Significance
The widespread use of recombinase-dependent adeno-associated virus (AAV) for targeting transgene expression to specific neural populations has made it a fundamental tool for neural manipulation and circuit dissection. Despite this, the well-known but oft-ignored issue of unwanted recombinase-independent “leak” expression from common recombinase-dependent DIO and FLEX AAV designs impedes certain applications and confounds data. Herein we discover that the leak expression from these constructs is due to both low-level expression of the inverted transgene and homology-dependent recombination in bacteria during plasmid production. Using this knowledge, we create a Cre-dependent AAV design that greatly decreases leak expression. This development will improve cell-type-specific transgene targeting for a wide spectrum of tools dependent on precise cell-type-specific targeting.
Keywords: AAV, leak, recombinase, Cre, Flp
Abstract
In combination with transgenic mouse lines expressing Cre or Flp recombinases in defined cell types, recombinase-dependent adeno-associated viruses (AAVs) have become the tool of choice for localized cell-type-targeted gene expression. Unfortunately, applications of this technique when expressing highly sensitive transgenes are impeded by off-target, or “leak” expression, from recombinase-dependent AAVs. We investigated this phenomenon and find that leak expression is mediated by both infrequent transcription from the inverted transgene in recombinant-dependent AAV designs and recombination events during bacterial AAV plasmid production. Recombination in bacteria is mediated by homology across the antiparallel recombinase-specific recognition sites present in recombinase-dependent designs. To address both of these issues we designed an AAV vector that uses mutant “cross-over insensitive” recognition sites combined with an “ATG-out” design. We show that these CIAO (cross-over insensitive ATG-out) vectors virtually eliminate leak expression. CIAO vectors provide reliable and targeted transgene expression and are extremely useful for recombinase-dependent expression of highly sensitive transgenes.
Site-specific recombinases, of which Cre and Flp are members, mediate recombination in double-stranded DNA (dsDNA) across pairs of recombinase-specific 34-base pair sequences. These sequences are comprised of an 8-base pair spacer sequence flanked by a 13-base pair inverted repeat (1–3). Besides the canonical loxp and frt sites recognized by Cre and Flp, respectively, numerous orthogonal sites containing small mutations to the spacer region have also been described (4–7). Depending on whether pairs of these recognition sites are in a parallel or antiparallel orientation, the recombinase will cause excision or inversion, respectively, of the intervening DNA. The ability to manipulate DNA in such a manner has led to diverse applications, including the development of many transgenic mouse lines expressing recombinases in restricted neuronal cell types (8). While these lines can be crossed with recombinase reporter lines to drive transgene expression in a target population globally (9), recombinase-dependent adeno-associated viruses (AAVs) allow for targeting of specific regions and expression of different transgenes without the comparatively cumbersome process of transgenic mouse line production.
AAV is a small relatively nontoxic unenveloped virus with a roughly 5-kb single-stranded DNA genome bookended by 145-bp GC-rich inverted terminal repeats (ITRs). These single-stranded ITRs form cruciform hairpins necessary for replication, packaging, episomal concatemerization of viral genomes, and host genome integration (10–13). While recombinase-dependent AAV designs using the lox-STOP-lox and frt-STOP-frt system have been used (14), double-inverted open reading frame (ORF) (DIO) and flip/excision (FLEX) constructs (15, 16), effectively identical in their design, have gained the most widespread use for their limited size and purported less leaky nature when using strong promoters (17). Briefly, the DIO and FLEX designs use 2 pairs of orthogonal recognition sites in an overlapping antiparallel orientation around the desired transgene that is, with respect to the rest of the expression cassette, inverted and thus transcriptionally repressed (Fig. 1A). When exposed to the appropriate recombinase, the transgene ORF is reverted and locked in-sense with the promoter and 3′ untranslated region (UTR), driving expression.
Fig. 1.
Recombinase-dependent AAV constructs carrying sensitive transgenes suffer from off-target “leak” expression and both their genomes and the plasmids used to make them contain reverted transgenes. (A) Overview of recombinant AAV production from cloning to injection. The plasmid diagram shows the typical Cre-dependent DIO or FLEX recombinase-dependent design utilizing 2 overlapping anti-sense pairs of orthogonal recombinase recognition sites. (B) Examples of leak expression from Flp-dependent Cre (Left) and Cre-dependent Flp (Right) AAVs injected into V1 of reporter mice in which Flp and Cre expression, respectively, is restricted to a specific interneuron population. White arrows indicate those neurons expressing the reporter tdTomato that are not labeled by antibody for the cell type in which recombinase expression is restricted. (C) PCR of recombinase-dependent AAV genomes from different core facilities using in-sense primers spanning recombinase recognition sites produce <1-kb doublets. Sequencing of these PCR fragments reveals amplicons containing transgenes in-sense with the transcriptional motifs of the vector (I and II). (D) PCR of recombinase-dependent AAV plasmids using in-sense and anti-sense primer pairs designed to detect recombination across both 5′ and 3′ lox pairs. Colored spheres above each lane indicate primer pairs. Labeled bands in the Right lane indicate lox pairs across which recombination occurred, as found by sequencing. See also SI Appendix, Fig. S1. Acronyms: RCF, “Rosa-CAG-Frt-STOP-Frt”; nEF, hybrid HTLV/EF1α promoter.
Our laboratory and others have noted evidence of transgene expression in off-target cell populations, termed “leak,” from DIO and FLEX constructs expressing sensitive transgenes, such as TVA for targeting EnvA-pseudotyped rabies virus to specific neural cell types (18), of which small quantities are sufficient for EnvA binding and viral entry (19). As this leak can confound or prohibit the use of these tools for certain applications, including rabies tracing of local circuits, efforts have been made to circumvent the issue, such as a TVA with decreased sensitivity (20, 21). Another strategy works on the assumption that low-level expression occurs from the inverted ORF. Sometimes called “ATG-out” or “split-transgene,” this design disrupts the ORF in DIO or FLEX constructs such that the kozak sequence and initiating codon of the transgene are placed outside the first set of recombinase recognition sites, leaving the transgene ORF to be reconstituted only following recombination (18, 22), a design that has been further optimized by the incorporating destabilized domains to rapidly degrade protein production resulting from leaky transcripts (23). While this design decreases leak expression, it still persists (18).
AAV production requires copious quantities of high concentration plasmid containing the recombinant genome (Fig. 1A). Due to the inverted homology of the ITRs, it is recommended that AAV plasmids are prepared in commercially available Escherichia coli containing mutations to the RecA or RecBJ genes (24), part of the RecBCD pathway mediating bacterial homologous recombination (25). However, previous work has shown that rates of recombination across short homologous regions of less than 50 base pairs in length are unaffected by RecA mutation and likely occur due to strand breaks at the replication fork (26, 27). Further, recombination resulting from homology across short inverted repeats spanning an inverted tetracycline resistance gene resulted in plasmid doubling events that reconstituted transcription and imparted resistance (26).
