Abstract
The data presented in this article are related to the research article entitled as “Targeted deletion of the BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: A promising approach for gene therapy of beta-thalassemia disease " [1]. BCL11A is a master regulator of γ-globin gene silencing, and suppresses fetal hemoglobin expression by association with other γ-globin suppressors, and also interacts with human beta-globin locus control region as well as intergenic region between the Aγ and δ-globin genes to reconfigure beta-globin cluster. Thus, HbF reactivation has been proposed to be an approach for the treatment of β-thalassemia through knockout of BCL11A. Accordingly, an erythroid enhancer sequence was identified that, when inactivated, led to repression of BCL11A and induction of γ-globin in the erythroid lineage [2–7]. This article describes data that obtained from BCL11A gene enhancer modification in KU812 and KG-1 cell lines using the CRISPR-Cas9 genome editing system in order to reactivate γ-globin gene expression.
Keywords: BCL11A, γ-globin, KU812, KG-1, CRISPR-Cas9 genome editing system
Specifications Table
| Subject area | Biochemistry, Genetics and Molecular Biology |
| More specific subject area | Biotechnology, Genome editing, Genetics, and Molecular Medicine |
| Type of data | Graph, figure |
| How data was acquired | Data were collected using PCR, Real-Time qPCR (QuantStudio™ 7) Flex Real-Time PCR Systems (Applied Biosystems) and StepOne Plus Real-Time PCR Systems (Applied Biosystems), Sanger sequencing, and GraphPad Prism 6 |
| Data format | Raw and analyzed |
| Experimental factors | BCL11A and -globin gene expression level were compared between wildtype conditions and conditions after erythroid-specific BCL11A enhancer gene editing through the CRISPR-Cas9 system in KU812 and KG-1 cells. |
| Experimental features | BCL11A-knockout KU812 and KG-1 cell lines were established using the CRISPR-Cas9 genome editing system. Single-cell cloning was performed by serial dilution, and then the expression level of BCL11A and -globin were analyzed in clones. |
| Data source location | Institute of Laboratory Animal Sciences - Institut für Labortierkunde (LTK) |
| Data accessibility | The data are available for this article |
| Related research article | M.A. Khosravi, M. Abbasalipour, J.-P. Concordet, J. Vom Berg, S. Zeinali, A. Arashkia, K. Azadmanesh, T. Buch, M. Karimipoor, Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: A promising approach for gene therapy of beta-thalassemia disease, Eur. J. Pharmacol. 854 (2019) 398–405. https://doi.org/10.1016/J.EJPHAR.2019.04.042 [1]. |
Value of the Data
|
1. Data
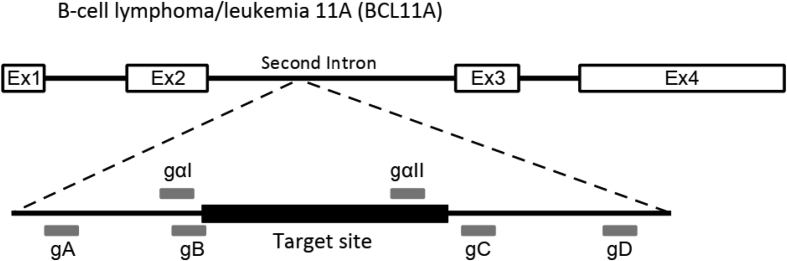
This article shows targeted inactivation of BCL11A gene expression for induction of γ-globin in the erythroid lineage [[2], [3], [4], [5], [6], [7]] using the CRISPR-Cas9 genome editing system through deletion of a 200 bp sequence encompassing the GATA1 motif within the BCL11A erythroid enhancer. Two sgRNAs based on SaCas9 (gα) and four sgRNA based on SpCas9 (g) were designed using an online CRISPR design tool (http://crispor.tefor.net/) (Fig. 1).
Fig. 1.
Schematic representation of single guide RNA (sgRNA) locations. Two sgRNAs based on SaCas9 (gα) and four sgRNAs based on SpCas9 (g) were designed within the erythroid enhancer of the BCL11A gene.
