Significance
The mechanisms of hepatocarcinogenesis are poorly understood. In this study, we interrogated the temporal gene expression profiles in mouse livers during progression of chronic fatty liver diseases to carcinogenesis. By establishing an analytical system that focuses on transcription factor (TF) clusters, we identified a sudden switch from healthy liver to tumor tissues in the transcriptomes at precancer stage, prior to detection of any tumor nodule. We further developed a platform to calculate tumorigenic index (TI) based on the transcriptome, especially the TF clusters, to measure tumorigenic signal strengths. This quantitative analytical tool of TI is powerful in assessing cancer risk of chronic liver disease patients and in predicting tumor stages and prognosis of liver cancer patients.
Keywords: liver cancer, tumorigenic index, quantitative analysis, transcription factor clusters
Abstract
Primary liver cancer develops from multifactorial etiologies, resulting in extensive genomic heterogeneity. To probe the common mechanism of hepatocarcinogenesis, we interrogated temporal gene expression profiles in a group of mouse models with hepatic steatosis, fibrosis, inflammation, and, consequently, tumorigenesis. Instead of anticipated progressive changes, we observed a sudden molecular switch at a critical precancer stage, by developing analytical platform that focuses on transcription factor (TF) clusters. Coarse-grained network modeling demonstrated that an abrupt transcriptomic transition occurred once changes were accumulated to reach a threshold. Based on the experimental and bioinformatic data analyses as well as mathematical modeling, we derived a tumorigenic index (TI) to quantify tumorigenic signal strengths. The TI is powerful in predicting the disease status of patients with metabolic disorders and also the tumor stages and prognosis of liver cancer patients with diverse backgrounds. This work establishes a quantitative tool for triage of liver cancer patients and also for cancer risk assessment of chronic liver disease patients.
The incidences and mortality of liver cancer, mainly hepatocellular carcinoma (HCC), are increasing rapidly worldwide (1). Diverse risk factors for primary liver cancer have been identified, including infection of hepatitis B virus and hepatitis C virus, alcohol abuse, nonalcoholic fatty liver disease (NAFLD) or nonalcoholic steatohepatitis (NASH), and intake of aflatoxin B1. Consistent with the complex and multifactorial etiologies, multiomics analyses of human liver tumor samples have identified vast genomic heterogeneity, molecular and cellular defects, metabolic reprogramming, and subtypes of tumors as well as altered tumor microenvironment in the liver (2–4).
However, it remains to be determined if any common molecular signatures in the transcriptomes exist for liver cancer, despite their considerable genomic heterogeneity. Furthermore, little is known about the kinetics and fashions, either gradual accumulation or dramatic transition, in generation of cell-intrinsic and cell-extrinsic signals that are intertwined to drive malignant transformation of hepatocytes and tumor initiation. To dissect the stepwise oncogenic signals and mechanisms at the precancer stages, we chose to work on mouse models that recapitulate key pathogenic features in human liver cancer. By pathological examination, RNA-sequencing (RNA-seq), and bioinformatic data analysis, we identified a sudden switch in transcription factor (TF) clusters at a precancer stage of hepatocarcinogenesis. Based on a multilayer analysis of the TF clusters and mathematic modeling, we developed a tumorigenic index (TI) calculation system for quantitative measurement of liver tumorigenesis. Although this platform was established based on the transcriptomic data derived from genetically modified mouse tumor models, we have found applicable effectiveness of the derived TI values in determination of disease status in other mouse models of chronic or malignant liver diseases with diverse backgrounds. Furthermore, using the TI method developed from animal models to interrogate the human patients’ data, we demonstrated the power of the TI tool in prognosis of liver cancer patients with diverse etiologies and also in diagnosis of precancer patients with steatosis, fibrosis, or cirrhosis. Upon further development and optimization, this TI platform may become a quantitative means for early detection of liver tumor initiation or risk assessment of chronic disease patients, based on comprehensive analysis of the whole transcriptome rather than a few biomarker molecules. We believe that the principle and rationale of this TI derivation method can be extended from HCC to other types of cancer, to develop a quantitative analytical tool for cancer prediction and prognosis in a given organ or tissue.
Results
Temporal Gene Expression Patterns in Liver Tumorigenesis.
