Abstract
The integrated stress response (ISR) is a conserved pathway which is activated by cells that are exposed to stress. In lung adenocarcinoma (LUAD), activation of the ATF4 branch of the ISR by certain oncogenic mutations has been linked to the regulation of amino acid metabolism. In the present study, we provide evidence for ATF4 activation across multiple stages and molecular subtypes of human LUAD. In response to extracellular amino acid limitation, LUAD cells with diverse genotypes commonly induce ATF4 in an eIF2α dependent manner, which can be blocked pharmacologically using the integrated stress response inhibitor (ISRIB). Although suppressing eIF2α or ATF4 can trigger different biological consequences, adaptive cell cycle progression and cell migration are particularly sensitive to inhibition of the ISR. These phenotypes require the ATF4 target gene asparagine synthetase (ASNS), which maintains protein translation independently of the mTOR/PI3K pathway. Moreover, NRF2 protein levels and oxidative stress can be modulated by the ISR downstream of ASNS. Finally, we demonstrate that ASNS controls the biosynthesis of select proteins, including the cell cycle regulator cyclin B1, which are associated with poor LUAD patient outcome. Our findings uncover new regulatory layers of the ISR pathway and its control of proteostasis in lung cancer cells.
Implications
We reveal novel regulatory mechanisms by which the integrated stress response controls selective protein translation and is required for cell cycle progression and migration of lung cancer cells.
Keywords: Lung adenocarcinoma (LUAD), integrated stress response (ISR), activating transcription factor 4 (ATF4), amino acid starvation, translation
Introduction
Non-small cell lung cancers (NSCLCs) are highly heterogeneous not only in terms of their cellular origins and molecular landscapes, but also in their abilities to adapt to cellular and metabolic stresses (1,2). Oncogenic mutations can induce intrinsic cellular stress (3). Moreover, lung tumor cells may be exposed to extrinsic cellular stressors in the lung tumor microenvironment (TME), which can include amino acid deprivation, glucose starvation, hypoxia, and acidosis at various stages of cancer progression (4). Malignant cells must therefore co-opt different adaptive responses to mitigate these detrimental effects as they expand and disseminate (5). The current standard of care for NSCLC includes immunotherapy and targeted therapies for tumors with a known oncogenic driver mutation (6). However, most NSCLC patients eventually become refractory to these treatments. Identifying and understanding the adaptive responses of lung cancer cells to metabolic stress may unveil additional therapeutic vulnerabilities of NSCLCs.
One highly conserved stress-induced pathway is the integrated stress response (ISR) (7). The ISR maintains proteostasis by halting global translation when environmental conditions are such that continued translation would be detrimental to cellular fitness. The ISR consists of four kinases which sense unique stressors and phosphorylate eIF2α, attenuating cap-dependent translation (8). The sensor PERK is activated by an accumulation of unfolded protein in the endoplasmic reticulum generated by metabolic stresses such as glucose deprivation and hypoxia, while the sensor GCN2 is activated by an accumulation of uncharged transfer RNAs (tRNAs) produced as a consequence of amino acid deprivation. Under nutrient stress, attenuation of global translation also allows for selective translation of the transcription factor ATF4 through bypass of an upstream open reading frame (uORF) (9,10). ATF4 activates diverse transcriptional programs to either regain homeostasis or, in the setting of chronic stress, trigger cell death (11,12). The adaptive consequences of ISR activation are highly context dependent and can be influenced by cell type, the nature of the stress, the sensor, and the amplitude and duration of the stress.
Several cancers types have been shown to be regulated by ATF4 and the ISR. For instance, genetic suppression of ATF4 in fibrosarcoma and prostate cancer results in tumor cell apoptosis (13,14). Conversely, PERK and ATF4 can inhibit tumor progression in melanoma (15). In NSCLC, tumors driven by KRAS mutations can activate ATF4 upon nutrient depletion (16). However, it remains unclear if ATF4 can regulate other molecular subtypes of lung cancer. Importantly, given the context dependent consequences of ISR activation, there remains a need to determine which of its effector functions are required for the fitness of lung cancer cells at different stages of tumor progression.
Materials and Methods
Cell lines and culture
Cell lines were cultured as recommended by ATCC and routinely tested for mycoplasma using the Universal mycoplasma detection kit (#30–1012k). Cells were cultured in RPMI 1640 (Thermo Fisher Scientific #11875093) containing 10% fetal bovine serum (Thermo Fisher Scientific #10437–028), 1% penicillin-streptomycin (Thermo Fisher Scientific #15140122), and 0.2% amphotericin B (Sigma Aldrich #A2942). Treatment media was prepared by adding back all constituents (Sigma #LAA21–1kt and #G7021), except those indicated, to RPMI 1640 without glucose and amino acids (US Biological #R9010–01). Clonogenic, cell viability, anoikis, bivariate cell cycle analysis, cleaved caspase-3 staining, CellROX, transwell migration assays, and scratch assays were performed as described in Supplementary Materials and Methods.
shRNA and cDNA expression
Independent shRNAs (Dharmacon) against ATF4 (a and b) or ASNS were subcloned into pINDUCER10 (17). See Supplementary Materials and Methods for sequences. ASNS (#OHS5897–202616233), ATF4 (#OHS5899–202616733), and RFP-expressing control (#OHS5832) constructs (pLOC) were purchased from Dharmacon. For both knockdown and overexpression, lentivirus was generated using standard calcium phosphate transfection protocols, concentrated, titered, and used to infect cell lines using standard methods (MOI = 0.5–4). Infected cells were selected using puromycin (for pINDUCER10) or blasticidin (for pLOC) over 3 or 7 days respectively. For shRNA induction, cells were incubated with 1 μg/mL doxycycline for 1–5 days.
Gene set enrichment analysis (GSEA) and pathway analysis
For GSEA analysis (18), a gene list consisting of 472 ATF4 target genes as defined by Han et al. (19) was used. Pre-ranked gene lists of ANOVA analysis for indicated comparisons were generated for either all TCGA LUADs (n = 489 tumors) (20), the TCGA Nature Core samples (n = 230 tumors and 45 matched normal tissues which include exome sequencing), or the Director’s Challenge Cohort of LUADs (n = 442) (21) where appropriate. DAVID analysis of leading edge genes from the GSEA analysis was performed as previously described (22). Additional details provided in Supplementary Materials and Methods.
Quantitative real time-PCR
Total RNA was extracted using an RNeasy kit (Qiagen #74106) and 1 μg used to generate cDNA with an iScript cDNA Synthesis Kit (Bio-Rad #1708890). cDNA was diluted 1:10, mixed with Fast SYBR Green master mix (Thermo Fisher Scientific #4385614), and technical quadruplicates were amplified and measured using a ViiA 7 Real-Time PCR machine (Thermo Fisher Scientific).
Western blotting
Cells were rinsed with PBS and lysed directly in the plate, using RIPA buffer, protease inhibitors (Roche # 11836170001), and phosphatase inhibitors (Sigma #P5726 and #P0044). Cells were incubated on ice for 30 min, vortexing every 10 min. Lysates were clarified by centrifugation for 15 min. Protein was quantified using the DC Protein Assay (Bio-Rad # 500–0112) and analyzed by SDS-PAGE using the Mini-PROTEAN system (Bio-Rad). Protein was transferred to either nitrocellulose or PVDF and membranes blocked using 5% milk in TBST (0.1% Tween20). Blots were incubated with primary antibodies at 4°C overnight, then HRP-secondary antibodies for 1 hr at room temperature. ECL was used to develop blots, and they were imaged using either a KwikQuant imaging system (Kindle Biosciences) or ChemiDoc Imaging System (Bio-Rad).
RNA sequencing and pathway analysis
RNA sequencing was performed by the Yale Center for Genome Analysis. Subsequent ANOVA analysis of all genes significantly changed (FDR < 0.05 by Benjamini–Hochberg step-up method) by at least 1.5 fold was performed using Partek Genomics Suite (Partek). All data are deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE126232. Ingenuity® Pathway Analysis software (Qiagen) was used to predict changes in upstream regulators and canonical pathways. Additional details provided in Supplementary Materials and Methods.
Translation assay
Cells were starved of L-methionine for 30 min and subsequently incubated with 50 μM homopropargylglycine (HPG) (Life Technologies #C10186) for 1–4 hr in treatment media. Cells were then trypsinized and a Click-IT kit (Life Technologies #C10269) used to label HPG. Labeled cells were analyzed using an LSR II flow cytometer (BD Biosciences) and data was processed using FlowJo® (FlowJo, LLC). Translation rates were determined based on the slope of HPG incorporation over time.
