Abstract
Mitochondria play a central role in bioenergetics, and fulfill a plethora of functions in cell signaling, programmed cell death, and biosynthesis of key protein cofactors. Mitochondria harbor their own genomic DNA, which encodes protein subunits of the electron transport chain and a full set of transfer and ribosomal RNAs. Mitochondrial DNA (mtDNA) is essential for cellular and organismal functions, and defects in mitochondrial genome maintenance have been implicated in common human diseases and mitochondrial disorders. mtDNA repair and degradation are known pathways to cope with mtDNA damage; however, molecular factors involved in this process have remained unclear. Such knowledge is fundamental to the understanding of mitochondrial genomic maintenance and pathology, because mtDNA degradation may contribute to the etiology of mtDNA depletion syndromes and to the activation of the innate immune response by fragmented mtDNA. This article reviews the current literature regarding the importance of mitochondrial DNA degradation in mtDNA maintenance and stress response, and the recent progress in uncovering molecular factors involved in mtDNA degradation. These factors include key components of the mtDNA replication machinery, such as DNA polymerase γ, helicase Twinkle, and exonuclease MGME1, as well as a major DNA-packaging protein, mitochondrial transcription factor A (TFAM).
Keywords: Mitochondrial DNA, DNA turnover, TFAM, DNA repair, DNA damage
1. Introduction
Mitochondrial DNA is a double-stranded circular molecule of 16,569 base pairs (bp) and is present in multiple copies in mitochondria [1–3]. mtDNA is essential for mitochondrial function and organismal health because it encodes 13 protein subunits of the electron transport chain system and a full set of tRNAs and rRNAs. Distinct from nuclear DNA (nDNA), the mtDNA genome exists as a heterogeneous population of molecules, known as heteroplasmy [4]. mtDNA copy number is specific to tissue type and to developmental stage and can number in thousands per cell. mtDNA is generally considered to be maternally inherited. In addition, the mtDNA genome contains no introns and very few noncoding intergenic nucleotides. It is replicated independently of the cell cycle and nDNA replication, although all the proteins involved in mtDNA replication are encoded by nDNA [3,5].
A distinctive property of the mitochondrial genome is that it has a much higher turnover rate than the nuclear genome [6,7]. Although this property has been known for decades, the basis of mtDNA degradation remains enigmatic. The identification of human diseases associated with mitochondrial DNA depletion has renewed interest in the topic [8,9], and inspired the development of mitochondrial-specific endonucleases targeting mutated mtDNA molecules [10–16]. Recently, mtDNA degradation has emerged as an important mechanism to counteract various insults to mtDNA. This review summarizes recent progress in understanding the molecular factors involved in mtDNA degradation in mammalian systems. For general discussion on mitochondrial genome maintenance and its relevance to human health, please see several excellent reviews [2,3,5,17].
2. Organization of mtDNA
2.1. Mitochondrial nucleoids and their DNA content
The mtDNA genome is organized in protein–DNA complexes known as nucleoids [18]. The term nucleoid was first used in 1959 in an electron microscopic study of rat oocytes [19], likely chosen in analogy to the organization of prokaryotic chromosomes. The understanding of the morphology and composition of mitochondrial nucleoids has advanced significantly over the past decade [18,20,21], driven primarily by the improved resolution of various imaging techniques. It is now known that nucleoid particles are located in the matrix between the cristae tubules, separated from the inner boundary membrane by cristae [22]. Historically, fluorescent DNA-binding probes, such as 4′,6-diamidino-2-phenylindole (DAPI), were used to visualize nucleoids. The early studies demonstrated that nucleoids exist as compact spherical or ovoid structures in yeast [23] and human cells [24]. The visualization was also achieved by tagging the green fluorescent protein to nucleoid-specific proteins, such as mitochondrial transcription factor A (TFAM) and mitochondrial single-stranded DNA-binding protein (mtSSB) [21]. The reported sizes of nucleoids vary considerably, ranging from an average diameter of 0.1 to 0.9μm, due to the lateral resolution limit of approximately 250nm of wide-field or confocal microscopy [25] and different sample preparation methods. The more recently developed stimulated emission depletion (STED) microscopy is capable of achieving a 30–80nm resolution range [25], which is more suited to determine the size of nucleoids and to resolve nucleoid clusters. Two recent studies using STED microscopy [22,26] obtained a uniform mean size of approximately 100nm in a panel of cultured mammalian cells, similar to the dimensions of nucleoids reconstituted in vitro using mtDNA and TFAM.
Determining the number of DNA molecules has proven challenging. Satoh and Kuriowa were the first to report a calculated average of 1.4 mtDNA per nucleoid, with 4.6 mtDNA molecules and 3.2 nucleoids per mitochondrion in human ovarian carcinoma cells [24]. Subsequent studies reported a range from about 2.4 to 7.8 mtDNA molecules per nucleoid, depending on the cell type and the methods employed. Using the super resolution STED microscopy, Kukat et al. found that a surprisingly large fraction (26%–54%) of all nucleoids exist as nucleoid clusters in a panel of mammalian cells [26]. When using bromodeoxyuridine to track newly synthesized DNA, the authors found 1.3 times more nucleoids using STED microscopy than with confocal microscopy, confirming the resolving power of STED microscopy. Notably, it was found that, on average, each mitochondrial nucleoid contains only a single mtDNA molecule in human and mouse fibroblasts [22,26], strikingly similar to the results by Satoh and Kuriowa almost 30 years ago [24]. It is conceivable that the DNA content could vary based on cell and tissue types, and the activity of DNA replication in nucleoids; additional research is required to clarify these questions.
2.2. Protein composition in mitochondrial nucleoids
The mtDNA genome is not naked but extensively coated with proteins. Early experiments showed that isolated mtDNA from HeLa [27] and rat cells [28] is associated with proteins and that certain regions of the Drosophila melanogaster genome are protected from trimethylpsoralen-induced DNA-interstrand crosslinking [29]. However, identifying nucleoid proteins is not as straightforward as it seems because of their dynamic composition and the low abundance of various proteins. The list of nucleoid proteins has been under constant expansion and revision, except for a list of core proteins that are essential for mtDNA maintenance and gene expression (for historical perspectives, please refer to previous reviews [18,20,30]). A layered structure of nucleoids was suggested by Bogenhagen and colleagues [31], which includes a set of core proteins for DNA replication and transcription, and peripheral proteins for translation, protein import, and metabolism.
Modern mass spectrometry-based methods have been the main driving force for identifying new nucleoid proteins. These methods generally involve two types of sample preparation methods. The first type involves formaldehyde cross-linking to covalently trap the interacting components followed by fractionation [31–33], and the second type employs known nucleoid proteins as bait to immunoprecipitate their interacting partners [33–35]. The former approach could produce false positives due to contamination from cytosolic and nuclear proteins, and the latter could generate false negatives owing to the failure of capturing transient or weak interactions. Consequently, there is a poor consensus among multiple data sets. Also, varying sensitivity of the instrumentation and different data processing methods may confound the analysis.
Recently, Han et al. used a proximity biotinylation method to identify a nucleoid proteome of 37 proteins [36], extending the specificity and coverage of previous mass spectrometry-based investigations [31,33] and those using immunoprecipitation-based approaches [34,35]. This method stands out, both in its sensitivity due to the ability to capture weak or transient interacting proteins in a cellular environment, and in its specificity due to avoidance of contaminants from cross-linking and fractionation steps. The selectivity of the data was further enhanced by filtering out by the nucleoid protein/matrix protein signal intensity ratios, although this data processing method could potentially miss some dual localized proteins [36]. Several new orphan nucleoid proteins were identified using this method, as summarized in Table 1. Interestingly, none of the published data sets identify any DNA repair factors in nucleoids, suggesting that these proteins exist in low abundance and are likely to interact transiently with DNA or other nucleoid proteins.
Table 1.
Summary of mitochondrial nucleoid proteins.
