Abstract
STUDY QUESTION
Is the combined use of fluorescence lifetime imaging microscopy (FLIM)-based metabolic imaging and second harmonic generation (SHG) spindle imaging a feasible and safe approach for noninvasive embryo assessment?
SUMMARY ANSWER
Metabolic imaging can sensitively detect meaningful metabolic changes in embryos, SHG produces high-quality images of spindles and the methods do not significantly impair embryo viability.
WHAT IS KNOWN ALREADY
Proper metabolism is essential for embryo viability. Metabolic imaging is a well-tested method for measuring metabolism of cells and tissues, but it is unclear if it is sensitive enough and safe enough for use in embryo assessment.
STUDY DESIGN, SIZE, DURATION
This study consisted of time-course experiments and control versus treatment experiments. We monitored the metabolism of 25 mouse oocytes with a noninvasive metabolic imaging system while exposing them to oxamate (cytoplasmic lactate dehydrogenase inhibitor) and rotenone (mitochondrial oxidative phosphorylation inhibitor) in series. Mouse embryos (n = 39) were measured every 2 h from the one-cell stage to blastocyst in order to characterize metabolic changes occurring during pre-implantation development. To assess the safety of FLIM illumination, n = 144 illuminated embryos were implanted into n = 12 mice, and n = 108 nonilluminated embryos were implanted into n = 9 mice.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Experiments were performed in mouse embryos and oocytes. Samples were monitored with noninvasive, FLIM-based metabolic imaging of nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) autofluorescence. Between NADH cytoplasm, NADH mitochondria and FAD mitochondria, a single metabolic measurement produces up to 12 quantitative parameters for characterizing the metabolic state of an embryo. For safety experiments, live birth rates and pup weights (mean ± SEM) were used as endpoints. For all test conditions, the level of significance was set at P < 0.05.
MAIN RESULTS AND THE ROLE OF CHANCE
Measured FLIM parameters were highly sensitive to metabolic changes due to both metabolic perturbations and embryo development. For oocytes, metabolic parameter values were compared before and after exposure to oxamate and rotenone. The metabolic measurements provided a basis for complete separation of the data sets. For embryos, metabolic parameter values were compared between the first division and morula stages, morula and blastocyst and first division and blastocyst. The metabolic measurements again completely separated the data sets. Exposure of embryos to excessive illumination dosages (24 measurements) had no significant effect on live birth rate (5.1 ± 0.94 pups/mouse for illuminated group; 5.7 ± 1.74 pups/mouse for control group) or pup weights (1.88 ± 0.10 g for illuminated group; 1.89 ± 0.11 g for control group).
LIMITATIONS, REASONS FOR CAUTION
The study was performed using a mouse model, so conclusions concerning sensitivity and safety may not generalize to human embryos. A limitation of the live birth data is also that although cages were routinely monitored, we could not preclude that some runt pups may have been eaten.
WIDER IMPLICATIONS OF THE FINDINGS
Promising proof-of-concept results demonstrate that FLIM with SHG provide detailed biological information that may be valuable for the assessment of embryo and oocyte quality. Live birth experiments support the method’s safety, arguing for further studies of the clinical utility of these techniques.
STUDY FUNDING/COMPETING INTEREST(S)
Supported by the Blavatnik Biomedical Accelerator Grant at Harvard University and by the Harvard Catalyst/The Harvard Clinical and Translational Science Center (National Institutes of Health Award UL1 TR001102), by NSF grants DMR-0820484 and PFI-TT-1827309 and by NIH grant R01HD092550-01. T.S. was supported by a National Science Foundation Postdoctoral Research Fellowship in Biology grant (1308878). S.F. and S.A. were supported by NSF MRSEC DMR-1420382. Becker and Hickl GmbH sponsored the research with the loaning of equipment for FLIM. T.S. and D.N. are cofounders and shareholders of LuminOva, Inc., and co-hold patents (US20150346100A1 and US20170039415A1) for metabolic imaging methods. D.S. is on the scientific advisory board for Cooper Surgical and has stock options with LuminOva, Inc.
Keywords: metabolism, mitochondria, embryo assessment, oocyte, noninvasive, spindle, fluorescence, nicotinamide adenine dinucleotide, flavin adenine dinucleotide
Introduction
The invention of precise, noninvasive methods of assessing oocyte and embryo quality has long remained a critical goal in ART (Gardner et al., 2015; Sanchez et al., 2017). Forty years after the first successful IVF procedures were performed, morphological assessment remains the primary method of evaluation, despite its well-known limitations (Wong et al., 2014). Morphological features, however, have no clear connection to the underlying biochemical factors that are essential for viability, such as metabolic function (Gardner et al., 2000, 2001; Leese, 2002; Babayev and Seli, 2015) and genetic integrity (Baart et al., 2006; Vanneste et al., 2009; Niakan et al., 2012).
Innovations in recent years have offered some promise, but failed to solve the problem of low overall success rates in IVF. Time-lapse imaging systems have been advanced in the hopes that embryo growth dynamics may reflect viability more accurately than single observations. Like the standard morphological assessment though, these systems can only sample gross morphological features, and clinical trials have not demonstrated efficacy in increasing success rates (Gardner and Sakkas, 2013; Armstrong et al., 2019).
The main advance has been the development of pre-implantation genetic testing for aneuploidy (PGT-A). These techniques have demonstrated some significant increase in success rates (Forman et al., 2013; Scott et al., 2013a;), mostly for older patients (Munne et al., 2019; Murphy et al., 2018). However, PGT is expensive and invasive, and concerns persist over accuracy, reproducibility between clinics and the role of mosaicism (Capalbo et al., 2017; Vega and Jindal, 2017). Perhaps most importantly, it conveys no information about metabolic competence. Among euploid embryos, 40% still fail (Scott et al., 2013a) due to nongenetic causes, including metabolic dysfunction.
Some studies have measured mitochondrial DNA copy number in biopsied cells in the hopes of gleaning some insight into metabolic health (Diez-Juan et al., 2015; Fragouli et al., 2016). Studies using this technique, however, have neither reported consistent results nor demonstrated predictive values (Diez-Juan et al., 2015; Treff et al., 2017; Cecchino and Garcia-Velasco, 2019). Additionally, high variation in the data remains a barrier for this technique (Treff et al., 2017).
It has long been known that embryo metabolism relates to viability (Renard et al., 1980; Van Blerkom et al., 1995; Gardner et al., 2000, 2011). Here, we investigate the potential utility of fluorescence lifetime imaging microscopy (FLIM)-based metabolic imaging of the electron carriers nicotinamide adenine dinucleotide (NADH) and flavin adenine dinucleotide (FAD) (Becker, 2012; Heikal, 2012) with second harmonic generation (SHG)-based spindle imaging (Hsieh et al., 2008; Yu et al., 2014). This approach has several benefits. First, NADH and FAD fluorescence can serve as a basis for imaging the spatial distribution of mitochondria within cells, as NADH is highly concentrated in the mitochondria relative to the cytoplasm (Stein and Imai, 2012) and nuclei (Cinco et al., 2016), and FAD is almost entirely localized to the mitochondria (Dumollard et al., 2004). Previous studies have shown strong co-localization of FAD fluorescence with mitochondrial fluorescence in mouse oocytes using mitochondrial dyes (Dumollard et al., 2004), and other studies showed incomplete overlap between NADH fluorescence with mitochondria in other cell types (Mujat et al., 2008; Tucker et al., 2016). Abnormal mitochondrial morphology has been associated with reduced developmental competence (Nagai et al., 2006). Hence, FLIM intensity images, alone, may have the potential to assist in oocyte/embryo screening. Fluorescence imaging of cytoplasmic NADH also contains some signal from NADPH, as their fluorescence spectra are almost identical (Ghukasyan and Heikal, 2014); however, NADH contributes the majority of the fluorescence signal in many cell types, as its concentration is typically several fold higher than that of NADPH (Klaidman et al., 1995). Therefore, for simplicity we adopt the convention of referring to this fluorescence signal as ‘NADH’ signal.
