Abstract
The human gastrointestinal tract harbors a dense and diverse microbial community, the makeup of which is intimately linked to health. Extrinsic factors such as diet and host immunity are insufficient to explain the constituents of this community, implicating direct interactions between co-resident microbes as an important driver of microbiome composition. The genomes of bacteria derived from the gut microbiome are replete with pathways that mediate contact-dependent interbacterial antagonism1–3. Many members of the Gram-negative order Bacteroidales encode the type VI secretion system (T6SS), which facilitates the delivery of toxic effector proteins into adjacent cells4,5. Here we report the occurrence of acquired interbacterial defense (AID) gene clusters in Bacteroidales residing within the human gut microbiome. These clusters encode arrays of immunity genes that protect against T6SS-mediated intra- and inter-species bacterial antagonism. Moreover, the clusters reside on mobile elements and we demonstrate that their transfer is sufficient to confer toxin resistance in vitro and in gnotobiotic mice. Finally, we identify and validate the protective capacity of a recombinase-associated AID subtype (rAID-1) present broadly in Bacteroidales genomes. These rAID-1 gene clusters have a structure suggestive of active gene acquisition and include predicted immunity factors of toxins deriving from diverse organisms. Our data suggest that neutralization of contact-dependent interbacterial antagonism via AID systems shapes human gut microbiome ecology.
Polymicrobial environments contain a plethora of biotic and abiotic threats to their inhabitants. Bacterial survival in these settings necessitates elaborate defensive mechanisms. Some of these are basal and protect against a wide range of threats, whereas others, for instance CRISPR-Cas, represent adaptations unique to the specific threats encountered by a bacterial lineage6,7. In densely colonized habitats such as the mammalian gut, overcoming contact-dependent interbacterial antagonism is likely a major hurdle to survival. The type VI secretion system (T6SS) is a pathway widely utilized by gut bacteria to mediate the delivery of toxic effector proteins to neighboring cells8–10. While kin cells are innately resistant to these effectors via cognate immunity proteins, it is unknown whether non-self cells in the gut can escape intoxication. Intriguingly, a number of recent studies have reported that bacteria from a range of environments can encode T6S immunity genes that lack an accompanying cognate effector gene2,9,11–13. It has been suggested that these genes play a role in interbacterial defense, but they have yet to be functionally investigated in a physiological context.
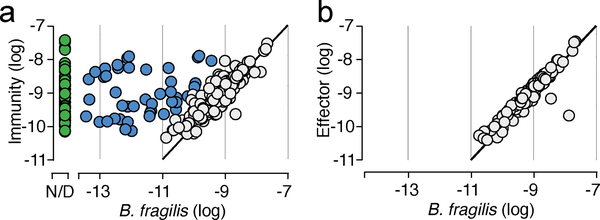
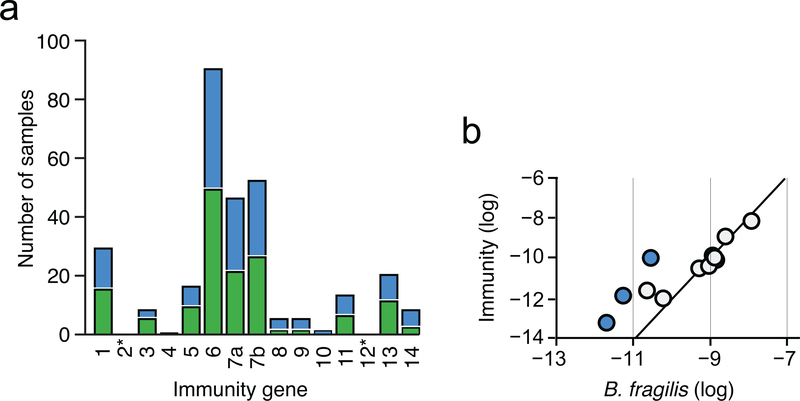
To identify potential mechanisms of defense against T6S-delivered interbacterial effectors in the human gut, we mined a large collection of shotgun metagenomic samples (n=553) from multiple studies of the human gut microbiome for sequences homologous to known immunity genes (Supplementary Table 1)14,15. We first focused our efforts on Bacteroides fragilis, which harbors a well described and diverse repertoire of effector and cognate immunity genes (Supplementary Table 2)3,8,9. As expected for genes predicted to reside within the B. fragilis genome, sequences mapping to these immunity loci were detected at an abundance similar to that of B. fragilis species-specific marker genes in many microbiome samples (Fig. 1a, grey; Supplementary Table 1). However, in a second subset of samples, immunity genes were detected at an abundance significantly higher (>10X) than expected given the abundance of B. fragilis (Fig. 1a, blue). Finally, we identified a third subset of samples wherein immunity gene sequences were detected in the absence of B. fragilis (Fig. 1a, green). These latter sequences include close homologs of 12 of 14 unique immunity genes (i1-i14) encoded by B. fragilis (Extended Data Fig. 1a). In contrast to the pattern observed for immunity genes, we found no samples in which the abundance of corresponding cognate effector genes exceeded that of B. fragilis by a significant margin (Fig. 1b, Supplementary Table 1).
Fig. 1 |. T6SS orphan immunity genes are found in human gut microbiomes.
Abundance comparison of B. fragilis-specific T6SS immunity genes a or effector genes b with B. fragilis marker genes in adult microbiome samples derived from the HMP and MetaHIT studies (Supplementary Table 1). Abundance values denote the number of reads mapped to the gene, normalized by gene length and total number of reads in the sample. Abundances are calculated as the average abundance of all immunity, effector, or B. fragilis species-specific marker genes. Samples with undetectable B. fragilis (green) and samples in which immunity gene abundance exceeds that of Bacteroides by over 10-fold (blue) are highlighted.
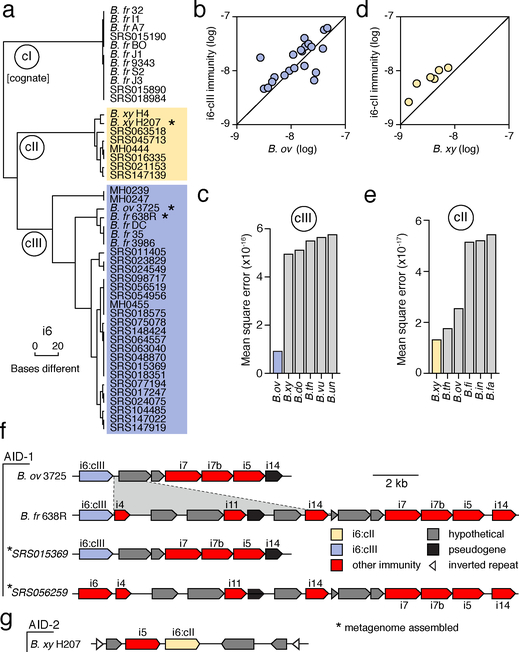
The detection of B. fragilis immunity gene homologs in samples in which we were unable to detect B. fragilis strongly suggests that these elements are encoded by other bacteria in the gut. Indeed, BLAST searches revealed the genomes of several Bacteroides spp. that harbor B. fragilis T6S immunity gene homologs, including B. ovatus (i6, i7, i5, i14), B. vulgatus (i8, i13), B. helcogenes (i1, i9, i10), and B. coprocola (i8, i13). To determine whether these bacteria could also account for the presence of immunity genes in the human gut microbiome, we assembled full-length predicted immunity genes from the metagenomic sequencing reads of individual microbiomes. We limited this assembly to homologs of i6, the most prevalent immunity gene detected in samples lacking B. fragilis (Extended Data Fig. 1a). Clustering of the recovered homologs showed that the majority of i6 sequences distribute into three discrete clades that differ by multiple nucleotide substitutions (i6:cI-cIII) (Fig. 2a and Supplementary Table 3). A comparison of these immunity sequences to available bacterial genomes revealed a clade matching cognate immunity genes in B. fragilis (i6:cI). Additionally, we found an i6 sequence homologous to i6:cIII in the genome of B. ovatus, which we previously found does not contain the cognate T6SS effector gene8.