| Protein name | Gene name | Biological function in mitochondria | Han et al. [36] | Bogenhagen et al. [31] | He et al. [35] | Rajala et al. [33] | Wang and Bogenhagen [34] |
|---|---|---|---|---|---|---|---|
| Transcription factor A, mitochondrial | TFAM | Transcription factor required for transcription [37,38] | √ | √ | √ | √ | √ |
| Single-stranded DNA-binding protein, mitochondrial | SSBP1 | Single-stranded DNA binding protein required for mtDNA replication [39] | √ | √ | √ | √ | √ |
| Lon protease homolog, mitochondrial | LONP1 | mtDNA binding and RNA binding, involved in degradation of other nucleoid proteins such as POLG, TFAM and PEOl [40–42] | √ | √ | × | √ | × |
| DNA-directed RNA polymerase, mitochondrial | POLRMT | RNA polymerase required for transcription [37] | √ | √ | √ | √ | √ |
| Serine hydroxymethyltransferase, mitochondrial | SHMT2 | Involved in thymidylate biosynthesis required to prevent uracil accumulation in mtDNA; also part of the mitochondrial folate pathway [43] | √ | √ | √ | √ | √ |
| Isoform 2 of putative ATP-dependent RNA helicase DHX30 | DHX30 | Known RNA binding protein that affects mtRNA abundance [44] | √ | √ | √ | √ | √ |
| GTPase Era, mitochondrial | ERAL1 | Binds to 12S-mtRNA [45,46] | √ | √ | √ | × | × |
| G-rich sequence factor 1 | GRSF1 | Associated with nascent transcript binding and processing [47,48] | √ | √ | × | × | × |
| Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial | DBT | Mutation causes mtDNA damage accumulation and affects mtDNA copy number [49] | √ | √ | √ | √ | × |
| Twinkle protein, mitochondrial | PEO1 | Helicase required for mtDNA replication [39,50] | √ | √ | × | √ | √ |
| DNA polymerase subunit gamma-1 | POLG | DNA polymerase required for mtDNA replication [51] | √ | √ | × | √ | √ |
| RNA pseudouridylate synthase domain-containing protein 3 | RPUSD3 | RNA modification enzyme [52] | √ | × | × | × | × |
| Mitochondrial ribonuclease P protein 1 | TRMT10C | Associated with nascent transcript binding and processing [53] | √ | × | × | × | × |
| 2-Oxoglutarate dehydrogenase-like, mitochondrial | OGDHL | Oxoglutarate dehydrogenase degrading glucose and glutamate [54] | √ | × | × | × | × |
| Aspartate aminotransferase, mitochondrial | GOT2 | Known RNA binding protein [55] | √ | × | × | × | × |
| Mitochondrial 10-formyltetrahy drofolate dehydrogenase | ALDH1L2 | Mitochondrial form of 10-formyltetrahydrofolate dehydrogenase [56–58] | √ | × | × | × | × |
| Monofunctional C1-tetrahydrofolate synthase, mitochondrial | MTHFD1L | Up regulated with mtDNA replication defects [59] | √ | × | × | × | × |
| Dimethyladenosine transferase 2, mitochondrial | TFB2M | Transcription factor required for transcription [37] | √ | √ | × | √ | √ |
| Peroxiredoxin-5, mitochondrial | PKDX5 | DNA binding, negative regulator of transcription [60] | √ | × | × | × | × |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | NDUFS6 | A subunit of the NADH:ubiquinone oxidoreductase for the transfer of electrons from NADH to the respiratory chain [61] | √ | × | × | × | × |
| Isovaleryl-CoA dehydrogenase, mitochondrial | IVD | Mitochondrial matrix enzyme for leucine catabolism [62–64] | √ | × | × | × | × |
| Isocitrate dehydrogenase [NADP], mitochondrial | IDH2 | Yeast homolog IDH2 interacts with TFAM and binds to mtRNA [65,66] | √ | × | × | × | × |
| Adenylate kinase 4, mitochondrial | AK4 | Nucleotides modification enzyme [67] | √ | × | × | × | × |
| Mycophenolic acid acyl-glucuronide esterase, mitochondrial | ABHD10 | Hydrolase removing glucuronide from mycophenolic acid acyl-glucuronide [68–70] | √ | × | × | × | × |
| Methionine-R-sulfoxide reductase B2, mitochondrial | MSRB2 | Predicted DNA binding, transcription factor activity [71] | √ | × | × | × | × |
| DNA polymerase subunit gamma-2, mitochondrial | POLG2 | DNA polymerase required for mtDNA replication [39,72] | √ | × | × | √ | √ |
| GrpE protein homolog 2, mitochondrial | GRPEL2 | Essential component of the PAM complex, a complex required for the translocation of transit peptide-containing proteins from the inner membrane into the mitochondrial matrix in an ATP-dependent manner [73,74] | √ | × | × | × | × |
| Mitochondrial genome maintenance exonuclease 1 | MGME1 | Mitochondrial 7S DNA exonuclease [75,76] | √ | × | × | × | × |
| Kynurenine—oxoglutarate transaminase 3 | CCBL2 | Known RNA binding protein that affects mtRNA abundance [55,77] | √ | × | × | × | × |
| FAST kinase domain-containing protein 1 | FASTKD1 | Predicted RNA binding protein [52] | √ | × | × | × | × |
| Carnitine O-acetyltransferase | CRAT | Transferase removing acyl groups from carnitine to coenzyme A (CoA) and regulates the acyl-CoA/CoA ratio [78,79] | √ | × | × | × | × |
| UPF0562 protein C7orf55 (Protein FMC1 homolog) | C70RF55 (FMC1) | Involved in the assembly/stability of the mitochondrial membrane ATP synthase [80] | √ | × | × | × | × |
| Probable glutamate—tRNA ligase, mitochondrial | EARS2 | tRNA binding for aminoacylation [81] | √ | × | × | × | × |
| Decaprenyl-diphosphate synthase subunit 2 | PDSS2 | Similar to oxoglutarate dehydrogenase (OGDH) of the OGDH complex, which degrades glucose and glutamate [82–84] | √ | × | × | × | × |
| Probable proline—tRNA ligase, mitochondrial | PARS2 | tRNA binding for aminoacylation [36] | √ | × | × | × | × |
| Polymerase delta-interacting protein 2 | POLDIP2 | Interacts with TFAM and SSBP1, DNA binding activity, cross-links to SSBP1, LRPPRC, LONP1, CLPX [85,86] | √ | × | × | × | × |
3. DNA turnover in mitochondrial DNA maintenance
3.1. Mitochondrial DNA turnover
In 1969, Gross et al. reported that mtDNA in different rat tissues has a much shorter half-life (typically a matter of days) than nDNA (approximately 30 days) [6]. The mtDNA copy number varies considerably between cell and tissue types [87]. Even in a given tissue, mtDNA copy number varies over a 2–10-fold range [88]; variation in the range of 40%–150% in the average mtDNA content is considered clinically normal [89]. Current understanding of how mtDNA copy number is regulated remains incomplete [8]. Intriguingly, mitochondria can be partially or entirely depleted of their mtDNA in certain cells; mammalian erythrocytes contain no mitochondria and hence no mtDNA, and auxotrophic ρ0 cells lose their mtDNA content upon the induction of mtDNA depletion [90,91]. The major noncoding region of many mitochondrial genomes across species contains a triple-stranded D-loop, formed by stable association of a third, short DNA strand known as 7S DNA [92]. Curiously, 7S DNA is turned over even more rapidly relative to other parts of the mitochondrial genome, with a half-life of about an hour in rodent cells [93]. The reason for such a rapid turnover remains under investigation. The observations that organelle DNA abandonment exists across the species had led to the proposal that DNA degradation serves as a protective mechanism against mutagenesis and to reduce the energy cost of repair [93a].
3.2. Mitochondrial DNA degradation under stress conditions
Similar to nDNA, mtDNA is susceptible to endogenous and exogenous chemicals [94–96]. It is well documented that a wide variety of environmental carcinogens and alkylating agents form covalent modifications preferentially with mtDNA relative to nDNA in mammalian cell culture and in experimental animals [94,95]. The ratio of the resulting mtDNA to nDNA lesion ratios ranges from several fold for alkylating agents [97–100] and aflatoxin B1 [101], 50–100-fold for peroxidation-derived DNA adducts [102], to 50–200-fold for polycyclic aromatic hydrocarbons [103,104]. The higher ratios for the latter compounds have been attributed to their lipophilicity, and the persistence of the resulting lesions due to the lack of nucleotide excision repair in mitochondria. Moreover, several types of oxidative DNA lesions have been shown to exist in higher amounts in mtDNA relative to nDNA (see [94] and references therein). Contradictory results exist in the literature concerning the relative abundance in the two compartments of the lesion 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG, a commonly considered marker for oxidative DNA damage), likely because of the probability of artificial formation of 8-oxodG during sample preparation. Aside from measuring specific DNA lesions, multiple laboratories have reported a higher level of mtDNA lesions relative to nDNA in a wide variety of cells and tissues, using quantitative polymerase chain reaction (QPCR) methodology [105,106]. Cumulatively, these data emphasize the importance of mtDNA as a target for endogenous and exogenous DNA damaging agents.
The biological consequence of mtDNA damage remains less well understood than for its nuclear counterpart, due to the multicopy and heterogeneous characteristics of mtDNA. Several known pathways are known to cope with mtDNA damage. First, mtDNA lesions can be repaired via several mitochondrial DNA repair pathways (see earlier reviews [2,5,107–112] and Chapter “Enzymology of mitochondrial DNA repair” by Alencar et al. of this volume), of which base excision repair (BER) is the most well understood. Second, during mtDNA replication, certain lesions can be bypassed by DNA polymerase γ (pol γ) or other mitochondrial DNA polymerases now reported in mitochondria [113–115], or by a replication restart mechanism likely facilitated by PrimPol [116–119]. Lastly, the damaged mtDNA molecules can undergo a mitochondria-specific degradation pathway to maintain DNA integrity, which will be the focus of later sections. Notably, these pathways are not mutually exclusive, and can occur in parallel to cope with mtDNA damage.
An early report pointing to the existence of degradation of damaged mtDNA by Suter and Richter [120] showed that fragmented mtDNA contains a 15-fold higher level of 8-oxodG than the intact circular mtDNA in rat liver mitochondria, which suggested that “an efficient mtDNA repair or degrading system” exists in mitochondria [120]. Data from experimental animals also support the notion that mtDNA degradation is an important response to cellular stress. In mice, acute intragastric ethanol administration leads to mtDNA degradation in different tissues, including liver, brain, heart, and skeletal muscle [121,122]. Loss of mtDNA content was also observed after ischemia/reperfusion injury, followed by the restoration of mtDNA to a normal level 24h after the reperfusion [123].
A body of studies by Alexeyev and colleagues have shown unequivocally that mtDNA degradation is an active mechanism to counteract DNA damage [87,124–127]. Shokolenko et al. showed that hypoxanthine/xanthine oxidase-induced oxidative stress can lead to mtDNA degradation, and that accumulation of linear mtDNA molecules proceeds degradation [124]. When BER activity was inhibited by methoxyamine treatment, the activity of mtDNA degradation increased, suggesting that BER and mtDNA degradation cooperate to cope with DNA damage [124]. Subsequently, the same group verified that mtDNA degradation is a direct and immediate consequence of mtDNA damage after the induction of mitochondrial-specific DNA single-strand breaks and abasic sites [125]. The rate of degradation varies based on the type of mtDNA damage [125] and cell type [126]. In particular, Kozhukhar et al. demonstrated that BER and mtDNA degradation are major pathways to counteract mitochondrial abasic (AP) sites (vide infra), and that translesion bypass of AP sites occurs much less frequently [127].
Data from other laboratories also support the importance of mtDNA degradation. Kai et al. observed a rapid reduction in the amount of mtDNA after treating rat hepatocytes with 2′,3′-dideoxycytidine or ethidium bromide [128]. Furda et al. observed mtDNA loss and persistent mtDNA lesions in mouse embryonic fibroblasts upon H2O2 treatment [129]. Although these authors did not observe a decrease in mtDNA level after treatment with methyl methanesulfonate in contrast to the results by Shokolenko et al. [124], the discrepancy can be explained by the different treatment times, the potential effect of compensatory DNA synthesis, and different methods in measuring the DNA copy number used in the two studies. Moretton et al. reported that rapid mtDNA loss occurs after the induction of double-strand breaks by expression of a mitochondria-targeted restriction enzyme [130]. Furthermore, that treatment of experimental animals with environmental carcinogens [131] or suppressing the expression of mitochondrial BER enzymes [132] did not cause an increase in the mtDNA mutation load also suggests the existence of specific mechanisms to eliminate damaged mtDNA molecules. mtDNA degradation has emerged as an important mechanism for mtDNA maintenance and stress response. Degradation is believed to be nonspecific with regard to DNA lesion type, and activated in response to difficult-to-repair DNA lesions or excessive DNA damage [126,130].
