Abstract
Introduction:
Many germline associations have been reported for urinary bladder cancer (UBC) outcomes and prognostic characteristics. It is unclear whether there are overlapping genetic patterns for various prognostic endpoints. We aimed to review contemporary literature on genetic associations with UBC prognostic outcomes and to identify potential overlap in reported genes.
Methods:
EMBASE, MEDLINE, and PubMed databases were queried for relevant articles in English language without date restrictions. The initial search identified 1346 articles. After exclusions, 112 studies have been summarized. Cumulatively, 316 single-nucleotide polymorphisms (SNPs) were reported across prognostic outcomes (recurrence, progression, death) and characteristics (tumor stage, grade, size, age, risk group). There were considerable differences between studied outcomes in the context of genetic associations. The most commonly reported SNPs were located in OGG1, TP53, and MDM2. For outcomes with the highest number of reported associations (ie, recurrence and death), functional enrichment annotation yields different terms, potentially indicating separate biological mechanisms.
Conclusions:
Our study suggests that all UBC prognostic outcomes may have different biological origins with limited overlap. Further validation of these observations is essential to target a phenotype that could best predict patient outcome and advance current management practices.
Keywords: Urinary bladder cancer, review, prognosis, characteristics
Introduction
Urothelial bladder cancer (UBC) results in considerable clinical input and necessitates ongoing research to reduce the burden of patients and health care providers.1 Current era of genomics offers new insights into UBC pathogenesis.2 However, due to the complex nature of genetics, many studies are difficult to summarize into clear recommendations for future research and clinical practice.
Urothelial bladder cancer is most frequently diagnosed as a non-muscle-invasive bladder cancer (NMIBC), accounting for 70% to 80% of all new cases.3 Management of NMIBC is complex with appropriate treatment dependent on multiple clinical and pathological components. Importantly, a significant proportion of patients are prone to tumor recurrence and/or progression, both events being difficult to predict. Previously developed multifactorial prognostic NMIBC tools4 have been useful to describe populations, but lack accuracy for individual outcomes and require further advances.5 Muscle-invasive bladder cancer (MIBC) cases are equally complex to treat with various permutations of chemotherapy, radiotherapy, and cystectomy,6 with an addition of recent initiatives in molecular-genomic subtyping.2
Although multiple studies have addressed the potential role of genetic variation in UBC prognosis, the findings are yet to be implemented into clinical practice. For the most part, genetic associations are often reported in small samples and their validity is difficult to establish.7 In addition, the interpretation of the biological relevance over many reports is challenging. Furthermore, it is not yet clear whether genetic associations overlap within and between the groups of direct (recurrence, progression, survival) and indirect (stage, grade, tumor size, age at the time of diagnosis) prognostic endpoints. Identifying existing genetic similarities between prognostic outcomes would help potentially decipher underlying pathological mechanisms and guide promising future directions in UBC research.
In this review, our objective is to summarize genetic associations for UBC prognostic phenotypes and to describe any overlap or existing patterns that would clarify their pertinence for future clinical practice.
Methods
The systematic review was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)8 (Supplementary Table 1).
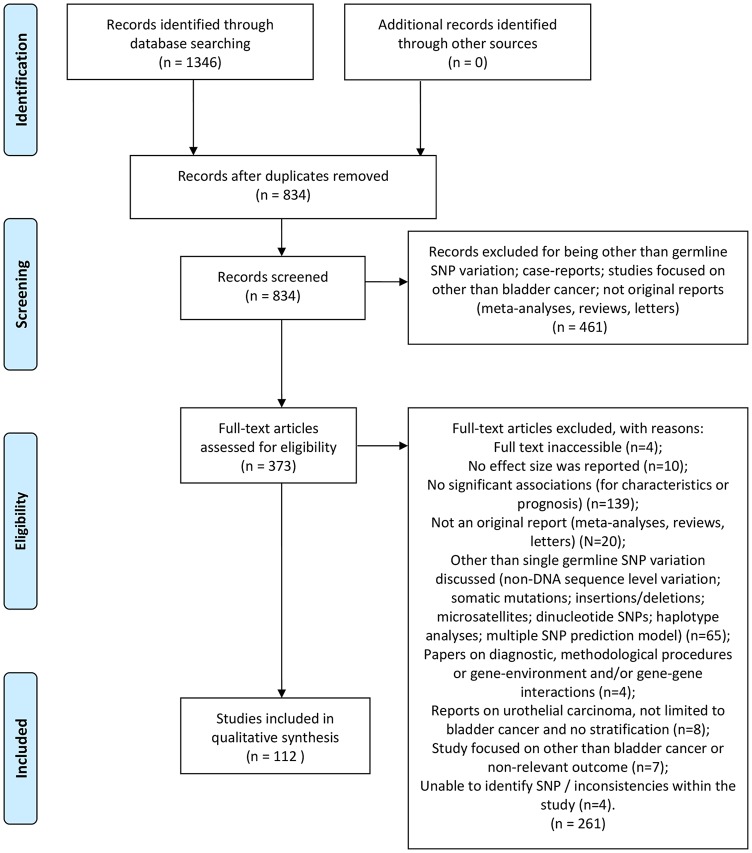
We queried EMBASE, Medline, and PubMed with the following search term: ((“urinary bladder neoplasms” OR “bladder cancer” OR “urothelial carcinoma”) AND (prognosis OR survival OR recurrence OR progression OR grade OR stage OR “tumour size” OR age) AND (polymorphi* OR SNP OR germline)). The search was limited to articles published prior to November 13, 2018, written in English, and describing human research only. A detailed flowchart on the selection and search process is presented in Figure 1. Reference lists of included manuscripts were checked for potentially missing reports. Study eligibility was determined by the 2 authors (N.L., A.W.).
Figure 1.
Flow diagram of study selection used in evidence synthesis. SNP indicates single-nucleotide polymorphism.
Inclusion criteria were as follows:
Studies assessing single germline single-nucleotide polymorphism (SNP) variants (not somatic mutations, insertion/deletions, microsatellites, haplotype analyses, dinucleotide polymorphism associations, multiple SNP prediction models);
Original reports (not meta-analyses, reviews, letters, case reports, others);
Studies focused on UBC or where UBC data are described distinctly from a broader urothelial carcinoma (UC) cohort;
Studies reporting an effect size;
Studies reporting significant associations (for characteristics or prognosis);
DNA sequence level variation described;
The described SNPs could be identified.
Studies describing diagnostic and methodological procedures and gene-gene and gene-environment interactions were excluded.
Each study was assessed for quality by evaluating reporting adequacy. Inconsistency was regarded as mismatching data within the study (eg, different SNP IDs reported in article sections). Data completeness was verified if all relevant data fields for a genetic association study9 were available to extract from the report. The quality criteria were part of the study selection process (eg, studies stating variant relevance for an outcome without providing an effect size were regarded as having low quality and excluded from further evaluation).
Data extraction
Further information was extracted from each eligible study: year of publication, first author, patient subgroup (UBC, MIBC, NMIBC, or other), cancer subtype (UC or other), ethnicity, sample size, SNP ID, locus, gene, effect allele, reference allele, effect allele frequency, effect size, corresponding 95% confidence intervals, and P value.
