Abstract
Serendipita (=Piriformospora) indica is a fungal endophytic symbiont with the capabilities to enhance plant growth and confer resistance to different stresses. However, the application of this fungus in the field has led to inconsistent results, perhaps due to antagonism with other microbes. Here, we studied the impact of individual bacterial isolates from the endophytic bacterial community on the in vitro growth of S. indica. We further analyzed how combinations of bacteria and S. indica influence plant growth and protection against the phytopathogens Fusarium oxysporum and Rhizoctonia solani. Bacterial strains of the genera Bacillus, Enterobacter and Burkholderia negatively affected S. indica growth on plates, whereas Mycolicibacterium, Rhizobium, Paenibacillus strains and several other bacteria from different taxa stimulated fungal growth. To further explore the potential of bacteria positively interacting with S. indica, four of the most promising strains belonging to the genus Mycolicibacterium were selected for further experiments. Some dual inoculations of S. indica and Mycolicibacterium strains boosted the beneficial effects triggered by S. indica, further enhancing the growth of tomato plants, and alleviating the symptoms caused by the phytopathogens F. oxysporum and R. solani. However, some combinations of S. indica and bacteria were less effective than individual inoculations. By analyzing the genomes of the Mycolicibacterium strains, we revealed that these bacteria encode several genes predicted to be involved in the stimulation of S. indica growth, plant development and tolerance to abiotic and biotic stresses. Particularly, a high number of genes related to vitamin and nitrogen metabolism were detected. Taking into consideration multiple interactions on and inside plants, we showed in this study that some bacterial strains may induce beneficial effects on S. indica and could have an outstanding influence on the plant-fungus symbiosis.
Keywords: Serendipita indica, bacterial endophytes, symbiosis, Mycolicibacterium, tripartite interactions, biocontrol, fungal stimulation
Introduction
In all terrestrial plants and natural ecosystems, multipartite interactions take place between plants and different kinds of microbes (Hardoim et al., 2015). While some bacteria can have a negative impact on the surrounding microflora (Barea et al., 2005; Deveau et al., 2018), other bacteria are known as boosting colonization and establishment of endophytic fungi such as in the case of mycorrhiza (van Overbeek and Saikkonen, 2016), acting therefore as helper bacteria (Frey-Klett et al., 2007). Some of these bacteria are also plant growth-promoting rhizobacteria (PGPR), capable of increasing plant growth and resistance against fungal pathogens by mobilizing nutrients, production of siderophores, auxins, ACC-deaminase, polyamines as well as antibiosis and chitinolytic compounds (Whipps, 2001; Glick, 2014). Most of the helper bacteria were isolated from the rhizosphere. However, some could thrive as endophytes in their host plants, crossing from the root surfaces to the inner plant tissues (Brader et al., 2017). These bacteria are particularly important for the development of biostimulant products, as those microbes with the ability to colonize roots are more prepared to survive and exert beneficial effects on the plant (Compant et al., 2005). Bacteria can further penetrate fungal hyphae and establish symbiosis, playing sometimes critical roles for the survival of the fungal hosts (Bertaux et al., 2005; Bonfante and Desirò, 2017; Guo et al., 2017). Fungal hyphae provide a nutrient-rich niche for bacterial growth (Linderman, 1992; Boer et al., 2005; Scherlach et al., 2013). The hyphae-associated bacteria may reciprocally promote hyphal development and root colonization through the supply of vitamins, nitrogen, phosphorus, sugars, and secondary metabolites to the fungal partner or by increasing ATP production (Hildebrandt et al., 2006; Ghignone et al., 2012), as exemplified by the mycorrhizal helper bacteria of arbuscular mycorrhizal fungi (AMF) (Frey-Klett et al., 2007). However, bacteria with suppressive effects on fungi (e.g. secretion of antifungal compounds, mycophagy) have also been widely reported (Bonfante and Anca, 2009; Kobayashi and Crouch, 2009). Understanding and exploiting the association between different kinds of beneficial microorganisms, together with the management and engineering of the plant microbiome, may lead to improving soil fertility, crop productivity, and biological control of plant pathogens (Berg, 2009; Collinge et al., 2019; Compant et al., 2019).
An example of beneficial fungi interacting with symbiotic bacteria is Serendipita indica, previously known as Piriformospora indica (Varma et al., 1999). It is a well-known root endophytic fungus that mimics the capabilities of AMF, but in contrast to AMF it can be cultured axenically (Varma et al., 1999). The fungus improves crop yield and confers resistance against biotic and abiotic stresses by triggering induced systemic resistance (ISR), boosting antioxidant capacity, mobilizing nutrients, and manipulating the hormone levels of the plant (Franken, 2012; Gill et al., 2016). Interestingly, it has been demonstrated that this fungus hosts an endobacterium, Rhizobium radiobacter (Sharma et al., 2008), which has endophytic as well as plant growth-promoting properties, although its functional role is not yet fully understood (Glaeser et al., 2016). Only few studies have demonstrated in vitro that particular bacterial strains can be detrimental for the growth of S. indica (Varma et al., 2013) or in contrast have stimulatory effects, like strain WR5 of Azotobacter chrococcum, which enhanced mycelial growth and sporulation of S. indica in vitro (Bhuyan et al., 2015). In the last few years, some researchers have developed co-inoculations (microbial consortia) of fungi and bacteria, searching synergisms between two beneficial microbes (Artursson et al., 2006; Collinge et al., 2019). Special focus has been laid on the combination of S. indica with PGPR. Kumar et al. (2012) detected increased plant growth by combining S. indica and pseudomonad R81. However, combinations of S. indica and bacteria have not always been successful (Sarma et al., 2011). Similarly, several combinations of different biocontrol agents have not improved the effect exerted by the most efficacious one, indicating no synergistic but more likely antagonistic interactions (Xu et al., 2011). This suggests that more research is needed in the exploration of compatible combinations. We hypothesized, nevertheless, that several bacterial taxa or strains can positively interact with the beneficial fungus S. indica and that these bacterial-fungal interactions could be exploited for crop enhancement and resistance against different phytopathogens.
The aim of this study was to identify bacterial taxa that stimulate the growth and effects of the beneficial fungus S. indica and to select the most promising “fungus-conducive” bacteria for dual-inoculation with S. indica. The effect of these combinations on plant growth and biocontrol activity against fungal pathogens was further assessed. In particular, we aimed at researching the tripartite interactions between S. indica, various bacterial strains and the plant pathogens F. oxysporum or R. solani. We further explored potential mechanisms involved using genome analysis of most promising bacterial endophytes showing positive interaction with S. indica.
Materials and Methods
Serendipita indica Cultivation
Serendipita (=Piriformospora) indica strain DSM 11,827 was provided by Pr. Philipp Franken and obtained from the “Deutsche Sammlung für Mikroorganismen und Zellkulturen,” Braunschweig, Germany (Varma et al., 1999). The fungus was maintained at −80°C in sterile Potato Dextrose Broth (PDB) (Carl Roth, Germany) amended with 25% glycerol and grown on Potato Dextrose Agar (PDA) plates or in liquid culture containing Aspergillus complete medium (Pontecorvo et al., 1953).
To produce inoculum, roughly fifty 2-mm agar plugs from a 2-week old culture of S. indica grown on PDA were transferred to 250 ml Erlenmeyer flasks containing 100 ml of Aspergillus CM and incubated for 3 weeks under constant shaking (150 rpm) at 26 ± 1°C. Mycelium and spores were collected by centrifugation (4,500 rpm, 5 min) and the remaining pellet was washed 3–5 times with sterile phosphate-buffered saline (PBS) of pH 6.5. The mixture of mycelium and spores, resuspended in PBS, was ground with a homogenizer Ultra Turrax T25 (IKA®, Staufen, Germany) for 3 min in intervals of 30 s. The number of spores + mycelium fragments was estimated with a hemocytometer (NanoEnTek, Seoul, Korea) and the viability of the CFU confirmed by plating on PDA. Final concentrations were adjusted with PBS.
Plant Assay for Isolation of Endophytic Bacteria
An agricultural soil was sampled in a field located in Meires, Lower Austria, Austria (48°46′43.8″N; 15°17′23.4″E) at a depth of 5–15 cm and stored at 4°C. To create an isolated microsystem, one potato tuber (Solanum tuberosum L. cv. Romina; NÖ. Saatbaugenossenschaft, Austria) and four tomato seeds (Solanum lycopersicum L. cv. Moneymaker; Austrosaat, Vienna, Austria) were grown in closed Magenta boxes (Sigma-Aldrich) containing ~200 g of the agricultural soil. The plants were kept in greenhouse with a Day/Night temperature of 22/21°C, a relative humidity 50/35% and a 12 h light/dark photoperiod. Plants were watered weekly with 20 ml of water. After 4 weeks, potato and tomato plants were inoculated by drenching a mixture of spores + fragmented mycelium of S. indica at 104 CFU/g of soil. Control treatment was mock-inoculated with PBS. To mitigate a possible big shift in the bacterial community caused by S. indica, an additional treatment with low-concentrated (102 CFU/g). S. indica inoculum was included for potato plants. In total five treatments were prepared with three replicates (boxes) per treatment (Supplementary Figure 1).
Isolation of Endophytic Bacteria
Three weeks after inoculation, 1.5 g of potato and 0.2 g of tomato roots were harvested from each magenta box to isolate endophytic bacteria. For this, roots were rinsed abundantly with tap water, then surface-sterilized with 70% ethanol for 10 s followed by 2.5% sodium hypochlorite for 3 min and then rinsed 3 times with sterile water. For each sample, 100 μl of the final wash were plated in triplicates on Nutrient Agar No2 (NA) (Sigma-Aldrich, St. Louis, USA) until 6 days of incubation at 26°C to confirm the surface sterilization. To isolate bacteria, sterile roots, macerated in 5 ml of sterile 0.85% NaCl, were smashed with a mortar and pestle and homogenized by vortexing for 30 s at maximum speed. Smashed roots from each sample were 10-fold diluted on PBS and 100 μl from each dilution were plated on NA and further incubated at 26°C for 6 days. Based on visual differences in morphology, single colonies were randomly picked only from the most diluted cultures to avoid contaminations. Selected bacteria were further purified by repeating streaking on NA plates and all the recovered isolates were stored at −80°C in Nutrient Broth (NB) (Difco, Detroit, MI) supplemented with 25% glycerol. At least 100 isolates were obtained from every treatment with potato plants, and 50 with tomato plants.
DNA Isolation and 16S rRNA Gene Sequencing
Bacterial DNA of each isolate was extracted using the UltraClean® Microbial DNA Isolation Kit (QIAGEN, Venlo, Netherlands) according to the manufacturer's instructions. The 16S rRNA genes were amplified using the primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) (Weisburg et al., 1991) and 1520R (5′-AAGGAGGTGATCCAGCCGCA-3′) (Edwards et al., 1989). A conventional PCR amplification of 20 μl PCR reaction mix containing 2.5 mM MgCl2, 0.2 mM dNTPs, 0.3 mM of each primer, 1–2 μl of DNA template, 1 U HOT FIREPol® DNA polymerase (Solis BioDyne) and 1 × PCR reaction buffer (Invitrogen) was carried out in a thermocycler peqSTAR 96X HPL (PEQLAB Biotechnologie GmbH). An initial denaturation step at 95°C for 5 min was followed by 30 cycles of denaturation at 95°C for 45 s, annealing at 54°C for 60 s and elongation at 72°C for 90 s, plus a final extension at 72°C for 10 min were performed. Sequencing of the PCR product was performed by LGC-Genomics (Teddington, UK) using the primers 8F and 1495r (5′-CTACGGCTACCTTGTTACGA-3′) (Lane, 1991). To remove duplicate sequences from the library, sequences were de-replicated and clustered at 100% similarity with the Avalanche NextGen Workbench (http://www.visualbioinformatics.com/bioinf/index.html). Strains showing the same partial 16S rRNA genes and the same phenotypic interaction with S. indica were removed. The identification of isolates was performed by BLAST search on a local installation of the complete NCBI's nt database (downloaded in October 2018), targeting the first more significant 50 hits. Taxonomic assignment of BLAST hits was then refined with the approach implemented in BlobTools (Laetsch and Blaxter, 2017). Sequence data are available at NCBI database and GenBank under the accession numbers MN180888–MN181366.
Effect of Endophytic Bacteria on Mycelial Growth of S. indica
To determine the effect of bacteria on S. indica, the growth of the fungus in interaction with each endophytic bacterium was expressed in terms of hyphae expansion on agar plates. For this, bacteria were pre-cultured for 4 days on NA and the fungus for 2 weeks on PDA. To study the interaction between bacteria and S. indica, each bacterium was then streaked on 1 cm2 in the center of a Petri dish (9 cm Ø, containing 15 ml PDA) and a 0.5-cm2 agar plug of active S. indica mycelium was placed inverted over the streaked bacterium. As a control, S. indica was grown alone. All the co-cultures were replicated four times. Aiming a confirmation of results, some selected isolates were additionally co-cultured with S. indica under different nutrient conditions (a mixture 1:1 of PDA+NA), and under longer bacterial growth phase (9 days of bacterial preculture).
Plates were incubated at 26°C in darkness. After 13 days of dual-culturing, the surface of the plate covered with S. indica mycelium was measured with ImageJ 1.48 software (https://imagej.nih.gov/ij/) and the average measurement of the four replicated plates was employed to determine the type of interaction. The difference in growth between S. indica co-cultured with a bacterium (dual-cultured S. indica) and control was calculated as relative increase/decrease of dual-cultured S. indica growth respect to the control. To characterize the type of interaction between fungus and bacteria, an arbitrary scale was further established. When the growth of dual-cultured S. indica was reduced by more than 90% in respect to the control, the interaction was considered as a complete inhibition. A reduction between 90 and 20% was determined as a negative interaction. If the growth of dual-cultured S. indica was decreased or increased up to 20% respect to the control, a neutral interaction was established. An increment of dual-cultured S. indica growth larger than 20% defined a positive interaction (Figure 1B).
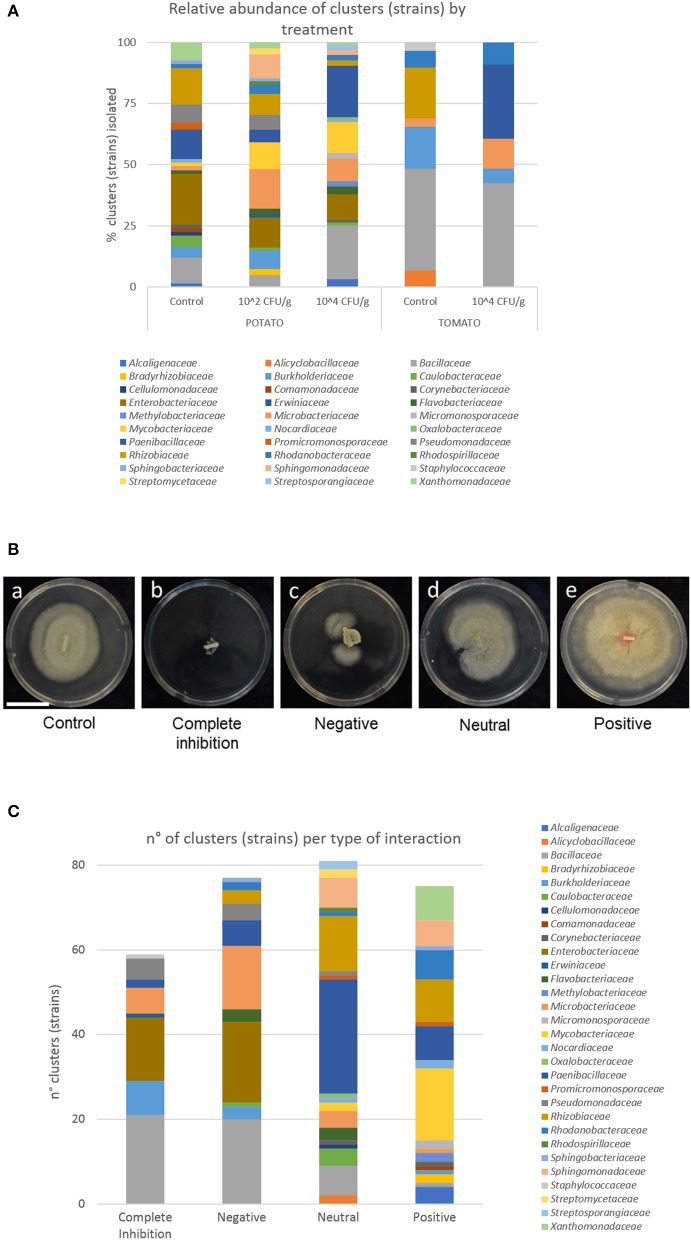
Figure 1.
