Abstract
Multidrug resistant organisms (MDROs) are a serious threat to human health1,2. Fast, accurate antibiotic susceptibility testing (AST) is a critical need in addressing escalating antibiotic resistance, since delays in identifying MDROs increase mortality3,4 and use of broad-spectrum antibiotics, further selecting for resistant organisms. Yet current growth-based AST assays, such as broth microdilution5, require several days before informing key clinical decisions. Rapid AST would transform the care of infected patients while ensuring that our antibiotic arsenal is deployed as efficiently as possible. Growth-based assays are fundamentally constrained in speed by doubling time of the pathogen, and genotypic assays are limited by the ever-growing diversity and complexity of bacterial antibiotic resistance mechanisms. Here, we describe a rapid assay for combined Genotypic and Phenotypic AST through RNA detection, GoPhAST-R, that classifies strains with 94–99% accuracy by coupling machine learning analysis of early antibiotic-induced transcriptional changes with simultaneous detection of key genetic resistance determinants to increase accuracy of resistance detection, facilitate molecular epidemiology, and enable early detection of emerging resistance mechanisms. This two-pronged approach provides phenotypic AST 24–36 hours faster than standard workflows, with <4 hour assay time on a pilot instrument for hybridization-based multiplexed RNA detection implemented directly from positive blood cultures.
Current gold standard AST assays that measure growth in the presence of an antibiotic, while slow, directly answer the key question of whether the antibiotic inhibits pathogen growth. By contrast, newer genotypic approaches6 fall short of universal AST because of our incomplete knowledge of the innumerable resistance-causing genes and mutations across all pathogens and antibiotics, and the interactions of these genetic factors with diverse genomic backgrounds within any given bacterial species7–9. While the genomics revolution has undeniably transformed our understanding of antibiotic resistance10–12, as a clinical diagnostic, WGS remains technically demanding, costly, and slow; and the complexity, variability, and continuing evolution of bacterial genomes under ongoing antibiotic exposure pose serious challenges to predicting susceptibility accurately enough to direct patient care9,13,14. These shortcomings have motivated several novel approaches that focus on faster phenotypic AST, including rapid automated microscopy15, ultrafine mass measurements16, and others17–19.
Among current MDROs, carbapenem resistant organisms are the most alarming, as their resistance to this broad-spectrum antibiotic class often leaves few to no treatment options20. Yet phenotypic carbapenem resistance detection can be challenging, as some carbapenemase-producing strains may be mistakenly identified as susceptible by current phenotypic assays21 while failing clinical carbapenem therapy22. Newer multiplexed PCR assays can detect several common carbapenemases in carbapenem-resistant Enterobactericeae (CRE)6, yet these genotypic approaches miss a significant fraction of CRE isolates (13–68%) with unknown or non-carbapenemase resistance mechanisms8,23. For non-Enterobacteriaceae, these alternative genetic resistance mechanisms account for the vast majority of resistance; just 1.9% of over 1000 carbapenem-resistant Pseudomonas surveyed in 2017 by the US Centers for Disease Control (CDC) harbored known carbapenemases23. These challenges have left clinical microbiology laboratories still seeking consensus on how to best apply the multiple possible workflows for detecting carbapenem resistance24,25, including phenotypic26, genetic24, and biochemical25 assays.
GoPhAST-R is a novel diagnostic approach that can detect both genotype and phenotype in a single assay, allowing integration of all information and simultaneously informing both resistance prediction and molecular epidemiology. GoPhAST-R detects specific mRNA expression signatures in bacteria after brief antibiotic exposure; susceptible cells that are stressed upon antibiotic exposure are transcriptionally distinct from resistant cells that are not, agnostic to resistance mechanism17. mRNA is uniquely informative in this regard, as it encodes genotypic information in its sequence and phenotypic information in its abundance: We show that multiplexed hybridization-based quantification of transcriptional responses within minutes of antibiotic exposure can distinguish susceptible from resistant organisms. We demonstrate this approach for three major antibiotic classes in common clinical use – fluoroquinolones, aminoglycosides, and carbapenems – in five pathogens with a propensity for multi-drug resistance through diverse mechanisms. We describe a generalizable process to extend this approach to any pathogen-antibiotic pair of interest, requiring only that an antibiotic elicit a differential transcriptional response in susceptible versus resistant isolates, a biological phenomenon that to date appears universal. For carbapenems, we incorporate simultaneous genotypic detection of key resistance determinants to improve accuracy of resistance detection and facilitate molecular epidemiology. Finally, we demonstrate GoPhAST-R directly on positive blood culture bottles, reporting phenotypic AST within hours of a positive culture. Together, this work establishes GoPhAST-R as a novel, accurate, rapid approach to AST that leverages the advantages of both phenotypic and genotypic assays.
To identify transcripts that robustly distinguish susceptible and resistant bacteria after brief antibiotic exposure, we used RNA-Seq to compare transcriptional timecourses of two susceptible and two resistant clinical isolates of K. pneumoniae, E. coli, and A. baumannii (Supplementary Table 1) treated with either meropenem (a carbapenem that inhibits cell wall biosynthesis), ciprofloxacin (a fluoroquinolone that targets DNA gyrase and topoisomerase), or gentamicin (an aminoglycoside that inhibits protein synthesis). Doses were chosen to match clinical breakpoint concentrations26, defined by the Clinical and Laboratory Standards Institute (CLSI) as the antibiotic concentration above which resistant strains (i.e. those at high risk of failing clinical therapy) can grow in broth microdilution assays. To enable these comparisons, we developed a library construction method optimized from RNAtag-Seq27 to include template switching, now termed RNAtag-Seq_TS, to dramatically decrease cost and increase throughput (see Supplementary Methods). For each pathogen, each antibiotic elicited a transcriptional response within 30–60 minutes in susceptible but not resistant organisms (Fig. 1a; Extended Data Figs. 1a and 2).
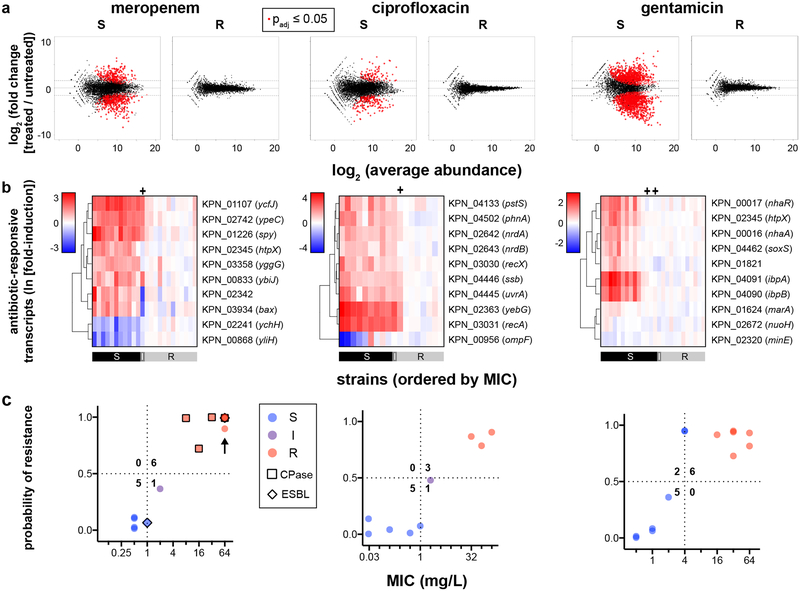
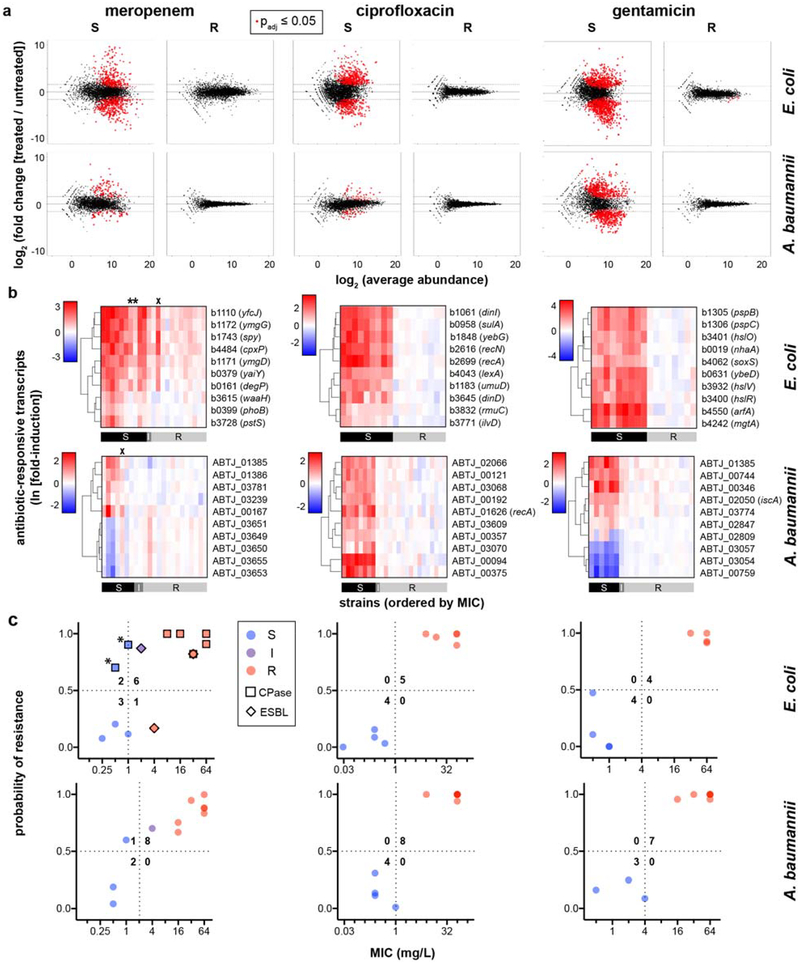
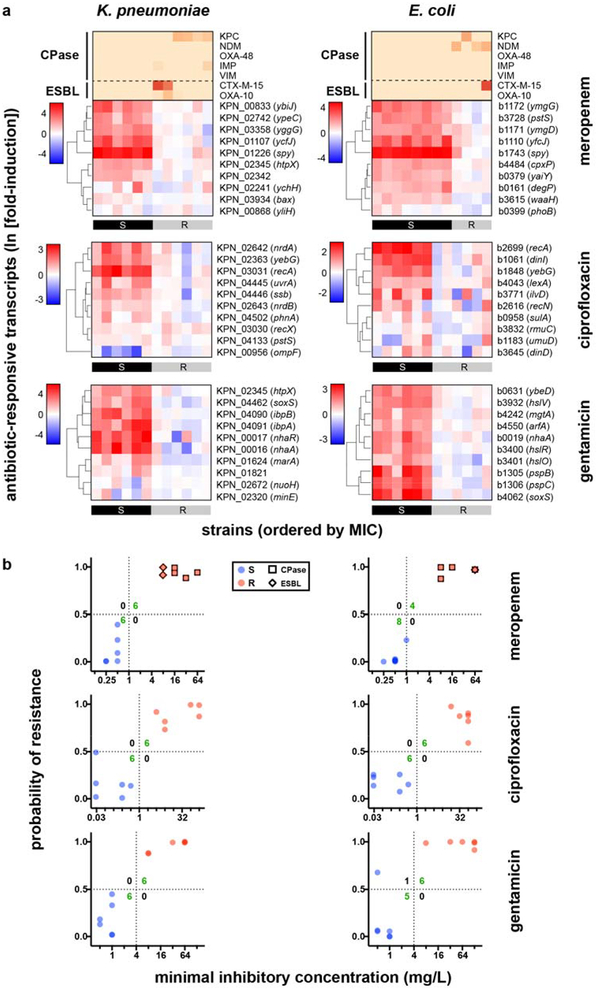
Fig. 1. Differential gene expression upon antibiotic exposure distinguishes susceptible and resistant strains.