4. Molecular factors involved in mtDNA degradation
Critical to the understanding of the mechanism by which mitochondria destruct damaged DNA molecules, several recent studies have identified protein factors involved. These include key components in the mtDNA replication machinery, such as pol γ, Twinkle, and MGME1, and the DNA packaging protein TFAM, as summarized below.
4.1. DNA polymerase γ
Encoded by the POLG gene, DNA polymerase γ has been considered historically as the sole DNA polymerase in human mitochondria [133,134]. It is now known that noncanonical DNA polymerases, such as PrimPol [116–119], pol β [113,114], and pol θ [115], can also localize to mitochondria and potentially participate in special replication and repair events. Nonetheless, pol γ remains the only replicative DNA polymerase, as it is responsible for the bulk of the mtDNA synthesis in human mitochondria [133,134]. Pol γ, mtSSB, and helicase Twinkle constitute the minimal mitochondrial replisome [39]. The human pol γ holoenzyme consists of a catalytic subunit and a dimeric form of its accessory subunit [134]. The catalytic subunit is an 140kDa polypeptide (p140 or pol γA) comprising an N-terminal exonuclease domain, a connecting linker region, and a C-terminal polymerase domain. The catalytic subunit has multiple enzymatic activities, including DNA polymerase activity, 3′ → 5′ exonuclease activity, and 5′ dRP lyase activity. The accessory subunit, a 55kDa protein (p55 or pol γB), is known to promote DNA binding and processive DNA synthesis [135].
Essential for high fidelity DNA synthesis, the exonuclease activity of pol γ proofreads the 3′-terminal nucleotide from both matched and mismatched primer termini, with a preference for the mismatched pairs. Moreover, the exonuclease activity cleaves single-stranded DNA, making it an excellent candidate for digesting linearized mtDNA fragments. Recently, results from several laboratories suggest the involvement of the exonuclease activity of pol γ in mtDNA degradation. Peeva et al. engineered human HEK-293 cells with mitochondrial-targeted restriction endonucleases [136]. Upon the induction of mtDNA double-strand breaks by restriction cleavage, progressive degradation occurs, to yield a mixture of DNA fragments ranging from a few hundred to several thousand base pairs downstream from the cleavage site. Ultra-deep sequencing revealed that mtDNA degradation occurs from both the 3′-end and 5′-end, suggesting the involvement of two types of exonucleolytic activities. Given the known specificities of mitochondrial nuclease MGME1 and pol γ, the authors verified the role of both enzymes in the process by generating MGME1-null cells and cells containing pol γ D274A (a known variant that is defective in its 3′ → 5′ exonuclease activity) using CRISPR–Cas9 technology. Results from Southern blotting and ultra-deep sequencing confirmed that the exonuclease activity of pol γ is required for removing linear mtDNA species. The accessory subunit did not appear to be involved in the degradation process as judged from experiments using siRNA-mediated knockdown of the POLG2 gene.
Nissanka et al. used the mtDNA mutator mice and the derived lung fibroblasts carrying an exonuclease-deficient pol γ to clarify the role of the exonuclease activity of pol γ in eliminating the fragmented mtDNA upon double-strand breaks [137]. In both cultured fibroblasts and the liver of the mutator mice, prolonged persistence of the mtDNA fragments was observed. Degradation did not depend on the DNA polymerase activity of pol γ or the origin of replication, suggesting that a separate population of pol γ molecules dock at the free double-stranded ends.
Currently, little is known regarding how the degradation mode of the replication machinery is regulated. Although the high-resolution crystal structures of apo pol γ and the pol γ ternary complexes have been solved [138] and revised [139–143], the existing structures do not shed light on the exonuclease mode of pol γ, or how the degradation mode is activated. It is conceivable that pol γ may adopt a different conformation in the degradative mode, and may cooperate with Twinkle and mtSSB to perform such functions [143]. Another potential factor regulating the pol γ degradative mode may be the level of deoxynucleoside triphosphates, reminiscent of the known activity of T4 DNA polymerase [144]. In addition, a switch between DNA synthesis and degradation by pol γ may depend on the homeostatic functions of autophagy, as has been observed in yeast [145]. All of these questions remain to be addressed.
4.2. MGME1
DNases play an essential role in mtDNA maintenance and repair (reviewed in [146]). Known mitochondrial DNases include APE1 [147], DNA2 [148,149], EXOG [150], ENDOG [151], FEN1 [152], MGME1 [76], and MRE11 [153,154]. EXOG, ENDOG, and MGME1 are localized exclusively to mitochondria, whereas the other enzymes are localized to both mitochondria and the nucleus. MGME1 (also known as Ddk1) belongs to the PD–(D/E)XK phosphodiesterase superfamily, which includes a broad spectrum of enzymes involved in DNA and RNA cleavage. MGME1 has a documented role in mtDNA degradation, evidenced from patients with loss-of-function MGME1 mutations [75] and MGME1-depleted cells [76]. Patients carrying MGME1 mutations develop multisystemic mitochondrial disorders characterized by external ophthalmoplegia, emaciation, and respiratory failure [75,155]. Muscle biopsies of the affected individuals show substantial mtDNA depletion and deletions. In fibroblast cultures from an affected patient, mtDNA repopulation is severely impaired after the nucleotide analog 2′,3′-dideoxycytidine (ddC) treatment and subsequent withdrawal. In addition, the mtDNA depletion rate upon ddC treatment is significantly slower in the MGME-null cells than that of the control fibroblasts, arguing a role for MGME1 in mtDNA degradation. Furthermore, MGME1 is involved in maintaining 7S DNA, the single-stranded DNA species formed by premature replication termination at the end of the control region of mtDNA. Patients carrying MGME1 mutations or MGME1-depleted cells exhibit an increase in 7S DNA levels [75,76]. The accumulated 7S DNA is elongated due to incomplete processing of 5′-ends by MGME1 [155]. Consistent with these findings, MGME1-knockout mice show mtDNA depletion, deletion of the minor arc of mtDNA, and increased stability of 7S DNA [156].
MGME1 is thought to be a component of the mitochondrial replisome, considering that MGME1 interacts with all three core components (pol γ, mtSSB and Twinkle) of the minimal mitochondrial replisome [155–157]. In the context of degrading flap DNA during Okazaki fragment processing or long-patch BER, MGME1 cooperates with the 3′ → 5′ exonuclease activity of pol γ to yield ligatable nicks in cells [158]. In cellular models containing restriction endonuclease-induced mtDNA double-strand breaks, MGME1, the 3′ → 5′ exonuclease activity of DNA polymerase γ, and Twinkle helicase together contribute to the removal of linear mtDNA [136]. The loss of MGME1 activity does not affect the efficiency of mtDNA cleavage, but causes a substantial accumulation of the linear mtDNA fragments induced by engineered mitochondrial endonucleases [136]. In MGME1−/− mice, tissue-specific replication stalling has been observed in addition to the aforementioned effects on 7S DNA and mtDNA deletion [156].
Biochemically, recombinant MGME1 cleaves single-stranded DNA (ssDNA) at the 5′- or 3′-terminus, and DNA flap substrates with a 5′- or 3′-flap [75,76]. MGME1 can also degrade the DNA segment in RNA-DNA chimeric substrates at a position two to five nucleotides downstream from the RNA-DNA junction [75]. MGME1 does not have endonucleolytic activity on a single-strand circular DNA [75,76]. In addition, MGME1 processes DNA 5′- and 3′-splayed-arm substrates by digesting the ssDNA segment but pauses at the ssDNA-dsDNA junction [75]. MGME1 is active in the presence of either Mg2+ or Mn2+, with a different optimal concentration for each metal ion [76]. Together, these enzymatic activities suggest that MGME1 is an excellent candidate for processing displaced DNA-containing Okazaki fragments on the lagging strand, and/or DNA flaps during long-patch BER [75].
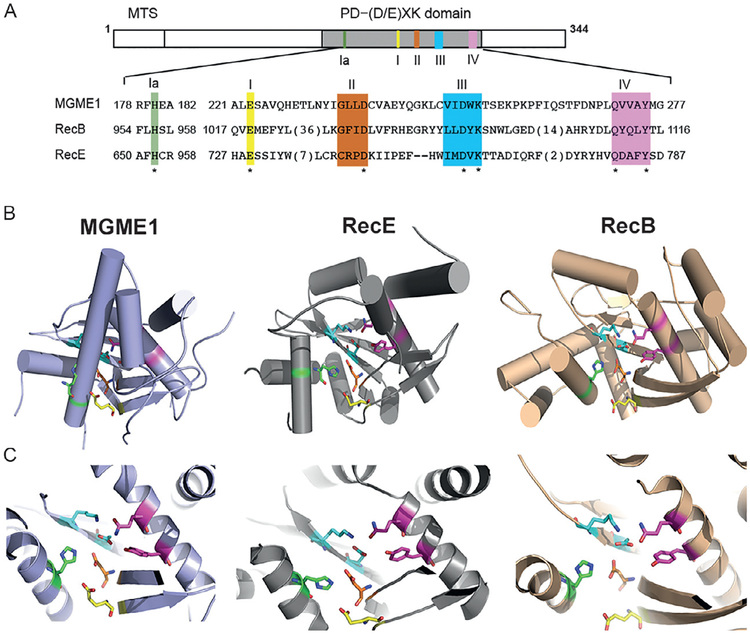
In the PD–(D/E)XK phosphodiesterase superfamily, MGME1 shares high sequence similarity with its orthologs from other species, and very low sequence similarity with other PD–(D/E)XK nucleases. Nonetheless, MGME1 contains several motifs (Ia, I, II, III, and IV, depicted in Fig. 1A) that are characteristic of RecB-type and RecE-type nucleases. Recently solved X-ray crystal structures of MGME1 have revealed the detailed architecture of the enzyme [159]. A Dali structural comparison search using the apo MGME1 (PDB:5ZYW) resulted in a list of structurally similar proteins, including RecE (PDB: 3H4R) and RecB (PDB: 1W36) nucleases (Fig. 1B). The conserved motifs share a considerable structural similarity when comparing the catalytic domains of these proteins (Fig. 1C). The structure of MGME1-ssDNA complex closely resembles that of apo MGME1, except that the connecting loop (aa 255–264) between β3 and α5 undergoes a large conformational change to allow DNA binding. The complex of MGME1 with a 3′-flap DNA also superimposes well with the MGME1-ssDNA complex, suggesting a common mechanism to cleave the ssDNA segment of different substrates. MGME1 uses a structural pin (F173) for duplex unwinding, as revealed in the structure of MGME1 with 3′-flap DNA [159].
