Abstract
Esophageal cancer is one of the leading malignancies worldwide, while around sixty percent of newly diagnosed cases are in China. In recent years, genome-wide sequencing studies and cancer biology studies show that Hippo signaling functions a critical role in esophageal squamous cell carcinoma (ESCC) progression, which could be a promising therapeutic targets in ESCC treatment. However, the detailed mechanisms of Hippo signaling dys-regulation in ESCC remain not clear. Here we identify SHARPIN protein as an endogenous inhibitor for YAP protein. SHARPIN depletion significantly decreases cell migration and invasion capacity in ESCC, which effects could be rescued by further YAP depletion. Depletion SHARPIN increases YAP protein level and YAP/TEAD target genes, such as CTGF and CYR61 in ESCC. Immuno-precipitation assay shows that SHARPIN associates with YAP, promoting YAP degradation possibly via inducing YAP K48-dependent poly-ubiquitination. Our study reveals a novel post-translational mechanism in modulating Hippo signaling in ESCC. Overexpression or activation of SHARPIN could be a promising strategy to target Hippo signaling for ESCC patients.
Abbreviations: SHARPIN, SHANK-associated RH domain interacting protein; ESCC, Esophageal squamous cell carcinoma; UBL, Ubiquitin-like domain; NZF, Npl4 zinc finger domain; EMT, Epithelial-mesenchymal transition; ATCC, American Type Culture Collection
Background
Esophageal cancer accounts for 3.4% of malignancy incidence and 2.6% in cancer-related mortality worldwide [1]. Among the cases, more than 50% newly diagnosed cases happen in China, while the major subtype of esophageal cancer is esophageal squamous cell carcinoma [2]. Although over 300,000 newly diagnosed cases each year in China, the incidence of esophageal carcinoma has high area variations with high incidence in certain district, such as Henan province [3]. Besides of the known environmental factors, such as smoking and alcohol, the alternation of genetic factors also play important role in carcinogenesis process [4], [5], [6]. Recent genomic-based sequencing and molecular biology studies reveal that the dysregulation of Hippo signaling is common in ESCC, while inhibition of Hippo signaling core factor YAP leads to decreased cell proliferation and invasion of ESCC [7], [8]. However, the detailed regulation of Hippo signaling in esophageal cancer is still not clear. Since the specific role of Hippo signaling could be a promising target for ESCC therapeutics, it is particularly important to elucidate the regulatory mechanism of YAP in ESCC.
Hippo signaling was firstly uncovered by genetic screening in Drosophila, which was further revealed as an evolutional conserved tumor suppressor pathway [9]. Hippo signaling controls tissue growth and organ size by a delicate balance between cell proliferation and cell death [10]. The core Hippo pathway consist of a kinase cascade: an upstream kinase MST1/2 phosphorylates and activates a downstream kinase LATS1/2, leading to phosphorylation and inactivation of a transcriptional co-activator YAP/TAZ. When YAP/TAZ is activated, they trans-locate into the nuclear and trans-activate several transcriptional factors, including TEADs and RUNX [10], [11]. The abnormality of Hippo signaling components was found in several cancers, including esophageal cancer [7], [12], [13]. For example, Hippo signaling is dys-regulated by mutations, such as FATs and AJUBA, and gene amplifications, such as YAP in esophageal cancer [7]. Besides, YAP expression level is elevated in esophageal cancer, while YAP protein level is correlated with tumor metastasis and later tumor stage [8], [14].
SHARPIN (SHANK-associated RH domain interacting protein), a linear ubiquitin chain-related protein, was firstly identified from the post-synaptic density of excitatory synapses in the brain [15]. SHAPRIN has essential roles in many aspects, including tissue development, inflammation and homeostasis [16], [17]. However, recent studies reported that SHARPIN was up-regulated in several cancers, such as breast cancer and lung cancer [18], [19]. Quite a few studies showed that SHAPRIN might function as an oncogenic role in cancer, through modulating NFKB signaling and PTEN signaling [20], [21]. In our previous studies, SHARPIN was identified as an oncogene in breast cancer, through promoting ER alpha signaling and suppressing P53 pathway [22], [23]. However, in this current study, SHARPIN functions the tumor-suppression role in esophageal cancer progression. SHARPIN promotes YAP protein poly-ubquitination, which subsequently inhibits the transcriptional regulation of YAP/TEAD target genes in esophageal carcinoma.
Materials and Methods
Cell Culture
EC109, KYSE105 and HEK293 cells were acquired form American Type Culture Collection (ATCC). HEK293 cells were cultured in Dulbecco’s Modified Eagle’s Medium that contains 4.5 g/L glucose and 4 mM L-glutamine (DMEM, 41965, Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS, 10270, Life Technologies). EC109 and KYSE150 cells grown in RPMI-1640 (42401, Life Technologies) supplemented with 2 mM L-glutamine (25030, Life Technologies) and 10% FBS. All cell lines were subject to cell line authentication. The cell line authentication via Short Tandem Repeat (STR) was performed via PowerPlex 21 system. The STR data of HEK293 and KYSE150 cell lines were found consistent with STR data in ATCC.
Plasmids and siRNA
The Flag-tag-SHARPIN plasmid was used in our previous study [22]. The HA-K48 and HA-K63 Ubi plasmids were acquired form our previous study [24]. The Lipofectamin 2000 (1662298, Invitrogen) was used for the plasmids transfection. Small interfering RNAs were used for specific gene knocking-down. The SHARPIN siRNA sequences were: CCUGGAAACUUGACGGAGAdTdT; CUGCUUUCCUCUACUUGCUdTdT. The YAP siRNA sequences were GCUCAUUCCUCUCCAGCUUdTdT. The negative control siRNA sequences were: UUCUCCGAACGUGUCACGUTT. The RNAiMAX reagent (13778150, invitrogen) was used for siRNA transfection.
RNA Extraction and qPCR Analysis
RNeasy plus mini kits were used to extract total RNA (Qiagen) [25]. Real-time PCR was performed as previously described. 36B4 was used for internal control. The primer sequences were shown here. SHARPIN: F: tag cag cca cca gag gtt ac; R: agc agt cag tag agg tcc cc. 36B4: F: ggc gac ctg gaa gtc caa ct; R: cca tca gca cca cag cct tc. CTGF: F: ctc gcg gct tac cga ctg; R: ggc tct gct tct cta gcc tg. CYR61: F: agc agc ctg aaa aag ggc aa; R: agc ctg tag aag gga aac gc.
