Abstract
Inorganic polyphosphates (polyP) consist of linear chains of orthophosphate residues, linked by high-energy phosphoanhydride bonds. They are evolutionarily old biopolymers that are present from bacteria to man. No other molecule concentrates as much (bio)chemically usable energy as polyP. However, the function and metabolism of this long-neglected polymer are scarcely known, especially in higher eukaryotes. In recent years, interest in polyP experienced a renaissance, beginning with the discovery of polyP as phosphate source in bone mineralization. Later, two discoveries placed polyP into the focus of regenerative medicine applications. First, polyP shows morphogenetic activity, i.e., induces cell differentiation via gene induction, and, second, acts as an energy storage and donor in the extracellular space. Studies on acidocalcisomes and mitochondria provided first insights into the enzymatic basis of eukaryotic polyP formation. In addition, a concerted action of alkaline phosphatase and adenylate kinase proved crucial for ADP/ATP generation from polyP. PolyP added extracellularly to mammalian cells resulted in a 3-fold increase of ATP. The importance and mechanism of this phosphotransfer reaction for energy-consuming processes in the extracellular matrix are discussed. This review aims to give a critical overview about the formation and function of this unique polymer that is capable of storing (bio)chemically useful energy.
1. Introduction
Any kind of chemical or biochemical reaction follows the thermodynamic laws. While chemical systems tend to reach an equilibrium between reactants and products, the reactants in living biochemical systems are usually in a nonequilibrium state.1 Living systems remain in the latter state due to thermodynamic processes that dissipate energy. This fact implies that in biological systems energy-generating/providing circuits are coupled to endergonic reactions. Intracellularly it is ATP that is capturing and transferring free energy.2,3 Under standard conditions, enzymatic hydrolysis of the terminal high-energy β–γ and α–β phosphoanhydride bonds in ATP to ADP and ADP to AMP, respectively, results in the release of −30.5 kJ mol–1 of Gibbs free energy change (ΔG0). This released free energy is used as fuel for driving energetically unfavorable processes, like channeling of molecules against a concentration gradient via ATP-powered pumps, movement of cilia, contraction of muscle or synthesis of biopolymers. In animal cells, and most nonphotosynthetic cells, fatty acids and sugars are metabolized during aerobic oxidation to CO2 and H2O, and the energy liberated is, at least partially, stored as biochemically usable energy in phosphoanhydride bonds of ATP. In eukaryotes, the final stages of aerobic oxidation occur in mitochondria, whereas in prokaryotes those reactions proceed on the plasma membrane. In addition, substrate-level phosphorylation steps take place during which ATP or GTP is formed directly from ADP or GDP. In plants, light energy is converted to chemical energy and stored in the chemical bonds of carbohydrates.
The intracellular cytosol has a sol consistency which thwarts especially the low-molecular weight molecules to diffuse freely. This highly dynamic and yet exquisitely organized cytosol undergoes sol–gel transition (gelation) in response to changing pH conditions and changing ions/molecules supplementation.4 Already this property allows a fairly stable regional organization of the cytosolic metabolic pathways. In the next higher hierarchical level of complexity, the cytosol in eukaryotes harbors insoluble suspended particles, like ribosomes and organelles. Semipermeable membranes around the organelles build a barrier and implement a further possibility for control of the different metabolic cycles, the intracellular transport or metabolic energy production/utilization.5 In addition, this compartmentalization builds a protection mechanism against the activity of lytic enzymes, like those of the lysosomes.
One prerequisite for the emergence of life was the separation of the cellular environment from the surrounding, external environment, which was accomplished by the cytoplasmic membrane with a width of ∼6–10 nm built of a phospholipid bilayer into which (glyco)proteins, glyoclipids, and cholesterol molecules are embedded too.6 These hydrophobic structures undergo self-assembly and exclude water interactions between the cell interior and the extracellular space.7 ATP-dependent as well as ATP-independent transporters establish and maintain a membrane asymmetry. The exchange of molecules between the cytoplasm and the extracellular milieu is enabled by both passive (without energy consumption) and active transport (energy-dependent transport against a concentration gradient or an electrochemical gradient) using a variety of channels and transporters.8 Mitochondria are the major intracellular ATP generator producing ∼32 mol of ATP from 1 mol of glucose which is metabolized via glycolysis and the citric acid cycle. ATP synthesized in mitochondria or during glycolysis is found to play parallel and/or separate, distinct roles during differentiation and migration.9,10
ATP cannot diffuse across the plasma membrane into the extracellular space unless it is channeled through, for example, the conductive ATP-releasing pathways.11,12 In addition, a series of ATP-permeable channels have been identified that are grouped to the connexin channels, hemichannels, pannexin 1, Ca2+ homeostasis modulator 1, volume-sensitive outwardly rectifying anion channels, and the maxi-anion channels. In spite of these varieties of ATP export channels, the intracellular ATP concentration is much higher and can reach up to 100 mM in the neuronal synaptic vesicles, compared to the extracellular environment with 10 nM (see refs (11 and 13)). In human blood, the ATP levels are in the range of 20 and 100 nM.13 In this context, it is interesting to mention that mammalian erythrocytes do not contain any mitochondria with the consequence that the level of ATP in those cells is low with 360 nM, compared to 8500 nM (average level for 106 cells) in T lymphocytes.14 It should be noted that precise measurements of the low, nanomolar ATP levels in the tissue interstitium are technically difficult, in particular, close to the cell surface where ATP seems to increase.13
While the energetics within cells is quite well understood the extracellular energetic cycle(s) are poorly addressed. In general, the extracellular space does not contain mitochondria, with the exception of the blood platelet environment during inflammatory responses. During this state extracellular mitochondria have been identified together with neutrophils in vivo, triggering neutrophil adhesion toward the endothelial wall.15 In most human tissues the volume occupied by cells is substantially smaller compared to the volume of the surrounding extracellular matrix (ECM), like in cartilage which contains about 1–2% of cells16 or calcified bone which comprises about 25% organic matrix (of which 2–5% contributes to cells), 5% water and 70% inorganic mineral.17 Now, the pressing question emerges, which addresses the fueling of the dynamic and complex ECM with metabolic energy, a space which does not contain ATP-generating mitochondria. Even more, the ECM is, as outlined above, poor in ATP, and if present in this compartment, this nucleotide is exposed to the extracellular or cell membrane-bound alkaline phosphatase (ALP [EC 3.1.3.1]) and the cell membrane glycoprotein-1 phosphodiesterase [EC 3.1.4.1]/nucleotide pyrophosphatase [EC 3.6.1.9].18 The diffusion level of ATP in the protein-rich ECM as well as in the cytosol is lower than the one in water;19 for example, the diffusion in the cytosol amounts to approximately 5 × 10–10 m2 s–1.20 An extracellular ADP/ATP carrier has not been identified. It is widely acknowledged that the maintenance of the organization of the ECM in general and especially in cell-poor organs is a high-energy-demanding process. It is very obvious that tissues which contain only a small number of cells, like cartilage or intervertebral discs, receive some of their metabolic energy from their cells by metabolic respiration during glycolysis and mitochondrial oxidative phosphorylation.21 Hence it must be assumed that nutrients/glucose can only be supplied by diffusion from blood vessels to the margins of the cartilage/discs through the dense ECM of that tissue. Quantitative determinations revealed that the values of the diffusion coefficients in cartilage vary within the range from 0.3 × 10–11 to 30 × 10–11 m2 s–1;22 the diffusion is higher in damaged/degenerated tissue compared to normal cartilage.
Until now, no quantitative determinations are available that support the view that ATP released from cells is sufficient to maintain the coordinated complex ECM structure. Perhaps one reason for this inappropriate situation is the lack of data on energy requiring and energy consuming reactions that proceed in the extracellular space. These, or at least some of those, will be outlined in this review. Furthermore, besides ATP, a new candidate physiological metabolic energy-storing compound in eukaryotes will be introduced, inorganic polyphosphate (polyP). This polymeric polyanion consists of unbranched chains of three up to 1000 phosphate (Pi) residues, linked by high-energy phosphoanhydride bonds. PolyP has been conserved in all biotic organisms from bacteria to animals and can reach in yeast vacuoles levels as high as 20% with respect to dry weight.23 It has been proposed that already in the prebiotic environment polyP was abundantly formed during volcanic eruptions. Emerging evidence is now available, which qualifies polyP, being an abundantly present polymer in metazoan tissue, due to the presence of high-energy phosphate bonds/acid anhydride linkages (reviewed in refs (24−27)), as a polymer that can transfer Gibbs free energy to AMP or ADP under formation of ATP after enzymatic cleavage of the polymer by ALP (reviewed in refs (28−30)).
2. Extracellular Matrix Organization
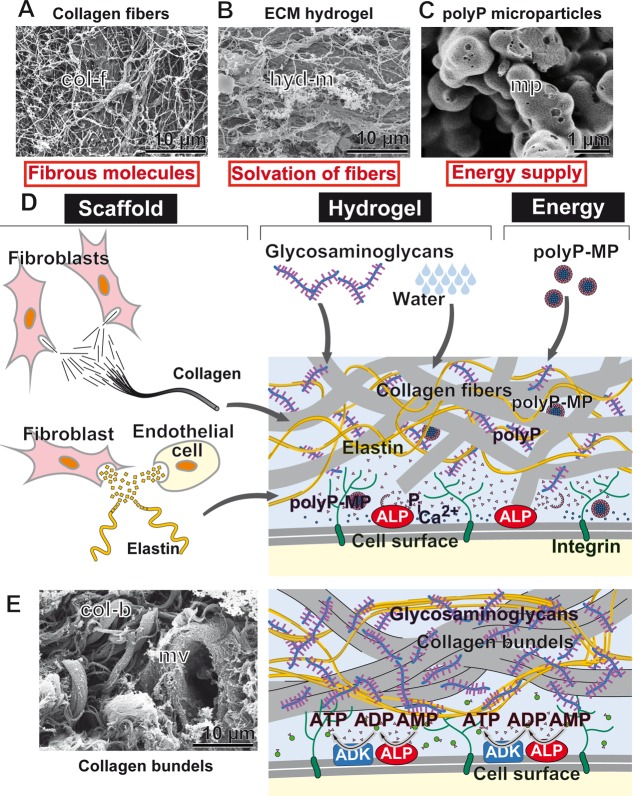
It is conceivable that the formation of the bulky ECM of metazoan tissues, most prominent with bone [70% contributes to the bone mineral and the remaining organic matrix, consisting of proteoglycans, glycosaminoglycans, and collagen, and only 25% is water],31 cartilage [<20% are cells],32 eye lens [<10% cells],33 and cornea [<10% cells],34 is a strongly energy-dependent process. The reason is that the (macro)molecular components of the ECM exist in a highly ordered, functional arrangement (Figure 1). These extracellular structural molecules are hierarchically composed and have a modular composition. The organization can be operationally grouped into three phases (Figure 1D). First, the synthesis of the macromolecules building the scaffold,35,36 such as collagen extruded from fibroblasts, elastin released from fibroblasts and endothelial cells, and fibronectin secreted from hepatocytes. These fiber-forming or rubber-like structural molecules of the ECM share the property of being dynamic and adapting to the physicochemical conditions of their environment. They are spiral-like and are loosely held together by noncovalent hydrophobic forces. Second, the elasticity of the ECM molecules is a prerequisite for the next step of complexity, the structural and functional organization of these components in the ECM. At this stage the macromolecules become embedded in a hydrogel consisting of proteoglycans and glycosaminoglycans (hyaluronan or heparin) and, to some extent, the fibrous proteins collagen, elastin, laminin, and fibronectin.37 Third, at least some of these organization processes are certainly driven by exergonic reactions, which are kinetically controlled by the flow of energy along metastable morphologies, assembled from the same or adjacent molecular building blocks.38 In addition, the self-assembly of these blocks into regional, tissue-specific, and distinct ECM tissue units requires metabolic energy, especially in the form of ATP. Exemplary highlighted: the transparent cornea with the stromal ECM as the major refractive element of the eye is composed of homogeneous small diameter collagen fibrils which are regularly packed in a highly ordered hierarchical way.39 Initially, in the developing cornea, collagen fibrillogenesis involves multiple molecules interacting in a sequential way, a process which is driven by a cell-directed deposition of aligned collagen fibrils. In the next step tuned interactions between keratocytes and stroma matrix components follow. In parallel, collagen fibrils nucleate and form bundles within cell surface channels or fuse to form fascicles of uniaxial collagen.40 While the mechanism of interaction, e.g., between the small leucine-rich proteoglycans and the collagen fibrils, has been well addressed also with respect to the temporal and spatial organization, the underlying energetics have only been poorly studied. It has been proven that ATP in the extracellular space promotes not only ECM biosynthesis, mediated by cell-based molecular pathways, but also increases the intracellular energy supply.41 In turn, the maintenance of the functional organization of the ECM requires metabolic energy, surely in the form of nucleoside triphosphates, mainly of ATP, as well. Some of those ATP-dependent processes will be mentioned (Figure 2).
Figure 1.
Extracellular matrix (ECM) as a hydrogel-scaffold into which structural fibrous molecules are embedded in a functionally organized manner. (A) Initially, the collagen fibrils/fibers (col-f) are quite randomly arranged, and (B) become incorporated into a hydrogel matrix (hyd-m), depicted as contracted globular deposits. (C) Microparticles (mp), either released from the blood platelets or added as artificial particles, like Ca-polyphosphate microparticles (polyP-MP), integrate into the scaffold and function as storage for and generators of metabolic energy. (D) The three phases of the hierarchical organization of the ECM scaffold molecules; first, synthesis of the structural macromolecules (scaffold) such as collagen from the fibroblasts and elastin from fibroblasts and endothelial cells, second, formation of the hydrogel molecules such as proteoglycans and glycosaminoglycans, and third, the compartmentation of these scaffold molecules into functional tissue units, which requires organization processes driven by supramolecular assembly/interaction and metabolic energy, like ATP, as stressed in this review. It is proposed that enzymatically controlled hydrolysis of Ca-polyP-MP via ALP generates inorganic phosphate (Pi), Ca2+, and metabolic energy that, in the presence of adenylate kinase (ADK), is stored in ADP and ATP. (E) Organized arrangement of collagen bundles (col-b) that are often associated with microvessels (mv). [(A–C,E) SEM; scanning electron microscopy].
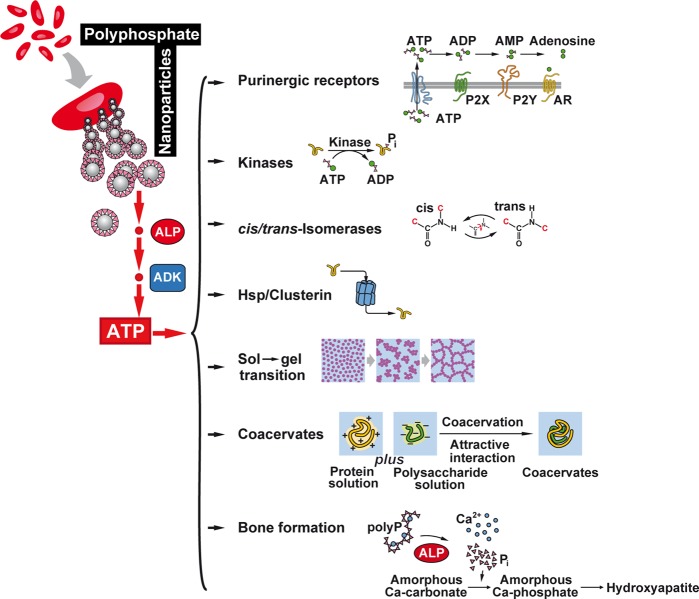
Figure 2.
ATP-requiring processes in the ECM. It is proposed that polyP in the ECM occurs as nano/microparticles that undergo hydrolytic cleavage via the ALP. In concert with the ADK, ATP (ADP, AMP) is synthesized that acts as a metabolic energy reservoir. In comparison, it is shown that ATP, exported from the cells, can act in the extracellular space as signal/ligand for purinergic receptors. During this reaction ATP is not used as metabolic fuel but activates purinergic receptor(s), P2X, P2Y, or adenosine receptors (AR). The ATP metabolizing systems in the ECM include the ECM kinases that control metabolic processes by phosphorylation of key molecules. ATP might also be required during peptidylprolyl cis–trans isomerase reactions. Surely, heat shock proteins (HSP), like clusterin, are also present in the ECM that are involved in the physiological folding of functional polymeric molecules. In addition, processes like sol to gel transitions during supramolecular polymer organization are critical organization principles in the ECM involving exergonic reactions. The transition processes during coacervation likewise follow an energy-favorable reaction pathway. Bone and cartilage formation in the extracellular space are prominent energy-requiring reactions. During bone mineralization Pi and ADP/ATP are generated by enzymatic hydrolysis of polyP via ALP. The released Pi is driving the transition of amorphous Ca-carbonate “bio-seeds”, initially formed during bone mineralization, to amorphous Ca-phosphate and the final deposition of hydroxyapatite.
It should be stressed here that ATP, as well as polyP, is expected to be associated, especially in the extracellular space, with binding proteins. However, only very fragmentary and rudimentary first data have been gathered in this field.42,43 ATP- and polyP-binding proteins could have the role of protecting these metabolites toward degrading enzymes, interfering with potential functional receptors, or allowing their transportation.
2.1. Purinergic Receptors
The plasma membrane comprises integrated purinergic receptors, purinoceptors, that respond to extracellular nucleosides (like adenosine) and nucleotides (ADP, ATP, UDP, or UTP), which act as signaling molecules.44,45 They are involved in learning and memory, locomotion/movement, feeding behavior, as well as in sleep.46 As typical signaling molecules these nucleosides and nucleotides react locally with the purinergic receptors.47 As an example, ATP released from aggregating blood platelets acts as a signaling molecule and elicits endothelium-dependent vasodilatation. During this process nucleosides and nucleotides are released from intracellular organelles and stores and act locally around the extracellularly exposed receptor(s).48 ATP acts as a signaling molecule on the purinergic receptors and, as such, needs to exist only at defined, usually low concentrations, triggering intracellular metabolic reactions.49
Important to note that polyP, present in the mammalian brain, acts in micromolar concentrations as a gliotransmitter between astrocytes P2Y1 purinergic receptors.50 The cells respond with an activation of phospholipase C, followed by a release of Ca2+ from the intracellular stores. Furthermore, besides neural cells, the P2Y purinergic receptors are also found at the surface of cardiomyocytes51 as well as on platelets and other hematopoietic and nonhematopoietic cells with, e.g., the subtype P2Y12.52
In addition to the function of ATP/ADP as a signaling molecule and a link in an autocrine signaling loop, the nucleotides are fed as substrates for metabolic energy-requiring processes in the ECM space. Some examples are given below.
2.2. Kinase Reactions
The intracellular signaling is dependent on amplifiers that potentiate extracellular signals (like hormones) via enzyme reactions (kinase reactions). Over 500 kinases have been described in humans53 and about 30% of the intracellularly existing proteins are phosphorylated.54 Initiated by analyses of the mammalian phosphoproteome, a series of secreted, extracellular proteins with phosphotyrosine units has been disclosed,55 like the vertebrate lonesome kinase (VLK). It is secreted in the ECM as a Tyr kinase which phosphorylates proteins both in the secretory pathway and outside the cell.56 Evidence has been presented that this kinase is physiologically regulated during platelet degranulation and enzymatically active.
2.3. Peptidylprolyl cis–trans Isomerases
The triple-helical protein collagen represents the most abundant ECM component. A series of enzymes, molecular chaperones, and post-translational modifiers facilitates the maturation of collagen. Among them are the peptidyl-prolyl cis–trans isomerases (PPIases) which catalyze an essential step during trans isomerization of the peptidylprolyl bonds, a rate limiting step in protein folding.57 The PPIases cover three families of structurally unrelated proteins, the cyclophilins, FK506-binding proteins, and parvulins. Spontaneous isomerization of peptidylprolyl bonds is a rather slow process at physiological pH conditions; the PPIases substantially accelerate this reaction.58 The first evidence that PPIases are present in the extracellular space came from studies with hensin, an ECM protein that forms 50- to 100 nm-long fibers.59 On the basis of theoretical consideration, it has been proposed that the cis–trans interconversion does not require ATP but instead is driven by energy derived from conformational changes in the protein.58,60 However, more experimental data are required to rule out that ATP is not indirectly involved in those PPIases processes, since for the FK506-binding protein binding of ATP to the domain II has been identified, which has been shown to be functionally active, while GTP is not.61 It remains also to be studied if a functional interplay between prolyl isomerases and ATP consuming HSPs, like with HSP60, during mitochondrial protein import exists, allowing a reduction of the number of ATP binding and release cycles and, in turn, making the folding more efficient and less energy-consuming.62
Interestingly, the formation/assembly of collagen type I monomers into higher organization complexes, into SLS (segment long spacing) crystallites63 with a length of ∼300 nm, is accelerated in the presence of ATP under in vitro conditions.64
2.4. Heat Shock Proteins/Clusterin
Heat shock proteins (HSPs) support and restore not only intracellularly but also extracellularly correct protein folding.65 The chaperones can be grouped into different families, according to their structure, complexity, and regulation.66 First, energy-independent chaperones, which function as monomers, dimers, or oligomers, and second, complex multichaperone machines, which bind to ATP, hydrolyze the nucleotide, and regulate cochaperone as well as substrate protein binding and a subsequent release (reviewed in ref (67)) are key regulators of proteostasis and include TRiC, HSP70, and HSP90. Surprisingly, it has been disclosed that polyP shares protein-like chaperone properties.68 The authors demonstrated that at micromolar concentrations, polyP efficiently prevent protein aggregation during stress conditions, like oxidative stress and higher temperature. Even more, it has been reported that in the presence of polyP, enzymes are stabilized as soluble micro-β-aggregates with amyloid-like properties.69 In continuation of this line, the authors showed that polyP prevents misfolding of the proteins by rendering them in a β-sheet-rich, amyloid-like, soluble conformation.69
Solid evidence exists that HSP70 is present in the extracellular space and released via the MyD88/NF-κB signal transduction pathway.70 Since HSP70 shows functional activity in the presence of ATP, it is more than hypothetical to suppose that HSP70 also binds ATP in the extracellular space, allowing energy-dependent protein folding.71
Perhaps more experimental data on the potential function of ATP toward the extracellularly present HSP clusterin are needed.72 For this protein it has been suggested that it functions independently of ATP hydrolysis,73 even though it comprises the ATP binding domain.74 Another example of an extracellular chaperone with ambiguous interaction with ATP is α-crystallin.72 This molecule protects vertebrate eye lens proteins from adverse protein aggregation. In addition, this protein exists also in extra-lenticular organs, including heart, kidney, and brain.75 In an experimental approach, it could be demonstrated that αB-crystallin, as a molecular chaperone, increases its chaperone functions in the presence of ATP.76 Further experiments are needed to clarify if those mentioned ATP-independent folding processes are not indirectly coupled with ATP consuming reactions.
2.5. Sol to Gel Transition: Supramolecular Polymers
Basically the ECM can be considered as a dense matrix which appears, at first glance, as an impermeable barrier for cells to infiltrate and to migrate into. In accordance with the gelation theory, the sol–gel transition theory,77 it is assumed that gelation starts when soluble polymer chains randomly bind together under formation of growing soluble clusters which assemble together and form a gel. This transition state is critical and fast and only depends upon a slight variation in the bond quantity.78 The directed navigation of cells through the ECM requires a series of intracellular biochemical signaling pathways which are probing the microenvironment through integrin-mediated adhesion circles. These cellular physiological processes must be coordinated with phase transition changes in the ECM during which the thermodynamical equilibrium shifts to a partition. This process requires free energy.79,80 It is known that within cells, ATP-driven processes cause fluctuations which usually result in random movements that can become directional if driven by motor proteins.81
Sol–gel transition processes often accelerate the coassembly of two components in supramolecular systems, like in hydrogels, the ECM, or in the cytoplasm. Supramolecular polymerization is caused and mediated by noncovalent interactions, which are usually weaker than covalent ones,38 but in contrast to covalent ones more wide-ranging and more specific. Even more, because of their property to be highly flexible in their organization, the molecules associated with functional units in the ECM are rarely used for a static stabilization of the cells. Specifically, the supramolecular polymers are prime candidates allowing a transitionally membranous scaffold formation in the ECM in order to set up extracellular functional compartments.82 In addition to those, some transport systems lined with epithelia, transcellular, pericellular, and paracellular transport pathways are characterized by a high flexibility in the ECM.83 Examples are the claudin–claudin cis interfacial structures that organize the dynamic and flexible nature of tight junction strands.84 Likewise, transcellular functional organization of aligned scaffolds is relevant for the formation of transcellular units and, by that, for an efficient transmission of information between neurons85 and basically between any other cell systems. Those flexible structures, based on supramolecular polymers, are energy requiring and energy consuming. If those structures are not fueled by metabolic energy, the transport systems should stall. In consequence, the mentioned self-adaptive supramolecular organizations and cycles are energy dissipating systems. As shown for micelles, the growth and breakdown of supramolecular associations are ATP-dependent.86 More generally, it can be proposed that a failure of ATP-driven transient supramolecular interactions in the ECM, like in the intracellular environment, results in a collapse of the metastable state of all living systems.
2.6. Coacervation
Related to the sol to gel transition process is the coacervate phase transition during which organic-rich droplets are formed via liquid–liquid phase separation, resulting in an association of oppositely charged polymeric molecules.87 This process requires the availability of free energy and entropy in the system.88
2.7. Bone and Cartilage
It is well-established that mechanical energy is stored as elastic energy in both nonmineralized and mineralized components in the ECM.89 Besides those energy-storing and dissipating systems in collagens and tendons,90 the less flexible human skeleton, contributing with about 15% to the total body weight, requires metabolic energy, like for released elastic strain energy, for the formation as well as continual maintenance and repair of this dynamic tissue. It is indicative that during the onset of osteoblast mineralization, ATP levels in those cells peak, a process which is paralleled with the accumulation of mitochondria with high-transmembrane potentials in those regions;91 subsequently, fully differentiated osteoblasts revert to glycolysis to maintain ATP production.92 These findings indicate that the development and the bioenergetic programs of the osteoblasts are coupled with the mineralization state. Furthermore, osteocalcin, secreted solely by osteoblasts, increases insulin sensitivity and modulates an array of genes involved in energy expenditure, like for Ucp2. Its protein product causes mitochondrial uncoupling of oxidative phosphorylation under dissipation of energy as heat.93
In addition, polyP turned out to act as a source of inorganic phosphate needed during bone mineralization for the transformation of the initially formed amorphous calcium carbonate deposits (“bio-seeds”) into amorphous calcium phosphate (for a review, see ref (30)). While the calcium carbonate bioseed formation is enzyme-catalyzed by a membrane-associated carbonic anhydrase (CA-IX), the exchange of carbonate by phosphate in the amorphous calcium carbonate proceeds nonenzymatically but requires prior hydrolysis of polyP mediated by ALP. The amorphous calcium phosphate subsequently undergoes a phase transition to hydroxyapatite (Figure 2).
During bone and cartilage formation, like during wound healing, vascularization is one important process to happen and is essential for tissue construction and repair. During the initiation of this process, endothelial cells have to form initial small vessels. During this initial phase, the orientation, guiding, and ring formation of endothelial cells is mediated by ATP which is released into the extracellular space and acts as a chemotactic gradient along which the cells migrate.10,94
In conclusion, among the above-mentioned processes in the ECM which are evoked by ATP, some of them are most likely dependent on metabolic energy, generated during the cleavage of the energy-rich bonds of the nucleotide. Examples are the ATP-requiring processes during the following reactions: phosphorylation reactions driven by ECM kinases, the peptidylprolyl cis–trans isomerase process during collagen packaging, some heat shock protein-mediated actions, coacervation, sol to gel transition processes during supramolecular polymer organization, and finally formation of bulky extracellular tissue, like in bone and cartilage.
In section 13 experimental evidence is given, underscoring that extracellular ATP is enzymatically generated from polyP.
3. Structure of polyP
Every pro- and eukaryotic cell contains inorganic polyphosphate (polyP) with a chain length that can reach up to several thousands of orthophosphate (Pi) residues (Figure 3; see refs (24, 25, 95−97)). The tetrahedral Pi units forming this polymer are linked together by high-energy phosphoanhydride bonds similar to those found in ATP. While microorganisms usually contain very long-chain polyP, ranging in size from hundreds to thousands of Pi residues, eukaryotic cells synthesize polyP chains which are much less heterodisperse and comprise polymer lengths of ∼60–100 Pi units.
Figure 3.
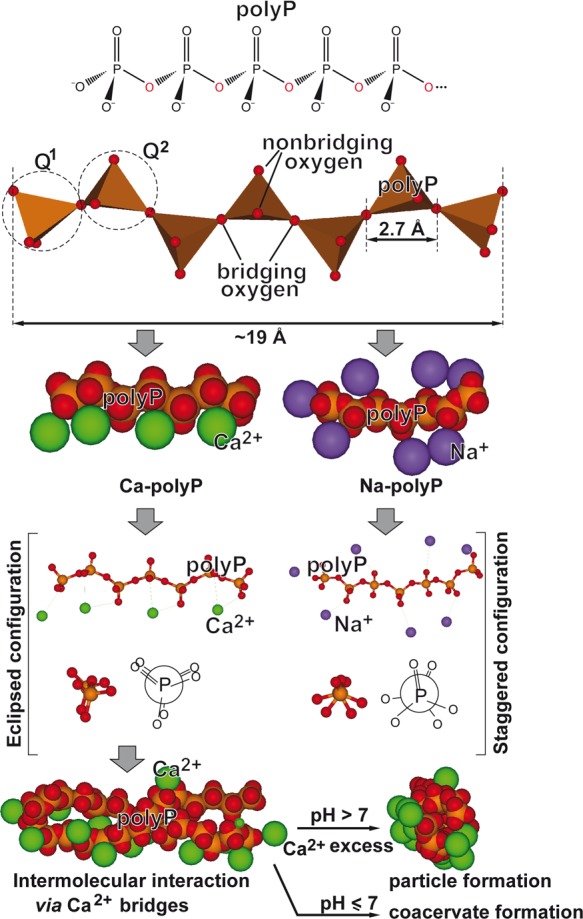
Structure of the polyP molecule composed of interconnected PO4 tetrahedra, linked via their bridging oxygen atoms. The linear polyP consists of both Q1 (one bridging oxygen) and Q2 (two bridging oxygens) tetrahedra. Because of the rotational flexibility of the P–O–P (phosphoanhydride) bonds, the polymer can occupy various steric conformations (staggered and eclipsed), in dependence on the counterion. The space-filling models and ball-and-stick models of Ca-polyP and Na-polyP are shown. The divalent Ca2+ salt of polyP assumes an eclipsed configuration, while for the monovalent Na+ salt of polyP a staggered configuration is found. Higher-ordered structures formed by an intermolecular Ca2+ bridge formation comprise both solid particles (at alkaline pH and Ca2+ excess) and coacervates (at lower pH values).
