Abstract
Background
Endocrine therapy reduces breast cancer mortality by 40%, but resistance remains a major clinical problem. In this study, we sought to investigate the impact of aromatase inhibitor (AI) therapy on gene expression and identify gene modules representing key biological pathways that relate to early AI therapy resistance.
Methods
Global gene expression was measured on pairs of core-cut biopsies taken at baseline and at surgery from 254 patients with ER-positive primary breast cancer randomised to receive 2-week presurgical AI (n = 198) or no presurgical treatment (control n = 56) from the POETIC trial. Data from the AI group was adjusted to eliminate artefactual process-related changes identified in the control group. The response was assessed by changes in the proliferation marker, Ki67.
Results
High baseline ESR1 expression associated with better AI response in HER2+ tumours but not HER2− tumours. In HER2− tumours, baseline expression of 48 genes associated with poor antiproliferative response (p < 0.005) including PERP and YWHAQ, the two most significant, and the transcription co-regulators (SAP130, HDAC4, and NCOA7) which were among the top 16 most significant. Baseline gene signature scores measuring cell proliferation, growth factor signalling (ERBB2-GS, RET/GDNF-GS, and IGF-1-GS), and immune activity (STAT1-GS) were significantly higher in poor AI responders. Two weeks of AI caused downregulation of genes involved in cell proliferation and ER signalling, as expected. Signature scores of E2F activation and TP53 dysfunction after 2-week AI were associated with poor AI response in both HER2− and HER2+ patients.
Conclusions
There is a high degree of heterogeneity in adaptive mechanisms after as little as 2-week AI therapy; however, all appear to converge on cell cycle regulation. Our data support the evaluation of whether an E2F signatures after short-term exposure to AI may identify those patients most likely to benefit from the early addition of CDK4/6 inhibitors.
Trial registration
ISRCTN, ISRCTN63882543, registered on 18 December 2007.
Keywords: Breast cancer, Oestrogen receptor, Aromatase inhibition, Resistance, Residual proliferation, Signatures
Background
Breast cancer (BC) is the most common malignancy in women worldwide [1]. Over 80% [2] of primary BCs express oestrogen receptor (ER) alpha. While tamoxifen is an effective agent for reducing recurrence and death from BC, its effectiveness is impeded by its partial agonist activity. Aromatase inhibitors (AIs) show greater efficacy than tamoxifen. They reduce BC mortality by c.40% and have become the preferred first-line agent in postmenopausal women [3–5]. While treatment with an AI is sufficient to control the disease in many patients, for others, additional treatment to target resistance pathways is necessary, but identifying the mechanisms of resistance is mandatory to optimise this strategy.
Identifying relevant mechanisms of resistance in individual patients presenting with ER+ primary disease and treated post-surgically with adjuvant AI is prohibitively difficult, because patients are clinically disease-free after surgery and the absence of recurrence may relate to the absence of subclinical micrometastases or to disease control by the AI. In contrast, in the presurgical setting, gene expression in an individual tumour may be assessed in relation to validated markers of response in the same tumour. Multiple clinical trials provide strong evidence to support change in the expression of the nuclear proliferation marker, Ki67, after only 2-week treatment with an endocrine agent to be a valid predictor of the long-term benefit from adjuvant endocrine therapy and to be a better predictor of such benefit than clinical response [6–9]. In addition, the residual level of Ki67 after short exposure to endocrine therapy provides better prognostic information than pre-treatment Ki67 [10]. Thus, change in Ki67 can be used to measure a tumour’s response to AI and to study the mechanisms underpinning this, while residual level of Ki67 after short-term AI may be used to identify patients whose tumours retain significant proliferative drive, who are thereby at high-risk of recurrence and merit additional treatment. Identifying the molecular pathways associated with the residual Ki67 may allow such additional treatment to be targeted at relevant resistance pathway(s).
While a small number of presurgical studies have the potential to identify pathways associated with response and early resistance in ER+ patient populations, including some by our group [11, 12], most reports have lacked adequate patient numbers to allow the identification of effects restricted to subgroups of patients. In addition, and importantly, previous reports have not included controls that can identify artefacts that result from the experimental design of pre-surgical studies; we have recently reported that the changes in gene expression of the greatest magnitude in AI-treated patients in a short-term presurgical study are entirely artefactual. This makes the inclusion of a control set of tumours critical for eliminating these artefacts [13]. In the following analyses, we have utilised a study design that avoids these limitations by accessing samples from the PeriOperative Endocrine Therapy-Individualising Care (POETIC, CRUK/07/015) trial [14]. The inclusion of the no-treatment group in POETIC allowed us to adjust our observation in order to eliminate the impact of pre-analytical artefacts.
The POETIC trial, randomised postmenopausal women with primary ER+ BC 2:1 receive perioperative AI (2 weeks pre- + 2 weeks post-surgery, termed AI-treated) or no perioperative treatment (termed control). We report analyses from the cohort of 254 (AI-treated = 198; control = 56) patients from whom samples in RNA-later were available and provided high-quality genome-wide expression data. This is the largest presurgical study of the mechanisms of response and resistance to AIs to date and has sufficient numbers for separate analyses of HER2− and HER2+ subsets (i) to determine the associations between the baseline expression of individual genes or biological pathways with the change in Ki67 and the residual on-treatment Ki67 and (ii) to investigate the early impact of AI on gene expression and gene signatures.
Methods
Detailed methods are described within the STAR file (Additional file 1)
Patients and samples
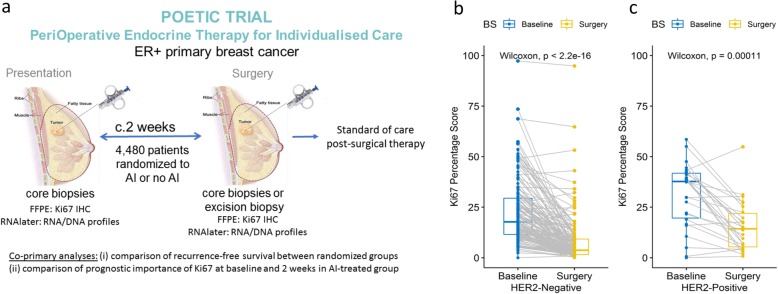
The patients studied were a subpopulation of the POETIC (PeriOperative Endocrine-Therapy for Individualised Care) [14] study. The study design is illustrated in Fig. 1a.
Fig. 1.
a POETIC schema, study design POETIC Trial PeriOperative Endocrine Therapy for Individualised Care. b Individual Ki67 changes in HER2− (n = 159) AI-treated groups. c Individual Ki67 changes in HER2+ (n = 26) AI-treated groups. The boxes indicate the median and interquartile ranges
RNA extraction
Total RNA was extracted using miRNeasy (Qiagen, Sussex, UK). RNA quality was checked using an Agilent Bioanalyser (Santa Clara, CA, USA), as previously described [15].
Ethics statement
Ethical approval for POETIC (Trial Number CRUK/07/015) was provided by NRES Committee London–South-East. All patients consented to molecular analysis of their samples for research purposes.
Gene expression analysis and data pre-processing
RNA amplification, labelling, and hybridization on HumanHT-12_V4 expression BeadChips (Illumina, San Diego, CA, USA) were performed, according to the manufacturer’s instructions. The raw data was extracted using GenomeStudio Software and was processed in R using lumi package (http://www.bioconductor.org) (Additional file 1).
Elimination of gene expression changes in the control group
To correct for potential artefactual changes in the gene expression that resulted from study procedures (10), the 2-week changes in the expression resulting from AI treatment were estimated for each gene by comparing the expression changes (log2(Surgery/Baseline)) in the AI-treated tumours and the expression changes (log2(Surgery/Baseline)) of the un-treated tumours. The relative (corrected) gene expression level in a given sample was calculated by subtracting the mean expression for the gene in the control samples from the expression of the given gene in the AI-treated tumour. All data shown that relate to either on-treatment expression/signature score or changes in the expression/signature score were corrected in this manner.
Biomarker analyses
Ki67% staining on formalin-fixed samples was carried out using anti-MIB-1 (M7240, DAKO UK), as previously described (7). HER2 status was measured locally using immunohistochemistry (IHC) and/or in situ hybridisation [16].
Published gene signatures
We determined the association of gene signatures representative of various biological processes for their association with the antiproliferative response to AI. In some cases, these signatures have been suggested as associated with resistance to endocrine therapy, and the work here can be considered an assessment of the validity of those findings: Inflammatory-GS [11], STAT1-GS [12], IGF1-GS [12], RBloss-GS and DiLeoRBloss-GS [17, 18], E2Factivation-GS [19], E2F4-GS [20], TP53-GS [21], and GDNF-GS [22]. For other signatures, our analyses were exploratory, and positive findings would need further validation. Many of the signatures have a predominance of known proliferation-associated genes (PAGs) that obscure the likely relationship with the signalling pathways per se; therefore, we conducted analyses that included and excluded PAGs from the respective signatures, as previously described [12] (Additional file 2: Table S1).
Immune or stromal score estimation
To allow comparison of the extent of immune or stromal admixture between samples, we used ESTIMATE [23].
Statistical analysis
Unpaired t tests were used to compare the mean changes in the gene expression (log2[Surgery/Baseline]) of tumours in the treated vs the control group using BRB-Array Tools (https://brb.nci.nih.gov/BRB-ArrayTools/). The Ingenuity Pathways Analysis (IPA) was conducted on the lists of genes that associated with the change in Ki67, or residual Ki67, or were differentially expressed to identify over-represented pathways. For individual pathways, the Benjamini-Hochberg procedure was used to calculate the false discovery rate (FDR) in order to adjust for multiple testing; the association between the two groups was considered to be statistically significant when p value < 0.005; the difference between the two sets of data was considered to be statistically significant when p value < 0.001. Reported p values are two-sided.
Endpoints
Four endpoints were used in this study: (i) change in Ki67 between baseline and 2 weeks as a continuous variable and (ii) responder or non-responder, defined as a reduction of > 60% or < 60%, respectively [24]; (iii) residual Ki67 as a continuous variable, and (iv) presence or absence of complete cell cycle arrest (CCCA or noCCCA), i.e. residual Ki67 < 2.7% or > 2.7%, respectively [25]. Each of the endpoints provides different information: (i) and (ii) reflect the antiproliferative response to AI treatment which relates to the benefit from the treatment, and endpoints (iii) and (iv) relate to the residual risk after AI therapy as described in a reference endpoints’ table (Additional file 2: Table S2). Patients with a baseline Ki67 value < 5% were excluded from (i) and (ii) because low pretreatment values can lead to highly aberrant estimates of proportional change.
Results
Patient demographics and changes in Ki67
There were 198 AI-treated patients with a baseline gene expression profile and paired Ki67 values (Additional file 8). Of these, 157 also had a gene expression profile at surgery. There were 56 controls with a gene expression profile at both baseline and surgery. Reasons for samples being excluded are shown in the consort diagram (Additional file 3: Figure S1). The demographics of the AI-treated patients are shown in Additional file 2: Table S3. Of the tumours, 81% were ductal and 61% were histologic grade 2. At surgery, 66% had a tumour diameter between 2 and 5 cm. All tumours were ER+, except 1 case which was found to be ER-negative after all analyses had been completed. Data on HER2 status, the individual changes in Ki67, and categorisation into responders or non-responders is shown in Additional file 2: Table S4.
Twenty-six (13.1%) of the AI-treated tumours and 8 (14.3%) of the control tumours were HER2+. Major between-patient heterogeneity in Ki67 change was evident in both the HER2− and HER2+ AI-treated groups, but there was significantly greater geometric mean suppression of Ki67 in the HER2− compared to the HER2+ cases (77.7% and 50.0%, respectively; p = 2.72E−04) (Fig. 1b, c). One hundred thirteen of 155 (72.9%) of the HER2− cases (with baseline Ki67 > 5%) were classed as good responders, compared with 9/23 (39.1%) HER2+ cases (Fisher’s exact test p = 2.90E−03). Furthermore, a higher proportion, 40.0% (66/161), of HER2− cases reached CCCA compared with 11.5% (3/26) of the HER2+ cases (Fisher’s exact test p = 4.00E−03) (Additional file 2: Table S5 a,b,c). This observation confirms previous studies indicating that the antiproliferative response to AIs is impeded in HER2+ tumours [26, 27]. As a consequence, all further analyses were conducted separately for HER2− and HER2+ subgroups.
HER2-negative tumours
Predictors of de novo antiproliferative response to AI
Association of individual genes and gene signatures with change in Ki67
Baseline expression of 123 genes correlated with the 2-week change in Ki67 with p value < 0.005 (Additional file 4: Figure S2; Additional file 2: Table S6). Of note, because the change is a reduction in Ki67, correlations with good response are negatively signed. High expression of 75 genes was associated with better response and 48 genes with poorer response. These 2 sets of genes segregated as the 2 major arms when the 123 genes were subjected to hierarchical clustering. The 6 genes with the strongest correlations were all genes associated with better response, but even for these, the absolute r values were all < 0.40 (Table 1; Additional file 2: Table S6). No further distinct groupings were apparent in the heatmap other than a tendency for non-luminal subtypes to show poorer Ki67 suppression.
