Abstract
Significance: Acute respiratory distress syndrome (ARDS) is a severe, highly heterogeneous critical illness with staggering mortality that is influenced by environmental factors, such as mechanical ventilation, and genetic factors. Significant unmet needs in ARDS are addressing the paucity of validated predictive biomarkers for ARDS risk and susceptibility that hamper the conduct of successful clinical trials in ARDS and the complete absence of novel disease-modifying therapeutic strategies.
Recent Advances: The current ARDS definition relies on clinical characteristics that fail to capture the diversity of disease pathology, severity, and mortality risk. We undertook a comprehensive survey of the available ARDS literature to identify genes and genetic variants (candidate gene and limited genome-wide association study approaches) implicated in susceptibility to developing ARDS in hopes of uncovering novel biomarkers for ARDS risk and mortality and potentially novel therapeutic targets in ARDS. We further attempted to address the well-known health disparities that exist in susceptibility to and mortality from ARDS.
Critical Issues: Bioinformatic analyses identified 201 ARDS candidate genes with pathway analysis indicating a strong predominance in key evolutionarily conserved inflammatory pathways, including reactive oxygen species, innate immunity-related inflammation, and endothelial vascular signaling pathways.
Future Directions: Future studies employing a system biology approach that combines clinical characteristics, genomics, transcriptomics, and proteomics may allow for a better definition of biologically relevant pathways and genotype–phenotype connections and result in improved strategies for the sub-phenotyping of diverse ARDS patients via molecular signatures. These efforts should facilitate the potential for successful clinical trials in ARDS and yield a better fundamental understanding of ARDS pathobiology.
Keywords: acute respiratory distress syndrome (ARDS), genome-wide association studies (GWAS), ARDS mortality, reactive oxygen species (ROS), inflammation, pathway analysis, candidate gene studies
Introduction
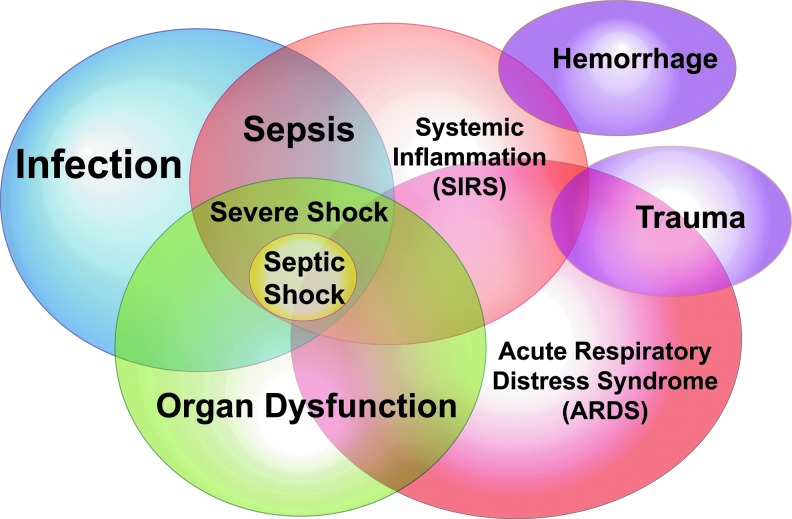
Critical illnesses, including infection, sepsis, trauma, pancreatitis, hemorrhage, and acute respiratory failure/acute respiratory distress syndrome (ARDS) (Fig. 1), account for 20% of health care costs in the United States ($90 billion annually) (152, 206). Estimates of the incidence of ARDS vary between ∼200,000 and 400,000 individuals annually in the United States with mortality rates of ∼30%–40% (23, 40, 49, 51, 64, 129, 176). The pathologic hallmarks of ARDS are marked increases in high permeability pulmonary edema related to acute endothelial cell activation/dysfunction resulting in paracellular gap formation (57, 58, 62), alveolar flooding, decreased lung compliance, severe hypoxemia, and a requirement for mechanical ventilation.
FIG. 1.
Schema of the overlapping syndromes and complications in the critically ill. Depicted are the major causes of critical care illnesses and associated complications that contribute to the staggering morbidity, mortality, and health care costs in the critically ill.
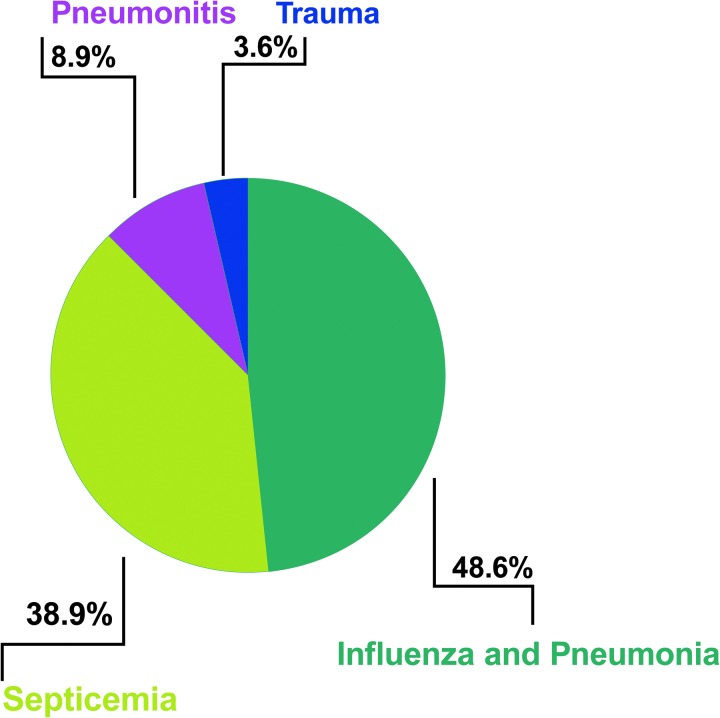
From a pathobiological standpoint, two categories of lung injury are recognized in ARDS. Direct pulmonary injury, which is to the local lung epithelium such as in pneumonia, aspiration, mechanical ventilation, inhalation injury, and lung contusion. Indirect pulmonary injury occurs secondary to vascular endothelial damage (156), with sepsis being the most common cause of indirect lung injury and the highest contributor to ARDS mortality (Fig. 2) (49). However, these broad categorizations and classifications of ARDS fail to capture the nuances of the disease and the substantial heterogeneity of phenotypes within ARDS has made interrogation of basic mechanisms and therapeutic development extremely challenging. This has contributed to the abysmal track record of Phase II/III trials of novel therapies in ARDS (70, 98, 152). Clinical factors alone have failed to predict which patients will develop ARDS or develop severe ARDS (122).
FIG. 2.
ARDS causes. An adaptation of the summary of Cochi et al. data from 15 years of ARDS comorbidity data (1999–2013) that identified four major comorbidities that occur with ARDS (49). ARDS, acute respiratory distress syndrome.
Although promising attempts exist to sub-phenotype ARDS cohorts via blood-derived biomarkers that are predictive of mortality in hope of identifying at-risk individuals, successful validation has been elusive (189–191). Scoring systems such as the acute physiology and chronic health evaluation II score (101, 152) or the injury severity score (130) in critically ill patients predict patient outcomes but can only be applied to general intensive care unit populations and do not provide consistent and accurate estimates of the risk of death in specific intensive care unit (ICU) patient populations. For example, mean lung injury scores were not significantly different between ARDS survivors and non-survivors (42, 104) and attempts to characterize predictors of death in ARDS by developing a prognostic index (189) remain controversial and without replication or validation.
Thus, there is a compelling unmet need to identify ARDS sub-phenotypes that risk-stratify patients for both accurate prognostication and clinical trial purposes. A risk score is a standardized metric for the likelihood that an individual will experience a particular outcome—in this instance, higher risk for ARDS mortality (32, 189). Risk stratification is the aggregation of multiple individual risk scores to create a broader, more complex profile of risk.