Fig. 2 |. T6SS orphan immunity gene clusters are encoded by multiple species in the human gut microbiome.
a, Dendrogram depicting hierarchical clustering of orphan immunity gene i6 sequences extracted from genomes (n=15) and metagenomes (n=32) derived from the indicated HMP (SRS) or MetaHIT (MH) samples. Sequence clades discussed in the text are denoted (cI-III). Asterisks indicate strains diagrammed in panels f and g. b-d Abundance comparison of genes from the indicated immunity clades (colors as in a) with B. ovatus b or B. xylanisolvens d marker genes in adult microbiome samples (Supplementary Table 1). c, e, Linear model error values for the six species best fitting i6:cIII c and i6:cII e gene abundances calculated as in Fig. 1. f, g, Representative AID-1 f and AID-2 g gene clusters containing homologs of the indicated B. fragilis T6S immunity genes. The i6 gene of SRS056259 did not meet our sequence depth coverage requirements for inclusion in i6:cIII. The B. xylanisolvens strain in g was sequenced as a part of this study (BioProject PRJNA484981). A region of difference between B. ovatus 3725 and B. fragilis 638R clusters is highlighted. All strain abbreviations defined in Supplementary Table 3.
To identify the species encoding these sequences in human gut microbiomes, we used a simplified linear model to identify Bacteroides spp. whose abundance in microbiome samples best fits that of each immunity sequence clade. We found that i6:cIII is best explained in gut metagenomes by B. ovatus (Fig. 2b,c), suggesting that although reference genomes of both B. fragilis and B. ovatus contain these sequences, it is most often harbored by the latter in natural populations. We could not confidently define a single species harboring i6:cII by this method (Fig. 2d,e); therefore, we employed the same analysis pipeline to an infant microbiome dataset for which matching stool samples were available16 (Extended Data Fig. 1b and Supplementary Table 4). Whole genome sequencing of isolates identified B. xylanisolvens as a bacterium harboring i6:cII in these samples. Notably, this species best fit the abundance of i6:cII genes in the large adult metagenomic datasets, albeit by a narrow margin (Fig. 2d). Based on these observations, we hypothesized that orphan immunity genes – encoded by B. fragilis and other species of Bacteroides – serve an adaptive role in the gut by providing defense during intra- and inter-species antagonism.
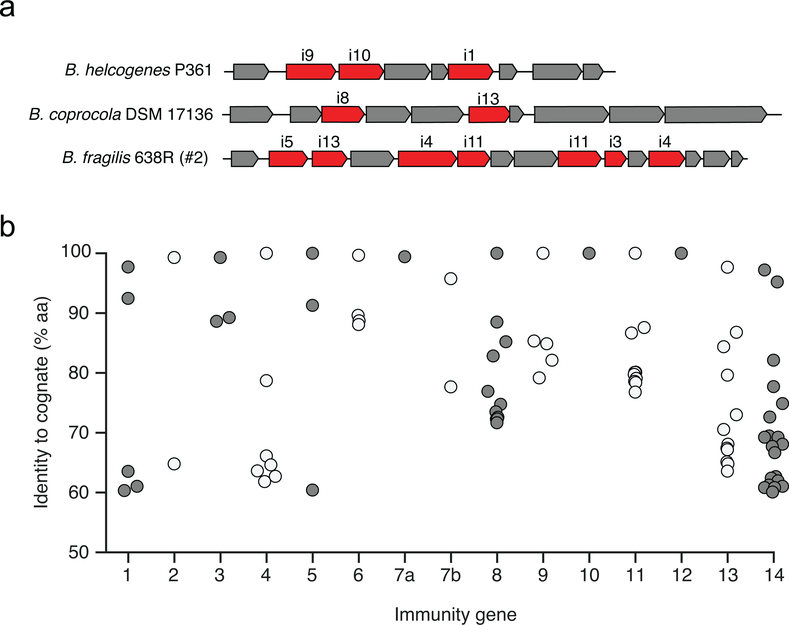
To gain insight into the function of orphan immunity genes, we examined their genomic context in available reference genomes and assembled sequence scaffolds from metagenomic data. We found that homologs of B. fragilis T6S immunity genes i6 and i7 are located together within discrete gene clusters, which we termed AID-1 (acquired interbacterial defense 1) systems, in several Bacteroides strains and in microbiome samples (Fig. 2f). Within AID-1, we identified distant homologs and pseudogenized remnants of additional B. fragilis immunity genes, including i4, i5, i11, and i14 (Fig. 2f and Supplementary Table 5). These findings prompted us to search for more distant homologs of B. fragilis T6S immunity genes. This revealed gene clusters containing orphan homologs of i1, i3, i8, i9, i10, and i13 in diverse Bacteroides genomes and we also identified distant homologs of these genes in a metagenomic gene catalog (Extended Data Fig. 2a,b). In B. xylanisolvens, we found that genes belonging to i6:cII are located in a unique, but analogous context adjacent to a homolog of i5 on an apparent transposable element (Fig. 2g)17. We designated this sequence AID-2.
