Abstract
Embryo assessment and selection is a critical step in an In-vitro fertilization (IVF) procedure. Current embryo assessment approaches such as manual microscopic analysis done by embryologists or semi-automated time-lapse imaging systems are highly subjective, time-consuming, or expensive. Availability of cost-effective and easy-to-use hardware and software for embryo image data acquisition and analysis can significantly empower embryologists towards more efficient clinical decisions both in resource-limited and resource-rich settings. Here, we report the development of two inexpensive (<$100 and <$5) and automated imaging platforms that utilize advances in artificial intelligence (AI) for rapid, reliable, and accurate evaluations of embryo morphological qualities. Using a layered learning approach, we have shown that network models pre-trained with high quality embryo image data can be re-trained using data recorded on such low-cost, portable optical systems for embryo assessment and classification when relatively low-resolution image data is used. Using two test sets of 272 and 319 embryo images recorded on the reported stand-alone and smartphone optical systems, we were able to classify embryos based on their cell morphology with >90% accuracy.
Introduction
Over 48 million couples are affected by infertility globally, making it one of the most underestimated problems in world 1, 2. Globally, in-vitro fertilization (IVF) has provided an expensive solution, costing tens of thousands of dollars, for a significant number of infertile couples 3–5. IVF involves cycles of culturing patient embryos, in-vitro, for a few days and transferring the best quality embryos back to the patient. The inefficient process has an average success rate of 30% and patients usually require multiple cycles for a positive outcome 5. Embryo selection based on quality is a critical factor that influences the eventual clinical outcome.
Traditionally, embryos are manually assessed by embryologists visually, which is a highly subjective procedure 6, 7. Time-lapse imaging (TLI) systems have enabled automated and uninterrupted continuous imaging of embryos over the 3 to 6 days of embryo development, however, studies have suggested that TLI systems may also be subjective and time-consuming due to input requirements from embryologists 8, 9. Furthermore, these systems are bulky and prohibitively expensive which limit the accessibility even to fertility centers in the developed nations 10. Numerous alternative methods of embryo analysis that involve phenotypical and genotypical analyses have also been available to embryologists, but owing to factors such as their lack of cost-effectiveness and clinical validation, traditional methods of visual assessments at specific time-points are still preferred by most clinical practices 11, 12. Therefore, manual morphological assessments using static images, regardless of the subjectivity, is the most widely used method of embryo analysis due to simplicity and cost-effectiveness 13.
Embryos are usually transferred to a patient’s uterus during either the cleavage or the blastocyst stage of development. Embryos reach the blastocyst stage 5–6 days after fertilization. Blastocysts have fluid filled cavities and two distinguishable cell types, the trophectoderm (TE) and the inner cell mass (ICM). Blastocysts are generally selected for transfer based on the expansion of the blastocoel cavity and the quality of the TE and ICM 14.
Most of the previously developed systems make use of traditional rule-based computer vision algorithms and are limited strictly by controlled imaging systems. These algorithms are also computationally intensive requiring additional expensive computing hardware and space. Deep neural networks on the other hand provide a suitable alternative to traditional computer vision-based approaches and have shown great potential in biomedical and diagnostic applications 15–17. Recently, deep learning-based approaches have been extensively explored in evaluating embryos using data acquired from time-lapse systems and have shown promise in automating embryo analysis 18–28. Unlike all prior computer-aided algorithms used for embryo assessment, the deep convolutional neural networks (CNN) allow for automated embryo feature selection and analysis at the pixel level without any interference by an embryologist. However, these networks are still limited to the data acquired using bulky and expensive (>$100,000 USD in most cases) imaging systems, which is not available to the majority of fertility clinics, even in resource-rich settings, where over 80% of clinics do not have access to such systems 12.