3.1. PolyP Chain
In accordance with the Qi nomenclature (i = number of bridging oxygens), the linear polyP is composed of both Q1 and Q2 tetrahedra with one (terminal Pi units) and two bridging oxygens (internal Pi units), respectively98 (Figure 3). Branched polyP chains (“ultraphosphates”) with Q3 tetrahedra (three bridging oxygens) are not found in living organisms. The P–O bond lengths within the Pi units are between 1.62 and 1.66 Å for the single bonds99,100 and 1.52 Å for the P=O double bond.101,102 The length of the P=O bond is much shorter than the sum of the two atomic single-bond radii (P: 1.07 Å and O: 0.66 Å)100 due to the difference in electronegativity (P = 2.1; O = 3.5). On the basis of these values and the bond angles of P–O–P and O–P–O bonds (P–O–P: 130° and O–P–O: 102°),103 the maximum length (longitudinal size) of the linear polyP molecules can be estimated from the size of each phosphate unit (2.7 Å); e.g., the maximum size of polyP40 (usually used for the formation of biomimetic polyP nanoparticles; see below) amounts to 108 Å, while the maximum size of the long chain polyP produced by bacteria (around 1000 Pi units), which accomplish structural properties, is in the range of ∼3 × 103 Å (∼300 nm). Since the bond angles within the Pi units can vary between ∼120° to 180°,104 and the polymer is physiologically complexed to divalent cations, a smaller value for the space-filling area is, however, more realistic.
3.2. PolyP Conformations
In addition and due to a high degree of rotational flexibility, the polyP chain can occupy various conformations, staggered and eclipsed,104 depending on the arrangement of the tetrahedral PO4 units (Figure 3). In this way, the polymer can adjust to the charge and the coordination requirements of the respective counterions which can be monovalent (e.g., Na+ or K+), divalent (e.g., Mg2+ or Ca2+), or trivalent (e.g., Gd3+) cations. The eclipsed configuration seems to be optimal for divalent Ca2+ ions, as confirmed by modeling studies with pyrophosphate,105 while the monovalent Na+ salt of polyP prefers a staggered configuration. The higher binding energy of the fully charged polyanionic polyP to divalent cations is the major factor that determines the preferential binding of Mg2+ and Ca2+ at physiological pH; at a lower pH/charge density of the polymer, monovalent cations increasingly bind to the polyP chain.106 Based again on modeling studies, Ca2+ ions are able to form with polyP molecules organized aggregates/particles whereby–in the presence of a small number of polyP chains–the divalent cations occupy a more peripheral position (Figure 3).
4. Difficulties and Challenges in Analysis of Cell-Associated polyP and in Identification of polyP Metabolizing Enzymes
The polyP polymer has been identified comparably late after the discovery of ATP, first described as adenyl pyrophosphoric acid, by Lohmann.107 Initially named RNA,108 the basophilic polymer was later identified as metaphosphate by Schmidt et al.109 and later as polyphosphate.110 The first suggestion that polyP can serve as an energy-rich phosphate came from Ebel in 1952 (see ref (111)). The first attempts to understand the polyP metabolism were performed by Liss and Langen.112
4.1. Chemical Analysis of polyP
Despite the increasing importance of polyP in medicine and biotechnology as well as in environmental research, the chemical and biochemical analytics of the polymer are not well developed.113,114 Starting with biological and environmental samples, a significant disadvantage is seen in the isolation procedure because during the extraction of the polymer other phosphate-containing compounds are coisolated. Initially, polyP was adsorbed to charcoal, followed by LiCl desorption.115 After delipidation and alkali treatment, the material was neutralized with perchloric acid. The polyP in the fractions was determined after hydrolysis in acid and corrected for the orthophosphate content.116 In addition, especially if an accurate chain length is attempted to be determined, an interference with the ALP is a serious pitfall since this enzyme readily hydrolyzes the polymer during and after the extraction process from living tissue, especially from eukaryotic cells.117
An improvement was reached by application of a sequential extraction procedure, by isolation first of acid-soluble polyP and second of long-chain polyP at neutral pH.118 After removal of proteins using phenol-chloroform and nucleic acids by precipitation of polyP at neutral pH, the polyP factions (short chain and long chain) were collected by ion exchange chromatography. The polymer was quantitatively assessed after charcoal adsorption, by a total phosphate colorimetric assay, including also the toluidine blue colorimetric reaction.118,119 This basic extraction concept is still applied today97,120 and includes extraction after breaking up the cells with a chaotropic agent, like guanidinium thiocyanate, followed by enzymatic digestion with proteinases, phenol/chloroform mixture extraction, and anion exchanger-based chromatography. The chain length of polyP was determined by thin layer chromatography (short- to medium-chain polyP), ion-exchange chromatography, gel-filtration, electrophoresis on urea-polyacrylamide gels or 31P NMR spectroscopy.119−123 The calibration of the systems was achieved with either fluorescently or radiolabeled polyP markers.124−126 Introduced is also the overall determination of polyP, using the Fourier transform spectroscopy (FTIR),30 a technique which can be used also quantitatively.113 A less practical but comparably sensitive method for the quantitation of polyP was introduced with the fluorescence Ca2+ indicator fura-2.127 The fluorescence intensity in this system was found to be abolished with increasing polyP concentrations. This method turned out also to be applicable for the determination of polyP catabolic enzymes.
4.2. Determination of polyP in Intact Cells
It is of serious disadvantage that for polyP a specific dye does not exist. Very often the fluorescent dye DAPI (4′,6-diamidino-2-phenylindole) is used for in situ detection of polyP within cells or tissues.128 However, this reaction is not specific since DAPI stains also nucleic acids with only slightly different characteristics. The dye toluidine blue is less frequently used.129
4.3. Assays for polyP Metabolizing Enzymes
Due to the lack of precise polyP quantification techniques, faithful methods for the quantification of enzymatic reactions of polyP metabolizing enzymes are also not available. As an example, the photometric determination of the polyP kinase both acting as a polyP synthesizing enzyme and as a polyP hydrolyzing enzyme might be mentioned.130,131 In the first system, an enzyme-coupled assay with pyruvate kinase and lactate dehydrogenase has been elaborated, while for the reverse reaction, the determination of NADH consumption is used as a changing parameter. Furthermore, a similar assay has been introduced for the polyP-AMP-phosphotransferase reaction132 during which ATP, ADP, and AMP in combination with luciferase and luciferase/adenylate kinase is quantified. Finally, the purine-nucleoside phosphorylase has been used as an enzyme indicator for the quantification of polyP in the reaction.119
In all these assay systems, inaccuracies can creep in especially during the preceding isolation of the polyP product/substrate and the indirect approaches used for quantification of the product. Therefore, more reliable and specific methods have to be developed in the future suitable for a more precise quantification of polyP in cells and also during the enzyme reactions. These remarks should also indicate that some of the existing enzymatic and functional data should be taken with some caution.
5. Physiological Roles of polyP
Because of the presence of multiple energy-rich phosphodiester bonds, long-chain polyP is able to store large amounts of metabolically utilizable energy, much more than an ATP molecule. The energy (ΔG0; standard conditions) released during hydrolysis of a single phosphoanhydride bond within this polymer is similar to that of hydrolysis of the β–γ or α–β phosphoanhydride bond in ATP (−30.5 kJ mol–1) or even higher (ΔG0≈ −40 kJ mol–1).133
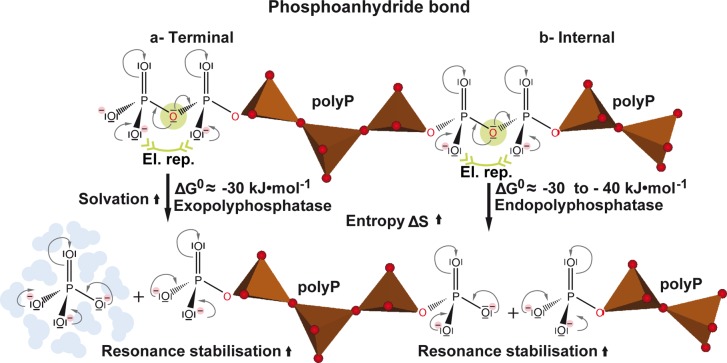
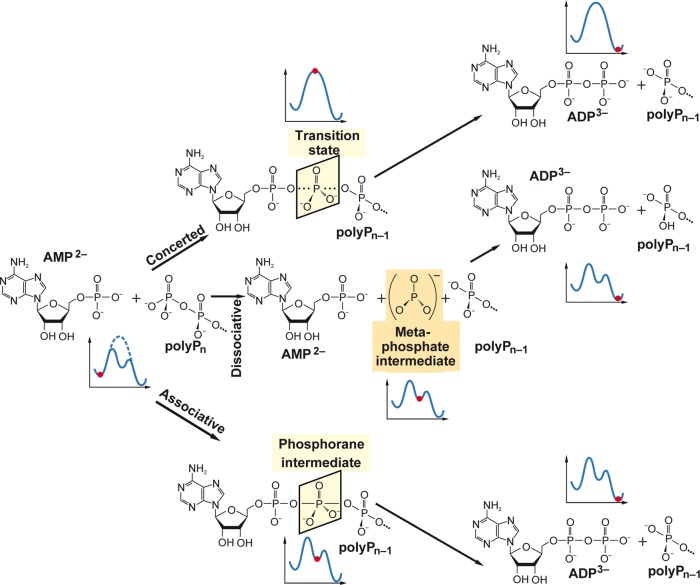
As sketched in Figure 4, several factors, similar to ATP hydrolysis, may contribute to the highly exergonic character of the hydrolytic cleavage of the phosphoanhydride bonds of polyP, which is catalyzed by polyP exo- and endopolyphosphatases, such as the extensive electrostatic repulsion of the negatively charged oxygen atoms on the phosphate groups,134 changes in entropy135 or resonance stabilization136,137 and a higher degree of solvation of the reaction products,135 compared to polyP (Figure 4).
Figure 4.
Various factors contributing to the large amount of Gibbs’ free energy released during polyP hydrolysis by exopolyphosphatases and endopolyphosphatases. El. rep., electrostatic repulsion.
It should be noted, however, that under physiological conditions, the actual ΔG values, as with ATP, may be influenced by a number of factors, such as pH, temperature, divalent cations, and ionic strength.138−143 For example, physiologically, polyP (like ATP)144 is coordinated with divalent cations, like Ca2+ and Mg2+.25 Therefore, it is expected that the actual ΔG values for polyP hydrolysis will be significantly different from ΔG0, similar to ATP hydrolysis. For example, in the case of ATP hydrolysis, the actual ΔG values, for example, in the liver, are between −59 and −53.5 kJ mol–1, and in the heart, between −61.7 and −59.5 kJ mol–1;141,145 for in vivo data (human brain and muscle), see ref (146).
In contrast to microorganisms, the biological and biochemical pathways of polyP in higher eukaryotes have been largely unknown for a long time, although this polymer is detectable throughout the entire animal kingdom. This situation changed more recently when it became obvious that polyP has a functional role during bone mineralization147 as well as affects the energy balance within the cells and also in the surrounding ECM.148−150 First indications came from an observation that the amount and turnover of polyP is considerably higher in tissues with high metabolic rates and requirements like the brain or heart.150−152 However, caution is advisible about the reliability of those data since the isolation, purification, and analysis methods used are sometime immature, as pointed out.153
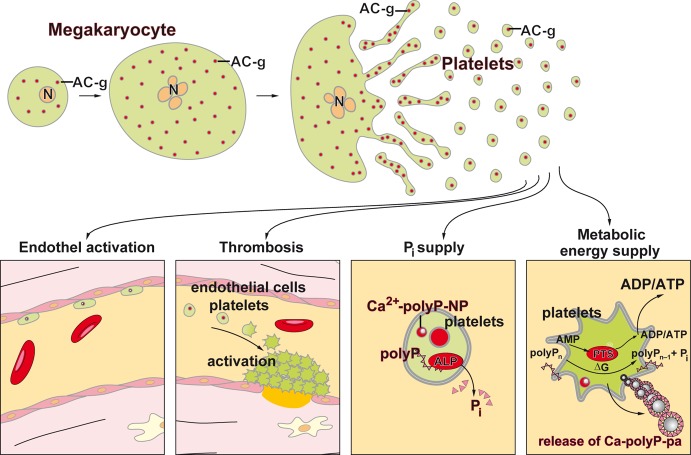
Microorganisms such as bacteria and yeast can accumulate large amounts of polyP. Multiple functions of polyP in these organisms have been demonstrated, e.g., being a source of energy, a donor for sugar and adenylate kinases, and also acting as a chelator for divalent cations and working as a functional buffer against alkaline stress or as a regulator of development. Likewise, the enzymes catalyzing these reactions have been identified (see refs (25 and 26)). However, also vertebrates contain significant amounts of polyP. This polymer has been identified in every animal tissue investigated so far, from mouse heart (∼114 μM), brain (95 μM), and lungs (91 μM) to kidney (64 μM). Lower amounts of polyP are found in the liver (38 μM), while high levels are present in mast cells and especially in blood platelets (up to 1.1 mM).121,154 It might be stressed that blood platelets comprise ∼5%, after erythrocytes (∼85%) but before bone marrow cells (2.5%) and lymphocytes (1.5%), the second most abundant cells in the human body.155 In human blood, a concentration of 1–3 μM polyP has been calculated.96 Also in other body fluids, like in synovial fluids, polyP exists.156 Within mammalian cells, this polymer has been identified in lysosomes,157 dense granules of the acidocalcisomes,121 mitochondria, and nuclei.158
Focusing on the polyP effect within the eukaryotic, mainly mammalian cells, it could be elucidated that polyP is likely to be involved in the following pathways.
5.1. Energy Production and Permeability Transition Pore Induction
In the mitochondria, polyP was found to increase in response to the activation of the metabolic respiration and be reduced by their inhibitors.159 Oligomycin, a known inhibitor of complex V/F1FO-ATP synthase, abolishes the proton gradient at the inner mitochondrial membrane and strongly interferes with mitochondrial polyP metabolism.159 The crucial role of polyP in mitochondrial energy metabolism has been demonstrated by enzymatic depletion of the mitochondrial polyP content using polyP-hydrolyzing yeast exopolyphosphatase PPX1; a significant decrease in mitochondrial membrane potential required for F1FO-ATP synthase-mediated ATP production was observed.160 The major role of mitochondrial polyP at certain pathological conditions associated with ischemic cell death like stroke and myocardial infarction161,162 has been intensively studied. These pathological conditions are associated with a Ca2+-induced activation of the mitochondrial permeability transition pore (mPTP), a large, voltage-dependent multiprotein channel complex in the inner mitochondrial membrane, causing opening of the channel and leading to increased membrane permeability, decreased mitochondrial membrane potential, abolition of ATP production, and ultimately cell death (for a review, see refs (154 and 163)). Especially reperfusion/reoxygenation therapy of ischemic tissue can result in excessive Ca2+ accumulation inside the mitochondria. Already in the first report on the effect of polyP, evidence has been presented that this polymer is closely associated or a component of the mPTP complex.164 In this study, the isolation of a polyP/Ca2+/poly-3-hydroxybutyrate (PHB) complex from mammalian (rat liver) mitochondria was reported,164 a complex which was first identified in bacteria.165 On the basis of its properties, it was concluded that this complex might act as the ion-conducting module of mPTP.164 The proposed involvement of the polyP/Ca2+/PHB complex in mPTP opening was supported by polyP depletion experiments that revealed a strong inhibition of mPTP function,159,166 resulting in prevention of mitochondrial Ca2+ accumulation and cell death (see refs (154 and 167)). The effects of polyP are correlated with the chain length of the polymer. While long chain polyP (polyP120) can induce/activate PTP and cell death as well as mitochondrial depolarization in astrocytes, medium- (polyP60) and short-chain polyP (polyP15) failed to cause this effect.168 This result has been supported by findings revealing that short and medium polyP does not cause apoptosis while long chain polyP activates the apoptotic cascade in neurons and astrocytes.168 In a more recent study, a highly purified preparation has been obtained from mammalian mitochondria that contained besides the polyP/Ca2+/PHB also the C-subunit of the ATP synthase.169 The properties of this isolated channel-forming complex were quite similar to the native mPTP. The data also showed that polyP, PHB, and the C-subunit are intimately associated as an integral component of the Ca2+-induced mPTP.169 This conclusion was also supported by the finding that the formation of this channel-forming complex is inhibited by cyclosporine A, an inhibitor of mPTP.169 Interestingly, polyP has also been found to be an modulator (activator) of mTORC1 (target of rapamycin complex 1),170 a complex that acts as an intracellular homeostatic ATP sensor by activating a series of anabolic metabolic pathways.171 Recently, it has been reported that a decrease of mitochondrial membrane potential by opening mPTP pores is associated with a rise in calcification of polyP-treated MC3T3-E1 osteoblastic cells.172
5.2. Cell Sensation
The receptor-activated nonselective cation channel TRPM8 (transient receptor potential melastatin subtype 8), a cold-activated channel, is involved in sensing of extracellular temperature, such as coolness, but is also activated by certain chemicals such as menthol. Whole-cell patch-clamp and fluorescent calcium measurements with human embryonic kidney and F-11 neuronal cells revealed that the TRPM8 protein complex forms a stable association with polyP and PHB resulting in a modulation of the sensitivity threshold.173 A related but mechanistically distinct function of polyP, acting on the nociceptive sensation, has been described.174 The TRPA1 receptor (transient receptor potential A1) is expressed in sensory afferent neurons and requires polyP as a soluble factor to become functionally active. In contrast to TRPM8 which copurifies with polyP, TRPA1 only weakly interacts with polyP.
5.3. Cell Metabolism
The overall cell metabolism of human fibroblast cells is activated by polyP.175 This effect has been deduced from the findings that the mitogenic activities of both the acidic fibroblast growth factor (FGF-1) and the basic fibroblast growth factor (FGF-2) are enhanced by this polymer. In addition, polyP increases the adhesion capacity of the cells via their cell surface receptors. Support came also from more recent studies with Caco2/BBE cells.176 These authors described that polyP induces HSP27 gene expression in vitro and causes in vivo a protection of mice against inflammation, elicited by Na-dextran sulfate. Finally, it should be mentioned that higher levels of polyP are present in myeloma plasma cells, which are assumed to modulate the transcriptional activity, catalyzed by RNA polymerase I.177
5.4. DNA Synthesis and Repair
Using yeast cells as a model, polyP was found to increase the intracellular pool of deoxynucleotide triphosphates (dNTPs). These “extra” dNTPs were identified to protect the cells against DNA damage and extend the longevity of those compromised cells.178 Subsequent studies with mammalian HEK293 cells and in human dermal primary fibroblasts confirmed this effect by demonstrating that after deprivation of those cells for polyP the cells become more prone to DNA damage.178 In turn, the authors claim that polyP increases the intracellular metabolic energy in the form of ATP needed during the DNA repair process. In the bacterial system Pseudomonas aeruginosa, a close coupling of polyP synthesis with the completion of cell cycle exit during starvation has been demonstrated.179 This aspect of the interaction of polyP with the DNA replication machinery is, at the present state, not completely understood.
5.5. Blood Clotting
It has been suggested that after release from activated blood platelets, synthetic polyP initiates clotting of plasma via the contact pathway on the level of factor XII.120 However, subsequent studies disclosed that when platelets become activated only physiologically occurring short-chain polyP is released, which has only a very low capacity of direct activation of factor XII (see ref (180)).
5.6. Bone Mineralization and Cartilage Formation
Among the most energy-consuming processes proceeding in the extracellular space in humans are the bone and cartilage anabolic processes. An extensive review on this subject has been very recently published,30 and it is also discussed here in the context of energy homeostasis during bone and cartilage formation in the body.
Increasing evidence indicates that polyP can also act as an important mediator of pro-inflammation and pro-coagulation.181 Both activities are beneficial in innate immune response and during at least the initial stages of wound healing, e.g., accelerating the sealing process for severe injuries.182 These authors identified in rat basophilic leukemia mast cells polyP with a chain length of 60 Pi units which they attribute to the serotonin-containing acidocalcisomes. It has been described that those polymeric molecules are secreted from platelets/acidocalcisomes upon activation.120,121
The supporting role of ATP and adenosine being an important metabolite that controls the activity of chondrocytes in rat tibia explants was demonstrated in mice lacking the adenosine A2A receptor.183 Animals with this defect develop spontaneous osteoarthritis, a degenerative joint disorder. Supplementation with adenosine via intra-articular injection prevented the development of the disease.
6. PolyP Metabolism in Prokaryotic and Eukaryotic Cells: Enzymes
Although polyP is an abundantly present inorganic polymer found in all cells from prokaryotes to eukaryotes, the way of synthesis and degradation of polyP in the different kingdoms of life appears to be distinctly different. This partially depends on the different functions of polyP in these organisms. In (eu)bacteria, polyP takes an active/functional role in survival at the stationary phase, stress responses, activation of ATP-dependent serine peptidases, Lon proteases, as well as during virulence184 (reviewed in ref (185)) but also acts extracellularly as an intercellular communication/signaling molecule during quorum sensing processes, and biofilm formation.186 Also in the budding yeast Saccharomyces cerevisiae and the social amoeba Dictyostelium discoideum, polyP acts as a signaling molecule during pH homeostasis187 and growth control.188 In animals, polyP is also released into the extracellular space, especially from the platelets, and functions there as a regulator of a series of regeneration processes, e.g., in wound healing,189,190 inflammation,189 and coagulation.180,191
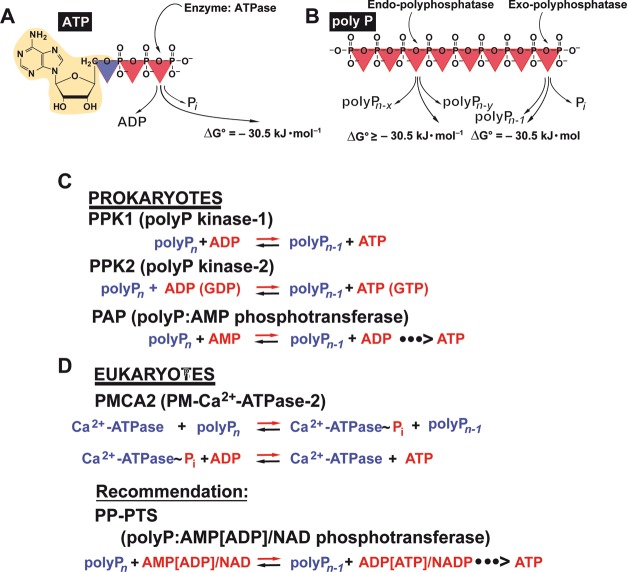
The chain length of the polyP has a decisive effect on the biological effects of the polymer. This has been reported both for intracellular (e.g., mitochondrial) and extracellular polyP. Therefore, the control of the size of the polymer seems to be of utmost importance. In principle, the polyP chain length can be controlled both on the level of polyP synthesis, catalyzed, e.g., by bacterial polyP kinases (PPK) [EC 2.7.4.1], and on the level of polyP degradation. In yeast and higher eukaryotes, different mechanisms for polyP synthesis have been developed, as discussed later.
polyP degradation can occur either by hydrolysis of the polyP chain, catalyzed by polyphosphatases, both by exopolyphosphatases [EC 3.6.1.11] which hydrolyze the polyP chain from the ends of the polymer under liberation of orthophosphate (Pi) and by polyP endopolyphosphatases [EC 3.6.1.10] which cleave polyP within the polymer chain, or by transfer of the terminal Pi groups to an acceptor molecule, mediated by a phosphotransferase. Examples of the latter group of enzymes are a polyP-AMP phosphotransferase [EC 2.7.4.B2] which catalyzes the phosphotransfer from polyP to AMP,192 or a polyP glucokinase [EC 2.7.1.63] which phosphorylates glucose by polyP.193 While in the first case, the energy of the energy-rich phosphoanhydride bond is liberated in the form of heat, this energy is, at least partially, conserved in the latter case in the newly formed phosphoanhydride or phosphoester bond at the acceptor molecule.
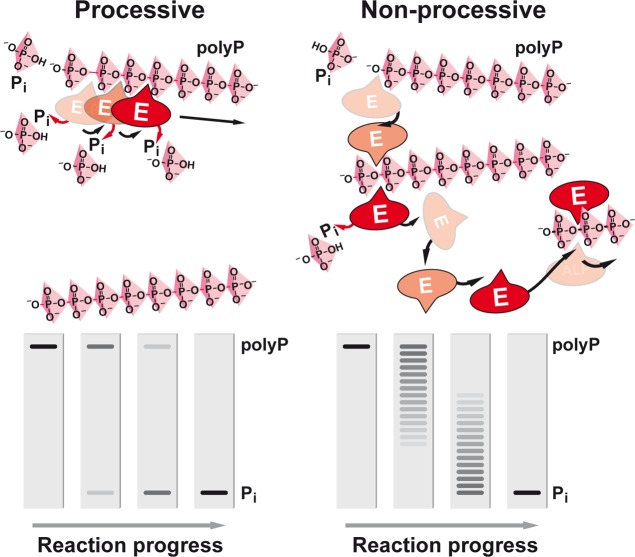
In addition, two principal mechanisms can be discerned through which degradation (but also synthesis) of polyP can proceed: processive or nonprocessive (Figure 5).24,25,117 In the processive reaction, the enzyme (here an exopolyphosphatase) remains bound to the polymer until the degradation of the polyP chain is completed. Analysis of the products on high-percentage polyacrylamide gels will reveal only two bands, the undegraded polyP substrate, which decreases during the reaction, and the product (e.g., Pi), which increases in the course of the reaction. No intermediate polyP chain lengths can be seen. In the nonprocessive mechanism, a successive association and dissociation of the enzyme from the polyP during each catalytic step takes place. In turn, the enzyme continually “jumps” from one polyP molecule to another until the substrate is completely turned over. Consequently, polyacrylamide gel analysis will show a ladder-like pattern reflecting the occurrence of shorter polyP chains between the polyP substrate and Pi product in the course of the reaction. It should be noted that some polyphosphatases break down the polyP to a certain chain length or change from a processive to a nonprocessive mechanism beyond a certain chain length; in the latter case, a laddering pattern is observed below that size of the polyP molecule.
Figure 5.
Processive versus nonprocessive degradation of polyP. In the processive mechanism (left), the enzyme (E) remains bound to the polyP substrate until the degradation is completed, while in the nonprocessive reaction (right), the enzyme dissociates from the substrate after each catalytic cycle and then reassociates either with the same or another polyP molecule.
6.1. Eubacterial Enzymes
The intracellular, enzyme-driven polyP anabolism and catabolism, especially with respect to the energy metabolism, is well understood in bacteria. In the center of the bacterial energy transfer reactions are the polyphosphate kinases (PPKs),26,194 which synthesize polyP by using ATP as a substrate194 (Figure 6). Besides in bacteria, the PPKs have been identified in archaea, fungi, yeast, and algae (reviewed in ref (185)) but not yet unequivocally in animals.159 The PPKs are reversible acting enzymes transferring energy-rich bonds in both directions.185
Figure 6.
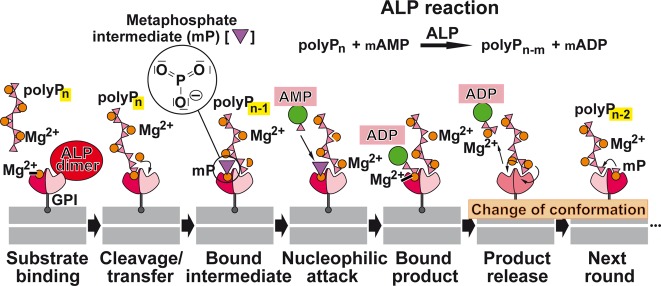
Enzymes involved in energy storage and energy supply by polyP. (A) ATP as the intracellular energy generator, and (B) polyP as an intra- and extracellular energy source and energy generator during the enzymatic hydrolysis by exo- or endopolyphosphatase(s). In both compounds, energy-rich bonds are present that comprise a ΔG0 of about −30.5 kJ mol–1 per bond. The polyP metabolizing enzymes that catalyze reversible reactions are present in both (C) prokaryotes and (D) eukaryotes. In prokaryotes, the initial ATP-dependent step of polyP synthesis is catalyzed by polyP kinases; the reactions catalyzed by polyP kinase 1 (PPK1) and polyP kinase 2 (PPK2) are shown. These reversible reactions can also serve for the formation of ATP (GTP) from polyP. Additionally, it has been suggested that polyP-AMP-phosphotransferases are using polyP as a substrate for the synthesis of ATP via AMP and ADP. For eukaryotes, it has been described that the plasma membrane associated Ca2+-ATPase (PMCA) acts as an ATP-polyphosphate transferase and polyphosphate-ADP transferase. Finally, an ALP is present, which is involved in both polyP hydrolysis and ADP formation from AMP, as well as a NAD-kinase which catalyzes the transfer of the terminally located energy-rich phosphate units of the polyP chain to NAD+ resulting in NADP+ production. We recommend to group these phosphotransferases which also includes the PPK (reversible reaction) as polyP-phosphotransferase system [PP-PTS].