Summarizing overlap in genetic associations and outcomes
To investigate whether previously reported genes may play a role across multiple UBC outcomes, results were put in a ranked table. Genes associated with many UBC endpoints were ranked high, whereas genes that were reported for one or few of the outcomes were ranked low. As such, we were able to suggest genes that are important for UBC prognosis overall (ie, ranked high) and which genes are more likely to be outcome specific (eg, only associated with cancer recurrence and ranked low).
The resulting ranking acted as a guideline for identifying genes that were commonly observed for most of the prognostic outcomes and characteristics. Outcomes with at least 20 genes were chosen and their functional roles were further described in additional detail.
Functional annotation
After summarizing the overlap, some outcomes have been associated with multiple genes. Every biological process is polygenic, and having bigger sets of identified genes helps elucidate biological pathways behind the studied phenotype. We chose outcomes with the largest number of reported genes and submitted those sets to the DAVID Functional Annotation Tool.10 This tool groups genes by their functional similarity, using information from well-known databases, such as Gene Ontology (GO) for biological mechanisms and KEGG (Kyoto Encyclopedia of Genes and Genomes) for pathways, among others. Gene clustering was performed with setting the highest level of classification stringency. A high level of stringency generates fewer clusters, but genes within them are associated more tightly. Moreover, to reduce the likelihood of describing false-positive clusters, only gene groups containing pathways with false discovery rates (FDRs) < 5% were interpreted as valid results.
Statistical analysis
Overall, our search has resulted in multiple genes corresponding to various outcomes. As such, the resulting data are very difficult to describe in a comprehensive manner. To reduce the dimensionality of current data, we performed a principal component analysis (PCA). Principal component analysis can be seen as a form of an exploratory analysis to identify group-level correlations in the sample. It is a useful tool for improving the interpretation of data, as it allows visualizing similarities between groups regarding chosen characteristics. In our analysis, we aimed to investigate the similarity between UBC outcomes regarding their genetic background. For example, clinical outcomes that share genes would plot more closely, whereas an outcome that does not share any genes with other endpoints would plot far from other groups. In the currently reviewed literature, some outcomes have been investigated more often (eg, recurrence and death), and hence we have adjusted the size of data points in a PCA plot to be relative to the number of associated genes. The first 2 principal components were plotted for all studied endpoints.
Results
For this review, 373 full-text articles were evaluated in depth, resulting in a final set of 112 articles for further summary (Figure 1). In total, 316 associations were extracted across all investigated outcomes as follows: age (N = 1211-22), stage (N = 7911,13,18,20,23-66), tumor size (N = 267,68), grade (N = 4911,18,27,29,32,33,35-37,42,47,50,54,58,62,66,67,69-75), risk groups (N = 1512,14,17,24,25,30,31,33,40,76,77), recurrence (N = 8114,23,25,30,31,40,43,49,50,52,53,69,71,78-107), progression (N = 2426,33,46,87-89,107-112), and death (N(cancer-specific) = 1234,43,46,101,107,113-117, N(overall) = 4234,43,50,56,89,90,111,112,118-122).
There was considerable heterogeneity across all associations, including assumed patterns of inheritance, studied ethnic populations, and outcome definitions.
Age was investigated using multiple year cut-offs, namely, 50,13 56,17 60,11,16,22 65,12,15,18-21 and once as a continuous variable14 (Supplementary Table 2).
Tumor size was investigated either as using a cut-off of ⩾3 cm67 or defined as a large tumor, corresponding to stages T1 to T468 (Supplementary Table 3).
Tumor stage was analyzed using multiple combinations. Broadly, we have differentiated between stages corresponding to NMIBC and MIBC cases. For studies reporting on NMIBC, the following endpoints were used: tumors of Tis,61,65 T1,37 Ta + T1,13,18,36,39,54,58,62-64,66 and Ta + T1 + Tis32,59,60 (Supplementary Table 4). As for MIBC, most studies have defined the primary outcome as T2+ staged tumors.11,20,23-32,34-57 However, some associations have been reported for a merged group of T2+ and T1 stages.33,34
Most reports on grade can be roughly categorized into containing either low- or high-grade UBC cases. Low-grade UBC definitions were as follows: G1,18,54,62,70 G2,35,58,62,70 G1 + G2,36 low-grade,69,71 and G1 + G2 + papilloma.32 High-grade UBC was usually defined as grade 3 UBC,11,27,29,32,36,37,47,50,67,70,72-75 a combination of G2 and G3 NMIBC,54,66 and some studies have reported estimates for grade 4 tumors, without a reference for the grading system used (G3 + G442 and G2 + G3 + G4)33 (Supplementary Table 5).
It was common for studies to classify UBC as a disease of low or high risk that corresponded to various combinations of clinical stage and grade. For low-risk tumors, researchers used the following definitions: TaG2,33,34 TaG1,33 and TaG1-2.12,14,17 In contrast, high-risk tumors were defined as TaG2-3 + T1G1-3,25,30,31,40,76 TaG3 + T1G2-3,24 G2-3 with T1-4,77 and TaG3 + T133 (Supplementary Table 6).
For genetic associations with tumor recurrence, studies mostly focused on NMIBC cases (except for few reports considering UBC group overall49 or MIBC).50,71 The NMIBC recurrence was investigated as an overall outcome,49,69,78,89,90,94,97,104,106 or in specific groups: patients younger than 64 years,98 patients not treated with Bacillus Calmette-Guérin (BCG) therapy40,43; BCG-treated patients23,25,30,31,40,43,52,53,79-83,85-87,93,95,96,99,100,102,103,107; patients treated only with transurethral urinary bladder tumor (TURBT) resection88,101; patients who have received both TURBT and BCG treatments91,92; patients having received treatments of TURBT and epirubicin105; and recurrence only among low-risk NMIBC14,84 (Supplementary Table 7).
Progression was defined as an increase of stage within NMIBC33,109,110 or UBC108 overall. Also, transition from NMIBC to MIBC or metastatic disease87-89 was considered a disease progression, sometimes expanding the latter definition to include cancer-specific death.107,112 In other cases, alternative definitions were considered, namely, occurrence of metastases26,46 and a confirmed relapse among MIBC111 (Supplementary Table 8).
Regarding death outcomes, there were 2 broad groups of overall33,43,56,89,90,112,118-121 and cancer-specific34,43,46,101,107,113-117 survival endpoints (Supplementary Table 9).
Retrieved data and detailed study characteristics, including outcome definition for each study, are presented in Supplementary Tables 2 to 9.
Overlap between outcomes
A summary table of existing overlap between outcomes and associated genes is presented in Table 1.
Table 1.
