Abstract
Missing data is almost always present in real datasets, and introduces several statistical issues. One fundamental issue is that, in the absence of strong uncheckable assumptions, effects of interest are typically not nonparametrically identified. In this article, we review the generic approach of the use of identifying restrictions from a likelihood-based perspective, and provide points of contact for several recently proposed methods. An emphasis of this review is on restrictions for nonmonotone missingness, a subject that has been treated sparingly in the literature. We also present a general, fully-Bayesian, approach which is widely applicable and capable of handling a variety of identifying restrictions in a uniform manner.
Keywords: missing data, MNAR, mixture models, multiple imputation, non-ignorable missingness, nonparametric Bayes
1. Introduction
Missing data is highly prevalent in real datasets. Within a likelihood-based framework, missing data can best be categorized as either ignorable or nonignorable (Rubin, 1976); the former does not require a model for the missingness process, while the latter does. Nonignorable missingness introduces fundamental identifiability issues because, by virtue of the fact that we did not observe the missing data, we have no data with which to estimate its distribution.
The literature is filled with approaches which resolve identifiability issues by making parametric modeling assumptions (see Section 2 for a review). Following Cox and Donnelly (2011, page 96), however, we believe that if an issue cannot be resolved nonparametrically given an infinite sample then it is “usually dangerous to resolve it parametrically.” While parametric approaches are useful, we argue that they should not indirectly resolve identifiability issues. An alternative approach is to incorporate non-identifiability into the analysis. The full-data distribution can be factored into two components: (1) the observed-data distribution, which is identified by the observed data; and (2) the conditional distribution of the missing data given the observed data, sometimes called the extrapolation distribution, which is not identified (Daniels and Hogan 2008, Section 8.2; Little 1995). Different assumptions about the missing data can be expressed in terms of identifying restrictions which allow the analyst to recover the full-data distribution from the observed data distribution. The most well-known identifying restriction is the missing at random (MAR) assumption (Rubin, 1976), but many alternatives exist.
The National Research Council (2010) recommends the routine use of sensitivity analysis to assess the impact of assumptions about the missing data on inference. Two approaches to sensitivity analysis are to first consider many different identifying restrictions (Thijs et al., 2002) and second (in the spirit of Rotnitzky et al. 1998 and Daniels and Hogan 2008, Chapter 9) to introduce an unidentified sensitivity parameter ξ which represents an interpretable deviation from a benchmark identifying restriction. The sensitivity parameter ξ should be such that (1) there is no information in the data to inform ξ and (2) upon specification of ξ, the effects of interest are identified.
Concerns about parametric assumptions have motivated frequentist semiparametric approaches (Robins et al., 1995; Scharfstein et al., 1999) which make minimal assumptions about the full-data distribution. These approaches posit a parametric model for the missing data mechanism and a semiparametric model for the outcome distribution, and produce estimates by solving inverse-probability-weighted (IPW) estimating equations. These procedures are frequently doubly-robust, requiring the analyst to specify one of the two models correctly to attain consistent estimation (Scharfstein et al., 1999; Rotnitzky et al., 1998; Tsiatis, 2007). Recently, there have been various likelihood-based approaches proposed which have the flexibility of semiparametric approaches and allow a flexible sensitivity analysis (Wang et al., 2010; Linero and Daniels, 2015; Linero, 2017). An advantage of the Bayesian approach is that it allows for uncertainty about the unidentified components of the model to be encoded in an informative prior, allowing the analyst to incorporate subject-matter expertise formally into the analysis.
This article has three goals. First, we provide a review of model-based approaches to nonignorable missingness, including parametric approaches which identify the full-data distribution (see National Research Council, 2010; Ibrahim and Molenberghs, 2009, for additional reviews of MNAR modeling strategies). Our second goal is to summarize and review existing identifying restrictions in the literature. A special emphasis is given to recent proposals for nonmonotone missingness, as this subject has received a sparser treatment in the literature. We highlight several recently proposed identifying restrictions and characterize them as generalizations of monotone restrictions.
Our third goal is to propose a flexible, fully-Bayesian, framework for incomplete outcome data. First, a flexible Bayesian nonparametric model is chosen for the observed data distribution. Second, we use an identifying restriction to identify the extrapolation distribution. The framework allows for many different restrictions to be used without needing to change the model used for the observed data, can accommodate both monotone and nonmonotone missingness, and allows for the introduction of sensitivity parameters. The proposed approach might be perceived as a competitor to the IPW approaches which are prevalent in the literature. However, it has several features which IPW approaches do not. First, the Bayesian framework allows for expert knowledge to be formally incorporated into the analysis by eliciting informative priors on sensitivity parameters. Second, the approach allows for simultaneous inference about functionals of the full-data distribution, rather than just a specifically chosen functional such as the mean; for example it is possible to make inferences about means and quantiles simultaneously. Third, we are not required to fit different models depending on the choice of identifying restriction, allowing for a more principled comparison of different restrictions.
To illustrate the necessity of conducting a principled sensitivity analysis, we analyze data from the Breast Cancer Prevention Trial (BCPT). A concern in this study was that the treatment tamoxifen might cause depression. We show that the evidence for this hypothesis is strongly influenced by the assumptions made about the missingness, and that seemingly similar assumptions can yield dramatically different results. This underscores the need for statisticians and subject-matter experts to work together in determining which assumptions about the missing data are most appropriate for a particular problem.
1.1. Notation
Let denote the measurement of variable j intended to be collected on subject i for i = 1, . . . ,N, and let . Let be a vector of missingness indicators such that or 0 according to whether is observed or not. For a given binary vector r ∈ {0,1}J, let yr = (yj : rj = 1) and y−r = (yj : rj = 0). The observed data on subject i is then given by , and the missing data is given by .
We assume the pairs (Y(i),R(i)) are iid with density p(y, r) with respect to some measure; implicitly, p(y, r) may depend on a parameter vector θ. We refer to p(y, r) as the full-data distribution. To lighten notation, we will often work with an iid copy (Y,R) of (Y(1),R(1)). For simplicity we omit covariates; in principle all distributions we discuss can be defined conditional on fully-observed covariates X = x.
We will abuse notation, for example writing p(y) for the marginal density of Y or p(r | y) for the probability of R = r given Y = y; it will always be clear from context what density is being referred. When specific arguments are required, we will write for example p(Rj = 1 | Y = y) for the probability of Rj = 1 given Y = y.
For a fixed r, let p(y, r) = p(yr, r)p(y−r | yr, r) denote the extrapolation factorization (Daniels and Hogan, 2008, Section 8.2) of p(y, r). This factors p(y, r) into the product of a term which is identified and a term which is unidentified. Note that p(yr, r) is the density of the observed data (YR,R) while p(y−r | yr, r) is the conditional density of the missing data Y−R. We refer to p(yr, r) as the observed-data distribution and to p(y−r | yr, r) as the extrapolation distribution.
Missingness is said to be monotone if Rj = 0 implies Rj+1 = 0. This commonly occurs in longitudinal trials when missingness is due to dropout. Missingness can then be summarized by the last time at which a subect is measured , which we refer to as the (index of the) dropout time. For longitudinal studies it is also useful to let denote the history of the response up-to time j, and let denote the future of the response strictly after time j. Thus, . We similarly define and .