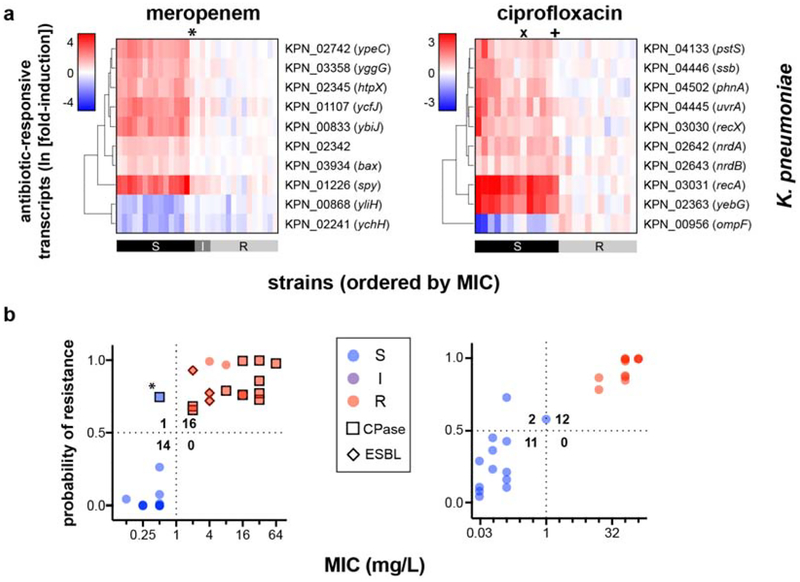
(a) RNA-Seq data from two susceptible (left panels) or two resistant (right panels) clinical isolates of K. pneumoniae treated with meropenem (60 min), ciprofloxacin (30 min), or gentamicin (60 min) at CLSI breakpoint concentrations are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package28. (b) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from K. pneumoniae treated at CLSI breakpoint concentrations with meropenem (left, 24 independent clinical isolates), ciprofloxacin (center, 18 independent clinical isolates), or gentamicin (right, 26 independent clinical isolates). Gene identifiers are listed at right, along with gene names if available. CLSI classifications of each strain based on broth microdilution are shown below. + = strains with one-dilution errors in classification. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on NanoString data from the derivation cohort and tested on the validation cohort (y-axis) are compared with standard CLSI classification based on broth microdilution MIC (x-axis) for K. pneumoniae isolates treated with meropenem, ciprofloxacin, and gentamicin. Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (i.e. intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on the meropenem plot. Arrow indicates a strain with high-level meropenem resistance, but no carbapenemase.
To identify transcripts that best distinguish susceptible from resistant strains for each pathogen-antibiotic combination, we initially selected 60–100 candidate antibiotic-responsive transcripts from these RNA-Seq datasets to evaluate in more clinical isolates. We used DESeq228 followed by Fisher’s combined probability test to identify transcripts whose expression changed more upon antibiotic treatment than under any phase of growth during the timecourse, thus enriching for genes directly affected by antibiotic exposure (see Supplementary Methods). Gene ontology enrichment analysis (Supplementary Table 2) revealed that meropenem affected lipopolysaccharide biosynthesis in both Enterobacteriaceae species, and induced a heat shock response in both E. coli and Acinetobacter. In all three species, ciprofloxacin induced the SOS response, while gentamicin induced the unfolded protein response and quinone binding. For normalization across samples, we also used DESeq2 to select 10–20 control transcripts for each pathogen-antibiotic pair that were most invariant to antibiotic treatment and growth phase (see Methods).
For each pathogen-antibiotic pair, we designed probesets for multiplexed detection of each candidate control and responsive transcript using NanoString, a simple, quantitative fluorescent hybridization platform that does not require nucleic acid purification or enzymology and thus works on crude lysates17,29. Each probeset comprised pairs of 50mer probes to conserved regions (see Methods) of the targeted transcripts (Supplementary Table 3). Using a protocol modified from the standard NanoString nCounter assay to accelerate detection (see Methods), we quantified these target transcripts in 18–24 diverse clinical isolates of each species collected from various geographic locations (Supplementary Table 1) and spanning the known phylogenetic landscape of each species (Extended Data Fig. 3). Because of a homology screening step in probe design (see Methods), each probe will uniquely hybridize to the target transcript from its cognate species, thereby enabling simultaneous species identification17. Normalized expression signatures of all responsive genes are shown as heatmaps (Extended Data Fig. 4) and summarized as one-dimensional projections (Extended Data Fig. 5).
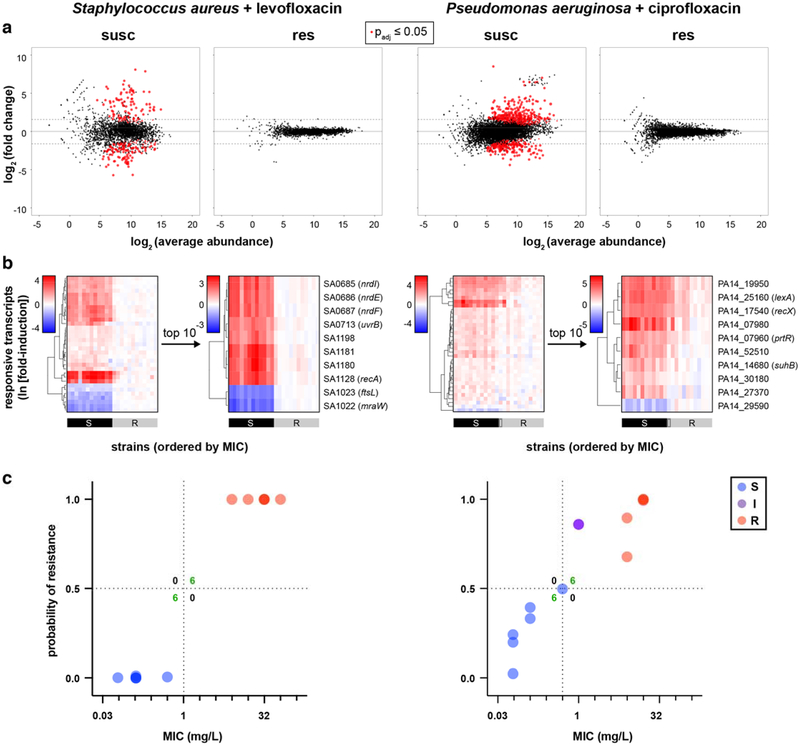
To further test the generalizability of this approach, we repeated these steps from RNA-Seq through NanoString detection of candidate responsive and control genes for two additional high-priority and frequently multidrug-resistant pathogens – S. aureus, a gram positive, and P. aeruginosa, another gram negative – each treated with a fluoroquinolone, levofloxacin (given its greater potency against gram positives) and ciprofloxacin, respectively (Extended Data Fig. 6). Each robustly induced the SOS response (Supplementary Table S2) in susceptible but not resistant clinical isolates.