The ability to leverage newly described systems biology and “omic” approaches and techniques (genetic and biomarker panels) hold promise for improving the capacity to characterize risk and prognosis for ARDS and in other ICU patients with critical illness and respiratory failure. ARDS genetic and genomic studies potentially provide the basis for identifying candidate genes, biomarker discovery, risk stratification, and novel ARDS therapeutic targets (83, 122). Although ARDS is not a known inheritable condition, the pattern of injury response-recovery has significant heritability (121, 122, 197). Unlike rare and high penetrance monogenetic diseases, ARDS risk and severity are influenced by multiple genes to a varying effect.
The potential to utilize genomic approaches to identify ARDS high inflammatory sub-phenotypes at higher risk for death is a currently untapped area of therapeutic stratification in severe lung injury (32, 65, 122, 149, 175). The diversity of potential genetic biomarkers in ARDS ranges from markers of epithelial injury (the receptor of advanced glycation end products [RAGE]) (62, 97), endothelial activation/injury (angiopoetin [ANGPT-1], intercellular adhesion molecule 1 [ICAM-1], vascular endothelial growth factor [VEGF]) (33, 60, 62, 133, 159), pro-inflammatory (interleukin [IL]-1B, IL-18, IL-6, IL-8) (55, 68, 79, 118, 120, 140, 166, 183), anti-inflammatory molecules (IL-10) (140), coagulation and fibrinolysis proteins (pediocin PA-1) (28, 37, 56, 144), and macrophage markers (high mobility group box 1 [HMGB1], macrophage migration inhibitory factor [MIF]) (27, 50, 71, 74, 131).
Significance
We (Oita et al., manuscript in preparation, 3, 23–26, 35, 46, 59, 76, 82, 90, 126, 132, 141, 157, 169, 187, 199, 202, 205) and others (2, 13, 19, 21, 22, 45, 47, 63, 69, 92, 122–124, 149, 153, 163, 173, 181, 194) have contributed to the notion that ARDS represents the ultimate in genetic stress, with a complex assortment of genes that contribute a limited overall effect size per gene (149). Individually, many of these loci have limited value for risk prediction, but their aggregated impact on lung injury phenotypes (effect size) in ARDS pathology is much greater (122).
Unlike other complex genetic diseases, ARDS has not benefited from family pedigree studies (149); however, using both candidate gene and genome-wide association study (GWAS) approaches, we have identified several novel genetic targets and biomarkers of ARDS risk and severity, including nicotinamide phosphoribosyl transferase (NAMPT) (108, 168, 199), toll-like receptor 4 (TLR4) (169, 200), encoding myosin light chain kinase (MYLK) (46, 47, 72, 73, 119, 187), iodothyronine deiodinase 2 (DIO2) (187, 194), growth arrest and DNA damage-inducible gene (GADD45a) (124, 126), MIF (71), and sphingosine 1-phosphate receptors 1 and 3 (S1P1, S1P3) (132, 169).
In this article, we have chosen to integrate studies utilizing peripheral blood mononuclear cells (PBMCs) for identification of genetic signature in ARDS, meta-analysis of ARDS risk, and mortality biomarker studies (175) with GWAS. We speculate that this strategy may expand the understanding of ARDS pathobiology and potentially identify genes associated with ARDS mortality that may serve as diagnostic makers and therapeutic targets.
We evaluated PubMed literature relevant to ARDS to identify a total of 201 dysregulated genes potentially associated with either ARDS risk or severity followed by pathway analysis. Pathway analysis included genes from both candidate and agnostic GWAS studies, genes from mRNA microarray and sequencing studies, and proteomic evaluations that identified putative candidate genes but without associated single nucleotide polymorphisms (SNPs) related to ARDS risk or ARDS mortality. Blood biomarkers without genomic/genetic evidence were not included.
We further summarize the current state of ARDS genetics in terms of susceptibility risk SNPs and those that confer increased mortality selected on the basis of adjusted significance in their respective study. We have also chosen to evaluate ARDS studies that use mortality as an end-point as this captures the most severe outcome for ARDS patients. Our bioinformatically derived results are consistent with the concept that evolutionarily conserved inflammatory networks comprising reactive oxygen species (ROS), innate immunity-related inflammation, and endothelial vascular signaling pathways are potent contributors to multiple organ dysfunction-related ARDS mortality and pathobiology (82, 83).
Recent Advances
A brief timeline of ARDS clinical and genetic literature
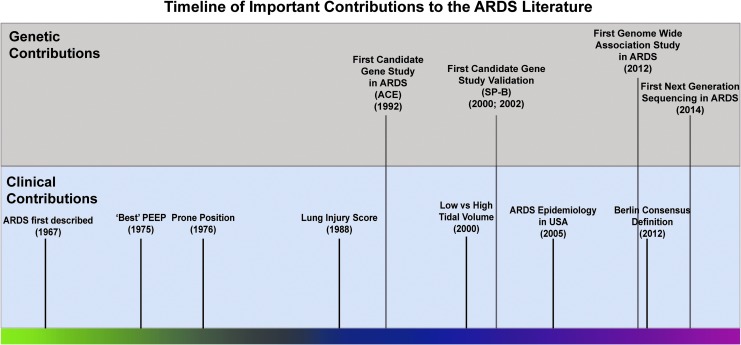
ARDS was first described in the classic Ashbaugh et al. report in 1967 (10). Clinical therapeutics in ARDS have been studied exclusively in ARDS for several decades (107). Figure 3 depicts a brief timeline of the initial important clinical studies for common therapeutics in ARDS. The two earliest clinical methods to manage ARDS were management of lung collapse utilizing positive end-expiratory pressure and the prone position to aid oxygenation (30a, 171). Ventilator-induced lung injury (VILI) or barotrauma was later described, and the further lung injury and alveolar rupture induced by the mechanical ventilator used to treat ARDS patients provides an additional challenge in the clinic (57). In 2000, the landmark ARDSNet clinical trial using lower tidal volumes to reduce VILI and ARDS mortality represented another significant clinical therapeutic advance (8) (Fig. 3), the sole ARDS clinical trial to succeed.
FIG. 3.
A timeline of ARDS clinical and genetic contributions. A selection of clinical contributions for ARDS (bottom) and genetic contributions to the ARDS literature (top). The relative recent history of genetic contributions to the ARDS literature should be noted compared with a much long history of clinical contributions to treatment. Clinical and genetic timelines adapted from Laffey et al. and Reilly et al. (107, 149).
In contrast, the first candidate gene study in ARDS involving angiotensin-converting enzyme (ACE) polymorphisms corresponding to higher ACE plasma levels in ARDS was reported in 1992. Candidate gene studies in ARDS, rare until the sequencing of the human genome (177), have subsequently significantly populated the ARDS literature (Oita et al., manuscript in preparation, 2, 3, 9, 11, 33, 36, 52, 61, 79, 89, 91, 98, 105, 106, 111, 112, 116, 120, 123, 127, 128, 131, 134, 140, 147, 150, 160, 168, 169, 173, 174, 183, 186, 187, 191–194, 198, 201, 203). The chronology of the discovery of the major candidate genes in ARDS as well as the transition into ARDS GWAS studies has been previously elegantly detailed (149); however, in this review, we have attempted to capture more recent genetic developments and reporting in ARDS (53, 107).
We integrated a variety of genetic studies into the ARDS literature, and excluding the initial ACE polymorphism study (177), all candidate gene studies were published after 2000 (Fig. 3). GWAS studies enter the ARDS literature starting in 2012, but compared with GWAS studies in other disorders and clinical arenas, ARDS GWAS studies generally include smaller cohorts (122). The literature of ARDS genetics is relatively small and recent compared with other academic fields, and the genes presented are a comprehensive list of all mapped genes in ARDS that were significant (irrespective of the individual study) for either (i) a specific polymorphism associated with ARDS or (ii) an overall gene expression level significant for ARDS risk.
