Abstract
Stem cells are a sub population of cell types that form the foundation of our body, and have the potential to replicate, replenish and repair limitlessly to maintain the tissue and organ homeostasis. Increased lifetime and frequent replication set them vulnerable for both exogenous and endogenous agents-induced DNA damage compared to normal cells. To counter these damages and preserve genetic information, stem cells have evolved with various DNA damage response and repair mechanisms. Furthermore, upon experiencing irreparable DNA damage, stem cells mostly prefer early senescence or apoptosis to avoid the accumulation of damages. However, the failure of these mechanisms leads to various diseases, including cancer. Especially, given the importance of stem cells in early development, DNA repair deficiency in stem cells leads to various disabilities like developmental delay, premature aging, sensitivity to DNA damaging agents, degenerative diseases, etc. In this review, we have summarized the recent update about how DNA repair mechanisms are regulated in stem cells and their association with disease progression and pathogenesis.
Keywords: DNA damage and repair, Stem cells, Disease, Gene Therapy
1. Introduction
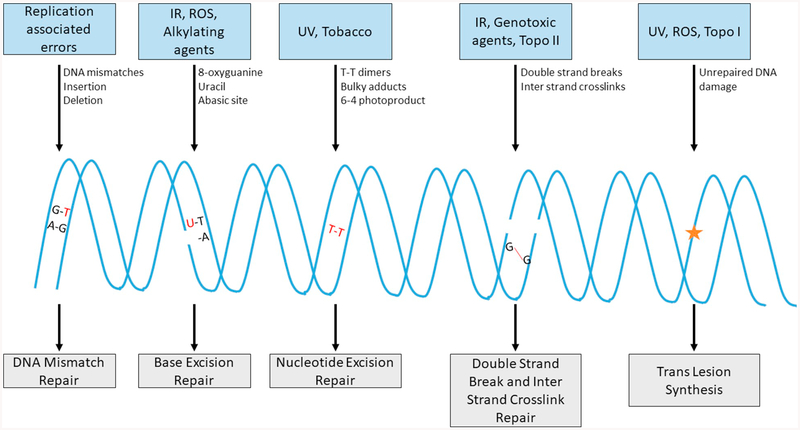
DNA damage response (DDR) and repair processes are dynamic mechanisms and plays important role in maintaining the integrity of genomic DNA, which is critical to pass the precise genomic details to next generation. In human body, each individual cell faces 1000 to 10,000 genotoxic insults from endogenous metabolic byproducts such as reactive oxygen species (ROS), lipid peroxidation, inflammation, estrogen metabolites, etc. [1,2]. In addition to the endogenous reactive metabolites, several exogenous agents like radiation, smoking and several organic and inorganic chemicals present in the environment can cause DNA lesions upon exposure [3]. In order to counter these potential injuries to DNA, cells have evolved with various repair mechanisms with the ability to detect and fix these damages (Fig. 1). Particularly, mismatches during DNA replication are fixed by mismatch repair (MMR) [4], damaged bases are restored by base excision repair (BER) [5], bulky base lesions are repaired by nucleotide excision repair (NER) [6], inter-strand cross-links (ICLs) are repaired by Fanconi anemia (FA) pathway [7] and highly lethal DNA double-strand breaks (DSBs) are repaired by non-homologous end joining (NHEJ) [8] and/or homologous recombination (HR) [9] and some of these damages are also bypassed by low fidelity trans-lesion DNA synthesis (TLS) [10]. Failure in any of these DNA damage sensing and repair mechanisms can cause various diseases including cancer [11].
Fig. 1.
Different types of DNA damage and repair mechanisms. Cells in human body undergoes various types of DNA damages like mismatch DNA during replication, damaged base, bulky adducts and potentially lethal double strand breaks (DSBs) which are recognized and repaired by mismatch repair, base excision repair, nucleotide excision repair and DSB repair (non-homologous end joining and/or homologous recombination) respectively. These damages are also bypassed by translesion synthesis during replication to be repaired later.
Stem cells are undifferentiated cells in multicellular organisms, which have the ability to differentiate into multiple lineages [12]. Stem cells possess unlimited potential to replenish when there is a need to repair the tissues and organs in our body. Thus, the maintenance and preservation of stem cells have been much of the focus for research to use them in treatment and prevention of certain diseases and to restore damaged tissues and organs, also called as stem cell therapy [13]. Various types of stem cells include, (1) embryonic stem cells (ESCs), which can be isolated from the inner mass of blastocyst, and pluripotent in nature, meaning, they can differentiate into any cell type of the body except placenta and umbilical cord, (2) adult stem cells (ADCs), which are mostly multipotent, meaning, they can differentiate into specific lineages of cells in which they are present, (3) induced pluripotent stem cells (iPSCs) are generated from fully differentiated cells by engineering them in the lab to behave like ESCs [14] and (4) multi lineage-differentiating stress-enduring (MUSE) cells, which are the subpopulation of mesenchymal stem cells (MSCs) and has the pluripotency ability to differentiate into endoderm, ectoderm and mesoderm lineages compared to rest of the non-MUSE MSCs subpopulation [15,16].
2. Association between DNA repair and stem cells
Unlike normal somatic cells, stem cells can remain quiescent and start to replicate limitlessly when needed and can survive for the long term. This increased survival stretch of stem cells puts them in an increased risk of accumulating more endogenous and exogenous-induced DNA damages compared to the short-lived normal cells [17]. Additionally, replication-associated mutations are more common in normal stem cells as they replicate more times than normal somatic cells [18]. Stem cells are also at increased risk of transferring acquired mutations to their daughter cells [17]. These challenges mark stem cells to evolve with dynamic and effective DNA repair mechanisms. For example, studies shown that mouse embryonic stem cells (mESCs) and human embryonic stem cells (hESCs) possess superior DNA repair efficiency compared to their respective somatic cells [19,20]. Interestingly, several DNA damage sensing and repair factors are overexpressed in stem/progenitor cells compared to non-stem cells (Table 1).
Table 1.
DDR genes/proteins that are upregulated in various types of stem/progenitor cells in comparison with the non-stem cells.
| DDR Pathway | Members of DDR | References |
|---|---|---|
| MMR | MSH2, MSH6, PMS2, MLH1, EXO1, MSH3 | [21–23] |
| BER | APE1, PARP1, XRCC1, DNA Ligase III, FEN1, NTHL1, UNG, POLE, POLE2, AAG | [20–24] |
| NER | XPA, XPC, XRCC1, RAD23B, GTF2H2, GTF2H3 | [20,22,24] |
| DSB/HR | RAD50, RAD51, RAD52, RAD54, BRCA1, BRCA2, NBS1, XRCC5, BLM, WRN | [20,22,23,25] |
| DSB/NHEJ | KU70, KU80, NHEJ1, XRCC6 | [22,25] |
| ICLR | FANCG, FANCL, FANCA | [20,22] |
| TLS | PCNA | [22] |
Furthermore, microarray based gene expression profiles indicate significant increase in the expression of genes associated with DNA repair in hESCs, when compared to differentiated cells [20]. In fact, as an additional layer of protection to genomic DNA, ESCs preferentially uses the efficient HR-mediated DSB repair rather than the inefficient NHEJ mediated DSB repair [25]. Not wanting to survive with unrepairable DNA damage, ESCs have constitutively active pro-apoptotic protein Bax in the Golgi, which translocate to mitochondria and initiates apoptosis-mediated cell death in a p53 dependent manner [26]. Thus, stem cell’s integrity is well preserved by DNA repair mechanisms, and in its fiasco, it affects the function and/or survival of stem cells, which in turn leads to the manifestation of numerous diseases, as summarized in Table 2. The role of DNA repair and how it influences cancer stem cells (CSCs) has already been revised in the previous issue of this journal [27]. Thus, we focused on the other diseases apart from cancer. In this review, we have summarized the recent findings of how DNA repair and signaling mechanisms are regulated in stem cells, and their association with human health and disease.
Table 2.
List of human diseases and disorders affected by deficiency in various DDR pathways.
| Disease or disorder | DDR pathway | Genes involved |
|---|---|---|
| Lynch syndrome | MMR | MLH1, MSH2, MSH6, PMS2 |
| Turcot syndrome | MMR | MSH2, MSH6, MLH1, PMS1, PMS2 |
| Muir-Torre syndrome | MMR | MSH2, MLH1 |
| Metabolic syndrome | BER | NEIL1 |
| Atherosclerosis | BER | OGG1 |
| Xeroderma pigmentosa | NER | XP (A–G) |
| Cockayne syndrome | NER | ERCC6, ERCC8 |
| Trichothiodystrophy | NER | Various XP and CS related |
| Ataxia telangiectasia | Global | ATM |
| Cerebro-oculo-facial-skeletal syndrome | NER | ERCC1, 2, 5 or 6 |
| AT-like disease | Global | MRE11 |
| Seckel syndrome 1 | Global | ATR |
| Nijmegen breakage syndrome | DSB/NHEJ | NBS |
| Ligase IV syndrome | DSB/NHEJ | LIGASE IV |
| Werner syndrome | DSB/HR | WRN |
| Rothmund–Thomson syndrome | DSB/HR | RECQL4 |
| Blooms syndrome | DSB/HR | BLM |
| Spinocerebellar ataxia with axonal neuropathy | DSB | TDP1 |
| Fanconi anemia | ICL | FA genes |
| Karyomegalic interstitial nephritis | ICL | FAN1 |
| Xeroderma pigmentosum variant | TLS | POLH |
3. MMR and stem cells
3.1. Mechanism of MMR
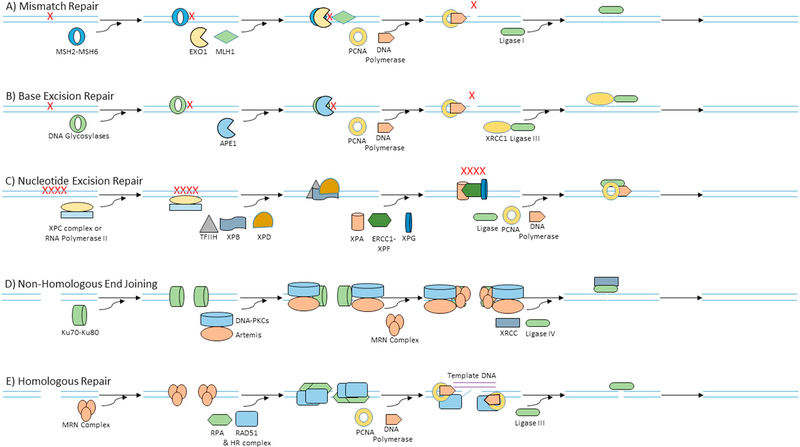
High fidelity DNA polymerases are typically involved in DNA synthesis during replication. Nonetheless, these polymerases still do make errors at a rate of about 1 per every 100,000 nucleotides. This means every time a diploid cell with 6 billion base pairs divides, it would make up to 120,000 mistakes. Most of these mistakes are immediately fixed by MMR mechanism (Fig. 2A). Mismatches are primarily detected by MSH2-MSH6, which then recruits MLH1 to facilitate incision of DNA on either side of the mismatch. EXO1 nucleases then remove the mismatch. DNA polymerase and PCNA resynthesize the DNA with the help of template strand to preserve the genetic information, which is finally ligated by Ligase I [5].
Fig. 2.
Different DNA repair mechanisms. A) Mismatch repair is initiated by MSH2-MSH6 complex, which recognizes the mismatched bases and recruits nuclease (MLH1 and EXO1) to incise and remove the mismatch. The abasic site is then resynthesized by DNA polymerase and PCNA, and the gap is ligated by Ligase I. B) Base excision repair recognizes damaged bases using DNA glycosylases, which then recruits APE1 to cleave the abasic site. DNA polymerase and PCNA then add the missing bases which is finally ligated by XRCC1-Ligase IIIα complex. C) Nucleotide excision repair recognizes DNA adducts in total genomic DNA using XPC-hHR23B complex (global genome repair) and in transcribed DNA strand of active genes (transcription-coupled repair) by the presence of RNA polymerase II stalled at the damage site. Transcription factor TFIIH and helicases XPB and XPD are then recruited to unwind the strands to give access to XPA-RPA complex to recruit nucleases ERCC1-XPF and XPG which then incise the damaged nucleotides. After incision, DNA polymerase and PCNA synthesizes the new nucleotides and ligated by ligases. D) DSBs before replication and also sometimes after replication is repaired by non-homologous end joining. Ku70-Ku80 complex recognizes DSB and recruits DNA-PKcs and Artemis, which then facilitates the DNA end processing by MRN (MRE11-RAD50-NBS1) complex. After DNA end processing, the broken strands are ligated by XRCC-Ligase IV complex. E) DSBs after replication are mostly repaired by homologous recombination. MRN complexes recognize and initiate the resection of DSB DNA ends. The exposed single strands are coated by RPA and replaced by RAD51, which then initiates the crossover. PCNA and polymerase synthesizes new nucleotides with the help of template DNA and the free ends are finally ligated using ligase.
