Abstract
microRNAs (miRNAs) are highly conserved, short (~ 21-nucleotide), endogenous, non-coding RNA molecules that play major roles in post-transcriptional silencing by guiding target mRNA cleavage or translational inhibition. In this study, applying high-stringent genome-wide computational-based approaches, a total of 28 putative miRNAs belonging to 17 miRNA families were identified from an antioxidant-rich medicinal plant passion fruit (Passiflora edulis). Inter-tissue (leaves and fruits) and inter-varietal (yellow and purple fruit varieties) quantitative study of six putative passion fruit miRNAs (ped-miR160, ped-miR164, ped-miR166, ped-miR393, ped-miR394, and ped-miR398) showed differential expression. Using psRNATarget tool, a total of 25 potential target proteins of the characterized passion fruit miRNAs were also identified. Most of the target proteins identified in this study, including SQUAMOSA promoter binding, Class III HD-Zip, NAC, Scarecrow, APETALA2, Auxin response factor, MYB, and superoxide dismutase, were found to be involved in development, metabolism, and defense/stress response signaling.
Electronic supplementary material
The online version of this article (10.1007/s13205-019-2000-5) contains supplementary material, which is available to authorized users.
Keywords: Passion fruit, microRNA (miRNA), Minimum folding free energy index (MFEI), microRNA expression, miRNA targets
Introduction
microRNAs (miRNAs) are small (~ 21-nucleotide) non-coding endogenous RNA molecules that play a vital post-transcriptional regulatory role in gene expression by target mRNA cleavage or translational inhibition (Jones-Rhoades et al. 2006). In plant, mature miRNAs are generated from the long stem-loop primary transcript (pre-miRNA) by a dicer-like RNA endonuclease via the intermediate step of pre-miRNA synthesis, and finally the RNA inducing silencing complex (RISC) guided by ARGONAUTE 1 (AGO1) protein directs the miRNA to the complementary target mRNA sequence (Voinnet 2009). Plant microRNAs are reported to possess important functions in several metabolic and biological pathways such as tissue development and differentiation, biotic and abiotic stress responses, phytohormones signaling, and secondary metabolite production (Wu 2013; Rajwanshi et al. 2014; Paul et al. 2011, 2017, 2018; Sharma et al. 2019; Gupta et al. 2017). Nonetheless, the evolutionary highly conserved nature of an extensive number of miRNAs simplified the process of characterization of novel miRNA orthologs in new plant species through homologs identification (Zhang et al. 2006b; Ye et al. 2013; Sharma et al. 2019). However, only the computational-based homology approach for the identification of miRNAs in new plant species can generate false-positive results, and hence several other parameters such as length, minimum free energy (MFE), and the minimum folding free energy index (MFEI) of the pre-miRNAs need to be studied to increase the precision of the prediction (Paul et al. 2018; Sharma et al. 2019). Moreover, experimental validation of the predicted miRNAs is highly recommended (Sharma et al. 2019).
Passiflora edulis, commonly known as passion fruit is a flowering vine native to Brazil and cultivated worldwide with distinctive aromas and flavors making it a popular additive to many tropical fruit juice blends (Talcott et al. 2003). The most common varieties of this fruit are with yellow (P. edulis var flavicarpa) and purple (P. edulis var edulis) skin (Xiaojing and Liuxin 1994). Both the varieties contain a yellow soft pulp and lots of seeds inside a hard rind (Supplementary file 1) (López-Vargas et al. 2013). The characteristic color of yellow passion fruit is due to provitamin A carotenes and xanthophylls, while the anthocyanins contribute to the spectacular deep purple color to the purple variety (Talcott et al. 2003). Because of their acidic flavor and higher juice yield, yellow passion fruits are most widely used for processed juices, while due to their sweeter taste, purple varieties are typically consumed fresh (Talcott et al. 2003). Passion fruit has recently gained a lot of attention worldwide due to it being a rich source of powerful antioxidants as well as for showing anti-inflammatory, anti-cancer, anti-diabetic, anti-hypertension, and anti-aging properties (Vera et al. 2019; Ayala-Zavala et al. 2011). On the other hand, the peel of passion fruit contains saponin, triterpenoid, flavonoids, and phenolic compounds that can help to inhibit elastase activity facilitating the production of anti-wrinkle cosmetic cream (Vera et al. 2019). Typical phytochemicals found in passion fruit are passiflorine, harmine, harman, harmol, and harmaline (Pereira et al. 2014). Moreover, the leaves of passion fruit also contain several flavonoids such as isoorientin, isovitexin, and vitexin which have been employed in several European and American countries as sedatives and anti-inflammatory agents (Ferreres et al. 2007). Despite the fact that miRNAs regulate several metabolic processes in plants, none of the miRNA-related studies have been performed so far in passion fruit, and none are listed in the miRBase. It has been confirmed that several conserved, as well as species-specific miRNAs, are very important for plants in different biological processes and hence, profiling miRNAs in a non-model plant is essential to understand the regulation of various biological phenomena. With the recent draft genome sequence available (Wu et al. 2019), the current study offers an insight of miRNAs and their respective targets in passion fruit using bioinformatics as well as wet-lab approach for better understanding the physiological processes in this plant. In summary, in this study for the first time, we have generated the miRNA and respective target profile of passion fruit and performed a comparative miRNA expression study between vegetative and reproductive tissues (leaves and fruits) as well as between two different varieties (yellow and purple skins).
