaction
- SRA
Sequence Read Archive
- TUCPs
Transcripts of unknown coding potential
Authors’ contributions
CL and ZX conceived, designed, and supervised this study. XZ, BP, and MC collected and compiled data from the literature and from public databases. XZ, WC, and JL designed and developed the database. XZ, JL, and CL prepared the draft of the manuscript. All authors reviewed, edited, and approved the manuscript.
Funding
Publication of this supplement was funded by the National Natural Science Foundation of China (No. 31970609, 31670612, 31870291 and 31370595), a Start-up Fund from Xishuangbanna Tropical Botanical Garden (XTBG), the Programme of the Chinese Academy of Sciences (kfj-brsn-2018-6-008 and 2017XTBG-T02) and the ‘Top Talents Program in Science and Technology’ of Yunnan Province.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuan Zhang and Bang-Zhen Pan contributed equally to this work.
Contributor Information
Xuan Zhang, Email: zhangxuan@xtbg.ac.cn.
Bang-Zhen Pan, Email: pbz@xtbg.org.cn.
Maosheng Chen, Email: chenms@xtbg.org.cn.
Wen Chen, Email: chenwen@biochen.com.
Jing Li, Email: lijing3@xtbg.ac.cn.
Zeng-Fu Xu, Email: zfxu@xtbg.ac.cn.
Changning Liu, Email: liuchangning@xtbg.ac.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6356-z.
References
- 1.Mazumdar P, Singh P, Babu S, Siva R, Harikrishna JA. An update on biological advancement of Jatropha curcas L.: new insight and challenges. Renew Sust Energ Rev. 2018;91:903–917. doi: 10.1016/j.rser.2018.04.082. [DOI] [Google Scholar]
- 2.Giwa A, Adeyemi I, Dindi A, Lopez CG-B, Lopresto CG, Curcio S, Chakraborty S. Techno-economic assessment of the sustainability of an integrated biorefinery from microalgae and Jatropha: a review and case study. Renew Sust Energ Rev. 2018;88:239–257. doi: 10.1016/j.rser.2018.02.032. [DOI] [Google Scholar]
- 3.Laviola BG, Rodrigues EV, Teodoro PE, Peixoto LA, Bhering LL. Biometric and biotechnology strategies in Jatropha genetic breeding for biodiesel production. Renew Sust Energ Rev. 2017;76:894–904. doi: 10.1016/j.rser.2017.03.116. [DOI] [Google Scholar]
- 4.Moniruzzaman M, Yaakob Z, Khatun R. Biotechnology for Jatropha improvement: a worthy exploration. Renew Sust Energ Rev. 2016;54:1262–1277. doi: 10.1016/j.rser.2015.10.074. [DOI] [Google Scholar]
- 5.Montes JM, Melchinger AE. Domestication and breeding of Jatropha curcas L. Trends Plant Sci. 2016;21(12):1045–1057. doi: 10.1016/j.tplants.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Abdelgadir HA, Van Staden J. Ethnobotany, ethnopharmacology and toxicity of Jatropha curcas L. (Euphorbiaceae): a review. S Afr J Bot. 2013;88:204–218. doi: 10.1016/j.sajb.2013.07.021. [DOI] [Google Scholar]
- 7.Maghuly F, Laimer M. Jatropha curcas, a biofuel crop: functional genomics for understanding metabolic pathways and genetic improvement. Biotechnol J. 2013;8(10):1172–1182. doi: 10.1002/biot.201300231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King AJ, He W, Cuevas JA, Freudenberger M, Ramiaramanana D, Graham IA. Potential of Jatropha curcas as a source of renewable oil and animal feed. J Exp Bot. 2009;60(10):2897–2905. doi: 10.1093/jxb/erp025. [DOI] [PubMed] [Google Scholar]
- 9.Islam AKMA, Yaakob Z, Anuar N. Jatropha: a multipurpose plant with considerable potential for the tropics. Sci Res Essays. 2011;6(13):2597–2605. [Google Scholar]
- 10.Ong HC, Mahlia TMI, Masjuki HH, Norhasyima RS. Comparison of palm oil, Jatropha curcas and Calophyllum inophyllum for biodiesel: a review. Renew Sust Energ Rev. 2011;15(8):3501–3515. doi: 10.1016/j.rser.2011.05.005. [DOI] [Google Scholar]
- 11.Kumar A, Sharma S. An evaluation of multipurpose oil seed crop for industrial uses (Jatropha curcas L.): a review. Ind Crop Prod. 2008;28(1):1–10. doi: 10.1016/j.indcrop.2008.01.001. [DOI] [Google Scholar]
- 12.Carvalho CR, Clarindo WR, Praça MM, Araújo FS, Carels N. Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci. 2008;174(6):613–617. doi: 10.1016/j.plantsci.2008.03.010. [DOI] [Google Scholar]