3.2. Regulation of MMR in stem cells
Individuals with MMR deficiency are susceptible to various types of cancers including leukemia and colorectal cancer (CRC). Additionally, mutations in the MMR associated genes often mark with microsatellite instability syndrome (MSI) and early onset of CRC. MMR deficient hematopoietic stem cells (HSCs) reveal increased hematopoietic repopulation defect and stem cell exhaustion in response to temozolomide-induced genomic insults [28]. Furthermore, mESCs that are deficient in Msh2 and/or Msh3 shows increased levels of dinucleotide repeat instability [29]. Analysis of MSI in ten different hESC lines at different passages revealed that MSI occurs at a very low frequency compared to the normal somatic cells [30]. The mechanism was not clear until recently, as follows. Lentiviral-mediated knockdown of MMR in hPSCs displays increased sensitivity towards DNA alkylation damage in an MMR-dependent manner. Notably, damaged somatic cells experience stable G2/M arrest only during the second cell cycle after DNA damage. On the other hand, pluripotent stem cells (PSCs) instantaneously activate apoptosis signaling during the first cell cycle. From these results, it is conceivable that DDR and cell death signaling mechanisms are differentially regulated in stem cells to avoid accumulation of mutations or MSI in their DNA [31].
Even though stem cells maintain these high standards of MMR, certain external factors like hypoxia can obliterate the integrity of MMR efficiency in stem cells. Prolonged exposure to hypoxic conditions in neural stem cells (NSCs) reduces the efficacy of MMR. Mechanistically, promoter regions of MMR associated genes MLH1 and MSH6 were found to be hypoacetylated and hypermethylated. Nevertheless, treatment with histone deacetylase (HDAC) inhibitors facilitates the re-expression of MMR associated genes [32]. These studies suggest that variations in physiologic conditions such as hypoxia could alter epigenetic milieu of gene regulation, and influence integrity of DDR and pose threat to genomic stability. Recent study demonstrates that inhibition of SIRT1 can lead to DNA damage and trigger cell death in hESCs, but not in differentiated cells. Systematically, it is also found that SIRT1 inhibition leads to decreased expression of MSH2 and MSH6 [33]. Furthermore, hESCs isolated from trinucleotide repeat expansions associated diseases (Huntington Disease and Myotonic Dystrophy 1) were evaluated for MMR efficiency before and after differentiation. MSH2, MSH3 and MSH6 levels were often found to be downregulated in association with increased genomic instability in differentiated cells [34]. Conversely, the role of MMR in irradiated HSC isolated from mice did not show a dependence on MMR status [35]. Increased replication rate in stem cells has put them on increased risk to develop mismatches. It is evident from the above results that various types of stem cells engage increased expression of MMR associated genes/proteins to efficiently repair errors during replication and to prevent mutations.
3.3. MMR deficiency in stem cells and diseases
Most of the disease or syndromes associated with deficient MMR results in a predisposition to cancers like Lynch syndrome, Turcot syndrome and Muir-Torre syndrome (MTS). Cells isolated from cutaneous and internal tumors from MTS exhibits increased MSI and also lacks MSH-2 staining, which confirms the role of MMR in MTS [36]. In order to demonstrate their pathologic significance, the two MSH2 gene variants found in Lynch Syndrome patients MSH2-Y165D and MSH2-Q690E were recreated in mESCs. Exposure of Msh2-Y165D and Msh2-Q690E mESCs revealed MMR deficiency in these cells in repair of ultraviolet (UV) irradiation induced DNA damages [37].
4. BER and stem cells
4.1. Mechanism of BER
BER primarily repairs the common base modifications like oxidized, deaminated, depurinated and alkylated bases (Fig. 2B). Apurination is one of the common damage that happens on an average of 10,000–100,000 times per day in a human cell. Mechanistically, altered bases are recognized and excised by DNA glycosylases. Then the abasic site is cleaved by apurinic/apyrimidinic endonuclease 1 (APE1). A single nucleotide replacement is done by short patch BER and 2–10 nucleotide replacement is done by long patch BER. In short batch BER, polymerase β adds the single nucleotide and the gaps are sealed by XRCC1-Ligase IIIα complex. In long batch BER, polymerase δ/ε adds 2 or more into the repair gap, the 5′-flap structure is excised by FEN-1, and finally, the remaining DNA single-strand break (SSB) ends are then sealed by Ligase I [5].
4.2. Regulation of BER in stem cells
BER repair plays a significant role in maintaining the genomic stability of hESCs during their adaptation to in-vitro culture conditions and also during the reprogramming of iPSCs [38]. Evaluation of human ESCs shows an increased expression of various BER associated genes compared to the somatic cells [20]. For example, 8-oxoguanine DNA glycosylase-1 (OGG1), a major DNA glycosylase responsible for the removal of ROS induced 8-oxoguanine was found to be highly expressed in NSCs. Correspondingly, a significant reduction in OGG1 expression was observed in the fully differentiated cells [39]. Similarly, NEIL3 a mammalian DNA glycosylase is expressed more in NSCs compared to the differentiated neurons [40]. Moreover, both of these glycosylases (OGG1 and NEIL3) are necessary for the survival of NSCs, and their knockdown by siRNAs affects the differentiation ability of NSCs as well as the expression of Musashi-1, an important stem cell marker [41].
Levels of BER proteins like Ligase III, APE1, and DNA polymerase β were also found to be overexpressed in ESCs and iPSCs compared to the differentiated cells. In line with this observation, ESCs and iPSCs exhibits enhanced resistance towards methyl methanesulfonate (MMS) induced mutagenesis [42]. Terminally differentiated muscle cells displays decreased BER activity and undetectable levels of key BER proteins DNA ligase I, XRCC1 and DNA ligase III [43].
DNA methylation is one of the critical epigenetic tools that controls the pluripotent status of stem cells [44]. Methylation-associated DNA damage induced by N-methyl-N-nitrosourea in HSCs are repaired by a broad range of DNA repair mechanisms including BER. Respectively, BER genes such as AAG, APE was found to be overexpressed in multipotent CD34 (+) HSCs compared to the mature CD34 (−) cells isolated from cord blood of the same donor [23]. Not only DNA methylation associated DNA damage, but active DNA methylation status in ESCs are also maintained by BER mechanism, regulated by PR domain-containing transcriptional regulator (PRDM14) protein [45].
In order to assess the efficacy of BER in long-term in-vitro culture conditions, efficiency of BER was evaluated in long-term in vitro cultures of adipose-tissue derived MSCs. These MSCs displays a prolonged efficacy of glycosylase activities and BER at least for 12 passages in-vitro [40]. Nevertheless, after a 55 month culturing of hESCs demonstrates diminished BER activity and reduced expression of APE1, the major nuclease required for BER [46].
4.3. BER deficiency in stem cells and diseases
Diseases or syndromes that are associated directly with BER proteins are not available. However, few studies suggest a link between BER deficiency and certain type of pathological conditions. For example, defective 8oxoG base excision repair by OGG1 in vascular smooth muscle cells leads to increased accumulation of oxidative damage and progression of atherosclerosis [47]. NEIL1 is another BER DNA glycosylase that is important for repairing the oxidatively damaged purines. In a transgenic mouse model, Neil1 deficient mouse develops metabolic syndrome displaying greater total weight, fat mass gains, and more severe fatty liver disease compared to Neil1 proficient mice [48].
5. NER and stem cells
5.1. Mechanism of NER
NER repairs bulky DNA adducts that are induced by external agents such as UV and chemicals (Fig. 2C). Especially, NER repairs UV radiation-induced lesions like cyclobutane pyrimidine dimers and 6–4 photoproducts. Both these lesions alter DNA structure and inhibit DNA transcription and replication. NER is classified into two models. (1) GG-NER (global genome repair) which preferentially removes lesions from total genomic DNA (coding parts of the genome and non-transcribed strands of active genes). (2) TC-NER (transcription-coupled repair) is restricted to removing lesions preferentially from the transcribed DNA strand of active genes. Mechanistically, only the damage recognition step differs between GG-NER and TC-NER. In GG-NER, the initial damage is recognized by XPC-hHR23В complex, whereas in TC-NER, the presence of RNA polymerase II stalled at the damage site is sufficient for recognition. The transcription factor TFIIH and helicases XPB and XPD are recruited to unwind the double helical structure. XPA-RPA complex is then loaded into the DNA that allows the nucleases ERCC1-XPF and XPG to introduce the 5′-and the 3′-incisions, respectively. After incision, DNA polymerase δ or ε and PCNA synthesizes the new strands which are then ligated by DNA ligase III or ligase I [6].
5.2. Regulation of NER in stem cells
NER deficient transgenic mouse model that lacks XPD demonstrates a progressive deterioration of HSCs with age and upon exposure to genotoxic stress [49]. Similarly, ERCC1 deficient ESCs were ten-fold hypermutable compared to the wild-type cells [50]. GG-NER-defective Xpa(−/−) and Xpc(−/−) ESCs shows hypersensitivity towards UV radiation. TC-NER defective Ercc6(−/−) ES cells exhibit only mild UV sensitivity. Furthermore, GG-NER-defective Xpa(−/−) and Xpc(−/−) ES cells display increased S-phase delay and increased apoptosis upon UV treatment compared to the differentiated Xpa(−/−) and Xpc (−/−) mouse embryonic fibroblasts (MEFs). However, mutation induction upon UV is similar in both the NER-defective ESCs and NER-defective MEF’s. These results suggest that a differential cell cycle response and apoptosis are indeed sufficient to protect against proliferation of damaged ESCs [51]. In addition, spectral karyotyping analysis of TC-NER defective Ercc6(−/−) ESCs exhibits increased complex exchanges that involves multiple chromosome interactions [52]. Moreover, ERCC6-mediated DNA repair is also important for the self-renewal of embryonic NSCs after UV induced DNA damage [53].
A recent study demonstrates that DNA damage induced increase in Xpg levels are necessary for cell cycle arrest in HSCs upon ionizing radiation (IR) treatment. Knocking down of Xpg causes abrogation of IR induced cell cycle arrest and increased number of HSCs [54]. Additionally, a study shows that ERCC1-XPF heterodimer endonuclease in NER mechanism is also important for the HR induced targeted gene replacement in ESCs [55].
Apart from their role in protecting stem cells from genotoxic lesions, XPC also serves as a critical component of the transcriptional co-activation of pluripotency gene regulatory network (NANOG, OCT4, and SOX2) [56]. Moreover, knockout of XPC results in compromised pluripotency in ESCs and somatic cell reprogramming of fibroblasts to iPSCs [57]. XAB2 levels also decreased in the differentiated cells during myeloid cell differentiation, a process by which stem cells develop into mature monocytes or granulocytes [58]. Overall, the NER mechanism shows a very promising role in the maintenance of the genome, as well as stemness in stem cells, and deficiency of NER results in various disorders as described below.
5.3. NER deficiency in stem cells and diseases
Xeroderma pigmentosum (XP) is an inherited condition with defective GG-NER, where the patients develop extreme sensitivity towards UV and other DNA damaging agents [59]. Recently, iPSCs derived from the fibroblasts of five different patients bear mutations in XPA, XPB, XPC, XPG, and XPV genes were examined for their NER efficiency. Comparatively, XPA-iPSCs shows decreased NER efficiency upon differentiation into neural cells and stimulates apoptosis in dividing NSCs [60]. Cockayne syndrome is a premature aging disorder with defective TC-NER and characterized by microcephaly (abnormal head size), developmental delay, and importantly, they have increased sensitivity to sunlight. At the molecular level, these patients have mutations in either ERCC6 or ERCC8 genes, which involves in NER [61]. ERCC6 mutated patient’s fibroblasts were successfully reprogrammed into iPSCs. However, these ERCC6-iPSCs show increased cell death and ROS [62]. The apparent connection between upregulated ROS and aging has already been established for decades [63]. Trichothiodystrophy (TTD) is an intellectual impairment disease with sensitivity towards radiation exposure and carry mutations in XPD, XPB, or GTF2H5 genes, which are primarily involved in TC-NER [64]. Studies using TTD mice models with mutant Xpd displays the developmental defect, accelerated bone aging and decreased MSCs/osteoprogenitors [65,66]. Cerebro-oculo-facio-skeletal (COFS) syndrome is another degenerative disorder that affects the individuals with an intellectual and developmental disability. Mutations in ERCC1, ERCC2, ERCC5, or ERCC6 have been documented for COFS syndrome [67].