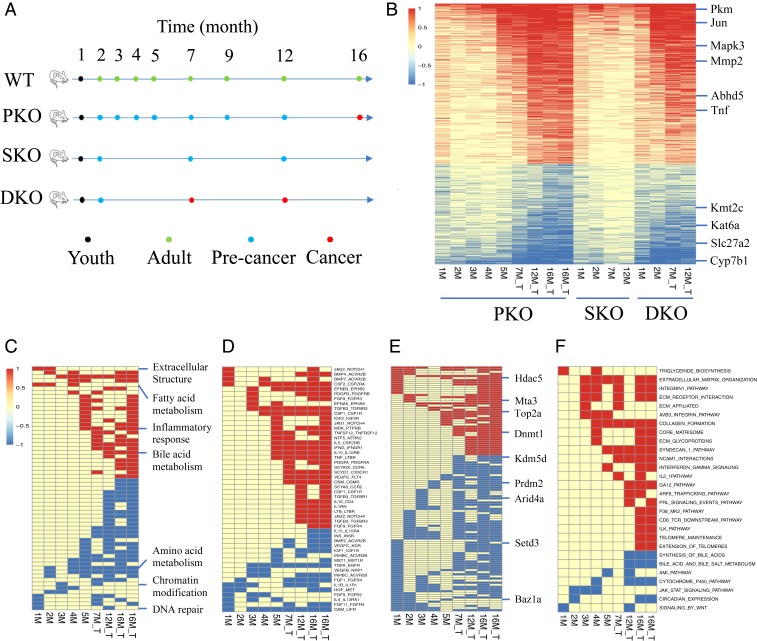
Pten is a tumor suppressor that counteracts the PI3K/Akt signaling pathway, and Pten deficiency has often been detected in liver cancer patients (5, 6). Targeted deletion of Pten in hepatocytes induced NAFLD and, subsequently, NASH, followed by tumor development in mice (7, 8). The NASH-driven pathogenic process in Pten-deficient liver was accelerated and aggravated by additional deletion of Shp2/Ptpn11 (9), a newly identified liver tumor suppressor (10, 11). These mutant mouse lines with defined genetic background and characterized tumor phenotype constitute an ideal group of animal models to dissect stepwise mechanisms underlying NASH-HCC development. Toward this goal, we isolated liver samples at multiple time points from mice with hepatocyte-specific deletion of Pten (PKO), Shp2 (SKO), Pten and Shp2 (DKO), and wild-type (WT) control (Fig. 1A). Based on the developmental or pathological features, these liver samples were divided into 4 groups: 1) youth, 2) adult, 3) precancer, and 4) cancer (Fig. 1A). Representative histological or pathological properties of the specimens were shown (SI Appendix, Fig. S1) and were also described in detail previously (9). RNA-seq was performed for all liver samples, and the data quality analysis showed high levels of correlation for biological replicates in each group (SI Appendix, Fig. S2A), with reduced expression of Shp2 and/or Pten in corresponding mutants as expected (SI Appendix, Fig. S2 B and C). By comparing expression levels in each mutant with WT control at the same time point, we identified genes that were significantly changed in mutant livers at different stages (Dataset S1). The resulting heatmap depicts the progressive changes in liver transcriptomes of the 3 mutant mouse lines (Fig. 1B), which correlated well with the kinetics and severity of tumor progression in SKO, PKO, and DKO livers (SI Appendix, Fig. S1). Highlighted in Fig. 1B are some significantly changed genes related to the MAPK pathway, fatty acid, glucose or bile acid metabolism, extracellular matrix, epigenetic machinery, and inflammatory response.
Fig. 1.
Temporal gene expression patterns during progression of chronic fatty liver disease to tumorigenesis. (A) Liver samples were collected from WT, PKO, SKO, and DKO mice and were classified into 4 groups based on developmental or pathological stages. The youth group represented the 1-mo-old (1M) livers of all 4 genotypes; the adult group included WT livers at 2 to 16M; the precancer group included PKO livers at 2 to 12M, SKO livers at 2, 7, and 12M, and DKO livers at 2M; and the cancer group included PKO livers at 16M and DKO livers at 7 and 12M. RNA-seq was performed for at least 3 mouse livers of each genotype at every time point shown. (B) Heatmap of differentially expressed genes in PKO, SKO, and DKO livers, relative to WT controls at the same time point (t test, false discovery rate (FDR) < 0.05). We selected genes that were significantly changed in at least 2/3 mutant livers. Highlighted here are genes related to MAPK pathway (Mapk3, Jun), fatty acid metabolism (Abhd5, Slc27a2), glucose metabolism (Pkm), bile acid and cholesterol metabolism (Cyp7b1), extracellular structure (Mmp2), epigenetic machinery (Kmt2c, Kat6a), and inflammatory response (Tnf). (C) GSEA was performed to identify significantly up-regulated (red) or down-regulated (blue) biological processes in PKO livers, relative to WT. Marked on the right are some examples with a detailed list provided in Dataset S2. (D–F) Comparative analysis of transcriptomes between PKO and WT livers identified differentially expressed ligands and receptors (D), epigenetic regulators (E), and biological pathways (F) at these time points (FDR < 0.05). A detailed list of significantly changed epigenetic regulators is provided in Dataset S4.
By comparing the transcriptomes between PKO and WT livers at multiple time points, we identified significantly changed biological processes in the mutant liver using gene set enrichment analysis (GSEA) (Fig. 1C and Dataset S2). Consistent with the development of NAFLD and NASH, changes in metabolism of fatty acids and lipids were detected very early in PKO liver starting from 1 mo (1M) (SI Appendix, Fig. S3A). Changes in epigenetic and DNA repair pathways (SI Appendix, Fig. S3B) and extracellular structure organization (SI Appendix, Fig. S3C) were also detected very early at 1 mo, followed by changes in amino acid metabolism at 2 mo (SI Appendix, Fig. S3D), in several immunological and inflammatory processes at 5 mo (SI Appendix, Fig. S3E), and the metabolic processes of bile acids and cholesterol at 12 mo (SI Appendix, Fig. S3F). By comparing expression levels of ligand and receptor genes between PKO and WT livers, we identified ligand and receptor pairs that were changed in a coordinated fashion in Pten-deficient livers (Fig. 1D and Dataset S3). Significantly changed ligands and receptors included Jag2-Notch1, TGFb3-TGFBR2, IFNG-IFNGR1, IL1-IL1RN, and TGFA-EGFR. We also identified significantly changed epigenetic regulators such as DNMT1, ARID4A, and SETD3 in PKO liver (Fig. 1E and Dataset S4). The GSEA approach identified pathways that were significantly altered, such as integrin1, syndecan1, IFNγ, IL-2, GA12, ARF6, P38, CD8-TCR, ILK, Telomere, AMI, JAK-STAT, and WNT pathways, in PKO liver (Fig. 1F and Dataset S5). In SKO liver, MAPK activity and cell cycle were down-regulated, and inflammatory response such as leukocyte proliferation was up-regulated, starting from 1M (SI Appendix, Fig. S4 A–C), followed by changes in epigenetics at 12M (SI Appendix, Fig. S4D). The DKO liver exhibited all of the changes detected in PKO and SKO livers in an expedited fashion (SI Appendix, Fig. S4 E–G), resulting in aggravated and accelerated tumorigenesis.