Overlap between reported outcomes and mapped genes.
| Outcomes | Number of overlapping genes | Mapped genes |
|---|---|---|
| Age/grade/recurrence/risk group/stage | 1 | OGG1 |
| Death/grade/recurrence/risk group/stage | 2 | TP53, MDM2 |
| Age/grade/risk group/stage | 1 | CCND1 |
| Age/recurrence/risk group/stage | 1 | XRCC7(PRKDC) |
| Age/death/recurrence/stage | 1 | XRCC1 |
| Grade/progression/risk group/stage | 1 | HRAS |
| Death/grade/recurrence/stage | 2 | PDCD6, XPD(ERCC2) |
| Age/grade/stage | 1 | H19 |
| Age/death/stage | 1 | EGFR |
| Grade/risk group/stage | 1 | MSH6 |
| Death/risk group/stage | 1 | NQO1 |
| Progression/recurrence/stage | 1 | MIR146A |
| Death/grade/recurrence | 1 | IL6 |
| Grade/recurrence/tumor size | 1 | TSP-1(THBS1) |
| Death/progression/recurrence | 1 | NOS3 |
| Age/recurrence/risk group | 1 | TACC3/FGFR3 |
| Death/grade/stage | 2 | RAD51, MTHFR |
| Recurrence/risk group/stage | 2 | CASP9, IL18 |
| Grade/recurrence/stage | 4 | CCR2, PPARG, GSTP1, XPC |
| Risk group/stage | 1 | XRCC5 |
| Progression/stage | 1 | IL4 |
| Grade/recurrence | 1 | IL31 |
| Progression/recurrence | 1 | RGS1 |
| Age/risk group | 2 | CASC11, TP63 |
| Death/stage | 2 | TLR10, IL27 |
| Death/recurrence | 3 | RGS2, GSTO1, XPF(ERCC4) |
| Grade/stage | 4 | LEPR, IGFBP3, XPG(ERCC5), PSCA |
| Recurrence/stage | 4 | IL17A, TNFA, GPX1, NAMPT |
| Death/progression | 4 | NOD2, BCL2, RGS5, ERCC1 |
| Tumor size | 1 | WISP1(CCN4) |
| Risk group | 2 | POLG2, BRCA2 |
| Age | 3 | PCAT1, POR, HOTAIR |
| Grade | 6 | MIR143_CARMN, TMEM129_TACC3_FGFR3, CLPTM1L, MYC, TNFRSF10A(TRAILR1, DR4), CCNE1 |
| Progression | 9 | DGCR8, NOS2, CDKN2A, TGFB1, RGS4, RGS7, IL10, UNG, RGS14 |
| Stage | 15 | CD44, SDF1(CXCL12), CXCR4, SLC23A1, MATR3, DNAJC18, C13ORF31(LACC1), CD4, CFH, XRCC3, IL22, MMP12, COX2(PTGS2), P21(CDKN1A), PMS2 |
| Death | 20 | IL8RB(CXCR2), RPTOR, RGS12, GSTO2, MRE11, RB1CC1, EPHX1, BCL2L1, GATA3, UGT1A1, XRCC4, PIK3R1, DRD4, RGS3, TERT, CD80, AURKA, AKT2, TGFBR1, GNB3 |
| Recurrence | 26 | VDR, Survivin(BIRC5), MMP2, GPX4, NFKBIA, CDH1, IGF1, GLI2, NEIL2, GLI3, RNASEN(DROSHA), IL8(CXCL8), ICAM1, IFN-G, SHH, RGS13, RGS16, RGS10, DDX20, GSS, CWC27, SOD1, ERCC6, NRAMP1(SLC11A1), ALDH2, TNFRSF10A(TRAILR1, DR4) |
OGG1 was the most commonly reported gene, having been associated with patient age,19 tumor stage,54 grade,54 recurrence,40,78 and risk group.40 Associations on OGG1 and UBC did not cluster within a clearly defined subgroup and instead showed relationships with various characteristics: increased age at diagnosis (>65 years)19 and elevated risks of the following: non-muscle-invasive and invasive UBC,54 low- and high-grade tumors,54 rate of recurrence,40,78 and high-risk tumors.40
A set of 2 genes (TP53 and MDM2) have also been reported for multiple endpoints, specifically UBC grade,42,69 stage,25,41,42 recurrence,25,69,101 survival,101,117 and risk group.25,76 For most outcomes (death, risk category, grade, stage), the associations for MDM2- and TP53-related variants were in opposite directions.
Regarding the number of genes corresponding to a single endpoint, tumor recurrence was the outcome with the highest sum of genes (N = 28) showing associations, followed by death (N = 21) (Table 1).
To elucidate any unifying pathways between these genes, gene sets for recurrence and death were submitted to the functional annotation tool DAVID.10
For recurrence, DAVID identified 2 gene clusters of similar functions that contained pathways with acceptable FDR values (Supplementary Table 10). The first group (enrichment score = 2.72) was formed entirely of RGS family genes (RGS10, RGS13, RGS16). The second cluster (enrichment score = 2.42) was formed by GLI2, GLI3, and SHH genes. Out of 10 functional terms within the cluster, 1 was of satisfactory FDR and reached Bonferroni-adjusted statistical significance < .05, termed “hindgut morphogenesis.”
For individual enriched pathways, 20 have yielded FDR < 5% and are listed in Supplementary Table 11. Three functional terms—“hindgut morphogenesis,” “pathways in cancer,” and “positive regulation of transcription from RNA polymerase II promoter”—have shown low FDR rates and were also below the conventional level of statistical significance (P < .05) after multiple-comparison adjustment.
For genes associated with UBC survival, the submitted set retrieved 6 functional clusters in total; however, no individual terms had acceptable FDR values.
Nonetheless, multiple individual functional pathways with FDR < 5% were identified instead (Supplementary Table 12). One term, “pancreatic cancer,” has reached a Bonferroni-adjusted statistical significance (P = .05).
Finally, a performed PCA analysis for previously reported genetic associations showed UBC recurrence to be the most distinct outcome (Figure 2), with tumor stage and grade also showing significant deviations from other endpoints.
Figure 2.
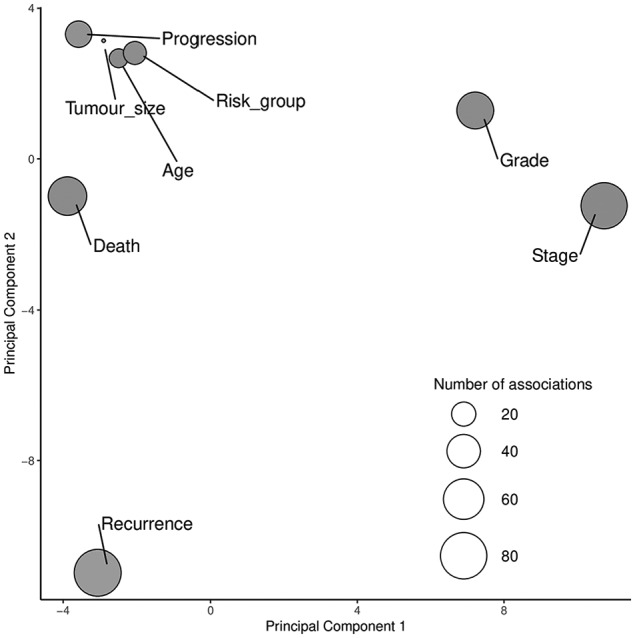
Principal component analysis for genetic associations with urinary bladder outcomes.
Data point sizes are indicative of the number of associated genes with each outcome. The first 2 plotted principal components include all genetic reports and explain most of the variance in all plotted outcomes.
Discussion
In this review, we have summarized existing evidence for single SNP genetic associations with UBC characteristics (tumor size, stage, grade, patient’s age) and prognostic outcomes (recurrence, progression, survival). There were multiple associations for considered endpoints with limited overlap. Based on these data, we have made several observations.