The high cost of such systems is one of the main reasons for its low penetration in fertility centres. Deep learning approaches, in general, are specific to the data domain that they are trained on and do not perform well on shifted domains, let alone on data with loss in quality 29. Therefore, deep learning networks developed using high-quality data cannot be directly adapted to inexpensive, portable optical systems. CNNs, generally require large annotated datasets of a target domain for effective learning. However, generation of such datasets is inherently difficult, let alone for medical devices and newer inexpensive, portable optical hardware with relatively low-resultion image data 30, 31. The development of an automated, low-cost system for embryo assessments is, thus, an unmet challenge.
Here, we report the development and validation of two portable, low-cost optical systems for human embryo assessment using a neural network developed through layered transfer learning technique (Fig.1A and B). We employed a layered learning scheme to achieve the best performance with our inexpensive lossy imaging system using a relatively small dataset (Fig S1). We first trained the algorithm, pre-trained on ImageNet weights, using a dataset of over 2,450 embryos imaged using a commercially available time-lapse imaging system. We then retrained the algorithm using embryo images recorded with our portable optical devices. The performance of the developed system systems in embryo classifications was similar to the performance of highly trained embryologists as well as CNNs trained and tested using high-quality clinical time lapse embryo image data.
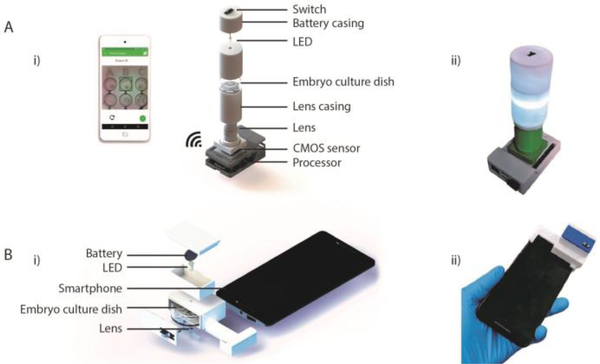
Figure 1. The AI-empowered portable imaging systems for embryo assessment.
(A) Stand-alone optical system for embryo assessment. (i) Schematic of exploded image of the stand-alone optical system and its various components. The system is wirelessly controlled using a smartphone to image embryos on a standard embryo culture dish. (ii) The actual image of the fabricated stand-alone optical system. (B) The optical accessory for embryo imaging and analysis on-phone. (i) Schematics of exploded image of the smartphone-based imaging system and its various components. (ii) The actual image of the fabricated smartphone-based optical system for embryo assessment. Both systems can acquire embryo images using standard embryo culture dishes.
Materials and Methods
Stand-alone optical system
The optical housing of the stand-alone system was designed using SolidWorks (Dassault Systèmes) and 3D-printed using an Ultimaker 2 Extended (Ultimaker) with polylactic acid (PLA). The overall dimensions of the system were 62 × 92 × 175 mm. The core system itself contained an electronic circuit with a white light-emitting diode (LED) (Chanzon HNPC-59042), a 3 V battery (Panasonic CR1620), and a single pole double-throw switch (Teenitor SS-12D00G3). The embryo sample was transilluminated as seen in Figure 1. The stand-alone optical system used a 10× Plan-Achromatic objective (Amscope PA10XK-V300) for image magnification and a complementary metal–oxide–semiconductor (CMOS) image sensor (Sony IMX219) for embryo image data acquisition. The CMOS sensor was connected to an inexpensive single-board computer (SBC; Raspberry Pi 3), which processed the captured images through a custom script (Fig. S2). In addition, the system was connected to a smartphone device via WIFI for system operation and data visualization (Fig. 1, S3). The captured images were stored on cloud for post verification and analysis. The total cost of the system was ~$85, which includes $5 for PLA, ~$30 for the objective, $0.4 for the battery, $0.1 for the LED, $19 for the CMOS, and ~$30 for the SBC and the switches and wires (Table S1). The application connected to the SBC in the stand-alone system wirelessly when recording images (Fig. S2). Furthermore, we developed a smartphone application which was running on Android, to connect and interface with the optical system.