Materials and methods
Computational prediction of miRNA
For the in silico prediction of potential passion fruit miRNAs, a reference set of mature plant miRNAs were retrieved from miRBase database (http://www.mirbase.org/cgi-bin/sequence_get.pl) and aligned against the whole genome sequence of passion fruit. The reference set consisted of a total 1370 mature miRNA sequences including Arabidopsis thaliana (428 mature sequences), Glycine max (756 mature sequences), and Vitis vinifera (186 mature sequences). BLASTn tool was used for the alignment and sequences with exact matches were chosen manually. The possible precursor (pre-miRNA) sequences of approximately 400 nucleotides (nt) (200 nt upstream and 200 nt downstream to the BLAST hit region) were mined and sequences coded for proteins were eliminated. To check the reliability of the possible precursors, secondary structures were predicted using MFOLD (http://unafold.rna.albany.edu/?q=mfold) webserver. Since an authentic secondary structure of the precursor is considered as one of the vital factors to be a miRNA candidate, some previously demonstrated strict filtering criteria were applied during prediction, such as: (1) the precursors must form a stem‐loop structure containing mature miRNA sequences within one arm, (2) the potential miRNA sequences should not be positioned at the terminal loop of the hairpin structures, (3) mature miRNAs should have fewer than nine mismatches with the opposite miRNA*sequence, and (4) the predicted secondary structures must have low MFE and high MFEI values, since it is required for distinguishing the miRNAs from other RNAs molecules (MFEIs of tRNAs, rRNAs or mRNAs candidates are 0.64, 0.59 and 0.62–0.66, respectively) (Zhang et al. 2006a). The MFE or ΔG (−kcal/mol) values generated from the MFOLD web server of the stem-loop structures were used for calculating the MFEI values using the following formula:
Prediction of miRNA targets
The precise or near-precise complementarity of plant miRNAs and their targets facilitated in silico miRNA target prediction in non-model plants. In this study, web tool psRNATarget (http://plantgrn.noble.org/psRNATarget/?dowhat=Help) was used to identify the potential miRNA targets of passion fruit. Due to unavailability of passion fruit protein database in the psRNATarget web server, target search was conducted against the protein database of Populus trichocarpa, which is considered as the evolutionary closest species of passion fruit (Wu et al. 2019). The parameters were adjusted manually, such as maximum expectation value of 3, translation inhibition ranges of 9–11 nt, number of top targets of 10, penalty for G:U pair of 0.5, and number of mismatches allowed in seed region of 1.5.
Collection of plant materials, RNA extraction, and miRNA expression analysis
To validate the predicted results, fresh ripe yellow and purple passion fruits and leaves were collected from the local field in Queretaro, Mexico. Total RNA including small RNA was extracted from leaves and both the fruit varieties using the miRNeasy Mini Kit (Qiagen) and pooled separately for each sample. The quality and quantity of RNA samples were measured with Nanodrop One (Thermo Scientific), and subsequently polyadenylated (using modified oligo dT primer) as well as reverse transcribed using mRQ Buffer (2 ×) and enzyme provided with Mir-X miRNA First-Stand Synthesis kit (Takara, Tokyo, Japan). The resulting cDNA was then amplified by T100 Thermal Cycler (Bio-Rad, CA, USA) using the entire predicted miRNA sequence as forward primer and the adapter-specific mRQ3′ primer provided with Mir-X miRNA qRT-PCR TB Green Kit (Takara, Tokyo, Japan) as the reverse primer. Selected six passion fruit miRNAs (miR160, miR164, miR166, miR393, miR394, and miR398) were experimentally validated in this study. The PCR was programmed as follows: initial denaturation at 94 °C for 3 min followed by 45 cycles of denaturation at 94 °C for 30 s and annealing at 60 °C for 30 s, extension at 72 °C for 25 s, and a final elongation step at 72 °C for 7 min. The resulted PCR products (~ 80 bp) were checked in 2% agarose gel. To study the quantitative differential expression pattern of the aforesaid passion fruit miRNAs between fruits and leaves as well as between two varieties, a quantitative real-time PCR (qPCR) was performed. The reaction was made in 12.5 μl volume containing: 3.8 μl of ddH2O, 6.25 μl of TB Green Advantage Premix (2 ×), 0.23 μl of ROX Dye (50 ×), 0.1 μl of miRNA-specific primer (10 μM), 0.1 μl of mRQ3′ primer (10 μM), and 2 μl of cDNA. Each sample was done with three technical replicates. Step One Real-Time PCR System (Applied Biosystems, Carlsbad, CA) and Mir-X miRNA TB Green qRT-PCR kit (Takara, Tokyo, Japan) were used, respectively for the qPCR experiment. The qPCR was programmed as follows: initial denaturation at 95 °C for 10 s followed by 45 cycles of denaturation at 95°C for 5 s and annealing at 60 °C for 20 s, dissociation curve at 95 °C for 30 s, 55 °C for 20 s, and 95 °C for 20 s. The detail of miRNA-specific primers used in this study is presented in Supplementary file 1. The comparative Ct (2−ΔΔCT) method was employed to determine the relative fold changes and U6 was employed as an internal reference for the miRNA qPCR analysis (Livak and Schmittgen 2001).