Beyond imaging, FLIM provides quantitative information on the local environment of NADH and FAD molecules, which is reflective of the biochemical processes they are engaged in (Ghukasyan and Heikal, 2014). FLIM measures not only the fluorescence intensity of a sample but also the rates at which fluorophores decay from their excited state (Becker, 2012). These rates, or ‘fluorescence lifetimes’, are affected by the microenvironment of the fluorophores and thus have been used to probe various intracellular processes such as protein binding via FLIM-FRET (Förster resonance energy transfer) (Yoo et al., 2018), viscosity (Parker et al., 2010) and temperature (Okabe et al., 2012). Because NADH and FAD are integral to cellular respiration, their microenvironment is affected by changes in metabolic function, and these changes are reflected in their FLIM signatures (Ghukasyan and Heikal, 2014). Thus, FLIM measurement of NADH and FAD is commonly referred to as metabolic imaging. Furthermore, metabolic imaging has the potential to probe metabolic processes with subcellular resolution if cytoplasmic and mitochondrial regions are analyzed separately. NADH in the cytoplasm is involved in a variety of pathways, including glycolysis and fermentation, while NADH and FAD in mitochondria are primarily associated with the tricarboxylic acid cycle and the electron transport chain (Berg et al., 2007). In total, a single metabolic measurement can generate up to 12 quantitative metrics, providing a detailed profile of egg/embryo metabolic state.
FLIM can be performed via one-photon or two-photon excitation of fluorophores: the former typically uses pulsed photodiode lasers (Becker, 2017), and the latter typically uses mode-locked lasers, such as a Ti-sapphire (Smith, 1970), which deliver ultra-short light pulses of 100–150 fs in duration. In two-photon excitation, fluorophores absorb two low-energy (long wavelength) photons simultaneously. This method has several benefits, including efficiency for deep-tissue imaging, intrinsic confocal imaging and lower phototoxicity (Potter, 1996).
The sensitivity of a metabolic assay can be investigated by probing known changes in metabolic state using chemical perturbations. Oxamate inhibits lactate dehydrogenase, which reduces NAD+ to NADH during the conversion of lactate into pyruvate (Dumollard et al., 2007). Rotenone inhibits the transfer of electrons from Complex I to ubiquinone, reducing the activity of the electron transport chain (Staniszewski et al., 2013). Embryo metabolism is known to undergo distinct shifts over the course of early development (embryonic days E1–E5) (Rieger, 1992; Gardner, 1998; Chason et al., 2011), including a gradual increase in pyruvate and glucose metabolism during the cleavage stage (E1–E3) (Gardner and Leese, 1986) and the commencement of aerobic glycolysis around compaction (morula) and blastocyst formation (Gardner and Harvey, 2015). Thus, comparing developmental time points can serve as an additional standard for testing assay sensitivity to detecting known metabolic differences.
In conjunction with FLIM measurements, we also simultaneously acquire high-fidelity images of meiotic spindles in oocyte and mitotic spindles via SHG (Hsieh et al., 2008; Campagnola, 2011; Yu et al., 2014). SHG is a nonlinear effect, in which photons are up-converted to a single scattered photon of exactly twice the frequency. Thus, it is commonly generated using the same kind of ultra-short pulsed light sources as two-photon fluorescence. Significant SHG signals are only produced by highly ordered, non-centrosymmetric materials. In biology, biofilament bundles, such as collagen and microtubules in spindles, are among the few materials that produce an SHG signal (Campagnola and Loew, 2003). This provides an endogenous source of contrast that allows for high-quality, noninvasive imaging of spindles (Hsieh et al., 2008; Campagnola, 2011; Yu et al., 2014). As chromosome segregation errors are associated with spindle defects (Battaglia et al., 1996), visualizing spindle morphology could also have clinical relevance.
As these methods probe physiological factors that are known to be important for embryo viability, they could potentially serve as a basis for assessing embryo quality and identifying embryos with the highest chance of success. Furthermore, they have the potential to serve as powerful new research tools to produce new insights into early embryo biology. We have already shown that FLIM parameters exhibit strong differentiation between a mitochondria protein [caseinolytic peptidase P (Clpp)]-knockout versus wild type, and old versus young mouse oocytes (Sanchez et al., 2018). FLIM measurements of mouse embryos at the compaction stage have also been reported to correlate with blastocyst development (Ma et al., 2018).
With any noninvasive, light-based assay, an assessment of safety must be performed before any clinical application is possible, as cell illumination has the capacity to cause photo damage (Masters and So, 2008). We present evidence supporting the technique’s safety for use on embryos: we implanted illuminated and nonilluminated embryos into surrogate female mice and measured both live birth rates and pup weights as indicators of possible embryo damage.
Materials and Methods
Imaging system and FLIM measurements
FLIM measurements were performed on a Nikon TE300 microscope with either a Nikon 20× objective (0.75 NA) or a Nikon 40× objective (1.25 NA), with a galvanometer scanner and a TCSPC module (SPC-150, Becker and Hickl GmbH, Germany). Two-photon excitation was supplied via a Ti:Sapphire pulsed laser (M Squared Lasers, UK) with an 80-MHz repetition rate and ~150-fs pulse width. A 750-nm illumination (3–60 mW) and a 447/60-nm bandpass filter (BrightLine from Semrock, USA) were used for imaging NADH fluorescence. For imaging FAD fluorescence, 890- or 845-nm illumination (12–80 mW) and a 550/88-nm bandpass filter were used. Illumination intensities were calibrated by measuring power output through the objective with a handheld power meter (Newport, USA). Fluorescence was detected in the epi direction with a hybrid detector (HPM-100-40, Becker and Hickl GmbH, Germany). SHG was detected simultaneous with FAD imaging by a single-photon counting detector (PMC-150, Becker-Hickl GmbH, Germany), placed in the forward direction, with combined 650 short-pass and 440/20-nm bandpass filters (BrightLine from Semrock, USA). Scans were acquired for 5–60 s, with either one or three separate Z-planes for metabolic measurements. A customized motorized stage (using CONEX TRA12CC actuators, Newport, USA) was used for multi-dimensional acquisition. Bright-field was performed using the same 20× and 40× objectives and an Amscope (USA) MU300 camera. Acquisitions were performed using custom LabVIEW software.
Embryo and oocyte imaging, metabolic perturbations and live birth experiments
Cryopreserved oocytes and one-cell mouse embryos (Embryotech, Haverhill, MA, USA), from crosses between B6C3F1 females and B6D2F1 males, were thawed and transferred to pre-equilibrated dishes for either culture or perturbation experiments. Research was conducted under a protocol approved by Harvard’s institution’s Animal Care and Use Committee (IACUC), which has a Letter of Assurance (File No. A3593-01) from the National Institutes of Health Office of Laboratory Animal Welfare. Embryos were imaged in either custom-fabricated microwell dishes with glass coverslip bottoms, commercial plastic microwell dishes (Primo Vision from Vitrolife AB, Sweden) or commercial glass bottom dishes (MatTek P35G-0.170-14-C, USA) in KSOM media (MR-121-D, MilliporeSigma, USA) in an on-stage incubator (10918 from ibidi GmbH, Germany), at 37°C, 5% CO2 and 5% O2. Prior to imaging, embryos were stored in a tabletop commercial incubator (MCO5MPA from Panasonic, Japan).
For MitoTracker experiments, embryos were incubated in advanced KSOM (aKSOM MR-101-D from Millipore-Sigma, USA) with 5 nM MitoTracker Red CMXRos (M7512 from Thermo Fisher, USA) for 20 min, then transferred directly to an aKSOM droplet in a glass-bottomed dish for imaging.
For metabolic perturbation experiments, oocytes were imaged in custom glass-bottomed microwell dishes to prevent movement of oocytes caused by pipetting in drugs. Oxamate and rotenone were pipetted in during acquisition to final concentrations of 10 mM and 1 μM, respectively.