Isolates of bacteria and interaction with S. indica. (A) Relative abundance of bacterial families isolated from potato and tomato roots, and from plants pre-inoculated or not with S. indica. (B) Growth of the fungus alone (a) and in combination with bacteria completely inhibiting fungal growth (b), interacting negatively (c), in a neutral way (d), and stimulating the growth of the fungus (e). Bar corresponds to 3 cm. (C) Number of bacterial clusters (strains) and their phylogeny per type of interaction.
Combining Selected Beneficial Endophytic Bacteria and S. indica for Tomato Growth Promotion
Since most of the isolates from one genus (Mycolicibacterium) stimulated S. indica growth, four isolates (P1-5, P1-18, P9-22, and P9-64) of this genus were selected for further experiments. With the aim of studying the effect of dual inoculations of S. indica and bacteria on tomato plants, a pot experiment was conducted in the greenhouse.
For the pot experiment, S. indica inoculum was produced as described above. In the case of bacteria, they were grown for 2.5 days in 10 ml bottom-rounded Falcon tubes containing 5 ml NB at 26°C and constant shaking (190 rpm). The cultures were centrifuged (4,600 rpm, 6 min, room T°C) and washed 3 times with sterile PBS to remove traces of media. Cell growth was determined by measuring the OD and CFU were estimated by standard serial dilution on NA.
Tomato seeds cv. Moneymaker were germinated for 4 days in a Whatman® filter paper (110 mm Ø) at room temperature on Petri dishes. Germinated seeds were transferred to 50 ml falcon tubes containing 15 ml of PBS and either (i) bacterial cells (5 × 107 CFU/ml), (ii) spores + hyphae fragments of S. indica (5 × 105 CFU/ml), or (iii) a mixture of bacterial cells (5 × 107 CFU/ml), and S. indica (5 × 105 CFU/ml). Falcon tubes were maintained in a Tube Roller RS-TR5 (Phoenix Instrument GmbH, Garbsen, Germany) for 30 min. For control treatment, the seeds were immersed in 1 × PBS.
Two seeds per pot were then sown at 1 cm depth in pots (1 l capacity) containing the substrate “Fruhstorfer Erde Typ Nullerde” (Hawita Gruppe, Vechta, Germany). The experiment had 10 treatments, ± S. indica and ± individual bacterial strains with 14 replicates (7 pots × 2 plants per pot) for each treatment. The plants were grown in the greenhouse (with conditions described before) and watered twice a week with tap water. Plants were harvested 6 weeks after planting and shoot fresh weight and leaf area were measured (using ImageJ). A confirmatory experiment was repeated with a richer soil in which pots were filled with a mixture (1:1:1 v/v) of perlite, sand, and the substrate “Tonsubstrat ED63 Special” (Einheitserde, Germany). After harvesting, shoot fresh and dry weight (after oven-drying for 3 days at 70°C) were measured.
In vitro Interaction Between Selected Bacteria and Plant Pathogens
In addition to determining the effect of selected bacteria on the growth of S. indica and tomato plants, they were also tested for their effects on pathogens such as Fusarium oxysporum f. sp. lycopersici Fol4287 (kindly provided by Maria E. Constantin, University of Amsterdam, Netherlands) (Di Pietro and Roncero, 1996) and Rhizoctonia solani AG-3 (kindly provided by Rosanna C. Hennessy, University of Copenhagen, Denmark). These fungi were maintained at −80°C in PDB amended with 25% glycerol. In vitro dual culture assays between these fungal pathogens and selected bacterial isolates were performed as described above for the beneficial fungus S. indica, but due to different growth rates, fungal preculture and final measurements were shortened to 3 days in case of R. solani, and 6 days for F. oxysporum.
Tomato Protection Against Fusarium oxysporum and Rhizoctonia solani Using Multipartite Interaction
Biocontrol of F. oxysporum was evaluated by pot experiment in which plants were infected with the pathogen and with single or dual inoculations of S. indica and selected bacteria. Inoculum production of S. indica and bacteria and seed inoculation were carried out as described above. The seeds were planted in 1 l pots containing a mixture (1:1:1 v/v) of perlite, sand and the substrate “Tonsubstrat ED63 Special” (Einheitserde, Germany) (2 seeds per pot, 5 pots per treatment).
For production of Fusarium, spores were obtained according to van der Does et al. (2019). Briefly, an agar plug from a 6-day old PDA culture of Fusarium was transferred to a 250-ml Erlenmayer flask containing 100 ml minimal media (3% sucrose, 0.17% yeast nitrogen base without amino acids or ammonia, and 100 mM KNO3), and incubated for 5 days at 26°C, 190 rpm. Spores were filtered through a Miracloth filter (Millipore), washed twice with sterile 1 × PBS and diluted to a concentration of 107 spores/ml.
Ten days after planting, tomato plants were infected with the solution of F. oxysporum described before according to the root dip method (Wellman, 1939). Seedlings were uprooted and trimmed leaving roughly 1 cm of root, to facilitate the penetration of Fusarium. Roots were placed for 30 min in the spore suspension of Fusarium, and directly repotted. Five weeks after inoculation, plant weight above the cotyledons was measured, and the extent of disease progression was scored according to de Lamo et al. (2018). Briefly, disease index was 0 = no symptoms, 1 = one brown vessel above the soil, 2 = one or two brown vascular bundles at the cotyledon level, 3 = at least three brown vessels and growth distortion, 4 = all vessels brown or the plant is small and wilted, 5 = dead plant.
The effect of dual-inoculation of S. indica and endophytic bacteria against the damping-off causative agent R. solani was analyzed, in parallel to F. oxysporum test, by a germination assay in closed boxes (Steri Vent Containers 107 × 94 × 96 mm, Duchefa Biochemie b.v, Haarlem, Netherlands). These boxes contained 120 g of a sterile (2 times, 121°C, 20 min) 1:4 mixture (w/w) of vermiculite (2–3 mm, Sigma-Aldrich) and distilled water. Tomato seeds cv. Moneymaker were surface-sterilized with 2.5% sodium hypochlorite for 5 min and rinsed 8 times with sterile water. Seeds were inoculated with either (i) S. indica, (ii) (x4) bacteria, or (iii) (x4) combination of fungus and bacteria as earlier described.
For this, Rhizoctonia was cultured on 1/5 PDA for 3 weeks. Five agar plugs of Rhizoctonia mycelium were placed in a row in the center of each box, and 2 rows of tomato seeds (5 seeds per row) were sown on both sides of the pathogen row at 2 cm distance. Both phytopathogen and seeds remained at 0.5 cm depth. Control was prepared with plugs of 1/5 PDA. Three replicated boxes were prepared per treatment and maintained in the greenhouse (see above). The number of germinated seedlings per box was monitored regularly for 4 months by scoring as follows: 1 = Plant germinated and no disease symptoms, 0.5 = Plant germinated, alive, with necrotic areas in leaves and stems, 0 = plant dead.
Bacterial Genome Sequencing and Analysis
Bacterial genomic DNA from 4 selected Mycolicibacterium strains were isolated using a phenol-chloroform based protocol according to Samad et al. (2016). Concisely, cells were grown on NB for 3 days and collected by centrifugation. Each bacterial pellet was resuspended in lysis buffer (5 mM EDTA pH8, 50 mM Tris-Cl, 1% SDS, 0.5 M NaCl, 0.2 mg/ml Proteinase K) and incubated at 65°C overnight, 400 rpm. DNA was extracted 2 times using 1 volume of phenol-chloroform-isoamylalcohol (25:24:1) and collected by centrifugation. Genomic DNA was further cleaned with Amicon Ultra 0.5 mL 30K Centrifugal Filter Units (Millipore, Cork, Ireland) and re-suspended in water. Whole-genome shotgun sequencing was performed on an Illumina HiSeq (GATC Biotech, Konstanz, Germany), producing 2 × 150 bp reads.
Illumina reads were checked for the presence of PhiX using Bowtie 2 (v2.3.4.3) (Langmead and Salzberg, 2012) and adapters were removed with fastp (v0.19.5) (Chen et al., 2018). Sequence quality and length distribution were checked via FastQC1 (Andrews, 2010). Genome assembly was carried out with SPAdes v3.13.0 (Bankevich et al., 2012) and short (<500 bp), low-abundant (<2×) contigs filtered out. The presence of contaminant contigs was assessed using BlobTools and alien contigs were eventually removed. Genome assembly quality was then inferred using QualiMap v2.2 (Okonechnikov et al., 2015) and QUAST v5.0.0 (Gurevich et al., 2013) and genome completeness reconstruction was evaluated with BUSCO v3.0 (Waterhouse et al., 2017). Gene annotation was performed using Prokka v1.12 (Seemann, 2014) and NCBI Prokaryotic Genome Annotation Pipeline (PGAP). Contigs were further screened for the presence of antimicrobial resistance or virulence genes with ABRicate v0.8.10. The presence of plasmids was ascertained by using Mash v2.1 against the PLSDB database (Galata et al., 2018).
Functional annotation was performed using EggNOG 4.5 (Huerta-Cepas et al., 2015) and the ClassicRAST (Rapid Annotation using Subsystem Technology) web server (http://rast.nmpdr.org) (Aziz et al., 2008). Prediction of biosynthetic gene clusters and secondary metabolites was additionally carried out using antiSMASH version 4.0.2 (Weber et al., 2015). CAZy families were identified with dbCAN2 according to the DIAMOND database. A cutoff of E-Value of 1e-102 was set for the output. When a gene contained a CBM with other CAZy classes, the gene was classified as CBM. Protein annotation was based on the CAZy database (Cantarel et al., 2008; Lombard et al., 2013).
The Average Nucleotide Identity (ANI) analysis was further used to determine the relatedness between the assembled genomes and affiliated genomes available in NCBI database classified as Mycobacterium or Mycolicibacterium. For this, 169 genomes were downloaded using the script available at https://github.com/kblin/ncbi-genome-download and ANI was calculated with the pyani Python module available at https://github.com/widdowquinn/pyani, using BLAST (ANIb) and TETRA methods. Based on the ANI pair-wise values, a distance matrix representing ANI-divergence (defined as 100% ANI data) (Chan et al., 2012) was compiled to display a heat map and compute a dendrogram using the hierarchical clustering adopting the complete linkage algorithm, with the software Morpheus (https://software.broadinstitute.org/morpheus/).
The draft genome sequences for the Mycolicibacterium strains P1-5, P1-18, P9-22, and P9-64 are available at NCBI, BioProject PRJNA393298, with the DDBJ/ENA/GenBank accession numbers NPKT00000000, NPKR00000000, NPKP00000000, and NPKO00000000, respectively.
Statistical Analysis
Statistical analysis of in vitro fungal growth, analysis of biomass and leaf area of samples from the growth enhancement experiments were performed in R 3.5.1 (R Core Team, 2019). Data distributions were checked using the fitdistrplus package (Delignette-Muller and Dutang, 2015) and linear or linear mixed-effects models (nlme R package) (Pinheiro et al., 2019), when applicable, were generated. After graphical verification of homogeneity assumption, ANOVA was applied on previously generated models, followed by pairwise comparisons (Tukey's method, P = 0.05) calculated using Estimated Marginal Means (emmeans R package) (Lenth, 2019). Quantitative data were processed with dplyr package (Wickham et al., 2019) and results visualized with boxplots using ggplot2 package (Wickham and Chang, 2016). The samples from the biocontrol experiment were analyzed with PRISM 8.0 (GraphPad). Concerning Fusarium, a non-parametric Mann-Whitney U-test was applied on the fresh weight and disease index data (de Lamo et al., 2018). The germination assay with Rhizoctonia was analyzed by one-way ANOVA and Tukey's test (P = 0.05).
Results
Isolation and Identification of Bacteria
In total, 479 isolates were recovered and identified at the genus level (Table 1). The most abundant families of isolates were Bacillaceae (21.09% of the total isolates), Enterobacteriaceae (13.78%), Rhizobiaceae (11.48%), and Paenibacillaceae (10.02%) (Table 1 and Supplementary Figure 2A). At the genus level, the overall top five genera were Bacillus (21.09% of isolates), Enterobacter (13.57%), Paenibacillus (8.56%), Burkholderia (7.52%), and Agrobacterium (6.05%). After de-replication with a 100% threshold, we obtained 260 different clusters (strains) with different 16S rRNA genes. The most abundant strains belonged to the families Bacillaceae (17.31%), Paenibacillaceae (15.0%), Enterobacteriaceae (11.15%), and Microbacteriaceae (9.23%). At the genus level, the most abundant strains belonged to Bacillus (17.31%), Paenibacillus (12.69%), Enterobacter (10.77%), and Mycolicibacterium (7.31%).
Table 1.