Fig. 1.
Domain illustration of MGME1 and structural comparison with RecE and RecB nucleases. (A) MGME1 contains characteristic of RecB-type and RecE-type nuclease motifs (Ia, I, II, III, and IV). (B) Overall view of MGME1 (PDB:5ZYW), RecE (PDB: 3H4R), and RecB (1W36) nucleases with their active sites highlighted in (C). Conserved residues at their active sites are highlighted.
In evaluating the superimposition of structural components of several crystal structures, a two-metal ion catalytic mechanism has been proposed for MGME1 [159]. The two-metal ion mechanism, initially adopted from that of the exonuclease activity of E. coli DNA polymerase I, has been shown to be a common mechanism for many nucleases. In this model, metal ion A coordinates a water molecule to lower its pKa, and K253 acts as a general base to deprotonate the water molecule and to produce a strong nucleophile for the phosphoryl transfer reaction. Metal ion B also contributes to stabilizing the transition state by chelating the phosphate backbone and a conserved aspartate residue (D238). The solved MGME1 structures unveil the basis of the MGME1 3′-nuclease activity, although the mechanism of its 5′-nuclease activity remains to be clarified. At present, how MGME1 interacts with other components of the mitochondrial replisome is unknown.
4.3. TFAM
TFAM is a core mitochondrial transcription factor, responsible for recruiting mitochondrial RNA polymerase and transcription factor T2BM to activate transcription. Additionally, TFAM is an abundant protein that coats and packages mtDNA into nucleoids by imposing a U-turn on mtDNA [38,160] and cross-strand interactions [22]. TFAM knockout mice show embryonic lethality, indicating that TFAM is a critical protein for mitochondrial genome maintenance. Furthermore, TFAM has been proposed to play a role in regulating mtDNA repair [161], though the underlying mechanism remains to be clarified. Recently, my laboratory has identified a novel role for TFAM in facilitating the degradation of damaged mtDNA containing abasic (AP) sites [162], which are ubiquitous DNA lesions and an important intermediate during BER.
The importance of AP sites lies in their abundance and chemical reactivity. AP sites exist at a steady-state level of approximately 30,000 AP lesions per cell [163,164], and can number in the hundreds in the mitochondria of each cell [165,166]. AP sites are chemically reactive and labile, existing as an equilibrating mixture of a cyclic hemiacetal and a ring-opened aldehyde [167,168]. The latter functionality renders AP sites susceptible to reactions with amino groups in DNA, nuclear DNA repair enzymes, histones, small peptides, and endogenous polyamines (e.g., spermine), which can lead to DNA interstrand cross-links, DNA-protein cross-link intermediates, or single-strand breaks [167,169–177]. Several oxidized forms of AP sites can also yield stable DNA-protein cross-links [178,179].
AP sites and their derivatives pose tremendous threats to nuclear and mitochondrial genome integrity. AP sites are cytotoxic and mutagenic and can block nuclear DNA replication and transcription [180–182]. The DNA-protein cross-links formed between oxidized AP sites and histone proteins have the potential to alter histone epigenetic marks [183]. In mitochondria of human cells, an elevated level of AP sites leads to rapid loss of mtDNA, suggesting that the DNA degradation is unlikely due to autophagy, mitophagy, or apoptosis [126,130]. By inducing AP sites at either T or C residues in mouse embryonic fibroblasts, Kozhukhar et al. analyzed the relative contribution of DNA lesion bypass synthesis versus the combined contribution of BER and mtDNA degradation [127], and found that AP sites increase only moderately the overall mutation load, and that BER and mtDNA degradation are the major pathways to cope with abasic DNA [127]. Evidently, mitochondria are capable of abandoning the abasic DNA as a way to alleviate its cytotoxic effects.
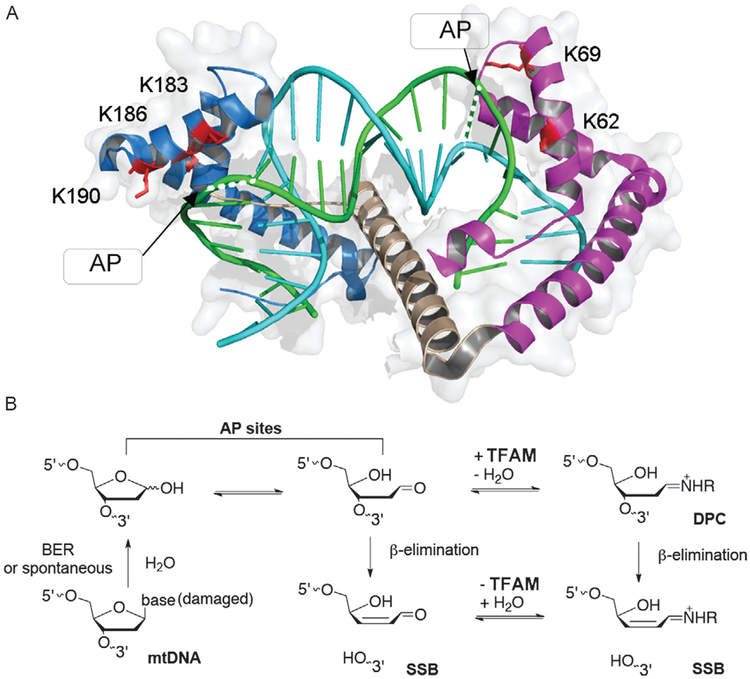
What protein factors could be involved in abasic mtDNA degradation? Considering the abundance of TFAM and its role as a histone-like protein in packaging mtDNA, it is inevitable that TFAM encounters the ubiquitous AP sites. Recently, my laboratory has shown that TFAM accelerates the degradation of AP lesion-containing DNA using biochemical assays and mitochondrial extracts from human cells [162]. Based on the proximity of the ε-NH2 group of several lysine residues and the C1′-carbon of the nearby nucleotide residue in the TFAM-DNA crystal structures [38,160], we hypothesized that several lysine residues might react with AP sites nearby to form single-strand breaks (Fig. 2A). Site-specifically modified oligodeoxynucleotides with an AP lesion were designed and synthesized using light strand promoter (LSP) and nonspecific DNA sequences. On the basis of structural analysis, an AP lesion was positioned near K69 in the high-mobility group (HMG) 1 domain, or in K183, K186, and K189 in the HMG 2 domain (Fig. 2A). In reconstituted TFAM-DNA complexes, we found that the half-life of AP sites is reduced by 2- to 3-orders of magnitude, depending on the position of the AP sites and the sequence of the oligodeoxynucleotides. We were able to trap the reaction intermediates (TFAM-DNA cross-links) using NaBH3CN, a reagent that selectively reduces imine to amine, indicating that the reaction occurs via Schiff base intermediates (the proposed mechanism is shown in Fig. 2B). The reaction was observed with either recombinant TFAM or mitochondrial extracts, suggesting that TFAM interacts strongly with AP-DNA in the presence of other mitochondrial proteins. The TFAM-induced reduction of AP lesion half-life resembles that of histone proteins in reconstituted core particles, although the extent of decrease in AP lesion half-life appears to be more dramatic in TFAM-DNA complexes [172,184].
Fig. 2.
Structural basis and proposed mechanism of TFAM-promoted abasic DNA strand scission. (A) Crystal structure of TFAM in complex with DNA containing the mitochondrial light-strand promoter (LSP) sequence (PDB: 3TQ6). The two high-mobility group (HMG) box domains of TFAM are in purple (HMG1) and blue (HMG2), and the inter-domain linker is in wheat. Potentially reactive (with AP sites) lysine residues within the two HMGs are highlighted in red. The heavy strand is in cyan, and the light strand is in green with the position of AP lesions shown in the dashed lines. (B) The proposed mechanism of TFAM-mediated DNA strand cleavage at AP sites via Schiff base intermediates (DNA-TFAM cross-links, DPC) to form single-strand breaks (SSB).
Remarkably, substituting putative lysine residues with alanine did not completely abolish TFAM-DNA cross-link formation, suggesting the potential contribution of other lysine or arginine residues in TFAM to promote strand scission. This observation is consistent with the dynamic nature of TFAM-DNA complexes, which could lead to transient interactions between additional lysine or arginine residues and AP sites, perhaps through a butterfly-like TFAM-DNA complex breathing [185]. In addition, considering the flexibility of the inter-domain linker [185], other Lys or Arg residues in the region could potentially contribute to the observed reaction with TFAM variants. The potential for multiple residues to contribute to the AP-DNA strand cleavage, and the fact that TFAM coats nearly-uniformly the entire mtDNA molecule, suggest a ubiquitous role for TFAM-promoted AP-DNA degradation. The role of TFAM in accelerating the strand cleavage of AP-DNA also suggests that TFAM could be involved in the rapid DNA depletion resulting from mitochondrial AP sites [127]. Additional research is required to establish the role of TFAM in DNA degradation in cellulo and in vivo.
APE1 is known to localize to mitochondria and particularly relevant to AP lesion repair. We found that TFAM competes actively with APE1-mediated DNA cleavage using mitochondrial extracts from HeLa and HEK-293 cells [162], suggesting that TFAM interacts strongly with AP lesions under physiological concentration ratios of TFAM to APE1, and TFAM could play a regulatory role in mtDNA repair, as suggested previously [161]. Because mtDNA degradation and repair may occur in parallel to alleviate abasic DNA damage [127], it is plausible that TFAM could act redundantly with APE1 to trigger mtDNA turnover or to regulate the partitioning between mtDNA degradation and repair. The cooperation between degradation and repair is likely to be regulated by additional factors, such as the total amount of TFAM protein (regulated by Lon protease [186,187]), the posttranslational modification of TFAM [187a,b], and the interactions between TFAM and its interacting partners.
4.4. Interactions of replisomal components
It is well established that interactions among replisome proteins are critically important for their functions [143]. Relevant to mtDNA degradation, it has been shown that mtSSB stimulates the DNA polymerase and exonuclease activities of pol γ [188,189], albeit moderate in the human system relative to the high level of stimulation observed in the Drosophila system [190,191]. Similarly, mtSSB stimulates the DNA unwinding activity of the human mtDNA helicase, Twinkle [188,189]. The importance of Twinkle helicase is supported by the accumulation of linear mtDNA fragments in human cells after siRNA knockdown [136]. It is reasonable to presume that functional interactions between mtSSB and both Twinkle and pol γ, are vital for their roles in mtDNA degradation.