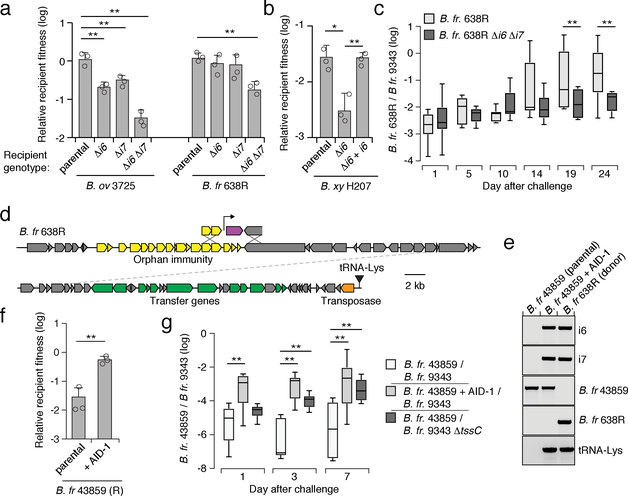
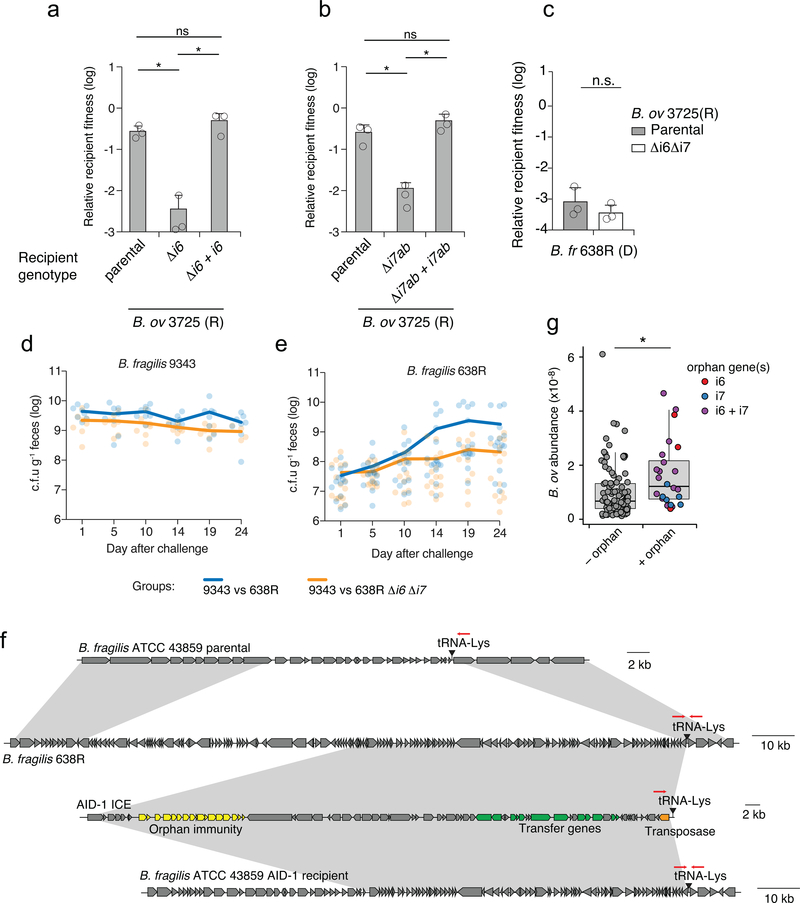
We next defined the phenotypic implications of orphan immunity genes of Bacteroides spp. during interbacterial competition. B. fragilis 9343 encodes the cognate effectors for i6 and i7, and prior data demonstrate that the corresponding toxins antagonize assorted Bacteroides spp. in vitro and in gnotobiotic mice9. We thus employed this strain in growth competition assays against Bacteroides spp. bearing orphan immunity genes, derivative strains containing deletions of these genes, or genetically complemented strains. These experiments showed that in both B. ovatus and B. fragilis, AID-1 system genes grant immunity against corresponding T6S effectors (Fig. 3a, Extended Data Fig. 3a,b). The i6 and i7 genes of B. ovatus did not influence the outcome of its competition with B. fragilis 638R, which possesses an orthogonal effector repertoire (Extended Data Fig. 3c). Finally, we also found that an i6:cII gene from a B. xylanisolvens AID-2 system affords this bacterium protection against e6 of B. fragilis 9343 (Fig. 3b). In total, these data show that the orphan immunity genes of multiple Bacteroides spp. – localized to AID systems – can confer protection against effectors delivered by the T6SS of B. fragilis.
Fig. 3 |. Orphan immunity genes are mobile and protect against T6S-delivered toxins.
a, b, Outcomes of growth competition assays between the indicated AID-1 a or AID-2 b harboring strains versus B. fragilis 9343. Relative recipient fitness was determined by calculating the ratio of final to initial colony forming units (c.f.u.) and normalizing to corresponding experiments with T6S-inactive B. fragilis 9343 (ΔtssC). Data represent mean ± s.d. of n=3 technical replicates indicative of at least three biological replicates,*P < 0.05, **P < 0.01, (unpaired two-tailed t-test). c, Outcome of pairwise competition between the indicated B. fragilis strains in germ-free mice. Mice were colonized with B. fragilis 9343 for one week and challenged with 638R (n=8 mice per group for each of two independent experiments). Boxplots represent the interquartile range with indicated means for each group, whiskers represent the minimum and maximum values, **P < 0.01 (Mann-Whitney U test for each time point). d, Schematic depicting a B. fragilis 638R ICE harboring the AID-1 cluster depicted in Fig. 2f. e, PCR analysis of an AID-1 transfer experiment. Schematic with primer locations provided in Extended Data Fig. 3f. Transfer data are representative of two independent biological replicates. f, Outcomes of growth competition assays between B. fragilis 43859 or a derivative AID-1 ICE recipient and B. fragilis 9343. Data presentation and statistics as in a,b. g, Results of pairwise competition between the indicated B. fragilis strains in germ-free mice (n=6 mice from two independent experiments). Statistics, and data presentation performed as in c.
B. fragilis is typically found as a clonal population in the human gut microbiome, and recent studies suggest that this is in part due to active strain exclusion via the T6SS8,18. However, in gnotobiotic mouse colonization experiments, certain B. fragilis strain pairs inexplicably co-exist10. We noted that one such pair corresponds to B. fragilis 9343 and B. fragilis 638R, the latter of which contains an AID-1 system harboring homologs of i6 and i7. To determine whether our in vitro results with these strains extend to a more physiological setting, we measured the fitness contribution of the orphan immunity genes encoded by B. fragilis 638R following pre-colonization of germ-free mice with B. fragilis 9343 (Fig. 3c, Extended Data Fig. 3d, e). Our results indicated that the cumulative protection afforded by orphan i6 and i7 genes underlies the capacity of B. fragilis 638R to persist during T6S-mediated antagonism in vivo.
Interestingly, we found that AID-1 resides on a predicted mobile integrative and conjugative element (ICE), potentially providing an explanation for its distribution (Fig. 3d)19. To test whether this element can be transferred between strains, we performed mobilization studies using B. fragilis 638R as a donor and B. fragilis 43859 as a recipient. An antibiotic resistance marker was inserted within AID-1 to facilitate the detection of its transfer. With this tool, we readily detected AID-1 transfer (Fig. 3e). This occurred at a frequency of approximately 5×10−6, in line with previous quantification of ICE mobility in Bacteroides spp.20. Next, we asked whether the transfer of AID-1 to B. fragilis 43859 is sufficient to confer resistance to T6S-mediated antagonism. In vitro growth competition assays against B. fragilis 9343 showed that AID-1 effectively neutralizes intoxication by e6 and e7 (Fig. 3f). The receipt of AID-1 also granted significant protection to B. fragilis 43859 against T6-mediated killing in germ-free mice pre-colonized with B. fragilis 9343 (Fig. 3g). Together, these findings indicate that the transfer of a mobile orphan immunity island to a naïve Bacteroides strain is sufficient to provide defense against T6S effectors.
Deciphering the contribution of individual gene products, or even whole pathways, to bacterial fitness in complex microbial communities is challenging. We reasoned that the ability to identify orphan immunity genes in human gut metagenomes, coupled with our capacity to infer their organismal source, provided an opportunity to measure the impact of these defensive factors on competitiveness in the gut. To this end, we compared the abundance of B. ovatus strains with and without i6 and i7 orphan immunity genes in human gut metagenome samples. Interestingly, we found that the average abundance of B. ovatus strains with orphan immunity genes significantly exceeds that of those without (Extended Data Fig. 3g). One interpretation of this finding is that the acquisition of i6 and i7 allows B. ovatus to increase its niche; however, there are several potential caveats inherent to these correlative data that cannot be ruled out. For instance, B. ovatus strains containing i6 and i7 might be related and enriched for other fitness determinants that account for their abundance.