6. DSB Repair and stem cells
6.1. Mechanism of DSB repair (DSBR)
DSB break is the most lethal type of DNA damage, where even 2–3 DSBs in a normal healthy cell will initiate apoptosis to avoid the detrimental consequences. Given its importance in maintaining genomic stability, DSBs are repaired by two fundamental sovereign mechanisms based on the accessibility to template DNA. DNA damages before replication are primarily repaired by error-prone-NHEJ (Fig. 2D), and DNA damage during or after replication is repaired by predominantly by error-free-HR (Fig. 2E) and to an extent by NHEJ. Exactly how cells choose between NHEJ and HR to fix its DSB is still unknown. Mechanistically, NHEJ is initiated by Ku70-Ku80 complex that binds to the damaged ends and recruits DNA-PKcs. DNA-PKcs holoenzyme and Artemis facilitates the DNA end processing by MRN (MRE11-RAD50-NBS1) complex that possesses exonuclease, endonuclease and helicase activity. After DNA end processing, the broken strands are ligated by XRCC-Ligase IV complex. On the other hand, HR is initiated by the resection of the DSB DNA ends by MRN complex. SSB protein RPA which then coats the single strands are replaced by nucleoprotein RAD51 with the help of BRCA2. This nucleofilament then initiates the homology search and recombinase activity which results in the synthesis of new DNA strands. Finally, the free ends are ligated using Ligase I or III [8,9].
6.2. Regulation of NHEJ in stem cells
Importance of NHEJ mediated DSB repair in stem cells is evident from the research conducted on zygotes, the initial stage of ESCs. During embryonic development, zygotes preferably choose NHEJ mediated DNA repair [68]. DNA repair during embryonic development is mostly a maternal trait, depending on proteins and mRNAs provided by the oocyte [69]. IR-induced DNA damage in sperms are repaired mostly by NHEJ mediated repair after fertilization. Specifically, DNA PKcs plays a vital role in repairing DSB breaks during the initial stages of zygotes [70]. Development of NSCs into differentiated neurons in-vivo is mainly dependent on the NHEJ repair mechanism [71]. NSCs are very sensitive to DNA damaging agents; even mild aphidicolin-induced replication stress prompted recurrent DSB clusters. Interestingly, 90% of this gene fragility locates within gene bodies [72].
Lipopolysaccharide-induced inflammation in dental pulp stem cells (DPSC) activates DSB associated repair proteins. In these DPSCs, knockdown of Ku70 increases DNA damage and apoptosis [73]. DPSCs treatment with hydroxyurea sensitizes the cells to undergo senescence. This increased senescence associates with decreased expression of NHEJ proteins (Ku70, Ligase IV and XRCC4). Furthermore, DPSCs derived from young donors exhibits increased resistant to apoptosis and increased NHEJ activity compared to old donors [74]. Similar results were observed in mammary stem cells isolated from mouse, which shows increased apoptosis and NHEJ activity [75].
DSB repair kinetics measured in mouse NSCs, neurons differentiated from NSCs in culture and MEFs derived from the same littermates. Outcomes show that DSB repair was quicker in NSC and neurons compared to MEFs. In parallel with these observations, increased expression and activity of DNA-PKcs was observed in both NSCs and neurons compared to MEFs [76]. A recent study presents that levels of DNA-PKcs dependent NHEJ in neurons derived from iPSCs were regulated by a scaffold protein Translin-associates protein X (TRAX) [77]. It is essential to note that TRAX protein has various functions and associated with mental illnesses, including schizophrenia. Furthermore, SIRT6, an HDAC has been shown to regulate NHEJ in iPSCs derived from old mice. Mechanistically, SIRT6 was found to be directly binding with Ku80 and aids the Ku80/DNA-PKcs interaction which in turns facilitates the DNA-PKcs phosphorylation at residue Serine-2056, leading to efficient NHEJ. Interestingly, NHEJ mediated DSB was only active in iPSCs derived from old mice, whereas in iPSCs derived from young mice HR plays a significant role [78]. In skeletal muscle stem cells, DNA-PKcs dependent NHEJ is more efficient compared to the differentiated cells [79].
Ligase IV is another essential member of NHEJ, which performs the end joining process of DSB. Mutations in Ligase IV results in Ligase IV syndrome, which is characterized by growth defects, microcephaly, reduced number of blood cells, increased predisposition to leukemia and variable degrees of immunodeficiency. hiPSCs that lack Ligase IV exhibits increased accumulation of DSBs and increased apoptosis. Additionally, Ligase IV-deficient hiPSCs show increased chromosomal aberrations and a significant decrease in reprogramming efficiency [80]. Furthermore, a hypomorphic Ligase IV (Y288C) mutant mouse model has been developed for human Ligase IV syndrome. Ligase IV (Y288C) mouse exhibits decreased DSB repair in association with progressive loss of HSCs, bone marrow cellularity during aging and severely impaired stem cell function in tissue culture and upon transplantation [81].
DSB DNA damage always does not induce NHEJ mediated repair in stem cells. For example, hair follicle stem cells in mouse epidermis show increased accumulation of DSB during the natural aging process. Conversely, aging-induced 53BP1 foci formation does not co-localize with NHEJ repair components suggesting entirely different DDR events [82]. NHEJ appears to be more important even in the G2 phase of human ESCs. Compared to the differentiated derivatives, hESCs displays increased radiation-induced chromatid breaks, a G2 phase associated damage. Remarkably, inhibition of DNA-PKcs results in a significant decrease in radiation-induced chromatid exchanges in hESCs but not in somatic cells. Thus, establishing a role for DNA-PKcs in mis-rejoining of chromosomes in pluripotent cells [83]. Polymerase micro (Polmicro) is a DNA polymerase and a member of NHEJ repair. Defective in hematopoietic development, as well as reduction in progenitors’ number and expansion potential were observed in Polmicro (−/−) mice [84].
MUSE cells comprises of 10–15% of all MSCs and play important role in tissue repair and in maintaining tissue homeostasis. Though the data is limited, but interesting DNA repair studies were done in the MUSE cells. Evaluation of MUSE cells ability to repair various types of DNA damage showed no significant difference in BER and NER activities among MSCs, non-MUSE cells, and MUSE cells. However, basal level expression of DNA damage sensing kinase ATM was elevated in MUSE cells when compared to non-MUSE cells. On the other hand, in response to H2O2 exposure, MUSE cells expressed increased levels of DNA-PKcs (NHEJ) compared to MSC and non-MUSE cells. Based on these observations, authors proposed that DNA-PKcs-mediated NHEJ might be the prominent mechanism involved in repair of H2O2-induced DSB in MUSE cells [85].
Even though the role of NHEJ in repairing DSBs is robust based on the literature as mentioned above, few types of damages are not successfully processed by NHEJ. mESCs were genetically modified to carry two I-SceI sites in cis separated by a distance of 9 kbp. Even though NHEJ repair was active in this system, it failed to efficiently restore breaks induced in two distant chromosome ends [86].
6.3. Regulation of HR in stem cells
Although NHEJ plays a significant role in stem cell DNA repair, most of the time HR is used by stem cells as an additional protection. Furthermore, whenever NHEJ repair fails, HR seems to take control of the repair process, provided if DNA replication has already occurred [87]. Even though the mechanism is not understood, RAD51 proteins are recruited at DSB sites throughout the mESCs cell cycle, including G1 [88]. This observation challenges the common assumption that HR can be active only during the S-phase of the cell cycle, and HR in mESC can be independent of DNA replication. Additionally, treating hESCs with replication inhibitors does not always activate CHK1 mediated S-phase arrest [89]. hESCs prefer elimination of cells with replication-associated DNA damage through apoptosis rather than DNA repair.
Though they are inactive during unperturbed conditions, HR proteins are highly expressed throughout different stages of the cell cycle in mESCs until their differentiation [90]. Possibly, stem cells might have this constant HR expression to immediately activate and fix the DNA damage, thereby avoiding delay in time for their transcriptional activation.
Maintenance of telomere length is a critical hallmark of stem cells and for their immortality. Accumulation of extrachromosomal telomeric repeats in the form of T-circles after excessive telomere elongation is a common problem in stem cells. HR proteins, specifically, Nijmegen breakage syndrome 1 (NBS1) and XRCC3 play a significant role in trimming the excessive telomere elongation [91].
The efficiency of HR has been applied in gene editing of stem cells. Recently, Artemis knockout (Art (−/−)) mouse has successfully reverted the Art (−/−) phenotype. Technically, I-Sce1 recognition site was replaced in the exon 12 of the Artemis gene and transfected along with the Artemis correction template, which got incorporated into the murine genome by HR [92]. Similarly, Sickle Cell disease pathology was reversed in mESCs using I-Sce1 mediated HR gene-knock-in [93]. Still, it is important to note that I-Sce1 mediated HR may be locus dependent in mammalian stem cells [94]. The success of these gene therapies mostly relies on the efficiency of HR mechanism. List of human diseases that have been successfully gene corrected in-vitro using HR in stem cells is given in table 3.
Table 3.
Homologous recombination-mediated gene correction of diseases in-vitro in human stem cells.
| Disease | Gene Targeted | Disease Characterization | Reference |
|---|---|---|---|
| Cystic fibrosis | CFTR | Progressive damage to the respiratory system and chronic digestive system problems | [95] |
| X-linked agammaglobulinemia | BTK | Deficiency in humoral immunity | [96] |
| Myotonic dystrophy type 1 | DMPK | Progressive muscle wasting and weakness | [97] |
| Huntington disease | HTT | Progressive brain disorder that causes uncontrolled movements, emotional problems, and loss of thinking ability (cognition) | [98] |
| Beta-thalassemia | HBB | Blood disorder that reduces the production of hemoglobin | [99] |
| Sickle cell disease | HBB | Blood disorder that distort red blood cells into a sickle, or crescent, shape | [100] |
| Duchenne muscular dystrophy | DMD | Progressive muscle weakness and wasting (atrophy) and a heart condition called dilated cardiomyopathy | [101] |
| Pulmonary alveolar proteinosis | CSF2 | Accumulation of proteins and phospholipids in the alveolar spaces | [102,103] |
| Wiskott-Aldrich syndrome | WAS | X-linked primary immunodeficiency disease | [104] |
| Gyrate atrophy | OAT | Progressive vision loss | [105] |
| Kostmann disease | HAX1 | Shortage (deficiency) in neutrophils leads to recurrent infections | [106] |
| Xeroderma pigmentosum | XPC | Hypersensitivity to UV light and a dramatic predisposition to skin neoplasms | [107] |
| Limb girdle muscular dystrophy | DYSF & SGCA | Weakness and wasting of the muscles in the arms and legs | [108] |
| X-linked chronic granulomatous disease | CYBB | A form of immunodeficiency | [109] |
| Laminopathy | LMNA | Associated with various degenerative diseases | [110] |
| Retinitis pigmentosa | RPGR | Progressive vision loss | [111] |
| Fanconi anemia | FANCA & FANCD2 | Progressive bone marrow failure and cancer predisposition | [112] |
| Dystrophic epidermolysis bullosa | COL7A1 | Associated with skin fragility and skin blisters | [113] |
Several new methods have been identified to enhance the efficiency of HR in stem cells. Combined inhibition of MEK and GSK3β pathways enriches the HR efficiency in ESCs [114]. Similarly, combined obstruction of FGF receptor, ERK, and GSK3 or dual impediment of Src and GSK3 also enhanced HR efficiency in mESCs [115,116].
BRCA proteins are potential tumor suppressors and have a significant role in DSB DNA repair. Brca1 mutated mouse develops hematopoietic defects in early adulthood that included reduced HSCs [117]. Expression of BRCA1 was observed in NSCs isolated from rat brain; however, this expression of BRCA1 decreased as soon as the NSCs differentiated [118]. Impaired Brca2 mESCs reveals increased genomic instability and hypersensitivity to genotoxic agents [119]. Both the human iPSCs and ESCs indicate increased expression of BRCA2 upon irradiation, which shows its importance in maintaining the integrity of the stem cell genome [120].
RAD51 is the final effector of HR mechanism and mainly involved in the strand exchange process [121]. Irradiation-induced ATM-mediated RAD51 foci formation was observed in hESCs but not in fully differentiated astrocytes [122]. Co-expression of RAD51 along with the four reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) increases the reprogramming efficiency of iPSCs compared with the expression of four reprogramming factors alone [123]. Conversely, inhibition of RAD51 in mESCs does not affect its self-renewal or differentiation capacity. However, RAD51 suppression slowed the proliferation of mESCs by arresting them at the G2/M phase of the cell cycle [124]. Combined ectopic expression of RAD51 and RAD52 results in efficient differentiation of ESCs [125]. Consistently, recent studies showed increased expression of RAD51 in response to UV exposure in MUSE cells suggests a role for HR in repair of UV-induced DSB. Together these studies indicate swift activation of DNA repair processes facilitate these cells resistance to senescence and apoptosis which otherwise is activated upon unrepairable DNA damage [85].
6.4. DSBR deficiency in stem cells and diseases
ATM kinase is an important signal transducer of DDR and its loss results in a neurodegenerative disorder called ataxia-telangiectasia (A-T). A-T patients typically show increased chromosomal instability and sensitivity towards IR. They also suffer from severe cellular degeneration due to Purkinje cell death [126]. iPSCs from A-T patient exhibits abnormal DNA repair mechanism, defective radiation-induced signaling, radiosensitivity, and cell cycle checkpoint defects [127]. NBS1 protein plays a significant role in the repair of DSB repair and its deficiency in humans’ results in NBS, characterized as growth retardation and cancer predisposition. Predictably, iPSCs derived from NBS patient’s fibroblast shows increased chromosomal instability, slower growth, mitotic inhibition, a reduced apoptotic response to stress, and abnormal cell-cycle-related gene expression. Especially, NBS neural progenitor cells show downregulation of neural developmental genes [128]. Ataxia-telangiectasia-like disorder (ATLD) is a disorder that has mutated MRE11 and shows similar features to that of A-T and NBS [129]. Conditional knockout of Mre11 in mESCs results in the inhibition of stem cells proliferation [130].