Table 1.
Genes whose baseline expression significantly correlated with the change in Ki67 (p < 0.005) based on 155 HER2− of the 178 AI-treated samples
| Gene symbol | Entrez gene name | Correlate with change in Ki67 | Correlate with residual Ki67 | ||||
|---|---|---|---|---|---|---|---|
| Rank by correlation coefficient (rho) | Rho | Parametric p value | Ranking by rho | Rho | Parametric p value | ||
| ESR1 | NA | − 0.11 | 1.70E−01 | NA | − 0.16 | 5.30E−02 | |
| Associated with good antiproliferative response | |||||||
| ACADVL | Acyl-CoA dehydrogenase very long | 1 | − 0.355 | 6.90E−06 | 1 | − 0.419 | 1.00E−07 |
| TRABD | TraB domain containing | 2 | − 0.333 | 2.51E−05 | 208 | − 0.241 | 2.58E−03 |
| CCND1 | Cyclin D1 | 3 | − 0.328 | 3.34E−05 | 294 | − 0.226 | 4.69E−03 |
| TRIP6 | Thyroid hormone receptor interactor 6 | 4 | − 0.299 | 1.71E−04 | 58 | − 0.286 | 3.18E−04 |
| CTTN | Cortactin | 5 | − 0.297 | 1.88E−04 | NA | NA | > 5.00E−3 |
| MRPL23 | Mitochondrial ribosomal protein L23 | 6 | − 0.295 | 2.04E−04 | 277 | − 0.228 | 4.44E−03 |
| CCNI | Cyclin I | 7 | − 0.289 | 2.81E−04 | 89 | − 0.271 | 6.79E−04 |
| EPHX2 | Epoxide hydrolase 2 | 7 | − 0.289 | 2.82E−04 | 8 | − 0.356 | 6.30E−06 |
| IMPDH2 | Inosine monophosphate | 9 | − 0.288 | 3.00E−04 | 114 | − 0.264 | 9.45E−04 |
| ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor 6 | 10 | − 0.283 | 3.70E−04 | NA | NA | > 5.00E−3 |
| ACY1 | Aminoacylase 1 | 10 | − 0.283 | 3.73E−04 | 86 | − 0.272 | 6.53E−04 |
| EIF3G | Eukaryotic translation initiation factor 3 subunit G | 12 | − 0.282 | 3.91E−04 | 122 | − 0.261 | 1.08E−03 |
| GLI3 | GLI family zinc finger 3 | 12 | − 0.282 | 3.97E−04 | 155 | − 0.252 | 1.59E−03 |
| TTC17 | Tetratricopeptide repeat domain 17 | 14 | − 0.279 | 4.70E−04 | 6 | − 0.366 | 3.30E−06 |
| SCUBE2 | Signal peptide, CUB domain and EGF-like domain containing 2 | 15 | − 0.276 | 5.24E−04 | 3 | − 0.393 | 5.00E−07 |
| MRPS27 | Mitochondrial ribosomal protein S27 | 16 | − 0.274 | 5.84E−04 | 245 | − 0.236 | 3.23E−03 |
| Associated with poor antiproliferative response | |||||||
| PERP | PERP, TP53 apoptosis effector | 1 | 0.291 | 2.53E−04 | 161 | 0.273 | 6.09E−04 |
| YWHAQ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta | 2 | 0.29 | 2.63E−04 | 86 | 0.328 | 3.47E−05 |
| SNIP1 | Smad nuclear interacting protein 1 | 3 | 0.275 | 5.47E−04 | 210 | 0.26 | 1.15E−03 |
| PNO1 | Partner of NOB1 homologue | 4 | 0.274 | 5.74E−04 | 214 | 0.258 | 1.21E−03 |
| SEC22B | SEC22 homologue B, vesicle trafficking protein (gene/pseudogene) | 5 | 0.268 | 7.64E−04 | NA | NA | > 5.00E−3 |
| GOLT1B | Golgi transport 1B | 5 | 0.268 | 7.92E−04 | 87 | 0.326 | 3.82E−05 |
| SAP130 | Sin3A-associated protein 130 | 7 | 0.264 | 9.56E−04 | 285 | 0.24 | 2.65E−03 |
| GPKOW | G-patch domain and KOW motifs | 8 | 0.261 | 1.08E−03 | 135 | 0.289 | 2.78E−04 |
| NUS1 | NUS1 dehydrodolichyl diphosphate synthase subunit | 9 | 0.259 | 1.19E−03 | 187 | 0.266 | 8.67E−04 |
| PLCB1 | Phospholipase C beta 1 | 10 | 0.257 | 1.28E−03 | NA | NA | > 5.00E−3 |
| MBIP | MAP3K12 binding inhibitory protein 1 | 11 | 0.256 | 1.34E−03 | NA | NA | > 5.00E−3 |
| VCAM1 | Vascular cell adhesion molecule 1 | 12 | 0.254 | 1.50E−03 | 132 | 0.29 | 2.62E−04 |
| DNAJC8 | DnaJ heat shock protein family (Hsp40) member C8 | 13 | 0.25 | 1.79E−03 | NA | NA | > 5.00E−3 |
| HENMT1 (C1orf59H) | EN methyltransferase 1 | 14 | 0.249 | 1.87E−03 | 201 | 0.261 | 1.08E−03 |
| MRPL50 | Mitochondrial ribosomal protein L50 | 15 | 0.246 | 2.07E−03 | NA | NA | > 5.00E−3 |
| ODF2 | Outer dense fibre of sperm tails 2 | 16 | 0.245 | 2.15E−03 | NA | NA | > 5.00E−3 |
| PIGA | Phosphatidylinositol glycan anchor biosynthesis class A | 16 | 0.245 | 2.16E−03 | 209 | 0.26 | 1.13E−03 |
| HDAC4 | Histone deacetylase 4 | 16 | 0.245 | 2.19E−03 | 311 | 0.236 | 3.21E−03 |
| NCOA7 | Nuclear receptor coactivator 7 | 16 | 0.245 | 2.21E−03 | NA | NA | > 5.00E−3 |
The genes of top rank 16 that associated with good antiproliferative response and the genes of top rank 16 that associated with poor antiproliferative response, plus ESR1. All the 123 genes showing a significant correlation with the change in Ki67 are shown in Additional file 2: Table S5
Among the 48 genes whose high expression associated with poorer response, PERP (a TP53 apoptosis effector) and YWHAQ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) were the top 2 best correlated genes (r = 0.291 and 0.290, respectively), while 3 transcription co-regulators, SAP130, HDAC4, and NCOA7, were among the top 16 most correlated with poor Ki67 repression (Table 1).
The most highly correlated of the genes associated with better response was ACADVL which is related to fatty acid degradation [28]. CCND1 and SCUBE2 which are known to be associated with better response to endocrine therapy [29, 30] were among the top 16 best correlated with good suppression of Ki67. ESR1 expression was not correlated with the change in Ki67 after 2 weeks of AI therapy (Table 1; Additional file 5: Figure S3a).
Pathway analysis of the 123 genes identified HIPPO signalling as the most significantly over-represented pathway together with others directly or indirectly related to cell cycle regulation including p53 and p70S6K signalling (Additional file 6: Figure S4).
Of the pre-selected baseline signature scores, only proliferation-based modules (Gene70-GS, GGI-GS, AURKA-GS, CIN70-GS) and Rbloss-GS were significantly correlated with poor Ki67 response and these only weakly so (r = 0.243 to r = 0.161, all p < 0.05). WntTarget34-GS score was significantly correlated with good response, while TP53-GS score (signature associated with functional TP53) and several previously defined oestrogen signalling signatures approached significance (Additional file 7: Figure S5a; Additional file 8: Table S18A).
When the Ki67 changes were dichotomised to responders and non-responders, most of the baseline GSs whose score significantly associated with poor response were proliferation-based modules and Rbloss signatures, which was similar to the above. However, four additional GS that are not directly associated with proliferation but rather represent growth factor signalling pathways were significantly higher in non-responder tumours: ERBB2-GS, IGF1-GS, STAT1-GS, GDNF-GS (Table 2; Additional file 2: Table S7). Furthermore, five genes (CCND1, EPHX2, TRIP6, IMPDH2, and ACADVL) showed baseline expression that was significantly higher in AI responder tumours (p ≤ 1.5E−4);
Table 2.
Unpaired t test of significance for the difference between the two group baseline gene expression means of (i) non-responders vs responders and (ii) noCCCAs vs CCCAs in HER2− group. The means of gene signatures that directly associated with proliferation and represent growth factor signalling pathways were significantly different between AI responder and non-responder tumours, and most of them were statistically different between CCCAs and noCCCAs
| Module name | Reference | Non-responders vs responders | noCCCAs vs CCCAs | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (non-responders) | Mean (responders) | t test p value | FDR | Mean (no CCCAs) | Mean (CCCAs) | t test p value | FDR | ||
| ERBB2-GS.NoPAGs | Desmedt et al. | 4.901 | 4.773 | 5.01E−03 | 3.78E−02 | 4.814 | 4.798 | 6.49E−01 | 6.49E−01 |
| ERBB2-GS | Desmedt et al. | 4.475 | 4.348 | 5.04E−03 | 3.78E−02 | 4.391 | 4.372 | 5.98E−01 | 6.35E−01 |
| GGI-GS.NoPAGs | Sotiriou et al. | 0.683 | 0.565 | 7.83E−03 | 3.78E−02 | 0.669 | 0.476 | 7.10E−07 | 3.02E−06 |
| AURKA-GS | Desmedt et al. | 2.323 | 2.217 | 1.10E−02 | 3.78E−02 | 2.311 | 2.139 | 4.82E−06 | 1.37E−05 |
| IGF1-GS | Creighton et al. | -0.365 | − 0.402 | 1.62E−02 | 3.78E−02 | − 0.371 | − 0.426 | 3.95E−04 | 8.39E−04 |
| Immune.2.STAT1.NoPAGs | Desmedt et al. | 7.720 | 7.526 | 1.83E−02 | 3.78E−02 | 7.608 | 7.526 | 2.27E−01 | 3.22E−01 |
| STAT1-GS | Desmedt et al. | 7.819 | 7.623 | 1.84E−02 | 3.78E−02 | 7.706 | 7.623 | 2.23E−01 | 3.22E−01 |
| Gene70-GS | van ‘t Veer et al. | 3.029 | 2.928 | 2.54E−02 | 3.78E−02 | 3.019 | 2.846 | 2.19E−06 | 7.45E−06 |
| GDNF-GS.NoPAGs | Morandi et al. | 1.586 | 1.475 | 2.61E−02 | 3.78E−02 | 1.520 | 1.474 | 2.74E−01 | 3.58E−01 |
| removedPAGs | Gao et al. | 0.555 | 0.507 | 2.65E−02 | 3.78E−02 | 0.552 | 0.467 | 7.46E−06 | 1.81E−05 |
| WntTarget34-GS | VerhaeghCR2014TableS1 | 8.091 | 8.191 | 2.78E−02 | 3.78E−02 | 8.153 | 8.181 | 4.70E−01 | 5.33E−01 |
| IGF1.NoPAGs | Creighton et al. | -0.603 | − 0.636 | 2.91E−02 | 3.78E−02 | − 0.608 | − 0.657 | 1.09E−03 | 2.06E−03 |
| GDNF-GS | Morandi et al. | 1.699 | 1.591 | 3.08E−02 | 3.78E−02 | 1.634 | 1.594 | 3.31E−01 | 4.02E−01 |
| E2Factivation-GS.NoPAGs | Miller et al. | 9.119 | 9.029 | 3.12E−02 | 3.78E−02 | 9.091 | 8.996 | 7.83E−03 | 1.33E−02 |
| JClinInvest2007RBloss-GS | Bosco et al. | 7.961 | 7.802 | 3.44E−02 | 3.82E−02 | 7.960 | 7.654 | 2.83E−07 | 1.60E−06 |
| GGI-GS | Sotiriou et al. | 5.364 | 5.219 | 3.59E−02 | 3.82E−02 | 5.372 | 5.068 | 1.55E−07 | 1.60E−06 |
| DiLeoRBloss-GS | Malorni et al. | 7.771 | 7.600 | 4.21E−02 | 4.21E−02 | 7.781 | 7.423 | 2.29E−07 | 1.60E−06 |
Association of baseline gene expression and pre-selected signatures with 2-week residual Ki67
Baseline expression of 678 genes correlated with residual Ki67 after AI treatment. High expression of 376 genes was associated with high residual proliferation, and 302 genes were associated with low residual proliferation (Additional file 2: Table S8). Consistent with its association with good Ki67 suppression, ACADVL was the gene whose baseline expression was most strongly associated with low residual Ki67 (r = 0.419) and SCUBE2 the third most strongly associated (Table 1). Interestingly, the baseline expression of ACADVL and SCUBE2 was significantly correlated (r = 0.27, p = 0.0006). ESR1 expression was not correlated with residual Ki67 (r = − 0.16, p = 5.3E−2; Table 1; Additional file 5: Figure S3b).