Due to the comprehensive studies of the Kornberg’s group, the enzymatic metabolism of polyP in prokaryotes has been resolved. The initial ATP-dependent step of polyP synthesis is mediated by the polyP kinase 1 (PPK1 [EC 2.7.4.1]), most likely the dominant enzyme in bacteria (reviewed in refs (195 and 196)). Experimental data indicate that the metabolic energy, stored in polyP, can be reutilized in bacteria to synthesize ATP. A second bacterial polyP kinase, the PPK2 [EC 2.7.4.1]197 has been described that shows similarity to the mammalian thymidylate kinase.198 In addition, several bacterial PPKs have been identified that use preferentially pyrimidine nucleotides as cosubstrates; they were termed PPK3 [EC 2.7.4.1].199
polyP degradation in bacteria is catalyzed both by the polyP kinases (reversible reaction of PPKs) and exopolyphosphatases which only hydrolyze the polymer chain.200,201 A well investigated polyP degrading exopolyphosphatase is the bacterial exopolyphosphatase (PPX [EC 3.6.1.11]) from Escherichia coli a highly processive enzyme that preferentially hydrolyzes long polyP chains (≥1000 Pi residues).125,201,202 The enzyme is a dimer201 which produces four discrete intermediates: polyP2, polyP3, polyP14, and polyP50. In a model proposed,125 each monomer of the PPX dimer contains two catalytically active N-terminal sites and three polyP binding sites located at a distance of 3, 14, and 50 Pi residues from the active site.
Two further examples of an enzyme that produces intermediates with a specific chain length during degradation are the E. coli polyP guanosine pentaphosphate phosphohydrolase (GppA) [EC 3.6.1.40]203 and the polyP glucokinase [EC 2.7.1.63] of Propionibacterium shermani which phosphorylates glucose by polyP.193,204 Both enzymes degrade long chain polyP by processive hydrolysis of the terminal phosphates until a specific chain length of 40 Pi units (polyP40; for GppA)203 or 100 Pi units (polyP100; for polyP glucokinase)193,204 is released.
Additionally, a bacterial polyP-AMP-phosphotransferase (PAP [EC 2.7.4.B2])192,205 has been described that apparently metabolizes polyP as a substrate for the synthesis of ADP and ATP via ADP (the latter reaction, ATP formation, proceeds together with an adenylate kinase),206,207 as well as an enzyme from P. aeruginosa that acts both as a polyP:ADP phosphotransferase and an exopolyphosphatase.208,209 The active site of latter enzyme is assumed to be involved in both enzyme activities.209
In contrast to yeast and animals, endopolyphosphatases which cleave polyP in the middle of the chain seem to be absent in bacteria.
Interestingly, the polyP glucokinase from P. shermanii can change from a processive to nonprocessive mechanism. The degradation of long chain polyP by this enzyme proceeds via a processive mechanism, while short polyP is hydrolyzed nonprocessively.193,204 This change of the type of reaction is paralleled by a change of the Km of the enzyme that first increases slowly with decreasing polymer size but then dramatically increases at a chain length of 30 (polyP30). After reaching this size, the enzyme dissociates from the polymer and further degradation proceeds by a nonprocessive mechanism.
6.2. Fungal Enzymes
Some phylogenetic data revealed that eukaryotic fungi share a close relationship with animals.210,211 In yeasts, polyP accumulates in large amounts inside the vacuole, an acidic, acidocalcisome-like organelle,212 where cellular compounds are recycled. This vacuole is a dynamic structure that can rapidly modify its morphology. There the polyP polymer accumulates and is additionally found in smaller levels in the cytosol, nucleus, and mitochondria (reviewed in ref (213)).
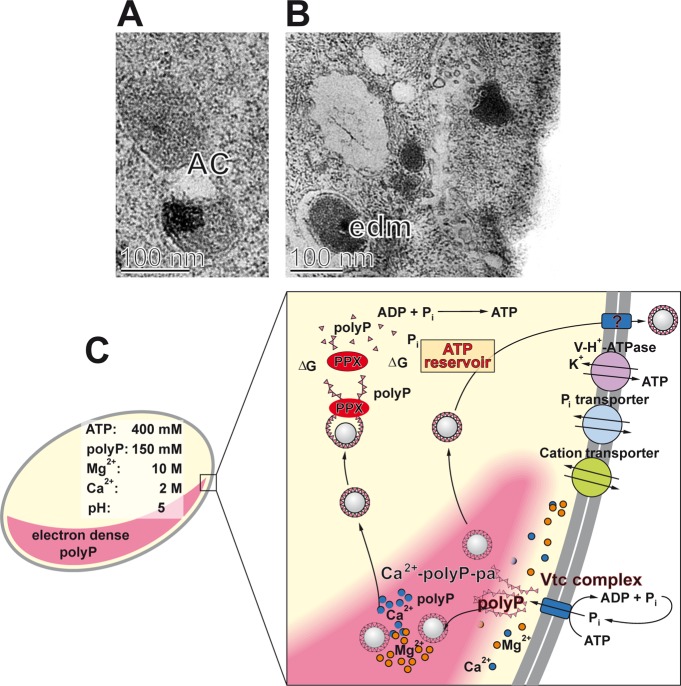
Using the yeast S. cerevisiae as a model, most knowledge on polyP metabolism in eukaryotes has been gathered. Among the yeast enzymes involved in polyP metabolism are the cytosolic exopolyphosphatase PPX1,214,215 the cytosolic endopolyphosphatase DDP1,216−218 and the vacuolar endopolyphosphatases PPN1,219−221 which can switch between exo- and endopolyphosphatase activity,222 and PPN2.223,224 In addition, a vacuole transporter chaperon complex (Vtc complex) has been identified that synthesizes polyP and acts as a main phosphate storage molecule in yeast. This polymerase uses ATP as a substrate and transfers the γ-phosphate to an acceptor phosphate under formation of a polyP chain.225 This complex is also involved in the translocation of polyP across the vacuolar membrane (see section 8).
Different reversibly acting PPKs have been identified in other fungal taxa, like in arbuscular mycorrhiza or in the funguslike slime mold D. discoideum.226,227 In addition, PPK-like enzymes have been identified that use other cosubstrates such as the 1,3-diphosphoglycerate:polyphosphate phosphotransferase.25,228
The S. cerevisiae polyphosphatases have been extensively studied. These enzymes differ in their substrate specificity (preference for long polyP or for short polyP chains), mode of action (processive or nonprocessive), dependence on metal ions, and response to inhibitors;126,229,230 for a recent review, see ref (213). The PPX1 which degrades long-chain polyP to PPi in a processive manner under the release of Pi230 is a member of the DHH family of phosphoesterases,231 like the mammalian protein h-prune which also acts as an exopolyphosphatase (see below).232 On the basis of the analysis of the crystal structure, the yeast enzyme is assumed to contain a positively charged tunnel-like structure with multiple arginine and lysine residues, which channels the entering polyP polymer from one end to the catalytic site at the other end of the protein.233 The existence of such a tunnel-like structure has also been proposed for certain polyP-binding proteins.234 Also an enzyme related to the yeast PPN1 has been partially purified from rat and bovine brain220 but not further studied. The DDP1 belongs to the Nudix hydrolase family and seems to be primarily involved in inositol pyrophosphate metabolism but can also act as a polyP-hydrolyzing endopolyphosphatase.216−218
6.3. Enzymes from Other Eukaryotes and Animals
In the trypanosomes, the acidocalcisomes have been first discovered and, in turn, used as a model for the understanding of the role and function of these organelles in polyP metabolism also in animals.235
6.3.1. Trypanosomes/Unicellular Eukaryotes
Several exopolyphosphates have been isolated, cloned, and expressed from the major human pathogenic unicellular eukaryotes Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. In contrast to the yeast enzymes, these exopolyphosphatases preferentially hydrolyze short-chain polyP, such as the exopolyphosphatase from T. cruzi,236 the cytoplasmic PPX1 from T. brucei (degradation of polyP3, but not PPi and long-chain polyP),237 and the exopolyphosphatase from L. major, which is localized in the acidocalcisomes and cytosol.238 In addition, some members of the Nudix family of enzymes were found to hydrolyze polyP, such as the T. brucei Nudix hydrolases TbNH2, an exopolyphosphatase located in peroxisome-like organelles (glycosomes) where glycolysis occurs, and TbNH4, an endo/exopolyphosphatase located in the cytosol and nucleus.239,240
6.3.2. Animals
As outlined, until today, an enzyme related to the PPKs has not been identified in higher eukaryotes. It should be granted that, within the cells, in the first place ATP should be considered as the cosubstrate for the enzymatic synthesis of polyP. This energy-carrying nucleotide is assumed to be synthesized intracellularly at two metabolic intersections, either within the mitochondria during the enzyme-controlled reduction/oxidation reactions or in the cytoplasm at the substrate-level phosphorylation steps, like in glycolysis during transfer of a phosphate group to ADP from 1,3-bisphosphoglycerate yielding 3-phosphoglycerate (phosphoglycerate kinase reaction) and from phosphoenolpyruvate producing pyruvate (pyruvate kinase reaction). In these two compartments, mitochondria and cytosol (membrane associated), polyP synthesizing enzymes have been identified.
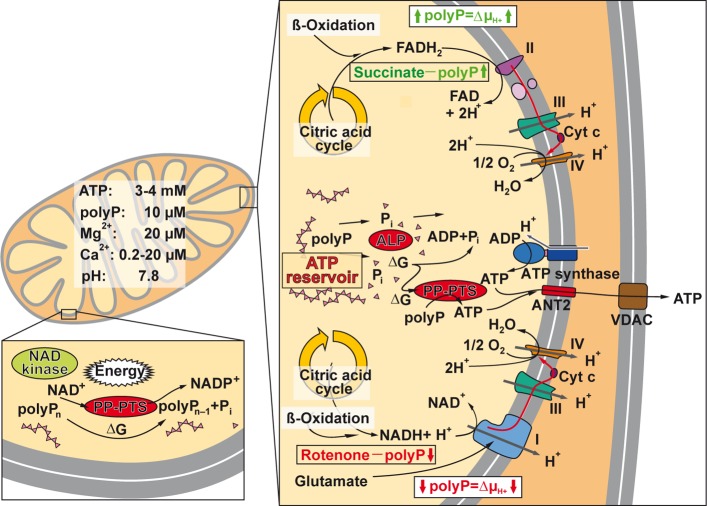
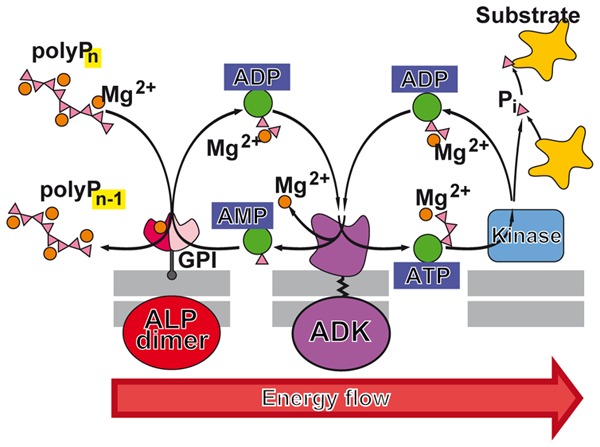
The mitochondrial ATP-synthase (F1FO-ATP synthase [EC3.6.1.34]) has been proposed to be involved in the synthesis of polyP.159 The authors conclude that polyP synthesis is directly linked with the ATP generation system of the mitochondria and with their energetic state; they propose that a feedback mechanism regulates the levels of polyP and of ATP and reciprocally (Figure 7). It has been suggested, based on the strong evidence that the energy level of the mitochondria is closely correlated with polyP level, that the synthesis of the polymer starts on pyrophosphate241 that becomes elongated by an unknown polyP-synthesizing enzyme. The level of polyP in mitochondria is ∼10 μM151 and that of ATP 3–4 mM.242 Enzymatic removal of polyP in mitochondria, via expression of yeast exopolyphosphatase (PPX), results in a decrease of the mitochondrial membrane potential and an impairment of the respiratory chain.243 This finding can be taken as an indication that polyP might act in the mitochondria as a reservoir for metabolic energy generation in the form of ATP (Figure 7).
Figure 7.
Mitochondria as storage of ATP and ATP buffering organelles. These organelles are the major intracellular ATP generator producing ATP from reducing equivalents during metabolism of glucose and fatty acids via glycolysis (cytosol) and β-oxidation and finally the citric acid cycle (mitochondrion). The reducing equivalents drive the electron transport chain in the inner mitochondrial membrane, which ends in the ATP generator, the ATP synthase. The adenine nucleotide translocase (ANT2) allows the ATP to cross the mitochondrial inner membrane, while the voltage-dependent ion channel (VDAC) is involved in the energetic flux and transport of ATP and ADP across the outer mitochondrial membrane. Within the mitochondrial matrix, polyP is proposed to be metabolized via the NAD kinase (insert), an enzyme that catalyzes the phosphorylation of NAD+ to NADP+ under consumption of ATP and other nucleoside triphosphates as well as of polyP as a phosphate source. Most likely many of these complex enzymatic reactions, including the ALP and the NAD kinase, act as PP-PTS. A reduced polyP level parallels with a decrease of the mitochondrial membrane potential (ΔμH)/proton gradient (ΔpH+) at the inner mitochondrial membrane and vice versa. The approximate concentrations of ATP, polyP, Mg2+, and Ca2+ as well as the intramitochondrial pH value are given.
In addition, experimental evidence has been presented demonstrating that the plasma membrane associated Ca2+-ATPase (PMCA) from erythrocytes might function as an ATP-polyphosphate transferase and polyphosphate-ADP transferase.244 The PMCA belongs to the group of PMCA2 [EC 3.6.3.8] since the enzyme forms a phosphate intermediate during the reaction cycle.245−247 Consequently, it has been postulated that the PMCA2 is associated with polyP allowing also a transient covalent autophosphorylation, followed by a transferase reaction and, by that, the formation of ATP.244,248 It would be interesting to study this enzyme in further detail.
Among the mammalian enzymes involved in polyP degradation, only one enzyme that corresponds to the yeast PPX1 has been identified, the exopolyphosphatase h-prune which belongs to the DHH family of phosphoesterases.232 However, this enzyme, in contrast to the yeast PPX1, is a short-chain exopolyphosphatase and inhibited by long polyP chains.232 The major exopolyphosphatase in mammalian organisms including humans, which also degrades long polyP chains, seems to be the alkaline phosphatase (ALP); see section 7. We propose that this enzyme also acts as a phosphotransferase, as discussed in section 13, like the NAD kinase [EC 2.7.1.23]. The latter enzyme has been traced in mitochondria, mediating NADP+ biosynthesis from NAD+ (Figure 7);249,250 this metabolite is required in mitochondria for a series of specific biosynthetic pathways and acts also as an antioxidant.251 The NAD kinase uses not only ATP but also polyP as phosphate donor for phosphorylation of NAD+ at the 2′-hydroxyl group of the adenosine ribose moiety of the molecule.252,249
Therefore, the NAD kinase, like the ALP, belongs to a group of enzymes capable of using energy-rich polyP instead of nucleotides/ATP as a phosphate donor in phosphotransferase reactions. We propose to operationally classify this group of functionally related enzymes (e.g., ALP and NAD kinase) that catalyze the transfer of energy-rich phosphate from polyP to a substrate (e.g., AMP, ADP, or NAD+) as a polyP phosphotransferase system (PP-PTS; EC number 2.7) (Figure 6).
7. Alkaline Phosphatase: Role in polyP Catabolism
The major exopolyphosphatase in animals and humans, which is able to degrade long chain polyP, is the alkaline phosphatase (ALP).117 This metalloenzyme, which can also act as a phosphotransferase, can therefore be grouped into the polyP phosphotransferase system PP-PTS and is functionally active only as a dimer.253 It contains two Zn2+ ions and one Mg2+ ion in each catalytic site of the dimer; these metal ions are involved in the catalytic mechanism of the enzyme.254 We have demonstrated that the ALP degrades polyP following a processive mechanism.117 Four isozymes are known, the intestinal ALP, the placental ALP, the germ cell ALP, and the tissue-nonspecific ALP or liver/bone/kidney ALP.254 The ALP dimers are allosteric enzymes.253 Studies on the human placental ALP which only requires Zn2+ for activity revealed that this isoform, in its fully metalated form, shows the characteristics of a noncooperative allosteric enzyme; both subunits of the ALP dimer function independently, but their catalytic activity is controlled by the conformation of the respective other subunit.253 On the other hand, the tissue-nonspecific ALP (liver/bone/kidney ALP; also present in brain), which like the intestinal ALP, needs both Zn2+ and Mg2+, shows a negative cooperativity with respect to Mg2+ binding, which is relevant for the mechanism of polyP degradation, as discussed in section 13. The ALP is characterized by a broad substrate specificity. Besides polyP, the ALP hydrolyzes pyrophosphate, AMP, ADP, ATP, glucose-1-phosphate, glucose-6-phosphate, and β-glycerophosphate.
Apart from the ALP, only two other human proteins have proven to be an exopolyphosphatase. The first protein, h-prune, is characterized by a high sequence homology to PPX but preferably hydrolyzes only short-chain polyP (≤25 Pi residues).232 The second protein is the osteoclast tartrate-resistant acid phosphatase (TRAP).255 This exopolyphosphatase hydrolyses, like h-prune, only shorter-chain polyP molecules; it is inhibited by long-chain polyP. On the basis of the facts that the chain length of the acidocalcisomal and platelet polyP is around 70–75 Pi units and the mammalian ALP, in contrast to h-prune and TRAP, hydrolyzes both long and short polyP molecules,117,256 it can be concluded that the ALP plays a predominant role at least in the control of long chain polyP in mammalian/human organisms. The ALP might function in close cooperation with the endopolyphosphatase described by Kumble and Kornberg.220 This enzyme which corresponds to the yeast PPN1 has been reported also to exist in mammalian tissue; it cleaves long chain polyP to polyP60 and has been partially purified from the rat brain and bovine brain.220 Unfortunately, this enzyme which might be responsible for the occurrence of polyP molecules of distinct, intermediate chain lengths during cell cycle/apoptosis has not been studied further. For example, analysis of the size of polyP in HL-60 cells revealed that both proliferating and nonproliferating cells contain two classes of polyP, long-chain polyP with a narrow size distribution of around 150 Pi residues and medium-chain polyP with 25–45 Pi residues. In apoptotic cells, the long-chain polyP but not the medium-chain polyP disappeared.257
With regard to the differential biological effects of different size classes of polyP, the processive mode of action of polyP degradation shown by ALP might be of particular importance. This mechanism allows the degradation of long polyP molecules to monomeric Pi without the intermediate release of polyP molecules of shorter, intermediate chain lengths. In this way, it is prevented that during degradation of long chain polyP shorter polyP molecules are formed with a size that might show different (possibly unwanted) activities. For example, very long polyP polymers, administered extracellularly, with chain length of ≥500, such as those present in microorganisms, have been shown to activate the contact pathway of the blood coagulation cascade, while medium size polyP molecules, with polymer lengths of about 100 phosphate units, such as those released by platelets, accelerate activation of factor V via factor Xa and thrombin and abrogate the anticoagulant function (inhibition of factor Xa) of the tissue factor pathway inhibitor (TFPI); polyP chains with a size of ≥250 are required to enhance fibrin polymerization122 (reviewed in refs (96 and 258)).
The mechanism by which the mammalian polyP degrading enzymes recognize specific chain lengths of polyP is not known. This is in particular intriguing for exopolyphosphatases, like the ALP, because the degradation of long polyP chains begins at the ends of the polymer. In contrast to the E. coli enzyme, this, also processive, exopolyphosphatase degrades long-chain polyP, e.g. polyP750–800, without formation of any intermediary product.117 It might be possible that, like seen for the E. coli exopolyphosphatase (see above), multiple binding sites on distant portions of the enzyme proteins exist. Both enzymes are functionally active as a dimer. The affinity of the ALP to the polyP substrate increases with increasing chain length of the polymer. Measurements with low and medium chain polyP at pH 9.5 (pH 7.5) and using the enzyme from calf intestine revealed a decrease in the Km value (in terms of polymer concentration) from 187 μM (218 μM) for polyP2 to 28 μM (0.5 μM) for polyP69–95.117 This increase in affinity is paralleled by a decrease in the maximum rate of the reaction, vmax, from 1025 × 10–3 (176 × 10–3) mol min–1 mg–1 to 60 × 10–3 (2.5 × 10–3) mol min–1 mg–1, in line with the processive type of reaction catalyzed by the enzyme (increase in reaction rate in the course of the processive degradation of the polyP chain).
8. PolyP Metabolism in Eukaryotic Cells: Organelles
There are two intracellular organelles which are rich in polyP, the mitochondria and the acidocalcisomes.168 Interestingly, within eukaryotic cells, exemplarily demonstrated in the eukaryotic parasite T. brucei, these two organelles are in close contact,259 a finding that is suggestive and indicative toward a functional interaction of the acidocalcisomes with the mitochondria during regulation of cellular bioenergetics.260
8.1. Mitochondria
It has been especially the Abramov group261 that disclosed polyP metabolism being correlated with the polymer chain-length in mitochondria. Short polyP (14 Pi residues) or medium-sized polyP (∼70 Pi) increases the respiratory coefficient by activation of complex III and inhibition of complex IV, compared to the control. Simultaneously, these two classes of polyP markedly reduce the oxidative phosphorylation (ADP/O ratio). Long polyP (130 Pi) causes cell death in primary neurons and astrocytes, while medium and short polyP, again, do not elicit this effect. However, it might be considered that any kind of polyP is prone to degradation by ALP, or ALP-like enzymes, a fact which complicates a strong conclusion.117,262
A hypothetical polyP synthase has been proposed to be located in the inner membrane of the mitochondria, presumably associated with the mitochondrial ATP synthase (Figure 7).159 Complex I-linked substrates, like pyruvate, malate and glutamate or the complex II-linked substrate succinate, which stimulate mitochondrial oxidative phosphorylation,263 are also activators of the mitochondrial polyP synthesis.159 Inhibition of the complex I system by Rotenone causes a drop of the polyP pool. This fact implies that alterations of the function of these complexes are correlated with the polyP level in the matrix of the mitochondria. This circuit is paralleled with a potential effect of polyP on the proton gradient across the inner mitochondrial membrane (ΔpH+); at a high potential, polyP concentration increases and vice versa.168 In turn, two circuits which control the level of polyP in the mitochondria can be formulated, one linked with the individual complexes of the respiratory chain and another linked with the overall proton gradient.
In addition to an ALP-like enzyme, a PPX has been described to be present in the mitochondria of animals (ticks).264 This enzyme acts as an energy sensor. It has been found that the mitochondrial PPX becomes activated and responds with a higher polyP hydrolysis if the mitochondria are rich in ADP.264
These data might suggest that the metabolic cycles of polyP in the mitochondria, perhaps both polyP synthesis and the catabolic pathway of polyP hydrolysis, control the ATP level in these organelles and function as a storage reservoir for ATP. The polyP synthesis requires the energy from ATP (for the formation of the phosphoric acid anhydride bond). Further studies are needed to clarify the metabolism of polyP in mitochondria.
8.2. Acidocalcisomes
As mentioned, the mitochondria are in close apposition235 or even directly attached to the acidocalcisomes.260 It has been found that the latter organelles, collectively referred to as acidocalcisomes,265,266 which occur in both prokaryotic and eukaryotic cells, from bacteria via yeasts to higher vertebrates/humans, store polyP, accumulate Ca2+, and show an acidic milieu. The acidocalcisomes are round-shaped organelles with an 8 nm thick membrane and have a size of 50–300 nm. They are filled with amorphous polyP that appears as electron-dense deposits267,268 (Figure 8A,B). These particles can be stained by DAPI, showing a green fluorescence.128 In the acidocalcisomes, the polyP level reaches millimolar concentrations.121 Furthermore, a series of proteins has been identified in these organelles that are involved in phosphate and polyP metabolic pathways, like the Vtc complex or a putative phosphate transporter and a putative acid phosphatase.269 The Ca2+ concentration in the acidocalcisomes reaches levels of 2 M, and their matrix pH is about 5.4.270 This pH value is maintained by an H+-ATPase, a pump which is different from F1FO- as well as the P-type ATPases271 (Figure 8C). Besides polyP and energy-rich nucleotides, the acidocalcisomes contain pyrophosphate and serotonin.180,270
Figure 8.
Acidocalcisomes as polyP-synthesizing organelles. (A and B) TEM images of acidocalcisomes (AC) from bone cells. The AC is an organelle which harbors inclusions of electron dense material (edm) that is composed of polyP. (C) The acidocalcisomes are rich in ATP, polyP, Mg2+, and Ca2+ and have a low pH of ∼5 which is maintained by an H+-ATPase. The organelle is surrounded by a membrane into which a series of transporters are embedded. Among them is the vacuolar transporter chaperone (Vtc) complex that has been identified in yeast and implicated in polyP synthesis. The polymer is released into the lumen of the organelle. There, polyP is cleaved by the exopolyphosphatase (PPX) at the energy-rich anhydride bonds under release of metabolic energy (ΔG), or, as proposed here, the energy-rich phosphate is transferred to AMP/ADP under formation of ATP by the PP-PTS. Again, the acidocalcisomes would act, like the mitochondria, as an ATP reservoir but with a much higher capacity.
A (putative) transport mechanism of polyP from the mitochondria to the acidocalcisomes and in continuation to the cytosol is not yet known. It is most likely that the polymeric polyP is not exported from the mitochondria to the acidocalcisomes via the mitochondrial ADP/ATP carrier, since this carrier is highly specific for ADP, ATP, and their deoxy-analogues.272 As an alternative, the export of polyP could occur in nanoparticles. If so, polyP must be complexed with Ca2+, which is present in mitochondria only in micromolar concentrations and is an essential cation functioning during the energy/ATP synthesis. A similar role plays Mg2+ which is also present in only small levels with 20–30 μM273 and is crucially important for the maintenance of the energy homeostasis in mitochondria.274 Therefore, based on quantitative consideration (next paragraph), it appears to be likely that the main intracellular organelles producing polyP are the acidocalcisomes.
Comparing the polyP level in mitochondria versus acidocalcisomes (∼10 μM – 150 mM), the ATP concentration of 3–4 mM versus 400 mM, and the Ca2+/Mg2+ level in mitochondria versus acidocalcisomes of ∼10 μM – ∼2 M/∼10 M,275 it can be assumed that the bulk of polyP, provided by the acidocalcisomes to the cells and the surrounding tissue, is synthesized in the acidocalcisomes (Figure 8C). In turn it can be deduced that polyP acts in the acidocalcisomes like in the mitochondria as a reservoir for the deposition of metabolic energy.159,168,241,276
The acidocalcisomes are surrounded by a membrane (Figure 8C) into which a series of transporters and channels is integrated (see ref (266)); the existence of some of those channels is still not yet clear. These organelles contain the electron-dense material, composed of polyP, that adheres to one side of the membrane of the organelle with the inositol 1,4,5-trisphosphate receptor channel (IP3R; not shown in Figure 8C), required for Ca2+ release from the acidocalcisomes, to the mitochondrial Ca2+ uniporter.260 Among the other transporters/channels within the acidocalcisome membrane are the vacuolar H+-ATPase and the vacuolar H+-pyrophosphatase and other transporters, which import Ca2+ and Pi and additionally Zn2+, Fe2+, and polyamines. These pumps keep the pH of the acidocalcisomes around 5.277 Strong evidence has been worked out that in trypanosomes and yeast it is the Vtc complex that mediates polyP synthesis.225 In the yeast S. cerevisiae, the Vtc complex consists of four subunits, one catalytic (Vtc4) and three accessory (Vtc1, Vtc2, and Vtc3).278 The synthesis of polyP from ATP depends on the presence of the electrochemical gradient across the acidocalcisome membrane and requires the formation of a tunnel-like structure by the central domain of Vtc4, which is assumed to be involved in the translocation of the polyP chain into the lumen of the vacuole.225,279 This tunnel-like structure with the polyP chain has been analyzed by X-ray crystallography of the catalytic domain in the presence of ATP.213,225 As presently assumed, the polyP formation driven by Vtc occurs at the cytoplasmic side of the complex in close association with the acidocalcisome membrane. It remains open from where the large amounts of ATP are coming from. As mentioned above, the acidocalcisomes are very rich in ATP (400 mM). The polyP formed from ATP then accumulates in the lumen of acidocalcisomes and is not present in a freely diffusible form.280 The formation of polyP particles is strongly favored because of the high concentration of Ca2+ (2 M) and Mg2+ (10 M) present in the acidocalcisomes.256 It is agreed that polyP released to the lumen of the acidocalcisomes is prone to hydrolysis by the surrounding exopolyphosphatase [PPX] (see refs (266 and 281)), which is opening the energy-rich anhydride bond under the release of metabolic energy (ΔG). Again, on this step metabolic energy is released from the polymer in addition to Pi which might contribute to the high level of ATP within the acidocalcisomes (Figure 8C). This catalysis might occur via an enzyme grouped, like the ALP proposed here, to the class of enzymes classified to the phosphotransferase system (PP-PTS), and polyP could act as a large reservoir for ATP production in the acidocalcisomes.
In addition to its biocatalytic function as a polyP polymerase, the yeast Vtc complex has been implicated in multiple processes such as membrane trafficking, membrane fusion, and microautophagy.282,283 The polyP in the Vtc has been reported to activate fusion with other vacuoles under stimulation of SNAREs (soluble NSF attachment protein receptors). The latter process, activation of SNAREs requires zippering supported by the release of the vesicular-fusion protein SEC18 and addition of ATP.284 Most likely adjacent to the Vtc transmembrane regions within the membranes of the acidocalcisomes, they release their granula.279
9. (Potential) ATP Transfer between Mitochondria and Acidocalcisomes
The concentration gradients for ATP and polyP from the mitochondria to the acidocalcisomes are substantial with amounts consisting of several orders of magnitude (Figure 7 and Figure 8). Certainly, this comparison does not necessarily mean that the primary place of intracellular ATP synthesis is in the acidocalcisomes. It might indicate that ATP and perhaps also polyP are actively transported from the mitochondria to the acidocalcisomes (Figure 9).
