Abstract
Psychiatric diseases are often accompanied by circadian disruptions, but the molecular underpinnings remain largely unclear. To address this, we screened genes that have been previously reported to be associated with psychiatric diseases and found that TRRAP, a gene associated with schizophrenia, is involved in circadian rhythm regulation. Knocking down Nipped‐A, the Drosophila homolog of human TRRAP, leads to lengthened period of locomotor rhythms in flies. Molecular analysis demonstrates that NIPPED‐A sets the pace of the clock by increasing the mRNA and protein levels of core clock genes timeless (tim) and Par domain protein 1ε (Pdp1ε). Furthermore, we found that NIPPED‐A promotes the transcription of tim and Pdp1ε possibly by facilitating deubiquitination of histone H2B via the deubiquitination module of the transcription co‐activator Spt‐Ada‐Gcn5 acetyltransferase complex. Taken together, these findings reveal a novel role for NIPPED‐A in epigenetic regulation of the clock.
Keywords: circadian clock, Drosophila, histone deubiquitination, NIPPED‐A
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Neuroscience; Signal Transduction
The fly homolog of the schizophrenia‐linked TRRAP gene acts via the deubiquitination module of the SAGA chromatin‐modifying complex to set the pace of the circadian clock.
Introduction
A myriad of our behavioral and physiological processes exhibit ~24‐h rhythms or circadian rhythms (Dibner, 2019). These rhythms exist in almost all organisms on the earth and are driven by a relatively conserved molecular clockwork consisting of several transcriptional and translational feedback loops (Li & Zhang, 2015). In Drosophila and many other animals including humans, these loops center around two transcription factors, CLOCK (CLK) and CYCLE (CYC; Hardin, 2011). As a heterodimer, CLK and CYC activate the transcription of period (per) and timeless (tim) via E‐box elements in the promoter regions of these two genes. PER and TIM proteins accumulate in the cytoplasm, form a complex, and translocate into the nucleus where they inhibit the transcriptional activities of CLK/CYC, thus repressing their own transcription. PER/TIM undergo a series of post‐translational modifications (PTMs) that ultimately lead to their degradation, thus enabling CLK/CYC to activate transcription and start a new cycle. CLK/CYC also activate the transcription of two additional transcription factors, vrille (vri) and PAR domain protein 1ε/δ (Pdp1ε/δ), with the former repressing while the latter activating clk transcription.
Although generally accepted by the field, this is an over‐simplified model of the clock. In vivo, DNA wraps around nucleosomes and together they are packaged into chromatin. Chromatin state, determined by histone PTMs, is a key factor regulating transcription. A number of studies in mammals have demonstrated daily oscillations of histone acetylations and methylations which modulate rhythmic circadian gene expression (Papazyan et al, 2016). Consistently, it has also been shown in flies that the acetylation of histone H3‐K9 and trimethylation of H3‐K4 occur in concert with CLK/CYC binding to E‐boxes and activation of per/tim transcription (Taylor & Hardin, 2008). One recent study reported circadian H2B monoubiquitination in the mouse liver which may influence the expression of clock genes (Tamayo et al, 2015). Overall, it is largely unclear how these chromatin modifications contribute to clock regulation and ultimately to rhythmic behavior and physiology.
Disruptions of circadian rhythms are associated with many diseases and disorders (Takahashi et al, 2008). Psychiatric diseases such as major depressive disorder, bipolar disorder, and schizophrenia make up approximately 20% of all illnesses and about 25% of the population is affected at some point during their lifetime (Zordan & Sandrelli, 2015). Most psychiatric disorders involve circadian disruptions including alterations of sleep/wake cycle, core body temperature rhythms, as well as rhythms in melatonin and cortisol secretion (Jones & Benca, 2015). Moreover, nearly every anti‐depressant and anti‐psychotic medication exerts effects on circadian rhythm, and chronobiological interventions demonstrate efficacy in improving mood‐related symptoms (Zhang et al, 2014; Jones & Benca, 2015). Although circadian abnormalities have long been hypothesized to play a role in the pathology of various psychiatric diseases, how circadian disruptions occur in these illnesses remains elusive (McClung, 2013).
In this study, we screened genes that have been reported to be associated with human psychiatric disorders to identify candidates involved in regulating circadian rhythms in flies. We discovered Nipped‐A, the fly homolog of TRRAP which is associated with schizophrenia, to play a role in regulating the circadian clock (Xu et al, 2012). TRRAP/NIPPED‐A is an evolutionarily conserved protein known to function as a histone acetyltransferase (HAT) cofactor by facilitating the recruitment of HAT to chromatin to regulate transcription and DNA repair (Murr et al, 2007). Here, we demonstrate that knocking down Nipped‐A leads to lengthened period of locomotor rhythm, likely due to decreased transcription of tim and Pdp1ɛ. Furthermore, we found that NIPPED‐A is physically associated with tim and Pdp1 loci, facilitating the deubiquitination of H2B by the deubiquitination (DUB) module of the Spt‐Ada‐Gcn5 acetyltransferase (SAGA) complex which could in turn enhance transcription.
Results
Knocking down Nipped‐A lengthens the period of locomotor rhythm
We initiated a RNAi screen of Drosophila homologs of genes reported to be associated with psychiatric conditions in humans to identify genes that are involved in circadian regulation. We knocked down the expression of these candidate genes in all clock cells (including neurons and glial cells) using a timGAL4 driver or mainly in circadian neurons using a cryptochrome (cry)GAL4‐16 driver (Emery et al, 1998, 2000), and assessed the effects of these manipulations on fly locomotor rhythm. So far we have tested 24 genes and have identified Nipped‐A, the Drosophila homolog of human TRRAP, to be involved in determining the period length of fly locomotor rhythm under constant darkness (DD). Knocking down Nipped‐A results in roughly 1‐ to 3‐h longer period with 6 RNAi lines (generated from 3 independent RNAi transgenes; Fig 1A–C; Appendix Table S1). When Nipped‐A is knocked down only in the PIGMENT‐DISPERSING FACTOR (PDF)‐expressing neurons, which are believed to be the major pacemaker neurons, the period is lengthened by ~1 h in 3 of the RNAi lines (Fig 1D; Appendix Table S1; Renn et al, 1999). We also observed significant reduction in the power of the rhythms in some of the lines and have verified that Nipped‐A mRNA levels are decreased in all RNAi lines used (Appendix Figs S1 and S2, and Table S1).
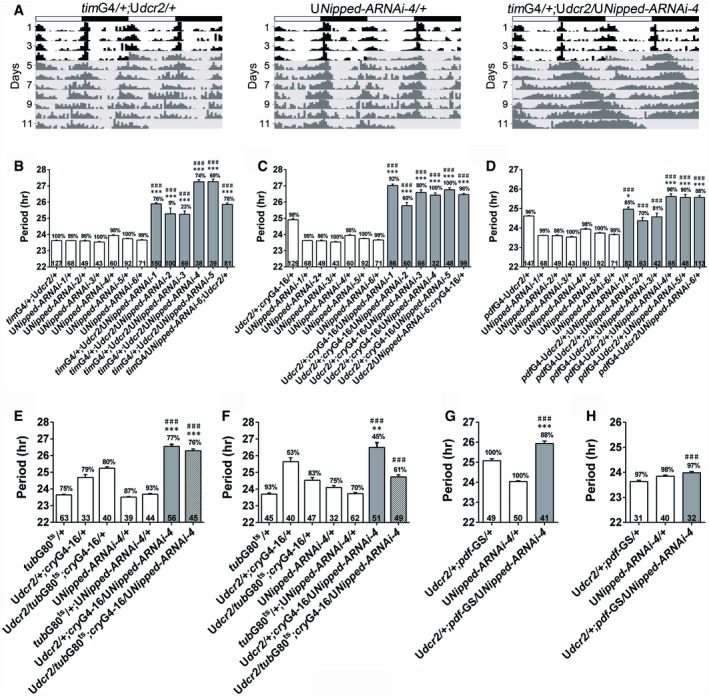
Figure 1. Knocking down Nipped‐A lengthens the period of fly locomotor rhythms.
-
ADouble‐plotted representative actograms of the indicated genotypes. Flies are monitored in LD for 4 days and then DD for 7 days. Dicer2 (dcr2) is co‐expressed to enhance the effects of RNAi. White boxes indicate the light phase, and black boxes indicate the dark phase. Gray shades indicate DD.
-
B–DThe period of DD locomotor rhythms of flies with Nipped‐A knocked down and controls.
-
E, FUsing temperature‐sensitive GAL80 system to control Nipped‐A RNAi expression. (E) The period of DD locomotor rhythms of flies raised at 18°C and tested at 29°C. (F) The period of DD locomotor rhythms of flies raised at 29°C and tested at 18°C.
-
G, HThe period of DD locomotor rhythms of flies treated with RU486 to activate the pdfG4‐geneswitch (pdf‐GS) driver (G) or vehicle control (H).