Results and discussions
Characterization of passion fruit miRNAs
Employing rigorous filtering approach, 28 conserved passion fruit miRNAs were identified in this study belonging to 17 miRNA families (miR156/miR157, miR160, miR162, miR164, miR166, miR167, miR169, miR171, miR172, miR319, miR393, miR394, miR395, miR397, miR398, miR399, and miR828) (Table 1). The majority of the identified passion fruit miRNAs are 21 nt long. The length of the predicted pre-miRNA varied from 60 nt to 185 nt with an average length of 102 nt. In addition, all the predicted pre-miRNAs produced secondary structures with stem loop and 50% of the mature miRNAs were found either at the 5′ or 3′ end corroborating Gorodkin et al. (2006) studies which exhibit an equal distribution of miRNAs in both arms in other plant species. Moreover, 75% of the predicted passion fruit miRNAs started with the nucleotide uracil (U) agreeing with the study of Zhang et al. (2008) that miRNA-mediated regulation is greatly dependent on the uracil present at the first position of the mature miRNA. The GC content of passion fruit miRNAs had an average of 46.25%. It is well established that low MFE values of stem-loop precursors attain more stable predictions (Bonnet et al. 2004); in this study, the MFE values of the precursors ranged from − 24.80 to − 79.40 with an average of − 44.02, while the MFEI values oscillated between 0.70 and 1.30 with an average of 0.95, excluding the possibility of being other small RNAs. Furthermore, as exhibited in Table 1, clusters of miRNA genes in passion fruit are abundant. Several clusters are compacted indicating some groups of miRNAs are expressed from specific transcription elements (polycistron) such as miR166a-5p, miR166b-3p, miR166h-3p, miR166h-5p; miR171b-3p, miR171j-3p, miR171k-5p; miR319b, miR319I, miR319p; miR399d, miR399e, and miR399i. However, in the current analysis, the maximum number of miRNA members was found to be present in miR166 family. The predicted secondary structures of passion fruit miRNA precursors with higher MFEI values (top 10) are shown in Fig. 1. The in silico prediction of passion fruit miRNAs was found accurate, as all the selected miRNAs displayed expected bands in the agarose gel (around ~ 80 bp) (Fig. 2).
Table 1.
Summary of the identified miRNAs from passion fruit
| Identified miRNAs | LM (nt) | Query miRNAs | miRNA sequences | Accession | Location | LP (nt) | GC % | MFEs (ΔG) | MFEI |
|---|---|---|---|---|---|---|---|---|---|
| ped-miR156a-5p | 20 | ath-miR156a-5p | UGACAGAAGAGAGUGAGCAC | MUZT01071036.1 | 5′ | 82 | 47.56 | – 51.10 | 1.31 |
| ped-miR157a-5p | 21 | ath-miR157a-5p | UUGACAGAAGAUAGAGAGCAC | MUZT01093399.1 | 5′ | 84 | 40.48 | – 42.90 | 1.26 |
| ped-miR160c-5p | 21 | ath-miR160c-5p | UGCCUGGCUCCCUGUAUGCCA | MUZT01072383.1 | 5′ | 79 | 58.23 | – 43.50 | 0.94 |