Species identified by sequencing the 16S rRNA gene and number of isolates and clusters (strains) assigned to species.
| Order | Family | Genus | Species | Clusters | Isolates | Potato | Tomato | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl. | 102 CFU | 104 CFU | Ctrl. | 104 CFU | ||||||
| Caulobacteriales | Caulobacteraceae | Brevundimonas | B. lenta | 1 | 2 | 1 | 0 | 1 | 0 | 0 |
| B. vesicularis | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Caulobacter | C. sp. | 2 | 2 | 2 | 0 | 0 | 0 | 0 | ||
| Rhizobiales | Bradyrhizobiaceae | Bosea | B. thiooxidans | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| Tardiphaga | T. robiniae | 1 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| Methylobacteriaceae | Methylobacterium | M. tardum | 2 | 2 | 0 | 0 | 2 | 0 | 0 | |
| Rhizobiaceae | Agrobacterium | A. rhizogenes | 5 | 12 | 2 | 3 | 0 | 7 | 0 | |
| A. tumefaciens | 5 | 17 | 3 | 7 | 2 | 5 | 0 | |||
| Neorhizobium | N. galegae | 1 | 1 | 1 | 0 | 0 | 0 | 0 | ||
| Rhizobium | R. alamii | 1 | 3 | 1 | 0 | 0 | 2 | 0 | ||
| R. gallicum | 1 | 4 | 0 | 4 | 0 | 0 | 0 | |||
| R. sullae | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| R. tibeticum | 2 | 4 | 4 | 0 | 0 | 0 | 0 | |||
| Shinella | S. zoogloeoides | 2 | 13 | 12 | 1 | 0 | 0 | 0 | ||
| Rhodospirillales | Rhodospirillaceae | Inquilinus | I. ginsengisoli | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| Sphingomonadales | Sphingomonadaceae | Novosphingobium | N. barchaimii | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| Sphingobium | S. japonicum | 1 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| S. sp. | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Sphingomonas | S. asaccharolytica | 3 | 3 | 0 | 3 | 0 | 0 | 0 | ||
| S. kyeonggiensis | 1 | 3 | 0 | 0 | 3 | 0 | 0 | |||
| S. melonis | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| S. paucimobilis | 1 | 3 | 0 | 3 | 0 | 0 | 0 | |||
| S. pituitosa | 1 | 2 | 0 | 0 | 2 | 0 | 0 | |||
| Burkholderiales | Alcaligenaceae | Achromobacter | A. sp. | 4 | 5 | 1 | 0 | 4 | 0 | 0 |
| Burkholderiaceae | Burkholderia | B. ambifaria | 8 | 35 | 3 | 10 | 0 | 22 | 0 | |
| B. sp. | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Paraburkholderia | P. phenazinium | 1 | 1 | 0 | 0 | 0 | 0 | 1 | ||
| P. soli | 1 | 1 | 0 | 0 | 0 | 0 | 1 | |||
| Ralstonia | R. pickettii | 1 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| Comamonadaceae | Variovorax | V. paradoxus | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |
| Oxalobacteraceae | Massilia | M. haematophila | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Enterobacterales | Enterobacteriaceae | Enterobacter | E. asburiae | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| E. cloacae | 17 | 51 | 19 | 18 | 14 | 0 | 0 | |||
| E. ludwigii | 9 | 11 | 2 | 6 | 3 | 0 | 0 | |||
| E. sp. | 1 | 2 | 2 | 0 | 0 | 0 | 0 | |||
| Lelliottia | L. amnigena | 1 | 1 | 1 | 0 | 0 | 0 | 0 | ||
| Erwiniaceae | Pantoea | P. ananatis | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |
| Pseudomonadales | Pseudomonadaceae | Pseudomonas | P. brassicacearum | 3 | 9 | 8 | 1 | 0 | 0 | 0 |
| P. graminis | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| P. koreensis | 2 | 2 | 0 | 2 | 0 | 0 | 0 | |||
| P. putida | 2 | 5 | 5 | 0 | 0 | 0 | 0 | |||
| P. sp. | 1 | 3 | 0 | 3 | 0 | 0 | 0 | |||
| Xanthomonadales | Rhodanobacteraceae | Dyella | D. marensis | 2 | 2 | 0 | 2 | 0 | 0 | 0 |
| Luteibacter | L. rhizovicinus | 4 | 10 | 1 | 1 | 2 | 6 | 0 | ||
| Rhodanobacter | R. lindaniclasticus | 1 | 1 | 0 | 0 | 0 | 0 | 1 | ||
| R. sp. | 2 | 2 | 0 | 0 | 0 | 0 | 2 | |||
| Xanthomonadaceae | Stenotrophomonas | S. maltophilia | 8 | 12 | 8 | 2 | 2 | 0 | 0 | |
| Flavobacteriales | Flavobacteriaceae | Chryseobacterium | C. daecheongense | 3 | 7 | 0 | 4 | 3 | 0 | 0 |
| C. taeanense | 1 | 2 | 1 | 0 | 1 | 0 | 0 | |||
| Sphingobacteriales | Sphingobacteriaceae | Mucilaginibacter | M. sp. | 2 | 2 | 1 | 1 | 0 | 0 | 0 |
| Corynebacteriales | Corynebacteriaceae | Corynebacterium | C. sp. | 2 | 2 | 1 | 0 | 1 | 0 | 0 |
| Mycobacteriaceae | Mycolicibacterium | M. frederiksbergense | 2 | 2 | 0 | 1 | 1 | 0 | 0 | |
| M. gilvum | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| M. hodleri | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| M. llatzerense | 3 | 4 | 1 | 2 | 1 | 0 | 0 | |||
| M. moriokaense | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| M. mucogenicum | 3 | 7 | 0 | 3 | 4 | 0 | 0 | |||
| M. neoaurum | 4 | 4 | 0 | 1 | 3 | 0 | 0 | |||
| M. pallens | 1 | 3 | 0 | 0 | 3 | 0 | 0 | |||
| M. peregrinum | 2 | 2 | 0 | 0 | 2 | 0 | 0 | |||
| M. smegmatis | 1 | 2 | 0 | 2 | 0 | 0 | 0 | |||
| Nocardiaceae | Rhodococcus | R. erythropolis | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |
| Nocardia | N. nova | 1 | 2 | 2 | 0 | 0 | 0 | 0 | ||
| Micrococcales | Cellulomonadaceae | Cellulomonas | C. hominis | 1 | 1 | 1 | 0 | 0 | 0 | 0 |
| Promicromonosporaceae | Cellulosimicrobium | C. cellulans | 2 | 3 | 3 | 0 | 0 | 0 | 0 | |
| Microbacteriaceae | Leifsonia | L. shinshuensis | 9 | 14 | 0 | 3 | 4 | 0 | 7 | |
| L. soli | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| L. xyli | 4 | 6 | 1 | 2 | 3 | 0 | 0 | |||
| Lysinimonas | L. sp. | 3 | 3 | 0 | 2 | 0 | 1 | 0 | ||
| Microbacterium | M. oxydans | 2 | 2 | 0 | 1 | 1 | 0 | 0 | ||
| Rathayibacter | R. agropyri | 4 | 7 | 0 | 7 | 0 | 0 | 0 | ||
| R. tritici | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| Micromonosporales | Micromonosporaceae | Micromonospora | M. sp. | 2 | 2 | 0 | 0 | 2 | 0 | 0 |
| Streptomycetales | Streptomycetaceae | Streptomyces | S. niveus | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| S. mirabilis | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| Streptosporangiales | Streptosporangiaceae | Microbispora | M. rosea | 1 | 1 | 0 | 0 | 1 | 0 | 0 |
| M. sp. | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| Bacillales | Alicyclobacillaceae | Tumebacillus | T. luteolus | 2 | 2 | 0 | 0 | 0 | 2 | 0 |
| Bacillaceae | Bacillus | B. altitudinis | 4 | 4 | 0 | 1 | 0 | 0 | 3 | |
| B. aryabhattai | 3 | 3 | 0 | 0 | 3 | 0 | 0 | |||
| B. cereus | 2 | 2 | 2 | 0 | 0 | 0 | 0 | |||
| B. drentensis | 2 | 2 | 0 | 0 | 0 | 0 | 2 | |||
| B. flexus | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |||
| B. megaterium | 5 | 7 | 3 | 0 | 3 | 1 | 0 | |||
| B. mycoides | 1 | 2 | 0 | 0 | 0 | 1 | 1 | |||
| B. niacini | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| B. pumilus | 6 | 15 | 1 | 0 | 1 | 6 | 7 | |||
| B. simplex | 3 | 5 | 0 | 0 | 2 | 2 | 1 | |||
| B. sp. | 7 | 41 | 4 | 1 | 14 | 7 | 15 | |||
| B. subtilis | 5 | 12 | 0 | 0 | 8 | 4 | 0 | |||
| B. thuringiensis | 3 | 3 | 0 | 0 | 2 | 1 | 0 | |||
| B. velezensis | 2 | 3 | 0 | 2 | 0 | 0 | 1 | |||
| Paenibacillaceae | Brevibacillus | B. brevis | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |
| Cohnella | C. plantaginis | 5 | 6 | 3 | 0 | 3 | 0 | 0 | ||
| Paenibacillus | P. aceris | 1 | 1 | 0 | 0 | 0 | 0 | 1 | ||
| P. agaridevorans | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. alginolyticus | 2 | 2 | 0 | 0 | 2 | 0 | 0 | |||
| P. amylolyticus | 1 | 1 | 0 | 0 | 0 | 0 | 1 | |||
| P. anaericanus | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. campinasensis | 1 | 2 | 0 | 2 | 0 | 0 | 0 | |||
| P. chondroitinus | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. elgii | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| P. glycanilyticus | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. graminis | 1 | 1 | 1 | 0 | 0 | 0 | 0 | |||
| P. mucilaginosus | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. polymyxa | 1 | 2 | 0 | 0 | 1 | 0 | 1 | |||
| P. provencensis | 3 | 3 | 0 | 0 | 0 | 0 | 3 | |||
| P. sp. | 3 | 4 | 2 | 0 | 2 | 0 | 0 | |||
| P. stellifer | 1 | 1 | 0 | 0 | 0 | 0 | 1 | |||
| P. taichungensis | 2 | 3 | 0 | 1 | 0 | 0 | 2 | |||
| P. terrigena | 1 | 1 | 0 | 1 | 0 | 0 | 0 | |||
| P. validus | 2 | 3 | 1 | 0 | 0 | 0 | 2 | |||
| P. vulneris | 1 | 1 | 0 | 0 | 1 | 0 | 0 | |||
| P. xylanexedens | 6 | 7 | 0 | 0 | 7 | 0 | 0 | |||
| P. xylanilyticus | 1 | 3 | 0 | 3 | 0 | 0 | 0 | |||
| Staphylococcaceae | Staphylococcus | S. warneri | 1 | 1 | 0 | 0 | 0 | 1 | 0 | |
| Total | 260 | 479 | 109 | 123 | 125 | 69 | 53 | |||
Ctrl stands for control plants.
Tomato roots were thinner than potato roots, thus fewer isolates were recovered from tomato plants (25.47% of the total isolates) using the sterilization procedure. After de-replication, only 15 strains were shared by tomato and potato plants (Supplementary Figure 2B). The bacterial community isolated from potato plants differed considerably to the tomato plants (Figure 1). Interestingly, strains belonging to Enterobacteriaceae (overall top second family) were only found in potato roots. Abundances of bacterial taxa isolated from the same plant species, but under different treatment, were more similar. Nevertheless, there were some differences between control plants and plants inoculated with S. indica (Table 1 and Figure 1A). Particularly, strains of the genus Burkholderia (9 strains, 36 isolates) were only found in control plants of tomato and potato, or in plants inoculated at 102 CFU/g, but never in the treatments of plants heavily (104 CFU/g) inoculated with S. indica (Table 1 and Supplementary Figure 2C).
Interaction Between S. indica and Bacteria
From the whole assemblage of bacteria co-cultured in vitro with S. indica, similar number of strains were found in each type of interaction. Twenty percentage of the total strains were completely inhibitory, 26% negative, 28% neutral, and 26% positive for S. indica growth (Figure 1C), revealing that S. indica must coexist with antagonistic microbes, but also with synergistic ones, during the process of root colonization by the fungus.
Bacillaceae, Enterobacteraceae, and Burkholderiaceae were the most detrimental families for S. indica growth (Figure 1C). All the strains from Enterobacteriaceae displayed an inhibitory or negative interaction with the beneficial fungus. Similar results were obtained with members of Bacillaceae, especially strains of B. subtilis, B. pumilus, B. velezensis, and B. thuringiensis, although strains of B. simplex displayed a neutral effect to S. indica growth. In the Burkholderiaceae family, all the strains of Burkholderia completely inhibited S. indica growth or were strongly negative, while one strain of Paraburkholderia displayed positive interactions (Supplementary Figure 2D). Likewise, the great majority of Leifsonia, Rathayibacter, and Pseudomonas (95%) strains showed an inhibitory or negative interaction with S. indica.
The most abundant families showing a positive interaction were Mycobacteriaceae (22.67% of the total positive strains), Rhizobiaceae (13.33%), Xanthomonadaceae (10.67%), Paenibacillaceae (10.67%), and Rhodanobacteraceae (9.33%). Considering the type of interaction within a family, every strain of Xanthomonadaceae, and 17 out of 19 for Mycobacteriaceae stimulated S. indica growth. At the genus level, several strains classified as Achromobacter and Sphingomonas were further positive for S. indica growth, although some others were just neutral.
Screening of Bacterial Strains for Further Experiments
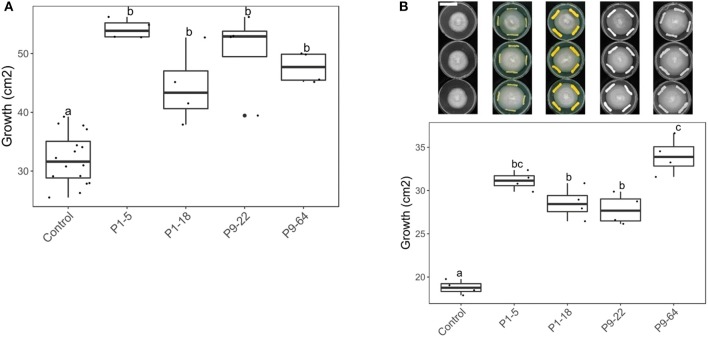
Since Mycobacteriaceae was the family that contains more strains stimulating fungal growth, four isolates of the genus Mycolicibacterium (P1-5, P1-18, P9-22, and P9-64) recovered from potato roots were selected for further experiments. These isolates strongly stimulated S. indica growth when co-cultured on PDA and after 4 days of bacterial pre-culture (Figure 2A). The four isolates further stimulated S. indica growth on different growing media (PDA+NA) and bacterial growth phase (9 days preculture) (Supplementary Figure 3), except for P9-22 that did not significantly increase fungal growth on PDA+NA. To rule out the hypothesis that the stimulating effect of bacteria on fungal growth is due to stressed hyphae running away from the bacterium, we further confronted S. indica with the selected isolates, but growing on different zones of the Petri dish. Concomitantly, S. indica growth was also stimulated when the bacteria were streaked few centimeters away from the fungus (Figure 2B).
Figure 2.
Effect of Mycolicibacterium strains on S. indica growth (in cm2). Co-cultured on PDA, with bacteria pre-cultured for 4 days. Same letters represent non-significantly different mean values, according to Tukey's test (P < 0.05), after ANOVA (n = 4). (A) With Serendipita indica and bacteria co-cultured at the same spot (direct contact). (B) Bacteria and fungus co-cultured at different spots. Bar in the picture corresponds to 4 cm.
Effect of Combined Inoculation of Endophytic Bacteria and S. indica on Tomato Growth
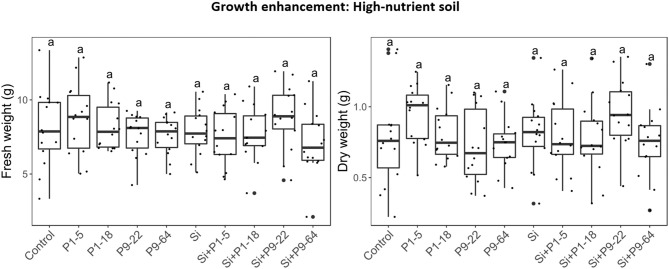
To determine effects of selected Mycolicibacterium strains and S. indica on plants, tomato plants were inoculated with single or dual inoculations. The fresh weight in all the inoculated treatments was never lower than control (untreated) plants (Figure 3), therefore none of these microbes seemed detrimental or pathogenic for plant growth. Apart from the strain P1-5, single inoculations of bacteria increased plant growth, but only P1-18 and P9-22 increased shoot fresh weight significantly. Inoculation of plants with S. indica increased shoot fresh weight (3.3-fold). Dual inoculations of S. indica+P1-5 and S. indica+P1-18 further enhanced the beneficial effect triggered by S. indica, but it resulted significant only for leaf area measurements of plants inoculated with S. indica+P1-5. Contrarily, dual inoculations of S. indica+P9-22 and S. indica+P9-64 displayed lower performance than inoculation of S. indica alone. To confirm the plant growth promotion triggered by these microbes, this experiment was repeated in soil with high content of nutrients but in this experiment no differences were obtained between treatments (Figure 4).
