SUMMARY
Csm, a type III-A CRISPR-Cas interference complex, is a CRISPR RNA (crRNA)-guided RNase that also possesses target RNA-dependent DNase and cyclic oligoadenylate (cOA) synthetase activities. However, the structural features allowing target RNA-binding-dependent activation of DNA cleavage and cOA generation remain unknown. Here, we report the structure of Csm in complex with crRNA together with structures of cognate or non-cognate target RNA bound Csm complexes. We show that depending on complementarity with the 5′ tag of crRNA, the 3′ anti-tag region of target RNA binds at two distinct sites of the Csm complex. Importantly, the interaction between the non-complementary anti-tag region of cognate target RNA and Csm1 induces a conformational change at the Csm1 subunit that allosterically activates DNA cleavage and cOA generation. Together, our structural studies provide crucial insights into the mechanistic processes required for crRNA-meditated sequence-specific RNA cleavage, RNA target-dependent non-specific DNA cleavage, and cOA generation.
Data Resources: 6IFN, 6IFL, 6IFY, 6IG0, 6IFR, 6IFK, 6IFU, 6IFZ
Graphical Abstract
In Brief
Structures of the type III-A CRISPR-Csm complex in the presence of target RNAs reveal the molecular basis for a unique mode of self/non-self discrimination.
INTRODUCTION
CRISPR-Cas (CRISPR-associated genes) are RNA-guided adaptive immune systems that protect most archaea and approximately half of bacteria against invading foreign nucleic acids (Barrangou et al., 2007). CRISPR response is commonly divided into three different phases: adaptation, CRISPR RNA (crRNA) biogenesis, and interference (Marraffini, 2015). During the interference phase, crRNA and specific Cas proteins form an effector complex. Cas proteins involved in CRISPR interference are extremely diverse. Based on effector complex composition, CRISPR-Cas systems are broadly divided into two classes (Makarova et al., 2015). Class 1 includes type I, III, and IV CRISPR-Cas systems whose effector complexes are composed of multiple Cas proteins. Class 2 (type II, V, and VI) effector complexes consist of a single Cas protein. Type III CRISPR-Cas systems are characterized by the presence of Cas10 protein and are subdivided into four (III A–D) subtypes. Types III-A and III-D systems comprise Csm effector complex, which is composed of five different Csm proteins (Csm1–Csm5) and crRNA. Types III-B and III-C systems comprise Cmr effector complex, which consists of six distinct subunits (Cmr1–Cmr6) and crRNA. Cas10 proteins in the Csm and Cmr complexes are also called Csm1 and Cmr2, respectively. The crRNA in both Csm and Cmr complexes has 8 nucleotides at its 5′ end (termed 5′ tag) originating from CRISPR repeat and 30–45 nucleotides in guide sequence, derived from one of CRISPR array spacers.
Type III CRISPR-Cas systems employ a unique target RNA-dependent mechanism to defend hosts against invading nucleic acids. The crRNA-guided complexes recognize complementary RNA targets and cleave them via the Csm3/Cmr4 subunit (Ramia et al., 2014; Staals et al., 2014; Tamulaitis et al., 2014). Importantly, the crRNA-dependent target RNA binding also activates non-specific single-stranded DNA cleavage by the Csm1/Cmr2 HD domain, and cOA generation by the Csm1/Cmr2 Palm domains (Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016, 2017; Niewoehner et al., 2017). Recognition of nascent transcripts targets the single-stranded DNA cleavage activity toward transcribed DNA, allowing specific defense from actively transcribed foreign genomes (Goldberg et al., 2014). In addition, the Palm domains convert ATP to cOAs, which act as second messengers allosterically regulating RNase activity of Csm6/Csx1 (Niewoehner and Jinek, 2016; Sheppard et al., 2016). Activation of DNA cleavage and cOA synthesis require the 5′ tag of crRNA and the 3′-sequence flanking the target sequence to be non-complementary (cognate). In contrast, if these sequences are complementary (non-cognate), both activities are repressed, presumably to prevent recognition of antisense CRISPR array transcripts and avoid autoimmune response (Kazlauskiene et al., 2016, 2017; Niewoehner et al., 2017). Previous structural studies revealed the overall architecture of the Csm and Cmr complexes (Osawa et al., 2015; Rouillon et al., 2013; Staals et al., 2014; Taylor et al., 2015). However, the exact molecular mechanism of crRNA-guided target RNA cleavage, target RNA-binding dependent non-specific single-stranded DNA (ssDNA) cleavage, and cOA generation remained unknown.
Here, we report the crystal structure of Streptococcus thermophilus (Sth) Csm and a series of cryo-EM structures of SthCsm in complex either with cognate or non-cognate RNA targets. Our structural analysis shows that complementarity between the 5′ tag of crRNA and 3′ anti-tag of the target affects the conformation of the Csm1 Linker and the Loop L1 region, allosterically regulating the nuclease and cOA synthesis activities of Csm1 subunit.
RESULTS
Overall Structure of the SthCsm Complex
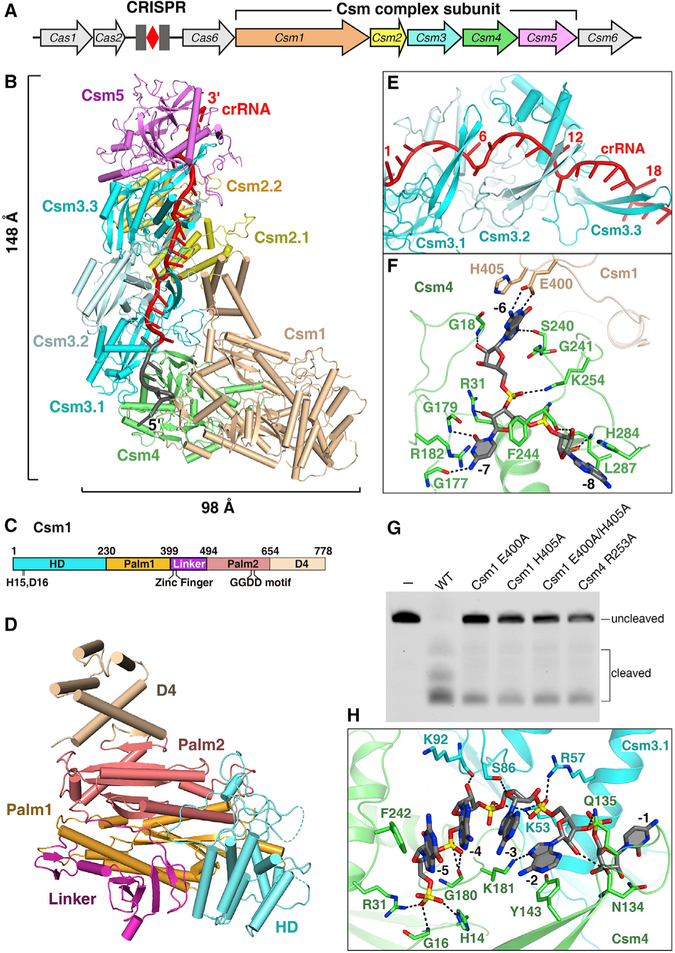
To understand the molecular basis of how Csm architecture determines its function, we purified recombinant SthCsm complex from E. coli cells containing a S. thermophilus repeat-spacer-repeat array with a single spacer and overproducing SthCsm1–SthCsm5 and SthCas6 proteins (Figure 1A). The Cas6 protein is required for crRNA processing. We determined the crystal structure of the Csm complex at 2.9 Å resolution (Figure 1B; Table S1). The Csm complex consists of a 9-subunit assembly composed of five Csm subunits (Csm1–Csm5) and one crRNA with a protein stoichiometry of Csm1122334151 (Figure S1A). The complex exhibits wormlike features in which the central double-helical core of three Csm3 and two Csm2 subunits is capped by the Csm1-Csm4 subcomplex at one end and by a Csm5 subunit at the opposite end.
Figure 1. Crystal Structure of the SthCsm Complex.
(A) Schematic view of a type III-A CRISPR-Cas system in S. thermophilus.
(B) The overall structure of the SthCsm complex associated with a crRNA. Color coding used for Csm1–Csm5 is identical to that used in (A). The spacer and 5′ tag regions of crRNA are in red and gray, respectively.
(C) The schematic representation of Csm1 domains. The HD, Palm1, Linker, Palm2, and D4 domains are shown in cyan, orange, purple, salmon, and wheat, respectively.
(D) Structure of Csm1 subunit in the same orientation as in (B).
(E) Close-up view of the crRNA spacer region and Csm3 β-thumb. The β-thumb of each Csm3 subunit folds over the top of the crRNA, creating a kink in the crRNA at 6-nt interval.
(F) Magnified view of the interactions between nucleotides (−6)–(−8) and the Csm1-Csm4 subcomplex illustrates that nucleotides (−6)–(−7) are sequence specifically recognized by Csm1 and Csm4 subunits.
(G) The target RNA dependent non-specific DNA cleavage with either Csm4 or Csm1 mutant form.
(H) Interactions between the nucleotides (−2)–(−5) of crRNA and Csm4-Csm3.1 subcomplex showing that these nucleotides are solvent exposed.
Three Csm3 subunits pack in a head-to-tail fashion with an azimuthal rotation between consecutive subunits, forming a helical stack. Two Csm2 subunits are also arranged in a head-to-tail fashion. Csm1 subunit contains one HD domain at the N-terminal, D4 domain at the C-terminal, Palm1 and Palm2 domains, and a zinc-finger containing Linker (Figures 1C and 1D). The HD-nuclease domain is positioned on the opposite side of the Csm4 binding surface of Csm1. The Linker, located between two Palm domains, contains a conserved zinc finger. Together with the Csm1 D4 domain, the two Csm2 subunits form the second helical stack. Like the Csm2 monomer, the Csm1-D4 domain consists of a five-α helix bundle, suggesting that Csm1-D4 and Csm2 play similar functions.
The crRNA passes through the entire Csm complex, suggesting that it plays an essential role in the Csm complex assembly. While the spacer in CRISPR array is designed 36 base pairs long, crRNA containing a 27-nt spacer fragment and an 8-nt repeat sequence (the 5′ tag) was found bound in the crystallized Csm complex (Figures S1B and S1C). The 27-nt spacer fragment is fastened tightly within the central channel formed by the Csm3 and Csm2 subunits. Three interlocking copies of Csm3 and the Csm5 monomer are lined up along the length of the crRNA spacer (Figure S1D). Long, thumb-like β strand protrusions emerge from the palm of each Csm3 and Csm4 subunit and are positioned close to its adjacent subunit (Figure 1E). As a result of this arrangement, every 6th base of cRNA spacer part is flipped. This structural feature is similar to that observed in crRNA within the Cmr complex (Osawa et al., 2015) and in type I Cascade complexes (Jackson et al., 2014; Zhao et al., 2014). The C(−1) nucleotide of the 5′ tag is also flipped out and stabilized by both Csm3.1 and the thumb of Csm4. This finding suggests that C(−1) cannot form base pairs with its complementary sequence in the target.