Given the short size of recombinase-specific sites, the fact that ORF disruption fails to abrogate leak expression, and the previous literature finding that recombination across inverted repeats can reconstitute an inverted transgene, we hypothesized that spontaneous recombinase-independent inversion occurs during the AAV production process in DIO or FLEX constructs. Our investigation found recombinants in all AAV preparations and AAV plasmids we tested and confirmed that intra- or interplasmid recombination indeed occurs across recombinase sites in RecA-mutant competent E. coli. In addition, we show that while this recombination occurs at a low rate in most AAV plasmids, its nature is such that some AAV preparations could be largely or entirely comprised of recombinants. By independently disrupting spontaneous inversion and the transgene ORF, we show that both must be disrupted to fully abrogate leak. Further, while leak expression from an intact ORF is only detectable in highly sensitive systems, spontaneous inversions can drive low but detectable levels of expression of fluorescent proteins. Finally, we show that the use of mutant recombinase recognition sites with reduced homology in AAV’s utilizing an ATG-out transgene design, which we dub CIAO (cross-over insensitive ATG-out), greatly reduces leak expression in the mouse brain of a recombinase reporter mouse.
Results
Commonly Used Recombinase-Dependent AAV Constructs Suffer from Off-Target Expression and Contain Reverted Transgenes.
To illustrate the phenomenon of leak expression, recombinase-dependent AAVs expressing an orthogonal recombinase were used, due to the widespread availability of reporter lines, the increased application of intersectional approaches requiring such viral designs (28–30), and the highly sensitive nature of the recombinase system, as only 4 peptide copies are required to mediate recombination (31, 32). These AAVs were then injected in primary visual cortex of transgenic mouse lines crossed such that transgene expression should be restricted to common interneuron subclasses. Injections of a Flp-dependent AAV vector expressing Cre (AAV8 nEF-fDIO-iCre) were made into the cerebral cortex of a Cre-dependent TdTomato reporter mouse line crossed to a somatostatin-Flp driver line (Ai14 x SomFlp). This should result in expression of TdTomato only in somatostatin-expressing neurons. Results show the expected sparse tdTomato labeling around the injection site (Fig. 1B); however, staining against somatostatin reveals many tdTomato+ cells unlabeled by the somatostatin antibody (white arrows). Similarly, leak is evident from injection of a Cre-dependent Flp AAV vector (AAV8 nEF-DIO-FlpO) into the cortex of a Flp-dependent reporter mouse crossed to a parvalbumin-Cre driver line (RCF-tdT x PvalbCre). Results show many tdTomato+ neurons unlabeled by the parvalbumin antibody (Fig. 1 B, Right). These results provide prime examples of typical leak expression from recombinase-dependent AAVs.
Knowing that removal of the start codon and kozak sequence from the inverted transgene does not fully abolish leak expression, we hypothesized that spontaneous inversion of the transgene sequence was occurring at some point during the sequential exposure of the recombinant AAV genome sequence to bacterial and mammalian cell environments during AAV production and application (Fig. 1A). To test if such recombinants were present in the final AAV product, prior to its application, we designed PCR primers to selectively amplify template DNA containing transgenes that had reverted to be in-sense with the promoter and downstream transcription termination motifs (Fig. 1C). We tested DNA extracted from AAV prepared by popular vector cores to ensure inversion events were not specific to the protocol used by a single core. In addition, both DIO and FLEX constructs were tested to assess if any inversions were specific to either design. All 3 AAVs produced a doublet of bands (Fig. 1C) which, when sequenced, invariably revealed reversion of the transgene to be in-sense with the expression-enhancing woodchuck hepatitis virus posttranscriptional regulatory element (wpre). The sequence difference differentiating these 2 bands was the presence of either a single or triplicate of recombinase sites in a pattern indicative of inversion across 1 set of orthogonal recognition sites (Fig. 1C, I and II and see SI Appendix, Fig. S1).
To see whether this same pattern of reversion was inherited from the plasmids used to produce AAV preparations, prior to transfection of HEK cells in the production process, we applied a similar strategy of parallel and antiparallel PCR primers (Fig. 1D) to the plasmid pAAV-nEF-DIO-FlpO-wpre-pA. Test conditions spanning either end of the transgene (Fig. 1D, lanes 3 and 6) produced doublet and triplicate bands like those seen in PCR of AAV genomes (Fig. 1C) and of the predicted size for reversions across orthogonal sites. Sequencing of the largest and smallest of these bands produced sequences identical to those found from AAV genomes. Based on these sequences, the lox sites across which recombination is expected to have occurred to have produced these bands is indicated (Fig. 1 C, Bottom Right). Interestingly, a third band containing 2 lox sites between the reverted transgene and wpre was also found, the sequence of which suggested recombination across orthogonal sites loxp and lox2722 (SI Appendix, Fig. S1).
A larger band, or bands, was typically, but not always, found in these and similar PCR experiments (for example, Fig. 1C, ∼2 kb). Sequencing attempts of this fragment using the wpre primer failed to return results. Sequencing using primers against the ends of the FlpO coding region revealed that this band contained the FlpO sequence flanked by inverted wpre sequences. Given these sequence results, the single band, and the difficulty of sequencing with the wpre plasmid, we reasoned that this band resulted from single-primer amplification across a region containing flanking inverted sequences, which suggested a duplicative nature of the recombinant plasmid.
Plasmid Doubling due to a Recombination Across Homologous Recombinase Sites Leads to Transgene Reversion.
Due to the presence of 2 nested sets of recombinase recognition sites in DIO and FLEX constructs, we developed a simplified model plasmid system to ascertain the nature of spontaneous transgene inversions. In these plasmids, the LacZ element of the common cloning plasmid pUC19 was inverted and flanked by antiparallel 34-base pair sequences (Fig. 2A). To assess whether the mechanism of inversion was specific to the structure of the site-specific recombinase recognition site or due to gross homology we designed these 34-bp flanking regions to be either the canonical common recombinase recognition sequences (loxp for Cre and frt for Flp) or shuffled loxp sequences lacking any of the sequence structure of typical recombinase binding motifs in either a homologous or nonhomologous setup (Fig. 2B).
Fig. 2.