Smartphone-based optical system
A low-cost, portable smartphone-based optical system was also developed for embryo assessment. The smartphone attachment was designed using Solidworks and printed using an Ultimaker 2 Extended 3D printer with PLA (Fig 1). The dimensions of the device were 82 × 34 × 48 mm. The acrylic lens used in this system was extracted from a DVD drive. The total material cost of the system was ~$3, which includes an estimated $1 for PLA, ~$1 for lens, $0.4 for the battery, and $0.1 for the LED and the switches and wires (Table S1). For this study, a Moto X Pure (XT1575) was used along with the attachment to collect embryo images.
Data collection and structure
Embryo images were collected for research after institutional review board approval by the Massachusetts General Hospital Institutional Review Board (IRB#2017P001339 and IRB#2019P002392). All of the experiments were performed in compliance with the relevant laws and institutional guidelines of Massachusetts General Hospital, Brigham and Women’s Hospital, and Partners Healthcare. Embryos donated at the Massachusetts General Hospital’s Fertility Center for research were imaged using the smartphone and stand-alone imaging systems. Only cells designated for discard or for research purposes were used in this study such as unfertilized oocytes (those that were designated to be discarded due to being unfertilized by 16–18 hours post insemination), abnormally fertilized oocytes (those that do not have 2 pronuclei by 16–18 hours post insemination), and embryos that arrest during pre-implantation development and those that are not selected for embryo transfer or cryopreservation. We have also included oocytes and embryos that have been frozen and consented to be discarded specifically for research purposes. A total of 157 and 385 embryo images were collected using the stand-alone and smartphone-based optical systems, respectively. The embryos were visually viewed under a standard desktop microscope by an embryologist for quality assessment and annotation. Embryos with normal fertilization were evaluated based on their morphology at the blastocyst stage. At the blastocyst stage, embryos are conventionally graded through 83 classes of blastocysts based on the combinations of (i) the degree of blastocoel expansion (grades 1–6), (ii) inner cell mass quality (grades 1–4), and (iii) trophectoderm quality (grades 1–4) along with 3 classes of non-blastocysts (Table S2). For the CNN classification algorithm, the grading system was simplified to encompass all 86 classes within a 2-level hierarchy of training and inference classes (Fig S4, Table S2). The non-blastocysts category included the training classes 1 and 2, and blastocysts category included the training classes 3, 4, and 5. The embryos were annotated under the 5-class categorization system and trained for the 5 classes. However, the inference was at a 2-class level (blastocyst and non-blastocysts) that is more universal, since these clinical blastocyst grading systems are center-specific and do apply to all clinics.
We used 1,968 embryo images including augmented data (flips and rotations), collected on the stand-alone optical device, along with 2,450 Embryoscope images in training the network developed for the stand-alone device (Fig S1 and S4). We also used 1,056 embryo images including augmented data (flips and rotations), collected on the smartphone optical device, along with 2,450 Embryoscope images in training the network developed for the smartphone device (Fig S1 and S4). We performed flips and 90° rotations to the images prior to training and testing, and the data-generator dynamically performed random rotations at every batch during data balancing procedures. Augmentation through brightness shifts was not used since our portable imaging systems collect data in controlled lighting conditions. Other morphological augmentations such as zoom and stretching were not used because changes to the morphology of the embryo may affect the overall assessment of embryos. The networks were evaluated using independent tests of containing 272 and 319 embryo images collected using the stand-alone and smartphone imaging systems, respectively (Fig S1).