Reverse phase protein arrays (RPPA)
RPPA was performed by the MD Anderson Reverse Phase Protein Array Core Facility. Of the antibodies plotted in Figure 5, FOXO3A antibodies were annotated as “use with caution.” Partek Genomic Suite® software (Partek, Inc.) was used for ANOVA analysis (Supplementary Table 4). Proteins whose expression changed by at least 1.5 fold with FDR < 0.05 by Benjamini–Hochberg step-up method were considered significantly changed. A heap map showing representative proteins that significantly changed was generated using Prism software (GraphPad). Supplementary Table 4 was filtered for targets whose protein levels changed in the RPPA, but whose corresponding mRNA levels from the RNA sequencing data remained unchanged. Independently, Spearman correlation of ASNS expression in the TCGA with corresponding RPPA levels was performed for all LUAD samples with available RPPA data (n = 181) (20) in order to determine proteins that clinically correlate with ASNS expression. Targets that changed at the protein but not RNA levels in our experiments were plotted against the corresponding correlation between protein levels and ASNS expression from the TCGA. Additional details are available in Supplementary Materials and Methods.
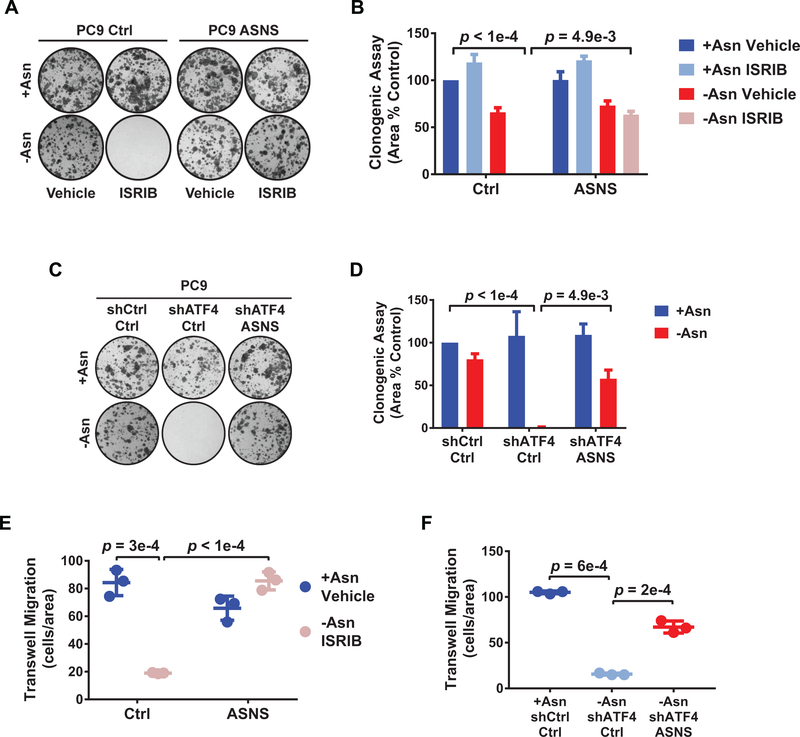
Figure 5. ASNS-mediated protein translation is sufficient for adaptive cell proliferation and migration.
A) Representative images of clonogenic assay of the indicated PC9 cell lines treated as shown. B) Quantification of A). Error bars show SEM. C) Representative images of clonogenic assay of the indicated PC9 cells in + Asn or -Asn conditions. D) Quantification of C). Error bars show SEM. E) Trans-well migration assay of PC9 cells expressing empty vector control (Ctrl) or ASNS in treatment media with 0.2% FBS and using treatment media with 10% FBS as a chemoattractant for 24 hr. Migrating cells stained for DAPI and quantified using ImageJ. F) Trans-well migration assay of the indicated PC9 cell lines treated as indicated for 24 hr and quantified as in E). n = 3 for all experiments, error bars show SD, and p-values by Unpaired t-test.
Animal studies
All work was done in accordance with Yale Institutional Animal Care and Use Committee policies (protocol #201611338). Isonitrogenous, amino acid-defined chow containing (#TD01084) or lacking (#TD160365) asparagine was purchased from Envigo. For experiments that involved knockdown, the mouse chow also contained 625 ppm doxycycline (#TD160774 and #TD160775). Trans-ISRIB (Tocris #5284) was sterilely dissolved using the following formulation: 5:2:20:73 v/v DMSO:Tween80:PEG400:D5W. Cells were suspended in sterile PBS and mixed 1:1 with growth factor reduced-matrigel (Corning #356231) and 100 μL, containing 5e4 cells, was injected subcutaneously in the flanks of 4–5 weeks old male athymic nu/nu mice (Charles River #088). Tumor volume was measured using calipers and calculated using the formula: tumor volume = (major axis)(minor axis2) x 0.52.
Statistical Analysis
Data was presented as mean ± SEM or SD, as indicated, with p-values calculated by two-tailed Student’s t-test for in vitro work and Mann-Whitney test for in vivo work. FDR (Benjamini–Hochberg step-up method) was used for multiple test correction where appropriate. Statistics were performed using Prism software (GraphPad) or Partek Genomics Suite (Partek).
Results
ATF4-regulated amino acid stress response is activated in advanced stage LUAD and can be inhibited by ISRIB
To understand the biological and mechanistic context of ISR regulation in human lung cancer, we compared the activation status of this pathway across different stages and subtypes of primary human lung adenocarcinoma (LUAD) (20), which is the most frequently diagnosed type of NSCLC. We inferred ISR pathway activation by performing gene set enrichment analysis (GSEA) using conserved transcriptional targets of ATF4 (18,19). We did not observe a bias in ATF4 target gene activation when comparing LUADs expressing several major driver mutations or as a function of mutation burden, although we note that KRAS mutant tumors had higher ATF4 target gene expression when compared to EGFR mutant tumors specifically (Fig. 1A). ATF4 targets genes were significantly enriched in late stage (III/IV) versus early stage (I/II) LUADs (Fig. 1A). ATF4 genes were also preferentially activated in transcriptional subtypes of LUAD with poor prognosis and a high probability for relapse (e.g. proximal inflammatory and distal alveolar stem cell-like tumors (20,23)) (Fig. 1A and Supplementary Fig. S1A).
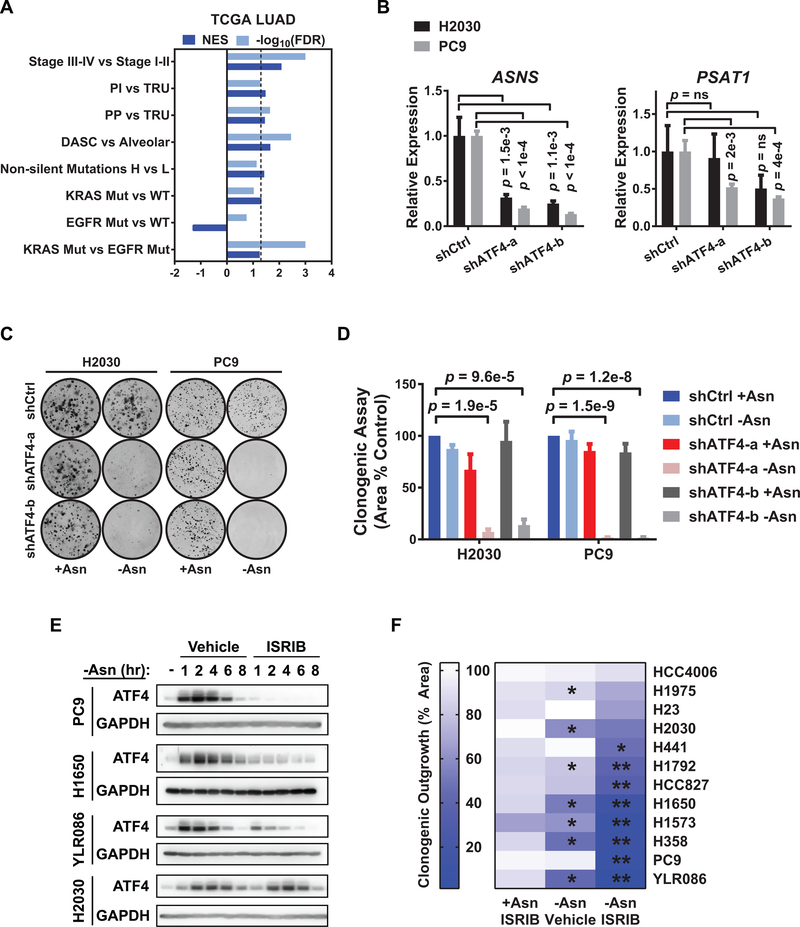
Figure 1. The ISR is activated in diverse molecular subtypes and stages of LUAD and can be inhibited by ISRIB.
