SUMMARY
In vitro differentiation of pluripotent cells into β cells is a promising alternative to cadaveric-islet transplantation as a cure for type 1 diabetes (T1D). During the directed differentiation of human embryonic stem cells (hESCS) by exogenous factors, numerous genes that affect the differentiation process are turned on and off autonomously. Manipulating these reactions could increase the efficiency of differentiation and provide a more complete control over the final composition of cell populations. To uncover in vitro autonomous responses, we performed single-cell RNA sequencing on hESCs as they differentiate in spherical clusters. We observed that endocrine cells and their progenitors exist beside one another in separate compartments that activate distinct genetic pathways. WNT pathway inhibition in the endocrine domain of the differentiating clusters reveals a necessary role for the WNT inhibitor APC during islet formation in vivo. Accordingly, WNT inhibition in vitro causes an increase in the proportion of differentiated endocrine cells.
In Brief
In vitro differentiation of pluripotent cells into β cells is a promising alternative to cadaveric islet transplantation as a cure for type 1 diabetes. Sharon et al. use scRNA-seq to identify the cell populations that form during the process and uncover a role for WNT pathway inhibition during endocrine differentiation.
Graphical Abstract
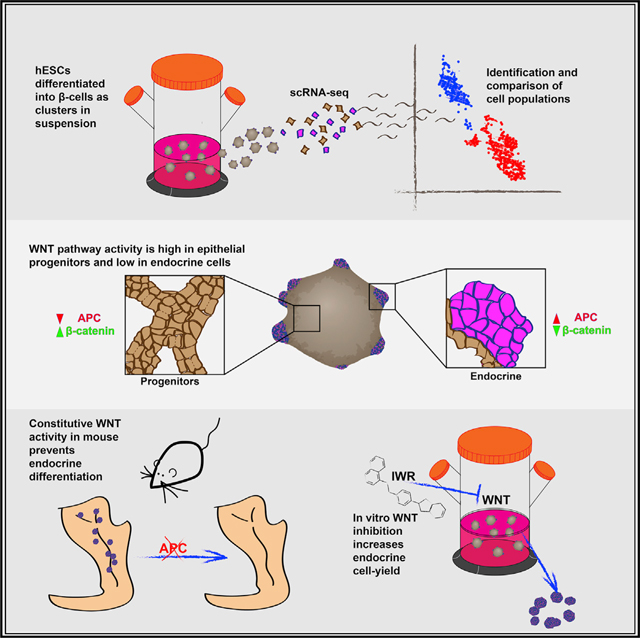
INTRODUCTION
Type 1 diabetes (T1D) is caused by autoimmune destruction of the insulin-producing β cells in the pancreatic islets. Transplantation of cadaveric islets can cure the disease (Shapiro et al., 2000), but donor scarcity and high cost limit its feasibility. In an attempt to develop a ready supply of β cells for transplantation, several protocols for the in vitro differentiation of pluripotent cells into β cells were developed in recent years (Pagliuca et al., 2014; Rezania et al., 2014; Russ et al., 2015). Our protocol directs differentiation of human embryonic stem cells (hESCs) into β cells that resemble cadaveric β cells in both gene expression and function, including the ability to secrete insulin in response to changing glucose levels (Pagliuca et al., 2014). Still, under these in vitro conditions, only about 30% of the generated cells are, in fact, β cells, and finding ways to increase the efficiency of the differentiation will be valuable.
An obstacle to protocol improvement is our incomplete understanding of the complex process of β cell differentiation. During normal embryonic development, the nascent pancreas contains a network of monolayered tubules composed of epithelial progenitors, called epithelial cords (Pan and Wright, 2011). As cells in the cords divide, some turn on NEUROG3 and form “peninsulas”—bud-like structures that grow and develop to become the islets (Sharon et al., 2019). Current protocols aim to recapitulate embryonic islet development by stepwise application of defined factors. Here, we use single-cell RNA sequencing (scRNA-seq) to characterize the cell populations that appear during the in vitro differentiation process and identify pathways that affect β cell yield.
RESULTS
Single-Cell RNA Sequencing of Differentiating β Cells
hESCs were differentiated into stem-cell-derived β cells as clusters in suspension using a six-stage protocol (Pagliuca et al., 2014) (Figure 1A). scRNA-seq was performed on undifferentiated cells and on 10 consecutive time points, representing the end of each of the differentiation stages and select intermediate points (Figures S1A and S1B). To analyze the relationships between the cells, we combined SIMLR analysis (single-cell interpretation via multikernel learning) with topic modeling (TM). SIMLR is a method that groups cells based on cell-to-cell similarity and then displays them in lower dimensional space (Wang et al., 2017) (Figure 1B). TM is a probabilistic unsupervised learning algorithm that, in the context of gene expression analysis, identifies groups of genes that are frequently expressed together in the same cell and gathers them into “expression profiles” (EPs) (Blei, 2012; Gerber et al., 2007; Teh et al., 2006). For each EP, every gene receives a “relevance value,” which describes the gene’s weight in the identification of this particular EP. While establishing which genes constitute an EP, the TM algorithm simultaneously quantifies how active each EP is within a particular cell via a “usage value.” Cells that tend to use genes from the same EPs (have high usage values for similar EPs) can be grouped together. Whereas customarily used clustering methods, such as hierarchical clustering, assume that the relationships between genes are strict (e.g., Euclidean distance, correlation), TM analyzes these relationships as probability distributions. This allows the clustering of cells and genes in a flexible arrangement. Rather than forcing each gene to one expression module, with TM, a gene can be relevant to several EPs, reflecting its possible expression in the context of different biological processes. Similarly, since each cell uses several biological processes, a single cell may use several EPs, to varying extents. Furthermore, since conventional clustering methods allow a gene to belong only to a single expression module, many genes can be lost to artificial modules caused by technical noise. However, the inherent flexibility of TM allows these genes to appear in biologically meaningful EPs as well. Altogether, the advantages of TM analysis over conventional clustering methods are especially relevant for discovering hidden structures in highly complex datasets, including scRNA-seq of heterogeneous populations.
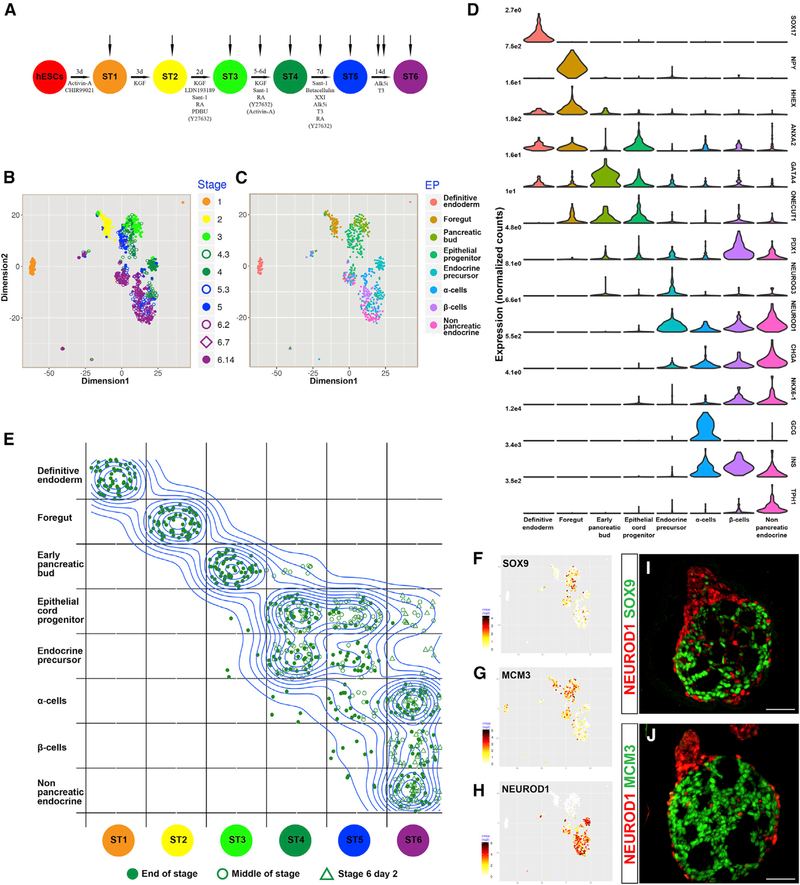
Figure 1. scRNA-Seq Analysis of the Directed Differentiation of β cells In Vitro.
(A) Overview of the protocol for the directed differentiation of hESCs into β cells(Pagliuca et al., 2014). Cells were collected from undifferentiated hESCs at the end of stages 1 through 5, on the 3rd day of stages 4 and 5, and on days 2,7, and 14 of stage 6.