Quantification of Cell Viability
EC109 and KYSE150 cells were transfected with siSHARPIN or siControl in 24-well plates. Twenty-Four hours after transfection, the cells number was countered and 4000 cells were seeded into 96-well plates. The relative cell viability was measured at indicated time points. Cell numbers were determined using the WST-1 cell proliferation reagent as previously described [5].
Wound Healing Assay
EC109 and KYSE 150 cells were transfected with 50 µM SHARPIN siRNA or sControl. After twenty-four hours, cells were seeded into 12-well plates with 1%FBS. The cells were 100% confluence. The yellow pipette tips were applied for straight scratch. The wound distance was measured at indicated time points and normalized with starting time point. The wound healing recovery was expressed as: [1 − (Width of the wound at a given time/width of the wound at t = 0)] × 100%.
Trans-Well Assay
Cell invasion capacity was measured using the modified two-chamber plates as before [5]. For invasion assay EC109 cells and KYSE105 cells were transfected with 50 µM SHARPIN siRNA or sControl. In order to stimulate invasion, the bottom wells were filled with complete medium, while the upper chambers were added with FBS-free medium. After 12 h, cells were carefully removed and the cells that invaded through the membrane were fixed and stained with Crystal Violet Staining solution. The cell numbers are counted by microscope.
Clone Formation Assay
EC109 and KYSE 150 cells were seeded in six-well plates overnight and treated with 50 nM SHARPIN siRNA or 50 nM siControl. Twenty-four hours post-transfection, the cells were washed with PBS, trypsinized and plated at low density (5000 cell/well in six-well plate). The cells were cultured for 10 days and the medium was refreshed every two days. The colonies were stained with crystal violet. The number of the clones in a given area was counted for each condition.
Western Blotting
Cells were harvested and lysed with RIPA buffer. Proteins were separated by electrophoresis on SDS-polyacrylamide gel electrophoresis (PAGE) and electro-transferred to PVDF membrane. The antibodies used in this study were listed here: Anti-SHARPIN alpha (Ab125188, Abcam); Anti-YAP (SC-101199, Santa Cruz); Anti-HA (MMS-101R, COVANCE); Anti-myc (9E10, ab32, Abcam); Anti-myc (Ab9106, Abcam); Anti-GAPDH (GB12002, Servicebio). Membranes were then washed with PBS for three times and incubated with secondary antibodies Peroxidase-Conjugated AffiniPure Goat Anti-Mouse IgG or Goat Anti-Rabbit IgG. Fluorescent signals were visualized with ECL system. (amersham imager 600, USA).
Co-Immunoprecipitation Assay
Immunoprecipitation was performed as described in previous study [23]. The EC109 cells total cell lysls were pre-cleared with rabbit IgG for 2 h and subsequently immunoprecipitated with SHARPIN antibody (Ab125188, Abcam) over night, while rabbit IgG (Santa Cruz) was used as the negative control. The bounded protein was analyzed by Anti-YAP antibody (SC-101199, Santa Cruz).
Protein Stability Assays
About 105 HEK293 cells were seeded into twenty-four well plates and transfected with 0.5ug Flag-SHARPIN or Flag-vector. After 48 h, cells were treated with 100uM cycloheximide (C7698, Sigma) for indicated time points. Samples were subject to western blot for YAP degradation. For EC109 cells, 105 cells were seeded into 24 well-plate and transfected with 50 nM siSHARPIN or siControl. After 24 h, cells were treated with 100 µM cycloheximide (C7698, Sigma) for indicated time points. Samples were subject to western blot for YAP degradation.
Poly-Ubiquitination Detection Assay
To directly detect the enriched K48-ubiquitinated and K63-ubiqutinated YAP from the cell extracts, HEK293 cells were transfected with 0.5 µg K48 Ubi or 4 µg K63 Ubi plasmids together with 0.5 µg Flag-SHARPIN or Flag-vector. After 48 h, total protein was extracted and pre-cleared with 20ul protein A (santa cruz, SC-2001) for 2 h. The supernatant was collected and immunoprecipitated by YAP antibody. Western blot with HA antibody was performed to detect K48 or K63 poly-ubiquitinated YAP.
Immunofluorescence Assay
EC109 cells were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.2% Triton X-100 for 5 min, and blocked by 5% BSA in PBS for 1 h. A rabbit anti-SHARPIN polyclonal antibody (Ab125188, Abcam) and mouse anti-YAP monoclonal antibodies (SC-101199, Santa Cruz) were used, followed by Alexa Flour 647 (Invitrogen) anti-rabbit antibody and FITC-conjugated anti-mouse antibodies (Jackson ImmunoResearch, West Grove, PA). As negative controls, the samples were incubated with the secondary antibodies without primary antibodies. Images were acquired under conditions fulfilling the Nyquist criterion using Nikon A+ laser scanning confocal system with a 60X oil NA1.4 objective and pinhole size of 1.0 Airy Unit. The acquired pictures were further processed and assembled using ImageJ.
Clinical Breast Tumor Samples
Two hundred and twenty-nine ESCC samples were collected from the first affiliated Hospital of Xinxiang Medical University. All the ESCC cancer samples were examined and the immuno-histochemistry of SHARPIN and YAP were carried out according to standard method. The IHC results of SHARPIN and YAP were examined through pathological specialists.
Statistics
Student’s t-test, Pearson correlation coefficient, and Cox regression analysis were used for comparisons. A P-value of <0.05 was considered to be significant.