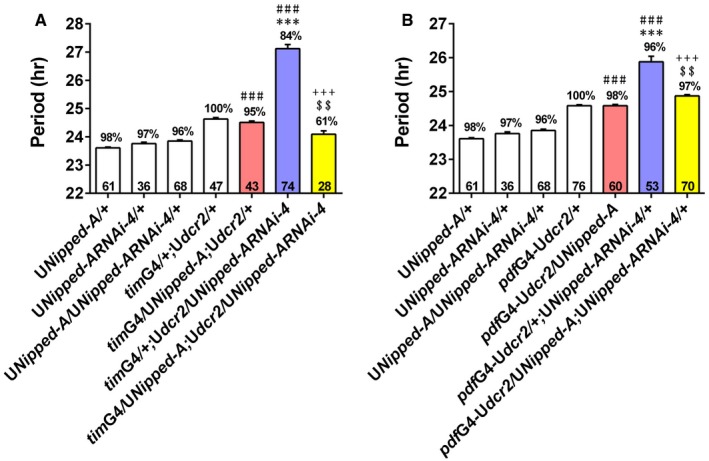
We employed a CRISPR/Cas9‐based transcriptional activation system, flySAM, to over‐express Nipped‐A (Jia et al, 2018). This manipulation does not significantly alter period length on WT background, but it rescues the long‐period phenotype in Nipped‐A RNAi flies, validating that the lengthened period is indeed caused by Nipped‐A deficiency (Fig EV1; Appendix Table S2). As a control, we expressed GFP and did not observe shortening of the period length in Nipped‐A RNAi flies (Appendix Table S3).
Figure EV1. Over‐expressing Nipped‐A rescues the long period induced by knocking down Nipped‐A .
-
A, BThe period of DD locomotor rhythm of Nipped‐A RNAi flies when over‐expressing Nipped‐A using timG4 (A) and pdfG4(B).Error bars represent SEM. Digits on the bar are the number of flies tested. Percentage of rhythmicity is indicated above the bars. Statistical difference is measured using one‐way ANOVA, P < 0.001, Tukey's multiple comparison test, $$ P < 0.01, ***/###/+++ P < 0.001, * compared with the G4 control; # compared with the UAS control, + compared with the Nipped‐A RNAi flies, $ compared with the over‐expression flies. White bar indicates UAS or GAL4 controls. Red bar indicates Nipped‐A over‐expressing flies. Blue bar indicates flies with Nipped‐A knocked down. Yellow bar indicates flies with Nipped‐A over‐expressing and knocked down at the same time. G4, GAL4; U, UAS.
To test whether NIPPED‐A functions in the adult circadian system, we used a temperature‐sensitive tubulin (tub)GAL80ts in combination with cryGAL4‐16 to knock down Nipped‐A specifically during the adult or developmental stage (McGuire et al, 2004). tubGAL80ts represses the transcriptional activities of GAL4 at permissive temperature (18°C), and thus, GAL4‐driven transcription can only occur under restrictive temperature (29°C). When RNAi is specifically expressed in adults, the period is lengthened by ~1 h (Fig 1E; Appendix Table S1; McGuire et al, 2004). On the other hand, when RNAi is expressed exclusively during the developmental stage, the period does not appear to be altered (Fig 1F; Appendix Table S1). We also employed a gene switch system to turn on RNAi expression specifically in the PDF neurons of adult flies a few days before the start of behavioral monitoring (Depetris‐Chauvin et al, 2011). Similarly, we observed ~1‐h longer period in drug‐treated flies which have RNAi expression activated, but not in the vehicle‐treated controls (Fig 1G and H; Appendix Table S1). Taken together, these results indicate that NIPPED‐A is involved in setting the pace of the adult clock.
NIPPED‐A sets the pace of the clock by promoting tim and Pdp1ε mRNA levels
We next sought to characterize the mechanism of how NIPPED‐A regulates the clock by examining the effects of knocking down Nipped‐A on clock gene expression. tim and Pdp1ε mRNA levels are significantly reduced in the heads of these flies compared to the control (Fig 2A). We furthered validated the effects by measuring TIM and PDP1ε protein levels and found these to be reduced as well (Fig 2B and C). Moreover, we observed a significant decrease of PER in flies with Nipped‐A knocked down, whereas per mRNA levels are not significantly altered (Fig 2A–C). This is likely caused by the decrease in TIM levels, as TIM is known to stabilize PER (Hardin, 2011). Since DD period is believed to be determined by the PDF‐expressing small ventral lateral neurons (s‐LNvs), we assessed the expression of core clock proteins in these cells (Stoleru et al, 2005). Consistent with changes occurring at the whole‐head level, TIM, PDP1ε, and PER protein levels are also reduced in the s‐LNvs (Fig 2D and E). We identified a similar role for NIPPED‐A in cultured Drosophila cell line. Knocking down Nipped‐A in S2 cells leads to substantial reduction of tim and Pdp1ε mRNA levels, and to a lesser extent, vri mRNA level (Fig EV2).
Figure 2. NIPPED‐A acts to increase the mRNA and protein levels of core clock gene tim and Pdp1ε .
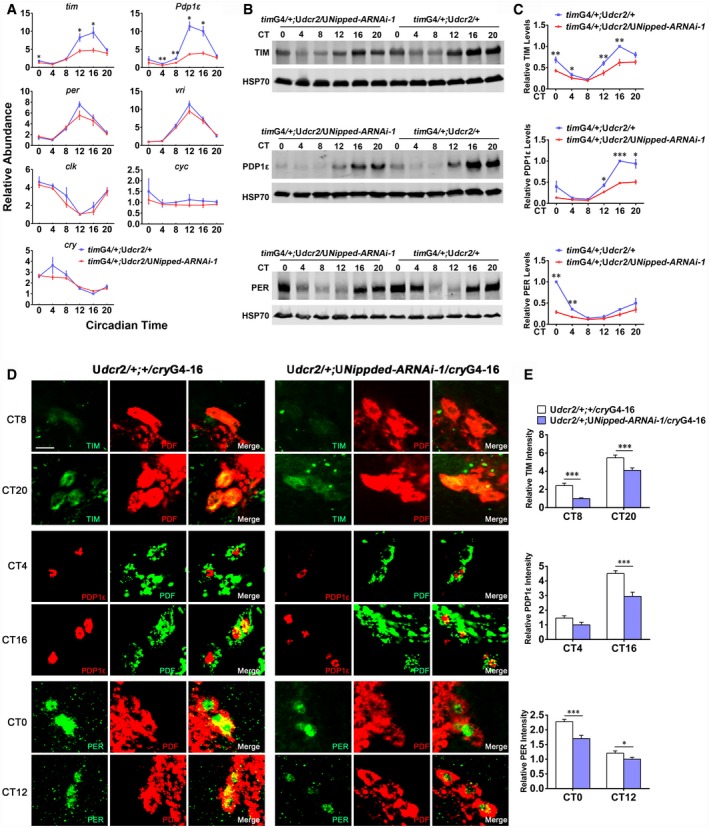
- Plots of relative mRNA abundance vs. circadian time (CT, CT0 is the time of subjective lights on) for clock genes determined by qRT–PCR in whole‐head extracts of Nipped‐A RNAi (timG4/+;Udcr2/UNipped‐ARNAi‐1/+) and control (timG4/+;Udcr2/+) collected on the first day of DD (DD1) (tim/cry, n = 5; Pdp1ε/per/clk/cyc, n = 3; vri, n = 6). For each time series, the value of the lowest time point was set to 1.
- Western blots of proteins from whole‐head extracts of Nipped‐A RNAi and control flies collected on DD1 and probed with TIM, PDP1ε, and PER antibodies.
- Quantification of TIM (n = 5), PDP1ε (n = 3), and PER (n = 3) protein levels of blots in (B). TIM, PDP1ε, and PER protein levels were normalized to that of HSP70. For each time series, the value of the control at the peak time point was set to 1.
- Brains from Udcr2/+;cryG4‐16/+ and Udcr2/+;cryG4‐16/UNipped‐ARNAi flies collected at CT0, 4, 8, 12, 16, 20 on DD1 were immunostained with TIM (green), PDP1ε (red), PER (green), and PDF (red or green) antisera. The scale bar represents 10 μm.
- Quantification of TIM, PDP1ε, and PER protein levels in the s‐LNvs of images in (D) (TIM: CT8: Udcr2/+;cryG4‐16/+, n = 89; Udcr2/+;cryG4‐16/UNipped‐ARNAi, n = 58; CT20: Udcr2/+;cryG4‐16/+, n = 130; Udcr2/+;cryG4‐16/UNipped‐ARNAi, n = 80; PDP1ε: CT4: Udcr2/+;cryG4‐16/+, n = 46; Udcr2/+;cryG4‐16/UNipped‐ RNAi, n = 26; CT16: Udcr2/+;cryG4‐16/+, n = 65; Udcr2/+;cryG4‐16/UNipped‐ARNAi, n = 41; PER: CT0: Udcr2/+;cryG4‐16/+, n = 90; Udcr2/+;cryG4‐16/UNipped‐ARNAi, n = 58; CT12: Udcr2/+;cryG4‐16/+, n = 86; Udcr2/+;cryG4‐16/UNipped‐ARNAi, n = 64).
Figure EV2. Knocking down Nipped‐A decreases tim, Pdp1ε, and vri mRNA levels in cultured cells.
