Abstract
Advanced prostate cancer is often treated with AR antagonists which target the androgen receptor (AR) on which the growth of the tumour depends. Prostate cancer often develops AR-antagonist resistance via a plethora of mechanisms, many of which are as yet unknown, but it is thought that AR upregulation or AR ligand-binding site mutations, may be responsible. Here we describe the production of cell lines based on LNCaP and VCaP, with acquired resistance to the clinically relevant AR antagonists, bicalutamide and enzalutamide. In these resistant cells, we observed, via RNA-seq, that new variants in the 3′UTR of the AR mRNA were detectable and that the levels were increased both with AR-antagonist treatment and with hormonal starvation. Around 20% of AR transcripts showed a 3 kb deletion within the 6.7 kb 3′UTR sequence. Actinomycin D and luciferase fusion studies indicated that this shorter mRNA variant was inherently more stable in anti-androgen-resistant cell lines. Of additional interest was that the AR UTR variant could be detected in the sera of prostate cancer patients in a cohort of serum samples collected from patients of Gleason grades 6–10, with an increasing level correlated to increasing grade. We hypothesise that the shorter AR UTR variant is a survival adaptation to low hormone levels and/or AR-antagonist treatment in these cells, where a more stable mRNA may allow higher levels of AR expression under these conditions.
Keywords: prostate, cancer, bicalutamide, enzalutamide, anti-androgen, therapeutics
Introduction
Targeting the androgen receptor (AR) via AR-antagonist therapies is still the mainstay of advanced prostate cancer (PCa) treatments (1, 2). AR antagonists, such as enzalutamide, flutamide and bicalutamide, target and competitively bind the AR, to the exclusion of testosterone (or DHT). Additionally AR antagonists may mediate a misfolding of AR helix 12 and cause the AR to become inactive (3). In either event, the starvation of prostate cancer cells of testosterone leads to a reduction in tumour cell growth. Often patients undergoing AR-antagonist therapy can become resistant to bicalutamide via many pathways – involving AR amplification, AR ligand-binding domain (LBD) mutations, cofactor/coregulator changes, and multidrug efflux pumps. Alternatively, in high-grade prostate cancers, tumours may synthesise their own androgens from precursor molecules as a mechanism for escape from such therapies (reviewed in (4)). Often PCa cells can acquire AR LBD mutations which can change the activity of an antagonist to an agonist, and in some cases patients may acquire PCa that proliferates in the presence of AR antagonists. Such patients may respond better (initially) when AR antagonists are withdrawn (5, 6). In addition to all these processes, prostate cancer cells have been shown to produce AR splice variants – truncated versions of the AR with missing ligand activation domain which remain constitutively active for example, truncated version known as ARv7 (7).
The LNCaP cell line represents a human PCa model derived from a patient who relapsed with flutamide therapy (8). The LNCaP cell line contains an AR mutation within the LDB (T877A), which allows a greater plethora of ligands to bind and activate the AR, including flutamide. However, the cell line remains sensitive to AR antagonists such as bicalutamide and enzalutamide, but several publications report that prolonged treatment of the LNCaP cell line with AR antagonists or prolonged hormonally starved cells can produce relevant cell lines that mimic clinically acquired AR-antagonists resistance.
The VCaP cell line was established in 1997 from a lumbar vertebral bone metastasis from a metastatic lesion from a patient with hormone refractory prostate cancer. The cell line has been reported to have WT AR but overexpresses it at high levels (9).
Recently, we generated bicalutamide and enzalutamide-resistant clones of the LNCaP and VCaP cell lines, as tools for screening novel AR-antagonist compounds (10, 11). The VCaPBicR / EnzR-resistant clones showed resistance to both bicalutamide and enzalutamide – showing cytostatic effects at high doses. However, the LNCaPBicR cell lines showed a proliferative response to bicalutamide, whilst LNCaP/EnzR cells showed a more cytostatic effect at high doses.
In an attempt to discover the potential mechanism of AR-antagonist resistance and to discover if any novel AR variants were being expressed or if novel LBD mutations were present, we subjected LNCaPBicR cells to RNA-seq. Here we discuss the finding that although no new coding variant or mutations were seen, we did discover an increase in an AR variant, in the 3′UTR region. Here we investigate how the variant conveys extra stability to the AR mRNA transcript and may be in part responsible for increased AR mRNA and protein expression in response to AR-antagonist-mediated hormone starvation.
Materials and methods
Cell culture
LNCaP cells were maintained at 37°C, 5% CO2 in RPMI medium with 10% foetal bovine serum (First Link Ltd, Brierley Hill, UK). PC3, VCaP and Du145 cells were maintained in DMEM medium (Sigma) with 10% foetal bovine serum (First Link UK). All media was supplemented with 2 mM l-glutamine, 100 units/mL penicillin, 100 mg/mL streptomycin (Sigma). For hormone depletion experiments, 72 h before androgen exposure, medium was replaced with ‘starvation medium’ consisting of phenol red-free RPMI (or DMEM) medium, supplemented with 5% charcoal-stripped foetal bovine serum (First Link UK). All cells were obtained from the ATCC cell bank in 2017, and kept in liquid N2 in aliquots thereafter. All cells were used within ten passages of the original stock. RNA-seq of the samples also verified these cell lines in terms of oncogenes and documented mutations.
Treatments
All AR-antagonist compounds were obtained from Sigma, and made up in DMSO stock at 10 mM and kept at −20°C. Actinomycin-D was made up in DMSO at 100 µg/mL, and kept at −20°C. Working solution was 1 µg/mL.
Generation of bicalutamide-resistant cells
LNCaP and VCaP cells were grown in increasing doses of bicalutamide or enzalutamide (0–30 µM) for up to 6 weeks at which time cells began to show proliferation. Resistant cell lines were expanded from these and resistant clones grew stably in medium containing 20 µM AR antagonist.
MTT assays
The MTT assay was used as a cell viability assay for the all the cell lines listed using the AR-antagonist compounds. Briefly, cells (5 × 104 cells/mL) were seeded into 96-well plates (200 µL/well), allowed to attach and grow for 24 h and subsequently treated with varying concentrations of AR antagonists (0–100 μM) for 96 h. Optimal seeding densities were premeasured for linear growth over the 96 h. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, 5 mg/mL in PBS) was added to a final concentration of 0.5 mg/mL for 4 h at 37°C. After 4 h, purple formazan crystals, formed by mitochondrial reduction of MTT, were solubilized in acidified isopropanol (200 μl/well) and the absorbance was read at 570 nm after 10 min incubation. Percent inhibition of viability was calculated as a % of untreated control and the cytotoxicity/cell growth inhibition was expressed as IC50.