Results
SHARPIN Depletion Facilitates Cell Invasion and Migration in Esophageal Squamous Cell Carcinoma
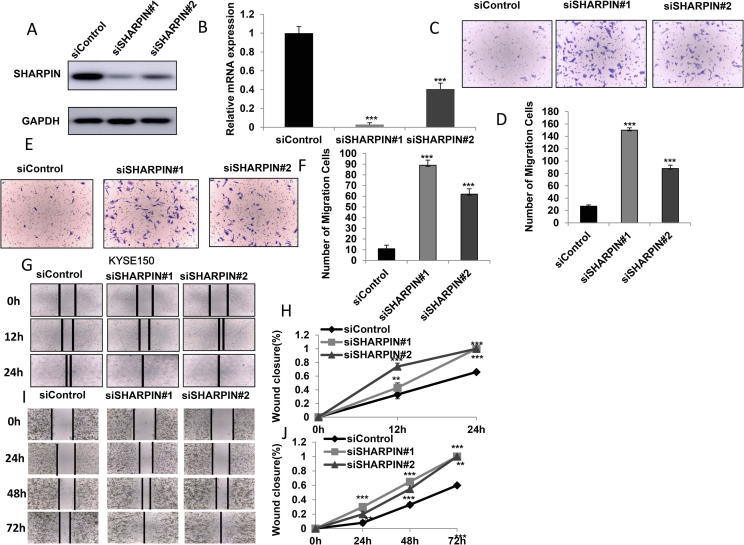
In order to investigate the role of SHARPIN in esophageal cancer cells, SHARPIN was depleted in EC109 and KYSE150 cells. Two independent siRNAs were used in the experiments. The SHARPIN depletion effects were shown in Figure 1A and B (protein and mRNA level). The trans-well assay indicated that SHARPIN depletion via two independent siRNAs significantly increased cell invasion capacity in EC109 and KYSE150 cells (Figure 1C–F). Besides, wound-healing assay showed that SHARPIN depletion promoted cell migration in both EC109 and KYSE150 cells (Figure 1G–J). The WST-1 assay indicated that depletion SHARPIN does not statistically affect cell proliferation and proliferation related gene expression in both EC109 and KYSE150 cells (Supplementary Figure 1A–D).
Figure 1.
SHARPIN depletion facilitates cell invasion and migration in esophageal squamous cell carcinoma. A and B: SHARPIN depletion effect by two different siRNA oligos. EC109 cells are transfected with siSHARPIN or siControl. After 48 h, SHARPIN mRNA and protein levels are determined by Western blot analysis. Actin was used as internal control. C and D: SHARPIN depletion promoted EC109 cell invasion capacity. EC109 cells were transfected with siControl or siSHARPIN. After 24 h, cells were seeded into the chamber for trans-well assay. The cell number was counted and Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). E and F: SHARPIN depletion promoted HYSE 150 cell invasion capacity. HYSE 150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were seeded into the chamber for trans-well assay. The cell number was counted and Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). G and H: Wound-healing assay of EC109 cells were transfected with indicated 50 nM SHARPIN siRNA (mix of #1 and #2) or 50 nM control siRNA. Quantification of wound closure at the indicated time points. Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). I and J: Wound healing assay of HYSE 150 cells were transfected with indicated 50 nM SHARPIN siRNA (mix of #1 and #2) or 50 nM control siRNA. Quantification of wound closure at the indicated time points. Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test).
SHARPIN Depletion Increases YAP Protein Level and Activates YAP/TEAD Signaling in Esophageal Squamous Cell Carcinoma
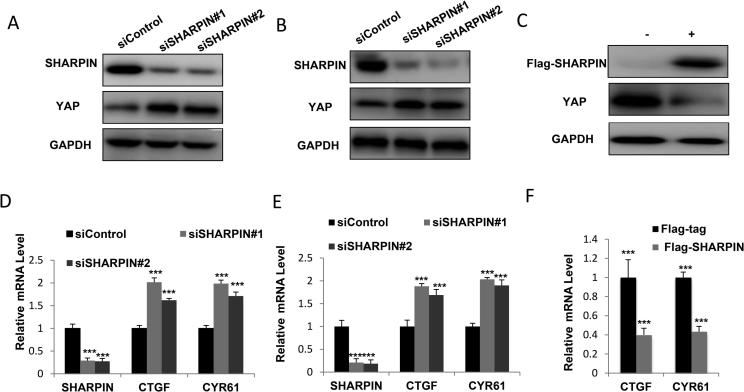
In order to confirm SHARPIN function in Hippo signaling, we depleted SHARPIN via two different siRNAs. SHARPIN depletion significantly increased YAP protein level in both EC109 and KYSE150 cells (Figure 2A and B). Besides, SHARPIN over-expression decreased YAP protein level in EC109 cells (Figure 2C). By examining of YAP/TEAD target genes, we found that SHARPIN depletion significantly increased YAP/TEAD target gene expression (CTGF and CYR61) in both EC109 and KYSE150 cells (Figure 2D and E). Consistently, SHARPIN over-expression inhibited YAP/TEAD target gene expression in EC109 cells (Figure 2F).
Figure 2.
SHARPIN depletion increases YAP protein level and activates YAP/TEAD signaling in esophageal squamous cell carcinoma. A: SHARPIN depletion increased YAP protein levels in ESCC. EC109 cells were transfected with siControl or siSHARPIN. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. B: SHARPIN depletion increased YAP protein levels in ESCC. HYSE150 cells were transfected with siControl or siSHARPIN. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. C: SHARPIN over-expression decreased YAP protein levels in ESCC. HYSE150 cells were transfected with Flag-SHARPIN or Flag plasmids. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. D: SHARPIN depletion increased YAP target gene expression in ESCC. EC109 cells were transfected with siControl or siSHARPIN. After 48 h, total RNA was extracted for gene expression analysis. Each group was done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for target gene expression comparison. E: SHARPIN depletion increased YAP target gene expression in ESCC. HYSE150 cells were transfected with siControl or siSHARPIN. After 48 h, total RNA was extracted for gene expression analysis. Each group was done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for target gene expression comparison. F: SHARPIN overexpression decreased YAP target gene expression in ESCC. EC109 cells were transfected with Flag-SHARPIN or Flag plasmids. After 48 h, total RNA was extracted for gene expression analysis. Each group was done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for target gene expression comparison.