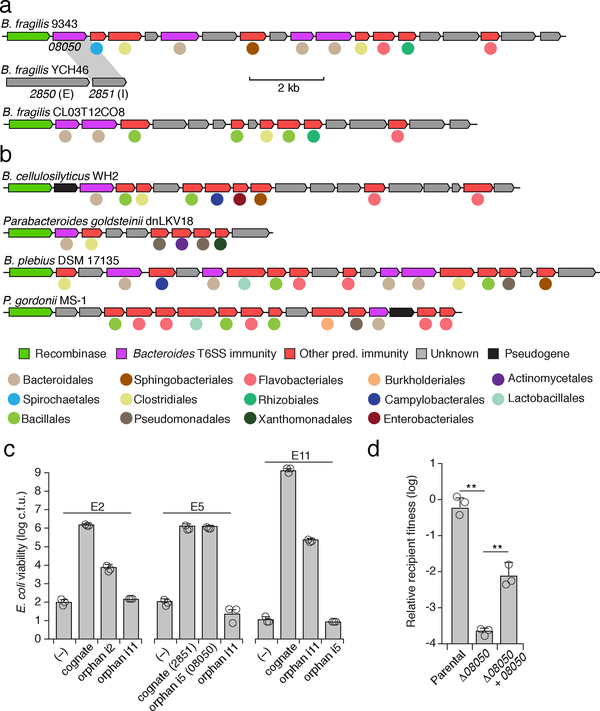
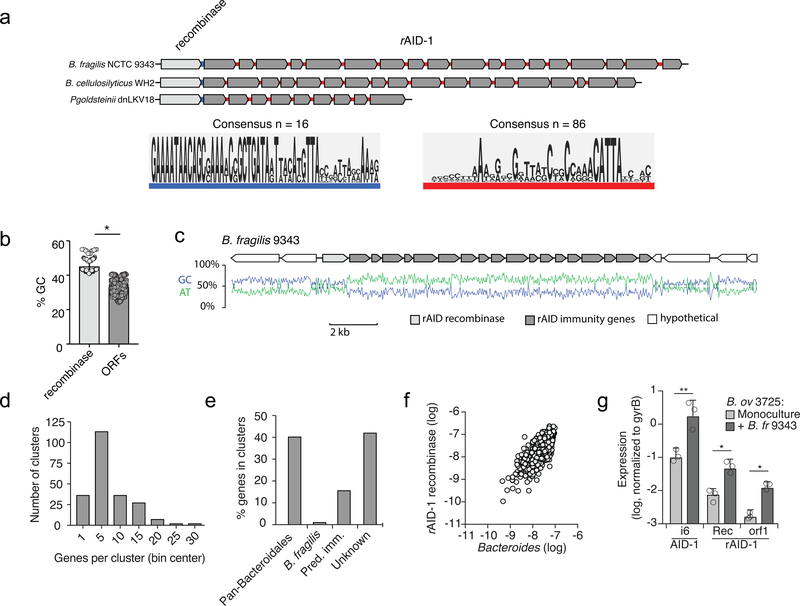
Given the benefit of orphan immunity genes against B. fragilis effectors, we hypothesized that this mechanism of inhibiting interbacterial antagonism should extend to effectors produced by other species. We previously reported evidence that B. fragilis is antagonized by other Bacteroides spp. in the human gut microbiome8. In addition to the T6SS present exclusively in B. fragilis, this species and other Bacteroides spp. can possess other T6SSs, referred to as GA1 (genetic architecture 1) and GA2, with a distinct and non-overlapping repertoire of effector and immunity genes3. The effectors of these systems exhibit hallmarks of antibacterial toxins and we demonstrated their capacity to mediate interbacterial antagonism (Fig 4, Extended Data Fig. 4)2,8. Therefore, we searched B. fragilis genomes for sequences homologous to the immunity genes corresponding to these systems. In 29 of the 122 available B. fragilis genomes, we identified apparent orphan homologs of these immunity genes grouped within gene clusters (Fig. 4a). While analogous to the AID-1 and −2 systems, these clusters have several unique characteristics including skewed GC content, conservation of a gene encoding a predicted XerD-family tyrosine recombinase, and repetitive intergenic sequences (Extended Data Fig. 5a–c)21. These often-large gene clusters, hereafter referred to as recombinase-associated AID (rAID-1) systems, can exceed 16 kb and contain up to 31 genes with varying degrees of homology to T6S immunity genes and predicted immunity genes associated with other interbacterial antagonism pathways (Fig. 4a, Extended Data Fig. 5d, e, Supplementary Table 6)2. Using the shared characteristics of B. fragilis rAID-1 systems, we searched for related gene clusters across sequenced Bacteroidales genomes. We found that over half of sequenced bacteria belonging to this order possess a rAID-1 system (226 of 423) (Fig. 4b, Supplementary Tables 6,7). In sum, these gene clusters contain 579 unique genes, encompassing homologs of 25 Bacteroidales T6S immunity genes.
Fig. 4 |. rAID systems encode toxin-neutralizing immunity genes and are prevalent in human gut microbiomes.
a, b, rAID-1 clusters from the indicated B. fragilis a, or Bacteroidales b, species. rAID-1 genes were assigned to functional immunity classes (indicated by gene coloring) via profile HMM scans and BLAST against a curated database of Bacteroidales T6SS immunity genes2,8. Colored circles indicate taxonomic association of the top non-rAID-1 homolog. Homology (70% a.a. identity) between gene 08050 of the B. fragilis 9343 rAID-1 cluster and a T6S cognate immunity gene from B. fragilis YCH46 (2851) is indicated. c, Viable E. coli cells recovered from cultures expressing indicated proteins (refer to Supplementary Table 10 for locus tags). Data indicate n=3 technical replicates representative of 3 independent biological replicates, bars indicate mean ± s.d. d, Outcomes of growth competition assays between the indicated Bacteroides strains (n=3 biologically independent samples). The relevant rAID-1 gene of B. fragilis 9343 and its corresponding effector within B. fragilis YCH46 are depicted in a. Statistics were performed using unpaired two-tailed t-tests, **P>0.01, bars indicate mean ± s.d.
The prevalence of rAID-1 genes in Bacteroidales genomes suggested that these elements may be common in the human gut microbiome. To investigate this, we searched metagenomic data for sequences mapping to Bacteroidales T6S orphan immunity genes found within rAID-1 systems. Remarkably, we found one or more rAID-1 immunity genes in 551 of 553 samples using a 97% sequence identity threshold to map reads (Supplementary Table 8). These rAID-1 immunity genes diverge significantly from corresponding cognate immunity genes (corresponding to 32–91% amino acid identity), suggesting that the latter are an unlikely source of significant false positives in this analysis. We also searched the same samples for rAID-1-associated recombinase sequences. Although recombinase genes are widely distributed across bacteria, close homologs (>50% amino acid identity) of those found associated with rAID-1 systems are restricted to this context and only found in Bacteroidales genomes. Consistent with this, we found the abundance of rAID-1 recombinase genes correlates strongly with the genus Bacteroides (Extended Data Fig. 5f).
Orphan immunity genes encoded within rAID-1 clusters diverge more significantly from cognate immunity than do those within AID systems. Thus, we sought to experimentally validate the capacity of rAID-1 immunity genes to protect bacteria from intoxication. Since most bacteria harboring rAID-1 systems have limited genetic tools, we employed E. coli to identify three Bacteroidales T6S effector genes that intoxicate cells in a manner that is neutralized by cognate immunity. In each case, we found that co-expression of these effector genes with corresponding rAID-1-associated orphan immunity genes, but not mismatched orphan immunity genes, restored E. coli growth (Fig. 4c). Both genes from one effector–orphan immunity pair we validated derive from genetically tractable strains: B. fragilis YCH46 (effector, 2850) and B. fragilis 9343 (orphan immunity, 08050). In vitro growth competition experiments with these strains, and mutant and genetically complemented derivatives, showed that an endogenous rAID-1 orphan immunity gene of B. fragilis 9343 can neutralize a T6S-delivered toxin (Fig. 4d).
