Abstract
Chromosome Conformation Capture (3C) has emerged as a powerful approach for revealing the conformation and features of three-dimensional (3D) genomic organization. Yet attainment of higher resolution in organisms with compact genomes presents a challenge. Here, we describe modifications in the 3C technique that substantially enhance its resolution and sensitivity when applied to the 3D genome of budding yeast. Keys to our approach include use of a 4 bp cutter, Taq I, for cleaving the genome and quantitative PCR for measuring the frequency of ligation. Most importantly, we normalize the percent digestion at each restriction site to account for variation in accessibility of local chromatin structure under a given physiological condition. This strategy has led to the detection of physical interactions between regulatory elements and gene coding regions as well as intricate, stimulus-specific interchromosomal interactions between activated genes. We provide an algorithm that incorporates these and other modifications and allows quantitative determination of chromatin interaction frequencies in yeast under any physiological condition.
Keywords: Chromosome conformation capture (3C), 3D genome, budding yeast, DNA looping, interchromosomal interactions, transcription factories, transcription hubs
1. Introduction
Chromatin is packaged into intricate folds or loops within the three-dimensional (3D) space of the nucleus. From yeast to mammals, chromatin loops serve to control gene expression by bringing distal regulatory elements of the genome into physical proximity. These loops can juxtapose enhancers and promoters located tens or hundreds of kilobases from each other [1], or bring gene promoters into close proximity with their corresponding terminators [2-6]. Furthermore, loops encompassing protein-coding genes tend to adopt distinct nuclear positions [7]. Genes can interact with the nuclear envelope and its associated components [8, 9], or cluster within nuclear substructures locally enriched in transcription machinery [10, 11], pre-mRNA splicing factors [12] or Polycomb proteins [13, 14]. Such interactions can either activate or dampen gene expression. Clearly transcription, as well as other DNA-templated processes such as DNA repair [15, 16], are profoundly dependent on the 3D architecture and nuclear organization of chromatin.
Two types of approaches are principally used for studying 3D genome architecture and organization: fluorescence microscopy-based techniques and molecular methods. Microscopy-based procedures such as FISH, LacO-LacI tagging and immunofluorescence have provided insights into chromosome positioning [17], gene re-localization [18, 19] and more recently, biomolecular condensates that may participate in the transcriptional control of cell identity genes [20, 21]. Microscopy makes the study of single and live cells possible; however, this method is limited in throughput as well as in resolution (~200 nm). Molecular techniques such as chromosome conformation capture (3C), in contrast, permit detection of chromatin contacts representative of a large population of cells, even highly transient ones [22]. 3C has a resolution of 1 to 5 nm [23], far greater than that attainable by microscopy. 3C and its derivative techniques have also been successfully applied in a variety of organisms.
3C enables the detection and characterization of chromatin contacts between any two genomic elements in physical proximity. Use of this technique in the yeast Saccharomyces cerevisiae has unveiled important features of the Rabl-like configuration of chromosomes [24, 25] as well as higher-order chromatin folding [26, 27]. It has also revealed interactions between gene promoters and terminators [5, 22, 28-30]; upstream activator sites (UAS’s) and promoters; and regulatory and coding regions [22, 31]. 3C has also uncovered activator-dependent intergenic interactions that take place between genes located on the same or different chromosomes [22, 31]. Application of 3C technology to higher eukaryotes has revealed long-range gene control via chromatin loops [32-34] and higher-order chromosomal structures such as topologically associating domains (TADs) [35, 36]. Such approaches have additionally revealed the existence of co-regulated gene clusters termed transcription factories or hubs [10, 11, 37].
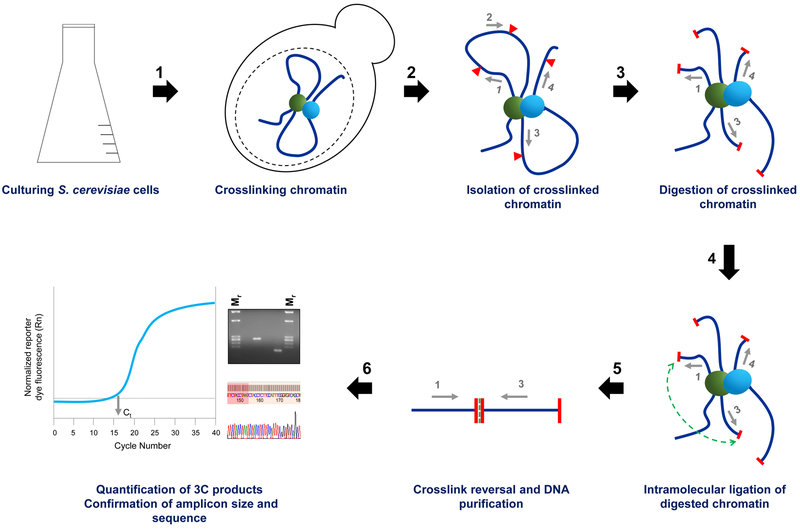
The 3C procedure typically incorporates four steps: (1) chemical crosslinking of spatially proximal chromatin segments; (2) fragmentation of the genome using appropriate restriction enzyme(s); (3) intra-molecular ligation of the linked chromatin segments; and (4) quantification of purified ligation products (schematically summarized in Figure 1). Each of these steps is subject to variability; therefore, incorporation of proper controls is necessary for ensuring high-quality data. For instance, the efficiency of restriction enzyme digestion can vary dramatically from one genomic locus to the next (or from one physiological state to another). In particular, location of the recognition site within a nucleosome [38] and protein density across these sites can dictate their accessibility and by extension, cleavability. Therefore, for accurate assessment of contact frequency between two chromatin loci, a normalization step that takes into account the efficiency of cleavage of each locus is necessary.
Figure 1. Technical workflow of chromosome conformation capture (3C).
S. cerevisiae cells are grown to mid-log density. (1) Whole cells are fixed in the presence of formaldehyde. This step of chemical crosslinking preserves native protein-protein and protein-DNA interactions inside the cell (proteins, green and cyan spheres; DNA, dark blue lines). (2) Cells are lysed, and the crosslinked chromatin is extracted (enzyme recognition sites, red triangles; primers abutting restriction sites, grey arrows numbered 1-4). (3) The chromatin is digested using a restriction enzyme of choice (restriction digestion, red capping of blue lines). (4) The digested ends are ligated in dilute conditions (proximity ligation event, green dotted line). (5) The crosslinks are reversed, and the cut and ligated (3C) DNA is purified. (6) The abundance of novel ligation joints is quantified by PCR-based methods. In addition, the size and sequence of the amplicon is confirmed. Ct, cycle threshold.
Moreover, attainment of enhanced resolution in organisms with more compact genomes presents a challenge. In yeast, UAS and promoter regions are typically located within a few hundred base pairs. Therefore, earlier 3C protocols designed to map long-range chromosomal interactions [39] or promoter-terminator proximity [40, 41] are inadequate to detect such short-range interactions. Here we describe a 3C procedure, Taq I-3C, that incorporates a normalization step that accounts for efficiency of digestion along with other important modifications, including the use of qPCR to quantify novel joint formation. Collectively, these improvements substantially enhance the resolution, sensitivity and reproducibility of 3C, and have led to unexpected insights into the dynamic structure and 3D organization of the yeast genome.
2. Development of Taq I-3C
A key feature of Taq I-3C is the use of a four base-pair (bp) cutter, as opposed to more commonly used 6 bp cutters [2, 28, 29, 42]. Taq I represented an attractive choice for several reasons. First, Taq I digestion yields cohesive ends that can ligate efficiently. Second, as a 4 bp cutter, Taq I cleaves frequently, roughly every 256 bp. Small fragments arising from Taq I cleavage could, in principle, resolve between regulatory versus coding regions of a gene. Indeed, in tests for candidate gene interactions, novel UAS-promoter and regulatory-coding region contacts were detected [22, 31], in addition to the previously reported promoter-terminator gene loops [5, 28-30], owing in part to the frequent and favorable distribution of Taq I within and around the genes.
