How the gut microflora influences the progression of bacterial infections is only beginning to be understood. Antibiotics are counterindicated for E. coli O157:H7 infections, limiting treatment options. An increased understanding of how the gut microflora directs O157:H7 virulence gene expression may lead to additional treatment options. This work identified E. coli strains that enhance the production of Shiga toxin by O157:H7 through the secretion of a proposed microcin. Microcins are natural antimicrobial peptides that target specific species, can act as alternatives to antibiotics, and mediate microbial competition. This work demonstrates another mechanism by which non-O157 E. coli strains may increase Shiga toxin production and adds to our understanding of microcins, a group of antimicrobials less well understood than colicins.
KEYWORDS: Escherichia coli, O157:H7, Shiga toxins, bacteriocins, microcins
ABSTRACT
Escherichia coli O157:H7 is a foodborne pathogen implicated in various multistate outbreaks. It encodes Shiga toxin on a prophage, and Shiga toxin production is linked to phage induction. An E. coli strain, designated 0.1229, that amplified Stx2a production when cocultured with E. coli O157:H7 strain PA2 was identified. Growth of PA2 in 0.1229 cell-free supernatants had a similar effect, even when supernatants were heated to 100°C for 10 min, but not after treatment with proteinase K. The secreted molecule was shown to use TolC for export and the TonB system for import. The genes sufficient for production of this molecule were localized to a 5.2-kb region of a 12.8-kb plasmid. This region was annotated, identifying hypothetical proteins, a predicted ABC transporter, and a cupin superfamily protein. These genes were identified and shown to be functional in two other E. coli strains, and bioinformatic analyses identified related gene clusters in similar and distinct bacterial species. These data collectively suggest that E. coli 0.1229 and other E. coli strains produce a microcin that induces the SOS response in target bacteria. Besides adding to the limited number of microcins known to be produced by E. coli, this study provides an additional mechanism by which stx2a expression is increased in response to the gut microflora.
IMPORTANCE How the gut microflora influences the progression of bacterial infections is only beginning to be understood. Antibiotics are counterindicated for E. coli O157:H7 infections, limiting treatment options. An increased understanding of how the gut microflora directs O157:H7 virulence gene expression may lead to additional treatment options. This work identified E. coli strains that enhance the production of Shiga toxin by O157:H7 through the secretion of a proposed microcin. Microcins are natural antimicrobial peptides that target specific species, can act as alternatives to antibiotics, and mediate microbial competition. This work demonstrates another mechanism by which non-O157 E. coli strains may increase Shiga toxin production and adds to our understanding of microcins, a group of antimicrobials less well understood than colicins.
INTRODUCTION
Escherichia coli O157:H7 is a notorious member of the enterohemorrhagic E. coli (EHEC) pathotype, which causes hemolytic colitis and hemolytic-uremic syndrome (HUS) through the production of virulence factors, including the locus of enterocyte effacement (LEE) and Shiga toxin (Stx) (1, 2). Stx is encoded on a lambdoid prophage (3). Induction of the prophage and subsequent upregulation of stx are tied to the activation of the bacterial SOS response (4). Therefore, DNA-damaging agents, including certain antibiotics, increase Stx synthesis and are typically counterindicated during treatment (5). There are two Stx types, referred to as Stx1 and Stx2 (6). Stx1 is further divided into three subtypes, Stx1a, Stx1c, and Stx1d (7). Stx2 also has multiple subtypes, designated Stx2a, Stx2b, Stx2c, Stx2d, Stx2e, Stx2f, Stx2g (7), Stx2h (8), and Stx2i (9). In general, infections caused by Stx1 and, interestingly, even those caused by both Stx1 and Stx2 (such as strains EDL933 [10] and Sakai [11]) are associated with less severe disease symptoms than Stx2-only-producing E. coli (12–14). Of the Stx2 subtypes, Stx2a is more commonly associated with clinical cases and instances of HUS (14–17). Indeed, the FAO and WHO consider STEC carrying stx2a to be of the greatest concern (18).
Stx2a levels can be affected in vitro and in vivo when E. coli O157:H7 is cultured along with other bacteria. Indeed, it was found that stx2a expression is downregulated by various probiotic species (19, 20) or in a medium conditioned with human microbiota (21). Conversely, nonpathogenic E. coli strains that are susceptible to infection by the stx2a-converting phage were reported to increase Stx2a levels (22, 23). This mechanism is O157:H7 strain dependent (23) and requires the expression of E. coli BamA, which is the phage receptor (24, 25).
Production of Stx2a by O157:H7 is mediated by quorum sensing (26) and can also increase in response to molecules secreted by other members of the gut microbiota (24, 27), such as bacteriocins and microcins. Bacteriocins are proteinaceous toxins produced by bacteria that inhibit the growth of closely related bacteria. For example, a colicin E9 (ColE9)-producing strain amplified Stx2a when grown together with Sakai to higher levels than a colicin E3 (ColE3)-producing strain (27). ColE9 is a DNase, while ColE3 has RNase activity, and this may explain the differences in SOS induction and Stx2a levels. In support of this, the addition of extracted DNase colicins to various E. coli O157:H7 strains increased Stx2a, but not Stx1, production (27). Additionally, microcin B17 (MccB17), a DNA gyrase inhibitor, was shown to amplify Stx2a production (24). It was hypothesized that nonpathogenic E. coli strains could secrete additional colicins and microcins capable of increasing Stx2a production by O157:H7.
RESULTS
0.1229 amplifies Stx2a production in a cell-independent manner.
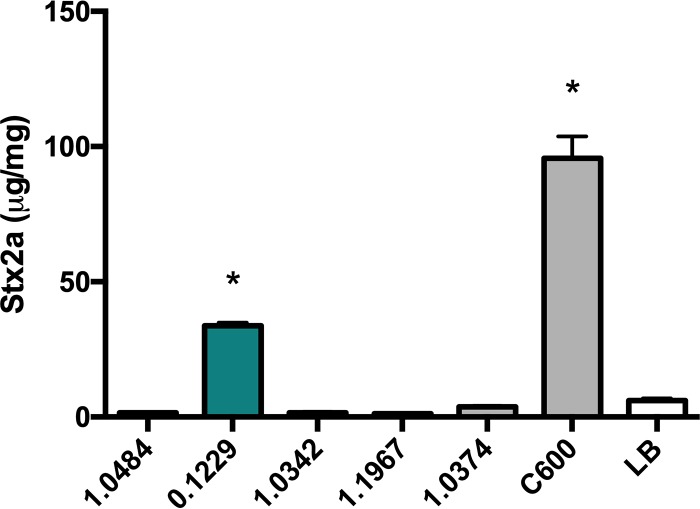
Twelve human-associated E. coli isolates were tested for their ability to enhance Stx2a production in coculture with O157:H7. One of four amplifying isolates (data not shown), strain 0.1229, significantly increased Stx2a production of PA2, compared to PA2 alone (Fig. 1). C600 was included as a positive control, as it was previously shown to increase Stx2a production when cocultured with O157:H7 (22, 23).
FIG 1.
PA2 was grown with various E. coli strains, and Stx2a levels were measured using an R-ELISA. LB refers to PA2 grown in monoculture. One-way analysis of variance (ANOVA) was used, and bars marked with an asterisk show values that were significantly higher than for LB (P < 0.05 by Dunnett’s test).
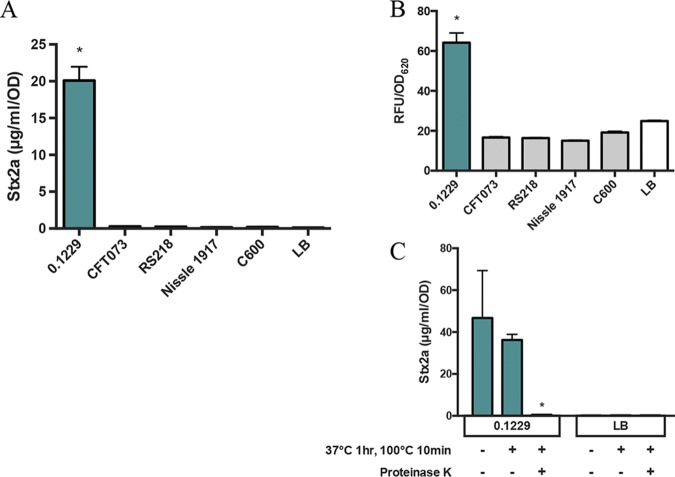
Growth of PA2 in cell-free supernatants of 0.1229 also amplified Stx2a production, indicating that this phenomenon does not require whole cells (Fig. 2A). Sequencing of the genome of 0.1229 using Illumina technology revealed that it belonged to the same sequence type (ST73) as E. coli strains CFT073 (28) and Nissle 1917 (29) and carried a plasmid similar to pRS218 in E. coli RS218 (30). However, supernatants harvested after growth of these strains failed to increase Stx2a production by PA2 (Fig. 2A). To test whether increased Stx2a production was dependent on recA, 0.1229 was cocultured with the W3110 ΔtolC PrecA-gfp reporter strain. As anticipated, we found that among this collection, only strain 0.1229 increased green fluorescent protein (GFP) expression in coculture (Fig. 2B). Treatment of 0.1229 supernatants revealed that this bioactivity was resistant to boiling and sensitive to proteinase K (Fig. 2C). This phenotype is not unique to O157:H7 strain PA2, as EDL933 and C600 carrying the 933W stx2a-converting phage also exhibit increased Stx2a production when grown in the supernatant of 0.1229 (see Fig. S1 in the supplemental material).