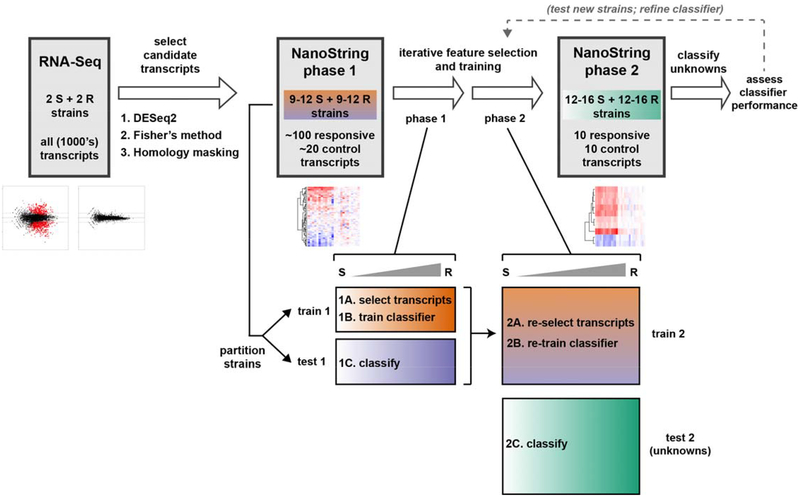
Importantly, expression signatures alone merely show that reliable differences occur in the transcriptional response in susceptible versus resistant organisms, while AST requires binary classification of a strain as susceptible or resistant. To address this general classification problem, we deployed machine-learning algorithms (Extended Data Fig. 7, Phase 1), first to identify the most informative transcripts, and then to use these select transcripts to classify unknown isolates. To avoid overtraining, we partitioned the tested strains into a training (derivation) cohort for both feature selection and classifier training, and a testing (validation) cohort as a naïve strain set for assessing classifier performance. We used reliefF to identify the 10 transcripts whose normalized expression best distinguished susceptible from resistant organisms among the training cohort (Fig. 1b; Extended Data Figs. 1b and 6b; Supplementary Table 3). Although fewer than 10 transcripts were required to robustly distinguish between the strains we have thus far tested, we opted to keep more genes in the refined signature to lessen the potential impact of unanticipated diversity in gene content, sequence, or regulation among clinical isolates.
We next trained an ensemble classifier using a random forest model for binary classification of isolates in the derivation cohort based solely on these selected features, then tested this trained classifier on the validation cohort. Across all 11 bacteria-antibiotic combinations, 109 isolates were used as derivation strains for training, and 108 isolates were tested as validation. The ensemble classifier correctly classified 100 of these 108 (93% categorical agreement, 95% confidence interval [CI] 87–96%), including 51 of 52 resistant isolates (1.9% very major error rate, 95% CI 0.21–8.6%) and 35 of 38 susceptible isolates (7.9% major error rate, 95% CI 2.3–20%), compared with standard broth microdilution (Fig. 1c, Extended Data Figs. 1c and 6c; Supplementary Table S4). Of note, these error rates are typically reported on a natural distribution of isolates. In contrast, for this study, we deliberately assembled a “challenge set” of strains, intentionally overrepresented for isolates near the clinical breakpoints, which will artificially inflate all errors, since discrepant classifications are more common for strains with minimal inhibitory concentrations (MICs) near the breakpoint – both due to possible errors in the assay and to one-dilution errors inherent in the gold standard broth microdilution assay30. Consistent with this, all major and very major errors in Phase 1 testing involved strains less than or equal to two dilutions away from the clinical breakpoint (Fig. 1c; Extended Data Figs. 1c and 6c).
To assess this approach to classification as it would be deployed on unknown isolates, and to further ensure against overtraining on the initial set of isolates, we performed a second, iterative round of training on all strains (from both derivation and validation cohorts) from Phase 1. We then tested a new set of K. pneumoniae isolates treated with meropenem and ciprofloxacin (Extended Data Fig. 7, Phase 2), this time measuring only the top 10 selected responsive transcripts (as we envision for the ultimate assay), rather than the 60–100 transcripts measured in Phase 1 (Extended Data Fig. 8a). Here, GoPhAST-R correctly classified 52 of 55 strains (95% categorical agreement, 95% CI 86–98%), including all 25 resistant isolates (0% very major error rate, 95% CI 0–9.5%) and 25 of 27 susceptible isolates (7.4% major error rate, 95% CI 1.6–22%), compared with broth microdilution (Extended Data Fig. 8b). All three discrepant isolates were less than or equal to two dilutions from the breakpoint.
Three isolates classified as meropenem-resistant by GoPhAST-R but susceptible by broth microdilution exhibited a large inoculum effect (ref). These three isolates, a K. pneumoniae (BAA2524; Fig. 1b–c) and two E. coli (BAA2523 and AR0104; Extended Data Fig. 1b–c), all had MICs of 0.5–1 mg/L on standard broth microdilution with an inoculum of 105 cfu/mL, but MICs of ≥32 mg/L with an inoculum of 107 cfu/mL. Each of these strains carried a carbapenemase gene: BAA2523 and BAA2524 contained blaOXA-48, and AR0104 contained blaKPC-4, as has been reported for other such strains with large inoculum effects31. While the clinical consequences of such large inoculum effects are uncertain, they may portend clinical failure, particularly in the setting of carbapenemase production22; detection of this phenomenon is a known gap in standard broth microdilution assays5,25,32. GoPhAST-R classified these strains as phenotypically resistant, perhaps because the assay is performed at higher cell density (>107 cfu/mL). Thus, the gold standard method likely misclassified these isolates as susceptible, while GoPhAST-R correctly recognized their resistance.
Importantly, classifier performance was independent of resistance mechanism, as exemplified for meropenem resistance. In total, 22 of 47 meropenem-resistant isolates, including 7 of 22 K. pneumoniae, 4 of 12 E. coli, and 11 of 13 A. baumannii, lacked carbapenemases (Supplementary Table 1), yet 46 of these 47 isolates were correctly recognized as resistant by GoPhAST-R. These results underscore the ability of GoPhAST-R to assess phenotypic resistance, agnostic to its genotypic basis.
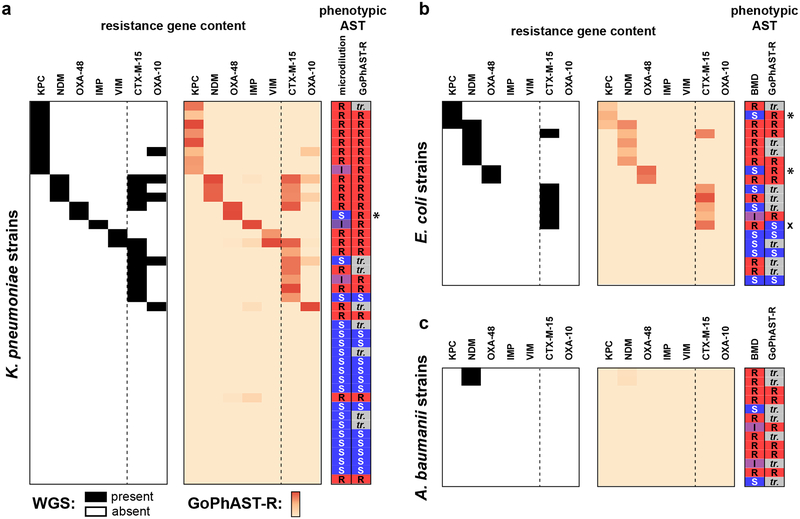
GoPhAST-R can readily accommodate simultaneous profiling of additional transcripts, including genetic resistance determinants such as carbapenemases. Such genotypic information can complement GoPhAST-R’s phenotype-based AST classification while providing valuable epidemiological data. For example, each of the three isolates with discrepant classifications and prominent inoculum effects carried a carbapenemase gene. Indeed, the most common known mechanism for carbapenem resistance among the Enterobacteriaceae involves acquisition of one of several carbapenemase genes23, most commonly the KPC, NDM, OXA-48, IMP, and VIM families33. We thus incorporated probes for these carbapenemases into the GoPhAST-R assay for meropenem AST, as well as two extended-spectrum beta-lactamase (ESBL) gene families that have been associated with carbapenem resistance when expressed in the context of porin loss-of-function, CTX-M-1534 and OXA-1035 (Supplementary Table 3).
GoPhAST-R correctly detected all 39 carbapenemase genes across 38 strains known to be present by WGS, including at least one member of each of the five targeted classes, and all 29 ESBL genes across 26 strains; no signal was detected in the 22 meropenem-resistant strains nor the 38 susceptible isolates known to lack these gene families, across all three species (Fig. 2). This included detection of OXA-48 or KPC in the three cases of discrepant phenotypic AST classification and prominent inoculum effects, reinforcing GoPhAST-R’s resistant phenotypic classification. Thus, in a single assay, GoPhAST-R can provide both phenotypic AST and genotypic information about resistance mechanism, which together result in the most accurate AST classification and valuable characterization of resistant isolates, including immediate recognition of unexplained resistance.
Fig. 2. GoPhAST-R detects carbapenemase and ESBL gene content from tested strains.
Known carbapenemase and select ESBL transcript content based on WGS data (left panels) are compared with heatmaps of GoPhAST-R results (right panels) for all (a) K. pneumoniae, (b) E. coli (, and (c) A. baumannii isolates tested for meropenem susceptibility for which WGS data was available (42, 20, and 13 independent clinical isolates, respectively). Heatmap intensity reflects normalized, background-subtracted, log-transformed NanoString data from probes for the indicated gene families. Vertical dashed line separates carbapenemases (left) from ESBL genes (right). Phenotypic AST classification by broth microdilution and GoPhAST-R is shown at right (“S” = susceptible, “I” = intermediate, “R” = resistant; “tr.” = strain used in training cohort, thus not classified by GoPhAST-R). * = strains with large inoculum effects in meropenem MIC; x = strain discordant by more than one dilution.