Figure 9.
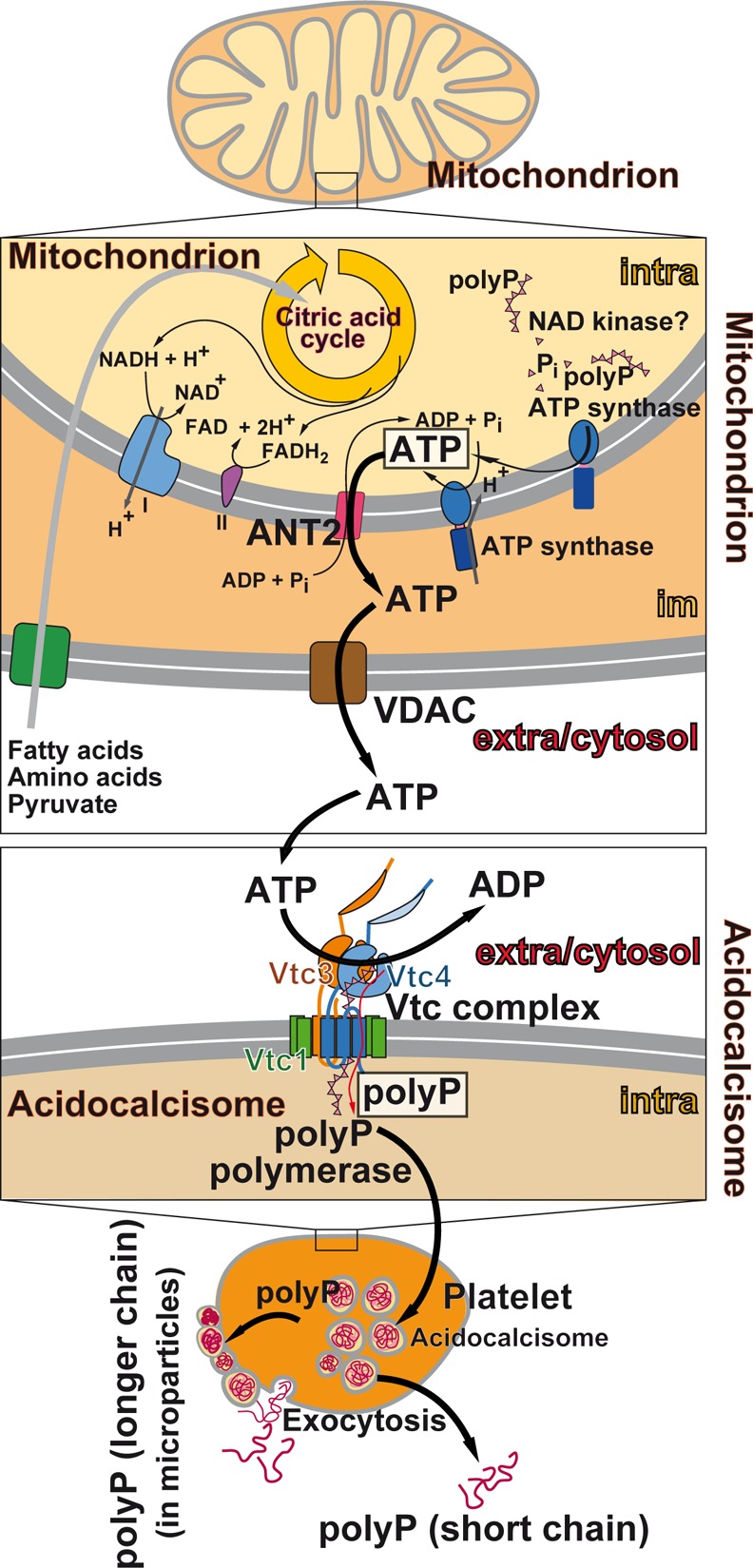
Most probable synthesis route of polyP in eukaryotic cells. The mitochondria take up fatty acids, amino acids, and pyruvate which fuel the citric acid cycle. Subsequently, the reduced coenzymes are channeled into the respiratory chain resulting in ATP synthesis by using the membrane potential to drive the ATP synthase. ATP is transported via ANT2 from the matrix to the intermembrane space and from there through the VDAC to the extramitochondrial cytosol. In the cytosol, ATP reaches the Vtc complex, at the acidocalcisomes, and there the polyP polymerase forms polyP and translocates the polymer into the acidocalcisomes. Subsequently, the polyP rich organelles, abundantly present in blood platelets, release polyP into the cytosol and then via exocytosis into the extracellular space as either short chain polyP or as longer chain polyP, packed into Ca2+ rich particles.
It is suggested, based on the existing close apposition of acidocalcisomes to the mitochondrial outer membrane, that a transfer or even a shuttle of ions or molecules between the two organelles exists, like postulated for Ca2+ from the acidocalcisomes to the mitochondria through the voltage-dependent anion channels (VDACs).260 It would be helpful for an understanding of the bioenergetical interaction between the two organelles if ATP is channeled from the mitochondria via their VDACs to the acidocalcisomes either directly or after bridging a small cytoplasmic gap. The VDACs are located in the mitochondrial outer membrane and are key regulators of the import of the respiratory substrates, ADP, and Pi, into mitochondria and simultaneously of the release of mitochondrial ATP to the cytosol (Figure 9). As observed, a blockade of these transport channels results in an inhibition of a series of cell functions, like cell proliferation.285 The flux of ATP through VDAC saturates only a concentration of 100 mM, a value which is much higher than the overall concentration of ATP in the mitochondria which is 3–4 mM.286
Interestingly, the VDAC can be blocked by the binding of tubulin to the channel. Experiments using carboxy-terminal tail peptides of the tubulin dimer revealed that only the detyrosinated but not the tyrosinated peptides close the channel.287 The post-translational modification of tubulin by tyrosination is catalyzed by an ATP-dependent tubulin-tyrosine ligase [EC 6.3.2.25] that adds a tyrosine residue at the carboxyl end of α-tubulin.288 It might be intriguing to study if this enzyme that might regulate the passage of molecules through the channel causes channel closure at high cytosolic ATP levels and opening (ATP efflux) at low levels of the nucleotide.
It has been briefly noted that within the VDAC, polyP can interact with the binding site of NADH.289 Thereafter, it has been discussed if polyP can cross the mitochondrial outer membrane through VDAC channel from inside to outside. However, the available experiments did not support this view.290 On the basis of the few experimental evidence, which is first of all indirect, it is most likely that polyP within the acidocalcisomes is synthesized de novo through the Vtc polyP polymerase, at least in the yeast model.280
In conclusion and according to the currently existing data, it appears that the bulk of polyP is synthesized in the acidocalcisomes and distributed from there to the cytoplasm and then to the extracellular space. This view is supported by the finding that ionized polyP, if present in the cytosol, is toxic if the expression of cytosolic PPX1 is suppressed.279 This adverse effect is most likely caused by the property of polyP to be a strong chelating and/or sequestering agent for metallic ions.291 It is reported in section 10 that in acidocalcisomes polyP exists as Ca2+ or Mg2+ salts and packed there into amorphous nano/microparticles. These particles are, due to their high zeta potential (−34 mV; at pH 7.4), less prone to ALP hydrolysis.87 This high value is found at pH 11 and a calcium (as CaCl2) to phosphorus (Na-polyP) weight ratio of >2. If the pH is lowered to 7 or even to 4, the zeta potential drops to ∼8 mV.
On the basis of the given experimental data, the most likely route along which polyP is synthesized from the mitochondrial ATP is summarized in Figure 9. ATP is synthesized in the mitochondria, tightly linked to the respiratory chain, and released from the matrix to the cytosol via ANT2 and VDAC. There, ATP generates via the Vtc complex in the acidocalcisomal membrane, polyP.213 Within the acidocalcisome polyP is deposited as amorphous polyP material (reviewed in refs (235 and 292)). Packed in those organelles within the cells, polyP is released, for example, from the blood platelets via exocytosis (see ref (293)).
The released polyP occurs in two forms as shorter polymers [60–100 Pi residues] that promote coagulation, and the longer polymers [>100 Pi] that are packed into microparticles, and initiate coagulation.180,294 The size of the particles is around 500 nm (Z-average) or ∼200 nm (SEM). The polyP molecules exist in these particles in an amorphous state,292 surely caused by the high stoichiometric surplus of Ca2+ over polyP.256 Associated with the outer cell membrane, the microparticles are assembled with other components of the blood clotting in polar caps, after platelet activation.295
10. PolyP Nano(micro)particles: Physiological Formation and Biomimetic Synthesis
It is conceivable to fabricate polyP for any kind of biomedical application in the same way, with a similar size and chain length, like the cells are preparing it. The chain length of polyP in metazoan cells is physiologically between 20 and 200 Pi units.96,97 The highest amounts of polyP are found in the blood platelets (thrombocytes),121 mast cells,181 and in mineralizing bone cells of the vertebrate skeleton.262 There, the polymer exists in the form of amorphous particles together with the divalent cations. The platelets are nucleus-free cell fragments, with a life-span of about 7–10 days, that originate from megakaryocytes.296,297 They are released within the bone marrow or during lung passage.298 During their maturation to the platelets, the megakaryocytes underwent an interlaced series of remodeling events; thousands of platelets are formed from a single megakaryocyte.296,299 Subsequently, the platelets circulate in the blood and primarily function as regulators of hemostasis (initiated by endothelial activation [attachment-adhesion-aggregation/signaling]) and thrombosis (formation of a blood clot with a central core [fibrin rich] and a loosely packed shell).300 After enzymatic hydrolysis of polyP with the ALP, the produced Pi residues contribute to the maintenance of the phosphate homeostasis301 and, bioenergetically, the polyP, released from platelets, also serves as a supply of metabolic energy28,29 (Figure 10). In the megakaryocytes, dense granules are formed which are embedded into acidocalcisomes,302 filled with polyP of a chain length of 70–75 Pi units and other molecules comprising energy-rich bonds.120,121 These organelles are subsequently directed to the platelets during their maturation process. Mast cells are filled with polyP entrapped into acidocalcisomes as well.303 These cells have a longer half-life with ∼40 d.304
Figure 10.
Formation of polyP nanoparticles (polyP-NP) in blood platelets. The platelets are released into the bloodstream by precursor cells, the megakaryocytes, residing within or originating from the bone marrow. The megakaryocytes undergo a series of remodeling events, resulting in the release/budding of thousands of platelets from a single megakaryocyte. Within the megakaryocytes, and especially in the platelets acidocalcisomes, granules (AC-g) accumulate that are rich in polyP complexed with Ca2+ or Mg2+. Prominent (patho)physiological roles of the platelets are their interactions with the coagulation system, the leukocytes, and the activated endothelium; their role in hemostasis and thrombosis; supply of ortho-phosphate, as a result of the ALP-catalyzed hydrolysis of Ca-polyP-NP; and, as suggested here, the generation of metabolically synthesized ATP via the PP-PTS system, either before or after the release of polyP, most likely in the form of Ca-polyP particles.
10.1. Synthesis within the Cells
As determined, and basically as expected, a major portion of polyP in the platelets is packed to nano/microparticles (size of around 200 nm) that are amorphous and exposed membrane-associated on their cell surface.180,305 The platelet polyP is present as medium-sized chains (∼100 Pi units) in the dense granules of these cells. The anionic polyP readily binds to cations, consisting of only single atoms, like Ca2+,256 or to longer chain polycations, as shown for synthetic polyethylene glycol linked to charged tertiary amine groups.306 As outlined above (section 8), the acidocalcisomes contain ∼150 mM polyP and 2 M Ca2+/10 M Mg2+. In turn, a large stoichiometric surplus of the cation over the polymeric anion (in terms of Pi units) exists in the acidocalcisomes (Figure 8), and the overall pH value in acidocalcisomes adjusts to ∼5.277 In contrast, the polyP synthesis at the Vtc complex proceeds at a pH of 7 or even higher, as might be present at the cytosolic side of the organelles280 (Figure 8). The pH environment in the acidocalcisomes shifts rapidly after alkaline stress, resulting in an increased mobilization of polyP.307 Consequently, our group prepared the Ca2+-polyP salt from CaCl2 and sodium-polyP (Na-polyP) at an excess of Ca2+ at pH 10, the third equivalence point of phosphoric acid.10,256
10.2. Biomimetics
Applying these reaction conditions, nano/microparticles are formed from Ca2+ and Na-polyP (Ca-polyP-MP), having a size of 100–300 nm256 (Figure 11). Importantly, these particles, obtained at a stoichiometric ratio between Ca2+ and polyP (based on Pi) of ≥ 2, are amorphous. Only particles in the amorphous phase are prone to a considerable dissolution process, compared to those in the crystalline state.308 During preparation of the particles, the pH was kept constant at 10 in the pH-Stat. After termination of the reaction, the particles were washed extensively with water and ethanol; they remain stable in the dry state. If suspended in nonbuffered water, the pH in the environment dropped down to a pH of ∼5 without dissolution of the particles.
Figure 11.
Biomimetically fabricated amorphous Ca-polyP microparticles (Ca-polyP-pa); SEM; (A and B) Particles formed after a short incubation period of 0.5 h and a longer reaction period (C and D) of 1 h and (E and F) of 4 h. Adapted with permission from ref (30). Copyright 2018 The Royal Society of Chemistry.
The size of the biomimetically fabricated particles can be controlled, perhaps like in the acidocalcisomes, by two parameters, first, by the duration of the addition of the CaCl2 solution to the Na-polyP solution and, second, by the chain length of the Na-polyP, present at the beginning of the reaction. In contrast to longer chain Ca-polyP-MP, Ca-polyP samples, prepared with CaCl2 from polymers with a chain length of ≤3, form crystalline Ca-polyP particles (Ca-polyP-pa).30 SEM (scanning electron microscopy) inspections of the amorphous polyP particles revealed that the size of the globular to irregular-ellipsoid particles, formed during a short reaction period of 30 min (Ca-polyP-pa: 0.5 h), is large and measures ∼1.2 μm (Figure 11A,B). These particles have a porous interior space transversed by channels of ∼40 nm (Figure 11B). An extension of the reaction time during the preparation of the particles to 1 h (Ca-polyP-pa: 1 h) results in much smaller globular particles with ∼60 nm (Figure 11C,D). A further prolongation to 4 h (Ca-polyP-pa: 4 h) causes a further reduction of the size of the particles to ∼25 nm (Figure 11E,F). Decreasing the chain length of the polyP reactant likewise causes a reduction of the size of the particles.30
Since in physiological acidocalcisomes the size of the polyP-containing electron-dense particles is around 50–300 nm, they are amorphous and have a chain length of ∼70 Pi units,180,305 we have adjusted the size of the particles for all our studies between 100 and 300 nm and used as a reactant polyP with a chain length of ∼40 Pi units.256 The amorphous state of the particles has been verified by XRD (X-ray powder diffraction).256
The FTIR recordings show the characteristic signals for polyP in the Ca-polyP-pa,30 such as the asymmetric stretching vibration of (PO2)− at ∼1259 cm–1, the asymmetric vibration of (PO3)2– at ∼1103 cm–1, the asymmetric vibration of (P–O–P) at ∼865 cm–1, as well as the symmetric vibration of (P–O–P) at 755 cm–1 (Figure 12). It is apparent that the intensity of the (PO2)− decreases with the duration of the reaction, a sign that the packaging of the particles increases with time. EDX analyses revealed that the Ca/P atomic ratio within the Ca-polyP microparticles is 0.62, reflecting that every ionic phosphate group is bound to the divalent Ca2+ in a substoichiometric way (based on phosphate).309
Figure 12.
FTIR spectral analysis of Ca-polyP microparticles, prepared during a short (0.5 h) incubation period and after the longer (1 and 4 h) fabrication time. Characteristic signals are highlighted, like the asymmetric stretching vibration of (PO2)− at ∼1259 cm–1, the asymmetric vibration of (PO3)2– at ∼1103 cm–1, as well as the two signals for P–O–P at ∼865 and 755 cm–1.
11. Applications of Biomimetic polyP Particles
Amorphous polyP particles have been fabricated with the cations Mg2+, Ca2+, Sr2+, Zn2+, and Fe2+/Fe3+. The Mg2+-polyP complex was prepared from Na-polyP and MgCl2 and found to form heterodisperse particles with an average size of 250 nm.310 The Na-polyP reactant could be partially substituted by the natural polyanionic polymer hyaluronic acid resulting in a product that comprises similar biomechanical properties like cartilage. Interestingly, this composite material was able to dissolve small calcium phosphate bone splinters, present in synovial fluid, and is prone to ALP hydrolytic degradation.156
11.1. Morphogenetic Activity
The Mg2+-polyP particles were found to elicit morphogenetic activity since they enhance growth of chondrocytes and induce gene expression in these cells, such as the expression of ALP, aggrecan, and several collagen genes. In addition, Ca2+-polyP particles have been prepared from CaCl2 and Na-polyP resulting in the fabrication of 100–300 nm particles that can be degraded by ALP.256 Again, these particles were able to induce a series of genes involved in bone formation (see ref (30)). Animal studies with normal and diabetic mice revealed that the Ca2+-polyP particles like the Mg2+-polyP particles strongly accelerate wound healing even in diabetic animals.311 Strontium has been proven to enhance bone formation and improve bone microarchitecture in osteoporosis.312 Considering this fact Sr2+-polyP particles have been prepared from SrCl2 and Na-polyP having a size of around 340 nm. Besides their in vitro morphogenetic activity, these particles strongly induced the regeneration process of bone defects in vivo.313 Zinc is an essential ion that is crucial for maintaining cell membrane integrity and cell intermediary metabolism. Zinc deficiency causes delayed wound healing and immune dysfunction.314 Therefore, Zn2+-polyP particles have been prepared from Na-polyP and ZnSO4 (unpublished). The ∼250 nm particles were found to accelerate migration of human endothelial cells in the in vitro wound healing model.
The polyanion polyP offers another option toward a biomedical application in host–guest chemistry. It has been shown that polyP can be partially substituted by other anions like zoledronic acid,315 hyaluronic acid,156 or adenine arabinoside monophosphate (araAMP) (unpublished). With the use of this technology, complexes could be fabricated that might be applicable for drug delivery in which the components are held together by noncovalent bonds without losing their biological activities. Advantageous is the fact that the host polyP nano/microparticles also display their beneficial functional properties, e.g., eliciting morphogenetic activity, in addition to the biomedical effect of the guest as inhibitor of tumor cell proliferation,315 stimulus of cartilage repair,156 or inhibitor of herpesvirus infection.316
11.2. Coacervation
Recently, it could be demonstrated that polyP particles do not undergo dissolution in water or culture medium unless they come into contact with peptides or proteins.87 During this process, the amorphous Ca2+-polyP particles, characterized by a high zeta potential, disintegrate and form a coacervate phase. This coacervate formed from the Ca2+-polyP particles is the functionally active form of the biomaterial and characterized by the property to be biodegradable; in vitro it becomes disintegrated with time.
12. Intracellular Energy: From polyP to ATP
It is the rule that all metabolically active tissues produce heat with the adipose tissue as the most prominent example.317 Most of the ATP is synthesized in mitochondria, tightly coupled to the electron transport chain that moves electrons from an electron donor to the terminal electron acceptor (O2) via a series of redox reactions. The degree of utilization of the energy of the formed proton gradient, the proton motive force, that is converted into biochemically utilizable energy in ATP during the reaction from ADP is not hundred percent; a portion of the energy generated during oxidation is lost as heat.318,319
12.1. Intracellular Generation of Energy As Heat
There are only a few tissues in which the energy produced during certain exergonic chemical processes is almost completely released as heat; it is primarily the brown adipose tissue that acts as a heat generator, while the white tissue functions as an energy storage. This uniqueness of the brown adipose tissue is due to the presence of the brown fat-specific uncoupling protein, thermogenin, that allows the dissipation of the electrochemical/chemical energy of the proton gradient formed at the inner mitochondrial membrane as heat and, by this, the uncoupling of the oxidative phosphorylation/ATP production.320 A second, enzymatic pathway that has been identified to expend energy proceeds through futile creatine cycling by a coordinated upregulation of genes controlling creatine metabolism in beige adipocytes.321,322 Thermogenin or the creatine cycling enzymes residing at the mitochondrial membrane(s) promote thermogenesis by dissipating the proton-motive force and/or increasing the rate of substrate flux through the mitochondrial respiratory chain.
12.2. Metabolic Energy: ATP
Most commonly ATP is used as a primary energy carrier that is storing and transferring the energy of the mitochondrial transmembrane electrochemical potential (ECP) into the energy of a chemical bond, a process that is mediated by the F1/FO-like ATP synthases. These enzymes contain two motor sectors: (i) the FO couples proton translocation to rotation, and F1 couples rotation to ATP synthesis.323 The rotation, the cyclic conformational changes in the catalytic sites within the F1 unit, is driving ATP synthesis and release. During this process, the Gibbs free energy of the ECP is first converted into the mechanical energy of the rotation of the centrally located γ-subunit and then into the chemical energy-rich phosphoanhydride bond. With the uncoupling protein UCP-1 (thermogenin), the electrochemical potential is completely and irreversibly converted to heat, possibly resulting in an entropy change,324 while with the ATP cycling via the ATP/ADP carrier between the F1/FO-like ATP synthases and the creatine cycling the energy is only partially dissipated and the rest is restored into ATP/creatine phosphate. Basically this partial reusage of chemical energy from ATP to the formation of phosphoanhydride and phosphate ester linkages describes the energy shuttle mediated by the large group of phosphotransferases [EC 2.7] enzymes.
Usually the phosphotransferases, like the hexokinase, use 1 mol of ATP per 1 mol of substrate (glucose) to synthesize 1 mol of phosphorylated substrate (glucose-6-phosphate) and 1 mol of ADP. In this reaction, a sum of ΔG0 of −30.5 kJ mol–1 is released during hydrolysis of ATP to ADP and Pi. From this sum of free energy (ΔG0), a portion of 13.7 kJ mol–1 is reused and stored in the phosphorylated product.325 The remaining energy ΔG0 ≈ – 17 kJ mol–1 is dissipated. This means that during the phosphotransferase reaction a major portion of the chemical energy originally present in the phosphoanhydride bond of ATP is not stored as metabolic energy. As outlined later, the experimental data available strongly suggest that, both in the extracellular space and intracellularly, polyP acts as a phosphate donor for the generation of ADP or ATP.148,149 In mammalian organisms, the reaction during which ΔG0 is released is primarily catalyzed by the enzyme ALP. Assuming that the phosphorylation of AMP to ADP or ADP to ATP is also catalyzed by a phosphotransferase, it is likely that also during this reaction a portion of the energy stored in the (terminal) phosphoanhydride bond of polyP undergoes dissipation, most likely in the form of heat. Consequently, more than one phosphoanhydride bond in the polyP would be needed to be invested for the formation of an energy-rich bond in the nucleotide. Moreover, such a reaction might require an intermediate prior to the formation of the energy-rich phosphoanhydride bond. A strong candidate for such an intermediate has been recently identified in studies on myosin. The myosin light chain kinase mediates the reversible phosphorylation of calmodulin.326 This enzyme has been found to hydrolyze ATP via formation of a stable intermediate with the γ-phosphate of ATP in the dissociated metaphosphate (PγO3–) state.327,328 Hence, we propose that in a close interaction of the two subunits of the membrane-bound ALP homodimer, the formed metaphosphate intermediates can react either with AMP under formation of ADP which is subsequently converted into ATP by adenylate kinase (ADK) reaction or with water under release of orthophosphate (see section 13).
13. PolyP, ALP, and ADK: An Extracellular ATP Generator Complex
The ALPs are ubiquitous membrane-bound glycoproteins.329 In vertebrates, the enzyme is attached, as an ecto-enzyme, to the plasma membrane by a GPI anchor.330 The formation of the functionally active ALP dimer at the cell membrane has been reported to depend on the redox state of the cell (NAD+/NADH ratio).331,332 Previously it has been shown that the ALP degrades polyP through a processive mechanism,117,262 i.e., without dissociation of the inorganic polymer from the enzyme after each catalytic cycle until degradation is completed (see section 7). The ALP has been considered as catalytically promiscuous;333,334 the enzyme has broad substrate specificity and accepts both polyP and AMP as substrate.117,335,336
13.1. Cell Biological Evidence for a Role of ALP and ADK in ATP Generation
Cell biological studies revealed that inhibitors of ALP and ADK severely block migration behavior and metabolic activity of some cells, especially of fibroblasts and epi/endothelial cells.10,149,337 An illustrative example is the migration process of endothelial cells during the initial stage of vascularization (Figure 13). During this migration process, the irregularly scattered cells onto the culture substrate (Figure 13A) join to 100 μm large ring-shaped cell circles during a surprisingly short incubation period in vitro, of about 4 to 8 h (Figure 13B,C). In the presence of Ca-polyP-MP, the organized tube-like arrangement comprising cell migration and pattern formation is strongly enhanced (Figure 13D,E). This increase can be significantly prevented by adding levamisole (LEV), an established inhibitor of ALP338 (Figure 13F,G). In addition, the accelerated process of tube-like formation can be severely suppressed by the addition of the natural ADK inhibitor P,1P5-di(adenosine-5′)pentaphosphate [Ap5A]339 (Figure 13H,I). On the basis of previously gathered data demonstrating that the levels of ADP and ATP increase following addition of polyP to cells in vitro, a more direct study was conducted to examine whether ATP is involved in this enhanced migration and accelerated tube-like pattern formation. The polyP-supplemented cell culture assays were coincubated with apyrase, an enzyme that hydrolyses nucleoside triphosphates to their corresponding di- and mononucleotides under Pi release.336 During this coincubation study, polyP with apyrase, the tube-like organization pattern of the cells was almost completely blocked (Figure 13J,K). These cell biological data were completed and corroborated by analytical data measuring the ATP level in the culture medium in the absence or presence of polyP (Figure 14A). Upon addition of polyP to the culture system, an over 3-fold increase in the ATP pool is measured. Addition of both the ALP inhibitor (levamisole) and the ADK inhibitor (Ap5A) strongly reduced the polyP-mediated extracellular ATP level. Interestingly, the phosphate trimeric Ca-polyP3-MP particles did not cause a significant change in the ATP level measured in controls (without polyP). A quantitative assessment of the effect of polyP on tube formation in the absence or presence of polyP is shown in Figure 14B. The strong increase in this pattern formation in assays with polyP is significantly reduced after coincubation with levamisole and Ap5A. The removal of ATP from the system with apyrase blocked the polyP stimulatory effect almost completely.
Figure 13.
Migration and pattern formation (tube-like formation assay) of endothelial cells in vitro during an incubation period of 4 and 8 h, respectively. The cells were stained with calcein and visualized by fluorescence microscopy. (A–C) Control assay in the absence of polyP and inhibitor. (D and E) Organization of the cells in the presence of 50 μg/mL of Ca-polyP-MP. In addition, coincubation experiments with 1 mM LEV, an ALP inhibitor (F and G), or with 40 μM of the ADK inhibitor Ap5A (H and I) were performed. Finally, the cultures were incubated with Ca-polyP-MP and 10 U/ml apyrase (J and K). Reprinted with permission from ref (10). Copyright 2018 Portland Press Limited on behalf of the Biochemical Society.
Figure 14.
(A) Quantitative assessment of the extracellular ATP level in endothelial cell culture in vitro, in the absence of polyP (control) or after addition of 50 μg/mL of Ca2+-containing microparticles, prepared from Na-tripolyphosphate (Ca–P3-MP) or from Na-polyP (40 phosphate units; Ca-polyP-MP) and CaCl2. The level of ATP in the assays with Ca-polyP-MP was set to 100% [corresponding to 600 pmol/106 cells]. After coincubation with either levamisole (LEV) or Ap5A, inhibitors of ALP or ADK, the ATP level drops significantly. (B) Reduction of the polyP-caused tubelike pattern formation after coaddition of levamisole (LEV) or Ap5A to values close to that measured in the control (without polyP). A complete removal of ATP from the culture system with the enzyme apyrase almost completely stopped the tubelike pattern formation. Adapted with permission from ref (10). Copyright 2018 Portland Press Ltd..
The results of the tube-like formation assays were confirmed in assays measuring the migration capability of the endothelial cells (Figure 15). For this series of experiments, the in vitro scratch assay was applied.340 Endothelial cells were seeded onto an ECM substrate. After incubation for 12 h, a “scratch” was performed in the cell monolayer, and the migration rate of the cells into the scratch area was monitored. Microscopic analysis revealed that after an incubation period of 12 h, cells in both the controls (without polyP) and in the polyP-treated cultures migrated into the initially removed cell strips. However, the density of the in-migrated cells within the previously cell-free strips was significantly higher in the polyP-containing assays (Figure 15C,D) as compared to the controls (Figure 15A,B).
Figure 15.
Analysis of endothelial cell migration onto ECM extract substratum by the in vitro scratch assay. The assays in A and B were free of polyP, while in C and D, 50 μg/mL of Ca-polyP-MP were added. The dotted lines mark the areas lacking cells at the beginning of the experiments; into these zones, the cells migrated in. The images were taken at 0 h (A,C) and 12 h (B,D) after scratch formation.
These experimental data strongly suggest that after enzymatic hydrolysis of polyP by ALP and the consecutive ADK reaction, an increase in ATP level in the extracellular space of the cultures occurs. The following theoretical consideration supports this empirical observation.
13.2. Model of Concerted ALP-ADK Action
As summarized in Figure 16, the ALP-mediated phosphoryl transfer reaction from polyP to AMP could, in principle, proceed through one of three different mechanisms: first, a SN2-type mechanism via a pentavalent transition state with simultaneous bond cleavage and new bond formation; second, a dissociative (elimination-addition) mechanism, characterized by the formation of a metaphosphate intermediate after cleavage of the leaving Pi group; and third, an associative (addition–elimination) mechanism during which a pentavalent phosphorane intermediate is formed by nucleophilic attack, followed by the release of the leaving group from polyP.341 In the first (concerted) mechanism, no intermediate is formed, while the dissociative and associative mechanisms are two-step processes that do involve an intermediate. Both mechanisms (dissociative and associative) are stepwise SN1-type processes, in contrast to the concerted mechanism, in which bond-breakage and new bond formation occur simultaneously. These reactions can also be described using a More O’Ferrall–Jencks diagram.342 In view of the results reported for the myosin light chain kinase (see section 12), as well as the results reported for model systems,342 we assume that the ALP reaction proceeds through a dissociative mechanism, involving the formation of a metaphosphate intermediate.