A) Gene set enrichment analysis (GSEA) of ATF4 targets in LUAD samples from the TCGA (either n = 489 or n = 275 with paired RNA seq and whole exome sequencing data. Comparisons are stage III-IV (late) versus I-II (early) disease, proximal inflammatory (PI) versus terminal respiratory unit (TRU) tumors, proximal proliferative (PP) versus TRU tumors, DASC-like versus alveolar-like tumors, non-silent mutation high versus low tumors, KRAS mutant versus wildtype tumors, EGFR mutant versus wildtype tumors, and KRAS versus EGFR mutant tumors. NES (normalized enrichment score) and -log10(FDR) are listed for each analysis. Dashed line is FDR < 0.05. B) qRT-PCR of ATF4 target genes involved in amino acid biosynthesis in the H2030 and PC9 cell lines with doxycycline-induced control shRNA or shRNAs against ATF4. Data for two independent shRNAs (a and b) are shown. Relative expression normalized to GUSB with upper and lower limits of a representative experiment shown; n = 2. ns = not significant. C) Representative images of clonogenic assay with the indicated cells plated in +Asn or -Asn media. D) Quantification of C) showing mean and SEM. E) Representative western blots for ATF4 and GAPDH (loading control) from PC9, H1650, YLR086, and H2030 cells following incubation in -Asn media with vehicle (DMSO) or 200 nM ISRIB for the indicated times. F) Heat map depicting quantification of clonogenic assay of a panel of LUAD cell lines treated as indicated. +Asn ISRIB: ISRIB treatment alone. -Asn Vehicle: asparagine deprivation alone. -Asn ISRIB: asparagine deprivation with ISRIB treatment (dual treatment). Asterisks indicate FDR < 0.05 for indicated condition versus (*) +Asn Vehicle or (**) -Asn Vehicle. Unless otherwise noted n = 3 and p-values by Unpaired t-test.
Increased levels of phosphorylated eIF2α (p-eIF2α) have been observed in human lung cancer tissue (24). Moreover, in late stage LUADs, activated ATF4 targets were enriched for genes involved in amino acid metabolism and, in particular, serine and asparagine biosynthesis (Supplementary Fig. S1B and Supplementary Table 1). These findings suggest that adaptive responses to amino acid supply may be a major determinant of ISR activation throughout tumor progression and across oncogenic subtypes of LUAD. Accordingly, we tested the role of the ISR in the H2030 and PC9 LUAD cell lines which are dependent on prototypical oncogenic mutant KRAS or mutant EGFR, respectively, for their viability. Reducing ATF4 using two independent shRNAs (a and b) (Supplementary Fig. S1C) robustly decreased the expression of ASNS, which drives asparagine biosynthesis, and more modestly reduced PSAT1, a gene in involved in serine biosynthesis (Fig. 1B). ATF4 knockdown did not affect the outgrowth of these cell lines under complete amino acid conditions (+Asn or +Ser), but concurrent removal of asparagine or serine from the media (-Asn or -Ser) significantly reduced their clonogenicity (Fig. 1C and 1D and Supplementary Fig. S1D and S1E), as has been shown in other types of cancers (13).
To ascertain the dependency of LUAD cells on the ATF4 regulator eIF2α, we tested the effects of the integrated stress response inhibitor (ISRIB) on cell outgrowth. ISRIB is a small molecule that targets eIF2α’s guanine exchange factor, eIF2B, stabilizing it in an activate state (25,26). ISRIB can maintain global translation, thus blocking ATF4 translation, in the presence of cellular stress (27). We confirmed that asparagine starvation increases eIF2α phosphorylation and ATF4 protein levels and that the latter can be blocked by ISRIB in many, but not all, cell lines (Supplementary Fig. S1F and Fig. 1E). We next evaluated the effects of ISRIB on the outgrowth of a panel of LUAD cell lines, including tumor cells derived from a treatment refractory LUAD patient (YLR086). Cells were cultured under ISRIB treatment in the presence or absence of asparagine. Whereas ISRIB treatment alone (+Asn ISRIB) (Fig. 1F, left column) had no significant effect, several lines demonstrated modest decreases in outgrowth upon asparagine deprivation alone (-Asn Vehicle) (Fig. 1F, middle column). Combining asparagine limitation with ISRIB treatment (-Asn ISRIB; dual treatment) inhibited the outgrowth of most of the lines tested (8 out of 12), with some lines responding additively or selectively (e.g. PC9, HCC827, and H441) to the dual treatment (Fig. 1F, right column).
ISRIB did not further sensitize PC9 cells to the deprivation of serine, glutamine, or glucose (Supplementary Fig. S1G–S1I), indicating that LUAD cells can be preferentially dependent on the ISR when extracellular asparagine is limiting. A smaller proportion of LUAD cell lines were partially or fully resistant to ISRIB under asparagine deprivation (e.g. H2030, H1975, H23, and HCC4006) (Fig. 1F). For the H2030 cell line, although its clonogenicity required ATF4 induction under asparagine starvation (Fig. 1C), ATF4 levels were not reduced by ISRIB (Fig. 1E), suggesting that the pharmacological activity of ISRIB or inhibition of its target may be abated in this line. Recent studies suggest that asparagine is an essential amino acid for some tumor cells specifically as extracellular glutamine levels become limiting (28,29). Hence, we also tested ISRIB on two other resistant lines, HCC4006 and H1975, when deprived of both asparagine and glutamine. In this setting, ISRIB had an additive effect on the growth inhibition of both cell lines (Supplementary Fig. S1J). In summary, despite variable adaptive responses to amino acid stress, most LUAD cells can be sensitized to asparagine limitation by targeting eIF2α and ATF4.
Pharmacological inhibition of the ISR arrests the proliferation and motility of LUAD cells during amino acid deprivation
Inhibition of the ISR causes a variety of cell biological effects in a context dependent manner. By using the well-characterized PC9 LUAD cell line, dual treatment was shown to significantly decrease viability in an ISRIB dose dependent manner (Fig. 2A and 2B). Bi-variate cell cycle analysis demonstrated that dual treatment caused an arrest in G2/M phases of the cell cycle by 72 hours (Fig. 2C). Apoptosis, as measured by cleaved caspase-3 staining, increased slightly, but only after 9 days of treatment (Fig. 2D). These data suggests that the proximal effects of dual treatment are cytostatic. Moreover, growth inhibition was only partially reversed by either halting ISRIB treatment (ISRIB removed) or replenishing the cells with extracellular asparagine (Asn added) (Fig. 2E). Clonogenic outgrowth under dual treatment could also be rescued by ectopic expression of ATF4 (Supplementary Fig. S2A and S2B).
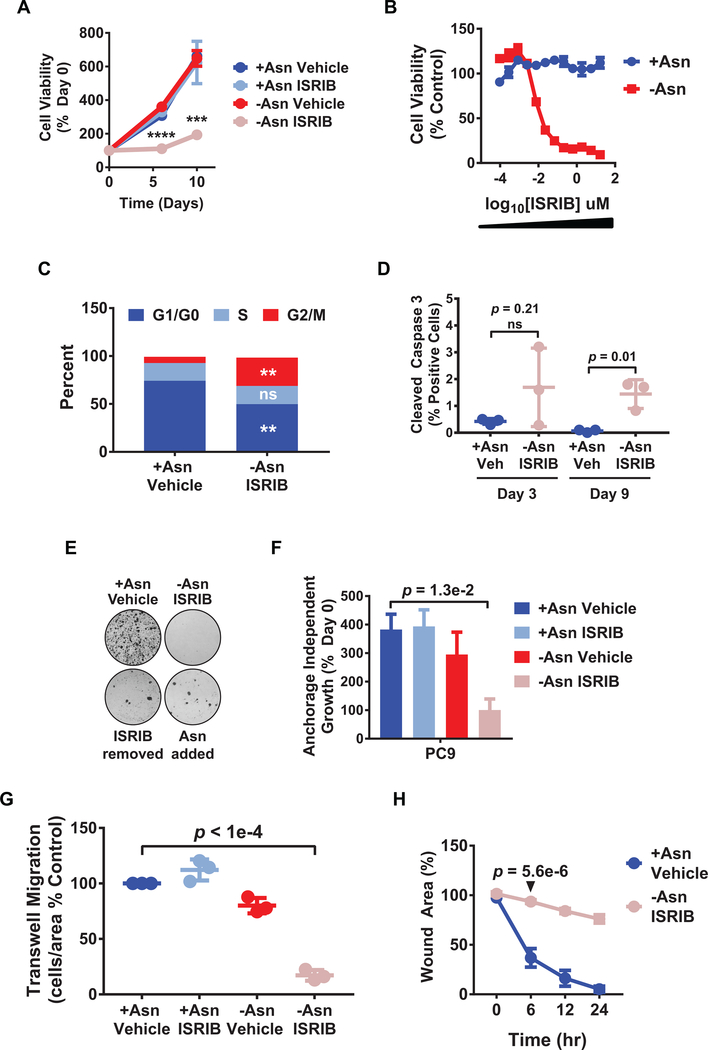
Figure 2. ISRIB arrests the adaptive proliferation and motility of LUAD cells under amino acid stress.