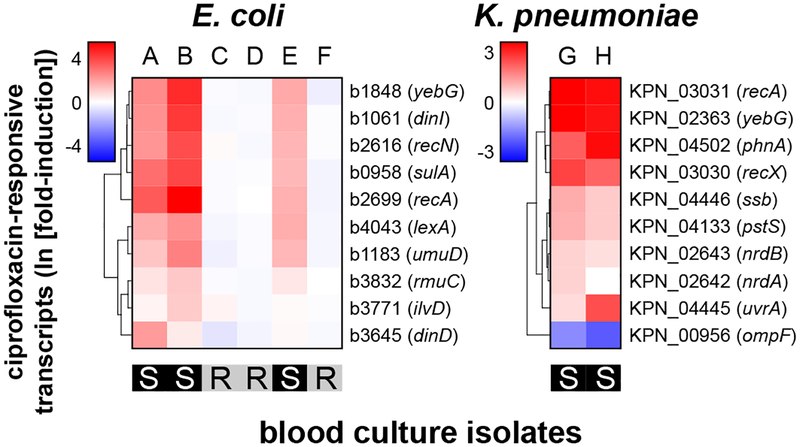
We previously demonstrated that a simulated positive blood culture bottle contains sufficient bacteria to permit mRNA detection36. To demonstrate one valuable clinical application, we used GoPhAST-R to rapidly determine ciprofloxacin susceptibility in blood culture bottles that grew gram-negative rods from the clinical microbiology laboratory at Massachusetts General Hospital (MGH). Ciprofloxacin was chosen because no rapid genotypic method exists for detection of fluoroquinolone resistance due to the diversity of genetic causes, and because the prevalence of fluoroquinolone resistance facilitated acquisition of both sensitive and resistant cases. Of six E. coli and two K. pneumoniae clinical positive blood cultures (Fig. 3), GoPhAST-R clearly distinguished three susceptible from three resistant E. coli; both K. pneumoniae species were susceptible. Given the relative scarcity of gentamicin and meropenem resistance at our institution, in order to test assay performance in this growth format, we generated simulated positive blood cultures by spiking in susceptible or resistant isolates of K. pneumoniae and E. coli. GoPhAST-R detected transcriptional signatures for each pathogen-antibiotic pair directly from these positive blood culture bottles, and blinded AST prediction using a random forest model and leave-one-out cross-validation correctly classified 71 of 72 blood cultures (99% categorical agreement with broth microdilution, 95% CI 94–100%), including 31 of 31 resistant isolates (0% very major error rate; 95% CI 0–7.7%) and 37 of 38 susceptible isolates (2.6% major error rate; 95% CI 0.29–11%), compared with broth microdilution (Extended Data Fig. 9).
Fig. 3. GoPhAST-R detects antibiotic-responsive transcripts directly from positive blood culture bottles.
Heatmaps of normalized, log-transformed fold-induction of the top 10 ciprofloxacin-responsive transcripts from 8 positive blood culture bottles that grew either E. coli (6 independent bottles, A-F) or K. pneumoniae (2 independent bottles, G-H). CLSI classifications of isolates performed by the clinical microbiology laboratory, which were blinded until analysis was complete, are displayed below each heatmap.
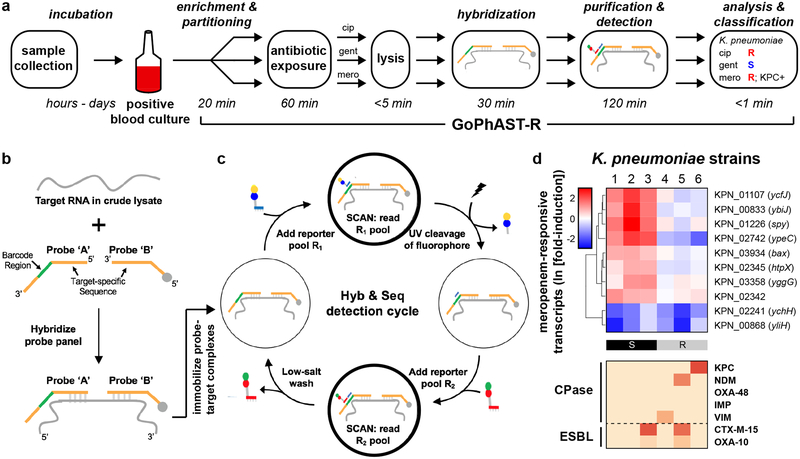
To decrease the time to answer, we deployed GoPhAST-R on a next-generation nucleic acid detection platform, NanoString Hyb & Seq™ (J.B., AGBT Precision Health 2017, unpublished abstract), whose accelerated detection technology enables AST in <4 hours from the time a blood culture turns positive (Fig. 4a). Relative to the nCounter detection platform, Hyb & Seq (Fig. 4b) accelerates hybridization by utilizing unlabeled reporter probes that are far smaller and thus equilibrate much faster than standard nCounter probes, which contain bulky fluorophores. Accelerated optical scanning rapidly quantifies these smaller reporter probes via sequential cycles of binding, detection, and removal of complementary barcoded fluorophores (Fig. 4c; see Methods). On a prototype Hyb & Seq instrument, GoPhAST-R measured meropenem susceptibility signatures and carbapenemase content in <4 hours (Fig. 4d). A head-to-head time trial on simulated blood culture bottles demonstrated GoPhAST-R results in <4 hours from the time of culture positivity, compared with 28–40 hours in the MGH clinical microbiology laboratory by standard methods (subculture followed by AST on a VITEK-2 instrument).
Fig. 4. GoPhAST-R workflow with the NanoString Hyb & Seq™ platform distinguishes phenotypically susceptible from resistant strains and detects genetic resistance determinants in <4 hours.
(a) The GoPhAST-R workflow on the Hyb & Seq detection platform begins once growth is detected in a blood culture bottle. Pathogen identification could either be done prior to this process, or in parallel by multiplexing mRNA targets from multiple organisms (see Supplementary Text). (b) Hyb & Seq hybridization scheme: probe pairs targeting each RNA transcript are hybridized in crude lysate. Each probe A contains a unique barcode sequence (green) for detection and a shared 3’ capture sequence; each probe B contains a biotin group (gray circle) for surface immobilization and a shared 5’ capture sequence. (c) Hyb & Seq detection strategy: immobilized, purified ternary probe-target complexes undergo sequential cycles of multi-step imaging for spatially resolved single-molecule detection. Each cycle consists of reporter probe binding and detection, UV cleavage, a second round of reporter probe binding and detection, and a low-salt wash to regenerate the unbound probe-target complex. 5 Hyb & Seq cycles were used to generate the data shown. See Supplemental Methods for details. (d) Pilot studies for accelerated meropenem susceptibility testing of 6 clinical K. pneumoniae isolates. Above: heatmaps of normalized, log-transformed fold-induction of top 10 meropenem-responsive transcripts measured using this Hyb & Seq workflow, with strains arranged in order of MIC for each antibiotic. CLSI classifications are shown below. Below: heatmaps of normalized, background-subtracted, log-transformed NanoString data from carbapenemase (“CPase”) and select ESBL transcripts measured in the same Hyb & Seq assay.
In summary, by quantifying a refined set of transcripts whose antibiotic-induced expression reflects susceptibility, GoPhAST-R provides a conceptually distinct approach to rapid phenotypic AST, agnostic to resistance mechanism and extendable to any pathogen and antibiotic class, while simultaneously providing select, complementary genotypic information. The machine learning approach to strain classification developed for GoPhAST-R provides actionable AST information in excellent categorical agreement with the gold standard broth microdilution assay and should continue to improve in accuracy as it is trained on additional strains. Omitting three carbapenemase-producing strains with ambiguous and likely errant susceptible classification by the gold standard assay, GoPhAST-R correctly classified 100 of 106 strains (94%) in Phase 1, 52 of 54 strains (96%) in Phase 2, and 71 of 72 (99%) simulated blood cultures, with 8 of the 9 discrepancies occurring on strains within two dilutions of the clinical breakpoint.
By integrating genotypic and phenotypic information in a single rapid multiplexed RNA detection assay, GoPhAST-R offers several advantages over the current gold standard that are unique among other rapid AST assays under development. First, like other phenotypic assays, it determines susceptibility agnostic to mechanism of resistance, a clear advantage over genotypic AST assays. Second, combining genotypic and phenotypic information enhances AST accuracy over conventional growth-based methods, improving sensitivity of resistance detection in carbapenemase-producing Enterobacteriaceae that test susceptible by standard methods but may exhibit resistance upon treatment21,22,25. Third, identification of carbapenem resistance determinants can inform antibiotic choice, as certain novel beta-lactamase inhibitors like avibactam or vaborbactam will overcome some classes of carbapenemases (e.g., KPC) but not others (e.g., metallo-beta-lactamases such as the NDM class)37,38. Solely phenotypic assays would require additional, serial testing to provide such guidance. Fourth, detecting resistance determinants together with a phenotypic assay enables molecular epidemiology for local, regional, national, or global tracking of the emergence and spread of resistance, without requiring additional testing. We demonstrate this advantage for one major class of high-value resistance determinants, the carbapenemases; this combined approach can readily extend to other critical emerging resistance determinants such as mcr genes, plasmid-borne colistin resistance determinants recently found in Enterobacteriaceae39, or key virulence factors such as Shiga toxin, in seamless conjunction with a phenotypic AST assay. Fifth, strains with unknown resistance mechanisms, such as CREs without carbapenemases (see Fig. 1 and Extended Data Fig. 8), can be immediately identified from a single assay and flagged for further study such as WGS if desired.