Increase Cell Invasion and Migration by SHARPIN Depletion Could be Rescued by YAP Knocking-Down in Esophageal Squamous Cell Carcinoma
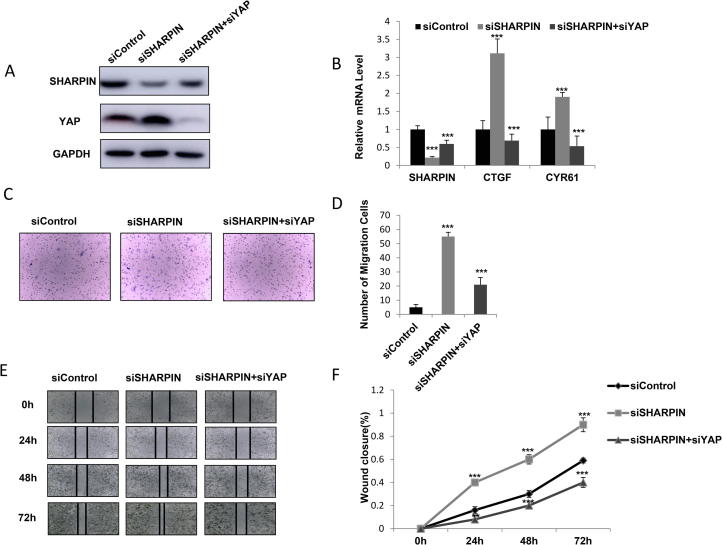
Our previous data showed that SHARPIN depletion could increase YAP signaling activity and dramatically increase cancer cell migration capacity, our further experiments aimed to provide the inner logic link between YAP signaling and cell phenotype in SHARPIN depletion condition. Our data indicated that SHARPIN depletion increased YAP protein level and YAP/TEAD target gene expression, while further depletion of YAP in the cells could bring back the YAP protein level and YAP/TEAD target gene expression in both EC109 and KYSE150 cells (Figure 3A and B; Supplementary Figure 2A). Interestingly, the trans-well assay showed that the increased invaded cell number by SHARPIN knocking-down could be at least partially rescued by further YAP depletion in EC109 and KYSE150 cells (Figure 3C and D; Supplementary Figure 2B and C). Besides, the wound-healing assay also indicated that the increased wound healing speed by SHARPIN knocking-down could be partially rescued by further YAP depletion in EC109 cells (Figure 3E and F; Supplementary Figure 2D and E).
Figure 3.
Increase cell invasion and migration by SHARPIN depletion could be rescued by YAP knocking-down in esophageal squamous cell carcinoma. A: SHARPIN depletion increased YAP protein level, which effect could be reversed by YAP knocking-down. EC109 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. B: SHARPIN depletion increased YAP/TEAD target gene expression, which effect could be reversed by YAP knocking-down. EC109 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After 48 h, total RNA was extracted for gene expression analysis. Each group was done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for target gene expression comparison. C and D: SHARPIN depletion increased ESCC cell invasion capacity, which effect could be reversed by YAP knocking-down. EC109 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After another 24 h, cancer cells were seeded into the chamber for trans-well assay. The cell number was counted and Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). E and F: Wound healing assay indicated that SHARPIN depletion increased ESCC cell migration capacity, which effect could be reversed by YAP knocking-down. EC109 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. Quantification of wound closure at the indicated time points. Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test).
SHARPIN Modulates YAP Protein Stability
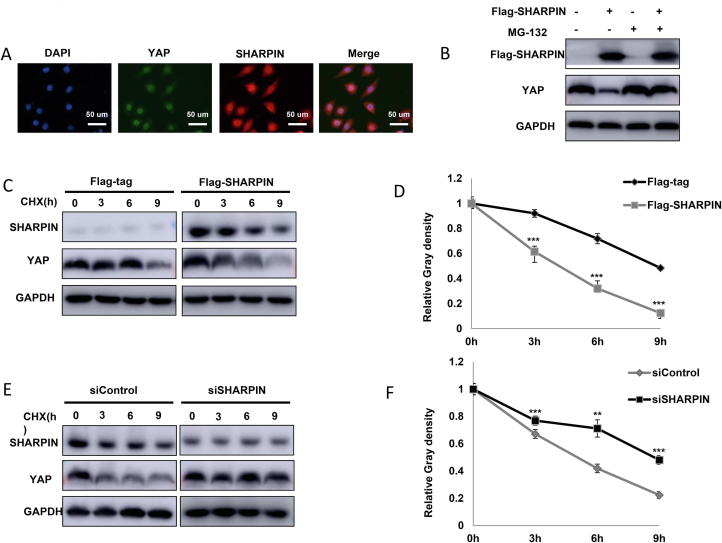
We further investigated the localization of SHARPIN and YAP in ESCC cell lines. Both SAHRPIN and YAP antibody were validated via siRNA knocking down (Supplementary Figure 3A and B). Immuno-staining indicated that both SHARPIN and YAP were mainly localized in the nuclear (Figure 4A). SHARPIN overexpression could suppress the endogenous YAP protein level, which effect could be reversed by inhibition of proteasome inhibitor MG132 in EC109 cells (Figure 4B). We infer that SHARPIN could modulate YAP through post-translational modifications. Up on inhibition of protein synthesis of cycloheximide, the presence of SHARPIN significantly decreased the half-life of YAP in HEK293 cells (Figure 4C and D). On the contrary, depletion of endogenous SHARPIN in EC109 cells dramatically prolonged the half-life of YAP (Figure 4E and F), but did not affect P53 half-life in EC109 cells (Figure 3C and D). Interestingly SHARPIN depletion also prolongs YAP half-life in MDAMB231 cells (Breast cancer cell line) (Supplementary Figure 4A and B). We further analyzed 229 ESCC tumors samples. The Immuno-histochemistry showed that SHARPIN protein level reversely correlated with YAP (P < 0.01) (Supplementary Figure 4C and D).
Figure 4.