Significantly Changed Transcription Factor Clusters in Liver Tumors.
To probe the driving force for the transcriptomic changes associated with liver tumorigenesis, we combined a given TF with its target genes as a TF cluster, as their coexpression controls specific cellular activities (SI Appendix, Fig. S5A). The TF cluster-based analysis is superior over a TF alone, which often showed high variations among samples. A total of 1,568 TF clusters have been collected and defined in the public datasets CellNet (12) and GeneFriends (13) (SI Appendix, Fig. S5B and Dataset S6). We confirmed the power of downstream genes to capture key features of specific TFs (SI Appendix, Fig. S5C and Dataset S7). Using the TF clusters as basic units, we compared the transcriptomic data between tumors (isolated from PKO livers at 16 mo and DKO livers at 7 and 12 mo) and WT adult liver samples collected at 2 to 16 mo (Fig. 1A). A total of 61 TF clusters were significantly changed in tumors relative to WT livers, with 36 up-regulated and 25 down-regulated (SI Appendix, Fig. S5D and Dataset S8), as exemplified by up-regulated RelA and down-regulated HNF4α clusters (SI Appendix, Fig. S5D). We summarized well-documented biological functions of some TFs from the literature in Dataset S8. Consistent with the overall gene expression profile changes, as above, the up-regulated TF clusters participate in immune responses and extracellular structure, while down-regulated TF clusters operate in metabolic activities and epigenetic regulation. We also identified some TFs, such as PRRX2, AEBP1, and SFPI1, which have not been reported in liver cancer.
A Transcriptomic Switch at the Precancer Stage and a Coarse-Grained Correlative Network.
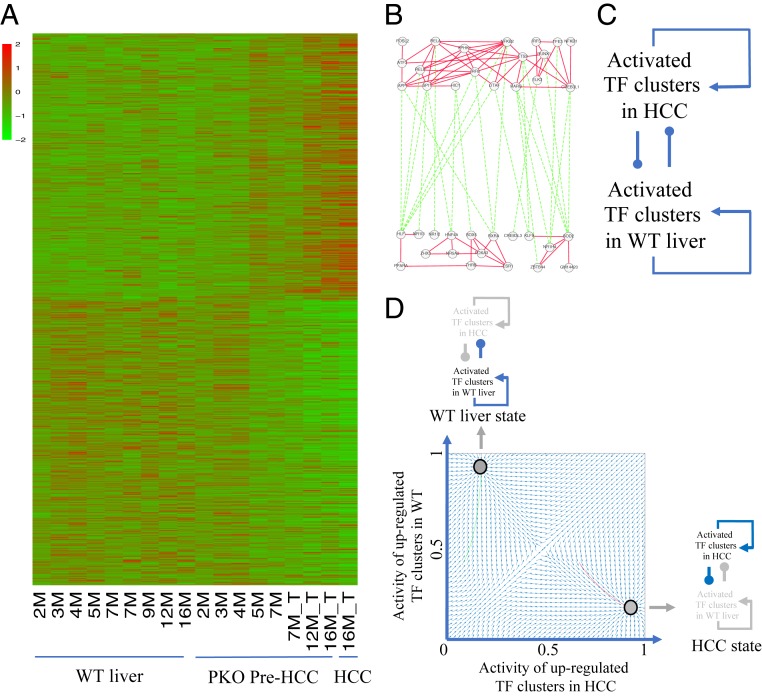
Having identified distinct TF clusters in tumors relative to healthy adult livers, we then compared expression of these TF clusters between WT and PKO livers at multiple time points, in order to trace their changes along the tumorigenic process. The overall expression patterns of the 61 TF clusters remained relatively stable in adult WT livers during a long period of 2 to 16 mo and did not change drastically in PKO livers at 2 to 4 mo. However, a sudden switch occurred in the PKO liver at 5 mo (Fig. 2A and SI Appendix, Fig. S6 A and B), prior to detection of tumor nodules. Of note, the dire transcriptomic change detected in PKO livers at 5 mo was observed in DKO livers even at 2 mo but was not visible in SKO livers until 12 mo (SI Appendix, Fig. S6 A and B), correlating well with the kinetics of tumor development in these 3 mutant mouse livers. Thus, instead of a gradual change, a sharp molecular transition of TF clusters occurred at a precancer stage. It is also interesting to note a high level of similarity of the transcriptomes between tumor tissues and the youth livers at age of 1 mo, reinforcing a notion of cell dedifferentiation during the tumorigenic process (SI Appendix, Fig. S6 A and B).
Fig. 2.
A transcriptomic switch at precancer stage and a coarse-grained correlative network. (A) The expression patterns of TF clusters in the WT adult livers at 2 to 16M, PKO livers at 2 to 16M, as well as tumor tissues as shown. (B) The TF clusters were divided into 2 groups according to their relative expression levels in WT adult livers and tumors. Correlations between TF clusters were inferred as a correlative network, in which nodes denote TF clusters, with solid and dashed lines indicating positive or negative relationships. (C) A coarse-grained network model shows the TF clusters positively stimulating within the same group and mutually inhibiting between the 2 groups. (D) The WT adult liver and tumor were regarded as 2 distinct attractors. Once the changes were accumulated to reach a threshold in Pten-deficient liver, the feedback regulation in the coarse-grained network ensued a dramatic switch to a tumorigenic fate.