It is widely accepted that complex disease genetic architecture is highly polygenic.123 However, the currently summarized list of associations for UBC outcomes and characteristics is far from exhaustive. It is essential to note that future studies with higher per-study power will contribute additional associations and will clarify the validity of those already reported.
Importantly, our review underscores the sensitivity of outcome definition in genetic studies. It has been demonstrated that genetic variants for UBC risk are unlikely to be relevant for prognosis,124 and our report implies that prognostic outcomes demonstrate further within-group heterogeneity. Interestingly, the PCA revealed differences for direct prognostic outcomes: UBC death and progression showed similar characteristics, whereas UBC recurrence significantly deviated from the group. From a biological perspective, cancer recurrence is not an equivalent to progression or death, and it is likely that the mechanisms involved are triggered and organized via different pathways. Similarly, tumor characteristics (grade, stage, size) and patient characteristics (age) are likely different entities in genetic contribution.
When trying to elucidate unifying pathways for multiple genes involved in certain outcomes, UBC recurrence was found to be associated with terms that relate to the formation of a new tissue (eg, “hindgut morphogenesis”). In contrast, functional pathway terms were different for death as an outcome and may indicate a separate biological mechanism. Interestingly, the most promising associated term for death was “pancreatic cancer,” which exhibits very low survival rates in comparison with cancers of any other site.125
In the light of our analyses, UBC prognosis may represent a complex phenotype, and this review indicates that different outcomes imply distinct genetic associations. The genetic relationships may overlap but, nonetheless, should be treated as independent endpoints.
Importantly, the review identifies a number of commonly reported genes, specifically OGG1, TP53, and MDM2. OGG1 encodes a protein involved in base excision repair (BER) pathways to protect cells from oxidative stress.126 Although having a clear role in mutagenic processes, OGG1-null mice showed only moderate increases in malignancy rate, likely due to effective alternative damage repair pathways.127 Evidence from multiple meta-analyses128-130 of OGG1 involvement in UBC cancerogenesis is contradictory, and, when having a genuine effect, is more likely to play a supporting role in a multi-stage process rather than being the main cause of it.127 It is also probable that the establishment of the type and direction of genetic associations requires larger populations (underscoring sufficient sample sizes for different ethnicities), not yet available to researchers.
In addition, the link between TP53 and MDM2 genes has been extensively reported in the literature, offering an attractive pharmacologic target in cancer treatment.131 P53 protein acts as a tumor suppressor, which is negatively regulated by MDM2 oncoprotein. The pattern is somewhat mirrored in observed associations, where a variation in SNPs of the 2 genes seemed to correspond to effects in opposite direction (eg, SNPs in TP53 increased the risk of T2+ stage, whereas alterations in MDM2 showed reduced risk of invasive tumors).
Collectively, OGG1, TP53, and MDM2 are relevant for multiple essential DNA-preserving cellular mechanisms and hence would be expected to have importance for a variety of UBC characteristics and outcomes, as observed in our review.
The limitations of our study are important to acknowledge. Many reports have analyzed different ethnicities, which alone does not undermine the reported associations, but makes inter-population relevance improbable due to differing allele frequencies.132 Moreover, assumed genetic patterns of inheritance (eg, recessive, dominant, additive) differed highly between the studies, without a clear preference for the chosen model. Usually, the reported model was chosen ad hoc as a consequence of being statistically significant, making it difficult to be confident that the reported model reflected true genetic architecture of the association. Because associations were highly heterogeneous, we were unable to perform a meta-analysis (which would have provided a preferred summary of these data). Furthermore, most of the included studies were of candidate-gene design; we would expect different results if all studies followed an agnostic genome-wide association approach. Finally, sample sizes were limited, and it is difficult to establish whether all reported associations are robust. The lack of external replication studies for genetic associations is detrimental to translating science into practice, as many genetic findings are likely to be false positives.7 Optimally, only validated variants would be included in review studies. We underscore the importance of validation efforts for future studies to summarize only unambiguous variants.
Finally, we interpret this report as exploratory in its nature, and although no clear guidelines can be drawn due to the heterogeneity of previous studies, it is nonetheless an important exercise in drawing research directions. First of all, as more independent research groups have access to genotype data of bladder cancer patients, this article can prove a useful resource to replicate already reported associations in a time-efficient manner. Second, reporting those results will help identify which associations and corresponding genetic regions are most promising to pursue in other studies. As such, the landscape of genetic bladder cancer investigations may be accelerated by collaborative contributions made by the wider research community and preserve resources for studies of higher likelihood to produce meaningful results.
To conclude, we have summarized existing genetic associations for tumor and patient characteristics and disease prognosis for UBC. Multiple loci have been identified that demonstrate little consensus and highlight the possibility of UBC prognostic outcomes being unique entities in the context of genetic contribution. We recommend that further replication of previously identified SNPs should be undertaken. Consecutive formal reviews of existing associations will help facilitate their potential use in clinical practice.
Supplemental Material
Supplemental material, Supplementary_Tables_1_12_Sep24_xyz268777d1d25d8 for Systematic Review: Genetic Associations for Prognostic Factors of Urinary Bladder Cancer by Nadezda Lipunova, Anke Wesselius, Kar K Cheng, Frederik J van Schooten, Jean-Baptiste Cazier, Richard T Bryan and Maurice P Zeegers in Biomarkers in Cancer
Footnotes
Funding:The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: NL designed the study, organized the data, performed statistical analyses, and wrote the first draft of the manuscript. All authors contributed to the manuscript and study design revision, read, and approved the submitted version.