A plasmid duplication event resulting from homologous recombination across recombinase-specific sites results in transgene reversion. (A) Design of an inverted LacZ expression plasmid based on the pUC19 expression vector. The LacZ coding sequence is inverted anti-sense to the Lac operon and start codon and bookended by 2 34-bp anti-sense recognition sites. (B) The sequences of 4 pairs of tested 34-bp “recognition sites,” both canonical and shuffled, designed to test if the mechanism of inversion was specific to the structure of site-specific recombinases or due to gross homology. Positions 1 and 2 are as denoted in A. For a full list of tested sequences and recombination rates, see SI Appendix, Fig. S3. (C) X-Gal/IPTG plate of colonies from a diluted bacterial miniprep grown from a white colony harboring the pUC19-loxp-inv_LacZ plasmid. Inset shows blue colonies among white colonies. (D) The frequency of blue colonies on X-Gal/IPTG plates grown from white colonies, from 3 minipreps grown across 3 plates. Horizontal lines signify mean value across minipreps. The average colony number per plate and the SD are shown below. (E) The relative amounts of reverted LacZ coding sequences to total LacZ copies in DNA minipreps grown from white colonies by qPCR. Horizontal lines signify the mean value across minipreps (n = 7, circles), and vertical lines are SD. (F) X-Gal/IPTG plate of colonies from a diluted bacterial miniprep grown from a blue colony found in plates shown in C from a transformation of the pUC19-loxp-inv_LacZ plasmid. Inset shows blue colonies. (G) The frequency of white colonies on X-Gal/IPTG plates grown from blue colonies, from 3 minipreps grown across 3 plates. Horizontal lines signify mean value across minipreps. The average colony number per plate and the SD are shown below. (H) The relative amounts of reverted LacZ coding sequences to total LacZ copies in DNA preparations grown from blue colonies by qPCR. Horizontal lines signify mean value across minipreps (n = 7, circles), and vertical lines are SD. (I) Schematic of proposed inter- or intraplasmid recombination events across anti-sense homologous recombinase sites at different relative positions resulting in transgene reversion and plasmid doubling. P and P′ denote mirrored plasmids. Approximate SacI site denoted by orange asterisks. For a schematic of plasmid doubling in recombinant-dependent AAVs, see SI Appendix, Fig. S4. (J) SacI restriction profile of minipreps grown from white and blue colonies from conditions in which blue colonies were present. First and last lanes are a 1-kb ladder.
To assess the rate of recombination in each condition, NEB stable E. coli were transformed with these plasmids and grown on plates coated with the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and lac operon inducer isopropyl β-D-1-thiogalactopyranoside (IPTG), from which a single white colony was grown in 2 mL LB media at 30 °C. This preparation was then diluted and plated on X-Gal/IPTG plates, revealing low numbers of spontaneous blue colonies (Fig. 2C, red arrows). Counting of both white and blue colonies revealed a recombination rate slightly greater than 1/1,000 colonies across the 3 conditions containing homologous regions, regardless of sequence (Fig. 2D). No blue colonies were found in the nonhomologous condition.
To increase the sensitivity with which reversion events were detected, we conducted qPCR in which the targeted amplicon was specific to plasmids in which the LacZ was in-sense with the upstream operon. Absolute quantification against a synthesized dsDNA standard indicated recombinant frequencies between 1/10,000 and 1/1,000 when compared to LacZ copy number (Fig. 2E). Similar frequencies were found for loxp and frt derivates lox2722, loxn, F3, and F5 (SI Appendix, Fig. S3). While all plasmid preparations used here were grown in competent cells at 30 °C, as recommended by the manufacturer, it should be noted that no significant difference in recombination rate was found in the competent cell lines used in any of the experiments presented herein when grown at 37 °C (SI Appendix, Fig. S2).
It was our hypothesis that transgene reversion events, at the molecular level, were similar in nature to those of typical recombinase reactions, in which the nucleotides within a set of antiparallel lox or frt sites are flipped while the surrounding genetic material remains unchanged. Based on this hypothesis, we surmised that the rate of recombination from white to blue colonies should be the same as those from blue to white. Therefore, we conducted the same set of experiments on blue colonies selected from those conditions in which they were found. Surprisingly, no white colonies were found on any X-Gal/IPTG plates from bacterial preparations started from a blue colony (Fig. 2 F and G). Further, qPCR of these plasmid preparations revealed a recombinant:LacZ ratio of roughly 1:1 (Fig. 2H).
Given these results, the pattern of sequences found in AAV genome and plasmid PCR experiments, and the previously published model of recombination between inverted repeats (26), we predicted that recombination across homologous sites at different positions within or between plasmids resulted in a plasmid doubling event leaving the inverted LacZ coding sequence in-sense with the operon (Fig. 2I). Given the nature of this doubling, we further predicted that DNA from blue colonies would produce unique bands when cut with a restriction enzyme expected to have a single recognition site on the original plasmid. SacI restriction of DNA from preparations grown from white and blue colonies revealed bands equivalent to those predicted by such a doubling (Fig. 2J), confirming this hypothesis and suggesting that DIO and FLEX AAV plasmid preparations are contaminated by 2 potential first-order plasmid duplicates (SI Appendix, Fig. S4B).
Both Intact ORF and Spontaneous Reversions Contribute to Leak from Recombinase-Dependent AAV Plasmids.