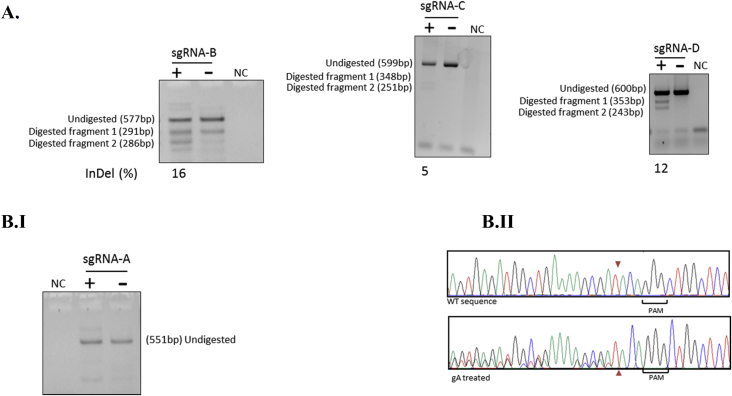
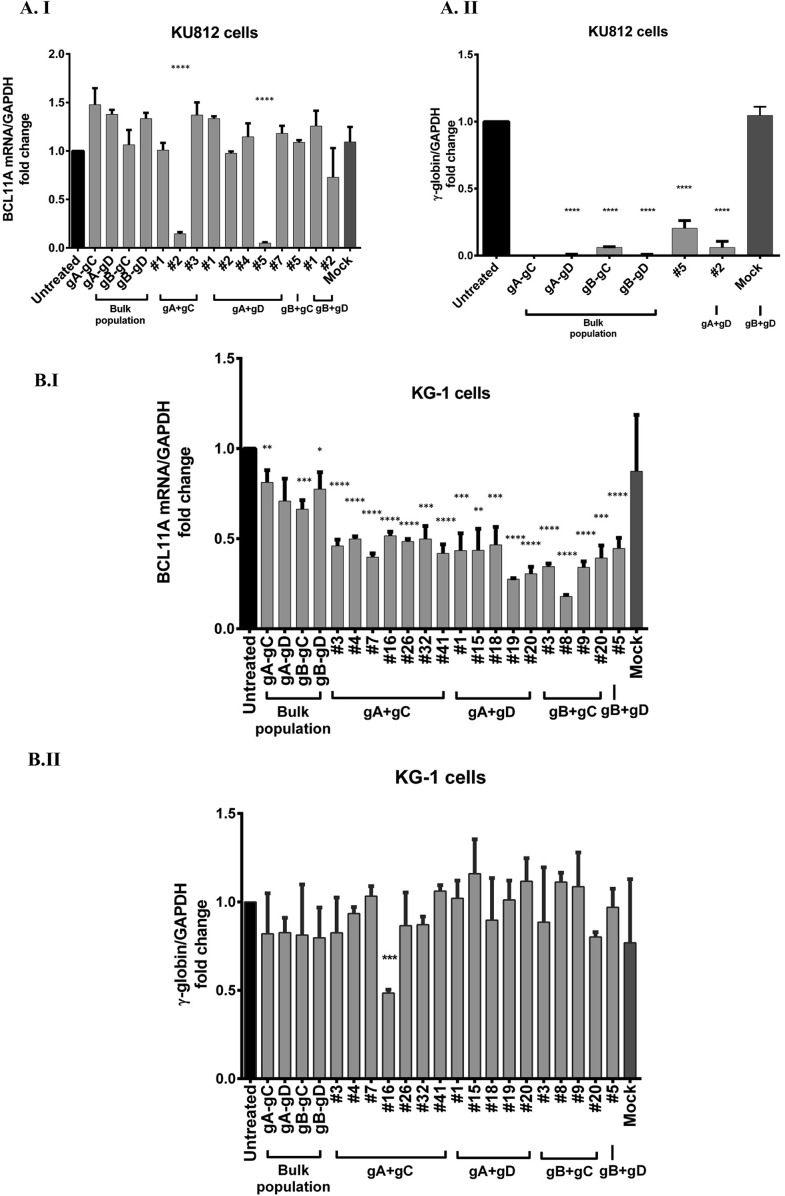
Cutting efficiency of each gRNA was determined by T7EI assay and Sanger sequencing in HEK293T cells. Cutting efficiencies of 16%, 5%, and 12% obtained for guide RNAs gB, gC, and gD, respectively. However, no cutting efficiency was determined for gRNAs gαI and gαII. Moreover, no cutting activity was observed for gA by T7EI assay, but it showed activity when the target site was analyzed by Sanger sequencing (Fig. 2, Fig. 3-Add sequencing files Fig. 2, Fig. 3 gαI, gαII, gA, gB, gC, gD, WT, and T7EI assay data for Fig. 2, Fig. 3). The constructs were subsequently transfected into KU812 and KG-1 cells. Following validation of targeted genomic deletion by pairs of sgRNA using PCR (Fig. 4), quantitative real-time PCR (qPCR) was performed on mRNAs purified from bulk cells, as well as on edited clones to assess the mRNA level of BCL11A and γ-globin (Fig. 5-Add qPCR data for Fig. 5).