This work represents an important demonstration of this new approach to AST, enabling rapid, accurate classification across a genetically diverse sampling (Extended Data Fig. 2) of five high-priority pathogens and three major antibiotic classes. Still, further development will be needed for translation to clinical practice. With wider testing, while the specific classifiers will improve, the general strategy and approach remains valid. Indeed, the capacity to learn through iterative retraining is one of the strengths of this approach as it is used more broadly. Likewise, extending this assay to more pathogen-antibiotic pairs will be critical for widespread clinical utility. Fortunately, the experimental and computational approach described here allows rapid and conceptually straightforward extension to all pathogens and antibiotic classes, including those with novel mechanisms of action and as-yet-unknown or newly emerging mechanisms of resistance. Underscoring the generalizability of this approach, we have generated preliminary RNA-Seq data for 50 additional pathogen-antibiotic pairs, spanning gram positive, gram negative, and mycobacteria, treated with bactericidal and bacteriostatic antibiotics, that demonstrate early differential transcriptional responses to antibiotics in all cases tested (Hung lab, unpublished results). Because GoPhAST-R is explicitly informed by MIC, it leverages decades of prior studies linking in vitro behavior to clinical outcomes26, facilitating extension to new pathogens or antibiotics. GoPhAST-R cannot, however, overcome all limitations of current diagnostics, including an initial delay for blood culture, driven by the low abundance of bacteria, and thus bacterial mRNA, in the bloodstream of infected patients. Still, considering the widespread adoption and clinical benefits40 of rapid pathogen identification by matrix-associated laser desorption and ionization / time-of-flight (MALDI-TOF) mass spectrometry in 2 hours from subcultured colonies streaked from blood culture bottles40, this comparatively more informative AST assay directly from blood culture bottles in <4 hours would be potentially transformative. We also anticipate the potential to expand GoPhAST-R to other clinical specimen types; some, like urine, contain enough bacteria without culture to enable direct mRNA profiling17. Additionally, characterizing mixed populations of susceptible and resistant strains within an individual sample remains challenging for GoPhAST-R, which integrates transcription across bulk populations, just as it is for existing gold-standard methods that undergo a single-colony bottleneck during subculture. Finally, GoPhAST-R cannot completely overcome the challenge of identifying delayed inducible resistance, though this would be true for any rapid phenotypic test. In fact, GoPhAST-R may accurately identify at least some of these cases through simultaneous genotypic detection of induced resistance determinants, if known.
We have herein demonstrated the application of a next-generation nucleic acid detection platform that can return AST answers in <4 hours. We have shown that the strategy is generalizable across a number of bacterial species and antibiotic classes, and will likely be widely applicable based on its conserved underlying biological principle. Notably, a reliable transcriptional signature of susceptibility is present in ≤1 hour for each of these key antibiotic classes. Thus, as RNA detection methods become faster and more sensitive, GoPhAST-R has the potential for even faster phenotypic AST on timescales that can inform early antibiotic decisions and thus transform infectious disease practice.
Methods
Strain acquisition and characterization:
All strains in this study (Supplementary Table 1) were obtained from clinical or reference microbiological laboratories, including both local hospitals and MDRO strain collections from the CDC’s Antibiotic Resistance Isolate Bank (https://wwwn.cdc.gov/ARIsolateBank/) and the New York State Department of Health. MICs reported from those laboratories were validated by standard broth microdilution assays5 in Mueller-Hinton broth (Difco); any discrepancies of >1 doubling from reported values were resolved by repeating in triplicate. Partners Health Care IRB 2015P002215 approved strain collection under waiver of patient consent, since bacterial isolates were obtained from clinical microbiology laboratories without any human material. Strains for which WGS data was available at the National Center for Biotechnology Information (NCBI) were visualized using the Interactive Tree of Life41.
RNA-Seq experimental conditions
For each bacteria-antibiotic pair, selected independent clinical isolates (Supplementary Table 1), two susceptible and two resistant, were grown at 37°C in Mueller-Hinton broth to early logarithmic phase, then treated with the relevant antibiotic at breakpoint concentrations set by the Clinical Laboratory Standards Institute (CLSI)26: 2 mg/L for meropenem, 1 mg/L for ciprofloxacin, and 4 mg/L for gentamicin. Total RNA was harvested from paired treated and untreated samples at 0, 10, 30, and 60 minutes. cDNA libraries were made using a variant of the RNAtag-Seq protocol we previously described27 and sequenced on either an Illumina HiSeq or NextSeq. Sequencing reads were aligned using BWA42 and tabulated as previously described27. See Supplementary Methods for further details.
Differential gene expression analysis and selection of responsive and control transcripts
Differentially expressed genes were determined using the DESeq2 package28, comparing treated vs untreated samples at each timepoint using a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing. Fisher’s combined probability test was used to select only those genes whose expression after antibiotic treatment is statistically distinguishable from its expression at any timepoint in the untreated samples. Gene ontology (GO) terms were assigned using blast2GO43 (version 1.4.4), with hypergeometric testing for enrichment. For each pathogen-antibiotic pair, the fold-change threshold in DESeq2 used to test statistical significance was increased to select 60–100 antibiotic-responsive transcripts with maximal stringency, a number readily accommodated by the NanoString assay format. Control transcripts were also determined with DESeq2 using an inverted hypothesis test as described28 to select genes whose expression we expect to be unaffected by antibiotic exposure or growth in both susceptible and resistant isolates, at all timepoints and treatment conditions. As with responsive genes, the fold-change threshold was varied in order to select the top 10–20 control transcripts. The resulting control and responsive gene lists for each pathogen-antibiotic pair, and the fold-change thresholds used to generate them, are shown in Supplementary Table 3. See Supplementary Methods for further details.
Targeted transcriptional response to antibiotic exposure
After using BLASTn to identify regions of targeted transcripts with maximal conservation across all RefSeq genomes from that species (see Supplemental Methods), NanoString probes were designed per manufacturer’s standard process29 to these conserved regions. Strains treated with antibiotic at the CLSI breakpoint concentration, and untreated controls, were lysed via bead-beating at the desired timepoint. The resulting crude lysates were used as input for standard NanoString (Seattle, WA) assays, which were performed on the nCounter® Sprint platform with variations on the manufacturer’s protocol to enhance speed, detailed in Supplementary Methods. Raw counts for each target were extracted and processed as described in Supplementary Methods. Briefly, for each sample, each responsive gene was normalized by control gene expression as a proxy for cell loading using a variation on the geNorm algorithm44, then converted to fold-induction in treated compared with untreated strains. Pilot NanoString Hyb & Seq™ assays (Fig. 4) were performed on a prototype Hyb&Seq instrument at NanoString, with 20 minute hybridization time and 5 imaging cycles to detect hybridization probes with two-segment 10-plex barcodes. See Supplementary Methods for more details.
Machine learning: feature selection and susceptibility classification
For each pathogen-antibiotic pair, we first partitioned the normalized data, grouping half the strains into a derivation cohort on which we trained our algorithm, reserving the other half for validation (Extended Data Fig. 5), ensuring equivalent representation of susceptible and resistant isolates in each cohort. For comparison of GoPhAST-R classifications with gold-standard results, confidence intervals for accuracy estimates were calculated using Jeffrey’s interval45.
In Phase 1, implemented for all pathogen-antibiotic pairs, normalized fold-induction data of responsive genes from strains in the training cohort, along with CLSI susceptibility classification for each training strain, were input to the ReliefF algorithm46 using the CORElearn package (version 1.52.0) to rank the top 10 responsive transcripts that best distinguished susceptible from resistant strains. These 10 features were then used to train a random forest classifier47 using the caret package (version 6.0–78) in R (version 3.3.3) on the same training strains. Performance of this classifier was then assessed on the testing cohort, to which the classifier had yet to be exposed.
In Phase 2, implemented for K. pneumoniae + meropenem and ciprofloxacin, all 18–24 strains from Phase 1 were combined into a single, larger training set. For each antibiotic, ReliefF was again used to select the 10 most informative responsive transcripts, which were then used to train a random forest classifier on the same larger training set. Transcriptional data were then collected on a test set of 25–30 new strains using a trimmed NanoString nCounter® Elements™ probeset containing only probes for these 10 selected transcripts, plus 8–13 control probes. Susceptibility of each strain in this test set was predicted using the trained classifier. See Supplemental Methods for further detail on machine learning strategy and implementation.
For classification of simulated blood cultures, NanoString data were collected for the top 10 transcripts (selected in Phase 1) from 12 strains for each pathogen-antibiotic pair, and analyzed using a leave-one-out cross-validation approach48, training on 11 strains and classifying the 12th, then repeating with each strain omitted once from training and used for prediction.