ORCID iD: Nadezda Lipunova  https://orcid.org/0000-0001-6478-9408
https://orcid.org/0000-0001-6478-9408
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Svatek RS, Hollenbeck BK, Holmang S, et al. The economics of bladder cancer: costs and considerations of caring for this disease. Eur Urol. 2014;66:253-262. [DOI] [PubMed] [Google Scholar]
- 2. Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540-556.e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet (London, England). 2016;388:2796-2810. [DOI] [PubMed] [Google Scholar]
- 4. Babjuk M, Bohle A, Burger M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: update 2016. Eur Urol. 2017;71:447-461. [DOI] [PubMed] [Google Scholar]
- 5. Sylvester RJ. How well can you actually predict which non-muscle-invasive bladder cancer patients will progress? Eur Urol. 2011;60:431-433; discussion 433-434. [DOI] [PubMed] [Google Scholar]
- 6. Alfred Witjes J, Lebret T, Comperat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71:462-475. [DOI] [PubMed] [Google Scholar]
- 7. Grotenhuis AJ, Dudek AM, Verhaegh GW, et al. Independent replication of published germline polymorphisms associated with urinary bladder cancer prognosis and treatment response. Bladder Cancer. 2016;2:77-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sagoo GS, Little J, Higgins JP. Systematic reviews of genetic association studies. Human Genome Epidemiology Network. PLoS Med. 2009;6:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang DW, Sherman BT, Tan Q, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hua Q, Lv X, Gu X, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. 2016;31:531-538. [DOI] [PubMed] [Google Scholar]
- 12. Wang M, Wang M, Zhang W, et al. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991-996. [DOI] [PubMed] [Google Scholar]
- 13. Kelsey KT, Park S, Nelson HH, Karagas MR. A population-based case-control study of the XRCC1 Arg399Gln polymorphism and susceptibility to bladder cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1337-1341. [PubMed] [Google Scholar]
- 14. Kiemeney LA, Sulem P, Besenbacher S, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 2010;42:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao X, Ma G, Li S, et al. Functional POR A503V is associated with the risk of bladder cancer in a Chinese population. Sci Rep. 2015;5:11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y, Ge Y, Wang Y, et al. The association of rs710886 in lncRNA PCAT1 with bladder cancer risk in a Chinese population. Gene. 2017;627:226-232. [DOI] [PubMed] [Google Scholar]
- 17. Stern MC, Van Den Berg D, Yuan JM, et al. Sequence variant on 3q28 and urinary bladder cancer risk: findings from the Los Angeles-Shanghai bladder case-control study. Cancer Epidemiol Biomarkers Prev. 2009;18:3057-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan L, Gu X, Shao J, et al. Cyclin D1 G870A polymorphism is associated with risk and clinicopathologic characteristics of bladder cancer. DNA Cell Biol. 2010;29:611-617. [DOI] [PubMed] [Google Scholar]
- 19. Ma L, Chu H, Wang M, et al. hOGG1 Ser326Cys polymorphism is associated with risk of bladder cancer in a Chinese population: a case-control study. Cancer Sci. 2012;103:1215-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu H, Wang M, Jin H, et al. EGFR 3’UTR 774T>C polymorphism contributes to bladder cancer risk. Mutagenesis. 2013;28:49-55. [DOI] [PubMed] [Google Scholar]
- 21. Wang SY, Peng L, Li CP, et al. Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int J Urol. 2008;15:534-539. [DOI] [PubMed] [Google Scholar]
- 22. Wang X, Wang W, Zhang Q, et al. Tagging SNPs in the HOTAIR gene are associated with bladder cancer risk in a Chinese population. Gene. 2018;664:22-26. [DOI] [PubMed] [Google Scholar]
- 23. Ahirwar DK, Mandhani A, Dharaskar A, Kesarwani P, Mittal RD. Association of tumour necrosis factor-alpha gene (T-1031C, C-863A, and C-857T) polymorphisms with bladder cancer susceptibility and outcome after bacille Calmette-Guerin immunotherapy. BJU Int. 2009;104:867-873. [DOI] [PubMed] [Google Scholar]
- 24. Guey LT, Garcia-Closas M, Murta-Nascimento C, et al. Genetic susceptibility to distinct bladder cancer subphenotypes. Eur Urol. 2010;57:283-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gangwar R, Mittal RD. Association of selected variants in genes involved in cell cycle and apoptosis with bladder cancer risk in North Indian population. DNA Cell Biol. 2010;29:349-356. [DOI] [PubMed] [Google Scholar]
- 26. Deng S, Wang W, Li X, Zhang P. Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J Surg Oncol. 2015;13:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Safarinejad MR, Shafiei N, Safarinejad SH. The association between bladder cancer and a single nucleotide polymorphism (rs2854744) in the insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) gene. Arch Toxicol. 2011;85:1209-1218. [DOI] [PubMed] [Google Scholar]
- 28. Lin HH, Ke HL, Hsiao KH, et al. Potential role of CCND1 G870A genotype as a predictor for urothelial carcinoma susceptibility and muscle-invasiveness in Taiwan. Chin J Physiol. 2011;54:196-202. [DOI] [PubMed] [Google Scholar]
- 29. Safarinejad MR, Safarinejad S, Shafiei N, Safarinejad S. Association of genetic polymorphism of glutathione S-transferase (GSTM1, GSTT1, GSTP1) with bladder cancer susceptibility. Urol Oncol. 2013;31:1193-1203. [DOI] [PubMed] [Google Scholar]
- 30. Gangwar R, Mandhani A, Mittal RD. Caspase 9 and caspase 8 gene polymorphisms and susceptibility to bladder cancer in North Indian population. Ann Surg Oncol. 2009;16:2028-2034. [DOI] [PubMed] [Google Scholar]
- 31. Jaiswal PK, Singh V, Srivastava P, Mittal RD. Association of IL-12, IL-18 variants and serum IL-18 with bladder cancer susceptibility in North Indian population. Gene. 2013;519:128-134. [DOI] [PubMed] [Google Scholar]
- 32. Ali SHB, Bangash KS, Rauf A, et al. Identification of novel potential genetic predictors of urothelial bladder carcinoma susceptibility in Pakistani population. Fam Cancer. 2017;16:577-594. [DOI] [PubMed] [Google Scholar]
- 33. Sanyal S, De Verdier PJ, Steineck G, et al. Polymorphisms in XPD, XPC and the risk of death in patients with urinary bladder neoplasms. Acta Oncol. 2007;46:31-41. [DOI] [PubMed] [Google Scholar]
- 34. Sanyal S, Ryk C, De Verdier PJ, et al. Polymorphisms in NQO1 and the clinical course of urinary bladder neoplasms. Scand J Urol Nephrol. 2007;41:182-190. [DOI] [PubMed] [Google Scholar]
- 35. Wang S, Tang J, Wang M, Yuan L, Zhang Z. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31:621-624. [DOI] [PubMed] [Google Scholar]
- 36. Safarinejad MR, Shafiei N, Safarinejad S. Genetic susceptibility of methylenetetrahydrofolate reductase (MTHFR) gene C677T, A1298C, and G1793A polymorphisms with risk for bladder transitional cell carcinoma in men. Med Oncol. 2011;28:S398-S412. [DOI] [PubMed] [Google Scholar]