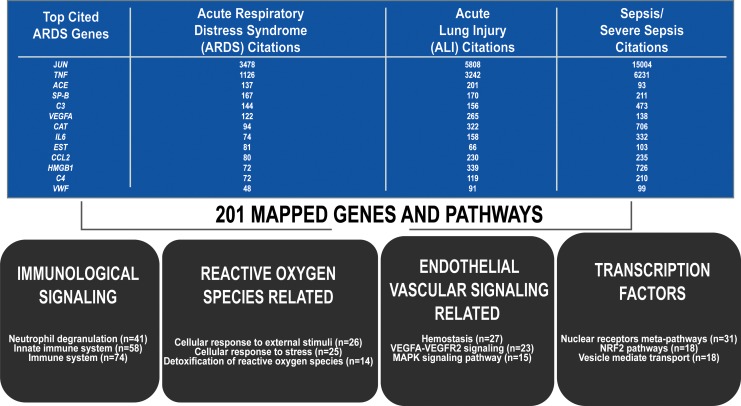
Later, the importance of gene expression studies and PBMC mortality risk genes are discussed as these genes potentially serve as novel therapeutic targets. The “Pathway analysis methods” section given next highlights our attempt to synthesize the recent and diverse genetic studies in the ARDS literature. A list of 201 mapped genes were identified via PMC/PubMed literature search of ARDS, acute lung injury (ALI), and “lung injury” studies that identified genes that were differentially expressed or conferred risk for ARDS, severe sepsis, or mortality (15) (Fig. 4 and Supplementary Table S1).
FIG. 4.
ARDS gene table and top pathways. Two hundred one genes identified through differential gene expression (mRNA sequencing, mRNA CHIP array, RNA microarray, proteomics, candidate gene sequencing, or GWAS) that mapped onto either the Reactome or Wikipathways databases for pathway analysis (5359 human pathways covered). The second, third and fourth columns reflect the number of citations for each gene (14). Four broad categories of enriched pathways (immunological signaling, ROS-related signaling, vascular signaling, and transcription factor-related pathways) and the number or genes represented in each (p < 0.01, five members minimum per pathway). GWAS, genome-wide association study; ROS, reactive oxygen species.
ARDS genes identified by dysregulated gene expression
The 201 ARDS genes were derived from studies with clinical populations, human-derived cell lines, or genes validated across multiple animal models with genetically conserved regions (29, 53, 82, 83, 149, 196). Cross-species analyses of VILI models (rat, mouse, canine) and human ARDS patients have yielded a list of genes that are conserved across species and of potential importance in the pathophysiology of ARDS and VILI (196). Eleven genes were “immune response” genes that were highly significant with Expression Analysis Systematic Explorer scores, and two genes (IL-1B, IL-6) were noted to harbor SNPs that were independently associated with ARDS risk or ARDS mortality (17, 180, 196).
Six genes were involved in “inflammatory response” and “innate immune response” pathways. Taken together, these data indicate that multiple genes fall into the evolutionary conserved inflammatory and immunological-related pathways across species and are potentially important in ARDS and VILI pathology (196). A limited mRNA study of ARDS and healthy controls yielded 12 upregulated genes in ARDS (106) with IL-1R2, a decoy receptor that dampens IL-1 signaling (106), identified as the top upregulated gene. Three genes (Arginase-1, MHC-DRB1, CCR2) are macrophage-specific genes expressed by activated macrophages (106).
Another strategy to study pathways incorporating genes of interest is to utilize genetically engineered preclinical murine models involving exposure to ARDS and VILI followed by genome-wide lung tissue gene expression and pathway analysis (90). For example, NAMPT, also known as PBEF, is an ARDS candidate gene (175) that harbors several promoter SNPs (−2422A/G, −948G/T) that are associated with an increased risk of ARDS and ARDS mortality (3, 12, 108, 135, 169).
We have shown that extracellular NAMPT (eNAMPT) directly interacts with TLR4 (62) and genomic comparisons of wild-type mice and NAMPT heterozygous mice exposed to eNAMPT, VILI, or lipopolysaccharide revealed significant NAMPT-influenced pathways involved in “acute phase response signaling,” “IL-10 signaling,” “IL-6 signaling,” “NF-кB signaling,” “LXR/RXR activation,” “Leukocyte Extravasation Signaling,” “PPAR signaling,” “Death Receptor Signaling,” “Apoptosis Signaling,” and “TLR signaling” (35, 202). Similar genomic-intensive studies independently identified TLR1 and interleukin-1 receptor-associated kinase (IRAK1) as ARDS risk genes (142, 155).
A complementary approach to identify pathways relevant to ARDS mortality is to utilize proteomic analyses to identify ARDS biomarkers that identify ARDS sub-phenotypes (21). Proteomic analysis of bronchoalveolar lavage fluid (BALF) in ARDS survivors and non-survivors (22) revealed differentially expressed proteins that fall within “acute phase signaling” and “FXR/RXR Activation” pathways, results remarkably similar to results from preclinical models of ARDS (22, 90).
The “oxidative ethanol degradation” and “fatty acid α-oxidation” pathways were significantly upregulated in BALF obtained from ARDS non-survivors (22). Utilization of the quantitative electrophoresis-based proteomics method (difference gel electrophoresis) identified 37 proteins differentially expressed between ARDS patients and healthy controls (39), with “Wounding” and “Inflammatory Response” being the top network pathways that included calgranulin A (S100A8), calgranulin B (S100A9), calgranulin C (S100A12), serum amyloid protein (SAA), complement C9 precursor (C9), hemopexin precursor (HPX), peroxiredoxin 5 mitochondrial (PDX5), complement C3 precursor (C3), annexin A1 (ANXA1), and alpha-1-antitrypsin (SERPINA1) (39).
PBMC gene expression in ARDS as predictors of mortality
PBMCs are an easily obtainable blood cell fraction that is broadly representative of innate immunity status. A meta-analysis of PBMC molecular biomarkers (54 distinct studies) attempted to validate ARDS gene biomarkers (175) and found two significant sets of biomarkers (175). One set consisted of ARDS risk genes and included Krebs von den Lugen-6 (KL-6), lactate dehydrogenase (LDH), soluble RAGE, and von Willebrand factor (vWF). A second set of genes associated with increased ARDS mortality (175) included IL-4, IL-2, angiopoetin 2 (Ang-2), and KL-6. IL-4 is also an ARDS candidate gene with SNPs associated with ARDS risk, possibly via regulation of lung repair in cellular and animal models of ARDS (52, 85, 120, 173).
