Abstract
Valency can be defined as the number of discrete interactions a biomolecule can engage in. Valency can be critical for function, such as determining whether a molecule acts as a scaffold for assembling large supramolecular complexes or it forms a functional dimer. Here, we highlight the importance of the role of valency in regulating immune responses with a focus on innate immunity. We discuss some of the ways in which valency itself is regulated through transcriptional, post-transcriptional, and post-translational modifications. Finally, we propose that the valency model can be applied at the whole cell level to study differences in individual cell responses with relevance to putative therapeutic applications.
Keywords: valency, NLRP3, inflammasome, pro-inflammatory cytokines, IL-1β, IL-1α, AIM2, cGAS, myddosome, MYD88, IRAK4, IL-18, pyroptosis, ASC, CASP1, DNA sensing, stress granules
Valency dictates physical interactions between molecules
The question “Can 1000 reviews be wrong?” was part of the title of a preview accompanying two articles published in Cell in 2005 [1]. The articles exposed an important limitation in the consensus model of the adherens junction-actin filament connection; it had been assumed that α-catenin, a molecule that can bind to both actin filaments and β-catenin, acted as a bridge between the adherens junction complex and actin filaments [1]. However, studies showed that α-catenin cannot bind to β-catenin and actin filaments simultaneously because the molecules shared the same binding site on α-catenin [2,3]. Essentially, α-catenin had a valency (see Glossary) of one, for physical interactions in this scenario. This was an important fact that, mainly due to the inadequacy of structural and biochemical data available at the time, was not considered in the original model. This highlights the need to consider the number of independent binding sites available when building models of biological processes that require physical interactions.
The physical interaction between biomolecules is a fundamental property for the function of all biological systems. These interactions can occur between molecules of the same type (homotypic interaction) or different types (heterotypic interaction), leading to the assembly of biomolecular complexes that are often the functional units controlling processes such as biochemical reactions or information transfer. Assembly of biomolecular complexes is regulated fundamentally in two ways, by the spatiotemporal localization of the interactors and by the availability of the interaction sites within them. Biochemists have borrowed the concept of valency from chemistry to model and explain the role of binding site availability in the regulation of biomolecular complex assembly [4]. Valency in this context can be defined as the number of independent binding sites on a molecule that allow homotypic or heterotypic physical interactions [4]. In this opinion, we focus on the regulation mediated by valency, mainly in the context of immune system regulation. Signaling downstream of innate immune sensors is regulated by biomolecular complexes that require homotypic and heterotypic interactions for their assembly. We propose that assembly of these biomolecular complexes is critically regulated by multivalent interactions. We further propose that the idea of valency can be extended to the whole cell level and define the concept of cellular valency. Additionally, we discuss examples of cells exploiting cellular valency to regulate their response.
Valency regulates the assembly of supramolecular structures and phase separation
The idea that the number of binding sites, or valency, plays a central role in the assembly of supramolecular structures such as stress granules and P-bodies has been extensively studied and tested [4–7]. The basic premise is that a valency of at least two is required to assemble a large complex: the valency of one would mean that complex formation would be limited to a dimer, with the valency of each molecule being satisfied by binding to the other (Fig. 1A). Moreover, the efficiency of assembly of large supramolecular structures has been shown to increase with an increase in valency, resulting in a higher probability of assembly (or phase separation) at lower concentrations of the components, as evidenced from in vitro experiments with the SRC homology 3 (SH3) domain and its proline-rich motif (PRM) ligand [4]. Phase separation is inhibited in the presence of high concentrations of monovalent molecules that can compete for binding sites on the multivalent scaffold molecules, further corroborating the important role valency plays in these processes [4]. Thus, multivalency is a fundamental property required for phase transitions of biomolecules.
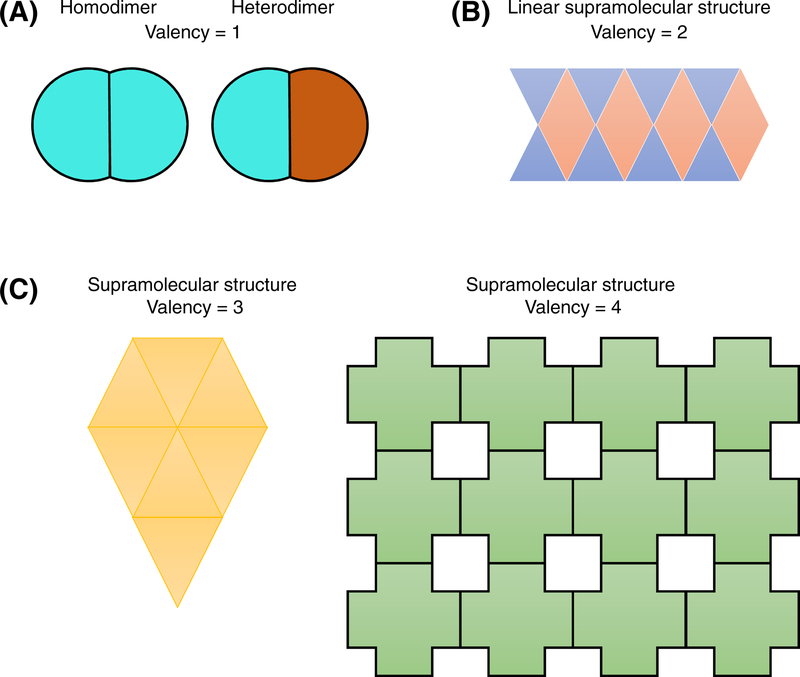
Figure 1. Valency can determine the architecture and size of a biomolecular complex.
(A) A valency of one limits the size of a complex to a dimer. (B) A valency of two can lead to formation of large, linear complexes. (C) A valency of three or more can allow assembly of complexes with varying architecture and sizes.
One of the earliest studies to use the concept of valency involved antibody-antigen binding [8]. Different antibody classes have different valency for their antigen based on the number of immunoglobulin molecules in the functional unit of the antibody class. Multivalency of antibodies is the fundamental property that gives rise to a property known as avidity, where higher valency leads to higher avidity, if the binding affinity remains the same. Another well-studied example is that of lectin-carbohydrate interactions [9]. If both the lectin and its ligand carbohydrate are bivalent, formation of the linear chain lectin-carbohydrate complex can occur (Fig. 1B). However, if they have a valency greater than two, formation of ordered lattice-like structures is possible (Fig. 1C) [9]. Taken together, this evidence shows that the valency of biomolecules can determine the size and topology of biomolecular complexes.
The valency of nucleic acids plays an important role in cellular homeostasis. Phase transitions involving repetitive DNA (for example, ribosomal DNA [rDNA] and telomeres) also exploit the multivalent nature of rDNA, telomeres, and their constituent proteins [10,11]. These phase transitions are critical for maintaining genome organization and regulating transcription [12–16]. Increases in valency can promote phase transitions that can have pathophysiological consequences [17]. In repeat expansion disorders, such as amyotrophic lateral sclerosis, myotonic dystrophy, and Huntington’s disease, the valency of repetitive DNA, or transcribed RNA, increases with the increasing number of repeats [18–20]. Working with chemically synthesized RNA with a defined number of repeats, the valency of the RNA has been shown to determine its phase separation kinetics [17]. Taken together, emerging evidence suggests that the valency of nucleic acids may play important roles in certain human diseases.