FIG 2.
(A and B) Stx2a levels (A) and fluorescence (B) of nonpathogenic E. coli, after PA2 growth in the cell-free supernatant or W3110 ΔtolC PrecA-gfp coculture, respectively. Samples were normalized to the OD600 or OD620 for Stx2a or fluorescence, respectively. One-way ANOVA was used, and levels marked with an asterisk were significantly higher than for LB (P < 0.05 by Dunnett’s test). (C) Stx2a levels of PA2 grown in the 0.1229 cell-free supernatant (left) or LB (right), with or without heat and proteinase K treatments. Two-way ANOVA was used, and bars marked with an asterisk show values that were significantly lower than for untreated 0.1229 or LB (P < 0.05 by Dunnett’s test).
The plasmids of 0.1229 may play a role in Stx2a amplification.
Further analysis of the Illumina sequence data revealed high sequence identity between the chromosome of 0.1229 and those of other E. coli isolates. CFT073 was 99.97% identical at the nucleotide level (97% query coverage), Nissle 1917 was 99.99% identical (96% query), and RS218 was 99.21% identical (93% query) to 0.1229, using BLAST. The most notable differences were in predicted plasmid content. To obtain a more complete picture, Pacific Biosciences (PacBio) long-read technology was used to sequence the genome of 0.1229.
The largest plasmid of 0.1229, designated p0.1229_1, was 114,229 bp and 99.99% identical with 100% query coverage to pUTI89 (31) and pRS218 (30) (Fig. S2A). A second plasmid, designated p0.1229_2, had five identifiable antimicrobial resistance genes, was 96,272 bp, and carried the operon for MccB17, a microcin that inhibits DNA gyrase (32). The plasmid p0.1229_2 shared high sequence identity with other known plasmids, being 99.96% identical with 57% query coverage to pRS218, 97.81% identical with 82% query coverage to pECO-fce (NCBI accession number CP015160), and 100% identical with 89% query coverage to pSF-173-1 (33) (Fig. S2B). A third plasmid was smaller than the cutoff for size selection used during PacBio library preparation; however, this sequence was identified within the Illumina sequence assemblies. This plasmid, designated p0.1229_3, was 12,894 bp and encoded ampicillin resistance (blaTEM-1b) (Fig. S2C). It was similar to pEC16II (NCBI accession number KU932034), being 99.85% identical with 61% query coverage, and to pHUSEC41-3 (34), being 98.89% identical with 61% query coverage.
As RS218 did not amplify Stx2a production (Fig. 2A), it was assumed that p0.1229_1 did not carry the genes responsible for this phenotype. Similarly, E. coli strain SF-173 did not increase GFP levels when cocultured with W3110 ΔtolC PrecA-gfp, suggesting that genes carried on p0.1229_2 were also not required (data not shown).
Strain 0.1229 encodes microcin B17, which is partially responsible for Stx2a amplification.
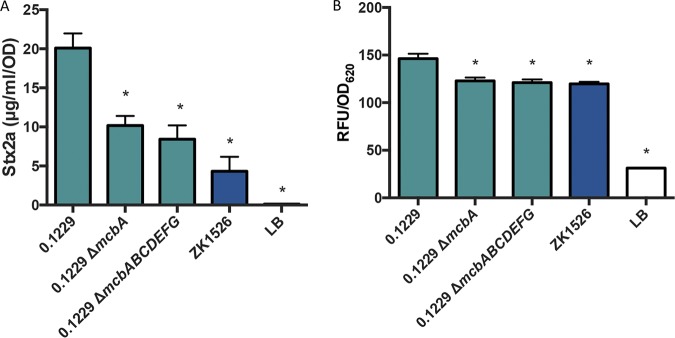
MccB17 is a 3.1-kDa (43-amino-acid) DNA gyrase inhibitor; its gene is found on a seven-gene operon, with mcbA encoding the 69-amino-acid microcin precursor (32). Although pSF-173-1 carries this operon, there was a 3-nucleotide deletion observed in mcbA in pSF-173-1, compared to a previously reported sequence (32). This deletion is predicted to shorten a 10-Gly homopolymeric stretch by 1 amino acid residue. Although this Gly-rich region is not important for interaction with the gyrase-DNA complex (35), it seemed prudent to confirm that the results reported above for strain SF-173 were not due to the production of a nonfunctional McbA. Therefore, knockouts of mcbA (ΔmcbA) and the entire operon (ΔmcbABCDEFG) were constructed in 0.1229. These mutations decreased Stx2a amplification by O157:H7 compared to wild-type 0.1229 (Fig. 3A); however, they did not ablate the Stx2a levels back to monoculture levels. Similar results were seen with the PrecA-gfp strain (Fig. 3B), although differences were less pronounced than those seen with the Stx assays.
FIG 3.
Stx2a levels (A) and fluorescence (B) of 0.1229, its MccB17 knockouts, and ZK1526, after PA2 growth in the cell-free supernatant or W3110 ΔtolC PrecA-gfp coculture, respectively. Samples were normalized to the OD600 or OD620 for Stx2a or fluorescence, respectively. One-way ANOVA was used, and bars marked with an asterisk show values that were significantly lower than for 0.1229 (P < 0.05 by Dunnett’s test).
Four ORFs carried by p0.1229_3 are necessary for the Stx2a amplification phenotype.
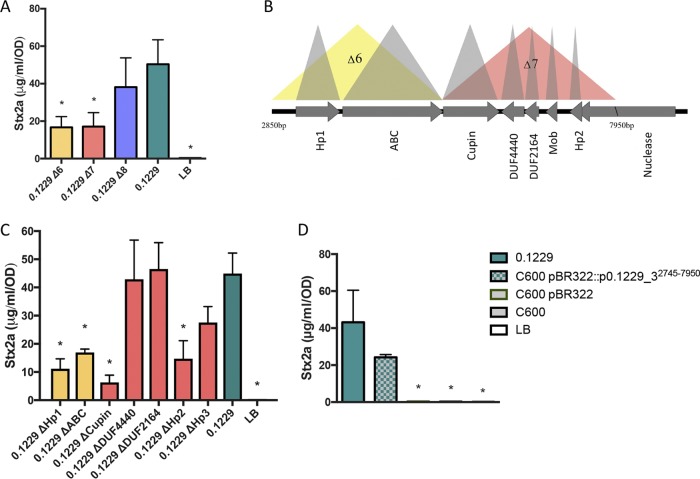
To identify the other factors encoded on p0.1229_3 that affect Stx2a amplification, p0.1229_3 was transformed into strain C600. Spent supernatants from C600(p0.1229_3) were found to amplify Stx2a production of PA2 (Fig. 4), confirming the importance of this plasmid. By systematically deleting portions of p0.1229_3, two regions were identified as being essential for increased Stx2a production (Fig. 5A). The genes annotated in these regions are referred to as those for hypothetic proteins (Hps), domains of unknown function (DUF), an ATP-binding cassette (ABC)-type transporter, and a member of the cupin superfamily of conserved barrel domains. The mutant, 0.1229Δ6 (bp 2850 to 5473), deleted two open reading frames (ORFs), referred to as Hp1 and ABC ORFs, and 0.1229Δ7 (bp 5426 to 7950) deleted cupin, DUF4440, DUF2164, Hp2, Hp3, and a portion of a predicted nuclease (Fig. 5B). These results were confirmed using coculture assays with the PrecA-gfp reporter (Fig. S3A). Insertional inactivation of individual ORFs in these regions identified four, Hp1, ABC, cupin, and Hp2 ORFs, that were necessary for enhanced Stx2a production (Fig. 5C). Similar results were shown for cocultures with PrecA-gfp, although 0.1229 Hp2 showed only a moderate decrease in GFP expression (Fig. S3B). Cloning of a 5.2-kb region of the plasmid, spanning upstream of HP1 through the beginning of the putative nuclease-encoding gene, confirmed that this activity is encoded within this region (Fig. 5D). Cloning of a similar region that ended after abc did not provide C600 the ability to increase GFP levels (data not shown).
FIG 4.
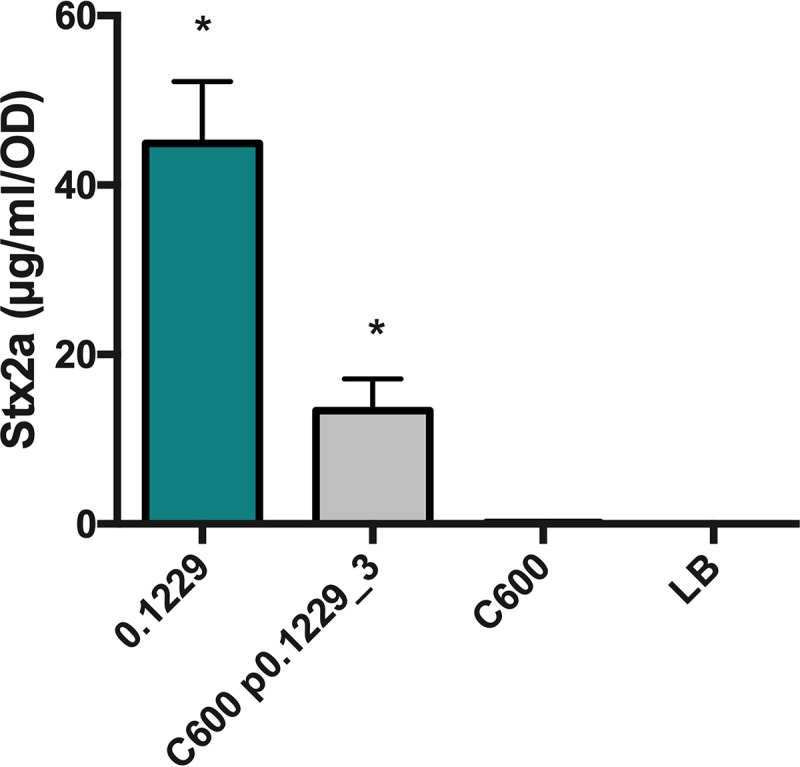
PA2 was grown in the cell-free supernatants of 0.1229, C600 containing p0.1229_3, and C600. Stx2a levels were measured using an R-ELISA. LB refers to PA2 grown in LB. One-way ANOVA was used, and bars marked with an asterisk show values that were significantly higher than for LB (P < 0.05 by Fisher’s least significant difference [LSD] test).