Blood culture processing
Under Partners Health Care IRB 2015P002215, bacteria were isolated from real or simulated blood cultures in the MGH clinical microbiology laboratory, isolated by differential centrifugation, resuspended in Mueller-Hinton broth, and immediately split for treatment with the indicated antibiotics. Lysis and targeted RNA detection were performed as above. Specimens were blinded until all data acquisition and analysis was complete. See Supplemental Methods for more detail. As approved by Partners IRB, samples were collected under waiver of patient consent due to experimental focus only on the bacterial isolates, not the patients from which they were derived.
Code availability
Custom scripts for transcript selection from RNA-Seq data are available at https://github.com/broadinstitute/GeneSelection/. Custom scripts for feature selection and strain classification from NanoString data are available at https://github.com/broadinstitute/DecisionAnalysis/. See Life Sciences Reporting Summary for additional detail.
Data availability
All RNA-Seq data generated and analyzed for this study, supporting the analyses in Fig. 1, and Extended Data Figs. 1, 2, and 5, have been deposited as aligned bam files in the NCBI Sequencing Read Archive under study ID PRJNA518730. All other datasets generated during the current study, including raw and processed NanoString data, are available from the corresponding author on reasonable request.
Extended Data
Extended Data Figure 1. Differential gene expression upon antibiotic exposure distinguishes susceptible and resistant strains.
(a) RNA-Seq data from two susceptible (left panels) or two resistant (right panels) clinical isolates of E. coli or A. baumannii treated with meropenem (60 min), ciprofloxacin (30 min), or gentamicin (60 min) at CLSI breakpoint concentrations are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package47. (b) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from 24 clinical isolates of E. coli or A. baumannii treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin. Gene identifiers are listed at right, along with gene names if available. CLSI classifications of each strain based on broth microdilution are shown below. * = strains with large inoculum effects in meropenem MIC; x = strains discordant by more than one dilution. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on NanoString data from the derivation cohort and tested on the validation cohort (y-axis) are compared with standard CLSI classification based on broth microdilution MIC (x-axis) for E. coli (top) or A. baumannii isolates treated with meropenem, ciprofloxacin, and gentamicin. Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (i.e. intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on the meropenem plot.
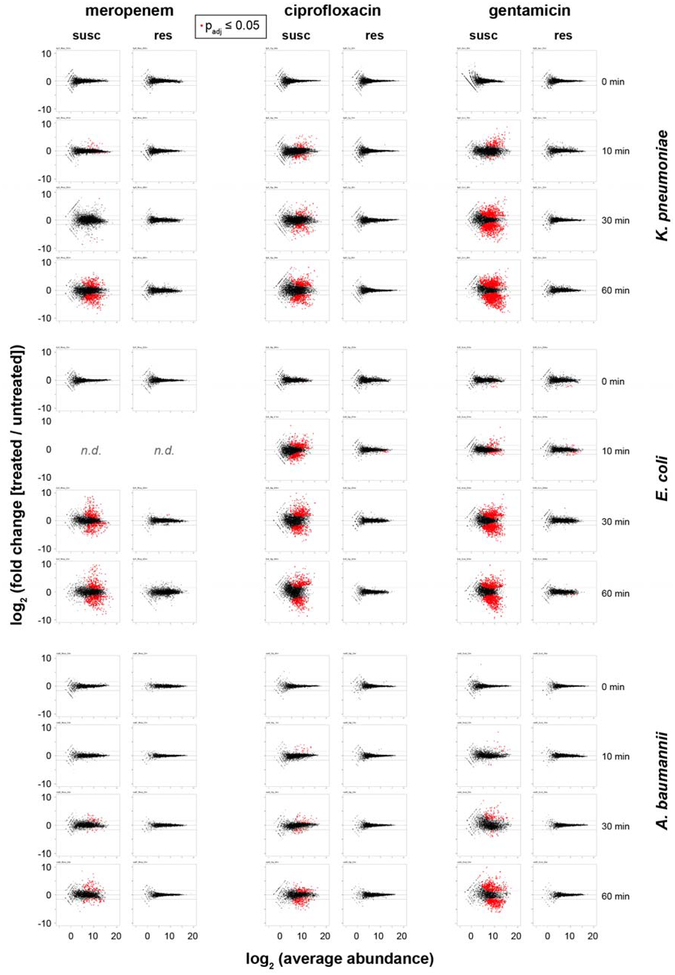
Extended Data Figure 2. Timecourse of RNA-Seq data upon antibiotic exposure reveals differential gene expression between susceptible and resistant clinical isolates.
Susceptible (left panels) or resistant (right panels) clinical isolates of K. pneumoniae, E. coli, or A. baumannii treated with meropenem, ciprofloxacin, or gentamicin at CLSI breakpoint concentrations for the indicated times. Data are presented as MA plots, Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package47.
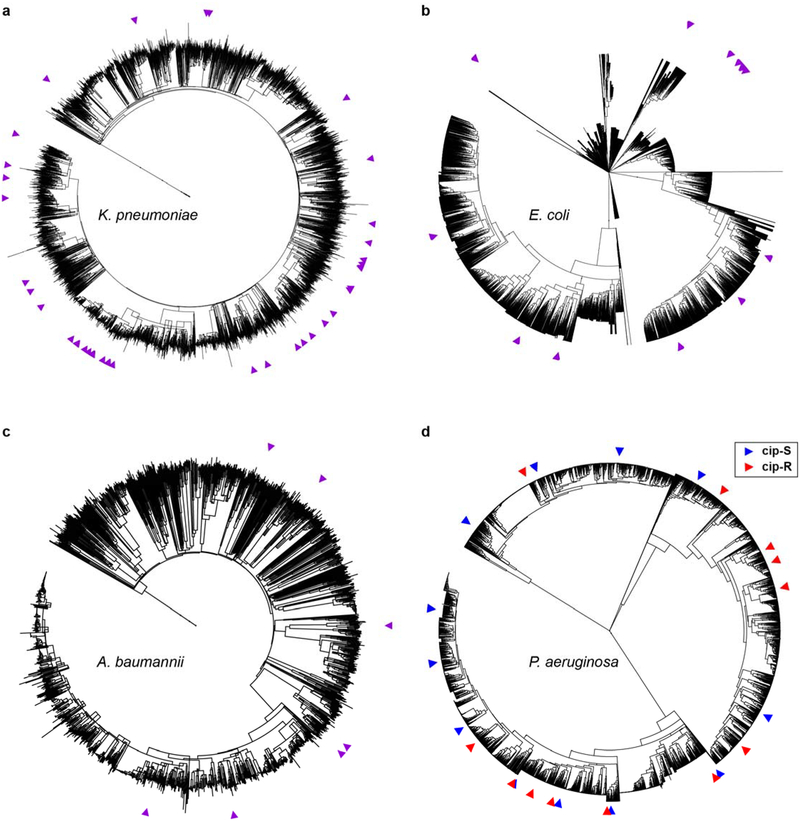
Extended Data Figure 3. Phylogenetic trees highlight the diversity of strains used in this study.
Phylogenetic trees of all sequenced isolates deposited in NCBI for (a) K. pneumoniae, (b) E. coli, (c) A. baumannii, and (d) P. aeruginosa, with all sequenced isolates used in this study indicated by colored arrowheads around the periphery. See Supplemental Methods for details.
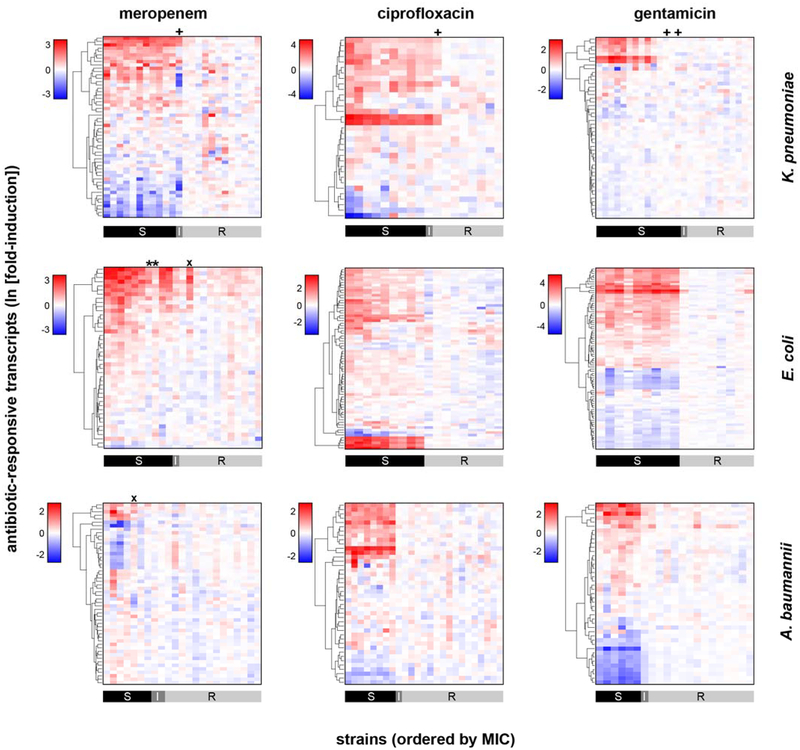
Extended Data Figure 4. NanoString data from dozens of antibiotic-responsive genes distinguish susceptible from resistant isolates.
Heatmaps of normalized, log-transformed fold-induction of antibiotic-responsive transcripts from clinical isolates of K. pneumoniae (24, 18, and 26 independent clinical isolates for the three antibiotics, respectively), E. coli (24 independent clinical isolates for each antibiotic), or A. baumannii (24 clinical isolates for each antibiotic) treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin. CLSI classifications are shown below. All antibiotic-responsive transcripts chosen as described from RNA-Seq data are shown here; the subset of these chosen by reliefF as the 10 most discriminating transcripts are shown in Fig. 1b or Supplemental Fig. 1b. * = strains with large inoculum effects in meropenem MIC; + = one-dilution errors; x = strains discordant by more than one dilution.
