Abstract
Prokaryotes deploy CRISPR-Cas–based RNA-guided adaptive immunity to fend off mobile genetic elements such as phages and plasmids. During CRISPR adaptation, which is the first stage of CRISPR immunity, the Cas1-2 integrase complex captures invader-derived prespacer DNA and specifically integrates it at the leader-repeat junction as spacers. For this integration, several variants of CRISPR-Cas systems use Cas4 as an indispensable nuclease for selectively processing the protospacer adjacent motif (PAM) containing prespacers to a defined length. Surprisingly, however, a few CRISPR-Cas systems, such as type I-E, are bereft of Cas4. Despite the absence of Cas4, how the prespacers show impeccable conservation for length and PAM selection in type I-E remains intriguing. Here, using in vivo and in vitro integration assays, deep sequencing, and exonuclease footprinting, we show that Cas1-2/I-E—via the type I-E–specific extended C-terminal tail of Cas1—displays intrinsic affinity for PAM containing prespacers of variable length in Escherichia coli. Although Cas1-2/I-E does not prune the prespacers, its binding protects the prespacer boundaries from exonuclease action. This ensures the pruning of exposed ends by exonucleases to aptly sized substrates for integration into the CRISPR locus. In summary, our work reveals that in a few CRISPR-Cas variants, such as type I-E, the specificity of PAM selection resides with Cas1-2, whereas the prespacer processing is co-opted by cellular non-Cas exonucleases, thereby offsetting the need for Cas4.
Keywords: CRISPR-Cas, protein-nucleic acid interaction, protein-DNA interaction, Escherichia coli (E. coli), molecular biology, microbiology, bacterial immunity, Cas1-Cas2, exonuclease, prespacer processing, CRISPR adaptation, protospacer-adjacent motif (PAM), Cas1, Cas2, prespacer integration
Introduction
Prokaryotes utilize an adaptive immune response mediated by CRISPR and CRISPR-associated proteins (Cas)2 to respond to infections by mobile genetic elements (MGE) (viz. phages and plasmids) (1–4). CRISPR encompasses a typical architecture that comprises an array of direct repeats (∼30–40 bp) (5), which are partitioned by short spacer sequences of viral origin (6, 7). The repository of spacers acts as a vaccination card, and this genetic memory acquired during pathogenic invasions guides the adaptive immune response by CRISPR-Cas (1, 2). The molecular choreography of CRISPR-Cas defense is trichotomized into (i) adaptation, (ii) maturation, and (iii) interference stages (3). Upon phage attack, the CRISPR adaptation machinery derives short stretches of nucleic acids (prespacers) from the invaders and incorporates them into the CRISPR array. This process generates an infection memory and immunizes the host (2, 4, 8–10). An A-T–rich leader region present upstream to the repeat-spacer array regulates the transcription of the CRISPR locus, and the subsequent Cas nuclease–mediated processing of this inert transcript generates short regulatory mature CRISPR RNA (11–14). In association with other Cas proteins, the CRISPR RNA forms a surveillance complex that detects the recurring infections by MGE via base complementarity. This event triggers the Cas nucleases to annihilate the MGE, thereby interfering with the spread of infection (1–3, 13).
The CRISPR adaptation machinery expands the spacer archive, thus bestowing on the host a compendium of immunological memory to counter the ever-evolving phages. Highly conserved Cas1 and Cas2 spearhead the spacer acquisition (9, 10, 15); however, previous investigations have also demonstrated the indispensable role of additional Cas proteins and host factors in the uptake of spacers that are derived from phage DNA and RNA (16–28). Although the spacer sequences are extremely variable, the majority of the organisms (CRISPR types I, II, and V) display conservation of a 2–5-nt protospacer adjacent motif (PAM) at its origin site in MGE (8, 10, 29–31). In addition to specifying the prespacer region for integration, PAM also guides the differentiation between self and nonself during the interference step (32–34). Mutations in the PAM region on MGE lead to impaired target recognition, thus evading CRISPR-Cas immune response. In such circumstances, the imperfectly paired surveillance complex at the target region directs the interference machinery and Cas1-2 to display an inflammatory immune response by rapidly acquiring more spacers by a process termed “primed adaptation” (24, 35–38).
The CRISPR adaptation pathway proceeds via two sequential events: the capture of the prespacer fragments from invading MGE and the site-specific integration of these captured fragments at the leader-repeat junction. The CRISPR adaptation machinery of Escherichia coli (type I-E) derives its spacers predominantly from the DNA debris generated during the action of a multisubunit RecBCD DNA repair complex (18). Modulation in helicase, exonuclease, and endonuclease activities of RecBCD results in the production of ssDNA fragments ranging from tens to thousands of nucleotides in length (39, 40). However, the legitimate spacers in E. coli are strictly 33 bp in length and abut a 5′-AAG-3′ PAM (where “G” is destined to be the first residue of the spacer) (10, 30, 33, 35, 41, 42). Additionally, structural studies have demonstrated that Cas1-2/I-E efficiently binds to partial duplex prespacers that are 33 bp in length (42, 43). The existence of such spacer-sized DNA fragments is infinitesimal among the RecBCD products. Moreover, a previous study also demonstrated the incorporation of 33-bp spacers that are directly acquired from electroporated longer DNA duplexes (63 bp) (44). These findings augment the involvement of an additional DNA-trimming step to generate befitting substrates for CRISPR adaptation.
Recent studies in Sulfolobus solfataricus (type I-A), Sulfolobus islandicus (type I-A), Bacillus halodurans (type I-C), Synechocystis sp. 6803 (type I-D), Pyrococcus furiosus (type I-G), and Geobacter sulfurreducens (type I-U) (25, 45–51) highlighted the indispensable role of Cas4 nuclease in PAM selection and prespacer processing. The occurrence of Cas4 is prevalent in type I CRISPR-Cas systems except in subtypes I-E and I-F (15). In one case, it was observed that an extended variant of Cas2 with an unorthodox C terminus DnaQ exonuclease domain assists Streptococcus thermophilus DGCC7710 (type I-E) in prespacer trimming (52). Cas2-DnaQ domain fusion is nonubiquitous, and even model organisms for spacer acquisition studies like E. coli (type I-E) do not harbor it. Although recent studies envisage the involvement of exonucleases during spacer acquisition in E. coli (53), the molecular events guiding PAM selectivity and prespacer processing remain obscure.
Upon production of legitimate prespacers, Cas1-2 integrase complex catalyzes the prespacer incorporation at the leader-adjoining repeat (10, 23, 24, 41, 54–56). This polarized mechanism records the chronology of infections by positioning the new spacer toward a promoter encompassing leader. During recurring infections, swift expression of the latest spacers ensures a productive fight by a rapid and robust immune response (54). In conjunction with Cas1-2, various conserved DNA motifs present in the leader and repeat regions mandate the fidelity of prespacer integration (16, 17, 54, 57–65). In E. coli (type I-E), upon interaction with these conserved regions, a sequence-specific genome architectural protein termed integration host factor (IHF) restructures the leader and generates a docking site for Cas1-2 integrase at the leader-repeat junction (16, 17, 66). In contrast, the presence of a Cas1-2–binding site abutting the leader proximal region obviates the involvement of host factors during spacer acquisition in the CRISPR type II-A system (54, 67, 68).
Unlike many of the type I CRISPR-Cas encompassing prokaryotes, E. coli (type I-E) lacks Cas nucleases like Cas4 and Cas2-DnaQ to generate processed prespacers for efficient homing at the CRISPR locus (15). Nevertheless, this inadequacy does not appear to hinder PAM selection or spacer size preference (10, 30, 33, 41, 44). Intrigued by these observations, we sought to understand how prespacers are selected and tailored to the appropriate size for CRISPR adaptation. Here, we demonstrate that the PAM-directed interactions with longer DNA fragments signals Cas1-2 to demarcate potential prespacer boundaries. Upon supplementing the reaction with exonucleases to mimic the cellular environment, we found that Cas1-2-DNA nucleoprotein complex could protect DNA fragments of ∼33 bp in length. Further, we show that these protected fragments could be efficiently integrated into the CRISPR locus. These findings demystify the mechanism by which E. coli efficiently scales the fragments of the foreign DNA to generate viable prespacers of desired length, in contrast to other CRISPR-Cas subtypes that possess dedicated prespacer-processing nucleases such as Cas4.
Results
Length of the prespacers dictates their integration at the CRISPR array
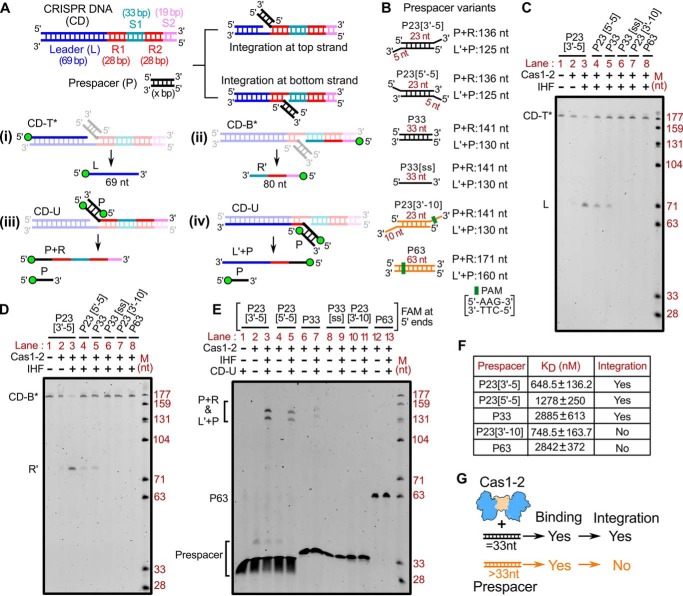
We utilized a previously established in vitro spacer integration assay (17) to understand the parameters that necessitate the uptake of prespacers during CRISPR adaptation. In this assay, we employed Cas1-2 integrase, IHF, prespacers, and linear CRISPR DNA (69-bp leader followed by two repeat-spacer units) (Fig. 1A and Fig. S1A). Spacers in E. coli are routinely derived from the remnant ssDNA fragments generated by the action of RecBCD-mediated double-stranded break repair (18). Upon reannealing to their complementary sequences, variably sized DNA fragments that encompass blunt ends, 3′- or 5′-overhangs could be generated. To simulate these conditions in vitro, we employed various types of DNA fragments, such as P33 (33-bp duplex), P33[ss] (33-nt ssDNA), P23[3′-5] (23-bp duplex with 5-nt 3′-overhangs), P23[5′-5] (23-bp duplex with 5-nt 5′-overhangs), P23[3′-10] (23-bp duplex with 10-nt 3′-overhangs), and P63 (63-bp blunt duplex) as prespacers in the integration assays (Fig. 1B).
Figure 1.
Prespacer length regulates the fate of spacer integration at CRISPR locus. A, schema of CRISPR DNA (CD) and prespacer (P) used in Cas1-2–mediated prespacer integration assay. Regions corresponding to 69-bp leader (L in blue), 28-bp repeats (R1-R2 in red), 33-bp spacer 1 (S1 in cyan), and 19-bp spacer 2 (S2 in magenta) of the CRISPR DNA are indicated. Two integration events resulted from 3′-OH nucleophilic attack of Cas1-2–bound prespacer at the top strand (L-R1 junction) and bottom strand (R1-S1 junction), and their respective denatured DNA fragments are shown. The design of integration assays to determine the positions of nucleophilic attack (top strand (i) and bottom strand (ii)) and prespacer ligation (top strand (iii) and bottom strand (iv)) are displayed. B, pictures depicting various prespacers that are employed in the integration assay. Prespacers with an overall length of 33 nt are colored black, whereas those of >33 nt are represented in orange. Sizes of the respective DNA fragments due to top strand integration (P+R) and bottom strand integration (L′+P) are indicated. C–E, representative denaturing polyacrylamide gels displaying the prespacer integration at the top strand (C) or at the bottom strand (D) or both (E). Absence (−) or presence (+) of each reaction component and the type of prespacer used in each sample are indicated at the top of the respective lanes. DNA molecular weight marker (M) positions are shown on the right. The positions of intermediate products of integration (L, R′, P+R, and L′+P) are displayed to the left of the respective gels. F, equilibrium disassociation constant values (KD) of Cas1-2 with each type of prespacer substrate. The success (Yes) or failure (No) of integration for each prespacer is shown. G, cartoon depicting the relationship between prespacer length (33 and >33 nt) and Cas1-2 (in blue and brown)-mediated binding and integration. The possibility of binding and integration of each prespacer is denoted as Yes or No.