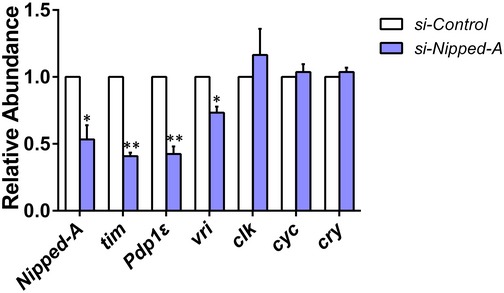
Relative mRNA abundance of clock genes determined by qRT–PCR in S2 cells transfected with small interfering RNA targeting Nipped‐A (si‐Nipped‐A; n = 3). Error bars represent SEM. Student's t‐test, *P < 0.05, **P < 0.01.
Based on our findings, we propose that NIPPED‐A regulates clock timing by promoting tim and Pdp1ε mRNA levels. To test this hypothesis, we first over‐expressed tim in flies with Nipped‐A knocked down and found this partially rescues the long‐period phenotype, whereas over‐expressing tim in control flies does not affect the period (Fig 3A; Appendix Table S4 and Fig S3A; McDonald et al, 2001). On the other hand, knocking down Nipped‐A in a tim 01 heterozygous mutant background dramatically reduces the power of the rhythm, while knocking down Nipped‐A in a wild‐type (WT) background does not substantially decrease the power (Sehgal et al, 1994; Fig 3B; Appendix Table S4). Next, we show that over‐expressing Pdp1ε in Nipped‐A RNAi flies completely reverts the lengthened period, whereas over‐expressing Pdp1ε in WT flies does not alter the period (Fig 3C; Appendix Table S5; Benito et al, 2007). We have validated that this manipulation not only increases Pdp1ε mRNA level, but also the mRNA levels of clk, per, and tim (Appendix Fig S3B). Heterozygous Pdp1ε 3135 mutation further enhances the long‐period phenotype caused by knocking down Nipped‐A, but exerts no effect in control flies (Zheng et al, 2009; Fig 3D; Appendix Table S5). Since knocking down Nipped‐A leads to reduced PER protein level, we over‐expressed per in Nipped‐A RNAi flies and found this manipulation fully rescues the long‐period phenotype, while a per L mutation synergistically enhances this phenotype (Fig 3E and F; Appendix Table S6 and Fig S3C; Kaneko & Hall, 2000; Konopka & Benzer, 1971). The genetic interactions appear to be relatively specific, as we do not detect prominent interaction between Nipped‐A and the core clock gene clock (clk; Appendix Fig S4 and Table S7; Zhao et al, 2003). Taken together, these data support a role for NIPPED‐A in determining the speed of the molecular clock by promoting tim (and consequently PER) and Pdp1ε expression.
Figure 3. Nipped‐A synergistically interacts with tim, Pdp1ε, and per to determine period length.
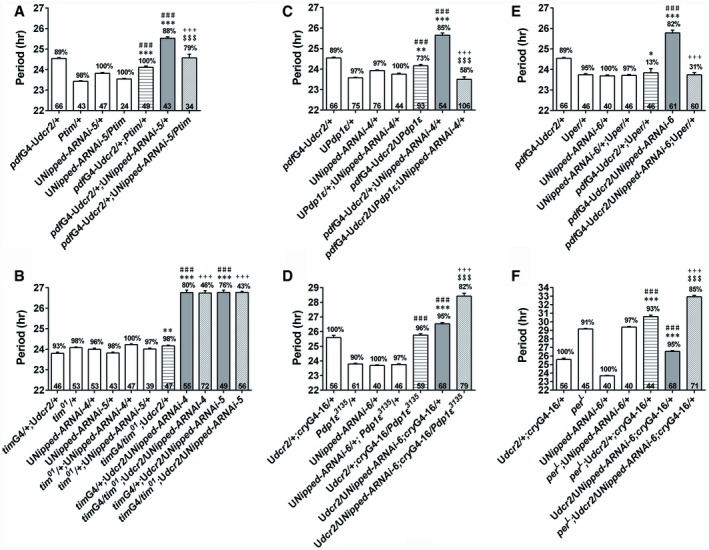
-
A, BThe period of DD locomotor rhythm of Nipped‐A RNAi flies over‐expressing tim (A) or carrying heterozygous tim 01 mutation (B). Ptim is a tim cDNA construct driven by tim promoter.
-
C, DThe period of DD locomotor rhythm of Nipped‐A RNAi flies over‐expressing Pdp1ε (C) or carrying heterozygous Pdp1ε 3135 mutation (D).
-
E, FThe period of DD locomotor rhythm of Nipped‐A RNAi flies over‐expressing per (E) or carrying per L mutation (F).
NIPPED‐A is physically associated with tim/Pdp1ε loci and does not exhibit significant oscillation
We next tested whether NIPPED‐A functions as an instructive or permissive signal to the clock by assessing its temporal expression pattern. Neither the mRNA nor the protein level of Nipped‐A displays significant oscillation under light–dark cycles (LD) or DD (Fig 4A and B). We detected binding of NIPPED‐A at tim and Pdp1 gene loci that is significantly above background, although the binding does not appear to cycle throughout the day, suggesting that NIPPED‐A plays a permissive rather than instructive role in timing the clock (Fig 4C; Appendix Fig S5). As a positive control, we examined the binding of CLK at tim and Pdp1ε promoters and observed significant changes throughout the day, consistent with previous studies that report cyclic binding of CLK at E‐box genes (Appendix Fig S6; Zhou et al, 2016). Notably, knocking down Nipped‐A significantly increases CLK binding at Pdp1ε and per promoters, while a trend of increase is observed at tim promoter. On the other hand, CLK binding at vri promoter is not significantly altered. We also observed binding of NIPPED‐A at per locus although NIPPED‐A does not appear to regulate per transcription (Fig 4C).
Figure 4. NIPPED‐A is physically associated with tim and Pdp1ε loci.
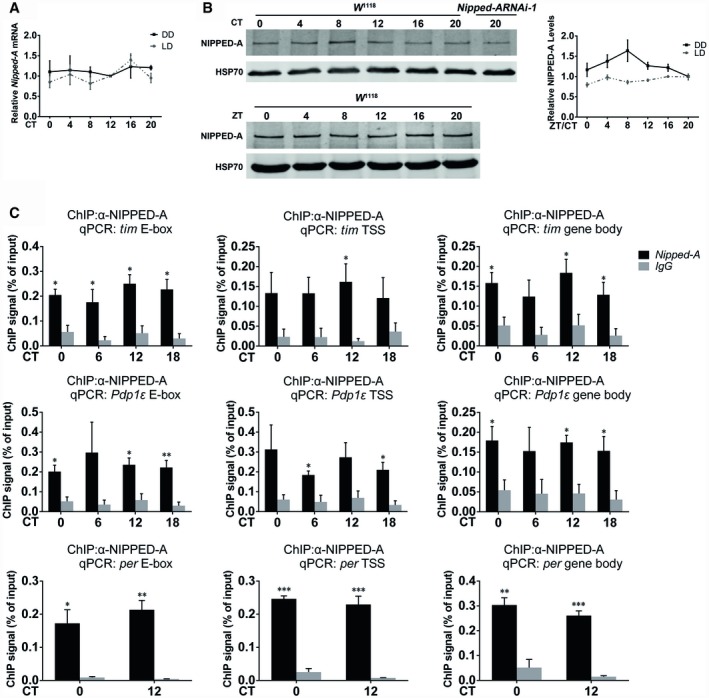
- Plots of relative mRNA abundance of Nipped‐A in whole‐head extracts of w 1118 flies in LD and DD1 determined by qRT–PCR (n = 3).
- Left panel: Western blots of proteins from whole heads of w 1118 and Nipped‐A RNAi flies collected during LD or DD1 and probed with NIPPED‐A antibody. Right panel: quantification of NIPPED‐A protein levels of blots in the left panel (LD, n = 3; DD1, n = 5). ZT, Zeitgeber time. ZT0 is the time of lights on.
- Chromatin immunoprecipitation (ChIP) assays to detect NIPPED‐A binding at E‐box, transcription start site (TSS), and gene body of tim, Pdp1ε, and per using w 1118 flies (n = 3).
Knocking down Nipped‐A reduces tim/Pdp1ε pre‐mRNA levels and increases H2B ubiquitination at tim/Pdp1ε loci
To address how NIPPED‐A enhances the mRNA levels of tim and Pdp1ε, we examined the pre‐mRNA levels of these two genes in Nipped‐A RNAi flies and observed significant reduction, suggesting that knocking down Nipped‐A down‐regulates the transcription of tim and Pdp1ε (Fig 5A). This is consistent with a role for NIPPED‐A/TRAPP in transcriptional activation (Murr et al, 2007).
Figure 5. NIPPED‐A promotes the transcription of tim and Pdp1ε and facilitates H2B deubiquitination.