SHARPIN modulates YAP protein stability. A: Intracellular localization analysis of SHARPIN and YAP by immunofluorescence assay. EC109 cells were cultured in normal medium before fixation. Intracellular localization of YAP (green) and SHARPIN (red) were shown. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). B: In the presence of the proteasome inhibitor MG132, the degradation effect of SHARPIN on YAP did not further increase YAP protein levels. HEK293 cells were transfected with 0.5 µg Flag-tag or Flag-SHARPIN plasmids. After 24 h, cells were treated with 10 µM MG132/vehicle for 6 h. Cell lysates were prepared for Western blot analysis. The results are representative for three independent experiments. C and D: SHARPIN decreased YAP half-life in HEK293 cells. HEK293 cells were transfected with 0.5 µg Flag-tag or Flag-SHARPIN plasmids. After 24 h, cells were treated with 100 µM cycloheximide/vehicle for indicated times. Cell lysates were prepared for Western blot analysis. The results are representative for three independent experiments. The YAP relative density was measured by Image J software. E and F: SHARPIN depletion increased YAP half-life in EC109 cells. EC109 cells were transfected with 50 µM siControl or siSHARPIN. After 24 h, cells were treated with 100 µM cycloheximide/vehicle for indicated times. Cell lysates were prepared for Western blot analysis. The results are representative for three independent experiments. The YAP relative density was measured by Image J software.
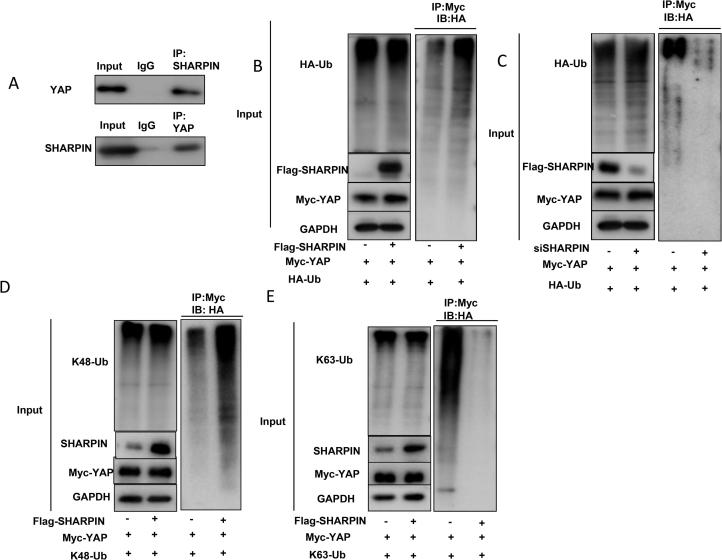
SHARPIN associates with YAP and Promotes YAP K48-Linked Poly-Ubquitination and Degradation
We obtained the further support of functional cooperation of YAP and from immuno-precipitation assay. Co-immunoprecipation (co-IP) of the endogenous proteins from EC109 cells showed that YAP associated with SHARPIN (Figure 5A). Further co-IP showed that SHARPIN associated with YAP through its UBL domain, while YAP interacted with SHARPIN by its WW domain (171AA-292 AA) (Supplementary Figure 5). As an ubiquitin-binding protein, SHARPIN possibly exerted its function through ubiquitin-based manner. We carried out ubiquitin-based immuno-precipitation assay in HEK293 cells, which indicated that SHARPIN overexpression could dramatically increased the overall YAP poly-ubiquitination (Figure 5B). We further detected the endogenous ubiquitin of YAP in EC109 cells. It showed that SHARPIN depletion decreased the endogenous YAP poly-ubiquitination (Figure 5C). Then, we examined SHARPIN ubiquitination activity on YAP in two common ubiquitination manners (K48-linked ubiquitination and K63-linked ubiquitination). Previous studies showed that K48-linked ubiquitination of YAP leaded to protein degradation, while K63-linked ubiquitination of YAP linked to non-proteolytic modification and promoted YAP co-activator function in the nuclear [26]. Interestingly, our ubiquitin-based immuno-precipitation assay showed that SHARPIN promoted K48-linked ubiquitination (proteolytic modification), but inhibited K63-linked ubiquitination (non-proteolytic modification).
Figure 5.
SHARPIN associates with YAP and promotes YAP K48-linked poly-ubquitination and degradation. A: Co-IP assay revealed association between endogenous SHARPIN and YAP protein in EC109 cells. EC109 cells were harvested with RIPA lysis buffer. CO-IP was performed using antibody as indicated. B: SHARPIN increased the overall poly-ubiquitination of YAP. HEK293 cells were transfected with 0.5 µg Flag-SHARPIN or Flag vector. After 24 h, cells were transfected with 1 µg HA-Ub plasmid. After another 24 h, the cell extracts were immunoprecipitated with HA antibody. The poly-ubiquitinated YAP was detected via western blotting analysis. C: SHARPIN depletion decreased the overall poly-ubiquitination of YAP. EC109 cells were transfected with 50 µM siControl or siSHARPIN. After 24 h, cells were transfected with 1 µg HA-Ub plasmid. After another 24 h, the cell extracts were immunoprecipitated with HA antibody. The poly-ubiquitinated YAP was detected via western blotting analysis. D: SHARPIN increases K48-linked poly-ubiquitination of YAP. HEK293 cells were transfected with 0.5 µg Flag-SHARPIN or Flag vector, together with 1 µg HA-K48 Ubi plasmid. The cell extracts were immunoprecipitated with HA antibody. The K48 specific poly-ubiquitinated YAP was detected via western blotting analysis. E: SHARPIN decreases K63-linked poly-ubiquitination of YAP. HEK293 cells were transfected with 0.5 µg Flag-SHARPIN or Flag vector, together with 1 µg HA-K63 Ubi plasmid. The cell extracts were immunoprecipitated with HA antibody. The K63 specific poly-ubiquitinated YAP was detected via western blotting analysis.
Discussion
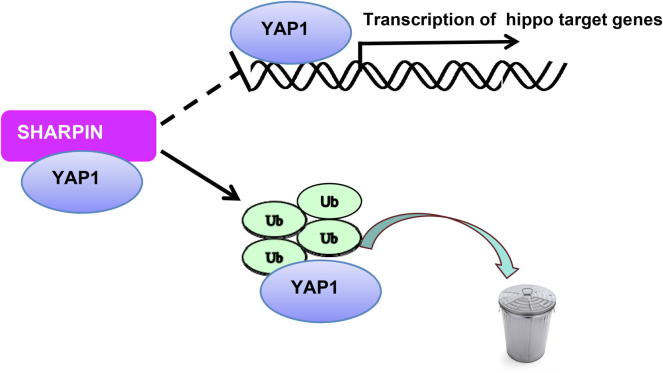
In this study, we report that the ubiquitin-binding protein SHARPIN associates with YAP and promotes YAP degradation in esophageal cancer, which subsequently lead to decreased YAP transcriptional activity and cancer cell progression capacity. Interestingly, SHARPIN could shift the ubiquitination manner of YAP from non-proteolytic to proteolytic dominant way and inhibits YAP nuclear function (Figure 6). On this basis, modulation SHARPIN expression level or activity could be a strategy to modulate YAP/TEAD signaling and subsequently inhibit cancer cell progression in ESCC.