Upon incubation with Cas1-2 and IHF, prespacer integration proceeds via a trans-esterification reaction, wherein 3′-OH of the Cas1-2–bound prespacer makes two nucleophilic attacks at the target sites to get itself ligated into the CRISPR array (L-R1 junction in the top strand and R1-S1 junction in the bottom strand) (Fig. 1A) (55, 56). To precisely identify the site of nucleophilic attack, we employed the CRISPR DNA substrates that were 5′-end–labeled with fluorescein (FAM) either on the top strand (CD-T* in Fig. 1A (i)) or on the bottom strand (CD-B* in Fig. 1A (ii)). Likewise, to monitor the prespacer ligation, we used an unlabeled CRISPR DNA (CD-U in Fig. 1A (iii and iv)) and prespacers with FAM at their 5′-ends. Here, we observed that P23[3′-5] or P23[5′-5] or P33 could alone make a successful nucleophilic attack at the integration sites and result in the generation of top strand (L) and bottom strand (R′) cleavage products from CD-T* and CD-B*, respectively (lanes 3–5 in Fig. 1 (C and D); Fig. S1 (B and C)). Further, utilizing 5′-FAM–labeled prespacers, we observed the ligation of P23[3′-5], P23[5′-5], and P33 at these nicked sites on the top strand (P + R) and bottom strand (L′ + P) (lanes 3, 5, and 7 in Fig. 1E; Fig. S1 (B and C)).
Interestingly, we did not observe bands corresponding to the integration products when we substituted the reaction mixtures with P33[ss], P23[3′-10], and P63 (lanes 6, 7, and 8 in Fig. 1 (C and D); lanes 9, 11, and 13 in Fig. 1E; Fig. S1 (B and C)). These findings suggest that either duplex (P33) or partial duplex (comprising 3′-overhang (P23[3′-5]) or 5′-overhang (P23[5′-5])) prespacers with an effective length of 33 nt are strictly required during CRISPR adaptation. This bias in prespacer size preference could possibly arise due to the weakening of Cas1-2 interaction with long substrate precursors (such as P23[3′-10] and P63 with an effective length of 43 and 63 nt, respectively) and/or inefficient integration of such DNA fragments at the target site in the CRISPR locus. To test whether longer prespacers (>33 nt) have weak interaction with Cas1-2, thereby leading to inefficient integration, we performed EMSA to assess the interaction between Cas1-2 integrase and various prespacers (Fig. S2 and Fig. 1F). This indicated that the affinity of Cas1-2 toward P23[3′-10] (KD = 748.5 ± 163.7 nm) is comparable with its affinity to P23[3′-5] (KD = 648.5 ± 136.2 nm). Similarly, the affinity of P63 (KD = 2.842 ± 0.372 μm) for Cas1-2 is on par with that of P33 (KD = 2.885 ± 0.613 μm). These experiments suggest that Cas1-2 can interact with DNA fragments of varying lengths; however, the integration at the target site could be achieved only in the presence of DNA fragments with an effective length of 33 nt (Fig. 1G).
Cas1-2 foothold protects potential prespacer regions during exonuclease action
CRISPR adaptation in E. coli mandates precisely sized prespacers (Fig. 1). To cater to this need, long DNA fragments generated during RecBCD activity have to be trimmed further by nuclease action. Of the multitude of proteins encoded by the Cas operon, only Cas1 and Cas2 contribute to naive spacer acquisition in E. coli. We could not notice processing of longer prespacers (P23[3′-10] and P63) even at higher Cas1-2 concentrations (Fig. S3A).
The type I-E system is devoid of prespacer processing exonuclease Cas4, and its deficit cannot be complemented by Cas1-2 alone (Fig. S3A). Hence, we predicted the involvement of cytoplasmic exonucleases in trimming the longer prespacer to a suitable length. Having established that Cas1-2 indeed binds DNA fragments of variable length (Fig. 1F and Fig. S2), we sought to test the fate of Cas1-2–bound DNA fragments against exonucleases. Here, Cas1-2-P63 nucleoprotein complex was treated with a mixture containing 5′→3′–acting T5 exonuclease (T5exo) and 3′→5′–acting exonuclease III (ExoIII) (Fig. 2A). To our surprise, we identified a smear of protected DNA fragments (P63exo+) that ranged from 30 to 40 nt in the sample containing both P63 and Cas1-2 (lane 8 in Fig. 2A and lanes 4–13 in Fig. S3B), whereas such protection was not observed when we treated P63 in the absence of Cas1-2 or in the presence of either Cas1 or Cas2 (lanes 5, 6, and 7 in Fig. 2A). Coincidentally, the length of the protected fragments corresponded to legitimate spacer size in E. coli (∼33 nt). Additionally, it was noted that this protection was absent when Cas1-2-P63 was treated with ExoIII alone (lane 11 in Fig. 2A). The nuclease generates 5′-overhangs with which Cas1-2 binds weakly (compare P23[3′-5] and P23[5′-5] in Fig. 1F and Fig. S2). Therefore, it appears that ExoIII seems to dislodge the weakly bound Cas1-2 from its position on the prespacer. In contrast, the treatment of Cas1-2-P63 with 5′→3′–acting T5exo resulted in incompletely digested fragments that were predominantly of greater length (∼45 nt) than that of P63exo+ (compare lanes 8 and 13 in Fig. 2A).
Figure 2.
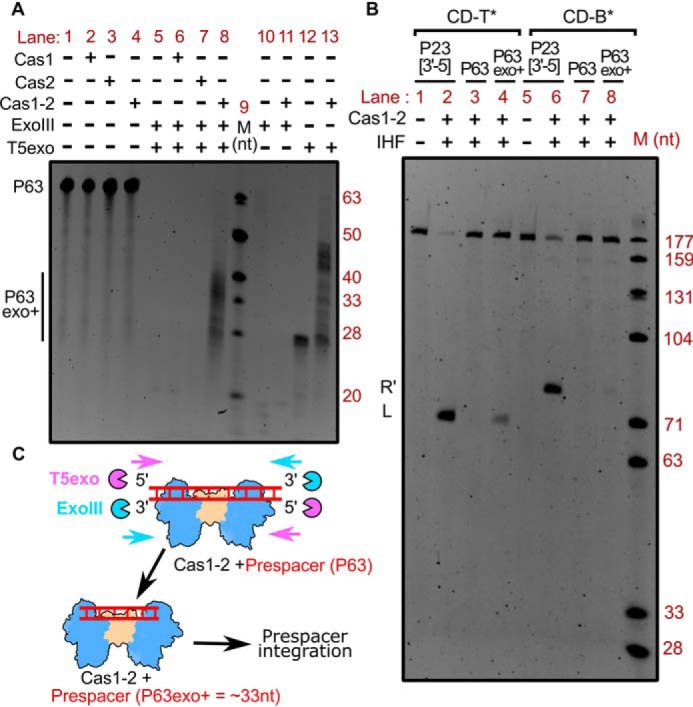
Tailoring of Cas1-2 bound large DNA fragments by exonucleases generates integration-competent prespacers. A, denaturing PAGE depicting the nuclease treatment of Cas1-2 bound P63 DNA fragments. The presence (+) or absence (−) of each reaction component is indicated at the top of each lane. Positions corresponding to the substrate (P63) and T5 exo/ExoIII-digested DNA fragments (P63exo+) are indicated on the left, whereas oligonucleotide marker (M) positions are shown on the right. B, denaturing PAGE displaying the integration reactions employing various prespacers (P23[3′-5] (lanes 1 and 2 and lanes 5 and 6), P63 (lanes 3 and 7), and Cas1-2 protected DNA fragments (P63exo+) (lanes 4 and 8)) and CRISPR DNA substrates (CD-T* (lanes 1–4) and CD-B* (lanes 5–8)) is shown. The presence (+) or absence (−) of each reaction component is indicated at the top of each lane. Positions corresponding to labeled DNA products that result from prespacer nucleophilic attack (L and R′) are displayed on the left. The DNA molecular weight marker (M) positions are shown on the right. C, schema illustrating the mechanism of Cas1-2–mediated protection of prespacer boundaries. Cas1-2 (in blue and brown), T5 exonuclease (magenta pie), exonuclease III (cyan pie), and prespacer P63 (red ladder) are portrayed.
As the Cas1-2–protected P63exo+ fragments were approximately of E. coli spacer size, we wondered whether they could act as potential prespacers for integration. To test this hypothesis, we purified and utilized P63exo+ DNA fragments as prespacers in a spacer integration assay. In line with the previous experiment (Fig. 1), we could not observe any integration events when we employed longer prespacer P63 (lanes 3 and 7 in Fig. 2B). To our surprise, integration was observed when P63exo+ was employed (lanes 4 and 8 in Fig. 2B; lane 7 in Fig. S3C). In this case, although we monitored efficient nucleophilic attack at the top strand (L in lane 4 of Fig. 2B), the integration at the bottom strand seemed to be sparse (R′ in lane 8 of Fig. 2B). By recapitulating these observations, we could suggest that Cas1-2–mediated binding of large DNA fragments secures the boundaries of suitable prespacers from the exonucleolytic action of cellular nucleases in E. coli (Fig. 2C).
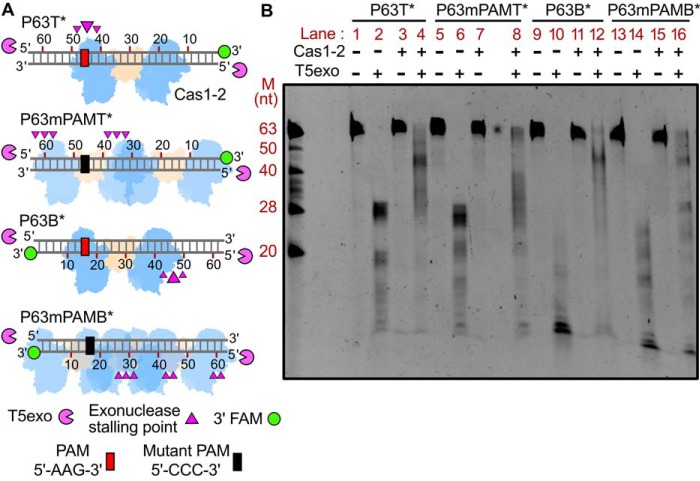
PAM-directed binding of Cas1-2 defines the boundary for prespacers
Because Cas1-2 binding was shown to mark the spacer boundaries (Fig. 2), we sought to identify and map these regions. As no protected DNA fragments appeared upon treatment of Cas1-2-P63 complex with ExoIII alone (lane 11 in Fig. 2A), we sought to utilize T5exo digestion (lane 13 in Fig. 2A) to demarcate the prespacer boundaries defined by Cas1-2. To accomplish this, we utilized variants of 63-bp blunt-ended prespacers that encompass fluorescein-labeled 3′-end (6-FAM) either on the top (P63T*) or on the bottom (P63B*) strand (Fig. 3A). Further, to identify the footprints of Cas1-2 on these 3′-end–labeled prespacers, we incubated Cas1-2–bound prespacer complex with T5exo. Here, the Cas1-2 binding on the prespacer acts as a roadblock that stalls the 5′→3′ progression of T5exo. The length of the resultant labeled fragments specifies the stalling points of the exonuclease, which, in turn, indicates the binding position of Cas1-2 on the prespacer. Utilizing this approach, we mapped the cleavage termination points on the top and bottom strands of the prespacer. After T5exo treatment of P63T*, we could observe an ∼28-nt labeled fragment. This is indicative of an inherent nuclease stalling point (in the absence of roadblocks such as Cas1-2) around the 28th nt position from the labeled end in P63T* (lane 2 in Fig. 3B; lane 12 in Fig. 2A). However, owing to complete exonucleolytic cleavage of the bottom strand, we did not observe such stalling on P63B* (lane 10 in Fig. 3B). Upon T5exo treatment of Cas1-2–bound P63T* complex, we noticed a shift in the nuclease stalling point to about the ∼45th nt position from the labeled end (lane 4 in Fig. 3B). This maps the Cas1-2–binding position to be around 45 nt from the labeled end (P63T* in Fig. 3A). Coincidentally, this binding position of Cas1-2 on P63T* is localized around a cognate PAM sequence (5′-AAG-3′ ranging from 47 to 49 nt upstream of the labeled position) (P63T* in Fig. 3A).