Csm1-Csm4 Subcomplex Sequence Specifically Recognizes the 5′ Tag
The conserved 8-nt 5′ tag of crRNA is held in the Csm1-Csm4 heterodimer. Csm4 and Csm1 subunits recognize the ((−6)–(−7)) nucleotides of 5′ tag in a sequence-specific manner. Notably, the Watson-Crick base edge of G(−6) forms two hydrogen bonds with the side-chain of Glu400 of Csm1, and the Watson-Crick edge of C(−7) base hydrogen bonds with Gly177 and Gly179 from Csm4 (Figure 1F). Nucleotides (−6)–(−8) are further stabilized by stacking interactions. Thus, nucleotides (−6) and (−7) are completely buried by the Csm4 and Csm1 subunits, and are not available for base pairing with other nucleic acids. In addition, Csm1 Glu400Ala and His405Ala mutants reduced target RNA-dependent DNA cleavage, suggesting these sequence-specific interactions contribute to optimal Csm effector function (Figure 1G).
Unlike the nucleotides (−6)–(−8), nucleotides (−2)–(−5) of 5′ tag are solvent-exposed. The segment consisting of nucleotides (−2)–(−5) adopts a regular single-stranded A-form, and is sandwiched by residues Tyr143 and Phe242 of the Csm4 subunit (Figure 1H). This segment is further stabilized by hydrogen bonds formed between their phosphate backbone and Csm. However, base edges of nucleotides (−2)–(−5) form no contact with Csm subunit and are solvent-exposed, suggesting that they are available for base pairing.
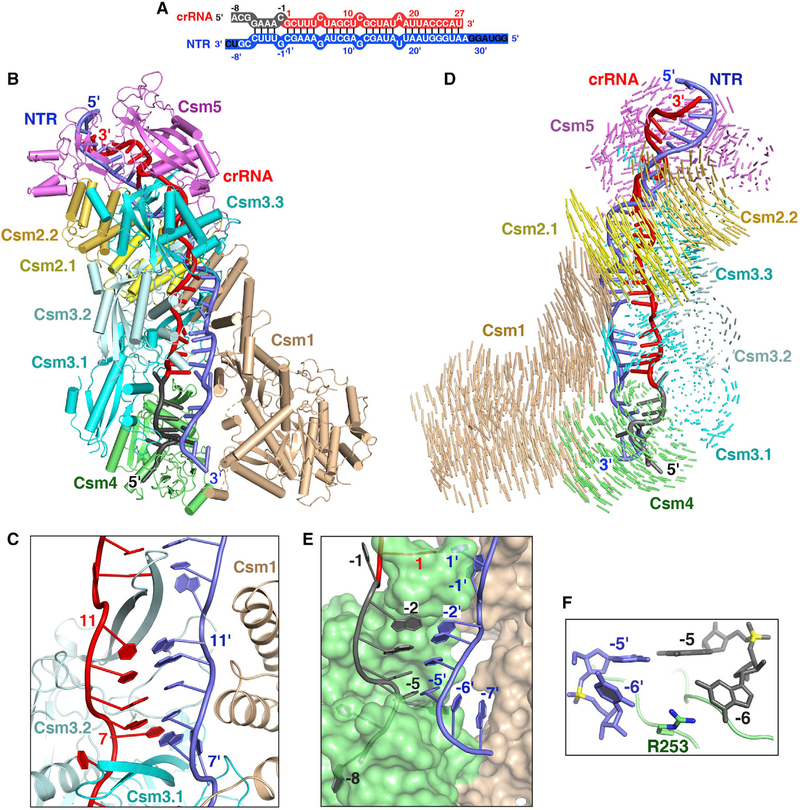
Overall Structure of Non-cognate Target RNA-Bound Csm Complex
To investigate how the crRNA recognizes target RNA, we determined the 3.16 Å cryo-EM structure of Csm complex bound to non-cognate target RNA (NTR) that is fully complementary to crRNA (Figures 2A and S2; Table S2). The complex contained Csm3 Asp33Asn mutant to prevent target RNA cleavage, as well as the mutation of Csm1 Asp16Asn. The guide-target RNA duplex adopts a discontinuous helical structure (Figure 2B). Two Csm2 subunits and the Csm1 D4 domain form a positively charged binding surface for target RNA (Figure S3A). In the spacer region, the target RNA base pairs with crRNA forming crRNA-target RNA duplex with every 6th base being flipped (Figure 2C), as is also observed in the cognate structure discussed further below. The crRNA-target RNA duplex is stabilized by the stacking interactions and hydrogen bonds formed between RNA’s phosphate groups and Csm subunits (Figure S3B). Notably, the flipped-out bases of target RNA stack on the side-chains of Csm2 Arg41 or Csm1 Tyr686 (Figure S3C).
Figure 2. Cryo-EM Structure of the Csm-NTR Complex.
(A) Schematic representation of the crRNA-NTR duplex. The base pairs observed in the NTR-bound Csm complex are depicted by lines. The non-cognate target RNA is shown in blue. The ordered and disordered nucleotides of NTR are shown in white and black, respectively.
(B) Overall structure of the NTR-bound Csm complex.
(C) The β-thumb of each Csm3 subunit inserts into the crRNA-NTR duplex, leading to the periodical nucleotide displacement of both RNA strands.
(D) Structural comparison between Csm-crRNA binary complex and Csm-crRNA-NTR ternary complex showing the conformational change upon target RNA binding. Vector length correlates with the domain motion scale (color-coded as defined in Figure 1A).
(E) Nucleotides at positions (−2)′–(−5)′ in the 3′ anti-tag of NTR base pair with nucleotides (−2)–(−5) in the 5′ tag of the crRNA.
(F) The side-chain of Arg253 of Csm4 subunit inserts into the bases at position (−6) of crRNA and target RNA, preventing base pairing at this position.
See also Figures S2 and S3 and Table S2.
The Csm complex undergoes conformational change upon target RNA binding. To analyze the rearrangement of the Csm complex upon target RNA binding, we aligned the Csm3 and crRNA of Csm binary and NTR-bound ternary Csm complex structures. We found that two Csm2 subunits rotate away from the Csm3 subunits, whereas Csm1 rotates toward the 5′ end of the crRNA, generating a wider binding channel that is large enough to harbor the crRNA-target RNA duplex (Figure 2D). Similar conformational changes were observed for the type I-E Cascade complex and type III-B Cmr complex upon target binding (Hayes et al., 2016; Taylor et al., 2015). In addition, upon NTR binding two Csm1 fragments (residues 393–417 and 257–265) adjacent to the 3′ anti-tag region of NTR become disordered. Thus, the base of G(−6) within 5′ tag does not form a pair of hydrogen bonds with Csm1 Glu400 as seen in the Csm binary complex.
The 3′ Anti-tag of NTR Forms Four Base Pairs with the 5′ Tag of the crRNA
To understand why complementarity between the 3′ anti-tag of the target RNA and the 5′ tag of the crRNA abrogates DNA cleavage, we analyzed the structure of the 5′ tag and the 3′ anti-tag region. A thumb-like β-hairpin of Csm4 protrudes and kinks the target RNA at position (−1)′, causing the nucleotide (−1)′ flip out and insert into a cleft formed by the Csm4 subunit. In contrast, nucleotides (−2)′ to (−5)′ of complementary NTR form four base pairs with nucleotides (−2) to (−5) within the 5′ tag of crRNA (Figure 2E). Upstream nucleotides ((−6)′–(−8)′) rotate away from crRNA, thus failing to base pairs with the 5′ tag. The 4-base pair 5′ tag-3′ anti-tag duplex is located in the cleft formed by Csm1 and Csm4. Notably, no direct interactions were found between nucleotides (−2)′ to (−5)′of NTR and the Csm1 subunit. Interestingly, upon NTR binding, Csm4 Arg253 undergoes a conformational change and inserts between G(−6) and C(−6)′ bases. The side chain of Arg253 contacts with the base of G(−6) of crRNA and C(−6)′ of target RNA (Figure 2F) and thus prevents duplex formation beyond nucleotide (−5).
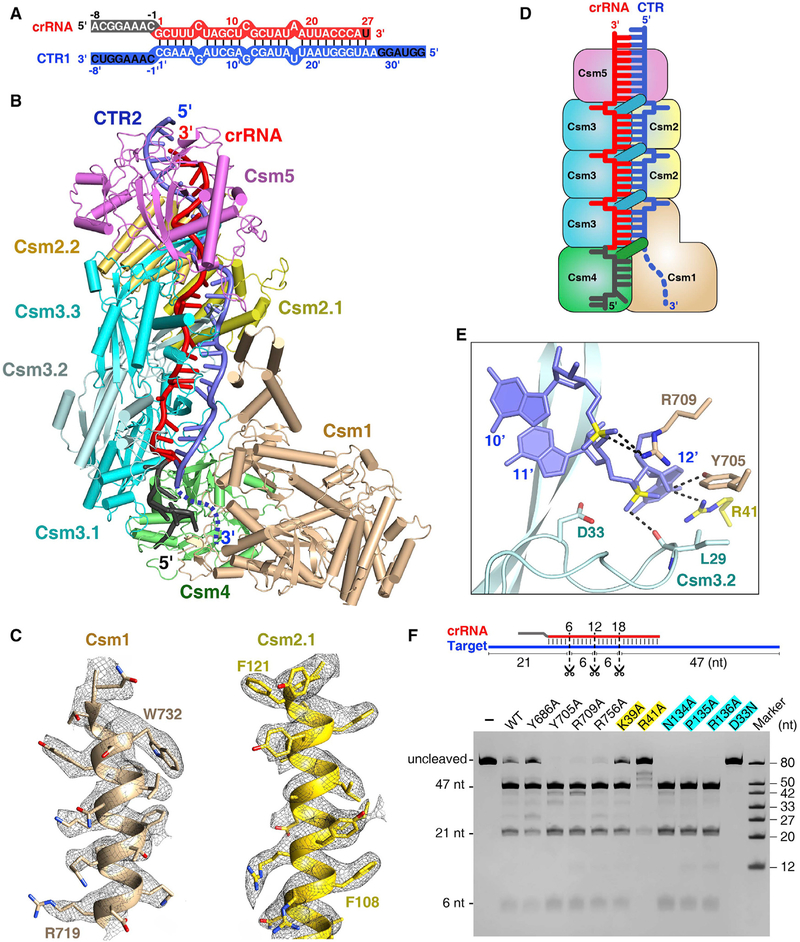
Structure of the Cognate Target RNA-Bound Csm Complex
To investigate how ssDNA cleavage is triggered by cognate target RNA, we determined the cryo-EM structure of Csm3 Asp33Asn and Csm1 Asp16Asn double mutant of Csm effector that lacks both RNase and DNase activities, bound to CTR1 at 3.9 Å resolution (Figures 3A and S4A). We next determined the cryo-EM structure of the Csm3 Asp33Asn, Csm1 His15Ala double mutant in complex with cognate target RNA (termed CTR2), and either with ssDNA or bubbled double-stranded DNA (dsDNA) at 3.5 Å and 3.05 Å resolution, respectively (Figures 3B, S4B, and S4C). These three CTR-bound Csm complexes have similar structures, further on we discuss the 3.05 Å Csm complex, whose electron density is well defined, allowing reliable modeling (Figure 3C). Unexpectedly, we failed to identify a clear DNA electron density map in either of these DNA-bound complexes, despite the presence of DNA in the sample (Figure S4D). In these CTR-bound Csm complexes, the spacer region forms base pairs with nucleotide displacement at 6-nt intervals similar to what is seen in the NTR-bound Csm complex (Figures S3A and S3B). However, the 3′ anti-tag region is disordered in the CTR-bound Csm complex, whereas it forms an ordered duplex with the 5′ tag in the NTR-bound Csm complex.