Figure 6.
The hypothetical model for SHARPIN regulating YAP/TEAD signaling in esophageal squamous cell carcinoma: SHARPIN protein associated with YAP and promoted YAP degradation via inducing YAP K48-linked poly-ubiquitination and inhibiting YAP K63-linked poly-ubiquitination.
There are accumulating evidences showing that Hippo signaling regulates tumorigenesis in several cancers [27], [28], [29], [30], [31]. For example, YAP gene is found to have amplification in several cancer types [7], [32]. Besides, several cancer biology studies showed that YAP could transduce several oncogenic pathways, including TEADs, RUNX, and AP1 [33], [34]. YAP could also promote cancer migration capacity and was regarded as key factor in epithelial-mesenchymal transition (EMT) in several cancers [11], [35]. When it comes to esophageal cancer, He et al reported that approximate 40% of esophageal tumors contain genomic abnormalities in hippo signaling, such as YAP gene amplification and FATs mutations [7]. YAP protein level was shown to correlate with later tumor state and poor prognosis in esophageal cancer [8]. Based on the importance of hippo signaling in esophageal cancer, targeting Hippo signaling could be a promising way to treat esophageal cancer.
YAP protein was firstly reported as WW domain containing protein, which is composed of three protein domains: TEAD interaction domain, WW domain and transcriptional activation domain [36]. YAP protein depends WW domain to recognize a specific motif called PPxY, which makes the correct subcellular localization. The TEAD binding domain functions to associate and trans-activate several transcriptional factors, such as TEADs [36]. YAP protein plays control roles in Hippo signaling transduction. When Hippo signaling is activated several serine and threonine kinases, such as MST1/2 (STE20-like protein kinase 1/2) and LATS1/2 (Large tumor suppressor 1/2), promote YAP phosphorylation, nuclear exporting and protein degradation. For example, LATS1/2 could promote YAP phosphorylation in multiple sites (S61, S109, S127 and S381), which subsequently promote YAP1 association with 14-3-3 proteins. This effect leads to YAP retaining in the cytoplasm and protein degradation. However, when Hippo signaling is turn-off, the un-phosphorylated form of YAP could translocate into the nuclear and trans-activate several transcriptional factors, such as TEADs, which lead to the activation of Hippo target gene and cancer cell progression [10]. However, recently studies showed that the YAP ubiquitination modification also played important role in hippo signaling activity. For example, SCFb-TRCP complex could associate with YAP protein and promote its proteasome-mediated degradation [37]. Beside, FBW7, which is a RING E3 ubiquitin ligase, could also induce YAP protein K48-linked ubiquitination and degradation [38]. In our current study, we identify a novel ubiquitin binding protein SHARPIN, which promotes K48-linked ubiquitination and degradation. We believe this not only help to understand the tiny modulation of YAP protein, but also increase the understanding of the ubiquitin binding protein in hippo signaling regulation.
SHARPIN protein is composed of several functional domains, including UBL domain (Ubiquitin-like domain) and NZF domain (Npl4 zinc finger domain) [17]. Based on the structure knowledge, it is more likely treated as a component in an ubiquitin assembly complex. One of the important finding is that SHARPIN forms a linear ubiquitin ligase complex with RNF31 and RBCK1, which promotes the linear ubiquitination of IKKr. This process is necessary for the activation of NFKB pathway [16]. SHARPIN knockout mice manifest with chronic proliferative dermatitis (CPDM), progressive multi-organ inflammation, such as splenic while pulp deficiency and immunoglobulin production deficiency in B/T cells [39], [40], [41], [42]. When it comes to cancer, SHARPIN is more regarded as an oncogene. Several studies reported SHARPIN protein is elevated in various cancers, such as lung cancer, prostate cancer and melanoma [18], [43], [44]. We, and others have shown that SHARPIN could inhibit several tumor-suppressive proteins, such as PTEN and P53 [21], [23]. Besides, SHARPIN could facilitate quite a few oncogenic pathways, including ER alpha signaling and NFKB signaling [22]. However, ER alpha signaling does not exist (https://www.proteinatlas.org/), while the majority of P53 is mutated in ESCC (about 75% mutation) [45], which indicates both of the pathways are not largely compromised in ESCC. Our current study indicated SHARPIN played a tumor-suppressor role in esophageal cancer, which was opposite to previous cancer studies. Depletion SHARPIN significantly promoted cancer invasion and migration capacity in ESCC. The interesting finds not only increase the understanding of SHARPIN role in YAP/TEAD signaling. Further on, it reveals the “multi-face” role of SHARPIN function in different cancer background.
This study identifies the first time, the ubiquitin binding protein SHARPIN as a modulator of YAP/TEAD signaling in human esophageal cancer cells. SHARPIN depletion promotes cancer cell progression and activates YAP/TEAD signaling in multiple esophageal cancer cell lines. As a novel discovered modulator for Hippo signaling, modulation of SHARPIN activity or expression level could be a promising approach to treat esophageal cancer.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
We thank all the members of Henan Key Laboratory of immunology and targeted therapy for sharing valuable material and research support.
Ethics approval and consent to participate
This study was reviewed and approved by the Ethical Board at Xinxiang Medical University.
Consent for publication
All authors consent for publication.
Availability of supporting data
Not applicable.