Figure 3.
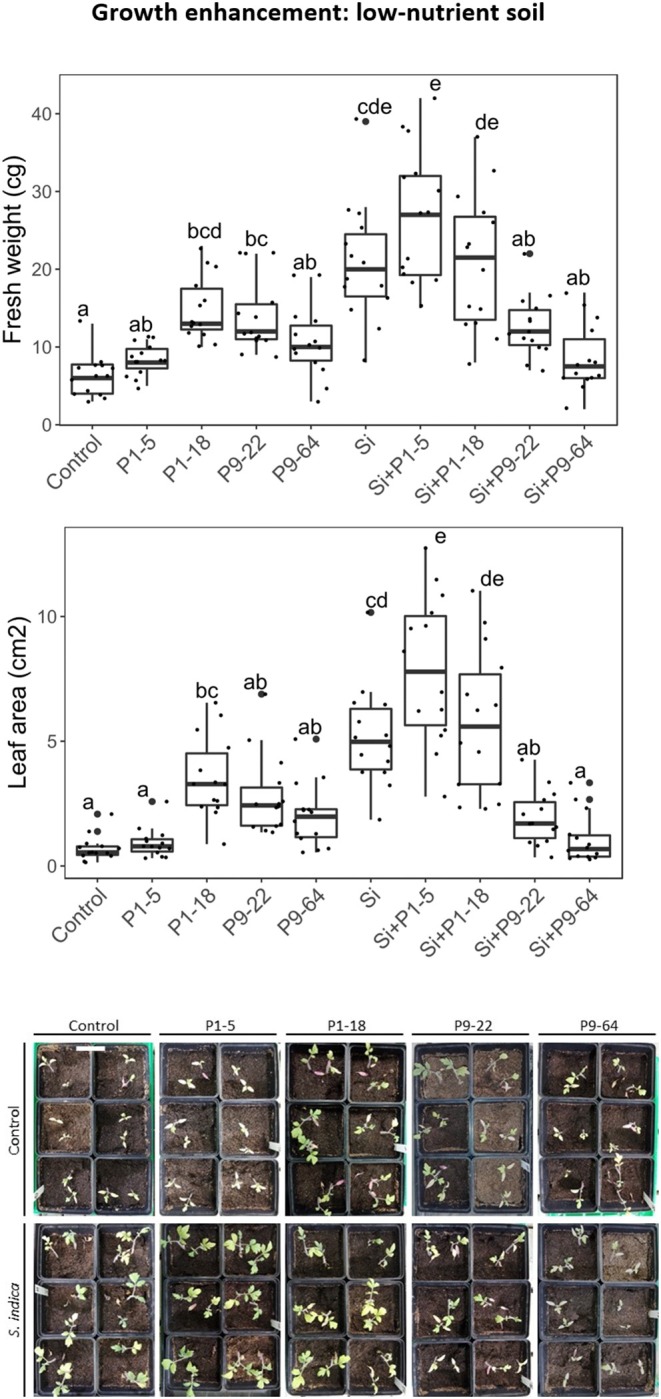
Effect of isolates alone or in combination with Serendipita indica on tomato plants under low-nutrient soil conditions. Same letters represent non-significantly different mean values, according to Tukey's test (P < 0.05) after ANOVA. n = 14. Bar in the picture corresponds to 6 cm.
Figure 4.
Effect of isolates alone or in combination with Serendipita indica on tomato plants under rich soil conditions. Same letters represent non-significantly different mean values, according to Tukey's test (P < 0.05) after ANOVA. n = 14.
In vitro Interaction Between Selected Bacterial Strains and Fungal Pathogens
By in vitro dual-culturing, the effect of selected Mycolicibacterium strains on Fusarium and Rhizoctonia growth was studied. In contrast to the beneficial interaction observed between S. indica and the selected Mycolicibacterium strains, none of these bacteria stimulated F. oxysporum growth. Interestingly, some strains significantly reduced hyphal growth respect to the control (Supplementary Figure 4) under certain growth conditions. In the case of Rhizoctonia solani, none of the four bacteria restrained fungal growth in vitro. Contrarily, the strains P9-22 and P9-64 slightly stimulated fungal growth under certain growth conditions.
Tomato Protection Against Fusarium oxysporum and Rhizoctonia solani Using Dual Inoculations
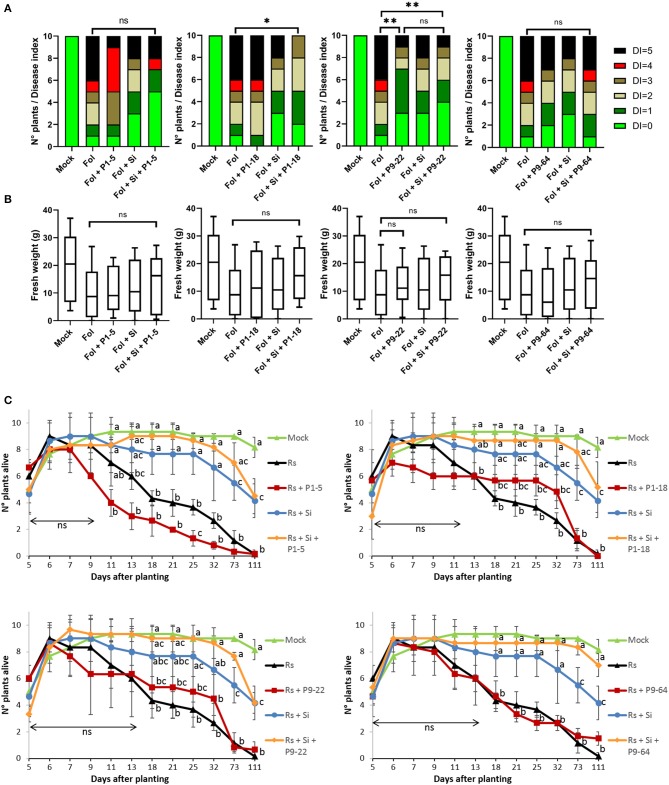
Tomato plants infected with the pathogen F. oxysporum (Fol) showed typical symptoms as leaf yellowing, necrotized vessels, wilting, and death (as described in van der Does et al., 2019). On average, fresh weights of Fusarium-treated plants were always reduced in comparison with mock-inoculated plants, although it was statistically significant uniquely for plants single-inoculated with P9-64 (Figure 5B). The extent of disease progression (i.e., yellowing, brown bundles, wilting) seemed to be alleviated when the plants were treated with the beneficial fungus S. indica but it was not statistically significant. Only during combined treatments of S. indica+P1-18 and S. indica+P9-22, the level of disease progression was significantly reduced (Figure 5A). Furthermore, single inoculation of P9-22 significantly reduced Fol symptoms to the same extent as the combination S. indica+P9-22.
Figure 5.
Effect of isolates in combination or not with Serendipita indica (Si) on tomato plants against fungal pathogens. (A,B) Biocontrol of Fusarium oxysporum (Fol). (A) Disease index (DI) score. (B) Fresh weight (FW). Boxes represent standard deviation with median. Whiskers represent the Min to Max of all the values. Analysis of FW and DI was performed using the non-parametric Mann–Whitney U-test (*P < 0.05, **P < 0.01, ns = non significant; n = 10); (C) biocontrol of Rhizoctonia solani (Rs). Points represent mean values, and whiskers standard deviations. Same letters represent non-significantly different mean values, according to Tukey's test (P < 0.05) after ANOVA (n = 3). ns, non significant.
Concerning the experiments with Rhizoctonia, the effect of seedlings germinated and alive was regularly monitored up to 111 days. The effects of the damping-off caused by Rhizoctonia first appeared 7 days after planting (dap), affecting only treatments in which S. indica was not inoculated (Figure 5C). Seedlings inoculated with S. indica as well as combinations of S. indica + bacteria were not affected by the pathogen until 11 dap. In contrast, single inoculation of Mycolicibacterium P1-5 accelerated the damping off caused by Rhizoctonia and the number of seedlings alive was significantly decreased 25 dap in comparison to Rhizoctonia control. In general, inoculation of seedlings with S. indica conferred resistance against the pathogen, but uniquely dual inoculations of S. indica + bacteria significantly maintained the number of seedlings alive for all the measurements from 18 to 111 dap, in comparison to Rhizoctonia control. The combinations of S. indica+P1-18 (at 73 dap) and S. indica+P9-64 (at 73 and 111 dap) further significantly increased the number of plants alive in comparisons to single inoculation of S. indica.
Genome Analysis
Genomic Features, ANI and Phylogeny of Sequenced Strains
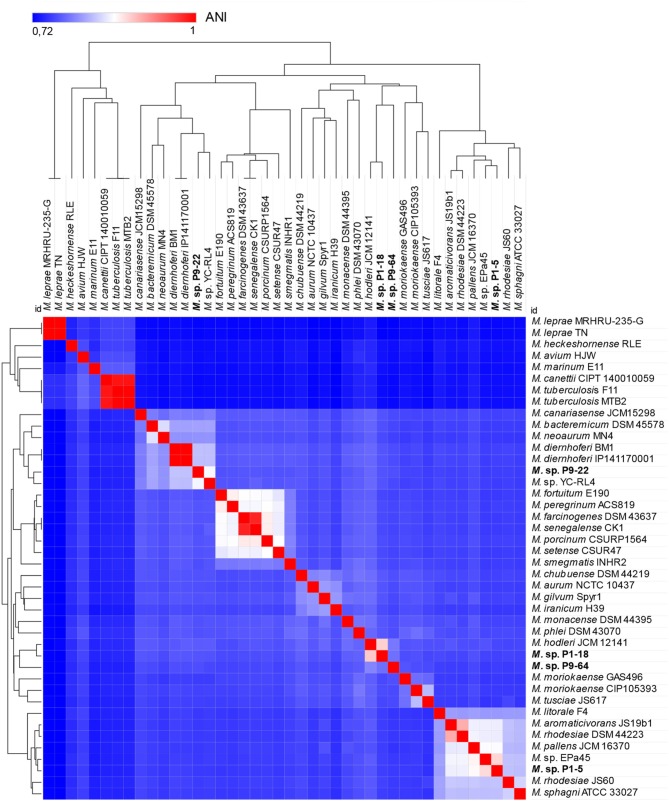
The genomes of the Mycolicibacterium strains P1-5, P1-18, P9-22, and P9-64 have a total of 5.47, 6.70, 6.79, and 7.34 Mb with an average G+C content of 65.95, 68.74, 66.89, and 66.27%, respectively. No evidence of plasmids was ascertained. The analysis of antimicrobial resistance or virulence genes detected few genes (Supplementary Table 1). In particular, strain P1-5 shows the presence of the rbpA gene, that can confer resistance to rifampin, and the strains P1-18 and P9-64 harbor the gene tet(V), possibly involved in tetracycline resistance. The genomic features of the four genomes are summarized in Table 2. To determine the relatedness of the sequenced strains to genomes publicly available at the NCBI database, ANI was calculated with the BLAST algorithm (ANIb), and with tetranucleotide frequency correlation coefficients (TETRA). The maximum values of ANIb for the four sequenced genomes were only in the range of 80–90% (Supplementary Table 2) and therefore the proposed threshold of ≈95% ANIb as the putative boundary for species circumscriptions was not reached (Konstantinidis and Tiedje, 2005; Richter and Rosselló-Móra, 2009). Contrarily, the TETRA values for the isolates P1-5 and P9-22 reached the 99% threshold (Supplementary Table 2) required to support the species circumscription (Richter and Rosselló-Móra, 2009). However, since TETRA values > 99% should agree with ANIb > 95–96% (Richter and Rosselló-Móra, 2009), we did not assign species names to these isolates.
Table 2.
Summary of the genomic features of the four Mycolicibacterium strains.
| Feature | P1-5 | P1-18 | P9-22 | P9-64 |
|---|---|---|---|---|
| Length (bp) | 5,470,684 | 6,702,551 | 6,794,647 | 7,340,553 |
| G + C content (%) | 65.95 | 68.74 | 66.89 | 66.27 |
| Contigs | 54 | 35 | 39 | 46 |
| Total genes | 5,292 | 6,515 | 6,609 | 7,061 |
| Predicted CDS | 5,213 | 6,430 | 6,522 | 6,974 |
| rRNA number | 2 | 2 | 3 | 2 |
| tRNA number | 51 | 54 | 54 | 51 |
| miscRNA number | 25 | 28 | 29 | 33 |
| tmRNA number | 1 | 1 | 1 | 1 |
| GenBank accession | NPKT00000000 | NPKR00000000 | NPKP00000000 | NPKO00000000 |
The dendrogram computed with the distance matrix of pairwise ANI values showed two separated groups (Figure 6). One group represents the clade of slow-growing mycobacteria designated as “Tuberculosis-Simiae” and depicts the emended genus Mycobacterium (Gupta et al., 2018; Oren and Garrity, 2018). This group includes well-known human pathogens (Gupta et al., 2018), most notably Mycobacterium leprae and Mycobacterium tuberculosis, causative agents of leprosy and tuberculosis, respectively (Magee and Ward, 2012; Lory, 2014). The second group, in which the four sequenced bacteria are included, encompasses species from the clade “Fortuitum-Vaccae,” recently transferred to a new genus, Mycolicibacterium gen. nov. (Oren and Garrity, 2018).
Figure 6.
Heat map and dendrogram of average nucleotide identity (ANI) values amongst different strains of Mycobacteriaceae showing separation of two groups. The group on the left include common human pathogens and are included in the genus Mycobacterium, clade “Tuberculosis-Simiae.” The group on the right consists of species of the new genus Mycolicibacterium, clade “Fortuitum-Vaccae.” The four sequenced strains P1-5, P1-18, P9-22, and P9-64 are included in the group of Mycolicibacterium (right side).
Genes and Proteins Predicted to Stimulate S. Indica Growth
Analysis of the genomes revealed the four Mycolicibacterium strains contain numerous genes predicted to be involved in the stimulation of S. indica growth. Some vitamins and cofactors are indispensable for fungal growth (van Overbeek and Saikkonen, 2016), and since some vitamins like cobalamin (B12) can only be synthesized by bacteria (Ghignone et al., 2012; Danchin and Braham, 2017), we hypothesized that production of vitamins could be one of the key factors in the stimulation of fungal growth. We identified several genes involved in the synthesis of six vitamins of the vitamin B complex: cobalamin (B12), biotin (B7), thiamin (B1), riboflavin (B2), pyridoxin (B6), and folate (B9), as well as menaquinone and phylloquinone of the vitamin K complex in all the four genomes. Moreover, genes related to nitrogen metabolism such as those for nitrate and nitrite reductase, ammonification, ammonium transporters, and glutamine synthase were detected in all four genomes (Table 3 and Supplementary Table 3).
Table 3.