Figure 3.
Cas1-2 complex is predominantly localized around the PAM region. A, schematic representation of various fluorescein-labeled prespacer substrates (P63T*, P63mPAMT*, P63B*, and P63mPAMB* as a gray ladder) used in the assay. Labeled 3′-end of each prespacer is highlighted as green circles, whereas PAM and its mutated variant are depicted as red and black boxes, respectively. Numbering on the DNA represents the distance (in nt) of a particular position from the labeled end. T5 exo (magenta pie) is positioned at susceptible 5′-ends of DNA substrate. Positions of T5exo stalling points (magenta triangles) and binding sites of Cas1-2 (blue and brown blobs) that are estimated from a nuclease footprinting assay performed in B are displayed. B, denaturing PAGE depicting the T5exo treatment of WT Cas1-2–bound fluorescein-labeled P63 variants (P63T*, P63mPAMT*, P63B*, and P63mPAMB*). The presence (+) or absence (−) of each reaction component is indicated at top of each lane. Positions corresponding to the DNA fragments of oligonucleotide marker (M) are shown on the left.
Prompted by this finding, we were interested in identifying the extent of protection that Cas1-2 could confer on the bottom strand upon its binding at the PAM region. To accomplish this, we treated the Cas1-2–bound P63B* complex with T5exo. Here, the resulting length of the protected fragments upon exonuclease treatment indicated that Cas1-2 complex interaction can guard a region spanning ∼45 nt from the labeled end in P63B* (lane 12 in Fig. 3B and P63B* in Fig. 3A). As the PAM residues are positioned at 14 nt from the labeled end of the P63B*, the effective length of the protected prespacer from the PAM is ∼30 nt. Overall, these results suggest a PAM-dependent mechanism by which Cas1-2 could selectively acquire 33-nt prespacers.
To test the role of PAM in defining the protospacer boundary, we employed mutated P63 DNA fragments (P63mPAMT* and P63mPAMB* in Fig. 3A) that are devoid of any cognate E. coli PAM sequence (5′-AWG-3′, where W represents A/T). These labeled fragments were incubated with Cas1-2 and later treated with T5exo. Here, we found extended smears and multiple bands upon employing P63mPAMT* (Fig. 3A and lane 8 in Fig. 3B) and P63mPAMB* (Fig. 3A and lane 16 in Fig. 3B). The varied length of the resultant labeled fragments is indicative of numerous stalling points on these P63 mutants (Fig. 3A) that occurred due to Cas1-2 binding. These results highlight that Cas1-2 gets specifically recruited toward a PAM-containing region that plays a crucial role in defining prespacer boundaries, whereas such specificity is lost when the DNA fragments lack PAM. Here the promiscuous interaction of Cas1-2 results in the generation of illicit prespacers that defy the productive length of the prespacers for integration (lanes 8 and 16 in Fig. 3B; P63mPAMT* and P63mPAMB* in Fig. 3A).
Intrinsic specificity of Cas1-2 circumvents the requirement of Cas4 during PAM selection in E. coli
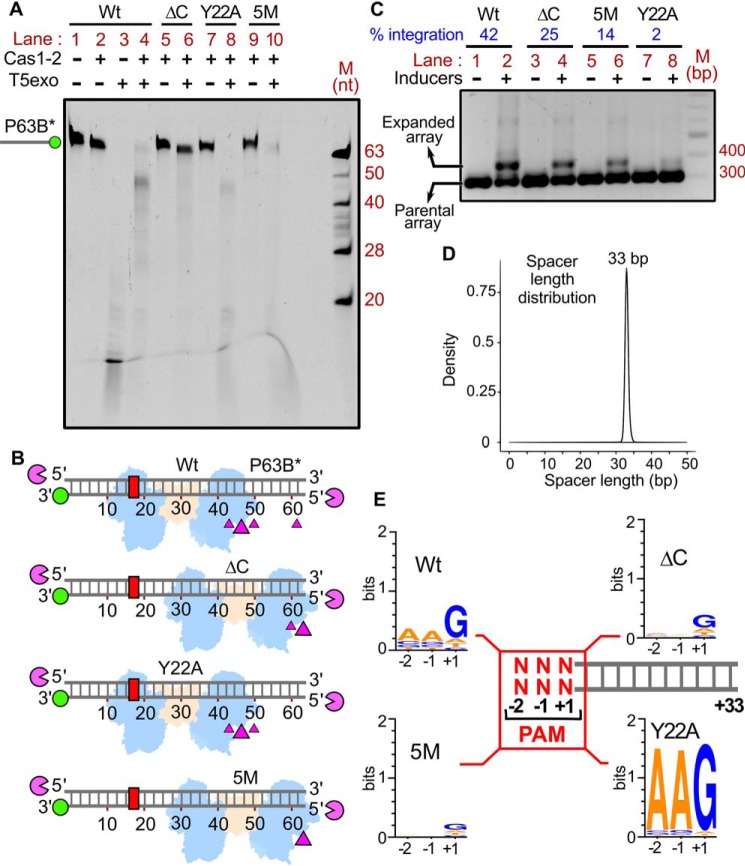
Having found the role of PAM-mediated interaction in selecting the prespacers for uptake, we attempted to understand the intrinsic molecular principles that confer precision to Cas1-2 in PAM selectivity and prespacer scaling. Previous structural studies of CRISPR adaptation complex in E. coli suggested that the extended Cas1 C-terminal tail of apoCas1-2 complex (69) gets organized around the PAM residues upon binding to prespacer DNA (Fig. 4) (42). In particular, the Gln-287 and Ile-291 residues of this proline-rich C-terminal tail make direct contacts with the nucleotides of PAM (Fig. 4B), thus possibly imparting the PAM specificity. In another striking feature, a pair of Tyr-22 residues that are derived from two different Cas1 protomers scales a 23-bp duplex region of prespacer via stacking interactions at either end (Fig. 4B) (42, 43). This gating mechanism at the Cas1-2 platform seems to scale the spacer length by facilitating the positioning of 3′-overhang at the catalytic groove for integration (Fig. 4B). To validate these observations, we employed Cas1-2 variants that encompass either deletion of Cas1 C-terminal tail (ΔC; ΔP279-S305) or a Cas1 point mutant, Y22A (Fig. S4A), and analyzed their footprints upon binding to P63B* and P63T* (Fig. 5A and Fig. S5D). As a control, we also used a Cas1 variant (5M; Q24H, P202Q, G241D, E276D, and L297Q) (Fig. 4B) that was previously shown to abrogate PAM selectivity (44).
Figure 4.
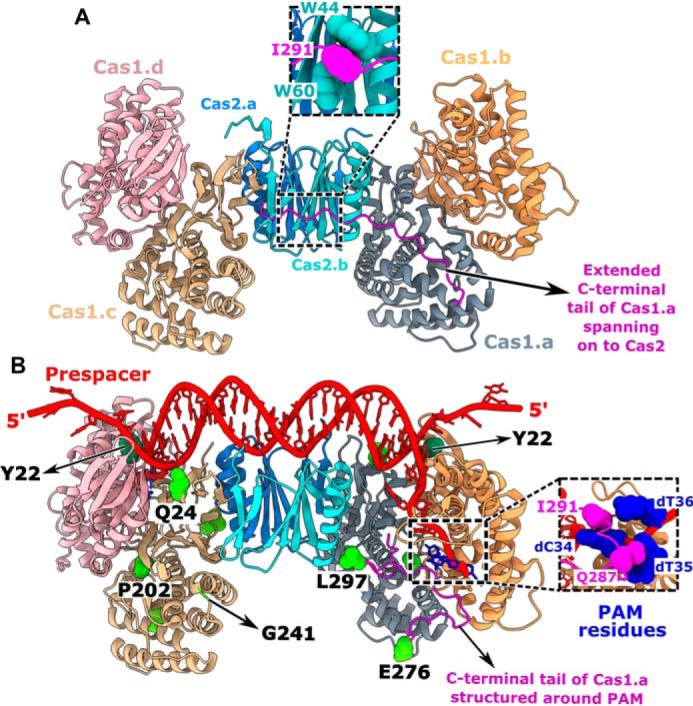
Structural features of Cas1-2 that determine the prespacer selection. A and B, structural comparison of apo-Cas1-2 integrase (PDB code 4P6I) (A) and Cas1-2-prespacer complex (PDB code 5DQZ) (B). Four protomers of Cas1 (indicated as Cas1.a to -d) and two protomers of Cas2 (indicated as Cas2.a and -b) are shown in different colors. The stacking of Cas2 tryptophan residues (Trp-44 and Trp-60 in cyan) with Ile-291 of the Cas1.a C-terminal tail (Gly-275 to Ser-305 in magenta) in apo-Cas1-2 (close-up view in A) and the interactions of Cas1.a Ile-291 and Gln-287 residues with PAM residues (in navy blue) cytosine 34 (dC34) and thymidine 35 (dT35) in Cas1-2-prespacer complex (close-up view in B) are shown. The Cas1 Tyr-22 residues (in dark green spheres) that stack the nucleic acid bases at the prespacer duplex ends are denoted (B). Amino acid residues corresponding to Cas1 mutations (Q24H, P202Q, G241D, E276D, and L297Q) in the 5M variant are displayed (green spheres) as part of the Cas1.a and Cas1.c protomers. For clarity, Glu-276 and Leu-297 of Cas1.a and Gln-24, Pro-202, and Gly-241 of Cas1.c are labeled at their respective positions.
Figure 5.