A typical 3C procedure fails to account for changes in efficiency of restriction digestion, which can be a confounding variable. The efficiency of a restriction enzyme to cleave its recognition sites depends on how accessible those sites are within chromatin. This accessibility is dictated by the local structure of chromatin, which is subject to variation due to changing gene expression patterns (among other things). In our experience, digestion efficiencies tend to vary – sometimes considerably – between conditions that are non-inducing, inducing or attenuating for gene transcription. As digestion efficiency would affect the frequency of ligation, this parameter is important to take into account.
Previous 3C studies in yeast have typically used a semi-quantitative method for assessment of interaction frequencies [2, 5, 28-30, 43, 44]. The Taq I-3C procedure utilizes a real-time qPCR-based quantification method that measures signals within the linear range of amplification, eliminating the need for titration controls. In addition, the method is cost-effective; offers high throughput analysis; and enables detection and comparison of a wide range of signals. Additionally, we use tandem primers for amplifying ligation products that minimizes the possibility of detecting PCR products resulting from crosslink-independent ligation events, a concern when convergent or divergent primers are used.
Using Taq I-3C, we have detected novel features of gene conformation and organization heretofore unappreciated in budding yeast, including the existence of chromatin contacts between the regulatory elements and other genomic loci separated by less than 500 bp. In addition, Taq I-3C has enabled detection of stimulus-specific interactions between genomic regions located across the genome.
3. Experimental Design
3.1. Crosslinking of chromatin
Crosslinking can be performed either on spheroplasts [39] or intact cells [40]. We use intact cells, not only because it streamlines the procedure but avoids an unintended consequence of spheroplasting: induction of the heat shock response [45, 46]. Formaldehyde, a potent yet reversible crosslinker, is ideal for Taq I-3C. It is extensively used for studying chromatin structure and composition [47]. It forms a methylene linkage between the amino or imino groups of neighboring proteins and nucleic acids [48]. Yeast cells are permeable to formaldehyde, unlike other crosslinkers that require that cells be first converted to spheroplasts. As formaldehyde-mediated crosslinks bridge short distances (2 Å) [47], crosslinking of indirect interactions is minimized. Typically, 1-3% of formaldehyde is used in 3C procedures for yeast [39, 40, 49]. However, optimal conditions should be determined, as overly crosslinked chromatin might trap indirect chromosomal interactions and be difficult to digest whereas inefficient crosslinking would result in a failure to capture legitimate chromatin interactions.
3.2. Restriction digestion
In previous 3C-based studies in yeast, restriction enzymes that recognize 6 bp sequences were most commonly used [2, 28, 29, 42] (summarized in Table B.1). Rarely employed were 4 bp cutters whose cleavage frequency is on average 16-fold more frequent, and thus the resultant mapping of higher resolution. In most cases where 4 bp cutters were employed, they generated blunt-ends that ligate less efficiently than cohesive ones (see Table B.1 for examples). A second important consideration is that the restriction sites should lie close to putatively interacting regions, as a failure to do so could lead to a loss of resolution. To avoid this problem, a combination of restriction enzymes that generate compatible ends could be used.
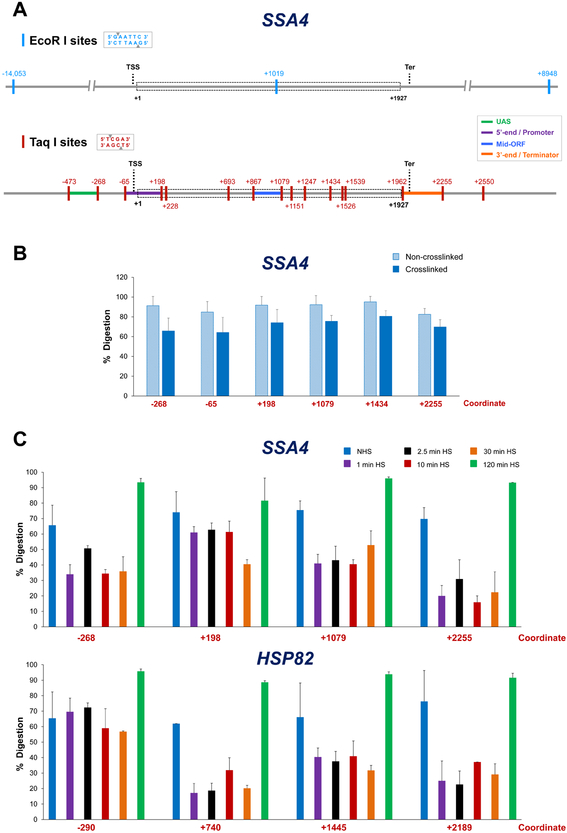
The importance of carefully assessing the distribution of recognition sites at a gene of interest is illustrated in Figure 2A. As can be seen, the 6 bp cutter EcoR I cleaves only once within the SSA4 gene and its associated regulatory sequences; the nearest 5’- and 3’-flanking sites are 14 kb and 9 kb away, respectively. By contrast, Taq I sites are frequent and distributed nearly uniformly across SSA4 and surrounding regions, yielding discrete regulatory and coding region fragments. The presence of multiple Taq I sites within the SSA4 coding region provides an opportunity of “walking down” the gene for mapping intragenic interactions, in contrast to EcoR I. While the distribution of Taq I sites was favorable at most genes that we have evaluated (Figures 2A and B.1A) [22, 31], any 4 bp cutter that exhibits a similarly favorable distribution could in principle be used.
Figure 2. Distribution of restriction sites at a representative Pol II gene and cleavage efficiency under different crosslinking and physiological conditions.
A) EcoR I and Taq I restriction maps of SSA4. Depicted is the genomic locus encompassing the SSA4 open reading frame (ORF; dashed rectangle) and varying lengths of flanking sequence. Coordinates correspond to the location of restriction sites (vertical colored lines) and are numbered relative to the ATG initiation codon (+1); transcription start site (TSS) and termination site (Ter) are indicated. Top: distribution of EcoR I (6-bp cutter) recognition sites. Bottom: Taq I (4-bp cutter) distribution. UAS, 5’-end/Promoter, Mid-ORF and 3’-end/Terminator regions are color-coded as indicated. For additional examples, see Figure B.1A.
B) Percent digestion of representative Taq I sites within SSA4 using templates isolated from non-heat-shocked (NHS) cells that were crosslinked (or not) prior to restriction digestion. Depicted are means + SD; N=2, qPCR=4.
C) As above, except percent digestion of Taq I sites within SSA4 and HSP82 is shown for crosslinked templates isolated from NHS cells or cells subjected to heat shock (HS) for the times indicated.
Nonetheless, the fact that Taq I cleaves optimally at 65°C makes it particularly useful in 3C. In a standard 3C procedure, a brief period of chromatin solubilization at 65°C in the presence of 1% SDS precedes restriction digestion [49]. This step ensures removal of loosely bound, non-crosslinked proteins from the DNA. However, since residual traces of SDS remain in the digestion mix, enzymes such as BamH I, Spe I, Pst I, and Nde I fail to work optimally [48]. The protocol for Taq I makes the addition of SDS unnecessary, as chromatin solubilization is achieved without it. A potential concern associated with incubation at elevated temperature is the reversal of formaldehyde-mediated crosslinks [50]. However, in our experience, a 7 h incubation at 60°C preserves many features of 3D chromosome topology, as evidenced by the fact that chromatin contacts identified using Taq I-3C are strictly formaldehyde-dependent (see Figure 4 below).