AR 3′UTR transfection and luciferase assays
AR UTR reporters were a kind gift from Dr Paivi Ostling (Science for Life Laboratory, Sweden). Cells were transfected with AR-UTR reporter vectors (12), and a constitutive expression Renilla (hRluc) vector (pGL4.75 Promega) alongside using Lipofectamine 3000 (Life Technologies). 24 h after transfection cells were washed and lysed in reporter lysis buffer (Promega). Lysate was mixed with d-luciferin substrate (Promega), and then with Renilla substrate (Promega). Light emission measured using the Promega Glomax multi luminometer. Luciferase activity was normalised to Renilla (hRluc) expression.
RNA extraction and RT-PCR
Total RNA samples were prepared using TRIzol reagent (Sigma) and converted to cDNA using the GoScript™ RT System (Promega).
Q-PCR
Reactions were performed in triplicate on cDNA samples in 96-well optical plates on an ABI Prism StepOne System (ThermoFisher). Reactions consisted of 2 μL cDNA, 7 μL PCR-grade water, 10 μL 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 1 μL TaqMan-specific assay probes (Applied Biosystems) for PSA, and RPL19 or SYBR green primers for AR, AR-UTR (variants), β-actin (ACTB) and GAPDH (for primer details, see Supplementary Table 2, see section on supplementary materials given at the end of this article). Parameters were 50°C for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 s and 60°C for 1 min. Data were recorded using Sequence Detector Software (SDS version 2.3; PE Applied Biosystems). Levels were normalised to GAPDH, β-actin and RPL19.
RNA-seq analysis
PolyA mRNA was isolated from total RNA using the Dynabeads mRNA DIRECT Kit (Life Technologies) and verified using a Bioanalyser-2100 (Agilent). RNA fragment libraries (150–200 bp) were generated using the Ion Total RNA-Seq kit (Life Technologies) and ligated to adapters for cDNA synthesis. cDNA was then amplified using IonXpress RNA-seq barcoded primers (5′) and quantified with a Qubit assay (Life Technologies).
cDNA libraries were clonally amplified by emulsion PCR on Ion Sphere Particles (ISP’s) using Ion PI template OT2 200 kit (Life Technologies) on an Ion OneTouch2 system (Life Technologies) as per manufacturer’s instructions. The template-positive ISP’s were recovered and enriched to remove non-template ISP’s on Ion One Touch ES (Life Technologies). The ISPs were processed using the Ion Proton 200 sequencing kit and loaded onto a P1 chip and sequenced on an Ion Proton (Life technologies) using default parameters (single-end, forward sequencing). Base calling, adaptor trimming, barcode deconvolution and alignment was performed on Torrent Suite, version 3.6 (Life Technologies) using the STAR RNA-seq aligner plugin. The Partek Genomic Suite 6.6 software was used for data analysis. The RPKM normalization method for RNA-seq 45 was used followed by a one-way ANOVA test for gene differential expression (from n = 4 samples per group). For transcript analysis, we utilised the RNA STAR aligner (or HISAT2 v2.1.0) followed by the Bowtie and TopHat splice aware mapping software (UCSC Galaxy). Finally, Cufflinks (version 2.2.1) and DESeq2 (version 1.12.4 running on R version 3.3.0) were used to build transcript information and visualised in IGV (Integrative Genomics Viewer) software (Broad Institute, University of California, USA).
For online GEO dataset analysis, we used an SRA download tool (UCSC Galaxy), followed by alignment as listed earlier, and analysis via StringTie (v1.3.6).
Serum collection
Clinical samples were obtained from the Wales Cancer Bank (WCB) in accordance with their ethics and patient consent, following application and approval (WCB application number 15/009). In total 124 samples of serum were used from patients with confirmed prostate cancer. The median patient age was 65 years and the median follow-up period was 4 years. Patient serum was collected at the point of first diagnosis based on pathological Gleason grade from biopsy, prior to any treatment. No TNM data were available. In addition we collected serum samples from normal healthy male volunteers, with an age range of 20–55 years (ethics number SMREG ref:13.69). 50 µL of serum was used and RNA was extracted using 150 µL of TRIzol LS.
Results
AR mRNA transcript and protein expression are increased with acquired AR-antagonist resistance
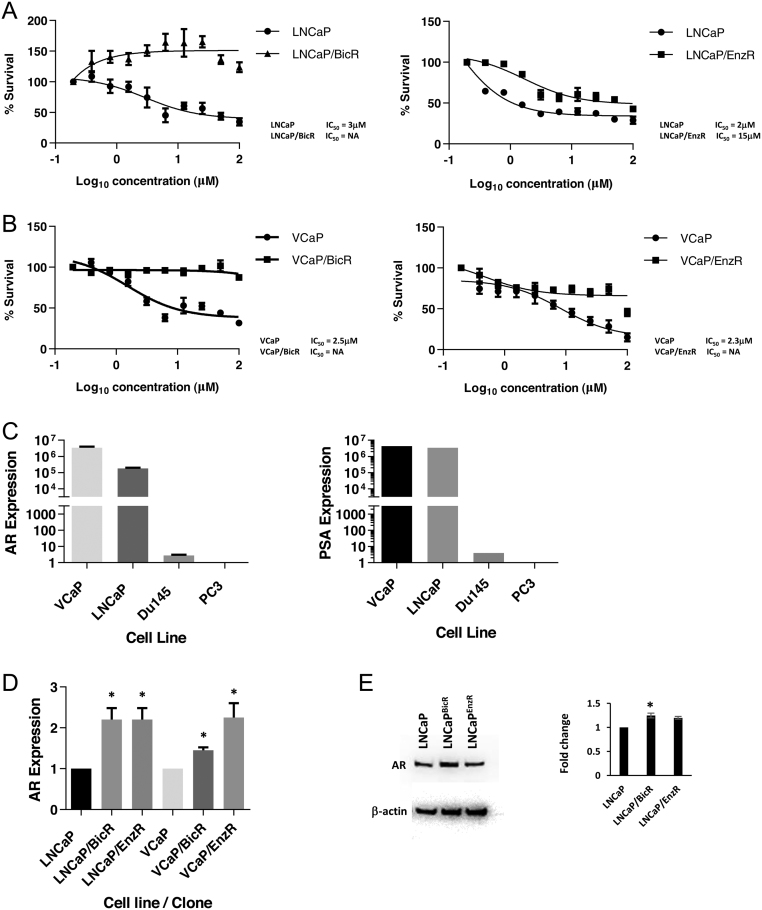
We set out to analyse and study the mechanism of action of acquired AR-antagonists resistance in prostate cancer cell models. The baseline sensitivity of LNCaP and VCaP cells to bicalutamide and enzalutamide was measured using the MTT assay for cell viability. Treating cells for 96 h in increasing doses of bicalutamide showed an IC50 of 3 µM for LNCaP cells and 2.5 µM for VCaP cells. Enzalutamide gave IC50 values of 2 and 2.3 µM respectively. After continual culture and growth in 20 µM bicalutamide or enzalutamide for 6 weeks approx., these cells began to grow and proliferate as normal, and cell lines were expanded – henceforth labelled LNCaPBicR / EnzR and VCaPBicR / EnzR. A further MTT analysis for the IC50 revealed that LNCaPBicR cells now proliferated in bicalutamide reaching maximal growth at approx. 20 µM (Fig. 1A). Enzalutamide still showed some activity in LNCaPEnzR cells, but the IC50 was now 15 µM (approx.) (Fig. 1A). In VCaP-resistant cell lines, both bicalutamide and enzalutamide failed to give an IC50 dose, but no dose-responsive proliferation was observed (Fig. 1B).