Protein encoding genes predicted to be involved in fungal growth stimulation of strains P1-5, P1-18, P9-22, and P9-64 determined by RAST.
| Category | Subsystem | Myco. P1-5 | Myco. P1-18 | Myco. P9-22 | Myco. P9-64 | ||||
|---|---|---|---|---|---|---|---|---|---|
| n°R | n°G | n°R | n°G | n°R | n°G | n°R | n°G | ||
| Vitamins/Cofactors | Biotin (vitamin B7) biosynthesis | 11 | 47 | 12 | 36 | 11 | 51 | 12 | 49 |
| Thiamin (vitamin B1) biosynthesis | 8 | 11 | 12 | 12 | 11 | 16 | 10 | 11 | |
| Menaquinone and Phylloquinone (vitamin K1 and K2) biosynthesis | 7 | 13 | 6 | 10 | 7 | 12 | 8 | 12 | |
| Cobalamin (vitamin B12) synthesis | 12 | 11 | 24 | 23 | 13 | 12 | 24 | 21 | |
| Heme and Siroheme biosynthesis | 14 | 15 | 14 | 24 | 14 | 18 | 14 | 19 | |
| Riboflavin (vitamin B2) metabolism | 11 | 10 | 12 | 9 | 12 | 10 | 12 | 11 | |
| Pyridoxin (vitamin B6) biosynthesis | 7 | 10 | 7 | 16 | 8 | 20 | 7 | 16 | |
| Folate (vitamin B9) biosynthesis | 21 | 27 | 21 | 29 | 21 | 28 | 21 | 29 | |
| Cell wall/Secretion | Lipoprotein releasing | 1 | 1 | ||||||
| Amino acid and peptide ABC transporter | 12 | 14 | 14 | 39 | 14 | 47 | 14 | 47 | |
| Nitrogen | Nitrate reductase | 5 | 6 | 1 | 2 | 1 | 2 | 5 | 5 |
| Nitrite reductase | 2 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | |
| Nitrate/Nitrite transporter | 1 | 3 | 3 | 3 | 4 | 11 | 3 | 9 | |
| Nitric oxide reductase | 2 | 4 | 3 | 5 | 1 | 1 | |||
| Ammonium transporter | 1 | 2 | 1 | 2 | 1 | 4 | 1 | 2 | |
| Glutamine synthetase | 2 | 4 | 3 | 6 | 3 | 8 | 4 | 9 | |
| Glutamate synthase | 3 | 5 | 3 | 7 | 3 | 8 | 3 | 5 | |
| Carbohydrates | Trehalose biosynthesis | 12 | 9 | 10 | 13 | 9 | 11 | 10 | 12 |
n°R, number of subsystem roles/proteins; n°G, number of protein encoding genes (peg).
Genes and Proteins Related to Plant Growth Promotion Traits
The RAST annotation and the functional annotation of proteins based on the eggNOG protein database detected various genes related to plant growth promotion (PGP) traits. In the genomes of the strains P1-18, P9-22, and P9-64, we identified key genes attributable to well-known plant growth-promoting compounds like siderophore synthesis and receptors as well as siderophore-interacting proteins, auxin biosynthesis and acetoin and butanediol metabolism (Table 4 and Supplementary Table 4). Regarding strain P1-5, the antiSMASH analysis detected a secondary metabolite cluster identified as mycobactin (Supplementary Table 5), a siderophore used by members of the genus Mycobacterium to shuttle free extracellular iron ions into the cytoplasm (McMahon et al., 2012). This cluster was also detected in the genomes of P9-22 and P9-64.
Table 4.
Protein encoding genes predicted to be involved in plant growth promotion and resistance of strains P1-5, P1-18, P9-22, and P9-64 determined by RAST.
| Category | Subsystem | Myco. P1-5 | Myco. P1-18 | Myco. P9-22 | Myco. P9-64 | ||||
|---|---|---|---|---|---|---|---|---|---|
| n°R | n°G | n°R | n°G | n°R | n°G | n°R | n°G | ||
| Siderophore/Fe uptake | Siderophore receptors/transport | 1 | 1 | 5 | 6 | 3 | 3 | ||
| Ferrous iron transporter | 3 | 3 | 4 | 6 | 2 | 2 | 3 | 3 | |
| Phosphate solubilization | Phosphatase | 2 | 3 | 3 | 5 | 4 | 6 | 4 | 6 |
| Pyrroloquinoline Quinone biosynthesis | 5 | 5 | |||||||
| Phosphate uptake and transport | Low-affinity inorganic phosphate transport system | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| High-affinity phosphate transport system | 8 | 12 | 8 | 13 | 8 | 12 | 8 | 14 | |
| Phosphonate ABC transporter | 3 | 3 | |||||||
| Plant hormone | Auxin biosynthesis | 3 | 5 | 4 | 7 | 5 | 10 | ||
| Polyamine | Putrescine/spermidine synthesis | 4 | 3 | 4 | 10 | 8 | 8 | 6 | 11 |
| Putrescine/spermidine transport | 4 | 4 | 8 | 20 | 7 | 7 | 8 | 23 | |
| Resistance to heavy metals | Cobalt-zinc-cadmium resistance | 2 | 5 | 3 | 10 | 3 | 12 | 3 | 7 |
| Copper homeostasis/tolerance | 6 | 9 | 8 | 11 | 6 | 12 | 6 | 14 | |
| Arsenic resistance | 4 | 9 | 4 | 15 | 4 | 11 | 4 | 10 | |
| Mercury resistance and detoxification | 1 | 2 | 2 | 3 | 1 | 1 | 2 | 5 | |
| Chromium compounds resistance | 2 | 2 | 1 | 1 | |||||
| Uptake of selenate/selenite | 2 | 4 | 5 | 11 | 3 | 6 | 5 | 12 | |
| VOC's PGP | Acetoin butanediol metabolism | 5 | 9 | 6 | 9 | 6 | 12 | ||
| Resistance to antibiotics | Fluoroquinolone resistance | 4 | 5 | 4 | 4 | 4 | 4 | 4 | 4 |
| Beta-lactamase | 3 | 9 | 3 | 9 | 3 | 10 | 3 | 10 | |
| Oxalate catabolism | 1 | 1 | 1 | 1 | |||||
| 4-hydroxybenzoate degradation | 1 | 1 | |||||||
| Antibiosis compounds | Clavulanic acid biosynthesis | 1 | 1 | 1 | 1 | ||||
| Chitinase & β-hexosaminidase | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Resistance to oxidative stress | Peroxidase | 5 | 6 | 6 | 9 | 6 | 8 | 6 | 8 |
| Catalase | 1 | 4 | 1 | 10 | 1 | 3 | 1 | 6 | |
| Superoxide dismutase | 1 | 2 | 3 | 3 | 4 | 4 | 4 | 4 | |
| Hydroperoxide reductase | 4 | 6 | 3 | 6 | 5 | 7 | 3 | 7 | |
| Glutathione-mediated detoxification | 2 | 3 | 2 | 3 | 3 | 7 | 2 | 3 | |
| Mycothiol | 10 | 9 | 10 | 9 | 10 | 9 | 10 | 12 | |
| Heat/cold shock | Heat shock protein/chaperone | 15 | 17 | 15 | 18 | 15 | 18 | 15 | 19 |
| Cold shock protein | 2 | 3 | 2 | 2 | 2 | 3 | 2 | 3 | |
| Salt tolerance | Choline/betaine uptake and biosynthesis | 7 | 7 | 13 | 33 | 11 | 18 | 10 | 23 |
| Ectoine biosynthesis | 4 | 4 | |||||||
| K+/Na+ transport | 15 | 22 | 19 | 27 | 19 | 27 | 19 | 28 | |
| Trehalose biosynthesis | 12 | 9 | 10 | 13 | 9 | 11 | 10 | 12 | |
| Protection from UV radiation and oxidative stress | Mycosporine synthesis | 4 | 4 | ||||||
| Carotenoids | 9 | 13 | |||||||
n°R, number of subsystem roles/proteins; n°G, number of protein encoding genes (peg).
The four genomes, and especially P9-64, contain also genes involved in phosphate solubilization (Table 4 and Supplementary Table 4). These genomes encode phosphatases and pyrroloquinoline quinone biosynthesis (pqqE genes) that catalyzes the synthesis of gluconic acid, considered as one of the major organic acids responsible for mineral phosphate solubilization (Wagh et al., 2014; Liu et al., 2016). Phosphate solubilization was additionally confirmed in vitro (data not shown). Furthermore, inorganic phosphate transport and uptake may be facilitated by low- and high-affinity phosphate transport systems, detected in these genomes. Similarly, iron is an essential nutrient for plant nutrition that is mainly absorbed by plants as ferrous iron (Morrissey and Guerinot, 2009). Genes coding for ferrous iron transporters are present in the four genomes analyzed, contributing to the provision of iron to the plants. Polyamines are phytohormone-like compounds with biological activity in processes like plant growth, development, and stress mitigation (Niemi et al., 2002; Kuznetsov et al., 2006). Furthermore, we identified several proteins involved in the transport and synthesis of the polyamines spermidine and putrescine in all genomes. Moreover, the strains P1-18 and P9-64 contain the gene acdS for 1-aminocyclopropane-1-carboxylate (ACC) deaminase (Supplementary Table 6), that enhances plant growth by lowering plant ethylene levels (Glick, 2014).
Genes and Proteins Related to Stress Tolerance
The four genomes encode genes implicated in protection against diverse stresses (Table 4 and Supplementary Table 4), and this protection may indirectly lead to growth promotion (Liu et al., 2016). We further found genes involved in resistance to heavy metals and metalloids including cobalt, zinc, cadmium, copper, arsenic, mercury, chromium, and selenite. We also identified numerous proteins and compounds that protect the cell from oxidative stress: peroxidases, catalases, hydroperoxide reductases, superoxide dismutases, glutathione S-transferases, and mycothiol (Newton et al., 2008). The four genomes also encode domain proteins of rhodanese (Supplementary Table 6), an enzyme that detoxifies cyanide (Cipollone et al., 2008), and nitrilases and cyanide hydratases, enzymes with critical roles in plant-microbe interactions for defense, nitrogen utilization, detoxification, and synthesis of plant hormones (Howden and Preston, 2009). Strain P1-18 further contains genes involved in the synthesis of mycosporines (Table 4), secondary metabolites considered to be amongst the strongest natural absorbers of UV radiation and with antioxidative capacities (Oren and Gunde-Cimerman, 2007).
The four strains might potentially confer tolerance to salt stress as they possess several copies of the genes encoding choline dehydrogenase and betaine aldehyde dehydrogenase (Table 4 and Supplementary Table 4), required to produce glycine betaine, one of the most important solutes to face osmolarity fluctuations (Nau-Wagner et al., 2012). The genomes contain also several genes involved in trehalose biosynthesis, a sugar with a protective effect under salt and drought stress (Garg et al., 2002). The strain P9-22 further harbors genes for ectoine (Tables 4, 5), an osmolyte that helps organisms survive extreme osmotic stress (Bernard et al., 1993). Complementarily, all strains also contain various K+ transport and Na+/H+ antiporters that contribute to resist hyperosmotic stress (Liu et al., 2016).
Table 5.
Distribution and number of biosynthetic gene clusters (BGCs) predicted by antiSMASH analysis.
| Cluster type | Myco. | Myco. | Myco. | Myco. | Description |
|---|---|---|---|---|---|
| P1-5 | P1-18 | P9-22 | P9-64 | ||
| Cf_saccharide | 6 | 7 | 4 | 4 | Possible saccharide cluster |
| Terpene | 1 | 2 | 2 | 2 | Terpene |
| Cf_fatty_acid | 4 | 1 | 2 | 3 | Possible fatty acid cluster |
| Cf_putative | 64 | 83 | 93 | 99 | Putative cluster of unknown type |
| T1pks | 4 | 3 | 1 | 3 | Type I Polyketide synthase (PKS) |
| T3pks | 2 | 2 | 1 | Type III Polyketide synthase | |
| Otherks | 2 | Other Polyketide synthase | |||
| T1pks-cf_saccharide-nrps | 1 | 1 | 1 | Type I PKS/saccharide/nrps | |
| T1pks-nrps | 1 | 1 | 2 | 2 | Type I PKS/saccharide |
| Nrps | 1 | 2 | 2 | Non-ribosomal peptide synthetase | |
| Arylpolyene | 1 | Aryl polyene cluster | |||
| Bacteriocin | 1 | 1 | 2 | 1 | Bacteriocin cluster |
| Ectoine | 1 | Ectoine cluster | |||
| Other | 2 | 5 | 3 | 4 | Other secondary metabolite protein |
Resistance to Antibiotics and Production of Antibiotic Compounds
Genes for antibiotic resistance may protect the plant against other pathogenic microbes. The four genomes encode the enzymes β-lactamases, that provide multi-resistance to β-lactam antibiotics such as penicillins (Neu, 1969), genes involved in resistance to the bactericide fluoroquinolone, and genes encoding multidrug resistance proteins (Table 4 and Supplementary Table 6). P1-18 and P9-64 have genes involved in the degradation of oxalate, a compound secreted by fungi to promote their growth and colonization of substrates (Dutton and Evans, 1996), which might contribute to plant defense against pathogenic fungi.
Similarly, we identified several genes involved in the production of antibiotics compounds (Tables 4, 5 and Supplementary Tables 4–6), including bacteriocins, clavulanic acid (Reading and Cole, 1977), aminoglycosides antibiotics (Davies and Wright, 1997) and type IV pili, a bacterial virulence mechanism that appears operational during pathogenesis of fungal hosts (Dörr et al., 1998). Polyketides are secondary metabolites that have antimicrobial properties, including the mycotoxins produced by fungi (Huffman et al., 2010). We identified in the four genomes several enzymes involved in the synthesis of polyketides (Table 5 and Supplementary Tables 5, 6). Among the biosynthetic gene clusters (BGCs) characterized as polyketides synthases (PKS) by antiSMASH analysis, several have been identified as alkylresorcinols, phenolic lipids with the ability to inhibit bacterial and fungal growth (Stasiuk and Kozubek, 2010). Others BGCs characterized as PKS were identified as the antibiotics rifamycin, FK520 (ascomycin), ansamitocin and tetrocarcin A, and as the siderophore griseobactin (Patzer and Braun, 2010) (Supplementary Table 5). However, the percentage of gene match was very low in comparison to their homologs BGCs, suggesting that these BGCs might encode novel antibiotic and siderophore biosynthetic pathways (de Los Santos-Villalobos et al., 2018). Other antibiotic BGCs detected by antiSMASH with antifungal properties, although with low percentage of gene match, include galbonolides (Fauth et al., 1986) identified in all the genomes, bacillomycin (Gu et al., 2017) in the genomes of P9-22 and P9-64, angucycline Sch 47554 (Basnet et al., 2006), pimaricin (Aparicio et al., 2016) in P1-18, and fengycin (Vanittanakom et al., 1986) in P1-5. We further identified proteins involved in the synthesis of phenazines (Supplementary Table 6), heterocyclic compounds that have been shown to control a wide range of plant pathogenic fungi (Chin-A-Woeng et al., 2003) and to elicit ISR (Pierson and Pierson, 2010).
In addition, these bacteria contain genes encoding enzymes involved in the degradation of the fungal cell-wall, like β-hexosaminidases and chitooligosacharide deacetylases, which have been proved to degrade chitin and chitooligosaccharides (Barber and Ride, 1989; Zhao et al., 2010). Furthermore, genes encoding chitinases are present in the genome of P1-5 (Tables 4, 6) and these enzymes can potentially contribute to biocontrol of fungal pathogens by disruption of fungal cell walls (Whipps, 2001).
Table 6.