For live birth safety experiments, illuminated and nonilluminated embryos were transported at 37°C in pre-equilibrated KSOM to the Harvard Genome Modification facility at the blastocyst stage, where they were transferred into pseudo-pregnant CD1 mice as previously described (Gardner and Sakkas, 1993). Embryos were transferred in groups of 12, with six embryos in each uterine horn. Pregnant mice were then monitored daily by the facility, and at birth, pups were counted and weighed.
Data analyses
We used supervised machine-learning-driven segmentation software (Sonka et al., 2015) to classify pixels in intensity images into three groups: mitochondrial and cytoplasmic NADH and mitochondrial FAD [the concentration of FAD in the cytoplasm is extremely low (Dumollard et al., 2004), so photons from that potential group were not analyzed]. The classification utilized a random forest algorithm (Breiman, 2001), which considered intensity, texture and edge properties. The algorithm was trained on representative embryo NADH and FAD intensity images, wherein the user manually draws over the regions to specify their classification (e.g. mitochondrial, cytoplasmic, background). For each embryo segment, the photon arrival time histogram was modeled as a bi-exponential model decay:
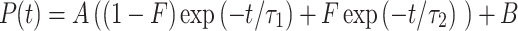 |
Here, A is a normalization factor, B is the background level from factors such as room light (fits typically gave B/A ~ 0.005, indicating low background levels), 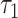 is the short lifetime,
is the short lifetime, 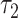 is the long lifetime and F is the fraction of molecule with long lifetime (the fraction engaged with enzymes for NADH and unengaged for FAD). This function was convolved with a measured instrument response function to model the experimental data, and least-square fitting yielded quantitative values for these three fit parameters. An additional parameter, I, the fluorescence intensity, was calculated for each region by dividing the total number of photons detected in the region by the area of the region. Thus, between NADH cytoplasm, NADH mitochondria and FAD mitochondria, a single metabolic measurement produces up to 12 quantitative parameters for characterizing the metabolic state of an embryo. Alternatively, if mitochondria and cytoplasm are not segmented and analyzed separately, then eight parameters can be measured: four from NADH and four from FAD.
is the long lifetime and F is the fraction of molecule with long lifetime (the fraction engaged with enzymes for NADH and unengaged for FAD). This function was convolved with a measured instrument response function to model the experimental data, and least-square fitting yielded quantitative values for these three fit parameters. An additional parameter, I, the fluorescence intensity, was calculated for each region by dividing the total number of photons detected in the region by the area of the region. Thus, between NADH cytoplasm, NADH mitochondria and FAD mitochondria, a single metabolic measurement produces up to 12 quantitative parameters for characterizing the metabolic state of an embryo. Alternatively, if mitochondria and cytoplasm are not segmented and analyzed separately, then eight parameters can be measured: four from NADH and four from FAD.
To evaluate the ability of FLIM to distinguish between different metabolic states, the three FLIM parameters with the largest separation between data groups were considered together. As a test condition, a separating plane was fit between data groups using a support vector machine algorithm (Cristianini and Shawe-Taylor, 2000). T tests on individual metabolic parameters were also performed to detect changes, and P < 0.05 was taken to be statistically significant.
Results
Mitochondria and spindle visualization and FLIM quantification
To investigate the potential utility of metabolic and SHG imaging for revealing mammalian egg and embryo physiology, we first explored the purely morphological data these techniques provide. Standard bright-field microscopy of mouse blastocysts produces a clear view of the overall morphology of the embryo, but gives limited information on the cellular-scale (Fig. 1A, left). In contrast, imaging NADH autofluorescence in mouse blastocysts provides a detailed view of subcellular structures, such as nuclei (Fig. 1A, center image, indicated by white arrow). Two-photon imaging provides intrinsic optical sectioning, with the 1.25 NA objective used here resulting in a theoretical lateral (XY) resolution of 230 nm and an axial (Z) resolution of 536 nm. We consecutively imaged Z planes two microns apart (Supplementary Movies S1 and S2) and combined the resulting data, enabling us to create 3D reconstructions of embryo morphology (Fig. 1A, right).
Figure 1.
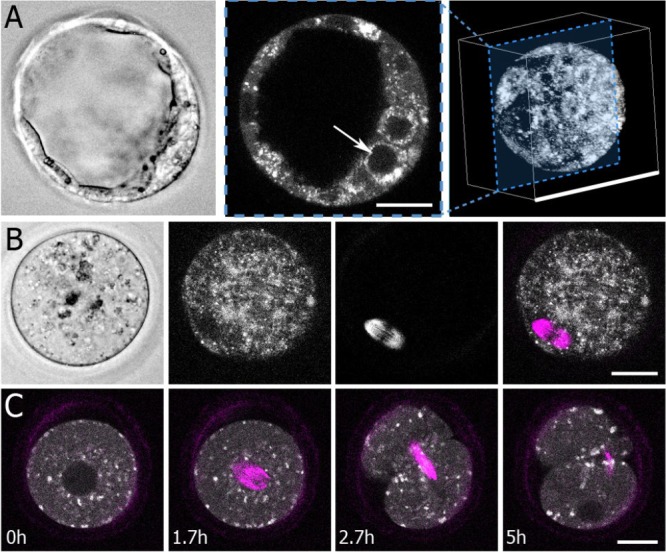
Morphological features imaged with fluorescence lifetime imaging microscopy and second harmonic generation imaging. (A) Left: a standard bright-field image of a mouse blastocyst shows gross morphological features. Middle: fluorescence lifetime imaging microscopy (FLIM) measurements of nicotinamide adenine dinucleotide (NADH) generate intensity images that visualize subcellular structures, including nuclei (white arrow, 25-μm scale bar). Right: 3D reconstruction from multiple focal planes (100-μm scale bar). (B) Left: Bright-field image of an oocyte. Second to left: a flavin adenine dinucleotide (FAD) FLIM image of the same oocyte. Second to right: a second harmonic generation (SHG) image of the same oocyte, which clearly shows the oocyte’s spindle. Right: Overlay image of SHG (magenta) and FAD (grey scale) (25-μm scale bar). (C) Time lapse FLIM (NADH) and spindle measurements capture key embryo dynamics, such as cell division (25-μm scale bar).
Similarly, bright-field images reveal the morphology of oocytes, but do not provide information on specific subcellular structures (Fig. 1B, left), while FAD, which is almost exclusively localized to mitochondria, shows a complex mitochondrial distribution inside oocytes (Fig. 1B, second to left). The same laser illumination that excites FAD autofluorescence can also nonlinearly interact with, and scatter from, the sample, enabling simultaneous imaging with SHG. The meiotic spindle and the zona pellucida are the only subcellular structures in mammalian oocytes that produce SHG, with the spindle generating by far the largest signal. This provides excellent intrinsic contrast (Fig. 1B, second panel from right) for spindle imaging. The combination of FLIM imaging and SHG allows detailed visualization of the internal structure of oocytes (Fig. 1B, right). Time lapse FLIM and SHG measurements provides information on dynamics. As the one-cell embryo proceeds to divide, the spindle assembles in the middle of the cell (Fig. 1C, 1.7 h) where the nucleus previously was, then elongates and thins during cytokinesis (Fig. 1C, 2.7 h). At later times, the spindle midbody is clearly visible, as the two-cell embryo’s nuclei form (Fig. 1C, 5 h) (Supplementary Movie S3).
NADH and FAD are highly enriched in mitochondria, arguing that bright regions and puncta within the cell contain high concentrations of mitochondria. To investigate this further, we used MitoTracker to specifically label the mitochondria and imaged one- and two-cell embryos with high resolution (40× 1.25-NA objective, 20-s integration time). We imaged MitoTracker (4 mW illumination power), then either NADH (7 mW) or FAD (30 mW) immediately afterward (27 s delayed), and found strong colocalization between the MitoTracker signal and the bright regions of both the NADH and the FAD intensity images (Fig. 2A and B). Machine-learning segmentations of the mitochondria in the NADH images had a 74.0 ± 0.65% (SEM) overlap with the mitochondria in the MitoTracker images. Segmentations based on FAD images had a 77.5 ± 3.6% (SEM) with mitochondria in the MitoTracker images. Thus, NADH/FAD autofluorescence imaging provides information on the localization, morphology and dynamics of mitochondria.