Figure 3. Cryo-EM Structure of the Csm-CTR Complex Either in the Absence or Presence of DNA.
(A) Schematic representation of the crRNA-CTR1 duplex. The color codes of crRNA and target RNA are identical to those used in Figure 2A.
(B) Overall structure of Csm1 H15A and Csm3 D33N catalytic mutant Csm in complex with CTR2 in the presence of bubble dsDNA at 3.05 Å resolution. The disordered the 3′ anti-tag of CTR2 is shown by a blue dashed line.
(C) Cryo-EM maps and fits for selected regions of Csm1 (residues 719–734) and Csm2.1 (residues 106–122) in the 3.05 Å CTR-bound Csm complex.
(D) Schematic representation of the Csm-CTR complex. Color-coding is identical to that used in Figure 1B.
(E) Close-up view of the scissile phosphate and the conserved catalytic residue Csm3 Asp33. The Csm3 D33N mutant was used for structural studies.
(F) Target RNA cleavage assay. Three scissors indicate the cleavage sites. The Csm1 mutants are in black, and Csm2 and Csm3 mutants are highlighted in yellow and cyan, respectively.
While the 3′ anti-tag region of target RNA remained disordered in these complexes, a weak continuous electron density was found between the nucleotide (+1)′ of the target RNA and the tip of the Csm1 Linker in the bubbled DNA containing structures, indicating the presence of a possible binding site for the 3′ anti-tag region of cognate RNA target (Figure S4E). Furthermore, disruption of this possible interaction by substituting Csm1 Linker amino acids 393–417 by a (GGS)4 sequence significantly reduced DNA cleavage and the cOAs synthesis.
In the CTR-bound Csm complex, each of the Csm3 subunits covers a 6-nt fragment of crRNA, while each of the Csm2 subunits and the Csm1 D4 domain covers a 6-nt fragment of target RNA (Figure 3D). Thus, one Csm3-Csm2 subcomplex covers 6 base pairs of the crRNA-target RNA duplex, and the number of Csm3 subunits always exceeds the number of Csm2 subunits by one. Because mature crRNAs contain a uniform 5′ tag and variable 3′ ends, a variable number of Csm2 and Csm3 subunits in the complex is expected, with one additional Csm3-Csm2 subcomplex assembled into the Csm complex with every 6-nt extension of the crRNA; in other words, the protein stoichiometry of Csm effector can be Csm112n3n+14151 (Figure S4F). This phenomenon explains why the Csm complexes frequently contain crRNAs that differ from each other by 6-nt increments.
Csm3 Cleaves Target RNA at 6-nt Intervals
The thumb-like β-hairpins of Csm3 go through the gap at each 6N positions in the crRNA-target RNA duplex, with each 5-base pair duplex bracketed by Pro135 and Arg136 originating from adjacent β-hairpins of Csm3 (Figures S3B and S3C). The flipped out target bases stack on the side chain of Csm2 Arg41 or Csm1 Tyr686. This nucleotide displacement of the target and interactions between target RNA and Csm complex periodically place the scissile bonds near the conserved Asp33 catalytic residues of Csm3 monomers (Figure 3E). As a result, the Csm3 subunits cleave complementary target RNAs at multiple sites with 6-nt intervals, explaining why the number of target RNA cleavage sites correlates with the number of Csm3 nuclease subunits (Tamulaitis et al., 2014). Our SthCsm complex cleaves the target RNA at three sites with 6-nt intervals, and the Csm3 Asp33Asn mutant abolishes the target RNA cleavage (Figure 3F). While Csm2 Arg41Ala mutant dramatically reduced cleavage activity, alanine substitution of Csm1 Tyr686 or Csm2 Lys39 reduced RNA cleavage only slightly. Other mutations of Csm1 or Csm3 residues that interact with the flipped out nucleotide or its downstream or upstream neighbors within the target RNA lacked any detectable effect on target RNA cleavage. In addition, previous studies revealed that Cmr4 also cleaves the target at multiple positions at exactly 6-nt intervals (Hale et al., 2009; Osawa et al., 2015; Staals et al., 2013). Thus, both the Csm and Cmr complexes cleave the target RNA by a 5′ ruler-like endonuclease mechanism.
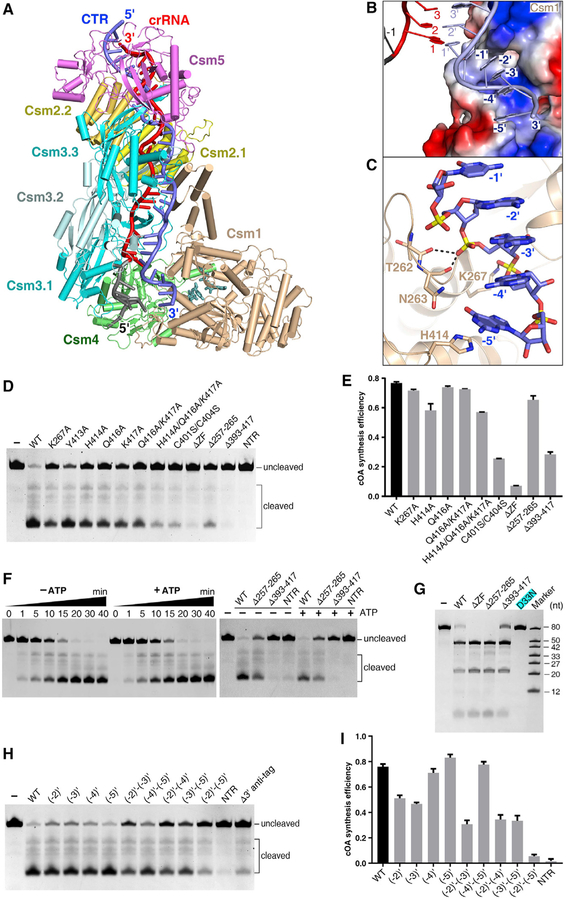
3′ Anti-tag of Target RNA Interacts with Csm1 Linker
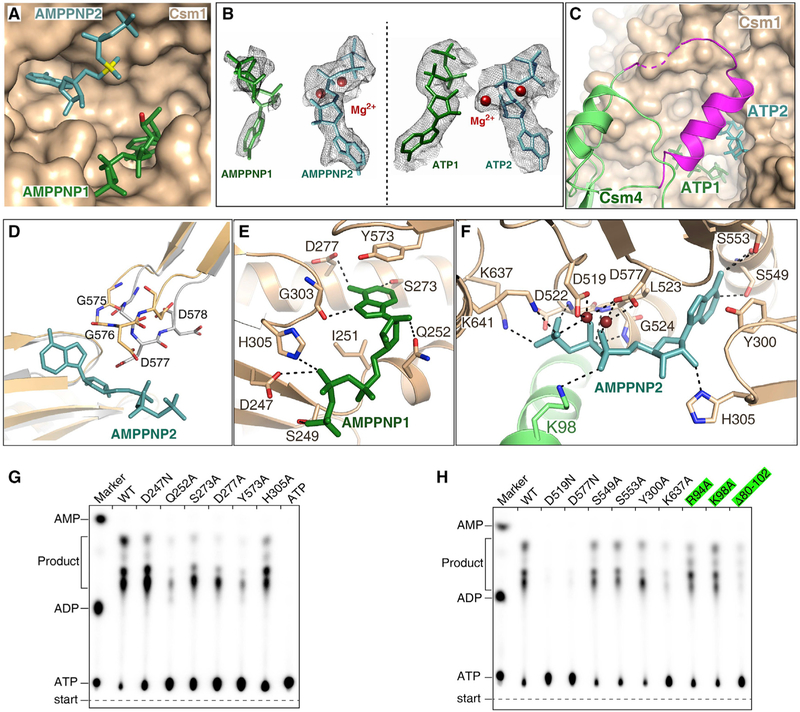
To investigate how the Csm1 Palm domains convert ATP into cOA, we incubated the Csm-CTR complex with AMPPNP or ATP and solved the cryo-EM structures of these complexes at 3.2 Å and 3.4 Å, respectively (Figures 4A and S5A; Table S3). We found that the overall structures of these two ternary complexes are similar. Therefore, below we discuss the 3.2 Å ternary structure containing AMPPNP. The nucleotides (−2)′–(−5) of the 3′ anti-tag are positioned inside a narrow positive-charged channel, which is formed by the Csm1 Linker region and a loop in the Palm1 domain (termed L1) (Figure 4B). The phosphate group of nucleotide (−4)′ interacts with conserved Lys267, anchoring the 3′ anti-tag (Figure 4C). In addition, the 3′ anti-tag also interacts with Csm1 Linker. The base of nucleotide (−5)′ interacts with Csm1 His414, which is positioned close to the zinc-finger.
Figure 4. Cryo-EM Structure of the Csm-CTR Complex in the Presence of ATP or AMPPNP.
(A) Overall structure of Csm1 D16N and Csm3 D33N catalytic mutant Csm in complex with cognate target RNA in the presence of AMPPNP.
(B) The non-complementary 3′ anti-tag region of the CTR binds in a positively charged groove of the Csm1 subunit, which is shown as a surface representation colored according to its electrostatic potential.
(C) Close-up view of the interaction between the 3′ anti-tag region of the CTR and Csm1 subunit.
(D) Nonspecific ssDNA cleavage with Csm1 mutants showing the impact of mutation in the Csm1 Linker region and Loop L1 on HD nuclease activity. WT, wild-type Csm; NTR, non-cognate target RNA bound wild-type Csm complex; ΔZF, Ser substitution of four Cys residues in the zinc finger. WT and mutant forms are CTR-bound Csm complexes containing either wild-type or mutant Csm1.
(E) Effect of mutations in Csm1 on the cOA synthesis. Averages with SDs are shown, n = 3 replicates.
(F) The DNA cleavage assay showing ATP has little impact on the DNA cleavage with either the wild-type (left) or mutant Csm complex (right).
(G) RNA cleavage assay showing the Csm1 mutation has little impact of on target RNA cleavage.
(H) DNA cleavage assay showing the impact of 3′ anti-tag mutation on the DNase activity of Csm1 HD domain.
(I) Effect of mutations of the 3′ anti-tag sequence in CTR on cOAs synthesis. Averages with SDs are shown, n = 3 replicates.
To investigate whether the interactions between the non-complementary 3′ anti-tag and Csm1 are important for both ssDNA cleavage and cOAs synthesis, we performed assays measuring these two activities. As shown in Figures 4D and 4E, following the substitution of Csm1 Loop L1 (residues 257–265) by a (GGS)3 sequence to block this interaction, ssDNA cleavage is greatly reduced, while cOA synthesis is only slightly reduced. We obtained similar results for the single mutation of Lys267Ala. Furthermore, the substitution of either the Csm1 Linker (residues 393–417) or the zinc finger exhibited dramatically reduced ssDNA cleavage and cOAs synthesis. Alanine substitution of Csm1 His414 and Gln416 reduced DNase activity, but had less effect on cOAs production. These results suggest that the interaction between 3′ anti-tag and the Csm1 Linker is crucial for both target RNA-dependent DNA cleavage and cOAs synthesis. In addition, the structural comparison between CTR-bound complexes with or without ATP suggests that ATP might stabilize the 3′ anti-tag. However, the presence of ATP lacks remarkable effect on the efficiency of DNA cleavage (Figure 4F). Thus, although both DNase and cOA synthetase activities depend on a non-complementary 3′ anti-tag sequence, these two activities show little interdependency.