Layered network training
We used Xception as the base network architecture to evaluate embryo images 32. For the networks designed to evaluate embryo images collected on the portable systems, we added a Dense classification layer (5×1) followed by Softmax activation layer (5×1) to Xception’s bottleneck section, which results in an addition of 10,245 trainable parameters to the network. The model was pre-trained with ImageNet followed by Embryoscope data using embryo images recorded 113 hours post-insemination (hpi) 18. Transfer learning is a process of transferring knowledge from a trained model to another in order to perform a similar task. A model trained on ImageNet has learned features associated over 1,000 categories of non-medical images. We optimized this model to work with high quality embryo images for the task of embryo classification. We further optimized the network to domain-shifted data using images from the portable systems. The advantages of using transfer learning from ImageNet and higher-quality clinical embryo images include, lower training time and possibly lower data requirements during the subsequent optimization procedures. Both networks trained for this study were trained using stochastic gradient descent optimization with a decay of 0.9. The network training was performed within the Keras framework on Tensorflow on a NVidia 1080Ti GPU. The network was trained with a batch size set at 64. For each batch the network maintained an overall equal distribution of images across all classes. The network’s learning rate was updated with a time-based decay at the end of each epoch. The models for the stand-alone and smartphone datasets trained with an initial learning rate of 0.00075 and 0.005, respectively. Training performance was evaluated by monitoring the cross-entropy loss. An early stoppage scheme was employed to avoid overfitting, where only the model that achieves the lowest validation cross-entropy loss was saved. The total number of epochs for training was set at 150.
For Embryoscope training and evaluations, Xception pre-trained on ImageNet, was trained and optimized similar to previous reports 18, 20–26. The 2,450 embryo images were split into 1,708 for training and 742 for testing (Fig S1). The best performing model, identified through the lowest validation loss, was tested with the 742 test set which resulted in 2-class accuracy of 90.97%. We used this as the clinical performance baseline to compare the two networks developed for the portable systems.
Data visualization techniques
Saliency maps and t-distribution stochastic neighbor embedding (t-SNE) were utilized in visualizing our data. We have also utilized a dot map to visualize true and false predictions more easily. The activations from the activation layer following the final fully connected convolution layer were used in combination with Smoothgrad to generate our saliency maps. Both the network trained with smartphone images and with stand-alone images were used to generate the respective saliency maps. Similarly, t-SNE plots were generated to visualize separability in a 2D space after learning for both datasets. For the sake of computational speed, 2,048 dimensions were reduced to 50 using a principal component analysis (PCA), following which t-SNE reduced the 50 dimensions to 2 for visualized plots 33.
Results
Hardware characterization
The smartphone and the stand-alone optical systems were designed to be simple and easy-to-use requiring minimal number of parts for operation (Fig. 1). Furthermore, these systems were designed as inexpensive alternatives (over x1000 cheaper). It was important to confirm the suitability of image quality prior to testing the portable systems with human embryos. The smartphone optical setup contained a plano-convex lens of 9 mm diameter with a focal length of 13.5 mm for magnification. We used a 10x standard objective in the stand-alone optical system. Resolution of the images was calculated by imaging an USAF 1951 resolution target and effective digital margination was measured using 1 mm stage micrometer. Each micron was represented by approximately 25 pixels in the images collected by the stand-alone device with a resolvable limit of 0.78 microns (Fig. S5). Similarly, the smartphone system was able to acquire images where each micron was represented by 2 pixels with a resolvable limit of 1.74 microns (Fig. S5). Images of embryos collected using these systems, therefore, possessed a significant shift in quality relative to the clinical Embryoscope systems (Fig S4).
Network training results
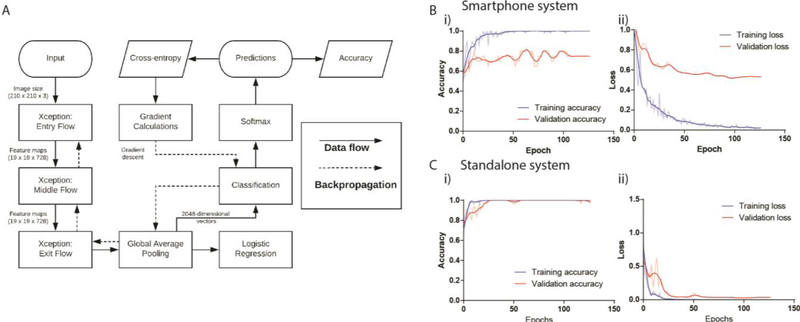
Depending on the complexity of the problem of interest, CNNs generally require large amounts of image data to accurately learn features and differentiate between the categories of classification. However, high-quality medical datasets are scarce and thus, we have transfer learned our networks over ImageNet weights followed by training with 2,450 embryo images recorded with an Embryoscope. Xception trained with high quality embryo image data was then re-trained using an augmented dataset of 1,968 and 1,056 embryo images acquired at 113 hpi collected using the stand-alone and smartphone imaging systems, respectively. The algorithm was then validated using 30% of the images from each dataset in order to optimize the system’s hyper-parameters. We implemented early stoppage rules during training that terminated training at the lowest validation loss to minimize overfitting. After fine-tuning hyperparameters based on performance with our validation data and training over 150 epochs, the models that achieved the lowest validation loss were saved. The validation accuracies for 5-class categorizations were 80%, and 100% for the smartphone-based and stand-alone imaging systems, respectively (Fig.2B i, 2C i ). The loss achieved by the best models were 0.5148 and 0.0259 at 96 epochs (Fig. 2B ii, 2C ii ), for the smartphone-based and stand-alone imaging systems, respectively.