The gene whose baseline expression was most strongly associated with high residual Ki67 was NEK2, a kinase involved in centrosome separation and bipolar spindle formation (r = 0.478). PTTG1 and the related PTTG3P were also among the top 5 most strongly correlated with residual Ki67 (r = 0.459 and 0.477, respectively). Both code for the members of the securin family that are homologues of yeast proteins that prevent separation of sister chromatid. Similarly, CDCA5, the third most strongly correlated gene, is also a regulator of sister chromatid cohesion, and all other genes strongly correlated at baseline with residual Ki67 are known to be associated with proliferation. Consistent with this, pathway analysis of the 678 genes showed p53, ATM, and EIF2 signalling pathways were among the most significantly over-represented (Additional file 2: Table S9), and of the pre-selected signatures, TP53-GS baseline score was the strongest inversely associated with residual Ki67 (r = − 0.46, p < 0.0001) (Additional file 7: Figure S5a; Additional file 8: Table S18A). The inverse correlation relates to high TP53-GS score being associated positively with TP53 wild-type status [21]. In contrast, baseline scores of Gene70-GS, GGI-GS, Rbloss-GS, DiLeoRBloss-GS, CIN70-GS, E2F4activation-GS, E2FmotifCellCycleAssociated-GS, AURKA-GS, PTEN-GS, and E2Factivation-GS score were positively correlated with residual Ki67 (all r ≥ 0.35, p < E−05).
As expected, higher baseline signature scores of PIK3CA-GS and modules measuring oestrogen signalling (ERGs-GS, ESR1-1-GS, ESR1-2-GS, SET-GS) were significantly associated with lower residual Ki67 (all p < 0.01). Higher STAT1-GS score was significantly but weakly correlated with higher residual Ki67 (r = 0.19, p = 1.57E−02) (Additional file 7: Figure S5a; Additional file 8: Table S18A).
Association of genes and pre-selected signatures with complete cell cycle arrest
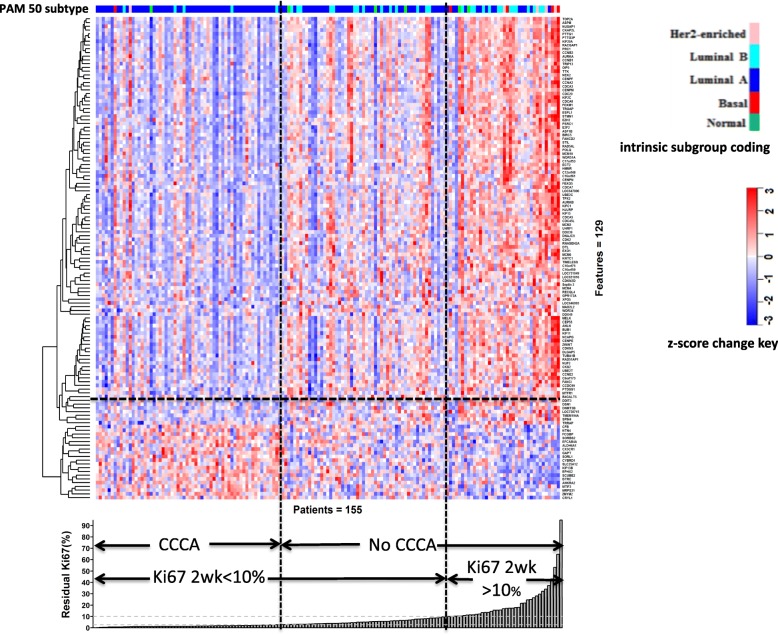
The baseline gene expression of 129 genes was significantly different between tumours reaching CCCA and noCCCA. Of the 109 genes whose baseline gene expression was significantly higher in the noCCCA tumours, 71.5% were proliferation-associated (Fig. 2; Additional file 2: Table S10). Similar to the above analysis of associations with residual proliferation, high baseline expression of PTTG1, PTTG3P, NEK2, and CDCA5 were prominent in being associated with noCCCA, but the most noticeable were TOP2A and UBE2C. High baseline NEK2 expression was also associated with poor antiproliferative response (Additional file 4: Figure S2). Notably, 5 genes (SCUBE2, FCGBP, EFCAB4A, EPHX2, and BTRC) whose baseline expression was significantly higher in tumours that achieved CCCA (Fig. 2; Additional file 2: Table S10) were also associated with good antiproliferative response (Additional file 4: Figure S2; Additional file 2: Table S6). Furthermore, ACADVL baseline expression was higher in CCCA tumours (p = 0.001).
Fig. 2.
Heatmap (Pearson, complete) of 129 genes whose baseline expression is significantly different (p < 0.001) between CCCA and noCCCA based on 155 HER2− of the 178 AI-treated samples. The gene expression across 155 samples was centred and scaled. Red denotes the gene expression in a sample is greater than the mean, blue denotes less than the mean. The tumours are ordered according to the residual level of Ki67
Of the pre-selected signatures, the baseline expression of TP53-GS, PIK3CA-GS, and ERGs-GS were significantly lower in noCCCA tumours. The lower TP53-GS score associated positively with dysfunctional TP53. In contrast, the expression of GGI-GS, DiLeoRBloss-GS, Rbloss-GS, CIN70-GS, E2FmotifCellCycleAssociated-GS, Gene70-GS, E2F4activation-GS, AURKA-GS, PTEN-GS, E2Factivation-GS, and IGF1-GS were significantly higher in the noCCCA tumours (all p < 0.0001) (Table 2; Additional file 2: Table S7).
One-dimensional clustering based on the relative baseline gene expression showed no distinct gene groups were apparent, and 5 of the 10 non-luminal tumours (excluding the normal-like) showed poorer than average Ki67 response to AI (Additional file 4: Figure S2). Of the 38 patients who had residual Ki67 (> 10%), 14 were from the original 33 (42%) luminal B tumours, 4 out of 5 (80%) were HER2-enriched, and 4 out of 5 (80%) were basal-like. Surprisingly, 13% of the original luminal A tumours (14 of the 106) were evident (Fig. 2).
Effects of oestrogen deprivation by AI treatment on gene expression and associated pathways
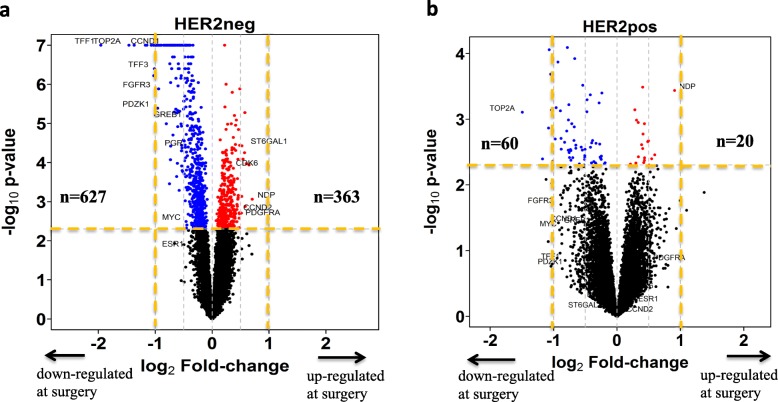
Oestrogen deprivation leads to profound effects on gene expression within 2 weeks. The expression of 902 genes was significantly changed: 560 downregulated and 342 upregulated (Fig. 3a; Additional file 2: Table S11). The most downregulated gene based on the amplitude of change was TFF1, followed by UBE2C and TOP2A, whose baseline expression was the most associated with noCCCA (both by > 60%). Similarly, NEK2 the gene most associated with residual Ki67 as a continuous variable was the ninth most downregulated gene.
Fig. 3.
Volcano plot highlighting the genes that were identified differentially expressed (p < 0.005) after AI treatment. Based on the difference of the expression mean changes (log2(Surgery/Baseline)) of paired samples between AI-treated and control. a Nine hundred ninety genes (n = 363 upregulated, n = 627 downregulated) in HER2− tumours (902 annotated genes). Number of AI-treated pairs, n = 135; control pairs, n = 46. b Eighty genes (n = 20 upregulated, n = 60 downregulated) in HER2+ tumours (71 annotated genes). Number of AI-treated pairs, n = 22; control pairs, n = 8. The p values range from 1 to a limited minimum value of 1.0E−07 was shown on the y-axis in a scale of −log10(p value)
Forty-nine of the top 50 genes that showed the greatest change in expression were downregulated by AI. The large majority of these were either related to proliferation or regulated by oestrogen. NDP was the only upregulated gene based on the amplitude of change (FC = 1.63, p = 8.69E−04). NDP is a norrin cystine knot growth factor, which activates the canonical Wnt signalling pathway through the frizzled family of receptors (FZD). Of note, FZD7, frizzled class receptor 7 was also upregulated (FC = 1.23, p = 0.0002) [31]. Furthermore, THRA, thyroid hormone receptor, was highly upregulated by AI (Additional file 2: Table S12).
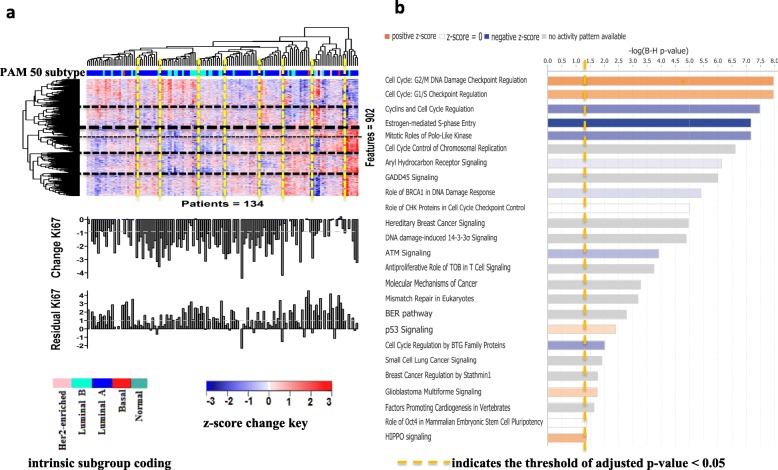
The heterogeneity of the changes in the gene expression between patients, irrespective of the change in Ki67, is illustrated in Fig. 4a. A large number of distinct groups of tumours were apparent, but these groups show a little distinct relationship with intrinsic subgrouping or both the change in Ki67 and residual Ki67 levels.
Fig. 4.
Unsupervised hierarchical clustering (Pearson, ward.D2) of 902 genes whose expression was significantly regulated after 2 weeks of treatment in HER2− tumours. And the overrepresented pathways (FDR < 5%) identified by pathway analysis (IPA). a The relative change in the gene expression across 134 HER2− tumours was standardised (centred and scaled). Red denotes the standardised z-score > 0, an increase in gene expression in a tumour after AI treatment compared to the average “relative changes” of the gene across all the 134 tumours; blue denotes the standardised z-score < 0, a decrease in gene expression in a tumour after AI treatment compared to the average “relative changes” of the gene across all the 134 tumours. b The 25 canonical pathways were significantly enriched (FDR < 5%). Positive z-score shown in orange colour specifies activated pathways; negative z-score shown in blue colour specifies inhibited pathways after AI treatment
Pathway analysis of the 902 genes that significantly changed with treatment revealed enrichment of 25 canonical pathways (adjusted p value < 0.05; Fig. 4b; Additional file 2: Table S13), the majority of which were proliferation-related. Cyclin-dependent kinases (CDK1, 2, and 6), CHEK1, cyclins (CCNE1, 2; CCND1, 2; and CCNB1, 2), and transcription factors E2F2 and E2F5, which were prominent in the majority of the 25 pathways, were also identified (Additional file 9: Figure S6).
Of particular note, CDK6 and CCND2 were significantly upregulated (p = 1.33E−04, p = 1.79E−03; Additional file 2: Table S12). In contrast to most of the cyclins and CDKs, CCND2 is a cell cycle regulator whose activity is dependent on its binding to CDK4/6 in G1 phase. Phosphorylation of Rb (retinoblastoma) by CDK4/6-CCND2 uncouples Rb from E2F allowing transcription of essential S-phase genes. Inhibition of CDK4/6-CCND2 in ER+ cells reduces cell proliferation and colony formation via a G1 cell cycle arrest [32]. The upregulation of CCND2 and CDK6 expression after AI therapy may be indicative of early tumour re-wiring that relates to residual proliferation.