Valency regulates innate immune sensing and signaling
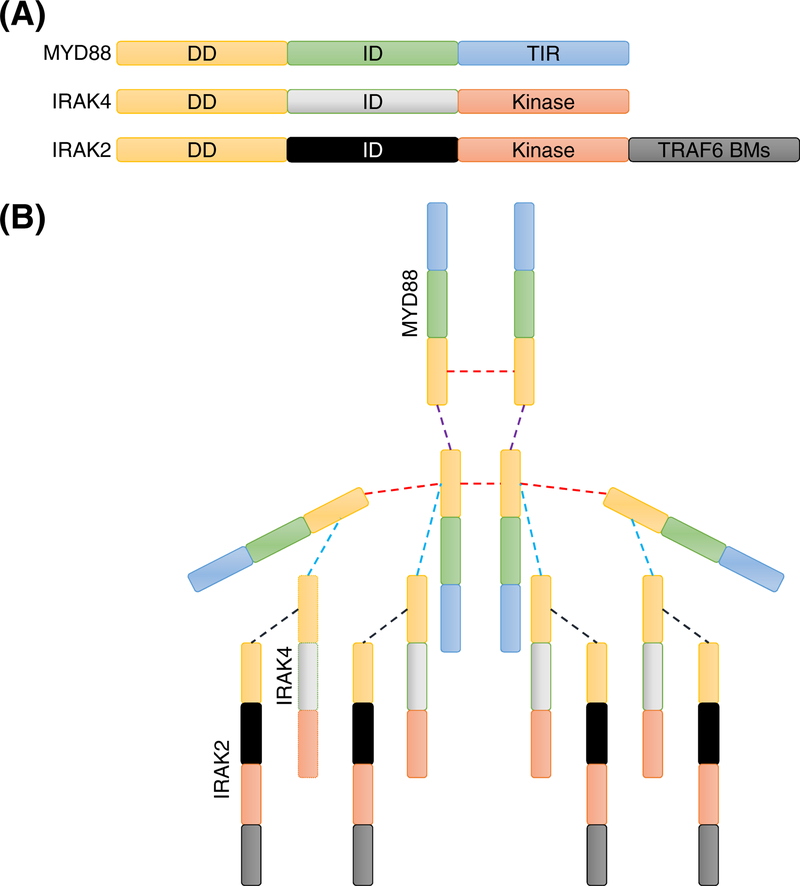
Innate immune sensors need to have at least two functionalities – sensing and signal transduction. This requires a valency of at least two, with distinct binding sites for the ligand that is sensed and the downstream adaptor/signal transducer. As a general principle, ligand binding activates or uninhibits the adaptor/transducer to increase its valency, allowing the sensor to activate downstream signaling. Assembly of an immune signaling complex called the myddosome depends on the valency of its component proteins (Fig. 2A) [21]. The complex is nucleated by activation of the sensor, Toll-like receptors (TLRs), through mechanisms such as the lipopolysaccharide (LPS)-mediated activation of TLR4 [21]. Working with the human myddosome complex, different types of homotypic interactions between the death domains (DD) of the MYD88 transducer have been found to allow assembly of the coordinating TLR sensor-mediated inflammatory signaling (Fig. 2B) [22]. The DD shows bivalency in the myddosome with one of the interaction interfaces in MYD88 stretching into its intermediate domain (ID) [22]. Disruption of the homotypic interactions through mutagenesis of critical amino acid residues in the transducer MYD88, which decreases the valency, disrupts the assembly of the myddosome [22]. Conversely, increasing the valency of the MYD88 transducer by adding a new interaction motif through genetic engineering allows novel signaling through the myddosome complex [23]. For example, using murine immortalized bone marrow-derived macrophages (BMDM), one study showed that genetically engineered MYD88 containing the pLxIS motif (p, hydrophilic residue; x, any residue; S, phosphorylation site) was able to activate interferon (IFN) signaling, while wild type MYD88 could not [23].
Figure 2. Mammalian myddosome assembly is driven by the multivalency of its components.
(A) Valencies of MYD88, interleukin-1 receptor-associated kinase (IRAK) 4, and IRAK2 help assemble the myddosome. DD is the death domain, ID is the intermediate domain, TIR is the Toll/interleukin-1 receptor (TIR) homology domain, and TRAF6 BMs is the TRAF6 binding motif. (B) Different types of interactions suggest distinct valencies in the DD domains of MYD88, IRAK4, and IRAK2 that allow assembly of the myddosome [22]. Domains are color-coded as in panel A. Dashed lines represent physical interactions between the DDs. Homotypic interactions between the different molecules representing distinct valencies in the DD are depicted with different colored dashed lines.
In addition to the role of valency in the adaptor/transducer, valency within the ligand is also important. A recent study suggested that the innate immune sensor of cytosolic DNA, cyclic GMP-AMP synthase (cGAS), phase transitions into a liquid-like state upon ligand binding, forming a large supramolecular structure that acts as a signaling platform to activate downstream molecules; this phase transition is dependent on the length of the DNA molecule and greatly enhances the innate immune response to cytosolic DNA relative to controls, by activating cGAS [24]. Since cGAS does not bind in a sequence-dependent manner, increases in the length of the ligand DNA can be interpreted as an increase in the valency of the ligand. Additionally, Influenza A virus (IAV) infection sensing by Z-DNA–binding protein 1 (ZBP1) also suggests that the multivalency of the ligand is important [25–27]. Specifically, studies in mouse models have shown that ZBP1 is a sensor of IAV infection [26,27]. In a recent study, super-resolution microscopy of the ZBP1-IAV complex indicated that multiple ZBP1 molecules were bound to the IAV ribonucleoprotein complex, suggesting that IAV ribonucleoprotein complexes are multivalent for ZBP1 binding [25]. These examples suggest that valency of these ligands is important in innate immune sensing.
Some cytokines such as IL-12 and IL-23 have a valency greater than one that allows them to bring together co-receptors for signaling [28]. The functional significance of multivalency of some cytokines remains a question that needs to be explored. IL-1 cytokines acting as ligands induce potent pro-inflammatory signaling upon binding to their receptor. IL-1β becomes activated after proteolytic processing and binds to its receptor (IL-1R) [29]. A great mystery in innate immunity is the function of another IL-1 cytokine, IL-1α [30,31]. IL-1α does not require proteolytic processing and has another domain that is not required for binding to its shared receptor IL-1R [29,32]. This suggests a hitherto undiscovered function of the second valency provided by the IL-1α N-terminal domain. Thus, these studies indicate that the valency of sensors, adaptors/transducers, and ligands can all contribute to regulation of innate immune sensing and signaling.