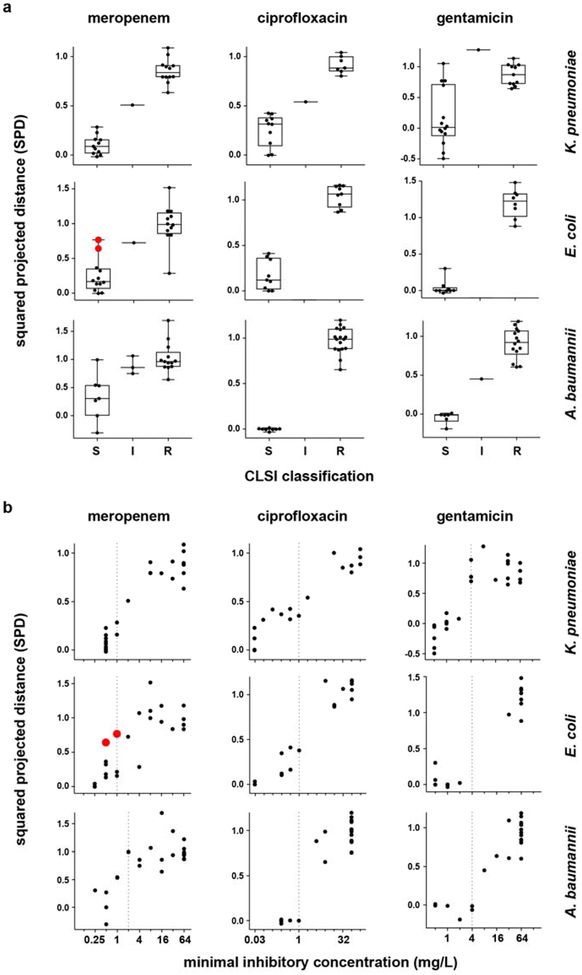
Extended Data Figure 5. One-dimensional projection of NanoString data distinguishes susceptible from resistant isolates and reflects MIC.
(a) Phase 1 NanoString data from Extended Data Fig. 2 (i.e., normalized, log-transformed fold-induction for each responsive transcript), analyzed as described to generate squared projected distance (SPD) metrics (y-axes) for each strain (see Supplemental Methods), are binned by CLSI classifications (x-axes), for clinical isolates of K. pneumoniae (24, 18, and 26 independent clinical isolates for the three antibiotics, respectively), E. coli (24 independent clinical isolates for each antibiotic), or A. baumannii (24 clinical isolates for each antibiotic) treated at CLSI breakpoint concentrations with meropenem, ciprofloxacin, or gentamicin (the same isolates shown in Fig. 1b–c and Extended Data Fig. 1b–c). By definition, an SPD of 0 indicates a transcriptional response to antibiotic equivalent to that of an average susceptible strain, while an SPD of 1 indicates a response equivalent to that of an average resistant strain. See Supplemental Methods for details. Data are summarized as box-and-whisker plots, where boxes extend from the 25th to 75th percentile for each category, with a line at the median, and whiskers extend from the minimum to the maximum. Note that for A. baumannii and meropenem, the clustering of the majority of susceptible strains by this simple metric (aside from one outlier which is misclassified as resistant by GoPhAST-R) underscores the true differences in transcription between susceptible and resistant isolates, despite the more subtle-appearing differences in heatmaps for this combination (Extended Data Fig. 1b), which is largely caused by one strain with an exaggerated transcriptional response (seen here as the strain with a markedly negative SPD) that affects scaling of the heatmap. (b) The same SPD data (y-axes) plotted against broth microdilution MICs (x-axes) reveal that the magnitude of the transcriptional response to antibiotic exposure correlates with MIC. In both (a) and (b), strains with a large inoculum effect upon meropenem treatment are displayed in red and enlarged. Vertical dashed line indicates the CLSI breakpoint between susceptible and not susceptible (i.e., intermediate or resistant).
Extended Data Figure 6. RNA-Seq and NanoString data reveal differential gene expression that distinguishes susceptible from resistant clinical isolates for S. aureus + levofloxacin and P. aeruginosa + ciprofloxacin.
(a) RNA-Seq data from two susceptible or two resistant clinical isolates of each species treated with the indicated fluoroquinolone at 1 mg/L for 60 minutes are presented as MA plots. Statistical significance was determined by a two-sided Wald test with the Benjamini-Hochberg correction for multiple hypothesis testing, using the DESeq2 package47. (b) Heatmaps of normalized, log-transformed fold-induction of antibiotic-responsive transcripts from 24 independent clinical isolates of each species treated with the indicated fluoroquinolone at 1 mg/L for 60 minutes. For each species, NanoString data from all candidate transcripts are shown at left, and top the 10 transcripts selected from Phase 1 testing are shown at right. (c) GoPhAST-R predictions of probability of resistance from a random forest model trained on Phase 1 NanoString data from the derivation cohort and tested on the validation cohort (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis). Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (i.e. intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution.
Extended Data Figure 7. Schematic of data analysis scheme, including “two-phase” machine learning approach to feature selection and strain classification.
Schematic representation of major data analysis steps in identifying antibiotic-responsive transcriptional signatures from RNA-Seq data, validating and optimizing these signatures using NanoString in two phases, and using these signatures to classify strains of unknown MIC, also in two phases. First, candidate antibiotic-responsive and control transcripts were chosen from RNA-Seq data using custom scripts built around the DESeq2 package47, and conserved regions of these transcripts were identified for targeting in a hybridization assay. In phase 1 (implemented for all pathogen-antibiotic pairs), these candidate transcripts were quantitated on the NanoString assay platform, and the resulting data were partitioned by strain into training and testing cohorts. Ten transcripts that best distinguish susceptible from resistant strains within the training cohort were then selected (step 1A) using the reliefF feature selection algorithm (implemented via the CORElearn package), then used to train an ensemble classifier (step 1B) on the same training cohort using a random forest algorithm (implemented via the caret package). This trained classifier was then used to predict susceptibilities of strains in the testing cohort (step 1C), and accuracy was assessed by comparing with broth microdilution results (Supplementary Table 4). In phase 2 (implemented for K. pneumoniae + meropenem and ciprofloxacin), the same process was repeated, but the phase 1 training and testing cohorts were combined into a single, larger training cohort for feature selection (step 2A) and classifier training (step 2B), and a new set of strains were obtained as a testing cohort. The 10 genes selected from the phase 2 training cohort were measured from this phase 2 testing cohort, and the trained classifier was used for AST on these new strains (step 2C), with accuracy again assessed by comparison with broth microdilution (Supplementary Table 4). See Supplemental Methods for detailed descriptions of each of these analysis steps.
Extended Data Figure 8. GoPhAST-R accurately classifies K. pneumoniae isolates tested in phase 2.
(a) Heatmaps of normalized, log-transformed fold-induction of top 10 antibiotic-responsive transcripts from K. pneumoniae treated at CLSI breakpoint concentrations with meropenem (31 independent clinical isolates) or ciprofloxacin (25 independent clinical isolates). CLSI classifications are shown below. * = strain with large inoculum effects in meropenem MIC; + = one-dilution error; x = strain discordant by more than one dilution. Note that the 10 responsive transcripts shown are the only 10 tested for this second phase of GoPhAST-R implementation. (b) GoPhAST-R predictions of probability of resistance from a random forest model trained on all Phase 1 NanoString data the independent Phase 2 cohort (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis). Horizontal dashed lines indicate 50% probability of resistance. Vertical dashed lines indicate the CLSI breakpoint between susceptible and not susceptible (i.e. intermediate/resistant). Numbers in each quadrant indicate concordant and discordant classifications between GoPhAST-R and broth microdilution. * = strain with large inoculum effects in meropenem MIC.
Extended Data Figure 9. GoPhAST-R accurately classifies AST and detects key resistance elements directly from simulated positive blood culture bottles in <4 hours.
(a) Heatmaps of normalized, log-transformed fold-induction NanoString data from the top 10 antibiotic-responsive transcripts directly from 12 simulated positive blood culture bottles for each indicated pathogen-antibiotic combination reveal antibiotic-responsive transcription in susceptible but not resistant isolates. For meropenem, results of carbapenemase / ESBL gene detection are also displayed as a normalized, background-subtracted, log-transformed heatmap above. CLSI classifications of isolates, which were blinded until analysis was complete, are displayed below each heatmap. (b) Probability of resistance from a random forest model trained by leave-one-out cross-validation on NanoString data from (a) (y-axis) compared with standard CLSI classification based on broth microdilution MIC (x-axis) for each isolate. Horizontal dashed lines indicate 50% chance of resistance based on random forest model. Vertical dashed lines indicate CLSI breakpoint between susceptible and resistant. Carbapenemase (square outline) and select ESBL (diamond outline) gene content as detected by GoPhAST-R are also displayed on meropenem plots. See Supplementary Methods for details of spike-in protocol.