The digestion efficiency of a restriction enzyme is dependent on several factors. First, as mentioned above, is the location of restriction sites within a nucleosome. The resistance to digestion can be up to 100-fold for DNA sequences located at the entry/exit points of the nucleosome and 100,000-fold for those located at its center [38]. Second, chemical-mediated stabilization of histones (or non-histone proteins) to DNA resists digestion. As shown for SSA4, crosslinked chromatin is digested 10-20% less efficiently than the non-crosslinked control (Figure 2B). This is in agreement with a study comparing digestion of EcoR I sites upstream, downstream and within the human FMR1 gene that showed a decrease in digestion efficiency (~25%) at each site, owing to the enrichment of crosslinked proteins [51].
As nucleosome position and stability (as well as occupancy of non-histone proteins) can vary dependent on a gene’s expression state [52, 53], we tested accessibility of Taq I sites within and flanking SSA4 under conditions of variable expression. We observed that the efficiency of Taq I digestion varied according to expression state (Figure 2C): typically highest when the gene was weakly expressed (either non-heat-shocked (NHS) or chronically induced (120 min heat shock [HS])) and lowest when the gene was most highly transcribed (1 - 30 min HS). Therefore, the efficiency of Taq I digestion dynamically changes during the course of heat shock, paralleling dynamic changes in the transcription rate of this gene [22]. Similar dynamic alterations in chromatin accessibility were observed at other thermal stress-responsive genes, both those induced by heat shock as well as those repressed by it (PGM2 and RPL10, respectively; Supplemental Figure B.1).
3.3. Primer design
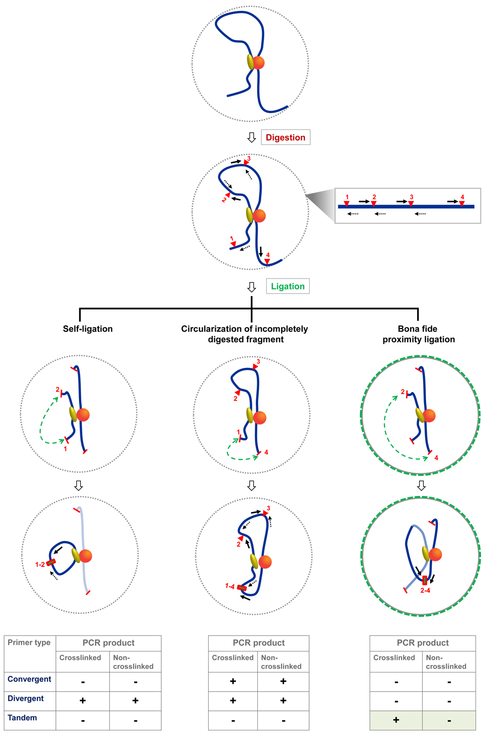
Primers must be designed against unique sequences in the genome (use BLAST in SGD [www.yeastgenome.org]). They should be 20-30 nts in length (ideally 24-27 nts) with a GC content of 40-50%. Ideally, they should correspond to sequences within 50 - 150 bp of the restriction sites of interest; this will keep amplicon sizes in the range of 100-300 bp, optimal for detection by qPCR. To avoid detection of false positives (i.e., signals not reflecting formation of crosslink-dependent ligation joints), we recommend using tandem primers. While divergent primers have been more commonly used (see Supplemental Table B.1), these can yield PCR products resulting from self-ligation of chromatin fragments as illustrated in Figure 3. Likewise, convergent primers can generate PCR products when the intervening restriction sites remain uncut. However, as shown in Figure 3, tandem primers specifically amplify crosslink-dependent ligation products, eliminating the non-informative background noise. It is nonetheless a good idea to confirm the size and sequence of each amplicon generated as there might be instances where incomplete digestion of restriction sites yields ligation products detectable by tandem primers. Under such circumstances, either multiple amplicons or a single amplicon of inappropriate length will be seen. Finally, since each primer pair amplifies with a different efficiency, it is critical to correct for this variable (see Section 3.4.2.5 below).
Figure 3. Tandem primers conclusively reflect crosslink-dependent proximity ligation.
Depicted are ligation events captured by convergent, divergent and tandem primers. A mono-molecular crosslinked complex is shown encompassing a set of proteins (shown as yellow oval and orange sphere) bound to a stretch of chromatin (blue line). Restriction sites are indicated by red triangles numbered 1 to 4. Primers abutting each restriction site are shown in forward and reverse orientations (solid and dotted black arrows, respectively); a map is shown on the right. Ligation events are indicated with green-dotted lines. PCR products from tandem primers are definitive of crosslink-dependent proximity ligation, as illustrated for sites 2 and 4 after their cleavage and ligation (right), whereas divergent and convergent primers yield products even in the absence of crosslinking (left and middle). In addition, the unidirectional primer design prevents the amplification of spurious ligation products, such as those resulting from self-ligation of digested fragments (left) detected by divergent primers. Also, convergent primers can yield PCR products under conditions of incomplete digestion (middle).
3.4. Detection and quantification of Taq I-3C signals
3.4.1. SYBR Green qPCR method
3C quantifies ligation products as a measure of the frequency of interaction between a given pair of genomic loci. Most previous 3C studies in budding yeast employed locus-specific end-point PCR [2, 5, 28-30, 43, 44], where a standard amount of genomic DNA template is used with a set of primer pairs for a certain number of PCR cycles. However, for this method to be quantitative, the template must be titrated to ensure linear amplification. On the other hand, real-time qPCR circumvents the need for any titration controls. It does so by identifying cycle threshold (Ct) values early within the exponential phase where PCR amplification occurs with >90% efficiency [54]. qPCR has a detection range vastly exceeding that of end-point PCR. It is also more sensitive; offers higher throughput; and requires smaller sample volume. We detect PCR products by using the SYBR Green fluorescent reporter, a convenient and cost-effective alternative to the previously used TaqMan probes [42, 55].
3.4.2. Controls and algorithm to calculate interaction frequencies
We developed an algorithim to quantify chromatin contacts using Taq I-3C. First, we divide crosslinked chromatin into three fractions: (i) undigested chromatin (“UND3C”), (ii) digested-only chromatin (“DO3C”) and (iii) digested and ligated chromatin (“Lig3C”). Second, we divide a naked genomic DNA (gDNA) control into three corresponding fractions: UNDgDNA, DOgDNA and LiggDNA(see also 3.4.2.5). For each template, two biological replicates are prepared. We then determine the cycle threshold value for all templates, and take an average of the replicates (see Table 1, equation 1). Additional controls incorporated into the algorithim are described below.
Table 1.
Step-by-Step Quantification of Taq I-3C
| Description | Terms and Equations | |
|---|---|---|
| 1. | Average cycle threshold (Ct) | a. Avg. Ct for DO3C
template = CtDO3C b. Avg.Ct for Lig3Ctemplate = CtLig3C |
| 2. | Net Ct values This control corrects for background noise due to primer dimers. |
a.
ΔCtDO3C = CtDO3C
– Ctno template b. ΔCtLig3C – CtLig3c, – Ctno template |
| 3. | Average fold signal above background | a. Avg. fold signal for
DO3C template =
−ΔCtDO3C b. Avg. fold signal for Lig3C template =2 −ΔCtLig3C |
| 4. | Amount of PCR product from ARS504 locus | a. ARS504 (in ng)
for DO3C template =
ARS504DO3C b. ARS504 (in ng) for Lig3C template = ARS5504Lig3c |
| 5. | Fold-over signals normalized to internal
control locus This control corrects for the variation in recovery of templates. |
a.
b. |
| 6. | Ligation-dependent 3C signal | |
| 7. | % digestion of each participating Taq I site | |
| 8. | Ligation-dependent 3C signals corrected for
variation in Taq I digestion efficiencies This control corrects for variation in Taq I digestion efficiencies. |
|
| 9. | Ligation-dependent signal corrected for
variation in Taq I digestion efficiencies for the gDNA
template Note: In our hands, digestion of gDNA is almost always close to 100%. |
|
| 10. | Normalized frequency of
interaction The control corrects for variation in primer pair efficiencies as well as the inherent tendency of any two DNA ends to be ligated. |
3.4.2.1. No-template control
SYBR Green intercalates within any duplex DNA, including primer dimers. Therefore, to correct for this background, we incorporate a ‘no-template’ control, which contains all qPCR reaction components except the template. For the analysis, we determine Ct of the no-template control and then take an average of the replicates, which we term “Ct no-template”. Next, we obtain ΔCt values for the DO3C and Lig3C templates by subtracting the Ct no-template value from the Ct values of DO3C and Lig3C (Table 1, equation 2).