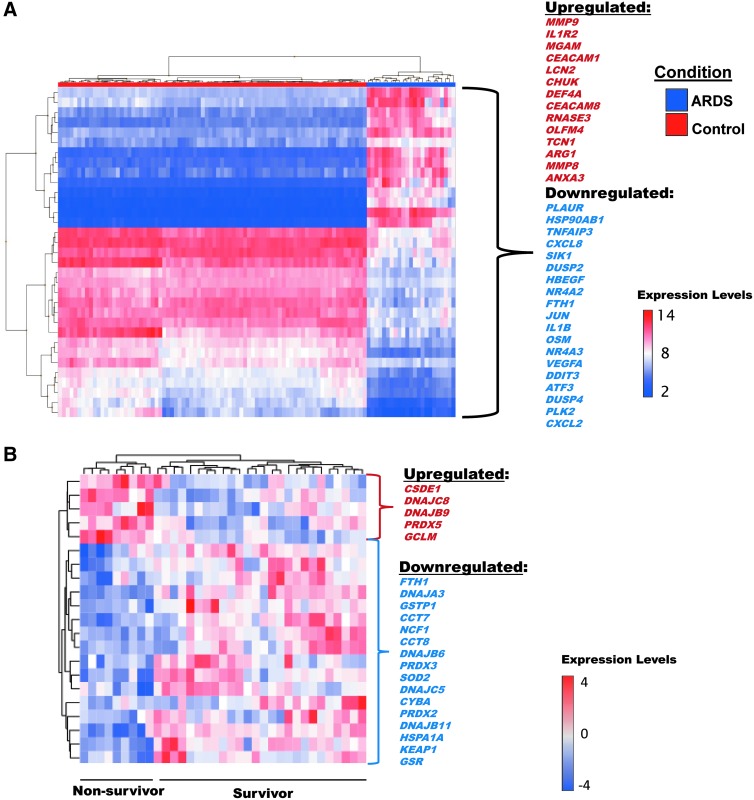
To further determine the utility of a PBMC-derived gene signature in the ICU setting, we interrogated differentially expressed PBMC genes in 55 ARDS survivors and non-survivors (Affymetrix GeneChip Human Exon 2.0 ST microarray). Figure 5A depicts heatmap-displayed results of bioinformatic analysis with 33 differentially expressed genes (DEGs) identified in a molecular signature in ARDS patients (n = 23) versus controls (n = 80), 19 genes downregulated, 14 upregulated (fold change >2 ± 4.41, p < 7.26e−23). Importantly, of the 215 genes (23 upregulated, 192 downregulated) predictive of survival with “Toll-like receptor signaling pathway,” the top enriched pathway, Figure 5B depicts DEGs in PBMCs from 23 ARDS patients that reflect survival with 16 downregulated genes and 5 upregulated genes (Fig. 5).
FIG. 5.
Heatmap of PBMC gene expression predicting ARDS susceptibility and ARDS mortality. (A) The 33 top genes identified in a molecular survival signature from PBMC mRNA in ARDS patients (n = 23) versus controls (n = 80, PMID:19222302). Nineteen genes are downregulated, and 14 are upregulated (Fold change >2 ± 4.41, p < 7.26e-23). (B) 22 genes in the ROS pathway were predictive of ARDS survival in ARDS patients. Five of these genes are upregulated, and 17 are downregulated (24). PBMC, peripheral blood mononuclear cell.
These gene lists includes genes previously reported as dysregulated in other ARDS studies (PLAUR, IL1B, VEGFA) (17, 83, 159). In addition, IL1R2 harbors SNPs that confer increased risk for ARDS and represents a biomarker potentially predictive of sepsis-induced ARDS mortality (123). The gene with the greatest magnitude of upregulation was matrix metallopeptidase 8 (MMP8) with >400-fold change. Although levels of plasma proteins reflecting the expression of these genes were previously reported, this is the first article of MMP8 and TIMP-1 as genomic markers among non-survivors in ARDS (91). This link between MMP8 and IL1B as molecular biomarkers in blood and gene expression biomarkers predicting survival in ARDS suggests that this approach may yield clinically- and biologically relevant ARDS biomarker candidates (14, 58, 85, 91, 109, 120).
Figure 5B results are also consistent with other reports that the TLR4 signaling pathway is a top pathway in predicting survival in ARDS patients (35, 92). eNAMPT, a validated ARDS blood biomarker whose NAMPT promoter SNPs confers risk of ARDS and ARDS mortality (3, 12, 108, 135, 169), is a TLR4 ligand (35) and both TLR4 and NAMPT are differentially expressed in animal models of ARDS (63, 173, 196). There is increasing interest in potentially therapeutically targeting the NAMPT pathway as a strategy to reduce ARDS mortality (Oita et al., manuscript in preparation). In addition, NAMPT genotypes and plasma protein levels represent an opportunity to develop a panel of biomarkers/genotypes that could be employed for clinical trial stratification based on ARDS mortality risk (27, 105).
Pathway analysis methods
High-throughput screenings have provided a wealth of valuable genome-wide data with pathway analysis; the logical next step will be to integrate these results to understand the biological phenomena that underpin these data and generate future hypotheses (29, 100). Database analysis is currently coding protein focused, and multiple database searches can be used to integrate GWAS, mRNA, and proteomic studies (100). We employed multiple genomic and biological pathway database searches to compile the results of ARDS genomic studies to facilitate a better understanding of the pathways involved in a complex genetic disease such as ARDS.
Although pathway analysis is not a meta-analysis of ARDS patients, pathway analysis using multiple databases allows the results of candidate gene, agnostic GWAS studies, and unclassified studies to be combined for analysis and for a better understanding of the cellular and molecular pathways that are involved in ARDS pathobiology. Pathway analysis has two major benefits: (i) It allows for thousands of genes to be reduced in complexity (29), and (ii) it allows for the development of active pathways that are significantly dysregulated in ARDS, potentially providing mechanistic insights that extend beyond creation of a simple gene list (29).
With a complex genetic disease such as ARDS, many genes with varying effect sizes will presumably be involved in ARDS pathobiology. Pathway analysis organizes the results of GWAS studies, candidate gene studies, and meta-analysis to understand the biological pathways that contribute significantly to disease progression. We used pathway analysis for gene sets (Gene Ontology terms), protein–protein interactions, and gene interactions (Reactome and Wikipathways; 5259 human pathways searched) to analyze our collected pool of 201 ARDS genes (100). These studies were performed on the Max Planck Institute for Molecular Genetics consensus pathway database (CPDB) across three pathway sources with the most relevance (Reactome and Wikipathways) (Table 1) (88, 102, 103).
Table 1.
Consensus Pathway Data Base Analysis: Thirty-Eight Enriched Acute Respiratory Distress Syndrome Pathways
| Pathway name | Candidates contained | p-Value | Q-Value | Pathway source | Genes contained |
|---|---|---|---|---|---|
| Immunological pathways | |||||
| Immune system | 74 (4.1%) | 7.69e-17 | 1.29e-15 | Reactome | ACTG1, AGER, ARG1, ARPC4, B2M, C3, C9, CAP1, CAT, CCT8, CEACAM1, CEACAM8, CHIT1, COTL1, CRISP3, CSF2, CSF2RB, CYBA, DEFA4, DNAJC5, FABP5, FGA, FTH1, FTL, GBP2, GHR, GPI, GSTP1, HBB, HMGB1, HSP90AB1, HSPA1A, HSPA8, IL13, IL1B, IL1R2, IL1RN, IL4, IL6, ISG15, JUN, KEAP1, LCN2, LGALS3, MAP3K1, MIF, MMP8, MMP9, MUC5AC, NCF1, NOS3, OLFM4, OSM, PDIA3, PGAM1, PI3, PLAUR, PPIA, PRDX6, RNASE3, S100A12, S100A8, S100A9, SAA1, SERPINA1, TCN1, TLR1, TNF, TNFAIP3, TNFRSF11A, TTR, TXN, VASP, YWHAZ |