Figure 2.
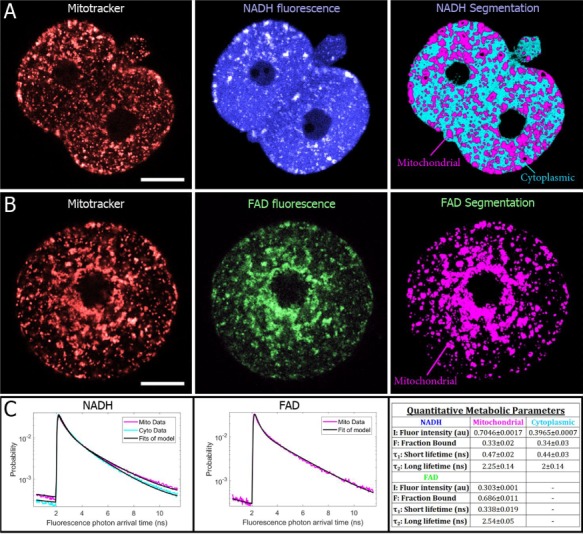
FLIM allows for mitochondrial imaging and separate segmentation of mitochondrial and cytoplasmic signals. (A) MitoTracker images (left) of a two-cell embryo shows co-localization with brighter regions of NADH (center) (25-μm scale bars). A machine-learning-based algorithm performed on autofluorescence images successfully segments mitochondria and cytoplasm (right). (B) MitoTracker and FAD autofluoresence images of a one-cell embryo also show co-localization. For FAD images, only mitochondrial regions were segmented. (C) For each region, all photon arrival times were binned into a separate histogram, which were normalized to produce a probability distribution, which represents the probability of being in an excited state after excitation. Fitting these curves to a two-exponential decay model produces a total of 12 parameters for characterizing the metabolic state of an embryo or oocyte (right).
Beyond mitochondrial visualization, FLIM provides a multi-parametric, quantitative characterization of metabolic state. We were able to probe metabolic processes with subcellular resolution by performing high-resolution imaging. This allowed us to separately segment mitochondrial and cytoplasmic regions, generating distinct arrival time histograms for each segment. Performing this segmentation on NADH and FAD intensity images (Fig. 2A and B, right panels), and fitting resultant histograms for NADH cytoplasm, NADH mitochondrial and FAD mitochondrial regions (Fig. 2C, left and middle panels), we obtained a total of 12 quantitative parameters (Fig. 2C, right panel). The arrival time histograms each contained ~100 000–200 000 photons, allowing the parameters to be measured with high precision: the 95% CI for every parameter was less than 10% of its mean.
Metabolic imaging enables in situ measurements of mitochondrial metabolism
To determine the extent to which FLIM parameters can be used to determine biologically relevant shifts in oocyte/embryo metabolism, we first investigated the impact of subjecting oocytes to the metabolic inhibitors, oxamate and rotenone. To obtain good spatial and temporal resolution, we performed these experiments in custom-fabricated, glass-bottomed microwell dishes with only one z-plane, 3 mW NADH, 12.5 mW FAD, 60 s integration time and a 40× objective.
Exposing oocytes to oxamate, a lactate dehydrogenase inhibitor, resulted in a readily visible decrease in cytosolic NADH intensity (Fig. 3A). Subsequent addition of rotenone caused a pronounced increase in NADH intensity and a decrease in FAD intensity (hence an increase in the redox ratio, NADH/FAD) in mitochondria. SHG images (Fig. 3B, overlaid in magenta) revealed that oxamate did not disrupt the meiotic spindle, but exposure to rotenone caused it to disappear (Supplementary Movie S4).
Figure 3.
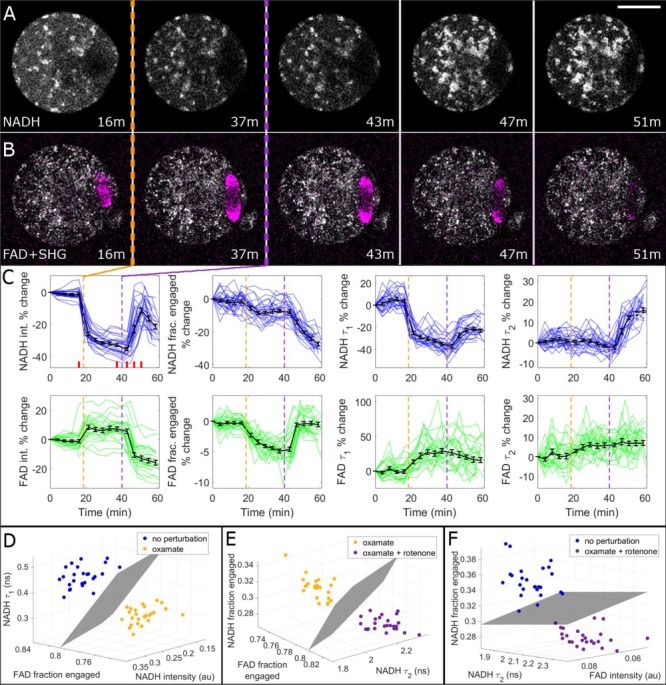
Metabolic and SHG imaging of oocytes detects effects of metabolic perturbations. Oocytes were exposed to 10 mM oxamate and then to 1 μM rotenone. (A) Oxamate exposure (indicated with a dashed orange line) caused a visible drop in cytosolic NADH FLIM intensity, whereas rotenone exposure (purple line) caused an increase in mitochondrial NADH intensity. Bar is 30 μm. (B) For the same oocyte, oxamate slightly increased FAD intensity, while rotenone decreased it. Simultaneous SHG imaging, overlaid in magenta, revealed that spindles were not disrupted by oxamate, but were disintegrated by rotenone. (C) Percentage-change time courses for all eight metabolic parameters. Colored traces represent individual oocyte trajectories (n = 25), and average curves are shown in black with SE bars. Vertical dashed lines indicate oxamate (orange) and rotenone (purple) exposures. Red bars in the NADH intensity panel correspond to the time stamps of the images shown in (A) and (B). (D) FLIM parameters were compared before and after oxamate exposure, and the three FLIM parameters with the largest separation were plotted on 3D plots, yielding complete separation. This comparison was also performed for (E) oxamate versus oxamate + rotenone, and (F) no perturbation versus oxamate + rotenone.
Averaging data from the 25 oocytes (Fig. 3C) revealed a significant change after oxamate exposure in six FLIM parameters: NADH intensity decreased by 33 ± 2% (P = 1.7e−9); NADH fraction engaged decreased by 7 ± 2% (P = 1.5e−6); NADH 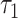 decreased by 35 ± 2% (P = 1.4e−16); FAD fraction engaged decreased by 4.9 ± 0.6% (P = 1.8e−11); FAD
decreased by 35 ± 2% (P = 1.4e−16); FAD fraction engaged decreased by 4.9 ± 0.6% (P = 1.8e−11); FAD 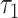 increased by 29 ± 7% (P = 1.7e−6); and FAD
increased by 29 ± 7% (P = 1.7e−6); and FAD 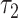 increased by 6 ± 2% (P = 3e−4). Oxamate is known to change cytosolic metabolic activity, and we detected highly significant changes in our measured FLIM parameters after exposure to the chemical. Thus, the results support our hypothesis that FLIM parameters can measure metabolic state and distinguish different states.
increased by 6 ± 2% (P = 3e−4). Oxamate is known to change cytosolic metabolic activity, and we detected highly significant changes in our measured FLIM parameters after exposure to the chemical. Thus, the results support our hypothesis that FLIM parameters can measure metabolic state and distinguish different states.