3.4.2.2. Internal recovery control
It is inevitable that variation in the recovery of samples between replicates or experiments will exist. An internal recovery control corrects for this variation, allowing accurate comparison between samples recovered from different cell types, states or genetic contexts [56]. Here, differences in the recovery of ‘DO’ and ‘Lig’ templates need to be controlled. For this, we measure the presence of a DNA sequence not affected by digestion and/or ligation in each template. We selected the ARS504 locus that lacks an internal Taq I site for this purpose.
In our analysis, we first determine the average fold-signal above background for the DO3C and Lig3C templates (Table 1, equation 3). Then, we quantify the amount of PCR product from the ARS504 locus deduced from interpolation of a standard curve (created using purified yeast gDNA as template; equation 4). Next, we derive fold-over signal normalized to the internal control locus (equation 5).
3.4.2.3. No-ligation control
Incorporation of this control in TaqI-3C ensures detection of a ligation-dependent 3C signal. This signal is calculated as the ratio of fold-over normalized signals of the Lig3C (digested and ligated chromatin) and DO3C templates (equation 6).
3.4.2.4. Digestion efficiency control
As discussed in Section 3.2, restriction site cleavage efficiency varies according to genomic location, genetic context and gene expression state. Therefore, for comparison of any two 3C signals, a control accounting for variation in digestion efficiencies is necessary. To determine percent digestion at each Taq I recognition site participating in the formation of a novel ligation joint, convergent primers are used. Such primers, corresponding to sequences on either side of the restriction site, amplify across the region. We determine the average Ct values for the UND3C and DO3C templates (termed “CtR”). Likewise, average Ct values are calculated for the non-cleaved locus, ARS504 (“CtARS504”). These are then incorporated into equation 7 (adapted and modified from [55]). We then derive ligation-dependent 3C signals corrected for variation in Taq I digestion efficiencies (equation 8). The normalized ligation-dependent signals for the gDNA control template are obtained in a similar manner (equation 9).
3.4.2.5. Primer-pair efficiency control
Different primer pairs amplify with different efficiencies; therefore, a control that corrects for this variation is necessary. For this we generate a gDNA control template that, in theory, has all possible ligation products in equimolar amounts [56]. The normalized frequency of interaction between the two tested loci is then determined by dividing normalized ligation-dependent signals for the 3C sample by those of the gDNA control (equation 10).
4. Results
4.1. Taq I - 3C detects ligation- and crosslink-dependent intragenic interactions within transcriptionally active genes
We initially tested the ability of Taq I-3C to detect ligation- and crosslink-dependent intragenic interactions. As shown in Figure 4A, the method readily identified looping between the promoter and terminator of a representative, heat shock-activated gene, HSP82 (3C assay, red bar). The frequency of this chromatin contact was >50-fold than that seen in the non-activated state (blue bar). Importantly, the promoter-terminator contacts within HSP82 were virtually undetectable in identically prepared, but non-crosslinked, chromatin (10 min HS, striped bar), thus verifying that the 3C signal was crosslink-dependent. We additionally validated that the primers used to detect gene looping of HSP82 gave rise to a single, ligation-dependent PCR product, and that its DNA sequence reflected the novel ligation joint formed between the promoter- and terminator-containing regions of the gene (Figures 4A and B.3). Our observations for HSP82 are consistent with earlier reports of ligation- and crosslink-dependent gene looping within constitutively active and other inducible genes using traditional 3C [2, 44].
As discussed above, Taq I, being a 4 bp cutter, cuts the genome on average once every 256 bp and its distribution within genes is typically favorable (e.g., see Figures 2A and B.1.A) [22, 31]. We exploited this to ask whether additional intragenic looping interactions might be detectable. As shown in Figure 4B, Taq I-3C readily detected contact between the UAS and coding region of the transcriptionally activated HSP104; no such contact was detectable in the non-induced state. This intragenic interaction between regulatory and coding sequences, which we term ‘crumpling’, was detectable in crosslinked but not in non-crosslinked chromatin. And as above, the 3C signal was ligation-dependent and the amplicon generated was confirmed by both gel electrophoresis and DNA sequencing (Figures 4B and 5A). Similar intragenic interactions, importantly including UAS-promoter contacts, have been detected at a variety of transcriptionally active genes [22, 31] and in different genetic backgrounds (Figure B.2, panels A-C), further validating the technique.
Figure 4. Taq I-3C detects ligation- and crosslink-dependent cis- and trans-interactions between genomic loci.
A) Left: Frequency of looping interactions detected between the indicated 5’ and 3’ regions of HSP82 (coordinates numbered relative to the ATG codon, +1). Chromatin was isolated from NHS and 10 min HS haploid cells (strain BY4741) as indicated, crosslinked (or not) prior to Taq I digestion and T4 DNA ligase treatment. Shown are means + SD; N=2, qPCR=4. **, P<0.01 (calculated by one-way ANOVA followed by post-hoc Tukey analysis). Right: Gel image of amplicon generated using primers corresponding to the 5’- and 3’-end of HSP82. Template was purified from crosslinked chromatin isolated from 10 min HS cells, digested with Taq I and then subject to proximity ligation (or not) by T4 DNA ligase (+L or −L, respectively). Expected size of amplicon: 276 bp.
B) Left: As above, except frequency of an intragenic interaction between the UAS and mid-coding regions of HSP104 is shown (means + SD; N=2, qPCR=4. **, P<0.01). Right: Gel image of amplicon arising from this primer combination and template purified from Taq I-digested chromatin that was subsequently ligated or not (+L or −L, respectively; expected size: 151 bp).
C) Left and Middle: As above, except interactions were detected between genes lying on the same or different chromosomes. Left: cis interactions between chromosomally linked genes in the presence or absence of crosslinking. Middle: trans interactions between the indicated regions of chromosomally unlinked genes in the presence or absence of crosslinking. Shown are means + SD; N=2, qPCR=4. **, P<0.01. Right: Gel image of the amplicons arising from the indicated primer combinations in the presence or absence of ligation.
4.2. Taq I - 3C detects intergenic interactions between chromosomally linked and unlinked genes
Previous 3C-based analyses in S. cerevisiae (including Hi-C) have provided scant evidence for interchromosomal interactions between natural, Pol II-transcribed genes beyond homologues [57-59]. Trans-interactions have been reported between Pol III-transcribed tRNA genes [24, 60], although these may be a consequence of the Rabl-like configuration of yeast chromosomes and the largely pericentric location of these genes [25]. We therefore asked whether Taq I - 3C could detect trans interactions between natural, Pol II transcribed genes. As shown in Figure 4C, interchromosomal interactions between HSP genes are readily seen (HSP12-HSP104), as are intrachromosomal interactions (3C assay, right and left panel, respectively). These interactions are transcription-dependent (Figure 4C; red bars); are distinct for one class of genes (they are not detected in genes under regulation of alternative, heat-responsive transcription factors [31]); and are quite intricate, as they involve sequences located within flanking regulatory regions as well as within coding regions (Figures 4C and 5B; [22, 31]). They are also ligation- and crosslink-dependent (Figure 4C) and seen in unrelated genetic backgrounds (Figure B.2D). DNA sequencing confirmed a novel intergenic interaction between HSP genes located on separate chromosomes (Figure 5B).