MGME1 interacts with all three replisome components: the catalytic subunit of pol γ, mtSSB and Twinkle [155,156]. Although MGME1 employs a structural pin for duplex DNA unwinding [159], structural comparison of MGME1 with RecBCD complex and the direct interactions between MGME1 and Twinkle have led to the hypothesis that Twinkle may mimic the helicase domains of RecBCD to assist the nuclease activities of MGME1 and pol γ [159]. The structural and biochemical basis of these interactions remain to be determined.
5. Conclusions and future perspectives
Prevailing evidence supports clearly the importance of mtDNA degradation as a unique mechanism for mtDNA maintenance and stress response. Studies of mtDNA turnover and the selective destruction of damaged mtDNA molecules have inspired the development of mitochondrial-specific endonucleases to target mutated mitochondrial molecules for therapeutic purposes [10–16]. Despite recent advances, a complete understanding of mtDNA degradation remains to be determined. Myriad questions remain to be addressed, including the additional factors involved, detailed molecular pathways, and regulatory mechanisms. The dynamics of mtDNA turnover require complex regulation on multiple levels, including, but not limited to, mtDNA replication and degradation, fission and fusion, and mitophagy. Additional complexity from tissue-specific factors also needs to be considered carefully. An understanding of these fundamental questions will provide additional insights into the development therapeutics for mitochondrial diseases.
Acknowledgments
We thank Dr. Wenyan Xu for preparing Table 1 and acknowledge funding support from the National Institutes of Health (NIH) Grant R35 GM128854. We apologize to the investigators whose important work was not cited because of space considerations.
References
- [1].Nunnari J, Suomalainen A, Mitochondria: in sickness and in health, Cell 148 (2012) 1145–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Alexeyev M, Shokolenko I, Wilson G, LeDoux S, The maintenance of mitochondrial DNA integrity—critical analysis and update, Cold Spring Harb. Perspect. Biol 5 (2013) a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gustafsson CM, Falkenberg M, Larsson N-G, Maintenance and expression of mammalian mitochondrial DNA, Annu. Rev. Biochem 85 (2016) 133–160. [DOI] [PubMed] [Google Scholar]
- [4].Wallace DC, Chalkia D, Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease, Cold Spring Harb. Perspect. Biol 5 (2013) a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Copeland WC, Longley MJ, Mitochondrial genome maintenance in health and disease, DNA Repair 19 (2014) 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gross NJ, Getz GS, Rabinowitz M, Apparent turnover of mitochondrial deoxyribonucleic acid and mitochondrial phospholipids in the tissues of the rat, J. Biol. Chem 244 (1969) 1552–1562. [PubMed] [Google Scholar]
- [7].Berk AJ, Clayton DA, Mechanism of mitochondrial DNA replication in mouse L-cells: asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence, J. Mol. Biol 86 (1974) 801–824. [DOI] [PubMed] [Google Scholar]
- [8].Moraes CT, What regulates mitochondrial DNA copy number in animal cells? Trends Genet. 17 (2001) 199–205. [DOI] [PubMed] [Google Scholar]
- [9].Rötig A, Poulton J, Genetic causes of mitochondrial DNA depletion in humans, Biochim. Biophys. Acta Mol. Basis Dis 1792 (2009) 1103–1108. [DOI] [PubMed] [Google Scholar]
- [10].Srivastava S, Moraes CT, Manipulating mitochondrial DNA heteroplasmy by a mitochondrially targeted restriction endonuclease, Hum. Mol. Genet 10 (2001) 3093–3099. [DOI] [PubMed] [Google Scholar]
- [11].Tanaka M, Borgeld H-J, Zhang J, Muramatsu S-I, Gong J-S, Yoneda M, et al. , Gene therapy for mitochondrial disease by delivering restriction endonucleaseSmaI into mitochondria, J. Biomed. Sci 9 (2002) 534–541. [DOI] [PubMed] [Google Scholar]
- [12].Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A, Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase, Proc. Natl. Acad. Sci. U. S. A 103 (2006) 19689–19694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bacman S, Williams SL, Hernandez D, Moraes CT, Modulating mtDNA heteroplasmy by mitochondria-targeted restriction endonucleases in a ‘differential multiple cleavage-site’model, Gene Ther. 14 (2007) 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alexeyev M, Venediktova N, Pastukh V, Shokolenko I, Bonilla G, Wilson G, Selective elimination of mutant mitochondrial genomes as therapeutic strategy for the treatment of NARP and MILS syndromes, Gene Ther. 15 (2008) 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M, Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations, EMBO Mol. Med 6 (2014) 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reddy P, Ocampo A, Suzuki K, Luo J, Bacman SR, Williams SL, et al. , Selective elimination of mitochondrial mutations in the germline by genome editing, Cell 161 (2015) 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pohjoismäki JL, Forslund JM, Goffart S, Torregrosa-Muñumer R, Wanrooij S, Known unknowns of mammalian mitochondrial DNA maintenance, Bioessays 40 (2018) 1800102. [DOI] [PubMed] [Google Scholar]
- [18].Chen XJ, Butow RA, The organization and inheritance of the mitochondrial genome, Nat. Rev. Genet 6 (2005) 815. [DOI] [PubMed] [Google Scholar]
- [19].Sotelo JR, Porter KR, An electron microscope study of the rat ovum, J. Cell Biol. 5 (1959) 327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bogenhagen DF, Mitochondrial DNA nucleoid structure, Biochim. Biophys. Acta Gene Regul. Mech 1819 (2012) 914–920. [DOI] [PubMed] [Google Scholar]
- [21].Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN, Composition and dynamics of human mitochondrial nucleoids, Mol. Biol. Cell 14 (2003) 1583–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kukat C, Davies KM, Wurm CA, Spåhr H, Bonekamp NA, Kühl I, et al. , Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T, Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae, J. Cell Sci. 88 (1987) 431. [DOI] [PubMed] [Google Scholar]
- [24].Satoh M, Kuroiwa T, Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell, Exp. Cell Res. 196 (1991) 137–140. [DOI] [PubMed] [Google Scholar]
- [25].Vangindertael J, Camacho R, Sempels W, Mizuno H, Dedecker P, Janssen K, An introduction to optical super-resolution microscopy for the adventurous biologist, Methods Appl. Fluoresc 6 (2018) 022003. [DOI] [PubMed] [Google Scholar]
- [26].Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson N-G, Jakobs S, Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Albring M, Griffith J, Attardi G, Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication, Proc. Natl. Acad. Sci. U. S. A 74 (1977) 1348–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Van Tuyle GC, McPherson ML, A compact form of rat liver mitochondrial DNA stabilized by bound proteins, J. Biol. Chem 254 (1979) 6044–6053. [PubMed] [Google Scholar]
- [29].Potter DA, Fostel JM, Berninger M, Pardue ML, Cech TR, DNA-protein interactions in the Drosophila melanogaster mitochondrial genome as deduced from trimethylpsoralen crosslinking patterns, Proc. Natl. Acad. Sci. U. S. A 77 (1980) 4118–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hensen F, Cansiz S, Gerhold JM, Spelbrink JN, To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins, Biochimie 100 (2014) 219–226. [DOI] [PubMed] [Google Scholar]
- [31].Bogenhagen DF, Rousseau D, Burke S, The layered structure of human mitochondrial DNA nucleoids, J. Biol. Chem 283 (2008) 3665–3675. [DOI] [PubMed] [Google Scholar]
- [32].Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA, In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins, Proc. Natl. Acad. Sci. U. S. A 97 (2000) 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Rajala N, Hensen F, Wessels HJ, Ives D, Gloerich J, Spelbrink JN, Whole cell formaldehyde cross-linking simplifies purification of mitochondrial nucleoids and associated proteins involved in mitochondrial gene expression, PLoS One 10 (2015) e0116726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang Y, Bogenhagen DF, Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane, J. Biol. Chem 281 (2006) 25791–25802. [DOI] [PubMed] [Google Scholar]
- [35].He J, Cooper H, Reyes A, Di Re M, Sembongi H, Litwin T, et al. , Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis, Nucleic Acids Res. 40 (2012) 6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Han S, Udeshi ND, Deerinck TJ, Svinkina T, Ellisman MH, Carr SA, et al. , Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells, Cell Chem. Biol 24 (2017) 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Litonin D, Sologub M, Shi Y, Savkina M, Anikin M, Falkenberg M, et al. , Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro, J. Biol. Chem 285 (2010) 18129–18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Rubio-Cosials A, Sidow JF, Jimenez-Menendez N, Fernandez-Millan P, Montoya J, Jacobs HT, et al. , Human mitochondrial transcription factor A induces a U-turn structure in the light strand promoter, Nat. Struct. Mol. Biol 18 (2011) 1281–1289. [DOI] [PubMed] [Google Scholar]
- [39].Korhonen JA, Pham XH, Pellegrini M, Falkenberg M, Reconstitution of a minimal mtDNA replisome in vitro, EMBO J. 23 (2004) 2423–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lu B, Yadav S, Shah PG, Liu T, Tian B, Pukszta S, et al. , Roles for the human ATP-dependent Lon protease in mitochondrial DNA maintenance, J. Biol. Chem 282 (2007) 17363–17374. [DOI] [PubMed] [Google Scholar]
- [41].Fu GK, Markovitz DM, The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner, Biochemistry 37 (1998) 1905–1909. [DOI] [PubMed] [Google Scholar]
- [42].Liu T, Lu B, Lee I, Ondrovicova G, Kutejova E, Suzuki CK, DNA and RNA binding by the mitochondrial Lon protease is regulated by nucleotide and protein substrate, J. Biol. Chem 279 (2004) 13902–13910. [DOI] [PubMed] [Google Scholar]
- [43].Anderson DD, Quintero CM, Stover PJ, Identification of a de novo thymidylate biosynthesis pathway in mammalian mitochondria, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Antonicka H, Shoubridge EA, Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis, Cell Rep. 10 (2015) 920–932. [DOI] [PubMed] [Google Scholar]