Figure 16.
Three possible mechanisms (concerted, dissociative, and associative) for the phosphoryl transfer reaction catalyzed by ALP. Comparing with model systems, the dissociative mechanism (formation of a metaphosphate intermediate; reaction scheme in the middle) is most likely. The concerted pathway (no intermediate, only a transition state is passed; upper reaction scheme) and the associative pathway with the formation of a phosphorane intermediate (lower reaction scheme) are less likely. The energy vs reaction coordinate plots given as inserts indicate the progress of the reaction (concerted pathway: one transition state; and dissociative or associate pathway: two transition states).
13.2.1. Mg2+ Ions
Mg2+ has a decisive role in the allosteric activation of the ALP; it enhances the reaction rate and binds to the ALP dimer with negative cooperativity.343,344 It is assumed that the particular ALP subunit, containing Mg2+, has the higher affinity for the substrate than the one without a bound Mg2+; consequently, both subunits exhibit an unequal affinity for the substrate/product.345 Here, we present a model that might allow an explanation of the phosphoryl-transfer from polyP to AMP, catalyzed by ALP. This model is based on, first, the findings that the functionally active ALP exists as an asymmetric homodimer,344,346 consisting of two ALP subunits that likely undergo alternating conformational changes, and changes in affinity to the substrate molecule, during the catalytic cycle,345 and, second, the assumption that the enzyme acts through a dissociative mechanism, involving the formation of a metaphosphate intermediate. In this model, depicted in Figure 17, we propose that the Mg2+ containing high affinity ALP subunit binds polyP. The product of the catalytic reaction, ADP, formed by phosphoryl-transfer from polyP to AMP, remains bound to this subunit and is released as soon as a conformational change occurs by binding of Mg2+ to the second low affinity subunit. The thereby activated second subunit (now present in the Mg2+ containing high affinity form) then binds the partially degraded (by one Pi) polyP product and performs the next cleavage reaction, and so forth. During this stepwise phosphoryl-transfer reactions, the degradation of the polyP proceeds.
Figure 17.
Proposed interaction of the ALP subunits during polyP degradation and ADP formation by membrane-bound ALP dimer. (Left to right) The ALP dimer consisting of a high-affinity subunit (red; containing a bound Mg2+) and a low-affinity subunit (pink; without Mg2+) is attached to the cell membrane via the GPI anchor of each subunit. After binding of the substrate, polyPn (n = number of Pi residues) to the high-affinity subunit and cleavage of the terminal Pi of the polyP chain, the resulting polyPn–1 is transferred to the low-affinity subunit, facilitated by the close proximity of the catalytic sites of both subunits. The cleaved terminal Pi, as a metaphosphate (mP) intermediate, noncovalently bound at the catalytic site of the high-affinity subunit becomes attacked by the second, AMP substrate. The ADP product formed by nucleophilic attack of AMP at the phosphorus of the metaphosphate intermediate remains tightly bound to the high-affinity subunit until a conformational change into the low-affinity form occurs, most likely induced by binding of Mg2+ to the low-affinity ALP subunit. Thereby the ADP is released, most likely as a Mg2+-ADP complex. The polyPn–1, bound to the second, now activated (by binding of Mg2+) high-affinity unit, then performs the next catalytic cycle (cleavage of polyPn–1 into polyPn–2 and enzyme-bound metaphosphate intermediate; release and transfer of polyPn–2 to the first subunit; formation of ADP by nucleophilic attack of AMP; and release of ADP after conformational change induced by Mg2+ binding to the other, first subunit).
13.2.2. Catalytic Reaction of ALP
In our model, the catalytic reaction involves the cleavage of the O–P bond at the phosphorus located at the terminal position of polyPn (n = number of Pi units) by one ALP molecule of the dimeric enzyme under formation of the metaphosphate intermediate (Figure 17). This metaphosphate intermediate is bound to the bimetallo Zn2+-center of active site of the ALP.347 Only after the release of the leaving group (polyP shortened by one Pi unit), the nucleophilic attack of the 2-fold negatively charged AMP at the enzyme-bound metaphosphate intermediate occurs (negatively charged but partially neutralized by interaction with positively charged functional groups in the active site) under formation and release of ADP from the enzyme. Simultaneously, the shortened polyP translocates to the second ALP molecule of the ALP dimer, where it is cleaved again under formation of an enzyme bound metaphosphate intermediate that is then transferred to a second AMP molecule, resulting in the formation and release of a further ADP. These steps are repeated until the polyP molecule is completely degraded. Consequently, the degradation of polyP follows an apparently processive mechanism that involves (i) an interchange of the ALP subunit conformations and (ii) the transfer of the progressively shortened polyP molecule from one subunit to the other and vice versa during each catalytic cycle. Thereby, the action of the subunits is not synchronous but time-lagged to each other. Of course, it must be taken into account that water which is present in excess amounts might compete with AMP in the ALP-catalyzed phosphoryl-transfer reaction. However, primarily the water hydroxide ions (OH–) act as a powerful nucleophile, and their concentration at neutral pH (with 10–7 M) is comparably low, in the same range or even lower than the concentration of AMP. Nevertheless, a possible coupling of both reactions, the strongly exergonic hydrolysis of the phosphoanhydride bond and the phosphoryl-transfer reaction, cannot be excluded, e.g., via alternating hydrolysis and transphosphorylation steps by the two ALP subunits.
13.2.3. Coupled ALP-ADK Reaction
As experimentally shown, exposure of cells in vitro, like SaOS-2 osteogenic sarcoma cells, to polyP elicits in those cells a 2- to 3-fold increase of the ADP/ATP level both in the intra- and the extracellular (medium) space.148,149 If the system is treated with levamisole (inhibitor of the ALP) and with Ap5A (inhibitor of the ADK) this increased nucleotide level was almost completely abolished.10 This finding suggested that the two enzymes in the cellular system are involved in the polyP-mediated generation of ADP/ATP.
Since ADP produced in the ALP reaction can subsequently be used as a substrate for the ADK in a cooperative reaction, the energy stored in polyP can be transformed into metabolically useful energy in the form of ATP.149 The ADK which is exposed, like the ALP, to the extracellular space at the outer cell membrane149 catalyzes the reversible interconversion of all three adenine nucleotides. The ATP generated in this transphosphorylation reaction, by the concerted action of the two enzymes, ALP and ADK (both enzymes are cell-membrane-bound), is formed extracellularly (Figure 18). Among the different isoforms of ADK known, the AK1β which is secreted into the extracellular space348,349 is the most likely isoform involved in this combined ALP-ADK reaction.
Figure 18.
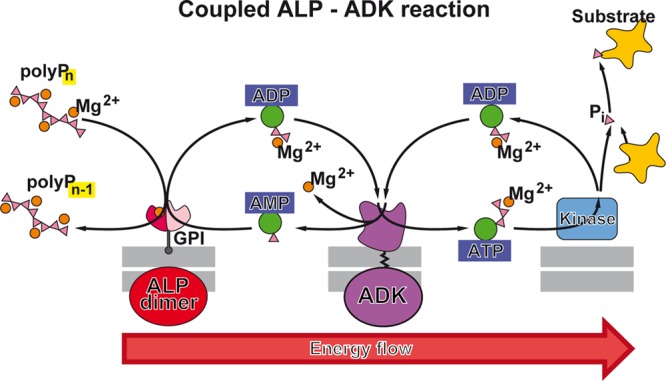
Energy channelling from membrane-bound ALP dimer via membrane-bound ADK to energy-consuming processes during ATP formation from polyP. The Mg2+-ADP complex released from the ALP dimer during each catalytic cycle, together with a second Mg2+-ADP, is converted by ADK into AMP and Mg2+-ATP which can be utilized for a variety of energy-consuming reactions in the extracellular space, e.g., as a Pi-donor for kinases. The concerted action of both enzymes (ALP and ADK) also allows the produced AMP to enter the next catalytic cycle of ALP via nucleophilic attack at the polyP metaphosphate intermediate bound at the catalytic site of the enzyme. It should be noted that only ADP and ATP but not AMP are complexed with a divalent cation (one Mg2+ ion each). Consequently, one Mg2+ is released during the ADK reaction, which could bind to the low-affinity ALP subunit to induce the conformational change of the high-affinity subunit to release the bound ADP product (see Figure 17).
The application of only the ADK inhibitor Ap5A (without levamisole) enabled a dissection of both enzyme activities of the ALP/ADK system. Cells exposed to polyP without this inhibitor showed an upregulation of the extracellular ATP with a high ATP/ADP ratio, while cells coincubated with polyP and Ap5A showed a relative increase in extracellular ADP (shift of the ATP/ADP ratio in favor of ADP).149 From these results, it can be concluded that the energy-rich phosphate liberated during the ALP-mediated cleavage of polyP is first transferred to AMP under formation of ADP which is subsequently interconverted to AMP and ATP through the ADK reaction. The final product, ATP, can then be used, among others, as a substrate in kinase reactions (Figure 18). Alternatively, the AMP molecule together with the ATP can undergo a series of subsequent catalytic cycles catalyzed by neighboring ADK molecules to channel the energy to more distant places along the membrane.
In contrast to the ALP, the phosphoryl transfer reaction catalyzed by the latter enzyme, the ADK, occurs via the concerted pathway, in which a negatively charged oxygen on AMP attacks, as a nucleophile, the terminal γ-Pi of ATP; bond formation to the nucleophile and bond cleavage to the leaving group proceed simultaneously. Only a single transition state with the pentavalently coordinated γ-Pi is formed that dissociates under the release of two ADP molecules.350
In the ADK reaction only ADP and ATP but not AMP is complexed with one Mg2+ ion each. Therefore, during the ADK equilibrium reaction (2 Mg2+-ADP ↔ AMP + Mg2+-ATP + Mg2+), one additional Mg2+ ion will be needed to obtain two Mg2+-ADP molecules from AMP and Mg2+-ATP (Figure 18). However, during polyP degradation/formation of Mg2+-ADP, only 0.5 Mg2+ ion is released per one phosphate unit of the polyP molecule, because, in the Mg2+-polyP, (maximally) one Mg2+ ion is bound to two Pi units of the polyP chain (maximum Mg:P ratio: 0.5). Consequently, the system will become depleted of free Mg2+ ions in the course of the reaction if not substituted by Mg2+ from other sources. It might be possible that the Mg2+ balance is restored by alternating hydrolysis- and phosphoryl-transfer reactions that could occur during degradation of Mg2+-polyP.
14. PolyP Transport and Energy Channeling in the Extracellular Space
As described in section 2, the extracellular space consists both of a fluid phase and a fibrillar ECM that provides structural support for the tissue.351 It is obvious that the structure and composition of the ECM can influence transport, stability, and degradation of polyP or polyP particles released into the extracellular space. The (charged) structural fibrous proteins of the ECM, collagen and, in particular, proteoglycans and glycosaminoglycans bind a large volume of water but can also interact electrostatically with polyanionic polyP or polyP particles. This could either facilitate or impair the transport and distribution of polyP in the extracellular space.
14.1. Potential Transport through the ECM
The potential transport ways of polyP through the ECM are not yet known. Principally, the extracellular conveyor system for polyP could be based on either a free polyP/polyP-nano/microparticle transport mechanism or a spreading of the polymer encapsulated into extracellular vesicles.351 Focusing on the first possibility, a transport of polyP could occur either by (i) free diffusion (following a concentration gradient), (ii) hindered diffusion (due to transient binding/immobilization of the molecules to ECM components), or (iii) facilitated diffusion (interaction with ECM components that facilitate the diffusion). Like the cell interior,352,353 the extracellular medium has a comparably high viscosity and its physical properties do not correspond to those of an ideal solution. This might even affect the diffusion of small molecules. In addition, the diffusion especially of larger molecules, of macromolecules/polymers, like polyP and polyP particles, might be hindered by steric restrictions or binding to ECM components. Generally speaking, the movement of particles through the ECM is triggered by Brownian random motion, a process that is influenced by steric, electrostatic, or hydrodynamic interactions.351 It has been shown that the transportability of the matrix is negatively correlated with the concentration of their fibrous elements, in particular glycosaminoglycans.354 Furthermore, the Brownian motion of particles is significantly impaired by electrostatic interactions between charged particles and charged components of the ECM.355
According to a model proposed by Stylianopoulos et al.,355 the effect of repulsive electrostatic interactions on the diffusion of nanoparticles in the ECM will only become significant at fiber diameters equivalent to the Debye length (a measure of a electrostatic effect of charged particles in a solution). Hence, electrostatic interactions should be relevant for glycosaminoglycans (negatively charged) with a size of a few nanometers but less relevant for collagen fibers (positively charged) with a size of up to a few micrometers. Moreover, neutral nanoparticles should diffuse faster than charged particles. Therefore, it can be expected that the diffusion of polyP-particles through the ECM can be influenced and modulated by changing the zeta potential of the particles,356 as described.87
14.2. Cell-Based Transport
It is reasonable to assume that the prevalent way of transport of polyP in the extracellular space likely occurs in form of platelet-packaged particles.357 Recently, it has been discovered that polyP is also present in exosomes. Exosomes are extracellular vesicles with a size of 30–100 nm that have attracted increasing interest as mediators of cell-to-cell communication.358 These vesicles are continuously released from cells into the extracellular fluid via exocytosis359 and can contain polyP, besides proteins, nucleic acids, and signaling molecules, as identified in so-called prostasomes that are secreted from prostate cancer cells and expose long chain polyP on their surface.360 The hydrodynamic size of exosomes can be markedly higher compared to the size of the membrane vesicles due to the formation of a proteinaceous “corona” which causes a retardation of their mobility.361 Exceptionally, polyP released from cells (D. discoideum) has also been reported to induce cell aggregation and to inhibit cytokinesis in an autocrine negative feedback loop.188,362
In addition to the transport of polyP in the extracellular space, the energy flow through the ECM in form of high-energy phosphate, released from polyP, is essential for the proper functioning of an extracellular energy supply system. As described in section 13, the ADKs play a decisive role in the phosphotransfer from polyP to adenine nucleotides. These enzymes are thought to form a phosphotransfer network both intra- and extracellularly that regulates a variety of energy-dependent processes and signaling events which are essential for cell growth and function, as well as differentiation.363
The mechanisms and advantages of the intra-cellular energy channeling has been intensively discussed.364 Because the diffusion of ATP and other energy carrying nucleoside triphosphates (NTP) through the cytosol is limited, it has been proposed that cellular energy metabolism involves multiple “microcompartments” where the production sites of the nucleotides are in close proximity to the sites of NTP consumption.364 This model enables a direct transfer of energy from the NTP-generating system to the NTP-consuming system.
On the basis of these considerations, we assume that polyP, together with the membrane-bound ALP and ADK, and an ATP consuming reaction can form such an energy transfer or energy channeling system in the extra-cellular space (Figure 18). Due to its processive mode of action, the ALP is able to produce large amounts of energy on the spot, and, in concert with the ADK, large amounts of ATP. The metabolic energy, in the form of ATP, could then be used by energy-consuming reactions, most likely occurring at or close to the cellular membrane. The advantage of this system: the close coupling of the two reactions at a specific site allows for a fast adaptation of ATP generation (via the ALP/ADK system using high-energy Pi from polyP) to the energy/ATP demand of those processes that utilize ATP in particular at rapidly changing or high turnover rates. Intracellularly, such enzymatic phosphotransfer networks can even couple spatially separated ATP-producing and ATP-consuming processes, as reviewed in Dzeja and Terzic.363
It should be mentioned that the site of polyP degradation/consumption within the interstitium/extracellular fluid or the cytosol is spatially separated from the site of polyP synthesis/storage within the platelet acidocalcisomes. This seems to be reasonable because in this way the formation of a futile cycle is avoided, i.e., a metabolic cycle that dissipates energy by hydrolysis of polyP without any meaningful anabolic reaction.365 The driving force of such a cycle would be the strongly exergonic hydrolytic cleavage of the polyP phosphoanhydride bonds. However, it might be possible that, e.g., in case of low availability of proteins/molecules acting as phosphoreceptors, polyP degradation can switch from ALP/ADK-mediated ATP generation to ALP-mediated hydrolysis under release of large amounts of energy in the form of heat. Under these conditions, polyP might play some role in extracellular thermogenesis, in particular in bradytrophic tissue with low cell numbers and vascularization.
15. Conclusion
It might be surprising that the importance of polyP, a purely inorganic biopolymer, as one of the oldest energy-delivering molecules in living systems has not been recognized for a long time. All the more so since there are no other molecules in living beings that can store as much energy in the form of energy-rich bonds as polyP. Compared to ATP, which is composed of both an inorganic and an organic part, polyP has a uniform, comparably simple structure. The discovery of the enzymes, enzyme systems, and mechanisms that enable one to convert the energy stored in the energy-rich phosphoanhydride bonds of that molecule into metabolic energy in the form of ATP has enhanced the understanding of the basic biochemical processes involved in energy storage and release. In particular, these discoveries make it possible to elucidate an aspect of energy metabolism that has hitherto not been considered or has received only little attention: energy supply and storage in the extracellular space. This extracellular space occupies a much larger volume in animals and humans than the intracellular space. For example, many tissues, such as cartilage and bone, consist of a huge extracellular matrix in which only a few cells are embedded. And this matrix does not contain the mitochondria which serve as energy factories of the cells.
The ECM, however, is rich in enzymes that metabolize extracellular nucleotides and nucleosides, most of them are involved in the regulation of purinergic signaling.366 Such a function implies that the concentration of the nucleotides is low and that they act only within a delimited region within the ECM, close to the surface of those cells that expose the respective nucleotide-sensitive receptors. Indeed, the concentration of ATP in the extracellular space only amounts to 10–9 to 10–7 M; only in the pericellular space, ATP levels from 10–6 to 10–5 M are reached. Therefore, in dependence on the adenylate energy charge (AEC)367 in the extracellular space, the levels of AMP can exceed those of the water OH– ions, making this nucleotide (AMP2–) an efficient nucleophile competing with water in the ALP-mediated phosphoryl-transfer reaction. The main factors that determine AEC in the intra-cellular space are (i) the F1FO ATP synthase reaction as the main cellular process that synthesizes ATP from ADP, (ii) the ADK reaction which catalyzes the interconversion of the three adenine nucleotides (ATP, ADP, and AMP), (iii) the enzymatic cleavage of the β,γ-phosphoanhydride bond in ATP releasing ADP like in phosphotransferase (kinase) or hydrolase (ATPase) reactions, and (iv) the enzymatic cleavage of α,β-phosphoanhydride bond in ATP yielding AMP like in many anabolic synthetase processes.368 In the extra-cellular compartment (interstitium or ECM), the phosphoryl-transfer from polyP, catalyzed by the cell membrane-bound ALP/ADK system, has to be added, which can be assumed to play a dominant role in determining AEC in the extracellular space as described in this review. In addition, a series of further enzymes are known to be involved in ecto-enzymatic interconversion reactions that control the level of the purinergic agonists like ATP or ADP, such as the nucleoside triphosphate diphosphohydrolases, the nucleotide pyrophosphatase/phosphodiesterases, and the ecto-5′-nucleotidase.366 Besides these degrading enzymes, the ATP regenerating nucleoside diphosphate kinases might be mentioned.367 Some of those enzymes are highly specific for nucleotides, while others degrade a variety of substrate molecules, like the ALP which hydrolyzes both ATP and the phosphoanhydride bonds in polyP.
In the model, proposed in this review, the ALP directly transfers the energy-rich phosphates from polyP, through a metaphosphate intermediate, to AMP which is subsequently converted to ATP via an ADK reaction. This reaction seems to be possible due to the broad substrate specificity of the ALP which accepts both polyP and AMP as a substrate. The ATP then transfers the phosphate to the final phosphate acceptor molecule. In this case, a highly exergonic reaction (hydrolysis of polyP) is linked with an endergonic reaction (formation of the phosphorylated substrate) via a shared intermediate product. Such a reaction scheme in which the product of the first (exergonic) reaction is used as a substrate for a second (endergonic) reaction is a typical example for a coupled reaction used by many reactions in biological systems.369
On the basis of our data, the ALP, like the PPX, which both release Pi processively from the termini of the substrate, acts as exopolyphosphatase. Evidence has been presented that this enzyme can also act as a phosphotransferase. If it is considered that the metabolic energy, which is released by cleavage of the P–O–P bond in the polyP molecule, is reused for ADP/ATP formation, the term polyP:AMP(ADP) phosphotransferase system (PP-PTS) is justified.
It will be an interesting task to elucidate the mechanism(s) of recognition of the diverse, possibly supramolecular structures of the complexes/particles formed by polyP and different counterions by the various proteins (enzymes and polyP-binding proteins) that interact with the polymer. Recently, in addition to the polyP-metabolizing enzymes described in this article, several polyP-binding proteins characterized by a CHAD (conserved histidine α-helical domain) motif have been identified, which are associated with the surface of the bacterial volutin granules.370 Later this group of proteins carrying the conserved CHAD domain has also been identified in archaea and eukaryotes (in the plant Ricinus communis).371 Structural analysis revealed that the CHAD domains are formed by helical bundles and a central pore surrounded by highly conserved basic amino acids. PolyP molecules but not nucleic acids are bound with high affinity (dissociation constants in the nanomicromolar range).371 The investigation of the potential function of these structurally interesting proteins in regulation of polyP metabolism, especially in higher eukaryotes, deserves further attention.
The elucidation of the function of polyP in extracellular energy metabolism is also of medical importance, especially for diseases of tissues that are composed of only a few cells and a large extracellular matrix and have poor vascularization. Thus, the discovery of the energy-supplying function of polyP has led to the development of first new approaches in regenerative medicine based on polyP, especially for the treatment of bone and cartilage diseases, but also for wound healing disorders.30,311 Even the application of polyP as a major component of a bioink, suitable for 3D printing of implant materials in the absence or presence of cells included, has been successfully shown.372
Nevertheless, many questions remain unanswered, for example, about the regulatory mechanisms that control the formation and degradation of polyP, its transport over greater distances via the bloodstream, the formation of the polyP-loaded platelets from the megakaryocytes, the release of polyP from the platelets, or its possible chaperone function and role in regulation of autophagy. PolyP is also a component of many materials used in bone replacement. However, in these cases, crystalline polyP is mostly used which, unlike the amorphous polyP and polyP MP described in this review, is not biologically active and cannot serve as an energy source.315 It can be expected that polyP, an inorganic polymer with a crucial function in biological systems, not only under physiological but also pathological conditions, will gain increasing attention besides the universal biochemical energy carrier ATP.
Acknowledgments
We thank Dr. Ingo Lieberwirth (Max Planck Institute for Polymer Research - Mainz) for helpful discussions and advise during the electron microscopic studies. W.E.G.M. is the holder of an ERC Advanced Investigator Grant (grant no. 268476). In addition, W.E.G.M. obtained three ERC-PoC grants (Si-Bone-PoC, grant no. 324564; MorphoVES-PoC, grant no. 662486; and ArthroDUR, grant no. 767234). Finally, this work was supported by grants from the European Commission (grant nos. 604036 and 311848), the International Human Frontier Science Program, and the BiomaTiCS research initiative of the University Medical Center, Mainz.
Biographies
Werner E.G. Müller is a Professor of Physiological Chemistry at the University Medical Center of the Johannes Gutenberg University Mainz, Germany. On the basis of his research, he has been awarded with one ERC Advanced Investigator Grant and three ERC Proof of Concept Grants in the field of enzyme-based biomineralization and regenerative medicine. His work focuses on processes running at the interfaces between molecular biology, (bio)chemistry, and tissue engineering. His achievements are, in enzymology, discovery and elucidation of the mode of action of a first antiviral agent (enzyme inhibitor) against herpes simplex virus, which is in use worldwide; in tissue engineering, (i) a novel biomaterial, amorphous polyphosphate nano/microparticles, a storage, and a generator of metabolic energy in the extracellular matrix and its application in regenerative medicine (wound healing and bone/cartilage repair) and (ii) discovery that enzymes are key drivers of biomineral formation, such as silicatein as the enzyme that forms an inorganic “biosilica” skeleton material in siliceous sponges and the two enzymes involved in human bone formation (alkaline phosphatase and carbonic anhydrase). His work has been recognized by 20 scientific awards, as well as the highest social award in Germany, the Federal Cross of Merit, first Class. He has more than 1100 publications (h-index: 84 [ISI-WOS]). In addition, he has been the coordinator of large EU projects, such as the Integrated Project BlueGenics.
Heinz C. Schröder is a chemist and physician and completed his doctorate in both disciplines with distinction. In 1982–1983, he did a Liebig fellowship at the Max Planck Institute for Biophysical Chemistry in Göttingen, Germany, and in 1988, he did a fellowship at the National Cancer Center Research Institute, Tokyo, Japan. Since 1985, he has been a professor at Mainz University/University Medical Center. His main scientific works include the description of polyphosphate in bone, as well as the disclosure of enzymes involved in polyadenylate and interferon 2-5A metabolism and of the role of HIV-Tat. His present research interests include the mechanisms of formation and application of biogenic nanoparticles. He has authored or coauthored more than 500 scientific articles (h-index: 59 [ISI-WOS]) and has received several awards. Together with Prof. Dr. W.E.G. Müller, he has been involved either as a coordinator (ENVRAD, UVTOX, BIOTOXmarin, BIO-LITHO, and BIOMINTEC) or partner in several large EU projects.
Xiaohong Wang is a chemist and material scientist. In 2005, she became a professor in inorganic chemistry. She has long-standing expertise in the development and characterization of regenerative-active materials and the molecular processes underlying their biological/morphogenetic activity. Her achievements include the elucidation of the mechanism of hardening of biogenetically formed silica and of the enzymatic formation of calcium carbonate “bio-seeds” in bone mineralization. Since 2006, she has had close collaboration with the group of W.E.G. Müller and she joined his team in Mainz in 2009. Her scientific work comprises over 300 publications (h-index: 34 [ISI-WOS]). In addition, she has coordinated/participated, as a PI, in several EU-funded projects, such as CoreShell, SPECIAL, MarBioTec, or the ongoing H2020-InnovaConcrete. In addition, she is the scientific coordinator of the German-Chinese “Joint Center” on Bioinspired Materials.
The authors declare no competing financial interest.
References
- van Esch J. H.; Klajn R.; Otto S. Chemical systems out of equilibrium. Chem. Soc. Rev. 2017, 46, 5474–5475. 10.1039/C7CS90088K. [DOI] [PubMed] [Google Scholar]
- Boyer P. D.; Cross R. L.; Momsen W. A new concept for energy coupling in oxidative phosphorylation based on a molecular explanation of the oxygen exchange reactions. Proc. Natl. Acad. Sci. U. S. A. 1973, 70, 2837–2839. 10.1073/pnas.70.10.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobel Prize. https://www.nobelprize.org/prizes/chemistry/1997/boyer/lecture/ (2019).