The valency of sensors, adaptors, and ligands can regulate inflammasome activation
Inflammasomes are cytoplasmic complexes that drive a pro-inflammatory programmed cell death process termed pyroptosis. Mammalian inflammasomes consist of three components: a sensor protein that senses pathogen- or damage-associated molecular patterns (PAMPs and DAMPs, respectively), a catalytic actuator caspase (caspase-1, CASP1) that processes the effector cytokines and pore-forming protein gasdermin D (GSDMD), and often an adaptor that brings the sensor and caspase together [33]. The adaptor protein apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC) contains a pyrin domain (PYD) and a caspase activation and recruitment domain (CARD) that allow assembly of the inflammasome complex through homotypic interactions with the PYD of the sensor and CARD of CASP1 [34]. Hypothetically, ASC seems to have a valency of two, which would limit the number of molecules in the inflammasome complex to three – one each for the sensor, CASP1, and ASC (Fig. 3A) because ASC needs to simultaneously interact with both the sensor and the catalytic actuator CASP1 [34]. However, the CARD domain of ASC is capable of multiple simultaneous homotypic interactions with other CARDs [35,36]. This multivalency in the adaptor molecule allows assembly of a larger supramolecular complex termed the ASC speck [37–39]. In addition, the ligand itself can act as a multivalent platform for assembly of the inflammasome in some cases [40,41]. For example, absent in melanoma 2 (AIM2) inflammasome activation requires 80 base pairs of double-stranded DNA [40,41]. However, the crystal structure of the AIM2 HIN domain in complex with double-stranded DNA has revealed that the ligand binding interface only spans 7–8 base pairs [40]. The size discrepancy between the length of double-stranded DNA required for inflammasome activation and the significantly shorter length of the ligand binding interface for AIM2 suggests that multiple AIM2 molecules need to bind to a ligand DNA molecule for inflammasome assembly, with the DNA molecule providing the required multivalency [40]. Additionally, the valency of the sensor molecule can play a role in NLRP3 and NLRC4 inflammasome formation. Specifically, the NLRP3 PYD alone appears to be a poor nucleator of ASC polymerization, but a truncated NLRP3 protein containing both the PYD and nucleotide binding domain (NBD) greatly enhanced ASC polymerization in mammalian cells relative to controls [35,42]. This suggested that the NBD domain could provide additional valency for homotypic interactions, thus leading to more ASC polymerization for efficient inflammasome assembly (Fig. 3B) [35].
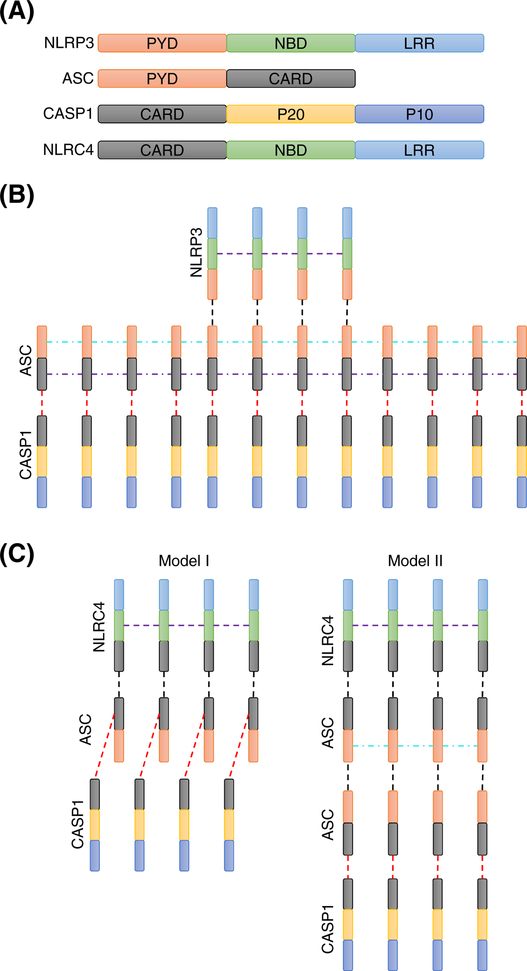
Figure 3. Valencies of the sensor, adaptor, and ligand are critical for mammalian inflammasome activation.
(A) Valency of the inflammasome core components NLRP3, apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC), caspase-1 (CASP1), and NLRC4. The ASC caspase activation and recruitment domain (CARD) is multivalent with respect to homotypic CARD-CARD interactions that allow it to polymerize into ASC filaments [36]. PYD is the pyrin domain. NBD is the nucleotide binding domain. LRR is the leucine rich repeat domain. The P20 fragment contains the caspase active site. The P10 fragment is auto-proteolytically cleaved for CASP1 activation. (B) NLRP3 PYD and NBD domains provide multivalency for efficient assembly of ASC filaments. Domains are color-coded as in panel A. Dashed lines represent physical interactions. Homotypic interactions between the different molecules representing distinct valencies are depicted with different colored dashed lines. (C) NLRC4 inflammasome assembly after ligand sensing can happen through two distinct molecular interaction pathways involving the CARD domains of ASC, CASP1, and NLRC4. In model I, the NLRC4 CARD binds to the ASC CARD, which can simultaneously recruit CASP1 through a CARD-CARD interaction. In model II, the NLRC4 CARD binds to the ASC CARD, leading to ASC polymerization through the ASC PYD. The ASC CARD domains in the ASC filament subsequently recruit CASP1 to facilitate its activation.
The NLRC4 inflammasome presents an interesting case where the adaptor ASC is recruited to the inflammasome through a CARD-CARD interaction between the NLRC4 CARD and ASC CARD domains [43]. This raises a question about the stoichiometry and structure of the NLRC4 inflammasome. If the ASC CARD is bound to the NLRC4 CARD, how does it recruit CASP1? We propose that two models are possible (Fig. 3C): In one model, the ASC CARD has a valency of two and contains distinct binding sites for the NLRC4 CARD and CASP1 CARD. In the second model, NLRC4 CARD-mediated recruitment of ASC can lead to ASC PYD-mediated polymerization of ASC filaments. The ASC CARD from another ASC molecule in the ASC filament is then free to recruit CASP1. It is interesting that ASC is dispensable for NLRC4 inflammasome activation because the NLRC4 CARD can directly bind to the CASP1 CARD, suggesting that the CARD domains of ASC and CASP1 might share a binding site on NLRC4 [44,45]. Although high-resolution cryo-electron microscopy (cryo-EM) structures are available for the NLRC4 inflammasome, these questions have not been resolved because the structures either lack the CARD or contain only the CARD domains [46–48]. Taken together, the evidence points towards multivalency of the adaptor ASC, ligands, and sensors playing critical roles in the assembly of a functional inflammasome complex, although further work is necessary to elucidate these roles and the putative mechanistic models that drive such functions. Understanding the role valency plays in inflammasome assembly is not just for academic interest; it can also help guide development of more potent drugs that target specific valency sites to modulate inflammasome activation. For example, targeting the ASC-NLRP3 valency for inhibition might be useful for treating patients with cryopyrin-associated periodic syndrome while leaving the NLRC4 and NLRP1 inflammasome functions intact to avoid systemic immunosuppression.