A) PC9 cell viability under the indicated treatments was measured over time by resazurin. ****p = 5.6e-6, ***p = 1.9e-4. B) Drug dose curve showing cell viability over a range of ISRIB concentrations in +Asn or -Asn media on day 9. C) Bivariate cell cycle analysis using BrdU and 7-AAD as analyzed by flow cytometry following 3 days of indicated treatment. Data was plotted as % of total tumor cells in a particular phase of the cell cycle. D) Staining for cleaved caspase-3 in PC9 cells treated as indicated with percent positive cells of total cells per sample shown. E) Clonogenic assays; top wells were treated for 9 days as indicated and harvested. Bottom wells were first treated for 9 days with –Asn/ISRIB and then either ISRIB was removed (ISRIB removed) or asparagine replenished (Asn added) for an additional 9 days before harvest. Representative images shown. F) Cells were cultured for 3 days under anchorage-independent growth conditions and indicated treatments. Cell viability was performed as in A); n = 2. G) Trans-well migration assay with cells plated in the indicated treatment media containing 0.2% FBS and using the corresponding treatment media containing 10% serum as a chemoattractant. After 24 hr, migrating cells were stained for DAPI, immunofluorescent images captured, and quantified. H) Scratch assay to measure cell migration over time under the indicated conditions. Values indicate remaining wound area as a percentage of initial wound area. p-value shown for 6 hr treatment. Unless indicated otherwise, n = 3, error bars show SEM, and p-values by Unpaired t-test.
Given that ATF4 activity may be increased in LUAD subtypes with a higher probability of metastatic relapse (Fig. 1A), we also evaluated the effects of dual treatment on anchorage independent growth and cell motility, two cellular phenotypes that are regulated by amino acid accessibility or metabolic stress (30,31) and are required for tumor cell dissemination. When PC9 cells were cultured in suspension, their growth was significantly inhibited by dual treatment (Fig. 2F). Moreover, a significant decrease in the migratory ability of PC9 cells was observed for dual treated cells (Fig. 2G and 2H and Supplementary Fig. S2C and S2D). We conclude that ISRIB also blocks adaptive anchorage independent tumor cell growth and motility in response to amino acid stress.
Direct and indirect molecular effects of ATF4 inhibition and ISRIB during asparagine starvation
Since eIF2α can regulate other stress response proteins in addition to ATF4 (7), we next comprehensively analyzed the molecular effects of ISRIB treatment. Based on the kinetics of ATF4 activation in PC9 cells starved of asparagine, we analyzed by RNA sequencing (RNAseq) the acute (6 hr) and long-term (24 hr) effects of ISRIB treatment, amino acid deprivation, or dual treatment. ISRIB alone did not affect the transcriptome of PC9 cells, consistent with its minimal biological effects in the absence of stress (Supplementary Table 2). Following 6 hr of asparagine deprivation, 42 genes were downregulated, while 284 genes were up-regulated by at least 1.5 fold (FDR < 0.05) of which 48 are known ATF4 targets (Fig. 3A). 24 hr following asparagine starvation alone, the expression of known ATF4 targets returned to baseline levels, with ASNS being the only target to remain relatively elevated. Of the acutely induced ATF4-targets, 42 were blocked by ISRIB (Fig. 3A). Pathway analysis identified ATF4 as well as the eIF2α kinases PERK and GCN2 as potential regulators of these targets (Fig. 3B and Supplementary Table 3), confirming that the ISR pathway is directly inhibited by ISRIB in LUAD cells. Genes that are acutely regulated by asparagine starvation and blocked by ISRIB include regulators of amino acid biosynthesis (ASNS), autophagy (MAP1LC3B), and amino acid transport (SLC7A11) (Fig. 3C). ASNS was also confirmed to be inhibited by ATF4 knockdown under similar conditions (Supplementary Fig. S3A).
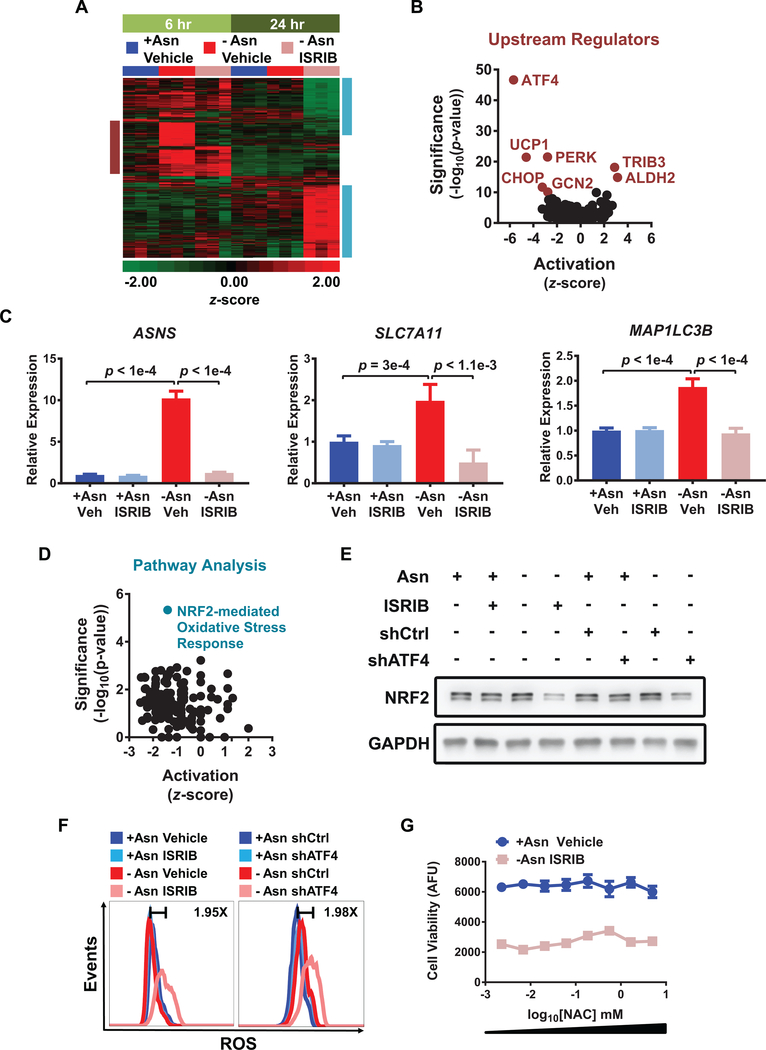
Figure 3. Molecular effects of ATF4 inhibition and ISRIB during asparagine starvation.