(B) SIMLR visualization of the 720 differentiating cells analyzed (excluding hESCs). Colors represent time of collection.
(C) SIMLR plot, with colors representing the most highly used cell-type EP.
(D) Violin plots represent gene expression distributions within the cell groups defined by cell-type EPs.
(E) A density-based topographic map is overlaid on top of a bins table to emphasize the changes that occur in the identity of the cells during in vitro differentiation. Cells are binned based on stage of collection (columns) and developmental identity (rows) and are randomly dispersed within each bin. For a numerical summary of the bins table, see Figure S1C.
(F-H) SIMLR map overlaid with the relative expression level of the indicated gene in each cell. (F) SOX9; (G) MCM3; and (H) NEUROD1.
(I and J) Immunofluorescent staining of clusters at the end of stage 4: (I) NEUROD1 and SOX9; (J) NEUROD1 and MCM3. Scale bars: 50 μm.
TM identified 36 EPs in the data (Figure S2). Of these, 8 EPs contain markers related to cell-type identity and, combined, are used by all differentiating cells (excluding undifferentiated hESCs). The identity of each cell could now be determined according to the EP it uses most (Figures 1C and 1D; Figures S1C and S1F). Cells collected in the first three stages correspond with the first three cell types, whereas cells collected at the 4th stage and beyond are heterogeneous and can be divided into five cell types (Figure 1E). An interactive database that combines SIMLR with TM and describes the major cell populations and their relative similarity to each other can be found at https://ifx.rc.fas.harvard.edu/invitrobetacells/.
Early In Vitro Differentiation Is Linear and Uniform, whereas Late Differentiation Gives Rise to Heterogeneous Populations
At the end of the first stage, marked by SOX17 expression (Pagliuca et al., 2014), all cells analyzed primarily use an EP comprising other definitive endoderm genes, including CER1 and LEFTY2. By the end of the second stage, cells primarily use an EP corresponding to foregut endoderm (HHEX, TTR) (Figures 1D and 1E; Figures S1C and S1F; Table S1) (Bort et al., 2004; Thomas et al., 1998). Almost all cells analyzed at the end of the third stage, when PDX1 is activated (Pagliuca et al., 2014), use an EP that contains additional pancreatic-bud markers such as GATA4, ONECUT1, and ONECUT2 (Figures 1D and 1E; Figure S1F; Table S1) (Decker et al., 2006; Jacquemin et al., 2003). The absence of NKX6.1 and expression of the foregut markers HOXA1, SHISA2, and HHEX suggest that these cells represent an early stage in pancreatic-bud development (Figure 1D; Table S1) (Filipe et al., 2006; Godwin et al., 1998; Jennings et al., 2013; Rodríguez-Seguel et al., 2013). Unlike the first two EPs, the early pancreatic-bud EP is also used by cells from later stages, albeit with low usage values and not as the primary EP (Figure 1E; Figures S3A and S3B). This suggests that many of the genes expressed at this point remain activated in later stages of development.
From stage 4 onward, the clusters contain heterogeneous populations that can be divided into two major cell groups (Figure 1E). The cells in one group express genes from an EP containing epithelial-cord markers, including SOX9 and ONECUT1 (HNF6), together with cell-cycle-related genes such as MCM3 and CCND2 (Figures 1E–1G; Table S1). A second group expresses genes from several EPs containing endocrine markers including CHGA, ISL1, and NEUROD1 (Figures 1D, 1E, and 1H; Table S1). The latter group also expresses lower levels of genes associated with cell proliferation (Figures 1G and 1H).
It was previously shown that treatment with Activin A and the ROCK inhibitor Y-27632 during stage 4 induces the appearance of buds on the clusters’ surface (Sharon et al., 2019). Immunostaining shows that NEUROD1+ non-proliferating endocrine cells reside primarily in peninsula-like buds that formed in stage 4, whereas SOX9+ proliferating epithelial progenitors are found in the cluster core (Figures 1I and 1J). Altogether, we conclude that the uniform population that formed in the first three stages of the protocol differentiates and splits into epithelial-cord-like progenitors and endocrine cells. These two cell types are physically separated within differentiating clusters.
The Endocrine Population Includes Four Distinct Cell Types
The endocrine EPs demarcate four major endocrine cell types (Figures 1C–1E). One group represents endocrine precursors and includes cells that express the endocrine initiating gene NEUROG3 (Figure 1D), with some cells co-expressing both progenitor markers (e.g., SOX11) and early-onset endocrine genes (e.g., RUNX1T1) (Osipovich et al., 2014) (Figure S1F). The hormones INS and GCG are expressed at low levels, and some cells express GHRL or SST. These endocrine precursors can be further classified into an early sub-group, collected mostly in the middle of stage 4, and a late sub-group, collected at the end of stages 4 and 5 (EPs 28 and 22, respectively, in Figure S2 and Table S1).
The remaining endocrine cells form three major types of hormonal cells. The first group expresses α cell markers including GCG, ETV1, and ARX, together with the non-α marker INS, and occasionally SST and PPY (Figures 1D and S1F; Table S1). Flow cytometry analysis indicates that, as cells progress into stage 6, these polyhormonal cells resolve into GCG+INS− α cells (Figures S1D and S1E). A second group is that of β cells, identified by high expression of INS, NKX6.1, PDX1, and LMO1 and scarcity of GCG transcripts (Figures 1D and S1F; Table S1). Finally, a group of CHGA+ endocrine cells express low levels of the pan-islet marker ISL1, serotonin-related genes such as TPH1, and non-islet genes (Segerstolpe et al., 2016; Uhlén et al., 2015) such as ADH6 and LMX1A (Figures 1D, 1E, and S1F; Table S1). These non-pancreatic endocrine cells may represent byproducts of the in vitro differentiation process that resemble enteroendocrine populations (Gunawardene et al., 2011; Veres etal., 2019).
Two Waves of Endocrine Differentiation
The presence of epithelial-progenitors side by side with endocrine cells observed from stage 4 onward is reminiscent of normal embryonic pancreas formation, wherein new endocrine cells emerge from epithelial-cord progenitors in a continouous, asynchronous flux (Johansson et al., 2007; Miyatsuka et al., 2009). To see whether such a flux occurs in vitro, we investigated the dynamics of endocrine differentiation during stages 4 and 5.
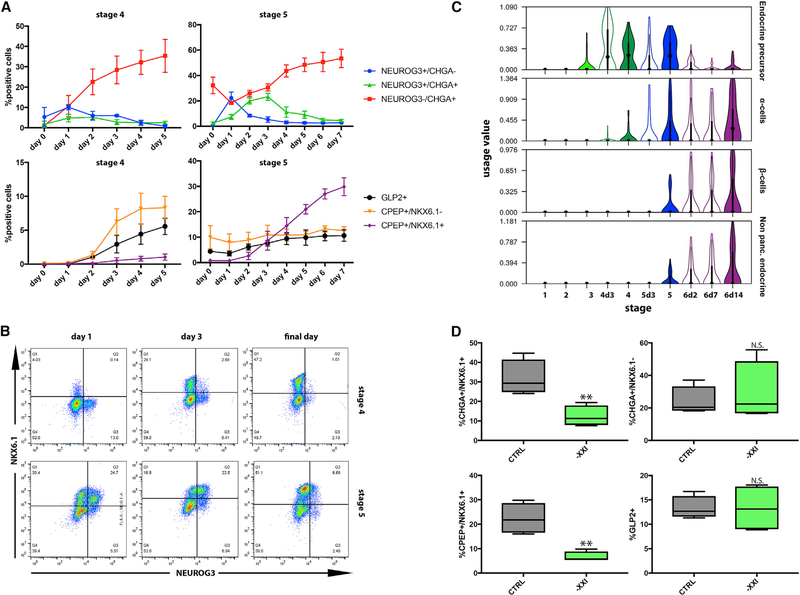
Flow cytometry reveals a wave of newly formed NEUROG3+ endocrine precursors during the first days of stage 4. A few precursors appear at the last day of stage 3, and within the first day of stage 4 their numbers peak to about 10% of the entire population (Figure 2A). In the following days, fewer NEUROG3+ cells appear, and as they differentiate, NEUROG3 is gradually replaced by the pan-endocrine gene CHGA. Less than 2% of cells are NEUROG3+CHGA− by the end of stage 4 (Figure 2A). During stage 5, we observed a second wave of endocrine precursor formation. Here, too, the wave peaks on the first day of the stage, when ~20% of the cells are NEUROG3+. Similar to stage 4, the initial rise is followed by a daily decline in the number of precursors, as NEUROG3+ is replaced with CHGA, until less than 3% of the cells are NEUROG3+ by the last day of stage 5 (Figure 2A).