- 37. Pandith AA, Shah ZA, Khan NP, Baba KM, Wani MS, Siddiqi MA. HRAS T81C polymorphism modulates risk of urinary bladder cancer and predicts advanced tumors in ethnic Kashmiri population. Urol Oncol. 2013;31:487-492. [DOI] [PubMed] [Google Scholar]
- 38. Zhou B, Zhang P, Wang Y, et al. Interleukin-17 gene polymorphisms are associated with bladder cancer in a Chinese Han population. Mol Carcinog. 2013;52:871-878. [DOI] [PubMed] [Google Scholar]
- 39. Chen WC, Wu HC, Hsu CD, Chen HY, Tsai FJ. p21 gene codon 31 polymorphism is associated with bladder cancer. Urol Oncol. 2002;7:63-66. [DOI] [PubMed] [Google Scholar]
- 40. Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Do DNA repair genes OGG1, XRCC3 and XRCC7 have an impact on susceptibility to bladder cancer in the North Indian population? Mutat Res. 2009;680:56-63. [DOI] [PubMed] [Google Scholar]
- 41. Lin HY, Huang CH, Yu TJ, Wu WJ, Yang MC, Lung FW. p53 codon 72 polymorphism as a progression index for bladder cancer. Oncol Rep. 2012;27:1193-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pandith AA, Shah ZA, Khan NP, et al. Role of TP53 Arg72Pro polymorphism in urinary bladder cancer predisposition and predictive impact of proline related genotype in advanced tumors in an ethnic Kashmiri population. Cancer Genet Cytogenet. 2010;203:263-268. [DOI] [PubMed] [Google Scholar]
- 43. Leibovici D, Grossman HB, Dinney CP, et al. Polymorphisms in inflammation genes and bladder cancer: from initiation to recurrence, progression, and survival. J Clin Oncol. 2005;23:5746-5756. [DOI] [PubMed] [Google Scholar]
- 44. Kader AK, Liu J, Shao L, et al. Matrix metalloproteinase polymorphisms are associated with bladder cancer invasiveness. Clin Cancer Res. 2007;13:2614-2620. [DOI] [PubMed] [Google Scholar]
- 45. Kucukgergin C, Sanli O, Amasyali AS, Tefik T, Seckin S. Genetic variants of MnSOD and GPX1 and susceptibility to bladder cancer in a Turkish population. Med Oncol. 2012;29:1928-1934. [DOI] [PubMed] [Google Scholar]
- 46. Guirado M, Gil H, Saenz-Lopez P, et al. Association between C13ORF31, NOD2, RIPK2 and TLR10 polymorphisms and urothelial bladder cancer. Hum Immunol. 2012;73:668-672. [DOI] [PubMed] [Google Scholar]
- 47. Kucukgergin C, Isman FK, Dasdemir S, et al. The role of chemokine and chemokine receptor gene variants on the susceptibility and clinicopathological characteristics of bladder cancer. Gene. 2012;511:7-11. [DOI] [PubMed] [Google Scholar]
- 48. Wen H, Feng CC, Fang ZJ, et al. Study on bladder cancer susceptibility and genetic polymorphisms of XPC, XPG, and CYP in smokers and non-smokers. Actas Urol Esp. 2013;37:259-265. [DOI] [PubMed] [Google Scholar]
- 49. Zhang K, Zhou B, Zhang P, et al. Genetic variants in NAMPT predict bladder cancer risk and prognosis in individuals from southwest Chinese Han group. Tumour Biol. 2014;35:4031-4040. [DOI] [PubMed] [Google Scholar]
- 50. Zhou B, Zhang P, Tang T, et al. Prognostic value of PDCD6 polymorphisms and the susceptibility to bladder cancer. Tumour Biol. 2014;35:7547-7554. [DOI] [PubMed] [Google Scholar]
- 51. Weng WC, Huang YH, Yang SF, et al. Effect of CD44 gene polymorphisms on risk of transitional cell carcinoma of the urinary bladder in Taiwan. Tumour Biol. 2016;37:6971-6977. [DOI] [PubMed] [Google Scholar]
- 52. Mittal RD, Singh R, Manchanda PK, et al. XRCC1 codon 399 mutant allele: a risk factor for recurrence of urothelial bladder carcinoma in patients on BCG immunotherapy. Cancer Biol Ther. 2008;7:645-650. [DOI] [PubMed] [Google Scholar]
- 53. Gangwar R, Mandhani A, Mittal RD. XPC gene variants: a risk factor for recurrence of urothelial bladder carcinoma in patients on BCG immunotherapy. J Cancer Res Clin Oncol. 2010;136:779-786. [DOI] [PubMed] [Google Scholar]
- 54. Ahmed T, Nawaz S, Noreen R, et al. A 3’ untranslated region polymorphism rs2304277 in the DNA repair pathway gene OGG1 is a novel risk modulator for urothelial bladder carcinoma. Ann Hum Genet. 2018;82:74-87. [DOI] [PubMed] [Google Scholar]
- 55. Gangwar R, Mandhani A, Mittal RD. Functional polymorphisms of cyclooxygenase-2 (COX-2) gene and risk for urinary bladder cancer in North India. Surgery. 2011;149:126-134. [DOI] [PubMed] [Google Scholar]
- 56. Zhou B, Zhang P, Tang T, et al. Polymorphisms and plasma levels of IL-27: impact on genetic susceptibility and clinical outcome of bladder cancer. BMC Cancer. 2015;15:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mittal RD, Mandal RK. Genetic variation in nucleotide excision repair pathway genes influence prostate and bladder cancer susceptibility in North Indian population. Indian J Hum Genet. 2012;18:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sobti RC, Kaur S, Sharma VL, Singh SK, Hosseini SA, Kler R. Susceptibility of XPD and RAD51 genetic variants to carcinoma of urinary bladder in North Indian population. DNA Cell Biol. 2012;31:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verhaegh GW, Verkleij L, Vermeulen SH, den Heijer M, Witjes JA, Kiemeney LA. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. 2008;54:1118-1126. [DOI] [PubMed] [Google Scholar]
- 60. Zhao T, Wu X, Liu J. Association between interleukin-22 genetic polymorphisms and bladder cancer risk. Clinics (Sao Paulo). 2015;70:686-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ito M, Habuchi T, Watanabe J, et al. Polymorphism within the cyclin D1 gene is associated with an increased risk of carcinoma in situ in patients with superficial bladder cancer. Urology. 2004;64:74-78. [DOI] [PubMed] [Google Scholar]
- 62. Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Influence of XPD and APE1 DNA repair gene polymorphism on bladder cancer susceptibility in North India. Urology. 2009;73:675-680. [DOI] [PubMed] [Google Scholar]
- 63. Wen H, Ding Q, Fang ZJ, Xia GW, Fang J. Population study of genetic polymorphisms and superficial bladder cancer risk in Han-Chinese smokers in Shanghai. Int Urol Nephrol. 2009;41:855-864. [DOI] [PubMed] [Google Scholar]
- 64. Ichimura Y, Habuchi T, Tsuchiya N, et al. Increased risk of bladder cancer associated with a glutathione peroxidase 1 codon 198 variant. J Urol. 2004;172:728-732. [DOI] [PubMed] [Google Scholar]
- 65. Lima L, Silva J, Amaro T, et al. IL-4 and TNF-alpha polymorphisms are associated with risk of multiple superficial tumors or carcinoma in situ development. Urol Int. 2011;87:457-463. [DOI] [PubMed] [Google Scholar]
- 66. Sakano S, Kumar R, Larsson P, et al. A single-nucleotide polymorphism in the XPG gene, and tumour stage, grade, and clinical course in patients with nonmuscle-invasive neoplasms of the urinary bladder. BJU Int. 2006;97:847-851. [DOI] [PubMed] [Google Scholar]
- 67. Gu J, Tao J, Yang X, et al. Effects of TSP-1-696 C/T polymorphism on bladder cancer susceptibility and clinicopathologic features. Cancer Genet. 2014;207:247-252. [DOI] [PubMed] [Google Scholar]
- 68. Lee HL, Chiou HL, Wang SS, et al. WISP1 genetic variants as predictors of tumor development with urothelial cell carcinoma. Urol Oncol. 2018;36:160.e15-160.e21. [DOI] [PubMed] [Google Scholar]