4.3. Taq I - 3C detects dynamic changes in both intra- and intergenic chromatin contacts during a heat shock time course
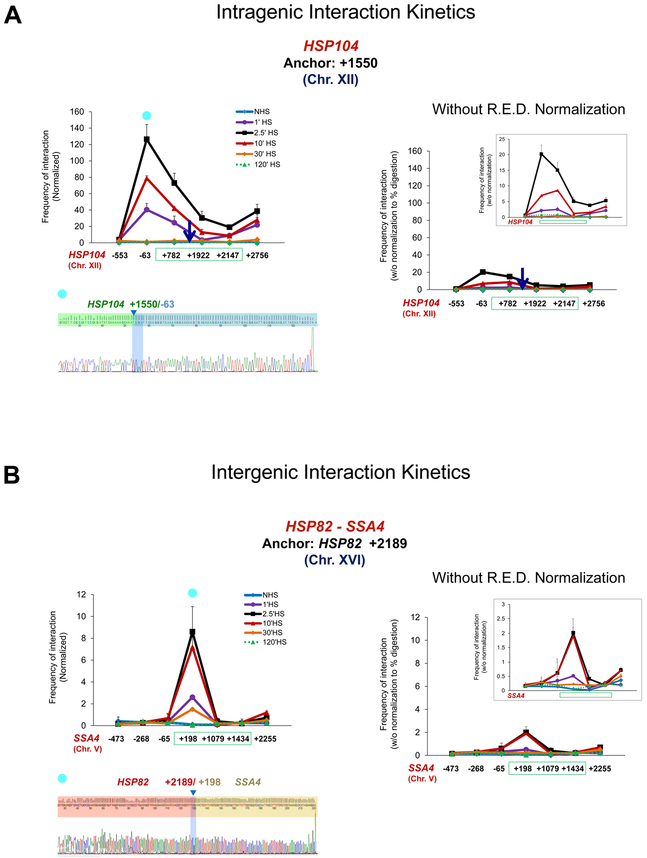
The foregoing examples demonstrate that Taq I-3C can differentiate transcription-associated changes in the 3D organization of genes. To further explore this idea and to assess the dynamic range of Taq I-3C, we subjected yeast cells to a heat shock time course during which the instantaneous rate of HSP gene transcription is dynamic: basally expressed under NHS conditions, induced within 1 min of thermal upshift (30°-39°C); peaking within 2.5-10 min; and attenuating within 30-60 min [22, 61]. Taq I-3C analysis of cells subjected to this treatment reveals a wide range of both intragenic and intergenic interactions occurring over the time course (Figure 5). We note that the dramatic changes detected in contact frequency were enabled, in part, by normalizing cleavage efficiency at each Taq I site for each time point (see Section 3.2 and Figure 2C above). Omission of this key control dramatically reduced the above-background signal and minimized detection of the dynamic chromatin contacts occurring during the transcriptional response to thermal stress (Figures 5A and 5B, compare 3C analysis performed with and without normalization control [left and right, respectively]).
Figure 5. Taq I-3C detects rapid and reversible interactions between genomic loci in cells subjected to heat shock.
A) Left: Kinetics of intragenic chromatin contacts within a representative heat shock-responsive gene, HSP104, in cells subjected to a heat shock time course. The mid-ORF anchor of HSP104 (+1550; arrow) was paired to the indicated loci upstream, within or downstream of the gene (HSP104 coding region spanned by green rectangle). Normalized 3C interaction frequencies were determined either prior to (NHS) or following HS for the times indicated. DNA sequence confirmation of a representative amplicon is shown below (10 min HS sample; primer paired with anchor is indicated by blue globe). Right: Same, except normalization to digestion efficiency was omitted from the 3C calculation. Data were plotted at same scale as on the left. Inset: Y-scale magnified 5X. R.E.D., restriction enzyme digestion. Shown are means + SD. N=2; qPCR=4 for each primer combination.
B) Left: As above, except kinetics of interchromosomal chromatin contacts between two heat shock-responsive genes, HSP82 and SSA4, as revealed by Taq I-3C, are depicted. DNA sequence confirmation of a representative amplicon is shown below (10 min HS sample). Right: Same, except normalization to digestion efficiency was omitted from the 3C calculation. Data were plotted at same scale as on the left. Inset: Y-scale magnified 3X.
4.4. Taq I - 3C is free of restriction fragment length and GC content bias
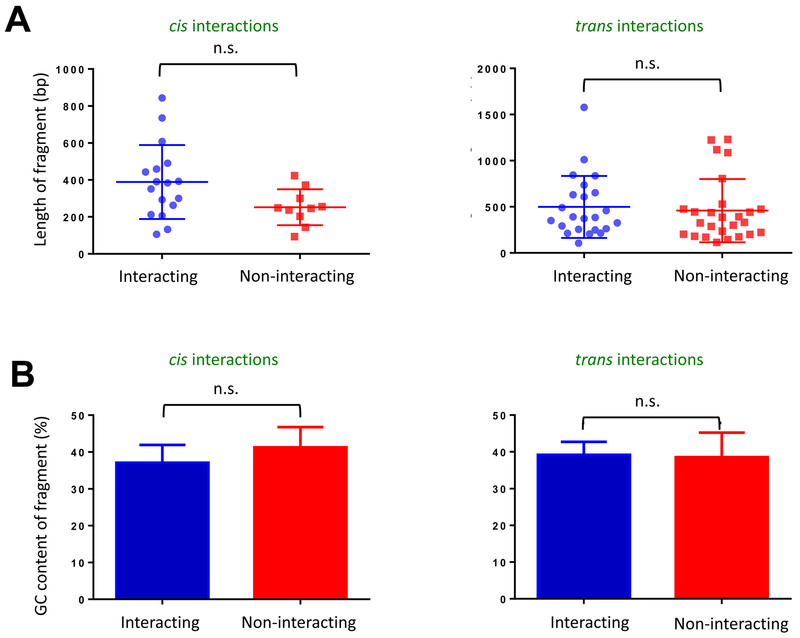
Finally, we investigated how two parameters, restriction fragment length and GC content of 3C products, contribute to the frequency of detection of 3C products. We did this given concerns that these properties can introduce bias into genome-wide chromosomal contact maps [62], including those of S. cerevisiae [24, 63]. To test for possible bias contributed by DNA fragment length, we compared lengths of interacting and non-interacting fragments revealed by TaqI-3C. As shown in Figure 6A, there is no significant difference in the length distribution of interacting DNA fragments (both cis- or trans-) versus non-interacting fragments. This observation is consistent with a previous point-to-point 3C analysis [51]. Similarly, to test for any possible bias contributed by GC content, we assessed this property in interacting versus non-interacting fragments. As shown in Figure 6B, there exists no significant difference in the GC content of either cis- or trans- interacting fragments when compared to their non-interacting counterparts. Thus, the two types of bias previously identified as substantially affecting the generation of genome-wide chromatin contact maps [62, 63], do not impact TaqI-3C analysis, and imply that the fragments we have tested are uniformly distributed with a GC content in the optimal range (~40%) for qPCR-based detection.
Figure 6. Analysis of potential bias in Taq I - 3C introduced by either restriction fragment length or restriction fragment GC content.
A) Test of Restriction Fragment Length Bias. Scatter plot illustrating the length distribution of Taq I fragments tested for cis- and trans-interactions as indicated. Depicted are means ± SD, n.s. (not significant), P>0.05 (calculated using unpaired t-test).
B) Test of GC Content Bias. Bar graph illustrating GC content of interacting and non-interacting Taq I restriction fragments tested as above. Depicted are means + SD. N= 18 (cis-interacting), 14 (trans-interacting), 10 (cis-non-interacting) and 17 (trans-non-interacting). n.s., P>0.05 (calculated as above).