The orphan immunity systems we defined consist of many genes and their expression could incur a substantial metabolic burden. As a first step toward understanding the regulation of AID systems, we performed quantitative RT-PCR to compare the expression of the systems in the presence and absence of a competitor strain. These studies provided evidence that transcription of both systems is induced by co-cultivation with a competitor strain (Extended Data Fig. 5g). We also examined metatranscriptomic data for evidence of AID expression22. Owing to a paucity of such data available for samples definitively containing AID-1 and AID-2, we could not systematically quantify expression of these systems. However, using conservative criteria for defining rAID-1-associated genes in metatranscriptomic data, we found evidence of the expression of this system in every sample derived from a large study (n=156) (Supplementary Table 9). In some samples, such as those with high levels of Bacteroides, rAID-1 genes accounted for nearly 1/10,000 of all metatranscriptomic reads. Taken together with our functional characterization of AID systems, these findings suggest that acquisition and maintenance of consolidated orphan immunity determinants is a common mechanism by which Bacteroidales defend against interbacterial antagonism in the human gut microbiome.
Mounting evidence suggests that competitive interactions between bacteria predominate in many environments23. This evolutionary pressure has undoubtedly led to the wide dissemination of idiosyncratically orphaned immunity genes predicted to provide resistance to diverse antagonistic pathways2,11,24,25. Modeling studies predict that interbacterial antagonism is a critical contributor to the maintenance of a stable gut community26. Our findings reveal that a corollary of the pervasiveness of antagonistic mechanisms is strong selective pressure for genes that can provide protection against attack, establishing a molecular arms race that has led to diversification and expansion of T6S effectors. Deciphering the linkage between orphan immunity genes and the bacteria harboring the cognate effectors has the promise of providing a window into the physical connectivity of bacteria in the gut microbiome.
It is now appreciated that phage defense mechanisms, including the adaptive system, CRISPR, are critical for bacteria to cope with the omnipresent threat and deleterious outcome of phage infection7. However, the ubiquity of interbacterial antagonistic systems suggests that in most habitats, bacteria are equally, or perhaps more likely to be subject to attack and potential cell death via the action of other bacteria2. Our characterization of AID systems encoded by prevalent members of the human gut microbiota appears to reconcile these observations and demonstrate that the neutralization of contact-dependent interbacterial antagonism can be a critical mechanism for survival in polymicrobial environments. Additionally, it suggests that analogous to the immune system of vertebrates, that of bacteria includes arms specialized in viral or bacterial defense.
Material and Methods
Microbiome Data
Metagenomic data from healthy adults were obtained from a number of large-scale sequencing projects. We specifically obtained 147 samples from the Human Microbiome Project 1.0, 100 samples from HMP 1.2, and 99 and 207 samples from two different MetaHIT datasets14,15,29,30. We further obtained paired metagenomic-metatranscriptomic data from a study of 156 individuals22. Finally, we obtained a database of genes identified from 1267 assembled metagenomes as part of the Integrated Gene Catalogue (IGC)27.
Analysis of gene and species abundances in microbiome samples
We previously compiled a list of T6SS immunity and effector genes8. We additionally compiled a list of species-specific marker genes for all Bacteroides species obtained from MetaPhlAn 2.031. In order to determine the abundance of a given immunity, effector, or marker gene in each metagenomic sample, single end metagenomic reads were aligned to gene sequences using bowtie2, allowing for one mismatch in the seed32. We counted the number of reads that aligned to each such gene with at least 80% nucleotide identity (to encompass divergent orphan immunity gene sequences) and minimum mapping quality of 20. The abundance of a gene was calculated as the number of reads aligned to this gene, normalized by the gene length and by the library size. For each species, the average gene level abundance of all species-specific marker genes was used to assess the species abundance. For the total Bacteroides abundance, we used the sum of all species-specific marker genes in the genus. Samples were only included in an analysis if they had at least 10 reads mapping to the type 6 genes in question (effectors, immunity or recombinases). Based on the abundance of GA3 immunity genes and B. fragilis we split samples into those where B. fragilis was not detectable, those where the immunity gene had >10X the B. fragilis marker gene abundance, and those where such discrepancy between the abundance of immunity genes and that of B. fragilis was not observed. Metatranscriptomics data was processed similarly to metagenomics data, except that abundance values were converted to an RPKM for familiarly with canonical RNA-Seq analysis.
Orphan immunity phylogenetic analysis
Filtered reads derived from human shotgun microbiome datasets were aligned using bowtie2 as described above and subsequently converted to a pileup using samtools with parameters --excl-flags UNMAP,QCFAIL,DUP -A -q0 -C0 -B32,33. A sequence corresponding to the most abundant version of the immunity gene in the sample was reconstructed from that pileup as follows. First, 50 bases from the start and end were trimmed due to a propensity for low coverage. Second, at all sites with at least 10X coverage the base was set to the major allele. Sites with <10X coverage were assigned an ambiguous base. Finally, we only kept the reconstructed sequence in metagenomic samples where at least 90% of the sequence had >10X coverage. The number of SNPs between all immunity sequences, both from metagenomic samples and from Bacteroides genomes, was calculated and used to populate a distance matrix. Since obtained distances were small (e.g., a single base difference), we used hierarchical clustering (with complete linkage), rather than standard phylogenetic reconstruction methods, to visualize the relatedness between different sequences. Sequence clades defined by hierarchical clustering are denoted (cI-III), as discussed in the text.
Assigning orphan immunity sequences to bacterial species
We aimed to identify the species most likely to encode the immunity gene in each cluster of identical sequences reconstructed from metagenomes. Only clusters with at least 3 sequences were used, to ensure statistical confidence. The abundance of each species was assessed based on species-specific marker genes as described above. We specifically employed a simple linear model that assumed that only a single species encodes the immunity gene. We further assumed a one to one relationship between species marker gene abundance and orphan immunity gene abundance, and accordingly fixed the intercept at zero and allowed a single species with a slope of one. The fit of the model for each species was calculated as the mean square error over all samples. The most likely species to encode the immunity gene was determined by the minimum mean squared error.
Assembly of orphan immunity sequences from metagenomes
Paired-end metagenomic sequencing data was assembled using SoapDeNovo2 with a kmer length of 63 and an average insert size of 20034. BLAST was used to identify the contig that contained the orphan immunity gene, and GeneMarkS was used to predict protein coding genes35.
Bacterial culture conditions
Anaerobic culturing procedures were performed either in an anaerobic chamber (Coy Laboratory Products) filled with 70% N2, 20% CO2 and 10% H2, or in Becton Dickson BBL EZ GasPak chambers. E. coli EC100D λ pir and S17–1 λ pir strains were grown aerobically at 37° C on lysogeny broth agar. Unless otherwise noted, Bacteroides strains were cultured under anaerobic conditions on brain heart infusion agar (BHI; Becton Dickinson) supplemented with 50 μg mL−1 hemin and 10% sodium bicarbonate36. Antibiotics and chemicals were added to media as needed at the following concentrations: trimethoprim 50 μg mL−1, carbenicillin 150 μg mL−1, gentamicin 15 μg mL−1 (E. coli), gentamicin 60 μg mL−1 (Bacteroides), erythromycin 12.5 μg mL−1, tetracycline 6 μg mL−1, chloramphenicol 12 μg mL−1, floxuridine (FUdR) 200 μg mL−1.
Genetic techniques
Standard molecular procedures were employed for creation, maintenance and E. coli transformation of plasmids. All primers used in this study were synthesized by Integrated DNA Technologies (IDT). Phusion polymerase, restriction enzymes, T4 DNA ligase, and Gibson Assembly Reagent were obtained from New England Biolabs (NEB). A comprehensive list of primers, plasmids, and strains are provided (Supplementary Table 10). Deletion of the gene encoding thymidine kinase in B. fragilis, B. ovatus, and B. xylanisolvens strains was performed by cloning respective genomic flanking regions into the vector pKNOCK as previously described37. Briefly, pKNOCK-tdk plasmids were mobilized into Bacteroides strains via overnight aerobic mating with E. coli. Integrants were isolated by plating on selective media, were passaged once without antibiotics to allow for plasmid recombination, and plated for counter selection on FUdR. Recovered single colonies were patched onto selective media to ensure loss of pKNOCK, and disruption of tdk confirmed by PCR. Subsequent deletion of orphan immunity genes was performed in Δtdk strains via a similar counter selection strategy, except employing the suicide plasmid pExchange in place of pKNOCK9. Genomic deletions were confirmed by PCR. Gene complementation was performed by cloning genes into pNBU2-erm_us1311 for constitutive expression38.