1.2. Running example: the Breast Cancer Prevention Trial
To make the concepts presented concrete, we will focus on applications to the Breast Cancer Prevention Trial (BCPT), a clinical trial which assigned women at high-risk of developing breast cancer to either a preventative drug, tamoxifen, or to a placebo. One aim of this study was to determine if tamoxifen causes depression. The response is 1 or 0 according to whether subject i is depressed or not at time j. Roughly N = 5000 subjects were assigned to each of tamoxifen (Z = 1) and control (Z = 0). Measurements were scheduled to be taken at baseline and 3, 6, 12, 18, 24, 30, and 36 months from baseline, for J = 8 intended measurements. There was a substantial amount of missingness at all time points, and missingness was highly nonmonotone. A concern is that depression at time j might be associated with missingness at time j, even after conditioning on other observables, resulting in MNAR missingness. Our primary interest is in the intention-to-treat effect ψ = E(YJ | Z = 1) − E(YJ | Z = 0).
To help illustrate concepts, we will also consider a simplified setting in which J = 2. We refer to this setting as the reduced Breast Cancer Prevention Trial (RBCPT). We assume that (Y1, Y2) represent continuous, rather than binary, measures of depression level (the actual binary responses were created from dichotomizing a quantitative score) to create more generality in the development.
2. Basic MNAR modeling strategies
We divide strategies for modeling p(y, r) into three categories: (1) selection models; (2) pattern mixture models; and (3) shared parameter models. In Section 2.4, we describe how any of these three approaches can be used to obtain a model for the observed data, without modeling the missing data.
2.1. Selection models
The selection modeling approach (Heckman, 1979) is based on the factorization p(y, r) = p(y)·p(r | y). The term p(r | y) is referred to as the missing data mechanism.
Example 1. Consider the RBCPT. With monotone missingness and Y1 always observed, following Diggle and Kenward (1994), we set
| (1) |
Selection models are attractive for their conceptual simplicity. In the context of the BCPT, the selection factorization suggests a causal mechanism in which depression causes missingness to occur. As p(y) is directly available, inference is usually straight-forward.
One drawback of parametric selection models is that they may “identify away” the missing data problem. Observe that ϕ2 = 0 corresponds to an MAR missing data mechanism in (1). One may be tempted to test for MNAR missingness by testing ϕ2 = 0. As we have stressed, testing for MAR cannot be done without recourse to parametric assumptions. As illustrated by Kenward (1998), inferences about MAR in this setup are extremely sensitive to parametric assumptions. When p(y) is a Gaussian density, (ϕ1, ϕ2) function as skewness parameters for p(y2 | y1, r) and can be estimated from the observed data. Hence, there are no sensitivity parameters which can be used as a basis of a sensitivity analysis. In practice, the likelihood of ϕ2 may be flat enough that it can be used as an approximate sensitivity parameter (Carpenter et al., 2002). This problem is mitigated to some extent when semiparametric or nonparametric models for Y are used, although this becomes more difficult as the dimension of the response increases. Note also that p(y−r | yr, r) is not available in closed form; consequently, it is difficult to describe on a conceptual level how missing values are imputed relative to the other approaches we describe.
2.2. Pattern mixture models
The pattern mixture approach (Little, 1994, 1993; Hogan and Laird, 1997) is based on the factorization p(y, r) = p(y | r)p(r). This characterizes p(y) as a mixture over missingness patterns . The pattern mixture factorization is closely related to the extrapolation factorization, with p(yr, r) = p(yr | r) · p(r). This makes the pattern mixture approach conducive to sensitivity analysis.
Example 2. Consider the RBCPT and assume monotone missingness with Y1 always observed. We set ϕ = p(R2 = 1), , and . The parameters are unidentified. One approach to identifying these parameters is to link them to the R2 = 1 pattern, setting for example and α(0) = α(1) + ξ. This implies that the influence of Y1 on Y2 and the conditional spread of Y2 do not depend on R2, while the conditional mean of Y2 does and is shifted by a fixed amount ξ. The parameter ξ is a sensitivity parameter, and can be varied as part of a sensitivity analysis.
Characteristic of pattern mixture models, the above model allows an interpretable sensitivity analysis and is transparent in how the it imputes missing values on a conceptual level. There are several shortcomings of the pattern mixture approach. Conceptually, it is typically not easy to interpret how the response Y influences the probability of missingness at time j. In the BCPT, a pattern mixture model suggests that those with missing values come from a distinct sub-population; an arguably more natural way to capture this intuition is through the use of latent class models (Roy, 2003) (though as constructed there, they do not allow sensitivity parameters). Pattern mixture models often possess a large number of unidentified parameters that the analyst must specify, with the situation becoming unwieldy in higher dimensions. Additionally, sparsity in the observed missing data patterns R(i) may necessitate further modeling of p(y | r) to share information across times.
2.3. Shared parameter approaches
The shared parameter approach captures dependence between Y(i) and R(i) through shared random effects (Wu and Carroll, 1988; Henderson et al., 2000), setting . The random effect distribution G(·) can be specified parametrically, usually as a multivariate Gaussian distribution, or nonparametrically.
Example 3. Consider the BCPT. We set (b1, b2) ~ Normal(μb,Σb) and assume that, conditional on b, all components of (Y,R) are mutually independent with and . For example, to get a random quadratic trend over time, we might set where tj is the time of measurement j. This type of shared parameter model is referred to as a correlated random effects model (Lin et al., 2010).
The shared parameter approach provides a highly flexible framework for analyzing nonignorable missingness, and is particularly effective for modeling complex data structures (Dunson and Perreault, 2001). Shared parameter models appeal strongly to intuition, suggesting that Y and R have a shared, unobserved, common cause. A drawback of the shared parameter approach is that it is difficult to separate p(yr, r) from p(y−r | yr, r), making it difficult to anchor a sensitivity analysis to an interpretable identifying restriction (see Section 3). Generally, it is not easy to see what assumptions about the missing data mechanism are encoded in a shared parameter model.
Methods for implementing a sensitivity analysis for shared parameter models have been developed by Creemers et al. (2010, 2011). In our example, one might set which gives an adjustment to the random effect b1 at the times for which rj = 0. One may then set, for example, δ ~ Normal(μδ,Σδ), with ξ = (μδ,Σδ) a sensitivity parameter. We feel that this is somewhat against the spirit of the shared parameter model, as Y and R are no longer conditionally independent and the causally suggestive motivation is stretched.
2.4. Observed data modeling
The models in Sections 2.1–2.3 have been presented as models for the joint density p(y, r). An alternative strategy is to model the observed data distribution p(yr, r) and leave the extrapolation distribution distribution p(y−r | yr, r) unspecified. One can then fit a model for p(yr, r) to the data and complete the model using one of the identifying restrictions described in Section 3.
Directly modeling p(yr, r) can be challenging to do in practice, as it requires a model for Yr for every pattern r. When missingness is monotone, one approach is to specify models for and . For examples of this approach, see Scharfstein et al. (2014) and Wang et al. (2010). Other approaches to directly modeling p(yr, r) often use the pattern mixture approach, specifying models for p(yr | r) while leaving p(y−r | yr, r) unspecified. See, for example, Little (1994) and Thijs et al. (2002).