- Munder M. C.; Midtvedt D.; Franzmann T.; Nüske E.; Otto O.; Herbig M.; Ulbricht E.; Müller P.; Taubenberger A.; Maharana S.; et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife 2016, 5, e09347 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallimann T.; Wyss M.; Brdiczka D.; Nicolay K.; Eppenberger H. M. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the ’phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 1992, 281, 21–40. 10.1042/bj2810021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson G. L. The fluid-mosaic model of membrane structure: still relevant to understanding the structure, function and dynamics of biological membranes after more than 40 years. Biochim. Biophys. Acta, Biomembr. 2014, 1838, 1451–1466. 10.1016/j.bbamem.2013.10.019. [DOI] [PubMed] [Google Scholar]
- Singer S. J. The structure and function of membranes-personal memoir. J. Membr. Biol. 1992, 129, 3–12. 10.1007/BF00232051. [DOI] [PubMed] [Google Scholar]
- Neverisky D. L.; Abbott G. W. Ion channel-transporter interactions. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 257–267. 10.3109/10409238.2016.1172553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa S.; Ueda S.; Imamura H.; Mori K.; Asanuma K.; Yanagita M.; Nakagawa T. Glycolysis, but not mitochondria, responsible for intracellular ATP distribution in cortical area of podocytes. Sci. Rep. 2016, 5, 18575. 10.1038/srep18575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Ackermann M.; Tolba E.; Neufurth M.; Ivetac I.; Kokkinopoulou M.; Schröder H. C.; Wang X. H. Role of ATP during the initiation of microvascularization. Acceleration of an autocrine sensing mechanism facilitating chemotaxis by inorganic polyphosphate. Biochem. J. 2018, 475, 3255–3273. 10.1042/BCJ20180535. [DOI] [PubMed] [Google Scholar]
- Taruno A. ATP release channels. Int. J. Mol. Sci. 2018, 19 (3), ii. 10.3390/ijms19030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marginedas-Freixa I.; Alvarez C. L.; Moras M.; Leal Denis M. F.; Hattab C.; Halle F.; Bihel F.; Mouro-Chanteloup I.; Lefevre S. D.; Le Van Kim C.; et al. Human erythrocytes release ATP by a novel pathway involving VDAC oligomerization independent of pannexin-1. Sci. Rep. 2018, 8, 11384. 10.1038/s41598-018-29885-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzoni S.; Donvito G.; Di Virgilio F. Detecting adenosine triphosphate in the pericellular space. Interface Focus 2013, 3, 20120101. 10.1098/rsfs.2012.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikirova N.; Riordan H. D.; Kirby R. K.; Klykov A.; Jackson J. A. Monitoring of ATP levels in red blood cells and T cells of healthy and Ill subjects and the effects of age on mitochondrial potential. J. Orthomolec. Med. 2004, 20, 50–58. [Google Scholar]
- Boudreau L. H.; Duchez A. C.; Cloutier N.; Soulet D.; Martin N.; Bollinger J.; Paré A.; Rousseau M.; Naika G. S.; Lévesque T.; et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood 2014, 124, 2173–2183. 10.1182/blood-2014-05-573543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T. M.; Häuselmann H. J.; Shintani N.; Hunziker E. B. Cell and matrix morphology in articular cartilage from adult human knee and ankle joints suggests depth-associated adaptations to biomechanical and anatomical roles. Osteoarthritis Cartilage 2013, 21, 1904–1912. 10.1016/j.joca.2013.09.011. [DOI] [PubMed] [Google Scholar]
- Upadhyay R. K. Role of calcium bio-minerals in regenerative medicine and tissue engineering. J. Stem Cell Res. Ther. 2017, 2, 00081. 10.15406/jsrt.2017.02.00081. [DOI] [Google Scholar]
- Johnson K. A.; Hessle L.; Vaingankar S.; Wennberg C.; Mauro S.; Narisawa S.; Goding J. W.; Sano K.; Millan J. L.; Terkeltaub R. Osteoblast tissue-nonspecific alkaline phosphatase antagonizes and regulates PC-1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 279, R1365–R1377. 10.1152/ajpregu.2000.279.4.R1365. [DOI] [PubMed] [Google Scholar]
- Kinsey S. T.; Locke B. R.; Dillaman R. M. Molecules in motion: influences of diffusion on metabolic structure and function in skeletal muscle. J. Exp. Biol. 2011, 214, 263–274. 10.1242/jeb.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley M. J.; Rosanske R. C.; Moerland T. S. Diffusion coefficients of ATP and creatine phosphate in isolated muscle: pulsed gradient 31P NMR of small biological samples. NMR Biomed. 1995, 8, 72–78. 10.1002/nbm.1940080205. [DOI] [PubMed] [Google Scholar]
- Urban J. P.; Smith S.; Fairbank J. C. Nutrition of the intervertebral disc. Spine 2004, 29, 2700–2709. 10.1097/01.brs.0000146499.97948.52. [DOI] [PubMed] [Google Scholar]
- Lee J. I.; Sato M.; Ushida K.; Mochida J. Measurement of diffusion in articular cartilage using fluorescence correlation spectroscopy. BMC Biotechnol. 2011, 11, 19. 10.1186/1472-6750-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. R.; Kornberg A. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16085–16087. 10.1073/pnas.0406909101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H. C.; Müller W. E. G. (Eds.) Inorganic Polyphosphates - Biochemistry, Biology, Biotechnology; Springer Press: Heidelberg, 1999. [Google Scholar]
- Kulaev I. S.; Vagabov V.; Kulakovskaya T.. The Biochemistry of Inorganic Polyphosphates; John Wiley: Chichester, 2004. [Google Scholar]
- Rao N. N.; Gómez-García M. R.; Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 2009, 78, 605–647. 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- Kulakovskaya T.; Pavlov E.; Dedkova E. N. (Eds.) Inorganic Polyphosphates in Eukaryotic Cells; Springer International Publishing: Basel, 2016. [Google Scholar]
- Müller W. E. G.; Tolba E.; Schröder H. C.; Wang X. H. Polyphosphate: a morphogenetically active implant material serving as metabolic fuel for bone regeneration. Macromol. Biosci. 2015, 15, 1182–1197. 10.1002/mabi.201500100. [DOI] [PubMed] [Google Scholar]
- Wang X. H.; Schröder H. C.; Müller W. E. G. Polyphosphate as a metabolic fuel in Metazoa: A foundational breakthrough invention for biomedical applications. Biotechnol. J. 2016, 11, 11–30. 10.1002/biot.201500168. [DOI] [PubMed] [Google Scholar]
- Wang X. H.; Schröder H. C.; Müller W. E. G. Amorphous polyphosphate, a smart bioinspired nano-/bio-material for bone and cartilage regeneration: Towards a new paradigm in tissue engineering. J. Mater. Chem. B 2018, 6, 2385–2412. 10.1039/C8TB00241J. [DOI] [PubMed] [Google Scholar]
- Alford A. I.; Kozloff K. M.; Hankenson K. D. Extracellular matrix networks in bone remodeling. Int. J. Biochem. Cell Biol. 2015, 65, 20–31. 10.1016/j.biocel.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Gentili C.; Cancedda R. Cartilage and bone extracellular matrix. Curr. Pharm. Des. 2009, 15, 1334–1348. 10.2174/138161209787846739. [DOI] [PubMed] [Google Scholar]
- Kwan K. M. Coming into focus: the role of extracellular matrix in vertebrate optic cup morphogenesis. Dev. Dyn. 2014, 243, 1242–1248. 10.1002/dvdy.24162. [DOI] [PubMed] [Google Scholar]
- Chen S.; Mienaltowski M. J.; Birk D. E. Regulation of corneal stroma extracellular matrix assembly. Exp. Eye Res. 2015, 133, 69–80. 10.1016/j.exer.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C.; Stewart K. M.; Weaver V. M. The extracellular matrix at a glance. J. Cell Sci. 2010, 123, 4195–4200. 10.1242/jcs.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer L. M.; Ma W.; Mosher D. F. Dynamic structure of plasma fibronectin. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 213–227. 10.1080/10409238.2016.1184224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivka P. F.; Dearth C. L.; Keane T. J.; Meng F. W.; Medberry C. J.; Riggio R. T.; Reing J. E.; Badylak S. F. Fractionation of an ECM hydrogel into structural and soluble components reveals distinctive roles in regulating macrophage behavior. Biomater. Sci. 2014, 2, 1521–1534. 10.1039/C4BM00189C. [DOI] [PubMed] [Google Scholar]
- Krieg E.; Bastings M. M.; Besenius P.; Rybtchinski B. Supramolecular polymers in aqueous media. Chem. Rev. 2016, 116, 2414–2477. 10.1021/acs.chemrev.5b00369. [DOI] [PubMed] [Google Scholar]
- Young R. D.; Knupp C.; Pinali C.; Png K. M.; Ralphs J. R.; Bushby A. J.; Starborg T.; Kadler K. E.; Quantock A. J. Three-dimensional aspects of matrix assembly by cells in the developing cornea. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 687–692. 10.1073/pnas.1313561110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starborg T.; Lu Y.; Huffman A.; Holmes D. F.; Kadler K. E. Electron microscope 3D reconstruction of branched collagen fibrils in vivo. Scand. J. Med. Sci. Sports 2009, 19, 547–552. 10.1111/j.1600-0838.2009.00907.x. [DOI] [PubMed] [Google Scholar]
- Gonzales S.; Wang C.; Levene H.; Cheung H. S.; Huang C. C. ATP promotes extracellular matrix biosynthesis of intervertebral disc cells. Cell Tissue Res. 2015, 359, 635–642. 10.1007/s00441-014-2042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber L.; Priel Z.; Aflalo C.; Shoshan-Barmatz V. Extracellular ATP binding proteins as potential receptors in mucociliary epithelium: characterization using [32P]3′-O-(4-benzoyl)benzoyl ATP, a photoaffinity label. J. Membr. Biol. 1995, 147, 83–93. 10.1007/BF00235399. [DOI] [PubMed] [Google Scholar]
- Smith S. A.; Collins J. N.; Choi S. H.; Morrissey J. H. Novel polyphosphate-binding proteins in human plasma and platelet releasates. Blood 2011, 118, 2270. 10.1182/blood.V118.21.2270.2270. [DOI] [Google Scholar]
- Burnstock G. Neural nomenclature. Nature 1971, 229, 282–283. 10.1038/229282d0. [DOI] [PubMed] [Google Scholar]
- Di Virgilio F.; Adinolfi E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 2017, 36, 293–303. 10.1038/onc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. Introduction to purinergic signalling in the brain. Adv. Exp. Med. Biol. 2013, 986, 1–12. 10.1007/978-94-007-4719-7_1. [DOI] [PubMed] [Google Scholar]
- Burnstock G. Purinergic signaling in the cardiovascular system. Circ. Res. 2017, 120, 207–228. 10.1161/CIRCRESAHA.116.309726. [DOI] [PubMed] [Google Scholar]
- Schwiebert E. M.; Fitz J. G. Purinergic signaling microenvironments: An introduction. Purinergic Signalling 2008, 4, 89–92. 10.1007/s11302-007-9091-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. Short- and long-term (trophic) purinergic signalling. Philos. Trans. R. Soc., B 2016, 371 (1700), 20150422. 10.1098/rstb.2015.0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmström K. M.; Marina N.; Baev A. Y.; Wood N. W.; Gourine A. V.; Abramov A. Y. Signalling properties of inorganic polyphosphate in the mammalian. Nat. Commun. 2013, 4, 1362. 10.1038/ncomms2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K.; Nakagawa H.; Oikawa T.; Noda M.; Ikeda S. Effects of 4(1H)-quinolinone derivative, a novel non-nucleotide allosteric purinergic P2Y2 agonist, on cardiomyocytes in neonatal rats. Sci. Rep. 2017, 7, 6050. 10.1038/s41598-017-06481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo M. The platelet P2Y12 receptor for adenosine diphosphate: congenital and drug-induced defects. Blood 2011, 117, 2102–2112. 10.1182/blood-2010-08-263111. [DOI] [PubMed] [Google Scholar]
- Milanesi L.; Petrillo M.; Sepe L.; Boccia A.; D’Agostino N.; Passamano M.; Di Nardo S.; Tasco G.; Casadio R.; Paolella G. Systematic analysis of human kinase genes: a large number of genes and alternative splicing events result in functional and structural diversity. BMC Bioinf. 2005, 6, S20. 10.1186/1471-2105-6-S4-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. Genomic overview of protein kinases. WormBook 2005, 13, 1–19. 10.1895/wormbook.1.60.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P. V.; Chabra I.; Kornhauser J. M.; Skrzypek E.; Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 2004, 4, 1551–1561. 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- Bordoli M. R.; Yum J.; Breitkopf S. B.; Thon J. N.; Italiano J. E.; Xiao J.; Worby C.; Wong S. K.; Lin G.; Edenius M.; et al. A secreted tyrosine kinase acts in the extracellular environment. Cell 2014, 158, 1033–1044. 10.1016/j.cell.2014.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H.; Vijayakumar S.; Schiene-Fischer C.; Li H.; Purkerson J. M.; Malesevic M.; Liebscher J.; Al-Awqati Q.; Schwartz G. J. Secreted cyclophilin A, a peptidylprolyl cis-trans isomerase, mediates matrix assembly of hensin, a protein implicated in epithelial differentiation. J. Biol. Chem. 2009, 284, 6465–6475. 10.1074/jbc.M808964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanghänel J.; Fischer G. Insights into the catalytic mechanism of peptidyl prolyl cis/trans isomerases. Front. Biosci., Landmark Ed. 2004, 9, 3453–3478. 10.2741/1494. [DOI] [PubMed] [Google Scholar]
- Watanabe S.; Tsuruoka S.; Vijayakumar S.; Fischer G.; Zhang Y.; Fujimura A.; Al-Awqati Q.; Schwartz G. J. Cyclosporin A produces distal renal tubular acidosis by blocking peptidyl prolyl cis-trans isomerase activity of cyclophilin. Am. J. Physiol. Renal. Physiol. 2005, 288, F40–F47. 10.1152/ajprenal.00218.2004. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu. Rev. Biophys. Biomol. Struct. 1993, 22, 123–142. 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- Le Bihan S.; Renoir J. M.; Radanyi C.; Chambraud B.; Joulin V.; Catelli M. G.; Baulieu E. E. The mammalian heat shock protein binding immunophilin (p59/HBI) is an ATP and GTP binding protein. Biochem. Biophys. Res. Commun. 1993, 195, 600–607. 10.1006/bbrc.1993.2088. [DOI] [PubMed] [Google Scholar]
- Schmid F. X. Protein folding. Prolyl isomerases join the fold. Curr. Biol. 1995, 5, 993–994. 10.1016/S0960-9822(95)00197-7. [DOI] [PubMed] [Google Scholar]
- Schmitt F. O.; Gross J.; Highberger J. H. A new particle type in certain connective tissue extracts. Proc. Natl. Acad. Sci. U. S. A. 1953, 39, 459–470. 10.1073/pnas.39.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. R.; Lewis R. J. The collagen type I segment long spacing (SLS) and fibrillar forms: Formation by ATP and sulphonated diazo dyes. Micron 2016, 86, 36–47. 10.1016/j.micron.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Calderwood S. K.; Gong J.; Murshid A. Extracellular HSPs: The complicated roles of extracellular HSPs in immunity. Front. Immunol. 2016, 7, 159. 10.3389/fimmu.2016.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–6342. 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L.; Jakob U. Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J. Biol. Chem. 2019, 294, 2180–2190. 10.1074/jbc.REV118.002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. J.; Wholey W. Y.; Wagner N. O.; Cremers C. M.; Mueller-Schickert A.; Hock N. T.; Krieger A. G.; Smith E. M.; Bender R. A.; Bardwell J. C.; et al. Polyphosphate is a primordial chaperone. Mol. Cell 2014, 53, 689–699. 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo N. G.; Dogra S.; Meinen B. A.; Tse E.; Haefliger J.; Southworth D. R.; Gray M. J.; Dahl J. U.; Jakob U. Polyphosphate stabilizes protein unfolding intermediates as soluble amyloid-like oligomers. J. Mol. Biol. 2018, 430, 4195–4208. 10.1016/j.jmb.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asea A.; Rehli M.; Kabingu E.; Boch J. A.; Bare O.; Auron P. E.; Stevenson M. A.; Calderwood S. K. Novel signal transduction pathway utilized by extracellular HSP70: role of toll-like receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. 10.1074/jbc.M200497200. [DOI] [PubMed] [Google Scholar]
- Trcka F.; Durech M.; Vankova P.; Chmelik J.; Martinkova V.; Hausner J.; Kadek A.; Marcoux J.; Klumpler T.; Vojtesek B.; et al. Human stress-inducible Hsp70 has a high propensity to form ATP-dependent antiparallel dimers that are differentially regulated by co-chaperone binding. Mol. Cell. Proteomics 2019, 18, 320–337. 10.1074/mcp.RA118.001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver J. A.; Rekas A.; Thorn D. C.; Wilson M. R. Small heat-shock proteins and clusterin: intra- and extracellular molecular chaperones with a common mechanism of action and function?. IUBMB Life 2003, 55, 661–668. 10.1080/15216540310001640498. [DOI] [PubMed] [Google Scholar]
- Poon S.; Easterbrook-Smith S. B.; Rybchyn M. S.; Carver J. A.; Wilson M. R. Clusterin is an ATP-independent chaperone with very broad substrate specificity that stabilizes stressed proteins in a folding-competent state. Biochemistry 2000, 39, 15953–15960. 10.1021/bi002189x. [DOI] [PubMed] [Google Scholar]
- Tsuruta J. K.; Wong K.; Fritz I. B.; Griswold M. D. Structural analysis of sulphated glycoprotein 2 from amino acid sequence. Relationship to clusterin and serum protein 40,40. Biochem. J. 1990, 268, 571–578. 10.1042/bj2680571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschek J.; Braun N.; Franzmann T. M.; Georgalis Y.; Haslbeck M.; Weinkauf S.; Buchner J. The eye lens chaperone α-Crystallin forms defined globular assemblies. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13272–13277. 10.1073/pnas.0902651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski P. J.; Clark J. I. ATP-enhanced molecular chaperone functions of the small heat shock protein human αB Crystallin. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 1004–1009. 10.1073/pnas.95.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam M.; Lairez D.. Sol-gel transition. In Cohen Addad J. P., Ed.; Physical properties of polymeric gels; John Wiley: Chichester, 1996; pp 87–142. [Google Scholar]
- Berry H.; Pelta J.; Lairez D.; Larreta-Garde V. Gel-sol transition can describe the proteolysis of extracellular matrix gels. Biochim. Biophys. Acta, Gen. Subj. 2000, 1524, 110–117. 10.1016/S0304-4165(00)00144-6. [DOI] [PubMed] [Google Scholar]
- Flory P. J. Molecular size distribution in three dimensional polymers. I. Gelation. J. Am. Chem. Soc. 1941, 63, 3083–3090. 10.1021/ja01856a061. [DOI] [Google Scholar]
- Stockmayer W. H. Theory of molecular size distribution and gel formation in branched polymers II. General cross linking. J. Chem. Phys. 1944, 12, 125–131. 10.1063/1.1723922. [DOI] [Google Scholar]
- Guo M.; Ehrlicher A. J.; Jensen M. H.; Renz M.; Moore J. R.; Goldman R. D.; Lippincott-Schwartz J.; Mackintosh F. C.; Weitz D. A. Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy. Cell 2014, 158, 822–832. 10.1016/j.cell.2014.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery L. D.; Gerth W. A.; Montgomery R. W.; Lew S. Q.; Klein M. M.; Stewart J. M.; Velasquez M. T. Monitoring intracellular, interstitial, and intravascular volume changes during fluid management procedures. Med. Biol. Eng. Comput. 2013, 51, 1167–1175. 10.1007/s11517-013-1064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günzel D. Claudins: vital partners in transcellular and paracellular transport coupling. Pfluegers Arch. 2017, 469, 35–44. 10.1007/s00424-016-1909-3. [DOI] [PubMed] [Google Scholar]
- Zhao J.; Krystofiak E. S.; Ballesteros A.; Cui R.; Van Itallie C. M.; Anderson J. M.; Fenollar-Ferrer C.; Kachar B. Multiple claudin-claudin cis interfaces are required for tight junction strand formation and inherent flexibility. Commun. Biol. 2018, 1, 50. 10.1038/s42003-018-0051-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T.; Kaeser P. S.; Blanpied T. A. Transcellular nanoalignment of synaptic function. Neuron 2017, 96, 680–696. 10.1016/j.neuron.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X.; Sang W.; Hu J.; Yan Q. Pulsating polymer micelles via ATP-fueled dissipative self-assembly. ACS Macro Lett. 2017, 6, 1151–1151. 10.1021/acsmacrolett.7b00649. [DOI] [PubMed] [Google Scholar]
- Müller W. E. G.; Wang S.; Tolba E.; Neufurth M.; Ackermann M.; Muñoz-Espí R.; Lieberwirth I.; Glasser G.; Schröder H. C.; Wang X. H. Transformation of amorphous polyphosphate nanoparticles into coacervate complexes: an approach for the encapsulation of mesenchymal stem cells. Small 2018, 14, 1801170 10.1002/smll.201801170. [DOI] [PubMed] [Google Scholar]
- Gupta A.; Reena; Bohidar H. B. Free-energy landscape of alcohol driven coacervation transition in aqueous gelatin solutions. J. Chem. Phys. 2006, 125, 054904. 10.1063/1.2219745. [DOI] [PubMed] [Google Scholar]
- Freeman J. W.; Silver F. H. Elastic energy storage in unmineralized and mineralized extracellular matrices (ECMs): a comparison between molecular modeling and experimental measurements. J. Theor. Biol. 2004, 229, 371–381. 10.1016/j.jtbi.2004.04.018. [DOI] [PubMed] [Google Scholar]
- Liu J.; Das D.; Yang F.; Schwartz A. G.; Genin G. M.; Thomopoulos S.; Chasiotis I. Energy dissipation in mammalian collagen fibrils: Cyclic strain-induced damping, toughening, and strengthening. Acta Biomater. 2018, 80, 217–227. 10.1016/j.actbio.2018.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle R. C.; Clemens T. L. Bone cell bioenergetics and skeletal energy homeostasis. Physiol. Rev. 2017, 97, 667–698. 10.1152/physrev.00022.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntur A. R.; Le P. T.; Farber C. R.; Rosen C. J. Bioenergetics during calvarial osteoblast differentiation reflect strain differences in bone mass. Endocrinology 2014, 155, 1589–1595. 10.1210/en.2013-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. K.; Sowa H.; Hinoi E.; Ferron M.; Ahn J. D.; Confavreux C.; Dacquin R.; Mee P. J.; McKee M. D.; Jung D. Y.; et al. Endocrine regulation of energy metabolism by the skeleton. Cell 2007, 130, 456–469. 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Ackermann M.; Wang S. F.; Neufurth M.; Muñoz-Espí R.; Feng Q. L.; Schröder H. C.; Wang X. H. Inorganic polyphosphate induces accelerated tube formation of HUVEC endothelial cells. Cell. Mol. Life Sci. 2018, 75, 21–32. 10.1007/s00018-017-2601-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A.; Rao N. N.; Ault-Riché D. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 1999, 68, 89–125. 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H.; Choi S. H.; Smith S. A. Polyphosphate: an ancient molecule that links platelets, coagulation, and inflammation. Blood 2012, 119, 5972–5979. 10.1182/blood-2012-03-306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Khong M. L.; Lui E. L. H.; Mebarek S.; Magne D.; Buchet R.; Tanner J. A. Long-chain polyphosphate in osteoblast matrix vesicles: Enrichment and inhibition of mineralization. Biochim. Biophys. Acta, Gen. Subj. 2019, 1863, 199–209. 10.1016/j.bbagen.2018.10.003. [DOI] [PubMed] [Google Scholar]
- Corbridge D. E. C.Phosphorus: An Outline of its Chemistry, Biochemistry, and Technology, 4th ed.; Elsevier: Amsterdam, 1990. [Google Scholar]
- Lager G. A.; Gibbs G. V. Effect of variations in O-P-O and P-O-P angles on P-O bond overlap populations for some selected ortho- and pyrophosphates. Am. Mineral. 1973, 58, 756–764. [Google Scholar]
- Cordero B.; Gómez V.; Platero-Prats A. E.; Revés M.; Echeverría J.; Cremades E.; Barragán F.; Alvarez S. Covalent radii revisited. Dalton Trans 2008, 2008, 2832–2838. 10.1039/b801115j. [DOI] [PubMed] [Google Scholar]
- Smith J. P.; Brown W. E.; Lehr J. R. Structure of crystalline phosphoric acid. J. Am. Chem. Soc. 1955, 77, 2728–2730. 10.1021/ja01615a013. [DOI] [Google Scholar]
- Gamoke B.; Neff D.; Simons J. Nature of PO bonds in phosphates. J. Phys. Chem. A 2009, 113, 5677–5684. 10.1021/jp810014s. [DOI] [PubMed] [Google Scholar]
- Rashchi F.; Finch J. A. Polyphosphates: A review. Their chemistry and application with particular reference to mineral processing. Miner. Eng. 2000, 13, 1019–1035. 10.1016/S0892-6875(00)00087-X. [DOI] [Google Scholar]
- Majling J.; Hanic F. Phase chemistry of condensed phosphates. Top. Phosphorus Chem. 1980, 10, 341–502. [Google Scholar]
- McCarthy W. J.; Adamowicz L.; Saint-Martin H.; Ortega-Blake I. A comparative ab initio study of the isomerizations and hydrolyses of neutral and anionic M-pyrophosphate complexes, with M = Ca, Zn. Rev. Soc. Quím. Méx. 2002, 46, 145–158. [Google Scholar]
- Reusch R. N. Transmembrane ion transport by polyphosphate/poly-(R)-3-hydroxybutyrate complexes. Biochemistry (Mosc.) 2000, 65, 280–295. [PubMed] [Google Scholar]
- Lohmann K. Darstellung der Adenylpyrophosphorsäure aus Muskulatur. Biochem. Z. 1931, 233, 460–466. [Google Scholar]
- Jenner R.; Brachet J. Recherches sur l’acide ribonucléique des levures. Enzymologia 1944, 11, 22–26. [Google Scholar]
- Schmidt G.; Hecht L.; Thannhauser S. J. The enzymatic formation and the accumulation of large amounts of a metaphosphate in bakers’ yeast under certain conditions. J. Biol. Chem. 1946, 166, 775–776. [PubMed] [Google Scholar]
- Wiame J. M. Etude d’une substance polyphosphoric, basophile et metachromatique chez les levures. Biochim. Biophys. Acta 1947, 1, 234. 10.1016/0006-3002(47)90137-6. [DOI] [Google Scholar]
- Ebel J. P.; Dirheimer G.; Stahl A.; Feller S.. Quelques aspects nouveaux de la biochimie des polyphosphates inorganiques. Composés organiques du phosphore; Coll. Nat. du CNRS; CNRS: Paris, 1966; pp 289–300. [Google Scholar]
- Liss E.; Langen P. Über ein hochmolekulares Polyphosphat der Hefe. Biochem. Z. 1960, 333, 193–201. [PubMed] [Google Scholar]
- Khoshmanesh A.; Cook P. L.; Wood B. R. Quantitative determination of polyphosphate in sediments using attenuated total reflectance-fourier transform infrared (ATR-FTIR) spectroscopy and partial least squares regression. Analyst 2012, 137, 3704–3709. 10.1039/c2an35289c. [DOI] [PubMed] [Google Scholar]
- Bru S.; Jiménez J.; Canadell D.; Ariño J.; Clotet J. Improvement of biochemical methods of polyP quantification. Microb. Cell 2017, 4, 6–15. 10.15698/mic2017.01.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Felter S.; Ebel J.-P. Séparation des acides ribonucléiques et des polyphosphates inorganiques. II. Mise au point d’une technique de séparation par adsorption différentielle sur charbon. Bull. Soc. Chim. Biol. (Paris) 1962, 44, 1175–1184. [PubMed] [Google Scholar]
- Lohmann K.; Langen P. Untersuchungen an den kondensierten Phosphaten der Hefe. Biochem. Z. 1956, 328, 1–11. [PubMed] [Google Scholar]
- Lorenz B.; Schröder H. C. Mammalian intestinal alkaline phosphatase acts as highly active exopolyphosphatase. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 2001, 1547, 254–261. 10.1016/S0167-4838(01)00193-5. [DOI] [PubMed] [Google Scholar]
- Clark J. E.; Beegen H.; Wood H. G. Isolation of intact chains of polyphosphate from ″Propionibacterium shermanii″ grown on glucose or lactate. J. Bacteriol. 1986, 168, 1212–1219. 10.1128/jb.168.3.1212-1219.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz B.; Schröder H. C. Methods for investigation of inorganic polyphosphates and polyphosphate-metabolizing enzymes. Prog. Mol. Subcell. Biol. 1999, 23, 217–239. 10.1007/978-3-642-58444-2_11. [DOI] [PubMed] [Google Scholar]
- Müller F.; Mutch N. J.; Schenk W. A.; Smith S. A.; Esterl L.; Spronk H. M.; Schmidbauer S.; Gahl W. A.; Morrissey J. H.; Renné T. Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 2009, 139, 1143–1156. 10.1016/j.cell.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F. A.; Lea C. R.; Oldfield E.; Docampo R. Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 2004, 279, 44250–44257. 10.1074/jbc.M406261200. [DOI] [PubMed] [Google Scholar]
- Smith S. A.; Choi S. H.; Davis-Harrison R.; Huyck J.; Boettcher J.; Rienstra C. M.; Morrissey J. H. Polyphosphate exerts differential effects on blood clotting, depending on polymer size. Blood 2010, 116, 4353–4359. 10.1182/blood-2010-01-266791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. Y. Study of polyphosphate metabolism in intact cells by 31-P nuclear magnetic resonance spectroscopy. Prog. Mol. Subcell. Biol. 1999, 23, 253–273. 10.1007/978-3-642-58444-2_13. [DOI] [PubMed] [Google Scholar]
- Choi S. H.; Collins J. N.; Smith S. A.; Davis-Harrison R. L.; Rienstra C. M.; Morrissey J. H. Phosphoramidate end labeling of inorganic polyphosphates: facile manipulation of polyphosphate for investigating and modulating its biological activities. Biochemistry 2010, 49, 9935–9941. 10.1021/bi1014437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolesch D. G.; Keasling J. D. Polyphosphate binding and chain length recognition of Escherichia coli exopolyphosphatase. J. Biol. Chem. 2000, 275, 33814–33819. 10.1074/jbc.M002039200. [DOI] [PubMed] [Google Scholar]
- Lorenz B.; Müller W. E. G.; Kulaev I. S.; Schröder H. C. Purification and characterization of an exopolyphosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 1994, 269, 22198–22204. [PubMed] [Google Scholar]
- Lorenz B.; Münkner J.; Oliveira M. P.; Leitão J. M.; Müller W. E. G.; Schröder H. C. A novel method for determination of inorganic polyphosphates using the dye fura-2. Anal. Biochem. 1997, 246, 176–184. 10.1006/abio.1996.9998. [DOI] [PubMed] [Google Scholar]
- Gomes F. M.; Ramos I. B.; Wendt C.; Girard-Dias W.; De Souza W.; Machado E. A.; Miranda K. New insights into the in situ microscopic visualization and quantification of inorganic polyphosphate stores by 4’,6-diamidino-2-phenylindole (DAPI)-staining. Eur. J. Histochem. 2013, 57, 34 10.4081/ejh.2013.e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullan A.; Quinn J. P.; McGrath J. W. A nonradioactive method for the assay of polyphosphate kinase activity and its application in the study of polyphosphate metabolism in Burkholderia cepacia. Anal. Biochem. 2002, 308, 294–299. 10.1016/S0003-2697(02)00249-X. [DOI] [PubMed] [Google Scholar]