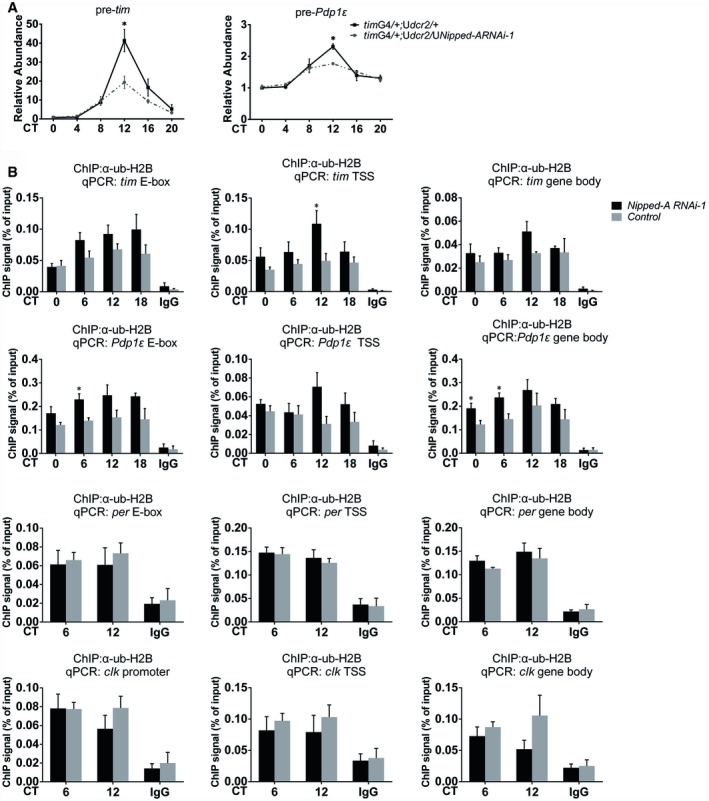
- Plots of relative pre‐mRNA abundance of tim (n = 5) and Pdp1ε (n = 3) determined by qRT–PCR in the whole‐head extracts of Nipped‐A RNAi (timG4/+;Udcr2/UNipped‐ARNAi‐1) and control (timG4/+;Udcr2/+) flies during DD1.
- ChIP assays to detect ub‐H2B binding at E‐box, TSS, and gene body of tim, Pdp1ε, per, and clk in Nipped‐A RNAi (timG4/+;Udcr2/UNipped‐ARNAi‐1) and control (timG4/+;Udcr2/+; n ≥ 3).
Since NIPPED‐A/TRRAP is known to facilitate transcription by recruiting HAT and thus promoting the acetylation of Histone H3 and H4, we measured the effects of knocking down Nipped‐A on H3 and H4 acetylation status at tim and Pdp1ε gene loci (Murr et al, 2007). In contrary to our expectation, we found there is either no significant change or in some cases even an increase in acetylation (H3K9 and H3K27), which is usually associated with elevated rather than diminished transcription (Fig EV3; Murr et al, 2007). This means reduction of tim and Pdp1ε transcription in Nipped‐A RNAi flies is probably not due to defects in histone acetylation.
Figure EV3. Knocking down Nipped‐A increases acetyl‐H3K9 and acetyl‐H3K27 at the promoters of tim and Pdp1ε .
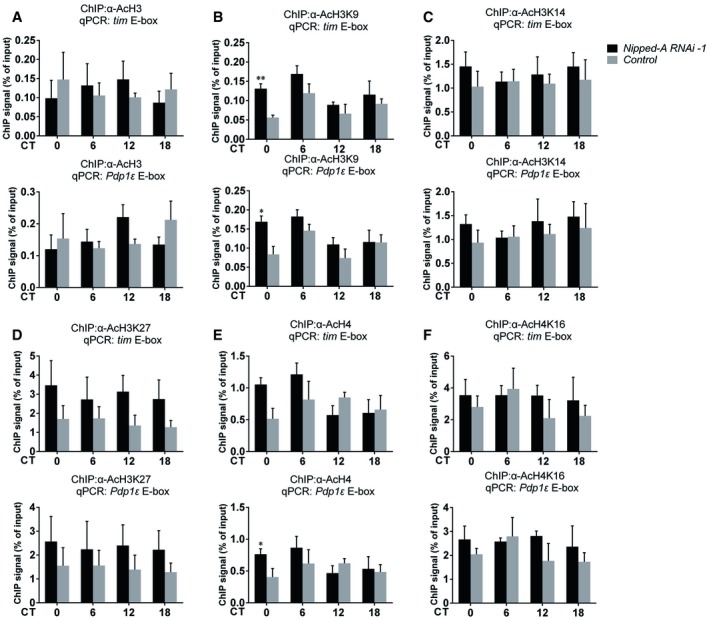
- ChIP assays to detect acetyl‐H3 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi (timG4/+;Udcr2/UNipped‐ARNAi‐1) and control (timG4/+;Udcr2/+) flies (n = 3).
- ChIP assays to detect acetyl‐H3K9 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi (timG4/+;Udcr2/UNipped‐ARNAi‐1) and control (timG4/+;Udcr2/+) flies (n = 3).
- ChIP assays to detect acetyl‐H3K14 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi and control flies (n = 4).
- ChIP assays to detect acetyl‐H3K27 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi and control flies (n = 4).
- ChIP assays to detect acetyl‐H4 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi and control flies (n = 4).
- ChIP assays to detect acetyl‐H4K16 binding at E‐box elements in tim and Pdp1ε promoters of Nipped‐A RNAi and control flies (n = 3).
It has been reported that NIPPED‐A/TRRAP is a member of the SAGA complex which contains both a HAT and a DUB module, and mutations in TRRAP can impair HAT activity of the SAGA complex (Koutelou et al, 2010; Helmlinger, 2012). Since TRRAP serves as a scaffold to interact with both enzymatic modules, we hypothesized that in our case here knocking down Nipped‐A interferes with the DUB activity of SAGA which in turn impairs transcription, as deubiquitination of histone H2B has been shown to promote transcription (Henry et al, 2003; Daniel et al, 2004; Wyce et al, 2007; Sharov et al, 2017). Indeed, we observed significantly enhanced H2B ubiquitination at the promoter and transcriptional start site of tim and Pdp1ε gene loci in Nipped‐A RNAi flies, which could account for reduced expression of these two genes (Fig 5B). On the other hand, H2B ubiquitination is not significantly altered at per and clk loci, which is consistent with the observation that the transcript levels of these two genes are not affected by Nipped‐A deficiency.
NIPPED‐A functions in synergy with SAGA DUB module to set the pace of the clock
To validate whether H2B deubiquitination by the SAGA complex plays a role in period determination, we knocked down not, the deubiquitinase in SAGA complex (Weake et al, 2008). The power of the rhythm is significantly reduced when cryGAL4‐16 is used to drive the expression of two independent RNAi lines, and we have confirmed that not mRNA level is indeed decreased (Fig 6A and B, and Appendix Fig S7A and Table S8). At the molecular level, we observed that knocking down not leads to significant reduction in the mRNA level of all six core clock genes and significant elevation of H2B ubiquitination at tim/Pdp1 loci (Fig 6C and D). Over‐expressing not shortens the long‐period phenotype caused by Nipped‐A deficiency (Fig 6E and F, and Appendix Fig S7B and Table S9). On the other hand, knocking down not synergistically enhances the period lengthening effect of Nipped‐A RNAi, while knocking down not alone does not substantially alter the period (Fig 6G; Appendix Table S9). These results indicate that NIPPED‐A and NOT function together to regulate the clock.
Figure 6. Nipped‐A synergistically interacts with not to determine period length.
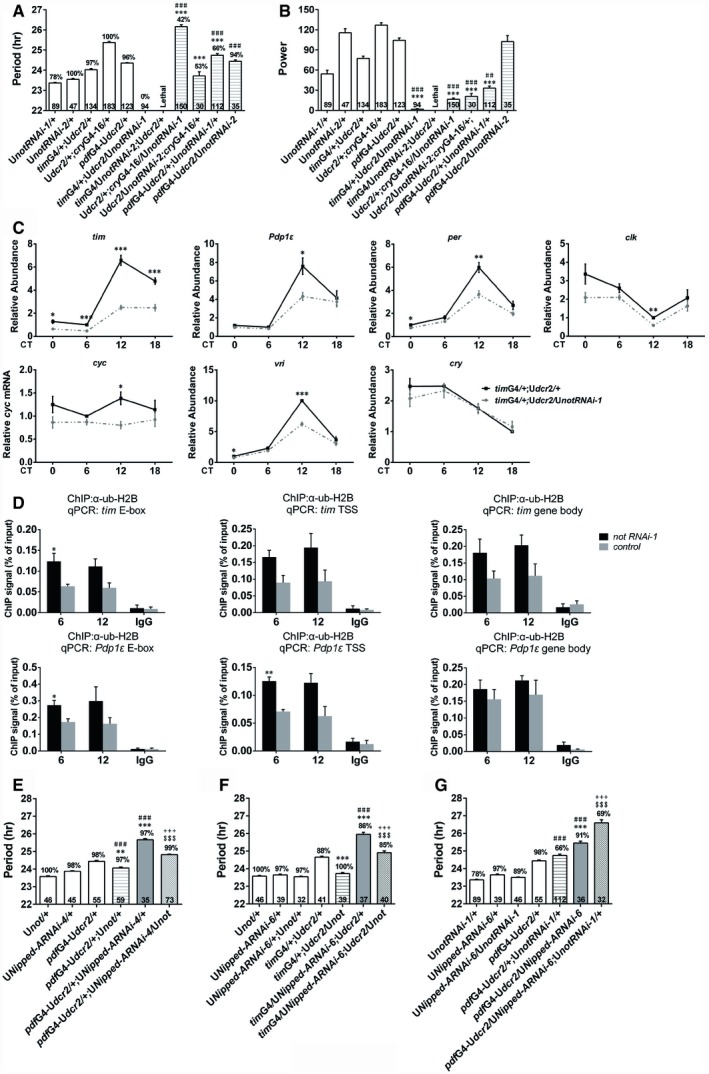
-
A, BThe period (A) and power (B) of DD locomotor rhythms of flies with not knocked down and controls.