Subsequent exposure of oocytes to rotenone, an inhibitor of the electron transport chain, also caused a significant, but distinct, set of parameter changes: NADH intensity increased by 32 ± 3% (P = 4e−6); NADH fraction engaged decreased by 14 ± 1.4% (P = 9e−20); NADH 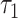 increased by 19 ± 4% (P = 1.6e−6); NADH
increased by 19 ± 4% (P = 1.6e−6); NADH 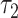 increased by 14 ± 2% (P = 3e−14); FAD intensity decreased by 19 ± 2% (P = 7e−8); and FAD fraction engaged decreased by 4.7 ± 0.6% (P = 7e−11). Rotenone is known to change mitochondrial metabolic activity, and we detected highly significant changes in our measured FLIM parameters after exposure to the chemical. Thus, the results again support our hypothesis that FLIM parameters can measure metabolic state and distinguish different states.
increased by 14 ± 2% (P = 3e−14); FAD intensity decreased by 19 ± 2% (P = 7e−8); and FAD fraction engaged decreased by 4.7 ± 0.6% (P = 7e−11). Rotenone is known to change mitochondrial metabolic activity, and we detected highly significant changes in our measured FLIM parameters after exposure to the chemical. Thus, the results again support our hypothesis that FLIM parameters can measure metabolic state and distinguish different states.
To further characterize the ability of metabolic imaging to distinguish between different metabolic states, we compared discrete time points by plotting the three metabolic parameters with the highest degree of separation on a 3D plot (Fig. 3D–F). Support vector machine planes were fit to the data as a test condition, and this comparison was performed for no perturbation versus oxamate (Fig. 3D), oxamate versus oxamate + rotenone (Fig. 3E) and no perturbation versus oxamate + rotenone (Fig. 3F). In all cases, the data sets were completely separated by FLIM.
These oxamate and rotenone experiments indicate that metabolic imaging can be used to readily resolve changes in metabolism induced by poisons. We next sought to determine if metabolic imaging is sensitive enough to also measure the changes in embryo metabolism that take place during pre-implantation development. For these experiments, we used multi-well plastic dishes with five embryos per well. Metabolic images were taken at three different Z planes every 2 h, over the course of 70 h, using 30 mW NADH, 50 mW FAD and 60 s integration time for each plane. Individual embryos were tracked from the one-cell stage to blastocyst (Fig. 4A, Supplementary Movie S5). To obtain a strong signal, photons from all three planes were binned into one histogram for each time point. We calculated metabolic parameters for each embryo, which underwent highly significant changes over the course of pre-implantation embryo development (Fig. 4B). Plotting data from 39 different embryos shows that changes over development were highly stereotyped (Fig. 4C). To further investigate the repeatability of these measurements, we split the data into two batches (with n = 20 and n = 19 embryos) from separately acquired time courses. The metabolic parameters from two batches were highly similar (Supplementary Fig. S1), arguing that these measurements are robust and repeatable.
Figure 4.
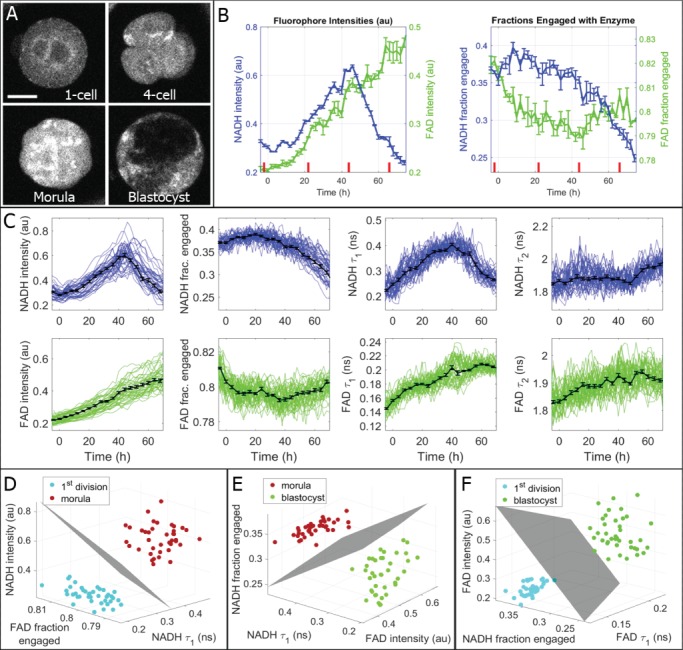
High time-resolution measurements of metabolic state during mouse embryo development. (A) Embryos were imaged together in 9-microwell dishes and individually tracked and analyzed to generate time plots for all FLIM parameters. Bar is 30 μm. (B) Time plots for NADH and FAD intensities and fractions engaged for the individual embryo shown in (A). Distinct changes are evident in multiple parameters, especially around the onset of blastocyst formation (~44 h after the 1st division, which is represented here as t = 0 h). Corresponding time points for the four images are displayed with red dashes on the plots. (C) The observed trends were robust and reproducible among all healthy embryos. These plots display individual embryo trajectories as thin-colored lines (n = 39), which were synchronized by 1st division. Averaged metabolic curves from all 39 embryos are displayed as thick black lines with SE bars. (D) FLIM parameters were compared between the 1st division and morula stages, and the three FLIM parameters with the largest separation were plotted as 3D plots, yielding complete separation. This comparison was also performed for (E) compaction versus expanded blastocyst and (F) 1st division versus expanded blastocyst.
Metabolic parameters changed monotonically between the period shortly after the first division (t = 0 in Fig. 4B and C) and compaction (44 h), and different monotonic changes were observed between compaction and expanded blastocyst stage. Comparing the one-cell embryo to morula revealed highly significant changes in four FLIM parameters (each with P < 10−6): NADH intensity, NADH 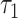 , FAD intensity and FAD
, FAD intensity and FAD 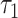 . Comparing morula to blastocyst revealed highly significant changes in four FLIM parameters (each with P < 0.002): NADH intensity, NADH fraction bound, NADH
. Comparing morula to blastocyst revealed highly significant changes in four FLIM parameters (each with P < 0.002): NADH intensity, NADH fraction bound, NADH 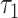 and FAD intensity. These observations reflect the known changes in metabolism, which occur over the course of embryo development. Using just three of the measured FLIM parameters, it is possible to completely separate one-cell embryos from morula (Fig. 4D), morula from blastocyst (Fig. 4E) and one-cell embryos from blastocyst (Fig. 4F). Thus, metabolic imaging is sensitive enough to measure natural changes in embryo metabolism.
and FAD intensity. These observations reflect the known changes in metabolism, which occur over the course of embryo development. Using just three of the measured FLIM parameters, it is possible to completely separate one-cell embryos from morula (Fig. 4D), morula from blastocyst (Fig. 4E) and one-cell embryos from blastocyst (Fig. 4F). Thus, metabolic imaging is sensitive enough to measure natural changes in embryo metabolism.
Live birth safety study
The ability of metabolic imaging and SHG to visualize mitochondria and spindles, and the obtained quantitative information on embryo metabolism, suggests that these techniques may be helpful in clinical embryology. These techniques are noninvasive in that they do not require exposing the embryos to any reagents; however, illumination light can damage embryos. Thus, we sought to determine if the illumination required for high signal-to-noise metabolic measurements damaged embryos. We exposed experimental groups of embryos to repeated acquisitions, every 2 h for 48 h, taking metabolic measurements with settings identical to those used in Fig. 4 (30 mW NADH, 50 mW FAD, 60 s integration time, three z-planes). Control groups of embryos were incubated in the same dishes as the experimental groups, but not exposed to any illumination. For the experimental group, n = 144 illuminated embryos were implanted into n = 12 mice, and for the control group, n = 108 nonilluminated embryos were implanted into n = 9 mice. Numbers of pups per mouse and individual pup weights were later measured when the mice gave birth.
There was no significant difference in either birth rate or pup weight between the illuminated and control groups (Fig. 5). For the birth rate measurement, the high labor cost per data point (pups/mouse) limited the feasibility of obtaining a very high sample size. As such, it was only possible to power this study to detect a 50% decrease in birth rate with an alpha of 0.05 and beta of 0.8. For pup weight, however, the sample size was much larger, such that the study was powered to detect a 7% decrease in weight. We observed an average experimental pup weight that differed by less than 1% from the control group.