Figure 1.
AR transcript expression is increased with acquired AR-antagonists resistance. (A) MTT cytotoxicity assays of LNCaP, LNCaPBicR and LNCaPEnzR cells treated with increasing concentrations of either bicalutamide (left hand side) and enzalutamide (right hand side), at 0–100 µM for 96 h. (B) MTT cytotoxicity assays of VCaP, VCaPBicR and VCaPEnzR cells treated with increasing concentrations of either bicalutamide (left hand side) and enzalutamide (right hand side), at 0–100 µM for 96 h. (C) qPCR analysis of AR and PSA expression levels in four prostate cancer cell lines, as indicated. (D) qPCR analysis of AR expression levels in parental and resistant prostate cancer cell lines as indicated. Data represent the mean and s.e. from three independent replicates. Data are normalised to housekeeping genes GAPDH, RPL19 and β-actin. (E) Western blot for AR levels from LNCaP and LNCaP/Bic/Enz cells, normalised to β-actin. Statistical analysis * (P = 0.05), ** (P = 0.01) from a t-test.
Analysis of AR and prostate-specific antigen (PSA) expression in these cell lines confirmed that VCaP and LNCaP were indeed AR +ve when compared to PC3 and Du145 – cell lines known to be androgen receptor negative (Fig. 1C). When we compared parental and resistant cell lines we observed that AR expression was increased two-fold in the resistant cell lines (Fig. 1D) – although the levels of AR transcripts was much higher in VCaP cells (×100-fold). We PCR-amplified and sequenced the AR LBD domain from these cell lines and did not observe any AR mutations apart from T877A, carried by the LNCaP cell line. Protein levels for AR also increased in the resistant LNCaP cell lines by 1.3-fold (Fig. 1E).
AR transcripts differ in the 3′UTR region in LNCaP with acquired bicalutamide resistance
We then analysed the changes in gene expression in the LNCaP vs LNCaPBicR cell lines by RNA-seq. When analysing the AR transcripts we did not observe any AR-coding region splice variants for example, ARv7, however, we did observe that the 3′UTR region of some AR transcripts showed an anomaly. Data are available at NCBI BioProjects accession PRJNA578661 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA578661).
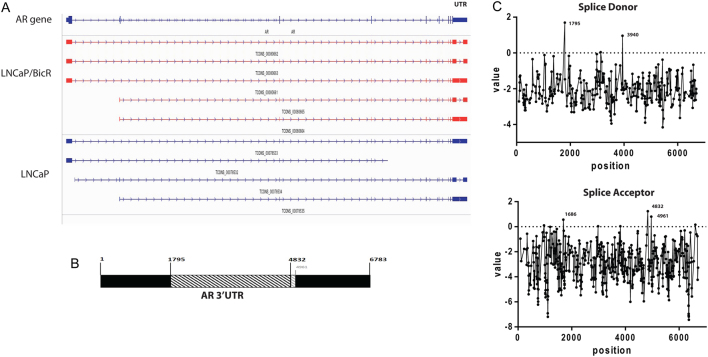
In the parental LNCaP cells, three distinct AR transcripts were identified. Two were complete matches of known transcripts (according to RefGene gene model) and one was a potentially novel transcript. However, in the LNCaPBicR cell line five distinct AR transcripts were identified, two of which were complete matches of known transcripts and three were potentially novel transcripts. Figure 2A showed the analysis schematic from IGV software, showing the Cuffmerge data output. Figure 2B showed a detailed schematic of the 3′UTR region of AR. The novel transcripts had a missing 3 kb region in the UTR sequence, from bases 1795 to 4832 of the UTR sequence. The coordinates are counted from +1 after the stop site in the AR mRNA sequence (Refseq: NM_000044) (Supplementary Results ‘AR mRNA UTR with primers’). The missing region corresponds to ChrX: position 66,945,478–66,948,515 (GRCh37/hg19 assembly). Additionally, we saw another transcript with a slightly larger missing region from bases 1795 to 4961 (Fig. 2B). The novel transcript will be henceforth referred to as AR 3.7kb-UTR, as opposed to AR 6.7kbUTR, for the normal full-length AR transcript.
Figure 2.
AR transcripts differ in the 3′UTR region with acquired AR-antagonists resistance. (A) IGV screenshot of the AR transcripts detected in RNAseq samples from LNCaP and LNCaPBicR cells. Upper panel indicated the genomic location of the AR intron and exon regions as well as the 5′ and 3′ UTR regions. (B) Schematic diagram of the full 3′ UTR region of AR (6.7 kb), with the missing 3 kb region indicated as a hatched box, and missing 3.1 kb regions (lighter hatched box). The numbers represent the bases in the UTR region at which the missing region are spanned by RNAseq reads. (C) Analysis results from the Spliceport splice analysis online tool. Left hand side indicates bases scoring highly as splice donor regions and right hand side shows bases scoring highly as splice acceptor regions.
3′UTR deletion correlates with splice sites
From the IGV analysis, reads mapping to the 3′UTR spanned the missing 3 kb region, indicating that a splicing or editing event was responsible. Therefore, we analysed the 3′UTR region of the AR mRNA via various online resources for splice site analysis. Utilising Spliceport (http://spliceport.cbcb.umd.edu/SplicingAnalyser2.html), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/) and RegRNA (http://regrna2.mbc.nctu.edu.tw/detection.html) analysis software we found that the bases 1795 and 4832 (and 4961) were predicted splice donor and acceptor regions respectively with the highest confidence value or free energy score. Figure 2B shows a schematic of the base numbers and deleted region of the 3′UTR of AR. Figure 2C shows the output from SplicePort for both donor and acceptor predicted sites. We did not detect the novel spliced region in genomic DNA from LNCaP cells, again indicating a post-transcriptional modification.