Among the upregulated genes after AI treatment (Tables 3, 4, and 5; Additional file 2: Table S11), several (SNAI2, TGFB3, TGFBR2, TWIST2, PDGFD, PDGFRA, and SMAD4) are known to contribute to the loss of E-cadherin, a key mechanism in the stabilisation of the mesenchymal state that plays a role in the epithelial-mesenchymal transition (EMT) [33]. In addition, the increasing expression of TGFBR2, ACVR1, TGFB3, SMAD4, and INHBB are all linked to the activation of TGF-β signalling (z-score = 2.236) (Additional file 2: Table S13); the TGF-β signalling pathway has an established role in promoting EMT by downregulating E-cadherin via a number of transcription factors, such as Twist and Slug [34]. Finally, FRMD6 and YAP1, members of the HIPPO pathway, were upregulated, while LATS1/2, known negative regulators of the pathway [35], were undetectable (Additional file 2: Table S11).
Table 3.
Relative changes in the expression of gene signatures in response to the 2-week AI treatment of the HER2− and HER2+ tumours.
| Module name | HER2-negative | HER2-positive | Significance of the difference between two changes in expression (HER2−vs HER2+) | ||
|---|---|---|---|---|---|
| %Δ of geometric mean of intensities of a module of pre- and post-treatment | 2-sided unadjusted p value of paired t test | Geometric mean of intensities of a module of pre- and post-treatment | Unadjusted p value of paired t test | ||
| ESR1 | − 23.06 | 1.54E−02 | 18.28 | 6.23E−01 | 9.18E−03 |
| ERGs-GS | − 30.79 | 8.58E−14 | − 21.96 | 2.85E−03 | 5.81E−02 |
| DiLeoRBloss-GS | − 27.61 | 9.81E−10 | − 30.64 | 1.41E−03 | 5.43E−01 |
| JClinInvest2007RBloss-GS | − 26.11 | 5.47E−10 | − 29.45 | 1.78E−03 | 4.72E−01 |
| CIN70-GS | − 25.92 | 3.26E−10 | − 28.69 | 1.81E−03 | 5.40E−01 |
| E2F4activation-GS | − 25.26 | 4.54E−09 | − 30.07 | 5.02E−03 | 3.13E−01 |
| GGI-GS | − 23.67 | 1.78E−09 | − 27.85 | 1.19E−03 | 3.40E−01 |
| ERTarget27-GS | − 17.80 | 1.32E−15 | − 11.41 | 3.32E−02 | 1.71E−02 |
| E2FmotifCellCycleAssociated-GS | − 16.75 | 5.71E−09 | − 21.72 | 2.00E−03 | 1.31E−01 |
| E2Factivation-GS | − 14.45 | 2.25E−07 | − 18.20 | 1.93E−02 | 2.62E−01 |
| AURKA-GS | − 13.91 | 1.08E−07 | − 15.45 | 3.94E−03 | 6.10E−01 |
| Gene70-GS | − 9.37 | 1.97E−07 | − 12.34 | 6.76E−03 | 1.98E−01 |
| SET-GS | − 9.34 | 1.55E−05 | − 2.08 | 5.69E−01 | 8.73E−03 |
| PTEN-GS | − 7.55 | 1.44E−06 | − 8.81 | 3.62E−02 | 5.38E−01 |
| ESR1.2-GS | − 5.82 | 3.01E−04 | − 0.46 | 8.49E−01 | 1.10E−02 |
| ESR1.1-GS | − 5.40 | 2.99E−03 | 0.29 | 9.29E−01 | 1.50E−02 |
| MYC-GS | − 4.36 | 1.98E−04 | − 3.54 | 2.24E−01 | 6.02E−01 |
| IGF1-GS | − 4.15 | 4.59E−04 | − 4.07 | 7.76E−02 | 9.58E−01 |
| VEGF-GS | − 3.85 | 3.33E−02 | − 0.21 | 9.48E−01 | 1.09E−01 |
| GDNF-GS | − 3.41 | 1.40E−01 | − 5.23 | 4.25E−01 | 5.71E−01 |
| obesity-GS | − 3.08 | 2.40E−02 | − 1.72 | 5.64E−01 | 4.55E−01 |
| E2F3-GS | − 2.89 | 5.01E−05 | − 3.79 | 2.62E−02 | 3.45E−01 |
| PI3K-GS | − 1.80 | 3.19E−03 | − 3.11 | 6.69E−02 | 1.23E−01 |
| SRC-GS | − 1.69 | 2.83E−01 | − 1.62 | 6.46E−01 | 9.74E−01 |
| AKT/mTOR-GS | − 1.62 | 4.57E−02 | − 1.95 | 3.31E−01 | 7.51E−01 |
| RAS-GS | − 1.47 | 1.93E−01 | − 0.47 | 8.51E−01 | 5.04E−01 |
| BetaCatenin-GS | − 1.28 | 3.83E−01 | 0.04 | 9.91E−01 | 5.07E−01 |
| WntTarget34-GS | − 0.30 | 8.90E−01 | − 2.56 | 6.50E−01 | 4.28E−01 |
| ERBB2-GS | 0.27 | 8.88E−01 | − 0.88 | 8.60E−01 | 6.55E−01 |
| PIK3CA-GS | 0.39 | 7.20E−01 | 1.77 | 3.99E−01 | 3.42E−01 |
| STAT1-GS | 0.79 | 8.27E−01 | − 7.73 | 2.98E−01 | 6.99E−02 |
| CASP3-GS | 1.37 | 5.57E−01 | − 1.18 | 8.48E−01 | 3.82E−01 |
| Bcell-GS | 1.71 | 6.92E−01 | − 7.38 | 3.51E−01 | 7.71E−02 |
| MacTh1-GS | 2.72 | 4.94E−01 | 0.20 | 9.79E−01 | 6.38E−01 |
| Tcell-GS | 2.83 | 5.38E−01 | − 6.13 | 4.73E−01 | 1.12E−01 |
| Inflammatory-GS | 2.86 | 4.92E−01 | − 4.80 | 5.15E−01 | 1.55E−01 |
| Immune.1-GS | 3.07 | 5.24E−01 | − 5.34 | 6.10E−01 | 1.49E−01 |
| MAPK-GS | 3.21 | 6.59E−03 | 1.94 | 5.02E−01 | 4.22E−01 |
| Stroma.2.PLAU-GS | 6.57 | 4.65E−02 | 4.54 | 5.14E−01 | 6.34E−01 |
| Stroma.1-GS | 20.71 | 1.17E−02 | 21.87 | 1.61E−01 | 9.18E−01 |
| TP53-GS | 29.90 | 6.45E−10 | 29.38 | 5.59E−03 | 9.44E−01 |
| PAGs | − 4.40 | 6.44E−06 | − 6.23 | 2.06E−02 | 1.37E−01 |
Significant difference test of the changes between HER2− and HER2+ tumours. The increase of TP53-GS score after AI treatment is associated with the TP53 wild-type status
Table 4.
Spearman rank correlation of surgery ESR1 expression/pre-selected gene signature scores and percentage of 2-week change in Ki67/residual Ki67 level in HER2− tumours
| Surgery gene signature scores | HER2− (n = 135) | ||||||
|---|---|---|---|---|---|---|---|
| Change in Ki67 (n = 121) | Residual Ki67 (n = 124) | ||||||
| Signature name | Reference | Rho | p value | FDR | Rho | p value | FDR |
| ESR1 | − 0.19 | 3.42E−02 | 4.62E−02 | − 0.17 | 6.53E−02 | 6.53E−02 | |
| TP53-GS | Coutant et al. | − 0.47 | 7.43E−08 | 6.50E−07 | − 0.54 | 6.61E−11 | 3.57E−10 |
| ERTarget27-GS | VerhaeghCR2014TableS2 | − 0.26 | 3.96E−03 | 8.22E−03 | − 0.25 | 5.25E−03 | 7.46E−03 |
| SET-GS | Symmans et al. | − 0.25 | 5.38E−03 | 1.04E−02 | − 0.3 | 7.82E−04 | 1.41E−03 |
| ESR1.2-GS | Desmedt et al. | − 0.24 | 8.58E−03 | 1.32E−02 | − 0.27 | 2.52E−03 | 4.00E−03 |
| ESR1.1-GS | Mackay et al. | − 0.23 | 1.15E−02 | 1.63E−02 | − 0.29 | 1.34E−03 | 2.26E−03 |
| ERGs-GS | Dunbier et al. | − 0.09 | 3.56E−01 | 3.70E−01 | − 0.21 | 1.96E−02 | 2.21E−02 |
| PIK3CA-GS | Loi et al. | − 0.05 | 5.53E−01 | 5.53E−01 | − 0.18 | 4.07E−02 | 4.23E−02 |
| MacTh1-GS | Iglesia et al. | 0.088 | 3.39E−01 | 3.66E−01 | 0.199 | 2.64E−02 | 2.85E−02 |
| Inflammatory-GS | Dunbier et al. | 0.116 | 2.05E−01 | 2.31E−01 | 0.22 | 1.43E−02 | 1.68E−02 |
| GDNF-GS | Morandi et al. | 0.116 | 2.04E−01 | 2.31E−01 | 0.254 | 4.40E−03 | 6.60E−03 |
| obesity-GS | Creighton et al. | 0.134 | 1.43E−01 | 1.76E−01 | 0.239 | 7.61E−03 | 9.34E−03 |
| Immune.2.STAT1-GS | Desmedt et al. | 0.158 | 8.26E−02 | 1.06E−01 | 0.247 | 5.64E−03 | 7.61E−03 |
| E2F3-GS | Bild et al. | 0.237 | 8.79E−03 | 1.32E−02 | 0.24 | 7.38E−03 | 9.34E−03 |
| IGF1-GS | Creighton et al. | 0.24 | 8.06E−03 | 1.32E−02 | 0.303 | 6.22E−04 | 1.20E−03 |
| AKT/mTOR-GS | Majumder et al. | 0.249 | 5.83E−03 | 1.05E−02 | 0.362 | 3.63E−05 | 7.54E−05 |
| PTEN-GS | Saale at al. | 0.286 | 1.49E−03 | 3.35E−03 | 0.392 | 6.68E−06 | 1.50E−05 |
| PI3K-GS | Creighton 2010 | 0.291 | 1.21E−03 | 2.97E−03 | 0.413 | 1.92E−06 | 4.71E−06 |
| AURKA-GS | Desmedt et al. | 0.32 | 3.53E−04 | 9.53E−04 | 0.427 | 7.36E−07 | 1.99E−06 |
| E2Factivation-GS | Miller et al. | 0.323 | 3.08E−04 | 9.24E−04 | 0.432 | 5.39E−07 | 1.62E−06 |
| E2FmotifCellCycleAssociated-GS | Miller et al. | 0.402 | 4.79E−06 | 1.62E−05 | 0.454 | 1.22E−07 | 4.12E−07 |
| Gene70-GS | van ‘t Veer et al. | 0.431 | 8.22E−07 | 3.17E−06 | 0.546 | 5.64E−11 | 3.57E−10 |
| CIN70-GS | Carter et al. | 0.442 | 3.89E−07 | 1.75E−06 | 0.529 | 2.79E−10 | 1.08E−09 |
| E2F4activation-GS | Guerrero-Zotano et al. | 0.454 | 1.73E−07 | 9.34E−07 | 0.535 | 1.51E−10 | 6.80E−10 |
| GGI-GS | Sotiriou et al. | 0.462 | 9.63E−08 | 6.50E−07 | 0.571 | 4.52E−12 | 6.10E−11 |
| JClinInvest2007RBloss-GS | Bosco et al. | 0.491 | 1.12E−08 | 1.51E−07 | 0.545 | 5.89E−11 | 3.57E−10 |
| DiLeoRBloss-GS | Malorni et al. | 0.526 | 5.61E−10 | 1.51E−08 | 0.584 | 1.08E−12 | 2.92E−11 |
TP53-GS surgery score was the strongest inversely associated with the change in Ki67 and residual Ki67. The inverse correlation relates to high TP53-GS score being associated positively with TP53 wild-type status
Table 5.