Werner syndrome (WS) is a premature aging disorder induced by mutations in WRN. WRN gene belongs to the RecQ family of helicases that are involved in the separation of double-stranded DNA. Telomere dysfunction and increased chromosomal instability are the hallmarks of WS in cells isolated from WS patients [131]. WRN−/− ESCs maintained standard karyotyping and successfully differentiated into all three germ layers. Nevertheless, WRN−/− ESCs differentiated into WRN−/− MSCs presents characteristics of premature aging like premature loss of pro-liferative potential, increased number of senescence-associated–β-galactosidase positive cells, up-regulation of aging-associated genes p16 and p21, activation of senescence-associated secretory phenotype and elevated expression of DDR proteins [132].
Similar to WRN protein, Bloom also belongs to RecQ family of helicases. Mutations in BLM leads to Bloom’s syndrome (BLMS) that impairs its functions and results in increased genomic instability, elevated sister chromatid exchanges and predisposition to cancer [133]. Mutation in murine Blm results in embryonic delay and decease by embryonic day 13.5. Increased DNA damage induced by chromosomal segregation results in elevated apoptosis of epiblast cells [134,135].
It is interesting to note that both WS and BLMS occur as a result of mutations in the RecQ family of helicases. Nonetheless, clinically they are different, where, WS is mostly an aging disorder, and BLMS is mostly a cancer predisposition disorder. Remarkably, few differences occur at the molecular level between these two disorders. First, telomere dysfunction/shortening happens only in WS but not in BLMS. Association between telomere dysfunction and aging has been well documented [136]. Secondly, HR is elevated in BS cells and depressed in WS cells. Finally, WRN is expressed in both dividing and non-dividing cells, whereas, BLM is expressed mostly in dividing cells. This situation might make the quiescent stem cells to accumulate more damage in WS but not in BLMS [137]. These observations show that mutations in genetically similar genes might have very different clinical characteristics.
Another disease that is associated with RecQ family is Rothmund–Thomson syndrome (RTS) and has mutations in RECQL4. RTS is clinically characterized as premature aging and cancer predisposition disorder [138]. Similar to other RecQ associated diseases, germline Recql4-deficient in mice is found to be embryonically lethal and somatic Recql4-deficient mice indicates increased genomic instability, apoptosis and defective hematopoiesis [139].
7. ICLR and stem cells
7.1. Mechanism of ICLR
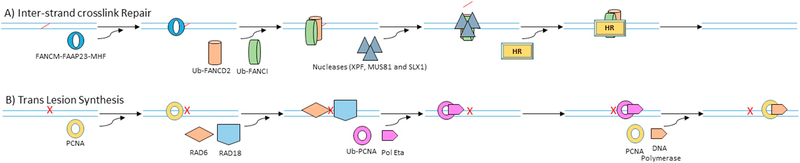
ICL repair is a complex DNA damage that employs a various type of DNA repair mechanisms including NER, HR, FA group and TLS (Fig. 3A). NER mostly repairs ICLs at G0/G1 phase. However, during replication, the stalled forks are recognized by FANCM-FAAP23-MHF 1/2 complex which in turn recruits the FA proteins and other accessory proteins to form a multi-subunit ubiquitin E3 ligase complex or the FA core complex. Within the core complex, an E3 ligase FANCL mono-ubiquitinates FANCD2 and FANCI at Lysine-561 and 523 respectively and activates them. Activated FANCD2 and I complex then localizes into the chromatin and interacts with the downstream components, which comprise nucleases (XPF, MUS81, and SLX1), HR proteins (BRCA2, BRIP1, PALB2, and RAD51C) and scaffold proteins (SLX4) to complete the repair.
Fig. 3.
Different DNA repair mechanisms. A) Inter-strand cross links during replication is repaired by Fanconi anemia (FA) pathway. Cross links induced stalled forks are recognized by FANCM-FAAP23-MHF ½ complex which then recruits FA core complex with E3 ligase activity that mono-ubiquitinates FANCD2 and FANCI. Mono-ubiquitination of FANCD2 and FANCI facilitates their localization to the damaged site, which in turn recruits nucleases (XPF, MUS81, and SLX1) to remove the cross link that results in DSB. DSB is then finally fixed by homologues repair. B) Unrepaired damages are sometimes bypassed by translesion synthesis during replication to avoid stalled forks. Upon experiencing adducts during replication, RAD6/RAD18 ubiquitin complex is recruited to the stalled fork site, which mono-ubiquitinates PCNA. Mono-ubiquitinated PCNA then undergoes confirmation change that facilitates the recruitment of low fidelity polymerase eta, which bypasses the damaged DNA. After bypassing, normal high fidelity DNA polymerase replaces the polymerase eta to continue replication.
7.2. Regulation of ICLR in stem cells
Deficiency in FA complementation group of proteins results in FA disease, characterized by bone marrow failure, developmental abnormalities and predisposition to cancer [140]. FANCD2 is a vital HR member that involves in DSB and ICL repair. Like any other HR proteins, defective FANCD2 regulation by Uap1 leads to increased chromosomal instability in mESCs and mouse embryonic lethality [141]. FancD2 deficient mice show defective HSCs including defective long-term in-vivo repopulating ability. Similar results were found in FancC knockout mice [142] and also in Usp1 deficient mice, an enzyme necessary for deubiquitinating/deactivating FANCD2 [143].
FancU deficient mouse was observed to be embryonically lethal and the embryos that were able to survive for a few days exhibit increased genomic instability [144]. FancB mutant mESCs exhibits hypersensitivity to the crosslinking agent mitomycin C (MMC) and did not facilitate the recruitment of FANCD2 to MMC induced foci [145]. FancA and FancG deficient mice display increased apoptosis of embryonic neural progenitors accompanied by NSCs exhaustion with aging. Additionally, NSCs displays a reduced capacity to self-renew in-vitro [146]. hiPSCs derived from somatic cells of FANCA mutant patients maintained pluripotency but underwent profound G2 arrest and apoptosis [147]. Additionally, lentiviral-mediated gene correction of FancA reversed phenotype of affected HSCs and promotes a healthy pattern of clonal turnover in-vivo [148]. FANCC mutant patient-derived iPSCs did not show any difference in their pluripotency. However, FANCC deficient iPSCs exhibits increased chromosomal abnormalities and unable to generate teratoma composed of all three germ layers in-vivo. Even though FANCC deficient iPSCs successfully differentiate into HSCs, the hematopoietic progenitors display increased apoptosis and reduced clonogenic potential [149].
7.3. ICLR deficiency in stem cells and diseases
FA is an inherited disease that leads to bone marrow failure, physical abnormalities, organ defects, and an increased risk of certain cancers. Currently, at least 22 genes were found to be associated with this disease. iPSCs derived from FANCA deficient patients display an early pathologically defective differentiation phenotype in both hematopoietic and endothelial lineages [150]. Similarly, in another study, iPSCs derived from FANCA deficient patients show decreased clonogenicity and increased sensitivity towards DNA crosslinking agents. FA-HPCs and FA-MSCs show similar deficiencies in maintenance and proliferation. Prominently, these FANCA deficient iPSCs exhibit defective differentiation into HPCs, MSCs, and NSCs [151]. Furthermore, FA deficient hHSCs reveal a higher rate of cytokinesis failure resulting in increased binucleated cells [152].
Karyomegalic Interstitial Nephritis (KIN) is a chronic interstitial nephropathy disorder that is characterized by tubulointerstitial nephritis and formation of enlarged nuclei in the kidneys and other tissues. Recent reports describe that mutations in the gene encoding FANCD2/FANCI-associated nuclease 1 (FAN1) causes KIN in humans. Mechanistically, FAN1 acts as a 5′−3′ exonuclease that cleaves DNA successively at every third nucleotide, allowing to excise an ICL from one strand through flanking incisions which are later taken care by FA pathway [153]. Chronic treatment of Fan1−/− mice with cisplatin exhibits renal failure within five weeks and defective MSCs [154]. Like-wise, karyomegaly became more prominent in kidneys and livers of Fan1−/− mice with age, and also develops liver dysfunction [155].
8. TLS and stem cells
8.1. Mechanism of TLS
Unrepaired DNA poses increased risk to the cells, especially during replication it stalls the DNA polymerases. Persistent stalling of replication forks leads to fork collapse, genomic instability, and cell death. These type of situations are handled by DNA damage tolerance (DDT) pathway, which facilitates to bypass the damage during replication and leaving the damage to get repaired at a later time (Fig. 3B). Mechanistically, TLS is a type of DDT that recruits RAD6/RAD18 ubiquitin complex that mono-ubiquitinates PCNA and results in the recruitment of specialized TLS polymerases like polymerase eta, which bypasses the damaged DNA.
8.2. Regulation of TLS in stem cells
Rev1, a core factor in TLS seems to be essential for normal stem cell replication and proliferation. Rev1 knockout hematopoietic stem and progenitor cells displays decreased proliferation and increased replication stress that is rescued with antioxidants. In addition to Rev1, Xpc knockout results in the perinatal loss of HSCs, progressive loss of bone marrow, and fatal aplastic anemia between three and four months of age. Further analysis reveals that this observation associates with increased replication stress, genomic breaks, DNA damage signaling, senescence, and apoptosis in bone marrow [156]. Recently, a detailed analysis of TLS pathway was studied in PcnaK164R/K164R mice as a unique DDT-defective mouse model. Both the HSCs and HSC derived multipotent progenitors show reduced numbers and stressed hematopoiesis was evident [157]. We have shown that RAD18, an essential member of TLS is critical for the functional interaction of HR proteins FANCD2, BRCA2 and RAD51 in cancer cells [158]. Rad18 knockout mESCs exhibits increased spontaneous sister chromatid exchange, hypersensitivity to multiple DNA damaging agents and a defect in postreplication repair [159]. Conversely, Rad18 deficient mice did not show any phenotype similar to that of Fanc deficient mice models. Nevertheless, Rad18−/− HSCs exhibits increased sensitivity towards in-vivo treatment with the non-cross linking and myelosuppressive agent 7 12-Dimethylbenz[a]anthracene [160]. RAD18, which primarily involves in the repair of crosslinking induced DNA damages in cancer cells seems to differ in HSCs. These results suggest that the stem cells depend on multiple DNA repair mechanism to protect its genomic stability.
8.3. TLS deficiency in stem cells and diseases
Xeroderma pigmentosum variant (XP-V) is a subtype of XP disease associated with typical pigmentation, sensitivity to radiation and types of cancer in the oral maxillofacial regions. Mutation in the POLH gene that encodes for TLS polymerase η (pol η) is documented for XP-V [161]. Mechanistically pol η can bypass sunlight induced TT-dimers with high accuracy. Polh−/− mice develops skin tumors upon UV irradiation, but not in control mice [162].
9. Conclusion
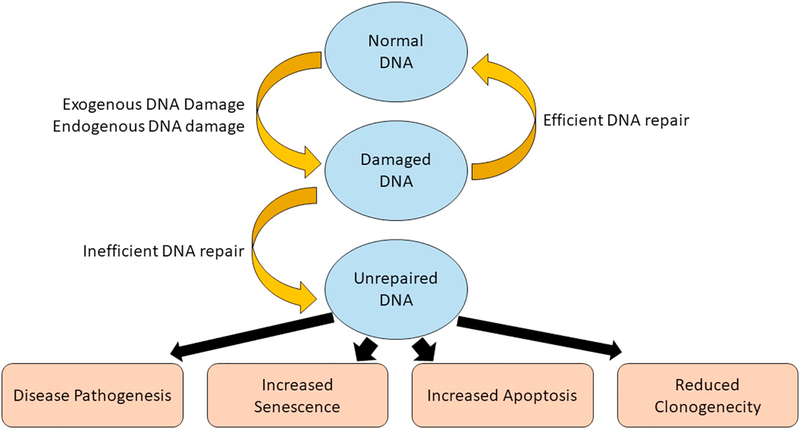
Overall, it is clear that stem cells can upregulate almost all types of DNA repair mechanisms to protect their genome from various types of genomic insults induced DNA lesions. It is also clear that failure of any of these DNA repair mechanism, stem cells undergo senescence or apoptosis in an accelerated manner to avoid the accumulation of damages in their genome. In some cases, failing of this process also makes them accumulate these damages and marks them predispose to aging and cancer (Fig. 4).
Fig. 4.