5. Discussion
Here we have described a 3C technique, Taq I-3C, that is both crosslink- and ligation-dependent and possesses sufficient sensitivity to detect features of S. cerevisiae chromosomal topology heretofore unknown. Its application has unveiled the fact that transcriptionally active genes engage in multiple intragenic interactions beyond the previously described gene looping [2, 5]. These include interactions between flanking regulatory regions and coding sequences, as well as between enhancer (UAS) and promoter regions (this study; [22, 31]). Both phenomena have been previously observed in metazoans (e.g., see [32, 36, 64]) but have not, to our knowledge, been seen at natural yeast loci. Even more significantly, Taq I-3C has revealed that a select subset of transcriptionally induced genes engages in robust cis- and trans-interactions. Certain of these intergenic interactions have been validated by fluorescence microscopy [22, 31] which revealed that interchromosomal interactions could be detected in up to 40% of all cells.
Additionally, Taq I-3C has revealed that the frequency of both intragenic and intergenic HSP gene interactions strongly correlates with their instantaneous rate of transcription [22], with concomitant mechanistic implications. We observe virtually identical looping, crumpling and coalescence interactions at HSP genes in unrelated strain backgrounds (Figure B.2), arguing that the observations reported using Taq I-3C are not peculiar to a particular genotype. While we use HSP genes as a model to illustrate the efficacy of Taq I-3C in this report, the technique described here should be suitable for all types of chromatin interactions in budding yeast.
The importance of two controls in the development and use of Taq I-3C cannot be overstated. First, is the availability of chromatin obtained from non-heat-shocked cells that served as a negative control for the heat shock-induced state. For HSP genes evaluated under NHS conditions, the signals detected likely represent random collisions since their frequency decays as a function of distance (1/L) [22], the expected outcome for non-specific contacts. By contrast, the signals detected at the same loci, but following a stimulus that strongly induces HSP gene transcription [61], are often >50-fold higher. We interpret these as representing bona fide long-range chromatin interactions. The second critical control is normalization of the 3C signal to the cleavage efficiency of each restriction site in chromatin, isolated from cells under distinct physiological conditions. At the heat-inducible SSA4 and HSP82 genes, for example, occupancies of Taq I sites within and flanking the coding region were reduced during the acute phases of heat shock relative to other phases (0 min and 120 min HS) (Figure 2C). These observations, which provide insight into dynamic protein occupancy under different physiological states, allow a sensitive detection of the long-range chromatin contacts taking place. When this step was omitted from the 3C calculation, chromatin contact frequencies were considerably muted (Figure 5, A and B, right panels). This is as expected, since sites that are dynamically occupied by DNA-binding proteins cannot be accurately evaluated without this normalization.
There are two additional key features of TaqI-3C. First is its high resolution, afforded by use of a 4 bp cutter, Taq I. Whether use of alternative 4 bp cutters would provide both the resolution and sensitivity of Taq I is unclear, although in theory they should. Second is its sensitivity. In the conventional protocol, 3C signal is quantified using linear PCR (amplicon visualized and quantified on a gel). The substantial increase in sensitivity afforded by qPCR represents a key facet underlying the sensitivity and dynamic range of quantification of TaqI-3C.
6. Conclusions
The TaqI-3C method has led to important new insights into features of yeast chromosome topology/ 3D nuclear architecture that have heretofore been unappreciated. The technique offers high resolution, high sensitivity and a dynamic range of quantitation. A key finding arising from its use, trans-interactions between nonallelic, transcriptionally active yeast genes, has been observed in unrelated genetic backgrounds and confirmed by live cell microscopy. Additionally, the strong correlation between frequency of intragenic contacts with nascent transcription of HSP, ribosomal protein (RPL) and perhaps other genes [22, 31,61], suggests that Taq I-3C can be a useful tool for revealing functional association between 3D topology and gene transcription. The detailed, step-by-step procedure provided here, along with accompanying equations used to calculate chromatin contact frequencies, should permit successful application of the technique to any organism in which crosslinked chromatin can be readily isolated.
7. Step-by-step protocol
7.1. Preparing TaqI-3C templates
7.1.1. Cell culture
Inoculate a 10 ml seed culture of S. cerevisiae in an appropriate growth medium. Incubate the culture overnight on a rotating or shaking platform at 30°C.
-
Inoculate 50 ml of fresh growth medium with the overnight culture to an A600 equivalent of 0.15. Allow cells to grow to a mid-log density (A600 =0.65-0.8) with continuous shaking at 30°C.
Note: When comparing strains in the presence or absence of drug treatments, such as Anchor-Away strains in +/−rapamycin conditions, one must start the treatment at a cell density lower than 0.8. This way cell densities of treated and non-treated strains will be roughly similar at the time of harvesting.
7.1.2. Crosslinking of cells
-
3.
Add 1.4 ml of 37% formaldehyde (final concentration = 1%) to the 50 ml culture. Mix thoroughly with continuous shaking for 15 min at 30°C.
Note: If crosslinking is performed at temperatures other than 30°C, appropriate controls must be incorporated to account for any temperature-dependent variation [22].
-
4.
To this, add 5.4 ml of 1.25 M glycine (final concentration = 135 mM) for quenching excess formaldehyde. Mix thoroughly with continuous shaking for 5 min at 30°C, then place the flask on ice.
-
5.
Transfer the cells to a 50 ml tube, and centrifuge at 3000 g for 10 min at 4°C.
-
6.
Discard the supernatant and wash the pellet once with 10 ml of ice-cold TBS containing 1% Triton X-100.
-
7.
Resuspend the pellet in 1 ml of ice-cold TBS containing 1% Triton X-100 and transfer the cell suspension to a microfuge tube.
-
8.
Pellet cells by centrifugation at 10,000 g for 10 min at 4°C. Discard the supernatant.
-
9.
Resuspend the pellet in 500 μl of FA lysis buffer by pipetting up and down. Do not vortex. Add PMSF to the FA lysis buffer just before use. For buffer recipes and reagents, see Appendix A.
-
10.
Store cells in −80°C or proceed to step 11.
7.1.3. Cell lysis and isolation of crosslinked chromatin
-
11.
Thaw cells on ice. To a 500 μl cell suspension, add an equal volume of glass beads (acid washed). Use ice-cold beads to prevent damage to the chromatin.
-
12.
Vortex mix (2300 rpm) for two cycles (20 min each) at 4°C, with an intermittent ice break of 10 min.
-
13.
Flip the tube upside down and pierce its bottom using a 23 G needle. Now place the tube on top of another microfuge tube. Centrifuge the nested tubes at 3000 g for 10 min at 4°C.
-
14.
Discard the tube containing glass beads. Resuspend the loose pellet by pipet mixing and centrifuge again at 13,000 g for 10 min at 4°C.
-
15.
Discard the supernatant and resuspend the pellet in 1 ml FA lysis buffer. Centrifuge again at 13,000 g for 10 min at 4°C.
-
16.
Discard the supernatant and resuspend the pellet in 500 μl of 10 mM Tris-Cl (pH 7.5).
-
17.
Distribute the chromatin suspension into ten microfuge tubes (50 μl each). Label four tubes as DO3C, four as Lig3C and the remaining two as UND3C. Store the aliquots in −80°C or proceed to step 18.
7.1.4. Restriction digestion of crosslinked chromatin
-
18.
Thaw eight tubes labeled as DO3C and Lig3C on ice. To each tube containing 50 μl of crosslinked chromatin suspension, add 10 μl of 10X Cutsmart buffer and 30 μl of ddH2O. Mix carefully by pipetting up and down. Then add 10 μl of Taq I restriction enzyme (20 U/ μl), pipet mix, and incubate the reaction mix at 60°C for 7h. Flip tubes intermittently.
-
19.