- 13.Sato S, Hirakawa H, Isobe S, Fukai E, Watanabe A, Kato M, Kawashima K, Minami C, Muraki A, Nakazaki N, et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18(1):65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirakawa H, Tsuchimoto S, Sakai H, Nakayama S, Fujishiro T, Kishida Y, Kohara M, Watanabe A, Yamada M, Aizu T, et al. Upgraded genomic information of Jatropha curcas L. Plant Biotechnol. 2012;29(2):123–130. doi: 10.5511/plantbiotechnology.12.0515a. [DOI] [Google Scholar]
- 15.Wu P, Zhou C, Cheng S, Wu Z, Lu W, Han J, Chen Y, Chen Y, Ni P, Wang Y, et al. Integrated genome sequence and linkage map of physic nut (Jatropha curcas L.), a biodiesel plant. Plant J. 2015;81(5):810–821. doi: 10.1111/tpj.12761. [DOI] [PubMed] [Google Scholar]
- 16.Ha J, Shim S, Lee T, Kang YJ, Hwang WJ, Jeong H, Laosatit K, Lee J, Kim SK, Satyawan D, et al. Genome sequence of Jatropha curcas L., a non-edible biodiesel plant, provides a resource to improve seed-related traits. Plant Biotechnol J. 2019;17(2):517–530. doi: 10.1111/pbi.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kancharla N, Jalali S, Narasimham JV, Nair V, Yepuri V, Thakkar B, Reddy VB, Kuriakose B, N, S A M. De Novo Sequencing and Hybrid Assembly of the Biofuel Crop Jatropha curcas L.: Identification of Quantitative Trait Loci for Geminivirus Resistance. Genes (Basel). 2019;10(1):96. [DOI] [PMC free article] [PubMed]
- 18.Qin X, Zheng X, Huang X, Lii Y, Shao C, Xu Y, Chen F. A novel transcription factor JcNAC1 response to stress in new model woody plant Jatropha curcas. Planta. 2014;239(2):511–520. doi: 10.1007/s00425-013-1993-y. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y, Yin Z, Ye J. Lipid biosynthesis and regulation in Jatropha, an emerging model for woody energy plants. 2017. pp. 113–127. [Google Scholar]
- 20.Tsuchimoto S, editor. The Jatropha genome: Volume 1st ed. 2017. Osaka University: Springer Japan; 2017.
- 21.Fresnedo-Ramirez J. The floral biology of Jatropha curcas L.-a review. Trop Plant Biol. 2013;6(1):1–15. doi: 10.1007/s12042-012-9113-x. [DOI] [Google Scholar]
- 22.Chen MS, Pan BZ, Fu Q, Tao YB, Martinez-Herrera J, Niu L, Ni J, Dong Y, Zhao ML, Xu ZF. Comparative transcriptome analysis between gynoecious and monoecious plants identifies regulatory networks controlling sex determination in Jatropha curcas. Front Plant Sci. 2016;7:1953. doi: 10.3389/fpls.2016.01953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Wu P, Zhang S, Song C, Chen Y, Li M, Jia Y, Fang X, Chen F, Wu G. Global analysis of gene expression profiles in developing physic nut (Jatropha curcas L.) seeds. PLoS One. 2012;7(5):e36522. doi: 10.1371/journal.pone.0036522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Zou Z, Wang S, Gong M. Global analysis of transcriptome responses and gene expression profiles to cold stress of Jatropha curcas L. PLoS One. 2013;8(12):e82817. doi: 10.1371/journal.pone.0082817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juntawong P, Sirikhachornkit A, Pimjan R, Sonthirod C, Sangsrakru D, Yoocha T, Tangphatsornruang S, Srinives P. Elucidation of the molecular responses to waterlogging in Jatropha roots by transcriptome profiling. Front Plant Sci. 2014;5:658. doi: 10.3389/fpls.2014.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Zhang C, Wu P, Chen Y, Li M, Jiang H, Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to salt stress. PLoS One. 2014;9(5):e97878. doi: 10.1371/journal.pone.0097878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C, Zhang L, Zhang S, Zhu S, Wu P, Chen Y, Li M, Jiang H, Wu G. Global analysis of gene expression profiles in physic nut (Jatropha curcas L.) seedlings exposed to drought stress. BMC Plant Biol. 2015;15:17. doi: 10.1186/s12870-014-0397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapeta H, Lourenco T, Lorenz S, Grumaz C, Kirstahler P, Barros PM, Costa JM, Sohn K, Oliveira MM. Transcriptomics and physiological analyses reveal co-ordinated alteration of metabolic pathways in Jatropha curcas drought tolerance. J Exp Bot. 2016;67(3):845–860. doi: 10.1093/jxb/erv499. [DOI] [PubMed] [Google Scholar]