Intrinsic specificity of Cas1-2 integrase directs the uniformity in spacer length and PAM preference during CRISPR adaptation. A, denaturing PAGE depicting the T5exo treatment of Cas1-2 (WT (lanes 1–4), ΔC (lanes 5 and 6), Y22A (lanes 7 and 8), or 5M (lanes 9 and 10))-bound fluorescein-labeled P63B* is displayed. The presence (+) or absence (−) of each reaction component is indicated at the top of each lane. The position of P63B*-labeled DNA fragment is shown on the left, whereas oligonucleotide marker (M) positions are indicated on the right. B, schematic illustration of the footprinting assay performed in A. DNA substrate P63B* (gray ladder), positions of 3′-fluorescein label (green circle), and PAM region (red rectangle) are represented. Numbering on the DNA represents the distance (in nt) of a particular position from the labeled end. T5 exo (magenta pie) is positioned at susceptible 5′-ends of DNA substrate. Positions of T5exo stalling points (magenta triangles) and binding sites of each variant of Cas1-2 (WT, ΔC, Y22A, or 5M in blue and brown blobs) that are estimated from the nuclease footprinting assay performed in A are displayed. C, agarose gel depicting the PCR products from the spacer acquisition assay performed in E. coli harboring the plasmids that express the Cas1-2 variants (WT (lanes 1 and 2), ΔC (lanes 3 and 4), 5M (lanes 5 and 6), and Y22A (lanes 7 and 8)). The absence (−) or presence (+) of inducers is indicated at the top of each lane. Positions corresponding to parental and expanded arrays (CRISPR 2.1 array) are indicated on the left. The percentage of integration is displayed at the top of the respective lanes (indicated in blue). DNA marker (M) positions are represented on the right. D, overlay of plots depicting length distribution of the newly acquired spacers that are incorporated into CRISPR 2.1 array by Cas1-2 variants (WT, ΔC, 5M, and Y22A) during the in vivo integration assay (C). The x axis depicts the length of the spacer (nt), whereas normalized frequency (density) is indicated on the y axis. E, illustration depicting the PAM preference of Cas1-2 variants (WT, ΔC, 5M, and Y22A) during the in vivo integration assay (C). +1 to +33 sequence of each spacer (gray ladder) was extracted from high-throughput sequencing data. Subsequently, sequence information of −1 and −2 positions of each spacer was derived from the respective plasmid/genome sequence. The conservation profile of PAM sequences (−2, −1, and +1 in red) corresponding to the respective Cas1-2 variant is shown as a sequence logo.
Similar to WT Cas1-2, T5exo treatment of Y22A-P63B* generated a nuclease stalling point at ∼45 nt, albeit with reduced efficiency (compare lanes 4 and 8 in Fig. 5A). These findings indicate that Cas1 Y22A does not result in the altered PAM specificity (WT and Y22A in Fig. 5B). Despite having a stronger affinity for P63 (Y22A KD = 1.781 ± 0.202 μm versus WT KD = 2.842 ± 0.372 μm) (Figs. S2E and S5C), the absence of Tyr-22–mediated stacking interaction seems to reduce the prespacer protection ability of Y22A against nuclease action. Additionally, a shift in the nuclease stalling point to 60 nt from the labeled ends of P63B* was observed for ΔC and 5M variants (lanes 6 and 10 in Fig. 5A). Due to its low affinity, 5M seems to display reduced protection of prespacers from nuclease action (ΔC KD = 2.675 ± 0.282 μm versus 5M KD = 17.08 ± 3.428 μm) (Fig. S5, A and B). The shift in this nuclease stalling point indeed indicates that the ΔC and 5M variants display an impaired PAM specificity and were randomly interacting at the ends of P63B* (ΔC and 5M in Fig. 5B). To fortify our observations, we sought to understand the impact of these mutations on the sequence composition of acquired spacers by expressing Cas1-2 variants in E. coli IYB5101. We observed that the mutations in ΔC and 5M have partly reduced the spacer incorporation efficacy of Cas1-2 (compare lanes 2, 4, and 6 in Fig. 5C). Surprisingly, Y22A displayed a drastic reduction of spacer uptake in vivo (compare lanes 2 and 8 in Fig. 5C), despite showing spacer integration in vitro (Fig. S4B).
Expanded CRISPR arrays corresponding to the expression of each mutant were purified, and the sequences of newly incorporated spacers were derived from high-throughput sequencing. In line with previous studies, we observed that the spacers originated from both genome and plasmid (44). Irrespective of the mutations in Cas1-2, the length of the incorporated spacers is strictly conserved (i.e. 33 nt) (Fig. 5D). This finding suggests that Tyr-22–mediated stacking interaction with prespacer or the Cas1 C-terminal restructuring is dispensable for the scaling of prespacers. These spacer sequences were mapped onto the plasmid and genome to identify the PAM. Despite the display of precise prespacer scaling by Cas1-2 variants, the specificity toward PAM region appears to be profoundly altered (Fig. 5E). In concurrence with previous studies (30, 44), we observed that most of the spacers acquired by WT Cas1-2 encompass a conserved PAM region (5′-AAG-3′, where “G” indicates the +1-position of the 33-nt spacer) (Fig. 5E). In line with the nuclease protection assay (Fig. 5A), we did not observe any preference toward the PAM region when we employed 5M or ΔC (Fig. 5E). This finding bolsters the involvement of Cas1 C-terminal tail in PAM selectivity. Despite the reduced efficiency in spacer acquisition in vivo (Fig. 5C), surprisingly, Y22A displayed a remarkable precision for PAM selectivity, suggesting that this mutation bestowed high fidelity with respect to PAM recognition (Fig. 5E).
Discussion
The CRISPR system in E. coli (type I-E) displays precise scaling of prespacer length and stringent selectivity of PAM (30, 41, 44). Here, we attempted to uncover the elusive molecular events that drive the generation of competent substrates for homing at the leader-repeat junction by a CRISPR adaptation complex. During naive adaptation, prespacers are predominantly foraged from the DNA fragments generated by RecBCD during DNA repair (18) or by Cas3 during primed adaptation (35, 36, 70). Because they are helicase-nuclease enzymes, RecBCD and Cas3 action generates varied-length ssDNA fragments (39, 71). Despite the association of ssDNA fragments with Cas1 in vivo (72), their integration at the CRISPR locus has been highly inefficient compared with duplex and partial-duplex DNA forms (Fig. 1) (56). This points to a certitude that duplex formation by annealing of complementary ssDNA strands upon helicase-nuclease action could potentially generate prespacers that elicit CRISPR adaptation.
We demonstrate that irrespective of being a duplex or partial duplex, the overall length of the prespacer must be ∼33 nt (i.e. E. coli spacer length) for successful integration (Fig. 1). A previous study has shown that an in vitro reconstituted Cas1-2 can solely process the prespacers with 10-nt 3′-overhangs to legitimate spacer size (42). In contrast, our experiments show the absence of such prespacer processing even with increasing Cas1-2 concentrations, despite the presence of PAM (P23[3′-10] in Fig. S3A). We posit that these variations could be due to differences in the purification strategies of Cas1-2 complex in both studies. Here, we employed Cas1-2 that was generated as a complex in vivo and was further purified extensively, whereas the previous study (42) utilized in vitro reconstituted Cas1-2 that potentially contained unassembled, free Cas1 protomers. The utilization of high concentrations of the in vitro reconstituted Cas1-2 and thus the increased presence of free Cas1, a known endonuclease (73), could have resulted in the cleavage of the prespacer overhangs (42). Alternatively, because the nuclease activity was only seen at micromolar concentration of in vitro reconstituted Cas1-2 (42), despite the fact that the integrase activity requires just nanomolar concentration (17, 56), it points to the possibility of trace cellular nuclease contamination.
Previous structural studies of E. coli Cas integrase complex have revealed that the 33-bp length of DNA can be exactly accommodated between two active-site regions of Cas1-2 (42, 43). This hints at the fact that the Cas1-2 foothold can only mask the 33-bp region and the rest is exposed to potential nuclease action. Mimicking such conditions, we incubated Cas1-2–bound longer DNA fragments (P63) with 3′→5′ (ExoIII) and 5′→3′ (T5exo) acting exonucleases. These reactions resulted in the generation of fragments that are in the range of cognate E. coli spacer size (Fig. 2A). Furthermore, these processed DNA fragments were readily utilized as substrates by Cas1-2 integrase (Fig. 2B). Likewise, in B. halodurans (type I-C) and S. thermophilus DGCC7710 (type I-E), the nuclease action of Cas4 and DnaQ (an auxiliary domain of Cas2), respectively, on Cas1-2–bound DNA, generates integration-competent prespacers (45, 52). Intriguingly, as observed in E. coli, when Cas1-2 of these systems was presented with legitimately sized prespacers, the need for nuclease processing was obviated (45, 52). This implies that Cas1-2 can solely catalyze spacer integration, but the generation of productive prespacers involves the action of an additional nuclease. The lack of a known prespacer-processing enzyme such as Cas4 in E. coli led us to hypothesize that the trimming action could be complemented by other cellular nucleases. Unlike in other CRISPR variants (25, 45–50), the productive pruning of prespacers by non-Cas nucleases is not sequence-specific (Fig. 2). Moreover, a parallel study on prespacer generation in E. coli also independently demonstrated that the generic nucleases (such as DNA polymerase III or exonuclease T) are sufficient for trimming the prespacers upon PAM recognition by Cas1-2 (74). These explain why the involvement of specific nucleases, such as Cas4, is precluded for prespacer processing in E. coli (see below).
In addition to spacer length conservation, most prokaryotes display selective uptake of phage-origin prespacers that are bordered by a PAM (8). Recent in vivo studies in various type I organisms (I-A, I-D, I-G, and I-U) have underscored the indispensable requirement of Cas4 in PAM selection as well as in prespacer processing (46–50). Moreover, in vitro studies performed with adaptation complex of B. halodurans (type I-C) and S. solfataricus (type I-A) revealed that Cas4 nuclease avoids the processing of free DNA ends that are devoid of PAM sequence (25, 45). This preferential activity of Cas4 seems to act as a critical checkpoint in ensuring the productive uptake of infection memory by Cas1-2 in the hosts.
A previous study in E. coli showed that upon expression of Cas1-2, a 33-nt prespacer bordered by PAM originated from the longer electroporated DNA (P63) (44). Interestingly, we observed the protection of the same region by Cas1-2 when we performed a nuclease footprinting assay on P63 (Fig. 3). These experiments highlight that the Cas1-2 complex alone is sufficient to recognize PAM in E. coli. The footprinting experiments also demonstrate binding of substrates at multiple points when PAM residues of P63 were mutated (Fig. 3). These nonspecific interactions of Cas1-2 could generate a heterogeneous population of protected prespacers. This explains how the adaptation complex of various type I organisms infrequently uptake prespacers with erroneous PAM (WT in Fig. 5E) (35, 75–80).
Structural analysis of Cas1-2-prespacer complex highlights the features that could lead to precise scaling and PAM selection of prespacers. A platform formed by the interaction of a Cas2 dimer with two Cas1 dimers on either side houses the 23-bp duplex region of prespacer (42, 43). Stationed at either end of this duplex is the aromatic ring of Tyr-22 residue that stacks the prespacer at the border of the Cas1 catalytic groove and directs the 3′-overhang to position its fifth nt at the catalytic site. Thus, Tyr-22–guided meticulous placement of DNA substrate seems to dictate the length of prespacer (42, 43). Furthermore, the flexible C-terminal tail of Cas1 is molded around the PAM region. The absence of such molecular architecture upon mutating the PAM hints at the role of C-terminal tail in PAM recognition (42). Deployment of Cas1-2 variants that encompass either deletion of Cas1 C-terminal tail (ΔC) or Y22A in spacer integration assays helped to unveil the role of these structural entities in determining the PAM selection (Fig. 5). As shown here, the deletion of C-terminal tail resulted in impaired PAM recognition (Fig. 5A) and led to an uptake of prespacers that were lacking PAM (ΔC in Fig. 5E).
A comparison of the structures of Cas1 from various type I organisms (Fig. 6, A–D) revealed a striking contrast between Cas1 C-terminal tail of type I-E and other subtypes. The C-terminal tail of E. coli Cas1 is noticeably longer with 31 amino acids (aa) than the shorter 12-aa tails of Archeoglobus fulgidus, Pyrococcus horikoshii, and B. halodurans (Fig. 6) (42, 81, 82). Additional comparative analysis reinforced these observations that type I-E encompasses the longest C-terminal tail with an average length of 29 aa (Fig. 6E and Figs. S6–S9). Coincidentally, CRISPR-Cas subtypes with shorter Cas1 C-terminal tails, such as type I-A, I-B, and I-C, encompass Cas4 (15), and previous studies suggest the indispensability of Cas4 in promoting PAM specificity (25, 45–47). In contrast to these systems, it appears that the extended C-terminal tail of Cas1 in E. coli (type I-E) compensates for the lack of Cas4 by guiding the PAM selection.