Figure 2. Stages 4 and 5 Produce Different Endocrine Cell Types.
(A) Daily flow cytometry analysis of endocrine Induction during stages 4 and 5. n = 3; data points represent mean ± SD.
(B) Representative flow cytometry plots show the relationship between NKX6.1 and NEUROG3+ expression during stages 4 and 5.
(C) Violin plots present the usage value distributions of endocrine EPs in cells collected in different stages.
(D) Flow cytometry analysis on cells at the end of stage 5, with (control) or without XXI. n = 4; p values, clockwise from top left: 0.0022,0.3933,0.1848, and 0.0066, Student’s t test.
These data show that, unlike embryonic development, in-vitro-generated endocrine cells form in two waves, one in stage 4 and the other in stage 5 of the protocol.
It should be noticed that, during stage 6, the proportion of endocrine cells continues to increase, but scRNA-seq shows that this is accompanied with a decline in the number of endocrine precursors detected (Figures 1E and S1C). Thus, this is most likely not the result of a third wave of endocrine differentiation but, rather, the result of existing endocrine precursors completing their differentiation, accompanied by a decline in the numbers of epithelial progenitors.
α Cells Start Forming in the First Wave of Endocrine Differentiation, whereas β cells Form in the Second Wave
Several findings suggest that the two waves of endocrine formation differ in the populations generated: (1) The α cell EP is utilized by cells already in the middle of stage 4, whereas the β cell and the non-pancreatic-endocrine programs are only used by cells from the end of stage 5 onward (Figure 2C). (2) NKX6.1, which is expressed together with INS in β cells (Figure 1D), is rarely detected together with either NEUROG3 or CHGA during stage 4. However, at the first day of stage 5, most of the newly emerging NEUROG3+ cells express NKX6.1 at medium levels and then gradually turn into CHGA+NKX6.1high endocrine cells (Figures 2B and S3C). (3) The GCG+ INS+ polyhormonal cells that appear at stage 4 express ARX (Table S1), indicative of an α cell state. Conversely, bona fide CPEP+NKX6.1+ β cells appear almost exclusively during stage (Figure 2A).
To test this hypothesis, we removed XXI from the media. XXI serves to inhibit the Notch pathway during stage 5, and induce general endocrine differentiation (Apelqvist et al., 1999; Beher et al., 2001; Murtaugh et al., 2003). Its removal is, therefore, expected to hamper endocrine differentiation during stage 5, without affecting cells that had differentiated earlier. Indeed, XXI removal during stage 5 dramatically reduced the number CPEP+NKX6.1+ β cells but did not significantly affect the number of GLP2+ α cells (Figure 2D). Thus, interference with endocrine differentiation at stage 5 limits β cell differentiation but does not affect the earlier formation of α cells. This confirms that the first wave of endocrine differentiation, during stage 4, produces primarily α cells, whereas the second wave, at stage 5, gives rise to β cells. This is reminiscent of normal mouse embryonic development, where α cell differentiation precedes that of β cells (Johansson et al., 2007; Sharon et al., 2019).
Identifying Pathways that Affect Pancreas Compartmentalization
The distinct fates of endocrine cells differentiating in the two waves suggests that controlling the timing of differentiation could affect the composition of the clusters. To control the timing of differentiation, we set out to find molecular mechanisms that act in the clusters autonomously rather than through the direct effect of factors already included in the protocol. Ligand-receptor interaction analysis suggested multiple potential pairs that could act either in an autoregulatory process or between different cell types (Figures S3D and S3E; Table S3).
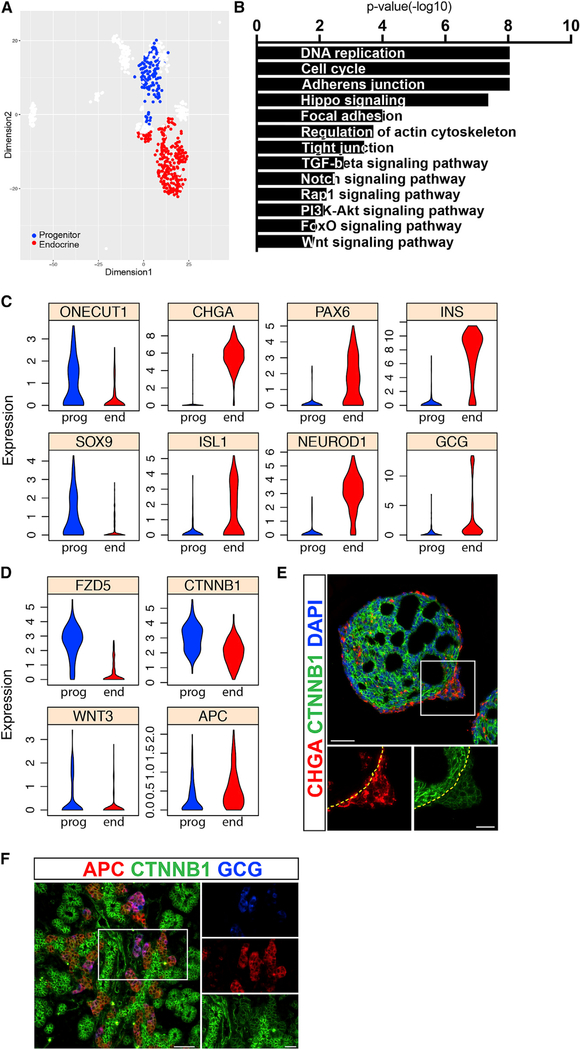
To focus on genetic pathways that control the overall balance between undifferentiated progenitors and endocrine cells, we compared cells with high usage values of the epithelial progenitors EP with cells that use the general endocrine EP (EPs 19 and 6, respectively) (Figure 3A). The expression of more than 4,800 genes is significantly different between the two groups (Figure S3F). As expected, the endocrine population is enriched for transcripts related to islet function and with maturity onset diabetes of the young (MODY), including PAX4, PDX1, PAX6, and NEUROD1; and it expresses higher levels of transcripts related to insulin or glucagon signaling (INS, GCG, ABCC8, GCK, FOXO1, and G6PC2) (Figure 3C; Figures S3F and S4A). Additional genes enriched in these cells are related with general mechanisms of secretion, including the endoplasmic reticulum, endocytosis, and calcium reabsorption (Table S2). The mechanisms shared by endocrine cells and neurons are also evident with the enrichment of transcripts related with synaptic transport (GABRB3 and CAMK2B). Lastly, in-vitro-generated endocrine cells are enriched for transcripts related to several molecular pathways that have a role in endocrine function, including cAMP (RYR2 and AKT3), cGMP/PKG (MAPK1 and MAPK3), AMPK (RAB2A and CREB3), mTOR (PTEN and PRKCA), VEGF (VEGFA and SHC2), and PPAR (PPARA) signaling (Table S2).
Figure 3. Identification of Genetic Pathways that Separate Progenitors and Endocrine Cells.
(A) SIMLR map presents the cells used for differential expression analysis. Cells with transitory intermediate characteristics were omitted to obtain clearer distinction between the populations.
(B) Representative gene categories enriched in the progenitor compartment.
(C) Violin plots show the expression distributions of various genes in progenitor (prog) and in endocrine (end) cells.
(D) Violin plots show the expression distributions of WNT-pathway related genes in progenitor (prog) and in endocrine (end) cells.
(E) Immunofluorescent staining of clusters at the end of stage 4. Dashed line marks the base of the endocrine bud. Scale bars: 50 μM.
(F) Immunofluorescent staining of a mouse embryonic pancreas at E15.5. Scale bars: 25 μm.
Conversely, the epithelial progenitors are enriched for transcripts related to cell adhesion (CDH1, CDC42, and ERBB2) and cell division (MCM7, CCND2, CHEK1, and PCNA). They also express genes that indicate a high activity of several signaling pathways known to have a role in progenitor-to-endocrine differentiation, including Notch (Apelqvist et al., 1999) (NOTCH2, NOTCH3, JAG1, and HES1), hippo-Yap (Gao et al., 2013; Rosado-Olivieri et al., 2019; Mamidi et al., 2018) (YAP1 and AMOT), activin (SMAD2/3, TGFBR1, and ACVR2A), and BMP (BMP2, BMP7, and BMPR1B) (Chung et al., 2010) (Figures S4C–S4F). The progenitor population also expressed higher levels of transcripts related with the PI3K (phosphatidylinositol 3-kinase)-AKT pathway (PIK3R1/3), possibly in context with fibroblast growth factor (FGF) activity and the enrichment of FGF receptors 1 and 2 (FGFR1 and FGFR2) in these cells (Figure S4G).