| ped-miR162a | 22 | ath-miR162a-5p | UGGAGGCAGCGGUUCAUCGAUC | MUZT01067906.1 | 5′ | 81 | 48.15 | – 28.50 | 0.73 |
| ped-miR164b-5p | 21 | ath-miR164b-5p | UGGAGAAGCAGGGCACGUGCA | MUZT01048418.1 | 3′ | 195 | 44.10 | – 60.40 | 0.70 |
| ped-miR166a-5p | 21 | gma-miR166a-5p | GGAAUGUUGUCUGGCUCGAGG | MUZT01089702.1 | 5′ | 131 | 45.80 | – 56.80 | 0.94 |
| ped-miR166b-3p | 21 | ath-miR166b-3p | UCGGACCAGGCUUCAUUCCCC | MUZT01068357.1 | 3′ | 80 | 48.75 | – 44.90 | 1.15 |
| ped-miR166h-3p | 21 | gma-miR166h-3p | UCUCGGACCAGGCUUCAUUCC | MUZT01172784.1 | 3′ | 88 | 45.45 | – 39.20 | 0.98 |
| ped-miR166h-5p | 21 | gma-miR166h-5p | GGAAUGUUGUUUGGCUCGAGG | MUZT01069089.1 | 5′ | 125 | 37.60 | – 44.50 | 0.94 |
| ped-miR167e | 21 | gma-miR167e | UGAAGCUGCCAGCAUGAUCUU | MUZT01041392.1 | 5′ | 67 | 44.78 | – 35.80 | 1.19 |
| ped-miR169a | 21 | gma-miR169a | CAGCCAAGGAUGACUUGCCGG | MUZT01219929.1 | 5′ | 179 | 43.58 | – 66.30 | 0.84 |
| ped-miR169b | 21 | gma-miR169b | CAGCCAAGGAUGACUUGCCGA | MUZT01009503.1 | 5′ | 99 | 46.46 | – 39.40 | 0.85 |
| ped-miR171b-3p | 21 | ath-miR171b-3p | UUGAGCCGUGCCAAUAUCACG | MUZT01091311.1 | 3′ | 84 | 44.05 | – 38.40 | 1.03 |
| ped-miR171j-3p | 21 | gma-miR171j-3p | UGAUUGAGCCGUGCCAAUAUC | MUZT01028930.1 | 3′ | 77 | 37.66 | – 37.80 | 1.30 |
| ped-miR171k-5p | 21 | gma-miR171k-5p | CGAUGUUGGUGAGGUUCAAUC | MUZT01137815.1 | 5′ | 79 | 46.84 | – 29.10 | 0.78 |
| ped-miR172b | 21 | vvi-miR172b | UGAAUCUUGAUGAUGCUACAC | MUZT01049317.1 | 3′ | 120 | 47.50 | – 42.20 | 0.74 |
| ped-miR319b | 21 | ath-miR319b | UUGGACUGAAGGGAGCUCCCU | MUZT01035900.1 | 3′ | 171 | 46.20 | – 74.80 | 0.94 |
| ped-miR319l | 22 | gma-miR319l | UUGGACUGAAGGGAGCUCCUUC | MUZT01112275.1 | 3′ | 185 | 46.49 | – 79.40 | 0.92 |
| ped-miR319p | 21 | gma-miR319p | UUUUGGACUGAAGGGAGCUCC | MUZT01179548.1 | 3′ | 76 | 47.37 | – 32.80 | 0.91 |
| ped-miR393a-5p | 22 | ath-miR393a-5p | UCCAAAGGGAUCGCAUUGAUCC | MUZT01078953.1 | 5′ | 64 | 46.88 | – 33.10 | 1.10 |
| ped-miR394a-5p | 20 | gma-miR394a-5p | UUGGCAUUCUGUCCACCUCC | MUZT01119677.1 | 5′ | 78 | 50.00 | – 33.40 | 0.85 |
| ped-miR395a | 21 | gma-miR395a | CUGAAGUGUUUGGGGGAACUC | MUZT01200730.1 | 3′ | 88 | 51.14 | – 50.20 | 1.11 |
| ped-miR397a | 21 | ath-miR397a | UCAUUGAGUGCAGCGUUGAUG | MUZT01096698.1 | 5′ | 79 | 51.90 | – 39.50 | 0.96 |
| ped-miR398d | 21 | gma-miR398d | UGUGUUCUCAGGUCGCCCCUG | MUZT01004242.1 | 3′ | 102 | 50.00 | – 42.30 | 0.82 |
| ped-miR399d | 21 | ath-miR399d | UGCCAAAGGAGAUUUGCCCCG | MUZT01051618.1 | 3′ | 102 | 49.02 | – 55.30 | 1.10 |
| ped-miR399e | 21 | gma-miR399e | UGCCAAAGGAGAUUUGCCCAG | MUZT01038065.1 | 3′ | 102 | 45.10 | – 35.80 | 0.77 |
| ped-miR399i | 21 | gma-miR399i | UGCCAAAGGAGAAUUGCCCUG | MUZT01092893.1 | 5′ | 60 | 45.00 | – 24.80 | 0.91 |
| ped-miR828b | 22 | gma-miR828b | UCUUGCUCAAAUGAGUAUUCCA | MUZT01146558.1 | 5′ | 123 | 39.84 | – 42.70 | 0.87 |
LM length of mature miRNAs, LP length of precursor
Fig. 1.