Impact of DNA repair in stem cells. Stem cells are under constant threat from both endogenous and exogenous DNA damage. Efficient DNA repair mechanisms are employed to fix these damages. However, failure to fix these damages in stem cells results in reduced clonogenic potential, increased senescence and apoptosis compared to damaged normal cells. However, in some occasions, stem cells can survive with DNA damages especially during development that can lead to various disease pathogenesis.
Deficiency of NER and DSBR mechanisms show a significant number of diseases or disorders compared to the MMR, BER and TLS. This phenomenon may be due to the fact that NER and DSBR are involved in repairing high magnitude DNA damages that are lethal by nature. Another reason might be that upon MMR and BER deficiency, mismatches and modified bases are repaired/compensated by NER. However, since there is no alternative for NER and DSBR, NER and DSBR deficiency is associated with various type of diseases.
Recent studies indicate that pluripotent MUSE stem cells possess increased DNA repair potential compared to the non-MUSE MSCs. However, further studies are needed to understand the regulation of different DNA repair processes in these cells, particularly during tissue repair and homeostasis.
Gene editing technologies using DNA repair has recently brought a tremendous potential for stem cells. Especially, HR in stem cells is used when dealing with knock-in, knock-out or precise mutagenesis using CRISPR CAS systems. Several disease models have already successfully reversed (gene corrected) in-vitro in patient-derived iPSCs using these gene editing techniques employing HR (Table 3). Even though there is still much need to be done for translating these technologies to in-vivo gene therapy, empowering stem cell potential with DNA repair is something phenomenal and vital for future gene therapy. Alternatively, specific mutations including the ones that can compromise DNA repair [29], and disease-specific mutations like Hemoglobin H [163], Lesch-Nyhan disease [164], etc., has also been achieved in stem cells using HR. Taking this research to another step ahead, mice models with defined genetic defects can now be generated using HR based gene therapy in mESCs with the potential for germline transmission [165]. Recently, several ways have been identified by researchers to improve the efficiency of HR in stem cells. For example, enrichment of hiPSCs in G2/M stage enhances HR-mediated gene editing with a 6-fold increase in targeting efficiency compared to the cells in the G1 phase [166]. These types of approaches that involve DNA repair mechanisms make gene editing in hPSCs and MUSE cells will be a more viable tool for disease modeling, regenerative medicine, and cell-based therapies. Further research about modifying DNA repair pathways will be un-doubtedly helpful to improve the therapeutic potential of stem cells.
Acknowledgments
We want to acknowledge funding sources for K. P. (NIH-R01GM098956; NIH-R01CA219187) and P. H. R. (NIH-R01AG42178, NIH-R01AG47812, NIH-R01NS105743 and NIH-R4160836). We greatly apologize to the colleagues whose works not cited due to space limitations.
Abbreviations:
- DDR
DNA damage response
- ROS
Reactive oxygen species
- MMR
Mismatch repair
- BER
Base excision repair
- NER
Nucleotide excision repair
- ICL
Interstrand crosslinks
- FA
Fanconi anemia
- DSB
Double strand breaks
- NHEJ
Non homologous end joining
- HR
Homologous recombination
- DSBR
Double-strand break repair
- ESC
Embryonic stem cell
- ADC
Adult stem cell
- PSC
Pluripotent stem cell
- hPSC
Human pluripotent stem cell
- iPSC
Induced pluripotent stem cell
- mESC
Mouse embryonic stem cell
- hESC
Human embryonic stem cell
- MUSE
Multi lineage-differentiating stress-enduring
- CSC
Cancer stem cell
- CRC
Colorectal cancer
- MSI
Microsatellite instability syndrome
- HDAC
Histone deacetylase
- MTS
Muir-Torre syndrome
- UV
Ultraviolet
- SSB
Single strand break
- NSC
Neural stem cell
- MSC
Mesenchymal stem cell
- COFS
Cerebro-oculo-facio-skeletal
- GG-NER
Global genome nucleotide excision repair
- TC-NER
Transcription-coupled nucleotide excision repair
- HSC
Hematopoietic stem cell
- MEF
Mouse embryonic fibroblast
- TTD
Trichothiodystrophy
- DPSC
Dental pulp stem cell
- TRAX
Translin-associated protein X
- NBS
Nijmegen breakage syndrome
- A-T
Ataxia-telangiectasia
- ATLD
Ataxia-telangiectasia like disorder
- WS
Werner syndrome
- BLMS
Bloom’s syndrome
- RTS
Rothmund–Thomson syndrome
- MMC
Mitomycin C
- KIN
Karyomegalic Interstitial Nephritis
- FAN1
FANCD2/FANCI-associated nuclease 1
- DDT
DNA damage tolerance
- TLS
Translesion synthesis
- OGG1
8-oxoguanine DNA glycosylase-1
Footnotes
Disclosures of potential conflict of interest
N/A
Transparency document
The Transparency document associated this article can be found, in online version.
References
- [1].Lindahl T, Barnes DE, Repair of endogenous DNA damage, Cold Spring Harb. Symp. Quant. Biol 65 (2000) 127–133. [DOI] [PubMed] [Google Scholar]
- [2].Tubbs A, Nussenzweig A, Endogenous DNA Damage as a Source of Genomic Instability in Cancer, Cell. 168 (2017) 644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jackson SP, Bartek J, The DNA-damage response in human biology and disease, Nature. 461 (2009) 1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jiricny J, The multifaceted mismatch-repair system, Nat. Rev. Mol. Cell Biol 7 (2006) 335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- [5].Wallace SS, Base excision repair: a critical player in many games, DNA Repair (Amst.) 19 (2014) 14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JHJ, Understanding nucleotide excision repair and its roles in cancer and ageing, Nat. Rev. Mol. Cell Biol 15 (2014) 465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- [7].Wang LC, Gautier J, The Fanconi anemia pathway and ICL repair: implications for cancer therapy, Crit. Rev. Biochem. Mol. Biol 45 (2010) 424–439. doi: 10.3109/10409238.2010.502166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Davis AJ, Chen DJ, DNA double strand break repair via non-homologous end-joining, Transl Cancer Res. 2 (2013) 130–143. doi: 10.3978/j.issn.2218-676X.2013.04.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jasin M, Rothstein R, Repair of strand breaks by homologous recombination, Cold Spring Harb Perspect Biol. 5 (2013) a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bi X, Mechanism of DNA damage tolerance, World J Biol Chem. 6 (2015) 48–56. doi: 10.4331/wjbc.v6.i3.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wiesmüller L, Ford JM, Schiestl RH, DNA Damage, Repair, and Diseases, J. Biomed. Biotechnol 2 (2002) 45. doi: 10.1155/S1110724302001985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yu J, Thomson JA, Pluripotent stem cell lines, Genes Dev. 22 (2008) 1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Biehl JK, Russell B, Introduction to stem cell therapy, J Cardiovasc Nurs. 24 (2009) 98–103; quiz 104–105. doi: 10.1097/JCN.0b013e318197a6a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chagastelles PC, Nardi NB, Biology of stem cells: an overview, Kidney Int Suppl (2011). 1 (2011) 63–67. doi: 10.1038/kisup.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kuroda Y, Kitada M, Wakao S, Nishikawa K, Tanimura Y, Makinoshima H, Goda M, Akashi H, Inutsuka A, Niwa A, Shigemoto T, Nabeshima Y, Nakahata T, Nabeshima Y-I, Fujiyoshi Y, Dezawa M, Unique multipotent cells in adult human mesenchymal cell populations, Proc. Natl. Acad. Sci. U.S.A 107 (2010) 8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dezawa M, Muse Cells Provide the Pluripotency of Mesenchymal Stem Cells: Direct Contribution of Muse Cells to Tissue Regeneration, Cell Transplant. 25 (2016) 849–861. doi: 10.3727/096368916X690881. [DOI] [PubMed] [Google Scholar]
- [17].Pazhanisamy SK, Stem cells, DNA damage, ageing and cancer, Hematol Oncol Stem Cell Ther. 2 (2009) 375–384. [DOI] [PubMed] [Google Scholar]
- [18].Vassilev A, DePamphilis ML, Links between DNA Replication, Stem Cells and Cancer, Genes (Basel). 8 (2017). doi: 10.3390/genes8020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saretzki G, Armstrong L, Leake A, Lako M, von Zglinicki T, Stress defense in murine embryonic stem cells is superior to that of various differentiated murine cells, Stem Cells. 22 (2004) 962–971. doi: 10.1634/stemcells.22-6-962. [DOI] [PubMed] [Google Scholar]
- [20].Maynard S, Swistowska AM, Lee JW, Liu Y, Liu S-T, Da Cruz AB, Rao M, de Souza-Pinto NC, Zeng X, Bohr VA, Human embryonic stem cells have enhanced repair of multiple forms of DNA damage, Stem Cells. 26 (2008) 2266–2274. doi: 10.1634/stemcells.2007-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tichy ED, Liang L, Deng L, Tischfield J, Schwemberger S, Babcock G, Stambrook PJ, Mismatch and base excision repair proficiency in murine embryonic stem cells, DNA Repair (Amst.) 10 (2011) 445–451. doi: 10.1016/j.dnarep.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liedtke S, Biebernick S, Radke TF, Stapelkamp D, Coenen C, Zaehres H, Fritz G, Kogler G, DNA damage response in neonatal and adult stromal cells compared with induced pluripotent stem cells, Stem Cells Transl Med. 4 (2015) 576–589. doi: 10.5966/sctm.2014-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Casorelli I, Pelosi E, Biffoni M, Cerio AM, Peschle C, Testa U, Bignami M, Methylation damage response in hematopoietic progenitor cells, DNA Repair (Amst.) 6 (2007) 1170–1178. doi: 10.1016/j.dnarep.2007.03.024. [DOI] [PubMed] [Google Scholar]
- [24].Bracker TU, Giebel B, Spanholtz J, Sorg UR, Klein-Hitpass L, Moritz T, Thomale J, Stringent regulation of DNA repair during human hematopoietic differentiation: a gene expression and functional analysis, Stem Cells. 24 (2006) 722–730. doi: 10.1634/stemcells.2005-0227. [DOI] [PubMed] [Google Scholar]
- [25].Tichy ED, Pillai R, Deng L, Liang L, Tischfield J, Schwemberger SJ, Babcock GF, Stambrook PJ, Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks, Stem Cells Dev. 19 (2010) 1699–1711. doi: 10.1089/scd.2010.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dumitru R, Gama V, Fagan BM, Bower JJ, Swahari V, Pevny LH, Deshmukh M, Human embryonic stem cells have constitutively active Bax at the Golgi and are primed to undergo rapid apoptosis, Mol. Cell 46 (2012) 573–583. doi: 10.1016/j.molcel.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Weeden CE, Asselin-Labat M-L, Mechanisms of DNA damage repair in adult stem cells and implications for cancer formation, Biochim Biophys Acta Mol Basis Dis. 1864 (2018) 89–101. doi: 10.1016/j.bbadis.2017.10.015. [DOI] [PubMed] [Google Scholar]
- [28].Reese JS, Liu L, Gerson SL, Repopulating defect of mismatch repair-deficient hematopoietic stem cells, Blood. 102 (2003) 1626–1633. doi: 10.1182/blood-2002-10-3035. [DOI] [PubMed] [Google Scholar]
- [29].Abuin A, Zhang H, Bradley A, Genetic analysis of mouse embryonic stem cells bearing Msh3 and Msh2 single and compound mutations, Mol. Cell. Biol 20 (2000) 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nguyen HT, Markouli C, Geens M, Barbé L, Sermon K, Spits C, Human embryonic stem cells show low-grade microsatellite instability, Mol. Hum. Reprod 20 (2014) 981–989. doi: 10.1093/molehr/gau059. [DOI] [PubMed] [Google Scholar]
- [31].Lin B, Gupta D, Heinen CD, Human pluripotent stem cells have a novel mismatch repair-dependent damage response, J. Biol. Chem 289 (2014) 24314–24, 324. doi: 10.1074/jbc.M114.570937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rodríguez-Jiménez FJ, Moreno-Manzano V, Lucas-Dominguez R, Sánchez-Puelles J-M, Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells, Stem Cells. 26 (2008) 2052–2062. doi: 10.1634/stemcells.2007-1016. [DOI] [PubMed] [Google Scholar]
- [33].Jang J, Huh YJ, Cho H-J, Lee B, Park J, Hwang D-Y, Kim D-W, SIRT1 Enhances the Survival of Human Embryonic Stem Cells by Promoting DNA Repair, Stem Cell Reports. 9 (2017) 629–641. doi: 10.1016/j.stemcr.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Seriola A, Spits C, Simard JP, Hilven P, Haentjens P, Pearson CE, Sermon K, Huntington’s and myotonic dystrophy hESCs: down-regulated trinucleotide repeat instability and mismatch repair machinery expression upon differentiation, Hum. Mol. Genet 20 (2011) 176–185. doi: 10.1093/hmg/ddq456. [DOI] [PubMed] [Google Scholar]