A generic approach to modeling the observed data is to specify a working model (Linero, 2017; Linero and Daniels, 2015; Daniels and Linero, 2015). One then implicitly obtains a model for the observed data . In principle, p⋆(y, r) may be a selection model, pattern mixture model, or shared parameter model. In Section 5 we will apply this approach using a nonparametrically modeled shared parameter to obtain a highly flexible model of the observed data.
A benefit of the working model approach is that it allows models which share information across missingness patterns and time, without identifying the extrapolation distribution. This allows one to avoid a common pitfall of pattern-mixture models; we can estimate p(yr, r) even when we do not observe some patterns or the amount of data in some patterns is sparse. Because the model p⋆(y, r) is used only to obtain a model for p(yr, r), and is not used as a basis for inference, we are allowed complete freedom in how to identify the extrapolation distribution. Conveniently, p⋆(y, r) can also be used as a basis for Markov chain Monte Carlo algorithms.
In practice, the working model framework has the drawback of being somewhat difficult to implement, in that one must be able to derive the conditional distributions p⋆(yr | R = r′). This places restrictions on which models can be tractably used; in particular, selection models and parametric shared parameter models are difficult to use. Fortunately, there are very flexible models that are tractable. An additional concern is that, when p(yr, r) is modeled parametrically, p(y, r) will usually fall outside of this parametric family. For example, when using identifying restrictions, if p(yr | r) is modeled with a Gaussian distribution, it will not typically be the case that p(y | r) is Gaussian (Wang and Daniels, 2011). Consequently, the joint model p(y, r) may not be easily interpretable, although causal effects may still be computed using MC integration (see Section 4).
3. Identifying restrictions
Identifying restrictions provide a useful starting point for identifying the extrapolation distribution and conducting a sensitivity analysis. Informally, an identifying restriction is an assumption about p(y, r) which links the observed data distribution p(yr,r) to the extrapolation distribution p(y−r | yr, r).
We remark that identifying assumptions differ subtlely throughout the literature; for example, Seaman et al. (2013) give several non-equivalent definitions of MAR. All restrictions we consider will be phrased in the form of conditional independencies, with (for example) MAR corresponding to the conditional independence statement for all patterns r.
The goal of specifying an identifying restriction is to nonparametrically identify the parameters of interest.
Definition 3.1. Let denote the set of observed data distributions q(yr, r), and let be some family of full-data distributions p(y, r). The family is said to nonparametrically identify a parameter ψ(p) if,
For every , there exists a such that q is the associated observed data density of p.
For every , if p, both marginalize to q, then ψ(p) = ψ(p′).
The family is said to be nonparametrically saturated (Robins, 1997; Vansteelandt et al., 2006) if, for each , there exists a unique which marginalizes to q.
In the absence of strong subject-matter knowledge, it is unwise to assume that a particular identifying restriction holds. Nevertheless, in practice it can be useful to specify a single identifying restriction as a benchmark assumption, and consider interpretable deviations from that benchmark. For example, one might “anchor” an analysis to MAR and consider smooth deviations from MAR. Considering several anchors, and deviations from these anchors, provides insight into how inferences are driven by our assumptions.
We differentiate three different types of identifying restrictions. Joint restrictions completely identify p(y, r); that is, they lead to nonparametrically saturated models. Marginal restrictions do not identify p(y, r), but identify the marginals p(yj); an example is the sequential explainability assumption (Vansteelandt et al., 2007) discussed later. Marginal restrictions do not lead to nonparametrically saturated models, but are sufficient to nonparametrically identify all marginal effects. Marginal restrictions can be useful because (i) they may be more readily interpretable than joint restrictions, and (ii) they may encode weaker assumptions. Marginal restrictions are special cases of partial restrictions, which are any restrictions which do not identify p(y, r).
3.1. Identifying restrictions under monotone missingness
The missing data problem becomes much simpler when missingness is monotone. In this case, the missing data pattern can be summarized by the dropout time S = max{j : Rj = 1}. Monotonicity occurs naturally when missingness is due to dropout in a longitudinal study. Techniques for monotone missingness can also be applied if there is a method of ordering the components of Y which makes missingness monotone.
Example 4 (NCMV). Consider the BCPT, and assume that missingness is monotone. We conjecture that the, if a subject drops out at time k < j, then their missing response at time j can reasonably be approximated using an equivalent individual who instead drops out at time j; so, we set . Thijs et al. (2002) refer to this as the neighboring case missing value (NCMV) restriction.
Example 5 (ACMV). Consider again the BCPT with monotone missingness. We conjecture that, if a subject drops out at time k < j, then their response at time j can reasonably be approximated by using an equivalent subject who dropped out after time j; so, we set . Little (1993) refers to this as the available case missing value (ACMV) restriction.
Example 6 (CCMV). In the BCPT, we decide to use the observations of those who complete the study to estimate the conditional distribution of the missing observations; so, we set ; Little (1993) refers to that as the complete case missing value (CCMV) restriction.
The goal of using these restrictions is to provide a starting point for a sensitivity analysis. In practice, when missingness is MNAR, none of the conditional independencies asserted above is realistic; in fact, ACMV is itself equivalent to MAR (Molenberghs et al., 1998)! In the BCPT, if the depression status of an individual at time j is a strong predictor of Rj = 0 then one may expect the conditional distribution of Yj to be stochastically larger than what is implied by ACMV, NCMV, or CCMV.
Under monotone missingness, ACMV is equivalent to MAR. This suggests that missingness at time j + 1 is causally linked only to the past values of . The NFD restriction (Kenward et al., 2003) generalizes this idea.
Example 7 (NFD). We posit that missingness at time j+1 is causally due to the past and present values of Y, so that , or equivalently . This is referred to as the non-future dependence (NFD) assumption.
Despite its causal motivation, we note that NFD is not a causal law; for example if (Y,R) share an unobserved common cause, NFD will usually be violated. Given that MAR implies NFD, but not vice-versa, NFD leads to an under-identified model (and thus is a partial restriction); in particular, the distribution is unidentified for j > 2. This is convenient, as it allows the analyst to consider families of restrictions, all of which satisfy the NFD restriction. For example, Linero and Daniels (2015) centers a sensitivity analysis on the MAR assumption by setting , with ξ = 0 corresponding to MAR.
The ACMV, NCMV, and CCMV restrictions are all joint restrictions. Birmingham et al. (2003) consider several partial restrictions, including the following marginal restriction which is implied by CCMV.
Example 8 (Last-occasion-pattern-mixture). We posit that the conditional distribution of YJ at the end of study, given and S = j, can reasonably approximated by the distribution of those who complete the study; hence, we set .
A general tool for extending the restrictions above to the nonmonotone settings is to assume that missingness is partially ignorable given S (Harel and Schafer, 2009). This sets p(R = r | Y = y, S = s) = p(R = r | Yr = yr, S = s), and assumes the parameters of p(r | y, s) are independent of the parameters of p(y, s). Analogously to ignorability, partial ignorability ensures that likelihood-based inferences for p(y, s) do not depend on how p(r | y, s) is modeled. See Wang et al. (2010) for an application of this assumption to the BCPT data.