We next performed a target RNA cleavage assay using Csm1 mutants. The independent substitution of Loop L1, Linker, or zinc finger exhibited little effect on target RNA cleavage (Figure 4G), suggesting that the interactions between 3′ anti-tag and Csm1 do not affect RNase activity of Csm3 subunits.
To evaluate the impact of changing the degree in non-complementarity of the 3′ anti-tag, we measured the effects of mutations within the 3′ anti-tag on both ssDNA cleavage and cOAs synthesis (Figures 4H and 4I). Single nucleotide mutation of nucleotides (−2)′ and (−3)′ reduced both ssDNA cleavage and cOAs synthesis. The double mutation at these positions resulted in greater reduction of both DNase and cOAs synthetase activities. In contrast, single or double nucleotide mutations at positions (−4)′ and (−5)′ showed considerably less effect on both activities. Nucleotides (−2)′–(−5)′ mutation dramatically reduced both activities. Furthermore, both activities were abolished in the presence of NTR. Together, these results confirm that non-complementarity between nucleotides (−2)′–(−5)′ of the 3′ anti-tag within target RNA and the 5′ tag of the crRNA is critical for activating both ssDNase and cOAs synthetase activities. Importantly, the first two nucleotides ((−2)′ and (−3)′) are likely to play more important roles in the activation of two activities and in defining the 3′ anti-tag region-binding channel.
Each Csm1 Palm Domain Binds One ATP
In order to understand how conserved Palm domains bind ATP and convert it into cOA, we analyzed ATP or AMPPNP bound Csm-CTR complex structures. Two AMPPNP or ATP molecules bind in the cleft formed by the Palm1 and Palm2 domains (Figures 5A and 5B). The positions of ATP or AMPPNP show that the 3′-OH of ATP1 is structurally ready to launch a nucleophilic attack on the α-phosphate group of ATP2, to form a 5′−3′ phosphodiester bond. Interestingly, in the presence of ATP or AMPPNP, Csm4 amino acids 82–104 (shown in magenta) form an α-helix and a loop (Figure 5C), forming a lid-shaped structure above the ATP binding pocket in Palm domains. In contrast, in the absence of ATP, these residues are disordered in target RNA-bound Csm complex. These results indicate that the flexibility of this Csm4 region keeps the ATP binding pocket open in the absence of ATP, and closes the pocket upon ATP binding. In addition, the AMPPNP2 or ATP2 binding triggers the conformational change at the GGDD motif within the Palm2 domain (Figure 5D).
Figure 5. Two ATP or AMPPNP Bind in the Palm Domains of Csm1 Subunit.
(A) Two AMPPNP molecules bind in the pocket formed by two Palm domains. AMPPNP1 (in green) and AMPPNP2 (in teal) bind to Palm1 and Palm2 domains, respectively. The a-phosphate group of AMPPNP2 and the O3′ atom of AMPPNP1 are shown in yellow and red, respectively.
(B) Cryo-EM map for two AMPPNP molecules in the CTR-bound Csm complex (left) and for two ATP in the NTR-bound Csm complex (right). Two Mg2+ are shown as brown spheres.
(C) Csm4 residues 82–104 (in magenta) cover the ATP binding pocket upon ATP binding, whereas they are disordered in the absence of ATP.
(D) Structural comparison the GGDD motif in the CTR-bound Csm complex either in the presence (wheat) or absence (gray) of AMPPNP, showing that the GGDD motif undergoes a conformational change upon the AMPPNP or ATP binding.
(E) Magnified view of the interactions between AMPPNP1 and the Palm1 domain.
(F) Expanded view of the interactions between AMPPNP2 and the Palm2 domain.
(G) Effect on cOAs synthesis of mutations of Csm1 residues that interact with the AMPPNP1.
(H) Effect on cOAs synthesis of the mutations of Csm1 and Csm4 residues that interact with the AMPPNP2. The Csm4 mutants are highlighted in green.
See also Figure S5.
AMPPNP1 binds to the Palm1 domain, with its adenylate base forming hydrogen bonds with the strictly conserved Ser273, Gly303, and modestly conserved Asp277 of Palm1. The base of AMPPNP1 is further stabilized by stacking on the side chain of Tyr573 (Figures 5E and S5B). AMPPNP2 binds to the Palm2 domain, and its base forms hydrogen bonds with the side-chain of conserved residues Ser549 and Ser553 (Figure 5F). These base contacts explain why Csm1 Palm domains bind ATP but not other nucleotides. Importantly, we also found that two Mg ions bind the GGDD motif and coordinate with three phosphate groups of AMPPNP2 and the side chains of Asp519 and Asp577. These two AMPPNP are further stabilized by the interactions between their phosphate groups and Csm1.
Next, we evaluated the effects of mutating ATP-binding amino acids of Csm1 on cOA synthesis (Figures 5G and 5H). Substitution of the AMPPNP1 base-contact residues Ser273, Asp277, and Tyr573 dramatically reduced cOAs synthesis (Figure 5G). In comparison, substitutions of AMPPNP2 base-contact residues Ser549, Ser553, and Tyr300 only slightly reduced cOAs synthesis (Figure 5H). Thus, mutations of the ATP1 base-contacts resulted in greater reduction compared to mutation of ATP2 base contacts. Substitution of any one of the phosphate-interacting amino acids substantially reduced cOAs synthesis. In contrast, the Asp519Asn and Asp577Asn mutants abolished cOA synthesis, suggesting that the interactions between the Mg2+ ions and the Asp residues play essential roles for the cOAs synthesis. Together, these results suggest that the interactions between the ATP and Palm domains are essential for cOA production.
To test whether complementarity between the 5′ tag and 3′ anti-tag affects ATP binding, we determined the 3.4 Å cryo-EM structure of Csm3 Asp33Asn Csm1 Asp16Asn mutant in complex with NTR and ATP (Figure S5C). This complex exhibits a similar overall structure to that bound to same NTR in the absence of ATP. The two ATP molecules bind to the Palm domains in a manner similar to those of AMPPNP in the CTR-bound Csm complex (Figures S5D and S5E). Thus, while complementarity between the 3′ anti-tag region and 5′ tag inhibits target RNA binding-dependent cOA synthesis, it has little effect on ATP binding.
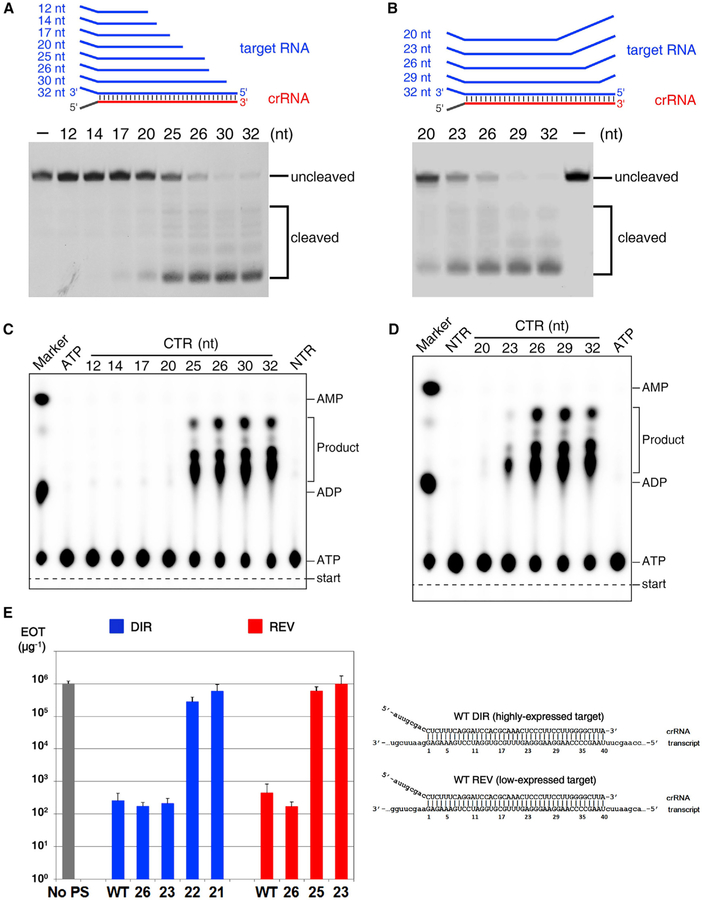
Determining the Functional Minimal Length of crRNA
Distinct from crRNAs in other CRISPR-Cas types, which have constant length, mature type III crRNAs contain variable 3′ ends. To investigate whether the crRNA length affects target RNA-dependent DNA cleavage, we performed a DNA cleavage assay using SthCsm complexes and CTR of different lengths. We found that when the crRNA-target duplex was shorter than 25 base pairs, DNase activity was greatly reduced (Figure 6A). To confirm this finding, we tested DNA cleavage using SthCsm effector complex and 32-nt cognate mutant target RNAs containing mismatches with crRNA 3′ end. We found that when the crRNA-target RNA duplex was 23 base pairs or less, DNA cleavage was greatly reduced (Figure 6B).
Figure 6. Determining the Minimal Length of crRNA Spacer Segment Required for DNase and cOA Synthetase Activities of the Csm Complex.
(A and B) DNA cleavage by various CTR-bound SthCsm complexes. The cognate target RNA is truncated from it 5′ end (A), and the target RNA contains mismatches from its 5′ end (B).
(C) cOAs synthesis assay with truncated cognate target RNA shown in (A).
(D) cOAs synthesis assay with the mismatch-containing cognate target RNA as shown in (B).
(E) Efficiency of transformation (EOT) of III-A+ T. thermophilus cells with plasmids carrying protospacers fully matched to (“WT”) or with multiple mismatches with the 3′ end of crRNA. Numbers indicate the length of crRNA-target duplex. Blue bars correspond to plasmids with protospacer cloned in direct orientation, which results in high level of transcripts targeted by crRNA, red bars correspond to plasmids with reverse protospacer orientation which results in reduced protospacer transcript levels. The gray bar indicates EOT with control plasmid without protospacer (“No PS”). The structures of crRNA-target duplexes for fully matching protospacers in both orientations are shown on the right. Mean values obtained from 3 independent experiments and SDs are shown.
See also Figure S6.
We next tested the cOAs synthesis with the shorter CTRs or mismatches containing CTR as above (Figures 6C and 6D). The CTR-crRNA duplexes with 25 base pairs or longer are able to activate the synthetase activity of the Palm domains, but the duplex shorter than 23 base pairs in length cannot. These data suggest that 25-base pair CTR-crRNA duplex is essential for the cOAs synthesis, and our SthCsm is the minimal functional Csm complex. Taken together, the 25-base pair CTR-crRNA duplex is the minimal requirement for activating both the DNase and cOAs synthetase activities. Notably, further investigations are needed to reveal the detailed molecular mechanism of regulation by the CTR-crRNA duplex length.