FIG 5.
(A and C) PA2 was grown in the cell-free supernatants of 0.1229 knockouts. (B) Depiction of a portion of p0.1229_3 with predicted open reading frames (ORFs). The colored triangles indicate the name of the regional knockout. (D) PA2 grown in the supernatant of a C600 strain containing a portion of p0.1229_3 (pBR322::p0.1229_32745–7950). Stx2a levels were measured using an R-ELISA. LB refers to PA2 grown in LB. One-way ANOVA was used, and bars marked with an asterisk show values that were significantly lower than for 0.1229 (P < 0.05 by Fisher’s LSD test).
In silico comparisons identified a nearly identical gene cluster in other species, including Shigella sonnei and Klebsiella pneumoniae (Fig. S4). The region of p0.1229_3 spanning bp 2745 to 7238 was >99.6% identical at the nucleotide level, comparing all the strains in Fig. S2. Similar gene clusters containing Hp1 at 36 to 68% amino acid identity were found in other species as well (Fig. S5). In these clusters, orthologs of Hp1, ABC, and cupin genes were commonly colocalized, and the genes were found in the same order.
The secreted molecule requires tolC for secretion and tonB for import into target strains.
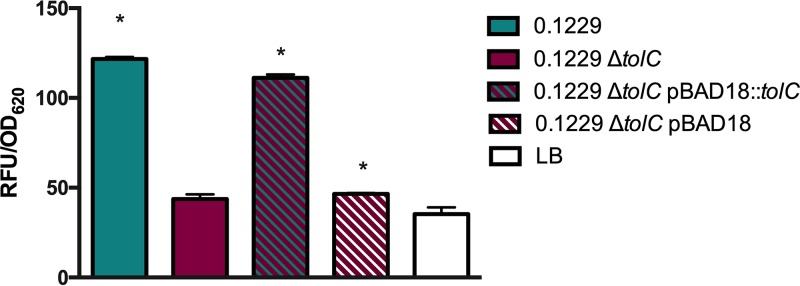
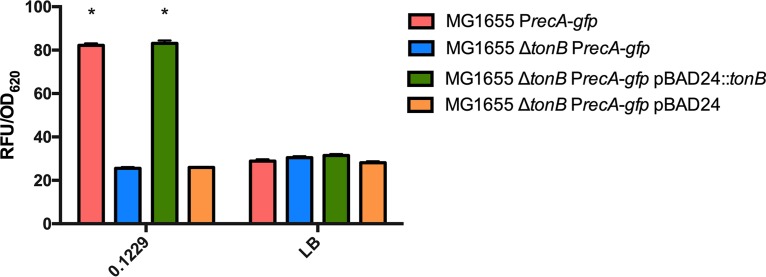
Some bacteriocins and microcins require genes located outside the main operon for secretion, such as the efflux protein TolC (36). The supernatant of 0.1229 ΔtolC did not increase Stx2a expression by strain PA2 to levels seen with wild-type 0.1229 supernatants (data not shown). Similar results were observed in coculture experiments using the PrecA-gfp-carrying strain (Fig. 6). The phenotype was restored when tolC was complemented on a plasmid but only when tested with the PrecA-gfp strain (Fig. 6). Similarly, numerous bacteriocins are translocated into target cells using the TonB system (37). A tonB knockout was constructed in the PrecA-gfp reporter strain, as we were unsuccessful in generating this in a O157:H7 background. In coculture with 0.1229, the MG1655 ΔtonB PrecA-gfp strain produced lower GFP levels than the MG1655 PrecA-gfp strain (Fig. 7). This phenotype was restored when pBAD24::tonB, but not pBAD24, was transformed into the mutant strain (Fig. 7).
FIG 6.
W3110 ΔtolC PrecA-gfp was grown in coculture with the 0.1229, 0.1229 ΔtolC, and 0.1229 ΔtolC pBAD18-containing strains, and fluorescence was measured. Samples were normalized to the OD620. One-way ANOVA was used, and bars marked with an asterisk show values that were significantly higher than for LB (P < 0.05 by Dunnett’s test).
FIG 7.
A plasmid expressing PrecA-gfp was electroporated into MG1655, MG1655 ΔtonB, MG1655 ΔtonB(pBAD24), and MG1655 ΔtonB(pBAD24::tonB). These strains were grown in coculture with 0.1229 or by themselves (LB). Two-way ANOVA was used, and bars marked with an asterisk show values that were significantly higher than for their respective monoculture controls (P < 0.05 by Dunnett’s test).
Identification of the gene cluster in additional strains.
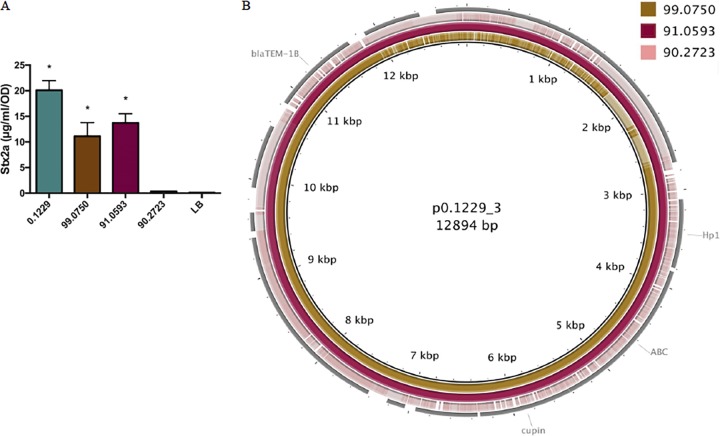
Finally, it was hypothesized that E. coli strains isolated from human feces would encode molecules similar to the ones identified here. A total of 101 human fecal E. coli isolates were obtained from Penn State’s E. coli Reference Center, and three of these were found to induce GFP production in the PrecA-gfp reporter assay (Fig. S6). Furthermore, the supernatants of two of these isolates, designated 91.0593 and 99.0750, increased Stx2a to levels similar to those in 0.1229; however, 90.2723 did not (Fig. 8A). Genome sequencing of these three organisms revealed that 91.0593 and 99.0750 carried plasmids similar to p0.1229_3; however, the latter plasmid had a deletion in the recombinase and transposon regions (Fig. 8B). Strain 99.0750 was of molecular serotype O36:H39, while 91.0593 could not be O typed but was identified as H10. Reads from 824 E. coli isolates (Table S1) were screened for the nucleotide sequence of the putative gene cluster, including Hp1, ABC, and cupin genes. However, none of the E. coli isolates tested carried this region.
FIG 8.
(A) PA2 was grown in the cell-free supernatants of 0.1229 and three human fecal E. coli isolates. Stx2a levels measured using an R-ELISA. LB refers to PA2 grown in LB. One-way ANOVA was used, and bars marked with an asterisk show values that were significantly higher than for LB (P < 0.05 by Dunnett’s test). (B) p0.1229_3 was compared to the contigs of 99.0750, 91.0593, and 90.2723 using BLAST and visualized using BRIG.
DISCUSSION
The concentration of E. coli in human feces ranges from 107 to 109 CFU (38, 39). Typically, there are up to five commensal E. coli strains colonizing the human gut at a given time (40, 41). As the human microbiota affects O157:H7 colonization and virulence gene expression (42–45), it is thought that community differences in the gut microflora may explain, in part, individual differences in disease symptoms (46). Indeed, commensal E. coli strains can be susceptible to stx2-converting phage and increase phage and Stx production (22, 23). In mice given a coculture of O157:H7 and phage-resistant E. coli, minimal toxin was recovered in the feces, but with E. coli strains that were phage susceptible, higher levels of toxin were found (47). However, it is clear that phage infection of susceptible bacteria is not the only mechanism by which the gut microflora affects Stx2 levels during infection (19, 20, 24, 27).
In this study, both whole cells and spent supernatants of E. coli 0.1229 enhanced Stx2a production by E. coli O157:H7 strain PA2. The latter strain is a member of the hypervirulent clade 8 (48) and was previously found to be a high-Stx2a producer in coculture with E. coli C600 (23) by a mechanism distinct from the one described here. E. coli 0.1229 produces at least two molecules capable of increasing Stx2a levels. The first is MccB17, a DNA gyrase inhibitor shown to activate Stx2a production in a previous study (24). The present study identified a second molecule localized to a 12.8-kb plasmid, and all genes necessary for production are found within a 5.2-kb region. Furthermore, gene knockouts identified four potential ORFs within this region, Hp1, ABC, cupin, and Hp2 ORFs, that are required for 0.1229-mediated Stx2a amplification. This gene cluster was also identified on pB51 (49), a plasmid similar to p0.1229_3; however, limited characterization was reported.