Unexpectedly, in spite of its known role in pancreas development, we did not observe enrichment of hedgehog pathway activity in either cell type (Kawahira et al., 2003, 2005). Upon examination of specific components of the pathway, we noticed that progenitors express significantly higher levels of both SHH and its signal transducer SMO (Alcedo et al., 2000), but neither of the pathway’s inhibitors PTCH1 or PTCH2 show a significant difference in RNA levels (Figure S4H). Accordingly, in situ hybridization showed that transcripts of PTCH1 are spread uniformly across the cluster, whereas SMO mRNA is dense in the non-endocrine region of the differentiating cluster and sparse in the endocrine bud (Figure S4I). Surprisingly, unlike its mRNA transcripts, high levels of PTCH1 protein were confined to the endocrine region both in clusters (Figure S4J) and in developing mouse islets (Figure S4K). This suggests that a post-translational mechanism acts to inhibit hedgehog signaling, specifically in the endocrine compartment.
Finally, transcripts upregulated in progenitors are enriched for factors related to the active form of WNT, including the ligand WNT3, the receptors FZD5 and −7, and the major pathway transducer β-catenin (CTNNB1), which shows a similar pattern on the protein level (Figures 3B, 3D, and 3E). Conversely, APC, which inhibits WNT activity, is upregulated in endocrine cells (Figure 3D). In mouse embryos, Apc protein is detected exclusively in the endocrine peninsula area, whereas β-catenin levels are high in the epithelial cords (Figure 3F).
WNT Pathway Inhibition Is Necessary for Endocrine Differentiation in Mice
High WNT pathway activity in progenitors and the opposing expression of APC, the WNT inhibitor, in the endocrine compartment suggest that WNT inhibition plays a role in endocrine differentiation. The Wnt pathway has a role in early murine pancreas formation and in the later expansion of the acinar compartment, but previous reports are inconclusive about a possible role for the WNT pathway in endocrine differentiation (Dessimoz et al., 2005; Heiser et al., 2006; Murtaugh, 2008; Murtaugh et al., 2005; Strom et al., 2007). Since our protocol does not involve manipulation of the pathway at this advanced stage, we decided to examine WNT pathway’s role in endocrine differentiation.
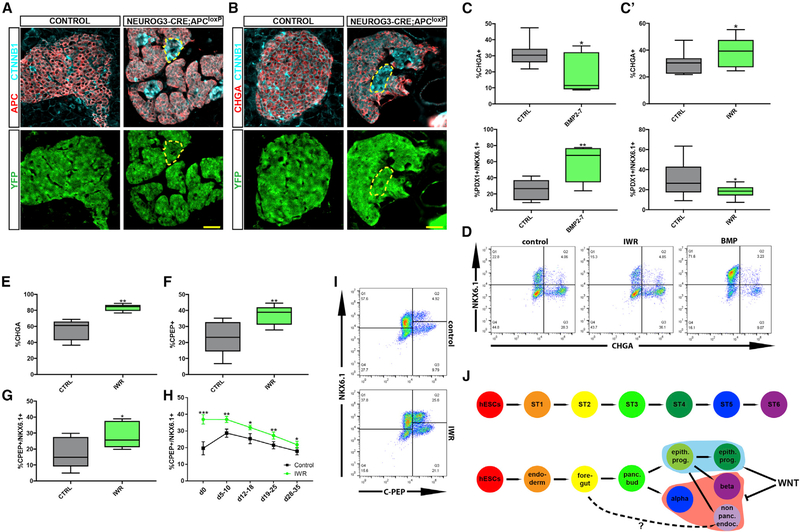
First, we examined the effect of Apc deletion on islet formation in vivo. Apc is a central component of the destruction mechanism that degrades Ctnnb1, and, in its absence, excess Ctnnb1 molecules accumulate in the nucleus and constitutively activate WNT pathway targets (MacDonald et al., 2009). In agreement with the importance of WNT inactivation during early pancreas development (Muñoz-Bravo et al., 2016), induced deletion of Apc in the pancreatic bud of mouse embryos resulted in the formation of a rudimentary pancreas (Figures S4L–S4N). To examine WNT’s later role in the formation of the endocrine compartment, we crossed mice carrying a conditional knockout of Apc (APCMIN ApcloxP/loxP) (Kuraguchi et al., 2006; Moser et al., 1990) with mice expressing CRE recombinase specifically in cells expressing Neurog3 (Schonhoff et al., 2004). Cells that efficiently deleted Apc presented nuclear Ctnnbl accumulation (Figure 4A) and failed to express either the pan-endocrine marker Chga (Figure 4B) or the hormones insulin and glucagon (Figure S4O). Since Neurog3 is also active during enteroendocrine cell differentiation (Jenny et al., 2002), APC was deleted in gut cells as well, leading to hyperplastic lesions (Figure S4P). Altogether, this shows that WNT pathway inhibition is necessary for proper endocrine differentiation in the mouse pancreas.
Figure 4. WNT Pathway Inhibition in Endocrine Differentiation.
(A and B) Immunofluorescent staining of pancreata from a 1-month-old Neurog3-cre female mouse (control) and a 2-month-old Neurog3-CRE;APCloxP/loxP (APC-endocrine-knockout [KO]) female mouse. Both mice bear a loxP-stop-loxP-YFP sequence in their ROSA26 locus to trace cells that activated CRE. Notice that APC was not deleted in all YFP+ cells. Scale bars: 25 μm. (A) APC− cells express high levels of β-catenin. (B) β-cateninhigh cells do not express CHGA.
(C) Cell-population proportions calculated using flow cytometry analysis at the end of stage 4, following treatment with BMP2–7. n = 7. Top: p = 0.0235. Bottom: p = 0.0098, Student’s t test. (C’) Flow cytometry analysis at the end of stage 4 following treatment with IWR1-endo. n = 9. Top: p = 0.0078. Bottom: p = 0.0314, Student’s t test.
(D) Representative flow cytometry plots at the end of stage 4.
(E-G) Flow cytometry analysis at the end of stage 5 following treatment with IWR1-endo. (E) n = 5; p = 0.0045. (F) n = 5; p = 0.0037. (G) n = 5; p = 0.0138. Student’s t test.
(H) Flow cytometry analysis of stage 6 cells following treatment with IWR1-endo during stage 5. Values represent mean ± SEM. d, days into stage 6; d0, last day of stage 5. 2-way ANOVA: p < 10−4 for change in β cell proportion over days and for overall difference in β cell proportion between conditions. Time-point-specific differences: d0, n = 8, p = 1.6 × 10−4; d5–10, n = 7, p = 0.002; d12–18, n = 7, p = 0.03; d19–25, n = 7, p = 0.001. d28–35, n = 7, p = 0.03, Student’s t test.
(I) Representative flow cytometry plots at the end of stage 5.
(J) An overview of the protocol (top) and the actual dynamics of cell differentiation in vitro (bottom).
WNT Inhibition and BMP Activation Modulate the Ratio between Progenitors and Endocrine Cells
Finally, we examined the potential of compartmentalizing pathways to regulate the ratio between progenitors and endocrine cells in vitro. We focused on WNT and BMP pathways, since they are not directly manipulated in the critical stages of our current protocol.
BMP is active in the pool of epithelial progenitors (Figures 3B and S4F), and it was previously shown to attenuate endocrine differentiation (Nostro et al., 2011; Russ et al., 2015). Accordingly, the addition of recombinant BMP2–7 at stage 4 resulted in the downregulation of endocrine genes (Figure S5D), and a nearly 2-fold decrease in the number of CHGA+ endocrine cells (Figures 4C and 4D). Concomitantly, the number of PDX1+/NKX6.1+ progenitors increased by more than 2-fold on average (Figures 4C and 4D). We confirmed this in an independent cell line (Figure S5C) but did not observe a similar effect when BMP2–7 was added during stage 5.