To show that in vitro DNA cleavage induced by Csm forming shortened target duplexes is biologically relevant, we investigated type III-A interference in vivo using Thermus thermophilus (Tth) as a model system. Tth cells with active III-A CRISPR-Cas system (III-A+) and a csm3 mutant (III-A−) were transformed with plasmids carrying a protospacer, a sequence matching one of Tth III-A spacers, cloned in different orientations. In direct orientation transcription of protospacer from upstream promoter was expected to generate RNA complementary to crRNA and promote interference (Figure S6A). Indeed, this plasmid was poorly transformed into III-A+ cells compared to plasmid with no protospacer. Both plasmids were efficiently transformed in III-A− cells. Unexpectedly, a plasmid with reverse protospacer orientation was also poorly transformed in III-A+ cells, seemingly contradicting transcription-dependence of Csm interference. This effect could have been due to transcription of both strands of plasmid DNA. Strand-specific sequencing of RNA extracted from cells transformed with empty plasmid vector revealed very strong “direct” orientation transcription across the site of protospacer cloning. While transcription in the opposite orientation was ~50 times less efficient, it was still 100 times higher than transcription of the essential rho gene (Figure S6B). When transcription terminator was introduced to decrease low-level counterclockwise transcription of the protospacer, efficiency of transformation of plasmid with reverse protospacer orientation increased by two orders of magnitude (Figures S6C and S6D). Thus, type III-A interference in Tth is transcription-dependent. Decreased transformation efficiency of protospacer plasmids must be due to RNA protospacer dependent cleavage of plasmid DNA by Tth Csm (TthCsm) complex.
We next created protospacer plasmids with multiple mismatches with the crRNA 3′ end and assayed their transformation efficiency. Reducing the crRNA-target duplex length for protospacer in direct orientation up to 23 base pairs had no effect on interference (Figure 6E). In contrast, a plasmid containing a protospacer that formed a 22-base pair crRNA-target duplex was transformed with high efficiency, indicating that interference was abolished. We conclude that transcription-dependent interference with plasmid transformation by TthCsm tolerates extensive shortening of the crRNA-target duplex. The threshold duplex length where in vivo interference is abolished corresponds well with in vitro data for SthCsm. With reverse protospacer orientation plasmids interference was abolished when RNA-target duplex length decreased to only 25 base pairs (Figure 6E), indicating that type III-A interference depends on target transcript abundance: higher abundance allows more extensive shortening of the duplex.
The 3′ Anti-tags of Cognate and Non-cognate Target RNAs Bind to Distinct Channels
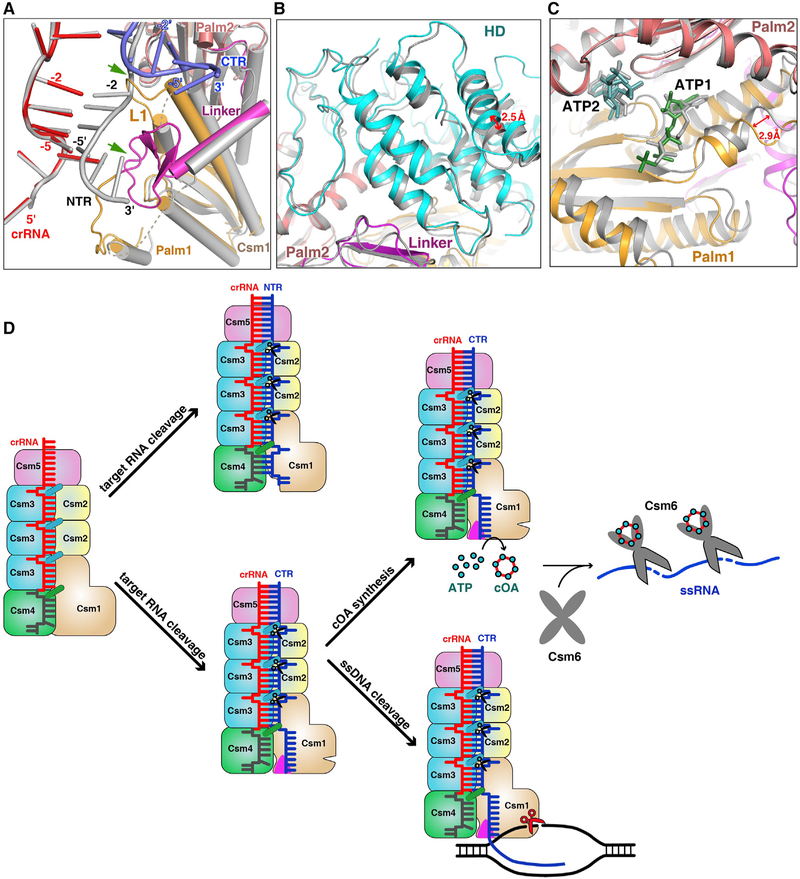
To understand how target RNA transcription activates the Csm1 nuclease and cOA synthetase, we compared the NTR-bound and CTR-bound Csm complex structures by aligning their Csm3 subunits and crRNA (Figure S7A). We found that the 3′ anti-tags of NTR and CTR bind in two distinct binding channels (Figure 7A). Nucleotides (−2)′–(−5)′ of NTR form four base pairs with crRNA and do not interact with Csm1 directly. Furthermore, most of the Csm1 Linker and Loop L1 are disordered (Figure S7B). In contrast, non-complementary nucleotides (−2)′–(−5)′ of CTR swing away from crRNA and are stabilized by the Csm1 Loop L1 and the Linker (Figure S7C). The Csm1 Linker and Loop L1 become ordered, suggesting that the interaction with CTR 3′ anti-tag anchors their position. The superposition of Csm1 in the two states clearly shows that the 3’ anti-tag of NTR clashes with the Csm1 Linker and Loop L1 of the CTR-bound complex (Figure 7A), suggesting that the Csm1 Linker and Loop L1 undergo a conformational change upon cognate target RNA binding. In addition, nucleotides 1′–2′ of CTR also undergo a conformational change (Figure S7D), suggesting the proper position of these nucleotides affects the target RNA-dependent DNase activity. This observation is consistent with previous mutant studies showing that mutations at positions 3–4 abrogate DNA cleavage (Kazlauskiene et al., 2016). The results explain how the complementarity between 5′ tag of crRNA and the 3′ anti-tag of target RNA prevents DNA cleavage (Marraffini and Sontheimer, 2010), without inhibiting target RNA cleavage (Kazlauskiene et al., 2016; Samai et al., 2015).
Figure 7. Non-complementarity of 3′ Anti-tag of Target RNA Activates the DNase and cOA Synthetase Activities.
(A) Structural comparison between NTR- and CTR-bound Csm complexes. Superposition of the NTR- and CTR-bound Csm complexes at the repeat region. The 5′ tag and 3′ anti-tag in the CTR-bound Csm complex are shown in red and blue, respectively. The Color-coding of Csm1 in the CTR-bound complex is identical to that used in Figure 1C. All components in the NTR-bound complex are shown in gray. Two green arrows highlight the clash region of between NTR and Csm1 within CTR-bound complex.
(B) Structural comparison of HD domains in the CTR- (cyan and purple) and NTR-bound (gray) Csm complexes by aligning the palm2 and D4 domains of the Csm1 subunit.
(C) Structural comparison of Palm domains in the CTR- (orange and salmon) and NTR- (gray) bound Csm complexes by aligning the palm2 and D4 domains of the Csm1 subunit.
(D) Model of type III-A interference. The crRNA associated with the Csm complex recognizes target RNA containing its complementary sequence forming the crRNA-target RNA duplex. The Csm3 subunit is an RNase that cleaves target RNA at 6-nt intervals. The 3′ anti-tag region of cognate target RNA binds to the Csm1 Linker and Loop L1 regions (shown in magenta triangles) and induces the rearrangement at the Linker and Loop L1 regions. This rearrangement of Csm1 allosterically activates the DNA cleavage and the production of cOAs, which activates the RNase activity of Csm6.
See also Figure S7.
To assess the conformational changes induced by 3′ anti-tag binding in HD and Palm catalytic pockets, we compared NTR-and CTR-bound Csm complex structures by aligning the Palm2 and D4 domains. The Palm2 domain is superposed well, whereas both HD and Palm1 domains undergo a positional shift within CTR-bound Csm complex (Figures 7B and 7C). It suggests that the binding of the non-complementary 3′ anti-tag region affects the dynamic properties of the HD and Palm1 domains. The fact that both the Csm1 HD-dependent ssDNA cleavage and Palm-dependent cOA synthesis depend on non-complementarity between the 5′ tag of crRNA and the 3′ anti-tag of target RNA suggests that a common conformational change activates both active sites of Csm1 upon cognate target binding. Thus, it is likely that the conformational changes within the Csm1 allosterically activate HD nuclease and Palm cOA synthetase.
DISCUSSION
Previous studies demonstrated that only type III CRISPR-Cas systems contain crRNA effectors that possess three entirely different enzymatic activities: target RNA cleavage, ssDNA cleavage and cOA synthesis. Importantly, RNA-dependent non-specific DNA cleavage and cOA synthesis are controlled by complementarity between the 5′ tag of crRNA and the 3′ anti-tag of target RNA. Here we show that nucleotides (−2)–(−5) of crRNA are solvent exposed and available for base-pairing with complementary target RNA. Our structural analysis of Csm complexes bound to different targets reveals that nucleotides (−2)′–(−5)′ of 3′ anti-tag are essential for activating both DNase and cOA synthetase activities. This mechanism provides the structural basis for control of target RNA-dependent DNA cleavage and cOA production. Structural similarities imply that the Cmr complex degrades foreign nucleic acids using a similar mechanism.
Formation of Csm/Cmr Complex
Our structural studies suggest that the protein stoichiometry of the Csm effector complex can be described as Csm112n3n+14151. Given the structural resemblance between the Csm and Cmr complexes, it is possible to predict that the copy number of the backbone subunits Cmr4 is one greater than that of the Cmr5 subunit, similar to Csm3 and Csm2 in the Csm complex. Thus, the stoichiometry of the Cmr complex can be presented as Cmr1121314n+15n61. Importantly, the stoichiometry of both Csm and Cmr complexes agrees with several high or medium resolution EM structures, including TtCmr complexes (Spilman et al., 2013; Staals et al., 2013; Taylor et al., 2015) and the Pyrococcus furiosus Cmr (PfCmr) complex (Ramia et al., 2014; Spilman et al., 2013), as well as the Cmr1-deficient chimeric Cmr-ssDNA complex (Osawa et al., 2015). In contrast, our stoichiometry does not agree with those obtained from low-resolution Csm complexes, including TthCsm complex (Staals et al., 2014), Sulfolobus solfataricus Csm (Rouillon et al., 2013), and Thermococcus onnurineus Csm (Park et al., 2017). Also, our stoichiometry differs from that for previously reported SthCsm, for which a stoichiometry was calculated on the basis of mass spectrometric data (Kazlauskiene et al., 2016). We note that the PfCmr complex was originally suggested to have three Cmr4 and three Cmr5 (Spilman et al., 2013), but later studies increased the number of Cmr4 subunits to four (Ramia et al., 2014), which is consistent with our present calculations. In addition, the 17 Å EM-map of TthCsm also fits the Csm1125364151 stoichiometry, instead of the formula suggested earlier (Csm1123364251) (Staals et al., 2014). It is possible that neither low-resolution EM nor mass spectrometry is sufficiently powerful to achieve accurate subunit assignments.