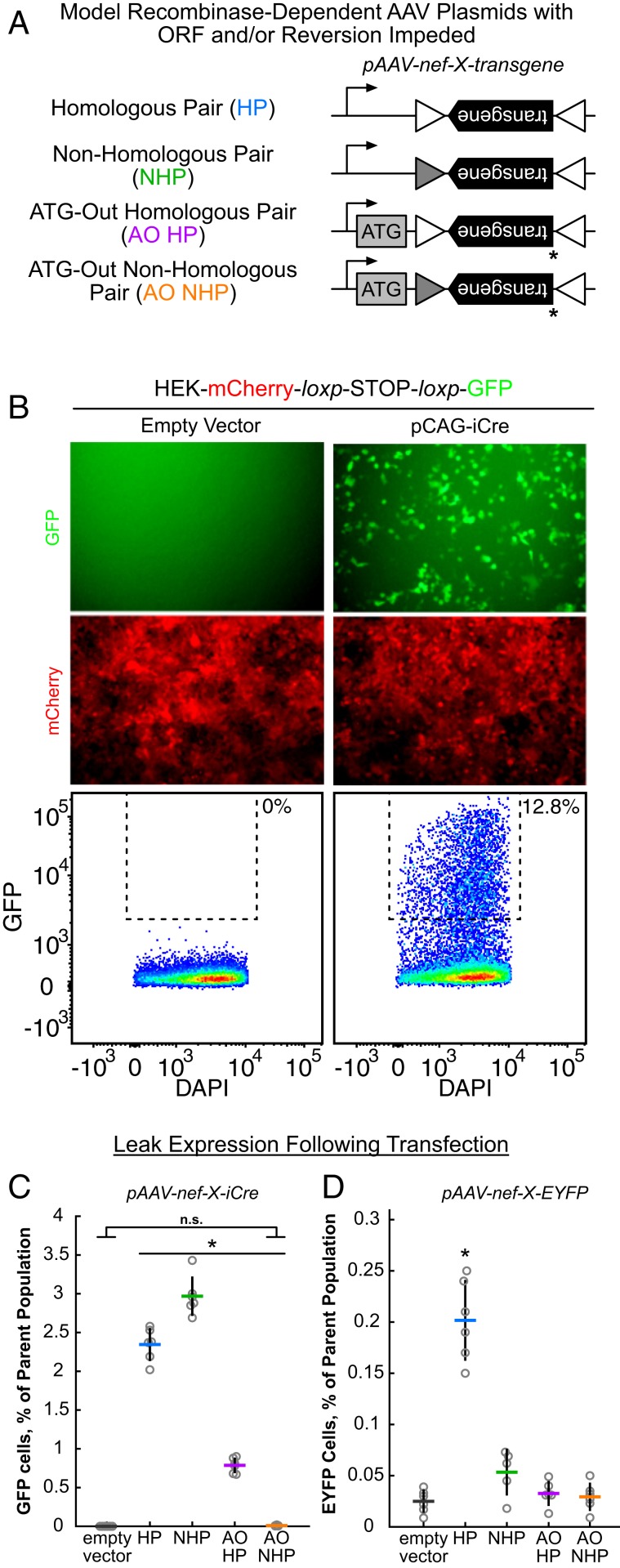
Having deduced the nature of these recombination events and knowing that disruption of the inverted transgene ORF leads to decreased leak, we hypothesized that previous strategies seeking to block leak by disrupting the ORF by moving the transgene start codon and kozak sequence outside of the recombinase recognition sites found persistent leak due to reconstitution of this ORF by recombination. To test this hypothesis we employed a model AAV plasmid design with a single pair of anti-sense 34-bp shuffled loxp sequences bookending the inverted transgene (Fig. 3A). To probe the role of spontaneous reversions in leak expression of Cre from these constructs we used either homologous (HS) or nonhomologous (NHS) shuffled loxp sequences, which left the plasmids sensitive or insensitive to spontaneous reversion, respectively. To ascertain the role of leak expression from the inverted ORF, the constructs were further modified by removal of the start codon (ATG-out [AO]) from the transgene and placing it outside the shuffle loxp sites and downstream from the promoter. Finally, a HEK-mCherry-loxp-STOP-loxp-GFP cell line was used as a reporter of leaky Cre expression, as flow cytometry of these cells when transfected with a pCAG-iCre vector (Fig. 3B) results in obvious and strong GFP expression. To measure the degree of leak expression in subsequent transfections of test constructs, a rectangular gate was constructed such that all GFP+ cells in the positive control were included, while all cells from the empty vector condition were avoided. Transfection of Cre leak test conditions revealed the highest degree of leak in conditions in which the ORF was maintained (Fig. 3C). When the ORF was disrupted but recombination was not, as is the case in previous attempts to mitigate leak from AAVs, leak expression was diminished but not extinguished. Finally, very few GFP+ cells were found in the condition in which both the ORF and spontaneous reversion were disrupted (P < 4 × 10−8 when compared to other modified plasmid conditions; P = 0.999 when compared to empty vector condition; 1-way ANOVA, Tukey–Kramer post hoc multiple comparison), confirming our hypothesis that near-complete mitigation of leak requires disruption of both recombination and the transgene ORF.
Fig. 3.
Disruption of both transgene ORF and spontaneous reversion are required to abolish leak expression. (A) Schematic of model AAV plasmid constructs designed to disrupt transgene ORF (AO plasmids) or reversion by recombination (NHP plasmids). Triangles denote 34-bp shuffled loxp sites; shadings denote heterologous sequences. Asterisks denote the removal of the ATG start codon from the ORF of the transgene. (B) The HEK293T line constitutively expressing mCherry and GFP under Cre control reports Cre expression. Example images show GFP (Top row) and RFP (Bottom row) channels of images of cell monolayers transfected 48 h earlier with the plasmids listed above. Example flow cytometry results showing degree of Cre leak in HEK-mcherry- loxp-STOP-loxp-GFP cells across Cre-expressing plasmids 48 h after transfection. Controls (Left column) show example GFP signal in empty vector and iCre transfection conditions. Dotted line denotes gate bounding GFP+ cells without any cells falling within it in empty vector condition. Percentages at the Upper Right of the doted gate signify percent of the total shown cell population contained within the gate. (C) Summary of leak expression from iCre plasmids in HEK-mCherry-loxp-STOP-loxp-GFP cells across conditions as measured by percent of cells falling within the dotted gate. Horizontal lines signify mean value across transfections (n = 6, circles) and vertical lines are SD. The AO NHP condition is significantly different (P < 4 × 10−8, 1-way ANOVA, Tukey–Kramer post hoc multiple comparison) when compared with other modified plasmid conditions (asterisk). No significance (n.s.) was found between AO NHP condition and empty plasmid. (D) Summary of leak from EYFP plasmids in HEK293T cells across conditions as measured by percent of cells falling within the dotted gate. EYFP positive control was omitted to maintain scale. Horizontal lines signify mean value across transfections (n = 6 transfections, except for NHS in which n = 5; circles) and vertical lines are SD. The HP condition is significantly different (P < 1.5 × 10−8, 1-way ANOVA, Tukey–Kramer post hoc multiple comparison) when compared to other conditions (asterisk).
To describe the role of leak expression in less-sensitive, or accumulative, transgenes, we repeated these experiments by transfecting HEK293T cells with the same plasmids with EYFP as the transgene instead of iCre (Fig. 3D). In these experiments, a gate was similarly drawn encompassing the EYFP+ cells in the positive control, but the bottom bounding line of the rectangular gate was drawn just above the bulk of cells in the empty vector condition. While no leak conditions showed expression levels as high as those seen in this positive control, as a percentage of parent population the homologous pair (HP) condition in which both ORF and spontaneous reversion are left unhampered showed a significant increase in the number of low-expressing GFP+ cells (P < 1.5 × 10−8, 1-way ANOVA, Tukey–Kramer post hoc multiple comparison), indicating that low, but present, levels of leak expression still occur from less-sensitive transgenes.
Use of Mutant Recombinase Recognition Sites with Decreased Homology in an ORF-Interrupted Design Mitigates Leak Expression.