Oxidizing agents, such as hydrogen peroxide (H2O2), and antibiotics targeting DNA replication, such as ciprofloxacin, mitomycin C, and norfloxacin, are known to induce stx-converting phage (5, 50, 51) and, subsequently, Stx2 production (5, 50). However, the Stx2-amplifying activity of the 0.1229 supernatant was abolished by proteinase K, suggesting that the inducing molecule is proteinaceous in nature. Colicins are bacteriocins found in E. coli (52) and are generally >30 kDa, and at least one member has been previously shown to enhance O157:H7 Stx2 production (27). While some colicins utilize TonB for translocation, they are not expected to be heat stable. The molecule produced by 0.1229 was resistant to 100°C for 10 min, strongly suggesting that it is not a colicin.
Microcins are bacteriocins that are generally smaller than 10 kDa (94). Their size and lack of secondary and tertiary structures make them more heat stable than colicins. Microcins are divided into three classes; members of class I and class IIa are plasmid encoded, while members of class IIb are chromosomally encoded. Members of class I and IIb are posttranslationally modified (53, 54), while members of class IIa are not. To date, all members of class II, but only one member of class I (microcin J25 [MccJ25]), use an ABC-type transporter in complex with TolC for export (36) and the TonB system for import into target cells (37). The putative microcin produced by 0.1229 is plasmid encoded, along with a predicted ABC transporter, and is TolC and TonB dependent. Therefore, this microcin appears to be more closely related to class IIa microcins. However, purification of the microcin to identify possible posttranslational modifications is necessary to confirm whether designating it as belonging to class I or IIa is more appropriate.
There are four known class IIa microcins, microcin V (MccV) (previously named colicin V) (55, 56), microcin N (MccN) (previously named Mcc24) (57), microcin L (MccL) (58), and microcin PDI (MccPDI) (59, 60). The operons encoding these microcins contain four or five genes, including the microcin precursor, immunity, and export genes. The MccN operon also encodes a putative regulator with a histone-like nucleoid domain (57). The microcin precursors possess leader sequences of approximately 15 amino acids, containing the signature sequence MRXI/LX9GG/A (where X is any amino acid), and are typically cleaved by the ABC transporters during export (61). A potential leader sequence with a double glycine was found in Hp2. Additionally, a small peptide (DHGSR) was identified in the supernatants of 0.1229 by mass spectroscopy (data not shown), corresponding to an ORF internal to the Hp2 ORF, encoded in the opposite direction. Future experiments will determine if one of these, or another region, encodes a secreted microcin.
One argument against designation as a class IIa microcin is the lack of an identifiable N-terminal proteolytic domain (62) in the predicted ABC transporter encoded on p0.1229_3. This domain is found in all other members of class IIa. Interestingly, the class I MccJ25 operon also encodes an ABC transporter lacking this domain. Unlike the other class I microcins, MccJ25 is TolC and TonB dependent for export and import, respectively. While the possibility that the system identified here is a class I microcin cannot be excluded, if so, it is more similar to MccJ25 than to other members of this group.
While the current mechanism of action is unknown, it is theorized that the putative microcin causes DNA damage through double-strand breaks, depurination, or inhibition of DNA replication. Such actions would lead to RecA-dependent phage induction and Stx2 production. The suspected mode of action would be divergent from those of the known class IIa microcins, which target the inner membrane (63), and MccJ25, which inhibits the RNA polymerase (64). Besides the predicted ABC transporter, the functions of the other ORFs are unclear. We anticipate that one of these may encode an ABC accessory protein known to be essential for these export complexes (65). One ORF encodes a cupin domain found in a functionally diverse set of proteins. An immunity gene protecting the host may also be expected in this region.
The genes encoding the putative microcin were additionally found in E. coli strains 99.0750 and 91.0593. Genome sequencing of these strains failed to identify genes encoding MccB17, which may explain the lower levels of Stx2a production seen in coculture with PA2 than those seen with 0.1229. Bioinformatic analyses also identified other E. coli strains that encode nearly identical regions. Interestingly, one of these was E. coli O104:H4 HUS, isolated in 2001 (34) and responsible for a large 2011 outbreak in Germany. However, a premature stop codon identified in the cupin gene suggests that it is nonfunctional. Homologs of the Hp1, ABC, and cupin genes were identified together in several other organisms distantly related to E. coli, suggesting that they encode a functional unit. The absence of the Hp2 ORF in most of these genetic clusters argues against this ORF encoding the antibacterial activity or may suggest that these organisms encode microcins distinct from Hp2. This putative microcin is more prevalent in human E. coli isolates than in environmental, animal, and food isolates, possibly explaining why it was only recently discovered.
In conclusion, a putative microcin was identified in E. coli, expanding our knowledge of this small group of antimicrobial peptides. This study also identifies another mechanism by which E. coli may enhance Stx2a production by E. coli O157:H7. Further studies may also provide new insights into the diverse genetic structures and functions of microcin-encoding systems.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
E. coli strains were grown in lysogeny broth (LB) at 37°C unless otherwise indicated, and culture stocks were maintained in 20% glycerol at −80°C. The following antibiotics were used at the indicated concentrations: ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), and tetracycline (10 μg/ml). All bacterial isolates, plasmids, and primers used in this study can be found in Tables 1 and 2. E. coli SF-173-1 was provided by Craig Stephens, Santa Clara University.
TABLE 1.
Bacterial isolates and plasmids used in this studya
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| E. coli strains | ||
| PA2 | stx2a; O157:H7; Pennsylvania | 66 |
| PA8 | stx2a; O157:H7; Pennsylvania | 66 |
| EDL933 | stx2a stx1a; O157:H7 | 86 |
| C600 | K-12 derivative | 87 |
| MG1655 | K-12 derivative | 88 |
| 1.0484 | A phylogroup; O147; Minnesota | ECRC |
| 0.1229 | B2 phylogroup; O18:H1; Ampr Tetr; ST73; California | ECRC |
| 1.0342 | D phylogroup; O11; Minnesota | ECRC |
| 1.1967 | B2 phylogroup; O21; Minnesota | ECRC |
| 1.0374 | D phylogroup; O77; Minnesota | ECRC |
| Nissle 1917 | Mutaflor; O6:H1; ST73 | 29 |
| CFT073 | UPEC; O6:H1; ST73 | 28 |
| RS218 | NMEC; O18:H7; ST95 | 89 |
| 99.0750 | O36:H39; Brazil | ECRC |
| 91.0593 | O?:H10; Mexico | ECRC |
| 90.2723 | O?:H12; New York | ECRC |
| ZK1526 | Microcin B17-producing strain; W3110 ΔlacU169 tna-2 pPY113; Ampr | 90 |
| Derivatives | ||
| 0.1229 ΔmcbA | 0.1229 ΔmcbA::cat; Catr Ampr Tetr | This study |
| 0.122 ΔmcbABCDEFG | 0.1229 ΔmcbABCDEFG::cat; Catr Ampr Tetr | This study |
| 0.1229Δ6 | 0.1229 Δp0.1229_32850–5473::cat; Catr Ampr Tetr | This study |
| 0.1229Δ7 | 0.1229 Δp0.1229_35426–7950::cat; Catr Ampr Tetr | This study |
| 0.1229Δ8 | 0.1229 Δp0.1229_38001–9950::cat; Catr Ampr Tetr | This study |
| 0.1229 Δhp1 | 0.1229 Δp0.1229_33084–3792::cat; Catr Ampr Tetr | This study |
| 0.1229 Δabc | 0.1229 Δp0.1229_33831–5423::cat; Catr Ampr Tetr | This study |
| 0.1229 Δcupin | 0.1229 Δp0.1229_35426–6319::cat; Catr Ampr Tetr | This study |
| 0.1229 ΔDUF4440 | 0.1229 Δp0.1229_36706–6344::cat; Catr Ampr Tetr | This study |
| 0.1229 ΔDUF2164 | 0.1229 Δp0.1229_36942–6703::cat; Catr Ampr Tetr | This study |
| 0.1229 Δhp2 | 0.1229 Δp0.1229_37227–7048::cat; Catr Ampr Tetr | This study |
| 0.1229 Δhp3 | 0.1229 Δp0.1229_39099–7546::cat; Catr Ampr Tetr | This study |
| 0.1229 ΔtolC | 0.1229 ΔtolC::cat; Catr Ampr Tetr | This study |
| 0.1229 ΔtolC(pBAD18::tolC) | 0.1229 ΔtolC::cat + pBAD18::tolC; Catr Ampr Tetr Kanr | This study |
| 0.1229 ΔtolC(pBAD18) | 0.1229 ΔtolC::cat + pBAD18; Catr Ampr Tetr Kanr | This study |
| W3110 ΔtolC PrecA-gfp | W3110 ΔtolC::tet + PrecA-gfp; Tetr Kanr | 67 |
| MG1655 PrecA-gfp | MG1655 PrecA-gfp; Kanr | This study |
| MG1655 ΔtonB PrecA-gfp | MG1655 ΔtonB::cat + PrecA-gfp; Catr Kanr | This study |
| MG1655 ΔtonB PrecA-gfp(pKP315) | MG1655 ΔtonB::cat + PrecA-gfp + pKP215; Catr Kanr Ampr | This study |
| MG1655 ΔtonB PrecA-gfp(pBAD24) | MG1655 ΔtonB::cat + PrecA-gfp + pBAD24; Catr Kanr Ampr | This study |
| C600(pBR322::p0.1229_32745–7950) | C600 + pBR322::p0.1229_32745–7950; Tetr | This study |
| C600(pBR322) | C600 + pBR322; Ampr Tetr | This study |
| C600(p0.1229_3) | C600 + p0.1229_3; Ampr | This study |
| Plasmids | ||
| p0.1229_1 | 114-kb plasmid of 0.1229 | This study |
| p0.1229_2 | 96-kb plasmid of 0.1229; Tetr | This study |
| p0.1229_3 | 13-kb plasmid of 0.1229; Ampr | This study |
| pKD3 | pKD3; Catr Ampr | 69 |
| pKD46-KanR | pKD46; ParaC-λ red recombinase; Kanr | Laboratory stocks |
| PrecA-gfp | pMSs201 + PrecA-gfp; Kanr | 67 |
| pBAD24 | pBAD24; araC; Ampr | 91 |
| pKP315 | pBAD24::tonB; ParaC; Ampr | 92 |
| pBAD18 | pBAD18; ParaC; Kanr | 91 |
| pBAD18::tolC | pBAD18::tolC; ParaC; Kanr | This study |
| pBR322 | pBR322; Ampr Tetr | 93 |
| pBR322::hp1end7 | pBR322::p0.1229_32850–7950; Tetr | This study |
ECRC, Penn State E. coli Reference Center; Ampr, ampicillin resistant; Catr, chloramphenicol resistant; Kanr, kanamycin resistant; Tetr, tetracycline resistant; stx2a, Shiga toxin 2a gene; stx1a, Shiga toxin 1a gene; ParaC, arabinose-inducible promoter; UPEC, uropathogenic E. coli; NEMC, neonatal meningitis Escherichia coli. Superscript numbers indicate regions that are knocked out.