- [45].Uchiumi T, Ohgaki K, Yagi M, Aoki Y, Sakai A, Matsumoto S, et al. , ERAL1 is associated with mitochondrial ribosome and elimination of ERAL1 leads to mitochondrial dysfunction and growth retardation, Nucleic Acids Res. 38 (2010) 5554–5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dennerlein S, Rozanska A, Wydro M, Chrzanowska-Lightowlers ZMA, Lightowlers RN, Human ERAL1 is a mitochondrial RNA chaperone involved in the assembly of the 28S small mitochondrial ribosomal subunit, Biochem. J 430 (2010) 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, et al. , GRSF1 regulates RNA processing in mitochondrial RNA granules, Cell Metab. 17 (2013) 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Antonicka H, Sasarman F, Nishimura T, Paupe V, Shoubridge EA, The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression, Cell Metab. 17 (2013) 386–398. [DOI] [PubMed] [Google Scholar]
- [49].Strand JM, Skinnes R, Scheffler K, Rootvelt T, Woldseth B, Bjoras M, et al. , Genome instability in Maple syrup urine disease correlates with impaired mitochondrial biogenesis, Metab. Clin. Exp 63 (2014) 1063–1070. [DOI] [PubMed] [Google Scholar]
- [50].Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, et al. , Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria, Nat. Genet 28 (2001) 2232001;29:100. [DOI] [PubMed] [Google Scholar]
- [51].Lestienne P, Evidence for a direct role of the DNA polymerase gamma in the replication of the human mitochondrial DNA in vitro, Biochem. Biophys. Res. Commun 146 (1987) 1146–1153. [DOI] [PubMed] [Google Scholar]
- [52].Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, et al. , Insights into RNA biology from an atlas of mammalian mRNA-binding proteins, Cell 149 (2012) 1393–1406. [DOI] [PubMed] [Google Scholar]
- [53].Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W, RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme, Cell 135 (2008) 462–474. [DOI] [PubMed] [Google Scholar]
- [54].Alters SE, McLaughlin B, Spink B, Lachinyan T, Wang CW, Podust V, et al. , GLP2–2G-XTEN: a pharmaceutical protein with improved serum half-life and efficacy in a rat crohn’s disease model, PLoS One 7 (2012) e50630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, et al. , The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts, Mol. Cell 46 (2012) 674–690. [DOI] [PubMed] [Google Scholar]
- [56].Strickland KC, Hoeferlin LA, Oleinik NV, Krupenko NI, Krupenko SA, Acyl carrier protein-specific 4’-phosphopantetheinyl rransferase activates 10-formyltetrahydrofolate dehydrogenase, J. Biol. Chem 285 (2010) 1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Strickland KC, Krupenko NI, Dubard ME, Hu CJ, Tsybovsky Y, Krupenko SA, Enzymatic properties of ALDH1L2, a mitochondrial 10-formyltetrahydrofolate dehydrogenase, Chem. Biol. Interact 191 (2011) 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA, ALDH1L2 Is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase, J. Biol. Chem 285 (2010) 23054–23061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Nikkanen J, Forsstrom S, Euro L, Paetau I, Kohnz RA, Wang LY, et al. , Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism, Cell Metab. 23 (2016) 635–648. [DOI] [PubMed] [Google Scholar]
- [60].Kropotov A, Sedova V, Ivanov V, Sazeeva N, Tomilin A, Krutilina R, et al. , A novel human DNA-binding protein with sequence similarity to a subfamily of redox proteins which is able to repress RNA-polymerase-III-driven transcription of the Alu-family retroposons in vitro, Eur. J. Biochem 260 (1999) 336–346. [DOI] [PubMed] [Google Scholar]
- [61].Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT, Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I, Mol. Cell. Biol 27 (2007) 4228–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Volchenboum SL, Vockley J, Mitochondrial import and processing of wild type and type III mutant isovaleryl-CoA dehydrogenase, J. Biol. Chem 275 (2000) 7958–7963. [DOI] [PubMed] [Google Scholar]
- [63].Mohsen AWA, Vockley J, Kinetic and spectral properties of isovaleryl-CoA dehydrogenase and interaction with ligands, Biochimie 108 (2015) 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AW, Vockley J, et al. , Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity, Biochemistry 36 (1997) 8455–8464. [DOI] [PubMed] [Google Scholar]
- [65].Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA, Mitochondrial nucleoids undergo remodeling in response to metabolic cues, J. Cell Sci. 121 (2008) 1861–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Elzinga SDJ, Bednarz AL, Vanoosterum K, Dekker PJT, Grivell LA, Yeast mitochondrial Nad(+)-dependent isocitrate dehydrogenase is an RNA-binding protein, Nucleic Acids Res. 21 (1993) 5328–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Panayiotou C, Solaroli N, Johansson M, Karlsson A, Evidence of an intact N-terminal translocation sequence of human mitochondrial adenylate kinase 4, Int. J. Biochem. Cell Biol. 42 (2010) 62–69. [DOI] [PubMed] [Google Scholar]
- [68].Iwamura A, Fukami T, Higuchi R, Nakajima M, Yokoi T, Human alpha/beta hydrolase domain containing 10 (ABHD10) is responsible enzyme for deglucuronidation of mycophenolic acid acyl-glucuronide in liver, J. Biol. Chem 287 (2012) 9240–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zuhl AM, Mohr JT, Bachovchin DA, Niessen S, Hsu KL, Berlin JM, et al. , Competitive activity-based protein profiling identifies aza-beta-lactams as a versatile chemotype for serine hydrolase inhibition, J. Am. Chem. Soc 134 (2012) 5068–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li Q, Vande Velde C, Israelson A, Xie J, Bailey AO, Dong MQ, et al. , ALS-linked mutant superoxide dismutase 1 (SOD1) alters mitochondrial protein composition and decreases protein import, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Huang W, Escribano J, Sarfarazi M, Coca-Prados M, Identification, expression and chromosome localization of a human gene encoding a novel protein with similarity to the pilB family of transcriptional factors (pilin) and to bacterial peptide methionine sulfoxide reductases, Gene 233 (1999) 233–240. [DOI] [PubMed] [Google Scholar]
- [72].Lim SE, Longley MJ, Copeland WC, The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance, J. Biol. Chem 274 (1999) 38197–38203. [DOI] [PubMed] [Google Scholar]
- [73].Konovalova S, Liu X, Manjunath P, Baral S, Neupane N, Hilander T, et al. , Redox regulation of GRPEL2 nucleotide exchange factor for mitochondrial HSP70 chaperone, Redox Biol. 19 (2018) 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Voisine C, Craig EA, Zufall N, von Ahsen O, Pfanner N, Voos W, The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70, Cell 97 (1999) 565–574. [DOI] [PubMed] [Google Scholar]
- [75].Kornblum C, Nicholls TJ, Haack TB, Scholer S, Peeva V, Danhauser K, et al. , Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease, Nat. Genet 45 (2013) 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Szczesny RJ, Hejnowicz MS, Steczkiewicz K, Muszewska A, Borowski LS, Ginalski K, et al. , Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels, Nucleic Acids Res. 41 (2013) 3144–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wolf AR, Mootha VK, Functional genomic analysis of human mitochondrial RNA processing, Cell Rep. 7 (2014) 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wu DH, Govindasamy L, Lian W, Gu YR, Kukar T, Agbandje-McKenna M, et al. , Structure of human carnitine acetyltransferase—molecular basis for fatty acyl transfer, J. Biol. Chem 278 (2003) 13159–13165. [DOI] [PubMed] [Google Scholar]
- [79].Drecourt A, Babdor J, Dussiot M, Petit F, Goudin N, Garfa-Traore M, et al. , Impaired transferrin receptor palmitoylation and recycling in neurodegeneration with brain iron accumulation, Am. J. Hum. Genet 102 (2018) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Li Y, Jourdain AA, Calvo SE, Liu JS, Mootha VK, CLIC, a tool for expanding biological pathways based on co-expression across thousands of datasets, PLoS Comput. Biol 13 (2017) e1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nagao A, Suzuki T, Katoh T, Sakaguchi Y, Suzuki T, Biogenesis of glutaminyl-mt tRNAGln in human mitochondria, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 16209–16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Saiki R, Nagata A, Kainou T, Matsuda H, Kawamukai M, Characterization of solanesyl and decaprenyl diphosphate synthases in mice and humans, FEBS J 272 (2005) 5606–5622. [DOI] [PubMed] [Google Scholar]
- [83].Chen P, Zhao SH, Chu YL, Xu K, Zhu L, Wu Y, et al. , Anticancer activity of PDSS2, prenyl diphosphate synthase, subunit 2, in gastric cancer tissue and the SGC7901 cell line, Anti-Cancer Drugs 20 (2009) 141–148. [DOI] [PubMed] [Google Scholar]
- [84].Chen P, Zhang Y, Polireddy K, Chen Q, The tumor-suppressing activity of the prenyl diphosphate synthase subunit 2 gene in lung cancer cells, Anti-Cancer Drugs 25 (2014) 790–798. [DOI] [PubMed] [Google Scholar]
- [85].Cheng XL, Kanki T, Fukuoh A, Ohgaki K, Takeya R, Aoki Y, et al. , PDIP38 associates with proteins constituting the mitochondrial DNA nucleoid, J. Biochem 138 (2005) 673–678. [DOI] [PubMed] [Google Scholar]
- [86].Liu L, Rodriguez-Belmonte EM, Mazloum N, Xie B, Lee MYWT, Identification of a novel protein, PDIP38, that interacts with the p50 subunit of DNA polymerase delta and proliferating cell nuclear antigen, J. Biol. Chem 278 (2003) 10041–10047. [DOI] [PubMed] [Google Scholar]
- [87].Shokolenko IN, Alexeyev MF, Mitochondrial DNA: a disposable genome? Biochim. Biophys. Acta 2015 (1852) 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Stringer HAJ, Sohi GK, Maguire JA, Côté HCF, Decreased skeletal muscle mitochondrial DNA in patients with statin-induced myopathy, J. Neurol. Sci 325 (2013) 142–147. [DOI] [PubMed] [Google Scholar]
- [89].Nakano Y, Murayama K, Tsuruoka T, Aizawa M, Nagasaka H, Horie H, et al. , Fatal case of mitochondrial DNA depletion with severe asphyxia in a newborn, Pediatr. Int 53 (2011) 240–242. [DOI] [PubMed] [Google Scholar]
- [90].Nass MM, Differential effects of ethidium bromide on mitochondrial and nuclear DNA synthesis in vivo in cultured mammalian cells, Exp. Cell Res. 72 (1972) 211–222. [DOI] [PubMed] [Google Scholar]
- [91].Kukat A, Kukat C, Brocher J, Schäfer I, Krohne G, Trounce IA, et al. , Generation of ρ 0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses, Nucleic Acids Res. 36 (2008) e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Nicholls TJ, Minczuk M, In D-loop: 40 years of mitochondrial 7S DNA, Exp. Gerontol 56 (2014) 175–181. [DOI] [PubMed] [Google Scholar]