-
CPlots of relative mRNA abundance vs. CT for clock genes determined by qRT–PCR in whole‐head extracts of not RNAi (timG4/+;Udcr2/UnotRNAi) and control (timG4/+;Udcr2/+) collected on DD1 (n = 5).
-
DChIP assays to detect ub‐H2B binding at E‐box, TSS, and gene body of tim and Pdp1ε in not RNAi (timG4/+;Udcr2/UnotRNAi) and control (timG4/+;Udcr2/+; n = 3).
-
E, FThe period of DD locomotor rhythm of flies when knocking down Nipped‐A and over‐expressing not.
-
GThe period of DD locomotor rhythm of flies co‐expressing Nipped‐A RNAi and not RNAi.
To further verify that deubiquitination by SAGA is involved in timing the clock, we tested flies lacking Sgf11, which is another subunit in the DUB module (Weake et al, 2008). Knocking down Sgf11 leads to significantly reduced power, and the phenotype is quite prominent with one of the RNAi lines but rather mild with the other RNAi line (Fig EV4A–C; Appendix Table S10). There is a significant lengthening of the period with one of the RNAi lines, but the extent of the change is quite small. Sgf11 mutants also do not show prominent changes in period or power (Fig EV4D; Appendix Table S11; Weake et al, 2008). Overall, Sgf11 deficiency does not appear to result in prominent locomotor rhythm phenotype. However, knocking down or mutating Sgf11 substantially lengthens the period in Nipped‐A RNAi flies (Fig EV4E and F; Appendix Table S11). This synergistic interaction between Nipped‐A and Sgf11 in determining period length further supports the idea that NIPPED‐A acts cooperatively with the DUB module of SAGA to set the pace of the clock, and consistent with the notion that the effects of NIPPED‐A on tim and Pdp1ε are mediated by H2B deubiquitination.
Figure EV4. Nipped‐A synergistically interacts with Sgf11 to determine period length.
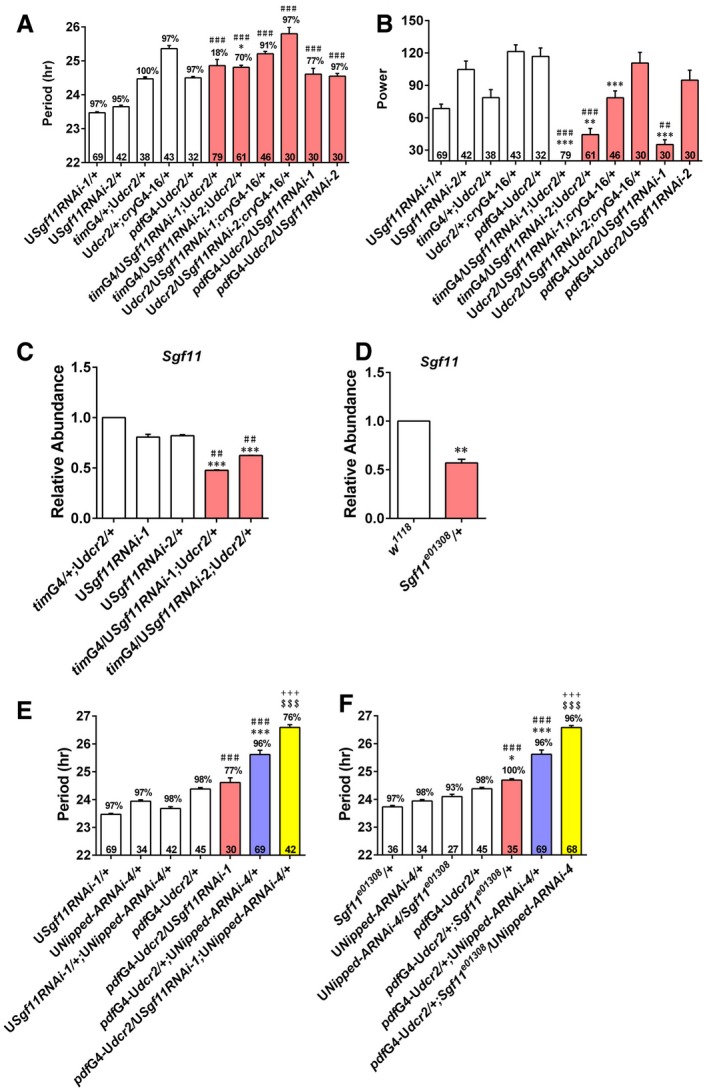
-
A, BThe period (A) and power (B) of DD locomotor rhythms of flies with Sgf11 knocked down and controls.
-
C, DPlots of relative mRNA abundance for Sgf11 determined by qRT–PCR in whole‐head extracts of Sgf11 RNAi (C) (sgf11RNAi‐1: n = 5; sgf11RNAi‐2: n = 3) and Sgf11 e01308 (D) flies (n = 3).
-
EThe period of DD locomotor rhythm of flies when knocking down Nipped‐A and Sgf11.
-
FThe period of DD locomotor rhythm of Sgf11 mutant flies with Nipped‐A knocked down.
Discussion
Our study identifies a role for TRRAP/Nipped‐A in setting the pace of the circadian clock in flies. A missense variant (Ile295Phe) and a splice site variant (c.7223.+6) in TRRAP have been reported to be associated with schizophrenia in humans, and up to 80% of schizophrenia patients experience circadian/sleep disruptions (Wulff et al, 2012; Xu et al, 2012; Cosgrave et al, 2018). The splice site mutation is predicted to affect the binding of SRP55 splicing factor within a splicing enhancer, and thus could severely disrupt the structure and function of the protein product (Xu et al, 2012). The missense variant is located within the Armadillo‐like helical motifs at the N terminal of the protein and may affect interactions with binding partners (Sharov et al, 2017). Therefore, it is possible these variants result in decreased TRRAP function, which can be mimicked by knocking down Nipped‐A in flies. Given the conservation of TRRAP protein and the molecular clockwork throughout evolution, we believe TRRAP may carry out similar functions in mammals (Murr et al, 2007; Li & Zhang, 2015).
We clearly demonstrate a role for NIPPED‐A in circadian period length determination in adults. However, UASdcr2/tubGAL80ts ;cryGAL4‐16/UASNipped‐ARNAi‐4 raised at the permissive temperature and tested at the restrictive temperature showed ~1 h lengthening of the period (compared to UASdcr2/tubGAL80ts ;cryGAL4‐16/+), whereas UASdcr2/+;cryGAL4‐16/UASNipped‐ARNAi‐4 flies in the same experiment showed nearly 2 h longer period (compared to UASdcr2/+;cryGAL4‐16/+). This implicates that knocking down Nipped‐A during development exerts influence on adult period, suggesting a role for NIPPED‐A in modulating the development of the clock.
Previous work reported a potential role for NIPPED‐A in mediating light resetting effects on the clock possibly by modulating light‐induced degradation of the blue light photoreceptor CRYPTOCHROME and subsequently TIM (Sathyanarayanan et al, 2008; Adewoye et al, 2015). However, the circadian changes we observed here are in the absence of light, and cry mutants do not show circadian defects in DD (Stanewsky et al, 1998). Therefore, we believe NIPPED‐A also functions in the clock under DD and the actions on TIM are likely independent of CRY. In line with this idea, we do not observe significant effects of knocking down Nipped‐A on cry mRNA level. Unfortunately, we were not able to obtain a working CRY antibody to examine whether there is any alteration of CRY protein.
The lengthened period observed in flies with Nipped‐A knocked down is at least in part due to decrease in TIM and consequently PER. This could lead to a delay in PER accumulation which ultimately slows down the pace of the molecular clockwork (Hardin, 2011). Consistently, over‐expressing tim or per rescues the lengthened period caused by knocking down Nipped‐A. As for whether alteration in PDP1ε levels can exert effects on the core clock is still an issue of debate (Cyran et al, 2003; Benito et al, 2007; Lim et al, 2007; Zheng et al, 2009). PDP1ε is known to activate clk transcription; however, in Nipped‐A RNAi flies, we observe reduced Pdp1ε but not clk mRNA level. This may be because the remaining PDP1ε is sufficient for maintaining normal clk expression. On the other hand, we are able to specifically rescue the period phenotype of Nipped‐A deficiency by over‐expressing Pdp1ε, and we have verified that over‐expressing Pdp1ε enhances clk mRNA level. This in turn leads to increased per and tim expression, thus reverting the long‐period phenotype. Pdp1 and per mutations interact with Nipped‐A to synergistically lengthen the period, whereas tim mutation appears to interact with Nipped‐A to impinge on the power of the rhythm. We reason that the effect of tim 01/+ on PER may not be strong enough to influence period length, but may act together with Nipped‐A to affect the power of the rhythm via other mechanisms that are yet unclear.