- 69. Xie L, Sun Y, Chen T, et al. Association between MDM2 SNP309 T>G polymorphism and the risk of bladder cancer: new data in a Chinese population and an updated meta-analysis. Onco Targets Ther. 2015;8:3679-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rothman N, Garcia-Closas M, Chatterjee N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li Q, Tang T, Zhang P, et al. Correlation of IL-31 gene polymorphisms with susceptibility and clinical recurrence of bladder cancer. Fam Cancer. 2018;17:577-585. [DOI] [PubMed] [Google Scholar]
- 72. Wu J, Huang Q, Meng D, Huang M, Li C, Qin T. A functional rs353293 polymorphism in the promoter of miR-143/145 is associated with a reduced risk of bladder cancer. PLoS ONE. 2016;11:e0159115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Timirci-Kahraman O, Ozkan NE, Turan S, et al. Genetic variants in the tumor necrosis factor-related apoptosis-inducing ligand and death receptor genes contribute to susceptibility to bladder cancer. Genet Test Mol Biomarkers. 2015;19:309-315. [DOI] [PubMed] [Google Scholar]
- 74. Wang L, Habuchi T, Takahashi T, et al. Cyclin D1 gene polymorphism is associated with an increased risk of urinary bladder cancer. Carcinogenesis. 2002;23:257-264. [DOI] [PubMed] [Google Scholar]
- 75. Gautam KA, Muktanand T, Sankhwar SN, Goel A, Sankhwar PL, Rajender S. Functional polymorphisms in the IL6 gene promoter and the risk of urinary bladder cancer in India. Cytokine. 2016;77:152-156. [DOI] [PubMed] [Google Scholar]
- 76. Jaiswal PK, Goel A, Mittal RD. Association of p53 codon 248 (exon7) with urinary bladder cancer risk in the North Indian population. Biosci Trends. 2011;5:205-210. [DOI] [PubMed] [Google Scholar]
- 77. Ratanajaraya C, Nishiyama H, Takahashi M, et al. A polymorphism of the POLG2 gene is genetically associated with the invasiveness of urinary bladder cancer in Japanese males. J Hum Genet. 2011;56:572-576. [DOI] [PubMed] [Google Scholar]
- 78. Kim EJ, Jeong P, Quan C, et al. Genotypes of TNF-alpha, VEGF, hOGG1, GSTM1, and GSTT1: useful determinants for clinical outcome of bladder cancer. Urology. 2005;65:70-75. [DOI] [PubMed] [Google Scholar]
- 79. Gu J, Zhao H, Dinney CP, et al. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11:1408-1415. [DOI] [PubMed] [Google Scholar]
- 80. Decobert M, Larue H, Bergeron A, et al. Polymorphisms of the human NRAMP1 gene are associated with response to Bacillus Calmette-Guerin immunotherapy for superficial bladder cancer. J Urol. 2006;175:1506-1511. [DOI] [PubMed] [Google Scholar]
- 81. Chiong E, Kesavan A, Mahendran R, et al. NRAMP1 and hGPX1 gene polymorphism and response to Bacillus Calmette-Guerin therapy for bladder cancer. Eur Urol. 2011;59:430-437. [DOI] [PubMed] [Google Scholar]
- 82. Gangawar R, Ahirwar D, Mandhani A, Mittal RD. Impact of nucleotide excision repair ERCC2 and base excision repair APEX1 genes polymorphism and its association with recurrence after adjuvant BCG immunotherapy in bladder cancer patients of North India. Med Oncol. 2010;27:159-166. [DOI] [PubMed] [Google Scholar]
- 83. Ahirwar DK, Agrahari A, Mandhani A, Mittal RD. Cytokine gene polymorphisms are associated with risk of urinary bladder cancer and recurrence after BCG immunotherapy. Biomarkers. 2009;14:213-218. [DOI] [PubMed] [Google Scholar]
- 84. Wang M, Wang M, Yuan L, et al. A novel XPF –357A>C polymorphism predicts risk and recurrence of bladder cancer. Oncogene. 2010;29:1920-1928. [DOI] [PubMed] [Google Scholar]
- 85. Chen M, Hildebrandt MA, Clague J, et al. Genetic variations in the sonic hedgehog pathway affect clinical outcomes in non-muscle-invasive bladder cancer. Cancer Prev Res (Phila). 2010;3:1235-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Srivastava P, Kapoor R, Mittal RD. Association of single nucleotide polymorphisms in promoter of matrix metalloproteinase-2, 8 genes with bladder cancer risk in Northern India. Urol Oncol. 2013;31:247-254. [DOI] [PubMed] [Google Scholar]
- 87. Wei H, Kamat A, Chen M, et al. Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guerin. PLoS ONE. 2012;7:e38533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ke HL, Chen M, Ye Y, et al. Genetic variations in micro-RNA biogenesis genes and clinical outcomes in non-muscle-invasive bladder cancer. Carcinogenesis. 2013;34:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lee EK, Ye Y, Kamat AM, Wu X. Genetic variations in regulator of G-protein signaling (RGS) confer risk of bladder cancer. Cancer 2013;119:1643-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Andrew AS, Gui J, Hu T, et al. Genetic polymorphisms modify bladder cancer recurrence and survival in a USA population-based prognostic study. BJU Int. 2015;115:238-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lima L, Oliveira D, Ferreira JA, et al. The role of functional polymorphisms in immune response genes as biomarkers of bacille Calmette-Guerin (BCG) immunotherapy outcome in bladder cancer: establishment of a predictive profile in a Southern Europe population. BJU Int. 2015;116:753-763. [DOI] [PubMed] [Google Scholar]
- 92. Ke HL, Lin J, Ye Y, et al. Genetic variations in glutathione pathway genes predict cancer recurrence in patients treated with transurethral resection and Bacillus Calmette-Guerin instillation for non-muscle invasive bladder cancer. Ann Surg Oncol. 2015;22:4104-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Deng X, Yang X, Cheng Y, et al. GSTP1 and GSTO1 single nucleotide polymorphisms and the response of bladder cancer patients to intravesical chemotherapy. Sci Rep. 2015;5:14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wang M, Li Z, Chu H, et al. Genome-wide association study of bladder cancer in a Chinese cohort reveals a new susceptibility locus at 5q12.3. Cancer Res. 2016;76:3277-3284. [DOI] [PubMed] [Google Scholar]
- 95. Williams SB, Kamat AM, Mmeje C, et al. Genetic variants in the inflammation pathway as predictors of recurrence and progression in non-muscle invasive bladder cancer treated with Bacillus Calmette-Guerin. Oncotarget. 2017;8:88782-88791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang Z, Lim YK, Lim HCC, et al. The role of Vitamin D receptor polymorphisms in predicting the response to therapy for nonmuscle invasive bladder carcinoma. J Urol. 2018;200:737-742. [DOI] [PubMed] [Google Scholar]
- 97. Yang X, Li P, Yang X, et al. TSP-1-1223 A/G polymorphism as a potential predictor of the recurrence risk of bladder cancer in a Chinese population. Int J Genomics. 2013;2013:473242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhao H, Liang D, Grossman HB, Wu X. Glutathione peroxidase 1 gene polymorphism and risk of recurrence in patients with superficial bladder cancer. Urology. 2005;66:769-774. [DOI] [PubMed] [Google Scholar]
- 99. Lin J, Dinney CP, Grossman HB, et al. E-cadherin promoter polymorphism (C-160A) and risk of recurrence in patients with superficial bladder cancer. Clin Genet. 2006;70:240-245. [DOI] [PubMed] [Google Scholar]