- [93].Bogenhagen D, Clayton DA, Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence, J. Mol. Biol 119 (1978) 49–68; [DOI] [PubMed] [Google Scholar]; (a) Bendich AJ, DNA abandonment and the mechanisms of uniparental inheritance of mitochondria and chloroplasts, Chromosome Res. 21, 2013, 287–296. [DOI] [PubMed] [Google Scholar]
- [94].Cline SD, Mitochondrial DNA damage and its consequences for mitochondrial gene expression, Biochim. Biophys. Acta Gene Regul. Mech 1819 (2012) 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Meyer JN, Leung MC, Rooney JP, Sendoel A, Hengartner MO, Kisby GE, et al. , Mitochondria as a target of environmental toxicants, Toxicol. Sci 134 (2013) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Roubicek DA, de Souza-Pinto NC, Mitochondria and mitochondrial DNA as relevant targets for environmental contaminants, Toxicology 391 (2017) 100–108. [DOI] [PubMed] [Google Scholar]
- [97].Wunderlich V, Schütt M, Böttger M, Graffi A, Preferential alkylation of mitochondrial deoxyribonucleic acid by N-methyl-N-nitrosourea, Biochem. J 118 (1970) 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Wunderlich V, Tetzlaff I, Graffi A, Studies on nitrosodimethylamine: preferential methylation of mitochondrial DNA in rats and hamsters, Chem. Biol. Interact 4 (1972) 81–89. [DOI] [PubMed] [Google Scholar]
- [99].Wilkinson R, Hawks A, Pegg AE, Methylation of rat liver mitochondrial deoxyribonucleic acid by chemical carcinogens and associated alterations in physical properties, Chem. Biol. Interact 10 (1975) 157–167. [DOI] [PubMed] [Google Scholar]
- [100].Myers KA, Saffhill R, O’Connor PJ, Repair of alkylated purines in the hepatic DNA of mitochondria and nuclei in the rat, Carcinogenesis 9 (1988) 285–292. [DOI] [PubMed] [Google Scholar]
- [101].Niranjan BG, Bhat NK, Avadhani NG, Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis, Science 215 (1982) 73. [DOI] [PubMed] [Google Scholar]
- [102].Wauchope OR, Mitchener MM, Beavers WN, Galligan JJ, Camarillo JM, Sanders WD, et al. , Oxidative stress increases M1dG, a major peroxidation-derived DNA adduct, in mitochondrial DNA, Nucleic Acids Res. 46 (2018) 3458–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Backer JM, Weinstein IB, Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene, Science 209 (1980) 297. [DOI] [PubMed] [Google Scholar]
- [104].Allen JA, Coombs MM, Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA, Nature 287 (1980) 244–245. [DOI] [PubMed] [Google Scholar]
- [105].Santos JH, Meyer JN, Mandavilli BS, Van Houten B, Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells, Methods Mol. Biol 314 (2006) 183–199. [DOI] [PubMed] [Google Scholar]
- [106].Furda A, Santos JH, Meyer JN, Van Houten B, Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells, Methods Mol. Biol 1105 (2014) 419–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Prakash A, Doublié S, Base excision repair in the mitochondria, J. Cell. Biochem 116 (2015) 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mandavilli BS, Santos JH, Van Houten B, Mitochondrial DNA repair and aging, Mutat. Res. Fundam. Mol. Mech. Mutagen 509 (2002) 127–151. [DOI] [PubMed] [Google Scholar]
- [109].Kazak L, Reyes A, Holt IJ, Minimizing the damage: repair pathways keep mitochondrial DNA intact, Nat. Rev. Mol. Cell Biol. 13 (2012) 659. [DOI] [PubMed] [Google Scholar]
- [110].Stein A, Sia EA, Mitochondrial DNA repair and damage tolerance, Front. Biosci 22 (2017) 920–943. [DOI] [PubMed] [Google Scholar]
- [111].Croteau DL, Bohr VA, Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells, J. Biol. Chem 272 (1997) 25409–25412. [DOI] [PubMed] [Google Scholar]
- [112].Croteau DL, Stierum RH, Bohr VA, Mitochondrial DNA repair pathways, Mutat. Res 434 (1999) 137–148. [DOI] [PubMed] [Google Scholar]
- [113].Sykora P, Kanno S, Akbari M, Kulikowicz T, Baptiste BA, Leandro GS, et al. , DNA polymerase beta participates in mitochondrial DNA repair, Mol. Cell Biol. 37 (2017). e00237–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Krasich R, Copeland WC, DNA polymerases in the mitochondria: a critical review of the evidence, Front. Biosci., Landmark Ed 22 (2017) 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Wisnovsky S, Sack T, Pagliarini DJ, Laposa RR, Kelley SO, DNA polymerase θ increases mutational rates in mitochondrial DNA, ACS Chem. Biol 13 (2018) 900–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, et al. , PrimPol, an archaic primase/polymerase operating in human cells, Mol. Cell 52 (2013) 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wan L, Lou J, Xia Y, Su B, Liu T, Cui J, et al. , hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity, EMBO Rep. 14 (2013) 1104–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Bailey LJ, Doherty AJ, Mitochondrial DNA replication: a PrimPol perspective, Biochem. Soc. Trans 45 (2017) 513–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Torregrosa-Muñumer R, Forslund JM, Goffart S, Pfeiffer A, Stojkovič G, Carvalho G, et al. , PrimPol is required for replication reinitiation after mtDNA damage, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 11398–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Suter M, Richter C, Fragmented mitochondrial DNA is the predominant carrier of oxidized DNA bases, Biochemistry 38 (1999) 459–464. [DOI] [PubMed] [Google Scholar]
- [121].Mansouri A, Gaou I, de Kerguenec C, Amsellem S, Haouzi D, Berson A, et al. , An alcoholic binge causes massive degradation of hepatic mitochondrial DNA in mice, Gastroenterology 117 (1999) 181–190. [DOI] [PubMed] [Google Scholar]
- [122].Mansouri A, Demeilliers C, Amsellem S, Pessayre D, Fromenty B, Acute ethanol administration oxidatively damages and depletes mitochondrial DNA in mouse liver, brain, heart, and skeletal muscles: protective effects of antioxidants, J. Pharmacol. Exp. Ther 298 (2001) 737–743. [PubMed] [Google Scholar]
- [123].Chen H, Hu C-J, He YY, Yang D-I, Xu J, Hsu CY, Reduction and restoration of mitochondrial DNA content after focal cerebral ischemia/reperfusion, Stroke 32 (2001) 2382–2387. [DOI] [PubMed] [Google Scholar]
- [124].Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF, Oxidative stress induces degradation of mitochondrial DNA, Nucleic Acids Res. 37 (2009) 2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Shokolenko IN, Wilson GL, Alexeyev MF, Persistent damage induces mitochondrial DNA degradation, DNA Repair 12 (2013) 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Shokolenko IN, Wilson GL, Alexeyev MF, The “fast” and the “slow” modes of mitochondrial DNA degradation, Mitochondrial DNA A DNA Mapp. Seq. Anal 27 (2016) 490–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kozhukhar N, Spadafora D, Fayzulin R, Shokolenko IN, Alexeyev M, The efficiency of the translesion synthesis across abasic sites by mitochondrial DNA polymerase is low in mitochondria of 3T3 cells, Mitochondrial DNA A DNA Mapp. Seq. Anal 27 (2016) 4390–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kai Y, Takamatsu C, Tokuda K, Okamoto M, Irita K, Takahashi S, Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture, Mitochondrion 6 (2006) 299–304. [DOI] [PubMed] [Google Scholar]
- [129].Furda AM, Marrangoni AM, Lokshin A, Van Houten B, Oxidants and not alkylating agents induce rapid mtDNA loss and mitochondrial dysfunction, DNA Repair 11 (2012) 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Moretton A, Morel F, Macao B, Lachaume P, Ishak L, Lefebvre M, et al. , Selective mitochondrial DNA degradation following double-strand breaks, PLoS One 12 (2017) e0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Valente WJ, Ericson NG, Long AS, White PA, Marchetti F, Bielas JH, Mitochondrial DNA exhibits resistance to induced point and deletion mutations, Nucleic Acids Res. 44 (2016) 8513–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Kauppila JH, Bonekamp NA, Mourier A, Isokallio MA, Just A, Kauppila TE, et al. , Base-excision repair deficiency alone or combined with increased oxidative stress does not increase mtDNA point mutations in mice, Nucleic Acids Res. 46 (2018) 6642–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kaguni LS, DNA polymerase γ, the mitochondrial replicase, Annu. Rev. Biochem 73 (2004) 293–320. [DOI] [PubMed] [Google Scholar]
- [134].Graziewicz MA, Longley MJ, Copeland WC, DNA polymerase γ in mitochondrial DNA replication and repair, Chem. Rev 106 (2006) 383–405. [DOI] [PubMed] [Google Scholar]
- [135].Lim SE, Longley MJ, Copeland WC, The mitochondrial p55 accessory subunit of human DNA polymerase γ enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance, J. Biol. Chem 274 (1999) 38197–38203. [DOI] [PubMed] [Google Scholar]
- [136].Peeva V, Blei D, Trombly G, Corsi S, Szukszto MJ, Rebelo-Guiomar P, et al. , Linear mitochondrial DNA is rapidly degraded by components of the replication machinery, Nat. Commun 9 (2018) 1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Nissanka N, Bacman SR, Plastini MJ, Moraes CT, The mitochondrial DNA polymerase gamma degrades linear DNA fragments precluding the formation of deletions, Nat. Commun 9 (2018) 2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lee Y-S, Kennedy WD, Yin YW, Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations, Cell 139 (2009) 312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Sohl CD, Szymanski MR, Mislak AC, Shumate CK, Amiralaei S, Schinazi RF, et al. , Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase γ, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 8596–8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Stumpf JD, Copeland WC, Mitochondrial DNA replication and disease: insights from DNA polymerase γ mutations, Cell. Mol. Life Sci. 68 (2011) 219–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Farnum GA, Nurminen A, Kaguni LS, Mapping 136 pathogenic mutations into functional modules in human DNA polymerase γ establishes predictive genotype–phenotype correlations for the complete spectrum of POLG syndromes, Biochim. Biophys. Acta, Gene Regul. Mech 1837 (2014) 1113–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Young MJ, Copeland WC, Human mitochondrial DNA replication machinery and disease, Curr. Opin. Genet. Dev 38 (2016) 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ciesielski GL, Oliveira MT, Kaguni LS, Animal Mitochondrial DNA Replication, The Enzymes, Elsevier, 2016, pp. 255–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Huang WM, Lehman I, On the exonuclease activity of phage T4 deoxyribonucleic acid polymerase, J. Biol. Chem 247 (1972) 3139–3146. [PubMed] [Google Scholar]