TABLE 2.
Primers used in this studya
| Target(s) | Primer | Sequence | Ta, variable time |
|---|---|---|---|
| ΔmcbA and ΔmcbABCDEFG | mcbA-KF | atactattcagatgtcataagcattaatttcccttaaaaaaggagtccttGTGTAGGCTGGAGCTGCTTC | 62°C, 100 s |
| mcbA-KR | ttttttaatatcagggagcaccatgctccctgaacggttaattcaacgtaCATATGAATATCCTCCTTAG | ||
| ΔmcbA | mcbA-VF | GGGGCTTAAAGGGGTAGTGT | 49°C, 45 s |
| mcbA-VR | AAGCGATTCGTCCAGTAGTTT | ||
| ΔmcbABCDEFG | mcbG-KR | gtccggttctgaggaggggcccgtccgggcaaccggcgggtctactcaccCATATGAATATCCTCCTTAG | 70.9°C, 2 min |
| mcbG-VR | CCTAACAACGCCACGACTTT | 49°C, 2 min | |
| Δ6 | 6-KF | acacatttcgtacagcctttacactcggtgaattagcggccctagatgcaGTGTAGGCTGGAGCTGCTTC | 67°C, 3 min |
| 6-KR | ttaaacctcatgttttgtgatatctataatctgtgctttaggtatattatCATATGAATATCCTCCTTAG | ||
| 6-VF | GAAGATATCGCACGCCTCTC | 54.5°C, 3 min | |
| 6-VR | CGCCTGTTTGGCTATATGTG | ||
| Δ7 | 7-KF | aatatacctaaagcacagattatagatatcacaaaacatgaggtttaaaaGTGTAGGCTGGAGCTGCTTC | 70°C, 90 s |
| 7-KR | tggagtttgtgcaggacgggagaaggaaatttctggttatcccgcaggggCATATGAATATCCTCCTTAG | ||
| 7-VF | TTCGATGAACCGACAAAAGG | 54°C, 2.5 min | |
| 7-VR | GGGTGAAAGAGGCGATGAT | ||
| Δ8 | 8-KF | tttaccgcagctgcctcgcacgcttcggggatgacggtgaaaacctctgaGTGTAGGCTGGAGCTGCTTC | 68°C, 90 s |
| 8-KR | agcagacaccgctcgccgcagccgaacgaccgagtgtagctagtcagtgaCATATGAATATCCTCCTTAG | ||
| 8-VF | CACGGAGGCATCAGTGACTA | 54.9°C, 2.5 min | |
| 8-VR | CAGCCTTTTCCTGGTTCTTG | ||
| ΔABC | ABC-KF | aattctagataacataaagcccgtaatatacgggctttaaggattataaaGTGTAGGCTGGAGCTGCTTC | 64.5°C, 90 s |
| ABC-KR | atatttcttaaatttttctatgatttccttttttataagattattcatttCATATGAATATCCTCCTTAG | ||
| ABC-VF | GCGAAAAGATGTTTGGAATGA | 52.7°C, 90 s | |
| ABC-VR | TCGGGAAAGTTGTCATTTGC | ||
| ΔHp1 | hp1-KF | ataaatgataactattctcatctacattcaaatatataattgggggtgttGTGTAGGCTGGAGCTGCTTC | 65.7°C, 90 s |
| hp1-KR | aataaaattcaatttataatccttaaagcccgtatattacgggctttatgCATATGAATATCCTCCTTAG | ||
| hp1-VF | ACTGGCTGCAAAAACCTTGT | 53.2°C, 75 s | |
| hp1-VR | TTTCTCCTATTGAATCTTTATTGTCA | ||
| ΔCupin | cupin KF | aatatacctaaagcacagattatagatatcacaaaacatgaggtttaaaaGTGTAGGCTGGAGCTGCTTC | 66.5°C, 3 min |
| cupin KR | agtttatatcgtatgaaaaaatctaaggggaagcccccttagattaatggCATATGAATATCCTCCTTAG | ||
| cupin VF | AAAGAGGAAAACAAGGAAAAGCA | 54°C, 2 min | |
| cupin VR | GCATTGCTTGTGTTTCAGGG | ||
| ΔDUF4440 | DUF4440 KF | aaaaaataaaacttgaacatatataaccattaatctaagggggcttccccGTGTAGGCTGGAGCTGCTTC | 68°C, 3 min |
| DUF4440 KR | aggaatgttggggatagattagaggaggaattagatatgaggaaggtagtCATATGAATATCCTCCTTAG | ||
| ΔDUF2164 | DUF2164 KF | tgcattataccattcttttcttattagatttaagtctgatttaaaattagGTGTAGGCTGGAGCTGCTTC | 68°C, 3 min |
| DUF2164 KR | tgataggaaaaatgttatattattaatttatttgtgaggcttcataaagaCATATGAATATCCTCCTTAG | ||
| ΔDUF4440 and ΔDUF2164 | DUF4440/2164 VF | GGCACAATGTTACGACTCAGA | 55°C, 90 s |
| DUF4440/2164 VR | GTTTCAGCGGTGCGTACAAT | ||
| ΔHp2 | hp2 KF | aaattacaactcaaccatactgcaacctggaatttcccaagcaagcatatGTGTAGGCTGGAGCTGCTTC | 68°C, 3 min |
| hp2 KR | tgtctctggctggcaattcctgcgtgattcacatggctgcatagctatgcCATATGAATATCCTCCTTAG | ||
| hp2 VF | TCCTCTGATTCAAACTGTCCAAG | 55°C, 90 s | |
| hp2 VR | TGTTGCTGTGTTTTGCCTCT | ||
| ΔHp3 | hp3 KF | aggcaaaacacagcaacaaaagacacaccagaatcgcgcccgtatgcgttGTGTAGGCTGGAGCTGCTTC | 68°C, 3 min |
| hp3 KR | acagcgagaacaggagataagggatgaacggctgatacaggaacgcgaacCATATGAATATCCTCCTTAG | ||
| hp3 VF | GAATTGCCAGCCAGAGACAG | 55°C, 90 s | |
| hp3 VR | GGTCATGCAGTTGAGTCAGC | ||
| ΔtonB | tonB-KF | tgcatttaaaatcgagacctggtttttctactgaaatgattatgacttcaGTGTAGGCTGGAGCTGCTTC | 68.6°C, 90 s |
| tonB-KR | ctgttgagtaatagtcaaaagcctccggtcggaggcttttgactttctgcCATATGAATATCCTCCTTAG | ||
| tonB-VF | AACATACAACACGGGCACAA | 54.9°C, 75 s | |
| tonB-VR | GACGACATCGGTCAGCATTA | ||
| ΔtolC | tolC-KF | aattttacagtttgatcgcgctaaatactgcttcaccacaaggaatgcaaGTGTAGGCTGGAGCTGCTTC | 64.5°C, 90 s |
| tolC-KR | atctttacgttgccttacgttcagacggggccgaagccccgtcgtcgtcaCATATGAATATCCTCCTTAG | ||
| tolC-VF | CCAAATGTAACGGGCAGGTT | 56°C, 2.5 min | |
| tolC-VR | GCGTGGCGTATGGATTTTGT | ||
| pBAD18::tolC | pBAD18 tolC L insert | GCTAGCGAATTCGAGCTCGGTACCCGGGGGAATCCGCAATAATTTTACAGTTTGATCGCG | 62°C, 2.5 min |
| pBAD18 tolC R insert | GCTTGCATGCCTGCAGGTCGACTCTAGAGGATAACCCGTATCTTTACGTTGCCTTACG | ||
| pBAD18 tolC R plasmid | CGTAAGGCAACGTAAAGATACGGGTTATCCTCTAGAGTCGACCTGCAGGCATGCAAGC | 62°C, 5 min | |
| pBAD18 tolC L plasmid | CGCGATCAAACTGTAAAATTATTGCGGATTCCCCCGGGTACCGAGCTCGAATTCGCTAGC | ||
| pBAD18 F | CTGTTTCTCCATACCCGTT | 45°C, 2.25 min | |
| pBAD18 R | CTCATCCGCCAAAACAG | ||
| pBR322::p0.1229_32745–7950 | pBR322 hp1 upstream L insert | GTATATATGAGTAAACTTGGTCTGACAGCATTAAAAGAGGCGTCAGAGGCAGAAAACG | 56°C, 4 min |
| pBR322 end 7 R insert | GCGGCATTTTGCCTTCCTGTTTTTGCGAAATCGGCAACGGTGATTCCCTATCAGGG | ||
| pBR322 end 7 R plasmid | CCCTGATAGGGAATCACCGTTGCCGATTTCGCAAAAACAGGAAGGCAAAATGCCGC | 62°C, 4 min | |
| pBR322 hp1 upstream L plasmid | CGTTTTCTGCCTCTGACGCCTCTTTTAATGCTGTCAGACCAAGTTTACTCATATATAC | ||
| pBR322-F | TTTGCAAGCAGCAGATTACG | 54.4°C, 2 min | |
| pBR322-R | GCCTCGTGATACGCCTATTT | ||
Ta, amplification temperature; KF, knockout forward; KR, knockout reverse; VF, verification forward; VR, verification reverse. Note that for the KF or KR primers, the lowercase letters indicate regions homologous to the target gene, and the uppercase letters indicate the primer for the antibiotic resistance cassette. Superscript numbers indicate regions knocked out.