- Robinson N. A.; Wood H. G. Polyphosphate kinase from Propionibacterium shermanii. Demonstration that the synthesis and utilization of polyphosphate is by a processive mechanism. J. Biol. Chem. 1986, 261, 4481–4485. [PubMed] [Google Scholar]
- Robinson N. A.; Clark J. E.; Wood H. G. Polyphosphate kinase from Propionibacterium shermanii. Demonstration that polyphosphates are primers and determination of the size of the synthesized polyphosphate. J. Biol. Chem. 1987, 262, 5216–5222. [PubMed] [Google Scholar]
- van Groenestijn J. W.; Deinema M. H.; Zehnder A. J. B. ATP production from polyphosphate in Acinetobacter strain 210A. Arch. Microbiol. 1987, 148, 14–19. 10.1007/BF00429640. [DOI] [Google Scholar]
- Matheja J.; Degens E. T.. Structural Molecular Biology of Phosphates; Gustav Fischer Verlag: Stuttgart, 1971. [Google Scholar]
- Hill T. L.; Morales M. F. On “high energy phosphate bonds” of biochemical interest. J. Am. Chem. Soc. 1951, 73, 1656–1660. 10.1021/ja01148a072. [DOI] [Google Scholar]
- George P.; Witonsky R. J.; Trachtman M.; Wu C.; Dorwart W.; Richman L.; Richman W.; Shurayh F.; Lentz B. Squiggle-H2O”. An enquiry into the importance of solvation effects in phosphate ester and anhydride reactions. Biochim. Biophys. Acta, Bioenerg. 1970, 223, 1–15. 10.1016/0005-2728(70)90126-X. [DOI] [PubMed] [Google Scholar]
- Kalckar H. M. The nature of energetic coupling in biological syntheses. Chem. Rev. 1941, 28, 71–178. 10.1021/cr60089a002. [DOI] [Google Scholar]
- Akola J.; Jones R. O. ATP hydrolysis in water – A density functional study. J. Phys. Chem. B 2003, 107, 11774–11783. 10.1021/jp035538g. [DOI] [Google Scholar]
- Alberty R. A. Effect of pH and metal ion concentration on the equilibrium hydrolysis of adenosine triphosphate to adenosine diphosphate. J. Biol. Chem. 1968, 243, 1337–1343. [PubMed] [Google Scholar]
- Alberty R. A. Change in the binding of hydrogen ions and magnesium ions in the hydrolysis of ATP. Biophys. Chem. 1998, 70, 109–119. 10.1016/S0301-4622(97)00114-2. [DOI] [PubMed] [Google Scholar]
- Alberty R. A. Thermodynamics of the hydrolysis of adenosine triphosphate as a function of temperature, pH, pMg, and ionic strength. J. Phys. Chem. B 2003, 107, 12324–12330. 10.1021/jp030576l. [DOI] [Google Scholar]
- Alberty R. A. Biochemical thermodynamics. Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol. 1994, 1207, 1–11. 10.1016/0167-4838(94)90045-0. [DOI] [PubMed] [Google Scholar]
- Romero P. J.; de Meis L. Role of water in the energy of hydrolysis of phosphoanhydride and phosphoester bonds. J. Biol. Chem. 1989, 264, 7869–7873. [PubMed] [Google Scholar]
- De Meis L. Role of water in the energy of hydrolysis of phosphate compounds—energy transduction in biological membranes. Biochim. Biophys. Acta, Bioenerg. 1989, 973, 333–349. 10.1016/S0005-2728(89)80440-2. [DOI] [PubMed] [Google Scholar]
- Ramirez F.; Marecek J. F. Coordination of magnesium with adenosine 5′-diphosphate and triphosphate. Biochim. Biophys. Acta, Bioenerg. 1980, 589, 21–29. 10.1016/0005-2728(80)90129-2. [DOI] [PubMed] [Google Scholar]
- Bergman C.; Kashiwaya Y.; Veech R. L. The effect of pH and free Mg2+ on ATP linked enzymes and the calculation of Gibbs free energy of ATP hydrolysis. J. Phys. Chem. B 2010, 114, 16137–16146. 10.1021/jp105723r. [DOI] [PubMed] [Google Scholar]
- Iotti S.; Frassineti C.; Sabatini A.; Vacca A.; Barbiroli B. Quantitative mathematical expressions for accurate in vivo assessment of cytosolic [ADP] and ΔG of ATP hydrolysis in the human brain and skeletal muscle. Biochim. Biophys. Acta, Bioenerg. 2005, 1708, 164–177. 10.1016/j.bbabio.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Leyhausen G.; Lorenz B.; Zhu H.; Geurtsen W.; Bohnensack R.; Müller W. E. G.; Schröder H. C. Inorganic polyphosphate in human osteoblast-like cells. J. Bone Miner. Res. 1998, 13, 803–812. 10.1359/jbmr.1998.13.5.803. [DOI] [PubMed] [Google Scholar]
- Müller W. E. G.; Tolba E.; Feng Q.; Schröder H. C.; Markl J. S.; Kokkinopoulou M.; Wang X. H. Amorphous Ca2+ polyphosphate nanoparticles regulate ATP level in bone-like SaOS-2 cells. J. Cell Sci. 2015, 128, 2202–2207. 10.1242/jcs.170605. [DOI] [PubMed] [Google Scholar]
- Müller W. E. G.; Wang S.; Neufurth M.; Kokkinopoulou M.; Feng Q.; Schröder H. C.; Wang X. H. Polyphosphate as a donor of high-energy phosphate for the synthesis of ADP and ATP. J. Cell Sci. 2017, 130, 2747–2756. 10.1242/jcs.204941. [DOI] [PubMed] [Google Scholar]
- Seidlmayer L. K.; Gomez-Garcia M. R.; Shiba T.; Porter G. A. Jr.; Pavlov E. V.; Bers D. M.; Dedkova E. N. Dual role of inorganic polyphosphate in cardiac myocytes: The importance of polyP chain length for energy metabolism and mPTP activation. Arch. Biochem. Biophys. 2019, 662, 177–189. 10.1016/j.abb.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumble K. D.; Kornberg A. Inorganic polyphosphate in mammalian cells and tissues. J. Biol. Chem. 1995, 270, 5818–5822. 10.1074/jbc.270.11.5818. [DOI] [PubMed] [Google Scholar]
- Gabel N. W.; Thomas V. Evidence for the occurrence and distribution of inorganic polyphosphates in vertebrate tissues. J. Neurochem. 1971, 18, 1229–1242. 10.1111/j.1471-4159.1971.tb00222.x. [DOI] [PubMed] [Google Scholar]
- Harold F. M. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol. Rev. 1966, 30, 772–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedkova E. N. Inorganic polyphosphate in cardiac myocytes: from bioenergetics to the permeability transition pore and cell survival. Biochem. Soc. Trans. 2016, 44, 25–34. 10.1042/BST20150218. [DOI] [PubMed] [Google Scholar]
- Sender R.; Fuchs S.; Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14 (8), e1002533 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. H.; Ackermann M.; Tolba E.; Neufurth M.; Wurm F.; Feng Q.; Wang S.; Schröder H. C.; Müller W. E. G. Artificial cartilage bio-matrix formed of hyaluronic acid and Mg2+-polyphosphate. Eur. Cell. Mater. 2016, 32, 271–283. 10.22203/eCM.v032a18. [DOI] [PubMed] [Google Scholar]
- Pisoni R. L.; Lindley E. R. Incorporation of [32P]orthophosphate into long chains of inorganic polyphosphate within lysosomes of human fibroblasts. J. Biol. Chem. 1992, 267, 3626–3631. [PubMed] [Google Scholar]
- Kornberg A. Inorganic polyphosphate: toward making a forgotten polymer unforgettable. J. Bacteriol. 1995, 177, 491–496. 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov E.; Aschar-Sobbi R.; Campanella M.; Turner R. J.; Gómez-García M. R.; Abramov A. Y. Inorganic polyphosphate and energy metabolism in mammalian cells. J. Biol. Chem. 2010, 285, 9420–9428. 10.1074/jbc.M109.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramov A. Y.; Fraley C.; Diao C. T.; Winkfein R.; Colicos M. A.; Duchen M. R.; French R. J.; Pavlov E. Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 18091–18096. 10.1073/pnas.0708959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A. P. Calcium, mitochondria and reperfusion injury: a pore way to die. Biochem. Soc. Trans. 2006, 34, 232–237. 10.1042/BST20060232. [DOI] [PubMed] [Google Scholar]
- Kwong J. Q.; Molkentin J. D. Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab. 2015, 21, 206–214. 10.1016/j.cmet.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solesio M. E.; Elustondo P. A.; Zakharian E.; Pavlov E. V. Inorganic polyphosphate (polyP) as an activator and structural component of the mitochondrial permeability transition pore. Biochem. Soc. Trans. 2016, 44, 7–12. 10.1042/BST20150206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov E.; Zakharian E.; Bladen C.; Diao C. T.; Grimbly C.; Reusch R. N.; French R. J. A large, voltage-dependent channel, isolated from mitochondria by water-free chloroform extraction. Biophys. J. 2005, 88, 2614–2625. 10.1529/biophysj.104.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N.; Sadoff H. L. Putative structure and functions of a poly-β-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc. Natl. Acad. Sci. U. S. A. 1988, 85, 4176–4180. 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidlmayer L. K.; Gomez-Garcia M. R.; Blatter L. A.; Pavlov E.; Dedkova E. N. Inorganic polyphosphate is a potent activator of the mitochondrial permeability transition pore in cardiac myocytes. J. Gen. Physiol. 2012, 139, 321–331. 10.1085/jgp.201210788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidlmayer L. K.; Blatter L. A.; Pavlov E.; Dedkova E. N. Inorganic polyphosphate--an unusual suspect of the mitochondrial permeability transition mystery. Channels 2012, 6, 463–467. 10.4161/chan.21939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova P. R.; Baev A. Y.; Berezhnov A. V.; Abramov A. Y. Role of inorganic polyphosphate in mammalian cells: from signal transduction and mitochondrial metabolism to cell death. Biochem. Soc. Trans. 2016, 44, 40–45. 10.1042/BST20150223. [DOI] [PubMed] [Google Scholar]
- Elustondo P. A.; Nichols M.; Negoda A.; Thirumaran A.; Zakharian E.; Robertson G. S.; Pavlov E. V. Mitochondrial permeability transition pore induction is linked to formation of the complex of ATPase C-subunit, polyhydroxybutyrate and inorganic polyphosphate. Cell Death Discov 2016, 2, 16070. 10.1038/cddiscovery.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Fraley C. D.; Faridi J.; Kornberg A.; Roth R. A. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 11249–11254. 10.1073/pnas.1534805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C. C.; Manning B. D. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat. Cell Biol. 2013, 15, 555–564. 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi K.; Sasase T. Cellular calcification induced by inorganic polyphosphate involves ATP depletion and opening of the mitochondrial permeability transition pore (mPTP). FEBS Open Bio 2019, 9, 1617–1622. 10.1002/2211-5463.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharian E.; Thyagarajan B.; French R. J.; Pavlov E.; Rohacs T. Inorganic polyphosphate modulates TRPM8 channels. PLoS One 2009, 4, e5404 10.1371/journal.pone.0005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Cavanaugh E. J. Requirement of a soluble intracellular factor for activation of transient receptor potential A1 by pungent chemicals: role of inorganic polyphosphates. J. Neurosci. 2007, 27, 6500–6509. 10.1523/JNEUROSCI.0623-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T.; Nishimura D.; Kawazoe Y.; Onodera Y.; Tsutsumi K.; Nakamura R.; Ohshiro M. Modulation of mitogenic activity of fibroblast growth factors by inorganic polyphosphate. J. Biol. Chem. 2003, 278, 26788–26792. 10.1074/jbc.M303468200. [DOI] [PubMed] [Google Scholar]
- Segawa S.; Fujiya M.; Konishi H.; Ueno N.; Kobayashi N.; Shigyo T.; Kohgo Y. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS One 2011, 6, e23278 10.1371/journal.pone.0023278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Nuñez M. D.; Moreno-Sanchez D.; Hernandez-Ruiz L.; Benítez-Rondán A.; Ramos-Amaya A.; Rodríguez-Bayona B.; Medina F.; Brieva J. A.; Ruiz F. A. Myeloma cells contain high levels of inorganic polyphosphate which is associated with nucleolar transcription. Haematologica 2012, 97, 1264–1271. 10.3324/haematol.2011.051409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru S.; Samper-Martín B.; Quandt E.; Hernández-Ortega S.; Martínez-Laínez J. M.; Garí E.; Rafel M.; Torres-Torronteras J.; Martí R.; Ribeiro M. P. C.; et al. Polyphosphate is a key factor for cell survival after DNA damage in eukaryotic cells. DNA Repair 2017, 57, 171–178. 10.1016/j.dnarep.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Racki L. R.; Tocheva E. I.; Dieterle M. G.; Sullivan M. C.; Jensen G. J.; Newman D. K. Polyphosphate granule biogenesis is temporally and functionally tied to cell cycle exit during starvation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2440–E2449. 10.1073/pnas.1615575114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoef J. J.; Barendrecht A. D.; Nickel K. F.; Dijkxhoorn K.; Kenne E.; Labberton L.; McCarty O. J.; Schiffelers R.; Heijnen H. F.; Hendrickx A. P.; et al. Polyphosphate nanoparticles on the platelet surface trigger contact system activation. Blood 2017, 129, 1707–1717. 10.1182/blood-2016-08-734988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Sanchez D.; Hernandez-Ruiz L.; Ruiz F. A.; Docampo R. Polyphosphate is a novel pro-inflammatory regulator of mast cells and is located in acidocalcisomes. J. Biol. Chem. 2012, 287, 28435–28444. 10.1074/jbc.M112.385823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landén N. X.; Li D.; Ståhle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell. Mol. Life Sci. 2016, 73, 3861–3885. 10.1007/s00018-016-2268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corciulo C.; Lendhey M.; Wilder T.; Schoen H.; Cornelissen A. S.; Chang G.; Kennedy O. D.; Cronstein B. N. Endogenous adenosine maintains cartilage homeostasis and exogenous adenosine inhibits osteoarthritis progression. Nat. Commun. 2017, 8, 15019. 10.1038/ncomms15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao N. N.; Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 1996, 178, 1394–1400. 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achbergerová L.; Nahálka J. Polyphosphate--an ancient energy source and active metabolic regulator. Microb. Cell Fact. 2011, 10, 63. 10.1186/1475-2859-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid M. H.; Rumbaugh K.; Passador L.; Davies D. G.; Hamood A. N.; Iglewski B. H.; Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 9636–9641. 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskes E.; Deprez M. A.; Wilms T.; Winderickx J. pH homeostasis in yeast; the phosphate perspective. Curr. Genet. 2018, 64, 155–161. 10.1007/s00294-017-0743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess P. M.; Watson J.; Chen W.; Gomer R. H. Extracellular polyphosphate signals through Ras and Akt to prime Dictyostelium discoideum cells for development. J. Cell Sci. 2017, 130, 2394–2404. 10.1242/jcs.203372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess P. M.; Chinea L. E.; Pilling D.; Gomer R. H. Extracellular polyphosphate promotes macrophage and fibrocyte differentiation, inhibits leukocyte proliferation, and acts as a chemotactic agent for neutrophils. J. Immunol. 2019, 203, 493–499. 10.4049/jimmunol.1801559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska E. M.; Poole A. W. Platelet secretion: From haemostasis to wound healing and beyond. Blood Rev. 2015, 29, 153–162. 10.1016/j.blre.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers R. J.; Smith S. A.; Morrissey J. H. Polyphosphate, platelets, and coagulation. Int. J. Lab. Hematol. 2015, 37, 31–35. 10.1111/ijlh.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishige K.; Noguchi T. Polyphosphate:AMP phosphotransferase and polyphosphate:ADP phosphotransferase activities of Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 2001, 281, 821–826. 10.1006/bbrc.2001.4415. [DOI] [PubMed] [Google Scholar]
- Pepin C. A.; Wood H. G. Polyphosphate glucokinase from Propionibacterium shermanii. Kinetics and demonstration that the mechanism involves both processive and nonprocessive type reactions. J. Biol. Chem. 1986, 261, 4476–4480. [PubMed] [Google Scholar]
- Kornberg A.; Kornberg S. R.; Simms E. S. Metaphosphate synthesis by an enzyme from Escherichia coli. Biochim. Biophys. Acta 1956, 20, 215–227. 10.1016/0006-3002(56)90280-3. [DOI] [PubMed] [Google Scholar]
- Brown M. R.; Kornberg A. Inorganic polyphosphate in the origin and survival of species. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 16085–16087. 10.1073/pnas.0406909101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. R.; Kornberg A. The long and short of it - polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 2008, 33, 284–290. 10.1016/j.tibs.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Ishige K.; Zhang H.; Kornberg A. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 16684–16688. 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead M. P.; Hooley P. W.; Brown M. R. Horizontal transfer of bacterial polyphosphate kinases to eukaryotes: implications for the ice age and land colonisation. BMC Res. Notes 2013, 6, 221. 10.1186/1756-0500-6-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahálka J.; Pätoprstý V. Enzymatic synthesis of sialylation substrates powered by a novel polyphosphate kinase (PPK3). Org. Biomol. Chem. 2009, 7, 1778–1780. 10.1039/b822549b. [DOI] [PubMed] [Google Scholar]
- Lindner S. N.; Knebel S.; Wesseling H.; Schoberth S. M.; Wendisch V. F. Exopolyphosphatases PPX1 and PPX2 from Corynebacterium glutamicum. Appl. Environ. Microbiol. 2009, 75, 3161–3170. 10.1128/AEM.02705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M.; Crooke E.; Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 1993, 268, 633–639. [PubMed] [Google Scholar]
- Alvarado J.; Ghosh A.; Janovitz T.; Jauregui A.; Hasson M. S.; Sanders D. A. Origin of exopolyphosphatase processivity: Fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog. Structure 2006, 14, 1263–1272. 10.1016/j.str.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Keasling J. D.; Bertsch L.; Kornberg A. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. U. S. A. 1993, 90, 7029–7033. 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepin C. A.; Wood H. G. The mechanism of utilization of polyphosphate by polyphosphate glucokinase from Propionibacterium shermanii. J. Biol. Chem. 1987, 262, 5223–5226. [PubMed] [Google Scholar]
- Bonting C. F.; Kortstee G. J.; Zehnder A. J. Properties of polyphosphate: AMP phosphotransferase of Acinetobacter strain 210A. J. Bacteriol. 1991, 173, 6484–6488. 10.1128/jb.173.20.6484-6488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishige K.; Noguchi T. Inorganic polyphosphate kinase and adenylate kinase participate in the polyphosphate:AMP phosphotransferase activity of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 14168–14171. 10.1073/pnas.011518098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick S. M.; Zehnder A. J. In vitro ATP regeneration from polyphosphate and AMP by polyphosphate:AMP phosphotransferase and adenylate kinase from Acinetobacter johnsonii 210A. Appl. Environ. Microbiol. 2000, 66, 2045–2051. 10.1128/AEM.66.5.2045-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T.; Shiba T.; Kameda A.; Ihara Y.; Munekata M.; Ishige K.; Noguchi T. The gene for an exopolyphosphatase of Pseudomonas aeruginosa. DNA Res. 1999, 6, 103–108. 10.1093/dnares/6.2.103. [DOI] [PubMed] [Google Scholar]
- Beassoni P. R.; Gallarato L. A.; Boetsch C.; Garrido M. N.; Lisa A. T. Pseudomonas aeruginosa exopolyphosphatase is also a polyphosphate: ADP phosphotransferase. Enzyme Res. 2015, 2015, 1–13. 10.1155/2015/404607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütze J.; Krasko A.; Custodio M. R.; Efremova S. M.; Müller I. M.; Müller W. E. G. Evolutionary relationship of Metazoa within the eukaryotes based on molecular data from Porifera. Proc. Royal Society London B 1999, 266, 63–73. 10.1098/rspb.1999.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S. B. The origin and evolution of model organisms. Nat. Rev. Genet. 2002, 3, 838–849. 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Li S. C.; Kane P. M. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim. Biophys. Acta, Mol. Cell Res. 2009, 1793, 650–663. 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimaitė R.; Mayer A. Enzymes of yeast polyphosphate metabolism: structure, enzymology and biological roles. Biochem. Soc. Trans. 2016, 44, 234–239. 10.1042/BST20150213. [DOI] [PubMed] [Google Scholar]
- Wurst H.; Shiba T.; Kornberg A. The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 1995, 177, 898–906. 10.1128/jb.177.4.898-906.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva N. A.; Kulakovskaya T. V.; Kulaev I. S. Purification and properties of exopolyphosphatase from the cytosol of Saccharomyces cerevisiae not encoded by the PPX1 gene. Biochemistry (Moscow) 2004, 69, 387–393. 10.1023/B:BIRY.0000026193.44046.29. [DOI] [PubMed] [Google Scholar]
- Trilisenko L. V.; Andreeva N. A.; Eldarov M. A.; Dumina M. V.; Kulakovskaya T. V. Polyphosphates and polyphosphatase activity in the yeast Saccharomyces cerevisiae during overexpression of the DDP1 gene. Biochemistry (Moscow) 2015, 80, 1312–1317. 10.1134/S0006297915100120. [DOI] [PubMed] [Google Scholar]
- Cartwright J. L.; McLennan A. G. The Saccharomyces cerevisiae YOR163w gene encodes a diadenosine 5′,5″’-P1,P6-hexaphosphate (Ap6A) hydrolase member of the MutT motif (Nudix hydrolase) family. J. Biol. Chem. 1999, 274, 8604–8610. 10.1074/jbc.274.13.8604. [DOI] [PubMed] [Google Scholar]
- Lonetti A.; Szijgyarto Z.; Bosch D.; Loss O.; Azevedo C.; Saiardi A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 2011, 286, 31966–31974. 10.1074/jbc.M111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman A.; Rao N. N.; Kornberg A. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 8542–8547. 10.1073/pnas.151269398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumble K. D.; Kornberg A. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J. Biol. Chem. 1996, 271, 27146–27151. 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- Andreeva N.; Trilisenko L.; Kulakovskaya T.; Dumina M.; Eldarov M. Purification and properties of recombinant exopolyphosphatase PPN1 and effects of its overexpression on polyphosphate in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2015, 119, 52–56. 10.1016/j.jbiosc.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Andreeva N.; Trilisenko L.; Eldarov M.; Kulakovskaya T. Polyphosphatase PPN1 of Saccharomyces cerevisiae: switching of exopolyphosphatase and endopolyphosphatase activities. PLoS One 2015, 10 (3), e0119594 10.1371/journal.pone.0119594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimaitė R.; Mayer A. Ppn2, a novel Zn2+-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J. Cell Sci. 2017, 130, 1625–1636. 10.1242/jcs.201061. [DOI] [PubMed] [Google Scholar]
- Andreeva N.; Ledova L.; Ryazanova L.; Tomashevsky A.; Kulakovskaya T.; Eldarov M. Ppn2 endopolyphosphatase overexpressed in Saccharomyces cerevisiae: Comparison with Ppn1, Ppx1, and Ddp1 polyphosphatases. Biochimie 2019, 163, 101–107. 10.1016/j.biochi.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Hothorn M.; Neumann H.; Lenherr E. D.; Wehner M.; Rybin V.; Hassa P. O.; Uttenweiler A.; Reinhardt M.; Schmidt A.; Seiler J.; et al. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 2009, 324, 513–516. 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- Tani C.; Ohtomo R.; Osaki M.; Kuga Y.; Ezawa T. ATP-dependent but proton gradient-independent polyphosphate-synthesizing activity in extraradical hyphae of an arbuscular mycorrhizal fungus. Appl. Environ. Microbiol. 2009, 75, 7044–7050. 10.1128/AEM.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-García M. R.; Kornberg A. Formation of an actin-like filament concurrent with the enzymatic synthesis of inorganic polyphosphate. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15876–15880. 10.1073/pnas.0406923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaev I. S.; Szymona O. V.; Bobyk M. A. The biosynthesis of inorganic polyphosphates in Neurospora crassa. Biokhimiya 1968, 33, 419–434. [PubMed] [Google Scholar]
- Shi X.; Kornberg A. Endopolyphosphatase in Saccharomyces cerevisiae undergoes post-translational activations to produce short-chain polyphosphates. FEBS Lett. 2005, 579, 2014–2018. 10.1016/j.febslet.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wurst H.; Kornberg A. A soluble exopolyphosphatase of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 1994, 269, 10996–11001. [PubMed] [Google Scholar]
- Tammenkoski M.; Moiseev V. M.; Lahti M.; Ugochukwu E.; Brondijk T. H.; White S. A.; Lahti R.; Baykov A. A. Kinetic and mutational analyses of the major cytosolic exopolyphosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 9302–9311. 10.1074/jbc.M609423200. [DOI] [PubMed] [Google Scholar]
- Tammenkoski M.; Koivula K.; Cusanelli E.; Zollo M.; Steegborn C.; Baykov A. A.; Lahti R. Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 2008, 47, 9707–9713. 10.1021/bi8010847. [DOI] [PubMed] [Google Scholar]
- Ugochukwu E.; Lovering A. L.; Mather O. C.; Young T. W.; White S. A. The crystal structure of the cytosolic exopolyphosphatase from Saccharomyces cerevisiae reveals the basis for substrate specificity. J. Mol. Biol. 2007, 371, 1007–1021. 10.1016/j.jmb.2007.05.066. [DOI] [PubMed] [Google Scholar]
- Wei Z. Z.; Vatcher G.; Tin A. H.; Teng J. L.; Wang J.; Cui Q. H.; Chen J. G.; Yu A. C. Positively-charged semi-tunnel is a structural and surface characteristic of polyphosphate-binding proteins: an in-silico study. PLoS One 2015, 10, e0123713 10.1371/journal.pone.0123713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo R.; de Souza W.; Miranda K.; Rohloff P.; Moreno S. N. Acidocalcisomes - conserved from bacteria to man. Nat. Rev. Microbiol. 2005, 3, 251–261. 10.1038/nrmicro1097. [DOI] [PubMed] [Google Scholar]
- Fang J.; Ruiz F. A.; Docampo M.; Luo S.; Rodrigues J. C.; Motta L. S.; Rohloff P.; Docampo R. Overexpression of a Zn2+-sensitive soluble exopolyphosphatase from Trypanosoma cruzi depletes polyphosphate and affects osmoregulation. J. Biol. Chem. 2007, 282, 32501–32510. 10.1074/jbc.M704841200. [DOI] [PubMed] [Google Scholar]
- Luginbuehl E.; Kunz S.; Wentzinger L.; Freimoser F.; Seebeck T. The exopolyphosphatase TbrPPX1 of Trypanosoma brucei. BMC Microbiol. 2011, 11, 4. 10.1186/1471-2180-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C. O.; Ruiz F. A.; Vieira M.; Hill J. E.; Docampo R. An acidocalcisomal exopolyphosphatase from Leishmania major with high affinity for short chain polyphosphate. J. Biol. Chem. 2002, 277, 50899–50906. 10.1074/jbc.M208940200. [DOI] [PubMed] [Google Scholar]
- Cordeiro C. D.; Ahmed M. A.; Windle B.; Docampo R. NUDIX hydrolases with inorganic polyphosphate exo- and endopolyphosphatase activities in the glycosome, cytosol and nucleus of Trypanosoma brucei. Biosci. Rep. 2019, 39 (5), 1. 10.1042/BSR20190894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matange N. Deorphanizing NUDIX hydrolases from Trypanosoma: tantalizing links with metabolic regulation and stress tolerance. Biosci. Rep. 2019, 39 (6), 1. 10.1042/BSR20191369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskaya T. V.; Lichko L. P.; Vagabov V. M.; Kulaev I. S. Inorganic polyphosphates in mitochondria. Biochemistry (Moscow) 2010, 75, 825–831. 10.1134/S0006297910070035. [DOI] [PubMed] [Google Scholar]
- Joyal J. L.; Aprille J. R. The ATP-Mg/Pi carrier of rat liver mitochondria catalyzes a divalent electroneutral exchange. J. Biol. Chem. 1992, 267, 19198–19203. [PubMed] [Google Scholar]
- Abramov A. Y.; Fraley C.; Diao C. T.; Winkfein R.; Colicos M. A.; Duchen M. R.; French R. J.; Pavlov E. Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 18091–18096. 10.1073/pnas.0708959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch R. N.; Huang R.; Kosk-Kosicka D. Novel components and enzymatic activities of the human erythrocyte plasma membrane calcium pump. FEBS Lett. 1997, 412, 592–596. 10.1016/S0014-5793(97)00863-6. [DOI] [PubMed] [Google Scholar]
- Chiesi M.; Zurini M.; Carafoli E. ATP synthesis catalyzed by the purified erythrocyte Ca-ATPase in the absence of calcium gradients. Biochemistry 1984, 23, 2595–2600. 10.1021/bi00307a009. [DOI] [PubMed] [Google Scholar]
- Kosk-Kosicka D.; Scaillet S.; Inesi G. The partial reactions in the catalytic cycle of the calcium-dependent adenosine triphosphatase purified from erythrocyte membranes. J. Biol. Chem. 1986, 261, 3333–3338. [PubMed] [Google Scholar]
- Brini M.; Carafoli E. The plasma membrane Ca2+ ATPase and the plasma membrane sodium calcium exchanger cooperate in the regulation of cell calcium. Cold Spring Harbor Perspect. Biol. 2011, 3 (2), a004168. 10.1101/cshperspect.a004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.; Vieira M.; Graves J.; Zhong L.; Moreno S. N. A plasma membrane-type Ca2+-ATPase co-localizes with a vacuolar H+-pyrophosphatase to acidocalcisomes of Toxoplasma gondii. EMBO J. 2001, 20, 55–64. 10.1093/emboj/20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi Y.; Yoshioka A.; Kawai S.; Murata K. Conferring the ability to utilize inorganic polyphosphate on ATP-specific NAD kinase. Sci. Rep. 2013, 3, 2632. 10.1038/srep02632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R. MNADK, a long-awaited human mitochondrion-localized NAD kinase. J. Cell. Physiol. 2015, 230, 1697–1701. 10.1002/jcp.24926. [DOI] [PubMed] [Google Scholar]