Figure 5.

Live birth safety study. To evaluate the safety of FLIM illumination on embryos, we implanted illuminated and nonilluminated embryos into pseudo-pregnant mice, later measuring birth rates and pup weights as metrics for possible damage. Sample numbers indicate number of mice and number of pups, respectively. No significant differences were observed.
Discussion
Here we have explored the utility of FLIM and SHG for imaging mammalian oocytes and embryos. These techniques allow noninvasive measurements of the location and morphology of mitochondria, nuclei and spindles. We also demonstrated that FLIM of NADH and FAD is robust and sensitive enough to detect metabolic changes in individual embryos associated with metabolic poisons and the natural changes that occur during embryo development. We found that this imaging does not significantly impair live births in mice. These results argue that FLIM and SHG can be used to noninvasively obtain relevant biological information on oocytes and embryos and thus are highly promising tools for ART.
Measuring metabolism of oocytes and embryos with FLIM
Proper metabolism is crucial for embryo development, and metabolic fluxes of glycolysis and oxidative phosphorylation, determined by nutrient uptake and oxygen consumption, have been found to correlate with embryo quality (Gardner and Leese, 1987; Gardner et al., 2011; Gardner and Wale, 2013). However, previous nontargeted metabolic assessments of embryo metabolism have not been clinically useful, likely because of their complexity and lack of sensitivity (Hardarson et al., 2012; Vergouw et al., 2012). Metabolic imaging via two-photon FLIM of NADH and FAD is a highly promising alternative approach for measuring embryo metabolism for ART, because it can provide detailed metabolic information on both the cytoplasm, related to glycolysis, and mitochondria, related to oxidative phosphorylation; it is highly quantitative and robust; it is noninvasive; and it does not require the use of any foreign stains or specialized media. Here, we have also shown that metabolic imaging with FLIM can be used to characterize the changes in metabolism that occur during pre-implantation embryo development. The observed large shifts in metabolism at the morula stage are consistent with expectations from previous studies (Houghton et al., 1996; Chason et al., 2011), but these measurements also yielded unexpected results that should be further investigated. For example, blastocysts engage in aerobic glycolysis (Gardner and Harvey, 2015), which would naively be expected to result in increases of NADH concentration (e.g. as in cancer cells (Yu and Heikal, 2009)). However, we observe an ~50% decrease in NADH intensity with blastocyst formation, suggesting that further study is needed to understand this metabolic transition. This could be due to the oxidation of cytosolic NADH during the conversion of pyruvate to lactate, which occurs as the blastocyst prepares to implant (Gardner, 2015). In addition to enabling new, fundamental insights, improved metabolic measures may also allow better refinement of embryo culture media.
Imaging spindles with SHG
A microscopy system for two-photon FLIM can also be used to simultaneously acquire SHG images, without the need for additional acquisition time or photo-exposure. SHG can produce high-quality images of meiotic and mitotic spindles (Campagnola and Loew, 2003; Hsieh et al., 2008; Campagnola, 2011; Yu et al., 2014) (Fig. 1B and C). Previous work has investigated the use of polarized light microscopy for imaging the spindle, and it has been reported that there are associations between meiotic spindle morphology and oocyte quality (Battaglia et al., 1996; Zeng et al., 2007; Tomari et al., 2011, 2018; Korkmaz et al., 2015; García-Oro et al., 2017; Guo et al., 2017). Thus, SHG imaging of meiotic spindles might also provide useful information for selecting high-quality oocytes. It will be interesting for future work to determine if imaging mitotic spindles in early embryos can provide additional information to aid in identifying aneuploid embryos.
Safety
It is paramount to investigate the safety of new technologies before attempts are made to apply them clinically. While metabolic imaging with FLIM does not entail exposing oocytes and embryos to foreign material, there is the potential concern that that illumination could cause harm. Excess light dosage from conventional microscopy is detrimental (Masters and So, 2008), but such effects can be minimized or eliminated by controlling light exposure, as performed in time lapse imaging systems (Nakahara et al., 2010). Similarly, the repeated laser pulse blastocysts conventionally receive during trophectoderm biopsy procedures do not appear to reduce embryo viability (Scott et al., 2013b), although when performed in excess they may lead to damage (Bradley et al., 2017). We thus investigated the safety of illumination during FLIM imaging of embryos and found that time lapse exposure of one measurement every 2 h for 48 h did not significantly impact live birth rates or pup weights. Previous work also demonstrated that this level of illumination did not cause a measurable difference in intracellular reactive oxygen species and blastocyst formation rates (Pedro et al., 2018). It is likely that in a clinical setting, far fewer measurements of metabolism would be sufficient to aid in selection, perhaps at only one to three time points.
Strengths and limitations
Because these experiments were performed in mouse, conclusions around the sensitivity and safety of these new methods may not generalize to human. Inbred mice used here are genetically homogenous, and the level of variation between human patients is not yet known. Performing these studies in human, where tracking individual embryos is possible, is the only way to measure the direct correspondence between FLIM measurements and birth outcome. True evaluation of this imaging system as an IVF tool will require clinical studies, including measurements on discarded embryos to obtain baseline data, expanded safety assessment, observational studies and clinical trials to demonstrate efficacy for predicting live birth. A limitation of the live birth data is also that although cages were routinely monitored, early morning and during the day, we could not preclude that some runt pups may have been eaten.
Supplementary Material
Acknowledgments
The authors would like to thank Becker Hickl GmbH for contributing a single-photon counting detector and time-correlated single photon counting (TCSPC) electronics to this research. We would also like to thank the Harvard Genome Modification Facility for performing the mouse transfer experiments.
Conclusion
Our results indicate that metabolic imaging is a highly sensitive assay for measuring changes in embryo metabolic function, which is known to be essential for viability. Taken together with simultaneous spindle imaging via SHG, we believe these proof-of-concept data and initial safety studies are encouraging indications that noninvasive metabolic imaging may be helpful for clinical embryo and oocyte assessment.
Authors’ roles
T.S.—contributed to the conception and design, acquisition, analysis and interpretation of experiments and was involved in drafting and completion of the article. He has approved the final version. M.V.—contributed to the design, acquisition, analysis and interpretation of the experiments. She was involved in completion of the article and has approved the final version. S.A.—co-designed and manufactured the custom glass-bottomed microwell dishes. He was involved in completion of the article and has approved the final version. X.Y.—Performed the segmentation analysis of the MitoTracker and NADH/FAD data. He was involved in the completion of the article and approved the final version. S.F.—was involved in the completion of the article and approved the final version. D.S.—contributed to the conception and design of experiments and to the interpretation of the data, and he was involved in the drafting and completion of the article. He has approved the final version. D.N.—contributed to the conception, design and interpretation of experiments, and he was involved in the drafting and completion of the article. He has approved the final version.
Funding
Blavatnik Biomedical Accelerator Grant at Harvard University; Harvard Catalyst|The Harvard Clinical and Translational Science Center (National Institutes of Health Award UL1 TR001102); National Science Foundation (DMR-0820484 and PFI-TT-1827309); National Institutes of Health (R01HD092550-01); National Science Foundation Postdoctoral Research Fellowship in Biology (1308878 T.S.); National Science Foundation (MRSEC DMR-1420382 to S.F. and S.A.). Becker and Hickl GmbH sponsored research with the loaning of equipment for FLIM.
Conflict of interest
T.S. is a co-founder and a shareholder and officer of LuminOva, Inc., and co-holds patent US20150346100A1 pending for metabolic imaging methods for assessment of oocytes and embryos and patent US20170039415A1 issued for nonlinear imaging systems and methods for assisted reproductive technologies. D.N. is a co-founder and a shareholder and officer of LuminOva and co-holds patent US20150346100A1 pending for metabolic imaging methods for assessment of oocytes and embryos and patent US20170039415A1 issued for nonlinear imaging systems and methods for assisted reproductive technologies. D.S. is on the scientific advisory board for Cooper Surgical and has stock options with LuminOva.