Mechanisms for regulating the valency of a biomolecule
Transcriptional regulation
Since valency may play an important role in innate immune signaling, it is not surprising that several mechanisms have evolved to regulate it. Valency can be controlled during gene expression as well as post-translationally. Expression-based regulation results in the synthesis of molecules that differ in their number or types of binding sites, and mechanisms of regulation include alternative transcription start site selection, alternative splicing, alternative polyadenylation, and alternative translation initiation [49–54]. MYD88, the central component of the myddosome [21], has two isoforms that are generated by alternative splicing [55,56]. The longer isoform forms the myddosome complex to activate NF-kB signaling [56]. The shorter isoform (MYD88s) lacks the intermediate domain that is required to recruit another component of the myddosome, interleukin-1 receptor-associated kinase 4 (IRAK4) [55,56]. Due to the loss of the binding capacity in the intermediate domain, MYD88s has a lower valency than the longer isoform and acts as a negative regulator of myddosome signaling in human and murine cell lines [56]. MYD88s is induced in response to LPS-mediated signaling, desensitizing HEK293T and Mf4/4 cells to LPS; it might perhaps act as a mechanism to control runaway inflammation [55,56]. Another myddosome component, IRAK4, also has two isoforms, IRAK4S and IRAK4L, which are regulated by alternative splicing and alternative translation initiation. Differential expression of these isoforms has been associated with disease; specifically, exon usage analysis of samples from The Cancer Genome Atlas found that human acute myeloid leukemia cells express IRAK4-L, while normal bone marrow-derived CD34+ hematopoietic cells and mononuclear cells express IRAK4S. IRAK4L contains the three domains required for myddosome assembly and signaling, while IRAK4S lacks the N-terminal death domain [57]. Studies with MYD88, IRAK4, and other molecules suggest that the cells utilize expression-based mechanisms for determining the valency of molecules to regulate biological processes. Cells can therefore exploit multivalency, or the lack thereof, to mount optimal responses that can maintain cellular and organismal homeostasis.
Post-transcriptional regulation
Molecular modifications of transcripts can also influence binding abilities and valency. Post-transcriptional modification of RNAs alters their binding specificity to proteins involved in epitranscriptomic regulation [58]. Similarly, DNA modifications play critical roles in gene expression regulation by altering the binding specificity of DNA for transcription factors and DNA-modifying enzymes [59]. Moreover, chromosomal translocations can lead to gene fusions that alter the regulation of gene expression or which generate chimeric proteins. Chimeric proteins might have additional valencies that can have profound effects on cell physiology and host homeostasis. Specifically, the Philadelphia chromosome generated by a chromosomal translocation of human chromosomes 9 and 22 is a common feature of chronic myelogenous leukemia [60]. This chromosomal translocation generates a gene fusion in which the oncogene ABL is fused to the (B-cell receptor) BCR gene [61]. The fused gene produces the BCR-ABL protein that has gained a valency for SH2 domain binding, which contributes to its oncogenicity in B cells [62]. This additional SH2 binding valency in the BCR-ABL chimeric protein then allows it to form a complex with the adaptor protein GRB-2 that leads to activation of Ras signaling [62]. A number of similar cases of gain of valency as well as loss of valency through gene truncations have been described [63].
Post-translational regulation
In addition to the impact of transcript expression and modifications on the valency of the resulting protein, the valency of protein molecules can also be regulated post-translationally. Proteolytic processing provides one post-translational mechanism to regulate valency in the innate immune system. For example, the mammalian inflammasome-dependent cytokines IL-1β and IL-18 are expressed as inactive monomers that cannot bind to their receptors [64,65]. Essentially, they have a valency of zero for their receptors. Removal of their autoinhibitory domains by inflammasome activation-dependent proteolytic cleavage unmasks the hidden valency increasing it to one and allowing them to bind to their receptors [34,66–68]. Another example of proteolysis-dependent regulation of valency is provided by a versatile kinase receptor-interacting serine/threonine kinase 1 (RIPK1). In its initial state, RIPK1 is fully functional; however, CASP8 proteolytically cleaves RIPK1, bifurcating its valency onto two fragments and inhibiting RIPK1 scaffold function-dependent processes [69,70]. Recent studies have shown that the valency of the inflammasome sensor NLRP1B is also regulated by proteolytic cleavage, with the proteasome removing the N-terminal inhibitory domain to control activation [71–74].
Post-translational modifications (PTMs) of proteins can also regulate valency. PTMs can induce conformational changes that expose or occlude binding sites, leading to an increase or decrease in valency, respectively. They can also change the surface properties of the protein, making it more or less prone to interact with its ligand. Intrinsically disordered regions (IDR) in proteins are promiscuous binding motifs that can drive the assembly of supramolecular structures in cells through IDR-IDR binding [75,76]. Phosphorylation in the IDRs can regulate their valency. For example, phosphorylation in the IDR of fused in sarcoma (FUS) inhibits assembly of FUS granules, likely by making IDR-IDR interactions energetically unfavorable due to electrostatic repulsion and decreasing valency [77]. Similarly, phosphorylation of the IDR of human GLE1, a stress granule component, triggers disassembly of stress granules in HeLa cells [78]. PTMs themselves can also act as binding sites, thereby increasing the valency of a protein [79]. K27 ubiquitination of stimulator of interferon genes (STING) increases its valency and allows it to bind the serine/threonine-protein kinase TBK1 and activate downstream signaling upon cytosolic DNA sensing [80]. M1-K63 hybrid ubiquitination of myddosome components can also increase valency, allowing recruitment of the transforming growth factor β-activating kinase 1 (TAK1) and IκB kinase (IKK) complex for their activation [81]. In another example, K63 ubiquitin chains can allow proteins to bind DNA, thereby modifying the valency of the K63-ubiquitinated proteins and facilitating the repair of damaged DNA [82]. Prenylation is another PTM that can expand the valency of proteins [83]. For example, prenylation of a protein can allow it to bind to a lipid membrane, increasing its valency for lipid membranes from zero to one [84–87]. Thus, post-translational modifications of proteins can increase or decrease their valencies.
Collectively, these examples illustrate different ways in which the valency of a diverse array of biomolecules can be regulated, and some cases in which such dysregulated valency status might contribute to, or result in, disease.
The valency of a cell might determine its response to external stimuli
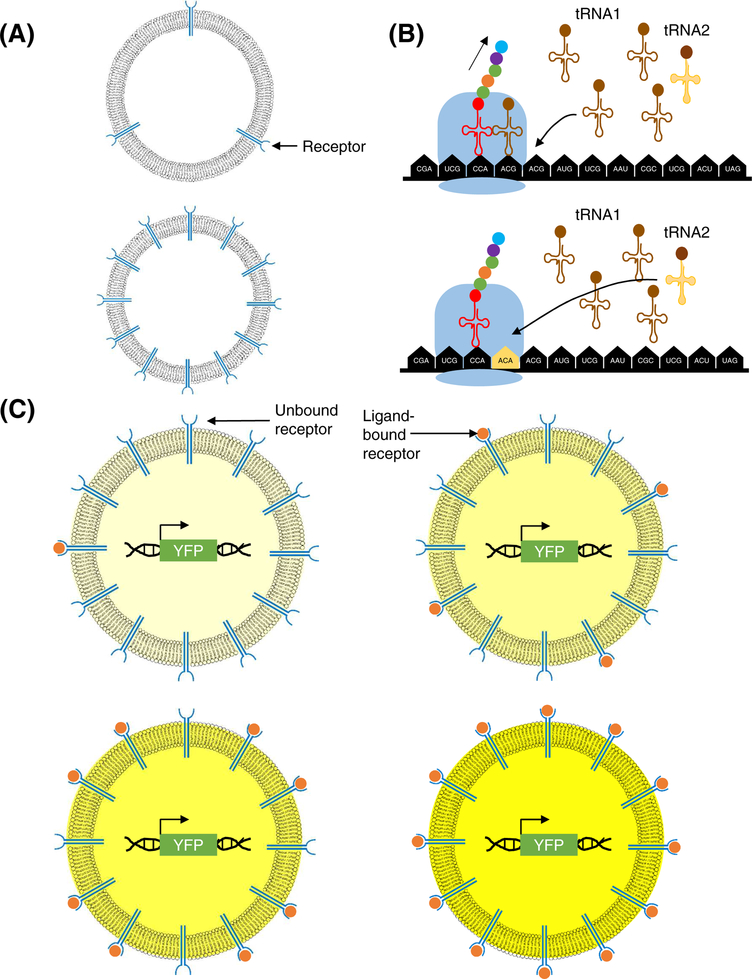
We have described many examples of the valency of a biomolecule determining its function and regulation. We propose that the idea of valency can also be applied at the whole cell level. For example, similarly to a biomolecule that has a defined number of binding sites for other molecules, or a valency, a cell also has an exact number of sensor molecules on its surface to recognize a particular PAMP; we can define this number as the “cell’s valency” for that PAMP (Fig. 4A). The same idea can be applied to any ligand-receptor combination, the number of adaptor and signal transducer molecules, the number of tRNA or mRNA molecules, the number of specialized ribosomes, gene copy numbers, or even the number of organelles, such as mitochondria and micronuclei (Fig. 4B) (See Outstanding Questions). Cells have evolved to utilize their valency in creative ways to regulate their functions. For example, 61 codon-tRNA pairs are used to code for 20 amino acids, meaning that some amino acids are coded by multiple redundant codon-tRNA pairs. The set of tRNAs possessed by a cell might be considered the tRNA valency of the cell. The tRNAs for certain codons are present in higher numbers than other redundant codons are, and the codons corresponding to those more abundant tRNAs are utilized more often by the cell [88–90]. These codons are considered ‘optimal codons’ because they allow the fastest possible translation rate [91,92]. However, cells might use suboptimal codons to regulate co-translational protein folding and even protein stability, modulating the speed of translation by utilizing the tRNA valency of a cell [93,94]. As an example, the use of a suboptimal codon in the γ-actin open reading frame exposes a lysine residue (K18) for ubiquitination and subsequent proteasome-mediated co-translational degradation in HEK293T cells [94].