For inactivation of Taq I, add 10 μl of 10% SDS to each of the eight tubes and incubate at 80°C for 20 min.
-
20.
To each tube, add 565 μl of ddH2O and 75 μl of 10% Triton X-100.
-
21.
Centrifuge tubes at 13,000 g for 20 min at room temperature. Discard the supernatant and resuspend pellets in 100 μl of 10 mM Tris-Cl (pH 7.5).
While it is important to remove the supernatant at this step, presumably because it has components that can interfere with ligation, we note that this fraction, which contains short chromatin fragments, may be impoverished in transcription-factor bound chromatin. Notably, Micro-C analysis, a 3C-based technique that uses micrococcal nuclease in lieu of a restriction enzyme, failed to detect gene looping interactions in the supernatant yet such interactions were present in the pellet [26, 65].
-
22.
Proceed to step 23 with four Lig3C tubes. To four DO3C tubes, add 600 μl of ddH2O and proceed to step 24. Thaw two UND3C tubes on ice, add 650 μl of ddH2O and proceed to step 24.
7.1.5. Intra-molecular ligation of crosslinked chromatin
-
23.
To each of the four Lig3C tubes containing 100 μl of digested chromatin, add 350 μl of the Quick ligase buffer, 245 μl of ddH2O and 5 μl of the Quick T4 DNA ligase enzyme (10,000 cohesive end units). Mix the tubes by gently pipetting up and down and incubate at 25°C for 2 h.
Note: We dilute the digested chromatin seven-fold before incubating with Quick T4 DNA ligase. This step is crucial as dilution will minimize inter-molecular contacts.
7.1.6. RNA removal, reversal of crosslinks and DNA purification
-
24.
To each of the four Lig3C tubes, add 2 μl of 10 mg/ml DNase free RNase (Sigma Aldrich; 30 ng/μl) and incubate at 37°C for 20 min. And, do likewise for DO3C and UND3C.
-
25.
To each of the ten tubes, add 7 μl of 10% SDS (final concentration = 0.1%) and 5 μl of 10 mg/ml Proteinase K (Sigma Aldrich; 70 ng/μl). Flip the tubes a few times and incubate overnight (12-14 h) at 65°C.
-
26.
To each tube, add an equal volume (700 μl) of the Phenol: Chloroform: Isoamyl alcohol (25:24:1; pH 8.0). Vortex tubes for 1-2 min, and centrifuge at 10,000 g for 10 min at room temperature.
-
27.
Collect the aqueous phase and repeat step 26. Do not discard the lower phase yet.
-
28.
To the aqueous phase, add Chloroform: Isoamyl alcohol (24:1) for removal of residual phenol. Vortex mix for 1-2 min and centrifuge at 10,000 g for 10 min at room temperature.
Note: Even after three rounds of organic extraction, there is DNA that is trapped within the crosslinked complexes. Therefore, we do an additional round of back-extraction.
-
29.
Pool aqueous phases for UND3C, DO3C or Lig3C into three separate tubes (15 ml each), and to each tube add, 1/10th volume of 3 M sodium acetate (pH 5.2) and 2 μl of glycogen (20 mg/ml). Mix well and add 2.5 volumes of ethanol. Mix gently by inverting tubes a few times and incubate for 30 min at room temperature.
-
30.
Centrifuge at 13,000 g for 20 min. Carefully aspirate the supernatant and remove any residual supernatant using a pipette. Air dry the pellet; do not over-dry it. Now dissolve the pellets from DO3C or Lig3C tubes in 450 μl TE (pH 8.0) and UND3C in 225 μl. The samples can be stored in −20°C for several months.
7.1.7. qPCR detection
-
31.
Thaw samples on ice. Prepare qPCR reaction master mix as below: 10 μl of 2X Power SYBR Green Master Mix + 0.5 μl Tandem Primer 1 + 0.5 μl Tandem Primer 2 + 7 μl ddH2O.
-
32.
To each well, add 18 μl of qPCR master mix and 2 μl of the 3C DNA template.
Note: Ideally, all 3C reactions should be prepared with the same qPCR master mix, but this may not be possible where multiple interactions are to be tested. However, we recommend running 3C and gDNA control templates on the same plate to minimize experimental variation. A minimum of two biological replicates and two technical replicates are recommended.
-
33.
Run the qPCR program for 40 cycles using standard cycling parameters. We use the standard settings on Applied Biosystems 7900HT. Use a compatible platform, if no access is available to this instrument.
Note: Add dissociation curve analysis step at the end of the qPCR run.
-
34.
Quantify interaction frequencies as described in section 3.4 above.
We note that many steps in Taq I-3C are common to the previously published yeast 3C protocol [40], including steps involved in generation of the genomic DNA control template.
Note: See Appendix A for buffer recipes and a troubleshooting guide.
Supplementary Material
Highlights.
Taq I-3C incorporates a novel normalization step to account for cleavage efficiency
Uses real-time qPCR to quantify interaction frequencies
Permits comparison of chromatin contacts in different physiological contexts
Has sufficient resolution to capture UAS-promoter interactions
Has led to the identification of evanescent intragenic and intergenic contacts
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Dekker J, Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture, Epigenetics Chromatin 7(1) (2014) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ansari A, Hampsey M, A role for the CPF 3'-end processing machinery in RNAP II-dependent gene looping, Genes Dev. 19(24) (2005) 2969–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tan-Wong SM, French JD, Proudfoot NJ, Brown MA, Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene, Proc Natl Acad Sci U S A 105(13) (2008) 5160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D, Genome-wide analysis of local chromatin packing in Arabidopsis thaliana, Genome Res 25(2) (2015) 246–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ, Gene loops juxtapose promoters and terminators in yeast, Nat. Genet 36(9) (2004) 1014–8. [DOI] [PubMed] [Google Scholar]
- [6].Rowley MJ, Lyu X, Rana V, Ando-Kuri M, Karns R, Bosco G, Corces VG, Condensin II Counteracts Cohesin and RNA Polymerase II in the Establishment of 3D Chromatin Organization, Cell Rep 26(11) (2019) 2890–2903 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cavalli G, Misteli T, Functional implications of genome topology, Nat Struct Mol Biol 20(3) (2013) 290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA, Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization, Cell 117(4) (2004) 427–39. [DOI] [PubMed] [Google Scholar]
- [9].Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H, Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development, Science 296(5565) (2002) 158–62. [DOI] [PubMed] [Google Scholar]
- [10].Cook PR, A Model for all Genomes: The Role of Transcription Factories, Journal of Molecular Biology 395(1) (2010) 1–10. [DOI] [PubMed] [Google Scholar]
- [11].Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P, Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells, Nat. Genet 42(1) (2010) 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N, Taylor S, Wood WG, Higgs DR, Iborra FJ, Buckle VJ, Association between active genes occurs at nuclear speckles and is modulated by chromatin environment, J Cell Biol 182(6) (2008) 1083–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B, Interactions among Polycomb domains are guided by chromosome architecture, PLoS Genet 7(3) (2011) e1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G, Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila, Cell 144(2) (2011) 214–26. [DOI] [PubMed] [Google Scholar]
- [15].Misteli T, Soutoglou E, The emerging role of nuclear architecture in DNA repair and genome maintenance, Nat Rev Mol Cell Biol 10(4) (2009) 243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hauer MH, Gasser SM, Chromatin and nucleosome dynamics in DNA damage and repair, Genes Dev 31(22) (2017) 2204–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cremer T, Cremer C, Chromosome territories, nuclear architecture and gene regulation in mammalian cells, Nat. Rev. Genet 2(4) (2001) 292–301. [DOI] [PubMed] [Google Scholar]