Isolation of Bacteroides strains from fecal samples
Fecal samples from healthy infants used for strain isolation were collected as part of a prior study approved by the Seattle Children’s Hospital Institutional Review Board16,39. Frozen stool samples stored at −80° C were manually homogenized, serially diluted in tryptone yeast glucose (TYG) broth, and plated under anaerobic conditions on Bacteroides bile esculin (BBE) agar plates (Oxyrase, Inc). Single colonies which exhibited esculin hydrolysis as indicated by the production of black pigment on BBE agar were sub-cultured in TYG broth with the addition of 60ug/mL gentamicin until stationary phase and then were frozen at −80° C following the addition of sterile glycerol to 20% final concentration. Single colonies isolated from these stocks were subsequently screened by PCR with primers targeting the orphan i6 gene as assembled from metagenomic short read sequence data16.
Genome sequencing
Genomic DNA used for Illumina sequencing was prepared by harvesting Bacteroides strains grown overnight on BHIS blood agar plates. Cells resuspended from plates were washed in phosphate buffered saline before DNA extraction with the Qiagen DNeasy Blood and Tissue Kit. Sequencing was performed on an Illumina MiSeq using the V3 Reagent kit at the Northwest Genomics Center sequencing facility at the University of Washington. AID clusters often appear in highly repetitive genomic contexts (e.g. mobile elements) and are often split into multiple scaffolds in reference genomes. To compensate for this, we additionally performed long read sequencing via PacBio on a subset of genomes. To this end, high-molecular weight DNA was extracted using the Qiagen Genomic-tip Kit and sequenced by SNPsaurus (Eugene, OR) using a PacBio Sequel. Hybrid long read-short read assemblies were conducted using Unicycler40. Species identification was performed by blast searches with species-specific marker genes31. Whole genome sequencing data generated in the course of this study has been deposited at the Sequence Read Archive under BioProject Accession PRJNA484981.
Interbacterial competition assays
Bacteroidales strains were grown on BHIS blood agar plates overnight at 37° C. Bacteria were resuspended from plates in BHIS broth and the optical density of each strain was adjusted to a 10:1 B. fragilis NCTC 9343 to competitor ratio (OD600 6.0 to 0.6) for competitions involving B. xylanisolvens and B. ovatus, or 1:1 ratio for competitions involving B. fragilis 638R (OD600 6.0). Equal volumes of each strain at the adjusted OD were mixed and 5ul of bacterial mixtures were spotted onto pre-dried BHIS blood agar plates, in triplicate spots. Competitions were allowed to proceed for 20–24 hours at 37° C under anaerobic conditions before spots were harvested into BHIS broth. Competition outcomes were quantified in one of two ways: 1) by serial dilution for enumeration of colony forming units after plating on BHIS selective plates containing either erythromycin or tetracycline, or 2) purification of total genomic DNA using the Qiagen DNeasy Blood and Tissue Kit and subsequent quantification by qPCR using strain-specific primers (see Supplementary Table 10). For antibiotic selection, B. fragilis 9343 was marked with erythromycin resistance by integration of pNBU2-erm at the att1 site38. Other strains were either naturally tetracycline resistant, or were marked by integration of pNBU2-tet-BCO1. Strains with insertions of pNBU2 were selected for matching integration sites by PCR with primers flanking att loci41. Interbacterial competitions between strains of B. fragilis occasionally exhibited T6SS-independent phenotypes that were dependent on the initial starting ratio of the strains used42.
Interbacterial mobile element transfer assays
Allelic exchange was used to engineer a high-expression chloramphenicol resistance cassette onto the AID-1 system of B. fragilis 638R, replacing BF638R_2056–205843. Chloramphenicol-resistant B. fragilis 638R cells were mixed on BHIS blood agar plates with erythromycin-resistant B. fragilis ATCC 43859 cells at a 1:1 ratio (OD600 6.0). Following overnight co-culture, bacterial mixtures were harvested and plated on BHIS plates containing either erythromycin alone (to quantify c.f.u. of total ATCC cells), or erythromycin and chloramphenicol (to quantify c.f.u of AID-1 recipient ATCC cells). Doubly-resistant colonies were screened individually by PCR to confirm strain identity, the presence of the AID-1 system, and the genomic integration site at a tRNA-Lys gene (see Supplementary Table 10 for primers used).
Gnotobiotic animal studies
Germ-free 6–12 week-old female Swiss Webster mice from multiple litters were randomized, housed simultaneously in pairs in single Techniplast cages with a 12-hour light/dark cycle, and fed a standard lab diet (Laboratory Autoclavable Rodent Diet 5010, LabDiet), in accordance with guidelines approved by the University of Washington Institutional Animal Care and Use Committee. Blinding was not performed, and no statistical methods were used to determine sample size. Reasonable numbers of animals were used considering limitations of housing and maintenance under gnotobiotic conditions.. Bacteroides fragilis strains were introduced into mice via oral gavage of 108 colony forming units (c.f.u.) suspended in 0.2mL of sterile 1X phosphate buffered saline with 20% glycerol. Challenge with B. fragilis 638R or B. fragilis ATCC strains occurred 7 days following pre-colonization with B. fragilis 9343 strains. Colonization levels by each strain in each mouse were tracked by collection of fecal pellets over a period of 4 weeks, plating on selective BHIS agar plates (B. fragilis 9343 on BHIS plus erythromycin; B. fragilis 638R and ATCC on BHIS plus tetracycline), and subsequent absolute quantification of c.f.u. by normalization of each sample to the initial pellet weight. Differences in the strain ratio of c.f.u between groups at each timepoint was assessed using Mann Whitney U tests. Non-parametric tests were used following Shapiro-Wilk analysis for normality of data at each time point. Mice were confirmed to be sterile prior to colonization by qPCR with primers targeting the 16S rRNA gene and free of non-Bacteroides contamination by plating fecal pellets on non-selective LB and BHIS plates incubated under either anaerobic and aerobic conditions44.
Bioinformatic analysis of rAID-1 clusters
The amino acid sequence of the B. fragilis NCTC 9343 polyimmunity-associated XerD-like tyrosine recombinase (BF9343_RS08045) was used as a query against a custom database of 423 Bacteroidales genomes downloaded from GenBank. rAID clusters in Bacteroidales genomes were identified based upon the following criteria: i) presence of a 5’ XerD-like tyrosine recombinase gene encoding a protein with amino acid identity exceeding 44% (corresponding to an e-value of 10−100), ii) 2 or more co-directionally oriented downstream genes which possessed iii) a GC content of 41% or lower. The end of the gene cluster was defined as the stop codon of the last co-directionally oriented gene in the cluster with similar GC content. To identify homologs of genes within rAID clusters, open reading frames within the clusters were translated and used as tblastn queries against the NCBI non-redundant nucleotide database. Top hits from these searches were often genes in other rAID clusters; therefore, these hits were discarded. The top non-rAID hit from tblastn searches with an e-value threshold of 10−30 was selected as the closest homolog. rAID cluster genes were assigned to interbacterial immunity gene families via hmm scans with profiles from Zhang et al.2 with an e-value cutoff of 10−3. rAID cluster genes were additionally compared via tblastn with forty-six Bacteroidales T6SS immunity genes from subtypes 1–33,8 with an e-value cutoff of 10−10. Percent amino acid identity with homologs was assessed if sequences could be aligned across ≥80% of their length. Motif enrichment analysis was performed on non-coding sequences within a subset of rAID-1 clusters (14 sequences immediately 3’ of the recombinase stop codon, and 86 intergenic sequences between rAID-1 ORFs), using MEME Suite 5.0.2 and default settings28.