Fig. 2.
Evaluation of SpCas9 cutting efficiency in HEK293T cells. (A) The mutation frequency of sgRNAs was evaluated using T7 endonuclease I assay in HEK293T cells transfected with the pMLM3636-gA, pMLM3636-gB, pMLM3636-gC, pMLM3636-gD, and hCas9 expression vector. Primers flanking the sgRNA recognition sequences were used to amplify regions around each sgRNA site, and the amplified products were cleaved by the T7 endonuclease enzyme. Expression vectors with sgRNA were indicated as (+), and control vectors without sgRNA were indicated as (−). The efficiency of cleavage has been indicated as % indels below each gel. (B) The mutation rate of sgRNA-A was assessed by (B. I) T7EI assay, and (B. II) Sanger sequencing on bulk cells. The reference sequence has been shown at the top, and the sequence derived from sgRNA-A transfected HEK293T cells has been shown below. NC: negative control.
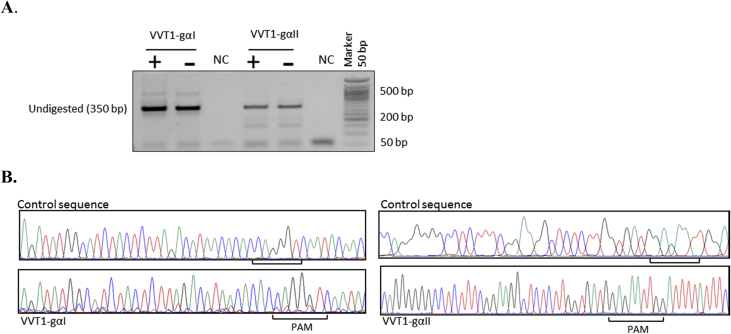
Fig. 3.
Evaluation of SaCas9 cutting efficiency in HEK293Tcells. The mutation frequency of sgRNAs was evaluated using T7 endonuclease I assay (A), and Sanger sequencing (B) in HEK293T cells transfected with the VVT1-αI, VVT1-αII, and BPK2139, a human expression vector for S.aureus Cas9, was a gift from Keith Joung (Addgene plasmid # 65776) [8]. The expected band sizes of gRNA-gI and gRNA-gII after digestion with T7 enzyme were 177 bp, 181 bp, and 226 bp, 127 bp, respectively. However, no difference was observed before and after digestion by T7 enzyme.