References
- Armstrong S, Bhide P, Jordan V, Pacey A, Marjoribanks J, Farquhar C. Time-lapse systems for embryo incubation and assessment in assisted reproduction. Cochrane Database Syst Rev [Internet] 2019; John Wiley & Sons, Ltd Available from: http://doi.wiley.com/10.1002/14651858.CD011320.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baart EB, Martini E, van den BI, Macklon NS, Galjaard RJH, Fauser BCJM, Van OD. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Hum Reprod 2006;21:223–233. [DOI] [PubMed] [Google Scholar]
- Babayev E, Seli E. Oocyte mitochondrial function and reproduction. Curr Opin Obstet Gynecol [Internet]2015;27:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia DE, Goodwin P, Klein NA, Soules MR. Influence of maternal age on meiotic spindle assembly in oocytes from naturally cycling women. Hum Reprod [Internet]1996;11:2217–2222. [DOI] [PubMed] [Google Scholar]
- Becker W. Fluorescence lifetime imaging – techniques and applications. J Microsc [Internet]2012;247:119–136. [DOI] [PubMed] [Google Scholar]
- Becker W. The bh TCSPC Handbook. Berlin, Germany: Becker & Hickl Gmbh, 2017 [Google Scholar]
- Berg J, Tymoczko J, Stryer L. Biochemistry. Biochemistry 2007. [Google Scholar]
- Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod [Internet]1995;10:415–424. [DOI] [PubMed] [Google Scholar]
- Bradley CK, Livingstone M, Traversa MV, McArthur SJ. Impact of multiple blastocyst biopsy and vitrification-warming procedures on pregnancy outcomes. Fertil Steril [Internet]2017;108:999–1006Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forrests. Mach Learn 2001;45:1–32. [Google Scholar]
- Campagnola P. Second harmonic generation imaging microscopy: applications to diseases diagnostics. Anal Chem [Internet]2011;83:3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol [Internet]2003;21:1356–1360. [DOI] [PubMed] [Google Scholar]
- Capalbo A, Ubaldi FM, Rienzi L, Scott R, Treff N. Detecting mosaicism in trophectoderm biopsies: current challenges and future possibilities. Hum Reprod 2017;32:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchino GN, Garcia-Velasco JA. Mitochondrial DNA copy number as a predictor of embryo viability. Fertil Steril [Internet]2019;111:205–211Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Chason RJ, Csokmay J, Segars JH, DeCherney AH, Armant DR. Environmental and epigenetic effects upon preimplantation embryo metabolism and development. Trends Endocrinol Metab [Internet]2011;22:412–420Elsevier Ltd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinco R, Digman MA, Gratton E, Luderer U. Spatial characterization of bioenergetics and metabolism of primordial to preovulatory follicles in whole ex vivo murine ovary. Biol Reprod [Internet]2016;95biolreprod:116–142141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristianini N, Shawe-Taylor J. An Introduction to Support Vector Machines: And Other Kernel-based Learning Methods. New York, NY, USA: Cambridge University Press, 2000 [Google Scholar]
- Diez-Juan A, Rubio C, Marin C, Martinez S, Al-Asmar N, Riboldi M, Diaz-Gimeno P, Valbuena D, Simon C. Mitochondrial DNA content as a viability score in human euploid embryos: less is better. Fertil Steril 2015;11:373–378. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Marangos P, Fitzharris G, Swann K, Duchen M, Carroll J. Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 2004;131:3057–3067. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development [Internet]2007;134:455–465. [DOI] [PubMed] [Google Scholar]
- Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, Treff NR, Scott RT. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril [Internet]2013;100:100–107.e1Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Spath K, Alfarawati S, Kaper F, Craig A, Michel CE, Kokocinski F, Cohen J, Munne S, Wells D. Altered levels of mitochondrial DNA are associated with female age, aneuploidy, and provide an independent measure of embryonic implantation potential. Obstet Gynecol Surv 2016;71:28–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Oro S, Rey MI, Rodríguez M, Durán Á, Devesa R, Valverde D. Predictive value of spindle retardance in embryo implantation rate. J Assist Reprod Genet 2017;34:617–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK. Changes in requirements and utilization of nutrients during mammalian preimplantation embryo development and their significance in embryo culture. Theriogenology 1998;49:83–102. [DOI] [PubMed] [Google Scholar]
- Gardner DK. Lactate production by the mammalian blastocyst: manipulating the microenvironment for uterine implantation and invasion? Bioessays 2015;37:364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner DK, Harvey AJ. Blastocyst metabolism. Reprod Fertil Dev 2015;27:638–654. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil Steril 2001;76:1175–1180. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Leese HJ. Non-invasive measurement of nutrient uptake by single cultured pre-implantation mouse embryos. Hum Reprod 1986;1:25–27. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Leese HJ. Assessment of embryo viability prior to transfer by the noninvasive measurement of glucose uptake. J Exp Zool 1987;242:103–105. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Meseguer M, Rubio C, Treff NR. Diagnosis of human preimplantation embryo viability. Hum Reprod Update 2015;21:727–747. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Pool TB, Lane M. Embryo nutrition and energy metabolism and its relationship to embryo growth, differentiation, and viability. Semin Reprod Med 2000;18:205–218. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Sakkas D. Mouse embryo cleavage, metabolism and viability: role of medium composition. Hum Reprod 1993;8:288–295. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Sakkas D. Human Gametes and Preimplantation Embryos[Internet] In: Gardner DK, Sakkas D, Seli E, Wells D (eds). New York, NY: Springer New York, 2013, Available from: http://link.springer.com/10.1007/978-1-4614-6651-2 [Google Scholar]
- Gardner DK, Wale PL. Analysis of metabolism to select viable human embryos for transfer. Fertil Steril [Internet]2013;99:1062–1072Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Wale PL, Collins R, Lane M. Glucose consumption of single post-compaction human embryos is predictive of embryo sex and live birth outcome. Hum Reprod 2011;26:1981–1986. [DOI] [PubMed] [Google Scholar]
- Ghukasyan VV, Heikal AA. Natural Biomarkers for Cellular Metabolism: Biology, Techniques, and Applications [Internet] CRC Press, 2014, Available from: https://www.crcpress.com/Natural-Biomarkers-for-Cellular-Metabolism-Biology-Techniques-and-Applications/Ghukasyan-Heikal/9781466509986 [Google Scholar]
- Guo Y, Liu W, Wang Y, Pan J, Liang S, Ruan J, Teng X. Polarization microscopy imaging for the identification of unfertilized oocytes after short-term insemination. Fertil Steril [Internet]2017;108:78–83Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Ahlström A, Rogberg L, Botros L, Hillensj T, Westlander G, Sakkas D, Wikland M. Non-invasive metabolomic profiling of day 2 and 5 embryo culture medium: a prospective randomized trial. Hum Reprod 2012;27:89–96. [DOI] [PubMed] [Google Scholar]
- Heikal A. A multiparametric imaging of cellular coenzymes for monitoring metabolic and mitochondrial activities. Rev Fluoresc 2010 [Internet]2012;2010:223–243. [Google Scholar]
- Houghton FD, Thompson JG, Kennedy CJ, Leese HJ. Oxygen consumption and energy metabolism of the early mouse embryo. Mol Reprod Dev 1996;44:476–485. [DOI] [PubMed] [Google Scholar]
- Hsieh C-S, Chen S-U, Lee Y-W, Yang Y-S, Sun C-K. Higher harmonic generation microscopy of in vitro cultured mammal oocytes and embryos. Opt Express [Internet]2008;16:11574–11588. [PubMed] [Google Scholar]
- Klaidman LK, Leung AC, Adams JD. High-performance liquid chromatography analysis of oxidized and reduced pyridine dinucleotides in specific brain regions. Anal Biochem [Internet]1995;228:312–317. [DOI] [PubMed] [Google Scholar]