Conversely, the role we found for WNT inhibition in islet formation suggests that it could be used to increase the number of endocrine cells generated in vitro. Indeed, treatment with the WNT-tankyrase inhibitor IWR1-endo (Chen et al., 2009) during both stages 4 and 5 increased the expression of endocrine markers and downregulated progenitor markers (Figure S5D). Similar results were obtained using XAV939, another WNT-tankyrase inhibitor (Figure S5D) (Huang et al., 2009). To estimate the effect of WNT pathway activity on the actual yield of the protocol, we measured the effect of its manipulation on cell proportions in vitro. Activation of the WNT pathway using the GSK3 inhibitor CHIR99021 (Ring et al., 2003) decreased the overall number of endocrine cells at stage 4 (Figure S5A). In contrast, inhibition of the pathway with IWR1-endo during stage 4 significantly raised the proportion of CHGA+ endocrine cells and reduced the proportion of PDX+NKX6.1+ progenitors (Figures 4C’ and 4D). IWR treatment during stage 5 caused an even higher relative increase in the proportion of CHGA+ endocrine cells (Figures 4E and S5C‘), and, specifically, the proportion of C-PEP+/NKX6.1+ β cells increased by more than 50% on average, relative to controls (Figures 4F and 4G). The higher proportion of β cells upon IWR addition in stage 5 was maintained during stage 6 of the protocol (Figure 4H), and overall cell numbers were slightly higher following treatment (Figure S5E), indicating that IWR increases the overall yield of in-vitro-generated β cells. IWR-treated cells maintain their ability to secrete insulin in response to high glucose levels (Figure S5F), but upon consecutive stimulations with high glucose, IWR-treated cells release less insulin in the second challenge, compared to controls (Figure S5G).
These experiments show that the ratio between the progenitor pool and the differentiated endocrine cells can be regulated by the activity of either the WNT pathway or the BMP pathway. The exogenous manipulation of the ratio between the two compartments at the right stage in the differentiation protocol affects β cell yield. Specifically, expanding the endocrine compartment through inhibition of the WNT pathway during β cell differentiation at stage 5 improves the yield of in vitro-generated β cells.
DISCUSSION
Protocols for in vitro-directed differentiation of hESCs into β cells are commonly described as linear processes that homogenously drive cells from one step to the next (Pagliuca et al., 2014; Petersen et al., 2017; Rezania et al., 2014; Russ et al., 2015). Using scRNA-seq, we found that differentiation is homogeneous only at the first three stages of the protocol, after which epithelial progenitors and endocrine cells reside side by side. A wave of endocrine differentiation at stage 4 tends to produce a cells, and a second wave, at stage 5, gives rise to β cells and non-pancreatic endocrine cells (Figure 4J).
These findings indicate that the in vitro differentiation protocol recapitulates multiple aspects of embryonic pancreas development. In both cases, the epithelial cord progenitors and the endocrine peninsulas that emerge from them exist side by side, and in both cases, α cell differentiation precedes that of β cells (Johansson et al., 2007; Pan and Wright, 2011; Sharon et al., 2019). However, we also noticed differences between the protocol and embryonic development, including the absence of early Cpa1+ multipotent progenitors that give rise to acinar, ductal, and endocrine cells (Zhou et al., 2007), and the formation of non-pancreatic enteroendocrine-like cells. It should be noted that the differentiating clusters may contain additional rare cell types, which have not been detected by our analysis.
Cell heterogeneity may be attributed to physical constraints such as the effect of cluster thickness on diffusion, but even planar differentiation protocols are often not homogeneous (Kelly et al., 2011). Here, we show that cells initiate autonomous molecular programs, and in the case of WNT and hedgehog pathways, for example, we observe opposing activity levels in the progenitor and endocrine compartments, suggesting a regulatory cross-talk between the various cell types. By intervening with potential regulatory mechanisms, we can manipulate the outcome of the differentiation process. Exogenous BMP activation increases the progenitor compartment in stage 4, whereas small-molecule WNT inhibitors increase the opposite, endocrine population.
The ultimate goal of in vitro differentiation protocols is to provide diabetic patients with β cells derived from pluripotent stem cells instead of cadaveric islets. However, it should be noticed that the currently practiced medical protocol involves whole islet transplantation, rather than β cells alone. In fact, there is no certainty that transplanting a single cell type will be able to regain the function of the entire islet. It is, therefore, sensible to aim efforts at generating whole islets in vitro, rather than pure populations of β cells. This work joins others (Petersen et al., 2017; Veres et al., 2019) in the attempt to characterize protocols for in vitro production of pancreatic endocrine cells at the single-cell level. Our use of scRNA-seq generates a large, unbiased dataset, which enabled the thorough characterization of the various cell types generated, the timing of their differentiation, and the identification of molecular pathways that affect cell fate. Looking ahead, the ability to finely tune the timing and balance of progenitor versus endocrine fates can be used to control the ratio between α and β cells and promote the in vitro formation of islets with proper cell composition.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Douglas A. Melton (dmelton@harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and animal handling
To generate mice lacking Apc in the pancreatic bud, we crossed heterozygote C57BL/6j-ApcMin/J (Moser et al., 1990) (Jackson laboratory, Bar Harbor, ME) mice, carrying a mutation in the gene (APCmin/+), with B6.Cg-Apctm2Rak/Nci (NCI/NIH) mice (Kuraguchi et al., 2006) (APCloxp) carrying a conditional knock out of Apc, and with Tg(Pdx1-cre)89.1Dam (Gu et al., 2002) mice that express CRE recombinase under a Pdx1 regulatory element. To delete Apc in endocrine cells, we used Tg(Neurog3-cre)C1Able/J (Jackson laboratory, Bar Harbor, ME) (Schonhoff et al., 2004) mice expressing CRE recombinase under a Neurog3 regulatory element with the previous Apc mice, to generate either heterozygotes that carry one mutated allele and one conditional allele, or mice that are homozygotes for the conditional allele. To validate CRE activity, these mice were also crossed with B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J(Jackson laboratory, Bar Harbor, ME) mice, that express YFP form the ROSA26 locus upon CRE mediated recombination. Both male and female mice were used for breeding all strains, except for C57BL/6j-ApcMin/J, where only heterozygote males were used for breeding and crossed with females from other strains. Sections of adult mice are from 1 and 2 month old females. Sex of the newborns analyzed was not determined. Mice were euthanized through CO2 inhalation prior to tissue isolation.
Animal studies were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocol number [16–05-269]. The protocol was approved by the Committee on the Use of Animals in Research and Teaching of Harvard University Faculty of Arts & Sciences (HU/FAS). The HU/FAS animal care and use program is AAALAC International accredited, has a PHS Assurance (A3593–01) on file with NIH’s Office of Laboratory Animal Welfare, and is registered with the USDA (14-R-0128).
HESC culture and differentiation
HUES8 (male) human embryonic stem cells and human induced pluripotent stem cells (hiPSCs) from line 1016 (male) were obtained from the Human Embryonic Stem Cell Facility and iPS Core Facility of the Harvard Stem Cell Institute. Cells were differentiated into beta like cells in suspension as described previously (Pagliuca et al., 2014). Briefly, 150Xe6 cells were suspended in 300ml of supplemented mTeSR1 and grown in spinning flasks (70rpm, 37°C, 5%CO2) for 48 hours prior to the beginning of a stepwise differentiation protocol:
Stage 1 – 24 hours in S1 medium supplemented with ActivinA(100ng/ml) and CHIR99021 (1.4μg/ml), followed by 48 hours without CHIR99021.
Stage 2 – 72 hours in S2 medium supplemented with KGF (50ng/ml).
Stage 3 – 48 hours in S3 medium supplemented with KGF (50ng/ml), LDN193189 (200nM), Sant1 (0.25μM), retinoic acid (2μM), and PDBU (500nM).
Stage 4 – 5 days in S3 medium supplemented with KGF (50ng/ml), Sant1 (0.25μM) and retinoic acid (0.1 μM).
Stage 5 – 7 days in BE5 medium supplemented with Beta Cellulin (20ng/ml), XXI (1μM), Alk5iII(10 μM) and T3 (1μM). Sant1 (0.25μM) was added in the first three days, and retinoic acid was added at 0.1 μM in the first three days, then at 0.025μM.
Stage 6 – CMRLS medium supplemented with Alk5iII (10μM) and T3 (1μM).
In many of the experiments following cell collection for scRNA-seq, the protocol was slightly modified to add Y-27632 (10μM) at stage 3–5, and ActivinA (5ng/ml) in stage 4. In some cases, an extra day at stage 4 was also added. For the presented functional and flow cytometry analysis in stage 6, CMRLS based media was replaced with S3 medium, without supplements.
Factors were tested in either 6-well plates placed on a rocker, or in Biott ABLE Bioreactors (Stemgent). IWR1-endo (Sigma-Aldrich) was used at 5μM. XAV939 (Santa Cruz, SC-296704A) was used at 5 μM. Recombinant human BMP2/BMP7 (Thermo-Fisher) was used at 100μg/ml. when added during stage 4, CHIR99021 was used at a concentration of 1.4–1.55μg/ml.