Spliced AR 3′UTR variant increases due to hormonal starvation or AR-antagonist treatment
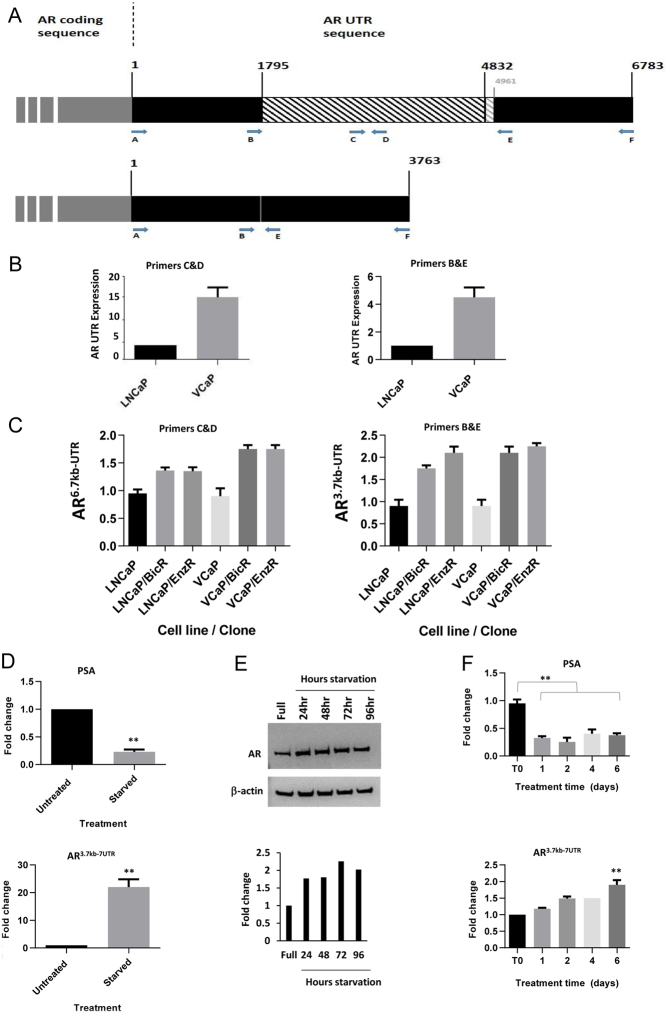
Utilising primers specific for various regions across the UTR (Fig. 3A), we analysed if the shorter UTR transcript could be detected by PCR. Utilising primers C&D for the long UTR variant and B&E for the shorter UTR variant (Fig. 3A), we carried out qPCR on the cell lines. The C&D primer set amplifies a region present only in the complete full-length AR 3′UTR. The B&E primer set only amplifies the AR 3′UTR across a small region (200 bp) of the shorter spliced variant.
Figure 3.
AR 3′UTR variant increases due to hormonal starvation or AR-antagonists treatment. (A) Schematic diagram indicating the binding location of qPCR primer sets used for detecting either a portion of the 6.7 kb 3′UTR of AR (upper panel) or the novel shorter AR 3 kb transcript (lower panel). Numbers represent base locations in the 3′UTR. (B) qPCR analysis of the AR UTR utilising the indicated primers sets in parental LNCaP or VCaP cells. (C) qPCR analysis of AR6.7kb-UTR variant and AR3.7kb-UTR variant in parental and resistant LNCaP and VCaP clones. (D) qPCR analysis of PSA (upper panel) and AR3.7kb-UTR (lower panel) in LNCaP cells grown in starvation medium for 72 h. (E) Western blot for AR protein levels in hormonally starved LNCaP cells for 0–96 h. Normalised to β-actin, with associated densitometry underneath. (F) qPCR analysis of PSA (upper panel) and AR3.7kb-UTR variant (lower panel) in LNCaP cells treated with bicalutamide (10 µM) for 6 days. Data represent the mean and s.e. from three independent replicates. Data are normalised to housekeeping genes GAPDH, RPL19 and β-actin. Statistical analysis * (P = 0.05), ** (P = 0.01) from a t-test.
Parental VCaP cells showed a 15-fold increase over LNCaP in AR-UTR expression (Fig. 3B). However, interestingly, parental VCaP cells also showed a four-fold increase in the novel shorter UTR variant than LNCaP cells (Fig. 3B). Further, we then analysed the levels of the normal UTR (primers C&D), and UTR variant (AR 3.7kb-UTR - primers B&E) in the resistant cell lines compared to the parental cell lines. This also showed increased expression of the shorter variant in all resistant cell lines (Fig. 3C), compared to their parental cells.
We then went on to study if the increase in levels of the 3′UTR variants in the parental cells changed rapidly following either hormone starvation or AR-antagonist treatment, or whether the AR variant was a product of a longer term adaptation. When LNCaP cells were hormonally starved for 72 h we saw a strong reduction in the levels of the androgen-inducible gene, PSA, as expected (Fig. 3D upper panel). Also, as expected, when starved, LNCaP cells upregulated their levels of AR mRNA, therefore, the AR-coding region and the normal UTR levels showed an increase of around 6–8 fold (Supplementary Fig. 1). However, the shorter UTR variant could be detected at approx. 20 times the level found in parental cells (Fig. 3D lower panel). Hormonal starvation for 96 h also increased the level of AR protein by wo-fold (Fig. 3E). When the parental LNCaP cells were treated with 20 µM bicalutamide for up to 6 days, we observed a decrease in PSA, as expected, but an increase in shorter UTR variant, which increased over time (Fig. 3F).