We attempted to determine a mechanism underlying such a sharp switch during hepatocarcinogenesis. In theory, the property of a biological process such as the transcriptomic change is dictated by the topology of a gene regulatory network. To define such topology in the context of liver cancer, we inferred correlations between the 61 TF clusters, based on the RNA-seq data (Dataset S9), and visualized the correlations as a network (Fig. 2B), in which the nodes denote individual TF clusters and the solid and dashed lines indicate positive and negative relationships, respectively. The TF clusters were divided into 2 groups, 1 that was activated in WT livers and the other up-regulated in tumors. The correlative network was further simplified by using a coarse-grained network model (Fig. 2C). The 2 TF cluster groups were assembled using 2 nodes, and a coarse-grained network between them was abstracted from the TF correlation network. In this network, activated TF clusters in WT livers or tumors showed positive feedback regulations that can maintain their distinctive expression patterns through reciprocal inhibition.
The coarse-grained network was further quantified using a set of nonlinear differential equations (SI Appendix) (14, 15), generating 2 robust attractors (Fig. 2D). In the first attractor, specific TF clusters were up-regulated in WT adult liver, with the expression patterns maintained by mutual positive regulation. However, the activated TF clusters in tumors were down-regulated due to inhibition by those unique to WT livers. The second attractor indicated the opposite trend, up-regulation of tumorigenic TF clusters and down-regulation of those in WT livers. These 2 attractors represented the states of WT livers and tumors, respectively, and Pten deficiency disturbed the stable expression of the TF clusters in adult livers. Once the changes were accumulated to reach a threshold, the feedback regulation in the coarse-grained network ensued a switch to a tumorigenic transcriptome.
A TI Is Defined by a Multilayer Computational Framework.
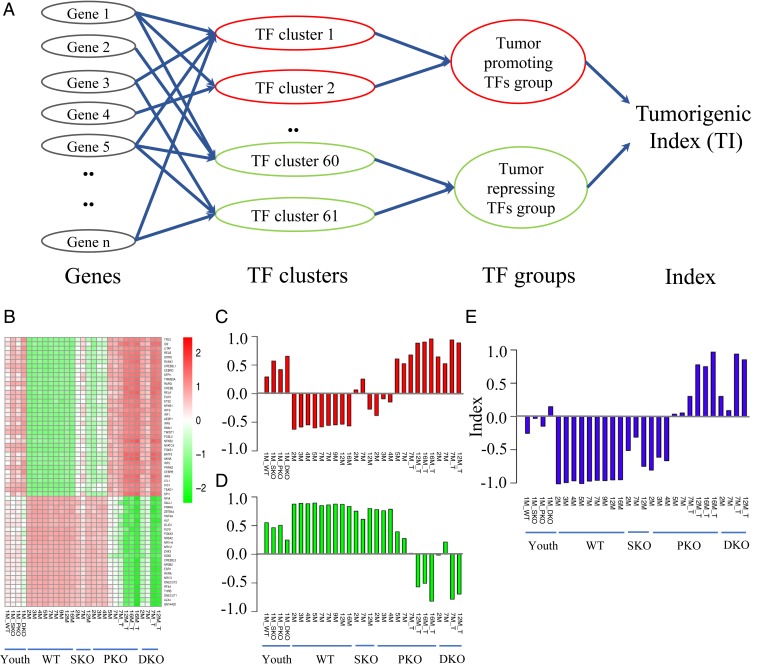
To develop a quantitative method for analysis of liver tumorigenesis, we defined a TI by formulating a multilayer computational model based on the transcriptomic data and the coarse-grained network (Fig. 2D). The first layer was the whole transcriptome (Fig. 3A), and the second layer included the 61 TF clusters changed significantly between WT livers and tumors. Each TF’s activity was determined by expression of its downstream target genes in the first layer. The TF clusters were divided into 2 groups based on their up- or down-regulated expression in tumors, relative to WT livers. The TF clusters up-regulated in tumors were assigned as protumorigenic, with down-regulated clusters assigned as antitumorigenic. The third layer represented the averaged activities of the TF clusters to calculate the tumor-promoting and tumor-inhibiting strengths. Finally, a TI value was defined ranging from −1 for healthy liver to +1 for tumor, to quantify tumorigenic signals in the last layer as output. The TI value of 0 marks the critical transition point from healthy or chronic liver diseases to irreversible tumorigenic fate.
Fig. 3.
A tumorigenic index (TI) for quantitative evaluation of liver tumorigenesis. (A) A multilayer computational framework. The whole transcriptomic profiles were used as input in the first layer. The second layer included the 61 TF clusters. The third layer represented protumorigenic and antitumorigenic TF clusters, with the averaged activities used to calculate the tumor-promoting and tumor-inhibiting strengths, respectively. Finally, a TI was derived ranging from −1 to +1 (from healthy liver to cancer), to quantify the progressive tumorigenic process in the last layer as output. (B) The activity of each TF cluster was derived from the transcriptomes in the first layer for youth, adult, precancer, and cancer samples. (C and D) The tumor-promoting (C) and tumor-inhibiting (D) strengths in the liver or tumor samples were calculated by averaging activities of protumorigenic and antitumorigenic TF clusters in the second layer. (E) The TI values were calculated from the tumor-promoting and tumor-inhibiting strengths of samples in the third layer.
The computational framework included multiple parameters to make quantitative connections between layers (SI Appendix). We first used the RNA-seq data of WT livers and tumor samples to train and optimize the values of these parameters. Using the trained computational framework, we analyzed all samples in Fig. 1A and calculated the activities of the 61 TF clusters (Fig. 3B), tumor-promoting or tumor-inhibiting strengths (Fig. 3 C and D), and a TI value (Fig. 3E). Indeed, the TI values were negative for all of the WT adult livers and positive for all tumor samples and even indicated the increasing severity of tumor phenotypes in SKO, PKO, and DKO livers (Fig. 3E). A switch from negative to positive TI was indeed found in PKO livers at 5 mo, in agreement with the heatmap (Fig. 2A). Of note, the index of the youth livers at 1 mo was higher than the adult livers at 2 to 16 mo, reflecting the decreased proliferative capacity of mature hepatocytes in adult liver. The TI value was positive for DKO liver even at 1 mo when it was negative for both SKO and PKO livers, indicating accelerated hepatooncogenesis driven by combined Shp2 and Pten deletion.