Plant and microbe cell-wall polysaccharide degrading enzymes (CE and GH classes) of strains P1-5, P1-18, P9-22, and P9-64 based on genome analysis.
| CAZy family | Substrate | Annotation | EC number | Copy number | |||
|---|---|---|---|---|---|---|---|
| P1-5 | P1-18 | P9-22 | P9-64 | ||||
| CE1 | Polysaccharide | diacylglycerol O-acyltransferase | 2.3.1.20 | 7 | 8 | 9 | 10 |
| esterase | 3.1.1.- | ||||||
| CE4 | Chitooligosaccharide | chitooligosaccharide deacetylase | 3.5.1.- | 1 | 2 | 2 | 2 |
| allantoinase | 3.5.2.5 | ||||||
| CE5 | Cutin | cutinase | 3.1.1.74 | 6 | 1 | 9 | |
| CE9 | Polysaccharides | N-acetylglucosamine 6-phosphate deacetylase | 3.5.1.25 | 1 | 1 | 1 | 1 |
| CE14 | Polysaccharides | N-acetyl-1-D-myo-inosityl-2-amino-2-deoxy-α-D-glucopyranoside deacetylase | 3.5.1.89 | 2 | 2 | 2 | 2 |
| GH1 | Cellulose | β-glucosidase | 3.2.1.21 | 1 | 1 | 2 | |
| Pectin (rhamnogalacturonan I) | β-galactosidase | 3.2.1.23 | |||||
| GH2 | Pectin (rhamnogalacturonan I) | β-glucuronidase | 3.2.1.31 | 2 | 1 | 2 | 2 |
| β-galactosidase | 3.2.1.23 | ||||||
| GH3 | Cellulose | β-glucosidase | 3.2.1.21 | 1 | 3 | 3 | 1 |
| GH4 | Polysaccharides | α-glucosidase | 3.2.1.20 | 1 | 2 | 3 | 4 |
| Hemicellulose (galactomannan) | α-galactosidase | 3.2.1.22 | |||||
| GH5 | Hemicellulose (xylan) | endo-β-1,4-xylanase | 3.2.1.8 | 1 | 2 | 3 | 2 |
| Glycosphingolipids | endoglycoceramidase | 3.2.1.123 | |||||
| GH6 | Cellulose | endo-β-1,4-glucanase | 3.2.1.4 | 1 | 1 | 1 | 1 |
| GH8 | Cellulose | cellulase | 3.2.1.4 | 1 | 1 | ||
| GH13 | Polysaccharides | α-amylase | 3.2.1.1 | 5 | 7 | 5 | 6 |
| α-glucosidase | 3.2.1.20 | ||||||
| trehalose synthase | 5.4.99.15 | ||||||
| α-1,4-glucan: phosphate α-maltosyltransferase | 2.4.99.16 | ||||||
| GH15 | Polysaccharides | glucoamylase | 3.2.1.3 | 3 | 3 | 2 | 2 |
| α,α-trehalase | 3.2.1.28 | ||||||
| GH16 | Polysaccharides | endo-1,3-β-glucanase | 3.2.1.39 | 6 | 2 | 1 | 3 |
| GH17 | Chitin | chitinase | 3.2.1.14 | 1 | |||
| GH23 | Peptidoglycans | peptidoglycan lyase | 4.2.2.1 | 3 | 2 | 3 | 4 |
| GH27 | Hemicellulose (galactomannan) | α-galactosidase | 3.2.1.22 | 1 | 1 | ||
| GH38 | Oligosaccharides | α-mannosidase | 3.2.1.24 | 1 | 1 | 1 | 1 |
| GH39 | Cellulose | cellulase | 3.2.1.4 | 3 | 1 | ||
| Pectin (rhamnogalacturonan I) | β-galactosidase | 3.2.1.23 | |||||
| GH57 | Polysaccharides | 1,4-α-glucan branching enzyme | 2.4.1.18 | 1 | 1 | 1 | 1 |
| GH65 | Polysaccharides | trehalose phosphorylase | 2.4.1.64 | 3 | 2 | 2 | 3 |
| maltose phosphorylase | 2.4.1.8 | ||||||
| GH77 | Polysaccharides | 4-α-glucanotransferase | 2.4.1.25 | 2 | 2 | 1 | 1 |
The numbers refer to the number of genes found in each CAZy family.
CAZy Analysis
We identified roughly 140 putative genes encoding carbohydrate-active enzymes (CAZy) in each genome analyzed (Table 7). CAZy were distributed unevenly among the six CAZy families and no enzymes belonged to the class polysaccharide lyases (PL). In the classes carbohydrate esterases (CE) and glycoside hydrolases (GH), we identified several plant cell-wall degrading enzymes (PCWDEs) related genes in all the genomes analyzed (Table 6 and Supplementary Table 7). The PCWDEs identified have the potential to degrade many plant cell-wall polymers, including cellulase, hemicellulose, pectin, and cutin. In addition, P1-18 encodes proteins involved in lignin degradation (Supplementary Table 6). Many other genes related to enzymes involved in the degradation of different plant intracellular polysaccharides were detected (Table 6), suggesting that the strains harbor traits for endophytic colonization.
Table 7.
Genes related to carbohydrate-active enzymes (CAZymes) in strains P1-5, P1-18, P9-22, and P9-64.
| Strain | CBM | CE | GH | GT | PL | AA |
|---|---|---|---|---|---|---|
| P1-5 | 4 | 18 | 46 | 76 | 0 | 0 |
| P1-18 | 4 | 15 | 43 | 72 | 0 | 3 |
| P9-22 | 5 | 25 | 54 | 59 | 0 | 1 |
| P9-64 | 4 | 17 | 49 | 71 | 0 | 2 |
CBM, Carbohydrate-Binding Module; CE, Carbohydrate Esterase; GH, Glycoside Hydrolase; GT, Glycosyl Transferase; PL, Polysaccharide lyases; AA, Auxiliary Activity.
Discussion
In our study, we confirm that the endophyte S. indica coexists with communities of deleterious, neutral and beneficial bacteria inside roots as revealed by in vitro assays, and there seems to be an ecological balance among these microbial communities (Varma et al., 2012). Bacillaceae, Enterobacteraceae, and Burkholderiaceae were the most detrimental families for S. indica growth. These results are congruent with the widely reported antifungal properties of several strains of Bacillus and Burkholderia (Compant et al., 2005; de Los Santos-Villalobos et al., 2018). In contrast, many Rhizobiaceae strains representing different species and genera stimulated S. indica growth in vitro. Intriguingly, it has been confirmed that S. indica hosts an endobacterium of Rhizobium radiobacter inside its hyphae and the bacterium increases host fitness (Sharma et al., 2008; Glaeser et al., 2016). Hence, it might be that this interaction is not specific and several Rhizobium relatives may promote S. indica growth. It is not known whether different rhizobia can colonize the fungus internally and it also remains unclear if the isolated Rhizobiaceae strains, rather than stimulating the fungus directly, stimulate the activity of the endobacterium of S. indica leading to improved growth. Likewise, several isolates identified as Paenibacillus exhibited a positive or neutral interaction with S. indica. These results are in line with Hildebrandt et al. (2006) who demonstrated that the bacterium Paenibacillus validus stimulates growth of the AMF Glomus intraradices. Moreover, one of the few endofungal bacteria detected in the Serendipita (= Sebacina) vermifera complex belongs to Paenibacillus (Sharma et al., 2008), suggesting a possible synergistic interaction between some strains of Paenibacillus and Serendipita. The fact that rhizobia and Paenibacillus inoculants are frequently applied as biofertilizers (Sessitsch et al., 2002; Grady et al., 2016), suggests that strains of these taxa could be further tested for application jointly with S. indica.
Most striking were the effects of Mycolicibacterium strains, which highly stimulated S. indica growth. This genus is only poorly understood regarding its interaction with plants, although some strains have been tested as bioinoculants owing to plant growth promotion effects (Egamberdiyeva, 2007). The predicted traits from the genomes of the four Mycolicibacterium strains revealed the presence of many genes responsible for vitamin production, which are potentially relevant for supplying vitamins to S. indica and thereby enhancing its growth. It has been reported that endosymbionts of mycorrhizal fungi are important for the provision of vitamin B12 (cobalamin) to their host (Ghignone et al., 2012), and vitamin B1 (thiamin) is implicated in the growth-promoting effect of Pseudomonas fluorescens on the ectomycorrhizal fungus L. bicolor (Deveau et al., 2010). Moreover, it has been described that S. indica possesses biotroph-associated genomic adaptations, such as lacking genes related to nitrogen metabolism and therefore suffering from some metabolic deficiencies. Congruent with this, S. indica barely grows on nitrate, but shows good growth on ammonium and glutamine as N source (Zuccaro et al., 2011). In accordance with these studies, we found several genes coding for nitrate and nitrate reductase as well as amino acid and peptide ABC transporters (genes families of proteins that have undergone contraction in S. indica genome) in all four Mycolicibacterium genomes. The strains also encode ammonium transporters and glutamine synthase, an enzyme that plays an essential role in the metabolism of nitrogen by catalyzing the condensation of glutamate and ammonia to form glutamine. These genomic features might be involved in increasing supply of glutamine and ammonium to the fungus, complementing the predicted metabolic deficiencies. Furthermore, it has been identified that trehalose is involved in the stimulation of hyphal growth of mycorrhizal fungus (Duponnois and Kisa, 2006; Hildebrandt et al., 2006). The Mycolicibacterium strains tested in this study encode genes for synthesis of trehalose, a compound that can be present as a disaccharide in the cytoplasm, but it is also present in the cell-wall glycolipids of Mycobacteria (Argüelles, 2000). The secretion of this sugar and the degradation of cell-wall glycolipids into oligosaccharides might also contribute to S. indica growth stimulation. To this point, there is no clear evidence, if only one, or a combination of the above-mentioned bacterial traits are responsible for the positive interactions with S. indica.
The four isolates contain many genes related to plant growth promotion traits. As predicted by the analysis of the genomes, strains P1-18, P9-22, and P9-64 enhanced plant growth, while strain P1-5 did not improve plant growth. A plausible reason is that, unlike the other strains, P1-5 lacks some of the most well-known genes involved in PGP, like those for biosynthesis of auxin, the volatile acetoin and ACC deaminase, besides harboring fewer number of genes involved in siderophore and phosphatases production as well as nitrilases, that have a potential role in the biosynthesis of indole-3-acetic acid (auxin) (Park et al., 2003). S. indica has been extensively shown to increase plant growth in different crops (reviewed in Franken, 2012). Consistent with this, in our study the inoculation of tomato plants with S. indica increased shoot fresh weight and leaf area.
Considering the PGP traits of the Mycolicibacterium strains tested in this study, a dual inoculation of S. indica with these bacteria can potentially increase plant growth. However, only dual inoculations of the strains P1-5 and P1-18 with S. indica moderately enhanced plant growth in comparison to single inoculations. These results are in agreement with Sarma et al. (2011) and Kumar et al. (2012), who observed plant growth promotion by combining pseudomonads and S. indica, and concomitantly with our results, the dual inoculation increased only slightly in comparison to single inoculations of each microbe. Interestingly, the strain P1-5 did not enhance plant growth when single-inoculated but was notably effective in plant growth promotion when applied in combination with S. indica. This synergistic effect can be ascribed to cooperation in the supply of phosphorus and nitrogen to the plant. These bacteria and S. indica encode genes involved in the provision of P to the plant, but it has been controversially discussed if, and how, S. indica supplies P to the plant. It has been shown that the fungus is able to solubilize phosphate from inorganic, but not from organic P sources (Ngwene et al., 2016), but also that S indica is not involved in the phosphate transfer to host plant (Achatz et al., 2010). The inconsistency of these results reveals how complex the interaction is, and the outcome might be dependent on abiotic factors, like adequate pH. In this regard, the combined effect of bacterial and fungal phosphatases and phosphate transport systems under certain pH levels triggered by the presence of both microbes might be the key factors to plant P uptake and growth promotion. Similarly, the fact that P1-5 and P1-18 possess genes for nitrate reductase, an enzyme which plays a key role in nitrate acquisition in plants (Gill et al., 2016), and for ferrous iron transport, might complement the nitrogen and iron supply to the host plant. Contrarily, co-inoculation of the strains P9-22 and P9-64 with S. indica displayed lower performance than single inoculations. These results coincide with Sarma et al. (2011), in which dual inoculation of S. indica and the pseudomonad R62 was more detrimental than R62 and S. indica inoculated singly. Some authors claim that the negative interaction might be due to niche competition for both space and nutrients as described by Whipps (2001). Similarly, this incompatibility might also be ascribed to alterations in the IAA (auxin) levels of the plants. Provision of low levels of IAA stimulate plant growth whereas high concentration of IAA may influence plant growth negatively (Sarwar and Frankenberger, 1994). S. indica can produce auxins, and these strains harbor the largest number of genes involved in auxin biosynthesis, therefore the dual inoculation might lead to auxin overproduction and the consequent imbalance of IAA levels, causing less growth promotion than each microbe inoculated singly.
Furthermore, our study reveals that the beneficial effect of the endophytes on plant growth is dependent on the cultivation substrate. Dual and single inoculation under nutrient-rich conditions did not exhibit shoot growth promotion 6 weeks after planting while on low-nutrient soil it did. Concomitant with our results, it has been shown that under certain nutrient conditions and plant stage S. indica and PGPR do not exert beneficial effects (Egamberdiyeva, 2007; Fakhro et al., 2010; Gill et al., 2016) and the outcome of fungal-bacteria interactions can be different from mutualistic to antagonistic. For example, Gorka et al. (2019) recently reported that the interaction between ectomycorrhiza and soil bacteria responded negatively to soil nitrogen application.
S. indica has been shown to confer resistance against several fungal pathogens (Waller et al., 2005; Qiang et al., 2012) including Fusarium (Deshmukh and Kogel, 2007; Sarma et al., 2011; Rabiey et al., 2015) and Rhizoctonia (Knecht et al., 2010), through activation of the antioxidant system (Prasad et al., 2013), defense related genes (such as PR, LOX2, and ERF1) (Zarea et al., 2012) and ISR. In agreement, S. indica always alleviated to some extent the symptoms caused by the fungal pathogens in these experiments. Bacteria employ many different mechanisms involved in biocontrol of fungal pathogens (Compant et al., 2005). Two of the most important mechanisms of biocontrol are the production of siderophores, depriving pathogenic fungi from iron acquisition (Kobayashi and Crouch, 2009), and production of antibiotics and fungal cell-wall degrading enzymes (Whipps, 2001). The genomes of Mycolicibacterium strains tested in this study encode genes for siderophore synthesis and receptors, as well as several antimicrobial compounds, including antibiotics, polyketides, phenolic lipids (alkylresorcinols), phenazines, and chitinolytic enzymes, confirming the genetic potential of these strains to exhibit biocontrol effects against pathogenic fungi. However, single inoculations of these bacteria did not protect the plants from pathogen attack. This might not be surprising considering the bacteria did not show in vitro direct antagonism to Fusarium and Rhizoctonia. Furthermore, inoculation of tomato seedlings with the strain P1-5 increased the negative effect caused by Rhizoctonia. A possible explanation is that in the same way the bacterium stimulates S. indica growth, it stimulates also the growth of Rhizoctonia enhancing damping off. Moreover, the bacterium P1-5 might help to detoxify the plant material after pathogen attack (Howden and Preston, 2009), by production of cyanide hydratases, carotenoids and antioxidants. Only strain P9-22 alleviated the symptoms of Fusarium wilt disease when single-inoculated. In reference to its genome, this biocontrol effect might stem from activation of ISR and competition for iron, as this strain encodes the highest number of genes implicated in siderophore synthesis and receptors (including BGCs identified as mycobactin, coelichelin, scabichelin) and numerous genes involved in triggering ISR (phenolic lipids—alkylresorcinols, cell wall-degrading enzymes, polyketides, antibiotics), as well as from direct antagonisms, like secreting the antifungal bacillomycin.