Multiple studies in yeast have indicated that Tra1, the yeast homolog of TRRAP, interacts with transcriptional activators and plays a crucial role in recruiting the SAGA complex to promoters (Weake & Workman, 2012). CLK and CYC are well‐characterized transcriptional activators of tim and Pdp1ε, binding rhythmically to the E‐box elements upstream of these genes (Li & Zhang, 2015). However, here we do not observe rhythmic association of NIPPED‐A at tim and Pdp1ε loci, suggesting that NIPPED‐A is not recruited by CLK/CYC. It has been shown that the SAGA complex may also be recruited and/or retained at the promoter region by interacting with the core transcription machinery or histone marks, which could potentially explain the constitutive association of NIPPED‐A at tim and Pdp1ε loci (Weake & Workman, 2012). Another possibility is that CLK/CYC recruits NIPPED‐A in clock cells, but this rhythmic recruiting event is masked by constitutive binding in non‐clock cells. More sensitive methods will be needed to reveal a potential temporal binding of NIPPED‐A to its targets. We found that knocking down Nipped‐A leads to enhanced (or a trend of enhanced) binding of CLK at Pdp1ε, per, and tim promoters but not at vri promoter, which may be due to some compensatory mechanism to make up for the lack of tim, Pdp1ε, and per.
Although TRRAP/NIPPED‐A has previously been characterized as a HAT cofactor, our results indicate that NIPPED‐A regulates the clock by facilitating H2B deubiquitination specifically at tim and Pdp1ε loci, thus promoting the transcription of these two genes. NIPPED‐A binding is not limited to tim and Pdp1ε loci, as NIPPED‐A also binds to the per locus and likely other sites. However, NIPPED‐A appears to specifically regulate H2B deubiquitination at tim/Pdp1ε loci possibly via the SAGA complex. Consistently, it has been shown in yeast that Tra1 only modulates the expression of a subset of the SAGA‐dependent genes and Tra1 mutation eliminates the recruitment of SAGA to some genes but not others (Helmlinger et al, 2011). Therefore, it is possible that tim and Pdp1ε transcription are particularly susceptible to NIPPED‐A deficiency, whereas the SAGA complex can still be assembled and recruited to the other clock genes.
Knocking down not reduces the mRNA level of several core clock genes, including but not limited to tim and Pdp1ε, which could contribute to the different behavioral phenotypes between not RNAi and Nipped‐A RNAi flies. Sgf11 RNAi and heterozygous mutant flies do not demonstrate prominent locomotor rhythm phenotypes, even though Sgf11 mRNA levels are substantially reduced. This is likely because the remaining SGF11 in these flies is sufficient to maintain circadian function. Despite the differences in circadian phenotypes caused by lack of Nipped‐A vs. not or Sgf11, both not and Sgf11 deficiency synergistically enhances the long‐period phenotype caused by knocking down Nipped‐A. Therefore, we believe NIPPED‐A functions together with the DUB module of SAGA to regulate the clock. On the other hand, the up‐regulated H3K9 and H3K27 acetylation at tim and Pdp1ε promoter regions may reflect some sort of compensatory mechanism in response to lack of tim and Pdp1ε. Interestingly, mutation of ATAXIN7, a protein that mediates the association of DUB module with the rest of the SAGA complex, has been shown to induce H3 hyperacetylation, but the genes that are hyperacetylated are transcriptionally down‐regulated, similar to what we observed here in Nipped‐A RNAi flies (Helmlinger et al, 2006; Koutelou et al, 2010).
Compared to other epigenetic modifications such as histone methylation and acetylation, ubiquitination of histones is much less studied and understood. A previous study in mouse liver demonstrates rhythmic H2B monoubiquitination of circadian E‐box genes including Per1 and Per2, which may be regulated by Ddb1‐Cullin‐4 ubiquitin ligase (Tamayo et al, 2015). Reducing H2B ubiquitination in fibroblast culture results in shortened period and enhanced Per1/2 mRNA levels. The authors propose a role for DDB1‐CULLIN‐4‐mediated H2B monoubiquitination during the transcriptional repression phase of the circadian cycle. Here, we showed that knocking down Nipped‐A leads to increased H2B ubiquitination accompanied by lengthened period and reduced tim/Pdp1ε mRNA levels, which is mediated by the DUB module of the SAGA complex. These results support a role for the SAGA complex and H2B deubiquitination in the transcriptional activation phase of the circadian cycle. However, we did not observe cycling of H2B ubiquitination, which could be masked by the constant levels of H2B ubiquitination in non‐clock cells in the fly head.
In summary, we propose a role for NIPPED‐A in promoting the transcription of tim and Pdp1ε by facilitating the deubiquitination of H2B, which adds another layer of epigenetic regulation to the clock (Fig 7). Moreover, environmental factors play critical roles in the etiology of schizophrenia, and epigenetic alterations including histone modifications have been implicated in this process (Thomas, 2017). Our findings here could thus provide a potential explanation for circadian disruptions associated with schizophrenia.
Figure 7. A model for the role of NIPPED‐A in setting the pace of the clock.
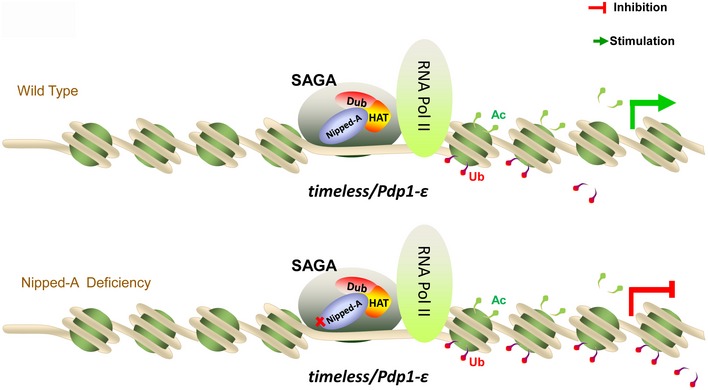
NIPPED‐A functions in the SAGA complex and promotes tim/Pdp1ε transcription by facilitating deubiquitination of H2B. When NIPPED‐A is deficient, ubiquitylation of H2B is enhanced, thus preventing the transcription of tim and Pdp1ε.
Materials and Methods
Fly strains
The following fly strains were used in this study: w 1118, yw, genetic background control lines for the RNAi lines (VDRC:60000, VDRC:60100, BDSC:36303), UASNipped‐ARNAi‐1 (THU5747), UASNipped‐ARNAi‐2 (THU3524), UASNipped‐ARNAi‐3 (THU0698), UASNipped‐ARNAi‐4 (VDRC:44781), Nipped‐ARNAi‐5 (VDRC:40789), UASNipped‐A RNAi‐6 (VDRC:40790), UASGFP, UASnotRNAi‐1 (THU3093), UASnotRNAi‐2 (v45776), UASnot (DGRC:206254), UASSgf11RNAi‐1 (v100581), UASSgf11RNAi‐2 (v17166), Sgf11 e01308 (BDSC:17941), timGAL4, timGAL4;UASdcr2, UASdcr2;cryGAL4‐16, pdfGAL4‐UASdcr2, tubGAL80ts, pdfGAL4‐geneswitch, tim 01, Ptim, UASPdp1ε, Pdp1ε 3135, UASper‐10, per L, and UASclk. UASflySAM2.0‐Nipped‐A is generated by Tsinghua Fly Center following previously published methods (Jia et al, 2018). sgRNA (GCAGTAAACATGCAAATAAG) targeting upstream sequence of Nipped‐A was cloned into flySAM2.0 vector. The construct was then injected into y sc v nanos‐integrase; attP40 embryos.
Drosophila activity monitoring and behavior analysis
Flies were reared on standard cornmeal–yeast–sucrose medium and kept in LD cycles at 25°C. Three‐ to four‐day‐old male flies were used to monitor locomotor activity levels using the Drosophila Activity Monitor system (Trikinetics) for 4 days of LD followed by 7 days of DD.
tubGAL80ts flies were raised at 18°C/29°C and transferred to 29°C/18°C 4 days before the start of behavioral assays. Behavioral assays are conducted at 29°C/18°C accordingly.
To monitor activities of strains containing the pdfG4‐geneswitch (pdf‐GS) driver, flies were raised on standard cornmeal–yeast–sucrose medium until pupation. After eclosion, flies were transferred to tubes with standard food containing RU486 (250 μM) for 2–3 days before monitoring locomotor activity. Food used during behavioral assay also contained 250 μM RU486. Ethanol used to solubilize RU486 was added to the food as vehicle control.
Cell culture and transient transfection
Drosophila S2 cells were maintained in Schneider's medium (Life Technologies) supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin (Life Technologies). The cells were plated in a 12‐well plate (2 × 105 cell/well) and incubated for 24 h, and si‐Nipped‐A was transiently transfected using Lipofectamine 2000 Transfection Reagent (Life Technologies) according to the protocol provided by the manufacturer. Cells were collected 48 h after the transfection.