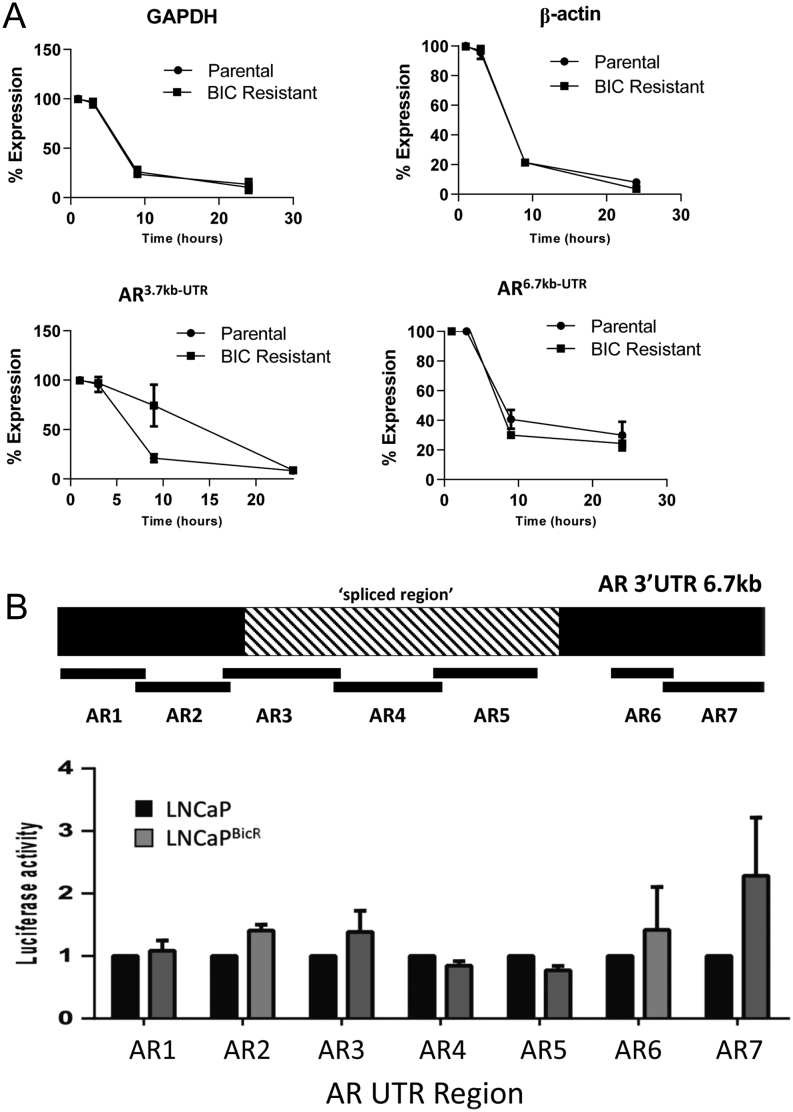
The spliced AR 3′UTR variant is more stable in cells with acquired AR-antagonist resistance
To monitor if the shorter AR UTR variant conferred an advantage to the stability of the AR mRNA strand we treated parental and AR-antagonist-resistant cells with actinomycin-D to halt RNA transcription. Total RNA was then extracted from cells at various timepoints. In LNCaP cells actinomycin-D significantly reduced expression of all transcripts over 24 h. The levels of GAPDH, β-actin and AR (coding region), AR 6.7kb-UTR did not show any significant differences between the cell lines. However, in the resistant cells the residual levels of AR 3.7kb-UTR transcript showed an increased level compared to the parental cell lines at 10 h (Fig. 4A), but was similarly reduced at 24 h. Similar results were seen in VCaP cells, but to a reduced level (Supplementary Fig. 2).
Figure 4.
Spliced AR 3′UTR variant is more stable in cells with acquired AR-antagonists resistance. (A) qPCR analysis of GAPDH, β-actin, AR3.7kb-UTR variant and AR6.7kb-UTR variant from LNCaP or LNCaPBicR cells treated with actinomycin D (1 µg/mL) for 0–24 h. Data represent the mean and s.e. from three independent replicates. Data are normalised to housekeeping genes GAPDH, RPL19 and β-actin, and then expressed as a % of expression at time 0. (B) Upper panel – schematic diagram of the location of the Luciferase-AR UTR reporter fusion constructs in the 6.7 kb region. Lower panel – luciferase activity of the reporter constructs in LNCaP or LNCaPBicR cells transfected with the firefly luciferase reporters and a constitutive renilla luciferase reporter. Data are expressed as firefly luciferase/Renilla luciferase. Statistical analysis * (P = 0.05), ** (P = 0.01) from a t-test.
We then tested the relative stability of different regions of the AR UTR using luciferase reporter constructs. Using seven AR UTR overlapping constructs (12) of approximately 1 kb each, were transfected into LNCaP or LNCaPBicR cells for 24 h, and measured luciferase activity over a constitutively expressed Renilla luciferase. Regions 1, 2, 3, 6 and 7 showed increased stability in the LNCaPBicR cells, but regions 4 and 5 which lie within the spliced region of the UTR showed a reduced stability (Fig. 4B).
The 3′UTR variant is present in online GEO datasets including LNCaP-abl cells, immortalised prostate epithelial cells and in patient samples prior to AR-antagonist therapy
Both the LNCaP and VCaP cell lines showed low levels of the AR UTR variant before the treatment with AR antagonist, but these cell lines represented cells taken from patients with advanced prostate cancer who had been treated with various endocrine therapies for example, flutamide (and/or other treatments). We, therefore, analysed several online datasets from the GEO database for the presence of the 3′UTR variant. In all studies analysed, the 3′UTR missing region was seen at some levels, albeit very low. Using online data from Olsen et al., GSE71797 (13), in hTert immortalised benign prostate epithelial cells we detected the presence of the transcript in some samples but not all (Supplementary Fig. 3A). Additionally, using data from Rajan et al. (GSE48403) (14), we again found the presence of the shorter AR UTR variant. The 3′UTR variant was seen to be elevated in prostate cancer patients post AR-antagonists therapy, but due to low sample numbers (n = 6), the data did not reach significance. Data are given in Supplementary Fig. 3B.
The AR UTR was also detected and was increased (P = 0.03) in a study by Knuuttila et al. (GSE95413) in orthotopically grown castration resistant VCaP xenografts treated with enzalutamide (Supplementary Fig. 4A). In a study by Shah et al. (GSE81796) using C4-2 cells (CRPC) the AR 3.7kb-UTR was present and was increased by enzalutamide treatment (Supplementary Fig. 4B). And in a study by Coleman et al. (GSE87153), the AR 3.7kb-UTR was seen to be increased in the LNCaP-derived MR49F cell line – grown in castrated mice and enzalutamide treated (Supplementary Fig. 4C). The variant was present in V16D cell line (LNCaP derived, castration resistant), but was not further elevated with enzalutamide. This missing region in the AR UTR was also observed in a study by Kohli et al. (GSE70380), in samples derived from metastatic deposits from a patient with disease progression whilst on androgen deprivation therapy (Supplementary Fig. 4D).
The LNCaP-abl cell line represents LNCaP cells grown in androgen-depleted medium for over 80 passages, and mimics castrate-resistant prostate cancer. A GEO dataset (GSE114708) utilising the LNCaP-abl cell line (15) showed the presence of the AR3.7kb-UTR variant (Supplementary Fig. 4E).
The AR 3′UTR variant is detectable in prostate cancer patient serum
Since prostate cancer cells and/or prostate cancer cellular DNA/RNA may circulate in the serum of prostate cancer patients, we evaluated whether the novel variant could be detected in their sera. We extracted total RNA from prostate cancer patients’ sera, collected from the Wales Cancer Bank (n = 124) ranging from Gleason grade 6–10, with associated PSA values. Additionally, serum was collected from apparently healthy male volunteers (n = 8). These were determined by clinical pathologists at the Heath Hospital Cardiff, UK. RNA was isolated and qPCR performed, for AR (coding region), AR 6.7kb-UTR (primers C&D), and for the AR3.7kb-UTR (primers B&E), along with GAPDH, β-actin (ACTB) and RPL19. Patient data and numbers can be seen in Supplementary Table 1.