Heterologous expression of Bacteroides toxin and immunity genes
To assess the ability of cognate immunity or orphan immunity to neutralize the toxicity of a Bacteroidales T6SS effector, genes were cloned into E. coli expression vectors pScrhab2-V (effectors) and pPSV39-CV (immunity). Immunity genes were fused with the P. aeruginosa ribosome binding site from hcp1 during the cloning process45. All cloning steps for effector genes involved growth of E. coli on media containing 0.1% glucose to ensure repression of expression. E. coli DH5a cells were co-transformed with pSchraB2 and pPSV39 plasmids bearing genes of interest. Overnight cultures were then grown from single co-transformed colonies to stationary phase in LB broth containing 50ug/mL trimethoprim, 15ug/mL gentamycin, and glucose. Cells were harvested from these cultures and washed to remove glucose before back-dilution to an OD600 of 0.05 into LB broth containing 50ug/mL trimethoprim, 15ug/mL gentamycin, 0.05% rhamnose, and 1mM isopropyl β-D-1-thiogalatopyranoside45,46. Cultures were then grown for 8 hours shaking at 37°C before plating to allow quantification of c.f.u. Experiments were performed with technical triplicates for each of at least two biological replicates.
Gene expression analysis of AID-1 and rAID-1 systems of B. ovatus 3725
To assess the level of expression of genes in the AID-1 and rAID-1 systems of B. ovatus 3725, bacterial cells were first grown overnight on BHIS blood agar plates containing gentamycin. Cells were then resuspended in BHIS to an OD600 of 3.0 for B. ovatus monocultures, or to an OD600 of 0.3 for mixed co-culture experiments with B. fragilis 9343 at 10-fold excess (OD600 of 3.0). 5uL volumes of bacterial mixtures were then spotted on BHIS blood agar plates. Plates were incubated at 37°C for 2 hours under anaerobic culture conditions before cells were harvested directly in Buffer RLT plus (twenty 5uL spots per condition per replicate, Qiagen RNeasy Micro Kit). Two separate rounds of DNase treatment were performed (Qiagen RNase-free DNase, Thermo Fisher Scientific Turbo DNase-free kit). RNA samples were confirmed to be free of genomic DNA by PCR with primers targeting the Bacteroides 16S rRNA gene. cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosciences). Following synthesis, cDNA was diluted 1:10. Quantitative PCR (primers listed in Supplementary Table 10) was performed using SSO Universal SYBR Green Supermix (Bio-Rad) on a CFX96 machine (Bio-Rad). Genomic DNA was used to generated standard curves47. Differences in gene expression between samples was performed by normalization to the expression level of B. ovatus 3725 gyrB. Primers targeting gyrB were designed to target regions of the genes that are highly polymorphic between B. fragilis and B. ovatus, and species-specificity for B. ovatus was confirmed by PCR using B. fragilis genomic DNA48.
Extended Data
Extended Data Fig. 1 |. Prevalence of B. fragilis-specific orphan immunity genes in adult and infant microbiomes.
a, Number of adult human gut microbiome samples in which the indicated immunity genes (1–14, GA3_i1–14 from ref8) can be detected at an 80% nucleotide identity threshold and an abundance >10-fold that of B. fragilis marker genes. Bars colored corresponding to Fig. 1a and asterisks indicate immunity genes without orphan representation. b, Abundance comparison of B. fragilis-specific T6SS immunity genes with B. fragilis species-specific marker genes in infant microbiome samples (Supplementary Table 4)16. Abundances are calculated as in Fig. 1a. Samples in which immunity gene abundance exceeds that of Bacteroides by over 10-fold (blue) are highlighted.
Extended Data Fig. 2 |. Diversity and genomic context of orphan immunity genes in human gut microbiomes and diverse Bacteroides species.
a, Representative AID-1 gene clusters containing homologs of the indicated B. fragilis T6S immunity genes from the indicated reference genomes. b, Data points indicate the amino acid identity of unique genes homologous to indicated B. fragilis-specific T6SS cognate immunity genes identified through BLAST analysis of the Integrated Gene Catalog (IGC) (n=88 genes, maximum E-value, 10−40; minimum percent identity, 60%)27.
Extended Data Fig. 3 |. Orphan immunity genes specifically enhance the fitness of Bacteroides strains in vitro and in vivo.
a, b, T6SS-dependent competitiveness of parental strains of B. ovatus 3725 and the indicated mutant and complemented derivatives during in vitro growth competition experiments with B. fragilis 9343. Relative recipient fitness was determined by calculating the ratio of final to initial colony forming units and normalizing to the corresponding experiment with B. fragilis 9343 lacking tssC (T6S-inactive). Data represent mean ± s.d. of n=3 independent biological replicates, unpaired two-tailed t-test, *P < 0.01. c, T6SS-dependent competitiveness of a parental strain of B. ovatus 3725 or a strain bearing in-frame deletions of indicated orphan immunity genes, during in vitro growth competition experiments with an orthogonal effector-bearing B. fragilis 638R parental strain or a derivative strain lacking tssC (T6-inactive). Relative recipient fitness and statistics were calculated as in a, b, n=3 independent biological replicates. d, e, Recovery of B. fragilis 9343 d, or 638R and the indicated orphan immunity mutant derivative e, from pairwise competitions of the strains in germ-free mice. Lines indicate the mean at each time point, (n=8 mice per group for each of two independent experiments). Alternating time points of these data are included in ratio form in Fig. 3c. f, Schematic depicting genomic loci for the B. fragilis ATCC 43859 parental strain, the B. fragilis 638R AID-1 donor strain, the AID-1 system, and the ATCC 43859 AID-1 recipient. Grey shading indicates homology, red arrows indicate the position of PCR primers used to infer insertion of the AID-1 element at the tRNA-Lys insertion site. g, Abundance of B. ovatus in samples lacking detected orphan immunity genes (−) and samples in which the indicated orphan immunity genes were assigned to B. ovatus (+). Abundances are calculated as in Fig. 1a. (Wilcoxon rank sum test, *P<0.001, n=128 non-orphan samples and n=24 for samples harboring orphan immunity, box plots indicate interquartile range with line at the median and whiskers indicating 1.5 times the IQR).
Extended Data Fig. 4 |. The GA2 system of Bacteroidales mediates interbacterial antagonism.
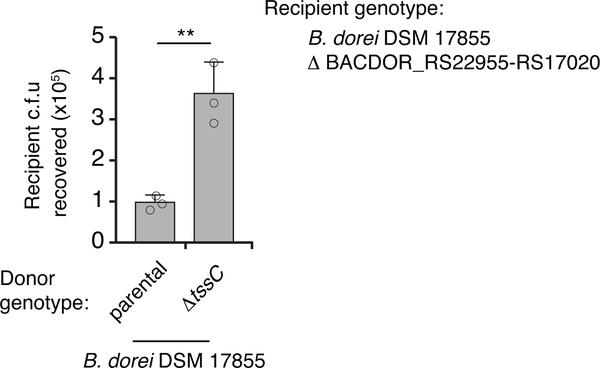
Recovery of B. dorei DSM 17855 cells lacking GA2_e14-i14 (BACDOR_RS22955–17020) from two-strain in vitro growth competition experiments with the indicated donor strains, n=3 technical replicates representative of three biological replicates, unpaired two-tailed t-test, **P < 0.01).