Figure 2. Graphical depiction of data flow with training and validation curves for accuracy and cross-entropy.
(A) The computational flow chart of the data through the Xception architecture. (B) The measured (i) accuracy and (i) cross-entropy loss during the training and validation phase of the network development for the smartphone imaging system. (C) The measured (i) accuracy and (ii) cross-entropy loss during the training and validation phases of the network development for the stand-alone imaging system. The bold lines indicate smoothed curves for easier visualization while the semi-transparent lines indicate their respective non-smoothened curves.
Performance of the deep learning model in assessing embryos using the stand-alone imaging system
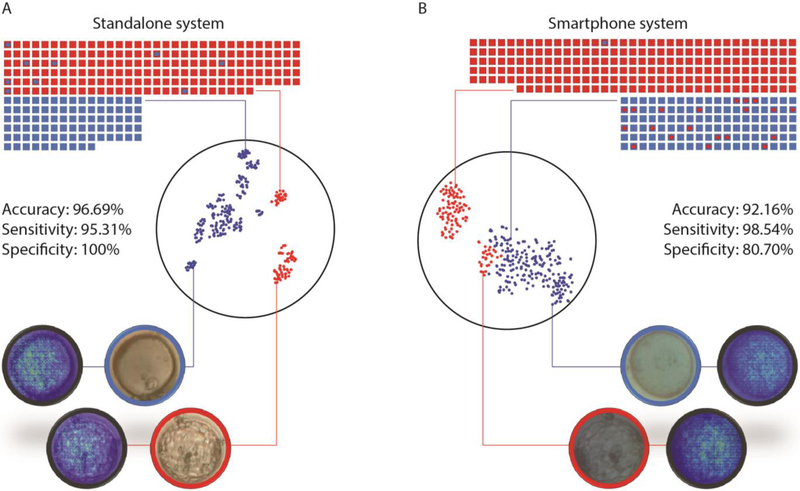
Our network performed well with unknown data collected from the inexpensive optical systems. We trained the CNN with embryo images collected using the stand-alone optical system and tested it with 272 embryo images. We evaluated the ability of the network in separating the two categories of images (blastocysts and non-blastocysts). The area under curve (AUC) as revealed by a receiver operator characteristic (ROC) of the system post-training was 0.98 (CI: 0.95 to 0.99). The accuracy of the neural network in classifying between blastocysts and non-blastocysts imaged using the stand-alone system was 96.69% (CI: 93.81 to 98.48%) (Fig. 3). Furthermore, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for the stand-alone system were 95.31% (CI: 91.29 to 97.83%), 100% (CI: 95.49 to 100%), 100%, and 89.89% (CI: 82.45 to 94.39%), respectively (Fig. 3). Saliency maps confirmed that the systems did use relevant morphological features of the embryos in these images for the classifications (Fig. 3).
Figure 3. System performance with images collected from the stand-alone and smartphone imaging systems along with TSNE and saliency visualizations.