- [35].Patel R, Qing Y, Kennedy L, Yan Y, Pink J, Aguila B, Desai A, Gerson SL, Welford SM, MMR Deficiency Does Not Sensitize or Compromise the Function of Hematopoietic Stem Cells to Low and High LET Radiation, Stem Cells Transl Med. 7 (2018) 513–520. doi: 10.1002/sctm.17-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Machin P, Catasus L, Pons C, Muñoz J, Conde-Zurita JM, Balmaña J, Barnadas M, Martí RM, Prat J, Matias-Guiu X, Microsatellite instability and immunostaining for MSH-2 and MLH-1 in cutaneous and internal tumors from patients with the Muir-Torre syndrome, J. Cutan. Pathol 29 (2002) 415–420. [DOI] [PubMed] [Google Scholar]
- [37].Wielders EAL, Hettinger J, Dekker R, Kets CM, Ligtenberg MJ, Mensenkamp AR, van den Ouweland AMW, Prins J, Wagner A, Dinjens WNM, Dubbink HJ, van Hest LP, Menko F, Hogervorst F, Verhoef S, te Riele H, Functional analysis of MSH2 unclassified variants found in suspected Lynch syndrome patients reveals pathogenicity due to attenuated mismatch repair, J. Med. Genet 51 (2014) 245–253. doi: 10.1136/jmedgenet-2013-101,987. [DOI] [PubMed] [Google Scholar]
- [38].Krutá M, Šeneklová M, Raška J, Salykin A, Zerzánková L, Pešl M, Bártová E, Franek M, Baumeisterová A, Košková S, Neelsen KJ, Hampl A, Dvořák P, Rotrekl V, Mutation frequency dynamics in HPRT locus in culture-adapted human embryonic stem cells and induced pluripotent stem cells correspond to their differentiated counterparts, Stem Cells Dev. 23 (2014) 2443–2454. doi: 10.1089/scd.2013.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hildrestrand GA, Diep DB, Kunke D, Bolstad N, Bjørås M, Krauss S, Luna L, The capacity to remove 8-oxoG is enhanced in newborn neural stem/progenitor cells and decreases in juvenile mice and upon cell differentiation, DNA Repair (Amst.) 6 (2007) 723–732. doi: 10.1016/j.dnarep.2006.12.008. [DOI] [PubMed] [Google Scholar]
- [40].Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjørås M, Luna L, Expression patterns of Neil3 during embryonic brain development and neoplasia, BMC Neurosci. 10 (2009) 45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Reis A, Hermanson O, The DNA glycosylases OGG1 and NEIL3 influence differentiation potential, proliferation, and senescence-associated signs in neural stem cells, Biochem. Biophys. Res. Commun 423 (2012) 621–626. doi: 10.1016/j.bbrc.2012.04.125. [DOI] [PubMed] [Google Scholar]
- [42].Cooper DJ, Chen I-C, Hernandez C, Wang Y, Walter CA, McCarrey JR, Pluripotent cells display enhanced resistance to mutagenesis, Stem Cell Res. 19 (2017) 113–117. doi: 10.1016/j.scr.2016.12.029. [DOI] [PubMed] [Google Scholar]
- [43].Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E, Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury, Proc. Natl. Acad. Sci. U.S.A 104 (2007) 17010–17, 015. doi: 10.1073/pnas.0701743104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Altun G, Loring JF, Laurent LC, DNA methylation in embryonic stem cells, J. Cell. Biochem 109 (2010) 1–6. doi: 10.1002/jcb.22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Okashita N, Kumaki Y, Ebi K, Nishi M, Okamoto Y, Nakayama M, Hashimoto S, Nakamura T, Sugasawa K, Kojima N, Takada T, Okano M, Seki Y, PRDM14 promotes active DNA demethylation through the ten-eleven translocation (TET)-mediated base excision repair pathway in embryonic stem cells, Development. 141 (2014) 269–280. doi: 10.1242/dev.099622. [DOI] [PubMed] [Google Scholar]
- [46].Krutá M, Bálek L, Hejnová R, Dobšáková Z, Eiselleová L, Matulka K, Bárta T, Fojtík P, Fajkus J, Hampl A, Dvořák P, Rotrekl V, Decrease in abundance of apurinic/apyrimidinic endonuclease causes failure of base excision repair in culture-adapted human embryonic stem cells, Stem Cells. 31 (2013) 693–702. doi: 10.1002/stem.1312. [DOI] [PubMed] [Google Scholar]
- [47].Shah A, Gray K, Figg N, Finigan A, Starks L, Bennett M, Defective Base Excision Repair of Oxidative DNA Damage in Vascular Smooth Muscle Cells Promotes Atherosclerosis, Circulation. 138 (2018) 1446–1462. doi: 10.1161/CIRCULATIONAHA.117.033249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS, The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase, Proc. Natl. Acad. Sci. U. S.A 103 (2006) 1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL, Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age, Nature. 447 (2007) 725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- [50].Van Sloun PP, Jansen JG, Weeda G, Mullenders LH, van Zeeland AA, Lohman PH, Vrieling H, The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage, Nucleic Acids Res. 27 (1999) 3276–3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].de Waard H, Sonneveld E, de Wit J, Esveldt-van Lange R, Hoeijmakers JHJ, Vrieling H, van der Horst GTJ, Cell-type-specific consequences of nucleotide excision repair deficiencies: Embryonic stem cells versus fibroblasts, DNA Repair (Amst.) 7 (2008) 1659–1669. doi: 10.1016/j.dnarep.2008.06.009. [DOI] [PubMed] [Google Scholar]
- [52].Griffin C, de Waard H, Deans B, Thacker J, The involvement of key DNA repair pathways in the formation of chromosome rearrangements in embryonic stem cells, DNA Repair (Amst.) 4 (2005) 1019–1027. doi: 10.1016/j.dnarep.2005.05.005. [DOI] [PubMed] [Google Scholar]
- [53].Sacco R, Tamblyn L, Rajakulendran N, Bralha FN, Tropepe V, Laposa RR, Cockayne syndrome b maintains neural precursor function, DNA Repair (Amst.) 12 (2013) 110–120. doi: 10.1016/j.dnarep.2012.11.004. [DOI] [PubMed] [Google Scholar]
- [54].Avila AI, Illing A, Becker F, Maerz LD, Morita Y, Philipp M, Burkhalter MD, Xpg limits the expansion of haematopoietic stem and progenitor cells after ionising radiation, Nucleic Acids Res. 44 (2016) 6252–6261. doi: 10.1093/nar/gkw376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Niedernhofer LJ, Essers J, Weeda G, Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH, Kanaar R, The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells, EMBO J. 20 (2001) 6540–6549. doi: 10.1093/emboj/20.22.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R, Architecture of the human XPC DNA repair and stem cell coactivator complex, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 14817–14,822. doi: 10.1073/pnas.1520104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R, A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells, Cell. 147 (2011) 120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Aoki Y, Sato A, Mizutani S, Takagi M, Hematopoietic myeloid cell differentiation diminishes nucleotide excision repair, Int. J. Hematol 100 (2014) 260–265. doi: 10.1007/s12185-014-1625-8. [DOI] [PubMed] [Google Scholar]
- [59].Black JO, Xeroderma Pigmentosum, Head Neck Pathol. 10 (2016) 139–144. doi: 10.1007/s12105-016-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fu L, Xu X, Ren R, Wu J, Zhang W, Yang J, Ren X, Wang S, Zhao Y, Sun L, Yu Y, Wang Z, Yang Z, Yuan Y, Qiao J, Izpisua Belmonte JC, Qu J, Liu G-H, Modeling xeroderma pigmentosum associated neurological pathologies with patients-derived iPSCs, Protein Cell. 7 (2016) 210–221. doi: 10.1007/s13238-016-0244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL, Bohr VA, Cockayne syndrome: Clinical features, model systems and pathways, Ageing Res. Rev 33 (2017) 3–17. doi: 10.1016/j.arr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Andrade L.N. de S, Nathanson JL, Yeo GW, Menck CFM, Muotri AR, Evidence for premature aging due to oxidative stress in iPSCs from Cockayne syndrome, Hum. Mol. Genet 21 (2012) 3825–3834. doi: 10.1093/hmg/dds211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wojas-Pelc A, Sułowicz J, Nastałek M, [Ultraviolet radiation, tobacco smoke and estrogens pathways of influence on skin aging; capabilities of prevention], Prz. Lek 65 (2008) 862–866. [PubMed] [Google Scholar]
- [64].Stefanini M, Botta E, Lanzafame M, Orioli D, Trichothiodystrophy: from basic mechanisms to clinical implications, DNA Repair (Amst.) 9 (2010) 2–10. doi: 10.1016/j.dnarep.2009.10.005. [DOI] [PubMed] [Google Scholar]
- [65].Diderich KEM, Nicolaije C, Priemel M, Waarsing JH, Day JS, Brandt RMC, Schilling AF, Botter SM, Weinans H, van der Horst GTJ, Hoeijmakers JHJ, van Leeuwen JPTM, Bone fragility and decline in stem cells in prematurely aging DNA repair deficient trichothiodystrophy mice, Age (Dordr). 34 (2012) 845–861. doi: 10.1007/s11357-011-9291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nicolaije C, Diderich KEM, Botter SM, Priemel M, Waarsing JH, Day JS, Brandt RMC, Schilling AF, Weinans H, Van der Eerden BC, van der Horst GTJ, Hoeijmakers JHJ, van Leeuwen JPTM, Age-related skeletal dynamics and decrease in bone strength in DNA repair deficient male trichothiodystrophy mice, PLoS ONE. 7 (2012) e35246. doi: 10.1371/journal.pone.0035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rafique M, Zia S, Cerebro-oculo-facio-skeletal syndrome, J Coll Physicians Surg Pak. 22 (2012) 607–609. doi:09.2012/JCPSP.607609. [PubMed] [Google Scholar]
- [68].Matsuda Y, Tobari I, Chromosomal analysis in mouse eggs fertilized in vitro with sperm exposed to ultraviolet light (UV) and methyl and ethyl methanesulfonate (MMS and EMS), Mutat. Res 198 (1988) 131–144. [DOI] [PubMed] [Google Scholar]
- [69].Generoso WM, Cain KT, Krishna M, Huff SW, Genetic lesions induced by chemicals in spermatozoa and spermatids of mice are repaired in the egg, Proc. Natl. Acad. Sci. U.S.A 76 (1979) 435–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Derijck A, van der Heijden G, Giele M, Philippens M, de Boer P, DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation, Hum. Mol. Genet 17 (2008) 1922–1937. doi: 10.1093/hmg/ddn090. [DOI] [PubMed] [Google Scholar]
- [71].Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW, A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis, Cell. 95 (1998) 891–902. [DOI] [PubMed] [Google Scholar]
- [72].Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B, Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells, Cell. 164 (2016) 644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Huang Y, Qiao W, Wang X, Gao Q, Peng Y, Bian Z, Meng L, Role of Ku70 in the apoptosis of inflamed dental pulp stem cells, Inflamm. Res (2018). doi: 10.1007/s00011-018-1167-2. [DOI] [PubMed] [Google Scholar]
- [74].Zhai Y, Wei R, Liu J, Wang H, Cai W, Zhao M, Hu Y, Wang S, Yang T, Liu X, Yang J, Liu S, Drug-induced premature senescence model in human dental follicle stem cells, Oncotarget. 8 (2017) 7276–7293. doi: 10.18632/oncotarget.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Chang C-H, Zhang M, Rajapakshe K, Coarfa C, Edwards D, Huang S, Rosen JM, Mammary Stem Cells and Tumor-Initiating Cells Are More Resistant to Apoptosis and Exhibit Increased DNA Repair Activity in Response to DNA Damage, Stem Cell Reports. 5 (2015) 378–391. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kashiwagi H, Shiraishi K, Sakaguchi K, Nakahama T, Kodama S, Repair kinetics of DNA double-strand breaks and incidence of apoptosis in mouse neural stem/progenitor cells and their differentiated neurons exposed to ionizing radiation, J. Radiat. Res 59 (2018) 261–271. doi: 10.1093/jrr/rrx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chien T, Weng Y-T, Chang S-Y, Lai H-L, Chiu F-L, Kuo H-C, Chuang D-M, Chern Y, GSK3β negatively regulates TRAX, a scaffold protein implicated in mental disorders, for NHEJ-mediated DNA repair in neurons, Mol. Psychiatry (2018). doi: 10.1038/s41380-017-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chen W, Liu N, Zhang H, Zhang H, Qiao J, Jia W, Zhu S, Mao Z, Kang J, Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice, Cell Rep 18 (2017) 2880–2892. doi: 10.1016/j.celrep.2017.02.082. [DOI] [PubMed] [Google Scholar]
- [79].Vahidi Ferdousi L, Rocheteau P, Chayot R, Montagne B, Chaker Z, Flamant P, Tajbakhsh S, Ricchetti M, More efficient repair of DNA double-strand breaks in skeletal muscle stem cells compared to their committed progeny, Stem Cell Res 13 (2014) 492–507. doi: 10.1016/j.scr.2014.08.005. [DOI] [PubMed] [Google Scholar]