3.2. Identifying restrictions for nonmonotone missingness
The topic of identifying restrictions under nonmonotone missingness was initiated by Robins (1997), who proposed the class of permutation missingness (PM) models. Let denote the observed data (including the Rj’s) up-to-and-including time j, and the data observed strictly after time j. The PM restriction assumes
| (2) |
possibly after applying an a-priori known permutation to Y. In words, (2) states that missingness at time j can depend on the “past” and the “observed future,” but not on the present, where the notion of time is determined by the given permutation. For longitudinal data, one can use (2) without a permutation, or use the reverse permutation to get which states that missingness depends on the “future” and the “observed past.”
Our opinion is that PM models are difficult to explain to practitioners. We review several alternative assumptions which have been introduced relatively recently.
Example 9 (Sequential explainability). For the BCPT, we believe that the observed depression levels prior to time j are sufficient to predict whether or not a subject will be measured at time j, while the outcome at time j is not predictive. We therefore impose the sequential explainability restriction (Vansteelandt et al., 2007) .
Example 10 (NIP). For the BCPT, we believe that, all other observed quantities being equal, missingness at time j is not predictive of depression at time j. We therefore posit the nearest identified pattern (NIP) (Linero, 2017) restriction, , where is equal to r, but with jth component fixed at 1.
Both NIP and sequential explainability are marginal restrictions. NIP appears similar to NCMV. A more direct analog is the itemwise conditional independence (ICIN) assumption, introduced independently by Sadinle and Reiter (2017a) and Shpitser (2016).
Example 11 (ICIN). For the BCPT, we believe that all other quantities (both observed and unobserved) being equal, missingness at time j is not predictive of depression at time j. We therefore posit the ICIN restriction where R−j = (Rk : k ≠ j) and Y−j = (Yk : k ≠ j) denote R and Y with the jth component removed.
ICIN and NIP differ in that (i) NIP conditions only on the observed components of Y and (ii) ICIN is a joint restriction. To the extent that conditioning on additional variables makes conditional independence more tenable, ICIN is very attractive. To our knowledge, practical algorithms for conducting inference under ICIN are lacking when J is moderately large. Results of Sadinle and Reiter (2017a) imply that ICIN is equivalent to NCMV when missingness is monotone. A proof of the following proposition is deferred to the supplementary material.
Proposition 3.2. ICIN is an extension of NCMV to nonmonotone missingness.
Tchetgen Tchetgen et al. (2016) introduced the pairwise missing at random assumption. The name is motivated by the observation that it corresponds to MAR when, for fixed r, we assume , where .
Example 12 (PMAR). For the BCPT, we believe that the distribution of the missing values of a subject can reasonably be approximated using an equivalent subject who was observed at all measurement times. We therefore posit the pairwise missing at random (PMAR) restriction, .
Just as ICIN is a joint restriction which generalizes NCMV, PMAR is a joint restriction which generalizes CCMV; the following proposition is immediate from the definition.
Proposition 3.3. PMAR is an extension of CCMV to nonmonotone missingness.
3.3. Sensitivity parameters for identifying restrictions
The identifying restrictions in Section 3.1 and Section 3.2 are phrased in terms of conditional independence relationships which, as we have noted, are not themselves particularly plausible when Yj is thought to directly influence Rj. We consider these assumptions not because we believe the conditional independencies they suggest, but rather to use as benchmark assumptions. These assumptions can be embedded in a family of restrictions indexed by a sensitivity parameter ξ ∈ Ξ such that (1) there is no information in the data to identify ξ and (2) upon specifying ξ, the effects of interest are identified. It is essential that the sensitivity parameter ξ be interpretable; our convention will be to associate the benchmark assumption with ξ = 0. The index ξ can then be thought of as a smooth deviation from our benchmark assumption.
Example 13. For the BCPT, we believe the NIP restriction is unreasonable because depression at time J should increase the risk of missingness, even after accounting for the observed data. We instead assume
Let ; using Bayes theorem it can be shown that
so that γ denotes the effect, on the log-odds scale, of YJ = 1 on missingness.
The exponential tilting strategy is very widely applicable, and we now outline it in a general form. Examples of works using this strategy include Birmingham et al. (2003); Wang et al. (2010); Scharfstein et al. (2014, 1999); Tchetgen Tchetgen et al. (2016); Vansteelandt et al. (2007). Consider a restriction of the form
| (3) |
where U is a subset of the missing data, W is a subset of the complete data distinct from U, and V is a subset of the missing data indicators. The values v and v′ are such that U is missing when V = v, while U is observed when V = v′. For example, under sequential explainability, one has while under PMAR one has . Let fv(u | w) and denote the densities of the distributions in (3). The exponential tilting approach sets
| (4) |
The function t(u,w) is a function-valued sensitivity parameter. By Bayes theorem,
Hence, t(·,w) determines the effect of a change in U on the log-odds of V = v versus V = v′.
Another option is to consider a transformation-based approach similar to Daniels and Hogan (2000). This is particularly useful when the underlying response is continuous.
Example 14. Consider the BCPT, but with Y instead representing a continuous measure of depression level. We believe the NIP restriction is unreasonable because we expect depression levels to be higher among those missing at time j, even after conditioning on the observed data. We instead assume , where ξj > 0 represents the expected increase in depression level when a subject is missing rather than observed.
More generally, starting from (3), one can specify a generic transformation
| (5) |
In practice we must specify to be interpretable by subject-matter experts. Location or location-scale transformations, such as , are popular (Daniels and Hogan, 2000; Wang and Daniels, 2011; Gaskins et al., 2016) and can be computationally advantageous. Non-affine choices for can be used to rescale the data before applying an affine transformation.
A meaningful sensitivity analysis requires serious engagement with subject-matter experts, and as such requires for Ξ to be low dimensional. A common approach that does not formally account for the effect of uncertainty in ξ is a “tipping point” approach. This identifies values, or regions of values, of ξ which result in substantively different conclusions for the effects of interest. If plausible values of ξ do not include any tipping points, then we can have confidence in our substantive conclusions; we note, however, that tipping point analyses do not incorporate uncertainty in ξ when quantifying uncertainty in treatment effects. For illustrations of tipping point analyses, see Scharfstein et al. (2014) and Liublinska and Rubin (2014). An option which formally incorporates uncertainty in ξ is to place an informative prior on ξ. As there is no information in the data about ξ, this prior for ξ will also be the posterior. An advantage of this approach is that it combines all restrictions under consideration to achieve a single, final, inference. For examples of this approach, see Daniels and Hogan (2008, Chapter 9, Case Study 2), Wang et al. (2010), and Gaskins et al. (2016).
3.4. A pattern mixture modeling example
We now show how one might combine the identifying restrictions described above with a model for the observed data for the RBCPT (using the original depression score). We specify a pattern mixture model
All parameters above can be estimated from the observed data using standard techniques; for example, we have . For convenience, we write
where (α, β, τ) is a function of (μ(1,1),Σ(1,1)). Suppose that interest is in the parameter ζ = E(Y1). We demonstrate how ζ is identified under the PMAR, sequential explainability, and NIP assumptions. First, by iterated expectation,
Observe that and . This leaves E(Y1 | R1 = 0, R2 = 0) and E(Y1 | R1 = 0, R2 = 1) to be identified.