Secondary stem-loop structures of the predicted passion fruit miRNA precursors/pre-miRNAs. Respective miRNAs are represented with red font
Fig. 2.
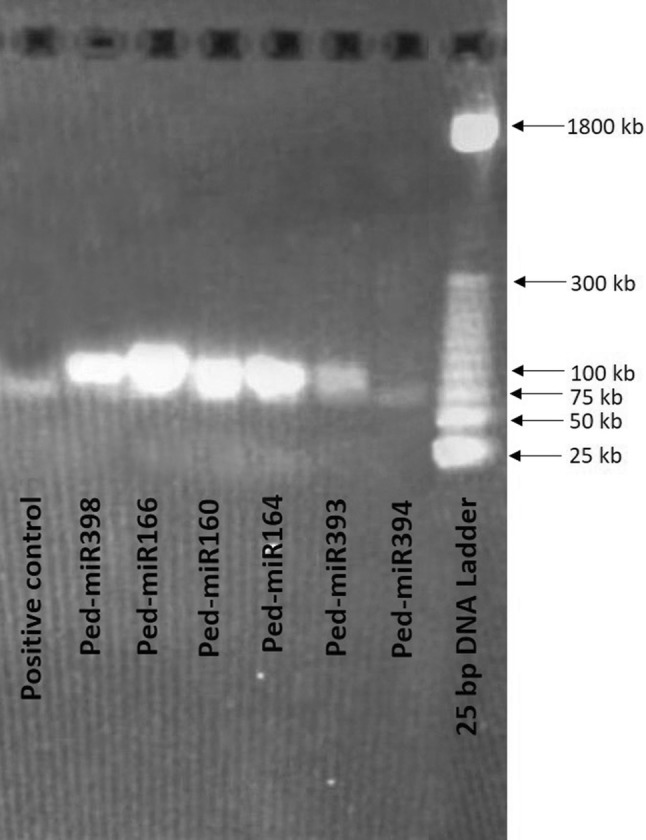
Validation of selected passion fruit miRNAs (ped-miR160, ped-miR164, ped-miR166, ped-miR393, ped-miR394, and ped-miR398) by semiquantitative reverse transcription PCR (fruit tissues). The resulting PCR products were checked in 2% agarose gel with EtBr staining. U6 was employed as a positive control
Identification of potential target transcripts of putative passion fruit microRNAs
In this study, a total of 25 possible target transcripts of passion fruit miRNAs were identified including some uncharacterized proteins and among those potential targets, several were found to be participating in signaling and metabolic pathways, defense mechanisms/stress response signaling, and cellular development (Table 2). Other targets were implicated in metal ion binding, ATP binding, DNA and RNA binding, and symporter activities. Therefore, target genes can be divided into three different groups: metabolism-related targets, stress-responsive targets, and transcription factors. However, the same miRNAs can have several distinct functions such as development as well as stress response signaling. Some miRNA: target pairs are well conserved among different plants species such as (1) transcription factors HD-ZIPs, participated in a variety of processes during plant growth and development, are principally targeted by miR166 family in poplar, rice, apple, and Arabidopsis; (2) Transcription factors NO APICAL MERISTEM (NAM) and NAC which were found to be involved in shoot development, fruit ripening, and also flavonoid biosynthesis are mostly targeted by miR164 family (Morishita et al. 2009; Zeng et al. 2015); (3) F-Box proteins, these are involved in many plants’ vegetative and reproductive growth and development are targeted by both miR393 and miR394 families in Arabidopsis, poplar, rice, and apple; (4) Auxin is a key regulator of virtually every aspect of plant growth and development from embryogenesis to senescence and auxin response factors (ARFs) which control the auxin-responsive genes as well as stress-responsive signals are often targeted by miR160 families; (5) superoxide dismutases (SODs), the major antioxidant defense systems in plants are the primary target of miRNA398 family in various plant species (Ye et al. 2013; Wang et al. 2004; Lu et al. 2005; Archak and Nagaraju 2007; Zhang et al. 2009; Colaiacovo et al. 2010; Sunkar et al. 2012; Bouzroud et al. 2018). Although few other miRNAs: target pairs are also conserved among different plant species in this study, we selected the six very important aforesaid miRNAs for the qPCR-based quantitative expression analysis.
Table 2.