METHOD DETAILS
Preparation of single-cell suspension
Clusters were collected prior to commencement of differentiation, at the last day of stages 1–5, on the 3rd day of stages 4 and 5, and on the 2nd, 7th and 14th days of stage 6. The clusters were allowed to settle, washed in PBS and dissociated by 5min incubation in TrypLE Express (GIBCO) at 37°C, followed by mechanical agitation through pipetting. TrypLE activity was terminated by washing with PBS+1% FCS.
Single cell library preparation and sequencing
Single-cell capture, cell lysis, reverse transcription of full-length mRNA and cDNA amplification were performed using the Fluidigm C1 system and the Smart-seq protocol (Ramsköld et al., 2012). Sequencing libraries were prepared using the Illumina Nextera XT DNA library preparation kit and samples were sequenced on an Illumina HiSeq.
DATA ANALYSIS
Alignment and processing of sequencing reads, data conversion and cell filtering
Reads were aligned to the human reference genome build hg19 with TopHat (v2.0.13), which was provided with gene annotations from GENCODE (v17). Gene expression profiles for each cell were computed using Cufflinks (2.2.0) as previously described in Trapnell et al., 2014. Per-cell fragments per kilobase million (FPKM) measures were converted into relative transcript counts using the Census algorithm from the Monocle package (Qiu et al., 2017a, 2017b; Trapnell et al., 2014). Cells with < 1k detected genes or with > 45k total mRNA were removed. Additional potential outliers were removed by defining lower and upper thresholds based on the appearance of the per-cell total mRNA distribution curve, separately for each sample. This approach automatically accounted for all cells lying outside of 2 standard deviations of each sample’s mean log-transformed per-cell total mRNA count and resulted in final number of 773 single-cell expression profiles (949 pre-filtering), with a median of 898,228 (mean: 945,907) usable paired end reads per cell (Figure S1B).
Inference of cellular gene expression programs via topic modeling
The topic model (TM) is a probabilistic unsupervised learning algorithm which is designed to discover latent topics present in a set of text documents, based solely on the observed usage of each word in each document (Blei, 2012). For text data, atopic is defined as set of (a priori unspecified) related words which probabilistically tend to co-occur. For example, statistical co-occurrence of words such as “banks,” “market,” and “GDP” within the same document provides strong evidence that a topic such as finance is being discussed. Once the topics have been inferred, a document can be more interpretably represented in terms of the relative presence of each topic rather than a massive collection of word frequencies. In our analysis, we utilize the topic model and inference algorithm used in the GeneProgram software tool (Gerber etal., 2007), which employs a Hierarchical Dirichlet Process (HDP) prior. This enables automatic inference of the number of latent topics (along with the topics themselves) from the data (Teh et al., 2006).
TM is adapted to scRNA-seq data by treating each cell as a document and each gene as a word. Within the cell, the unit amount of mRNA from the gene is therefore the genomic equivalent of a word’s frequency within a document. In this context, we refer to a latent topic as an Expression Program (EP), which represents a set of genes that behave coordinately in particular cells. Unlike traditional clustering techniques, our TM analysis allows many EPs to be active within a given cell, and a single gene may also be used by different EPs (one gene can regulate multiple different biological processes depending on the context in which it is expressed). For each EP, a gene receives a relevance value (quantifying how active the gene is within the EP) and a cell is assigned a usage value (quantifying how active the EP is within the cell). After inferring the EPs underlying our scRNA-seq measurements via Markov-Chain Monte Carlo (Gerber et al., 2007), we can interpretably represent and classify each cell in terms of its relative usage of different EPs, rather than the raw expression values of many thousand genes. Similarly, each EP is itself represented as a collection of genes along with their “relevance” in defining the EP. The biological significance of an EP can thus be elucidated by investigating which genes are the most actively used (Gerber et al., 2007).
The TM software used to infer the EPs in our analysis is available at http://groups.csail.mit.edu/cgs/geneprogram.html, and was run with the following settings: 4 levels of discretization for the expression values, 100 posterior samples (taken every 1,000 steps of the MCMC-chain with an initial burn-in of 30,000 steps), minimum of 2 genes per EP with a minimum usage value of 0.1, with the default prior HDP concentration and minimum-similarity/merging parameters used in (Gerber et al., 2007). See (Gerber et al., 2007) for detailed descriptions of these parameters.
SIMLR analysis
HESCs were removed from the 773 cleaned cells, retaining 720 cells for all subsequent analyses. A polynomial trend curve was fitted to the mean-variance plot of gene FPKM counts, and used as an estimate for technical variability. Altogether 3453 genes showed higher variance, and were used to cluster the cells using the SIMLR method implemented in the SIMLR R library (Wang et al., 2017). SIMLR calculates a cell-cell similarity metric based on parametrized Gaussian Kernels and uses the similarity values as input to the stochastic neighbor embedding method to plot the cell data in reduced dimensions. K-means clustering was subsequently used to assign the cells to different numbers of clusters, eventually settling on 10 predetermined clusters.
Differential expression was determined using the ROTS algorithm (Suomi et al., 2017). P values were corrected for multiple testing using the false discovery rate (FDR), and 4863 genes with FDR of less or equal to 0.05 were further characterized using Enrichr pathway analysis (Chen et al., 2013). The KEGG database (Kanehisa et al., 2017) was used as a source of curated lists of pathway genes.
Ligand-receptor interaction analysis
Ligands and receptors from Ramilowski etal. (Ramilowski etal., 2015) were introduced into the stage-specific analysis if expressed in 20% or more of the cells analyzed from that stage. The expression of these genes was compared between the cells belonging to the EPs analyzed on a given stage, and those that were significantly differentially expressed between EPs were retained and assigned to the EP where their average expression level is highest. This analysis was performed separately for stages 4 and 5.
Flow cytometry analysis
Differentiated cell clusters were dispersed into single-cell suspension by incubation in TrypLE Express at 37°C for 10–30 minutes, quenched with PBS+2% FBS, fixed with 4% PFA for at least 30 min at 4°C, washed once in PBS, and resuspended in PBS+ 2% FBS. Staining was done in 96-well plates. Cells were incubated in blocking buffer (PBS+0.1% Triton X-100+5% donkey serum) at 4°C for 30–60 min. Cells were then re-suspended in blocking buffer with primary antibodies and incubated at room temperature for 1 hour or at 4°C overnight. Cells were washed twice in blocking buffer and incubated in blocking buffer with secondary antibodies at 4°C for 1 hour. Cells were then washed three times in PBS+5% Donkey serum and analyzed using either the BD Accuri c6 flow cytometer or the LSR-II flow cytometer (BD Biosciences). Analysis of the results was performed using FlowJo software.
Nano-string gene expression analysis
Cells were collected for RNA extraction in RLT Lysis Buffer (QIAGEN) and frozen at −80°C. RNA was extracted using the RNeasy Mini Kit (QIAGEN), according the manufacturer’s instructions - including DNA digestion, and 100ng of RNA was used for each reaction. Nanostring set up was performed according to the protocol for the nCounter XT Gene Expression Assay. Data was analyzed in the nSolver 2.5 Software. All genes were normalized to housekeeping genes (ITCH, RPL15, RPL19, TCEB1, UBE2D3). Analysis of changes in gene expression of markers for endocrine and progenitor cells was done using R.
Immunofluorescence staining and In situ hybridization
Isolated tissue was fixed overnight in 4% paraformaldehyde, washed in PBS, embedded in paraffin and sectioned on a microtome. Paraffin was removed using Histo-Clear (National Diagnostics) and serial washes in declining ETOH concentrations. After antigen retrieval, samples were blocked in PBS with 5% donkey serum and 0.1% Triton x-100 for 1 hour in room temperature. Primary antibody was diluted in the blocking solution and incubated in room temperature for 1hr, followed by washing. Secondary antibody was incubated for 1 hour in room temperature, washed, and DAPI staining was added.
In situ hybridization was performed using ViewRNAISH tissue assay kit. For detection of human SMO and PTCH1 transcripts, VA1–14719 and VA6–20367 probes were used, respectively.