TI Is Applicable to Other Mouse Models with Liver Diseases or Tumors.
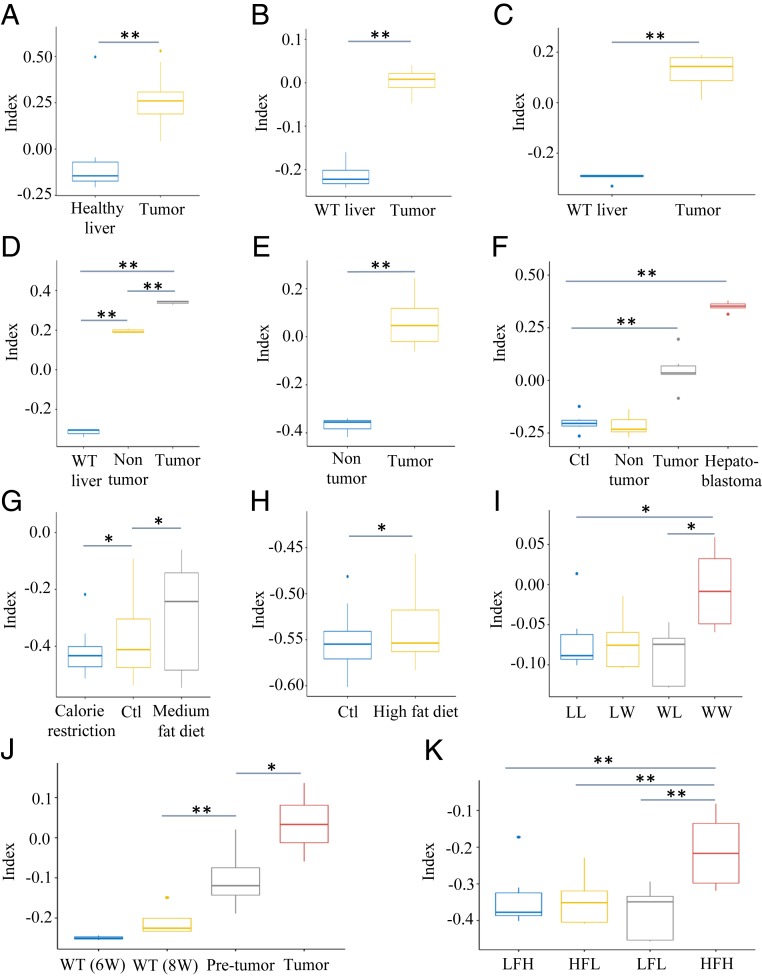
We tested if the TI platform, established using genetically modified mouse models, could be applied to evaluate and predict the pathogenic stages of other mouse models with chronic liver diseases or tumors of different etiologies. From the National Center for Biotechnology Information public databases, we selected 11 datasets that include transcriptomes of WT livers or liver tumors developed in mad2 and p53 null livers, ctnnb1 or gnmt knockout backgrounds, hepatoblastomas, and NAFLD and NASH livers induced by high-fat diet (Dataset S10). The TI values derived using the multilayer framework correctly predicted liver diseases and tumors in 11 out of the 12 datasets tested, based on the documented phenotypes of these samples. For example, the indexes of WT livers were negative (Fig. 4), and indexes of HCCs induced by different gene deletion (Fig. 4 A–D) or toxic agents (Fig. 4 E, F, and J) were positive. The TI values for precancer steatosis, NAFLD or NASH induced by toxic agents (Fig. 4J), or different diet models (Fig. 4 G–I and K) increased relative to WT livers. Most of the dietary models of NASH had negative TIs, indicating that their pathogenic processes did not pass the critical switch point yet in the course of tumorigenesis. Together, these analyses demonstrated the general applicability of the TI in predicting premalignant and malignant diseases in a variety of mouse models.
Fig. 4.
TI values of mouse livers with chronic metabolic disorders or tumors of diverse backgrounds. (A) TI values of healthy livers (n = 10) and Mad2 and p53 null liver tumors (n = 29) in dataset GSE63687. (B) WT livers and tumors of ctnnb1 knockout livers in dataset GSE43628 (n = 4). (C) WT livers and tumors of gnmt knockout mice in dataset GSE63027 (n = 5). (D) WT, tumor, and adjacent nontumor samples collected from SART+/− mice in dataset GSE71057 (n = 3). (E) Nontumor and tumor samples from dataset GSE29813 (n = 6). Tumors were spontaneously developed or induced by a transitional Chinese medicine, Ginkgo biloba leaf extract. (F) WT liver, nontumor, tumor, and hepatoblastoma samples from dataset GSE67316. All samples were collected from mice in a single NTP chronic bioassay (n = 6). (G) Liver samples fed calorie restriction diet, control diet, and medium fat diet from dataset GSE84495 (n = 4). (H) Liver samples with control diet or 21-d high-fat diet from dataset GSE43106 (n = 36). (I) Liver samples from dataset GSE44901. Female mice were fed western (W) or low-fat control (L) semisynthetic diet before and during gestation and lactation. At weaning, male offsprings were assigned either W or L diet, generating 4 groups: WW, WL, LW, and LL offsprings (n = 5–6). WW offsprings showed steatohepatitis. (J) Dataset GSE83596 contained WT liver samples at age 6 or 8 wk (n = 3), pretumor (n = 10) and tumor (n = 4) samples collected from a mouse NASH-associated HCC model (STAMTM model). (K) High- or low-fat-diet liver samples from dataset GSE24031. Mice were chronically fed a high-fat (HF) diet to induce NAFLD and compared with mice fed low-fat (LF) diet. Based on histological scoring, mice were divided into 4 subgroups. LF-low (LFL) responders (n = 4) showed normal liver morphology, LF-high (LFH) (n = 6) had benign hepatic steatosis, HF-low (HFL) (n = 4) exhibited pre-NASH, and HF-high (HFH) (n = 4) developed overt NASH. *P < 0.05; **P < 0.01 (Student’s t test).