Potential targets of identified passion fruit miRNAs
| miR family | Name of the target transcript | Molecular function | Biological process |
|---|---|---|---|
| miR157 | SQUAMOSA promoter binding-like protein | DNA-binding transcription factor, metal ion binding | Regulation of transcription |
| miR160 | Auxin response factor | DNA binding | Auxin-activated signaling pathway, leaf senescence, negative regulation of transcription, flavonoid biosynthesis |
| miR164 | NAC domain protein | DNA binding | Regulation of transcription, defense response, flavonoid biosynthesis |
| miR166 | Class III HD-zip protein | DNA and lipid binding | Development of shoot apical meristem |
| AG-motif binding protein-2 | Sequence-specific DNA and zinc ion binding | Regulation of transcription | |
| Chaperone protein ClpB3 | ATP binding | Protein metabolic process and protein refolding | |
| miR169 | Nuclear transcript factor Y subunit A | DNA-binding, protein heterodimerization activity | Abscisic and gibberellic acid signaling pathway, regulation of gene expression, positive regulation of photomorphogenesis |
| miR171 | Serine-rich protein | RNA binding | mRNA cis splicing via spliceosome |
| Scarecrow-like protein 6 | Sequence-specific DNA binding | Regulation of transcription | |
| Ethylene-responsive transcription factor RAP2-7 | DNA binding | Ethylene-activated signaling pathway | |
| Floral homeotic protein APETALA2 | DNA binding | Flower, seed, and plant ovule development, cell differentiation | |
| Transcription factor AHAP2 | DNA binding | Not found | |
| Inositol phosphate kinase | ATP and metal ion binding | Defense response, lateral root development | |
| Transcription factor GAMYB-like | Sequence-specific DNA binding | Anther development, cell, and pollen sperm cell differentiation, regulation of transcription, response to ethylene | |
| miR393 | Auxin response factor | DNA binding | Auxin-activated signaling pathway, leaf senescence, negative regulation of transcription |
| Protein transport inhibitor response 1 | Auxin binding and auxin receptor activity | Cell cycle, auxin activated and ethylene signaling pathway, defense response, response to auxin | |
| miR394 | F-box only protein | Photoreceptor activity | Circadian rhythm. Flower development, protein ubiquitination |
| miR395 | ATP-sulfurylase | ATP binding | Response to cytokinin and cadmium ion, sulfate assimilation |
| Sulfate adenylyltransferase | Nucleotide binding | Purine ribonucleotide and sulfur compound metabolic process | |
| miR397 | Laccase | Copper ion binding | Lignin biosynthetic and catabolic pathway |
| miR398 | Superoxide dismutase | Copper ion binding | Cellular response to light intensity, oxidative stress, ozone, and salt stress |
| Protein LAS1 | Terpenes’ synthase activity and magnesium ion binding | Defense response and metabolic process | |
| Dissimilatory sulfite reductase beta subunit | Metal ion binding | Sulfur compound metabolic process | |
| Zinc metalloprotease EGY2 | Metallopeptidase activity | Not found | |
| miR828 | Myb-like transcription factor | Sequence-specific DNA binding | Circadian regulation, histone H acetylation, photoperiodism, response to auxin, ethylene, abscisic acid, gibberellin, jasmonic acid, and salt stress |
Expression analysis of selected passion fruit miRNAs
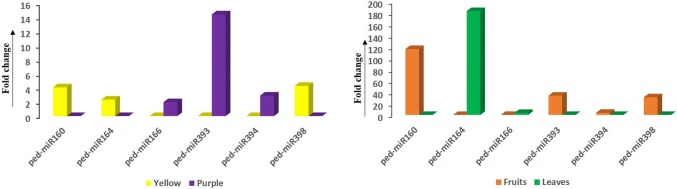
The qRT-PCR results showed a differential expression pattern of the selected miRNAs between fruits and leaves (both from yellow varieties) as well as between two different fruit varieties. The expression of miR160, and miR164, and miR398 was found to be higher in the yellow fruit variety than the purple one with a fold change of 4.05, 2.32, and 4.25, respectively. However, in the purple fruit variety miRNAs, miR166, miR393, and miR394 were found to be upregulated compared to the yellow one with the respective fold change of 1.96, 14.35, and 2.89. Furthermore, the miRNAs miR160, miR393, miR394, and miR398 showed higher expression in the fruit compared to leaves with a fold change of 116.16, 33.94, 3.77, and 31.12, respectively, while only miR164 and miR166 were found to be overexpressed in the leaves with a fold change of 183.01 and 3.53, respectively, compared to fruits. The fold change can be explained since the overexpression of certain miRNAs is linked to a decrease in the target transcript expression in the tissue and vice versa (Neutelings et al. 2012). For example, auxin is essential for plant development and high levels of auxin can repress the expression of the MYB–bHLH–WD (MBW) complex and thereby suppress the biosynthesis of anthocyanin. On the other hand, ARFs, target transcripts of miR-160 can control the expression of auxin-inducible genes by binding to auxin response elements in their promoters and therefore indirectly regulate the anthocyanin biosynthesis (Sun et al. 2017). Similarly, a relation between the overproduction of the NAC domain transcription factor (target of miR-164) and anthocyanin accumulation was recorded in blood‐fleshed peaches (Zhou et al. 2015). In this study, low expression of both ped-miR160 and miR-164 in the purple variety as compared to yellow variety indicated the accumulation of anthocyanin in purple variety and thus corroborates with the previous reports. Nevertheless, this differential expression pattern of passion fruit microRNAs obtained from the qPCR data demonstrates that a cell can control the target expression/functions in certain types of tissues or samples by manipulating the miRNA expression (Farh et al. 2005). The results of the qRT-PCR-based differential expression of the current study are shown in Fig. 3.