Key Figure, Figure 4. Model: The valency of a cell might be determined by the number of molecules it possesses.
(A) Cell surface receptor/sensor valency is the number of receptors/sensors, or a product of valency and number of receptors/sensors if the receptors/sensors can bind to more than one molecule of the ligand. (B) The number of tRNAs available may be another example of valency in the cell. The rate of mRNA translation can be modulated by the usage of redundant codons (tRNA1 and tRNA2 which code for the same amino acid here). For example, the use of a codon whose cognate tRNA abundance is low (tRNA2 here) will slow the rate of translation. (C) The fraction of receptors bound to their ligands can determine the response of individual cells. A higher fraction of bound receptors can lead to stronger cellular response. In the figure, we use an example of yellow fluorescent protein (YFP) expression as a readout of cellular response. There is an increase in YFP fluorescence corresponding with the increase in the fraction of valency being satisfied by ligand binding.
Outstanding Questions.
How do we determine the exact valency of a biomolecule? Homotypic and heterotypic interactions between well-studied domains are easy to predict. For example, interactions between PYRIN, CARD, RHIM, and DED domains play critical roles in regulating immune responses. However, post-translational modifications and interactions between under-studied proteins make it hard to accurately predict biophysical interactions. There is an important need to develop sets of tools to accurately predict such interactions.
How do we determine the exact valency of a cell? The number of a specific cellular component can determine the valency of a cell with respect to that component. However, in many cases multiple ligands can signal through the same receptor to induce different cell fates. IL-1α/IL-1β and IFNα/IFNβ are examples of pairs of ligands that share receptors. Alternatively, the ligand can signal through multiple receptors, with TNF signaling through TNFR1 and TNFR2 as an example. We need tools that allow us to count the numbers of each cellular component at the single-cell level. We also need a computational framework enabling the interrogation of diverse datasets (for example, proteomic and transcriptomic data) to build accurate models.
Can the valency of a cell determine its response to stimuli? We hypothesized that the valency of a cell and the extent to which the valency is satisfied might determine the responses of individual cells. Although it makes intuitive sense and agrees with the published literature, the hypothesis still needs to be directly tested.
How do we account for the valency of intrinsically disordered regions (IDRs) in proteins? IDRs are capable of engaging in promiscuous interactions. They drive the assembly of many cellular supramolecular structures. However, IDRs are also prone to inducing irreversible phase transitions to the insoluble phase, with potential pathophysiological consequences. IDRs are present in a vast number of proteins, including proteins involved in immune signaling, such as DDX3X. We need to find a way to clearly understand how IDR-IDR interactions work so that we can better predict which of the IDRs are capable of interacting with each other. Alternatively, is there any specificity to IDR-IDR interactions? What are the different ways in which IDR-IDR interactions are regulated?
The set of ribosomes present constitutes another type of valency in cells. A number of studies over the last decade have suggested that all ribosomes are not created equal [95,96]. Ribosomes differing in composition have different translational efficiency for specific mRNAs and act as a ribosome filter to fine-tune gene expression programs [95]. For instance, mice lacking one of the ribosomal proteins, RPL38, exhibit developmental defects linked to inefficient translation of certain Hox mRNAs [97]. Moreover, working with Saccharomyces cerevisiae, researchers have shown that cells can regulate the ribosome composition based on external stimuli, thereby regulating their ribosomal valency [98,99].
The valency of membrane-bound organelles can also play important roles in regulating cellular processes. In a recent study, mitochondrial replication increased mitochondrial valency and was found to be important for NLRP3 inflammasome activation [100]. In another report, NLRP3 binding to a dispersed trans-Golgi network was required for the activation of the NLRP3 inflammasome [101]. Furthermore, enzymatic activity potential can also be considered a type of valency. This can account for redundancies between different enzymes that act on the same substrate. For example, CASP1 and CASP8 can both process IL-1β and have been shown to play a role in a mouse model of osteomyelitis [102]. In this murine model, IL-1β–driven autoinflammatory osteomyelitis was inhibited with the loss of both Casp1 and Casp8. Mice lacking only one of these caspases had increased susceptibility to disease compared with the double knockout animals [102]. Overall, we propose that the number of cellular components or the activity potential of a cell can determine cellular valency with respect to that component or activity.
Using this concept, cellular valency might determine the cell’s response to external stimuli (See Outstanding Questions). For example, in the innate immune system, LPS stimulation leads to strong concentration-dependent kinetics for myddosome assembly, with higher concentrations of LPS leading to a more rapid assembly of the myddosome relative to stimulation with lower concentrations [103]. According to our model, this concentration dependence might be explained by the fraction of TLR4 molecules on the cell surface that are LPS-bound at a given concentration. At the highest LPS concentration tested in the study, most if not all of the TLR4 molecules on the cell surface might be expected to be LPS-bound, allowing maximal signaling and faster myddosome assembly. Similarly, under lower LPS concentrations, a fraction of TLR4 molecules would be expected to be free, potentially leading to lower downstream signal strength and slower kinetics of myddosome assembly relative to the signal strength and kinetics observed at higher concentrations [103]. We hypothesize that this dose-dependence could be generalized to explain the observed differences in the responses of individual cells to stimuli. Such differences might be expected to be greater under conditions when the components of a cell involved in responding to a stimulus or a ligand are present in limiting amounts (Fig. 4C). Since ligand-receptor binding is an equilibrium process, it is reasonable to suppose that the strength of an interaction might also be a critical parameter in determining the fraction of satisfied receptor valency. For example, when a population of cells is stimulated with less than the saturating concentration of a ligand, there might be differences in the fractions of ligand-bound and free receptors between cells. This might in turn lead to differential responses in individual cells.