| Innate immune system | 58 (5.4%) | 1.05e-18 | 5.29e-17 | Reactome | ACTG1, AGER, ARG1, ARPC4, B2M, C3, C9, CAP1, CAT, CCT8, CEACAM1, CEACAM8, CHIT1, COTL1, CRISP3, CYBA, DEFA4, DNAJC5, FABP5, FGA, FTH1, FTL, GPI, GSTP1, HBB, HMGB1, HSP90AB1, HSPA1A, HSPA8, IL1B, ISG15, JUN, LCN2, LGALS3, MAP3K1, MIF, MMP8, MMP9, MUC5AC, NCF1, NOS3, OLFM4, PGAM1, PI3, PLAUR, PPIA, PRDX6, RNASE3, S100A12, S100A8, S100A9, SAA1, SERPINA1, TCN1, TLR1, TNFAIP3, TTR, TXN |
| Neutrophil degranulation | 41 (8.5%) | 7.87e-20 | 7.94e-18 | Reactome | ARG1, B2M, C3, CAP1, CAT, CCT8, CEACAM1, CEACAM8, CHIT1, COTL1, CRISP3, CYBA, DEFA4, DNAJC5, FABP5, FTH1, FTL, GPI, GSTP1, HBB, HMGB1, HSP90AB1, HSPA1A, HSPA8, LCN2, LGALS3, MIF, MMP8, MMP9, OLFM4, PGAM1, PLAUR, PPIA, PRDX6, RNASE3, S100A12, S100A8, S100A9, SERPINA1, TCN1, TTR |
| Cytokine signaling in the immune system | 21 (4.6%) | 5.64e-06 | 1.73e-05 | Reactome | AGER, B2M, CSF2, CSF2RB, GBP2, GHR, HMGB1, IL13, IL1B, IL1R2, IL1RN, IL4, IL6, ISG15, JUN, OSM, S100A12, SAA1, TNF, TNFRSF11A, YWHAZ |
| Signaling by interleukins | 15 (5.9%) | 6.95e-06 | 2.06e-05 | Reactome | AGER, CSF2, CSF2RB, HMGB1, IL13, IL1B, IL1R2, IL1RN, IL4, IL6, JUN, OSM, S100A12, SAA1, YWHAZ |
| Interleukin-4 and 13 signaling | 15 (15.3%) | 1.67e-11 | 1.3e-10 | Wikipathways | ANXA1, CCL2, CDKN1A, CEBPD, CXCL8, HSPA8, IL1B, IL6, LCN2, MMP9, OSM, PTGS2, S1PR1, SAA1, TNF |
| Interleukin 10 signaling | 10 (26.3%) | 1.57e-10 | 1.06e-09 | Wikipathways | CCL2, CSF2, CXCL2, CXCL8, IL1B, IL1R2, IL1RN, IL6, PTGS2, TNF |
| Interleukin 1 signaling | 7 (11.7%) | 3.12e-05 | 7.32e-05 | Reactome | AGER, HMGB1, IL1B, IL1R2, IL1RN, S100A12, SAA1 |
| Development and heterogeneity of the ILC family | 7 (21.9%) | 4.02e-07 | 2.51e-06 | Wikipathways | AHR, AREG, IL13, IL1B, IL4, IL6, TNF |
| Reactive oxygen species (ROS)-related pathways | |||||
| Cellular response to external stimuli | 26 (6.3%) | 6.14e-10 | 3.65e-09 | Reactome | CAT, CDKN1A, CEBPB, CXCL8, CYBA, CYCS, DNAJA1, DNAJB6, EPAS1, GSR, GSTP1, HSP90AB1, HSPA1A, HSPA8, IL6, JUN, NCF1, NOX4, PRDX2, PRDX3, PRDX5, PRDX6, PRKAG2, SOD2, SOD3, TXN |
| Cellular responses to stress | 25 (7.3%) | 6.59e-11 | 4.75e-10 | Reactome | CAT, CDKN1A, CEBPB, CXCL8, CYBA, CYCS, DNAJA1, DNAJB6, EPAS1, GSR, GSTP1, HSP90AB1, HSPA1A, HSPA8, IL6, JUN, NCF1, NOX4, PRDX2, PRDX3, PRDX5, PRDX6, SOD2, SOD3, TXN |
| Detoxification of reactive oxygen species | 14 (38.9%) | 5.52e-17 | 1.29e-15 | Reactome | CAT, CYBA, CYCS, GSR, GSTP1, NCF1, NOX4, PRDX2, PRDX3, PRDX5, PRDX6, SOD2, SOD3, TXN |
| Focal adhesion | 13 (6.3%) | 1.51e-05 | 4.01e-05 | Wikipathways | ACTG1, COL1A1, COL3A1, COL5A1, EGF, EGFR, FLNA, FLT1, ITGA1, JUN, MYLK, VASP, VWF |
| Toll-like receptor cascades | 10 (6.5%) | 0.000112 | 0.000242 | Reactome | AGER, FGA, HMGB1, JUN, MAP3K1, S100A12, S100A8, S100A9, SAA1, TLR1 |
| Oxidative stress | 9 (30.0%) | 3.63e-10 | 2.29e-09 | Wikipathways | CAT, CYBA, CYP1A1, GCLC, GSR, NFE2L2, NOX4, SOD2, SOD3 |
| Cardiovascular and signaling pathways | |||||
| Hemostasis | 27 (4.0%) | 2.58e-06 | 9.29e-06 | Reactome | ANXA5, APOA1, CAP1, CEACAM1, CEACAM8, EGF, F3, FGA, FLNA, GYPA, GYPB, HRG, ITGA1, JMJD1C, MIF, NOS3, PFN1, PLAUR, PPIA, PRKAR2A, SELPLG, SERPINA1, SERPINE1, TGFB2, TMSB4X, VWF, YWHAZ |
| VEGFA-VEGFR2 signaling pathway | 23 (9.7%) | 1.01e-12 | 9.31e-12 | Wikipathways | ANXA1, CCL2, CXCL8, DNAJB9, EZR, F3, FGA, GJA1, HBEGF, HSPA1A, JUN, MYH9, NCF1, NOS3, NR4A1, NR4A2, NR4A3, PFN1, PLAUR, PTGS2, SOD2, TXN |
| MAPK signaling pathway | 15 (6.1%) | 4.71e-06 | 1.59e-05 | Wikipathways | EGF, EGFR, FLNA, GADD45A, HSPA1A, HSPA8, IL1B, IL1R2, JUN, MAP3K1, MAP3K6, NR4A1, STMN1, TGFB2, TNF |
| Platelet activation, signaling, and aggregation | 15 (5.8%) | 9.2e-06 | 2.58e-05 | Reactome | ANXA5, APOA1, CAP1, EGF, FGA, FLNA, HRG, PFN1, PPIA, SERPINA1, SERPINE1, TGFB2, TMSB4X, VWF, YWHAZ |
| Platelet degranulation | 14 (10.9%) | 8.35e-09 | 4.68e-08 | Reactome | ANXA5, APOA1, CAP1, EGF, FGA, FLNA, HRG, PFN1, PPIA, SERPINA1, SERPINE1, TGFB2, TMSB4X, VWF |
| Response to elevated platelet cytosolic Ca2+ | 14 (10.4%) | 1.37e-08 | 7.28e-08 | Reactome | ANXA5, APOA1, CAP1, EGF, FGA, FLNA, HRG, PFN1, PPIA, SERPINA1, SERPINE1, TGFB2, TMSB4X, VWF |
| Adipogenesis | 13 (9.9%) | 8.49e-08 | 4.08e-07 | Wikipathways | ADIPOQ, AGT, AHR, CDKN1A, CEBPB, CEBPD, EPAS1, GADD45A, IL6, MIF, NAMPT, OSM, SERPINE1, TNF |
| Focal adhesion-PI3K-Akt-mTOR-signaling pathway | 13 (4.3%) | 0.000645 | 0.00113 | Wikipathways | CDKN1A, COL1A1, COL3A1, COL5A1, EGF, EGFR, EPAS1, FLT1, GHR, HSP90AB1, NOS3, OSM, VWF |
| PI3K-Akt signaling pathway | 13 (3.8%) | 0.0019 | 0.00309 | Wikipathways | CDKN1A, COL1A1, EGF, EGFR, FLT1, GHR, HSP90AB1, IL4, IL6, ITGA1, NOS3, OSM, VWF |
| AGE-RAGE | 9 (13.6%) | 5.88e-07 | 3.5e-06 | Wikipathways | AGER, CYCS, EGFR, EZR, JUN, LGALS3, MMP9, NCF1, NOS3 |
| Complement and coagulation cascades | 7 (11.9%) | 2.79e-05 | 7.04e-05 | Wikipathways | C3, C9, F3, PLAUR, SERPINA1, SERPINE1, VWF |
| Transcription factor and signaling pathways | |||||
| Nuclear receptors meta-pathway | 31 (9.8%) | 6.42e-17 | 1.29e-15 | Wikipathways | ABCB1, AGER, AHR, APOA1, CCL2, CYP1A1, EGFR, FTH1, FTL, GCLC, GCLM, GSR, GSTP1, HBEGF, HSP90AB1, HSPA1A, IL1B, JUN, KEAP1, NFE2L2, PDE4B, PLK2, PRDX6, PTGS2, SERPINA1, SOD3, TGFB2, TNF, TNFAIP3, TXN, UGT2B7 |