Extended Data Fig. 5 |. rAID-1 systems include conserved and repetitive intergenic sequences and bear hallmarks of horizontal gene transfer.
a, Left - Motif enrichment analysis from the intergenic sequences immediately 3’ of the recombinase stop codon to the start codon of the first downstream open reading frame within 16 randomly selected rAID-1 gene clusters. This region is highlighted in blue in three representative rAID-1 systems shown above. Right - Motif enrichment analysis from all 86 intergenic sequences between the ORFs of six rAID-1 clusters (B. fragilis NCTC 9343, B. cellulosilyticus WH2, B. ovatus 3725, Paraprevotella clara YIT 11840, Parabacteroides goldsteinii dnLKV18, and Parabacteroides gordonii MS-1)28. This region is highlighted in red in three representative rAID-1 systems shown above. b, Average G+C nucleotide content of rAID-1-associated recombinase versus rAID-1 predicted ORFs, n=226, unpaired two-tailed t-test, ***P>0.0001. c, Schematic depicting the G+C and A+T nucleotide content across a representative rAID-1 system from B. fragilis 9343. d, Frequency distribution of gene number in rAID-1 clusters (n=1247 genes in 226 clusters). Bin width is 5 genes. e, Composition of genes in rAID-1 clusters (n=226 clusters) as determined by profile HMM scans and BLAST analysis against a curated database of Bacteroidales T6SS immunity genes2,8. f, Comparison of the total abundances of rAID-1-associated predicted recombinases and the Bacteroides genus in adult microbiome samples derived from the HMP and MetaHIT studies (Supplementary Table 8). Abundance values are calculated as in Fig 1; genus abundance corresponds to the sum of all Bacteroides spp. (calculated individually as the average of species-specific marker gene abundances). g, Results of qRT-PCR analyses for the indicated B. ovatus 3725 genes belonging to AID-1 (i6, M088_1971) or rAID-1 clusters (Rec, recombinase, M088_1401; orf1, M088_1400) under conditions of growth in mono- or co-culture with B. fragilis 9343 for 2 hrs. Data represent mean ± s.d. of n=3 independent biological replicates, Wilcoxon two-tailed sign-rank test, **P < 0.01, *P < 0.05.
Supplementary Material
Acknowledgements
We thank the UW GNAC for assistance with gnotobiotic experiments. We thank Cynthia Sears, Andy Goodman, Tomomi Kuwahara, and Eric Martens for generously providing Bacteroides strains. Funding: This work was supported by National Institutes of Health grants AI080609 (to JDM), P30DK089507 (to LRH as pilot study PI), R01DK095869 (to LRH), K99GM129874 (to BDR), R01GM124312 (to EB), and New Innovator Award DP2AT00780201 (to EB), and the Burroughs Wellcome Fund (to JDM). AJV was supported by a postdoctoral fellowship from the Natural Sciences and Engineering Research Council of Canada. BDR was supported by a Simons Foundation-sponsored Life Sciences Research Foundation postdoctoral fellowship. EB is a Faculty Fellow of the Edmond J. Safra Center for Bioinformatics at Tel Aviv University. JDM is an HHMI Investigator.
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and materials availability: Bacteroides strains acquired from Johns Hopkins University were obtained under an MTA. All data required to assess the conclusion of this research is available in the main text and supplemental materials, or has been deposited at the Sequence Read Archive under BioProject Accession PRJNA484981.
Code availability: Python and R scripts used in this work are available for download (http://borensteinlab.com/download.html).
References
- 1.Whitney JC et al. A broadly distributed toxin family mediates contact-dependent antagonism between gram-positive bacteria. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, de Souza RF, Anantharaman V, Iyer LM & Aravind L Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coyne MJ, Roelofs KG & Comstock LE Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell AB et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hood RD et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornforth DM & Foster KR Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol 11, 285–93 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Hille F et al. The Biology of CRISPR-Cas: Backward and Forward. Cell 172, 1239–1259 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Verster AJ et al. The Landscape of Type VI Secretion across Human Gut Microbiomes Reveals Its Role in Community Composition. Cell Host Microbe 22, 411–419 e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wexler AG et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113, 3639–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht AL et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17, 1281–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchberger PC, Unterweger D, Provenzano D, Pukatzki S & Boucher Y Sequential displacement of Type VI Secretion System effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci Rep 7, 45133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steele MI, Kwong WK, Whiteley M & Moran NA Diversification of Type VI Secretion System Toxins Reveals Ancient Antagonism among Bee Gut Microbes. MBio 8(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ting SY et al. Bifunctional Immunity Proteins Protect Bacteria against FtsZ-Targeting ADP-Ribosylating Toxins. Cell (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manor O et al. Metagenomic evidence for taxonomic dysbiosis and functional imbalance in the gastrointestinal tracts of children with cystic fibrosis. Sci Rep 6, 22493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siguier P, Gourbeyre E & Chandler M Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev 38, 865–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao S et al. Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host Microbe 25, 656–667 e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wozniak RA & Waldor MK Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8, 552–63 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Stevens AM, Shoemaker NB & Salyers AA The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unliked chromosomal DNA might also be involved in transfer of the element. Journal of Bacteriology 172, 4271–4279 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castillo F, Benmohamed A & Szatmari G Xer Site Specific Recombination: Double and Single Recombinase Systems. Front Microbiol 8, 453 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abu-Ali GS et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nature Microbiology 3, 356–366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foster KR & Bell T Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol 22, 1845–50 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Poole SJ et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7, e1002217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drider D, Fimland G, Hechard Y, McMullen LM & Prevost H The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70, 564–82 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coyte KZ, Schluter J & Foster KR The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–6 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Li J et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32, 834–41 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Bailey TL et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature 486, 207–14 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin J et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12, 902–3 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo R et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Besemer J, Lomsadze A & Borodovsky M GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29, 2607–18 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bacic MK & Smith CJ Laboratory maintenance and cultivation of bacteroides species. Curr Protoc Microbiol Chapter 13, Unit 13C 1 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koropatkin NM, Martens EC, Gordon JI & Smith TJ Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–15 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Degnan PH, Barry NA, Mok KC, Taga ME & Goodman AL Human gut microbes use multiple transporters to distinguish vitamin B(1)(2) analogs and compete in the gut. Cell Host Microbe 15, 47–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman LR et al. Escherichia coli dysbiosis correlates with gastrointestinal dysfunction in children with cystic fibrosis. Clin Infect Dis 58, 396–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wick RR, Judd LM, Gorrie CL & Holt KE Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS computational biology 13, e1005595–e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martens EC, Chiang HC & Gordon JI Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–57 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Bayona L & Comstock LE Bacterial antagonism in host-associated microbial communities. Science 361(2018). [DOI] [PubMed] [Google Scholar]
- 43.Lim B, Zimmermann M, Barry NA & Goodman AL Engineered Regulatory Systems Modulate Gene Expression of Human Commensals in the Gut. Cell 169, 547–558.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paik J et al. Potential for using a hermetically-sealed, positive-pressured isocage system for studies involving germ-free mice outside a flexible-film isolator. Gut Microbes 6, 255–65 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silverman JM et al. Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51, 584–93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardona ST & Valvano MA An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid 54, 219–28 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM & Kramer MF High-throughput real-time quantitative reverse transcription PCR. Curr Protoc Mol Biol Chapter 15, Unit 15 8 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Caro-Quintero A & Ochman H Assessing the Unseen Bacterial Diversity in Microbial Communities. Genome Biol Evol 7, 3416–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.