Cells can exploit their finite valencies with respect to their components to make cell fate decisions. If there is a common essential factor required for two different cellular processes, there might be a competition between the two processes for this factor. For example, when stress granule and NLRP3 inflammasome assembly are competing for the same common essential factor – e.g. DDX3X protein molecules-- the DDX3X valency might allow the cell to make a ‘live-or-die’ cell fate decision [104]. In murine BMDM, induction of stress granules inhibited NLRP3 inflammasome activation by sequestering DDX3X, preventing the cells from undergoing pyroptosis [104]. The concept of valency at the whole-cell level might also have important pathophysiological consequences and might lead to interesting future applications. An example would be assessing the optimal dose of a drug. Indeed different cell types might have different valencies for a drug. Hypothetically, the dose might be tailored based on the valency of the cell type being targeted. Valency could also be an important consideration in mathematical modeling of single-cell responses. Accordingly, studies of single-cell response have revealed differences in the responses of individual cells, even though all cells in a given population might be expected to be of the same genotype [105–107]. Presumably, such differences might be due to stochastic differences in the valencies of various types of cells. Furthermore, cells in a population can communicate with each other through cell-cell contacts and secreted signaling molecules, such as cytokines and chemokines. Thus, the amounts of cytokines and chemokines in the extracellular space might be expected to be lower than the amount needed to ensure that all the receptors are bound by their corresponding ligands. In such cases and as mentioned above, these differences in the fractions of ligand-bound receptors might also be expected to contribute to the differences in the responses of individual cells. Overall, such examples suggest that the concept of valency might be helpful in the study of immune responses.
Concluding remarks
In summary, we propose that valency is a fundamental property of biomolecules and cells. The valency of biomolecules is critical for the assembly of functional biomolecular complexes. Cells have evolved multiple mechanisms to regulate the valency of biomolecules to ensure homeostasis. We extend the concept of valency to whole cells and propose that it might be used to explain variations in the responses of individual cells. Testing the valency hypothesis to explain the responses of individual cells would require the development of more accurate and precise quantification techniques that can allow multiparametric measurements at the single cell level (see Outstanding Questions). Techniques such as Cy-TOF and multi-color flow cytometry analysis are already being used to study individual cells in suspension [108,109]. Nevertheless, there is still a need to develop techniques that allow these measurements to be performed on adherent cells. We posit that considering valency as an important parameter might lead to a better understanding of biological processes, which in turn could potentially lead to the development of better therapeutic interventions for a variety of immune-related pathologies. Indeed, the idea of using valency to modulate drug efficacy is already being exploited through the development of multivalent drugs, highlighting the potential applications of this exciting concept [110–112].
Highlights.
The valency of a biomolecule determines its biophysical interactions.
Innate immune signaling can require multivalent interactions for the proper functioning of signaling platforms such as the myddosome.
Inflammasome activation can exploit the multivalency of sensors, adaptors, and ligands.
We posit that the concept of valency might be applied at the whole-cell level to study differences in individual cell responses.
Acknowledgments
The authors acknowledge many investigators in the field whose primary data could not be cited in this review because of space limitations. Scientific editing and writing support were provided by Rebecca Tweedell, PhD. This work was supported by the National Institutes of Health grants CA163507, AR056296, AI124346, and AI101935 and by ALSAC to T‐DK. The authors have no financial conflicts of interest to disclose.
Glossary
- AIM2
cytosolic DNA sensor activated upon DNA binding to assemble the AIM2 inflammasome and initiate pro-inflammatory cell death through pyroptosis
- ASC
adaptor protein required for inflammasome assembly that bridges the sensor and the catalytic actuator caspase-1
- Avidity
measure of the accumulated strength of an interaction between biomolecules depending on the combination of all non-covalent interactions
- cGAS
cytosolic DNA sensor that catalyzes the synthesis of cyclic AMP-GMP (cGAMP) to induce the innate immune response by activating STING-mediated signaling
- Cy-TOF
a flow cytometry-based mass spectrometry technique to study single cells using cells labeled with lanthanide metal-tagged antibodies
- DAMP
molecules generated from cell and tissue injury
- FUS
nucleic acid binding protein that can undergo phase transitions that have pathophysiological consequences for amyotrophic lateral sclerosis disease
- IRAK4
component of the myddosome complex
- IDR
region that does not form a defined secondary structure in a protein; critical for the assembly of large protein complexes
- Inflammasome
protein complex formed in the cytoplasm after sensing intracellular damage and pathogen-associated molecular patters that contains a sensor, executioner caspase-1 or caspase-4/5/11, and often the adaptor ASC
- Lattice-like structures
biomolecules with a valency of 3 or more can form supramolecular assemblies that contain these structures
- Myddosome
signaling platform formed upon stimulation of toll-like receptors containing MYD88 and other proteins
- NLRC4
inflammasome sensor activated in response to intracellular bacteria by sensing components of the flagella and type III secretion system
- NLRP1B
inflammasome sensor activated by proteolytic processing of its inhibitor domains by bacterial toxins
- NLRP3
inflammasome sensor activated downstream of potassium efflux, dispersed Golgi network, mitochondrial DNA replication, and mitochondrial reactive oxygen species
- Optimal codons
codons within the cell that allow for the fastest possible translation rate; these are generally the most abundant codon for a given amino acid
- PAMP
molecular signatures of infection
- Phase separation
property of liquids where two liquids with sufficiently different physiochemical properties become immiscible
- PTM
molecular modifications, such as phosphorylation or ubiquitination, of proteins that occur after translation
- P-bodies
membraneless cytoplasmic compartments involved in RNA processing; formed in response to stress and which contain RNase enzymes
- Pyroptosis
pro-inflammatory cell death program activated downstream of inflammasome activation; characterized by cleavage of the pore-forming proteins gasdermin D/E and release of IL-1β and IL-18
- Repeat expansion disorders
class of human diseases characterized by an increase in the number of times a repeated pattern of nucleic acids appears
- STING
five transmembrane protein that plays a critical role in the innate immune response downstream of cyclic AMP-GMP synthase
- Stress granules
membraneless cytoplasmic compartments formed in response to stressors; may act as storage compartments for translation initiation complexes allowing cells to survive stress
- TBK1
serine/threonine kinase activated in response to innate immune sensing of bacterial and viral infections
- tRNA
type of RNA cofactor involved in protein translation
- TLR
type of pattern recognition receptor; helps cells sense damage- and pathogen-associated molecular patterns
- Valency
number of distinct binding sites for a specific interaction on a biomolecule. In the case of cellular valency, it is the effective number of a cellular component (product of the number of binding sites on a component and the total number of that component present in/on the cell); e.g. if a receptor can bind two ligand molecules, and there are three copies of that receptor on the cell, the cellular valency for that receptor will be six
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gates J and Peifer M (2005) Can 1000 Reviews Be Wrong? Actin, α-Catenin, and Adherens Junctions. Cell 123, 769–772 [DOI] [PubMed] [Google Scholar]