| NFR2 pathway | 18 (12.7%) | 3.98e-12 | 3.35e-11 | Wikipathways | AGER,FTH1, FTL, GCLC, GCLM, GSR, GSTP1, HBEGF, HSP90AB1, HSPA1A, KEAP1, NFE2L2, PRDX6, SERPINA1, SOD3, TGFB2, TXN, UGT2B7 |
| Vesicle-mediated transport | 18 (2.9%) | 0.00588 | 0.00843 | Reactome | AGTR1, APOA1, AREG, ARPC4, COL1A1, COL3A1, EGF, EGFR, FTH1, FTL, GJA1, HBB, HPX, HSPA8, PRKAG2, SAA1, SERPINA1, YWHAZ |
| G alpha (i) signaling events | 16 (4.0%) | 0.000347 | 0.000674 | Reactome | AGT, ANXA1, APOA1, C3, CXCL2, CXCL8, CXCR4, GRM3, HSPG2, PDE4B, PRKAR2A, RBP4, S1PR1, S1PR3, SAA1, TTR |
| Transcriptional regulation by TP53 | 13 (3.5%) | 0.00403 | 0.00598 | Reactome | BTG2, CDKN1A, CYCS, GADD45A, GPI, GSR, JUN, PLK2, PRDX2, PRDX5, PRKAG2, TXN, YWHAZ |
| Photodynamic therapy-induced NF-kB survival signaling | 8 (22.9%) | 3.83e-08 | 1.93e-07 | Wikipathways | CSF2, CXCL2, CXCL8, IL1B, IL6, MMP9, PTGS2, TNF |
| Other pathways | |||||
| Selenium micronutrient network | 19 (22.9%) | 1.1e-17 | 3.69e-16 | Wikipathways | APOA1, CAT, CBS, CCL2, GSR, HBB, IL1B, IL6, MTHFR, PRDX2, PRDX3, PRDX5, PTGS2, SAA1, SERPINE1, SOD2, SOD3, TNF, TXN |
| Spinal cord injury | 20 (17.1%) | 6.47e-16 | 9.33e-15 | Wikipathways | ANXA1, AQP1, ARG1, BTG2, CCL2, CXCL2, CXCL8, EGFR, GADD45A, GJA1, IL1B, IL4, IL6, LGALS3, MIF, MMP9, NOX4, NR4A1, PTGS2, TNF |
| Lung fibrosis | 14 (21.9%) | 4.82e-13 | 5.41e-12 | Wikipathways | CCL2, CEBPB, CSF2, CXCL2, CXCL8, EGF, IL13, IL1B, IL4, IL6, MMP9, NFE2L2, SERPINA1, TNF |
| Folate metabolism | 14 (21.2%) | 7.59e-13 | 7.67e-12 | Wikipathways | APOA1, CAT, CBS, CCL2, HBB, IL1B, IL4, IL6, MTHFR, SAA1, SERPINE1, SOD2, SOD3, TNF |
| Vitamin B12 metabolism | 13 (25.5%) | 3.99e-13 | 5.04e-12 | Wikipathways | APOA1, CBS, CCL2, HBB, IL1B, IL6, MTHFR, SAA1, SERPINE1, SOD2, SOD3, TCN1, TNF |
| Sudden infant death syndrome (SIDS) susceptibility pathways | 12 (7.5%) | 5.04e-06 | 1.63e-05 | Wikipathways | CEBPB, CXCL8, GJA1, HTR2A, IL13, IL1B, IL1RN, IL6, JUN, PRKAR2A, TNF, YWHAZ |
HMGB1, high mobility group box 1; KEAP1, Kelch-like ECH associated protein 1; MIF, macrophage migration inhibitory factor; MYLK, myosin light chain kinase; NAMPT, nicotinamide phosphoribosyl transferase; PRDXs, peroxiredoxin gene family; S1Ps, sphingosine 1-phosphate receptors.
Enriched pathways were defined as containing more than five genes represented and a p-value of <0.01, which resulted in 72 total enriched pathways. In addition, 38 relevant and/or highly enriched pathways were chosen based on clustering of genes (Fig. 4; Table 1). Of the enriched pathways, 17 resided in Reactome and 21 in Wikipathways (Table 1) and were further divided into ROS pathways (n = 6), immune and inflammatory pathways (n = 9), cardiovascular signaling pathways (n = 11), transcription factor signaling pathways (n = 6), and other pathways (n = 6).
ROS pathways
ROS play an important role in sepsis and ARDS, and they contribute to the severe disruption of the endothelial barrier and the resulting inflammation and inflammatory cascade in the lower airway (54). Neutrophils migrate across the endothelial barrier in response to endothelial-secreted cytokines and chemoattractants. In addition to endothelial-secreted pro-inflammatory signaling, neutrophils are further sources of released pro-inflammatory cytokines, ROS, proteolytic enzymes, nitrogen species, cationic proteins, and lipid mediators (54). Each inflammatory cell type in the lung generates and releases distinct profiles of ROS molecules (45). Leukocytes express NADPH oxidase and nitric oxidase synthases, which together generate peroxynitrite and other ROS species (45).
In ARDS, polymorphonuclear neutrophils and macrophages initiate prolific ROS activation (45). Several isoforms of NADPH oxidase (NOX1, NOX2, NOX4, NOX5) are expressed in the endothelium, and increased expression of NOX1, NOX2, and NOX4 drives endothelial and epithelial barrier dysfunction and generates substantial amounts of secondary ROS (19, 45, 81, 86). The main role of NOX is to catalyze the reduction of molecular oxygen (O2) to superoxide (O2−) (19). Pulmonary endothelial cells express both NOX2 and NOX4 that generate ROS under hypoxic conditions as well on exposure to mechanical stress caused by VILI (81, 86).
ROS generation is linked to survival in sepsis patients (2, 11, 25, 69, 73, 74, 96, 99, 105, 148, 169, 197), and we recently reported that a 21-gene ROS gene signature was significantly linked to survival in sepsis (25) with “Oxidative phosphorylation” being the top enriched pathway. Oxidative phosphorylation is the major pathway of ATP generation in eukaryotic cells, including the vascular endothelium (139). Endothelial mitochondria are also a major source of ROS under aerobic conditions, which include encode complex organelles, including multiple peroxisomes (the P450 complex, xanthine oxidases, and nicotinamide adenine dinucleotide [NADPH] oxidase complexes) that are encoded by a large number of genes (139).
Mitochondria and many genes involved in ATP production create ROS byproducts. The list of ROS genes (Fig. 5B) include chaperon proteins (HSP40, HSP70), which were not commented on in the original paper (25) but are included in the Kyoto Encyclopedia of Genes and Genomes (KEGG) oxidative phosphorylation pathway, and these chaperon proteins are of potential mechanistic interest to understanding the role of oxidative phosphorylation in ARDS. The 21 ROS gene signature (CSDE1, DNAJC8, DNAJB9, PRDX5, GCLM, FTH1, DNAJA3, GSTP1, CCT7, NCF1, CCT8, DNAJB6, PRDX3, SOD2, DNAJC5, CYBA, PRDX2, DNAJB11, HSPA1A, KEAP1, GSR) was used to create a sepsis risk score (25) (Fig. 5B) and significantly outperformed a thousand randomly picked genes in predicting survival among ARDS patients (25).