Coculture with PA2.
Coculture with E. coli O157:H7 PA2 was performed using methods similar to ones described previously (23). PA2 and commensal E. coli strains were grown overnight at 37°C (with shaking at 250 rpm). LB (2.5 ml) was added to 6-well plates (BD Biosciences Inc., Franklin Lakes, NJ) and allowed to solidify. PA2 and commensal strains were each diluted to an optical density at 600 nm (OD600) of 0.05 in 1 ml of LB and added to the 6-well plates. A monoculture of PA2 (at an OD600 of 0.05 in 1 ml) served as a negative control. The plates were incubated without shaking at 37°C. After 16 h, cultures were collected, cells were lysed with 6 mg/ml polymyxin B at 37°C for 5 min, and supernatants were collected. Samples were immediately tested with a receptor-based enzyme-linked immunosorbent assay (R-ELISA), as described below, or stored at −80°C. The total protein concentration was calculated using the Bradford assay (VMR Life Science, Philadelphia, PA) and used to calculate the concentration of Stx2 in micrograms per milligram.
R-ELISA for Stx2a detection.
Detection of Shiga toxin was performed using a sandwich ELISA approach previously described by Xiaoli et al. (24). Briefly, 25 μg/ml of ceramide trihexosides (bottom spot) (Matreya Biosciences, Pleasant Gap, PA) dissolved in methanol was used for coating of the plate. Washes were performed between each step by using phosphate-buffered saline (PBS) and 0.05% Tween 20. Stx2a-containing samples were diluted in PBS as necessary to obtain final readings in the linear range. Samples were added to the wells in duplicate and incubated with shaking for 1 h at room temperature. Supernatants of E. coli PA11, a high-Stx2a producer (66), were used as a positive control. Anti-Stx2 monoclonal mouse antibody (Santa Cruz Biotech, Santa Cruz, CA) was added to the plate at a concentration of 1 μg/ml, and the plate was then incubated for 1 h. Anti-mouse secondary antibody (MilliporeSigma, Burlington, MA) conjugated to horseradish peroxidase (1 μg/ml) was added to the plate, and the plate was incubated for 1 h. For detection, one-step Ultra-TMB (Thermo Fisher, Waltham, MA) was used, and 2 M H2SO4 was added to the wells to stop the reaction. The plate was read at 450 nm using a DU 730 spectrophotometer (Beckman Coulter, Atlanta, GA). A standard curve was generated from 2-fold serially diluted PA11 samples and used to quantify the concentration (in micrograms per milliliter) of Stx2a present in each sample.
Cell-free supernatant assay with PA2.
E. coli O157:H7 strain PA2 and nonpathogenic E. coli strains were individually grown with shaking at 37°C for 16 h. Cultures of the nonpathogenic strains grown overnight were centrifuged, and supernatants were filtered through 0.2-μm cellulose filters (VWR International, Radnor, PA). LB agar (2.5 ml) was added to the wells of 6-well plates (BD Biosciences Inc., Franklin Lakes, NJ) and allowed to solidify. PA2 was added to wells at a final OD600 of 0.05 in 1 ml of the spent supernatant. For the negative control, PA2 was resuspended in fresh LB to the same cell density, and 1 ml was added to a well. The plates were statically incubated at 37°C for 8 h, after which the cell density (OD600) was recorded. Cells were lysed with 6 mg/ml polymyxin B at 37°C for 5 min, and the supernatant was recovered. Samples were immediately tested for Stx2a by an R-ELISA or stored at −80°C. Data are reported as micrograms per milliliter per OD600 unit.
Detection of SOS-inducing agents using PrecA-gfp.
E. coli expressing PrecA-gfp, which encodes green fluorescent protein (GFP) under the control of the recA promoter (67), was purchased from Dharmacon (Lafayette, CO). The plasmid was transformed into E. coli W3110 ΔtolC. The tolC deletion reduces the potential efflux of recA-activating molecules. W3110 ΔtolC PrecA-gfp and commensal strains were individually grown overnight with shaking at 37°C. LB agar (2.5 ml) was added to 6-well plates and allowed to solidify. W3110 ΔtolC PrecA-gfp and one commensal strain were each diluted to a final OD600 of 0.05 in LB, and 1 ml was added to the 6-well plates. The negative control included only W3110 ΔtolC PrecA-gfp at a final OD of 0.05 in 1 ml LB. The plates were statically incubated at 37°C. After 16 h, 100 μl was removed from each well and added to black 96-well clear-bottom plates (Dot Scientific Inc., Burton, MI), and the OD620 was read using a DU 730 spectrophotometer. Relative fluorescence units (RFU) were measured at an excitation wavelength of 485 nm and an emission wavelength of 538 nm on a Fluoroskan Ascent FL instrument (Thermo Fisher Scientific, Waltham, MA) (68). RFU values were normalized to the cell density.
One-step recombination for E. coli knockouts.
Mutants of 0.1229 and MG1655 were constructed using one-step recombination (69). Primers contained either 50 bp upstream or downstream of the gene of interest, followed by sequences annealing to the P1 and P2 priming sites from pKD3. PCR was performed with the following settings: an initial denaturation step at 95°C for 30 s; 10 cycles of 95°C for 30 s, 49°C for 60 s, and 68°C for 100 s; 24 cycles of 95°C for 30 s, variable amplification temperatures (Ta) for 60 s, and 68°C at variable times; and a final extension step at 68°C for 5 min. Ta and variable times for each set of primers are reported in Table 2. The derivative pKD46-KanR was used instead of pKD46, as 0.1229 is resistant to ampicillin. Electroporation was used to construct E. coli 0.1229(pKD46) and MG1655(pKD46), using a Bio-Rad Gene Pulser II instrument according to protocols recommended by the manufacturer. Colonies containing pKD46-KanR were selected on LB plates with kanamycin. Strains containing pKD46 were grown to an OD600 of 0.3, and l-arabinose was added to a final concentration of 0.2 M. After incubation for 1 h, cells were washed and electroporated with the pKD3-derived PCR product. Transformants were selected on LB plates with chloramphenicol. Knockouts were confirmed by PCR using primers ∼200 bp upstream and downstream of the gene, using standard PCR settings (initial denaturation step at 95°C for 30 s; 35 cycles of 95°C 30 s, variable Ta for 60 s, and 68°C at variable times; and a final extension step at 68°C for 5 min). This strategy was followed for all the knockouts, including primers and temperatures specific for each gene (Table 2).
Gibson cloning.
The region spanning bp 2745 to 7950 of p0.1229_3 was cloned into pBR322 (pBR322::p0.1229_32745–7950), using Gibson cloning as previously described (70). Briefly, primer pairs containing 30 bp annealing to the pBR322 insert site and 30 bp that would anneal to p0.1229_3 were constructed. DNA from 0.1229 and pBR322 was amplified at these sites using standard PCR settings, and amplicons were cleaned up using a PCR purification kit (Qiagen, Germantown, MD) and subjected to assembly at 50°C using the Gibson cloning kit (New England Biosciences, Ipswich, MA). Assembled plasmids were propagated in DH5α competent cells (New England Biosciences, Ipswich, MA). Verification PCR was performed using primers 200 bp upstream and downstream of the insert site (Table 2) and confirmed using Sanger sequencing. Successful constructs were transformed into C600 electrocompetent cells. A similar process was used to clone tolC in pBAD18 (Kanr).
Whole-genome sequencing and bioinformatics.
For whole-genome sequencing of 0.1229, genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega, Madison, WI). Whole-genome sequencing was performed at the Penn State Genomics Core facility using the Illumina MiSeq platform. A PCR-free DNA kit was used for library preparation. The sequencing run produced 2- by 150-bp reads.
For whole-genome sequencing of 99.0750, 91.0593, and 90.2723, genomic DNA was isolated using the Qiagen DNeasy blood and tissue kit (Qiagen Inc., Germantown, MD). Whole-genome sequencing was performed using the NexTera XT DNA library prep kit and run on an Illumina MiSeq platform. The sequencing run produced 2- by 250-bp reads.