Model for Type III-A Interference
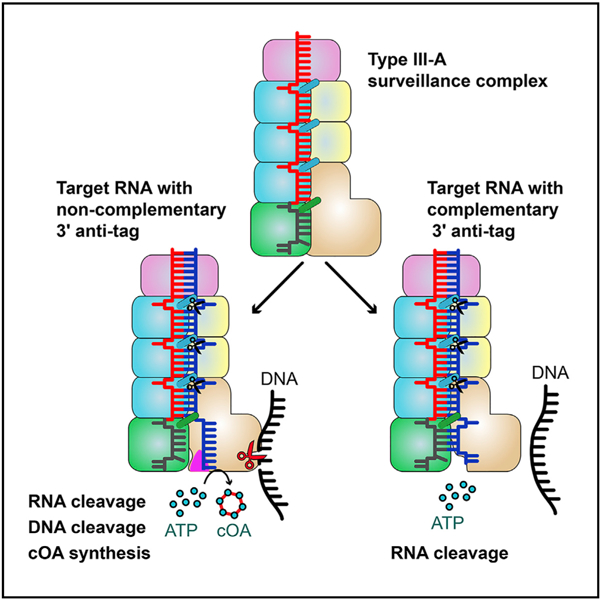
On the basis of the results presented here, we propose a possible mechanistic model of type III-A immunity against genetic invaders (Figure 7D). Type III immunity requires target transcription, which produces target nascent RNAs while generating a transcription bubble in transcribed DNA. First, crRNA specifically recognizes target RNA transcripts that contain sequences complementary to the spacer region. The Csm3 subunits periodically cleave the target transcripts. Second, the cognate target RNA binding activates both DNase and synthetase activities, under the condition that the 3′ anti-tag is not complementary to the 5′ tag of the crRNA. The non-complementary 3′ anti-tag, which fails to base pair with the 5′ tag of the crRNA, is positioned in the cleft of the Csm1 subunit. Although the 3′ anti-tag of the CTR is flexible and forms few interactions with the Csm1 subunit, the 3′ anti-tag binding drives the structural rearrangement of Csm1 Linker and Loop L1 (Figure 7A). These structural rearrangements of the Csm1 Linker and the interaction between the non-complementary 3′ anti-tag and the Csm1 Linker may stimulate the DNase and synthetase activities of the Csm1 subunit. In contrast, the complementary 3′ anti-tag does not trigger these two activities. The activated Csm1 HD domain cleaves non-template ssDNA within the transcription bubble of the template from which the target RNA is being transcribed (Elmore et al., 2016; Estrella et al., 2016; Samai et al., 2015). The activated Palm domains convert ATP to cOAs, which act as second messengers that allosterically regulates RNase activity of Csm6 (Kazlauskiene et al., 2017; Niewoehner et al., 2017). The presence of ATP or its analog may stabilize the 3′ anti-tag of CTR, without affecting the efficiency of DNA cleavage. Finally, both DNase and synthetase activities are rapidly de-activated as target RNA is cleaved and dissociates from the Csm complex and/or from the transcription elongation complex (Rouillon et al., 2018). In addition, the activated Csm6 then nonspecifically cleaves single-stranded RNA (ssRNA), thus temporally limiting both DNase and cOA synthetase activities (Rouillon et al., 2018). Taken together, this concerted three-pronged attack via type III CRISPR-Cas systems defends the host against actively transcribed genetic material of invading phages, thus minimizing the risk of parasite-induced cell death.
Interactions between the 3′ anti-tag region of CTR and Csm1 may allosterically activate the ssDNase and synthetase activities. In agreement with our results, one previous study revealed that the lack of a 3′ anti-tag region abolishes ssDNA cleavage (Kazlauskiene et al., 2016). The interactions between the 3′ anti-tag of CTR and Csm1 induce structural rearrangement at the Linker and Loop L1 regions. This rearrangement likely affects the dynamic properties of HD and Palm active sites. While the conformational changes of the HD and Palm active sites appear to be minimal, the resulting overall structural changes are likely to be important for both HD and Palm activities. It is therefore possible that the interactions of the 3′ anti-tag of the CTR with both Linker and loop L1 of the Csm1 subunit allosterically regulate the activities of the HD and Palm domains. As has been shown before for other protein complexes, allostery occurs even in the absence of measurable conformational change (Cooper and Dryden, 1984; Kornev and Taylor, 2015; Tsai et al., 2008). For instance, many protein kinases as well as G-coupled receptors (GPCR) are allosterically regulated; however, no significant conformational changes have ever been detected (Kornev and Taylor, 2015; Nussinov and Tsai, 2015). To provide an atomic-level description and an improved understanding of the allosteric activation and structural communication in the Csm complex, further studies that include molecular simulations and biochemical analysis are required.
Self and Non-self Discrimination
To distinguish non-self from self DNA, type I, II, and V CRISPR-Cas systems recognize short protospacer adjacent motif (PAM) sequences, which are present in invading nucleic acids, but are absent in the CRISPR array of the host (Anders et al., 2014; Hayes et al., 2016; Zetsche et al., 2015). In contrast, instead of depending on PAM recognition, type III CRISPR-Cas systems rely on non-complementarity between the 5′ tag of crRNA and 3′ anti-tag of RNA target (Elmore et al., 2016; Estrella et al., 2016; Kazlauskiene et al., 2016; Samai et al., 2015). The nucleotides (−2)′–(−5)′ within the 3′ anti-tag of the target RNA are essential for discriminating non-self from self nucleic acids. The non-complementarity between the 5′ tag of crRNA and 3′ anti-tag of the target RNA separates the crRNA and target RNA at the repeat region. As a consequence, the 3′ anti-tag region binds to the Csm1 Linker and Loop L1, activating the HD-nuclease and Palm-cOA synthetase activities. This finding agrees with the previous study that type III effectors are activated by non-complementarity rather than deactivated by complementarity between 3′ anti-tag region within target RNA and the 5′ tag of crRNA. A similar separation of crRNA and target RNA is found in type VI systems, where the nucleotide (−1)′ of target RNA is flipped outward, facing away from the crRNA-target RNA duplex (Liu et al., 2017). For RNA cleavage by Cas13a, it is essential that the non-complementary photospacer adjacent site (PFS) be flipped out, preventing base-pairing with crRNA (Liu et al., 2017).
Together with findings from previous studies, our results clearly show that complementarity between 5′ tag of crRNA and 3′ anti-tag region of target RNA plays a crucial role in regulating both HD-dependent nuclease activity of the Csm complex, together with its Palm-dependent cOAs production. Our data not only provide hitherto unknown structural details of the mechanisms responsible for crRNA-guided interference, but also show how subsequent target RNA-dependent nonspecific ssDNA and ssRNA cleavage are regulated by target RNA binding to Csm1.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Yanli Wang (ylwang@ibp.ac.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains
Csm complexes and mutant forms were expressed in E. coli BL21 (DE3) cells cultured at 37°C and 16°C in LB medium (OXOID), supplemented with 50 μg/ml kanamycin (AMRESCO), 34 μg/ml chloramphenicol (AMRESCO), 50 mg/ml streptomycin (AMRESCO), and 100 mg/ml ampicillin (AMRESCO).
Thermus thermophilus HB27 (GenBank: AE017221.1, AE017222.1) cells were grown in TBM medium (0.8% w/v tryptone, 0.4% w/v NaCl, 0.2% w/v yeast extract in Vittel mineral water supplemented with 0.5 mM MgSO4 and 0.5 mM CaCl2 at 70°C with aeration. For cultivation on plates, 2% w/v agar was added and cultivation temperature was decreased to 65°C. Transformants were selected on TBM plates containing 30 μg/ml of kanamycin. Plates containing 50 μg/ml hygromycin or 50 μg/ml hygromycin and 15 mg/ml bleomycin were used for selection of III-A+ and III-A− T. thermophilus mutants, respectively.
The III-A+ and III-A− strains were constructed from wild-type T. thermophilus HB27 by replacement of cmr4 (in III-A+) with hygromycin resistance genes or both cmr4 and csm3 (in III-A−) with hygromycin and bleomycin resistance genes via natural homologous recombination. To this end, the corresponding antibiotic resistance genes from plasmids pMH184 (a kind gift of Dr. J. Berenguer) and pWUR112 (a kind gift of Dr. J. van der Oost) were amplified with primers containing flanking areas of cmr4 or csm3 genes, the flanking areas were amplified from T. thermophilus HB27 genomic DNA and all fragments were cloned into the pT7blue (Nova-gene) plasmid using Gibson Assembly Mix (NEB). T. thermophilus cells were transformed with resulting plasmids according to protocol described previously (de Grado et al., 1999) and recombinant clones were selected on antibiotic-supplied medium and confirmed by PCR and sequencing.
METHOD DETAILS
Protein Expression and Purification
The genes csm1, csm2, csm3, csm4, csm5 and cas6 were amplified from Streptococcus thermophilus ND03 genomic DNA (GenBank accession number NC_017563) by polymerase chain reaction (PCR) and cloned into Duet expression vectors (Novagen). csm1 and csm4 were inserted into the pCDFDuet vector in two sub-cloning steps. csm3 and csm2 were inserted into the pRSFDuet vector with an N-terminal His6-tag of Csm3. csm5 was cloned into the pETDuet vector. The CRISPR sequence was synthesized (Sangon Biotech) and cloned into the pACYCDuet vector, together with cas6.
To express the Csm complexes (containing Csm1–5 and crRNA), the plasmids were co-transformed into the E. coli BL21 (DE3) cells (Novagen) that were induced with 0.2 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at OD600 of 0.6 for 15 h at 16°C. Cells were harvested and lysed by high-pressure cell disrupter in buffer containing 20 mM Tris-HCl, pH 7.5, 500 mM NaCl. After centrifugation, the supernatant was applied to Ni Sepharose (GE Healthcare). Bound proteins were eluted with buffer containing 200 mM imidazole and further purified on a Heparin column (GE Healthcare), eluting with buffer containing 20 mM Tris-HCl, pH 7.5 and 1 M NaCl. Proteins were collected and concentrated to a final concentration of 12 mg/ml.
All mutant forms were generated by site-directed mutagenesis and verified by sequencing, before being expressed and purified as described above.
Crystallization, Data Collection and Structure Determination
Crystals of the different Csm complexes were grown using the sitting drop vapor diffusion method at 16°C. Crystals were obtained by mixing 0.5 μL Csm complex solution and 0.5 μL of reservoir solution containing 0.1 M sodium citrate tribasic dihydrate, pH 5.0, 20% w/v polyethylene glycol 20,000. All crystals were cryo-protected by using corresponding reservoir buffers supplemented with 20% v/v glycerol and then flash frozen in liquid nitrogen.
All diffraction datasets were collected at beamline BL-19U1 at National Center for Protein Sciences Shanghai (NCPSS), and processed with HKL2000 (Otwinowski and Minor, 1997). The phases of Csm complex were solved by molecular replacement method with the cryo-EM map of Csm complex using PHENIX Phaser (Adams et al., 2002). The model was manually built and adjusted using the program COOT (Emsley et al., 2010). Iterative cycles of crystallographic refinement were performed using PHENIX. All data processing and structure refinement statistics for Csm complex are summarized in Table S1. All structure figures were prepared using PyMOL (http://www.pymol.org/).