A) RNAseq of PC9 cells treated as indicated for either 6 hr (light green) or 24 hr (dark green). The heat map depicts the z-scores for genes that were significantly changed (FDR < 0.05 and fold change < −1.5 or > 1.5) in any treatment comparisons (6 hr or 24 hr). The burgundy box highlights acute changes activated by the ISR at 6 hr, while the aqua box highlights changes associated with chronic asparagine starvation at 24 hr. Scale narrowed to −2 to +2 for increased contrast. B) Volcano plot of IPA upstream regulators analysis for -Asn ISRIB versus -Asn Vehicle at 6 hr. C) qRT-PCR of representative ATF4 targets in PC9 cells treated as indicated for 6 hr; veh = vehicle. Relative expression normalized to GUSB with upper and lower limits shown as representative experiment. D) Volcano plot of pathway analysis for -Asn ISRIB versus +Asn Vehicle at 24 hr. E) Western blotting for NRF2 and GAPDH (loading control) in PC9 cells treated as indicated. F) Flow cytometry analysis of ROS generation using CellROX probe in PC9 cells treated as indicated for 48 hr. Fold changes between controls (dark blue) and dual treatments (light pink) are indicated. Representative experiment depicted; n = 2. G) Cell viability of cells treated as indicated for 6 days with the addition of increasing concentrations of NAC. AFU is arbitrary fluorescence units. Error bars show SEM; n = 2. Unless otherwise noted, n = 3 and p-values by Unpaired t-test.
Long-term dual treatment increased the number of differentially expressed genes (604 up-regulated and 396 down-regulated) (Fig. 3A) in LUAD cells and was predicted to inhibit several signaling pathways, the most significant of these being the NRF2-mediated oxidative stress response (Fig. 3D and Supplementary Table 3). Previous studies suggest that NRF2 functions upstream or in conjunction with ATF4 to activate genes that control reactive oxygen species (ROS) and amino acid metabolism (32,33). Given the decrease in NRF2 targets by dual treatment, we evaluated mRNA and protein levels of NFE2L2/NRF2 itself. Although NFE2L2 was induced by asparagine limitation, this induction was not affected by ISRIB or ATF4 knockdown (Supplementary Fig. S3B). However, under stress, NRF2 protein levels were decreased by ISRIB or ATF4 knockdown (Fig. 3E), and this reduction correlated with decreased expression of the NRF2 target gene HMOX1 (Supplementary Fig. S3C). These results indicate that, upon amino acid starvation, NRF2 protein levels and activity can be indirectly regulated downstream of ATF4.
Oxidative stress and decreased autophagy are two potential consequences of perturbed ATF4 and NRF2 activity (34,35). Indeed, in PC9 cells, asparagine deprivation combined with genetic or ISRIB-mediated reduction of ATF4 led to increases in intracellular ROS (Fig. 3F). However, although n-acetylcysteine (NAC) treatment reduced ROS back to baseline levels, it was not sufficient to restore cell proliferation (Supplementary Fig. S3D and Fig. 3G). We also measured autophagic flux by monitoring the accumulation of microtubule-associated proteins 1A/1B light chain 3B (LC3-II) in the presence of lysosomal protease inhibitors. Neither ISRIB treatment nor ATF4 knockdown consistently affected LC3-II accumulation under the conditions tested (Supplementary Fig. S3E and S3F). As such, the regulation of ROS may be indirectly affected by long-term eIF2α or ATF4 inhibition, but neither ROS accumulation nor autophagy are required for the inhibition of LUAD cell growth during asparagine starvation.
ASNS-mediated protein translation is necessary and sufficient for adaptive cell proliferation and migration
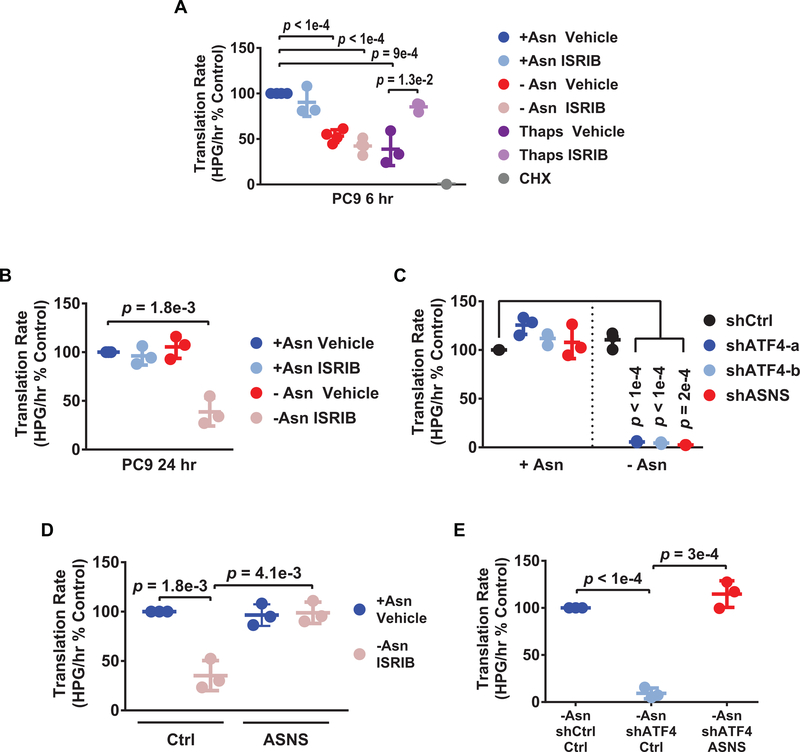
Another hallmark of the ISR is attenuation of global protein translation, through phosphorylation of eIF2α (7). We thus ascertained the effects of asparagine starvation and ISRIB on translation in LUAD cells. Protein translation rates were decreased following 6 hr of asparagine deprivation in the PC9 cell line, but were not rescued by ISRIB (Fig. 4A). This is in contrast to the ability of ISRIB to restore translation in cells undergoing ER stress via thapsigargin treatment (Fig. 4A). By 24 hr, translation rates returned to baseline following asparagine starvation alone, whereas translation rates remained inhibited in dual treated cells (Fig. 4B). Knockdown of ATF4 with asparagine deprivation also led to more pronounced decreases in translation (Fig. 4C). Finally, similar findings were observed in other cell lines, where ATF4 was reduced pharmacologically using ISRIB (H1650 and YLR086) or by gene knockdown (H2030) (Supplementary Fig. S4A–S4C). This suggests that sustained inhibition of translation in LUAD cells is a broader consequence of ISR inhibition downstream of eIF2α and ATF4.
Figure 4. ASNS mediates protein translation during asparagine deprivation.
A-E) Translation rates as measured by incorporation of the methionine analog homopropargylglycine (HPG) over time and evaluated by flow cytometry. Each data point represents the slope of HPG incorporation for at least four time points using median fluorescence intensity (MFI) from an independent experiment. A) PC9 cells following 6 hr treatment as indicated. Thaps = 100 nM thapsigargin. CHX = 100 μg/mL cycloheximide, used as a control to completely block de novo protein translation. B) PC9 cells following 24 hr treatment as indicated. C) PC9 cells expressing the indicated shRNAs in +Asn or -Asn conditions for 24 hr. D) PC9 cells expressing control shRNA (Ctrl) or ectopic ASNS (ASNS) treated as indicated for 24 hr. E) PC9 cells expressing the indicated shRNAs with control or ectopic ASNS expression cultured in -Asn for 24 hr. n = 3 for all experiments, error bars show SD, and p-values by Unpaired t-test.
Mammalian cells do not catabolize asparagine, but instead use asparagine primarily for protein translation (28,36). We therefore tested the dependence of LUAD cells on the sole asparagine biosynthetic enzyme ASNS, which is a target of ATF4 and regulated by stress (37). Knockdown of ASNS in the PC9 cells similarly decreased their translation rates during asparagine deprivation (Fig. 4C), phenocopied the decreased clonogenicity caused by ATF4 knockdown, and recapitulated the increase in intracellular ROS (Supplementary Fig. S4D–S4F). Importantly, forced expression of ASNS (Supplementary Fig. S4G and S4H) could restore protein translation when ATF4 was reduced by ISRIB treatment or ATF4 knockdown in the PC9 cells (Fig. 4D and 4E). Rescue of protein translation by ASNS expression was also sufficient to restore the clonogenicity (Fig. 5A–5D) and motility (Fig. 5E and 5F) of PC9 cells under asparagine deprivation. Finally, ASNS was also required by the H2030 cell line during asparagine starvation and ASNS expression downstream of ATF4 was also necessary and sufficient for adaptive protein translation and proliferation in this model system (Supplementary Fig. S5A–S5E). We conclude that asparagine bioavailability and protein translation are the primary ATF4 effector functions which mediate adaptive cell migration and proliferation by LUAD cells in response to amino acid stress.