(A) The dot plot illustrates the system’s performance in evaluating embryos imaged with the stand-alone system (n=272). (B) The dot plot illustrates the system’s performance in evaluating embryos imaged with the smartphone system (n=319). The squares represent true labels and the circles within them represent the system’s classification. Blue represents non-blastocysts and red represents blastocysts. t-SNE generated scatter plots are also presented to help visualize the separation of blastocyst and non-blastocyst embryo images obtained through (A) the stand-alone system and (B) the smartphone system. Each point represents an embryo image acquired through the respective imaging system. Saliency maps provided here were extracted from the networks to highlight the highest weighted features of the embryo images.
We also compared the performance of the developed model in evaluating embryos imaged using the stand-alone optical system with the performance of the model trained with Embryoscope data in evaluating embryos imaged with an Embryoscope. A chi-squared test revealed that the stand-alone optical system performed with an efficiency that was not inferior to an Embryoscope, with a difference of 5.72% (CI: 2.24 to 8.63%; P<0.05).
Performance of the deep learning model in assessing embryos using the smartphone optical system
Similarly, we tested our network with data collected from the smartphone system. We re-trained the CNN with embryo images collected using the smartphone optical system and tested it with 319 embryo images. We evaluated the ability of the network in separating the two categories of images (blastocysts and non-blastocysts). The AUC as revealed by a ROC analysis of the system post-training was 0.94 (CI: 0.90 to 0.96). The accuracy of the neural network in classifying between blastocysts and non-blastocysts imaged using the smartphone was 92.16% (CI: 88.65 to 94.86%) (Fig. 3). The sensitivity, specificity, PPV, and NPV of the smartphone-based optical system were 98.54% (CI: 95.78 to 99.70%), 80.70% (CI: 72.25 to 87.49%), 90.18% (CI: 86.31% to 93.04%), and 96.84% (CI: 90.86% to 98.95%), respectively (Fig. 3). Also, the saliency maps confirmed that the system did use relevant morphological features of the embryos in these images for the classifications (Fig. 3). An evaluation with the neural network trained on Embryoscope data through a chi-squared test for comparison revealed that the difference in performance between the two systems were non-differentiable (P=0.61; P>0.05), with a difference of 1.19% (CI: −2.8 to 4.7%; P>0.05).
Discussion
Embryo morphology is clinically analyzed at various stages of its development to identify and sort embryos based on their quality. Usually, embryos are assessed to check for fertilization during the first day of development, at the cleavage stage (2–3 days), and at the blastocyst (5–6 days) stage to evaluate the best embryo(s) for transfer. The most important timepoint for evaluation, arguably, remains to be at the 5th/6th day of development owing to the growing popularity of blastocyst transfers. Although, the devices presented in this work evaluate embryos at the 5th day of embryo development with >90% accuracies, they can be adapted to any of the embryo development stages for assessment. The primary goal of the study was to evaluate whether portable, low-cost optical systems, such as the ones presented here, can provide images of suitable quality to enable AI-mediated automated embryo assessments equivalent to approaches that are currently being researched with state-of-the-art imaging systems. Our results with the inexpensive stand-alone (<$100) and smartphone-based (<$5) imaging systems indicate that such platforms can indeed evaluate embryos with high accuracy and potentially act as support systems at a clinical level in both resource-limited and resource-rich settings.
One of primary limitations in access to care is high costs associated with IVF. In the recent years, technologies such as intracytoplasmic sperm injection (ICSI), time-lapse imaging, and pre-implantation genetic screening have been introduced in fertility centers that have helped in improving clinical outcomes. However, it is important to note that with the introduction of new technologies, costs associated with IVF have also dramatically increased. Furthermore, costs associated with setting up IVF labs can range upwards of $3 million dollars and with every additional procedure that is introduced new equipment and consumables are also added to the costs for the proper functioning of the lab. Therefore, newly developed technologies that demand expensive equipment are not available to most clinics. Unsurprisingly, the adoption of time-lapse imaging systems is limited to <17% of embryology labs even in a resource-rich setting such as the United States 12. Currently, IVF exists as a solution only to the economically well-off. Our deep learning-based approach on the other hand, does not rely on expensive hardware for both computation and data acquisition. We envision that these systems may improve the access to IVF procedures to many patients in need. Our presented systems here do not offer some of the features that time-lapse imaging systems possess such as a built-in incubator. However, the portability and low-cost of these systems can outweigh such features by allowing for availability of multiple imaging stations located at various key locations within an embryology lab, which does not hinder embryologists workflow within the lab while providing a rapid verification and assessment aid. Such systems can potentially improve the objectivity of assessment and reduce time spent per embryo. However, the evaluation of embryo morphologies at the blastocyst stage is just one of factors that dictate the clinical outcome in patients and other factors such as the male factor, medication, and patient prognosis and history also need to be taken into consideration by the deep learning model to potentially improve clinical outcomes directly. Future work involving the use the additional parameters is warranted.