Spearman rank correlation of change in ESR1 expression/pre-selected gene signature scores and percentage of 2-week change in Ki67/residual Ki67 level in (i) HER2− tumours, (ii) HER2+ tumours, and (iii) significance of the difference between the two correlation coefficients (HER2− vs HER2+).
| Change in gene signature scores | HER2− (n = 135) | HER2+ (n = 22) | Significance of the difference between two correlation coefficients (HER2− vs HER2+) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Change in Ki67 (n = 121) | Residual Ki67 (n = 124) | Change in Ki67 (n = 21) | Residual Ki67 (n = 22) | ||||||||
| Signature name | Reference | Rho | p value | Rho | p value | Rho | p value | Rho | p value | Change in Ki67 | Residual Ki67 |
| ESR1 | − 0.08 | 3.58E−01 | 0.03 | 7.37E−01 | 0.41 | 6.70E−02 | 0.48 | 2.20E−02 | 3.69E−02 | 4.61E−02 | |
| E2F4activation-GS | Guerrero-Zotano et al. | 0.38 | 2.12E−05 | 0.13 | 1.58E−01 | 0.42 | 5.84E−02 | 0.18 | 4.22E−01 | 8.47E−01 | 8.36E−01 |
| DiLeoRBloss-GS | Malorni et al. | 0.36 | 4.29E−05 | 0.09 | 3.12E−01 | 0.40 | 7.55E−02 | 0.23 | 2.94E−01 | 8.50E−01 | 5.60E−01 |
| JClinInvest2007RBloss-GS | Bosco et al. | 0.35 | 1.05E−04 | 0.08 | 3.87E−01 | 0.38 | 8.53E−02 | 0.22 | 3.31E−01 | 8.89E−01 | 5.62E−01 |
| GGI-GS | Sotiriou et al. | 0.34 | 1.30E−04 | 0.08 | 3.75E−01 | 0.41 | 6.28E−02 | 0.24 | 2.77E−01 | 7.42E−01 | 5.05E−01 |
| CIN70-GS | Carter et al. | 0.33 | 2.16E−04 | 0.09 | 3.26E−01 | 0.43 | 5.18E−02 | 0.23 | 3.04E−01 | 6.36E−01 | 5.60E−01 |
| E2FmotifCellCycleAssociated-GS | Miller et al. | 0.31 | 4.37E−04 | 0.04 | 6.91E−01 | 0.53 | 1.38E−02 | 0.09 | 6.84E−01 | 2.75E−01 | 8.39E−01 |
| Gene70-GS | van ‘t Veer et al. | 0.31 | 5.01E−04 | 0.05 | 5.90E−01 | 0.44 | 4.65E−02 | 0.14 | 5.46E−01 | 5.39E−01 | 7.13E−01 |
| E2Factivation-GS | Miller et al. | 0.29 | 1.49E−03 | 0.13 | 1.40E−01 | 0.42 | 5.92E−02 | 0.04 | 8.59E−01 | 5.46E−01 | 7.14E−01 |
| ERGs-GS | Dunbier et al. | 0.25 | 5.44E−03 | 0.28 | 1.96E−03 | 0.11 | 6.30E−01 | 0.49 | 2.07E−02 | 5.58E−01 | 3.15E−01 |
| PTEN-GS | Saale at al. | 0.23 | 1.16E−02 | 0.04 | 6.24E−01 | 0.46 | 3.43E−02 | 0.20 | 3.79E−01 | 2.87E−01 | 5.10E−01 |
| PI3K-GS | Creighton 2010 | 0.23 | 1.31E−02 | 0.12 | 1.95E−01 | 0.50 | 1.99E−02 | 0.37 | 9.24E−02 | 2.02E−01 | 2.79E−01 |
| AURKA-GS | Desmedt et al. | 0.21 | 2.32E−02 | 0.01 | 8.70E−01 | 0.46 | 3.37E−02 | 0.38 | 7.72E−02 | 2.50E−01 | 1.15E−01 |
| E2F3-GS | Bild et al. | 0.20 | 3.20E−02 | 0.07 | 4.34E−01 | 0.43 | 5.18E−02 | 0.33 | 1.36E−01 | 2.98E−01 | 2.70E−01 |
| WntTarget34-GS | VerhaeghCR2014TableS1 | 0.17 | 6.11E−02 | 0.16 | 6.99E−02 | 0.11 | 6.26E−01 | − 0.09 | 6.91E−01 | 8.04E−01 | 3.09E−01 |
| AKT/mTOR-GS | Majumder et al. | 0.15 | 9.44E−02 | 0.14 | 1.20E−01 | 0.29 | 2.03E−01 | 0.25 | 2.64E−01 | 5.51E−01 | 6.43E−01 |
| IGF1-GS | Creighton et al. | 0.13 | 1.61E−01 | − 0.05 | 5.80E−01 | 0.32 | 1.54E−01 | 0.42 | 5.41E−02 | 4.16E−01 | 4.41E−02 |
| ERTarget27-GS | VerhaeghCR2014TableS2 | 0.12 | 1.86E−01 | 0.19 | 3.26E−02 | 0.29 | 1.97E−01 | 0.41 | 5.85E−02 | 4.72E−01 | 3.25E−01 |
| MacTh1-GS | Iglesia et al. | 0.10 | 2.57E−01 | 0.09 | 3.23E−01 | − 0.17 | 4.51E−01 | − 0.14 | 5.33E−01 | 2.71E−01 | 3.50E−01 |
| CASP3-GS | Desmedt et al. | 0.09 | 3.06E−01 | 0.12 | 2.00E−01 | 0.17 | 4.64E−01 | − 0.08 | 7.32E−01 | 7.42E−01 | 4.17E−01 |
| STAT1-GS | Desmedt et al. | 0.07 | 4.50E−01 | 0.02 | 8.06E−01 | 0.11 | 6.30E−01 | − 0.22 | 3.18E−01 | 8.70E−01 | 3.24E−01 |
| GDNF-GS | Morandi et al. | 0.06 | 4.80E−01 | 0.04 | 6.99E−01 | 0.20 | 3.94E−01 | 0.03 | 8.91E−01 | 5.64E−01 | 9.68E−01 |
| MYC-GS | Bild et al. | 0.06 | 5.30E−01 | − 0.10 | 2.60E−01 | 0.40 | 7.14E−02 | 0.43 | 4.41E−02 | 1.41E−01 | 2.34E−02 |
| Inflammatory-GS | Dunbier et al. | 0.06 | 5.34E−01 | 0.03 | 7.56E−01 | − 0.19 | 3.97E−01 | − 0.28 | 2.04E−01 | 3.07E−01 | 1.99E−01 |
| Bcell-GS | Iglesia et al. | 0.05 | 5.55E−01 | 0.02 | 7.97E−01 | − 0.01 | 9.55E−01 | − 0.54 | 9.42E−03 | 8.08E−01 | 1.16E−02 |
| Tcell-GS | Iglesia et al. | 0.05 | 5.61E−01 | − 0.02 | 8.09E−01 | − 0.41 | 6.46E−02 | − 0.33 | 1.36E−01 | 4.95E−02 | 1.92E−01 |
| obesity-GS | Creighton et al. | 0.05 | 5.67E−01 | − 0.12 | 1.91E−01 | − 0.22 | 3.33E−01 | 0.23 | 3.06E−01 | 2.68E−01 | 1.51E−01 |
| VEGF-GS | Desmedt et al. | 0.03 | 7.41E−01 | − 0.02 | 8.34E−01 | 0.46 | 3.43E−02 | 0.63 | 1.57E−03 | 5.87E−02 | 2.07E−03 |
| Stroma.2.PLAU-GS | Desmedt et al. | 0.02 | 8.14E−01 | 0.01 | 9.55E−01 | − 0.21 | 3.51E−01 | − 0.17 | 4.46E−01 | 3.46E−01 | 4.62E−01 |
| BetaCatenin-GS | Bild et al. | 0.02 | 8.50E−01 | − 0.07 | 4.09E−01 | − 0.02 | 9.29E−01 | 0.33 | 1.39E−01 | 8.71E−01 | 9.48E−02 |
| Stroma.1-GS | Farmer at al. | 0.00 | 9.71E−01 | 0.04 | 6.27E−01 | − 0.20 | 3.88E−01 | − 0.26 | 2.51E−01 | 4.12E−01 | 2.16E−01 |
| MAPK-GS | Creighton et al. | − 0.01 | 9.24E−01 | − 0.07 | 4.24E−01 | − 0.27 | 2.36E−01 | − 0.30 | 1.70E−01 | 2.80E−01 | 3.33E−01 |
| Immune.1-GS | Teschendorff at al. | − 0.01 | 9.12E−01 | − 0.03 | 7.16E−01 | − 0.16 | 4.78E−01 | 0.03 | 8.79E−01 | 5.40E−01 | 8.08E−01 |
| PIK3CA-GS | Loi et al. | − 0.01 | 9.00E−01 | 0.02 | 8.03E−01 | − 0.23 | 3.22E−01 | − 0.15 | 5.13E−01 | 3.64E−01 | 4.89E−01 |
| SRC-GS | Bild et al. | − 0.03 | 7.43E−01 | − 0.13 | 1.58E−01 | − 0.06 | 7.97E−01 | − 0.14 | 5.46E−01 | 9.03E−01 | 9.67E−01 |
| ESR1.1-GS | Mackay et al. | − 0.05 | 6.16E−01 | 0.07 | 4.16E−01 | 0.31 | 1.71E−01 | 0.42 | 4.99E−02 | 1.34E−01 | 1.27E−01 |
| SET-GS | Symmans et al | − 0.05 | 5.70E−01 | 0.09 | 3.21E−01 | 0.22 | 3.28E−01 | 0.45 | 3.36E−02 | 2.68E−01 | 1.11E−01 |
| RAS-GS | Bild et al. | − 0.06 | 4.99E−01 | − 0.13 | 1.39E−01 | 0.40 | 7.44E−02 | 0.11 | 6.29E−01 | 5.04E−02 | 3.29E−01 |
| ESR1.2-GS | Desmedt et al. | − 0.10 | 2.81E−01 | 0.05 | 5.69E−01 | 0.29 | 1.97E−01 | 0.62 | 2.21E−03 | 1.07E−01 | 6.32E−03 |
| ERBB2-GS | Desmedt et al. | − 0.24 | 9.08E−03 | − 0.22 | 1.20E−02 | − 0.04 | 8.80E−01 | 0.15 | 5.06E−01 | 4.08E−01 | 1.29E−01 |
| TP53-GS | Coutant et al. | − 0.37 | 2.38E−05 | − 0.12 | 1.82E−01 | − 0.32 | 1.63E−01 | − 0.32 | 1.43E−01 | 8.18E−01 | 3.93E−01 |
| PAGs | Gao et al. | 0.27 | 3.11E−03 | 0.07 | 4.68E−01 | 0.47 | 2.99E−02 | 0.11 | 6.12E−01 | 3.45E−01 | 8.70E−01 |
Change in TP53-GS score was the strongest inversely associated with the change in Ki67 after 2-week AI treatment. The inverse correlation relates to the increase of TP53-GS score being associated positively with TP53 wild-type status
We next assessed the dynamic changes in the pre-selected signature response to 2-week AI treatment. ESR1 gene expression and ER-regulated/targeted genes (ERG-GS, ERTarget27-GS, and several proliferation-associated GSs were profoundly reduced by AI (%∆ of geometric mean > 10%)), but none to the same magnitude as the single IHC marker Ki67 (Table 3; Additional file 2: Table S5). Module scores of Gene70-GS, SET-GS, MYC-GS, PTEN-GS, and IGF1-GS were also all significantly suppressed but to a lesser degree. In contrast, the scores of Stroma.1-GS and TP53-GS had largely increased due to oestrogen deprivation. The increased TP53-GS score associated positively with TP53 wild-type status.
Association of 2-week pre-selected gene signature scores with changes in Ki67 and residual Ki67
On-treatment gene expression may be at least as important a determinant of resistance to AI therapy and a potential target for additional treatment as pre-treatment gene expression. We therefore assessed the association of on-treatment scores of the pre-selected signatures with the change in Ki67 and residual Ki67 (Table 4; Additional file 10: Figure S7a; Additional file 8: Table S19A). Significant correlations were found with several of the signatures and residual Ki67, and most of these were also significant for change in Ki67. Those correlations significant for both endpoints were (i) the two RB loss signatures [17, 18], (ii) proliferation-related signatures (GGI-GS, CIN70-GS, Gene70-GS, AURKA-GS), (iii) modules measuring oestrogen signalling (SET-GS, ESR1.1-GS, ESR1.2-GS, ERTarget27-GS), (iv) E2F signatures [19, 20], and (v) TP53-GS, PI3K-GS, PTEN-GS, AKT/mTOR-GS, and IGF1-GS. Of note, while high on-treatment oestrogen signalling module scores associated with lower residual proliferation and better antiproliferative response, high TP53-GS score that reflects wild-type TP53 function showed the highest correlation.
We found no significant relationship between the change in Ki67 and immune response gene signatures including Inflammatory-GS and the immune and stromal scores estimated by ESTIMATE. However, high STAT1-GS treatment score showed a significant association with high residual Ki67 (r = 0.25, p = 5.64E−03), as did the Inflammatory-GS and MacTh1-GS (Table 4).
Association of the change in pre-selected gene signature scores with changes in Ki67 and residual Ki67
Unsurprisingly, ten of the changes in signature scores that were significantly directly correlated with Ki67 change were proliferation-associated GSs. However, of particular note, reduction in the expression of the ERGs-GS was also directly associated with greater Ki67 suppression and low residual Ki67. In addition, increase in ERBB2-GS score was significantly associated with both greater Ki67 suppression and lower residual Ki67 after AI therapy, possibly as an immediate compensatory resistance mechanism (Table 5; Additional file 2: Table S14; Additional file 11: Figure S8a; Additional file 8: Table S20A). The change in ESR1 expression was significantly associated with the change in all the modules measuring oestrogen signalling (SET-GS, r = 0.72; ESR1-1-GS, r = 0.69; ESR1-2-GS, r = 0.59; ERTarget27-GS, r = 0.39; ERGs-GS, r = 0.36; all p < 0.0001).