Figure 6.
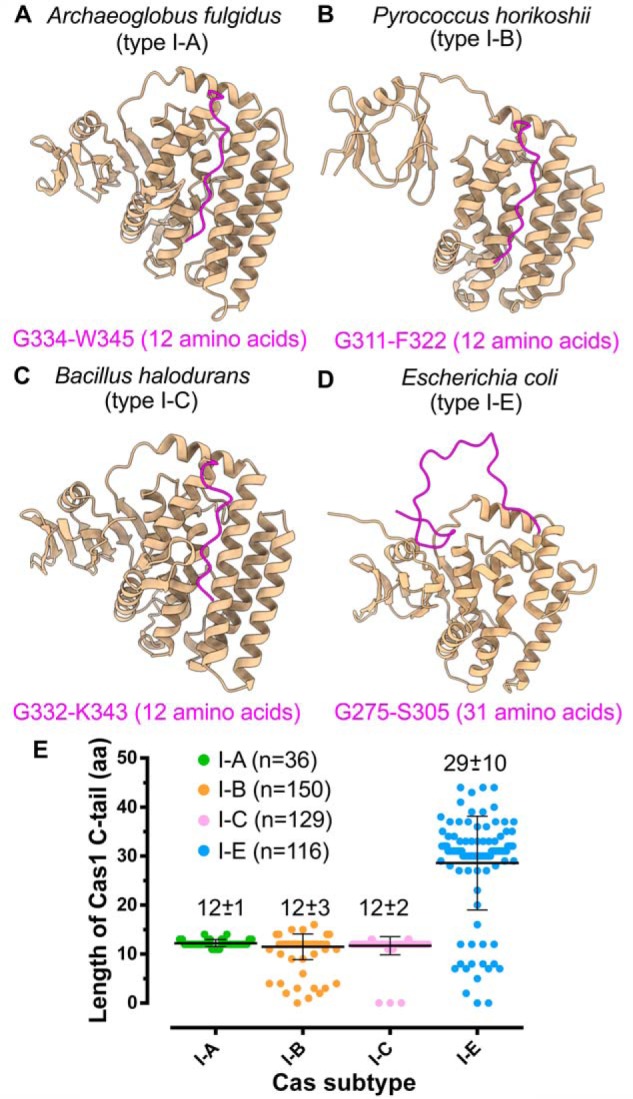
Cas1/I-E harbors extended C-terminal tail. A–D, structures highlighting Cas1 C-terminal tail (in magenta) of A. fulgidus (type I-A; PDB code 4N06) (A), P. horikoshii (type I-B; PDB code 4WJ0) (B), B. halodurans (type I-C; predicted model) (C), and E. coli (type I-E; PDB code 5DQZ) (D). The amino acids corresponding to the start and end positions of the C-terminal tail are displayed at the bottom of the respective structures. E, scatter plot representing the length differences among Cas1 of type I-A, I-B, I-C, and I-E. Each Cas subtype is shown in a different color, and the average length of amino acids in the C-terminal tail (mean ± S.D. (error bars)) of each subtype is indicated at the respective position. n corresponds to the number of Cas1 sequences from each subtype that are considered for the analysis (see “Experimental procedures”).
In the case of Y22A variant, although the prespacer integration is unaffected in vitro (Fig. S4B), in line with the previous observations (43), a rampant decrease in spacer integration efficiency was observed in vivo (Fig. 5C). Y22A conferred reduced prespacer protection against the nucleases compared with WT (Fig. 5A). This observation highlights the critical role of Tyr-22 residue in providing the WT with a better grip on bound DNA (Fig. 5A). As Y22A could lack such interactions with its substrates, nucleases might seamlessly dislodge it from the bound prespacer (Fig. 5A). This action appears to limit the substrate availability and impede spacer integration in vivo (Fig. 5C). Despite the reduction in spacer acquisition potential, Y22A showed high fidelity toward “AAG” PAM selection than WT (Fig. 5E). As discussed above, Y22A displays a reduced grip on the prespacers during nuclease-mediated processing (Fig. 5A). This weak prespacer binding could be further disrupted in the absence of PAM due to the loss of interactions with the Cas1 C-terminal tail. Therefore, only the presence of cognate PAM (AAG) is likely to allow Y22A to retain the hold on DNA during prespacer processing leading to selective enrichment (Fig. 5E). The strategic position of Cas1 Tyr-22 in the adaptation complex appears to have a key role in defining the prespacer length (Fig. 4B). Interestingly, our experiments with Y22A resulted in the uptake of prespacers that were predominantly 33 bp in length (Fig. 5D). These findings negate the involvement of Tyr-22–stacking interactions in deciding the prespacer boundary. Recent studies in type V-C demonstrated that a mini-integrase complex constituted by Cas1 tetramer prefers short (18-bp) spacers (83); likewise, in E. coli, the Cas1-2 structural framework alone appears to be a critical parameter in gauging the length of spacers (42, 43).
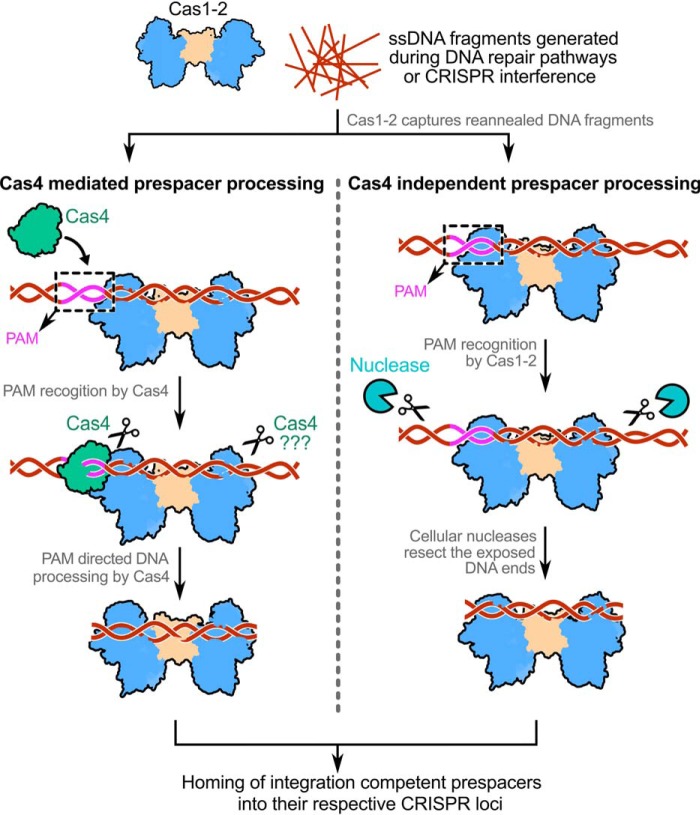
Our work in conjunction with the previous studies allows us to propose an updated model for prespacer capture in type I systems (Fig. 7). During CRISPR adaptation, the dispensability of sequence-specific auxiliary nucleases such as Cas4 seems to be contingent on the type of molecular players that are involved in PAM selection. Although Cas1-2 integrase catalyzes the prespacer homing, in the majority of type I systems, PAM selection and prespacer processing require Cas4 (25, 45–47). In contrast, in the type I-E system, the intrinsic affinity of Cas1-2 integrase alone is sufficient to recognize cognate PAM. This lineage-specific remarkable adaptation of Cas1-2/I-E offsets the requirement of PAM-specifying Cas4 nuclease. Instead, generic cellular non-Cas nucleases are co-opted to trim the exposed DNA ends of Cas1-2-prespacer complex for generating the legitimate prespacers for integration.
Figure 7.
Model depicting the mechanism of Cas4-dependent and -independent prespacer processing in type I CRISPR-Cas systems. The prespacer production in CRISPR systems encompasses a subset of two events: (i) PAM directed prespacer capture and (ii) processing of selected prespacer to a defined length for integration at the CRISPR locus. Generally, Cas1-2 captures long dsDNA fragments that are produced by reannealing of ssDNA products (in brown) derived from DNA repair pathways (such as RecBCD in E. coli) or CRISPR interference. Although in type I-E it is clear that the DNA capture by Cas1-2 precedes the prespacer processing event, the order of DNA capture and processing is yet to be understood in other CRISPR-Cas subtypes. The Cas4 (in green) in CRISPR-Cas subtypes I-A, I-C, I-D, and I-G or the Cas4 domain of Cas4-Cas1 fusion in type I-U trims the DNA upon recognizing the PAM region (in magenta) (25, 45–51). A second copy of Cas4 in type I-G was shown to trim the non-PAM end upon recognizing a short motif (47), whereas in other subtypes, it is not clear whether Cas4 processes this end. Here, the dual role of PAM recognition and prespacer processing by Cas4 propels CRISPR adaptation. Unlike this, in the type I-E system, the intrinsic affinity of Cas1-2 integrase toward PAM itself is sufficient to define the potential prespacer regions. This aspect of Cas1-2/I-E precludes the involvement of any PAM-specific Cas nucleases (such as Cas4) in prespacer selection. As the Cas1-2/I-E protects the prespacer boundaries efficiently upon recognizing the PAM, any common cellular non-Cas nucleases (in cyan) could trim the exposed ends to generate aptly sized prespacers for integration into the CRISPR locus.
Experimental procedures
Construction of plasmids
Lists of plasmids, strains, and oligonucleotides used in this study are detailed in the Tables S1–S3.
Genes encoding IHFα, IHFβ, Cas1, and Cas2 were amplified using E. coli K-12 MG1655 genomic DNA as template. To generate p1R-IHFαβ, a bicistronic cassette encoding IHFα and IHFβ was amplified and inserted at the SspI site of p1R. p13SR-Cas1 was generated by inserting an amplicon encoding Cas1 at the SspI site of p13SR, and pMS-Cas2 was created by introducing an amplicon encoding Cas2 between the BamHI/HindIII sites of pMS (84), respectively. Bicistronic cassettes (cas1-cas2) expressing His6-tagged WT and 5M Cas1 were amplified using pCas1-2[K] (85) and pMut89 (44) as templates, respectively. Amplified fragments encoding WT and 5M Cas1-2 were inserted between NcoI/NotI sites of pCas1-2[K] to generate pCas1-2H and p5M, respectively. PCR-based mutagenesis was used to generate pY22A and pΔC that express Y22A and ΔC variants of Cas1, respectively. All constructs were verified by Sanger sequencing.