TI Is Powerful in Predicting Tumor Stages and Prognosis of HCC Patients.
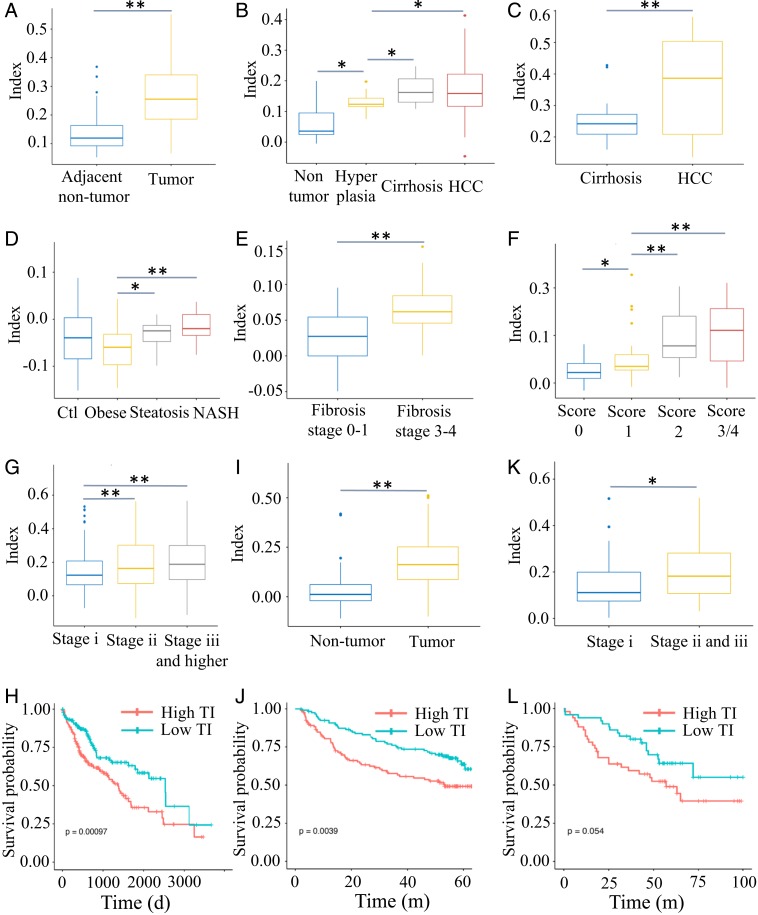
Having demonstrated the TI platform in predicting pathogenic stages in mouse models for liver diseases of diverse backgrounds, we tested the power of the TI in determining disease progression in human patients with HCC or chronic liver diseases. A total of 15 transcriptomic datasets were interrogated, including nontumor liver diseases such as steatosis, alcoholic hepatitis, steatohepatitis, NAFLD, NASH, fibrosis, and cirrhosis, and HCCs with diverse etiologies (Dataset S11). As expected, all of the HCC samples had positive indexes (Fig. 5 and SI Appendix, Fig. S7), and the TIs were negative for most of the NAFLD and NASH samples, with increasing values, as compared to the healthy controls (Fig. 5D). The TIs of livers with fibrosis or cirrhosis (Fig. 5 B, C, E, and F) were higher, some of which had positive values, passing the critical transition point in the tumorigenic process. Thus, the TI derived from a multilayer framework is powerful in predicting human HCC and assessing cancer risk of precancer liver diseases.
Fig. 5.
TI values of human HCC and precancer patients with steatosis, fibrosis, or NASH. (A) A total of 193 adjacent nontumor liver samples and 240 liver tumor samples were from the GSE36376 dataset. (B) A total of 10 nontumor liver samples, 17 dysplastic liver samples, 13 cirrhotic liver samples, and 35 HCC samples in GSE6764. (C) Cirrhotic liver (n = 34) and HCC (n = 35) samples in GSE56140. (D) Liver samples for control (n = 14), obese (n = 27), steatosis (n = 14), and NASH (n = 18) patients in GSE48452. (E) A total of 72 fibrotic liver samples (fibrosis stages 0–1, n = 40; fibrosis stages 3–4, n = 32) in GSE49541. (F) Hepatitis B virus-related liver fibrosis samples (n = 124) in GSE84044. The pathological Scheuer score of each sample was evaluated, score 0 (n = 43), score 1 (n = 20), score 2 (n = 33), and score 3 and 4 (n = 28). (G) A total of 371 liver tumor samples in the TCGA dataset. Of these, 347 tumor samples had well-documented tumor stages (stage I, n = 171; stage II, n = 86; stage III and more advanced, n = 90). (H) The 371 liver tumor samples in the TCGA dataset were subgrouped at the median TI value (high TI, n = 186; low TI, n = 185), and Kaplan–Meier analysis was done to compare survival for patients with high (red) and low (blue) TIs (log-rank test, P < 0.001). (I) TIs of 445 tumor and nontumor liver samples in GSE14520 (tumor, n = 221; nontumor, n = 224). (J) The 221 tumor samples in GSE14520 were subgrouped at the median TI value (high TI, n = 111; low TI, n = 110), and Kaplan–Meier analysis was done to compare survival for patients with high (red) and low (blue) TIs (log-rank test, P < 0.004). (K) TIs of 100 liver tumor samples in GSE16757. These tumor samples had well-documented tumor stages (stage I, n = 35; stage II and III, n = 65). (L) The 100 tumor samples in GSE16757 were subgrouped at the median TI (high TI, n = 50; low TI, n = 50), with Kaplan–Meier analysis done to compare survival for patients with high (red) and low (blue) TIs (log-rank test, P < 0.05). *P < 0.05; **P < 0.01.