Supplementary Material
Acknowledgments
We thank T. Abeel for valuable early discussions about the approach to machine learning for feature selection and strain classification; E. Lander, B. Birren, C. Nusbaum, and C. Russ for input on RNA-Seq experiments and overall strategy; and G. Giannoukos and D. Ciulla for valuable contributions to RNA-Seq methods development. This publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under awards 1R01AI117043-05 (DTH) and 1K08AI119157-04 (RPB), and contract No. HHSN272200900018C. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
R.P.B., P.M., J.Livny, and D.T.H. are co-inventors on subject matter in US provisional application No. 62/723,417 filed by the Broad Institute directed to RNA signatures for AST, as described in this manuscript. L.W., R.B., R.K., and J.B. are employees at NanoString, Inc., the company that manufactures the RNA detection platforms used in this manuscript. NanoString, Inc. has licensed the intellectual property for RNA-based AST from the Broad Institute. V.M.P. received research funds from SeLux Diagnostics, Inc. for work on an unrelated project.
References
- 1.Fauci AS & Morens DM The perpetual challenge of infectious diseases. N Engl J Med 366, 454–461, doi: 10.1056/NEJMra1108296 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Organization, W. H. Antimicrobial resistance: global report on surveillance 2014. (2014). [Google Scholar]
- 3.Kumar A et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34, 1589–1596, doi: 10.1097/01.CCM.0000217961.75225.E9 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Kadri SS et al. Difficult-to-Treat Resistance in Gram-negative Bacteremia at 173 US Hospitals: Retrospective Cohort Analysis of Prevalence, Predictors, and Outcome of Resistance to All First-line Agents. Clin Infect Dis 67, 1803–1814, doi: 10.1093/cid/ciy378 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiegand I, Hilpert K & Hancock RE Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3, 163–175, doi: 10.1038/nprot.2007.521 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Evans SR et al. Rapid Molecular Diagnostics, Antibiotic Treatment Decisions, and Developing Approaches to Inform Empiric Therapy: PRIMERS I and II. Clin Infect Dis 62, 181–189, doi: 10.1093/cid/civ837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arzanlou M, Chai WC & Venter H Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem 61, 49–59, doi: 10.1042/EBC20160063 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Cerqueira GC et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 114, 1135–1140, doi: 10.1073/pnas.1616248114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milheirico C, de Lencastre H & Tomasz A Full-Genome Sequencing Identifies in the Genetic Background Several Determinants That Modulate the Resistance Phenotype in Methicillin-Resistant Staphylococcus aureus Strains Carrying the Novel mecC Gene. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.02500-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnham CD, Leeds J, Nordmann P, O’Grady J & Patel J Diagnosing antimicrobial resistance. Nat Rev Microbiol 15, 697–703, doi: 10.1038/nrmicro.2017.103 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Consortium CR et al. Prediction of Susceptibility to First-Line Tuberculosis Drugs by DNA Sequencing. N Engl J Med 379, 1403–1415, doi: 10.1056/NEJMoa1800474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia B et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45, D566–D573, doi: 10.1093/nar/gkw1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya RP, Grad YH & Hung DT in Harrison’s Principles of Internal Medicine (eds Jameson JL et al. ) Ch. 474, 3491–3504 (McGraw-Hill Education, 2018). [Google Scholar]
- 14.Ellington MJ et al. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23, 2–22, doi: 10.1016/j.cmi.2016.11.012 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Charnot-Katsikas A et al. Use of the Accelerate Pheno System for Identification and Antimicrobial Susceptibility Testing of Pathogens in Positive Blood Cultures and Impact on Time to Results and Workflow. J Clin Microbiol 56, doi: 10.1128/JCM.01166-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cermak N et al. High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol 34, 1052–1059, doi: 10.1038/nbt.3666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barczak AK et al. RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities. Proc Natl Acad Sci U S A 109, 6217–6222, doi: 10.1073/pnas.1119540109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quach DT, Sakoulas G, Nizet V, Pogliano J & Pogliano K Bacterial Cytological Profiling (BCP) as a Rapid and Accurate Antimicrobial Susceptibility Testing Method for Staphylococcus aureus. EBioMedicine 4, 95–103, doi: 10.1016/j.ebiom.2016.01.020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Belkum A, Welker M, Pincus D, Charrier JP & Girard V Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues? Ann Lab Med 37, 475–483, doi: 10.3343/alm.2017.37.6.475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonomo RA et al. Carbapenemase-Producing Organisms: A Global Scourge. Clin Infect Dis 66, 1290–1297, doi: 10.1093/cid/cix893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lutgring JD & Limbago BM The Problem of Carbapenemase-Producing-Carbapenem-Resistant-Enterobacteriaceae Detection. J Clin Microbiol 54, 529–534, doi: 10.1128/JCM.02771-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weisenberg SA, Morgan DJ, Espinal-Witter R & Larone DH Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis 64, 233–235, doi: 10.1016/j.diagmicrobio.2009.02.004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodworth KR et al. Vital Signs: Containment of Novel Multidrug-Resistant Organisms and Resistance Mechanisms - United States, 2006–2017. MMWR Morb Mortal Wkly Rep 67, 396–401, doi: 10.15585/mmwr.mm6713e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMullen AR, Yarbrough ML, Wallace MA, Shupe A & Burnham CD Evaluation of Genotypic and Phenotypic Methods to Detect Carbapenemase Production in Gram-Negative Bacilli. Clin Chem 63, 723–730, doi: 10.1373/clinchem.2016.264804 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Humphries RM CIM City: The game continues for a better carbapenemase test. J Clin Microbiol, doi: 10.1128/JCM.00353-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th edn, CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; (2018). [Google Scholar]
- 27.Shishkin AA et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12, 323–325, doi: 10.1038/nmeth.3313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geiss GK et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26, 317–325, doi: 10.1038/nbt1385 (2008). [DOI] [PubMed] [Google Scholar]
- 30.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 11th edn, CLSI Supplement M07. Wayne, PA: Clinical and Laboratory Standards Institute; (2018). [Google Scholar]
- 31.Adler A, Ben-Dalak M, Chmelnitsky I & Carmeli Y Effect of Resistance Mechanisms on the Inoculum Effect of Carbapenem in Klebsiella pneumoniae Isolates with Borderline Carbapenem Resistance. Antimicrob Agents Chemother 59, 5014–5017, doi: 10.1128/AAC.00533-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith KP & Kirby JE The Inoculum Effect in the Era of Multidrug Resistance: Minor Differences in Inoculum Have Dramatic Effect on MIC Determination. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.00433-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordmann P, Dortet L & Poirel L Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med 18, 263–272, doi: 10.1016/j.molmed.2012.03.003 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Cubero M et al. Carbapenem-resistant and carbapenem-susceptible isogenic isolates of Klebsiella pneumoniae ST101 causing infection in a tertiary hospital. BMC Microbiol 15, 177, doi: 10.1186/s12866-015-0510-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma P, Laibinis HH, Ernst CM & Hung DT Carbapenem Resistance Caused by High-Level Expression of OXA-663 beta-Lactamase in an OmpK36-Deficient Klebsiella pneumoniae Clinical Isolate. Antimicrob Agents Chemother 62, doi: 10.1128/AAC.01281-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou HW, Bhattacharyya RP, Hung DT & Han J Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 15, 2297–2307, doi: 10.1039/c5lc00311c (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lomovskaya O et al. Vaborbactam: Spectrum of Beta-Lactamase Inhibition and Impact of Resistance Mechanisms on Activity in Enterobacteriaceae. Antimicrob Agents Chemother 61, doi: 10.1128/AAC.01443-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall S et al. Can Ceftazidime-Avibactam and Aztreonam Overcome beta-Lactam Resistance Conferred by Metallo-beta-Lactamases in Enterobacteriaceae? Antimicrob Agents Chemother 61, doi: 10.1128/AAC.02243-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caniaux I, van Belkum A, Zambardi G, Poirel L & Gros MF MCR: modern colistin resistance. Eur J Clin Microbiol Infect Dis 36, 415–420, doi: 10.1007/s10096-016-2846-y (2017). [DOI] [PubMed] [Google Scholar]
- 40.Florio W, Tavanti A, Barnini S, Ghelardi E & Lupetti A Recent Advances and Ongoing Challenges in the Diagnosis of Microbial Infections by MALDI-TOF Mass Spectrometry. Front Microbiol 9, 1097, doi: 10.3389/fmicb.2018.01097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Letunic I & Bork P Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47, W256–W259, doi: 10.1093/nar/gkz239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H & Durbin R Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gotz S et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36, 3420–3435, doi: 10.1093/nar/gkn176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandesompele J et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3, RESEARCH0034 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown LD, Cai TT & DasGupta A Interval Estimation for a Binomial Proportion. Statist Sci 16, 101–133, doi: 10.1214/ss/1009213286 (2001). [DOI] [Google Scholar]
- 46.Robnik-Šikonja M & Kononenko I Theoretical and Empirical Analysis of ReliefF and RReliefF. Machine Learning 53, 23–69, doi: 10.1023/a:1025667309714 (2003). [DOI] [Google Scholar]
- 47.Liaw A & Wiener M Classification and Regression by RandomForest. Vol. 23 (2001). [Google Scholar]
- 48.Efron B & Gong G A Leisurely Look at the Bootstrap, the Jackknife, and Cross-Validation. American Statistician 37, 36–48, doi:Doi 10.2307/2685844 (1983). [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-Seq data generated and analyzed for this study, supporting the analyses in Fig. 1, and Extended Data Figs. 1, 2, and 5, have been deposited as aligned bam files in the NCBI Sequencing Read Archive under study ID PRJNA518730. All other datasets generated during the current study, including raw and processed NanoString data, are available from the corresponding author on reasonable request.