ASNS mediates the synthesis of cell cycle regulators
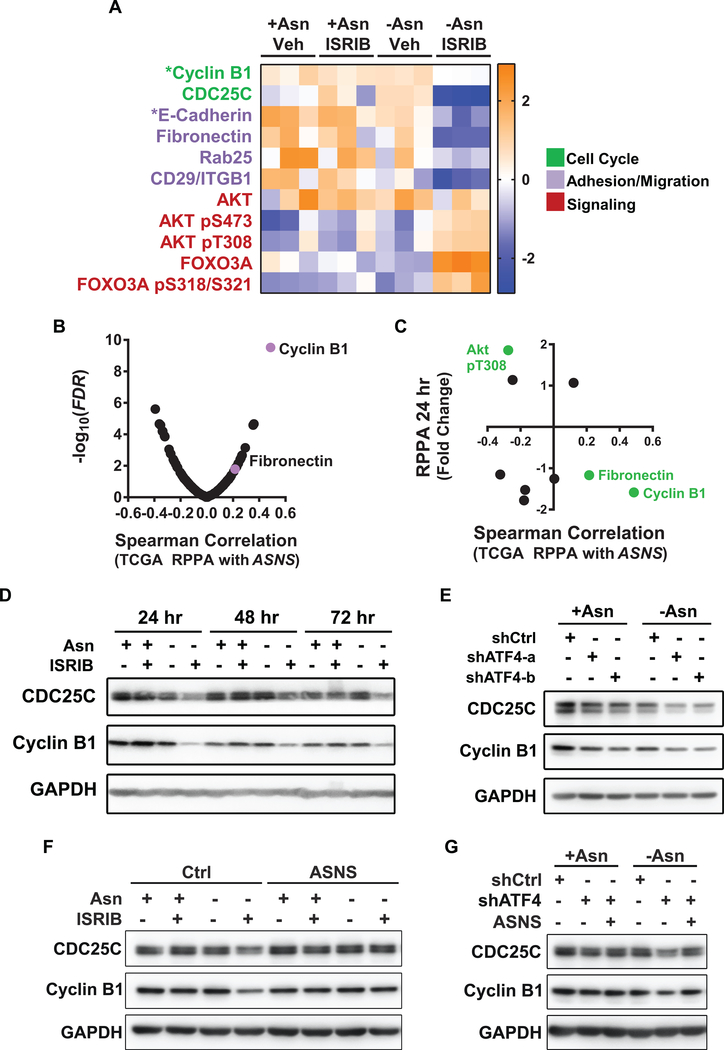
To identify steady state changes in protein and phospho-protein levels resulting from dual treatment, we performed Reverse Phase Protein Array (RPPA). No significant changes were observed in either the ISRIB or asparagine deprived conditions alone. However, upon dual treatment, subsets of 26 or 54 proteins (out of 239) and 10 or 18 phospho-proteins (out of 65) were increased or decreased by 1.5 fold (FDR p < 0.05) at 24 hr or 72 hr respectively (Supplementary Table 4).
Several of these proteins are involved in cell cycle regulation, cell adhesion/migration, and signal transduction. Specifically, activation of AKT and its target FOXO3A were increased upon dual treatment (Fig. 6A; red). Western blotting confirmed increased phosphorylation of AKT under acute asparagine deprivation that returned to baseline by 24 hr, but was maintained in the presence of dual treatment (Supplementary Fig. S6A). While no changes in mTOR phosphorylation or its downstream target 4E-BP1 were observed, phosphorylation of p70S6K, which phosphorylates S6 downstream of mTORC1, was sustained under dual treatment (Supplementary Fig. S6A). These changes correlated with inhibition of TRIB3 and DDIT4 (Supplementary Fig. S6B), two ATF4 targets which are known to inhibit AKT and mTORC1, respectively, during cellular stress (38,39). Nevertheless, alleviation of these ATF4 feedback mechanisms and activation of the PI3K/mTOR pathway did not restore protein translation under dual treatment (Fig. 4B).
Figure 6. Cyclin B1 protein synthesis is regulated by ASNS in LUAD cells under stress.
A) Heat map of reverse phase protein array (RPPA) for PC9 cells treated as indicated for 24 hr (indicated by asterisk) or 72 hr. Representative proteins that were significantly increased or decreased by dual treatment (FDR < 0.05 and 1.5 fold change) are plotted as z-scores. Functional categories indicated as cell cycle (green), cell adhesion/migration (purple), and signaling (red). B) Volcano plot of Spearman correlation between ASNS expression and protein levels as evaluated by RPPA, using LUADs from the TCGA with matching RPPA and RNA seq data (n = 181). C) Proteins that were differentially expressed in -Asn ISRIB versus +Asn Vehicle groups at 24 hr, but whose corresponding mRNA levels remained unchanged were identified. Fold change for these proteins were plotted against corresponding Spearman correlations between TCGA RPPA and ASNS from B). Green data points have a Spearman correlation with FDR < 0.05. D-G) Representative western blots for CDC25C, cyclin B1, and GAPDH (loading control) in PC9 cells lines modified and treated as indicated for D) 24, 48, or 72 hr, E) 72 hr, or F-G) 24 hr. Representative westerns shown, n = 3 for all experiments.
To further identify proteins whose translation and/or synthesis are regulated by the ISR, we integrated RPPA and RNA-seq data, focusing on proteins that were significantly altered by dual treatment independently of changes in their mRNA levels. This included key regulators of cell cycle progression (CDC25C and cyclin B1 (40)) as well as cell adhesion and migration (Rab25 (41), E-Cadherin, fibronectin and integrin beta 1/CD29 (42)) (Fig. 6A). To determine if these proteomic alterations correlated with ASNS expression in human LUAD, we identified proteins that were significantly altered under dual treatment and most positively associated with ASNS mRNA levels in human LUAD from TCGA. Among these, phosphorylation of AKT was negatively correlated with ASNS, whereas cyclin B1 and fibronectin (FN) levels were positively correlated with ASNS in human LUADs (Fig. 6B and 6C). As CDC25C and cyclin B1 are required for progression through the G2/M phase of the cell cycle and cyclin B1 expression in human lung cancers correlates with poor patient outcome (43), western blotting was used to confirm their expression in the PC9 cells. Asparagine deprivation and/or inhibition of ATF4 alone had mild effects on CDC25C and cyclin B1 levels (Fig. 6D and 6E). When combined with asparagine deprivation, ISRIB treatment, ATF4 knockdown or ASNS knockdown more significantly decreased both proteins (Fig. 6D and 6E and Supplementary Fig. S6C). mRNA levels of CCNB1 remained unchanged following ATF4 knockdown (Supplementary Fig. S6D). In addition, the rates of cyclin B1 turn-over did not correlate with the reduction in steady-state levels caused by dual treatment (Supplementary Fig. S6E). Finally, under asparagine deprived conditions with ISRIB or ATF4 knockdown, ectopic re-expression of ASNS was sufficient to restore levels of CDC25C and cyclin B1 (Fig. 6F and 6G). Our results demonstrate that the ISR maintains the levels of several key mediators of tumor cell proliferation via adaptive protein biosynthesis.
Dietary asparagine limitation combined with ATF4 inhibition delays LUAD growth in vivo
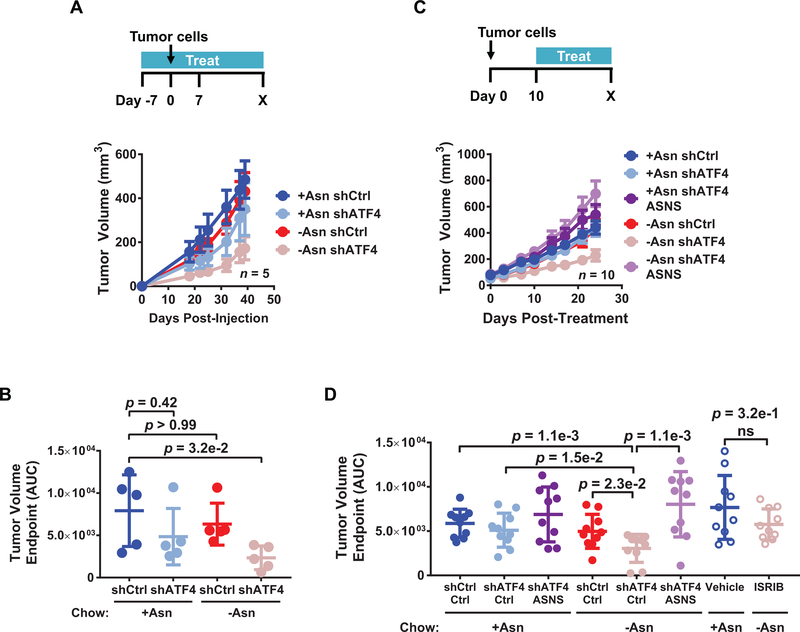
Because combining ISR inhibition with amino acid deprivation limits the fitness of LUAD cells in vitro, we tested the efficacy of this approach in vivo. First, PC9 cells expressing a control shRNA or shATF4 were injected into the flank of mice that were placed on an isonitrogenous amino acid-defined diet either containing or lacking asparagine. Analysis of tumor tissue by liquid chromatography-mass spectrometry (LC-MS/MS) confirmed a reduction in asparagine levels for mice on diets lacking asparagine (Supplementary Fig. S7A). Although ATF4 knockdown alone caused a minor reduction in tumor growth, this did not reach statistical significance (Fig. 7A and 7B). However, as compared to the control group, mice injected with cells expressing shATF4 had significantly delayed tumorigenic progression and decreased tumor volume when also fed a diet lacking asparagine (Fig. 7A and 7B). ASNS levels in vivo were effectively reduced by 80% in these residual tumor tissues (Supplementary Fig. S7B).