- [80].Tilgner K, Neganova I, Moreno-Gimeno I, Al-Aama JY, Burks D, Yung S, Singhapol C, Saretzki G, Evans J, Gorbunova V, Gennery A, Przyborski S, Stojkovic M, Armstrong L, Jeggo P, Lako M, A human iPSC model of Ligase IV deficiency reveals an important role for NHEJ-mediated-DSB repair in the survival and genomic stability of induced pluripotent stem cells and emerging haematopoietic progenitors, Cell Death Differ 20 (2013) 1089–1100. doi: 10.1038/cdd.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ, DNA repair is limiting for haematopoietic stem cells during ageing, Nature 447 (2007) 686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- [82].Schuler N, Rübe CE, Accumulation of DNA damage-induced chromatin alterations in tissue-specific stem cells: the driving force of aging?, PLoS ONE. 8 (2013) e63932. doi: 10.1371/journal.pone.0063932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Bogomazova AN, Lagarkova MA, Tskhovrebova LV, Shutova MV, Kiselev SL, Error-prone nonhomologous end joining repair operates in human pluripotent stem cells during late G2, Aging (Albany NY). 3 (2011) 584–596. doi: 10.18632/aging.100336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lucas D, Escudero B, Ligos JM, Segovia JC, Estrada JC, Terrados G, Blanco L, Samper E, Bernad A, Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair, PLoS Genet. 5 (2009) e1000389. doi: 10.1371/journal.pgen.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Alessio N, Squillaro T, Özcan S, Di Bernardo G, Venditti M, Melone M, Peluso G, Galderisi U, Stress and stem cells: adult Muse cells tolerate extensive genotoxic stimuli better than mesenchymal stromal cells, Oncotarget. 9 (2018) 19328–19, 341. doi: 10.18632/oncotarget.25039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Boubakour-Azzouz I, Ricchetti M, Low joining efficiency and non-conservative repair of two distant double-strand breaks in mouse embryonic stem cells, DNA Repair (Amst.) 7 (2008) 149–161. doi: 10.1016/j.dnarep.2007.09.005. [DOI] [PubMed] [Google Scholar]
- [87].Delacôte F, Han M, Stamato TD, Jasin M, Lopez BS, An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells, Nucleic Acids Res. 30 (2002) 3454–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Serrano L, Liang L, Chang Y, Deng L, Maulion C, Nguyen S, Tischfield JA, Homologous recombination conserves DNA sequence integrity throughout the cell cycle in embryonic stem cells, Stem Cells Dev. 20 (2011) 363–374. doi: 10.1089/scd.2010.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Desmarais JA, Hoffmann MJ, Bingham G, Gagou ME, Meuth M, Andrews PW, Human embryonic stem cells fail to activate CHK1 and commit to apoptosis in response to DNA replication stress, Stem Cells. 30 (2012) 1385–1393. doi: 10.1002/stem.1117. [DOI] [PubMed] [Google Scholar]
- [90].Choi E-H, Yoon S, Park K-S, Kim KP, The Homologous Recombination Machinery Orchestrates Post-replication DNA Repair During Self-renewal of Mouse Embryonic Stem Cells, Sci Rep. 7 (2017) 11610. doi: 10.1038/s41598-017-11,951-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Rivera T, Haggblom C, Cosconati S, Karlseder J, A balance between elongation and trimming regulates telomere stability in stem cells, Nat. Struct. Mol. Biol 24 (2017) 30–39. doi: 10.1038/nsmb.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rivière J, Hauer J, Poirot L, Brochet J, Souque P, Mollier K, Gouble A, Charneau P, Fischer A, Pâques F, de Villartay J-P, Cavazzana M, Variable correction of Artemis deficiency by I-Sce1-meganuclease-assisted homologous recombination in murine hematopoietic stem cells, Gene Ther. 21 (2014) 529–532. doi: 10.1038/gt.2014.20. [DOI] [PubMed] [Google Scholar]
- [93].Wu L-C, Sun C-W, Ryan TM, Pawlik KM, Ren J, Townes TM, Correction of sickle cell disease by homologous recombination in embryonic stem cells, Blood. 108 (2006) 1183–1188. doi: 10.1182/blood-2006-02-004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Fenina M, Simon-Chazottes D, Vandormael-Pournin S, Soueid J, Langa F, Cohen-Tannoudji M, Bernard BA, Panthier J-J, I-SceI-mediated double-strand break does not increase the frequency of homologous recombination at the Dct locus in mouse embryonic stem cells, PLoS ONE. 7 (2012) e39895. doi: 10.1371/journal.pone.0039895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sargent RG, Suzuki S, Gruenert DC, Nuclease-mediated double-strand break (DSB) enhancement of small fragment homologous recombination (SFHR) gene modification in human-induced pluripotent stem cells (hiPSCs), Methods Mol. Biol 1114 (2014) 279–290. doi: 10.1007/978-1-62,703-761-7_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Yamamoto H, Ishimura M, Ochiai M, Takada H, Kusuhara K, Nakatsu Y, Tsuzuki T, Mitani K, Hara T, BTK gene targeting by homologous recombination using a helper-dependent adenovirus/adeno-associated virus hybrid vector, Gene Ther. 23 (2016) 205–213. doi: 10.1038/gt.2015.91. [DOI] [PubMed] [Google Scholar]
- [97].Xia G, Gao Y, Jin S, Subramony SH, Terada N, Ranum LPW, Swanson MS, Ashizawa T, Genome modification leads to phenotype reversal in human myotonic dystrophy type 1 induced pluripotent stem cell-derived neural stem cells, Stem Cells. 33 (2015) 1829–1838. doi: 10.1002/stem.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM, Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells, Cell Stem Cell. 11 (2012) 253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wang Y, Zheng C-G, Jiang Y, Zhang J, Chen J, Yao C, Zhao Q, Liu S, Chen K, Du J, Yang Z, Gao S, Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice, Cell Res. 22 (2012) 637–648. doi: 10.1038/cr.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zou J, Mali P, Huang X, Dowey SN, Cheng L, Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease, Blood. 118 (2011) 4599–4608. doi: 10.1182/blood-2011-02-335,554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Popplewell L, Koo T, Leclerc X, Duclert A, Mamchaoui K, Gouble A, Mouly V, Voit T, Pâques F, Cédrone F, Isman O, Yáñez-Muñoz RJ, Dickson G, Gene correction of a duchenne muscular dystrophy mutation by meganuclease-enhanced exon knock-in, Hum. Gene Ther 24 (2013) 692–701. doi: 10.1089/hum.2013.081. [DOI] [PubMed] [Google Scholar]
- [102].Lachmann N, Happle C, Ackermann M, Lüttge D, Wetzke M, Merkert S, Hetzel M, Kensah G, Jara-Avaca M, Mucci A, Skuljec J, Dittrich A-M, Pfaff N, Brennig S, Schambach A, Steinemann D, Göhring G, Cantz T, Martin U, Schwerk N, Hansen G, Moritz T, Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis, Am. J. Respir. Crit. Care Med 189 (2014) 167–182. doi: 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- [103].Kuhn A, Ackermann M, Mussolino C, Cathomen T, Lachmann N, Moritz T, TALEN-mediated functional correction of human iPSC-derived macrophages in context of hereditary pulmonary alveolar proteinosis, Sci Rep. 7 (2017) 15195. doi: 10.1038/s41598-017-14,566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Laskowski TJ, Van Caeneghem Y, Pourebrahim R, Ma C, Ni Z, Garate Z, Crane AM, Li XS, Liao W, Gonzalez-Garay M, Segovia JC, Paschon DE, Rebar EJ, Holmes MC, Kaufman D, Vandekerckhove B, Davis BR, Gene Correction of iPSCs from a Wiskott-Aldrich Syndrome Patient Normalizes the Lymphoid Developmental and Functional Defects, Stem Cell Reports. 7 (2016) 139–148. doi: 10.1016/j.stemcr.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Howden SE, Gore A, Li Z, Fung H-L, Nisler BS, Nie J, Chen G, McIntosh BE, Gulbranson DR, Diol NR, Taapken SM, Vereide DT, Montgomery KD, Zhang K, Gamm DM, Thomson JA, Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy, Proc. Natl. Acad. Sci. U. S.A 108 (2011) 6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Pittermann E, Lachmann N, MacLean G, Emmrich S, Ackermann M, Göhring G, Schlegelberger B, Welte K, Schambach A, Heckl D, Orkin SH, Cantz T, Klusmann J-H, Gene correction of HAX1 reversed Kostmann disease phenotype in patient-specific induced pluripotent stem cells, Blood Adv. 1 (2017) 903–914. doi: 10.1182/bloodadvances.2016003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dupuy A, Valton J, Leduc S, Armier J, Galetto R, Gouble A, Lebuhotel C, Stary A, Pâques F, Duchateau P, Sarasin A, Daboussi F, Targeted gene therapy of xeroderma pigmentosum cells using meganuclease and TALEN™, PLoS ONE. 8 (2013) e78678. doi: 10.1371/journal.pone.0078678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Turan S, Farruggio AP, Srifa W, Day JW, Calos MP, Precise Correction of Disease Mutations in Induced Pluripotent Stem Cells Derived From Patients With Limb Girdle Muscular Dystrophy, Mol. Ther 24 (2016) 685–696. doi: 10.1038/mt.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Dreyer A-K, Hoffmann D, Lachmann N, Ackermann M, Steinemann D, Timm B, Siler U, Reichenbach J, Grez M, Moritz T, Schambach A, Cathomen T, TALEN-mediated functional correction of X-linked chronic granulomatous disease in patient-derived induced pluripotent stem cells, Biomaterials. 69 (2015) 191–200. doi: 10.1016/j.biomaterials.2015.07.057. [DOI] [PubMed] [Google Scholar]
- [110].Liu G-H, Suzuki K, Qu J, Sancho-Martinez I, Yi F, Li M, Kumar S, Nivet E, Kim J, Soligalla RD, Dubova I, Goebl A, Plongthongkum N, Fung H-L, Zhang K, Loring JF, Laurent LC, Izpisua Belmonte JC, Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs, Cell Stem Cell. 8 (2011) 688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bassuk AG, Zheng A, Li Y, Tsang SH, Mahajan VB, Precision Medicine: Genetic Repair of Retinitis Pigmentosa in Patient-Derived Stem Cells, Sci Rep. 6 (2016) 19969. doi: 10.1038/srep19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E, González F, Tiscornia G, Garreta E, Aasen T, Veiga A, Verma IM, Surrallés J, Bueren J, Izpisúa Belmonte JC, Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells, Nature. 460 (2009) 53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sebastiano V, Zhen HH, Haddad B, Derafshi BH, Bashkirova E, Melo SP, Wang P, Leung TL, Siprashvili Z, Tichy A, Li J, Ameen M, Hawkins J, Lee S, Li L, Schwertschkow A, Bauer G, Lisowski L, Kay MA, Kim SK, Lane AT, Wernig M, Oro AE, Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa, Sci Transl Med. 6 (2014) 264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lin Z, Zhang Y, Gao T, Wang L, Zhang Q, Zhou J, Zhao J, Homologous recombination efficiency enhanced by inhibition of MEK and GSK3β, Genesis. 52 (2014) 889–896. doi: 10.1002/dvg.22821. [DOI] [PubMed] [Google Scholar]
- [115].Kiyonari H, Kaneko M, Abe S, Aizawa S, Three inhibitors of FGF receptor, ERK, and GSK3 establishes germline-competent embryonic stem cells of C57BL/6 N mouse strain with high efficiency and stability, Genesis. 48 (2010) 317–327. doi: 10.1002/dvg.20614. [DOI] [PubMed] [Google Scholar]
- [116].Shimizu T, Ueda J, Ho JC, Iwasaki K, Poellinger L, Harada I, Sawada Y, Dual inhibition of Src and GSK3 maintains mouse embryonic stem cells, whose differentiation is mechanically regulated by Src signaling, Stem Cells. 30 (2012) 1394–1404. doi: 10.1002/stem.1119. [DOI] [PubMed] [Google Scholar]
- [117].Mgbemena VE, Signer RAJ, Wijayatunge R, Laxson T, Morrison SJ, Ross TS, Distinct Brca1 Mutations Differentially Reduce Hematopoietic Stem Cell Function, Cell Rep. 18 (2017) 947–960. doi: 10.1016/j.celrep.2016.12.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Korhonen L, Brännvall K, Skoglösa Y, Lindholm D, Tumor suppressor gene BRCA-1 is expressed by embryonic and adult neural stem cells and involved in cell proliferation, J. Neurosci. Res 71 (2003) 769–776. doi: 10.1002/jnr.10546. [DOI] [PubMed] [Google Scholar]
- [119].Marple T, Kim TM, Hasty P, Embryonic stem cells deficient for Brca2 or Blm exhibit divergent genotoxic profiles that support opposing activities during homologous recombination, Mutat. Res 602 (2006) 110–120. doi: 10.1016/j.mrfmmm.2006.08.005. [DOI] [PubMed] [Google Scholar]