We further tested the TI approach in predicting tumor stages and prognosis of human HCC patients using 3 HCC datasets (TCGA, GSE14520, GSE16757), which contained both transcriptomic and clinical data of the patients (Dataset S11). We evaluated the correlation between the TI values and the clinical outcomes. For 371 liver cancer patients in the TCGA dataset, the TI values predicted the tumor stages, with advanced HCCs having higher indexes (Fig. 5G). By dividing the patients into 2 groups based on the median TI value, Kaplan–Meier plotting showed shorter survival and poorer prognosis for the high-TI group than for the low-TI group (Fig. 5H). The TI values clearly distinguished the 221 tumors from 224 nontumor tissues (Fig. 5I), and also correctly predicted the prognosis of the 221 HCC patients, in the GSE14520 dataset (Fig. 5J) as well as the 100 tumor samples in the GSE16757 dataset (Fig. 5 K and L). As the public datasets were generated from different RNA-seq or microarray platforms, with gene expression levels varied at different orders of magnitude, the TI approach provides a platform-independent tool to accurately evaluate the clinical status and prognosis of HCC patients.
We also used 2 other methods, LASSO (16) and Random Forests (17), to calculate the TI values for these 3 sets of data in TCGA, GSE14520, and GSE16757 (SI Appendix, Figs. S8 and S9). With the identical samples used in the multilayer framework for model training, the indexes derived by LASSO did separate the stage I and II–III tumors for the data in GSE16757 (SI Appendix, Fig. S8E), but failed to distinguish between stage I and II liver cancer for the data in TCGA (SI Appendix, Fig. S8A), and the TI values were even higher for nontumor than tumor samples (SI Appendix, Fig. S8C). Kaplan–Meier survival analysis of the 100 samples in GSE16757 showed significantly shorter survival for patients with high TIs (SI Appendix, Fig. S8F), with no significant difference of survival between high and low TIs for the 371 patients in TCGA (SI Appendix, Fig. S8B) or the 221 tumor samples in GSE14520 (SI Appendix, Fig. S8D). Using Random Forests, the derived TI values did separate the nontumor from tumor samples in the GSE14520 dataset (SI Appendix, Fig. S9C), but did not distinguish the tumor stages for the 347 patients in TCGA (SI Appendix, Fig. S9A) or the 100 liver tumor samples in GSE16757 (SI Appendix, Fig. S9E). Kaplan–Meier analysis showed shorter survival for patients with high TIs in TCGA (SI Appendix, Fig. S9B) and GSE14520 (SI Appendix, Fig. S9D), but not for the patients in GSE16757 (SI Appendix, Fig. S9F). Thus, the TIs derived using LASSO and Random Forests were not sufficiently robust to predict liver tumor progression and prognosis. The better performance of our multilayer framework modeling is likely due to its ability to obtain robust results against noises on individual genes in processing transcriptomic data collected from different platforms.
Discussion
Primary liver cancer is characterized by diverse etiologies, genomic heterogeneity, and complex clinical presentations. Molecular dissection of hepatocarcinogenesis based on exome- and genome-sequencing data of patient samples is challenging because of the difficulty to distinguish the driver mutations from the vast majority of passenger mutations. To decipher the common oncogenic mechanisms in liver cancer, we chose to search and identify transcriptomic signatures that drive malignant transformation and tumor initiation, using a group of mouse models with clearly defined genetic defects and pathological properties. Intensive analysis of temporal gene expression profiles has revealed several important aspects in the liver. First, the transcriptomes in adult livers were relatively stable from 2 to 16 mo, suggesting a stringent control and maintenance of gene expression required for functional homeostasis as a major metabolic organ in mammals. Second, the transcriptomes in tumor tissues were very similar to the young liver at 1 mo, reinforcing a concept of dedifferentiation during the tumorigenic process. However, although a set of up-regulated genes were shared between tumors and the premature liver, genes down-regulated in tumors relative to adult liver were not down-regulated in the youth liver, especially those that are involved in maintaining liver-specific functions and epigenetic regulation. Thus, tumorigenesis is apparently not a simple dedifferentiation process but also involves loss of physiological functions in healthy organ or tissue. Third, by intensive month-by-month comparative analysis of gene expression between WT and PKO livers, we identified changes in multiple biological pathways, metabolic processes, and epigenetic factors at precancer stages, which happened in a progressive and accumulating manner. These changes were accelerated in DKO but delayed in SKO livers, relative to PKO liver, correlating well with the kinetics of tumor initiation and progression in these 3 mutants.
To determine the molecular mechanisms underlying these numerous changes, we then focused the analysis on TF clusters as basic units. This targeted analysis revealed interestingly an abrupt switch, rather than a gradual change, during the oncogenic progression from normal adult liver to tumor tissue. By comparing the WT adult livers of 2 to 16 mo with tumor tissues dissected from PKO livers at 16 mo and DKO livers at 7 and 12 mo, we identified a total of 61 significantly changed TF clusters, either up- or down-regulated. Using the 61 TF clusters as proxy to probe oncogenic mechanisms, we inferred correlations between TF clusters from the temporal transcriptomic data and viewed them as a correlative network. Quantitative analysis of the network together with mathematical modeling indicated that normal liver and cancer were associated with 2 distinct attractors, with their own basins of attraction. When perturbation-triggered changes were accumulated to reach a threshold, a sudden transition occurred from the normal liver attractor to the tumor attractor.