In the wider ARDS literature, 27 other genes were identified in pathways related to oxidative and cellular stress (Table 1). NOX4 represented a link across multiple pathways (“detoxification of ROS,” “Cellular stress,” “Cellular response to external stimuli,” and “oxidative stress”). Both the superoxide dismutase (SOD) family (SOD2, SOD3) and the peroxiredoxin gene family (PRDX2, PRDX3, PRDX5, PRDX6) were also represented across multiple pathways (“Detoxification of ROS,” “Cellular stress,” “Cellular response to external stimuli”) (Table 1) and exhibited common variants associated with ARDS in case–control studies (87, 112). These genes, together with those that comprise the previously identified 21 gene signature (25), are relevant to a genetic ROS risk and survival signature in ARDS.
“Oxidative stress” pathways join “inflammation” and “apoptosis” as pathways implicated as being important to ARDS pathology (22, 44, 90, 110, 151), with several “Oxidative stress” pathway genes exhibiting SNPs that are significantly linked to ARDS (NAMPT, IL-6, IL4, IL-13) and to vascular signaling pathways (44). Important redox-sensitive pathways in ARDS are the mitogen-activated protein kinase (MAPK) and signal transducer and activator of transcription (STAT) pathways that regulate several ARDS candidate genes (18, 135a, 182, 185, 203). Another redox-sensitive pathway involved in signaling and fibrotic proliferation is the PI3K/Akt pathway (36). Individually, many genes (NOX1, NOX4, STAT4, STAT5) in the MAPK/STAT and PI3K/Akt pathways exhibit mechanistic roles in ARDS pathology in animal models (36).
One highly ROS-related pathway is the NRF2 (NFE2L2) transcription factor and signaling pathway. NRF2 is a ubiquitous master transcription factor that regulates antioxidant response elements (ARE) and mediates cytoprotective and antioxidant protein expression (Fig. 5A) (43). In the healthy lung, NRF2 has a protective effect against hyperoxia, mechanical stress, and VILI (43, 150); it regulates NOX4 in the human and mouse lung, and ROS signaling in ARDS (141, 150).
NRF2 binds to Kelch-like ECH associated protein 1 (KEAP1), another differentially upregulated gene in the ROS ARDS survival gene signature (25, 150). The Keap1–Nrf2 complex translocates Nrf2 to the matrix that binds to AREs and transcribes heme oxygenase-1, NAD(P)H:quinone oxidoreductase 1, catalase, and SOD (41). Interestingly, NRF2 was recently shown to uniquely repress the expression of another ARDS candidate gene, MYLK, via a novel mechanism involving the AREs (113, 125, 141, 172), again highlighting the involvement of NRF2 in ARDS.
Immune-linked and inflammation-linked pathways
Many innate immunity genes are implicated in severe lung injury, contributing to neutrophil infiltration into the alveolar space and “cytokine storm,” an ARDS hallmark (90). In sepsis-induced ARDS, neutrophil-related genes (OLFM4, CD24, LCN2, BPI, RBP7, UTS2) are significantly expressed compared with sepsis patients alone (105). The most highly enriched pathways in our analysis were related to immune signaling (Table 1), with the caveat that many genes were shared between pathways.
A strong 37-gene signature (ARG1, CAP1, CAT, CCT8, CEACAM1, CEACAM8, CHIT1, COTL1, CRIPS3, CYBA, DEFA4, DNAJC5, FABP5, FTH1, FTL, GPI, GSTP1, HBB, HSP90AB1, HSPA1A, HSPA8, LCN2, LGALS3, MIF, MMP8, MMP9, OFLM4, PGAM1, PLAUR, PPIA, RNASE3, S100A8, S100A9) was shared between “neutrophils,” “innate immune system,” and “immune system signaling” pathways (Fig. 6). The strong neutrophil/immune system signature highlights dysregulation in neutrophil cellular biology as of key importance in ARDS risk and survival (84).
FIG. 6.
Inflammation pathways. Using the entirety of 201 genes identified by our eGWAS approach, we identified the top five enriched inflammation pathways: neutrophils, cytokine signaling, immune system, innate immune system, leukocytes. Shown are the individual genes in each pathway with significant overlap. eGWAS, expression genome-wide association study.
MMP8 and MMP9, two genes present in the ARDS survival signature depicted in Figure 6 (Manuel Gonzales-Garay, title unknown), are released by neutrophils at the site of acute inflammation (7). Both MMP8 and MMP9 had increased protein levels in models of lung injury (6, 7, 91). Absence of MMP8 and MMP9 in MMP8−/− and MMP9−/− mice exposed to VILI models shows decreased risk for ALI (6, 7). Both MMP levels in BALF correlate with increased lung injury (84).
HMGB1 is represented across all five of the top immune system pathways and in the IL-1 signaling pathway (Table 1; Figs. 6 and 7). HMGB1 was identified as a cytokine in a murine model of endotoxin-mediated lethality (184) and is upregulated in vitro under 18% cyclic stretch conditions of high mechanical stress (194). Similar to eNAMPT, HMGB1 binds TLR4 as well as RAGE, the primary receptor of HMGB1 (133, 161). HMGB1 protein expression correlates with ARDS severity, 28-day mortality, and 90-day mortality (34, 94–96, 179).
FIG. 7.
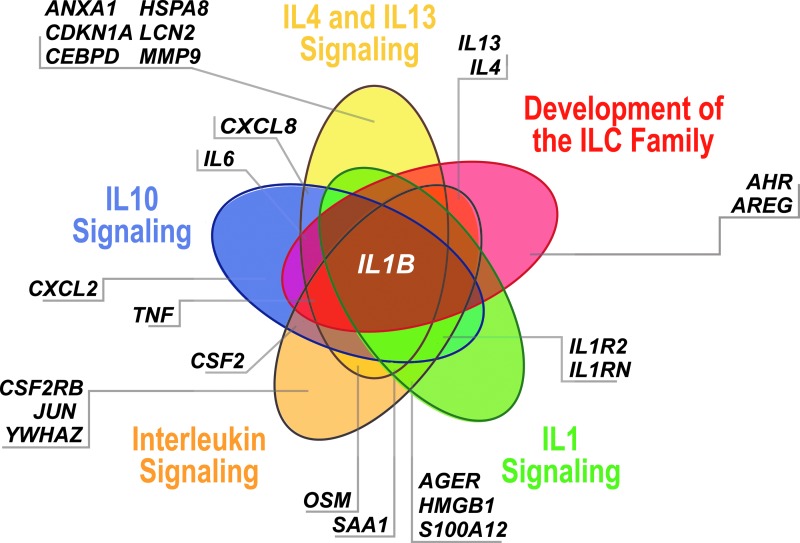
Five interleukin-associated signaling-specific pathways. (IL-10 signaling, IL-4 and IL-13 signaling, development of the ILC family, IL-1 signaling, leukocytes) and the genes that overlap. IL, interleukin.
Another important immunological cytokine in ARDS is MIF, first described in animal models of ARDS (20) and as a product of activated T cells that inhibit macrophage migration (71). In humans, MIF levels in BAL were elevated in both ARDS and septic patients, and we identified two SNPs (rs755622 and rs2070767) in MIF to be associated with African American ARDS patients (71). In pathway analysis, MIF is present in three of the immune pathways (neutrophils, immune system signaling, innate immune system signaling), hemostasis, and adipogenesis (Table 1).
IL-6 and IL-4 are well established biomarkers in ARDS (14, 25, 44, 79, 120, 175), and enrichment of IL signaling pathways in ARDS merits commentary (Fig. 7). Pathways for “IL-4 and IL-13 signaling,” “IL-1B signaling,” “IL-10 signaling,” development of the “ILC” family, and “leukocytes (general)” were enriched in pathway analysis (Table 1). IL1B is involved in all pathways, and IL-1B is an early candidate biomarker whose serum levels correlate with endothelial cellular injury (120, 143).
eNAMPT-driven gene pathways after TLR4 ligation include “IL10-signaling,” “IL-6 signaling,” “leukocyte extravasation signaling,” and “toll-like receptor signaling” (90). A total of 28 genes are involved in at least one IL-related signaling pathway (AGER, AHR, ANXA1, AREG, CCL2, CDKN1A, CEBPD, CSF2, CSF2RB, CXCL2, CXCL8, HMGB1, HSPA8, IL13, IL1B, IL1R2, IL1RN, IL4, IL6, JUN, LCN2, MMP9, OSM, PTGS2, S100A12, SAA1, TNF, YWHAZ) (Table 1; Fig. 7).