Knowing that recombination is mediated by sequence homology, we hypothesized that the use of recombination recognition sites with decreased homology would in turn decrease recombination. While canonical recombinase recognition sites are homologous, and mutations to the 8-bp spacer region lead to site incompatibility, mutations to the left and right ends of the flanking 13-bp repeat regions have yielded functional recognition sites that result in biased reaction kinetics in the presence of the appropriate recombinase (Fig. 4A; ref. 4). Given their decreased homology, several versions of mutant loxp pairs (Fig. 4B) were tested using the inverted LacZ assay previously described. Quantification by qPCR of recombinant frequency in the plasmid population revealed a sharp decrease in recombination in all mutants (Fig. 4C). Further, by comparing the recombination rate with the degree of sequence homology, (measured as 1 minus the normalized Levenshtein distance, the number of single character insertions, substitutions, or deletions between pairs of strings normalized by their length, such that a measurement of 1 is complete homology and 0 is no homology) we found that recombination rate decreased logarithmically with decrease in homology (Fig. 4D).
Fig. 4.
Mutated loxp sites with decreased homology mitigate spontaneous recombination and leak expression of sensitive transgenes from an ORF-disrupted AAV design. (A) Schematic of the directionally biased Cre-dependent recombination reaction when using mutant lox sites. Asterisks denote locations of mutations. (B) Candidate canonical and mutant lox sites with known compatibility and decreased homology. Nonhomologous bases within pairs are shown in red. (C) The frequency of spontaneous recombination in the pUC19-inv_LacZ test when using canonical and mutant loxp sequences as measured by qPCR. Circles indicate individual measurements, mean shown by a vertical line, and horizontal lines signify SD. n = 7. For a full list of tested sequences and recombination rates, see SI Appendix, Fig. S3. (D) Sequence homology, as measured by 1 minus the normalized Levenshtein distance between within-pair sequences, plotted against mean recombinant frequency as measured by qPCR in the pUC19_inv_LacZ test. Line of best fit was calculated using loxp derivatives (blue crosses). R2 = 0.9632. NHS condition shown for comparison (pink cross). (E) Images of brain tissue slices following injections of equal quantities of AAV8 nef-DIO-FlpO (Left column) and AAV8 nef-CIAO-Flp (Right column) into primary visual cortex of RCF-tdTomato x PvalbCre (Top) and RCF-tdTomato (Bottom). Asterisk in AAV model denotes removal of ATG start codon from Flp ORF. Flp-dependent tdTomato expression is in red and parvalbumin antibody in green. (Scale bars, 100 um.) (F) Quantification of total tdTomato+ neurons in RCF-tdTomato mice injected with AAV8 nef-DIO-FlpO (n = 5) and AAV8 nef-CIAO-Flp (n = 6). Error bars indicate SD; P = 0.0225, 2-tailed independent t test (asterisk).
Given these data, we set out to design a Cre-dependent AAV construct expressing Flp recombinase with minimized leak expression. Based on our finding that ORFs play a role in leak expression in recombinase-dependent AAVs based on an inverted transgene design, we opted to use an ATG-out lox-STOP-lox design to avoid any unintended transcription. While the pair did not return the highest degree of resistance to recombination in the LacZ assay, we used the loxp mutant pairs loxp66/71 based on their decreased recombination rate in the LacZ assay and known efficient Cre reactivity. Finally, we removed the start codon and SV40 nuclear locating sequence containing a second in-frame start site from the optimized FlpO coding sequence (33) and the wpre sequence from the vector. The resulting construct, AAV8 nEF-AO-loxp66-STOP-loxp71-Flp, or AAV8 nEF-CIAO-Flp, was injected in V1 of RCF-tdTomato × PvalbCre mice and compared with identical injections using an AAV8 nEF-DIO-Flp (Fig. 4 E, Top). Results from these injections showed efficient Flp-dependent expression of tdTomato in the target population for both vectors. To quantify degree of leak expression, viruses were titer matched and equal volumes were injected into V1 of RCF-tdTomato mice (not crossed to a Cre driver; Fig. 4 E, Bottom). Due to the absence of Cre in these mice, any Flp-dependent expression of tdTomato must be the result of leak expression of Flp from the AAV vector. Counting of tdTomato+ cells across the entire brain of these mice revealed a mean decrease of ∼20× in tdTomato leak expression in the AAV8 nEF-CIAO-Flp condition (Fig. 4F; AAV8 nEF-DIO-Flp mean 613.2 ± 526.46, AAV8 nEF-AO-CIAO-Flp mean 29 ± 15.86, P = 0.0225, t test).
Discussion
Recombinase-dependent AAVs have rightly become a popular tool, particularly in systems neuroscience. Coupled with transgenic mice, they allow for the targeted expression of transgenes to specific neuron populations. However, we’ve shown that current DIO and FLEX designs are not immune from off-target expression. To measure the extent of leak expression, we chose to use recombinases as transgenes due to the highly sensitive nature of Cre-lox and Flp-frt systems. These experiments showed clear leak expression in those plasmids in which the ORF was intact or spontaneous homologous recombination could occur between anti-sense recombination recognition sites. In contrast, the experiments conducted in vitro using EYFP revealed levels of leak expression which, though detectable by flow cytometry (Fig. 3D), are likely of negligible concern in typical fluorescent protein applications, and at worst are easily negated by ORF or recombination disruption. It remains unclear whether this increased expression is due to reversion of the transgene or from transcription of the inverted transgene, as we did not detect elevated levels of EYFP expression in either the AO HP or nonhomologous pair (NHP) conditions. One hypothesis is that the extra amino acids on the N-terminal of EYFP following read-through of the loxp sequence in the AO HP condition leads to decreased expression of florescence. (Indeed, the addition of extra amino acids is present in all ATG-out designs. The read-through of the loxp site following recombination results in the addition of 15 amino acids [MGTVRIAYIIRTVAS] to the N-terminal of the following peptide, an important caveat for genes that may be sensitive to such additions.) Regardless, these experiments confirm that DIO and FLEX designs leak when containing enzymatically or accumulatively detected transgenes. Whether an investigator should be concerned about AAV leak from these constructs depends on the intended application and the nature of the transgene being expressed.
We show here that the presence of recombinant plasmids in a model system, as measured by spontaneous blue colony frequency and absolute quantification by qPCR, to be between 1/1,000 and 1/10,000 recombinants per nonrecombinant (Fig. 4 D and E). Due to the overlapping nature of the 2 sets of recombinase recognition sites in DIO and FLEX designs, the duplicative nature of the recombinant plasmid, and the low GC content of the interstitial sequences flanking the recombinase recognition sites, we were unable to design a reliable method for quantifying the number of reverted genomes in AAV DIO or FLEX preparations or the plasmids used to make them. Given the size restrictions on genome packaging and potential differences in replication kinetics when multiple genomes of varying sizes are present in the same host cell, it is hard to estimate if the frequency of recombinants among AAV genomes is comparable to that seen in the plasmid populations used to make them. Previous work has shown that recombination between inverted repeats decreases as intervening sequence size increases (26), suggesting variance in recombination frequency in AAV designs harboring different sized transgenes. Of potentially greater concern are the rare cases in which entire AAV preparations contain genomes from recombinant plasmids, which would occur if the wrong bacterial colony was unwittingly selected for plasmid production and if quality control checks, typically based on Xma1 and SmaI restriction at the ITRs, were not stringent enough to detect the issue. Indeed, in our blue/white screens we found colonies that were only partially blue, suggesting recombination events after plating, which in an AAV preparation would lead a large percentage of the AAVs to contain genomes with reverted transgenes.