HER2-positive tumours
Class comparison of the mean changes between the 26 AI-treated HER2+ tumours and 8 HER2+ control tumours identified 71 annotated genes, which were significantly changed by AI therapy (n = 19 upregulated, n = 52 downregulated). (Fig. 3b; Additional file 2: Table S15). Pathway analysis of the 71 genes identified 7 canonical pathways as being significantly enriched (adjusted p value< 0.05; Additional file 12: Figure S9). Activation of the top pathway, mitotic roles of Polo-like kinase, was indicated as being significantly reduced by oestrogen deprivation consistent with the partial reduction in Ki67 for almost all of the HER2+ tumours and with the changes in proliferation-related genes in the HER2− cohort.
To identify any significant differences between HER2+ and HER2− tumours in their molecular response to AIs, we compared the AI-induced gene changes between the two groups (Additional file 2: Table S12). Seven of the 10 top downregulated genes in the HER2+ group were in the top 13 downregulated genes in HER2− tumours. The top upregulated gene NDP in the HER2− group was also the top upregulated in HER2+ tumours. Proliferation-associated and cell cycle genes were suppressed to a similar extent in both cohorts despite the difference in Ki67 suppression.
The classical oestrogen-regulated genes were suppressed to a significantly lesser extent by AI treatment in the HER2+ tumours, for example, downregulation of TFF1, TFF3, CCND1, and PGR was significantly less (p’s for difference = 0.0027, 0.0001, 0.035, and 0.0034, respectively). In contrast to the decrease in ESR1 levels seen in the HER2− tumours, in HER2+ tumours, ESR1 gene expression was not significantly changed (p = 0.009 for the difference between the groups). The GSs that measure oestrogen signalling (ERTarget27-GS, SET-GS, ESR1.2-GS, ESR1.1-GS) were also significantly less suppressed by AI in HER2+ tumours (Table 3). Again, in contrast with HER2− tumours, ESR1 expression was significantly correlated with the change in Ki67 (r = − 0.61, p = 2.57E−03) being among the 25 genes whose baseline expression correlated with better Ki67 response (Additional file 5: Figure S3c; Additional file 2: Table S16). ESR1 was among the 54 genes whose high baseline expression correlated with low residual proliferation in HER2+ tumours (r = − 0.62, p = 2.19E−03) while there were no such significant relationships with ESR1 in the HER2− group (Additional file 5: Figure S3d; Additional file 2: Table S17).
Association of genes and pre-selected signatures in HER2+ tumours
Analysis of the pre-selected signatures in the HER2+ cohort showed similar results to those observed in the HER2− tumours (Additional file 7: Figure S5b, Additional file 10: Figure S7b, Additional file 11: Figure S8b; Additional file 8: Table S18B, S19B, S20B). In those cases where there were differences between the relationships in HER2+ and HER2−, the p values were only ever moderately significant. Given the size of the HER2+ group and the multiple tests conducted, we did not pursue these further.
Discussion
AIs are well-established as the most effective and therefore most frequently used endocrine agents for treating ER+ BC in postmenopausal women [5]. Despite the efficacy of AIs, many patients recur with either de novo or acquired AI-resistant disease. Molecular characterisation of the resistance phenotype(s) is critical for enhanced control of the disease. In this study, we report the largest sample set describing the genome-wide transcriptional and related antiproliferative effects of AIs. In addition, for the first time, we have been able to correct for artefactual transcriptional changes that occurred in the control group in the absence of any treatment. We have described those artefactual changes in detail elsewhere and discussed the likely causes [13, 15]. Most importantly, had we been unable to correct for them; the most significantly and most quantitatively changed genes in the AI-treated group would have been accepted in error as due to AI treatment while they were in fact entirely related to tissue processing [13]. Our analyses applied rigorous statistical methods using Benjamini-Hochberg procedure where appropriate to calculate the FDR in order to adjust for multiple testing.
Our assessment of the biologic response/resistance of the tumours to AI was based on Ki67. It is important to appreciate the significance of the different endpoints and their validity for that purpose. Proportional or percentage change in Ki67 has been validated as reflecting the degree of benefit (or response) to AI [6–9] and is the relevant parameter for considering mechanisms of response/resistance to AI therapy. The 2-week (residual) value of Ki67 is determined in part by the proportional response to the AI but also by the pretreatment value; the value correlates with the residual risk of recurrence on the endocrine therapy [10] and is relevant as a marker of the value/need to apply additional adjuvant therapy, irrespective of whether or not there has been a good or poor proportional antiproliferative response to the AI. Continued or altered (rewired) signalling in the residual tissue may be more relevant to the targeting of the additional agents than baseline expression. It should be noted that while our sample set was drawn from the POETIC trial, we make no claims for it being a representative subset (although it may be). Rather, the design of POETIC and availability of the set of RNAlater-stored samples provided the opportunity for us to undertake the molecular analyses described; the relevance of our observations to ER+ breast cancer in general may be considered by reference to the demographics of this subset.
In the HER2− group, we confirmed that high baseline signature scores of IGF1-GS, STAT1-GS, and GDNF-GS were associated with poor antiproliferative response when Ki67 change was dichotomised [12, 22]. Although we validated STAT1-GS, which represents features of immune activity as being associated with AI resistance, there was no significant association between Inflammatory-GS and the change in Ki67. This is somewhat in contrast to our previous report in a smaller mixed HER2−/HER2+ set in which both these signatures were predictive of poor AI response of tumours [11, 12]. The significantly high baseline ERBB2-GS in AI non-responders suggests tumours with high HER2 signalling activity even in HER2− tumours were predictive of poor response. This is consistent with the observation of poor response to letrozole alone and improved outcome with added lapatinib in the HER2-enriched subtype of HER2− metastatic BC [36]. Further assessment of the interaction between this subtype and response to endocrine therapy is now underway in the much larger formalin-fixed set of tissues from POETIC. The strong correlations between several baseline signature scores and the residual Ki67 confirmed the high proliferation (AURKA-GS, PTEN-GS, Gene70-GS, GGI-GS, CIN70-GS), RB-loss (Rbloss-GS, DiLeoRBloss-GS), high E2F activation (E2F4activation-GS, E2Factivation-GS), and TP53 dysfunction (TP53-GS) were associated with high-oestrogen independent residual proliferation irrespective of whether the tumour showed an antiproliferative response to AI [12, 17–21].
We and others have previously described that HER2 positivity impedes the antiproliferative effect of endocrine therapy [26, 27]. The resultant major difference in the changes in Ki67 suppression seen here between the HER2+ and HER2− group led us to consider the HER2 subgroups separately. This allowed us to describe the substantial differences in oestrogen signalling that occurred between them. In HER2+ but not HER2− tumours, baseline ESR1 expression was significantly correlated with the change in Ki67 levels, and while those ESR1 levels were suppressed in HER2− tumours, they were not significantly changed in HER2+ tumours; expression of oestrogen-regulated genes and ER-related gene modules was also changed less in HER2+ than in HER2− tumours. This difference could be explained by the decrease in oestrogen signalling in the HER2− tumours being in part dependent on the lower ESR1 levels on-treatment and not just by the oestrogen deprivation with the AI. Alternatively, or in addition, the apparent persistent oestrogen signalling in HER2+ tumours might result from ligand-independent activation of ER by HER2. This highlights the complex crosstalk between HER2 and ER [37].
Assessment of the gene expression at baseline in the HER2− cohort to identify de novo biomarkers of resistance revealed a very marked heterogeneity between tumours with no new patterns of expression being associated with changes in Ki67. However, intrinsic subgrouping did reveal that luminal B and particularly the small number of non-luminal tumours showed less Ki67 suppression and greater residual Ki67 levels than luminal A tumours. This is somewhat in contrast to our report in a smaller earlier set of tumours in which the proportional change in Ki67 was found to be similar between luminal A and B tumours although the residual level of Ki67 was higher in the latter [11, 38].
Unsurprisingly, it is clear that proliferation and cell cycle-associated pathways dominated the gene signatures found to change with AI and also to be associated with residual Ki67. However, we also found that the baseline expression of several proliferation-related gene signatures was also related to the change in Ki67. This is consistent with the greater preponderance of luminal B and non-luminal tumours with a poor change in Ki67.
In contrast to the many cell cycle genes that were decreased in activity in parallel with the decrease in proliferation, CCND2 and CDK6 were increased. CDK4/6 inhibitors are now in widespread use in the treatment of ER+ metastatic BC and are in large clinical trials in primary BC, in each case in combination with endocrine therapy. It was also notable that the most prominent genes in the canonical pathways were CDK2 and CCNEs which are critical to triggering the G1- to S-phase transition. As previously reported, on-treatment E2F signatures were among those most strongly associated with residual Ki67 [19, 39], and the TP53-GS that reflects wild-type TP53 function showed the highest correlation with lower residual Ki67. Assessment of the early impact by AIs on each of these factors may be relevant to the success or not of CDK4/6 inhibition when combined with an AI. This argues for an initial treatment with an AI before the introduction of the CDK4/6 inhibitor. We are pursuing this concept in the design of a new clinical trial of the adjuvant use of CDK4/6 inhibition in high-risk ER+ disease (POETIC-A).
A particularly novel finding was that ACADVL baseline expression was the best predictor of both decrease in Ki67 and of low residual Ki67, and its expression was significantly higher in responder and CCCA tumours. In silico analysis of the BC dataset reveals that the lower baseline expression of ACADVL was associated with poor relapse-free survival in ER+ patients [40]. The gene encodes a very long chain-specific acyl-CoA dehydrogenase, mitochondrial (VLCAD) enzyme, a key enzyme of the mitochondrial fatty acid β-oxidation (FAO) pathway. A recent study [41] revealed that VLCAD interacts with the BH3 domain of MCL-1 via a non-canonical mechanism, which is associated with chemoresistance in human cancer and merits further study.
Two other novel findings were the high baseline expression of (i) PERP and YWHAQ as the most significantly associated with poor AI response, and (ii) NEK2 was most strongly associated with high residual proliferation. In silico analysis of the BC dataset reveals that high baseline expression of YWHAQ and NEK2 have been reported to be associated with poor relapse-free survival in an ER+/HER2− setting for the patients receiving endocrine therapy and no chemotherapy [40]. Furthermore, the expression of YWHAQ and NEK2 was significantly higher in luminal B compared to luminal A tumours in TCGA ER+/HER2− tumours [42]. Together, these findings suggest that the poor prognosis associated with these two genes may be at least partly due to an association with endocrine resistance. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22 family. A recent study [43] revealed that PERP is lost in more aggressive sparsely granulated human growth hormone pituitary tumours, and its loss and associated desmosomal instability may be an early driver of tumour progression. However, its significant association with poor antiproliferative response to AIs in ER+/HER2− tumours has not been previously reported and requires validation prior to further study.
Conclusions
It is clear from the above that our work identifies the possible involvement of multiple pathways in de novo resistance to AIs, some but not all of which have previously been described. However, there are other pathways whose baseline activity is unrelated to resistance but whose expression is modified or rewired within the first 2 weeks and at that stage is related to residual proliferation.
While the number of cases described is the largest reported to date and is sufficient to identify the possible involvement of each of the pathways described, their relative importance will require assessment in a yet larger population.
Overall, we conclude that there is a high degree of heterogeneity between tumours in their adaptive response to oestrogen deprivation; however, in this study, all appeared to converge on cell cycle regulation. Our data highlighting the relationship between the E2F signature and residual Ki67 along with the earlier proposal by Miller et al. [19] that on-treatment evaluation of this signature could indicate enhanced sensitivity to CDK4/6 inhibition suggests that it merits prospective evaluation in a clinical setting. This is a hypothesis that we will be testing in a major new national adjuvant trial, POETIC-A, in which patients with early ER+ breast cancer whose tumour continues to show high Ki67 expression after 2 weeks AI will be randomised to additional CDK4/6 inhibition or not.
Supplementary information
Additional file 1. Supplementary information. Additional description of the materials and methods.
Additional file 2. Table S1. Published gene-signatures. Table S2. Study endpoints. Table S3. Study demographics. Table S4. List of patients and their status of HER2, Ki67 and CCCA. Table S5. Ki67 data summary. (A) Percentage change in Ki67 expression after 2-weeks AI-treatment. (B) Anti-proliferative response summary. (C) Summary of number of tumours that reached CCCA. Table S6. Genes whose baseline expression significantly correlated with Change.Ki67 (p < 0.005) in HER2- group. Table S7. Unpaired t-test of significance for the difference between two-group means: i) non-responders vs responders; ii) noCCCAs vs CCCAs in HER2- group. Table S8. Genes whose baseline expression significantly correlated with residual Ki67 (p < 0.005) in HER2- group. Table S9. Pathway analysis of genes in Table S8. Table S10. Genes whose baseline expression is significantly different (p < 0.001) between CCCA and noCCCA in HER2- group. Table S11. Genes significantly regulated by AI with p < 0.005 by univariate Two-sample T-test in HER2- group. Table S12. Top 20 AI-downregulated / AI-upregulated genes, and additional AI-regulated genes of interest identified from HER2- paired tumours. Genes were ranked by fold-change (FC). The Significantly different (p-value) derived from Mann-Whitney test comparing the relative change in expression of a given gene in the HER2- group versus the HER2+ group. ns: p >= 0.05. Table S13. Enriched canonical pathways of the significantly AI-regulated genes in Table S11. Table S14. Correlations of the change in ESR1 / gene-signatures and change in Ki67 / residual-Ki67 in i) HER2- group, ii) HER2+ group, iii) Significance of the difference between the 2 correlation-coefficients. Table S15. Genes significantly regulated by AI with p < 0.005 by univariate Two-sample T-test in HER2+ group. Table S16. Genes whose baseline expression significantly correlated with Change.Ki67 (p < 0.005) in HER2+ group. Table S17. Genes whose baseline expression significantly correlated with residual-Ki67 (p < 0.005) in HER2+ group.