Glucose Stimulated Insulin Secretion (GSIS)
Differentiated cells were washed twice in Krebs buffer (Krb) containing 2.8mM glucose, and then distributed into 24-well plates containing Millicell Cell culture inserts for easy transfer between conditions. Each well received 0.5ml buffer with approximately 106 cells. After fasting in 2.8mM glucose for 1 hour in 37°C, the analysis of the cells’ response to changing glucose levels was performed by a series of 1 hour incubations in 37°C, in the following order: 1) 2.8mM (low) glucose. 2) 20mM (high) glucose. 3) brief wash in 2.8mM glucose. 4) 1 hour incubation in 2.8mM glucose. 5) 20mM glucose. 6) 2.8mM glucose with 30mM KCL. At the end of each step (excluding the wash), the incubation buffer was collected and the amount of insulin secreted during the hour’s incubation was determined using human insulin ELISA kit (ALPCO diagnostics; 80-INSHUU-E01.1). Clusters were then dispersed into single cells using TrypLE Express (Life Technologies), and cell number was counted automatically by a Vi-Cell (Beckman Coulter).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical methods used are described in the legend to the relevant figures. n marks the number of times an experiment was performed on independent differentiation batches, except for Figure S5B, which describes a triplicate of samples taken from the same differentiation batch. Data was analyzed and plotted using Prism software from GraphPad. When boxplots are presented, the bottom and top box borders represent the 25th and 75th percentiles, respectively. The line inside the box represents the median, and the whiskers represent the minimum and maximum values observed.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti C-peptide | DSHB | Cat#GN-ID4; RRID: AB_2255626 |
| Rabbit anti Chaga | Abcam | Cat#Ab15160; RRID: AB_301704 |
| Chicken anti GFP | Aves Labs inc. | Cat#GFP-1020; RRID: AB_10000240 |
| Goat anti glp2 | Santa Cruz Biotechnology | Cat#sc-7781; RRID: AB_2107346 |
| Mouse anti Glucagon | Santa Cruz Biotechnology | Cat#sc-514592; RRID: AB_2629431 |
| Goat anti Neurod1 | R&D systems | Cat#AF2746; RRID: AB_2149217 |
| Sheep anti Neurogenin3 | R&D systems | Cat#AF3444; RRID: AB_2149527 |
| Mouse anti Nkx6.1 | DSHB | Cat#F55A12; RRID: AB_532379 |
| Rabbit anti Sox9 | Abcam | Cat#Ab-5535; RRID: AB_2239761 |
| Goat anti PDX1 | R&D systems | Cat#AF2419; RRID: AB_355257 |
| Mouse anti somatostatin | Santa Cruz Biotechnology | Cat#sc-55565; RRID: AB_831726 |
| Rabbit anti MCM3 | Cell Signaling Technology | Cat#4012s; RRID: AB_2235150 |
| Mouse anti Ctnnb1 | ThermoFisher Scientific | Cat#MA1–2001; RRID: AB_326078 |
| Goat anti Patched1 | Santa Cruz Biotechnology | Cat#sc-6147; RRID: AB_2253263 |
| Rat anti PTCH | R&D systems | Cat#MAB41051; RRID: AB_2174045 |
| Rabbit anti APC | Abcam | Cat#Ab-15270; RRID: AB_301806 |
| Mouse anti E-cadherin | BD Transduction Laboratories | Cat#610181; RRID: AB_397580 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ActivinA | R&D Systems | Cat#338-AC |
| Rock Inhibitor Y-27632 | DNSK | Cat#DNSK-Kl15–02 |
| Chir99021 | Stemgent | Cat#04–0004-10 |
| KGF | Peprotech | Cat#100–19 |
| Sant1 | Sigma-Aldrich | Cat#S4572 |
| PDBu | EMD Millipore | Cat#524390 |
| XXI | EMD Millipore | Cat#565790 |
| Alk5i II | Axxora | Cat#ALX-270–445 |
| T3 | EMD Millipore | Cat#642511 |
| Betacellulin | ThermoFisher Scientific | |
| IWR1-endo | Sigma-Aldrich | Cat#I0161 |
| XAV939 | Santa Cruz Biotechnology | Cat#sc-296704A |
| Recombinant Human BMP2/BMP-7 Heterodimer | ThermoFisher Scientific | Cat#3229-BM |
| Critical Commercial Assays | ||
| ViewRNA ISH Tissue Assay Kit (2-plex) | ThermoFisher | Cat#QVT0012 |
| Anti human SMO ViewRNA probeset | ThermoFisher | Cat#VA1 −14719 |
| Anti human PTCH1 ViewRNA probeset | ThermoFisher | Cat#VA6–20367 |
| SMARTer® Ultra Low Kit for the Fluidigm® C1 System | Clontech | Cat#634833 |
| C1 Single-Cell Auto Prep Array for mRNA Seq (17–25 μm) | Fluidigm | Cat#100–5761 |
| Deposited Data | ||
| Raw and analyzed single cell sequencing data | This paper | SRA BioProject PRJNA532884 |
| Experimental Models: Cell Lines | ||
| HUES8hESCS | HSCI | hES Cell line: HUES-8 |
| hiPSC 1016 | HSCI | hiPSC 1016 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6j-ApcMin/J | Jackson Laboratories | RRID:IMSR_JAX:002020 |
| Mouse: B6.Cg-Apctm2Rak/Nci | NCI Mouse Repository | RRID:IMSR_NCIMR:01XAA |
| Mouse: Tg(Pdx1-cre)89.1Dam | In house | RRID:MGI:5902759 |
| Mouse: Tg(Neurog3-cre)C1Able/J | Jackson Laboratories | RRID:IMSR_JAX:005667 |
| Mouse: B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J | Jackson Laboratories | RRID:IMSR_JAX:006148 |
| Software and Algorithms | ||
| Topic modeling | This paper | http://groups.csail.mit.edu/cgs/geneprogram.html |
| Interactive database | This paper | https://ifx.rc.fas.harvard.edu/invitrobetacells/ |
Highlights.
scRNA-seq of hESC differentiation into β cells in vitro
Progenitor and endocrine compartments express different WNT pathway components
APC deletion in mouse endocrine precursors prevents endocrine differentiation
Small molecule inhibition of the WNT pathway in vitro increases endocrine yield
ACKNOWLEDGMENTS
N.S. is partly supported by the Gruss-Lipper Charitable Foundation. D.A.M. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the Harvard Stem Cell Institute, Helmsley Charitable Trust and the JPB Foundation. This research was performed using resources and/or funding provided by the NIDDK-supported Human Islet Research Network (HIRN; RRID:SCR_014393, UC4 DK104165, and UC4 DK104159).
We thank Prof. Aviv Regev from the Broad Institute and Dr. Alex Shalek, currently at MIT, for help with technical aspects of scRNAseq; and Mrs. Cathy MacGillvary and Mrs. Diane Faria for histological preparations.
Footnotes
DECLARATION OF INTERESTS
D.A.M. is a founder and SAB member for Semma Therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.04.083.
DATA AND SOFTWARE AVAILABILITY
Single-cell RNA-sequencing data was deposited to the NCBI Sequence Read Archive (SRA). The accession number for the raw sequences is: PRJNA532884.