Fig. 3.
Graphical representation of differential expression pattern (fold change) of selected passion fruit miRNAs (ped-miR160, ped-miR164, ped-miR166, ped-miR393, ped-miR394, and ped-miR398) between vegetative and reproductive tissues (leaves and fruits) as well as between two different varieties (yellow and purple). U6 was employed as an internal reference
Conclusion
To the best of our knowledge, this is the first report of the characterization of passion fruit microRNAs and their targets. In this report using strict filtering criteria, a total of 28 conserved passion fruit miRNAs belonging to 17 miRNA families as well as 25 corresponding targets were computationally identified. Among the predicted passion fruit miRNAs, six selected miRNAs (ped-miR160, ped-miR164, ped-miR166, ped-miR393, ped-miR394, and ped-miR398) were validated by semiquantitative RT-PCR and their quantitative expression was measured by qPCR in leaves and fruits as well as between two different fruit varieties. All the aforesaid miRNAs displayed significantly differential expression between the samples. Among the predicted passion fruit miRNA targets, several were found to be involved in metabolism, defense/stress response signaling, development, and flavonoid biosynthesis. Nevertheless, the identification of miRNAs and their targets is a key step towards the initiation of the miRNA-related study in a non-model plant and we believe that our current study will be helpful for strengthening the research on miRNA-mediated regulation in herbal plants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The research facility provided by the Departamento Regional de Bioingenieria, Region Centro is thankfully acknowledged.
Author contributions
SP conceived and designed the study; JL and CGM performed the experiments; SP and JL wrote the paper; AS reviewed and edited the manuscript. All authors read and approved the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Sujay Paul and Fuente Jiménez contributed equally.
Contributor Information
Sujay Paul, Email: spaul@tec.mx.
Ashutosh Sharma, Email: asharma@tec.mx.
References
- Archak S, Nagaraju J. Computational prediction of rice (Oryza sativa) miRNA targets. Genom Proteom Bioinform. 2007;5:196–206. doi: 10.1016/S1672-0229(08)60007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala-Zavala J, Vega-Vega V, Rosas-Domínguez C, Palafox-Carlos H, Villa-Rodriguez J, Siddiqui MW, Dávila-Aviña J, González-Aguilar G. Agro-industrial potential of exotic fruit byproducts as a source of food additives. Food Res Int. 2011;44:1866–1874. doi: 10.1016/j.foodres.2011.02.021. [DOI] [Google Scholar]
- Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, Smouni A, Bouzayen M, Zouine M. Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum) PLoS One. 2018;13:e0193517. doi: 10.1371/journal.pone.0193517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiacovo M, Subacchi A, Bagnaresi P, Lamontanara A, Cattivelli L, Faccioli P. A computational-based update on microRNAs and their targets in barley (Hordeum vulgare L.) BMC Genom. 2010;11:595. doi: 10.1186/1471-2164-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK-H, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Ferreres F, Sousa C, Valentão P, Andrade PB, Seabra RM, Gil-Izquierdo Á. New C-deoxyhexosyl flavones and antioxidant properties of Passiflora edulis leaf extract. J Agr Food Chem. 2007;55:10187–10193. doi: 10.1021/jf072119y. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Havgaard JH, Ensterö M, Sawera M, Jensen P, Öhman M, Fredholm M. MicroRNA sequence motifs reveal asymmetry between the stem arms. Comput Biol Chem. 2006;30:249–254. doi: 10.1016/j.compbiolchem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Gupta OP, Karkute SG, Banerjee S, Meena NL, Dahuja A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Front Plant Sci. 2017;8:374. doi: 10.3389/fpls.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. microRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-Vargas JH, Fernández-López J, Pérez-Álvarez JA, Viuda-Martos M. Chemical, physico-chemical, technological, antibacterial and antioxidant properties of dietary fiber powder obtained from yellow passion fruit (Passiflora edulis var. flavicarpa) co-products. Food Res Int. 2013;51(2):756–763. doi: 10.1016/j.foodres.2013.01.055. [DOI] [Google Scholar]