Funding
The project were supported by the National Science Foundation for Young Scientists of China (No. 81702725, Ting Zhuang), the Joint Fund of the National Natural Science Foundation of China (U1704169, Xiumin Li), the Foundation of Henan Educational Committee (No. 17A310025, Ting Zhuang), and the Program for Ph.D. starting research funding from Xinxiang Medical University (Ting Zhuang), The project of Science and Technology Department of Henan Province (182102310126, Xiumin Li), The National Natural Science Foundation of China (81872032, U1804262, Lidong Wang), The Major Science and Technology Projects of Henan Province (161100311300, Lidong Wang) and The National Key R&D program “Precision Medicine” of China (2016YFC0901403, Lidong Wang). This study is funded by graduate innovative practice base for clinical medicine of Xinxiang Medical University.
Authors’ contributions
AJ. Z., WL. W., ZJ. C, D. P., and XF. Z. performed most of the bench work. T. Z., XM. L., and LD. W. supervised the process of the study and performed the manuscript writing. K. L., JH. H., SJ. W., C. G., BJ. L., and ZY. Y. participated in western blot, real time PCR work. Z. C. performed the data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2019.12.001.
Contributor Information
Lidong Wang, Email: ldwang2007@126.com.
Ting Zhuang, Email: 77090993@qq.com.
Xiumin Li, Email: lxm3029981@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure 1A and B: SHARPIN depletion does not affect cancer cell proliferation. EC109 and HYSE 150 cells were transfected with siControl or siSHARPIN. After 24 h, the WST assay was used to determine the cellar metabolic activity at indicated time points after infection. Experiments were done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for cell growth comparison. Figure 1C and D: SHARPIN depletion does not affect growth-related gene expression. C109 and HYSE 150 cells were transfected with siControl or siSHARPIN. After 24 h, total RNA was extracted and expression was detected through QPCR. Figure 2A: SHARPIN depletion increased YAP protein level, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. Figure 2B and C: SHARPIN depletion increased ESCC cell invasion capacity, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After another 24 h, cancer cells were seeded into the chamber for trans-well assay. The cell number was counted and Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). Figure 2D and E: Wound healing assay indicated that SHARPIN depletion increased ESCC cell migration capacity, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. Quantification of wound closure at the indicated time points. Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). Figure 3A and B: YAP and SHARPIN antibody validation-YAP and SHARPIN were knocking down via siRNAs. After 24 h, Intracellular localization analysis of SHARPIN and YAP by immunofluorescence assay. EC109 cells were cultured in normal medium before fixation. Intracellular localization of YAP (green) and SHARPIN (red) were shown. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). Figure 3C and D: SHARPIN knocking down does not affect mutant P53 half-life in EC109 cells. Figure 4A and B: SHARPIN knocking increases YAP half-life in MDAMB231 cells. Figure 4C: Example tumor cases showing that SHARPIN and YAP protein in IHC. Figure 4D: Statistical analysis of SHARPIN correlation with YAP in ESCC tumor samples. Figure 5A: UBL domain is required for SHARPIN to associate with YAP protein. Figure 5B: WW domain (171-292 aa) is required for YAP to associate with SHARPIN protein.
References
- 1.Disease G.B.D., Injury I., Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100):1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng H. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thoracic Cancer. 2016;7(2):232–237. doi: 10.1111/1759-7714.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W. Cancer statistics: updated cancer burden in China. Chinese J Cancer Res. 2015;27(1):1. doi: 10.3978/j.issn.1000-9604.2015.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennathur A., Gibson M.K., Jobe B.A., Luketich J.D. Oesophageal carcinoma. Lancet. 2013;381(9864):400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhai S. PLCE1 promotes esophageal cancer cell progression by maintaining the transcriptional activity of snail. Neoplasia. 2017;19(3):154–164. doi: 10.1016/j.neo.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L.D. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42(9):759–763. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 7.Gao Y.B. Genetic landscape of esophageal squamous cell carcinoma. Nat Genet. 2014;46(10):1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J. Effect of YAP1 silencing on esophageal cancer. Onco Targets Therapy. 2016;9:3137–3146. doi: 10.2147/OTT.S102338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng Z., Moroishi T., Guan K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30(1):1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulkarni M. RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget. 2018;9(18):14175–14192. doi: 10.18632/oncotarget.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin H.D. Genomic characterization of esophageal squamous cell carcinoma reveals critical genes underlying tumorigenesis and poor prognosis. Am J Hum Genet. 2016;98(4):709–727. doi: 10.1016/j.ajhg.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawada G. Genomic landscape of esophageal squamous cell carcinoma in a Japanese population. Gastroenterology. 2016;150(5):1171–1182. doi: 10.1053/j.gastro.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 14.Wang L. Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene. 2019;38(12):2042–2055. doi: 10.1038/s41388-018-0476-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim S. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci. 2001;17(2):385–397. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda F. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature. 2011;471(7340):637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga F. SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature. 2011;471(7340):633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 18.De Melo J., Tang D. Elevation of SIPL1 (SHARPIN) increases breast cancer risk. PLoS ONE. 2015;10(5) doi: 10.1371/journal.pone.0127546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu T., Lv X., Kong Q., Yuan C. A novel SHARPIN-PRMT5-H3R2me1 axis is essential for lung cancer cell invasion. Oncotarget. 2017;8(33):54809–54820. doi: 10.18632/oncotarget.18957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang H. Elevation of SHARPIN protein levels in prostate adenocarcinomas promotes metastasis and impairs patient survivals. Prostate. 2017;77(7):718–728. doi: 10.1002/pros.23302. [DOI] [PubMed] [Google Scholar]