RNA extraction and quantitative real‐time PCR (qRT–PCR)
Flies were entrained in LD for 3 days, collected on the fourth day of LD or DD1 at the indicated time points (ZT or CT) and frozen immediately on dry ice. Fly heads or S2 cells were isolated and homogenized in TRIzol reagent (Ambion, Life Technologies). Chloroform was subsequently added and centrifuged at 12,000 g for 15 min at 4°C. Aqueous top layer was collected, and ethanol was added to precipitate RNA. The precipitates were collected by centrifuging at 12,000 g for 10 min at 4°C. RNA pellets were washed with 75% ethanol. After air dry, the pellets were dissolved in RNAse‐free water. Contaminating genomic DNA was removed by RQ1 DNase (Promega) digestion, and total RNA was directly amplified with TransScript Green One‐Step qRT–PCR SuperMix (TransGen Biotechnology). All qPCRs were carried out on a Step One Plus Real‐Time PCR System (Applied Biosystems, Life Technologies). The templates were reverse‐transcribed at 45°C for 5 min and denatured at 95°C for 30 s, followed by forty cycles with 5 s at 95°C, 15 s at 60°C, 30 s at 72°C, and 15 s at 75°C for data acquisition. Primers used for expression analysis are listed in Appendix Table S12.
Western blot
Flies were entrained in LD for 3 days and collected during LD or DD1 at the indicated time points (ZT or CT) and frozen immediately on dry ice. Heads were separated and homogenized in RIPA buffer (20 mM Tris at pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.05 mM EGTA, 10% glycerol, 1% Triton X‐100, 0.4% sodium deoxycholate, 0.1% SDS) containing protease inhibitor mixture (Roche) and phosphatase inhibitor mixture (Roche). This homogenate was sonicated 3–5 times for 8 s each time and then centrifuged at 16,200 g for 15 min at 4°C to remove cell debris. Supernatant was collected, transferred to new tubes, and centrifuged again at 16,200 g for 10 min at 4°C. Supernatant was collected, and protein lysates were prepared in SDS–PAGE loading buffer. Equal amounts of protein were run on 7.5% SDS–PAGE gels and then transferred to nitrocellulose membrane. After incubation with primary antibodies at 4°C overnight, membranes were incubated with secondary antibodies at room temperature for 1 h. The primary antibodies used were rat anti‐TIM (1:1,000; a gift from Joanna Chiu), guinea pig anti‐PDP1ε (1:5,000; a gift from Paul Hardin), guinea pig anti‐PER (1:1,000; a gift from Joanna Chiu), and mouse anti‐HSP70 (1:5,000; Sigma). Secondary antibodies used were conjugated either with IRDye 680 or IRDye 800 (LI‐COR Biosciences) and incubated at a concentration of 1:1,000. Blots were visualized with Odyssey Infrared Imaging System (LI‐COR Biosciences). Antibodies used in this study are listed in Appendix Table S13.
Immunostaining
Male flies were entrained for 3 days in LD and collected on DD1. Flies were anesthetized with CO2 and dissected in 3.7% formaldehyde diluted in PBS. After fixing for 30 min at room temperature, the brains were rinsed three times in PBS and incubated in PBS with 1% Triton for 15 min at room temperature. The brains were then incubated with 5% goat serum diluted in PBT (PBS with 0.3% triton) for 1 h at room temperature, followed by overnight incubation of 1:200 rat anti‐TIM (a gift from Joanna Chiu), 1:500 rabbit anti‐PER (a gift from Michael Rosbash), 1:1,000 guinea pig anti‐PDP1ε (a gift from Paul Hardin), and 1:50 mouse anti‐PDF (Drosophila Studies Hybridoma Bank) in PBT at 4°C. After 6 × 20 min PBT rinses, the brains were incubated with secondary antibodies: donkey anti‐mouse Alexa Fluor‐488 (1:1,000, Molecular Probes), donkey anti‐mouse Alexa Fluor‐594 (1:1,000, Life Technologies), donkey anti‐rat Alexa Fluor‐488 (1:500, Abcam), donkey anti‐guinea pig Alexa Fluor‐594 (1:1,000, Jackson ImmunoResearch Laboratories), and donkey anti‐rabbit Alexa Fluor‐488 (1:1,000, Abcam). After incubating overnight at 4°C, the brains were rinsed 6 × 20 min in PBS, and then mounted and imaged using an Olympus FV1000 confocal microscope with 20× or 60× oil lens (Olympus). Images were acquired using the same settings (power, gain, offset) for each experiment. Antibodies used in this study are listed in Appendix Table S13.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was conducted following a published protocol (Kwok et al, 2015). Flies were entrained in LD at 25°C for 3 days and collected at four time points (CT) on DD1. 600 μl of fly heads were mixed with 2 ml of NEB buffer [10 mM Tris–HCl at pH 8.0, 10 mM NaCl, 0.1 mM EGTA at pH 8.0, 0.5 mM EDTA at pH 8.0, 1 mM DTT, 0.5% tergitol NP‐10, 0.5 mM spermidine, 0.15 mM spermine, 1× protease inhibitor mixture (Roche), and phosphatase inhibitor mixture (Roche)]. The homogenates were centrifuged at 4,700 g for 10 min to remove cell debris. Pellets were resuspended in 1 ml of sucrose gradient in NEB (0.6 ml of 1.6 M sucrose in NEB, 0.4 ml of 0.8 M sucrose in NEB) and centrifuged at 13,800 g for 20 min. Pellets were subsequently resuspended in 1 ml of NEB with 1% formaldehyde and crosslinked for 15 min at room temperature with rotation. 150 μl of 1 M glycine was added to stop the crosslinking with rotation for 10 min at room temperature. Pellets were collected by centrifugation at 4,700 g for 5 min, washed two times with 1 ml NEB buffer each time, and resuspended in 500 μl of sonication buffer [10 mM Tris–HCl at pH 7.5, 2 mM EDTA, 1% SDS, 0.2% Triton X‐100, 0.5 mM spermidine, 0.15 mM spermine, 1× protease inhibitor mixture (Roche), and 1× phosphatase inhibitor mixture (Roche)]. Samples were sonicated 6 × 30 s on 30% of high setting and then centrifuged at 13,800 g for 10 min. Supernatants were collected and centrifuged again at 16,200 g for 10 min at 4°C. 200 μl of the supernatants were used for IP/IgG, respectively, and 20 μl for input. IP buffer [50 mM Tris–HCl at pH 7.6, 2 mM EDTA, 1% Triton X‐100, 0.1% DOC, 150 mM NaCl, 0.5 mM EGTA, 1× protease inhibitor mixture (Roche), and 1× phosphatase inhibitor mixture (Roche)] was added to IP reactions to a total of 800 μl per sample. Antibodies were then added and incubated with rotation overnight at 4°C. Antibodies used for ChIP are as follows: acetyl‐histone H3 (Millipore; 06‐599), acetyl‐histone H3‐K9 (Abcam; ab4441), acetyl‐histone H3‐K14 (Millipore; 07‐353), acetyl‐histone H3‐K27 (Abcam; ab4729), acetyl‐histone H4 (Millipore; 06‐866), acetyl‐histone H4‐K16 (Millipore; 07‐329), TRRAP (Sigma, PLA0167), ubiquityl‐histone H2B (Millipore; 05‐1312‐I), and mouse/rabbit anti‐IgG (Santa Cruz). For each IP/IgG, 50 μl of Protein G or Protein A magnetic beads (Bio‐Rad) were washed twice in 1 ml of CW buffer [50 mM Tris–HCl at pH 7.6, 1 mM EDTA, 1% Triton X‐100, 0.1% DOC, 0.1% BSA, 0.5 M KCl in PBS, 150 mM NaCl, 0.5 M EGTA, 0.1% SDS, 1× protease inhibitor mixture (Roche), and 1× phosphatase inhibitor mixture (Roche)]. The beads were then added to the samples and incubated for 2 h with rotation at 4°C. After that, beads were captured and washed for 30 min for two times in 1 ml of CW buffer, once in LW buffer (10 mM Tris–HCl at pH 8.0, 0.25 M LiCl, 0.5% NP40, 0.5% DOC, 1 mM EDTA), and once in TE buffer for 10 min. Supernatants were removed, and 150 μl of CE buffer (50 mM Tris–HCl at pH 8.0, 10 mM EDTA, 1% SDS, 1 mM DTT, 0.1 mg/ml proteinase K, 50 mM NaCl, and 0.05 mg/ml RNase A) was added. Equal amount of CE buffer was added to input samples as well. All samples were incubated for 2 h at 37°C. Beads were then removed from IP samples, and supernatants were de‐crosslinked overnight at 65°C. DNA was extracted using phenol:chloroform:iso‐amyl alcohol (25:24:1) and subjected to qPCR (primers used are listed in Appendix Table S12).Three technical replicates of qPCR were performed for each biological replicate, and at least three biological replicates were performed. Antibodies used in this study are listed in Appendix Table S13.