Figure 7. Dietary asparagine limitation combined with ISR inhibition delays LUAD growth in vivo.
A) Mice were placed on an amino acid-defined diet containing doxycycline (DOX) with or without asparagine. After 7 days, 5e4 PC9 cells expressing the indicated shRNAs were injected into the flanks of each mouse and tumor volume measured over time; n = 5 tumors. Error bars show SEM. B) Tumor volume as measured by area under the curve (AUC) until endpoint from A). C) 5e4 PC9 cells expressing the indicated shRNAs and vector control (no label) or ectopic ASNS were injected into the flanks of mice. Once tumors were palpable (10 days), mice were placed on an amino acid-defined diet containing DOX with or without asparagine and tumor volume was measured over time; n = 10 tumors. Error bars show SEM. D) Tumor volume as measured by area under the curve (AUC) until endpoint from C) and Supplementary Fig. S7D. Unless otherwise indicated error bars show SD and p-values by Mann-Whitney test. ns = not significant.
To test if inhibition of the ISR in combination with dietary asparagine limitation reduces the growth of established tumors, LUAD cells injected into mice were allowed to grow for 10 days at which point they generated palpable tumors. Tumor-bearing mice were then fed diets containing or lacking asparagine and treated with DOX to induce control shRNA, shATF4, or shATF4 with ectopic ASNS re-expression. Similarly, tumor-bearing mice were treated with 5 mg/kg ISRIB or vehicle ISRIB treatment. All treatments were well tolerated in mice over the duration of the experiment, as illustrated by endpoint mouse weights (Supplementary Fig. S7C). Knockdown of ATF4 in combination with asparagine limitation significantly reduced the progression of established tumors (Fig. 7C and 7D, solid pink circles). ISRIB treatment in combination with asparagine limitation also initially delayed tumor growth, but this effect was not maintained over time (Supplementary Fig. S7D and Fig. 7D, open pink circles). ISRIB treatment did not significantly reduce ASNS expression (Supplementary Fig. S7E) suggesting that the pharmacokinetics of ISRIB and/or targeting of eIF2α may be less efficacious in vivo under these conditions. Importantly, ectopic expression of ASNS was able to rescue the outgrowth of tumors with deficient ISR caused by ATF4 knockdown (Fig. 7C and 7D; light purple), confirming that this enzyme is a major effector of LUAD tumorigenesis under stress in vivo.
Discussion
Activation of the ISR has been observed in several types of cancers, but the underlying mechanisms and biological consequences of this are often unclear (11). In this study, we used genetic, pharmacological, and metabolic perturbations to understand the dependency of LUAD cancer cells on several effectors of the ISR, including eIF2α and ATF4. Moreover, we performed comprehensive transcriptomic as well as proteomic analysis to uncover novel regulatory layers of the ISR in LUAD cancer cells.
Regulation of amino acid stress response by the ISR is a common feature of LUADs
Our biological experiments confirm that a major role of the ISR in LUAD cells is to co-opt amino acid metabolism. We further demonstrate that pharmacological inhibition of eIF2α in multiple LUAD cell lines can be achieved in vitro using ISRIB, which sensitizes cancer cells to exogenous amino acid availability. Lung cancer cell lines expressing different oncogenic mutations require eIF2α and ATF4 to maintain their fitness in response to amino acid stress, particularly when extracellular asparagine levels are limited. Thus, although this phenomenon may be influenced by genetic alterations in LUAD, it is not necessarily restricted to a particular oncogenic driver.
Inhibition of eIF2α and ATF4 impacts not only LUAD cell proliferation, but also anchorage independent growth and cell motility, suggesting that the ISR may be required for multiple steps of LUAD progression. Accordingly, while knockdown of ATF4 alone did not significantly affect tumorigenesis, it was effective at blocking the growth of PC9 cells in vivo when combined with dietary asparagine limitation. This effect could be rescued by ectopic ASNS re-expression demonstrating that this ATF4 target is rate limiting for LUAD progression in vivo. Asparaginase mediated depletion of systemic asparagine can be used to inhibit tumorigenesis in childhood leukemia and pre-clinical models of breast cancer and NSCLC (16,30,44). Notably, ASNS overexpression confers asparaginase resistance in leukemia (45). Recent studies indicate that ISRIB can be used as a monotherapy in prostate cancer models (14). In our study, ISRIB initially delayed the growth of LUAD tumors in combination with asparagine limitation, but continuous treatment was not effective, likely due to insufficient reduction of ASNS under the conditions tested herein. Nevertheless, we conclude that ATF4 inhibition paired with exacerbation of cellular stress is a viable therapeutic paradigm for the treatment of LUAD and that pharmacological optimizations of ISRIB formulation and/or localized delivery are warranted for further pre-clinical testing in this cancer type.
Molecular link between ATF4 mediated transcriptional response and protein translation
Inhibition of eIF2α can affect global as well as selective protein translation. Our results suggest that the impact of ISRIB during amino acid deprivation of LUAD cells occurs mainly through ATF4 and its target ASNS. ATF4 can activate inhibitors of the AKT and mTOR pathways to suppress translation during nutrient deprivation (38,39,46). Consistent with signaling cross-talk, both AKT and mTOR activity were increased by dual treatment. Nevertheless, the net rates of translation in LUAD cells remained decreased upon ISRIB treatment or ATF4 knockdown because of the suppression of ASNS. In mammalian cells, asparagine is primarily used for protein biosynthesis, either through direct protein translation or indirectly as a co-transporter for amino acid uptake (28,36). ASNS can be activated downstream of the PI3K/mTOR pathways to enhance protein translation in KRAS mutant NSCLC (16). By performing epistasis experiments, we demonstrated that ASNS expression is also regulated independently of mTORC1 activation and is required for protein translation downstream of eIF2α and ATF4 in LUAD cell lines expressing various oncogenic mutations.
ASNS-mediated protein translation drives adaptive cell proliferation and migration
NRF2 can either independently activate ATF4 transcription or cooperate with ATF4 to induce target genes involved in anti-oxidant responses (16,32). Our data reveal an additional molecular link between proteostasis and the control of ROS. Specifically, NRF2 can be regulated downstream of ATF4, via ASNS mediated protein synthesis. Interestingly, while inhibition of ATF4 causes a decrease in NRF2 protein and accumulation of intracellular ROS, this level of oxidative stress was not responsible for the phenotypes that were incurred under dual treatment. Also, while ATF4 can upregulate autophagy genes (35), this did not correlate with the stress induced growth arrest of LUAD cells tested here. Alternatively, inhibition of eIF2α, ATF4, or ASNS under asparagine deprivation reduces the expression of a subset of proteins, several of which are important mediators of cell cycle progression and cell motility. In breast cancer, asparagine bioavailability can also regulate proteins involved in epithelial to mesenchymal transition (30). In LUAD cells, ASNS can regulate the G2/M mediator cyclin B1 and cell adhesion/extracellular matrix molecules such as FN. Importantly, ASNS expression is necessary and sufficient for adaptive lung cancer cell proliferation and migration in independent cell lines.