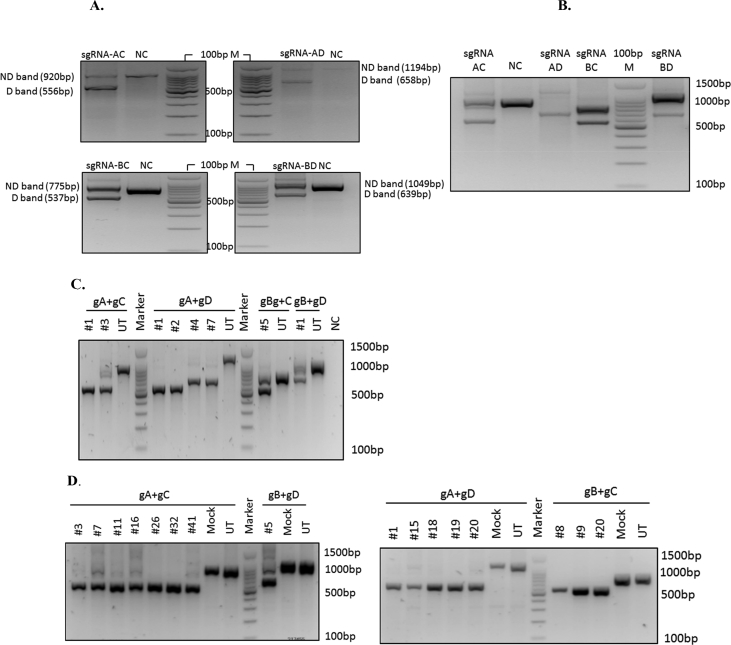
Fig. 4.
Targeted deletion of the BCL11A enhancer with CRISPR-Cas9. Targeted deletion of the core sequence of the +58DHS within the erythroid-specific enhancer of the BCL11A gene using the combination of sgRNAs AC, AD, BC, and BD was confirmed by conventional PCR. PCR was performed (A) on the bulk of the KU812 cells, and (B) on the bulk of the KG-1 cells. (C) Homozygously deleted (Biallelic deletion) clones of KU812 cells, and (D) KG-1 cells. Expected band sizes for gA + gC were as ND: 920 bp, and D: 556 bp; for gA + gD were as ND: 1194 bp, and D: 658 bp; for gB + gC were as ND: 775 bp, and D: 537 bp; for gB + gD were as ND: 1049 bp, and D: 639 bp. Abbreviations: D: deleted band. ND: Non-deleted band. NC: negative control. BenchTop 100 bp DNA Ladder, Promega, Cat.: #G8291 was used as the marker. All experiments were performed twice.
Fig. 5.
qRT-PCR Analysis of BCL11A and-globin gene expression. Real-time PCR was performed on mRNA purified from bulk cells and edited clones to assess BCL11A and γ-globin mRNA levels. The qRT-PCR was normalized to the level of human GAPDH in cells treated with four different pairs of sgRNA in two independent experiments. Evaluation of BCL11A (A. I), and γ-globin (A. II) expression in bulk and edited clones of KU812 cells treated by all combination of sgRNAs. Clone #2 from the combination of sgRNAs A and C, and clone #5 from transfection of sgRNAs A and D, indicated significant differences (p-value < 0.0001). Unpredictably, these clones showed a significant reduction in γ-globin expression despite the deletion. Evaluation of BCL11A expression in bulk and edited clones of KG-1 cells treated by all combination of sgRNAs (B.I), statistically, a significant difference was found compared with untreated control (B.II). Results from RT-qPCR have been depicted as relative fold change, which was calculated using the ddCT method [9,10], in which fold change data have been represented as 2−ddCT. Data were analyzed with GraphPad Prism software v6.0 and expressed as mean ± S.E.M. Statistically significant differences were identified by Student t-test compared to untreated cells and were indicated as follows: ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
2. Experimental design, materials and methods
2.1. Designing and cloning of sgRNAs
Single-guide RNAs were designed using the online CRISPR design tool (http://crispor.tefor.net/) [11,12]. Oligonucleotides encoding the sgRNAs were annealed and cloned into the BsmBI-digested pMLM363, VVT1, and pMAK vectors as previously described [1].