Consider first the PMAR assumption. This implies . Using iterated expectation, PMAR also implies
This gives
Next, we consider NIP. The derivations under NIP are exactly the same as those under PMAR, with the exception that . Therefore, under NIP we have
Hence, ζNIP will be larger than ζPMAR when , and vice versa. Lastly, we consider sequential explainability. At time j = 1 there is no observed history, so sequential explainability implies the marginal independence Y1 ⊥ R1. Consequently,
Sequential explainability differs fundamentally from NIP and PMAR as, due to its sequential nature, it does not use the distribution of (Y2, R2) to identify ζ.
We now incorporate sensitivity parameters under sequential explainability. Note that if , then ξ = 0 is consistent with sequential explainability. Under this assumption we have
Sensitivity analysis may now proceed either by eliciting an informative prior on ξ, or by identifying values of ξ which lead to substantively different inferences.
4. Inference and computation
We discuss two approaches to computation. First, we describe a fully-Bayesian approach, which can be computationally demanding. Second, we describe multiple imputation, which is a computationally simpler approximation. Let θ denote the parameters of the model of p(y, r), π(θ) a prior for θ, and the observed data. We first obtain samples of θ from its posterior distribution , usually by Markov chain Monte Carlo. When the working model framework described in Section 2.4 is used, samples of θ can be obtained by fitting the working model by data augmentation, taking advantage of the fact that . A benefit of this approach is that sampling θ can often be accomplished using general-purpose software for fitting Bayesian models. Software packages, such as JAGS and WinBUGS, allow for fast fitting of custom models, and accommodate missing values.
Fully-Bayesian inference then proceeds by computing effects of interest directly from the sampled θ’s and the chosen identifying restriction. Multiple imputation, by contrast, uses the sampled θ’s to impute completed datasets some number of times using the identifying restriction. Practically, these approaches are operationally quite similar. We begin by describing fully-Bayesian inference, and describe the changes required to perform multiple imputation.
4.1. Fully-Bayesian inference
Given sampled values , fully-Bayesian inference requires computing the desired effects. These will typically not be available in closed form, but can be computed by Monte Carlo integration. For illustrative purposes, we present the algorithm for sequential explainability in Algorithm 1. In the supplementary material we provide Monte Carlo integration algorithms for PMAR and NIP as well. While we do not pursue this here, Monte Carlo integration can also be implemented using IPW methods (see Robins, 1997; Birmingham et al., 2003; Shpitser, 2016, for such schemes). The number of Monte Carlo samples should be large relative to the sample size; in Section 5, we use 100 times the sample size. This appeal to Monte Carlo to estimate causal effects was initially proposed by Robins (1986) to implement G-computation. While computationally intensive, post-processing of the MCMC output is parallelizable and our experience is that the Monte Carlo integration is not a computational bottleneck. We can avoid repeating these computations for each ξ by using an informative prior, providing another advantage to the fully-Bayesian approach.
Algorithm 1.
Monte Carlo integration for sequential explainability
| 1: procedure GComp(θ T, j) | ▷ Approximates μj by simulating T samples from pθ(y) |
| 2: for t = 1,..., T do | |
| 3: Sample | |
| 4: if then | |
| 5: Sample | |
| 6: end if | |
| 7: end for | |
| 8: Set . | |
| 9: return μj | |
| 10: end procedure |
4.2. Multiple imputation
Multiple imputation (MI) proceeds by specifying two, potentially different, models. First, we use the sampled values to impute the missing data from pθ(y−r | yr, r) some number M > 1 times. The model pθ(y, r) is referred to as the imputation model. Next, an analysis model is specified to compute a point estimate and standard error from each of the m = 1, . . . ,M completed datasets . Rubin’s rules (see Harel and Zhou, 2007, for a review) are then used to produce a point estimate and standard error . The imputation is referred to as congenial (Meng, 1994) when and , in which case MI-based inference approximates fully-Bayesian inference. MI inference may be valid in the absence of congeniality, particularly when the analysis model is a sub-model of the imputation model. For further discussion of this issue, see Rubin (1996). For textbook level treatments of multiple imputation, see Rubin (1987) or Carpenter and Kenward (2012). For an exploration of impact of uncongeniality, see Daniels and Luo (2017).
Algorithm 2.
Multiple imputation algorithm for sequential explainability
| 1: procedure | ▷ Inference for μj using multiple imputation |
| 2: for m = 1,..., M do | |
| 3: Sample | |
| 4: for i = 1,..., N do | |
| 5: if then | |
| 6: Sample | |
| 7: end if | |
| 8: end for | |
| 9: Compute . | |
| 10: end for | |
| 11: Compute and using the rules for combining inferences under MI. | |
| 12: end procedure |
The imputation step for MI is operationally similar to the Monte Carlo integration used in Section 4.1, as it requires simulating from the same conditional distributions. Unlike Monte Carlo integration, MI only requires imputation of the missing data. Additionally, imputations can be used with different analysis models. MI is much more practical for large datasets than fully-Bayesian inference, at the cost of using an approximation. An MI-based algorithm for estimating μj = E(Yj) under sequential ignorability is given in Algorithm 2
Extreme caution is required in using MI with partial restrictions in terms of what analysis models can be used. A minimal condition for MI to be valid is that the analysis model is a submodel of the imputation model. Hence, when a partial restriction is used, the analysis model should not identify any part of the joint distribution which is unidentified by the imputation model. For example, if a marginal restriction identifies the marginals p(yj) but not the joints p(yj, yk), then the analysis model may also identify p(yj) but must not identify p(yj, yk).
We remark that there are other approaches to sensitivity analysis which are applied with multiple imputation. One approach is the so-called “δ-adjustment” (Leacy et al., 2017; Van Buuren, 2012, Section 3.9.1) in which imputations are adjusted, say, by a location shift δ. This approach is ad-hoc and somewhat lacking in transparency regarding what assumptions it encodes about the missing data, but is highly appealing due to its simplicity. Graphical methods for conducting a tipping-point analysis are given by Liublinska and Rubin (2014).
5. Application to the Breast Cancer Prevention Trial data
We apply the working model approach described in Section 2.4, using an infinite product-multinomial mixture (Dunson and Xing, 2009) which is implicitly stratified by treatment,
| (6) |
In the context of missing data, Si and Reiter (2013) applied this model to conduct multiple imputation in large-scale survey data under MAR. For longitudinal responses, various improvements are possible. One shortcoming of this model is that it does not incorporate temporal structure; additionally, a model with dependence within the mixture components would likely perform better (Murray and Reiter, 2016).
We give the stick-breaking prior associated with the Dirichlet process (Sethuraman, 1994) , . We approximate this by setting VK = 1 so that πk = 0 for k > K. For the BCPT data, we set K = 50 and α = 1. We view this truncation as a computational concession, leading to an approximation of inference under K = ∞; as pointed out by a referee, one may instead view the truncated model as a model in its own right, which is parametric rather than nonparametric. We model and . For ργj and βγj we specify independent Uniform(0,1) priors. Finally, for aγj and aβj we use a uniform shrinkage prior, with density fσ(a) = σ/(σ + a)2 with scale σ = 15. Larger values of σ encourage heavier shrinkage of the βkj’s and γkj’s towards their means. See Daniels (1999) and Wang et al. (2010) for motivation and details for the choice of this uniform shrinkage prior.