- 2.Drees F et al. (2005) α-Catenin Is a Molecular Switch that Binds E-Cadherin-β-Catenin and Regulates Actin-Filament Assembly. Cell 123, 903–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamada S et al. (2005) Deconstructing the Cadherin-Catenin-Actin Complex. Cell 123, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li P et al. (2012) Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banjade S and Rosen MK (2014) Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 3, e04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banani SF et al. (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin Y et al. (2015) Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 60, 208–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg RJ (1952) A Theory of Antibody—Antigen Reactions. I. Theory for Reactions of Multivalent Antigen with Bivalent and Univalent Antibody2. J. Am. Chem. Soc 74, 5715–5725 [Google Scholar]
- 9.Brewer CF et al. (2002) Clusters, bundles, arrays and lattices: novel mechanisms for lectin–saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol 12, 616–623 [DOI] [PubMed] [Google Scholar]
- 10.Tang S-J (2017) Potential Role of Phase Separation of Repetitive DNA in Chromosomal Organization. Genes 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall AC et al. (2019) Phase Separation as a Melting Pot for DNA Repeats. Trends Genet. DOI: 10.1016/j.tig.2019.05.001 [DOI] [PubMed] [Google Scholar]
- 12.Hnisz D et al. (2017) A Phase Separation Model for Transcriptional Control. Cell 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabari BR et al. (2018) Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boija A et al. (2018) Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larson AG et al. (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strom AR et al. (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain A and Vale RD (2017) RNA Phase Transitions in Repeat Expansion Disorders. Nature 546, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meola G and Cardani R (2015) Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim. Biophys. Acta BBA - Mol. Basis Dis 1852, 594–606 [DOI] [PubMed] [Google Scholar]
- 19.Pringsheim T et al. (2012) The incidence and prevalence of Huntington’s disease: A systematic review and meta-analysis. Mov. Disord 27, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 20.Zarei S et al. (2015) A comprehensive review of amyotrophic lateral sclerosis. Surg. Neurol. Int 6, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motshwene PG et al. (2009) An Oligomeric Signaling Platform Formed by the Toll-like Receptor Signal Transducers MyD88 and IRAK-4. J. Biol. Chem 284, 25404–25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S-C et al. (2010) Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Y and Kagan JC (2019) Innate Immune Signaling Organelles Display Natural and Programmable Signaling Flexibility. Cell 177, 384–398.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du M and Chen ZJ (2018) DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 361, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kesavardhana S et al. (2017) ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med DOI: 10.1084/jem.20170550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thapa RJ et al. (2016) DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 20, 674–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuriakose T et al. (2016) ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol 1, aag2045–aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teng MWL et al. (2015) IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med 21, 719–729 [DOI] [PubMed] [Google Scholar]
- 29.Lukens JR et al. (2012) IL-1 family cytokines trigger sterile inflammatory disease. Front. Immunol 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukens JR et al. (2013) RIP1-driven autoinflammation targets IL-1α independently of inflammasomes and RIP3. Nature 498, 224–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinarello CA (2009) Immunological and Inflammatory Functions of the Interleukin-1 Family. Annu. Rev. Immunol 27, 519–550 [DOI] [PubMed] [Google Scholar]
- 32.Werman A et al. (2004) The precursor form of IL-1α is an intracrine proinflammatory activator of transcription. Proc. Natl. Acad. Sci 101, 2434–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Place DE and Kanneganti T-D (2018) Recent advances in inflammasome biology. Curr. Opin. Immunol 50, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinon F et al. (2002) The Inflammasome: A Molecular Platform Triggering Activation of Inflammatory Caspases and Processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 35.Lu A et al. (2014) Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 156, 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahillioglu AC et al. (2014) Structural and Dynamics Aspects of ASC Speck Assembly. Structure 22, 1722–1734 [DOI] [PubMed] [Google Scholar]
- 37.Masumoto J et al. (1999) ASC, a Novel 22-kDa Protein, Aggregates during Apoptosis of Human Promyelocytic Leukemia HL-60 Cells. J. Biol. Chem 274, 33835–33838 [DOI] [PubMed] [Google Scholar]
- 38.Franklin BS et al. (2014) The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat. Immunol 15, 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venegas C et al. (2017) Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature 552, 355–361 [DOI] [PubMed] [Google Scholar]
- 40.Jin T et al. (2012) Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor. Immunity 36, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang B and Yin Q (2017) AIM2 inflammasome activation and regulation: A structural perspective. J. Struct. Biol 200, 279–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X et al. (2014) Prion-like Polymerization Underlies Signal Transduction in Antiviral Immune Defense and Inflammasome Activation. Cell 156, 1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamkanfi M and Dixit VM (2014) Mechanisms and Functions of Inflammasomes. Cell 157, 1013–1022 [DOI] [PubMed] [Google Scholar]
- 44.Broz P et al. (2010) Differential Requirement for Caspase-1 Autoproteolysis in Pathogen-Induced Cell Death and Cytokine Processing. Cell Host Microbe 8, 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Case CL et al. (2009) Asc and Ipaf Inflammasomes Direct Distinct Pathways for Caspase-1 Activation in Response to Legionella pneumophila. Infect. Immun 77, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenthorey JL et al. (2017) The structural basis of flagellin detection by NAIP5: A strategy to limit pathogen immune evasion. Science 358, 888–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L et al. (2015) Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y et al. (2018) Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl. Acad. Sci. U. S. A 115, 10845–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuvpilo S et al. (1999) Alternative Polyadenylation Events Contribute to the Induction of NF-ATc in Effector T Cells. Immunity 10, 261–269 [DOI] [PubMed] [Google Scholar]
- 50.Jia X et al. (2017) The role of alternative polyadenylation in the antiviral innate immune response. Nat. Commun 8, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian B and Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol 18, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pal S et al. (2011) Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res 21, 1260–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carpenter S et al. (2014) Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol 14, 361–376 [DOI] [PubMed] [Google Scholar]
- 54.Lichtinger M et al. (2007) Transcription Factor PU.1 Controls Transcription Start Site Positioning and Alternative TLR4 Promoter Usage. J. Biol. Chem 282, 26874–26883 [DOI] [PubMed] [Google Scholar]
- 55.Janssens S et al. (2002) Regulation of Interleukin-1- and Lipopolysaccharide-Induced NF-κB Activation by Alternative Splicing of MyD88. Curr. Biol 12, 467–471 [DOI] [PubMed] [Google Scholar]
- 56.Janssens S et al. (2003) MyD88S, a splice variant of MyD88, differentially modulates NF-κB- and AP-1-dependent gene expression. FEBS Lett 548, 103–107 [DOI] [PubMed] [Google Scholar]
- 57.Smith MA et al. (2019) U2AF1 mutations induce oncogenic IRAK4 isoforms and activate innate immune pathways in myeloid malignancies. Nat. Cell Biol 21, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.(2018) Epitranscriptomic Code and Its Alterations in Human Disease. Trends Mol. Med DOI: 10.1016/j.molmed.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 [DOI] [PubMed] [Google Scholar]