Additional file 3: Figure S1. Consort diagram showing derivation of samples for microarray analysis and Ki67 measurement. In total, RNA was extracted from 861 RNAlater stored core-cuts and 605 RNA samples with RNA integrity number (RIN) >4 and RNA >500 ng were sent for profiling. Samples were excluded due to lack of adequate estradiol suppression, RIN < 4 when profiling, or due to gene expression data of poor quality. “Pairs” indicates a tumour with matched baseline and surgery expression data; B (baseline) indicates a tumour with baseline expression data or baseline Ki67 value; S (surgery) indicates a tumour with surgical expression data or on-treatment Ki67. *Expression QC (quality control): samples with fraction of detection rate <30% or detected by lumi.outlier function from lumi R package as outlier were excluded.
Additional file 4: Figure S2. Heatmap (Pearson, complete) of 123 genes whose baseline expression significantly correlated with Change.Ki67 (p<0.005) based on 155 HER2- of the 178 AI-treated samples. The gene expression across 155 samples was centred and scaled. Red denotes the gene expression in a sample is greater than the mean, blue denotes less than the mean. The tumours are ordered according to the degree of change in Ki67.
Additional file 5: Figure S3. Scatter plot of baseline gene expression and Ki67 values. (a) ESR1 baseline expression and change in Ki67 of HER2- tumours; (b) ESR1 baseline expression and residual Ki67 of HER2- tumours; (c) ESR1 baseline expression and change in Ki67 of HER2+ tumours; (d) ESR1 baseline expression and residual Ki67 of HER2+ tumours.
Additional file 6: Figure S4. Pathway analysis of the list of 123 genes whose baseline expression correlated with change in Ki67 in the HER2- tumours by Spearman correlation at a p-value of <0.005.
Additional file 7: Figure S5. Heatmap of Spearman Rank Correlation Matrix (r-value and p -value) of baseline gene signature scores and percentage of 2-week change in Ki67 and residual Ki67 expression. r-values bottom, p-values top. (a) HER2- tumours, n=155. (b) HER2+ tumours, n=23.
Additional file 8: Matrices of correlation coefficient and p value. Table S18. Spearman Rank Correlation Matrix (r-value and p -value) of baseline gene signature scores and percentage of two-week change in Ki67 and residual Ki67 expression. r-values bottom, p-values top. (a) HER2- tumours, n=155 (the Signature name was sorted based on the rho-values of correlations between GS-score and residual.Ki67; (b) HER2+ tumours, n=23. Table S19. Spearman correlations (r-value and p -value) between on-treatment gene signature scores and i) percentage of two-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135; (b) HER2+ tumours, n=22. Table S20. Spearman correlations (r-value and p -value) between change in gene signature scores and i) percentage of two-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135; (b) HER2+ tumours, n=22.
Additional file 9: Figure S6. The 41 of the 902 significantly regulated genes after 2-week of treatment in HER2- tumours, that occurred in at least 3 of the significantly enriched 25 canonical pathways (FDR < 5%). Black-color and grey-color indicates presence and absence of a gene in a pathway. The numbers at the second row on the top of the black and grey image indicated number of pathways that a gene occurred. The numbers at the second column on the left-hand side of the black and grep image indicated number of genes occurred in a pathway.
Additional file 10: Figure S7. Heatmap of Spearman correlations (r-value and p -value) between on-treatment gene signature scores and i) percentage of 2-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135. (b) HER2+ tumours, n=22.
Additional file 11: Figure S8. Heatmap of Spearman correlations (r-value and p -value) between change in gene signature scores and i) percentage of 2-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135. (b) HER2+ tumours, n=22.
Additional file 12: Figure S9. Overrepresented pathways (FDR < 5%) identified by pathway analysis (IPA) of the 71 differentially expressed and annotated genes derived from HER2+ tumours. Negative z-score shown in blue-colour specifies inhibited pathway after AI-treatment. The yellow line indicates the threshold of adjusted p-value < 0.05.
Acknowledgements
We thank all participating patients and staff at POETIC centres and ICR-CTSU who contributed to the trial management and data and sample collection, and the independent data monitoring committee and trial steering committee for their oversight of the trial. This work was supported by the Mary-Jean Mitchell Green Foundation, Breast Cancer Now, working in partnership with Walk the Walk and the National Institute for Health Research (NIHR) Biomedical Research Centre at the Royal Marsden NHS Foundation Trust and The Institute of Cancer Research, London. The POETIC trial (C1491/A8671/CRUK/07/015, C1491/A15955, C406/A8962), from which samples were obtained for this study, was supported by Cancer Research UK as is ICR-CTSU through its core programme grant.
Authors’ contributions
QG analysed the data and drafted the manuscript. ELK extracted the RNA. MC provided the subtypes. JM provided the data and composed Additional file 2: Table S2. VM and AD sectioned and reviewed the histopathology of the samples. KS and DE recorded the samples for the study. AS, HC, EM, EA, and JR were involved in the sample acquisition. MD, IS, and JB were involved in the conception and design of POETIC. LAM, RR, and MD drafted the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
Global gene expression data supporting the finding from this manuscript was deposited at the NCBI gene expression omnibus (GEO) (http://ncbi.nlm.nih.gov/geo/) with accessions of GSE105777 and GSE126870.
Ethics approval and consent to participate
Ethical approval for POETIC (Trial Number CRUK/07/015) was provided by NRES Committee London–South-East. All patients consented to molecular analysis of their samples for research purposes.
Consent for publication
All authors approved the final version of this manuscript.
Competing interests
MD and LAM receive academic funding from Pfizer, Puma Biotechnology Inc., and AstraZeneca. MD receives honoraria from Myriad Genetics and speaker’s bureau of Roche; is a consultant and advisory board member of Radius, GTx, and Orion Pharma; and has received remuneration from the ICR rewards to Inventors Schemes. MCUC has a patent: US Patent No. 9,631,239 with royalties paid. All other authors declare that they have no competing interests.
Footnotes
James Morden is deceased. This paper is dedicated to his memory.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qiong Gao, Email: Alice.Gao@icr.ac.uk.
Elena López-Knowles, Email: Elena.LopezKnowles@icr.ac.uk.
Maggie Chon U. Cheang, Email: Maggie.Cheang@icr.ac.uk
James Morden, Email: James.Morden@icr.ac.uk.
Ricardo Ribas, Email: Ricardo.ribas@icr.ac.uk.
Kally Sidhu, Email: Kally.Sidhu@icr.ac.uk.
David Evans, Email: David.Evans@icr.ac.uk.
Vera Martins, Email: Vera.Martins@icr.ac.uk.
Andrew Dodson, Email: Andrew.Dodson@icr.ac.uk.
Anthony Skene, Email: Anthony.Skene@rbch.nhs.uk.
Chris Holcombe, Email: chris.holcombe@rlbuht.nhs.uk.
Elizabeth Mallon, Email: Elizabeth.Mallon2@ggc.scot.nhs.uk.
Abigail Evans, Email: abigail.evans@poole.nhs.uk.
Judith M. Bliss, Email: Judith.Bliss@icr.ac.uk
John Robertson, Email: John.Robertson@Nottingham.ac.uk.
Ian Smith, Email: ian.smith@rmh.nhs.uk.
Lesley-Ann Martin, Email: Lesley-Ann.Martin@icr.ac.uk.
Mitch Dowsett, Email: Mitchell.Dowsett@icr.ac.uk.
on behalf of the POETIC Trial Management Group and Trialists, Email: poetic-icrctsu@icr.ac.uk.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13058-019-1223-z.
References
- 1.Tao ZiQi, Shi Aimin, Lu Cuntao, Song Tao, Zhang Zhengguo, Zhao Jing. Breast Cancer: Epidemiology and Etiology. Cell Biochemistry and Biophysics. 2014;72(2):333–338. doi: 10.1007/s12013-014-0459-6. [DOI] [PubMed] [Google Scholar]
- 2.Dodson A, Parry S, Ibrahim M, Bartlett JM, Pinder S, Dowsett M, et al. Breast cancer biomarkers in clinical testing: analysis of a UK national external quality assessment scheme for immunocytochemistry and in situ hybridisation database containing results from 199 300 patients. J Pathol Clin Res. 2018;4(4):262–273. doi: 10.1002/cjp2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348(24):2431–2442. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 4.Chia YH, Ellis MJ, Ma CX. Neoadjuvant endocrine therapy in primary breast cancer: indications and use as a research tool. Br J Cancer. 2010;103(6):759–764. doi: 10.1038/sj.bjc.6605845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative G Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386(10001):1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 6.Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, Griffith C, et al. Short-term changes in Ki-67 during neoadjuvant treatment of primary breast cancer with anastrozole or tamoxifen alone or combined correlate with recurrence-free survival. Clin Cancer Res. 2005;11(2 Pt 2):951s–958s. [PubMed] [Google Scholar]
- 8.Ellis MJ, Suman VJ, Hoog J, Goncalves R, Sanati S, Creighton CJ, et al. Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: results from the American College of Surgeons Oncology Group Z1031 Trial (Alliance) J Clin Oncol. 2017;35(10):1061–1069. doi: 10.1200/JCO.2016.69.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype--ACOSOG Z1031. J Clin Oncol. 2011;29(17):2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowsett M, Smith IE, Ebbs SR, Dixon JM, Skene A, A’Hern R, et al. Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99(2):167–170. doi: 10.1093/jnci/djk020. [DOI] [PubMed] [Google Scholar]
- 11.Dunbier A. K., Ghazoui Z., Anderson H., Salter J., Nerurkar A., Osin P., A'hern R., Miller W. R., Smith I. E., Dowsett M. Molecular Profiling of Aromatase Inhibitor-Treated Postmenopausal Breast Tumors Identifies Immune-Related Correlates of Resistance. Clinical Cancer Research. 2013;19(10):2775–2786. doi: 10.1158/1078-0432.CCR-12-1000. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q, Patani N, Dunbier AK, Ghazoui Z, Zvelebil M, Martin LA, et al. Effect of aromatase inhibition on functional gene modules in estrogen receptor-positive breast cancer and their relationship with antiproliferative response. Clin Cancer Res. 2014;20(9):2485–2494. doi: 10.1158/1078-0432.CCR-13-2602. [DOI] [PubMed] [Google Scholar]
- 13.ELo-K QG, Cheang MCU, Morden J, Ribas R, Sidhu K, Evans D, Martins V, Dodson A, Anthony Skene CH, Mallon E, Evans A, Bliss JM, Robertson J, Smith I, Martin L-A, Dowsett M, on behalf of the POETIC Trial Management Group and Trialists Major impact of sampling methodology on gene expression in estrogen receptor–positive breast cancer. JNCI Cancer Spectrum. 2018;2(Issue 2):pky005. doi: 10.1093/jncics/pky005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowsett M, Smith I, Robertson J, Robison L, Pinhel I, Johnson L, et al. Endocrine therapy, new biologicals, and new study designs for presurgical studies in breast cancer. J Natl Cancer Inst Monogr. 2011;2011(43):120–123. doi: 10.1093/jncimonographs/lgr034. [DOI] [PubMed] [Google Scholar]
- 15.Lopez-Knowles E, Gao Q, Cheang MC, Morden J, Parker J, Martin LA, et al. Heterogeneity in global gene expression profiles between biopsy specimens taken peri-surgically from primary ER-positive breast carcinomas. Breast Cancer Res. 2016;18(1):39. doi: 10.1186/s13058-016-0696-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 17.Bosco EE, Wang Y, Xu H, Zilfou JT, Knudsen KE, Aronow BJ, et al. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J Clin Invest. 2007;117(1):218–228. doi: 10.1172/JCI28803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malorni L, Piazza S, Ciani Y, Guarducci C, Bonechi M, Biagioni C, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget. 2016;7(42):68012–68022. doi: 10.18632/oncotarget.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller TW, Balko JM, Fox EM, Ghazoui Z, Dunbier A, Anderson H, et al. ERalpha-dependent E2F transcription can mediate resistance to estrogen deprivation in human breast cancer. Cancer Discov. 2011;1(4):338–351. doi: 10.1158/2159-8290.CD-11-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guerrero-Zotano AL, Stricker TP, Formisano L, Hutchinson KE, Stover DG, Lee KM, et al. ER(+) breast cancers resistant to prolonged neoadjuvant letrozole exhibit an E2F4 transcriptional program sensitive to CDK4/6 inhibitors. Clin Cancer Res. 2018;24(11):2517–2529. doi: 10.1158/1078-0432.CCR-17-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coutant C, Rouzier R, Qi Y, Lehmann-Che J, Bianchini G, Iwamoto T, et al. Distinct p53 gene signatures are needed to predict prognosis and response to chemotherapy in ER-positive and ER-negative breast cancers. Clin Cancer Res. 2011;17(8):2591–2601. doi: 10.1158/1078-0432.CCR-10-1045. [DOI] [PubMed] [Google Scholar]
- 22.Morandi A, Martin LA, Gao Q, Pancholi S, Mackay A, Robertson D, et al. GDNF-RET signaling in ER-positive breast cancers is a key determinant of response and resistance to aromatase inhibitors. Cancer Res. 2013;73(12):3783–3795. doi: 10.1158/0008-5472.CAN-12-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gellert P, Segal CV, Gao Q, Lopez-Knowles E, Martin LA, Dodson A, et al. Impact of mutational profiles on response of primary oestrogen receptor-positive breast cancers to oestrogen deprivation. Nat Commun. 2016;7:13294. doi: 10.1038/ncomms13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Cynthia X., Gao Feng, Luo Jingqin, Northfelt Donald W., Goetz Matthew, Forero Andres, Hoog Jeremy, Naughton Michael, Ademuyiwa Foluso, Suresh Rama, Anderson Karen S., Margenthaler Julie, Aft Rebecca, Hobday Timothy, Moynihan Timothy, Gillanders William, Cyr Amy, Eberlein Timothy J., Hieken Tina, Krontiras Helen, Guo Zhanfang, Lee Michelle V., Spies Nicholas C., Skidmore Zachary L., Griffith Obi L., Griffith Malachi, Thomas Shana, Bumb Caroline, Vij Kiran, Bartlett Cynthia Huang, Koehler Maria, Al-Kateb Hussam, Sanati Souzan, Ellis Matthew J. NeoPalAna: Neoadjuvant Palbociclib, a Cyclin-Dependent Kinase 4/6 Inhibitor, and Anastrozole for Clinical Stage 2 or 3 Estrogen Receptor–Positive Breast Cancer. Clinical Cancer Research. 2017;23(15):4055–4065. doi: 10.1158/1078-0432.CCR-16-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowsett M, Harper-Wynne C, Boeddinghaus I, Salter J, Hills M, Dixon M, et al. HER-2 amplification impedes the antiproliferative effects of hormone therapy in estrogen receptor-positive primary breast cancer. Cancer Res. 2001;61(23):8452–8458. [PubMed] [Google Scholar]
- 27.Ellis MJ, Tao Y, Young O, White S, Proia AD, Murray J, et al. Estrogen-independent proliferation is present in estrogen-receptor HER2-positive primary breast cancer after neoadjuvant letrozole. J Clin Oncol. 2006;24(19):3019–3025. doi: 10.1200/JCO.2005.04.3034. [DOI] [PubMed] [Google Scholar]
- 28.NIH, U.S. National Library of Medicine. https://ghr.nlm.nih.gov/gene/ACADVL. Accessed 26 Nov 2019.