2.2. Cell culture and transfection
HEK293T cell line was cultured in Dulbecco's minimal essential medium (high glucose) (DMEM, Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Sigma). The KU812 cell line (chronic myelogenous leukemia, CML) and KG-1 as a human hematopoietic progenitor cell line model were cultured in RPMI 1640 medium (high glucose) (Sigma) supplemented with 2.0 mM l-glutamine (Life Technologies Gibco), 1% penicillin/streptomycin (Pen-Strep; Life Technologies Gibco, #15140–122), and 20% FBS for KU812, and 10% FBS for KG-1 cell lines. Transfection of HEK293T cells with pMLM3636 and hCas9 plasmids for CRISPR/SpCas9 system, as well as VVT1 and BPK2139 for CRISPR/SaCas9 system were performed using Lonza Nucleofector® Kit V (program CM-130), as previously described [1].
KU812 and KG-1 cell lines were transfected with pMAK-gA + gC, pMAK-gA + gD, pMAK-gB + gC, and pMAK-gB + gD vectors using the Neon® Transfection System 100 μl Kit (Thermo Fisher Scientific), as previously described [1].
2.3. Cleavage activity of sgRNAs in cultured cells
CRISPR-Cas9 editing efficiency with the different sgRNAs was assessed by T7 Endonuclease assay (New England Biolabs). Forty-eight hours after transfection, genomic DNA (gDNA) was extracted using the E.Z.N.A.® Tissue DNA Kit (Omega Bio-tek), and 50 ng of the gDNA was amplified by target PCR primer pairs flanking the sgRNA-targeted region with Phusion® High-Fidelity DNA Polymerase (New England Biolabs), based on the manufacturer's instructions. Following amplification, PCR amplicons were melted, annealed (5 min at 95 °C, 95 °C–25 °C at −0.5 °C/30 sec, and 15 min at 4 °C), and were treated by T7EI enzyme (New England Biolabs) for 20 min at 37 °C and run on a 2% agarose gel. To assess the gene modification efficiency, DNA band intensities were quantified using ImageJ software (http://imagej.nih.gov/ij/) using the following formula, as previously described:
% gene modification = 100 × (1 – (1- fraction cleaved)1/2) [13].
2.4. Generation of genomic deletions using the CRISPR-Cas9 system
Forty-eight hours post electroporation, transfected cells were plated at limiting dilution with 30 cells per 96-well plate to find homozygously deleted clones. After either 14 or 21 days of clonal expansion, genomic DNA was extracted from bulk cells as well as edited clones using DNA lysis buffer (Direct PCR–Tail PEQGOLD, USA). Single-cell clones were screened for monoallelic deletion via PCR to amplify the targeted sequence using a primer set flanking the two sgRNA; one primer for upstream of sgRNA-A and sgRNA-B, and another primer for downstream of sgRNA-C and sgRNA-D to amplify across the deletion sequence. Biallelic deleted clones were identified via the absence of non-deleted PCR band, and presence of deleted PCR band.
2.5. Measurement of BCL11A and γ -globin gene expression in modified cells
Total RNAs were isolated from the bulk cells as well as edited colonies using RNeasy Mini Kit (Qiagen) and were DNase-treated with Ambion® TURBO DNA-free™ DNase (Invitrogen™), according to the manufacturer's instructions. One microgram of extracted RNA was used for the generation of cDNA using the iScript™ cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's specifications. Levels of BCL11A, γ -globin, and GAPDH as an internal control were quantified by qPCR using the following primers: BCL11A primer (fw): 5′-TTGCCCCAAACAGGAACACA-3′, BCL11A primer (rv): 5′-CGGGGCATATTCTGCACTCA-3′, HbF primer (fw): 5′-CTTCAAGCTCCTGGGAAATGT-3′, γ -globin primer (rv): 5′-GTCACCATCTTCTGCCAGGA-3′, GAPDH primer (fw): 5′-TCCACCACCCTGTTGCTGTAG-3′, GAPDH primer (rv): 5′-ACACCCACTCCTCCACCTTTG-3′. Quantitative RT-PCR was performed in duplicate or triplicate using the QuantStudio™ 7 Flex Real-Time PCR Systems (Applied Biosystems) with SsoFast EvaGreen Supermix (Bio-Rad) or StepOne Plus Real-Time PCR Systems (Applied Biosystems) with the RealQ Plus 2× Master Mix Green High ROX™ (Ampliqon) following the manufacturer's procedures. SsoFast EvaGreen Supermix PCR conditions were as follows: 5 min at 98 °C, 40 cycles of 10 s at 98 °C and 20 s at 60 °C. RealQ Plus 2× Master Mix Green PCR conditions were as the following: 15 min at 95 °C, 40 cycles of 20 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. The relative values of the transcripts were normalized to that of GAPDH gene and analyzed using the 2-ddCT method. A standard curve for each gene was made from serial dilutions of the cDNA. Duplicate negative controls (no template cDNA) were also run with every experimental plate to assess the specificity, and to rule out contamination.