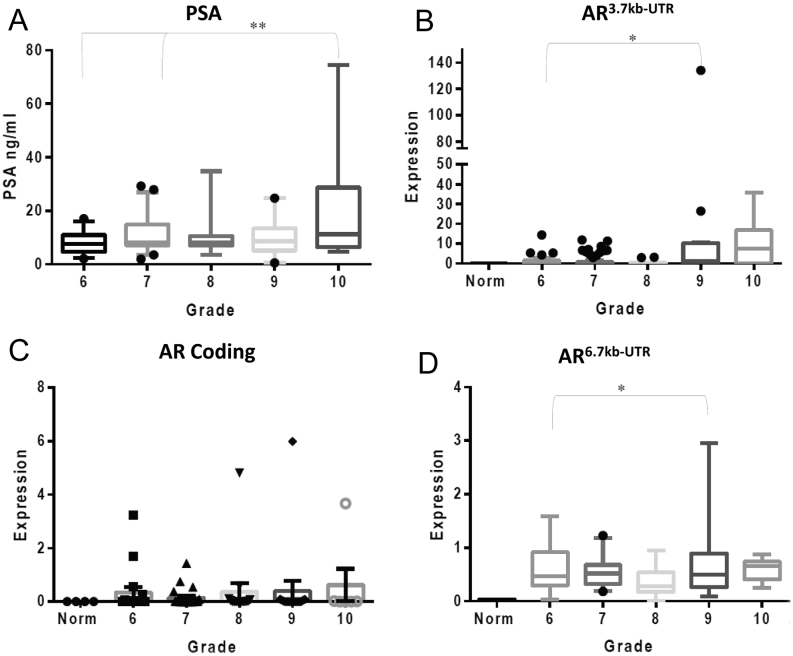
When analysed according to Gleason grade, we found a positive correlation with PSA as expected, but additionally we saw a positive correlation with increasing grade and the quantity of AR 3.7kb-UTR variant in the serum (Fig. 5A and B), and statistically significant differences between Gleason grade 6 and 9 (P ≤ 0.05). No correlation was seen between the levels of AR-coding region and Gleason grade; however, a positive correlation with grade was also seen for the full-length AR 6.7kb-UTR (Fig. 5C and D).
Figure 5.
Spliced AR 3′UTR variant is detectable in prostate cancer patient serum. (A) Serum PSA measurements from the prostate cancer patient cohort – data from the Wales Cancer Bank. Data are separated according to Gleason grade 6–10, assigned to each patient at pathological diagnosis. (B) qPCR analysis of the AR3.7kb-UTR variant from RNA extracted from patient serum samples. (C) qPCR analysis of the AR-coding region from RNA extracted from patient serum samples. (D) qPCR analysis of the AR6.7kb-UTR variant from RNA extracted from patient serum samples. Data represent the mean and s.e. from three independent measurements. Data are normalised to housekeeping genes GAPDH, RPL19 and β-actin. Statistical analysis * (P = 0.05), ** (P = 0.01) from a one-way ANOVA test (Kruskal–Wallis test).
When analysed for patient follow-up and survival, both PSA and AR3.7kb-UTR were elevated in the deceased group (P ≤ 0.05 and P ≤ 0.02) respectively. However, due to very small numbers we treat these results with caution. AR6.7kb-UTR and AR-coding region levels did not correlate with survival.
Discussion
During prostate cancer treatment AR antagonists are often used to reduce tumour growth and prolong life. Initially, AR antagonists are effective but frequently these become ineffective and patients relapse with prostate cancer which is resistant to further AR-antagonist treatment. Additionally, AR antagonists may bind and activate the AR causing a stimulatory effect. In instances such as these, prostate cancer patients may benefit from AR-antagonist withdrawal. We set out to investigate the mechanism behind acquired resistance to AR antagonist in prostate cancer cells by generating cells resistant to the two most clinically relevant AR antagonists, namely bicalutamide and enzalutamide.
For LNCaP and VCaP cells, the acquisition of AR-antagonist resistance was established at 20 µM, whereas at 30 µM no growth or very slow cell growth was observed. However, when these resistant cells were then exposed to a range of drug concentrations 0–100 µM we saw different effects dependent on the drug and cell line. VCaP showed resistance to enzalutamide and bicalutamide with no apparent cell killing up to 50–100 mM. LNCaP showed similar properties with enzalutamide. However, LNCaP cells showed a proliferation response with bicalutamide, peaking at 20 µM. This effect is probably due to the steric fit of bicalutamide in the mutant AR ligand-binding domain (T877A) carried by the LNCaP cell line causing a more promiscuous ligand binding. This may also mimic the clinical situation where patients sometimes respond better – albeit briefly – when AR antagonists are removed.
When analysing AR mRNA levels in these resistant cells, we found expression to be higher at the mRNA and protein levels. The increased expression of AR has been previously reported in cells either starved of hormone or inhibited by AR antagonists. VCaP cells show a much higher baseline expression of AR and has been reported previously, but even in these cells AR expression was still higher in the resistant clones.
When we analysed the AR by RNA-seq we did not discover any new or additional mutations in any of these cells lines, neither did we find any novel AR coding variants for example, ARv7. However, the RNA-seq and associated Cufflinks analysis did reveal an increase in transcript number and variation. Three isoforms were seen in parental LNCaP cells. The first being AR variant 1 (refseq: NM_000044) and the other being AR variant 2 (refseq: NM_001011546), which are previously identified variants. Variant 2 has an alternate start codon in the 5′ region, with an encoded isoform (variant 2) which has a distinct and shorter N-terminus than isoform 1. However, even in these parental cells another isoform was seen with a shorter 5′ UTR sequence and a missing 3 kb region from the 3′UTR. In the LNCaPBicR cell line five transcripts were seen – the three variants seen previously in the parental cell lines and two more variants which were the AR variant 2 but with a missing 3 kb region in the 3′UTR and an alternate variant 1 with a missing slightly larger 3.1 kb region.
Since RNAseq reads spanned this area and were aligned and mapped as genuine reads to the AR gene, we had to surmise that the missing 3–3.1 kb were caused by ‘splicing’ or ‘stitching’ events. Splice site analysis matched up precisely with donor and acceptor sites on several Web-based platforms. The novel variant region could not be PCR amplified from genomic DNA, also indicating a post-transcriptional splicing event.