- 21.De Melo J. SIPL1-facilitated PTEN ubiquitination contributes to its association with PTEN. Cell Signal. 2014;26(12):2749–2756. doi: 10.1016/j.cellsig.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang T. SHARPIN stabilizes estrogen receptor alpha and promotes breast cancer cell proliferation. Oncotarget. 2017;8(44):77137–77151. doi: 10.18632/oncotarget.20368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang H. SHARPIN facilitates p53 degradation in breast cancer cells. Neoplasia. 2017;19(2):84–92. doi: 10.1016/j.neo.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue M. Regulation of estrogen signaling and breast cancer proliferation by an ubiquitin ligase TRIM56. Oncogenesis. 2019;8(5):30. doi: 10.1038/s41389-019-0139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H. SMURF1 facilitates estrogen receptor a signaling in breast cancer cells. J Exp Clin Cancer Res: CR. 2018;37(1):24. doi: 10.1186/s13046-018-0672-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao F. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat Commun. 2018;9(1):2269. doi: 10.1038/s41467-018-04620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salem O., Hansen C.G. The hippo pathway in prostate cancer. Cells. 2019:8(4). doi: 10.3390/cells8040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J., Yu F.X. GPCR-hippo signaling in cancer. Cells. 2019;8:5. doi: 10.3390/cells8050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y. Analysis of the role of the Hippo pathway in cancer. J Transl Med. 2019;17(1):116. doi: 10.1186/s12967-019-1869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calses P.C., Crawford J.J., Lill J.R., Dey A. Hippo pathway in cancer: aberrant regulation and therapeutic opportunities. Trends Cancer. 2019;5(5):297–307. doi: 10.1016/j.trecan.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Ansari D. The hippo signaling pathway in pancreatic cancer. Anticancer Res. 2019;39(7):3317–3321. doi: 10.21873/anticanres.13474. [DOI] [PubMed] [Google Scholar]
- 32.Liu A.M., Xu M.Z., Chen J., Poon R.T., Luk J.M. Targeting YAP and Hippo signaling pathway in liver cancer. Expert Opin Therapeutic Targets. 2010;14(8):855–868. doi: 10.1517/14728222.2010.499361. [DOI] [PubMed] [Google Scholar]
- 33.Zanconato F. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17(9):1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santucci M. The hippo pathway and YAP/TAZ-TEAD protein-protein interaction as targets for regenerative medicine and cancer treatment. J Med Chem. 2015;58(12):4857–4873. doi: 10.1021/jm501615v. [DOI] [PubMed] [Google Scholar]
- 35.Santoro R. MEKK3 sustains EMT and stemness in pancreatic cancer by regulating YAP and TAZ transcriptional activity. Anticancer Res. 2018;38(4):1937–1946. doi: 10.21873/anticanres.12431. [DOI] [PubMed] [Google Scholar]
- 36.Li Z. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24(3):235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B., Li L., Tumaneng K., Wang C.Y., Guan K.L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24(1):72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu K. Fbxw7 is an independent prognostic marker and induces apoptosis and growth arrest by regulating YAP abundance in hepatocellular carcinoma. Mol Cancer. 2014;13:110. doi: 10.1186/1476-4598-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potter C.S. Chronic proliferative dermatitis in Sharpin null mice: development of an autoinflammatory disease in the absence of B and T lymphocytes and IL4/IL13 signaling. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seymour R., Shirley B.J., Hogenesch H., Shultz L.D., Sundberg J.P. Loss of function of the mouse Sharpin gene results in Peyer’s patch regression. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0055224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pouwels J. SHARPIN regulates uropod detachment in migrating lymphocytes. Cell Rep. 2013;5(3):619–628. doi: 10.1016/j.celrep.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z., Potter C.S., Sundberg J.P., Hogenesch H. SHARPIN is a key regulator of immune and inflammatory responses. J Cell Mol Med. 2012;16(10):2271–2279. doi: 10.1111/j.1582-4934.2012.01574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka Y. Sharpin promotes hepatocellular carcinoma progression via transactivation of Versican expression. Oncogenesis. 2016;5(12) doi: 10.1038/oncsis.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J. SHARPIN overexpression induces tumorigenesis in human prostate cancer LNCaP, DU145 and PC-3 cells via NF-kappaB/ERK/Akt signaling pathway. Med Oncol. 2015;32(2):444. doi: 10.1007/s12032-014-0444-3. [DOI] [PubMed] [Google Scholar]
- 45.Okuda E. Detection of p53 gene mutations in human esophageal squamous cell carcinomas using a p53 yeast functional assay: possible difference in esophageal carcinogenesis between the young and the elderly group. Clin Cancer Res. 2001;7(3):600–606. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1A and B: SHARPIN depletion does not affect cancer cell proliferation. EC109 and HYSE 150 cells were transfected with siControl or siSHARPIN. After 24 h, the WST assay was used to determine the cellar metabolic activity at indicated time points after infection. Experiments were done in triplicates. *P < 0.05; **P < 0.01; ***P < 0.001 for cell growth comparison. Figure 1C and D: SHARPIN depletion does not affect growth-related gene expression. C109 and HYSE 150 cells were transfected with siControl or siSHARPIN. After 24 h, total RNA was extracted and expression was detected through QPCR. Figure 2A: SHARPIN depletion increased YAP protein level, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After 48 h, cells were harvested for western blot analysis. SHARPIN and YAP protein levels were determined by Western blot. GAPDH was used as internal control. Figure 2B and C: SHARPIN depletion increased ESCC cell invasion capacity, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. After another 24 h, cancer cells were seeded into the chamber for trans-well assay. The cell number was counted and Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). Figure 2D and E: Wound healing assay indicated that SHARPIN depletion increased ESCC cell migration capacity, which effect could be reversed by YAP knocking-down. KYSE150 cells were transfected with siControl or siSHARPIN. After 24 h, cells were transfected with siYAP or siControl. Quantification of wound closure at the indicated time points. Data are presented as ±SD. **, P < 0.01, ***, P < 0.001 (student’s t-test). Figure 3A and B: YAP and SHARPIN antibody validation-YAP and SHARPIN were knocking down via siRNAs. After 24 h, Intracellular localization analysis of SHARPIN and YAP by immunofluorescence assay. EC109 cells were cultured in normal medium before fixation. Intracellular localization of YAP (green) and SHARPIN (red) were shown. Nuclei (blue) were stained with 4′,6-diamidino-2-phenylindole (DAPI). Figure 3C and D: SHARPIN knocking down does not affect mutant P53 half-life in EC109 cells. Figure 4A and B: SHARPIN knocking increases YAP half-life in MDAMB231 cells. Figure 4C: Example tumor cases showing that SHARPIN and YAP protein in IHC. Figure 4D: Statistical analysis of SHARPIN correlation with YAP in ESCC tumor samples. Figure 5A: UBL domain is required for SHARPIN to associate with YAP protein. Figure 5B: WW domain (171-292 aa) is required for YAP to associate with SHARPIN protein.
Data Availability Statement
Not applicable.