- 29.Ni J, Gao C, Chen MS, Pan BZ, Ye K, Xu ZF. Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2015;56(8):1655–1666. doi: 10.1093/pcp/pcv089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan BZ, Chen MS, Ni J, Xu ZF. Transcriptome of the inflorescence meristems of the biofuel plant Jatropha curcas treated with cytokinin. BMC Genomics. 2014;15:974. doi: 10.1186/1471-2164-15-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang M, Wu Y, Jin S, Hou J, Mao Y, Liu W, Shen Y, Wu L. Flower bud transcriptome analysis of Sapium sebiferum (Linn.) Roxb. and primary investigation of drought induced flowering: pathway construction and G-quadruplex prediction based on transcriptome. PLoS One. 2015;10(3):e0118479. doi: 10.1371/journal.pone.0118479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakurai N, Ogata Y, Ara T, Sano R, Akimoto N, Hiruta A, Suzuki H, Kajikawa M, Widyastuti U, Suharsono S, et al. Development of KaPPA-View4 for omics studies on Jatropha and a database system KaPPA-loader for construction of local omics databases. Plant Biotechnol. 2012;29(2):131–135. doi: 10.5511/plantbiotechnology.12.0508a. [DOI] [Google Scholar]
- 33.Leinonen R, Sugawara H, Shumway M. International nucleotide sequence database C: the sequence read archive. Nucleic Acids Res. 2011;39(Database issue):D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35(Web Server issue):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41(17):e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(Database issue):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 42.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi X, Zhang Z, Ling Y, Xu W, Su Z. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015;43(Database issue):D982–D989. doi: 10.1093/nar/gku1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arabidopsis Interactome Mapping C Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333(6042):601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333(6042):596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones AM, Xuan Y, Xu M, Wang RS, Ho CH, Lalonde S, You CH, Sardi MI, Parsa SA, Smith-Valle E, et al. Border control--a membrane-linked interactome of Arabidopsis. Science. 2014;344(6185):711–716. doi: 10.1126/science.1251358. [DOI] [PubMed] [Google Scholar]
- 49.Li P, Zang W, Li Y, Xu F, Wang J, Shi T. AtPID: the overall hierarchical functional protein interaction network interface and analytic platform for Arabidopsis. Nucleic Acids Res. 2011;39(Database issue):D1130–D1133. doi: 10.1093/nar/gkq959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brandao MM, Dantas LL, Silva-Filho MC. AtPIN: Arabidopsis thaliana protein interaction network. BMC Bioinformatics. 2009;10:454. doi: 10.1186/1471-2105-10-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin M, Shen X, Chen X. PAIR: the predicted Arabidopsis interactome resource. Nucleic Acids Res. 2011;39(Database issue):D1134–D1140. doi: 10.1093/nar/gkq938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poole RL. The TAIR database. Methods Mol Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- 53.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38(Database issue):D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buels R, Yao E, Diesh CM, Hayes RD, Munoz-Torres M, Helt G, Goodstein DM, Elsik CG, Lewis SE, Stein L, et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng W, Nickle DC, Learn GH, Maust B, Mullins JI. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user’s datasets. Bioinformatics. 2007;23(17):2334–2336. doi: 10.1093/bioinformatics/btm331. [DOI] [PubMed] [Google Scholar]
- 56.Heijmans K, Morel P, Vandenbussche M. MADS-box genes and floral development: the dark side. J Exp Bot. 2012;63(15):5397–5404. doi: 10.1093/jxb/ers233. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto N, Okada K. A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 2001;15(24):3355–3364. doi: 10.1101/gad.931001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yokoyama Y, Kobayashi S, Kidou S-i. PHD type zinc finger protein PFP represses flowering by modulating FLC expression in Arabidopsis thaliana. Plant Growth Regul. 2019;88:49.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The transcriptome data sources of Jatropha curcas.
Additional file 2. The entity relationship diagram of JCDB.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.