Although AR coding and AR UTR levels were elevated in AR-antagonists-resistant cells, PCR analysis confirmed that the missing UTR variant elevated in increasing amounts, indicating that additional AR transcripts with the missing region were present in these cells. Even though the high levels of AR 3.7kb-UTR were seen in fully resistant cells, we saw that the adaptive response was rapid, being detectable after 24 h of AR-antagonist treatment and after 6 days of hormone starvation.
Additionally, to test whether the AR ‘splicing’ event was unique to AR pathway inhibition or was a more general stress response, we subjected LNCaP cells to a variety of cellular stresses including hypoxia (mimicked by CoCl2), DNA damaging stress (by doxorubicin, cisplatin and paclitaxel) and environmental stress such as oxygen radicals (by H2O2) and extracellular low pH. We observed that both AR variants (3.7 and 6.7 kb UTRs) were upregulated in response to environmental stresses for example, oxygen or pH, but were not significantly altered in response to DNA-damaging agents (Supplementary Fig. 5). All stress treatments reduced cellular proliferation as measured by the expression of the MCM5 DNA replication gene. Oxygen level changes and low pH are significant factors within tumours, and may together with AR antagonism, play a role in prostate cancer. The ‘splicing’ events observed here may be part of a more generic stress-induced novel or aberrant splicing pathway.
We hypothesised that the AR variant was a result of an adaptive response to lack of androgen where the AR mRNA levels increased, and as is the case for alternative ‘splicing’, the mRNA strand itself may have an altered stability. Splicing of the coding region of the AR has been studied extensively, but UTR variants have not (7, 16). For example if the shorter UTR variant was more stable, then this would give the AR mRNA a longer half-life, and a higher expression which would give cells a survival advantage. We tested this hypothesis using actinomycin-D to stop cellular transcription of all transcripts and measured the half-life of the remaining transcripts. 24 h of actinomycin treatment reduced all transcripts measured including GAPDH, β-actin (ACTB), AR 6.7kb-UTR and AR 3.7kb-UTR. However, the AR 3.7kb-UTR showed a higher level and reduced rate of degradation. Additional analysis using AR UTR reporters fused to luciferase showed that two regions (4 and 5) showed a reduced activity/stability in the LNCaPBicR cell line, whereas all other regions showed an increased activity. Regions 4 and 5 were in the missing 3 kb region of the novel transcripts. Taken together, we hypothesise that in the adaptation to AR-antagonist resistance the 3 kb RNA region missing in AR 3.7kb-UTR may be detrimental for stability of the mRNA strand and may be spliced out to increase stability as a survival advantage for the cancer cells. Although a hypothetical explanation at this stage, we would suggest that this splicing or stitching of the UTR sequence may be a mechanism to avoid or circumvent microRNA or noncoding RNA downregulation of the AR strand. miR analysis software indicated at least 40 potential binding sites in the 3 kb were missing from the AR3.7kb-UTR mRNA strand. For example, if miRNA upregulation was a part of the adaptive AR-antagonist resistance response, then the shortening of the UTR would lessen the chance of AR itself being targeted by these miRs. Studies by Ostling et al. (12) found several active miRNAs which target the 3′UTR of AR in the deleted region for example, miR-185, miR-371-3p and miR-135. miR-185 has been shown to downregulate AR levels and is itself downregulated in prostate cancer samples (17, 18).
Splicing dysregulation has been hypothesised to be associated with cancer and survival mechanisms for example, in PI3kinase/Akt pathways (19, 20). 3′UTR splicing events have been found for other genes and is associated with transcript stability (21, 22, 23, 24). Often cells utilise alternative cleavage and polyadenylation sites to reduce UTR length and increase mRNA stability (25), but in the enzalutamide-resistant cells studied here, although the UTR length is indeed shortened the terminal polyadenylation site is unchanged. This would again imply a novel mRNA ‘splicing’ and editing event has occurred for the AR.
The AR 3.7kb-UTR variant was present in the parental LNCaP and VCaP cell lines, but was more prominent in the resistant cells. We then looked at several online GEO dataset to determine if the variant was common or cancer specific. In immortalised (hTERT) prostate epithelial cells and in epithelial cells undergoing mesenchymal transition, the variant was present but at very low levels; however, in cancer samples and cancer samples from AR-antagonist-resistant refractory patients we found the levels to be higher, with even higher levels in refractory patients – although not significant due to low numbers (P = 0.3). Therefore, the variant may be a naturally occurring transcript present at low levels, which is stabilised or upregulated at times of cellular stress.
Patients’ plasma and serum have been used in the other studies to measure circulating AR levels (26, 27). In prostate cancer patients’ sera, we tested whether the AR variant could be detected, as well as the normal AR UTR and the AR-coding region. Additionally, we compared the patient Gleason grade and PSA levels as indicators of disease. All three AR regions were detectable, but it was only the UTR regions that correlated with grade across the patients’ samples, and showed significant differences between grades 6 and 9 and 10. Additionally, both PSA and AR 3.7kb-UTR showed a significantly higher level in those patients who had died of prostate cancer in the interim follow-up. This was a surprising finding as numbers were low, but no significant correlation could be seen between PSA and AR 3.7kb-UTR. To our knowledge, these patients were naive to any endocrine treatment protocols, therefore taken together with the online GEO datasets, the appearance of the variant is not caused by AR-antagonist treatments but is upregulated by it. Unfortunately, follow-up data on how patients went on to respond to AR-antagonist therapies were unavailable at the time of this study. Previously, the detection of AR-v7 in whole blood did not predict effectiveness of AR-antagonist for castration-resistant prostate cancer (27).
In conclusion, we report that an AR 3.7kb-UTR variant levels increase in prostate cancer cells which have acquired resistance to bicalutamide/enzalutamide or indeed shorter-term hormonal starvation. This variant is present at low levels in most cells studied, but the levels are increased in the resistant cells, and the novel variant may be associated with a splicing event. We hypothesise that this variant increases AR mRNA stability, thus may increase protein levels and may provide prostate cancer cells a survival advantage.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector.
Acknowledgements
The authors would like to thank our funders – The Cardiff University – Peking University Cancer Institute and Cancer Research UK. Tissue (serum) samples were obtained from the Wales Cancer Bank which is funded by the Wales Assembly Government and Cancer Research Wales. Other investigators may have received specimens from the same subjects.