Expression and purification of proteins
IHF and Cas1 were purified as described before (17). To purify Cas2, E. coli BL21(DE3) harboring pMS-Cas2 was grown in autoinduction LB medium supplemented with 100 μg/ml kanamycin at 37 °C, 180 rpm. Upon reaching 0.6 A600, the temperature was shifted to 16 °C, and thereafter the growth and induction were continued for 16 h. Subsequently, cells were harvested and washed two times with Buffer 1A (20 mm HEPES-NaOH, pH 7.4, 500 mm KCl, and 10% glycerol). Bacterial pellet was resuspended in Buffer 1A containing 1 mm phenylmethylsulfonyl fluoride, and the cells were lysed by sonication. Here, Cas2 encompasses His6-tagged MBP-SUMO as an N-terminal fusion and a Strep-II tag on the C-terminal end. The clarified fraction of the lysate was applied to a 5-ml MBPTrap HP column (GE Healthcare) and was followed by a washing step with Buffer 1A. Thereafter, the bound proteins were eluted with Buffer 1A containing 10 mm maltose. Eluted fractions were mixed with SUMO protease (Ulp1403–621) (in a 400:1 ratio of His-MBP-SUMO-Cas2-strep/Ulp1403–621) (84), and the incubation was continued for 60 min at 25 °C. Following this, the mixture was loaded onto a 5-ml HiTrap IMAC HP column (GE Healthcare) five times to facilitate binding of histidine-tagged MBP-SUMO-Cas2-strep, MBP-SUMO, and Ulp1403–621. Column flow-through containing Cas2-strep was concentrated using a centrifugal membrane filter (Sartorius). To remove any trace protein contaminants, the concentrated sample was loaded onto a HiLoad Superdex 200 pg gel filtration column (GE Healthcare), that was pre-equilibrated with Buffer 1B (20 mm HEPES-NaOH, pH 7.4, 150 mm KCl, and 10% glycerol). Eluted fractions containing Cas2-strep were pooled, concentrated, snap-frozen in liquid nitrogen, and stored at −80 °C until required.
Integrase complex comprising untagged Cas1 and C-terminal His6-tagged Cas2 was expressed and purified as described before (61) with minor modifications. Here, E. coli BL21(DE3) transformed using pCas1-2H was grown in 2XYT broth supplemented with 100 μg/ml spectinomycin at 37 °C, 180 rpm until 0.6 A600. Thereafter, the protein expression was induced by the addition of 0.7 mm IPTG, and the growth was continued at 25 °C for 24 h. Simultaneously, cells were harvested and washed two times with Buffer 2A (20 mm HEPES-NaOH, pH 7.4, 150 mm KCl, 10% glycerol, and 30 mm imidazole). The pellet was resuspended in Buffer 2A containing 1 mm phenylmethylsulfonyl fluoride, and cells were lysed by sonication. Thereafter, the lysate was clarified and loaded onto a 5-ml HiTrap IMAC HP column (GE Healthcare) and was followed by a washing step with Buffer 2A. A linear gradient of imidazole (0.03–0.5 m) in Buffer 2A was applied to elute the proteins that were bound to the column resin. The purified fractions that contain the complex of Cas1-2 were pooled and concentrated using a centrifugal membrane filter (Sartorius). To remove trace protein contaminants and uncomplexed Cas2, the concentrate was further purified using a HiLoad Superdex 200 pg gel filtration column (GE Healthcare) that was pre-equilibrated with Buffer 2B (20 mm HEPES-NaOH, pH 7.4, 150 mm KCl, and 10% glycerol). Eluted fractions containing Cas1-2 integrase were pooled, concentrated, snap-frozen in liquid nitrogen, and stored at −80 °C until required. A similar procedure was implemented to purify 5M, ΔC, and Y22A Cas1 variants of Cas1-2 from the IPTG-induced E. coli BL21(DE3) cells that harbor p5M, pΔC, and pY22A, respectively.
In vitro integration assay
A 177-bp CRISPR DNA substrate that encompasses 69-bp leader and two repeat-spacer units of the CRISPR 2.1 locus of E. coli was amplified using pCSIR-T (85) as a template. CRISPR DNA substrates labeled with 5′-FAM at the top strand of the leader end (CD-T*) or at the bottom strand of the second spacer end (CD-B*) were prepared using PCR. To generate various prespacers (P33, P23[3′-5], P23[5′-5], P23[3′-10], P63, P63mPAM, and their 5′-FAM labeled variants), respective oligonucleotides (Table S3) were mixed in a buffer containing 10 mm Tris-Cl, pH 8.5. These mixtures were heated to 95 °C and gradually allowed to cool to room temperature to facilitate the formation of duplex and partial-duplex prespacers. In the case of P33ss, a 33-nt-long single-stranded oligonucleotide was used as a prespacer.
The in vitro integration assays were performed as described previously (17) with minor modifications. Briefly, 210 nm Cas1, Cas2, or Cas1-2 (WT, ΔC, Y22A, and 5M) was mixed with a 550 nm concentration of the desired prespacer and incubated at room temperature for 5 min. To this mixture, 0.5 μm IHF and 21 nm CRISPR DNA substrate were supplemented, and incubation was continued at 37 °C for 60 min in integrase buffer (20 mm HEPES-NaOH, pH 7.4, 25 mm KCl, 10 mm MgCl2, and 1 mm DTT). Subsequently, the reaction mixtures were supplemented with an equal volume of stopping solution (95% formamide, 5 mm EDTA, and 0.025% SDS) followed by heating at 95 °C for 20 min. These samples were loaded onto preheated 12% denaturing acrylamide gels that were maintained at 50 °C and electrophoresed in 1× TBE. Subsequently, gels were stained with EtBr and visualized using a gel documentation system (Bio-Rad), whereas in the assays that involve FAM-labeled CRISPR DNA or prespacers, gels were imaged without any post-staining step. All of the integration experiments were independently repeated at least twice, and the representative gel pictures were shown.
Electrophoretic mobility shift assays
The binding of Cas1-2 with various prespacers was monitored using electrophoretic mobility shift assays. Here, a 100 nm concentration of the desired 5′-FAM–labeled prespacers (P23[3′-5], P23[5′-5], P33, and P23[3′-10]) was incubated with increasing concentrations of WT Cas1-2 (0.1, 0.15, 0.2, 0.25, 0.45, 0.6, 0.8, 1, 1.5, 2, and 3 μm) in prespacer-binding buffer (20 mm HEPES-NaOH, pH 7.4, 125 mm KCl, 10 mm MgCl2, and 1 mm DTT) for 30 min at 37 °C. Subsequently, all of the samples were directly loaded onto 0.8% agarose gel and electrophoresed in 1× TAE at 4 °C. Bound fraction for each sample in the gel was estimated by quantifying the amount of DNA at each band using densitometric analysis (bound fraction of prespacer (%) at X μm Cas1-2 = ((amount of DNA in the absence of Cas1-2 − amount of unbound DNA at X μm Cas1-2)/(amount of DNA in the absence of Cas1-2)) × 100. To estimate KD values, the resulting plots of bound fraction (%) against Cas1-2 concentration were fitted to a nonlinear equation, y = Bmax × x/(KD + x) (where x, y, Bmax, and KD represent Cas1-2 concentration (μm), bound fraction (%), maximum concentration of Cas1-2 bound to prespacer, and dissociation constant, respectively). In EMSA that involves 5′-FAM–labeled P63 or P63mPAM prespacers, 100 nm DNA was incubated with 0.2–5 μm WT/ΔC/Y22A or 0.2–20 μm 5M Cas1-2 variants (Figs. S2 (E and F) and S5 (A–C)). All of the binding experiments were independently repeated three times, and the representative gel pictures were displayed.
To further verify the formation of Cas1-2 nucleoprotein complex, the release of prespacers was monitored upon Proteinase K treatment of Cas1-2 in each assay. To achieve this, an aliquot of sample containing prespacer and 3 μm Cas1-2 was mixed with 1 mg/ml Proteinase K and incubated at 37 °C for 15 min.
Exonuclease treatment of Cas1-2–bound DNA fragments
Exonuclease treatment was performed to identify the extent of protection conferred by binding of Cas1-2 onto a long DNA fragment. 40 μl of 0.5 μm P63 and a 6 μm concentration of either Cas1 or Cas2 or Cas1-2 in prespacer-binding buffer were incubated at 37 °C for 45 min. Subsequently, 20-μl aliquots of these samples were supplemented with 3 units of either T5 exonuclease (New England Biolabs) or exonuclease III (New England Biolabs) or 3 units of the mixture containing both exonucleases, and incubation was continued for 60 min at 37 °C. Thereafter, all of the samples were mixed with an equal volume of denaturation buffer that contains 200 mm Tris-Cl, pH 8.3, 200 mm boric acid, 20 mm EDTA, 0.05% SDS, and 8 m urea, followed by heating at 95 °C for 15 min. These samples were loaded onto preheated 20% denaturing acrylamide gels that were maintained at 50 °C and electrophoresed in 1× TBE. Subsequently, gels were stained with EtBr and visualized using a gel documentation system. This experiment was independently repeated three times, and a representative gel picture is shown.
Exonuclease footprinting
T5 exonuclease-mediated footprinting was performed to identify the interaction boundaries of Cas1-2 on longer prespacer DNA fragments. Here, 40 μl of 0.5 μm desired fluorescein-labeled P63 variant (P63T*, P63B*, P63mPAMT*, and P63mPAMB*) was mixed with 6 μm WT or one of the mutant variants of Cas1-2 (Y22A, ΔC, and 5M) in prespacer-binding buffer and incubated for 45 min at 37 °C. Subsequently, 20-μl aliquots of these samples were supplemented with 3 units of T5 exonuclease, and incubation was continued for 60 min at 37 °C. Thereafter, all of the samples were mixed with an equal volume of denaturation buffer that contains 200 mm Tris-Cl, pH 8.3, 200 mm boric acid, 20 mm EDTA, 0.05% SDS, and 8 m urea followed by heating at 95 °C for 15 min. These samples were loaded onto preheated 20% denaturing acrylamide gels that were maintained at 50 °C and electrophoresed in 1× TBE. Subsequently, gels were directly visualized using a gel documentation system. All of the footprinting experiments were repeated at least twice, and representative gel pictures are shown.
Spacer acquisition assays
The in vivo spacer acquisition assays were performed as described previously (10, 17) with minor modifications. After transformation using plasmids (pCas1-2H, p5M, pY22A, and pΔC), E. coli IYB5101 that expresses WT or a mutant of Cas1-2 was subjected to three cycles of growth and induction in LB medium supplemented with 100 μg/ml spectinomycin, 0.2% l-arabinose, and 0.1 mm IPTG for 16 h at 37 °C. After each cycle, cultures were diluted to 1:300 with fresh LB medium containing the aforementioned supplements, and the growth was continued for 16 h. Thereafter, genomic DNA was isolated according to the manufacturer's protocol (HiPurA bacterial genomic DNA purification kit, Himedia), and this was used as a template for PCR to monitor the spacer integration at CRISPR 2.1. All of the PCR-amplified samples were resolved on 1.5% agarose gels to identify the DNA bands corresponding to parental and expanded arrays (parental array + n × 61 bp, where n is a positive integer). DNA quantities corresponding to parental and expanded array were quantified by densitometric analysis. Utilizing these values, the percentage of spacer integration for each Cas1-2 variant was estimated (% integration = ((amount of expanded array)/(Amount of parental array + Amount of expanded array)) × 100).
High-throughput sequencing and analysis
To understand the effect of Cas1-2 mutants on prespacer scaling and PAM selectivity, high-throughput sequencing was performed to derive the sequences of newly incorporated prespacers. Expanded CRISPR arrays corresponding to the expression of each Cas1-2 variant were extracted from the agarose gels (QIAquick Gel Extraction Kit, Qiagen). Approximately 200 ng of each PCR product was further purified using HighPrep magnetic beads (MAGBIO). These purified samples were subjected to DNA end repair and adaptor ligation using the Illumina-compatible NEXTflex Rapid DNA sequencing kit (BIOO Scientific, Austin, TX). Subsequently, the ligated DNA products were purified with HighPrep magnetic beads, and further enrichment was achieved by eight cycles of PCR with Illumina-compatible primers (NEXTFlex DNA-sequencing kit). These amplicons were subjected to an additional step of purification with HighPrep magnetic beads and were sequenced on a Miseq 300 paired-end platform.