- Korkmaz C, Tekin YB, Sakinci M, Ercan CM. Effects of maternal ageing on ICSI outcomes and embryo development in relation to oocytes morphological characteristics of birefringent structures. Zygote 2015;23:550–555. [DOI] [PubMed] [Google Scholar]
- Leese HJ. Quiet please, do not disturb: a hypothesis of embryo metabolism and viability. Bioessays 2002;24:845–849. [DOI] [PubMed] [Google Scholar]
- Ma N, Mochel NR de, PDA P, Yoo TY, Ken, Cho MAD. Label-free assessment of preimplantation embryo quality by the fluorescence lifetime imaging microscopy (FLIM)-phasor approach. bioRxiv [Internet]2018; Available from: 10.1101/286682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters BR, So P. Handbook of biomedical nonlinear optical microscopy [Internet] J Biomed Opt [Internet]2008;14: Oxford University Press. [Google Scholar]
- Mujat C, Greiner C, Baldwin A, Levitt JM, Tian F, Stucenski LA, Hunter M, Kim YL, Backman V, Feld M et al. . Endogenous optical biomarkers of normal and human papillomavirus immortalized epithelial cells. Int J Cancer 2008;122:363–371. [DOI] [PubMed] [Google Scholar]
- Munne S, Kaplan B, Frattarelli JL, Gysler M, Child TJ, Nakhuda G, Shamma FN, Silverberg K, Kalista T, Oliver K et al. . Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertil Steril 2019 Sep 21. pii: S0015-0282(19)31979-X. doi: 10.1016/j.fertnstert.2019.07.1346 [DOI] [PubMed] [Google Scholar]
- Murphy LA, Seidler EA, Vaughan DA, Resetkova N, Penzias AS, Toth TL, Thornton KL, Sakkas D. To test or not to test? A framework for counselling patients on preimplantation genetic testing for aneuploidy (PGT-A). Hum Reprod [Internet]2018;1–8Available from: https://academic.oup.com/humrep/advance-article/doi/10.1093/humrep/dey346/5219186. [DOI] [PubMed] [Google Scholar]
- Nagai S, Mabuchi T, Hirata S, Shoda T, Kasai T, Yokota S, Shitara H, Yonekawa H, Hoshi K. Correlation of abnormal mitochondrial distribution in mouse oocytes with reduced developmental competence. Tohoku J Exp Med 2006;210:137–144. [DOI] [PubMed] [Google Scholar]
- Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, Kobayashi H, Takikawa S, Manabe S, Kikkawa F et al. . Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet [Internet]2010;27:93–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C, Pera RAR. Human pre-implantation embryo development. Development [Internet]2012;139:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe K, Inada N, Gota C, Harada Y, Funatsu T, Uchiyama S. Intracellular temperature mapping with a fluorescent polymeric thermometer and fluorescence lifetime imaging microscopy. Nat Commun [Internet]2012;3:705–709Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker WC, Chakraborty N, Vrikkis R, Elliott G, Smith S, Moyer PJ. High-resolution intracellular viscosity measurement using time-dependent fluorescence anisotropy. Opt Express [Internet]2010;18:16607–16617. [DOI] [PubMed] [Google Scholar]
- Pedro MV, Sanchez T, Seidler EA, Sakkas D, Needleman D. Safety of fluorescence lifetime imaging microscopy (FLIM) as a non-invasive assessment of embryo metabolism. Eur Soc Hum Reprod Embryol [Internet]2018;193.Available from: https://www.eshre.eu/Books/ESHRE2018/Programme2018/files/basic-html/page337.html. [Google Scholar]
- Potter SM. Vital imaging: two photons are better than one. Curr Biol 1996;6:1595–1598. [DOI] [PubMed] [Google Scholar]
- Renard J, Philippon A, Menezo Y. In-vitro uptake of glucose by bovine blastocysts. J Reprod 1980;58:161–164. [DOI] [PubMed] [Google Scholar]
- Rieger D. Relationships between energy metabolism and development of early mammalian embryos. Theriogenology 1992;37:75–93. [Google Scholar]
- Sanchez T, Seidler EA, Gardner DK, Needleman D, Sakkas D. Will noninvasive methods surpass invasive for assessing gametes and embryos? Fertil Steril [Internet]2017;108:730–737Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Sanchez T, Wang T, Venturas Pedro M, Zhang M, Esencan E, Sakkas D, Needleman D, Seli E. Metabolic imaging with the use of fluorescence lifetime imaging microscopy (FLIM) accurately detects mitochondrial dysfunction in mouse oocytes. Fertil Steril. 2018;110:1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RT, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, Tao X, Treff NR. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril [Internet]2013a;100:697–703Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Scott RT, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril [Internet]2013b;100:624–630Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Smith PW. Mode-locking of lasers. Proc IEEE 1970;58:1342–1355. [Google Scholar]
- Sonka M, Hlavac V, Boyle R. Image Processing, Analysis, and Machine Vision. Cengage Learning, 2015 [Google Scholar]
- Staniszewski K, Audi SH, Sepehr R, Jacobs ER, Ranji M. Surface fluorescence studies of tissue mitochondrial redox state in isolated perfused rat lungs. Ann Biomed Eng [Internet]2013;41:827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LR, Imai SI. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 2012;23:420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari H, Honjo K, Kunitake K, Aramaki N, Kuhara S, Hidaka N, Nishimura K, Nagata Y, Horiuchi T. Meiotic spindle size is a strong indicator of human oocyte quality. Reprod Med Biol 2018;17:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari H, Honjou K, Nagata Y, Horiuchi T. Relationship between meiotic spindle characteristics in human oocytes and the timing of the first zygotic cleavage after intracytoplasmic sperm injection. J Assist Reprod Genet 2011;28:1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff NR, Zhan Y, Tao X, Olcha M, Han M, Rajchel J, Morrison L, Morin SJ, Scott RT. Levels of trophectoderm mitochondrial DNA do not predict the reproductive potential of sibling embryos. Hum Reprod 2017;32:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KR, Cavolo SL, Levitan ES. Elevated mitochondria-coupled NAD(P) H in endoplasmic reticulum of dopamine neurons. Mol Biol Cell [Internet]2016;27:3214–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le CC, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F et al. . Chromosome instability is common in human cleavage-stage embryos. Nat Med 2009;15:577–583. [DOI] [PubMed] [Google Scholar]
- Vega M, Jindal S. Mosaicism: throwing the baby out with the bath water? J Assist Reprod Genet [Internet]J Assist Reprod Genet 2017;34:11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergouw CG, Kieslinger DC, Kostelijk EH, Botros LL, Schats R, Hompes PG, Sakkas D, Lambalk CB. Day 3 embryo selection by metabolomic profiling of culture medium with near-infrared spectroscopy as an adjunct to morphology: a randomized controlled trial. Hum Reprod 2012;27:2304–2311. [DOI] [PubMed] [Google Scholar]
- Wong KM, Repping S, Mastenbroek S. Limitations of embryo selection methods. Semin Reprod Med 2014;32:127–133. [DOI] [PubMed] [Google Scholar]
- Yoo TY, Choi JM, Conway W, Yu CH, Pappu RV, Needleman DJ. Measuring NDC80 binding reveals the molecular basis of tension-dependent kinetochore-microtubule attachments. Elife 2018;7:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CH, Langowitz N, Wu HY, Farhadifar R, Brugues J, Yoo TY, Needleman D. Measuring microtubule polarity in spindles with second-harmonic generation. Biophys J [Internet]2014;106:1578–1587Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Heikal A a . Two-photon autofluorescence dynamics imaging reveals sensitivity of intracellular NADH concentration and conformation to cell physiology at the single-cell level. J Photochem Photobiol B [Internet]2009;95:46–57Elsevier B.V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Ren Z, Yeung WSB, Shu Y, Xu Y, Zhuang G, Liang X-Y. Low mitochondrial DNA and ATP contents contribute to the absence of birefringent spindle imaged with PolScope in in vitro matured human oocytes. Hum Reprod [Internet]2007;22:1681–1686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