In vitro Transcription and Purification of Target RNA
All target RNAs were synthesized by in vitro transcription by T7 RNA polymerase using a linearized plasmid DNA as template. In vitro transcription reactions were performed at 37°C for 4 h in buffer containing 0.1 M HEPES-KOH, pH 7.9, 30 mM MgCl2, 30 mM DTT, 2.5 mM each NTP, 2 mM spermidine, 0.1 mg/ml T7 RNA polymerase, and 50 ng/μl linearized plasmid DNA template. Target RNAs were then purified by gel electrophoresis on a 20% denaturing (8 M urea) polyacrylamide gel (PAGE) and electroelution System. The target RNAs were resuspended in diethy pyrocarbonate (DEPC) H2O.
Csm-Target RNA Complex and Csm-Target RNA-DNA Complex Reconstitution
The Csm-RNA (NTR or CTR) complex was reconstituted at 4°C for 30 min by incubating Csm complex and target RNA at a molar ratio of 1:1.2. The complex was then further purified on a Superdex 200 Increase gel filtration column (GE Healthcare) in buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl. Fractions were collected for cryo-EM.
Csm-RNA-DNA complex was reconstituted by incubating purified Csm-NTR2 complex and DNA were mixed at a molar ratio of 1:1.2 and incubated at 4°C for 30 min. The complex was further purified on a Superdex 200 Increase gel filtration column (GE Healthcare) in buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM NaCl. Fractions were collected for cryo-EM.
For Csm-CTR1-ATP/AMPPNP complex reconstitution, purified Csm-CTR1 complex was mixed with 2 mM MgCl2 and 1 mM ATP or 0.5 mM AMPPNP and incubated at 4°C for 30 min. 0.5 mM ATP and 2 mM MgCl2 were added to Csm-NTR complex for Csm-NTRA-TP complex reconstitution.
Cryo-EM Specimen Preparation and Imaging
3 μL of each purified complex samples at a concentration of ~0.5mg/ml was spotted onto freshly glow-discharged holey carbon grids (GIG-C322). After incubation for 10 s, excess sample was blotted with filter paper for 2 s and the grid was flash-frozen in liquid ethane using a Leica EMGP device. All cryo-EM samples were prepared using the same procedure at 10°C and 60% relative humidity. The images were collected on a 300-kV FEI Titan Krios electron microscope equipped with Gatan K2 Summit direct electron detector positioned post a GIF quantum energy filter with energy filtered mode (the zero loss peak sits in the middle of a slit with the width of 20 eV), the camera was in super-resolution mode and the physical pixel size is 1.04 Å (0.52 Å super-resolution pixel size). The defocus range was set between 1.0 and 3.0 μm. Each image was exposed for 8 s, resulting in a total dose of ~60 e− Å−2 and 32 frames per movie stack.
Image Processing
Each movie stack was binned (1.04 Å pixel size) and then subjected to motion correction by the software MotionCor2 (Zheng et al., 2017). This procedure produced two images for each movie stack by summing all frames with or without dose-weighting. The contrast transfer function (CTF) parameters were measured by CTFFIND4 (Rohou and Grigorieff, 2015) using the summed image without dose-weighting. The summed image with dose-weighting was used for further data processing in Relion2.0 (Kimanius et al., 2016).
A subset of the Csm-NTR complex particles were manually picked and processed with reference-free 2D classification. Five 2D class average images were selected as references for automatic particle picking of the complete dataset. A total of 611,889 particles were picked in 1,541 micrographs and processed by reference free 2D classification. 247,202 particles were kept for further 3D classification which were classified into eight classes using the initial model created in RELION2.0. One class containing 133,127 particles were selected for further 3D auto-refinement, which resulted in a 3.16 Å density map estimated based on the gold-standard Fourier shell correlation with 0.143 criterion. To further improve the density, focused refinement was used for Csm1 and the remaining part of the complex. Focused refinement was performed by applying soft masks to the region of interest, with final alignments from previous global refinements used with local searches. Focus refinement performed on Csm1 and the remaining part resulted in resolutions of 3.60 Å and 3.13 Å, respectively. The remaining complexes were processed in the same procedure. Particles were automatic selected using the references used in Csm-NTR complex, and the final density map of Csm-NTR complex was used as initial model for 3D classification. For Csm-CTR2-dsDNA complex, 684,150 particles were picked in 1,218 micrographs and 413,326 particles were used for 3D classification. 75,887 particles were selected for further 3D auto-refinement and resulted in a 3.05 Å density map. For Csm-CTR1-AMPPNP complex, 364,667 particles were picked in 893 micrographs and 194,274 particles were used for 3D classification. 61,538 particles were selected for further 3D auto-refinement and resulted in a 3.20 Å density map. For Csm-CTR1-ATP complex, 598,192 particles were picked in 1,525 micrographs and 193,353 particles were used for 3D classification. 58,587 particles were selected for further 3D auto-refinement and resulted in a 3.37 Å density map. The local resolution of the final maps were calculated using ResMap (Kucukelbir et al., 2014).
Model Building and Refinement
The model of Csm-NTR complex was built de novo in Coot and the 3.15 Å electron density map of Csm-NTR complex was of high quality, allowing the tracing of most of the amino-acid residues. The models of the rest complexes (Csm-CTR2-dsDNA, Csm-CTR1-ATP, Csm-CTR1-AMPPNP, Csm-NTR-ATP) were generated by docking the refined Csm-NTR model into cryo-EM maps using UCSF Chimera (Pettersen et al., 2004), and manually fitting the model to the map in Coot. All models were improved iteratively by cycles of real-space refinement using PHENIX (Adams et al., 2002) and manual modification in Coot until no further improvement could be obtained. The refinement statistics of the structural models are summarized in Tables S2 and S3.
In vitro Target RNA Cleavage Assays
An 80-nt length target RNA was used for in vitro cleavage assays. 10 μM target RNA and 2 μM Csm complex were mixed in buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl. Reactions were initiated by adding 10 mM MgCl2 and incubated at 37°C for 45 min. Reactions were stopped by adding 2 × loading buffer. Cleavage products were analyzed by 20% denaturing (8M Urea) PAGE with TBE buffer, and visualized by toluidine blue staining.
In vitro DNA Cleavage Assays
Synthesized 5′-Cy3-labeled DNA (Sangon Biotech) was used for in vitro cleavage assays. 500 nM Csm, 500 nM RNA, and 50 nM 5′-Cy3-labeled DNA were mixed in 20 mM Tris-HCl, pH 7.5, 200 mM NaCl. Reactions were initiated by addition of 10 mM MnCl2 and performed at 37°C for 30 min. Reactions were stopped by adding 2 × loading buffer and were then quenched at 95°C for 5 min. Cleavage products were resolved by 20% denaturing (8M Urea) PAGE, and visualized by using FluorChem system (Proteinsimple).
In vitro cOA Synthesis Assays
200 nM Csm complex was mixed with 200 nM target RNA, 50 μM ATP and 10 μCi of [α−32P] ATP in reaction the buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl. The cOA synthesis reactions were initiated by addition of 10 mM MnCl2 and carried out at 37°C for 30 min. Reactions were stopped by adding 20 mM EDTA. Products were separated by Thin layer chromatography (TLC) on PEI-Cellulose F plate (Merck) in a buffer containing acetic acid 1N/ethanol 80/20 v/v with addition of 0.5 M LiCl and detected by phosphorimaging using a Typhoon FLA7000 imager (GE). ADP and AMP, used as a TLC marker, were generated by heating [α−32P] ATP at 95°C for 4 h. Products were quantified using densitometry by ImageJ software. Statistical analyses were performed using GraphPad Prism 7 software. Assays were performed in triplicate and error bars were calculated as standard deviation.
In vivo Interference Assays
To construct protospacer-containing plasmid, a oligonucleotide (5′-CTCTTTCAGGATCCACGCAAACTCCCTTCCTTGGGGCTTA-3′) corresponding to first spacer in CRISPR array #8 (Henne et al., 2004) and complementary oligonucleotide were annealed and cloned into the pMK18 E. coli – T. thermophilus shuttle vector (de Grado et al., 1999, a kind gift of Dr. J. Berenguer) between the HindIII and EcoRI restriction sites utilizing standard molecular cloning protocols. Protospacer plasmids carrying mismatched with crRNA spacer part was constructed in the same way using appropriate double-stranded oligonucleotides. For construction of a terminator-carrying plasmid, the t6 terminator of T. thermophilus phage FYS40 that efficiently terminates transcription by T. thermophilus RNA polymerase in vitro was cloned in appropriate orientation into the EcoRI restriction site of pMK18.
For in vivo interference assays, T. thermophilus cells were transformed with protospacer-bearing plasmids according to protocol described previously (de Grado et al., 1999). 10 μL drops of 10-fold dilutions of transformation mixtures were deposited on the surface of TBM agar plates supplemented with kanamycin and results were recorded after growth for 18–24 hours.
Strand-specific RNA-seq analysis
25 mL of OD600 = 0.4 cultures of T. thermophilus cells transformed with pMK18 were centrifuged at 3500 g for 3 minutes. Total RNA was extracted from pellets with ExtractRNA (Evrogen) solution according to manufacturer’s recommendations. rRNA depletion, cDNA libraries preparation (with fragmented and non-fragmented RNA), high-throughput RNA 75 cycles sequencing and primary data analysis were performed by the Core Unit Systems Medicine, University of Würzburg, Germany using Illumina kits and NextSeq500 platform. To estimate the transcription levels of different genomic and plasmid regions/strands, read counts for region of interest were counted using Samtools package (Li et al., 2009). RPKM values were calculated and plotted.
QUANTIFICATION AND STATISTICAL ANALYSIS
Biochemical assays were repeated three times, and representative results are shown. Crystallographic data collection and refinement statistics are listed in Table S1. Cryo-EM data collection and refinement statistics for the overall dataset and datasets used for focused refinement are listed in Tables S2 and S3.
DATA AND SOFTWARE AVAILABILITY
Data Resources
The accession numbers for the atomic coordinates reported in this paper are PDB: 6IFN (crystal structure of Csm binary complex), 6IFL (Csm-NTR complex), 6IFY (Csm-CTR1 complex), 6IG0 (Csm-CTR1-ATP complex), 6IFR (Csm-NTR-ATP complex), 6IFK (Csm-CTR1-AMPPNP complex), 6IFU (Csm-CTR2-dsDNA complex) and 6IFZ (Csm-CTR2-ssDNA complex).