Overall, in this study dual inoculations of S. indica and bacteria enhanced the protective effect conferred by S. indica against the two pathogens. The strains that better performed were P1-18 and P9-22 against Fusarium, and P9-22 and P9-64 against Rhizoctonia. Similar results showing synergism by dual-inoculations have been earlier reported (Whipps, 2001). For instance, dual inoculation of S. indica and pseudomonad R81 improved biocontrol of Fusarium compared to single inoculations (Sarma et al., 2011). This could be explained by cooperation in triggering the plant ISR. According to its biotrophic lifestyle, S. indica lacks also genes potentially involved in biosynthesis of toxic secondary metabolites and cyclic peptides, and family proteins involved in PKS and NRPS are contracted (Zuccaro et al., 2011). This deficiency could be ameliorated by these bacterial helpers, as they possess several genes involved in secondary metabolite production, including PKS and NRPS. These secondary metabolites might supplement S. indica metabolism, bioenergetic capacity, activation of defense related genes and production of antibiosis compounds (Bonfante and Anca, 2009; Bhuyan et al., 2015; Salvioli et al., 2016), that ultimately raises the plant ISR. These strains possess also genes that might cooperate in restraining pathogen expansion, like strains P1-18 and P9-64 that might degrade oxalate produced by pathogens. Moreover, competition for niche and nutrients (carbon, nitrogen, and iron) has been shown to be a mechanism associated with biocontrol or suppression of Fusarium wilt in several systems (Whipps, 2001). All in all, perhaps the combined effect of bacterial and S. indica-mediated ISR, bacterial production of siderophores and antimicrobial compounds [e.g., polyketides, non-ribosomal peptides, phenazines, chitinolytic enzymes, bacillomycin (P9-22 and P9-64), angucycline and pimaricin (P1-18), galbonolides] and the nutrient and niche competition between the pathogen and beneficial microbes might explain the enhanced resistance of plants inoculated with S. indica + Mycolicibacterium.
These results demonstrate the potential of Mycolicibacterium-S. indica combinations for biocontrol of plant pathogens, but the safety of these bacteria should be carefully addressed. As the dendrogram shows, these strains are separated from the clade “Tuberculosis-Simiae,” and included in the clade “Fortuitum-Vaccae,” primarily comprised of environmental species (Gupta et al., 2018). Besides, the analysis of antimicrobial resistance or virulence genes detected very few antimicrobial resistance genes of concern and the most related strains were classified as risk 1 according to the German classification TRBA. Nevertheless, the fact that these isolates are not related to the well-known human pathogens does not imply that they are completely safe. Moreover, future studies must consider how the application of fungal and bacterial inoculants affect soil microbial communities. For example, several studies provided evidence that mycorrhizal fungi modify the bacterial communities in the rhizosphere (Nuccio et al., 2013). Similarly, Meena et al. (2010) showed that S. indica affected population dynamics of pseudomonads in chickpea and also Nautiyal et al. (2010) observed changes in the microbial community structure in soil inoculated with S. indica. Most probably, the previously reported shifts in the bacterial communities might be attributed to the modified plant physiology, altered composition of root exudates and changes in the pH (Linderman, 1992; Barea et al., 2005; Svenningsen et al., 2018). Future research is needed in culture independent analysis to study the effect of S. indica in the native soil populations, and in exploring the interaction between S. indica and other positive strains belonging to different taxa that were isolated in this study. This might help to better understand bacteria-fungal interactions, and the selection of compatible microbial strains for field application, aiming at crop enhancement and biocontrol of fungal pathogens.
Data Availability Statement
The datasets generated for this study can be found in the Sequence data are available at NCBI database and GenBank under the accession numbers MN180888–MN181366. The draft genome sequences for the Mycolicibacterium strains P1-5, P1-18, P9-22, and P9-64 are available at NCBI, BioProject PRJNA393298, with the DDBJ/ENA/GenBank accession numbers NPKT00000000, NPKR00000000, NPKP00000000, and NPKO00000000, respectively.
Author Contributions
AB-D carried out the experiments and wrote the manuscript. JL and ASa helped for analysis. LA was responsible of genome assemblage and statistics. AB-D, ASe, and SC designed the experiments. All authors gave intellectual input and critically revised the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Maria E. Constantin for sharing expertise in scoring Fusarium wilt disease, Philipp Franken and Rosanna C. Hennessy for providing S. indica and R. solani, respectively, and Negar Ghezel Sefloo for laboratory support.
Funding. This project has received funding from the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 676480.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02888/full#supplementary-material
References
- Achatz B., von Rüden S., Andrade D., Neumann E., Pons-Kühnemann J., Kogel K. H., et al. (2010). Root colonization by Piriformospora indica enhances grain yield in barley under diverse nutrient regimes by accelerating plant development. Plant Soil 333, 59–70. 10.1007/s11104-010-0319-0 [DOI] [Google Scholar]
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed August 15, 2019).
- Aparicio J. F., Barreales E. G., Payero T. D., Vicente C. M., de Pedro A., Santos-Aberturas J. (2016). Biotechnological production and application of the antibiotic pimaricin: biosynthesis and its regulation. Appl. Microbiol. Biotechnol. 100, 61–78. 10.1007/s00253-015-7077-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüelles J. C. (2000). Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174, 217–224. 10.1007/s002030000192 [DOI] [PubMed] [Google Scholar]
- Artursson V., Finlay R. D., Jansson J. K. (2006). Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ. Microbiol. 8, 1–10. 10.1111/j.1462-2920.2005.00942.x [DOI] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genom. 9:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber M. S., Ride J. P. (1989). Purification and properties of a wheat leaf N-acetyl-β-D-hexosaminidase. Plant Sci. 60, 163–172. 10.1016/0168-9452(89)90162-3 [DOI] [Google Scholar]
- Barea J. M., Pozo M. J., Azcón R., Azcón-Aguilar C. (2005). Microbial co-operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. 10.1093/jxb/eri197 [DOI] [PubMed] [Google Scholar]
- Basnet D. B., Oh T. J., Vu T. T., Sthapit B., Liou K., Lee H. C., et al. (2006). Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC-2136: cloning, sequencing, and characterization. Mol. Cells 22, 154–162. [PubMed] [Google Scholar]
- Berg G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. 10.1007/s00253-009-2092-7 [DOI] [PubMed] [Google Scholar]
- Bernard T., Jebbar M., Rassouli Y., Himdi-Kabbab S., Hamelin J., Blanco C. (1993). Ectoine accumulation and osmotic regulation in Brevibacterium linens. J. Gen. Microbiol. 139, 129–136. 10.1099/00221287-139-1-129 [DOI] [Google Scholar]
- Bertaux J., Schmid M., Hutzler P., Hartmann A., Garbaye J., Frey-Klett P. (2005). Occurrence and distribution of endobacteria in the plant-associated mycelium of the ectomycorrhizal fungus Laccaria bicolor S238N. Environ. Microbiol. 7, 1786–1795. 10.1111/j.1462-2920.2005.00867.x [DOI] [PubMed] [Google Scholar]
- Bhuyan S. K., Bandyopadhyay P., Kumar P., Mishra D. K., Prasad R., Kumari A., et al. (2015). Interaction of Piriformospora indica with Azotobacter chroococcum. Sci. Rep. 5:13911 10.1038/srep13911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer W. D., Folman L. B., Summerbell R. C., Boddy L. (2005). Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 29, 795–811. 10.1016/j.femsre.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Anca I. A. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu. Rev. Microbiol. 63, 363–383. 10.1146/annurev.micro.091208.073504 [DOI] [PubMed] [Google Scholar]
- Bonfante P., Desirò A. (2017). Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in mucoromycota. ISME J. 11, 1727–1735. 10.1038/ismej.2017.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brader G., Compant S., Vescio K., Mitter B., Trognitz F., Ma L. J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. 10.1146/annurev-phyto-080516-035641 [DOI] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2008). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucl. Acids Res. 37, D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Z., Halachev M. R., Loman N. J., Constantinidou C., Pallen M. J. (2012). Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol. 12:302. 10.1186/1471-2180-12-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-A-Woeng T. F., Bloemberg G. V., Lugtenberg B. J. (2003). Phenazines and their role in biocontrol by Pseudomonas bacteria. New Phytol. 157, 503–523. 10.1046/j.1469-8137.2003.00686.x [DOI] [PubMed] [Google Scholar]
- Cipollone R., Ascenzi P., Tomao P., Imperi F., Visca P. (2008). Enzymatic detoxification of cyanide: clues from Pseudomonas aeruginosa rhodanese. J. Mol. Microbiol. Biotechnol. 15, 199–211. 10.1159/000121331 [DOI] [PubMed] [Google Scholar]
- Collinge D. B., Jørgensen H. J. L., Latz M., Manzotti A., Ntana F., Rojas Tayo E. C., et al. (2019). Searching for novel fungal biological control agents for plant disease control among endophytes, in Endophytes for a Growing World, eds Hodkinson T., Doohan F., Saunders M., Murphy B. (Cambridge: Cambridge University Press, 25–51. [Google Scholar]
- Compant S., Duffy B., Nowak J., Clément C., Barka E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. 10.1128/AEM.71.9.4951-4959.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: ecology, functions and emerging trends in microbial application. J. Adv. Res. 19, 29–37. 10.1016/j.jare.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danchin A., Braham S. (2017). Coenzyme B12 synthesis as a baseline to study metabolite contribution of animal microbiota. Microb. Biotechnol. 10, 688–701. 10.1111/1751-7915.12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Wright G. D. (1997). Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 5, 234–240. 10.1016/S0966-842X(97)01033-0 [DOI] [PubMed] [Google Scholar]
- de Lamo F. J., Constantin M. E., Fresno D. H., Boeren S., Rep M., Takken F. L. W. (2018). Xylem sap proteomics reveals distinct differences between R gene- and endophyte-mediated resistance against fusarium wilt disease in tomato. Front. Microbiol. 9:2977. 10.3389/fmicb.2018.02977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos-Villalobos S., Kremer J. M., Parra-Cota F. I., Hayano-Kanashiro A. C., García-Ortega L. F., Gunturu S. K., et al. (2018). Draft genome of the fungicidal biological control agent Burkholderia anthina strain XXVI. Arch. Microbiol. 200, 803–810. 10.1007/s00203-018-1490-6 [DOI] [PubMed] [Google Scholar]
- Delignette-Muller M. L., Dutang C. (2015). Fitdistrplus: an R package for fitting distributions. J. Stat. Softw. 64, 1–34. 10.18637/jss.v064.i04 [DOI] [Google Scholar]
- Deshmukh S. D., Kogel K. H. (2007). Piriformospora indica protects barley from root rot caused by Fusarium graminearum. J. Plant Dis. Protect. 114, 263–268. 10.1007/BF03356227 [DOI] [Google Scholar]
- Deveau A., Bonito G., Uehling J., Paoletti M., Becker M., Bindschedler S., et al. (2018). Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 42, 335–352. 10.1093/femsre/fuy008 [DOI] [PubMed] [Google Scholar]
- Deveau A., Brulé C., Palin B., Champmartin D., Rubini P., Garbaye J., et al. (2010). Role of fungal trehalose and bacterial thiamine in the improved survival and growth of the ectomycorrhizal fungus Laccaria bicolor S238N and the helper bacterium Pseudomonas fluorescens BBc6R8. Environ. Microbiol. Rep. 2, 560–568. 10.1111/j.1758-2229.2010.00145.x [DOI] [PubMed] [Google Scholar]
- Di Pietro A., Roncero M. I. G. (1996). Endopolygalacturonase from Fusarium oxysporum f. sp. lycopersici: purification, characterization, and production during infection of tomato plants. Phytopathology 86, 1324–1330. [Google Scholar]
- Dörr J., Hurek T., Reinhold-Hurek B. (1998). Type IV pili are involved in plant–microbe and fungus–microbe interactions. Mol. Microbiol. 30, 7–17. 10.1046/j.1365-2958.1998.01010.x [DOI] [PubMed] [Google Scholar]
- Duponnois R., Kisa M. (2006). The possible role of trehalose in the mycorrhiza helper bacterium effect. Botany 84, 1005–1008. 10.1139/b06-053 [DOI] [Google Scholar]
- Dutton M. V., Evans C. S. (1996). Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 42, 881–895. 10.1139/m96-114 [DOI] [Google Scholar]
- Edwards U., Rogall T., Blöcker H., Emde M., Böttger E. C. (1989) Isolation direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17, 7843–7853. 10.1093/nar/17.19.7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdiyeva D. (2007). The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Agric. Ecosyst. Environ. Appl. Soil Ecol. 36, 184–189. 10.1016/j.apsoil.2007.02.005 [DOI] [Google Scholar]
- Fakhro A., Andrade-Linares D. R., von Bargen S., Bandte M., Büttner C., Grosch R., et al. (2010). Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza 20, 191–200. 10.1007/s00572-009-0279-5 [DOI] [PubMed] [Google Scholar]
- Fauth U., Zähner H., Mühlenfeld A., Achenbach H. (1986). Galbonolides A and B-two non-glycosidic antifungal macrolides. J. Antibiot. 39, 1760–1764. 10.7164/antibiotics.39.1760 [DOI] [PubMed] [Google Scholar]
- Franken P. (2012). The plant strengthening root endophyte Piriformospora indica: potential application and the biology behind. Appl. Microbiol. Biotechnol. 96, 1455–1464. 10.1007/s00253-012-4506-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey-Klett P., Garbaye J. A., Tarkka M. (2007). The mycorrhiza helper bacteria revisited. New Phytol. 176, 22–36. 10.1111/j.1469-8137.2007.02191.x [DOI] [PubMed] [Google Scholar]
- Galata V., Fehlmann T., Backes C., Keller A. (2018). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202. 10.1093/nar/gky1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A. K., Kim J. K., Owens T. G., Ranwala A. P., Do Choi Y., Kochian L. V., et al. (2002). Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. PNAS 99, 15898–15903. 10.1073/pnas.252637799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghignone S., Salvioli A., Anca I., Lumini E., Ortu G., Petiti L., et al. (2012). The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J. 6, 136–145. 10.1038/ismej.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S. S., Gill R., Trivedi D. K., Anjum N. A., Sharma K. K., Ansari M. W., et al. (2016). Piriformospora indica: potential and significance in plant stress tolerance. Front. Microbiol. 7:332. 10.3389/fmicb.2016.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaeser S. P., Imani J., Alabid I., Guo H., Kumar N., Kämpfer P., et al. (2016). Non-pathogenic Rhizobium radiobacter F4 deploys plant beneficial activity independent of its host Piriformospora indica. ISME J. 10, 871–884. 10.1038/ismej.2015.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. 10.1016/j.micres.2013.09.009 [DOI] [PubMed] [Google Scholar]