- 100. Ahirwar D, Kesarwani P, Manchanda PK, Mandhani A, Mittal RD. Anti- and proinflammatory cytokine gene polymorphism and genetic predisposition: association with smoking, tumor stage and grade, and Bacillus Calmette-Guerin immunotherapy in bladder cancer. Cancer Genet Cytogenet. 2008;184:1-8. [DOI] [PubMed] [Google Scholar]
- 101. Horikawa Y, Nadaoka J, Saito M, et al. Clinical implications of the MDM2 SNP309 and p53 Arg72Pro polymorphisms in transitional cell carcinoma of the bladder. Oncol Rep. 2008;20:49-55. [PubMed] [Google Scholar]
- 102. Ahirwar DK, Mandhani A, Mittal RD. IL-8 –251 T > A polymorphism is associated with bladder cancer susceptibility and outcome after BCG immunotherapy in a northern Indian cohort. Arch Med Res. 2010;41:97-103. [DOI] [PubMed] [Google Scholar]
- 103. Jaiswal PK, Goel A, Mandhani A, Mittal RD. Functional polymorphisms in promoter survivin gene and its association with susceptibility to bladder cancer in North Indian cohort. Mol Biol Rep. 2012;39:5615-5621. [DOI] [PubMed] [Google Scholar]
- 104. Wang M, Chu H, Li P, et al. Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer Res. 2012;72:6173-6182. [DOI] [PubMed] [Google Scholar]
- 105. Deng X, Zhang X, Cheng Y, et al. XRCC1 polymorphisms associated with survival among Chinese bladder cancer patients receiving epirubicin and mitomycin C. Tumour Biol. 2015;36:4591-4596. [DOI] [PubMed] [Google Scholar]
- 106. Li P, Zhang X, Deng X, et al. Pharmacogenetic association between XRCC1 polymorphisms and improved outcomes in bladder cancer patients following intravesical instillation of epirubicin. Int J Clin Exp Med. 2015;8:11167-11173. [PMC free article] [PubMed] [Google Scholar]
- 107. Ryk C, Koskela LR, Thiel T, et al. Outcome after BCG treatment for urinary bladder cancer may be influenced by polymorphisms in the NOS2 and NOS3 genes. Redox Biol. 2015;6:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ryk C, Wiklund NP, Nyberg T, De Verdier PJ. Ser608Leu polymorphisms in the nitric oxide synthase-2 gene may influence urinary bladder cancer pathogenesis. Scand J Urol Nephrol. 2011;45:319-325. [DOI] [PubMed] [Google Scholar]
- 109. Sakano S, Berggren P, Kumar R, et al. Clinical course of bladder neoplasms and single nucleotide polymorphisms in the CDKN2A gene. Int J Cancer. 2003;104:98-103. [DOI] [PubMed] [Google Scholar]
- 110. Basturk B, Yavascaoglu I, Oral B, Goral G, Oktay B. Cytokine gene polymorphisms can alter the effect of Bacillus Calmette-Guerin (BCG) immunotherapy. Cytokine. 2006;35:1-5. [DOI] [PubMed] [Google Scholar]
- 111. Xu ZC, Cai HZ, Li X, et al. ERCC1 C118T polymorphism has predictive value for platinum-based chemotherapy in patients with late-stage bladder cancer. Genet Mol Res. 2016;15:1-9. [DOI] [PubMed] [Google Scholar]
- 112. Hess J, Stelmach P, Eisenhardt A, et al. Impact of BCL2 polymorphisms on survival in transitional cell carcinoma of the bladder. J Cancer Res Clin Oncol. 2017;143:1659-1670. [DOI] [PubMed] [Google Scholar]
- 113. Castillejo A, Rothman N, Murta-Nascimento C, et al. TGFB1 and TGFBR1 polymorphic variants in relationship to bladder cancer risk and prognosis. Int J Cancer. 2009;124:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Teo MT, Dyrskjot L, Nsengimana J, et al. Next-generation sequencing identifies germline MRE11A variants as markers of radiotherapy outcomes in muscle-invasive bladder cancer. Ann Oncol. 2014;25:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Eisenhardt A, Siffert W, Rosskopf D, et al. Association study of the G-protein beta3 subunit C825T polymorphism with disease progression in patients with bladder cancer. World J Urol. 2005;23:279-286. [DOI] [PubMed] [Google Scholar]
- 116. Teo MT, Landi D, Taylor CF, et al. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis. 2012;33:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Shinohara A, Sakano S, Hinoda Y, et al. Association of TP53 and MDM2 polymorphisms with survival in bladder cancer patients treated with chemoradiotherapy. Cancer Sci. 2009;100:2376-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Chen M, Gu J, Delclos GL, et al. Genetic variations of the PI3K-AKT-mTOR pathway and clinical outcome in muscle invasive and metastatic bladder cancer patients. Carcinogenesis. 2010;31:1387-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Andrew AS, Gui J, Sanderson AC, et al. Bladder cancer SNP panel predicts susceptibility and survival. Hum Genet. 2009;125:527-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mason RA, Morlock EV, Karagas MR, et al. EGFR pathway polymorphisms and bladder cancer susceptibility and prognosis. Carcinogenesis. 2009;30:1155-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sacerdote C, Guarrera S, Ricceri F, et al. Polymorphisms in the XRCC1 gene modify survival of bladder cancer patients treated with chemotherapy. Int J Cancer. 2013;133:2004-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Djukic TI, Savic-Radojevic AR, Pekmezovic TD, et al. Glutathione S-transferase T1, O1 and O2 polymorphisms are associated with survival in muscle invasive bladder cancer patients. PLoS ONE. 2013;8:e74724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Wray NR, Wijmenga C, Sullivan PF, Yang J, Visscher PM. Common disease is more complex than implied by the core gene omnigenic model. Cell. 2018;173:1573-1580. [DOI] [PubMed] [Google Scholar]
- 124. Grotenhuis AJ, Dudek AM, Verhaegh GW, et al. Prognostic relevance of urinary bladder cancer susceptibility loci. PLoS ONE. 2014;9:e89164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694-9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ba X, Boldogh I. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1-8. [DOI] [PubMed] [Google Scholar]
- 128. Zou H, Li Q, Xia W, Liu Y, Wei X, Wang D. Association between the OGG1 Ser326Cys polymorphism and cancer risk: evidence from 152 case-control studies. J Cancer. 2016;7:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhou PT, Li B, Ji J, Wang MM, Gao CF. A systematic review and meta-analysis of the association between OGG1 Ser326Cys polymorphism and cancers. Med Oncol. 2015;32:472. [DOI] [PubMed] [Google Scholar]
- 130. Wenjuan C, Jianzhong L, Chong L, et al. The hOGG1 Ser326Cys gene polymorphism and susceptibility for bladder cancer: a meta-analysis. Int Braz J Urol. 2016;42:883-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hellwege JN, Keaton JM, Giri A, Gao X, Velez Edwards DR, Edwards TL. Population stratification in genetic association studies. Curr Protoc Hum Genet. 2017;95:1.22.1-1.22.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_Tables_1_12_Sep24_xyz268777d1d25d8 for Systematic Review: Genetic Associations for Prognostic Factors of Urinary Bladder Cancer by Nadezda Lipunova, Anke Wesselius, Kar K Cheng, Frederik J van Schooten, Jean-Baptiste Cazier, Richard T Bryan and Maurice P Zeegers in Biomarkers in Cancer