References
- 1.Brinkmann AO, Trapman J. Prostate cancer schemes for androgen escape. Nature Medicine 2000. 628–629. ( 10.1038/76194) [DOI] [PubMed] [Google Scholar]
- 2.Brinkmann AO. Molecular mechanisms of androgen action – a historical perspective. Methods in Molecular Biology 2011. 3–24. ( 10.1007/978-1-61779-243-4_1) [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto Y, Miyachi H. Nuclear receptor antagonists designed based on the helix-folding inhibition hypothesis. Bioorganic and Medicinal Chemistry 2005. 5080–5093. ( 10.1016/j.bmc.2005.03.027) [DOI] [PubMed] [Google Scholar]
- 4.Penning TM. Androgen biosynthesis in castration-resistant prostate cancer. Endocrine-Related Cancer 2014. T67–T78. ( 10.1530/ERC-14-0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hara T, Miyazaki J, Araki H, Yamaoka M, Kanzaki N, Kusaka M, Miyamoto M. Novel mutations of androgen receptor: a possible mechanism of bicalutamide withdrawal syndrome. Cancer Research 2003. 149–153. [PubMed] [Google Scholar]
- 6.Brooke GN, Bevan CL. The role of androgen receptor mutations in prostate cancer progression. Current Genomics 2009. 18–25. ( 10.2174/138920209787581307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. New England Journal of Medicine 2014. 1028–1038. ( 10.1056/NEJMoa1315815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, Mirand EA, Murphy GP. LNCaP model of human prostatic carcinoma. Cancer Research 1983. 1809–1818. [PubMed] [Google Scholar]
- 9.Korenchuk S, Lehr JE, MClean L, Lee YG, Whitney S, Vessella R, Lin DL, Pienta KJ. VCaP, a cell-based model system of human prostate cancer. In Vivo 2001. 163–168. [PubMed] [Google Scholar]
- 10.Dart DA, Kandil S, Tommasini-Ghelfi S, Serrano de Almeida G, Bevan CL, Jiang W, Westwell AD. Novel trifluoromethylated Enobosarm analogues with potent anti-androgenic activity in vitro and tissue selectivity in vivo. Molecular Cancer Therapeutics 2018. 17 1846–1858. ( 10.1158/1535-7163.MCT-18-0037) [DOI] [PubMed] [Google Scholar]
- 11.Koushyar S, Economides G, Zaat S, Jiang W, Bevan CL, Dart DA. The prohibitin-repressive interaction with E2F1 is rapidly inhibited by androgen signalling in prostate cancer cells. Oncogenesis 2017. e333 ( 10.1038/oncsis.2017.32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Östling P, Leivonen SK, Aakula A, Kohonen P, Mäkelä R, Hagman Z, Edsjö A, Kangaspeska S, Edgren H, Nicorici D, et al Systematic analysis of microRNAs targeting the androgen receptor in prostate cancer cells. Cancer Research 2011. 1956–1967. ( 10.1158/0008-5472.CAN-10-2421) [DOI] [PubMed] [Google Scholar]
- 13.Olsen JR, Azeem W, Hellem MR, Marvyin K, Hua Y, Qu Y, Li L, Lin B, Ke X, Øyan AM, et al Context dependent regulatory patterns of the androgen receptor and androgen receptor target genes. BMC Cancer 2016. 377 ( 10.1186/s12885-016-2453-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajan P, Sudbery IM, Villasevil ME, Mui E, Fleming J, Davis M, Ahmad I, Edwards J, Sansom OJ, Sims D, et al Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. European Urology 2014. 32–39. ( 10.1016/j.eururo.2013.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bader DA, Hartig SM, Putluri V, Foley C, Hamilton MP, Smith EA, Saha PK, Panigrahi A, Walker C, Zong L, et al Mitochondrial pyruvate import is a metabolic vulnerability in androgen receptor-driven prostate cancer. Nature Metabolism 2019. 70–85. ( 10.1038/s42255-018-0002-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho Y, Dehm SM. Androgen receptor rearrangement and splicing variants in resistance to endocrine therapies in prostate cancer. Endocrinology 2017. 1533–1542. ( 10.1210/en.2017-00109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu F, Cui X, Hong Y, Wang J, Li Y, Chen L, Liu Y, Gao Y & Xu D. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Molecular and Cellular Biochemistry 2013 377 121–130. ( 10.1007/s11010-013-1576-z) [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Chen Z, Hu X, Wang L, Li C, Xue J, Zhang P, Chen W & Jiang A. MicroRNA-185 downregulates androgen receptor expression in the LNCaP prostate carcinoma cell line. Molecular Medicine Reports 2015 11 4625–4632. ( 10.3892/mmr.2015.3332) [DOI] [PubMed] [Google Scholar]
- 19.Graham JR, Hendershott MC, Terragni J, Cooper GM. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Molecular and Cellular Biology 2010. 5295–5305. ( 10.1128/MCB.00303-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.da Silva MR, Moreira GA, Gonçalves da Silva RA, de Almeida Alves Barbosa É, Pais Siqueira R, Teixera RR, Almeida MR, Silva Júnior A, Fietto JL, Bressan GC. Splicing regulators and their roles in cancer biology and therapy. BioMed Research International 2015. 150514 ( 10.1155/2015/150514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaagura M, Taal K, Koppel I, Tuvikene J, Timmusk T, Tamme R. Rat NEURL1 3′UTR is alternatively spliced and targets mRNA to dendrites. Neuroscience Letters 2016. 71–76. ( 10.1016/j.neulet.2016.10.041) [DOI] [PubMed] [Google Scholar]
- 22.Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3′untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biology 2012. 563–576. ( 10.4161/rna.20231) [DOI] [PubMed] [Google Scholar]
- 23.Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends in Cell Biology 2009. 465–474. ( 10.1016/j.tcb.2009.06.001) [DOI] [PubMed] [Google Scholar]
- 24.Yokoi S, Udagawa T, Fujioka Y, Honda D, Okado H, Watanabe H, Katsuno M, Ishigaki S, Sobue G. 3′UTR length-dependent control of SynGAP isoform α2 mRNA by FUS and ELAV-like proteins promotes dendritic spine maturation and cognitive function. Cell Reports 2017. 3071–3084. ( 10.1016/j.celrep.2017.08.100) [DOI] [PubMed] [Google Scholar]
- 25.Mayr C & Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009 138 673–684. ( 10.1016/j.cell.2009.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanel A, Gasi Tandefelt D, Conteduca V, Jayaram A, Casiraghi N, Wetterskog D, Salvi S, Amadori D, Zafeiriou Z, Rescigno P, et al Plasma AR and abiraterone-resistant prostate cancer. Science Translational Medicine 2015. 312re10 ( 10.1126/scitranslmed.aac9511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi T, Okuno Y, Hattori-Kato M, Zaitsu M, Mikami K. Detection of AR-V7 mRNA in whole blood may not predict the effectiveness of novel endocrine drugs for castration-resistant prostate cancer. Research and Reports in Urology 2016. 21–25. ( 10.2147/RRU.S98877) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a