- [145].Medeiros TC, Thomas RL, Ghillebert R, Graef M, Autophagy balances mtDNA synthesis and degradation by DNA polymerase POLG during starvation, J. Cell Biol. 217 (2018) 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Bruni F, Lightowlers RN, Chrzanowska-Lightowlers ZM, Human mitochondrial nucleases, FEBS J 284 (2017) 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, et al. , Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells, Nucleic Acids Res. 34 (2006) 2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Copeland WC, Longley MJ, DNA2 resolves expanding flap in mitochondrial base excision repair, Mol. Cell 32 (2008) 457–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, et al. , Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates, Mol. Cell 32 (2008) 325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, et al. , Apoptosis induced by persistent single-strand breaks in mitochondrial genome critical role of EXOG (5′-EXO/endonuclease) in their repair, J. Biol. Chem 286 (2011) 31975–31983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ohsato T, Ishihara N, Muta T, Umeda S, Ikeda S, Mihara K, et al. , Mammalian mitochondrial endonuclease G: digestion of R-loops and localization in inter-membrane space, Eur. J. Biochem 269 (2002) 5765–5770. [DOI] [PubMed] [Google Scholar]
- [152].Liu P, Qian L, Sung J-S, de Souza-Pinto NC, Zheng L, Bogenhagen DF, et al. , Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria, Mol. Cell. Biol 28 (2008) 4975–4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Dmitrieva NI, Malide D, Burg MB, Mre11 is expressed in mammalian mitochondria where it binds to mitochondrial DNA, Am. J. Phys. Regul. Integr. Comp. Phys 301 (2011) R632–R640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC, Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions, Mol. Biol. Cell 27 (2016) 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Nicholls TJ, Zsurka G, Peeva V, Schöler S, Szczesny RJ, Cysewski D, et al. , Linear mtDNA fragments and unusual mtDNA rearrangements associated with pathological deficiency of MGME1 exonuclease, Hum. Mol. Genet 23 (2014) 6147–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Matic S, Jiang M, Nicholls TJ, Uhler JP, Dirksen-Schwanenland C, Polosa PL, et al. , Mice lacking the mitochondrial exonuclease MGME1 accumulate mtDNA deletions without developing progeria, Nat. Commun 9 (2018) 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Liyanage SU, Coyaud E, Laurent EM, Hurren R, Maclean N, Wood SR, et al. , Characterizing the mitochondrial DNA polymerase gamma interactome by BioID identifies Ruvbl2 localizes to the mitochondria, Mitochondrion 32 (2017) 31–35. [DOI] [PubMed] [Google Scholar]
- [158].Uhler JP, Thörn C, Nicholls TJ, Matic S, Milenkovic D, Gustafsson CM, et al. , MGME1 processes flaps into ligatable nicks in concert with DNA polymerase γ during mtDNA replication, Nucleic Acids Res. 44 (2016) 5861–5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Yang C, Wu R, Liu H, Chen Y, Gao Y, Chen X, et al. , Structural insights into DNA degradation by human mitochondrial nuclease MGME1, Nucleic Acids Res. 46 (2018) 11075–11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ngo HB, Kaiser JT, Chan DC, The mitochondrial transcription and packaging factor Tfam imposes a U-turn on mitochondrial DNA, Nat. Struct. Mol. Biol 18 (2011) 1290–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Canugovi C, Maynard S, Bayne A-CV, Sykora P, Tian J, de Souza-Pinto NC, et al. , The mitochondrial transcription factor A functions in mitochondrial base excision repair, DNA Repair 9 (2010) 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Xu W, Boyd RM, Tree MO, Samkari F, Zhao L, Mitochondrial transcription factor A promotes DNA strand cleavage at abasic sites, Proc. Natl. Acad. Sci. U. S. A 116 (2019) 17792–17799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Swenberg JA, Lu K, Moeller BC, Gao LN, Upton PB, Nakamura J, et al. , Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment, Toxicol. Sci 120 (2011) S130–S145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Nakamura J, Mutlu E, Sharma V, Collins L, Bodnar W, Yu R, et al. , The endogenous exposome, DNA Repair 19 (2014) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hegler J, Bittner D, Boiteux S, Epe B, Quantification of oxidative DNA modifications in mitochondria, Carcinogenesis 14 (1993) 2309–2312. [DOI] [PubMed] [Google Scholar]
- [166].Mishra PK, Raghuram GV, Jain D, Jain SK, Khare NK, Pathak N, Mitochondrial oxidative stress-induced epigenetic modifications in pancreatic epithelial cells, Int. J. Toxicol 33 (2014) 116–129. [DOI] [PubMed] [Google Scholar]
- [167].Greenberg MM, Abasic and oxidized abasic site reactivity in DNA: enzyme inhibition, cross-linking, and nucleosome catalyzed reactions, Acc. Chem. Res 47 (2014) 646–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Greenberg MM, Looking beneath the surface to determine what makes DNA damage deleterious, Curr. Opin. Chem. Biol 21 (2014) 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Dutta S, Chowdhury G, Gates KS, Interstrand cross-links generated by abasic sites in duplex DNA, J. Am. Chem. Soc 129 (2007) 1852–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS, Interstrand DNA–DNA cross-link formation between adenine residues and abasic sites in duplex DNA, J. Am. Chem. Soc 136 (2014) 3483–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Guan L, Greenberg MM, Irreversible inhibition of DNA polymerase β by an oxidized abasic lesion, J. Am. Chem. Soc 132 (2010) 5004–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM, Rapid DNA–protein cross-linking and strand scission by an abasic site in a nucleosome core particle, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 22475–22480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Guan L, Bebenek K, Kunkel TA, Greenberg MM, Inhibition of short patch and long patch base excision repair by an oxidized abasic site, Biochemistry 49 (2010) 9904–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Lindahl T, Andersson A, Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid, Biochemistry 11 (1972) 3618–3623. [DOI] [PubMed] [Google Scholar]
- [175].Luke AM, Chastain PD, Pachkowski BF, Afonin V, Takeda S, Kaufman DG, et al. , Accumulation of true single strand breaks and AP sites in base excision repair deficient cells, Mutat. Res. Fundam. Mol. Mech. Mutagen 694 (2010) 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Piersen CE, McCullough AK, Lloyd RS, AP lyases and dRPases: commonality of mechanism, Mutat. Res./DNA Repair 459 (2000) 43–53. [DOI] [PubMed] [Google Scholar]
- [177].Male R, Fosse VM, Kleppe K, Polyamine-induced hydrolysis of apurinic sites in DNA and nucleosomes, Nucleic Acids Res. 10 (1982) 6305–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].DeMott MS, Beyret E, Wong D, Bales BC, Hwang J-T, Greenberg MM, et al. , Covalent trapping of human DNA polymerase β by the oxidative DNA lesion 2-deoxyribonolactone, J. Biol. Chem 277 (2002) 7637–7640. [DOI] [PubMed] [Google Scholar]
- [179].Quiñones JL, Thapar U, Yu K, Fang Q, Sobol RW, Demple B, Enzyme mechanism-based, oxidative DNA–protein cross-links formed with DNA polymerase β in vivo, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 8602–8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Loeb LA, Preston BD, Mutagenesis by apurinic/apyrimidinic sites, Annu. Rev. Genet 20 (1986) 201–230. [DOI] [PubMed] [Google Scholar]
- [181].Simonelli V, Narciso L, Dogliotti E, Fortini P, Base excision repair intermediates are mutagenic in mammalian cells, Nucleic Acids Res. 33 (2005) 4404–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Yu S-L, Lee S-K, Johnson RE, Prakash L, Prakash S, The stalling of transcription at abasic sites is highly mutagenic, Mol. Cell. Biol 23 (2003) 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Weng L, Greenberg MM, Rapid histone-catalyzed DNA lesion excision and accompanying protein modification in nucleosomes and nucleosome core particles, J. Am. Chem. Soc 137 (2015) 11022–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Zhou C, Sczepanski JT, Greenberg MM, Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles, J. Am. Chem. Soc 134 (2012) 16734–16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Rubio-Cosials A, Battistini F, Gansen A, Cuppari A, Bernadó P, Orozco M, et al. , Protein flexibility and synergy of HMG domains underlie U-turn bending of DNA by TFAM in solution, Biophys. J 114 (2018) 2386–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Matsushima Y, Goto Y-I, Kaguni LS, Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM), Proc. Natl. Acad. Sci. U. S. A 107 (2010) 18410–18415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Lu B, Lee J, Nie X, Li M, Morozov YI, Venkatesh S, et al. , Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease, Mol. Cell 49 (2013) 121–132; [DOI] [PMC free article] [PubMed] [Google Scholar]; (a) Lu B, et al. , Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease, Molecular Cell 49 (1) (2013) 121–132; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) King GA, et al. , Acetylation and phosphorylation of human TFAM regulate TFAM–DNA interactions via contrasting mechanisms, Nucleic Acid Res. 46 (7) (2018) 3633–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Oliveira MT, Kaguni LS, Functional roles of the N-and C-terminal regions of the human mitochondrial single-stranded DNA-binding protein, PLoS One 5 (2010) e15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Oliveira MT, Kaguni LS, Reduced stimulation of recombinant DNA polymerase γ and mitochondrial DNA (mtDNA) helicase by variants of mitochondrial single-stranded DNA-binding protein (mtSSB) correlates with defects in mtDNA replication in animal cells, J. Biol. Chem 286 (2011) 40649–40658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Thömmes P, Farr CL, Marton RF, Kaguni LS, Cotterill S, Mitochondrial Single-stranded DNA-binding protein from Drosophila embryos. Physical and biochemical characterization, J. Biol. Chem 270 (1995) 21137–21143. [DOI] [PubMed] [Google Scholar]
- [191].Farr CL, Wang Y, Kaguni LS, Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis, J. Biol. Chem 274 (1999) 14779–14785. [DOI] [PubMed] [Google Scholar]