We considered several approaches for abolishing leak expression. The simplest, given the apparent duplicative nature of the recombination event, is size exclusion of the offending recombinant by gel electrophoresis. Given that we are unaware of a technique by which plasmid size exclusion could be performed with yields high enough to produce high titer virus, the fact that we could not definitively show that nondoubling within-plasmid recombination events were also occurring (while unaware of any known mechanism by which this could be occurring), and the availability of other simple design-based solutions, we did not test whether this would be a suitable method for excluding the production of leaky genomes from recombined plasmids.
It may also be suggested that, given that it is the anti-sense nature of the homologous recombinase recognition sites that drives reversion of the inverted transgene in the DIO/FLEX system, a simple design change to the lox-STOP-lox system would abrogate leak. While we did not test it here directly, we see no reason why recombination and plasmid doubling could not also occur in this system and would predict from the data shown here that AAV preparations from such designs would harbor genomes containing lox and lox-STOP-lox-STOP-lox regions in place of the lox-STOP-lox motif. Indeed, we anticipate any remaining leak expression we see with application of the improved AAV8 nEF-CIAO-Flp virus described here to be the result of such recombination. Nor is the lox-STOP-lox system alone resistant to leaky transcription without further disruption of the ORF (17, 19).
Our finding that recombination decreased with homology led to the use of mutant loxp pairs. Given their decreased homology and equivalent or greater recombination frequency in the forward direction, we chose to use the previously published loxp66/71 pair (4). While this design proved effective at minimizing leak expression, use of the other mutant loxp sites should further decrease leak levels. Indeed, the pair loxp43/44 showed mean recombination rates lower than the nonhomologous condition. Though it should be noted that the exact accuracy of the nonhomologous reading may be skewed by differences between the qPCR amplicon from any recombinant in this condition (the sequence of which is unpredictable) and the loxp-based standard against which it is compared to generate this measurement, this finding strongly suggests the loxp43/44 pair is approaching the limits of recombination mitigation. However, the original description of these recognition sites detailed their decreased sensitivity to Cre (4), a drawback that informed our decision to use the loxp66/71 pair, which has been used previously. Indeed, even beyond improving reaction efficiency, there remains the need for the development of novel recombinase recognition sites with limited homology, a task that should be easily met with recent advances in prediction of sequence specificity for recombinases (34). Nowhere is this more needed than with the Flp-frt system, in which we found little effective difference in recombination rate between frt and frtRE/LE, to our knowledge the only previously described right end/left end mutant pair for frt (6, 35), likely due to the very minimal decrease in homology in this mutant pair (SI Appendix, Fig. S4). Further investigation will be needed to develop recognition sites with decreased homology for the Flp-frt system.
In the course of this work we attempted many potential solutions to abolish leak expression from recombinase-dependent AAVs, including the design of recombinant genomes resulting in the blockade of genomes carrying reverted transgenes (SI Appendix, Fig. S4) and the reversion-induced expression of bacterial toxin ccdb. These techniques did not prove successful, though further investigation may find more effective solutions. We also considered targeted mutation of bacterial competent cell lines to decrease recombination across short homologous sequences, a solution that would require minimal redesign of extant AAVs. However, it is our understanding that the RecA-independent recombination event is not dependent on a single gene product or pathway, but is the unwanted consequence of single-strand breaks occurring during plasmid replication. As such, this process may be difficult to target by mutation of a single gene or pathway. However, there may yet be a bacterial mechanism upon which manipulation would produce a competent strain with decreased RecA-independent recombination which would allow for leakless production of recombinase-dependent AAVs without plasmid modification.
While we believe the design described herein will greatly reduce leak expression from Cre-dependent AAVs in the majority of circumstances, it is important to note that there are other potential sources of leak from AAV constructs. Spontaneous or injection-induced changes in gene expression patterns may lead to transient off-target recombinase expression. Further, viral serotype, promoter strength, titer, and injection volumes could directly or indirectly effect the degree of leak expression. Indeed, such variability is evident in the DIO condition of Fig. 4F.
The expanding application of recombinase-dependent AAVs in neuroscience demands increasingly precise and replicable systems for neural circuit dissection. The issue of off-target transgene expression investigated here provides details regarding a problem that has long plagued an established technique. It is our hope that continued investigation will build on the results and solutions presented here. Nevertheless, the AAV-CIAO systems presented here represent a significant advance that can be readily applied to the viral toolkit used by the modern neuroscientist.
Methods
Plasmid Production.
NEB stable cells (NEB C3040) were used for DNA preparations and all blue/white assays. Chemical transformations were followed as described by the manufacturer. All minipreps were grown in a shaker at 220 rpm in 2 mL of LB containing 0.1 mg/mL ampicillin at 30 °C for 20 h, unless otherwise noted.
Blue/White Screening.
To produce minipreps of DNA, 2 mL of LB containing 100 μg/mL ampicillin were inoculated with a single colony and grown at 30 °C for 12 h. The bacterial solution was the diluted to 10 colony-forming units (cfu)/μL from an estimated starting concentration of 106 cfu/μL. A total of 50 μL of this dilution was plated on 100 μg/mL carbenicillin plates made with Chromomax IPTG/X-Gal solution (Fisher BP4200). Three plates were grown for each miniprep. Plates were grown at 37 °C overnight and moved to 4 °C before being imaged. For colony counting, a matlab script was used to identify blue and white colonies in each image before manual correction.
Plasmids and Cloning.