We use MCMC to draw samples of θ = (π, γ, β) from the posterior; details are provided in the supplementary material. We will focus our inference on the effect ψ = p(YJ = 1 | Z = 1) − p(YJ = 1 | Z = 0), where recall that Z = 1 corresponds tamoxifen and Z = 0 corresponds to the control. We consider four assumptions which identify ψ; the conditional distributions and algorithms needed are given in the supplementary material. First, we consider MAR by fitting the Y -marginal of (6) under ignorability. We also consider PMAR, sequential explainability, and the assumption
| (7) |
Assumption (7) is a nonmonotone, exponentially-tilted, variant of the last-occasion restriction of Birmingham et al. (2003). We refer to it as the tilted-last-occasion restriction. In addition to the interpretation of the exponential tilting strategy in Section 3.3, the parameter ξ can be interpreted as a location-shift on the logit-scale,
where ξ represents the log-odds ratio of [YJ = 1] relative to equivalent individuals with [R = r] and . We posit independent priors for ξ for each treatment; this has the effect of making the posterior variance of ψ large relative to dependent priors. Alternatively, one might take ξ constant across treatments to encode the belief that the effect of depression on missingness does not interact with treatment. To account for the fact that depression is expected to be positively correlated with missingness, we set ξ ~ Uniform(0,B). We set B = 0.8, corresponding to the belief that it is unlikely that the odds ratio of depression exceeds e0.8 ≈ 2.2. The above specification is made for illustrative purposes and is highly stylized. For a more realistic specification which seriously engages with subject-matter expertise, see Wang et al. (2010), who elicited informative priors from four subject-matter experts about analogous sensitivity parameters ξ; none posited values of ξ larger than 0.8.
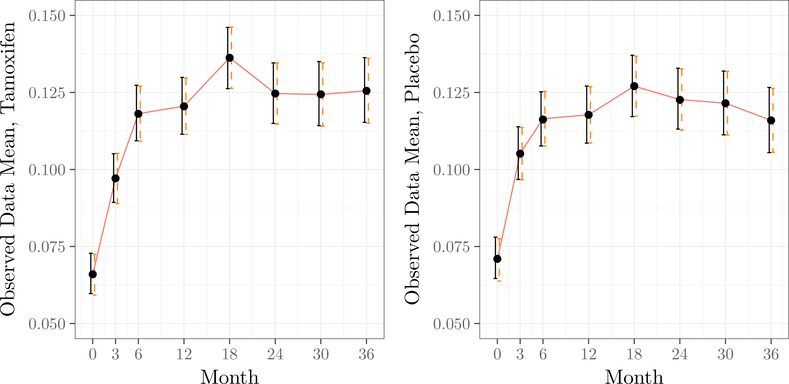
As a sanity check on the model, it is useful to verify that the posterior gives inferences which are consistent with the empirical distribution of the observed data. Let μobs,j = E(Yj | Rj = 1). In Figure 1, we compare the inferences based on the posterior distribution of the μobs,j’s to the inferences that would be obtained from the standard model-free estimates . We see that the posterior means are essentially identical to the the ’s, and the posterior credible intervals agree with the model-free intervals.
Figure 1:
Observed data means over time for the tamoxifen and placebo arms of the study. Dots correspond to the posterior mean using the prior outlined in this section. The line corresponds to the empirical mean of the observed data for each time point. Solid error bars give the 95% credible interval for the observed data mean; dashed error bars given the usual 95% confidence interval based on asymptotic normality of the observed-data means.
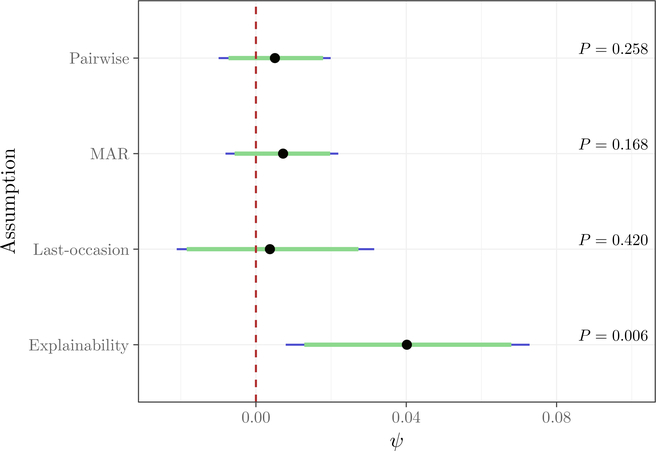
We report inferences for ψ obtained using the fully–Bayesian approach in Figure 2; results using multiple imputation with a nonparametric analysis model for p(yJ) are similar, and are given in the supplementary material, along with exact numerical results. The most striking feature is that inferences obtained under sequential explainability are very different from inferences obtained under either PMAR or MAR. First, the magnitude of the effect of tamoxifen on depression is much larger under sequential explainability; second, the posterior uncertainty is large. This is surprising, as one would expect the additional uncertainty in ξ to cause the tilted-last-occasion model to have the most posterior uncertainty.
Figure 2:
Posterior credible intervals for ψ under different assumptions. Dots give the posterior mean, green bars give two-sided 90% credible intervals, blue bars give two-sided 95% credible intervals. On the right, the posterior probability P = Pr(ψ < 0) is given for each assumption.
The additional posterior uncertainty can be explained from the fact that most of the missingness in the data was monotone. As a result, there is little information about for most missingness patterns. On the other hand, there are many fully observed individuals, so there is ample data to estimate for all patterns.
The fact that sequential explainability produces a larger effect size and leads to substantively different conclusions is concerning, and nessecitates an explanation. Further investigation revealed that, among those who were observed at the end of study, but who missed at least one visit (roughly 650 individuals per treatment), the difference in depression levels was a massive 6%. Moreover, this difference was highly significant, with Fisher’s exact test giving a P-value of 0.002. Under sequential explainability, those who were not observed at the end of the study are associated to this group, whereas under PMAR and the tilted-last-occasion model these individuals are associated to fully observed individuals. As there was no evidence of a difference in depression levels for fully observed subjects (P-value > 0.5 using Fisher’s exact test) the estimate of ψ is much smaller.
Whether PMAR or sequential explainability is more appropriate depends on subject matter considerations, as well as the causes of missingness. Regardless, the sensitivity analysis led us to find a treatment effect in a sub-population (those who were observed at the end of the study, but missed at least one prior visit) which is perhaps itself of interest. Hence, in addition to determining the robustness of our inferences, a sensitivity analysis can give substantive insight into the relationship between the missingness and the response.
6. Discussion and Open problems
In this paper we reviewed identifying restrictions with a focus on recent proposals for nonmonotone missingness. We also combined a flexible modeling approach for the observed data with a variety of identifying restrictions to analyze data from the BCPT.