The accession numbers for the cryoEM density maps reported in this paper are EM Data Bank: EMD-9654 (Csm-NTR complex), EMD-9658 (Csm-CTR1 complex), EMD-9660 (Csm-CTR1-ATP complex), EMD-9656 (Csm-NTR-ATP complex), EMD-9653 (Csm-CTR1-AMPPNP complex), EMD-9657 (Csm-CTR2-dsDNA complex) and EMD-9659 (Csm-CTR2-ssDNA complex).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Urea | AMRESCO | Cat# 0378–5KG |
| TRIS | AMRESCO | Cat# 0497–5KG |
| Toluidine Blue | AMRESCO | Cat# 0672–25G |
| HEPES | Sigma-Aldrich | Cat# H3375–250G |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | Cat# C8532–1KG |
| PEG 20,000 | Sigma-Aldrich | Cat# 81300–1KG |
| Glycerol | Sigma-Aldrich | Cat# G5516–1L |
| T7 RNA polymerase | Home-made | N/A |
| Spermindine | Sigma-Aldrich | Cat# S2626–1G |
| [α-32P] ATP | PerkinElmer | Cat# NEG503H250UC |
| ATP | Sigma-Aldrich | Cat# A2383 |
| UTP | Sigma-Aldrich | Cat# U6625 |
| CTP | Sigma-Aldrich | Cat# C1506 |
| GTP | Sigma-Aldrich | Cat# G8877 |
| AMPPNP | Sigma-Aldrich | Cat# A2647–25MG |
| AxyPrep Plasmid Maxiprep Kit 25-prep | Axygen | Cat# AP-MX-P-25G |
| Critical Commercial Assays | ||
| Chelating Sepharose Fast Flow | GE Healthcare | Cat# 17-0575-02 |
| HiTrap Heparin | GE Healthcare | Cat# 17-1152-01 |
| Superdex 200 Increase, 10/300 GL | GE Healthcare | Cat# 28-9909-44 |
| Deposited Data | ||
| The coordinates of SthCsm binary complex | This study | PDB: 6IFN |
| The coordinates of SthCsm-NTR complex | This study | PDB: 6IFL |
| The coordinates of SthCsm-CTR1 complex | This study | PDB: 6IFY |
| The coordinates of SthCsm-CTR2-bubbled DNA complex | This study | PDB: 6IFU |
| The coordinates of SthCsm-CTR2-ssDNA complex | This study | PDB: 6IFZ |
| The coordinates of SthCsm-CTR1-AMPPNP complex | This study | PDB: 6IFK |
| The coordinates of SthCsm-CTR1-ATP complex | This study | PDB: 6IG0 |
| The coordinates of SthCsm-NTR-ATP complex | This study | PDB: 6IFR |
| The density map of SthCsm-NTR complex | This study | EMDB: EMD-9654 |
| The density map of SthCsm-CTR1 complex | This study | EMDB: EMD-9658 |
| The density map of SthCsm-CTR2-bubbled DNA complex | This study | EMDB: EMD-9657 |
| The density map of SthCsm-CTR2-ssDNA complex | This study | EMDB: EMD-9659 |
| The density map of SthCsm-CTR1-AMPPNP complex | This study | EMDB: EMD-9653 |
| The density map of SthCsm-CTR1-ATP complex | This study | EMDB: EMD-9660 |
| The density map of SthCsm-NTR-ATP complex | This study | EMDB: EMD-9656 |
| Experimental Models: Organisms/Strains | ||
| Escherichia coli BL21 (DE3) | Novagen | Cat# 69450 |
| Thermus thermophilus HB27 | Henne et al., 2004 | N/A |
| Thermus thermophilus HB27 III-A+ | This study | N/A |
| Thermus thermophilus HB27 III-A− | This study | N/A |
| Recombinant DNA | ||
| pCDFDuet-Csm1-Csm4, various mutants | This study | N/A |
| pRSFDuet-His6-Csm3-Csm2, various mutants | This study | N/A |
| pETDuet-Csm5 | This study | N/A |
| pACYCDuet-Cas6-pre-crRNA | This study | N/A |
| Oligonucleotides | ||
| RNA and DNA | This study | Table S4 |
| Software and Algorithms | ||
| PHENIX | Adams et al., 2002 | http://www.phenix-online.org/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | Molecular Graphics System, Version 1.8 Schrodinger | https://pymol.org/2/ |
| MotionCor2 | Zheng et al., 2017 | http://msg.ucsf.edu/em/software/motioncor2.html |
| CTFFind4 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctffind4 |
| RELION2.0 | Kimanius et al., 2016 | http://www2.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| USCF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| ResMap | Kucukelbir et al., 2014 | http://resmap.sourceforge.net/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Amicon Ultra Centrifugal Filters Ultracel-30K | Merck Millipore Ltd | Cat# UFC903096 |
| 3 Well Midi Crystallization Plate (Swissci) | HAMPTON RESEARCH | Cat# HR3–125 |
| GiG-C322 grids (2 mm holes and spacing) | Beijing Zhongjingkeyi Technology | Cat# GIG-2020–300 |
Highlights.
Crystal structure of the type III-A CRISPR-RNA-guided surveillance complex
Cryo-EM structures of cognate and non-cognate target RNA-bound Csm complexes
Complementary and non-complementary 3′ anti-tag binds at two distinct sites
Target RNA binding allosterically activates ssDNA cleavage and cOA synthesis
ACKNOWLEDGMENTS
We thank the staff of the BL-17U1 and BL-19U1 beamlines at Shanghai Synchrotron Radiation Facility. We thank B. Zhu, X. Huang, G. Ji, D. Fan, F. Sun, and the other staff at the Center for Biological Imaging (IBP, CAS) for their support in cryo-EM data collection. We thank Matvey Kolesnik for assistance with RNA-seq data analysis and Hongjie Zhang at IBP for the guidance in handling radiolabeled chemicals. We thank Dr. Torsten Juelich (Peking University, China) for critical reading of the manuscript and linguistic assistance and Prof. Ruiming Xu for scientific discussion. This work was supported by the Chinese Ministry of Science and Technology (2017YFA0504203 and 2017YFA0504700), the Natural Science Foundation of China (31630015, 91440201, 31571335, 31700662, and 31400640), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010203 and XDB08030204), and the Opening Project of the State Key Laboratory of Microbial Resources. X.Z. received scholarships from the National Thousand (Young) Talents Program from the Office of Global Experts Recruitment in China. Work in K.S. laboratory is supported by NIH (R01 GM10407) and Russian Science Foundation (14-14-00988). D.A. was supported by Foundation for Assistance to Small Innovative Enterprises program “UMNIK” under the contract N 9188GU/2015 (0019877) of 12/24/2015.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and four tables and can be found with this article online at https://doi.org/10.1016/j.cell.2018.10.052.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, and Terwilliger TC (2002). PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr 58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, and Jinek M (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Cooper A, and Dryden DT (1984). Allostery without conformational change. A plausible model. Eur. Biophys. J 11, 103–109. [DOI] [PubMed] [Google Scholar]
- de Grado M, Castán P, and Berenguer J (1999). A high-transformation-efficiency cloning vector for Thermus thermophilus. Plasmid 42, 241–245. [DOI] [PubMed] [Google Scholar]
- Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, and Terns MP (2016). Bipartite recognition of target RNAs activates DNA cleavage by the type III-B CRISPR-Cas system. Genes Dev. 30, 447–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrella MA, Kuo FT, and Bailey S (2016). RNA-activated DNA cleavage by the type III-B CRISPR-Cas effector complex. Genes Dev. 30, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg GW, Jiang W, Bikard D, and Marraffini LA (2014). Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514, 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, and Terns MP (2009). RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139, 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RP, Xiao Y, Ding F, van Erp PB, Rajashankar K, Bailey S, Wiedenheft B, and Ke A (2016). Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature 530, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne A, Brüggemann H, Raasch C, Wiezer A, Hartsch T, Liesegang H, Johann A, Lienard T, Gohl O, Martinez-Arias R, et al. (2004). The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol 22, 547–553. [DOI] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PB, Carter J, Westra ER, Brouns SJ, van der Oost J, Terwilliger TC, Read RJ, and Wiedenheft B (2014). Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č, and Siksnys V (2016). Spatiotemporal control of type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306. [DOI] [PubMed] [Google Scholar]
- Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G, and Siksnys V (2017). A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609. [DOI] [PubMed] [Google Scholar]
- Kimanius D, Forsberg BO, Scheres SH, and Lindahl E (2016). Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, and Taylor SS (2015). Dynamics-driven allostery in protein kinases. Trends Biochem. Sci 40, 628–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A, Sigworth FJ, and Tagare HD (2014). Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li X, Ma J, Li Z, You L, Wang J, Wang M, Zhang X, and Wang Y (2017). The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 170, 714–726. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. (2015). An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Micro-biol. 13, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA (2015). CRISPR-Cas immunity in prokaryotes. Nature 526, 55–61. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, and Sontheimer EJ (2010). Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, and Jinek M (2016). Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Garcia-Doval C, Rostøl JT, Berk C, Schwede F, Bigler L, Hall J, Marraffini LA, and Jinek M (2017). Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548. [DOI] [PubMed] [Google Scholar]
- Nussinov R, and Tsai CJ (2015). The role of allostery in the termination of second messenger signaling. Biophys. J 109, 1080–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Sato C, and Numata T (2015). Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol. Cell 58, 418–430. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Park KH, An Y, Jung TY, Baek IY, Noh H, Ahn WC, Hebert H, Song JJ, Kim JH, Oh BH, and Woo EJ (2017). RNA activation-independent DNA targeting of the Type III CRISPR-Cas system by a Csm complex. EMBO Rep. 18, 826–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Ramia NF, Spilman M, Tang L, Shao Y, Elmore J, Hale C, Cocozaki A, Bhattacharya N, Terns RM, Terns MP, et al. (2014). Essential structural and functional roles of the Cmr4 subunit in RNA cleavage by the Cmr CRISPRCas complex. Cell Rep. 9, 1610–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, and White MF (2013). Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 52, 124–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Athukoralage JS, Graham S, Grüschow S, and White MF (2018). Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 7, e36734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, and Marraffini LA (2015). Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell 161, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard NF, Glover CV 3rd, Terns RM, and Terns MP (2016). The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosinespecific endoribonuclease. RNA 22, 216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, and Stagg S (2013). Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell 52, 146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. (2013). Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell 52, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, et al. (2014). RNA targeting by the type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol. Cell 56, 518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas Č, Nwokeoji AO, Dickman MJ, Horvath P, and Siksnys V (2014). Programmable RNA shredding by the type III-A CRISPR-Cas system of Streptococcus thermophilus. Mol. Cell 56, 506–517. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Zhu Y, Staals RH, Kornfeld JE, Shinkai A, van der Oost J, Nogales E, and Doudna JA (2015). Structural biology. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 348, 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, del Sol A, and Nussinov R (2008). Allostery: absence of a change in shape does not imply that allostery is not at play. J. Mol. Biol 378, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, and Wang Y (2014). Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature 515, 147–150. [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data Resources
The accession numbers for the atomic coordinates reported in this paper are PDB: 6IFN (crystal structure of Csm binary complex), 6IFL (Csm-NTR complex), 6IFY (Csm-CTR1 complex), 6IG0 (Csm-CTR1-ATP complex), 6IFR (Csm-NTR-ATP complex), 6IFK (Csm-CTR1-AMPPNP complex), 6IFU (Csm-CTR2-dsDNA complex) and 6IFZ (Csm-CTR2-ssDNA complex).
The accession numbers for the cryoEM density maps reported in this paper are EM Data Bank: EMD-9654 (Csm-NTR complex), EMD-9658 (Csm-CTR1 complex), EMD-9660 (Csm-CTR1-ATP complex), EMD-9656 (Csm-NTR-ATP complex), EMD-9653 (Csm-CTR1-AMPPNP complex), EMD-9657 (Csm-CTR2-dsDNA complex) and EMD-9659 (Csm-CTR2-ssDNA complex).