The blue/white screening plasmid pUC19-X-inv_LacZ was made by restricting pUC19 (NEB N3041) with BsmBI, EcoRI, and HindIII and collected both the backbone and LacZ fragments. Recombinase-specific and shuffled 34-bp LacZ flanking sites were designed as oligos to be annealed such that the final product contained overhangs matching those left by restriction. Sequences of these 34-bp sites can be found in Figs. 2B and 4B. To anneal, oligos were resuspended in molecular grade water to 100 μM, and 2 μL of each oligo solution was added to the reaction mix with 1 μL of T4 polynucleotide kinase (PNK; NEB M0201), 2 μL of 10× T4 PNK buffer (NEB BS0201), and 13 μL of molecular water. The PNK reaction was conducted as follows: 37 °C for 10 min, 65 °C for 10 min, 95 °C for 30 s, 60 °C for 10 min, and cooling at 0.1 °C/s to 20 °C. The reaction solution was diluted 20-fold and 1 μL of this annealed oligo solution was added to each T4 ligation (NEB M0202) along with the pUC19 backbone and LacZ fragment. Shuffled loxp sequences used in both model pUC19 and pAAV plasmids were generated by shuffling the loxp sequence, choosing sequences without more than 3 consecutive nucleotide repeats, and using Thermo Fisher’s Multiple Primer Analyzer to ensure there was no homology between the sequences. Prior to experiments all plasmids were sequenced across the cloned regions.
AAV PCR.
AAV preparations were treated with DNaseI (Epicentre D9905K) at a final concentration of 0.1 U/μL for 10 min at 37 °C. The solution was then immediately heated to 98 °C for 10 min to denature the DNaseI and capsid proteins. A total of 1 μL of this solution was used as a PCR template in a standard 50-μL PCR.
qPCR.
For experiments conducted to detect reversion events of LacZ in the pUC19 plasmid, the forward and reverse primers for the lacz amplicon used to measure total copy number were GCCTCTTCGCTATTACGCCA and TCACTGGCCGTCGTTTTACA, respectively. Forward and reverse primers used to assay for recombinant plasmids were GTGCTGCAAGGCGATTAAGT and TGTGGAATTGTGAGCGGATA, respectively. Primer3 (www.primer3plus.com) was used to design all qPCR primers. All qPCR reactions were carried out with SYBR Green (Thermo Scientific K0251) on a Lightcycler 480II for 45 cycles with an annealing temperature of 60 °C.
qPCR standards were 500 ng 500 bp gBlocks made by Integrated DNA Technologies and based upon the expected plasmid sequence following recombination. Separate gBlocks were used for loxp-and frt-based sequences. These sequences contained templates for both the lacZ and recombinant amplicons.
The template sequence used for all loxp-based sequences was as follows:
GTTGGCGGGTGTCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGATGCGTAAGGAGAAAATACCGCATCAGGCGCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGATAACTTCGTATAATGTATGCTATACGAAGTTATCAAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCC.
The template sequence used for all frt-based sequences was as follows:
GTTGGCGGGTGTCGGGGCTGGCTTAACTATGCGGCATCAGAGCAGATTGTACTGAGAGTGCACCATATGCGGTGTGAAATACCGCACAGATGCGTAAGGAGAAAATACCGCATCAGGCGCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAATTCGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGCAAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGCTCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTG CCCGCTTTCCAGTCGGGAAACCTGTCGTGCC.
Mammalian Cells and Transfections.
All cells were grown in 10% FBS (HyClone SH 30070.03) DMEM media (Gibco 11995-040) at 37 °C and 5% CO2. Cre-reporter HEK cells were made by lentiviral infection of a CAG-loxp-TdTomato-loxp-GFP vector and selection of the top 1% of RFP+ cells. Potentially due to multiple integrated copies per genome of the selected cells, these cells rarely switch from green to red as the expression cassette suggests.
For each microgram of DNA to be transfected, a 3× volume of 1 mg/mL polyethylenimine (PEI) was added to a 6× volume of Opti-MEM (Gibco 51985-043) and the appropriate volume of DNA was added to the same 6× volume of Opti-MEM. These 2 solutions were mixed and left at room temperature for 15 min, after which they were applied dropwise to the media of each target cell plate or well. After 4 to 6 h, the media were replaced.
Flow Cytometry.
Forty-eight hours following transfection, cells were trypsinated, collected in 2-mL Eppendorf tubes, and spun at 500 relative centrifugal force (rcf) for 3 min. The media were aspirated and the cellular pellet was resuspended in 1 mL of cold 1% FBS in dPBS (Gibco 14190-144). The cells were spun down again, the media aspirated, and cells resuspended in 1 mL of cold 10 μm DAPI in 1% FBS in dPBS. Cells were spun down, the media aspirated, and cells resuspended in 500 μL of cold 1% FBS in dPBS. This solution was moved to filter-topped tubes (Falcon 352235) on ice and run on a LSRII flow cytometer.
Viruses.
AAV plasmids were produced in NEB stable cells grown for 20 h in 250 mL of LB containing the appropriate antibiotic at 30 °C. These plasmids were provided to the Salk Vector Core for AAV production. All plasmids were restricted with XmaI to test for gross recombination prior to submission. Titer for AAV8 nEF-DIO-FlpO was 2.32 × 1012 genome copies (GC)/mL and for AAV8 nEF-AO-loxp66-STOP-loxp71-Flp (AAV8 nEF-CIAO-Flp) it was 7.79E + 13 GC/mL. AAV8 nEF-fDIO-iCre was an untitered crude prep. For producing equivalent titered viral dilutions for comparative injections, the higher titered virus was diluted in 0.22 μm filtered Hank’s Balanced Salt Solution.
To produce these crude preps, 10 μg of the AAV plasmid DNA and 15 μg of the helper plasmid pDP8 (36) were transfected by PEI into a 70 to 80% confluent 15-cm plate of HEK293T cells. Three days later, the media were aspirated, the cells washed with 10 mL of cold dPBS, and collected with a cell scraper into a 1.5-mL Eppendorf tube. Cells were then spun at 510 rcf for 3 min, the supernatant was aspirated, and the cells were resuspended in 1 mL of cold gradient buffer (10 mM Tris pH 7.6, 150 mM NaCl, 10 mM MgCl2, 100 mL, 0.22 μm filtered). Three rounds of freezing in liquid nitrogen, thawing in a 37 °C water bath, and vortexing were performed to lyse the cells. Finally, the lysed cells were spun at 14,000 rpm for 10 min at 4 °C and the supernatant was collected and filtered (0.22 μm; Millipore SLGV033RS) into a sterile 2-mL freezer tube. The crude prep was then aliquoted and stored at −80 °C. These crude preps, typically for small batch use, were not titered.
Mice.
All procedures using live animals were approved by the Salk Institute Institutional Animal Care and Use Committee. RCF-tdT mice, a gift from the laboratory of Sam Pfaff, Salk Institute for Biological Studies, La Jolla, CA, are Flp-reporter mice containing the Rosa-CAG-frt-Stop-frt-tdTomato allele and were made from crossing the Flp and Cre reporter mouse line Ai65 (RCFL-tdT; The Jackson Laboratory 021875) with a CMV-Cre mouse line. Ai14 (The Jackson Laboratory 007914) expresses tdTomato in a Cre-dependent manner from a Rosa-CAG-lox-Stop-lox-tdTomato allele. The Som-ires-Flp (The Jackson Laboratory 028579) and PV-ires-Cre (termed herein PvalbCre, The Jackson Laboratory 008069) were crossed with the aforementioned strains to specifically target the somatostatin and parvalbumin interneuron populations, respectively.