ADDITIONAL RESOURCES
An interactive website which presents gene expression and EP usage in the cells is accessible at https://ifx.rc.fas.harvard.edu/invitrobetacells/
REFERENCES
- Alcedo J, Zou Y, and Noll M. (2000). Posttranscriptional regulation of smoothened is part of a self-correcting mechanism in the Hedgehog signaling system. Mol. Cell 6, 457–465. [DOI] [PubMed] [Google Scholar]
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, and Edlund H. (1999). Notch signalling controls pancreatic cell differentiation. Nature 400, 877–881. [DOI] [PubMed] [Google Scholar]
- Beher D, Wrigley JD, Nadin A, Evin G, Masters CL, Harrison T, Castro JL, and Shearman MS (2001). Pharmacological knock-down of the presenilin 1 heterodimer by a novel gamma-secretase inhibitor: implications for presenilin biology. J. Biol. Chem. 276, 45394–45402. [DOI] [PubMed] [Google Scholar]
- Blei DM (2012). Probabilistic topic models. Commun. ACM 55, 77–84. [Google Scholar]
- Bort R, Martinez-Barbera JP, Beddington RS, and Zaret KS (2004). Hex homeobox gene-dependent tissue positioning is required for organogenesis of the ventral pancreas. Development 131, 797–806. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan CW, Wei S, Hao W, Kilgore J, Williams NS, et al. (2009). Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Andersson O, Row R, Kimelman D, and Stainier DY (2010). Suppression of Alk8-mediated Bmp signaling cell-autonomously induces pancreatic beta-cells in zebrafish. Proc. Natl. Acad. Sci. USA 107, 1142–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker K, Goldman DC, Grasch CL, and Sussel L. (2006). Gata6 is an important regulator of mouse pancreas development. Dev. Biol. 298, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessimoz J, Bonnard C, Huelsken J, and Grapin-Botton A. (2005). Pancreas-specific deletion of beta-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr. Biol. 15, 1677–1683. [DOI] [PubMed] [Google Scholar]
- Filipe M, GonÇalves L, Bento M, Silva AC, and Belo JA (2006). Comparative expression of mouse and chicken Shisa homologues during early development. Dev. Dyn. 235, 2567–2573. [DOI] [PubMed] [Google Scholar]
- Gao T, Zhou D, Yang C, Singh T, Penzo-Mendez A, Maddipati R, Tzatsos A, Bardeesy N, Avruch J, and Stanger BZ (2013). Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology 144, 1543–1553.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber GK, Dowell RD, Jaakkola TS, and Gifford DK (2007). Automated discovery of functional generality of human gene expression programs. PLoS Comput. Biol. 3, e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin AR, Stadler HS, Nakamura K, and Capecchi MR (1998). Detection of targeted GFP-Hox gene fusions during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 95, 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G, Dubauskaite J, and Melton DA (2002). Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development 129, 2447–2457. [DOI] [PubMed] [Google Scholar]
- Gunawardene AR, Corfe BM, and Staton CA (2011). Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 92, 219–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser PW, Lau J, Taketo MM, Herrera PL, and Hebrok M. (2006). Stabilization of beta-catenin impacts pancreas growth. Development 133, 2023–2032. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620. [DOI] [PubMed] [Google Scholar]
- Jacquemin P, Pierreux CE, Fierens S, van Eyll JM, Lemaigre FP, and Rousseau GG (2003). Cloning and embryonic expression pattern of the mouse Onecut transcription factor OC-2. Gene Expr. Patterns 3, 639–644. [DOI] [PubMed] [Google Scholar]
- Jennings RE, Berry AA, Kirkwood-Wilson R, Roberts NA, Hearn T, Salisbury RJ, Blaylock J, Piper Hanley K, and Hanley NA (2013). Development of the human pancreas from foregut to endocrine commitment. Diabetes 62, 3514–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny M, Uhl C, Roche C, Duluc I, Guillermin V, Guillemot F, Jensen J, Kedinger M, and Gradwohl G. (2002). Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21, 6338–6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson KA, Dursun U, Jordan N, Gu G, Beermann F, Gradwohl G, and Grapin-Botton A. (2007). Temporal control of neurogenin3 activity in pancreas progenitors reveals competence windows for the generation of different endocrine cell types. Dev. Cell 12, 457–465. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, and Morishima K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45 (D1), D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahira H, Ma NH, Tzanakakis ES, McMahon AP, Chuang PT, and Hebrok M. (2003). Combined activities of hedgehog signaling inhibitors regulate pancreas development. Development 130, 4871–4879. [DOI] [PubMed] [Google Scholar]
- Kawahira H, Scheel DW, Smith SB, German MS, and Hebrok M. (2005). Hedgehog signaling regulates expansion of pancreatic epithelial cells. Dev. Biol. 280, 111–121. [DOI] [PubMed] [Google Scholar]
- Kelly OG, Chan MY, Martinson LA, Kadoya K, Ostertag TM, Ross KG, Richardson M, Carpenter MK, D’Amour KA, Kroon E, et al. (2011). Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat. Biotechnol. 29, 750–756. [DOI] [PubMed] [Google Scholar]
- Kuraguchi M, Wang XP, Bronson RT, Rothenberg R, Ohene-Baah NY, Lund JJ, Kucherlapati M, Maas RL, and Kucherlapati R. (2006). Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2, e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, and He X. (2009). Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi A, Prawiro C, Seymour PA, de Lichtenberg KH, Jackson A, Serup P, and Semb H. (2018). Mechanosignalling via integrins directs fate decisions of pancreatic progenitors. Nature 564, 114–118. [DOI] [PubMed] [Google Scholar]
- Miyatsuka T, Li Z, and German MS (2009). Chronology of islet differentiation revealed by temporal cell labeling. Diabetes 58, 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, and Dove WF (1990). A dominant mutation that pre-disposes to multiple intestinal neoplasia in the mouse. Science 247, 322–324. [DOI] [PubMed] [Google Scholar]
- Muñoz-Bravo JL, Flores-Martínez A, Herrero-Martin G, Puri S, Taketo MM, Rojas A, Hebrok M, and Cano DA (2016). Loss of pancreas upon activated Wnt signaling is concomitant with emergence of gastrointestinal identity. PLoS ONE 11, e0164714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC (2008). The what, where, when and how of Wnt/β-catenin signaling in pancreas development. Organogenesis 4, 81–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, and Melton DA (2003). Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA 100, 14920–14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Law AC, Dor Y, and Melton DA (2005). Beta-catenin is essential for pancreatic acinar but not islet development. Development 132, 4663–4674. [DOI] [PubMed] [Google Scholar]
- Nostro MC, Sarangi F, Ogawa S, Holtzinger A, Corneo B, Li X, Micallef SJ, Park IH, Basford C, Wheeler MB, et al. (2011). Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development 138, 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovich AB, Long Q, Manduchi E, Gangula R, Hipkens SB, Schneider J, Okubo T, Stoeckert CJ Jr., Takada S, and Magnuson MA (2014). Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development 141, 2939–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, and Melton DA (2014). Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, and Wright C. (2011). Pancreas organogenesis: from bud to plexus to gland. Dev. Dyn. 240, 530–565. [DOI] [PubMed] [Google Scholar]
- Petersen MBK, Azad A, Ingvorsen C, Hess K, Hansson M, Grapin-Botton A, and Honoré C. (2017). Single-cell gene expression analysis of a human ESC model of pancreatic endocrine development reveals different paths to b-cell differentiation. Stem Cell Reports 9, 1246–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Hill A, Packer J, Lin D, Ma YA, and Trapnell C. (2017a). Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner H, and Trapnell C. (2017b). Reversed graph embedding resolves complex single-cell developmental trajectories. Nat. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, and Forrest AR (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun. 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, Ma ST, Reeder JW, Samuels I, Slabiak T, et al. (2003). Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes 52, 588–595. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Seguel E, Mah N, Naumann H, Pongrac IM, Cerdá-Esteban N, Fontaine JF, Wang Y, Chen W, Andrade-Navarro MA, and Spagnoli FM (2013). Mutually exclusive signaling signatures define the hepatic and pancreatic progenitor cell lineage divergence. Genes Dev. 27, 1932–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado-Olivieri EA, Anderson K, Kenty JH, and Melton DA (2019). YAP inhibition enhances the differentiation of functional stem cell-derived insulin-producing β cells. Nat. Commun. 10, 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. (2015). Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J. 34, 1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhoff SE, Giel-Moloney M, and Leiter AB (2004). Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 270, 443–454. [DOI] [PubMed] [Google Scholar]
- Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. (2016). Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro AM, Lakey JR, Ryan EA, Korbutt GS, Toth E, Warnock GL, Kneteman NM, and Rajotte RV (2000). Islet transplantation in seven patients with type 1 diabetes mellitus using a glucocorticoid-free immunosuppressive regimen. N. Engl. J. Med. 343, 230–238. [DOI] [PubMed] [Google Scholar]
- Sharon N, Chawla R, Mueller J, Vanderhooft J, Whitehorn LJ, Rosenthal B, Gürtler M, Estanboulieh RR, Shvartsman D, Gifford DK, et al. (2019). A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell 176, 790–804.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom A, Bonal C, Ashery-Padan R, Hashimoto N, Campos ML, Trumpp A, Noda T, Kido Y, Real FX, Thorel F, and Herrera PL (2007). Unique mechanisms of growth regulation and tumor suppression upon Apc inactivation in the pancreas. Development 134, 2719–2725. [DOI] [PubMed] [Google Scholar]
- Suomi T, Seyednasrollah F, Jaakkola MK, Faux T, and Elo LL (2017). ROTS: an R package for reproducibility-optimized statistical testing. PLoS Comput. Biol. 13, e1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh YW, Jordan MI, Beal MJ, and Blei DM (2006). Hierarchical dirichlet processes. J. Am. Stat. Assoc. 101, 1566–1581. [Google Scholar]
- Thomas PQ, Brown A, and Beddington RS (1998). Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development 125, 85–94. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Veres A, Faust AL, Bushnell HL, Engquist EN, Hyoje-Ryu Kenty J, Harb G, Poh Y-C, Sintov E, Gürtler M, Pagliuca FW, et al. (2019). Charting cellular identity during human in vitro β-cell differentiation. Nature, Published online May 8, 2019. 10.1038/s41586-019-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhu J, Pierson E, Ramazzotti D, and Batzoglou S. (2017). Visualization and analysis of single-cell RNA-seq data by kernel-based similarity learning. Nat. Methods 14, 414–416. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, and Melton DA (2007). A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.