Based on the transcriptomic data and the 61 TF clusters, we established a TI derivation system as a quantitative tool to measure tumorigenic signal strength and tumor progression in the liver. The derived TI values of WT, SKO, PKO, and DKO liver samples (Fig. 1A) accurately indicated the phenotypic severity of these mouse tumor models at various time points and captured a critical transition from negative to positive TI in PKO livers from 4 to 5 mo, which was accelerated in DKO livers. Using independent public datasets, we demonstrated the power of the TI calculation in predicting the disease status of precancer samples or the tumor stages of cancer samples in different mouse models (Fig. 4). Of note, these mouse models were generated by targeted gene deletion or were induced by high-fat diet. Therefore, although it was established by using genetically engineered mouse lines, this analytical platform was effectively applied to other mouse liver tumor models and also dietary models of NAFLD and NASH, independent of genetic backgrounds and etiologies.
We extended the quantitative analysis from mouse models to human patients. A large set of human patient samples were collected, including steatosis, fibrosis, cirrhosis, and HCC of diverse etiologies (Fig. 5). Indeed, we obtained negative TI values for these precancer samples, including healthy steatosis and NASH, etc., with all HCC samples being positive. The TI values accurately indicated the tumor stages and even predicted HCC patient survival in prognostic analysis. Furthermore, some cirrhosis patients without clinical detection of tumor nodules yet were found to have positive TI values, suggesting that the pathogenic process likely crossed the critical transition point in oncogenesis. Thus, this TI approach can be developed into a risk assessment or early diagnostic tool of HCC development for a huge population of chronic liver disease patients, especially those with cirrhosis. By quantifying the contribution of each TF cluster to TI, this model can be further used to predict the determining TFs in precision medicine.
This TI platform derived from analysis of tumorigenic TF clusters displayed superior accuracy in predicting tumor progression and prognosis of HCC patients, as compared to the 2 other state-of-the-art machine learning methods, LASSO and Random Forests. However, at this moment, the TI method is effectively applicable to liver tumors, mainly HCC. Using the same rationale to identify targeted TF clusters specific to various organs or tissues, similar approaches may be developed for quantitative analysis of other cancer types. With the rapidly evolving techniques of multiomics analysis, computational biology, and bioinformatics, we believe that quantitative analytical tools, either generic or specialized, will eventually be developed for all types of cancer.
Materials and Methods
Animal Protocols.
Hepatocyte-specific Shp2 KO mice (SKO), Pten KO mice (PKO), and Shp2 and Pten double-knockout (DKO) mice were generated and characterized as described previously (9, 10). All animal experimental protocols (S09108) have been approved by the Institutional Animal Care and Use Committee of the University of California San Diego, following NIH guidelines.
RNA-Sequencing and Data Analysis.
Total RNAs were extracted from liver tissues using QIAGEN RNeasy columns, and RNA-sequencing (RNA-seq) was performed using the multiplex analysis of polyA-linked sequence and the Illumina Hiseq2000 machine. Raw reads generated by RNA-seq experiments were mapped to the mm9 mouse reference genome using Star (2.3.0). The expression level of each gene under different conditions was obtained using cuffdiff.
Data and Code Availability.
The RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under ID code GEO: GSE123427. Codes have been deposited in the GitHub (https://github.com/wanyewang1/Index_model).
More materials and methods in this study are detailed in SI Appendix and Datasets S1–S11.
Supplementary Material
Acknowledgments
We thank our colleagues in the G.-S.F. laboratory for helpful discussion. This work was supported by R01CA188506 and R01CA176012 (to G.-S.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) database under ID code GEO: GSE123427. Codes have been deposited in GitHub (https://github.com/wanyewang1/Index_model).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911193116/-/DCSupplemental.
References
- 1.Llovet J. M., et al. , Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2, 16018 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zucman-Rossi J., Villanueva A., Nault J. C., Llovet J. M., Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239.e4 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network , Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169, 1327–1341.e23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaisaingmongkol J., et al. , Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 32, 57–70.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantley L. C., The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Worby C. A., Dixon J. E., Pten. Annu. Rev. Biochem. 83, 641–669 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Galicia V. A., et al. , Expansion of hepatic tumor progenitor cells in Pten-null mice requires liver injury and is reversed by loss of AKT2. Gastroenterology 139, 2170–2182 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horie Y., et al. , Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest. 113, 1774–1783 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X., et al. , Dual Shp2 and pten deficiencies promote non-alcoholic steatohepatitis and genesis of liver tumor-initiating cells. Cell Rep. 17, 2979–2993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bard-Chapeau E. A., et al. , Ptpn11/Shp2 acts as a tumor suppressor in hepatocellular carcinogenesis. Cancer Cell 19, 629–639 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng G. S., Conflicting roles of molecules in hepatocarcinogenesis: Paradigm or paradox. Cancer Cell 21, 150–154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cahan P., et al. , CellNet: Network biology applied to stem cell engineering. Cell 158, 903–915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dam S., Craig T., de Magalhães J. P., GeneFriends: A human RNA-seq-based gene and transcript co-expression database. Nucleic Acids Res. 43, D1124–D1132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Zhu X., Gu J., Ao P., Quantitative implementation of the endogenous molecular-cellular network hypothesis in hepatocellular carcinoma. Interface Focus 4, 20130064 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyson J. J., Chen K. C., Novak B., Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Friedman J., Hastie T., Tibshirani R., Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
- 17.Breiman L., Random Forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under ID code GEO: GSE123427. Codes have been deposited in the GitHub (https://github.com/wanyewang1/Index_model).
More materials and methods in this study are detailed in SI Appendix and Datasets S1–S11.