Immunoprecipitation
Flies were entrained in LD at 25°C for 3 days and collected at one time point (CT 12) on DD1. Heads were collected on dry ice, and 1 ml RIPA buffer was added for each reaction. Using a motorized plastic pestle, fly heads were homogenized and then sonicated for 3 × 5 s with 10‐s pauses. Debris were centrifuged at 17,000 g for 15 min at 4°C. Supernatants were transferred into a new 1.5‐ml tube, and pellets were discarded. Samples were centrifuged for another 10 min at 17,000 g (4°C), and then, supernatants were collected. Protein concentration was quantified. 50 μl of the supernatant was used as input, and the remaining sample was divided equally into two parts. 7 μg NIPPED‐A antibody and IgG were added, respectively. The samples were incubated at 4°C overnight, and 50 μl of Protein A magnetic beads (Bio‐Rad) were added followed by incubation for 2 h with rotation at 4°C. Beads were then captured and washed for 10 min for two times in 1 ml RIPA buffer. SDS–PAGE loading buffer was added subsequently. Samples were incubated at 95°C for 10 min. Equal amounts of protein were run on 7.5% SDS–PAGE gels.
Quantification and statistical analysis
Behavior analysis
Analyses of period, power, and rhythmicity during DD were carried out using ClockLab (Actimetrics) software. For DD rhythmicity, rhythmic flies were defined as those with chi‐squared power‐significance ≥ 10. Period calculations considered all flies with power‐significance ≥ 10. One‐way ANOVA and Tukey's multiple comparison test (Prism GraphPad) were used to compare the differences between different genotypes.
Immunostaining quantification
The intensity of TIM, PDP1ε, and PER signals was quantified by ImageJ software. Student's t‐test (Microsoft Excel) was used to compare the average intensity values between different genotypes.
Quantitative RT–PCR
Beta‐actin or rp49 (for mRNA analysis) and cbp20 (for pre‐mRNA analysis) were used as normalization control. The delta–delta CT method was used for quantification. The value of the control genotype was set to 1. Student's t‐test (Microsoft Excel) was used to compare the differences between genotypes.
Western blot quantification
Protein levels of TIM, PDP1ε, PER, NIPPED‐A, and HSP70 were quantified by the software image studio for Odyssey (LI‐COR Biosciences). TIM/PDP1ε/PER/NIPPED‐A levels were normalized to HSP70. The value of the control genotype was set to 1. Student's t‐test (Microsoft Excel) was used to compare the differences between genotypes.
ChIP assay quantification
ANOVA (Prism GraphPad) and Student's t‐test (Microsoft Excel) were used to compare the differences between genotypes and conditions.
Author contributions
BB, LC, and LZha designed the experiments; BB, LC, LZhe, and WH conducted the experiments; and LZha wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 2
Source Data for Figure 4
Acknowledgements
This work is supported by grants from the Natural Science Foundation of China (31471125 and 31671215) and 1000 Talents Program. We would like to thank Drs. Joanna Chiu, Yong Zhang, and Paul Hardin for helpful advice and discussions.
The EMBO Journal (2020) 39: e101259
References
- Adewoye AB, Kyriacou CP, Tauber E (2015) Identification and functional analysis of early gene expression induced by circadian light‐resetting in Drosophila . BMC Genom 16: 570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin P (2007) PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci 27: 2539–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrave J, Wulff K, Gehrman P (2018) Sleep, circadian rhythms, and schizophrenia: where we are and where we need to go. Curr Opin Psychiatry 31: 176–182 [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J (2003) vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341 [DOI] [PubMed] [Google Scholar]
- Daniel JA, Torok MS, Sun ZW, Schieltz D, Allis CD, Yates JR III, Grant PA (2004) Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J Biol Chem 279: 1867–1871 [DOI] [PubMed] [Google Scholar]
- Depetris‐Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF (2011) Adult‐specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol 21: 1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C (2019) The importance of being rhythmic: living in harmony with your body clocks. Acta Physiol (Oxf): e13281 [DOI] [PubMed] [Google Scholar]
- Emery P, So WV, Kaneko M, Hall JC, Rosbash M (1998) CRY, a Drosophila clock and light‐regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679 [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich‐Forster C, Emery‐Le M, Hall JC, Rosbash M (2000) Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26: 493–504 [DOI] [PubMed] [Google Scholar]
- Hardin PE (2011) Molecular genetic analysis of circadian timekeeping in Drosophila . Adv Genet 74: 141–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Hardy S, Abou‐Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L et al (2006) Glutamine‐expanded ataxin‐7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol 4: e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villen J, Swaney DL, Gygi SP, Bahler J, Winston F (2011) Tra1 has specific regulatory roles, rather than global functions, within the SAGA co‐activator complex. EMBO J 30: 2843–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D (2012) New insights into the SAGA complex from studies of the Tra1 subunit in budding and fission yeast. Transcription 3: 13–18 [DOI] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL (2003) Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA‐associated Ubp8. Genes Dev 17: 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Xu RG, Ren X, Ewen‐Campen B, Rajakumar R, Zirin J, Yang‐Zhou D, Zhu R, Wang F, Mao D et al (2018) Next‐generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc Natl Acad Sci USA 115: 4719–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SG, Benca RM (2015) Circadian disruption in psychiatric disorders. Sleep Med Clin 10: 481–493 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422: 66–94 [DOI] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S (1971) Clock mutants of Drosophila melanogaster . Proc Natl Acad Sci USA 68: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Hirsch CL, Dent SY (2010) Multiple faces of the SAGA complex. Curr Opin Cell Biol 22: 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok RS, Li YH, Lei AJ, Edery I, Chiu JC (2015) The catalytic and non‐catalytic functions of the Brahma chromatin‐remodeling protein collaborate to fine‐tune circadian transcription in Drosophila . PLoS Genet 11: e1005307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhang L (2015) Circadian control of global transcription. Biomed Res Int 2015: 187809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Lee J, Koo E, Choe J (2007) Targeted inhibition of Pdp1epsilon abolishes the circadian behavior of Drosophila melanogaster . Biochem Biophys Res Commun 364: 294–300 [DOI] [PubMed] [Google Scholar]
- McClung CA (2013) How might circadian rhythms control mood? Let me count the ways. Biol Psychiatry 74: 242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M, Emery P (2001) Wild‐type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol 21: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene‐switch systems in Drosophila . Sci STKE 2004: pl6 [DOI] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z (2007) Orchestration of chromatin‐based processes: mind the TRRAP. Oncogene 26: 5358–5372 [DOI] [PubMed] [Google Scholar]
- Papazyan R, Zhang Y, Lazar MA (2016) Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 23: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, Taghert PH (1999) Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J Neurobiol 41: 189–207 [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Kumar S, Chen CH, Chen D, Hay B, Sehgal A (2008) Identification of novel genes involved in light‐dependent CRY degradation through a genome‐wide RNAi screen. Genes Dev 22: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal A, Price JL, Man B, Young MW (1994) Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless [see comments]. Science 263: 1603–1606 [DOI] [PubMed] [Google Scholar]
- Sharov G, Voltz K, Durand A, Kolesnikova O, Papai G, Myasnikov AG, Dejaegere A, Ben Shem A, Schultz P (2017) Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA. Nat Commun 8: 1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R, Kaneko M, Emery P, Beretta B, Wagner‐Smith K, Kay SA, Rosbash M, Hall JC (1998) The cry b mutation identifies cryptochrome as a circadian photoreceptor in Drosophila . Cell 95: 681–692 [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M (2005) A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438: 238–242 [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL (2008) The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet 9: 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo AG, Duong HA, Robles MS, Mann M, Weitz CJ (2015) Histone monoubiquitination by Clock‐Bmal1 complex marks Per1 and Per2 genes for circadian feedback. Nat Struct Mol Biol 22: 759–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Hardin PE (2008) Rhythmic E‐box binding by CLK‐CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol 28: 4642–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA (2017) Histone posttranslational modifications in schizophrenia. Adv Exp Med Biol 978: 237–254 [DOI] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL (2008) SAGA‐mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J 27: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2012) SAGA function in tissue‐specific gene expression. Trends Cell Biol 22: 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff K, Dijk DJ, Middleton B, Foster RG, Joyce EM (2012) Sleep and circadian rhythm disruption in schizophrenia. Br J Psychiatry 200: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyce A, Xiao T, Whelan KA, Kosman C, Walter W, Eick D, Hughes TR, Krogan NJ, Strahl BD, Berger SL (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA‐related complex. Mol Cell 27: 275–288 [DOI] [PubMed] [Google Scholar]
- Xu B, Ionita‐Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M (2012) De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 44: 1365–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA 111: 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R (2003) Drosophila clock can generate ectopic circadian clocks. Cell 113: 755–766 [DOI] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A (2009) An isoform‐specific mutant reveals a role of PDP1 epsilon in the circadian oscillator. J Neurosci 29: 10920–10927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yu W, Hardin PE (2016) CLOCKWORK ORANGE enhances PERIOD mediated rhythms in transcriptional repression by antagonizing E‐box binding by CLOCK‐CYCLE. PLoS Genet 12: e1006430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan MA, Sandrelli F (2015) Circadian clock dysfunction and psychiatric disease: could fruit flies have a say? Front Neurol 6: 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Source Data for Figure 2
Source Data for Figure 4