Both male and female mice were used. Injections were conducted at ages between 80 and 150 d. Stereotactic coordinates used for all injections targeting primary visual cortex were, from bregma: anterior/posterior −3.28, medial/lateral 2.5, dorsal/ventral −0.5. All injections delivered 200 nL of virus.
Tissue Processing and Immunohistochemistry.
Mice were anesthetized with euthasol and intracardially perfused using 50 mL of PBS and 100 mL of 4% paraformaldahyde (PFA). Following perfusion, brains were dissected and placed in 5 mL of a 2% PFA and 15% sucrose in PBS mixture at 4 °C. Having been sunk in this solution, brains were transferred to 5 mL of 30% sucrose in PBS at 4 °C. Following sinking, brains were sliced on a microtome at 50 μm and stored in solution of 1% NaN3 in PBS.
Tissue staining started with a 5-min wash of 0.1% Triton X-100 in PBS followed by a 2- to 3-h block comprised of 5% normal donkey serum in 0.5% Triton X-100 in PBS at room temperature. Primary staining occurred overnight at 4 °C. Primary antibodies including rabbit anti-somatostatin (Peninsula Labs T-4103) and rabbit anti-parvalbumin (Swant PV27) were applied at 1:1,000 in block. Following primary staining, tissue was washed for 10 min in PBS 3 times. The secondary antibody Alexa-Fluor 647 donkey anti-rabbit (Invitrogen A31573) was diluted at 1:500 in PBS and applied to the tissue for 2 to 3 h at room temperature. Following secondary staining, tissue was washed thrice for 10 min in PBS. A total of 10 μm DAPI was applied for 10 min following by 3 5-min PBS washes. Tissue was stored in 1% NaN3 in PBS at 4 °C prior to mounting.
Data Availability.
All data necessary to support the paper's conclusions are present in the main text and SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Science Foundation (IOS-1707261), the National Institutes of Health (EY022577, MH063912), and the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915974116/-/DCSupplemental.
References
- 1.Voziyanov Y., Pathania S., Jayaram M., A general model for site-specific recombination by the integrase family recombinases. Nucleic Acids Res. 27, 930–941 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoess R. H., Ziese M., Sternberg N. A. T., P1 site-specific recombination: Nucleotide sequence of the recombining sites. Proc. Natl. Acad. Sci. U.S.A. 79, 3398–3402 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee G., Saito I., Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216, 55–65 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Albert H., Dale E. C., Lee E., Ow D. W., Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 7, 649–659 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Arakil K., Araki M., Yamamura K., Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 25, 868–872 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Senecoff J. F., Rossmeissl P. J., Cox M. M., DNA recognition by the FLP recombinase of the yeast 2 μ plasmid. A mutational analysis of the FLP binding site. J. Mol. Biol. 201, 405–421 (1988). [DOI] [PubMed] [Google Scholar]
- 7.Schlake T., Bode J., Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33, 12746–12751 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi H., et al. , A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013 (2011). Correction in: Neuron72, 1091 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling C., et al. , Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J. Virol. 89, 952–961 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini J. A., et al. , Sequence requirements for stable binding and function of Rep68 on the adeno-associated virus type 2 inverted terminal repeats. J. Virol. 68, 7448–7457 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang C. C., et al. , Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 71, 9231–9247 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., et al. , Concatamerization of adeno-associated virus circular genomes occurs through intermolecular recombination. J. Virol. 73, 9468–9477 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhlman S. J., Huang Z. J., High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One 3, e2005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnütgen F., et al. , A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 21, 562–565 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Atasoy D., Aponte Y., Su H. H., Sternson S. M., A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 28, 7025–7030 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z. J., Taniguchi H., He M., Kuhlman S., Cre-dependent adeno-associated virus preparation and delivery for labeling neurons in the mouse brain. Cold Spring Harb. Protoc. 2014, 190–194 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Wall N. R., Wickersham I. R., Cetin A., De La Parra M., Callaway E. M., Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proc. Natl. Acad. Sci. U.S.A. 107, 21848–21853 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seidler B., et al. , A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc. Natl. Acad. Sci. U.S.A. 105, 10137–10142 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rong L., et al. , Characterization of determinants for envelope binding and infection in tva, the subgroup A avian sarcoma and leukosis virus receptor. J. Virol. 72, 4552–4559 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamichi K., et al. , Dissecting local circuits: Parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron 80, 1232–1245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohara K., et al. , Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 17, 269–279 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung H., et al. , Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 10, 314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jungmann A., Leuchs B., Rommelaere J., Katus H. A., Müller O. J., Protocol for efficient generation and characterization of adeno-associated viral vectors. Hum. Gene Ther. Methods 28, 235–246 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Dillingham M. S., Kowalczykowski S. C., RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bi X., Liu L. F., DNA rearrangement mediated by inverted repeats. Proc. Natl. Acad. Sci. U.S.A. 93, 819–823 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovett S. T., Hurley R. L., Sutera V. A. Jr, Aubuchon R. H., Lebedeva M. A., Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics 160, 851–859 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Capelli P., Pivetta C., Soledad Esposito M., Arber S., Locomotor speed control circuits in the caudal brainstem. Nature 551, 373–377 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Penzo M. A., et al. , The paraventricular thalamus controls a central amygdala fear circuit. Nature 519, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., et al. , Hypothalamic circuits for predation and evasion. Neuron 97, 911–924.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Guo C., Yang W., Lobe C. G., A Cre recombinase transgene with mosaic, widespread tamoxifen-inducible action. Genesis 32, 8–18 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Chen Y., Narendra U., Iype L. E., Cox M. M., Rice P. A., Crystal structure of a Flp recombinase-Holliday junction complex: Assembly of an active oligomer by helix swapping. Mol. Cell 6, 885–897 (2000). [PubMed] [Google Scholar]
- 33.Raymond C. S., Soriano P., High-efficiency FLP and phiC31 site-specific recombination in Mammalian cells. PLoS One 2, e162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bessen J. L., et al. , High-resolution specificity profiling and off-target prediction for site-specific DNA recombinases. Nat. Commun. 10, 1937 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Branda C. S., Dymecki S. M., Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Grimm D., Kay M. A., Kleinschmidt J. A., Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 7, 839–850 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary to support the paper's conclusions are present in the main text and SI Appendix.