The paired-end reads were subjected to several preprocessing steps as described below. First, both F and R reads with Phred score less than 20 were removed by utilizing fastq_quality_trimmer from the FASTX-toolkit-version-0.0.13. The remaining F and R reads were trimmed in paired-end mode to remove F (5′-AGATCGGAAGAGCACACGTCTGAACTCCAGTCA-3′) and R (5′-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT-3′) adapter sequences using Cutadapt-1.18 (86). Following these, the leader-proximal spacer sequence (S0) was selectively retrieved in FASTA format. These S0 sequences that were derived from E. coli expressing WT and mutants of Cas1-2 were searched against plasmid (pCas1-2[K]) (85) and E. coli K-12 MG1655 genome (GenBankTM assembly accession: GCA_000005845.2), respectively, using BLASTN (87). From the BLAST hits, we identified the location of spacer sequences on plasmid and E. coli K-12 MG1655 genome, respectively, and extracted the triplet sequence corresponding to PAM. The conservation of PAM was analyzed using WEB-LOGO (88). For all sequence manipulations, including the extraction of S0 and PAM sequences, we employed custom-written Python codes utilizing the Biopython library (89).
Analysis of Cas1 C-terminal tail across CRISPR-Cas type I systems
Locus IDs corresponding to Cas1 genes of type I-A, I-B, I-C, and I-E were derived from the previous study (90). Utilizing these identifiers, we extracted Cas1 protein sequences of 36 type I-A, 150 type I-B, 129 type I-C, and 116 type I-E organisms from the NCBI protein database. Subsequent to this, we performed multiple sequence alignments in the T-COFFEE web server (91) for Cas1 proteins of each subtype separately (Files S1–S4). Utilizing Cas1 crystal structures of A. fulgidus (type I-A; PDB code 4N06), P. horikoshii (type I-B; PDB code 4WJ0), and E. coli (type I-E; PDB code 5DQZ) as a reference, C-terminal tail residues of each Cas1 were extracted from their multiple-sequence alignments. Owing to the absence of a type I-C Cas1 crystal structure in the PDB, the Cas1 (BH0341) structure of B. halodurans was predicted by I-TASSER web server (82) using structures corresponding to PDB entries 3LFX, 4N06, and 2YZS as threading templates. I-TASSER predicted five different models for B. halodurans Cas1. Among these, the model with the highest confidence score (C-score = 1.25) was used as a structural reference for predicting the C-terminal tail residues from a multiple-sequence alignment of 129 type I-C Cas1 proteins.
Utilizing ESpript server (92), various secondary structural element positions were mapped onto the multiple-sequence alignments of type I-A, I-B, I-C, and I-E Cas1 proteins (Figs. S6–S9). All of the protein structural representations were generated using ChimeraX (93).
Author contributions
K. N. Y. and B. A. conceptualization; K. N. Y. data curation; K. N. Y. and B. A. formal analysis; K. N. Y. validation; K. N. Y., M. M., and S. N. investigation; K. N. Y. visualization; K. N. Y., M. M., and S. N. methodology; K. N. Y. and B. A. writing-original draft; K. N. Y., M. M., and B. A. writing-review and editing; B. A. supervision; B. A. funding acquisition; B. A. project administration.
Supplementary Material
Acknowledgments
E. coli IYB5101 strain was a kind gift from Dr. Udi Qimron (Tel Aviv University, Israel); vectors pET StrepII TEV LIC cloning vector (p1R) (Addgene #29664) and pET StrepII TEV co-transformation cloning vector (p13SR) (Addgene #48328) were a kind gift from Dr. Scott Gradia (QB3 MacroLab, University of California); pLJSRSF7 (pMS) (Addgene #64693) and pFGET19_Ulp1 (Addgene #64697) were a kind gift from Dr. Hideo Iwai (University of Helsinki, Finland); pWUR 1+2 tetO mut89 (pMut89) (Addgene #80102) was a kind gift from Dr. George M. Church (Harvard University); and pCSIR-T and pCas1-2[K] were a kind gift from Dr. F. J. M. Mojica (University of Alicante, Spain). We sincerely acknowledge the gracious gesture of the aforementioned scientists for sharing bacterial strains and plasmids. We also thank all of the members of the Mechanistic Approaches to Biology lab for suggestions regarding the experiments and critical comments on the manuscript.
This work was supported by Department of Biotechnology (DBT) Grants BT/PR15925/NER/95/141/2015 and BT/08/IYBA/2014/05 and Science and Engineering Research Board (SERB) Grant YSS/2014/000286. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1–S3, Figs. S1–S9, and Files S1–S4.
Reads obtained from high-throughput sequencing have been deposited in the Sequence Read Archive (SRA) under accession number PRJNA527928.
- Cas
- CRISPR-associated protein(s)
- MGE
- mobile genetic element(s)
- PAM
- protospacer adjacent motif
- IHF
- integration host factor
- nt
- nucleotide(s)
- T5exo
- T5 exonuclease
- ExoIII
- exonuclease III
- SUMO
- small ubiquitin-like modifier
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- LB
- lysogeny broth
- PDB
- Protein Data Bank.
References
- 1. Marraffini L. A., and Sontheimer E. J. (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D. A., and Horvath P. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 3. Hille F., Richter H., Wong S. P., Bratovič M., Ressel S., and Charpentier E. (2018) The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259 10.1016/j.cell.2017.11.032 [DOI] [PubMed] [Google Scholar]
- 4. Deveau H., Barrangou R., Garneau J. E., Labonté J., Fremaux C., Boyaval P., Romero D. A., Horvath P., and Moineau S. (2008) Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunin V., Sorek R., and Hugenholtz P. (2007) Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8, R61 10.1186/gb-2007-8-4-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bolotin A., Quinquis B., Sorokin A., and Ehrlich S. D. (2005) Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 7. Mojica F. J., Díez-Villaseñor C., García-Martínez J., and Soria E. (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 8. McGinn J., and Marraffini L. A. (2019) Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat. Rev. Microbiol. 17, 7–12 10.1038/s41579-018-0071-7 [DOI] [PubMed] [Google Scholar]
- 9. Jackson S. A., McKenzie R. E., Fagerlund R. D., Kieper S. N., Fineran P. C., and Brouns S. J. (2017) CRISPR-Cas: adapting to change. Science 356, eaal5056 10.1126/science.aal5056 [DOI] [PubMed] [Google Scholar]
- 10. Yosef I., Goren M. G., and Qimron U. (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 10.1093/nar/gks216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hochstrasser M. L., and Doudna J. A. (2015) Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 40, 58–66 10.1016/j.tibs.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 12. Punetha A., Yoganand K. N. R., Nimkar S., and Anand B. (2018) Cutting it right: plasticity and strategy of CRISPR RNA specific nucleases. Proc. Indian Natl. Sci. Acad. B Biol. Sci. 84, 455–477 10.16943/ptinsa/2017/49241 [DOI] [Google Scholar]
- 13. Brouns S. J., Jore M. M., Lundgren M., Westra E. R., Slijkhuis R. J., Snijders A. P., Dickman M. J., Makarova K. S., Koonin E. V., and van der Oost J. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carte J., Wang R., Li H., Terns R. M., and Terns M. P. (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 22, 3489–3496 10.1101/gad.1742908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makarova K. S., Wolf Y. I., and Koonin E. V. (2018) Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1, 325–336 10.1089/crispr.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nuñez J. K., Bai L., Harrington L. B., Hinder T. L., and Doudna J. A. (2016) CRISPR immunological memory requires a host factor for specificity. Mol. Cell 62, 824–833 10.1016/j.molcel.2016.04.027 [DOI] [PubMed] [Google Scholar]
- 17. Yoganand K. N. R., Sivathanu R., Nimkar S., and Anand B. (2017) Asymmetric positioning of Cas1-2 complex and integration host factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res. 45, 367–381 10.1093/nar/gkw1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levy A., Goren M. G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., and Sorek R. (2015) CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 10.1038/nature14302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pul U., Wurm R., Arslan Z., Geissen R., Hofmann N., and Wagner R. (2010) Identification and characterization of E. coli CRISPR-Cas promoters and their silencing by H-NS. Mol. Microbiol. 75, 1495–1512 10.1111/j.1365-2958.2010.07073.x [DOI] [PubMed] [Google Scholar]
- 20. Mohr G., Silas S., Stamos J. L., Makarova K. S., Markham L. M., Yao J., Lucas-Elío P., Sanchez-Amat A., Fire A. Z., Koonin E. V., and Lambowitz A. M. (2018) A reverse transcriptase-Cas1 fusion protein contains a Cas6 domain required for both CRISPR RNA biogenesis and RNA spacer acquisition. Mol. Cell 72, 700–714.e8 10.1016/j.molcel.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silas S., Mohr G., Sidote D. J., Markham L. M., Sanchez-Amat A., Bhaya D., Lambowitz A. M., and Fire A. Z. (2016) Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science 351, aad4234 10.1126/science.aad4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ivančić-Baće I., Cass S. D., Wearne S. J., and Bolt E. L. (2015) Different genome stability proteins underpin primed and naive adaptation in E. coli CRISPR-Cas immunity. Nucleic Acids Res. 43, 10821–10830 10.1093/nar/gkv1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wei Y., Terns R. M., and Terns M. P. (2015) Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev. 29, 356–361 10.1101/gad.257550.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li M., Wang R., Zhao D., and Xiang H. (2014) Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492 10.1093/nar/gkt1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rollie C., Graham S., Rouillon C., and White M. F. (2018) Prespacer processing and specific integration in a Type I-A CRISPR system. Nucleic Acids Res. 46, 1007–1020 10.1093/nar/gkx1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fagerlund R. D., Wilkinson M. E., Klykov O., Barendregt A., Pearce F. G., Kieper S. N., Maxwell H. W. R., Capolupo A., Heck A. J. R., Krause K. L., Bostina M., Scheltema R. A., Staals R. H. J., and Fineran P. C. (2017) Spacer capture and integration by a type I-F Cas1-Cas2–3 CRISPR adaptation complex. Proc. Natl. Acad. Sci. U.S.A. 114, E5122–E5128 10.1073/pnas.1618421114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Westra E. R., Pul U., Heidrich N., Jore M. M., Lundgren M., Stratmann T., Wurm R., Raine A., Mescher M., Van Heereveld L., Mastop M., Wagner E. G., Schnetz K., Van Der Oost J., Wagner R., and Brouns S. J. (2010) H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77, 1380–1393 10.1111/j.1365-2958.2010.07315.x [DOI] [PubMed] [Google Scholar]
- 28. Ka D., Jang D. M., Han B. W., and Bae E. (2018) Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2. Nucleic Acids Res. 46, 9805–9815 10.1093/nar/gky702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heler R., Samai P., Modell J. W., Weiner C., Goldberg G. W., Bikard D., and Marraffini L. A. (2015) Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature 519, 199–202 10.1038/nature14245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yosef I., Shitrit D., Goren M. G., Burstein D., Pupko T., and Qimron U. (2013) DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc. Natl. Acad. Sci. U.S.A. 110, 14396–14401 10.1073/pnas.1300108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zetsche B., Gootenberg J. S., Abudayyeh O. O., Slaymaker I. M., Makarova K. S., Essletzbichler P., Volz S. E., Joung J., van der Oost J., Regev A., Koonin E. V., and Zhang F. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gleditzsch D., Pausch P., Müller-Esparza H., Özcan A., Guo X., Bange G., and Randau L. (2019) PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol. 16, 504–517 10.1080/15476286.2018.1504546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mojica F. J., Díez-Villaseñor C., García-Martínez J., and Almendros C. (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- 34. Sashital D. G., Wiedenheft B., and Doudna J. A. (2012) Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell 46, 606–615 10.1016/j.molcel.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Datsenko K. A., Pougach K., Tikhonov A., Wanner B. L., Severinov K., and Semenova E. (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945 10.1038/ncomms1937 [DOI] [PubMed] [Google Scholar]