Our reported data using a layered training approach showed that such systems may help reducing the required data for training. We pre-trained our models using clinical embryo images recorded with an Embryoscope. Clinical datasets used in this study was collected using time-lapse imaging systems that costs >$100k. While current applications of AI have focused on evaluating the state-of-the-art imaging systems, we have shown that these systems can also be adapted to low-cost optical devices. The layered training approach reported in this study can also be used in other non-fertility-related medical applications where image processing of low-quality image data is required.
Such low-cost, portable systems empowered with AI can shift the paradigm in fertility management in both resource-limited and resource-rich settings by providing an assistive tool to embryologists for consistent and reliable embryo assessment. Our work presents the first implementation of such a cost-effective alternative approach that is intended to assist embryologists and improve the quality of care provided to patients.
Supplementary Material
Acknowledgments
The authors would like to thank the Center for Clinical Data Science for their support and fruitful contributions. This work was supported by the Brigham and Women’s Hospital (BWH) Precision Medicine Developmental Award (BWH Precision Medicine Program) and Partners Innovation Discovery Grant (Partners Healthcare). It was also partially supported through 1R01AI118502 and R01AI138800 Awards (National Institute of Health).
Footnotes
Conflicts of interest
Hadi Shafiee, Charles Bormann, Manoj Kumar Kanakasabapathy, and Prudhvi Thirumalaraju are co-inventors of the presented AI technology that has been filed by Brigham and Women’s Hospital and Partners Healthcare.
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Turchi P, in Clinical Management of Male Infertility, eds. Cavallini G. and Beretta G, Springer International Publishing, Cham, 2015, DOI: 10.1007/978-3-319-08503-6_2, pp. 5–11. [DOI] [Google Scholar]
- 2.Mascarenhas MN, Flaxman SR, Boerma T, Vanderpoel S and Stevens GA, PLoS Med., 2012, 9, e1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birenbaum-Carmeli D, Sociol Health Illn, 2004, 26, 897–924. [DOI] [PubMed] [Google Scholar]
- 4.Toner JP, Fertil Steril, 2002, 78, 943–950. [DOI] [PubMed] [Google Scholar]
- 5.CDC, Fertility Clinic Success Rates Report, 2015. [Google Scholar]
- 6.Storr A, Venetis CA, Cooke S, Kilani S and Ledger W, Hum. Reprod., 2017, 32, 307–314. [DOI] [PubMed] [Google Scholar]
- 7.Baxter Bendus AE, Mayer JF, Shipley SK and Catherino WH, Fertil. Steril., 2006, 86, 1608–1615. [DOI] [PubMed] [Google Scholar]
- 8.Sundvall L, Ingerslev HJ, Breth Knudsen U and Kirkegaard K, Hum. Reprod., 2013, 28, 3215–3221. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y-G, Lazzaroni-Tealdi E, Wang Q, Zhang L, Barad DH, Kushnir VA, Darmon SK, Albertini DF and Gleicher N, Reprod. Biol. Endocrinol., 2016, 14, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M, Wei S, Hu J, Yuan J and Liu F, PLoS One, 2017, 12, e0178720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirkegaard K, Dyrlund TF and Ingerslev HJ, in Hum. Reprod., ed. Schatten H, 2016, DOI: 10.1002/9781118849613.ch7. [DOI] [Google Scholar]
- 12.Dolinko AV, Farland LV, Kaser DJ, Missmer SA and Racowsky C, J. Assist. Reprod. Genet., 2017, 34, 1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter RYL, Pyle AD, Tchieu J, Sridharan R, Clark AT, and Plath K, The National Academy of Sciences of the USA, 2008, vol. 105 2883–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardarson T, Van Landuyt L and Jones G, Hum. Reprod., 2012, 27, i72–i91. [DOI] [PubMed] [Google Scholar]