- 60.Nowell P and Hungerford D (1960) Minute Chromosome in Human Chronic Granulocytic Leukemia. Science 132, 1497–1497 [Google Scholar]
- 61.Shtivelman E et al. (1985) Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature 315, 550–554 [DOI] [PubMed] [Google Scholar]
- 62.Pendergast AM et al. (1993) BCR-ABL-induced oncogenesis is mediated by direct interaction with the SH2 domain of the GRB-2 adaptor protein. Cell 75, 175–185 [PubMed] [Google Scholar]
- 63.Mertens F et al. (2015) The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer 15, 371–381 [DOI] [PubMed] [Google Scholar]
- 64.Man SM and Kanneganti T-D (2015) Regulation of inflammasome activation. Immunol. Rev 265, 6–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karki R and Kanneganti T-D (2019) Diverging inflammasome signals in tumorigenesis and potential targeting. Nat. Rev. Cancer 19, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thornberry NA et al. (1992) A novel heterodimeric cysteine protease is required for interleukin-1βprocessing in monocytes. Nature 356, 768–774 [DOI] [PubMed] [Google Scholar]
- 67.Cheneval D et al. (1998) Increased Mature Interleukin-1β (IL-1β) Secretion from THP-1 Cells Induced by Nigericin Is a Result of Activation of p45 IL-1β-converting Enzyme Processing. J. Biol. Chem 273, 17846–17851 [DOI] [PubMed] [Google Scholar]
- 68.Perregaux D and Gabel CA (1994) Interleukin-1 beta maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem 269, 15195–15203 [PubMed] [Google Scholar]
- 69.Lin Y et al. (1999) Cleavage of the death domain kinase RIP by Caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13, 2514–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang L et al. (2015) RIP1 Cleavage in the Kinase Domain Regulates TRAIL-Induced NF-κB Activation and Lymphoma Survival. Mol. Cell. Biol 35, 3324–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandstrom A et al. (2019) Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu H et al. (2019) The N-end rule ubiquitin ligase UBR2 mediates NLRP1B inflammasome activation by anthrax lethal toxin. EMBO J 38, e101996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hellmich KA et al. (2012) Anthrax Lethal Factor Cleaves Mouse Nlrp1b in Both Toxin-Sensitive and Toxin-Resistant Macrophages. PLOS ONE 7, e49741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levinsohn JL et al. (2012) Anthrax Lethal Factor Cleavage of Nlrp1 Is Required for Activation of the Inflammasome. PLOS Pathog. 8, e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shin Y and Brangwynne CP (2017) Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- 76.Protter DSW et al. (2018) Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep 22, 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monahan Z et al. (2017) Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J 36, 2951–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aditi A et al. (2018) MAPK- and glycogen synthase kinase 3–mediated phosphorylation regulates the DEAD-box protein modulator Gle1 for control of stress granule dynamics. J. Biol. Chem DOI: 10.1074/jbc.RA118.005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akutsu M et al. (2016) Ubiquitin chain diversity at a glance. J. Cell Sci 129, 875–880 [DOI] [PubMed] [Google Scholar]
- 80.Wang Q et al. (2014) The E3 Ubiquitin Ligase AMFR and INSIG1 Bridge the Activation of TBK1 Kinase by Modifying the Adaptor STING. Immunity 41, 919–933 [DOI] [PubMed] [Google Scholar]
- 81.Emmerich CH et al. (2013) Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proc. Natl. Acad. Sci 110, 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu P et al. (2018) K63-linked polyubiquitin chains bind to DNA to facilitate DNA damage repair. Sci Signal 11, eaar8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang M and Casey PJ (2016) Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol 17, 110–122 [DOI] [PubMed] [Google Scholar]
- 84.Alexandrov K et al. (1994) Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J 13, 5262–5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muntz KH et al. (1992) Influence of gamma subunit prenylation on association of guanine nucleotide-binding regulatory proteins with membranes. Mol. Biol. Cell 3, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nantais DE et al. (1996) Prenylation of an interferon-γ-induced GTP-binding protein: the human guanylate binding protein, huGBP1. J. Leukoc. Biol 60, 423–431 [DOI] [PubMed] [Google Scholar]
- 87.Zeng Q et al. (2000) Prenylation-dependent Association of Protein-tyrosine Phosphatases PRL-1, −2, and −3 with the Plasma Membrane and the Early Endosome. J. Biol. Chem 275, 21444–21452 [DOI] [PubMed] [Google Scholar]
- 88.Dong H et al. (1996) Co-variation of tRNA Abundance and Codon Usage inEscherichia coliat Different Growth Rates. J. Mol. Biol 260, 649–663 [DOI] [PubMed] [Google Scholar]
- 89.Emilsson V and Kurland CG (1990) Growth rate dependence of transfer RNA abundance in Escherichia coli. EMBO J 9, 4359–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Berg OG and Kurland CG (1997) Growth rate-optimised tRNA abundance and codon usage11Edited by J. H. Miller. J. Mol. Biol 270, 544–550 [DOI] [PubMed] [Google Scholar]
- 91.Hanson G and Coller J (2018) Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol 19, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim CH et al. (1997) Codon optimization for high-level expression of human erythropoietin (EPO) in mammalian cells. Gene 199, 293–301 [DOI] [PubMed] [Google Scholar]
- 93.Kimchi-Sarfaty C et al. (2007) A “Silent” Polymorphism in the MDR1 Gene Changes Substrate Specificity. Science 315, 525–528 [DOI] [PubMed] [Google Scholar]
- 94.Zhang F et al. (2010) Differential Arginylation of Actin Isoforms Is Regulated by Coding Sequence–Dependent Degradation. Science 329, 1534–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mauro VP and Edelman GM (2002) The ribosome filter hypothesis. Proc. Natl. Acad. Sci 99, 12031–12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Komili S et al. (2007) Functional Specificity among Ribosomal Proteins Regulates Gene Expression. Cell 131, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kondrashov N et al. (2011) Ribosome-Mediated Specificity in Hox mRNA Translation and Vertebrate Tissue Patterning. Cell 145, 383–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Slavov N et al. (2015) Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep 13, 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Samir P et al. (2018) Identification of Changing Ribosome Protein Compositions using Mass Spectrometry. PROTEOMICS 18, 1800217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhong Z et al. (2018) New mitochondrial DNA synthesis enables NLRP3 inflammasome activation. Nature DOI: 10.1038/s41586-018-0372-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen J and Chen ZJ (2018) PtdIns4P on dispersed trans -Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gurung P et al. (2016) NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β–mediated osteomyelitis. Proc. Natl. Acad. Sci 113, 4452–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Latty SL et al. (2018) Activation of Toll-like receptors nucleates assembly of the MyDDosome signaling hub. eLife 7, e31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Samir P et al. (2019) DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Elowitz MB et al. (2002) Stochastic Gene Expression in a Single Cell. Science 297, 1183–1186 [DOI] [PubMed] [Google Scholar]
- 106.Neuert G et al. (2013) Systematic Identification of Signal-Activated Stochastic Gene Regulation. Science 339, 584–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bendall SC et al. (2011) Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science 332, 687–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vorobjev IA and Barteneva NS (2017) Multi-parametric imaging of cell heterogeneity in apoptosis analysis. Methods 112, 105–123 [DOI] [PubMed] [Google Scholar]
- 109.Diggins KE et al. (2015) Methods for discovery and characterization of cell subsets in high dimensional mass cytometry data. Methods 82, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Waring MJ et al. (2016) Potent and selective bivalent inhibitors of BET bromodomains. Nat. Chem. Biol 12, 1097–1104 [DOI] [PubMed] [Google Scholar]
- 111.Tanaka M et al. (2016) Design and characterization of bivalent BET inhibitors. Nat. Chem. Biol 12, 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lehar SM et al. (2015) Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328 [DOI] [PubMed] [Google Scholar]