- [18].Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D, Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei, J Cell Biol 166(6) (2004) 815–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brickner JH, Walter P, Gene recruitment of the activated INO1 locus to the nuclear membrane, PLoS Biol. 2(11) (2004) e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA, Coactivator condensation at super-enhancers links phase separation and gene control, Science 361(6400) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II, Mediator and RNA polymerase II clusters associate in transcription-dependent condensates, Science 361(6400) (2018) 412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chowdhary S, Kainth AS, Gross DS, Heat Shock Protein Genes Undergo Dynamic Alteration in Their Three-Dimensional Structure and Genome Organization in Response to Thermal Stress, Mol Cell Biol 37(24) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dekker J, Mirny L, The 3D Genome as Moderator of Chromosomal Communication, Cell 164(6) (2016) 1110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS, A three-dimensional model of the yeast genome, Nature 465(7296) (2010) 363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rutledge MT, Russo M, Belton JM, Dekker J, Broach JR, The yeast genome undergoes significant topological reorganization in quiescence, Nucleic Acids Res 43(17) (2015) 8299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ, Mapping nucleosome resolution chromosome folding in yeast by Micro-C, Cell 162(1) (2015) 108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Eser U, Chandler-Brown D, Ay F, Straight AF, Duan Z, Noble WS, Skotheim JM, Form and function of topologically associating genomic domains in budding yeast, Proc Natl Acad Sci U S A 114(15) (2017) E3061–E3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Laine JP, Singh BN, Krishnamurthy S, Hampsey M, A physiological role for gene loops in yeast, Genes Dev. 23(22) (2009) 2604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Singh BN, Hampsey M, A transcription-independent role for TFIIB in gene looping, Mol Cell 27(5) (2007) 806–16. [DOI] [PubMed] [Google Scholar]
- [30].Tan-Wong SM, Wijayatilake HD, Proudfoot NJ, Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex, Genes Dev. 23(22) (2009) 2610–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chowdhary S, Kainth AS, Pincus D, Gross DS, Heat Shock Factor 1 Drives Intergenic Association of Its Target Gene Loci upon Heat Shock, Cell Rep 26(1) (2019) 18–28 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W, Looping and interaction between hypersensitive sites in the active beta-globin locus, Mol. Cell 10(6) (2002) 1453–65. [DOI] [PubMed] [Google Scholar]
- [33].Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA, Mediator and cohesin connect gene expression and chromatin architecture, Nature 467(7314) (2010) 430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y, Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation, Cell 148(1-2) (2012) 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Bluthgen N, Dekker J, Heard E, Spatial partitioning of the regulatory landscape of the X-inactivation centre, Nature 485(7398) (2012) 381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL, A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping, Cell 159(7) (2014) 1665–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Papantonis A, Kohro T, Baboo S, Larkin JD, Deng B, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, Li G, Poh HM, Ruan X, Aburatani H, Ruan Y, Kodama T, Wada Y, Cook PR, TNFalpha signals through specialized factories where responsive coding and miRNA genes are transcribed, EMBO J. 31(23) (2012) 4404–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Polach KJ, Widom J, Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation, J. Mol. Biol 254 (1995) 130–149. [DOI] [PubMed] [Google Scholar]
- [39].Dekker J, Rippe K, Dekker M, Kleckner N, Capturing chromosome conformation, Science 295(5558) (2002) 1306–11. [DOI] [PubMed] [Google Scholar]
- [40].Singh BN, Ansari A, Hampsey M, Detection of gene loops by 3C in yeast, Methods 48(4) (2009) 361–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].El Kaderi B, Medler S, Ansari A, Analysis of interactions between genomic loci through Chromosome Conformation Capture (3C), Curr Protoc Cell Biol Chapter 22 (2012) Unit22 15. [DOI] [PubMed] [Google Scholar]
- [42].Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ, Gene loops enhance transcriptional directionality, Science 338(6107) (2012) 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Medler S, Ansari A, Gene looping facilitates TFIIH kinase-mediated termination of transcription, Sci Rep 5 (2015) 12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].El Kaderi B, Medler S, Raghunayakula S, Ansari A, Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries, J Biol Chem 284(37) (2009) 25015–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Adams CC, Gross DS, The yeast heat shock response is induced by the conversion of cells to spheroplasts and by potent transcriptional inhibitors, J. Bacteriol 173 (1991) 7429–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA, Remodeling of yeast genome expression in response to environmental changes, Mol Biol Cell 12(2) (2001) 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jackson V, Formaldehyde cross-linking for studying nucleosomal dynamics, Methods 17(2) (1999) 125–39. [DOI] [PubMed] [Google Scholar]
- [48].Splinter E, Grosveld F, de Laat W, 3C technology: analyzing the spatial organization of genomic loci in vivo, Methods Enzymol 375 (2004) 493–507. [DOI] [PubMed] [Google Scholar]
- [49].Belton JM, Dekker J, Chromosome Conformation Capture (3C) in Budding Yeast, Cold Spring Harb Protoc 2015(6) (2015) 580–6. [DOI] [PubMed] [Google Scholar]
- [50].Solomon MJ, Larsen PL, Varshavsky A, Mapping protein-DNA interactions in vivo with formaldeyde: evidence that histone H4 is retained on a highly transcribed gene, Cell 937-947 (1988) 937–947. [DOI] [PubMed] [Google Scholar]
- [51].Gheldof N, Tabuchi TM, Dekker J, The active FMR1 promoter is associated with a large domain of altered chromatin conformation with embedded local histone modifications, Proc Natl Acad Sci U S A 103(33) (2006) 12463–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zhao J, Herrera-Diaz J, Gross DS, Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density, Mol Cell Biol 25(20) (2005) 8985–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Svaren J, Horz W, Transcription factors vs. nucleosomes: regulation of the PHO5 promoter in yeast, Trends Biochem. Sci 22(3) (1997) 93–97. [DOI] [PubMed] [Google Scholar]
- [54].Fraga D, Meulia T, Fenster S, Real-Time PCR, Current Protocols Essential Laboratory Techniques. 00 (2008) 10.03.01–10.03.34. [Google Scholar]
- [55].Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T, Quantitative analysis of chromosome conformation capture assays (3C-qPCR), Nat. Protoc 2(7) (2007) 1722–33. [DOI] [PubMed] [Google Scholar]
- [56].Dekker J, The three 'C' s of chromosome conformation capture: controls, controls, controls, Nat Methods 3(1) (2006) 17–21. [DOI] [PubMed] [Google Scholar]
- [57].Zhang D, Bai L, Interallelic interaction and gene regulation in budding yeast, Proc Natl Acad Sci U S A 113(16) (2016) 4428–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kim S, Liachko I, Brickner DG, Cook K, Noble WS, Brickner JH, Shendure J, Dunham MJ, The dynamic three-dimensional organization of the diploid yeast genome, Elife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mirkin EV, Chang FS, Kleckner N, Dynamic trans interactions in yeast chromosomes, PLoS One 8(9) (2013) e75895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Thompson M, Haeusler RA, Good PD, Engelke DR, Nucleolar clustering of dispersed tRNA genes, Science 302(5649) (2003) 1399–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Pincus D, Anandhakumar J, Thiru P, Guertin MJ, Erkine AM, Gross DS, Genetic and epigenetic determinants establish a continuum of Hsf1 occupancy and activity across the yeast genome, Mol Biol Cell 29(26) (2018) 3168–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yaffe E, Tanay A, Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture, Nat Genet 43(11) (2011) 1059–65. [DOI] [PubMed] [Google Scholar]
- [63].Cournac A, Marie-Nelly H, Marbouty M, Koszul R, Mozziconacci J, Normalization of a chromosomal contact map, BMC Genomics 13 (2012) 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee K, Hsiung CC, Huang P, Raj A, Blobel GA, Dynamic enhancer-gene body contacts during transcription elongation, Genes Dev. 29(19) (2015) 1992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hsieh TS, Fudenberg G, Goloborodko A, Rando OJ, Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome, Nat Methods 13(12) (2016) 1009–1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.