- 36. Semenova E., Savitskaya E., Musharova O., Strotskaya A., Vorontsova D., Datsenko K. A., Logacheva M. D., and Severinov K. (2016) Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex. Proc. Natl. Acad. Sci. U.S.A. 113, 7626–7631 10.1073/pnas.1602639113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li M., Wang R., and Xiang H. (2014) Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 42, 7226–7235 10.1093/nar/gku389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Musharova O., Sitnik V., Vlot M., Savitskaya E., Datsenko K. A., Krivoy A., Fedorov I., Semenova E., Brouns S. J. J., and Severinov K. (2019) Systematic analysis of Type I-E Escherichia coli CRISPR-Cas PAM sequences ability to promote interference and primed adaptation. Mol. Microbiol. 111, 1558–1570 10.1111/mmi.14237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dillingham M. S., and Kowalczykowski S. C. (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 10.1128/MMBR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muskavitch K. M., and Linn S. (1982) A unified mechanism for the nuclease and unwinding activities of the recBC enzyme of Escherichia coli. J. Biol. Chem. 257, 2641–2648 [PubMed] [Google Scholar]
- 41. Swarts D. C., Mosterd C., van Passel M. W., and Brouns S. J. (2012) CRISPR interference directs strand specific spacer acquisition. PLoS One 7, e35888 10.1371/journal.pone.0035888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J., Li J., Zhao H., Sheng G., Wang M., Yin M., and Wang Y. (2015) Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163, 840–853 10.1016/j.cell.2015.10.008 [DOI] [PubMed] [Google Scholar]
- 43. Nuñez J. K., Harrington L. B., Kranzusch P. J., Engelman A. N., and Doudna J. A. (2015) Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527, 535–538 10.1038/nature15760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shipman S. L., Nivala J., Macklis J. D., and Church G. M. (2016) Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175 10.1126/science.aaf1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee H., Zhou Y., Taylor D. W., and Sashital D. G. (2018) Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59.e5 10.1016/j.molcel.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kieper S. N., Almendros C., Behler J., McKenzie R. E., Nobrega F. L., Haagsma A. C., Vink J. N. A., Hess W. R., and Brouns S. J. J. (2018) Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 22, 3377–3384 10.1016/j.celrep.2018.02.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shiimori M., Garrett S. C., Graveley B. R., and Terns M. P. (2018) Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824.e6 10.1016/j.molcel.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu T., Liu Z., Ye Q., Pan S., Wang X., Li Y., Peng W., Liang Y., She Q., and Peng N. (2017) Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 45, 8978–8992 10.1093/nar/gkx612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z., Pan S., Liu T., Li Y., and Peng N. (2019) Cas4 nucleases can effect specific integration of CRISPR spacers. J. Bacteriol. 201, e00747–18 10.1128/JB.00747-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Almendros C., Nobrega F. L., McKenzie R. E., and Brouns S. J. J. (2019) Cas4–Cas1 fusions drive efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res. 47, 5223–5230 10.1093/nar/gkz217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee H., Dhingra Y., and Sashital D. G. (2019) The Cas4-Cas1-Cas2 complex mediates precise prespacer processing during CRISPR adaptation. Elife 8, e44248 10.7554/eLife.44248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drabavicius G., Sinkunas T., Silanskas A., Gasiunas G., Venclovas Č., Siksnys V. (2018) DnaQ exonuclease-like domain of Cas2 promotes spacer integration in a type I-E CRISPR-Cas system. EMBO Rep. 19, e45543 10.15252/embr.201745543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Radovcic M., Killelea T., Savitskaya E., Wettstein L., Bolt E. L., and Ivancic-Bace I. (2018) CRISPR-Cas adaptation in Escherichia coli requires RecBCD helicase but not nuclease activity, is independent of homologous recombination, and is antagonized by 5′ ssDNA exonucleases. Nucleic Acids Res. 46, 10173–10183 10.1093/nar/gky799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. McGinn J., and Marraffini L. A. (2016) CRISPR-Cas systems optimize their immune response by specifying the site of spacer integration. Mol. Cell 64, 616–623 10.1016/j.molcel.2016.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rollie C., Schneider S., Brinkmann A. S., Bolt E. L., and White M. F. (2015) Intrinsic sequence specificity of the Cas1 integrase directs new spacer acquisition. Elife 4, e08716 10.7554/eLife.08716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nuñez J. K., Lee A. S., Engelman A., and Doudna J. A. (2015) Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198 10.1038/nature14237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wei Y., Chesne M. T., Terns R. M., and Terns M. P. (2015) Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 43, 1749–1758 10.1093/nar/gku1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang R., Li M., Gong L., Hu S., and Xiang H. (2016) DNA motifs determining the accuracy of repeat duplication during CRISPR adaptation in Haloarcula hispanica. Nucleic Acids Res. 44, 4266–4277 10.1093/nar/gkw260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Goren M. G., Doron S., Globus R., Amitai G., Sorek R., and Qimron U. (2016) Repeat size determination by two molecular rulers in the type I-E CRISPR array. Cell Rep. 16, 2811–2818 10.1016/j.celrep.2016.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Arslan Z., Hermanns V., Wurm R., Wagner R., and Pul Ü. (2014) Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 42, 7884–7893 10.1093/nar/gku510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Moch C., Fromant M., Blanquet S., and Plateau P. (2017) DNA binding specificities of Escherichia coli Cas1-Cas2 integrase drive its recruitment at the CRISPR locus. Nucleic Acids Res. 45, 2714–2723 10.1093/nar/gkw1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kieper S. N., Almendros C., and Brouns S. J. J. (2019) Conserved motifs in the CRISPR leader sequence control spacer acquisition levels in Type I-D CRISPR-Cas systems. FEMS Microbiol. Lett. 366, fnz129 10.1093/femsle/fnz129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Grainy J., Garrett S., Graveley B. R., and P Terns M. (2019) CRISPR repeat sequences and relative spacing specify DNA integration by Pyrococcus furiosus Cas1 and Cas2. Nucleic Acids Res. 47, 7518–7531 10.1093/nar/gkz548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim J. G., Garrett S., Wei Y., Graveley B. R., and Terns M. P. (2019) CRISPR DNA elements controlling site-specific spacer integration and proper repeat length by a Type II CRISPR-Cas system. Nucleic Acids Res. 47, 8632–8648 10.1093/nar/gkz677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Plateau P., Moch C., and Blanquet S. (2019) Spermidine strongly increases the fidelity of Escherichia coli CRISPR Cas1-Cas2 integrase. J. Biol. Chem. 294, 11311–11322 10.1074/jbc.RA119.007619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wright A. V., Liu J. J., Knott G. J., Doxzen K. W., Nogales E., and Doudna J. A. (2017) Structures of the CRISPR genome integration complex. Science 357, 1113–1118 10.1126/science.aao0679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xiao Y., Ng S., Nam K. H., and Ke A. (2017) How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature 550, 137–141 10.1038/nature24020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wright A. V., and Doudna J. A. (2016) Protecting genome integrity during CRISPR immune adaptation. Nat. Struct. Mol. Biol. 23, 876–883 10.1038/nsmb.3289 [DOI] [PubMed] [Google Scholar]
- 69. Nuñez J. K., Kranzusch P. J., Noeske J., Wright A. V., Davies C. W., and Doudna J. A. (2014) Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 10.1038/nsmb.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Künne T., Kieper S. N., Bannenberg J. W., Vogel A. I. M., Miellet W. R., Klein M., Depken M., Suarez-Diez M., and Brouns S. J. J. (2016) Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol. Cell 63, 852–864 10.1016/j.molcel.2016.07.011 [DOI] [PubMed] [Google Scholar]
- 71. Sinkunas T., Gasiunas G., Fremaux C., Barrangou R., Horvath P., and Siksnys V. (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 30, 1335–1342 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Musharova O., Klimuk E., Datsenko K. A., Metlitskaya A., Logacheva M., Semenova E., Severinov K., and Savitskaya E. (2017) Spacer-length DNA intermediates are associated with Cas1 in cells undergoing primed CRISPR adaptation. Nucleic Acids Res. 45, 3297–3307 10.1093/nar/gkx097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Babu M., Beloglazova N., Flick R., Graham C., Skarina T., Nocek B., Gagarinova A., Pogoutse O., Brown G., Binkowski A., Phanse S., Joachimiak A., Koonin E. V., Savchenko A., Emili A., et al. (2011) A dual function of the CRISPR–Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79, 484–502 10.1111/j.1365-2958.2010.07465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim S., Loeff L., Colombo S., Brouns S. J. J., and Joo C. (2019) Selective prespacer processing ensures precise CRISPR-Cas adaptation. bioRxiv 10.1101/608976 [DOI] [PubMed] [Google Scholar]
- 75. Savitskaya E., Semenova E., Dedkov V., Metlitskaya A., and Severinov K. (2013) High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 10, 716–725 10.4161/rna.24325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rao C., Chin D., and Ensminger A. W. (2017) Priming in a permissive type I-C CRISPR-Cas system reveals distinct dynamics of spacer acquisition and loss. RNA 23, 1525–1538 10.1261/rna.062083.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shmakov S., Savitskaya E., Semenova E., Logacheva M. D., Datsenko K. A., and Severinov K. (2014) Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 42, 5907–5916 10.1093/nar/gku226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li M., Gong L., Zhao D., Zhou J., and Xiang H. (2017) The spacer size of I-B CRISPR is modulated by the terminal sequence of the protospacer. Nucleic Acids Res. 45, 4642–4654 10.1093/nar/gkx229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jackson S. A., Birkholz N., Malone L. M., and Fineran P. C. (2019) Imprecise spacer acquisition generates CRISPR-Cas immune diversity through primed adaptation. Cell Host Microbe 25, 250–260.e4 10.1016/j.chom.2018.12.014 [DOI] [PubMed] [Google Scholar]
- 80. Musharova O., Vyhovskyi D., Medvedeva S., Guzina J., Zhitnyuk Y., Djordjevic M., Severinov K., and Savitskaya E. (2018) Avoidance of trinucleotide corresponding to consensus protospacer adjacent motif controls the efficiency of prespacer selection during primed adaptation. mBio 9, e02169–18 10.1128/mBio.02169-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kim T. Y., Shin M., Huynh Thi Yen L., and Kim J. S. (2013) Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochem. Biophys. Res. Commun. 441, 720–725 10.1016/j.bbrc.2013.10.122 [DOI] [PubMed] [Google Scholar]
- 82. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wright A. V., Wang J. Y., Burstein D., Harrington L. B., Paez-Espino D., Kyrpides N. C., Iavarone A. T., Banfield J. F., and Doudna J. A. (2019) A functional mini-integrase in a two-protein-type V-C CRISPR system. Mol. Cell 73, 727–737.e3 10.1016/j.molcel.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Guerrero F., Ciragan A., and Iwaï H. (2015) Tandem SUMO fusion vectors for improving soluble protein expression and purification. Protein Expr. Purif. 116, 42–49 10.1016/j.pep.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 85. Díez-Villasenor C., Guzmán N. M., Almendros C., García-Martínez J., and Mojica F. J. (2013) CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 10, 792–802 10.4161/rna.24023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 87. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Crooks G. E., Hon G., Chandonia J. M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cock P. J. A., Antao T., Chang J. T., Chapman B. A., Cox C. J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., and de Hoon M. J. L. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Makarova K. S., Wolf Y. I., Alkhnbashi O. S., Costa F., Shah S. A., Saunders S. J., Barrangou R., Brouns S. J., Charpentier E., Haft D. H., Horvath P., Moineau S., Mojica F. J., Terns R. M., Terns M. P., et al. (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 13, 722–736 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Notredame C., Higgins D. G., and Heringa J. (2000) T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- 92. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Goddard T. D., Huang C. C., Meng E. C., Pettersen E. F., Couch G. S., Morris J. H., and Ferrin T. E. (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.