After Illumina sequencing, Fastq files were checked using FastQC v0.11.5 (71) and assembled using SPAdes v3.10 (72). SPAdes assemblies were subjected to the Quality Assessment Tool for Genome Assemblies v4.5 (QUAST) (73), and contig number, genome size, N50, and percent GC content (GC%) were noted.
Strain 0.1229 was also sequenced at the Center for Food Safety and Nutrition, Food and Drug Administration, using the PacBio RS II sequencing platform, as previously reported (74). For library preparation, 10 μg genomic DNA was sheared to 20-kb fragments by using g-tubes (Covaris Inc., Woburn, MA) according to the manufacturer’s instructions. The SMRTbell 20-kb template library was constructed using DNA template prep kit 1.0 (Pacific Biosciences, Menlo Park, CA). BluePippin (Sage Science, Beverly, MA) was used for size selection, and sequencing was performed using P6/C4 chemistry on two single-molecule real-time (SMRT) cells, with a 240-min collection protocol along with stage start. SMRT Analysis 2.3.0 was used for read analysis, and de novo assembly was performed using the PacBio Hierarchical Genome Assembly Process (HGAP3.0) program. The assembly output from HGAP contained overlapping regions at the end, which can be identified using dot plots in Gepard (75). The genome was checked manually for even sequencing coverage. Afterwards, the improved consensus sequence was uploaded in SMRT Analysis 2.3.0 to determine the final consensus and accuracy scores using the Quiver consensus algorithm (76). The assembled genome was annotated using the NCBI’s Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP) (77).
Plasmid sequences were visualized using BLAST Ring Image Generator v0.95 (BRIG) (78). The Center for Genomic Epidemiology website was used for ResFinder v3.1.0 (90% identity and 60% length) (79), SerotypeFinder v2.0.1 (85% identity and 60% length) (80), and MLSTFinder v2.0.1 (81), using the multilocus sequence typing (MLST) scheme reported by Wirth et al. (82). The Integrated Microbial Genomics and Microbiomes website of the DOE’s Joint Genome Institute was utilized to perform BLAST analysis of the amino acid sequence of Hp1 against other genomes, and matches that were between 36 and 68% identical from various species were selected and then visualized using the gene neighborhood function (83).
Fastq files for 824 E. coli isolates were obtained using the BLAST+ (84) tool Fastq-dump. Strains were aligned to the nucleotide sequence of hp1-abc-cupin (bp 3094 to 6319 of p0.1229_3) using SRST2 (85). A cutoff of 90% nucleotide identity was used. Isolates 0.1229 and 91.0593 were used as positive controls.
Data analysis.
MS Excel (Microsoft Corporation, Albuquerque, NM) was used to calculate the means, standard deviations, and standard errors, and Prism 6 (GraphPad Software, San Diego, CA) was used for generating figures. Error bars report standard errors of the means from at least three biological replicates.
Data availability.
Nucleotide and SRA files for 0.1229 can be found in the NCBI database under BioSample accession number SAMN08737532. SRA files for 99.0750, 91.0593, and 90.2723 can be found under BioSample accession numbers SAMN11457477, SAMN11457478, and SAMN11457479, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Craig Stephens at Santa Clara University for providing strain SF-173, Roberto Kolter at Harvard Medical School for providing strain ZK1526, and Erin Nawrocki for manuscript proofreading.
H.M.M. was supported by USDA National Needs grant 2014-38420-21822. This work was supported by grant 1 R21 AI130856-01A1 through the National Institute of Allergy and Infectious Diseases and USDA National Institute of Food and Agriculture Federal Appropriations under project PEN04522s and accession number 0233376.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev 13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen Y, Sperandio V, Padola NL, Starai VJ. 2012. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol 2:90. doi: 10.3389/fcimb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han C-G, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Waldor MK, Friedman DI. 2005. Phage regulatory circuits and virulence gene expression. Curr Opin Microbiol 8:459–465. doi: 10.1016/j.mib.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 181:664–670. doi: 10.1086/315239. [DOI] [PubMed] [Google Scholar]
- 6.Scotland S, Smith HR, Rowe B. 1985. Two distinct toxins active on Vero cells from Escherichia coli O157. Lancet ii:885–886. doi: 10.1016/s0140-6736(85)90146-1. [DOI] [PubMed] [Google Scholar]
- 7.Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O’Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai X, Fu S, Zhang J, Fan R, Xu Y, Sun H, He X, Xu J, Xiong Y. 2018. Identification and pathogenomic analysis of an Escherichia coli strain producing a novel Shiga toxin 2 subtype. Sci Rep 8:6756. doi: 10.1038/s41598-018-25233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FAO/WHO STEC Expert Group. 2019. Hazard identification and characterization: criteria for categorizing Shiga toxin-producing Escherichia coli on a risk basis. J Food Prot 82:7–21. doi: 10.4315/0362-028X.JFP-18-291. [DOI] [PubMed] [Google Scholar]
- 10.Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O’Brien AD. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsushiro A, Sato K, Miyamoto H, Yamamura T, Honda T. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J Bacteriol 181:2257–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostroff S, Tarr P, Neill MA, Lewis JH, Hargrett-Bean N, Kobayashi JM. 1989. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis 160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 13.Donohue‐Rolfe A, Kondova I, Oswald S, Hutto D, Tzipori S. 2000. Escherichia coli O157:H7 strains that express Shiga toxin (Stx) 2 alone are more neurotropic for gnotobiotic piglets than are isotypes producing only Stx1 or both Stx1 and Stx2. J Infect Dis 181:1825–1829. doi: 10.1086/315421. [DOI] [PubMed] [Google Scholar]
- 14.Orth D, Grif K, Khan AB, Naim A, Dierich MP, Würzner R. 2007. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn Microbiol Infect Dis 59:235–242. doi: 10.1016/j.diagmicrobio.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 15.Persson S, Olsen KEP, Ethelberg S, Scheutz F. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J Clin Microbiol 45:2020–2024. doi: 10.1128/JCM.02591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shringi S, Schmidt C, Katherine K, Brayton KA, Hancock DD, Besser TE. 2012. Carriage of stx2a differentiates clinical and bovine-biased strains of Escherichia coli O157. PLoS One 7:e51572. doi: 10.1371/journal.pone.0051572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawano K, Okada M, Haga T, Maeda K, Goto Y. 2008. Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. Eur J Clin Microbiol Infect Dis 27:227–232. doi: 10.1007/s10096-007-0420-3. [DOI] [PubMed] [Google Scholar]
- 18.FAO/WHO. 2018. Shiga toxin-producing Escherichia coli (STEC) and food: attribution, characterization, and monitoring. FAO/WHO, Rome, Italy. [Google Scholar]
- 19.Carey CM, Kostrzynska M, Ojha S, Thompson S. 2008. The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods 73:125–132. doi: 10.1016/j.mimet.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Thévenot J, Cordonnier C, Rougeron A, Le Goff O, Nguyen HTT, Denis S, Alric M, Livrelli V, Blanquet-Diot S. 2015. Enterohemorrhagic Escherichia coli infection has donor-dependent effect on human gut microbiota and may be antagonized by probiotic yeast during interaction with Peyer’s patches. Appl Microbiol Biotechnol 99:9097–9110. doi: 10.1007/s00253-015-6704-0. [DOI] [PubMed] [Google Scholar]
- 21.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. 2009. Human microbiota-secreted factors inhibit Shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun 77:783–790. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamage SD, Strasser JE, Chalk CL, Weiss AA. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect Immun 71:3107–3115. doi: 10.1128/iai.71.6.3107-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goswami K, Chen C, Xiaoli L, Eaton KA, Dudley EG. 2015. Coculture of Escherichia coli O157:H7 with a nonpathogenic E. coli strain increases toxin production and virulence in a germfree mouse model. Infect Immun 83:4185–4193. doi: 10.1128/IAI.00663-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiaoli L, Figler HM, Goswami K, Dudley EG. 2018. Nonpathogenic E. coli enhance Stx2a production of E. coli O157:H7 through bamA-dependent and independent mechanisms. Front Microbiol 9:1325. doi: 10.3389/fmicb.2018.01325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith DL, James CE, Sergeant MJ, Yaxian Y, Saunders JR, McCarthy AJ, Allison HE. 2007. Short-tailed Stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J Bacteriol 189:7223–7233. doi: 10.1128/JB.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toshima H, Yoshimura A, Arikawa K, Hidaka A, Ogasawara J, Hase A, Masaki H, Nishikawa Y. 2007. Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl Environ Microbiol 73:7582–7588. doi: 10.1128/AEM.01326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mobley HLT, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nissle A. 1919. More about the Mutaflor treatment with special consideration of the chronic dysentery. Munch Med Wochenschr 25:678–681. [Google Scholar]
- 30.Wijetunge DSS, Karunathilake KHEM, Chaudhari A, Katani R, Dudley EG, Kapur V, DebRoy C, Kariyawasam S. 2014. Complete nucleotide sequence of pRS218, a large virulence plasmid, that augments pathogenic potential of meningitis-associated Escherichia coli strain RS218. BMC Microbiol 14:203. doi: 10.1186/s12866-014-0203-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen SL, Hung C-S, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meye RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davagnino J, Herrero M, Furlong D, Moreno F, Kolter R. 1986. The DNA replication inhibitor microcin B17 is a forty-three-amino-acid protein containing sixty percent glycine. Proteins Struct Funct Genet 1:230–238. doi: 10.1002/prot.340010305. [DOI] [PubMed] [Google Scholar]