- [120].Suchorska WM, Augustyniak E, Łukjanow M, Comparison of the early response of human embryonic stem cells and human induced pluripotent stem cells to ionizing radiation, Mol Med Rep. 15 (2017) 1952–1962. doi: 10.3892/mmr.2017.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Baumann P, West SC, Role of the human RAD51 protein in homologous recombination and double-stranded-break repair, Trends Biochem. Sci 23 (1998) 247–251. [DOI] [PubMed] [Google Scholar]
- [122].Adams BR, Golding SE, Rao RR, Valerie K, Dynamic dependence on ATR and ATM for double-strand break repair in human embryonic stem cells and neural descendants, PLoS ONE. 5 (2010) e10001. doi: 10.1371/journal.pone.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Lee J-Y, Kim D-K, Ko J-J, Kim KP, Park K-S, Rad51 Regulates Reprogramming Efficiency through DNA Repair Pathway, Dev Reprod. 20 (2016) 163–169. doi: 10.12717/DR.2016.20.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Yoon S-W, Kim D-K, Kim KP, Park K-S, Rad51 regulates cell cycle progression by preserving G2/M transition in mouse embryonic stem cells, Stem Cells Dev. 23 (2014) 2700–2711. doi: 10.1089/scd.2014.0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Choi E-H, Yoon S, Kim KP, Combined Ectopic Expression of Homologous Recombination Factors Promotes Embryonic Stem Cell Differentiation, Mol. Ther 26 (2018) 1154–1165. doi: 10.1016/j.ymthe.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Rothblum-Oviatt C, Wright J, Lefton-Greif MA, McGrath-Morrow SA, Crawford TO, Lederman HM, Ataxia telangiectasia: a review, Orphanet J Rare Dis. 11 (2016) 159. doi: 10.1186/s13023-016-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Nayler S, Gatei M, Kozlov S, Gatti R, Mar JC, Wells CA, Lavin M, Wolvetang E, Induced pluripotent stem cells from ataxia-telangiectasia recapitulate the cellular phenotype, Stem Cells Transl Med. 1 (2012) 523–535. doi: 10.5966/sctm.2012-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Halevy T, Akov S, Bohndorf M, Mlody B, Adjaye J, Benvenisty N, Goldberg M, Chromosomal Instability and Molecular Defects in Induced Pluripotent Stem Cells from Nijmegen Breakage Syndrome Patients, Cell Rep. 16 (2016) 2499–2511. doi: 10.1016/j.celrep.2016.07.071. [DOI] [PubMed] [Google Scholar]
- [129].Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NG, Raams A, Byrd PJ, Petrini JH, Taylor AM, The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder, Cell. 99 (1999) 577–587. [DOI] [PubMed] [Google Scholar]
- [130].Xiao Y, Weaver DT, Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells, Nucleic Acids Res. 25 (1997) 2985–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Oshima J, Sidorova JM, Monnat RJ, Werner syndrome: Clinical features, pathogenesis and potential therapeutic interventions, Ageing Res. Rev 33 (2017) 105–114. doi: 10.1016/j.arr.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu G-H, Belmonte JCI, Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging, Science. 348 (2015) 1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cunniff C, Bassetti JA, Ellis NA, Bloom’s Syndrome: Clinical Spectrum, Molecular Pathogenesis, and Cancer Predisposition, Mol Syndromol. 8 (2017) 4–23. doi: 10.1159/000452082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Chester N, Kuo F, Kozak C, O’Hara CD, Leder P, Stage-specific apoptosis, developmental delay, and embryonic lethality in mice homozygous for a targeted disruption in the murine Bloom’s syndrome gene, Genes Dev. 12 (1998) 3382–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Chester N, Babbe H, Pinkas J, Manning C, Leder P, Mutation of the murine Bloom’s syndrome gene produces global genome destabilization, Mol. Cell. Biol 26 (2006) 6713–6726. doi: 10.1128/MCB.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Opresko PL, Shay JW, Telomere-associated aging disorders, Ageing Res. Rev 33 (2017) 52–66. doi: 10.1016/j.arr.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].de Renty C, Ellis NA, Bloom’s syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases, Ageing Res. Rev 33 (2017) 36–51. doi: 10.1016/j.arr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wang LL, Plon SE, Rothmund-Thomson Syndrome, in: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Stephens K, Amemiya A(Eds.), GeneReviews®, University of Washington, Seattle, Seattle (WA), 1993. https://www.ncbi.nlm.nih.gov/books/NBK1237/ (accessed December 4, 2018). [Google Scholar]
- [139].Smeets MF, DeLuca E, Wall M, Quach JM, Chalk AM, Deans AJ, Heierhorst J, Purton LE, Izon DJ, Walkley CR, The Rothmund-Thomson syndrome helicase RECQL4 is essential for hematopoiesis, J. Clin. Invest 124 (2014) 3551–3565. doi: 10.1172/JCI75334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Gluckman E, Improving survival for Fanconi anemia patients, Blood. 125 (2015) 3676. doi: 10.1182/blood-2015-04-639,476. [DOI] [PubMed] [Google Scholar]
- [141].Park E, Kim JM, Primack B, Weinstock DM, Moreau LA, Parmar K, D’Andrea AD, Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice, Mol. Cell. Biol 33 (2013) 4360–4370. doi: 10.1128/MCB.00870-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kamimae-Lanning AN, Goloviznina NA, Kurre P, Fetal origins of hematopoietic failure in a murine model of Fanconi anemia, Blood. 121 (2013) 2008–2012. doi: 10.1182/blood-2012-06-439,679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Parmar K, Kim J, Sykes SM, Shimamura A, Stuckert P, Zhu K, Hamilton A, Deloach MK, Kutok JL, Akashi K, Gilliland DG, D’andrea A, Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1, Stem Cells. 28 (2010) 1186–1195. doi: 10.1002/stem.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Deans B, Griffin CS, Maconochie M, Thacker J, Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice, EMBO J. 19 (2000) 6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kim TM, Ko JH, Choi YJ, Hu L, Hasty P, The phenotype of FancB-mutant mouse embryonic stem cells, Mutat. Res 712 (2011) 20–27. doi: 10.1016/j.mrfmmm.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Sii-Felice K, Etienne O, Hoffschir F, Mathieu C, Riou L, Barroca V, Haton C, Arwert F, Fouchet P, Boussin FD, Mouthon M-A, Fanconi DNA repair pathway is required for survival and long-term maintenance of neural progenitors, EMBO J. 27 (2008) 770–781. doi: 10.1038/emboj.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Chlon TM, Ruiz-Torres S, Maag L, Mayhew CN, Wikenheiser-Brokamp KA, Davies SM, Mehta P, Myers KC, Wells JM, Wells SI, Overcoming Pluripotent Stem Cell Dependence on the Repair of Endogenous DNA Damage, Stem Cell Reports. 6 (2016) 44–54. doi: 10.1016/j.stemcr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Molina-Estevez FJ, Nowrouzi A, Lozano ML, Galy A, Charrier S, von Kalle C, Guenechea G, Bueren JA, Schmidt M, Lentiviral-Mediated Gene Therapy in Fanconi Anemia-A Mice Reveals Long-Term Engraftment and Continuous Turnover of Corrected HSCs, Curr Gene Ther. 15 (2015) 550–562. [DOI] [PubMed] [Google Scholar]
- [149].Yung SK, Tilgner K, Ledran MH, Habibollah S, Neganova I, Singhapol C, Saretzki G, Stojkovic M, Armstrong L, Przyborski S, Lako M, Brief report: human pluripotent stem cell models of fanconi anemia deficiency reveal an important role for fanconi anemia proteins in cellular reprogramming and survival of hematopoietic progenitors, Stem Cells. 31 (2013) 1022–1029. doi: 10.1002/stem.1308. [DOI] [PubMed] [Google Scholar]
- [150].Suzuki NM, Niwa A, Yabe M, Hira A, Okada C, Amano N, Watanabe A, Watanabe K-I, Heike T, Takata M, Nakahata T, Saito MK, Pluripotent cell models of fanconi anemia identify the early pathological defect in human hemoangiogenic progenitors, Stem Cells Transl Med. 4 (2015) 333–338. doi: 10.5966/sctm.2013-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Liu G-H, Suzuki K, Li M, Qu J, Montserrat N, Tarantino C, Gu Y, Yi F, Xu X, Zhang W, Ruiz S, Plongthongkum N, Zhang K, Masuda S, Nivet E, Tsunekawa Y, Soligalla RD, Goebl A, Aizawa E, Kim NY, Kim J, Dubova I, Li Y, Ren R, Benner C, Del Sol A, Bueren J, Trujillo JP, Surralles J, Cappelli E, Dufour C, Esteban CR, Belmonte JCI, Modelling Fanconi anemia pathogenesis and therapeutics using integration-free patient-derived iPSCs, Nat Commun. 5 (2014) 4330. doi: 10.1038/ncomms5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Vinciguerra P, Godinho SA, Parmar K, Pellman D, D’Andrea AD, Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells, J. Clin. Invest 120 (2010) 3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lachaud C, Slean M, Marchesi F, Lock C, Odell E, Castor D, Toth R, Rouse J, Karyomegalic interstitial nephritis and DNA damage-induced polyploidy in Fan1 nuclease-defective knock-in mice, Genes Dev. 30 (2016) 639–644. doi: 10.1101/gad.276287.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Airik R, Schueler M, Airik M, Cho J, Porath JD, Mukherjee E, Sims-Lucas S, Hildebrandt F, A FANCD2/FANCI-Associated Nuclease 1-Knockout Model Develops Karyomegalic Interstitial Nephritis, J. Am. Soc. Nephrol 27 (2016) 3552–3559. doi: 10.1681/ASN.2015101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Thongthip S, Bellani M, Gregg SQ, Sridhar S, Conti BA, Chen Y, Seidman MM, Smogorzewska A, Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction, Genes Dev. 30 (2016) 645–659. doi: 10.1101/gad.276261.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Martín-Pardillos A, Tsaalbi-Shtylik A, Chen S, Lazare S, van Os RP, Dethmers-Ausema A, Fakouri NB, Bosshard M, Aprigliano R, van Loon B, Salvatori DCF, Hashimoto K, Dingemanse-van der Spek C, Moriya M, Rasmussen LJ, de Haan G, Raaijmakers MHGP, de Wind N, Genomic and functional integrity of the hematopoietic system requires tolerance of oxidative DNA lesions, Blood. 130 (2017) 1523–1534. doi: 10.1182/blood-2017-01-764,274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Pilzecker B, Buoninfante OA, van den Berk P, Lancini C, Song J-Y, Citterio E, Jacobs H, DNA damage tolerance in hematopoietic stem and progenitor cells in mice, Proc. Natl. Acad. Sci. U.S.A 114 (2017) E6875–E6883. doi: 10.1073/pnas.1706508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Tripathi K, Mani C, Clark DW, Palle K, Rad18 is required for functional interactions between FANCD2, BRCA2, and Rad51 to repair DNA topoisomerase 1-poisons induced lesions and promote fork recovery, Oncotarget. 7 (2016) 12537–12, 553. doi: 10.18632/oncotarget.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Tateishi S, Niwa H, Miyazaki J-I, Fujimoto S, Inoue H, Yamaizumi M, Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells, Mol. Cell. Biol 23 (2003) 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Yang Y, Poe JC, Yang L, Fedoriw A, Desai S, Magnuson T, Li Z, Fedoriw Y, Araki K, Gao Y, Tateishi S, Sarantopoulos S, Vaziri C, Rad18 confers hematopoietic progenitor cell DNA damage tolerance independently of the Fanconi Anemia pathway in vivo, Nucleic Acids Res. 44 (2016) 4174–4188. doi: 10.1093/nar/gkw072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Guo J, Zhou G, Zhang W, Song Y, Bian Z, A novel POLH mutation causes XP-V disease and XP-V tumor proneness may involve imbalance of numerous DNA polymerases, Oncol Lett. 6 (2013) 1583–1590. doi: 10.3892/ol.2013.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, Kucherlapati R, Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice, Cancer Res. 66 (2006) 87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- [163].Chang J, Lu RH, Xu SM, Meneses J, Chan K, Pedersen R, Kan YW, Inactivation of mouse alpha-globin gene by homologous recombination: mouse model of hemoglobin H disease, Blood. 88 (1996) 1846–1851. [PubMed] [Google Scholar]
- [164].Urbach A, Schuldiner M, Benvenisty N, Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells, Stem Cells. 22 (2004) 635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- [165].Fung-Leung WP, Mak TW, Embryonic stem cells and homologous recombination, Curr. Opin. Immunol 4 (1992) 189–194. [DOI] [PubMed] [Google Scholar]
- [166].Yang D, Scavuzzo MA, Chmielowiec J, Sharp R, Bajic A, Borowiak M, Enrichment of G2/M cell cycle phase in human pluripotent stem cells enhances HDR-mediated gene repair with customizable endonucleases, Sci Rep. 6 (2016) 21264. doi: 10.1038/srep21264. [DOI] [PMC free article] [PubMed] [Google Scholar]