Several interesting avenues of research exist. Auxiliary covariates are often used to impute missing outcomes under an assumption that MAR holds only conditional on these additional covariates. This is sometimes called A-MAR missingness. The inclusion of such covariates can create parameter interpretation problems (Daniels et al., 2014) for categorical outcomes. A proposal similar to that introduced here for continuous outcomes and auxiliary covariates can be found in Zhou et al. (working paper).
This paper has focused on missing outcome data. Handling missing covariate data is also of general importance (see, e.g., Ibrahim et al., 1999; Xu et al., 2016; Murray and Reiter, 2016). One approach to addressing this would be to specify joint Bayesian nonparametric models, along with identifying restrictions for the combined vector of outcomes and covariates. An interesting problem here is how to specify a parsimonious set of sensitivity parameters which will correspond to conditional distributions of both missing outcomes and missing covariates. Multiple identifying restrictions could be used for such analyses, similar to what was used in Linero and Daniels (2015) for different types of dropouts (see also Sadinle and Reiter, 2017b). Nonignorable missingness for more complex data-structures, such as longitudinal images or networks, remains an underdeveloped area. Much of what has been proposed here could also be used for causal inference. Kim et al. (2017) and Roy et al. (2016) propose Bayesian nonparametric approaches similar to ours in the context of causal mediation and marginal structural models respectively. We are also intrigued by the ICIN restriction as an anchoring assumption, and believe practical methods for performing inference under ICIN would be valuable.
There are few software implementations for conducting sensitivity analysis using identifying restrictions, especially when missingness is nonmonotone. The primary challenge lies in imputing the missing data from the appropriate conditional distributions, as this requires model-specific software. Our R implementation of the multinomial mixture model is available at www.github.com/arlinero/NiNBayes. Beyond this, we mention several tools available for sensitivity analysis. Bunouf et al. (2015) provide SAS and R code for implementing pattern-mixture models under monotone missingness and a Gaussian assumption. Scharfstein et al. (2017) provide the R/SAS package SAMON for implementing semiparametric models under monotone missingness. Outside of our proposed framework, proc MI in SAS now supports δ-adjustments using the MNAR option, and the SOLAS software package implements the tipping-point strategy of Liublinska and Rubin (2014).
Supplementary Material
Acknowledgments
This work was partially supported by NIH grant R01CA183854 and NSF grant NSF-DMS 1712870. The BCPT data was collected under NIH grants U10-CA37377, U10-CA69974, R01AI078835, P30MH086043, and R01HL79457.
References
- Birmingham J, Rotnitzky A, and Fitzmaurice GM (2003). Pattern-mixture and selection models for analysing longitudinal data with monotone missing patterns. Journal of the Royal Statistical Society, Series B., 65:275–297. [Google Scholar]
- Bunouf P, Molenberghs G, Grouin J-M, and Thijs H (2015). A SAS program combining R functionalities to implement pattern-mixture models. Journal of Statistical Software, 68(8). [Google Scholar]
- Carpenter J and Kenward M (2012). Multiple Imputation and its Application. John Wiley & Sons. [Google Scholar]
- Carpenter J, Pocock S, and Johan Lamm C (2002). Coping with missing data in clinical trials: A model-based approach applied to asthma trials. Statistics in Medicine, 21(8):1043–1066. [DOI] [PubMed] [Google Scholar]
- Cox D and Donnelly CA (2011). Principles of Applied Statistics. Cambridge University Press, first edition. [Google Scholar]
- Creemers A, Hens N, Aerts M, Molenberghs G, Verbeke G, and Kenward MG (2010). A sensitivity analysis for shared-parameter models for incomplete longitudinal outcomes. Biometrical Journal, 52(1):111–125. [DOI] [PubMed] [Google Scholar]
- Creemers A, Hens N, Aerts M, Molenberghs G, Verbeke G, and Kenward MG (2011). Generalized shared-parameter models and missingness at random. Statistical modelling, 11(4):279–310. [Google Scholar]
- Daniels MJ (1999). A prior for the variance in hierarchical models. Canadian Journal of Statistics, 27(3):567–578. [Google Scholar]
- Daniels MJ and Hogan JW (2000). Reparameterizing the pattern mixture model for sensitivity analyses under informative dropout. Biometrics, 56(4):1241–1248. [DOI] [PubMed] [Google Scholar]
- Daniels MJ and Hogan JW (2008). Missing Data In Longitudinal Studies. Chapman and Hall/CRC, first edition. [Google Scholar]
- Daniels MJ and Linero AR (2015). Bayesian nonparametrics for missing data in longitudinal clinical trials In Nonparametric Bayesian Inference in Biostatistics, pages 423–446. Springer. [Google Scholar]
- Daniels MJ and Luo X (2017). A note on “congeniality” for missing data in the presence of auxiliary covariates. Technical report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels MJ, Wang C, and Marcus BH (2014). Fully Bayesian inference under ignorable missingness in the presence of auxiliary covariates. Biometrics, 70(1):62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle P and Kenward MG (1994). Informative drop-out in longitudinal data analysis. Applied Statistics, 43:49–73. [Google Scholar]
- Dunson DB and Perreault SD (2001). Factor analytic models of clustered multivariate data with informative censoring. Biometrics, 57(1):302–308. [DOI] [PubMed] [Google Scholar]
- Dunson DB and Xing C (2009). Nonparametric Bayes modeling of multivariate categorical data. Journal of the American Statistical Association, 104(487):1042–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskins J, Daniels M, and Marcus B (2016). Bayesian methods for nonignorable dropout in joint models in smoking cessation studies. Journal of the American Statistical Association, 111(516):1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel O and Schafer JL (2009). Partial and latent ignorability in missing-data problems. Biometrika, 96:37–50. [Google Scholar]
- Harel O and Zhou X-H (2007). Multiple imputation: Review of theory, implementation and software. Statistics in Medicine, 26(16):3057–3077. [DOI] [PubMed] [Google Scholar]
- Heckman JJ (1979). Sample selection bias as a specification error. Econometrica, 47:153–161. [Google Scholar]
- Henderson R, Diggle PJ, and Dobson A (2000). Joint modelling of longitudinal measurements and event time data. Biostatistics (Oxford), 1:465–480. [DOI] [PubMed] [Google Scholar]
- Hogan JW and Laird NM (1997). Mixture models for the joint distribution of repeated measures and event times. Statistics in Medicine, 16:239–257. [DOI] [PubMed] [Google Scholar]
- Ibrahim JG, Lipsitz SR, and Chen M-H (1999). Missing covariates in generalized linear models when the missing data mechanism is non-ignorable. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 61(1):173–190. [Google Scholar]
- Ibrahim JG and Molenberghs G (2009). Missing data methods in longitudinal studies: a review. Test, 18(1):1–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenward MG (1998). Selection models for repeated measurements with non-random dropout: an illustration of sensitivity. Statistics in Medicine, 17(23):2723–2732. [DOI] [PubMed] [Google Scholar]
- Kenward MG, Molenberghs G, and Thijs H (2003). Pattern-mixture models with proper time dependence. Biometrika, 90:53–71. [Google Scholar]