Cyclin B1 expression is associated with poor prognosis in human NSCLC (43), and we found cyclin B1 protein levels, but not CCNB1 mRNA, to correlate with ASNS in human LUADs. As such, the control of cell cycle progression via cyclin B1 translation is one likely mechanism by which the ISR and ASNS maintain human LUAD progression. Cyclin B1 is not preferentially enriched for asparagine residues, suggesting that the abundance of this amino acid does not necessarily determine which proteins are influenced by ASNS mediated translation. Alternatively, proteins that have short half-lives may be more sensitive to the limitation of asparagine and ISRIB treatment. Finally, stress granules have been shown to sequester CCNB1 mRNA (47) and could also inhibit protein translation of particular transcripts in response to unresolved amino acid stress and a maladaptive ISR. This potential selective control of proteostasis may provide novel therapeutic opportunities to target lung cancer cells as they attempt to overcome metabolic barriers over the course of treatment and tumor progression.
Supplementary Material
Acknowledgments
This study was funded by grants from the NIH/NCI to D.X.N. (R01CA166376 and R01CA191489) and M.M. (R01GM11738601). A.E.A. was supported by Training Grants from the NIH (T32GM007205) and the Yale Cancer Center. The MD Anderson Reverse Phase Protein Array Core is funded by NCI #CA16672. We thank Dr. Katerina Politi for comments on the manuscript and for providing the patient-derived NSCLC cell line YLR086. We thank Kevin B. Huang for his work related to this project.
Footnotes
Conflict of interest statement: D.X.N. has received research funding from AstraZeneca, Inc and Leidos.
References
- 1.Davidson SM, Papagiannakopoulos T, Olenchock BA, Heyman JE, Keibler MA, Luengo A, et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab 2016. doi 10.1016/j.cmet.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14(8):535–46 doi 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: Mechanisms and targets for therapy. Semin Cancer Biol 2015. doi 10.1016/j.semcancer.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolpaw AJ, Dang CV. Exploiting Metabolic Vulnerabilities of Cancer with Precision and Accuracy. Trends in cell biology 2018;28(3):201–12 doi 10.1016/j.tcb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Senft D, Ronai ZA. Adaptive Stress Responses During Tumor Metastasis and Dormancy. Trends in cancer 2016;2(8):429–42 doi 10.1016/j.trecan.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553(7689):446–54 doi 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 7.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016;17(10):1374–95 doi 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci 2013;70(19):3493–511 doi 10.1007/s00018-012-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 2004;101(31):11269–74 doi 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somers J, Poyry T, Willis AE. A perspective on mammalian upstream open reading frame function. The international journal of biochemistry & cell biology 2013;45(8):1690–700 doi 10.1016/j.biocel.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandewynckel YP, Laukens D, Geerts A, Bogaerts E, Paridaens A, Verhelst X, et al. The paradox of the unfolded protein response in cancer. Anticancer research 2013;33(11):4683–94. [PubMed] [Google Scholar]
- 12.Han J, Kaufman RJ. Physiological/pathological ramifications of transcription factors in the unfolded protein response. Genes & development 2017;31(14):1417–38 doi 10.1101/gad.297374.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. The EMBO journal 2010;29(12):2082–96 doi 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen HG, Conn CS, Kye Y, Xue L, Forester CM, Cowan JE, et al. Development of a stress response therapy targeting aggressive prostate cancer. Sci Transl Med 2018;10(439) doi 10.1126/scitranslmed.aar2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pytel D, Gao Y, Mackiewicz K, Katlinskaya YV, Staschke KA, Paredes MC, et al. PERK Is a Haploinsufficient Tumor Suppressor: Gene Dose Determines Tumor-Suppressive Versus Tumor Promoting Properties of PERK in Melanoma. PLoS Genet 2016;12(12):e1006518 doi 10.1371/journal.pgen.1006518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwinn DM, Lee AG, Briones-Martin-Del-Campo M, Conn CS, Simpson DR, Scott AI, et al. Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase. Cancer Cell 2018;33(1):91–107 e6 doi 10.1016/j.ccell.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America 2011;108(9):3665–70 doi 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nature cell biology 2013;15(5):481–90 doi 10.1038/ncb2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511(7511):543–50 doi 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Director’s Challenge Consortium for the Molecular Classification of Lung A, Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med 2008;14(8):822–7 doi 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 2009;4(1):44–57 doi 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Cheung WK, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell 2013;23(6):725–38 doi 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen E, Stinson A, Shan L, Yang J, Gietl D, Albino AP. Cigarette smoke induces endoplasmic reticulum stress and the unfolded protein response in normal and malignant human lung cells. BMC cancer 2008;8:229 doi 10.1186/1471-2407-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai JC, Miller-Vedam LE, Anand AA, Jaishankar P, Nguyen HC, Renslo AR, et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science 2018;359(6383) doi 10.1126/science.aaq0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zyryanova AF, Weis F, Faille A, Alard AA, Crespillo-Casado A, Sekine Y, et al. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science 2018;359(6383):1533–6 doi 10.1126/science.aar5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halliday M, Radford H, Sekine Y, Moreno J, Verity N, le Quesne J, et al. Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis 2015;6:e1672 doi 10.1038/cddis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pavlova NN, Hui S, Ghergurovich JM, Fan J, Intlekofer AM, White RM, et al. As Extracellular Glutamine Levels Decline, Asparagine Becomes an Essential Amino Acid. Cell Metab 2018;27(2):428–38 e5 doi 10.1016/j.cmet.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang J, Pavlova NN, Zhang J. Asparagine, a critical limiting metabolite during glutamine starvation. Mol Cell Oncol 2018;5(3):e1441633 doi 10.1080/23723556.2018.1441633.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knott SRV, Wagenblast E, Khan S, Kim SY, Soto M, Wagner M, et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature 2018;554(7692):378–81 doi 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009;461(7260):109–13 doi 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 2015. doi 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dey S, Sayers CM, Verginadis II, Lehman SL, Cheng Y, Cerniglia GJ, et al. ATF4-dependent induction of heme oxygenase 1 prevents anoikis and promotes metastasis. The Journal of clinical investigation 2015;125(7):2592–608 doi 10.1172/jci78031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Zhang L, Zhou L, Lei Y, Zhang Y, Huang C. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol 2018. doi 10.1016/j.redox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B’Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic acids research 2013;41(16):7683–99 doi 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krall AS, Xu S, Graeber TG, Braas D, Christofk HR. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nat Commun 2016;7:11457 doi 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balasubramanian MN, Butterworth EA, Kilberg MS. Asparagine synthetase: regulation by cell stress and involvement in tumor biology. American journal of physiology Endocrinology and metabolism 2013;304(8):E789–99 doi 10.1152/ajpendo.00015.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salazar M, Lorente M, Garcia-Taboada E, Perez Gomez E, Davila D, Zuniga-Garcia P, et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ 2015;22(1):131–44 doi 10.1038/cdd.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin HO, Seo SK, Woo SH, Kim ES, Lee HC, Yoo DH, et al. SP600125 negatively regulates the mammalian target of rapamycin via ATF4-induced Redd1 expression. FEBS Lett 2009;583(1):123–7 doi 10.1016/j.febslet.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 40.Harashima H, Dissmeyer N, Schnittger A. Cell cycle control across the eukaryotic kingdom. Trends in cell biology 2013;23(7):345–56 doi 10.1016/j.tcb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Developmental cell 2012;22(1):131–45 doi 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reticker-Flynn NE, Malta DF, Winslow MM, Lamar JM, Xu MJ, Underhill GH, et al. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nat Commun 2012;3:1122 doi 10.1038/ncomms2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper WA, Kohonen-Corish MR, McCaughan B, Kennedy C, Sutherland RL, Lee CS. Expression and prognostic significance of cyclin B1 and cyclin A in non-small cell lung cancer. Histopathology 2009;55(1):28–36 doi 10.1111/j.1365-2559.2009.03331.x. [DOI] [PubMed] [Google Scholar]
- 44.Pieters R, Hunger SP, Boos J, Rizzari C, Silverman L, Baruchel A, et al. L-asparaginase treatment in acute lymphoblastic leukemia: a focus on Erwinia asparaginase. Cancer 2011;117(2):238–49 doi 10.1002/cncr.25489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomelino CL, Andring JT, McKenna R, Kilberg MS. Asparagine synthetase: Function, structure, and role in disease. The Journal of biological chemistry 2017;292(49):19952–8 doi 10.1074/jbc.R117.819060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye J, Palm W, Peng M, King B, Lindsten T, Li MO, et al. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes & development 2015;29(22):2331–6 doi 10.1101/gad.269324.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. The Journal of cell biology 2013;202(7):1041–55 doi 10.1083/jcb.201302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.