- Lu S, Sun Y-H, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress–responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Kojima Y, Maruta T, Yokoi AN, Yabuta Y, Shigeoka S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009;50:2210–2222. doi: 10.1093/pcp/pcp159. [DOI] [PubMed] [Google Scholar]
- Neutelings G, Fénart S, Lucau-Danila A, Hawkins S. Identification and characterization of miRNAs and their potential targets in flax. J Plant Physiol. 2012;169:1754–1766. doi: 10.1016/j.jplph.2012.06.011. [DOI] [PubMed] [Google Scholar]
- Paul S. Identification and characterization of microRNAs and their targets in high-altitude stress-adaptive plant maca (Lepidium meyenii Walp) 3 Biotech. 2017;7:103. doi: 10.1007/s13205-017-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kundu A, Pal A. Identification and validation of conserved microRNAs along with their differential expression in roots of Vigna unguiculata grown under salt stress. Plant Cell Tiss Org. 2011;105:233–242. doi: 10.1007/s11240-010-9857-7. [DOI] [Google Scholar]
- Paul SC, Sharma A, Mehta R, Paul S. Genome wide computational identification of tuna (Thunnus orientalis) microRNAs and their targets. Ocean Sci J. 2018;53:727–734. doi: 10.1007/s12601-018-0041-z. [DOI] [Google Scholar]
- Pereira CAM, Rodrigues TR, Yariwake JH. Quantification of harman alkaloids in sour passion fruit pulp and seeds by a novel dual SBSE-LC/Flu (stir bar sorptive extraction-liquid chromatography with fluorescence detector) method. J Braz Chem Soc. 2014;25:1472–1483. [Google Scholar]
- Rajwanshi R, Chakraborty S, Jayanandi K, Deb B, Lightfoot DA. Orthologous plant microRNAs: microregulators with great potential for improving stress tolerance in plants. Theor Appl Genet. 2014;127:2525–2543. doi: 10.1007/s00122-014-2391-y. [DOI] [PubMed] [Google Scholar]
- Sharma A, Bejarano P, Castillo-Maldonado I, Capote M, Madariaga-Navarrete A, Paul S. Genome wide computational prediction and experimental validation of quinoa (Chenopodium quinoa) microRNAs. Can J Plant Sci. 2019 doi: 10.1139/CJPS-2018-0296. [DOI] [Google Scholar]
- Sun Y, Qiu Y, Duan M, Wang J, Zhang X, Wang H, Song J, Li X. Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Mol Genet Genom. 2017;292:215–229. doi: 10.1007/s00438-016-1268-y. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17:196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Talcott ST, Percival SS, Pittet-Moore J, Celoria C. Phytochemical composition and antioxidant stability of fortified yellow passion fruit (Passiflora edulis) J Agr Food Chem. 2003;51:935–941. doi: 10.1021/jf020769q. [DOI] [PubMed] [Google Scholar]
- Vera K, Raif A, Ikhtiari R. Antioxidant and anti-elastase activity of seed and peel extract of P. edulis. ASRJETS. 2019;53:43–48. [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang X-J, Reyes JL, Chua N-H, Gaasterland T. Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 2004;5:R65. doi: 10.1186/gb-2004-5-9-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. Plant microRNAs and development. J Genet Genomics. 2013;40:217–230. doi: 10.1016/j.jgg.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Wu Y, Tian Q, Liu J, Huang Y, Huang W, Xia X, Mou H, Yang X (2019) The genomic sequence and comparative genomic analysis of cultivated passion fruit (Passiflora edulis L.). bioRxiv 522128
- Xiaojing C, Liuxin L (1994) Karyotype analysis of two major cultivated forms of passiflora L.[J]. J Fujian Agric Univ. http://en.cnki.com.cn/Article_en/CJFDTOTAL-FJND401.006.htm
- Ye K, Chen Y, Hu X, Guo J. Computational identification of microRNAs and their targets in apple. Genes Genom. 2013;35:377–385. doi: 10.1007/s13258-013-0070-z. [DOI] [Google Scholar]
- Zeng S, Liu Y, Pan L, Hayward A, Wang Y. Identification and characterization of miRNAs in ripening fruit of Lycium barbarum L. using high-throughput sequencing. Front Plant Sci. 2015;6:778. doi: 10.3389/fpls.2015.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Stellwag EJ. Identification of soybean microRNAs and their targets. Planta. 2008;229:161–182. doi: 10.1007/s00425-008-0818-x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chia JM, Kumari S, Stein JC, Liu Z, Narechania A, Maher CA, Guill K, McMullen MD, Ware D. A genome-wide characterization of microRNA genes in maize. PLoS Genet. 2009;5:e1000716. doi: 10.1371/journal.pgen.1000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wang KL, Wang H, Gu C, Dare AP, Espley RV, He H, Allan AC, Han Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–121. doi: 10.1111/tpj.12792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.