- 33.Stephens CM, Skerker CM, Sekhon JM, Arkin MS, Riley AP. 2015. Complete genome sequences of four Escherichia coli ST95 isolates from bloodstream infections. Genome Announc 3:e01241-15. doi: 10.1128/genomeA.01241-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Künne C, Billion A, Mshana SE, Schmiedel J, Domann E, Hossain H, Hain T, Imirzalioglu C, Chakraborty T. 2012. Complete sequences of plasmids from the hemolytic-uremic syndrome-associated Escherichia coli strain HUSEC41. J Bacteriol 194:532–533. doi: 10.1128/JB.06368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson RE, Collin F, Maxwell A, Jolliffe KA, Payne RJ. 2014. Synthesis of full length and truncated microcin B17 analogues as DNA gyrase poisons. Org Biomol Chem 12:1570–1578. doi: 10.1039/c3ob42516a. [DOI] [PubMed] [Google Scholar]
- 36.Delgado MA, Solbiati JO, Chiuchiolo MJ, Farías RN, Salomón RA. 1999. Escherichia coli outer membrane protein TolC is involved in production of the peptide antibiotic microcin J25. J Bacteriol 181:1968–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braun V, Patzer SI, Hantke K. 2002. Ton-dependent colicins and microcins: modular design and evolution. Biochimie 84:365–380. doi: 10.1016/s0300-9084(02)01427-x. [DOI] [PubMed] [Google Scholar]
- 38.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 39.Slanetz LW, Bartley CH. 1957. Numbers of enterococci in water, sewage, and feces determined by the membrane filter technique with an improved medium. J Bacteriol 74:591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apperloo-Renkema HZ, Van Der Waaij BD, Van Der Waaij D. 1990. Determination of colonization resistance of the digestive tract by biotyping of Enterobacteriaceae. Epidemiol Infect 105:355–361. doi: 10.1017/s0950268800047944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson JR, Owens K, Gajewski A, Clabots C. 2008. Escherichia coli colonization patterns among human household members and pets, with attention to acute urinary tract infection. J Infect Dis 197:218–224. doi: 10.1086/524844. [DOI] [PubMed] [Google Scholar]
- 42.Leatham MP, Banerjee S, Autieri SM, Conway T, Cohen PS, Mercado-Lubo R. 2009. Precolonized human commensal Escherichia coli strains serve as a barrier to E. coli O157:H7 growth in the streptomycin-treated mouse intestine. Infect Immun 77:2876–2886. doi: 10.1128/IAI.00059-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A 96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sperandio V, Torres AG, Gir NJA, Kaper JB. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 183:5187–5197. doi: 10.1128/jb.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao T, Doyle MP, Harmon BG, Brown CA, Mueller PO, Parks AH. 1998. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. J Clin Microbiol 36:641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell BP, Griffin PM, Lozano P, Christie DL, Kobayashi JM, Tarr PI. 1997. Predictors of hemolytic uremic syndrome in children during a large outbreak of Escherichia coli O157:H7 infections. Pediatrics 100:E12. doi: 10.1542/peds.100.1.e12. [DOI] [PubMed] [Google Scholar]
- 47.Gamage SD, Patton AK, Strasser JE, Chalk CL, Weiss AA. 2006. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect Immun 74:1977–1983. doi: 10.1128/IAI.74.3.1977-1983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amigo N, Mercado E, Bentancor A, Singh P, Vilte D, Gerhardt E, Zotta E, Ibarra C, Manning SD, Larzábal M, Cataldi A. 2015. Clade 8 and clade 6 strains of Escherichia coli O157:H7 from cattle in Argentina have hypervirulent-like phenotypes. PLoS One 10:e0127710. doi: 10.1371/journal.pone.0127710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Micenková L. 2016. Bacteriocinogeny in pathogenic and commensal Escherichia coli strains. Masarykova Univerzita Lékařská Fakulta Biologický Ústav, Brno, Czechia. [Google Scholar]
- 50.Bielaszewska M, Idelevich EA, Zhang W, Bauwens A, Schaumburg F, Mellmann A, Peters G, Karch H. 2012. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob Agents Chemother 56:3277–3282. doi: 10.1128/AAC.06315-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Łoś JM, Łoś M, Węgrzyn A, Węgrzyn G. 2010. Hydrogen peroxide-mediated induction of the Shiga toxin converting lambdoid prophage ST2-8624 in Escherichia coli O157:H7. FEMS Immunol Med Microbiol 58:322–329. doi: 10.1111/j.1574-695X.2009.00644.x. [DOI] [PubMed] [Google Scholar]
- 52.Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 54.Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. 2003. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology 149:2557–2570. doi: 10.1099/mic.0.26396-0. [DOI] [PubMed] [Google Scholar]
- 55.Gilson L, Mahanty K, Kolter R. 1987. Four plasmid genes are required for colicin V synthesis, export, and immunity. J Bacteriol 169:2466–2470. doi: 10.1128/jb.169.6.2466-2470.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chehade H, Braun V. 1988. Iron-regulated synthesis and uptake of colicin V. FEMS Microbiol Lett 52:177–181. doi: 10.1111/j.1574-6968.1988.tb02591.x. [DOI] [Google Scholar]
- 57.Corsini G, Karahanian E, Tello M, Fernandez K, Rivero D, Saavedra JM, Ferrer A. 2010. Purification and characterization of the antimicrobial peptide microcin N. FEMS Microbiol Lett 312:119–125. doi: 10.1111/j.1574-6968.2010.02106.x. [DOI] [PubMed] [Google Scholar]
- 58.Morin N, Lanneluc I, Connil N, Cottenceau M, Pons AM, Sablé S. 2011. Mechanism of bactericidal activity of microcin L in Escherichia coli and Salmonella enterica. Antimicrob Agents Chemother 55:997–1007. doi: 10.1128/AAC.01217-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberhart LJ, Deringer JR, Brayton KA, Sawant AA, Besser TE, Call DR. 2012. Characterization of a novel microcin that kills enterohemorrhagic Escherichia coli O157:H7 and O26. Appl Environ Microbiol 78:6592–6599. doi: 10.1128/AEM.01067-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Z, Orfe LH, Liu J, Lu S-Y, Besser TE, Call DR. 2017. Microcin PDI regulation and proteolytic cleavage are unique among known microcins. Sci Rep 7:42529. doi: 10.1038/srep42529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Havarstein LS, Holo H, Nes IF. 1994. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology 140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 62.Havarstein LS, Diep DB, Nes IF. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol 16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 63.Yang CC, Konisky J. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J Bacteriol 158:757–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuzenkova J, Delgado M, Nechaev S, Savalia D, Epshtein V, Artsimovitch I, Mooney RA, Landick R, Farias RN, Salomon R, Severinov K. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J Biol Chem 277:50867–50875. doi: 10.1074/jbc.M209425200. [DOI] [PubMed] [Google Scholar]
- 65.Gilson L, Mahanty HK, Kolter R. 1990. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J 9:3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hartzell A, Chen C, Lewis C, Liu K, Reynolds S, Dudley EG. 2011. Escherichia coli O157:H7 of genotype lineage-specific polymorphism assay 211111 and clade 8 are common clinical isolates within Pennsylvania. Foodborne Pathog Dis 8:763–768. doi: 10.1089/fpd.2010.0762. [DOI] [PubMed] [Google Scholar]
- 67.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods 3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 68.Fan J, de Jonge BLM, MacCormack K, Sriram S, McLaughlin RE, Plant H, Preston M, Fleming PR, Albert R, Foulk M, Mills SD. 2014. A novel high-throughput cell-based assay aimed at identifying inhibitors of DNA metabolism in bacteria. Antimicrob Agents Chemother 58:7264–7272. doi: 10.1128/AAC.03475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Datsenko K, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 71.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 72.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yao K, Roberts RJ, Allard MW, Hoffmann M. 2017. Complete genome and methylome sequences of Salmonella enterica subsp. enterica serovars Typhimurium, Saintpaul, and Stanleyville from the SARA/SARB collection. Genome Announc 5:e00031-17. doi: 10.1128/genomeA.00031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 76.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 77.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. 2009. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res 37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res 47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med 308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 87.Appleyard RK. 1954. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39:440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 89.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun 39:315–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yorgey P, Lee J, Kördel J, Vivas E, Warner P, Jebaratnam D, Kolter R. 1994. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc Natl Acad Sci U S A 91:4519–4523. doi: 10.1073/pnas.91.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guzman L-M, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Larsen RA, Thomas MG, Postle K. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol Microbiol 31:1809–1824. doi: 10.1046/j.1365-2958.1999.01317.x. [DOI] [PubMed] [Google Scholar]
- 93.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. 1977. Construction and characterization of new cloning vehicle. II. A multipurpose cloning system. Gene 2:95–113. doi: 10.1016/0378-1119(77)90000-2. [DOI] [PubMed] [Google Scholar]
- 94.Rebuffat S. 2012. Microcins in action: amazing defence strategies of enterobacteria. Biochem Soc Trans 40:1456–1462. doi: 10.1042/BST20120183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide and SRA files for 0.1229 can be found in the NCBI database under BioSample accession number SAMN08737532. SRA files for 99.0750, 91.0593, and 90.2723 can be found under BioSample accession numbers SAMN11457477, SAMN11457478, and SAMN11457479, respectively.