- 15.Thirumalaraju P, Bormann C, Kanakasabapathy M, Doshi F, Souter I, Dimitriadis I and Shafiee H, Fertil. Steril., 2018, 110, e432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potluri V, Kathiresan PS, Kandula H, Thirumalaraju P, Kanakasabapathy MK, Pavan SKS, Yarravarapu D, Soundararajan A, Baskar K and Gupta R, Lab Chip, 2019, 19, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM and Thrun S, Nature, 2017, 542, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitriadis I, Bormann CL, Thirumalaraju P, Kanakasabapathy M, Gupta R, Pooniwala R, Souter I, Hsu JY, Rice ST, Bhowmick P and Shafiee H, Fertil. Steril., 2019, 111, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thirumalaraju P, Hsu JY, Bormann CL, Kanakasabapathy M, Souter I, Dimitriadis I, Dickinson KA, Pooniwala R, Gupta R, Yogesh V and Shafiee H, Fertil. Steril., 2019, 111, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thirumalaraju P, Kanakasabapathy MK, Gupta R, Pooniwala R, Kandula H, Souter I, Dimitriadis I, Bormann CL and Shafiee H, Fertil. Steril., 2019, 112, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hariton E, Dimitriadis I, Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Souter I, Rice ST, Bhowmick P, Ramirez LB, Curchoe CL, Swain JE, Boehnlein LM, Bormann CL and Shafiee H, Fertil. Steril., 2019, 112, e77–e78. [Google Scholar]
- 22.Hariton E, Thirumalaraju P, Kanakasabapathy MK, Gupta R, Pooniwala R, Souter I, Dimitriadis I, Veiga C, Bortoletto P, Bormann CL and Shafiee H, Fertil. Steril., 2019, 112, e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimitriadis I, Bormann CL, Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Souter I, Rice ST, Bhowmick P and Shafiee H, Fertil. Steril., 2019, 112, e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bortoletto P, Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Souter I, Dimitriadis I, Dickinson K, Hariton E, Bormann CL and Shafiee H, Fertil. Steril., 2019, 112, e272–e273. [Google Scholar]
- 25.Thirumalaraju P, Bormann CL, Kanakasabapathy MK, Kandula H and Shafiee H, Fertil. Steril., 2019, 112, e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanakasabapathy MK, Thirumalaraju P, Gupta R, Pooniwala R, Kandula H, Souter I, Dimitriadis I, Bormann CL and Shafiee H, Fertil. Steril., 2019, 112, e70–e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran D, Cooke S, Illingworth PJ and Gardner DK, Hum. Reprod., 2019, 34, 1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khosravi P, Kazemi E, Zhan Q, Malmsten JE, Toschi M, Zisimopoulos P, Sigaras A, Lavery S, Cooper LAD, Hickman C, Meseguer M, Rosenwaks Z, Elemento O, Zaninovic N and Hajirasouliha I, NPJ Digit Med, 2019, 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couzin-Frankel J, Science, 2019, 364, 1119–1120. [DOI] [PubMed] [Google Scholar]
- 30.Hoo-Chang S, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D and Summers RM, IEEE Trans. Med. Imaging, 2016, 35, 1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiens J, Guttag J and Horvitz E, Journal of the American Medical Informatics Association : JAMIA, 2014, 21, 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chollet F, Journal, 2016, 1610. [Google Scholar]
- 33.van der Maaten L and Hinton G, Journal of Machine Learning Research, 2008, 9, 2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.