- Kim C, Daniels MJ, Marcus BH, and Roy JA (2017). A framework for Bayesian nonparametric inference for causal effects of mediation. Biometrics, 73:401–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leacy FP, Floyd S, Yates TA, and White IR (2017). Analyses of sensitivity to the missing-at-random assumption using multiple imputation with delta adjustment: Application to a tuberculosis/hiv prevalence survey with incomplete hiv-status data. American journal of epidemiology, 185(4):304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Liu D, and Zhou X-H (2010). A correlated random-effects model for normal longitudinal data with nonignorable missingness. Statistics in medicine, 29(2):236–247. [DOI] [PubMed] [Google Scholar]
- Linero AR (2017). Bayesian nonparametric analysis of longitudinal studies in the presence of informative missingness. Biometrika, 104(2):327–341. [Google Scholar]
- Linero AR and Daniels MJ (2015). A flexible Bayesian approach to monotone missing data in longitudinal studies with informative dropout with application to a schizophrenia clinical trial. Journal of the American Statistical Association, 110(1):45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little RJ (1995). Modeling the drop-out mechanism in repeated-measures studies. Journal of the American Statistical Association, 90(431):1112–1121. [Google Scholar]
- Little RJA (1993). Pattern-mixture models for multivariate incomplete data. Journal of the American Statistical Association, 88:125–134. [Google Scholar]
- Little RJA (1994). A class of pattern-mixture models for normal incomplete data. Biometrika, 81:471–483. [Google Scholar]
- Liublinska V and Rubin DB (2014). Sensitivity analysis for a partially missing binary outcome in a two-arm randomized clinical trial. Statistics in medicine, 33(24):4170–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X-L (1994). Multiple-imputation inferences with uncongenial sources of input. Statistical Science, pages 538–558. [Google Scholar]
- Molenberghs G, Michiels B, Kenward MG, and Diggle PJ (1998). Monotone missing data and pattern-mixture models. Statistica Neerlandica, 52:153–161. [Google Scholar]
- Murray JS and Reiter JP (2016). Multiple imputation of missing categorical and continuous values via bayesian mixture models with local dependence. Journal of the American Statistical Association, 111(516):1466–1479. [Google Scholar]
- National Research Council (2010). The Prevention and Treatment of Missing Data in Clinical Trials. The National Academies Press. [PubMed] [Google Scholar]
- Robins J (1986). A new approach to causal inference in mortality studies with a sustained exposure periodapplication to control of the healthy worker survivor effect. Mathematical Modelling, 7(9–12):1393–1512. [Google Scholar]
- Robins JM (1997). Non-response models for the analysis of non-monotone non-ignorable missing data. Statistics in medicine, 16(1):21–37. [DOI] [PubMed] [Google Scholar]
- Robins JM, Rotnitzky A, and Zhao LP (1995). Analysis of semiparametric regression models for repeated outcomes in the presence of missing data. Journal of the American Statistical Association, 90(429):106–121. [Google Scholar]
- Rotnitzky A, Robins JM, and Scharfstein DO (1998). Semiparametric regression for repeated outcomes with non-ignorable non-response. Journal of the American Statistical Association, 93:1321–1339. [Google Scholar]
- Roy J (2003). Modeling longitudinal data with nonignorable dropouts using a latent dropout class model. Biometrics, 59:441–456. [DOI] [PubMed] [Google Scholar]
- Roy J, Lum KJ, and Daniels MJ (2016). A Bayesian nonparametric approach to marginal structural models for point treatments and a continuous or survival outcome. Biostatistics, 18(1):32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DB (1976). Inference and missing data. Biometrika, 63:581–592. [Google Scholar]
- Rubin DB (1987). Multiple Imputation for Nonresponse in Surveys. Wiley. [Google Scholar]
- Rubin DB (1996). Multiple imputation after 18+ years. Journal of the American statistical Association, 91(434):473–489. [Google Scholar]
- Sadinle M and Reiter JP (2017a). Itemwise conditionally independent nonresponse modeling for incomplete multivariate data. Biometrika, 104(1):207–220. [Google Scholar]
- Sadinle M and Reiter JR (2017b). Sequential identification of nonignorable missing data mechanisms. Statistica Sinica. To appear. [Google Scholar]
- Scharfstein D, Mcdermott A, Diaz I, Carone M, Lunardon N, and Turkoz I (2017). Global sensitivity analysis for repeated measres studies with informative dropout: A semiparametric approach. Biometrics. To appear. [DOI] [PubMed] [Google Scholar]
- Scharfstein D, McDermott A, Olson W, and Wiegand F (2014). Global sensitivity analysis for repeated measures studies with informative dropout: A fully parametric approach. Statistics in Biopharmaceutical Research, 6(4):338–348. [Google Scholar]
- Scharfstein DO, Rotnitzky A, and Robins JM (1999). Adjusting for nonignorable dropout using semiparametric nonresponse models. Journal of the American Statistical Association, 94. [Google Scholar]
- Seaman S, Galati J, Jackson D, and Carlin J (2013). What is meant by missing at random? Statistical Science, 28(2):257–268. [Google Scholar]
- Sethuraman J (1994). A constructive definition of Dirichlet priors. Statistica Sinica, 4:639–650. [Google Scholar]
- Shpitser I (2016). Consistent estimation of functions of data missing non-monotonically and not at random. In Advances in Neural Information Processing Systems, pages 3144–3152. [Google Scholar]
- Si Y and Reiter JP (2013). Nonparametric Bayesian multiple imputation for incomplete categorical variables in large-scale assessment surveys. Journal of Educational and Behavioral Statistics, 38(5):499–521. [Google Scholar]
- Tchetgen Tchetgen EJ, Wang L, and Sun B (2016). Discrete choice models for nonmonotone nonignorable missing data: Identification and inference. [DOI] [PMC free article] [PubMed]
- Thijs H, Molenberghs G, Michiels B, Verbeke G, and Curran D (2002). Strategies to fit pattern-mixture models. Biostatistics, 3:245–265. [DOI] [PubMed] [Google Scholar]
- Tsiatis AA (2007). Semiparametric Theory and Missing Data. Springer. [Google Scholar]
- Van Buuren S (2012). Flexible imputation of missing data.
- Vansteelandt S, Goetghebeur E, Kenward MG, and Molenberghs G (2006). Ignorance and uncertainty regions as inferential tools in a sensitivity analysis. Statistica Sinica, 16:953–979. [Google Scholar]
- Vansteelandt S, Rotnitzky A, and Robins JM (2007). Estimation of regression models for the mean of repeated outcomes under nonignorable nonmonotone nonresponse. Biometrika, 94(4):841–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C and Daniels MJ (2011). A note on MAR, identifying restrictions, model comparison, and sensitivity analysis in pattern mixture models with and without covariates for incomplete data. Biometrics, 67:810–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Danies MJ, Scharfstein DO, and Land S (2010). A Bayesian shrinkage model for incomplete longitudinal binary data with application to the breast cancer prevention trial. Journal of the American Statistical Association, 105:1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC and Carroll RJ (1988). Estimation and comparison of changes in the presence of informative right censoring by modeling the censoring process. Biometrics, 45:175–188. [PubMed] [Google Scholar]
- Xu D, Daniels MJ, and Winterstein AG (2016). Sequential BART for imputation of missing covariates. Biostatistics, 17(3):589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.