- 29.Bièche I, Olivi M, Noguès C, Vidaud M, Lidereau R. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86(4):580–586. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaklamani V. A genetic signature can predict prognosis and response to therapy in breast cancer: Oncotype DX. Expert Rev Mol Diagn. 2006;6(6):803–809. doi: 10.1586/14737159.6.6.803. [DOI] [PubMed] [Google Scholar]
- 31.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Lynsey McKenzie NM-S, Draper J, Nakjang S, Blair HJ, Wichmann C, Vormoor J, Bonifer C, Lacaud G, Heidenreich O. Identification of CCND2 as a RUNX1/ETO target Required for leukaemic propagation. Blood. 2016;128(22):835. doi: 10.1182/blood.V128.22.835.835. [DOI] [Google Scholar]
- 33.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 34.Mallini P, Lennard T, Kirby J, Meeson A. Epithelial-to-mesenchymal transition: what is the impact on breast cancer stem cells and drug resistance. Cancer Treat Rev. 2014;40(3):341–348. doi: 10.1016/j.ctrv.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 35.Britschgi A, Duss S, Kim S, Couto JP, Brinkhaus H, Koren S, et al. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERalpha. Nature. 2017;541(7638):541–545. doi: 10.1038/nature20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prat A, Cheang MC, Galvan P, Nuciforo P, Pare L, Adamo B, et al. Prognostic value of intrinsic subtypes in hormone receptor-positive metastatic breast cancer treated with letrozole with or without lapatinib. JAMA Oncol. 2016;2:1287–1294. doi: 10.1001/jamaoncol.2016.0922. [DOI] [PubMed] [Google Scholar]
- 37.Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2(2):101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- 38.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 39.Giltnane Jennifer M., Hutchinson Katherine E., Stricker Thomas P., Formisano Luigi, Young Christian D., Estrada Monica V., Nixon Mellissa J., Du Liping, Sanchez Violeta, Ericsson Paula Gonzalez, Kuba Maria G., Sanders Melinda E., Mu Xinmeng J., Van Allen Eliezer M., Wagle Nikhil, Mayer Ingrid A., Abramson Vandana, Gόmez Henry, Rizzo Monica, Toy Weiyi, Chandarlapaty Sarat, Mayer Erica L., Christiansen Jason, Murphy Danielle, Fitzgerald Kerry, Wang Kai, Ross Jeffrey S., Miller Vincent A., Stephens Phillip J., Yelensky Roman, Garraway Levi, Shyr Yu, Meszoely Ingrid, Balko Justin M., Arteaga Carlos L. Genomic profiling of ER+ breast cancers after short-term estrogen suppression reveals alterations associated with endocrine resistance. Science Translational Medicine. 2017;9(402):eaai7993. doi: 10.1126/scitranslmed.aai7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 41.Escudero S, Zaganjor E, Lee S, Mill CP, Morgan AM, Crawford EB, et al. Dynamic regulation of long-chain fatty acid oxidation by a noncanonical interaction between the MCL-1 BH3 helix and VLCAD. Mol Cell. 2018;69(5):729–743. doi: 10.1016/j.molcel.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiseljak-Vassiliades K, Mills TS, Zhang Y, et al. Elucidating the role of the desmosome protein p53 apoptosis effector related to PMP-22 (PERP) in growth hormone tumors. Endocrinology. 2017;158:1450–1460. doi: 10.1210/en.2016-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary information. Additional description of the materials and methods.
Additional file 2. Table S1. Published gene-signatures. Table S2. Study endpoints. Table S3. Study demographics. Table S4. List of patients and their status of HER2, Ki67 and CCCA. Table S5. Ki67 data summary. (A) Percentage change in Ki67 expression after 2-weeks AI-treatment. (B) Anti-proliferative response summary. (C) Summary of number of tumours that reached CCCA. Table S6. Genes whose baseline expression significantly correlated with Change.Ki67 (p < 0.005) in HER2- group. Table S7. Unpaired t-test of significance for the difference between two-group means: i) non-responders vs responders; ii) noCCCAs vs CCCAs in HER2- group. Table S8. Genes whose baseline expression significantly correlated with residual Ki67 (p < 0.005) in HER2- group. Table S9. Pathway analysis of genes in Table S8. Table S10. Genes whose baseline expression is significantly different (p < 0.001) between CCCA and noCCCA in HER2- group. Table S11. Genes significantly regulated by AI with p < 0.005 by univariate Two-sample T-test in HER2- group. Table S12. Top 20 AI-downregulated / AI-upregulated genes, and additional AI-regulated genes of interest identified from HER2- paired tumours. Genes were ranked by fold-change (FC). The Significantly different (p-value) derived from Mann-Whitney test comparing the relative change in expression of a given gene in the HER2- group versus the HER2+ group. ns: p >= 0.05. Table S13. Enriched canonical pathways of the significantly AI-regulated genes in Table S11. Table S14. Correlations of the change in ESR1 / gene-signatures and change in Ki67 / residual-Ki67 in i) HER2- group, ii) HER2+ group, iii) Significance of the difference between the 2 correlation-coefficients. Table S15. Genes significantly regulated by AI with p < 0.005 by univariate Two-sample T-test in HER2+ group. Table S16. Genes whose baseline expression significantly correlated with Change.Ki67 (p < 0.005) in HER2+ group. Table S17. Genes whose baseline expression significantly correlated with residual-Ki67 (p < 0.005) in HER2+ group.
Additional file 3: Figure S1. Consort diagram showing derivation of samples for microarray analysis and Ki67 measurement. In total, RNA was extracted from 861 RNAlater stored core-cuts and 605 RNA samples with RNA integrity number (RIN) >4 and RNA >500 ng were sent for profiling. Samples were excluded due to lack of adequate estradiol suppression, RIN < 4 when profiling, or due to gene expression data of poor quality. “Pairs” indicates a tumour with matched baseline and surgery expression data; B (baseline) indicates a tumour with baseline expression data or baseline Ki67 value; S (surgery) indicates a tumour with surgical expression data or on-treatment Ki67. *Expression QC (quality control): samples with fraction of detection rate <30% or detected by lumi.outlier function from lumi R package as outlier were excluded.
Additional file 4: Figure S2. Heatmap (Pearson, complete) of 123 genes whose baseline expression significantly correlated with Change.Ki67 (p<0.005) based on 155 HER2- of the 178 AI-treated samples. The gene expression across 155 samples was centred and scaled. Red denotes the gene expression in a sample is greater than the mean, blue denotes less than the mean. The tumours are ordered according to the degree of change in Ki67.
Additional file 5: Figure S3. Scatter plot of baseline gene expression and Ki67 values. (a) ESR1 baseline expression and change in Ki67 of HER2- tumours; (b) ESR1 baseline expression and residual Ki67 of HER2- tumours; (c) ESR1 baseline expression and change in Ki67 of HER2+ tumours; (d) ESR1 baseline expression and residual Ki67 of HER2+ tumours.
Additional file 6: Figure S4. Pathway analysis of the list of 123 genes whose baseline expression correlated with change in Ki67 in the HER2- tumours by Spearman correlation at a p-value of <0.005.
Additional file 7: Figure S5. Heatmap of Spearman Rank Correlation Matrix (r-value and p -value) of baseline gene signature scores and percentage of 2-week change in Ki67 and residual Ki67 expression. r-values bottom, p-values top. (a) HER2- tumours, n=155. (b) HER2+ tumours, n=23.
Additional file 8: Matrices of correlation coefficient and p value. Table S18. Spearman Rank Correlation Matrix (r-value and p -value) of baseline gene signature scores and percentage of two-week change in Ki67 and residual Ki67 expression. r-values bottom, p-values top. (a) HER2- tumours, n=155 (the Signature name was sorted based on the rho-values of correlations between GS-score and residual.Ki67; (b) HER2+ tumours, n=23. Table S19. Spearman correlations (r-value and p -value) between on-treatment gene signature scores and i) percentage of two-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135; (b) HER2+ tumours, n=22. Table S20. Spearman correlations (r-value and p -value) between change in gene signature scores and i) percentage of two-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135; (b) HER2+ tumours, n=22.
Additional file 9: Figure S6. The 41 of the 902 significantly regulated genes after 2-week of treatment in HER2- tumours, that occurred in at least 3 of the significantly enriched 25 canonical pathways (FDR < 5%). Black-color and grey-color indicates presence and absence of a gene in a pathway. The numbers at the second row on the top of the black and grey image indicated number of pathways that a gene occurred. The numbers at the second column on the left-hand side of the black and grep image indicated number of genes occurred in a pathway.
Additional file 10: Figure S7. Heatmap of Spearman correlations (r-value and p -value) between on-treatment gene signature scores and i) percentage of 2-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135. (b) HER2+ tumours, n=22.
Additional file 11: Figure S8. Heatmap of Spearman correlations (r-value and p -value) between change in gene signature scores and i) percentage of 2-week change in Ki67 protein expression and ii) residual KI67. r-values bottom, p-values top. (a) HER2- tumours, n=135. (b) HER2+ tumours, n=22.
Additional file 12: Figure S9. Overrepresented pathways (FDR < 5%) identified by pathway analysis (IPA) of the 71 differentially expressed and annotated genes derived from HER2+ tumours. Negative z-score shown in blue-colour specifies inhibited pathway after AI-treatment. The yellow line indicates the threshold of adjusted p-value < 0.05.
Data Availability Statement
Global gene expression data supporting the finding from this manuscript was deposited at the NCBI gene expression omnibus (GEO) (http://ncbi.nlm.nih.gov/geo/) with accessions of GSE105777 and GSE126870.