- Gorka S., Dietrich M., Mayerhofer W., Gabriel R., Wiesenbauer J., Martin V., et al. (2019). Rapid transfer of plant photosynthates to soil bacteria via ectomycorrhizal hyphae and its interaction with nitrogen availability. Front. Microbiol. 10:168. 10.3389/fmicb.2019.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady E. N., MacDonald J., Liu L., Richman A., Yuan Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Fact. 15:203. 10.1186/s12934-016-0603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q., Yang Y., Yuan Q., Shi G., Wu L., Lou Z., et al. (2017). Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 83:e01075–17. 10.1128/AEM.01075-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Glaeser S. P., Alabid I., Imani J., Haghighi H., Kämpfer P., et al. (2017). The abundance of endofungal bacterium Rhizobium radiobacter (syn. Agrobacterium tumefaciens) increases in its fungal host Piriformospora indica during the tripartite sebacinalean symbiosis with higher plants. Front. Microbiol. 8:629. 10.3389/fmicb.2017.00629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., Lo B., Son J. (2018). Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front. Microbiol. 9:67. 10.3389/fmicb.2018.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardoim P. R., Van Overbeek L. S., Berg G., Pirttilä A. M., Compant S., Campisano A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. 10.1128/MMBR.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt U., Ouziad F., Marner F. J., Bothe H. (2006). The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 254, 258–267. 10.1111/j.1574-6968.2005.00027.x [DOI] [PubMed] [Google Scholar]
- Howden A. J., Preston G. M. (2009). Nitrilase enzymes and their role in plant–microbe interactions. Microbial Biotechnol. 2, 441–451. 10.1111/j.1751-7915.2009.00111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M. C., et al. (2015). eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. 10.1093/nar/gkv1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman J., Gerber R., Du L. (2010). Recent advancements in the biosynthetic mechanisms for polyketide-derived mycotoxins. Biopolymers 93, 764–776. 10.1002/bip.21483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht K., Seyffarth M., Desel C., Thurau T., Sherameti I., Lou B., et al. (2010). Expression of BvGLP-1 encoding a germin-like protein from sugar beet in Arabidopsis thaliana leads to resistance against phytopathogenic fungi. Mol. Plant Microbe Int. 23, 446–457. 10.1094/MPMI-23-4-0446 [DOI] [PubMed] [Google Scholar]
- Kobayashi D. Y., Crouch J. A. (2009). Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 47, 63–82. 10.1146/annurev-phyto-080508-081729 [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. PNAS 102, 2567–2572. 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Sarma M. V. K., Saharan K., Srivastava R., Kumar L., Sahai V., et al. (2012). Effect of formulated root endophytic fungus Piriformospora indica and plant growth promoting rhizobacteria fluorescent pseudomonads R62 and R81 on Vigna mungo. World J. Microbiol. Biotechnol. 28, 595–603. 10.1007/s11274-011-0852-x [DOI] [PubMed] [Google Scholar]
- Kuznetsov V. V., Radyukina N. L., Shevyakova N. I. (2006). Polyamines and stress: biological role, metabolism, and regulation. Russ. J. Plant Physiol. 53, 583–604. 10.1134/S1021443706050025 [DOI] [Google Scholar]
- Laetsch D. R., Blaxter M. L. (2017). BlobTools: interrogation of genome assemblies. F1000Research 6:1287 10.12688/f1000research.12232.1 [DOI] [Google Scholar]
- Lane D. J. (1991). 16S/23S rRNA sequencing, in Nucleic Acid Techniques in Bacterial Systematics, eds Stackebrandt E., Goodfellow M. (New York, NY: John Wiley and Sons, 115–175. [Google Scholar]
- Langmead B., Salzberg S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenth R. (2019). emmeans: Estimated Marginal Means, aka Least-Squares Means. R package version 1.3.5.1. Available online at: https://CRAN.R-project.org/package=emmeans
- Linderman R. G. (1992). Vesicular-arbuscular mycorrhizae and soil microbial interactions, in Mycorrhizae in Sustainable Agriculture, eds Bethlenfalvay G. J., Linderman R. G. (Madison, WI: ASA Special Publication; ), 45–70. [Google Scholar]
- Liu W., Wang Q., Hou J., Tu C., Luo Y., Christie P. (2016). Whole genome analysis of halotolerant and alkalotolerant plant growth-promoting rhizobacterium Klebsiella sp. D5A. Sci. Rep. 6:26710. 10.1038/srep26710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Ramulu H. G., Drula E., Coutinho P. M., Henrissat B. (2013). The carbohydrate-active enzymes database (CAZy). Nucleic Acids Res. 42, D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lory S. (2014). The family Mycobacteriaceae, in The Prokaryotes-Actinobacteria, eds Rosenberg E., DeLong E., Lory S., Stackebrandt E., Thompson F. (New York, NY: Springer, 571–575. [Google Scholar]
- Magee G. M., Ward A. C. (2012). Genus I. Mycobacterium Lehmann and Neumann 1896, 363AL, in Bergey's Manual of Systematic Bacteriology, Vol. 5, Actinobacteria, eds Goodfellow M., Kampfer P., Busse H. J., Trujillo M. E., Suzuki K., Ludwig W., Whitman W. (New York, NY: Springer, 312–375. [Google Scholar]
- McMahon M. D., Rush J. S., Thomas M. G. (2012). Analyses of MbtB, MbtE, and MbtF suggest revisions to the mycobactin biosynthesis pathway in Mycobacterium tuberculosis. J. Bacteriol. 194, 2809–2818. 10.1128/JB.00088-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meena K. K., Mesapogu S., Kumar M., Yandigeri M. S., Singh G., Saxena A. K. (2010). Co-inoculation of the endophytic fungus Piriformospora indica with the phosphate-solubilising bacterium Pseudomonas striata affects population dynamics and plant growth in chickpea. Biol. Fert. Soils 46, 169–174. 10.1007/s00374-009-0421-8 [DOI] [Google Scholar]
- Morrissey J., Guerinot M. L. (2009). Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109, 4553–4567. 10.1021/cr900112r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal C. S., Chauhan P. S., DasGupta S. M., Seem K., Varma A., Staddon W. J. (2010). Tripartite interactions among Paenibacillus lentimorbus NRRL B-30488, Piriformospora indica DSM 11827, and Cicer arietinum L. World J. Microbiol. Biotechnol. 26, 1393–1399. 10.1007/s11274-010-0312-z [DOI] [Google Scholar]
- Nau-Wagner G., Opper D., Rolbetzki A., Boch J., Kempf B., Hoffmann T., et al. (2012). Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor. J. Bacteriol. 194, 2703–2714. 10.1128/JB.06642-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C. (1969). Effect of β-lactamase location in Escherichia coli on penicillin synergy. Appl. Environ. Microbiol. 17, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton G. L., Buchmeier N., Fahey R. C. (2008). Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 72, 471–494. 10.1128/MMBR.00008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngwene B., Boukail S., Söllner L., Franken P., Andrade-Linares D. R. (2016). Phosphate utilization by the fungal root endophyte Piriformospora indica. Plant Soil 405, 231–241. 10.1007/s11104-015-2779-8 [DOI] [Google Scholar]
- Niemi K., Häggman H., Sarjala T. (2002). Effects of exogenous diamines on the interaction between ectomycorrhizal fungi and adventitious root formation in Scots pine in vitro. Tree Physiol. 22, 373–381. 10.1093/treephys/22.6.373 [DOI] [PubMed] [Google Scholar]
- Nuccio E. E., Hodge A., Pett-Ridge J., Herman D. J., Weber P. K., Firestone M. K. (2013). An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 15, 1870–1881. 10.1111/1462-2920.12081 [DOI] [PubMed] [Google Scholar]
- Okonechnikov K., Conesa A., García-Alcalde F. (2015). Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 32, 292–294. 10.1093/bioinformatics/btv566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A., Garrity G. (2018). List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 68, 1411–1417. 10.1099/ijsem.0.002711 [DOI] [PubMed] [Google Scholar]
- Oren A., Gunde-Cimerman N. (2007). Mycosporines and mycosporine-like amino acids: UV protectants or multipurpose secondary metabolites? FEMS Microbiol. Lett. 269, 1–10. 10.1111/j.1574-6968.2007.00650.x [DOI] [PubMed] [Google Scholar]
- Park W. J., Kriechbaumer V., Müller A., Piotrowski M., Meeley R. B., Gierl A., et al. (2003). The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol. 133, 794–802. 10.1104/pp.103.026609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzer S. I., Braun V. (2010). Gene cluster involved in the biosynthesis of griseobactin, a catechol-peptide siderophore of Streptomyces sp. ATCC 700974. J. Bacteriol. 192, 426–435. 10.1128/JB.01250-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson L. S., Pierson E. A. (2010). Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 86, 1659–1670. 10.1007/s00253-010-2509-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D., R Core Team (2019). nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1-140. Available online at: https://CRAN.R-project.org/package=nlme
- Pontecorvo G., Roper J. A., Chemmons L. M., MacDonald K. D., Bufton A. W. J. (1953). The genetics of Aspergillus nidulans. Adv. Genet. 5, 141–238. 10.1016/S0065-2660(08)60408-3 [DOI] [PubMed] [Google Scholar]
- Prasad R., Kamal S., Sharma P. K., Oelmüller R., Varma A. (2013). Root endophyte Piriformospora indica DSM 11827 alters plant morphology, enhances biomass and antioxidant activity of medicinal plant Bacopa monniera. J. Basic Microbiol. 53, 1016–1024. 10.1002/jobm.201200367 [DOI] [PubMed] [Google Scholar]
- Qiang X., Weiss M., Kogel K. H., Schäfer P. (2012). Piriformospora indica—a mutualistic basidiomycete with an exceptionally large plant host range. Mol. Plant Pathol. 13, 508–518. 10.1111/j.1364-3703.2011.00764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019). A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing 2012. Available online at: https://www.R-project.org
- Rabiey M., Ullah I., Shaw M. W. (2015). The endophytic fungus Piriformospora indica protects wheat from fusarium crown rot disease in simulated UK autumn conditions. Plant Pathol. 64, 1029–1040. 10.1111/ppa.12335 [DOI] [Google Scholar]
- Reading C., Cole M. (1977). Clavulanic acid: a beta-lactamase-inhibiting beta-lactam from Streptomyces clavuligerus. Antimicrob. Agents Chemother. 11, 852–857. 10.1128/AAC.11.5.852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. PNAS 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvioli A., Ghignone S., Novero M., Navazio L., Bagnaresi P., Bonfante P. (2016). Symbiosis with an endobacterium increases the fitness of a mycorrhizal fungus, raising its bioenergetic potential. ISME J. 10, 130–144. 10.1038/ismej.2015.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A., Trognitz F., Antonielli L., Compant S., Sessitsch A. (2016). High-quality draft genome sequence of an endophytic Pseudomonas viridiflava strain with herbicidal properties against its host, the weed Lepidium draba L. Genome Announc. 4, e01170–e01216. 10.1128/genomeA.01170-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma M. V. R. K., Kumar V., Saharan K., Srivastava R., Sharma A. K., Prakash A., et al. (2011). Application of inorganic carrier-based formulations of fluorescent pseudomonads and Piriformospora indica on tomato plants and evaluation of their efficacy. J. Appl. Microbiol. 111, 456–466. 10.1111/j.1365-2672.2011.05062.x [DOI] [PubMed] [Google Scholar]
- Sarwar M., Frankenberger W. T. (1994). Influence of L-tryptophan and auxins applied to the rhizosphere on the vegetative growth of Zea mays L. Plant Soil 160, 97–104. 10.1007/BF00150350 [DOI] [Google Scholar]
- Scherlach K., Graupner K., Hertweck C. (2013). Molecular bacteria-fungi interactions: effects on environment, food, and medicine. Annu. Rev. Microbiol. 67, 375–397. 10.1146/annurev-micro-092412-155702 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sessitsch A., Howieson J. G., Perret X., Antoun H., Martinez-Romero E. (2002). Advances in Rhizobium research. Crit. Rev. Plant Sci. 21, 323–378. 10.1080/0735-260291044278 [DOI] [Google Scholar]
- Sharma M., Schmid M., Rothballer M., Hause G., Zuccaro A., Imani J., et al. (2008). Detection and identification of bacteria intimately associated with fungi of the order sebacinales. Cell. Microbiol. 10, 2235–2246. 10.1111/j.1462-5822.2008.01202.x [DOI] [PubMed] [Google Scholar]
- Stasiuk M., Kozubek A. (2010). Biological activity of phenolic lipids. Cell. Mol. Life Sci. 67, 841–860. 10.1007/s00018-009-0193-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenningsen N. B., Watts-Williams S. J., Joner E. J., Battini F., Efthymiou A., Cruz-Paredes C., et al. (2018). Suppression of the activity of arbuscular mycorrhizal fungi by the soil microbiota. ISME J. 12, 1296–1307. 10.1038/s41396-018-0059-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does H. C., Constantin M. E., Houterman P. M., Takken F. L. W., Cornelissen B. J. C., Haring M. A., et al. (2019). Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol. 154, 55–65. 10.1007/s10658-018-1596-3 [DOI] [Google Scholar]
- van Overbeek L. S., Saikkonen K. (2016). Impact of bacterial–fungal interactions on the colonization of the endosphere. Trends Plant Sci. 21, 230–242. 10.1016/j.tplants.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Vanittanakom N., Loeffler W., Koch U., Jung G. (1986). Fengycin-a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J. Antibiot. 39, 888–901. 10.7164/antibiotics.39.888 [DOI] [PubMed] [Google Scholar]
- Varma A., Bakshi M., Lou B., Hartmann A., Oelmueller R. (2012). Piriformospora indica: a novel plant growth-promoting mycorrhizal fungus. Agric. Res. 1, 117–131. 10.1007/s40003-012-0019-5 [DOI] [Google Scholar]
- Varma A., Fekete A., Srivastava A., Saxena A. K., Frommberger M., Li D., et al. (2013). Inhibitory interactions of rhizobacteria with the symbiotic fungus Piriformospora indica, in Piriformospora Indica, eds Varma A., Kost G., Oelmüller R. (Berlin; Heidelberg: Springer, 201–219. [Google Scholar]
- Varma A., Verma S., Sahay N., Bütehorn B., Franken P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 65, 2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh J., Shah S., Bhandari P., Archana G., Kumar G. N. (2014). Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to Herbaspirillum seropedicae Z67. Appl. Microbiol. Biotechnol. 98, 5117–5129. 10.1007/s00253-014-5610-1 [DOI] [PubMed] [Google Scholar]
- Waller F., Achatz B., Baltruschat H., Fodor J., Becker K., Fischer M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. PNAS 102, 13386–13391. 10.1073/pnas.0504423102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P., Klioutchnikov G., et al. (2017). BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., et al. (2015). antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Barns S. M., Pelletier D. A., Lane D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. 10.1128/jb.173.2.697-703.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellman F. L. (1939) A technique for studying host resistance and pathogenicity in tomato Fusarium wilt. Phytopathology 29, 945–956. [Google Scholar]
- Whipps J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. 10.1093/jxb/52.suppl_1.487 [DOI] [PubMed] [Google Scholar]
- Wickham H., Chang W. (2016). ggplot2: An Implementation of the Grammar of Graphics. Available online at: https://cran.r-project.org/web/packages/ggplot2/index.html
- Wickham H., François R., Henry L., Müller K. (2019). dplyr: A Grammar of Data Manipulation. R package version 0.8.3. Available online at: https://CRAN.R-project.org/package=dplyr
- Xu X. M., Jeffries P., Pautasso M., Jeger M. J. (2011). Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 101, 1024–1031. 10.1094/PHYTO-08-10-0216 [DOI] [PubMed] [Google Scholar]
- Zarea M. J., Hajinia S., Karimi N., Goltapeh E. M., Rejali F., Varma A. (2012). Effect of Piriformospora indica and Azospirillum strains from saline or non-saline soil on mitigation of the effects of NaCl. Soil Biol. Biochem. 45, 139–146. 10.1016/j.soilbio.2011.11.00631671710 [DOI] [Google Scholar]
- Zhao Y., Park R. D., Muzzarelli R. A. (2010). Chitin deacetylases: properties and applications. Mar. Drugs 8, 24–46. 10.3390/md8010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccaro A., Lahrmann U., Güldener U., Langen G., Pfiffi S., Biedenkopf D., et al. (2011). Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. PLoS Pathog. 7:e1002290. 10.1371/journal.ppat.1002290 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Sequence data are available at NCBI database and GenBank under the accession numbers MN180888–MN181366. The draft genome sequences for the Mycolicibacterium strains P1-5, P1-18, P9-22, and P9-64 are available at NCBI, BioProject PRJNA393298, with the DDBJ/ENA/GenBank accession numbers NPKT00000000, NPKR00000000, NPKP00000000, and NPKO00000000, respectively.