2.6. Statistical analysis
Results from RT-qPCR were depicted as relative fold change, which was calculated using the ddCT method, in which fold change data represented as 2-ddCT. Statistical analysis was performed using 2-tailed student t-test (Prism software v6.0, GraphPad Software).
Acknowledgments
This work was performed at the Institute of Laboratory Animal Science, University of Zurich, Switzerland. The authors would like to thank Stanislav Pantelyushin in the lab of Dr. Johannes vom Berg for his support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.104974.
Contributor Information
Mohammad Ali Khosravi, Email: mkhosravi96@gmail.com.
Thorsten Buch, Email: thorsten.buch@uzh.ch.
Morteza Karimipoor, Email: mortezakarimi@pasteur.ac.ir, mortezakarimi@yahoo.com.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
This work was supported by grants from Pasteur Institute of Iran [grant number: BP-9035].
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Khosravi M.A., Abbasalipour M., Concordet J.-P., Vom Berg J., Zeinali S., Arashkia A., Azadmanesh K., Buch T., Karimipoor M. Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: a promising approach for gene therapy of beta thalassemia disease. Eur. J. Pharmacol. 2019;854:398–405. doi: 10.1016/j.ejphar.2019.04.042. [DOI] [PubMed] [Google Scholar]
- 2.Bauer D.E., Orkin S.H. Hemoglobin switching's surprise: the versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr. Opin. Genet. Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brendel C., Guda S., Renella R., Bauer D.E., Canver M.C., Kim Y.J., Heeney M.M., Klatt D., Fogel J., Milsom M.D., Orkin S.H., Gregory R.I., Williams D.A. Lineage-specific BCL11A knockdown circumvents toxicities and reverses sickle phenotype. J. Clin. Investig. 2016;126:3868–3878. doi: 10.1172/JCI87885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavazzana M., Antoniani C., Miccio A. Gene therapy for ?-hemoglobinopathies. Mol. Ther. 2017;25:1142–1154. doi: 10.1016/j.ymthe.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoban M.D., Orkin S.H., Bauer D.E. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127:839–848. doi: 10.1182/blood-2015-09-618587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinjamur D.S., Bauer D.E., Orkin S.H. Recent progress in understanding and manipulating haemoglobin switching for the haemoglobinopathies. Br. J. Haematol. 2018;180:630–643. doi: 10.1111/bjh.15038. [DOI] [PubMed] [Google Scholar]
- 7.Wilber A., Nienhuis A.W., Persons D.A. Transcriptional regulation of fetal to adult hemoglobin switching: new therapeutic opportunities. Blood. 2011;117:3945–3953. doi: 10.1182/blood-2010-11-316893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., V Topkar V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao X., Huang X., Zhou Z., Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma. Biomath. 2013;3:71–85. http://www.ncbi.nlm.nih.gov/pubmed/25558171 [PMC free article] [PubMed] [Google Scholar]
- 10.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Canver M.C., Haeussler M., Bauer D.E., Orkin S.H., Sanjana N.E., Shalem O., Yuan G.C., Zhang F., Concordet J.P., Pinello L. Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat. Protoc. 2018;13:946–986. doi: 10.1038/nprot.2018.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J.-S., Concordet J.-P. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J. Humana Press; Totowa, NJ: 2010. A Rapid and General Assay for Monitoring Endogenous Gene Modification; pp. 247–256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.