- Outten C. E.; Culotta V. C. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J. 2003, 22, 2015–2024. 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K.; Kawai S.; Murata K. Identification and characterization of a human mitochondrial NAD kinase. Nat. Commun. 2012, 3, 1248. 10.1038/ncomms2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoylaerts M. F.; Manes T.; Millán J. L. Mammalian alkaline phosphatases are allosteric enzymes. J. Biol. Chem. 1997, 272, 22781–22787. 10.1074/jbc.272.36.22781. [DOI] [PubMed] [Google Scholar]
- Millan J. L. Alkaline phosphatases - Structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signalling 2006, 2, 335–341. 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K.; Itoh H.; Kawazoe Y.; Miyazaki S.; Doi K.; Kubo T.; Akagawa Y.; Shiba T. Polyphosphate-mediated inhibition of tartrate-resistant acid phosphatase and suppression of bone resorption of osteoclasts. PLoS One 2013, 8 (11), e78612 10.1371/journal.pone.0078612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Tolba E.; Schröder H. C.; Wang S.; Glaßer G.; Muñoz-Espí R.; Link T.; Wang X. H. A new polyphosphate calcium material with morphogenetic activity. Mater. Lett. 2015, 148, 163–166. 10.1016/j.matlet.2015.02.070. [DOI] [Google Scholar]
- Lorenz B.; Münkner J.; Oliveira M. P.; Kuusksalu A.; Leitão J. M.; Müller W. E. G.; Schröder H. C. Changes in metabolism of inorganic polyphosphate in rat tissues and human cells during development and apoptosis. Biochim. Biophys. Acta, Gen. Subj. 1997, 1335, 51–60. 10.1016/S0304-4165(96)00121-3. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H.; Smith S. A. Polyphosphate as modulator of hemostasis, thrombosis, and inflammation. J. Thromb. Haemost. 2015, 13, S92–S97. 10.1111/jth.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo R. The origin and evolution of the acidocalcisome and its interactions with other organelles. Mol. Biochem. Parasitol. 2016, 209, 3–9. 10.1016/j.molbiopara.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan S.; Asady B.; Docampo R. Acidocalcisome-mitochondrion membrane contact sites in Trypanosoma brucei. Pathogens 2018, 7, 1. 10.3390/pathogens7020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baev A. Y.; Holmstrom K. M.; Levitskaya Y. V.; Abramov A. Y. Role of polyphosphate in mitochondria: From modification of energy metabolism to induction of the cell death. Biophys. J. 2013, 104 (2), 660a. 10.1016/j.bpj.2012.11.3641. [DOI] [Google Scholar]
- Omelon S.; Georgiou J.; Henneman Z. J.; Wise L. M.; Sukhu B.; Hunt T.; Wynnyckyj C.; Holmyard D.; Bielecki R.; Grynpas M. D. Control of vertebrate skeletal mineralization by polyphosphates. PLoS One 2009, 4 (5), e5634 10.1371/journal.pone.0005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D.; Hirata N.; Miyashita R.; Kawaguchi R.; Yamakage M. Substrate-dependent modulation of oxidative phosphorylation in isolated mitochondria following in vitro hypoxia and reoxygenation injury. Exp. Clin. Cardiol. 2013, 18, 158–160. [PMC free article] [PubMed] [Google Scholar]
- Campos E.; Façanha A. R.; Costa E. P.; Fraga A.; Moraes J.; da Silva Vaz I. Jr.; Masuda A.; Logullo C. A mitochondrial membrane exopolyphosphatase is modulated by, and plays a role in, the energy metabolism of hard tick Rhipicephalus (Boophilus) microplus embryos. Int. J. Mol. Sci. 2011, 12, 3525–3535. 10.3390/ijms12063525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo R.; Moreno S. N. Acidocalcisomes. Cell Calcium 2011, 50, 113–119. 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander N.; Cordeiro C.; Huang G.; Docampo R. Polyphosphate and acidocalcisomes. Biochem. Soc. Trans. 2016, 44, 1–6. 10.1042/BST20150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I.; Gomes F.; Koeller C. M.; Saito K.; Heise N.; Masuda H.; Docampo R.; de Souza W.; Machado E. A.; Miranda K. Acidocalcisomes as calcium- and polyphosphate-storage compartments during embryogenesis of the insect Rhodnius prolixus Stahl. PLoS One 2011, 6 (11), e27276 10.1371/journal.pone.0027276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues C. O.; Ruiz F. A.; Rohloff P.; Scott D. A.; Moreno S. N. Characterization of isolated acidocalcisomes from Toxoplasma gondii tachyzoites reveals a novel pool of hydrolyzable polyphosphate. J. Biol. Chem. 2002, 277, 48650–48656. 10.1074/jbc.M208990200. [DOI] [PubMed] [Google Scholar]
- Huang G.; Ulrich P. N.; Storey M.; Johnson D.; Tischer J.; Tovar J. A.; Moreno S. N.; Orlando R.; Docampo R. Proteomic analysis of the acidocalcisome, an organelle conserved from bacteria to human cells. PLoS Pathog. 2014, 10, e1004555 10.1371/journal.ppat.1004555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmsen H.; Weiss H. J. Secretable storage pools in platelets. Annu. Rev. Med. 1979, 30, 119–134. 10.1146/annurev.me.30.020179.001003. [DOI] [PubMed] [Google Scholar]
- Dean G. E.; Fishkes H.; Nelson P. J.; Rudnick G. The hydrogen ion-pumping adenosine triphosphatase of platelet dense granule membrane. Differences from F1F0- and phosphoenzyme-type ATPases. J. Biol. Chem. 1984, 259, 9569–9574. [PubMed] [Google Scholar]
- Mifsud J.; Ravaud S.; Krammer E. M.; Chipot C.; Kunji E. R.; Pebay-Peyroula E.; Dehez F. The substrate specificity of the human ADP/ATP carrier AAC1. Mol. Membr. Biol. 2013, 30, 160–168. 10.3109/09687688.2012.745175. [DOI] [PubMed] [Google Scholar]
- Günther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+. Magnes. Res. 2006, 19, 225–236. [PubMed] [Google Scholar]
- Yamanaka R.; Tabata S.; Shindo Y.; Hotta K.; Suzuki K.; Soga T.; Oka K. Mitochondrial Mg2+ homeostasis decides cellular energy metabolism and vulnerability to stress. Sci. Rep. 2016, 6, 30027. 10.1038/srep30027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott D. A.; Docampo R.; Dvorak J. A.; Shi S.; Leapman R. D. In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J. Biol. Chem. 1997, 272, 28020–28029. 10.1074/jbc.272.44.28020. [DOI] [PubMed] [Google Scholar]
- Livermore T. M.; Chubb J. R.; Saiardi A. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 996–1001. 10.1073/pnas.1519440113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch-Tewfik J. L.; Flaumenhaft R. Platelet granule exocytosis: a comparison with chromaffin cells. Front. Endocrinol. (Lausanne) 2013, 4, 77. 10.3389/fendo.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller O.; Neumann H.; Bayer M. J.; Mayer A. Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 2003, 116, 1107–1115. 10.1242/jcs.00328. [DOI] [PubMed] [Google Scholar]
- Gerasimaitė R.; Sharma S.; Desfougères Y.; Schmidt A.; Mayer A. Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J. Cell Sci. 2014, 127, 5093–5104. 10.1242/jcs.159772. [DOI] [PubMed] [Google Scholar]
- Desfougères Y.; Gerasimaitė R. U.; Jessen H. J.; Mayer A. Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 2016, 291, 22262–22275. 10.1074/jbc.M116.746784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K.; de Souza W.; Plattner H.; Hentschel J.; Kawazoe U.; Fang J.; Moreno S. N. Acidocalcisomes in Apicomplexan parasites. Exp. Parasitol. 2008, 118, 2–9. 10.1016/j.exppara.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Müller O.; Bayer M. J.; Peters C.; Andersen J. S.; Mann M.; Mayer A. The Vtc proteins in vacuole fusion: coupling NSF activity to V0trans-complex formation. EMBO J. 2002, 21, 259–269. 10.1093/emboj/21.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttenweiler A.; Schwarz H.; Neumann H.; Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell 2007, 18, 166–175. 10.1091/mbc.e06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfougères Y.; Neumann H.; Mayer A. Organelle size control - increasing vacuole content activates SNAREs to augment organelle volume through homotypic fusion. J. Cell Sci. 2016, 129, 2817–2828. 10.1242/jcs.184382. [DOI] [PubMed] [Google Scholar]
- Shoshan-Barmatz V.; Ben-Hail D.; Admoni L.; Krelin Y.; Tripathi S. S. The mitochondrial voltage-dependent anion channel 1 in tumor cells. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 2547–2575. 10.1016/j.bbamem.2014.10.040. [DOI] [PubMed] [Google Scholar]
- Rostovtseva T.; Colombini M. VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys. J. 1997, 72, 1954–1962. 10.1016/S0006-3495(97)78841-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon K. L.; Gurnev P. A.; Bezrukov S. M.; Sackett D. L. Tubulin tail sequences and post-translational modifications regulate closure of mitochondrial voltage-dependent anion channel (VDAC). J. Biol. Chem. 2015, 290, 26784–26789. 10.1074/jbc.M115.678854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder H. C.; Wehland J.; Weber K. Purification of brain tubulin-tyrosine ligase by biochemical and immunological methods. J. Cell Biol. 1985, 100, 276–281. 10.1083/jcb.100.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. The VDAC channel: Molecular basis for selectivity. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863, 2498–2502. 10.1016/j.bbamcr.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Baev A. Y.; Negoda A.; Abramov A. Y. Modulation of mitochondrial ion transport by inorganic polyphosphate - essential role in mitochondrial permeability transition pore. J. Bioenerg. Biomembr. 2017, 49, 49–55. 10.1007/s10863-016-9650-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott R. P.; Straka R. P.; Garibaldi J. A. Polyphosphate inhibition of growth of Pseudomonads from poultry meat. Appl. Microbiol. 1964, 12, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelon S.; Habraken W.. Influence of condensed phosphates on the physical chemistry of calcium phosphate solids. In Kulakovskaya T.; Pavlov E.; Dedkova E. N., Eds.; Inorganic Polyphosphates in Eukaryotic Cells; Springer Press: Cham, 2016; pp 177–205. [Google Scholar]
- Angelova P. R.; Iversen K. Z.; Teschemacher A. G.; Kasparov S.; Gourine A. V.; Abramov A. Y. Signal transduction in astrocytes: Localization and release of inorganic polyphosphate. Glia 2018, 66, 2126–2136. 10.1002/glia.23466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz J. I.; Fredenburgh J. C. Platelet polyphosphate: the long and the short of it. Blood 2017, 129, 1574–1575. 10.1182/blood-2017-01-761593. [DOI] [PubMed] [Google Scholar]
- Podoplelova N. A.; Sveshnikova A. N.; Kotova Y. N.; Eckly A.; Receveur N.; Nechipurenko D. Y.; Obydennyi S. I.; Kireev I. I.; Gachet C.; Ataullakhanov F. I.; et al. Coagulation factors bound to procoagulant platelets concentrate in cap structures to promote clotting. Blood 2016, 128, 1745–1755. 10.1182/blood-2016-02-696898. [DOI] [PubMed] [Google Scholar]
- Patel S. R.; Hartwig J. H.; Italiano J. E. Jr. The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Invest. 2005, 115, 3348–3354. 10.1172/JCI26891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlus K. R.; Italiano J. E. Jr. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 2013, 201, 785–796. 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrançais E.; Ortiz-Muñoz G.; Caudrillier A.; Mallavia B.; Liu F.; Sayah D. M.; Thornton E. E.; Headley M. B.; David T.; Coughlin S. R.; et al. The lung is a site of platelet biogenesis and a reservoir for haematopoietic progenitors. Nature 2017, 544, 105–109. 10.1038/nature21706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malara A.; Abbonante V.; Di Buduo C. A.; Tozzi L.; Currao M.; Balduini A. The secret life of a megakaryocyte: emerging roles in bone marrow homeostasis control. Cell. Mol. Life Sci. 2015, 72, 1517–1536. 10.1007/s00018-014-1813-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holinstat M. Normal platelet function. Cancer Metastasis Rev. 2017, 36, 195–198. 10.1007/s10555-017-9677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penido M. G. M. G.; Alon U. S. Phosphate homeostasis and its role in bone health. Pediatr. Nephrol. 2012, 27, 2039–2048. 10.1007/s00467-012-2175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio A. L.; Di Pietro S. M. Storage pool diseases illuminate platelet dense granule biogenesis. Platelets 2017, 28, 138–146. 10.1080/09537104.2016.1243789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arelaki S.; Arampatzioglou A.; Kambas K.; Sivridis E.; Giatromanolaki A.; Ritis K. Mast cells co-expressing CD68 and inorganic polyphosphate are linked with colorectal cancer. PLoS One 2018, 13, e0193089 10.1371/journal.pone.0193089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerbäck L.; Löwhagen G. B. Long term increase of mucosal mast cells in the rat induced by administration of compound 48/80. Cell Tissue Res. 1979, 198, 209–215. 10.1007/BF00232005. [DOI] [PubMed] [Google Scholar]
- Donovan A. J.; Kalkowski J.; Smith S. A.; Morrissey J. H.; Liu Y. Size-controlled synthesis of granular polyphosphate nanoparticles at physiologic salt concentrations for blood clotting. Biomacromolecules 2014, 15, 3976–3984. 10.1021/bm501046t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafi A.; Abbina S.; Kalathottukaren M. T.; Morrissey J. H.; Haynes C.; Kizhakkedathu J. N.; Pfaendtner J.; Chou K. C. Design of polyphosphate inhibitors: A molecular dynamics investigation on polyethylene glycol-linked cationic binding groups. Biomacromolecules 2018, 19, 1358–1367. 10.1021/acs.biomac.8b00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F. A.; Rodrigues C. O.; Docampo R. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 2001, 276, 26114–26121. 10.1074/jbc.M102402200. [DOI] [PubMed] [Google Scholar]
- Weiner S.; Mahamid J.; Politi Y.; Ma Y.; Addadi L. Overview of the amorphous precursor phase strategy in biomineralization. Front. Mater. Sci. China 2009, 3, 104–108. 10.1007/s11706-009-0036-x. [DOI] [Google Scholar]
- Ackermann M.; Tolba E.; Neufurth M.; Wang S.; Schröder H. C.; Wang X. H.; Müller W. E. G. Biomimetic transformation of polyphosphate microparticles during restoration of damaged teeth. Dent. Mater. 2019, 35, 244–256. 10.1016/j.dental.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Müller W. E. G.; Ackermann M.; Tolba E.; Neufurth M.; Wang S.; Schröder H. C.; Wang X. H. A bio-imitating approach to fabricate an artificial matrix for cartilage tissue engineering using magnesium-polyphosphate and hyaluronic acid. RSC Adv. 2016, 6, 88559–88570. 10.1039/C6RA17043A. [DOI] [Google Scholar]
- Müller W. E. G.; Relkovic D.; Ackermann M.; Wang S.; Neufurth M.; Paravic-Radicevic A.; Ushijima H.; Schröder H. C.; Wang X. H. Enhancement of wound healing in normal and diabetic mice by topical application of amorphous polyphosphate - Superior effect of the host-guest composite material composed of collagen (host) and polyphosphate (guest). Polymers 2017, 9, 300. 10.3390/polym9070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdy N. A. Strontium ranelate improves bone microarchitecture in osteoporosis. Rheumatology (Oxford) 2009, 48, iv9–iv13. 10.1093/rheumatology/kep274. [DOI] [PubMed] [Google Scholar]
- Müller W. E. G.; Tolba E.; Ackermann M.; Neufurth M.; Wang S.; Feng Q.; Schröder H. C.; Wang X. H. Fabrication of amorphous strontium polyphosphate microparticles that induce mineralization of bone cells in vitro and in vivo. Acta Biomater. 2017, 50, 89–101. 10.1016/j.actbio.2016.12.045. [DOI] [PubMed] [Google Scholar]
- Kogan S.; Sood A.; Garnick M. S. Zinc and wound healing: A review of zinc physiology and clinical applications. Wounds 2017, 29, 102–106. [PubMed] [Google Scholar]
- Müller W. E. G.; Neufurth M.; Wang S.; Ackermann M.; Muñoz-Espí R.; Feng Q.; Lu Q.; Schröder H. C.; Wang X. H. Amorphous, smart, and bioinspired polyphosphate nano/microparticles: A biomaterial for regeneration and repair of osteo-articular impairments in-situ. Int. J. Mol. Sci. 2018, 19, 427. 10.3390/ijms19020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Zahn R. K.; Bittlingmaier K.; Falke D. Inhibition of herpesvirus DNA-synthesis by 9-ß-D-arabinofuranosyladenosine in vitro and in vivo. Ann. N.Y. Acad. Sci. 1977, 284, 34–48. 10.1111/j.1749-6632.1977.tb21935.x. [DOI] [PubMed] [Google Scholar]
- Hull D. Heat production in adipose tissue. Proc. R. Soc. Med. 1966, 59, 1278–1280. 10.1177/003591576605901221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartree W.; Hill A. V. The recovery heat-production in muscle. J. Physiol. 1922, 56, 367–381. 10.1113/jphysiol.1922.sp002019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof O. F.Energy conversions in muscle. Nobel Lecture, 11.12.1923, https://old.nobelprize.org/nobel_prizes/medicine/laureates/1922/meyerhof-lecture.html.
- Palou A.; Picó C.; Bonet M. L.; Oliver P. The uncoupling protein, thermogenin. Int. J. Biochem. Cell Biol. 1998, 30, 7–11. 10.1016/S1357-2725(97)00065-4. [DOI] [PubMed] [Google Scholar]
- Kazak L.; Chouchani E. T.; Jedrychowski M. P.; Erickson B. K.; Shinoda K.; Cohen P.; Vetrivelan R.; Lu G. Z.; Laznik-Bogoslavski D.; Hasenfuss S. C.; et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 2015, 163, 643–655. 10.1016/j.cell.2015.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S. N.; Seale P. Transcriptional control of brown and beige fat development and function. Obesity 2019, 27, 13–21. 10.1002/oby.22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubi J. M.; Naspreda M.; Kjelstrup S.; Bedeaux D. Energy transduction in biological systems: A mesoscopic non-equilibrium thermodynamics perspective. J. Non-Equilib. Thermodyn. 2007, 32, 351–378. 10.1515/JNETDY.2007.027. [DOI] [Google Scholar]
- Lucia U.; Grisolia G.; Ponzetto A.; Silvagno F. An engineering thermodynamic approach to select the electromagnetic wave effective on cell growth. J. Theor. Biol. 2017, 429, 181–189. 10.1016/j.jtbi.2017.06.029. [DOI] [PubMed] [Google Scholar]
- Meurer F.; Do H. T.; Sadowski G.; Held C. Standard Gibbs energy of metabolic reactions: II. Glucose-6-phosphatase reaction and ATP hydrolysis. Biophys. Chem. 2017, 223, 30–38. 10.1016/j.bpc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- Adelstein R. S.; Conti M. A.; Pato M. D. Regulation of myosin light chain kinase by reversible phosphorylation and calcium-calmodulin. Ann. N. Y. Acad. Sci. 1980, 356, 142–150. 10.1111/j.1749-6632.1980.tb29607.x. [DOI] [PubMed] [Google Scholar]
- Kiani F. A.; Fischer S. Stabilization of the ADP/metaphosphate intermediate during ATP hydrolysis in pre-power stroke myosin: quantitative anatomy of an enzyme. J. Biol. Chem. 2013, 288, 35569–35580. 10.1074/jbc.M113.500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiani F. A.; Fischer S. Catalytic strategy used by the myosin motor to hydrolyze ATP. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, E2947–E2956. 10.1073/pnas.1401862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U.; Pal D.; Prasad R. Alkaline phosphatase: an overview. Indian J. Clin. Biochem. 2014, 29, 269–278. 10.1007/s12291-013-0408-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y.; Kanzawa N.; Saito K.; Krawitz P. M.; Mundlos S.; Robinson P. N.; Karadimitris A.; Maeda Y.; Kinoshita T. Mechanism for release of alkaline phosphatase caused by glycosylphosphatidylinositol deficiency in patients with hyperphosphatasia mental retardation syndrome. J. Biol. Chem. 2012, 287, 6318–6125. 10.1074/jbc.M111.331090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy I.; Butterworth P. J. The inhibition of pig kidney alkaline phosphatase by oxidized or reduced nicotinamide-adenine dinucleotide and related compounds. Biochem. J. 1973, 131, 359–367. 10.1042/bj1310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor M. B.; Ramakrishnan S. K.; Legrand F.; Dolder S.; Siegrist M.; Durussel F.; Centeno G.; Firsov D.; Hynes N. E.; Hofstetter W.; et al. Redox-dependent bone alkaline phosphatase dysfunction drives part of the complex bone phenotype in mice deficient for Memo1. JBMR Plus 2018, 2, 195–205. 10.1002/jbm4.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O.; Tawfik D. S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- Pabis A.; Kamerlin S. C. Promiscuity and electrostatic flexibility in the alkaline phosphatase superfamily. Curr. Opin. Struct. Biol. 2016, 37, 14–21. 10.1016/j.sbi.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Millan J. L. Alkaline phosphatases - Structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signalling 2006, 2, 335–341. 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann H.; Zebisch M.; Sträter N. Cellular function and molecular structure of ecto-nucleotidases. Purinergic Signalling 2012, 8, 437–502. 10.1007/s11302-012-9309-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Wang S.; Wiens M.; Neufurth M.; Ackermann M.; Relkovic D.; Kokkinopoulou M.; Feng Q.; Schröder H. C.; Wang X. H. Uptake of polyphosphate microparticles in vitro (SaOS-2 and HUVEC cells) followed by an increase of the intracellular ATP pool size. PLoS One 2017, 12 (12), e0188977 10.1371/journal.pone.0188977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis T.; Engel A. M.; Bendiksen C. D.; Larsen L. S.; Houen G. Influence of levamisole and other angiogenesis inhibitors on angiogenesis and endothelial cell morphology in vitro. Cancers 2013, 5, 762–785. 10.3390/cancers5030762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard G. E.; Secemski I. I. P1,P5-Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J. Biol. Chem. 1973, 248, 1121–1123. [PubMed] [Google Scholar]
- Liang C. C.; Park A. Y.; Guan J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Lassila J. K.; Zalatan J. G.; Herschlag D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerlin S. C.; Sharma P. K.; Prasad R. B.; Warshel A. Why nature really chose phosphate. Q. Rev. Biophys. 2013, 46, 1–132. 10.1017/S0033583512000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappelet-Tordo D.; Iwatsubo M.; Lazdunski M. Negative cooperativity and half of the sites reactivity. Alkaline phosphatases of Escherichia coli with Zn2+, Co2+, Cd2+, Mn2+, and Cu2+ in the active sites. Biochemistry 1974, 13, 3754–3762. 10.1021/bi00715a022. [DOI] [PubMed] [Google Scholar]
- Cathala G.; Brunel C.; Chappelet-Tordo D.; Lazdunski M. Bovine kidney alkaline phosphatase. Catalytic properties, subunit interactions in the catalytic process, and mechanism of Mg2+ stimulation. J. Biol. Chem. 1975, 250, 6046–6053. [PubMed] [Google Scholar]
- Orhanović S.; Pavela-Vrančič M. Dimer asymmetry and the catalytic cycle of alkaline phosphatase from Escherichia coli. Eur. J. Biochem. 2003, 270, 4356–4364. 10.1046/j.1432-1033.2003.03829.x. [DOI] [PubMed] [Google Scholar]
- Malhotra O. P.; Singh L. R.; Srivastava D. K. Molecular asymmetry in alkaline phosphatase of Escherichia coli. Arch. Biochem. Biophys. 1983, 220, 519–529. 10.1016/0003-9861(83)90443-5. [DOI] [PubMed] [Google Scholar]
- Zalatan J. G.; Fenn T. D.; Herschlag D. Comparative enzymology in the alkaline phosphatase superfamily to determine the catalytic role of an active-site metal ion. J. Mol. Biol. 2008, 384, 1174–1189. 10.1016/j.jmb.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo H. J.; Kim B. W.; Kwon O. B.; Lee C. S.; Choi J. S.; Ko Y. G. Secretion of adenylate kinase 1 is required for extracellular ATP synthesis in C2C12 myotubes. Exp. Mol. Med. 2008, 40, 220–228. 10.3858/emm.2008.40.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja P.; Terzic A. Adenylate kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. 10.3390/ijms10041729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisliakov A. V.; Cao J.; Kamerlin S. C.; Warshel A.. Enzyme millisecond conformational dynamics do not catalyze the chemical step. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 17359–17364, 10.1073/pnas.0909150106(See also Supporting Information). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan D.; Creemers E. E.; Kassiri Z. Matrix as an interstitial transport system. Circ. Res. 2014, 114, 889–902. 10.1161/CIRCRESAHA.114.302335. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K. Cytoarchitecture and physical properties of cytoplasm: volume, viscosity, diffusion, intracellular surface area. Int. Rev. Cytol. 1999, 192, 189–221. 10.1016/S0074-7696(08)60527-6. [DOI] [PubMed] [Google Scholar]
- Swaminathan R.; Bicknese S.; Periasamy N.; Verkman A. S. Cytoplasmic viscosity near the cell plasma membrane: translational diffusion of a small fluorescent solute measured by total internal reflection-fluorescence photobleaching recovery. Biophys. J. 1996, 71, 1140–1151. 10.1016/S0006-3495(96)79316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levick J. R. Flow through interstitium and other fibrous matrices. Q. J. Exp. Physiol. 1987, 72, 409–437. 10.1113/expphysiol.1987.sp003085. [DOI] [PubMed] [Google Scholar]
- Stylianopoulos T.; Poh M. Z.; Insin N.; Bawendi M. G.; Fukumura D.; Munn L. L.; Jain R. K. Diffusion of particles in the extracellular matrix: the effect of repulsive electrostatic interactions. Biophys. J. 2010, 99, 1342–1349. 10.1016/j.bpj.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkus Z. B.; Nazir I.; Jalil A.; Tribus M.; Bernkop-Schnürch A. Zeta potential changing polyphosphate nanoparticles: a promising approach to overcome the mucus and epithelial barrier. Mol. Pharmaceutics 2019, 16, 2817–2825. 10.1021/acs.molpharmaceut.9b00355. [DOI] [PubMed] [Google Scholar]
- Gaertner F.; Ahmad Z.; Rosenberger G.; Fan S.; Nicolai L.; Busch B.; Yavuz G.; Luckner M.; Ishikawa-Ankerhold H.; Hennel R.; et al. Migrating platelets are mechano-scavengers that collect and bundle bacteria. Cell 2017, 171, 1368–1382. 10.1016/j.cell.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Rackov G.; Garcia-Romero N.; Esteban-Rubio S.; Carrión-Navarro J.; Belda-Iniesta C.; Ayuso-Sacido A. Vesicle-mediated control of cell function: The role of extracellular matrix and microenvironment. Front. Physiol. 2018, 9, 651. 10.3389/fphys.2018.00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannetta D.; Dragovic R.; Alyahyaei Z.; Southcombe J. Extracellular vesicles and reproduction-promotion of successful pregnancy. Cell. Mol. Immunol. 2014, 11, 548–563. 10.1038/cmi.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel K. F.; Ronquist G.; Langer F.; Labberton L.; Fuchs T. A.; Bokemeyer C.; Sauter G.; Graefen M.; Mackman N.; Stavrou E. X.; et al. The polyphosphate-factor XII pathway drives coagulation in prostate cancer-associated thrombosis. Blood 2015, 126, 1379–1389. 10.1182/blood-2015-01-622811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skliar M.; Chernyshev V. S.; Belnap D. M.; Sergey G. V.; Al-Hakami S. M.; Bernard P. S.; Stijleman I. J.; Rachamadugu R. Membrane proteins significantly restrict exosome mobility. Biochem. Biophys. Res. Commun. 2018, 501, 1055–1059. 10.1016/j.bbrc.2018.05.107. [DOI] [PubMed] [Google Scholar]
- Suess P. M.; Gomer R. H. Extracellular polyphosphate inhibits proliferation in an autocrine negative feedback loop in Dictyostelium discoideum. J. Biol. Chem. 2016, 291, 20260–20269. 10.1074/jbc.M116.737825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzeja P. P.; Terzic A. Phosphotransfer networks and cellular energetics. J. Exp. Biol. 2003, 206, 2039–2047. 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- Zala D.; Schlattner U.; Desvignes T.; Bobe J.; Roux A.; Chavrier P.; Boissan M. The advantage of channeling nucleotides for very processive functions. F1000Research 2017, 6, 724. 10.12688/f1000research.11561.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer J.; Schuster S.; de Figueiredo L. F.; Kaleta C. Detecting and investigating substrate cycles in a genome-scale human metabolic network. FEBS J. 2012, 279, 3192–3202. 10.1111/j.1742-4658.2012.08700.x. [DOI] [PubMed] [Google Scholar]
- Yegutkin G. G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. 10.3109/10409238.2014.953627. [DOI] [PubMed] [Google Scholar]
- Atkinson D. E.; Walton G. M. Adenosine triphosphate conservation in metabolic regulation. Rat liver citrate cleavage enzyme. J. Biol. Chem. 1967, 242, 3239–3241. [PubMed] [Google Scholar]
- De la Fuente I. M.; Cortés J. M.; Valero E.; Desroches M.; Rodrigues S.; Malaina I.; Martínez L. On the dynamics of the adenylate energy system: homeorhesis vs homeostasis. PLoS One 2014, 9 (10), e108676 10.1371/journal.pone.0108676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman F.Metabolic generation and utilization of phosphate bond energy. In Advances in Enzymology and Related Areas of Molecular Biology; John Wiley & Sons, Inc., 2006; Vol. 1, pp 99–162, 10.1002/9780470122464.ch4. [DOI] [Google Scholar]
- Tumlirsch T.; Jendrossek D. Proteins with CHADs (conserved histidine α-helical domains) are attached to polyphosphate granules in vivo and constitute a novel family of polyphosphate-associated proteins (phosins). Appl. Environ. Microbiol. 2017, 83 (7), 1. 10.1128/AEM.03399-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo-Orts L.; Hohmann U.; Zhu J.; Hothorn M. Molecular characterization of CHAD domains as inorganic polyphosphate-binding modules. Life Sci. Alliance 2019, 2 (3), e201900385. 10.26508/lsa.201900385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E. G.; Tolba E.; Schröder H. C.; Neufurth M.; Wang S.; Link T.; Al-Nawas B.; Wang X. H. A new printable and durable N,O-carboxymethyl chitosan-Ca2+-polyphosphate complex with morphogenetic activity. J. Mater. Chem. B 2015, 3, 1722–1730. 10.1039/C4TB01586J. [DOI] [PubMed] [Google Scholar]