Endothelial vascular and cellular signaling pathways
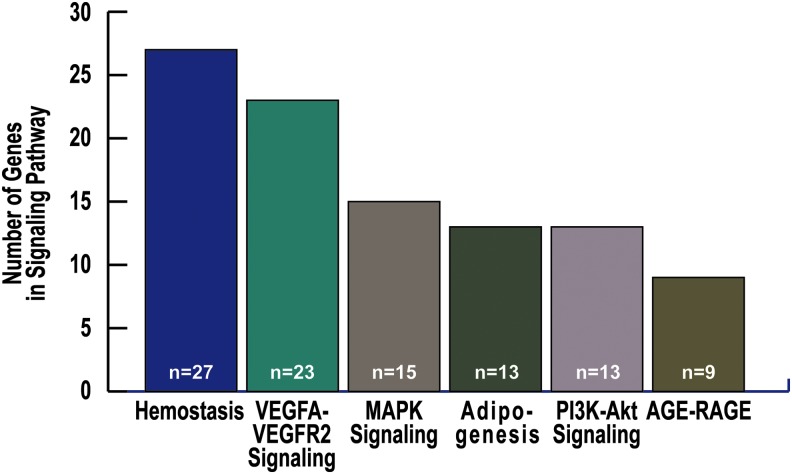
One of the most studied and diverse groups of genes responsible for the pathology of ARDS are the highly conserved vascular signaling genes (196). Our ARDS pathway analysis identified 11 pathways involved with vascular biology relevant to ARDS, with the 6 most highly enriched vascular and cellular signaling pathways shown in Table 1 and Figure 8 and genes from platelet-specific pathways shown in Figure 9. Both growth factors and coagulation factors are upregulated in mRNA studies across species (mouse, rat, canine, human) (196). Of the 37 genes upregulated in this cross-species study, 5 were related to cell proliferation, 6 were related to wound healing, 5 were related to extracellular spaces, and all were related to pro-fibrinolytic processes associated with poor outcomes in ARDS (167, 196). One of these genes, SERPINE1, encodes PAI-1, a potential biomarker for ARDS (137, 196).
FIG. 8.
Genes represented in top enriched endothelial vascular pathways. Shown are the top six cardiovascular signaling pathways and the number of genes in each pathway: Hemostasis, VEGFA-VEGFR2, MAPK signaling, PI3K-Akt signaling, AGE-RAGE. RAGE, receptor of advanced glycation end products; MAPK, mitogen-activated protein kinase; VEGF, vascular endothelial growth factor.
FIG. 9.
Platelet and coagulation pathways. Fourteen genes are shared among the top four enriched platelet and coagulation pathways.
Signaling by VEGF (KEGG pathway map04370) is associated with ARDS in a large genomic ARDS study that yielded 44 significant genes of interest (87), with 13 being specifically involved in VEGF signaling, a critical pathway for cellular proliferation in vascular signaling and ALI (87). Expression of the RAGE is correlated with severity in ARDS patients (97). RAGE is predominantly expressed in epithelial cells, and several RAGE SNPs are potential ARDS risk SNPs (30). NOS3, IL-1B, NOX4, SERPINE1, and IL6 all have a history of associations with ARDS pathology, risk, and severity (17, 24, 86). The VEGF signaling pathway also triggers the downstream activation of many transcription factors such as SP1, which regulates the key cytoskeleton protein, non-muscle myosin light chain kinase (160).
Genes associated with platelet count and coagulation have been discovered to be indirect mediators of endothelial damage in ARDS (193). Five genes associated with platelet counts (BAD, LRRC16A, CD36, JMJD1C, SLMO2) in a meta-analysis were studied in a larger population of ARDS and at-risk controls (146, 192, 193). Five pathways (“hemostasis,” “platelet activation,” “signaling and aggregation,” “platelet degranulation, response to elevated platelet cytosolic Ca2+,” “complement and coagulation cascades”) were involved with platelet signaling or coagulation (Table 1; Fig. 9). In a study with a canine model of lung injury, 7.4% of the differentially regulated genes were in blood coagulation pathways (163).
CPDB pathway analysis identified four separate significant enrichment pathways involving platelets and coagulation (“Hemostasis,” “Platelet activation, signaling and aggregation,” “Platelet degranulation,” “Response to elevated platelet cytosolic Ca2+”) (Table 1; Fig. 9). These pathways share 14 common genes that drive this platelet and coagulation pathway signal (Fig. 9). Many of these coagulation genes (ANXA1, APOA1, FGA, PPIA, SERPINA1) were identified in ARDS proteomic studies as well (39). Genes involved in the sphingolipid generation and signaling pathway, such as S1PR1 and S1PR3, are highly abundant in platelets and are also potential novel biomarkers and risk SNPs (132, 155, 169, 170).
Other transcription factor and signaling pathways
Several important transcription factor and signaling pathways emerged from our pathway analysis (Table 1). Nuclear receptors can directly interact with DNA as a ligand, and the “Nuclear receptors meta-pathways” represent a diverse group of genes that point to the overall importance of DNA regulation and transcription in ARDS. Nuclear receptor meta-pathways are a nebulous category but they present a large and highly significant pathway identified in this study (Table 1; p = 6.42e-17). Of the 201 mapped ARDS genes, 31 represent either nuclear receptors or their interacting genes (Table 1) (ABCB1, AGER, AHR, APOA1, CCL2, CYP1A1, EGFR, FTH1, FTL, GCLC, GCLM, GSR, GSTP1, HBEGF, HSP90AB1, HSPA1A, IL1B, JUN, KEAP1, NFE2L2, PDE4B, FLK2, PRDX6, PTGS2, SERPINA1, SOD3, TGFB2, TNF, TNFAIP3, TXN, UGT2B7).
Critical Issues
ARDS genetic variants/genes identified by GWAS
The recent advances (149) in identifying risk SNPs for ARDS not only present new therapeutic opportunities for ARDS therapies but also present challenges for validation and replication across multiple cohorts in a heterogeneous genetic disease such as ARDS (122, 175). Attempts to address the challenge of defining genetic risk factors involved in the development of ARDS and the severity of the ARDS phenotype largely relies on two approaches: candidate gene studies and genome-wide association studies (149). Candidate gene studies focus on specific gene(s) with probable biological and mechanistic links to vascular permeability, cytoskeletal protein dysregulation, apoptosis pathways, or pro-inflammatory cascades (185, 203).
GWAS focus on genotyping the entire genome without requiring an a priori hypothesis regarding specific genes or their biological significance (5). GWAS studies benefit from not requiring an understanding of mechanisms of gene involvement, allowing for the discovery of novel genes in ARDS (149). Candidate gene studies have several limitations that GWAS studies overcome but have the potential advantage of facilitating defining SNP functionality. In complex diseases such as ARDS with a myriad of environmental and genetic causes, GWAS studies are valuable as they may be performed without a complete mechanistic understanding of the many biological pathways involved (5), with this agnostic approach allowing for the discovery of unique genotype–phenotype relationships (149).
GWAS studies exploring ARDS risk are primarily divided between European populations (80%) and African populations (20%) (24, 116, 123, 162, 173). Together, 5 GWAS studies have yielded 11 genes with 15 independent SNPs associated with ARDS susceptibility in GWAS case studies (Table 2).
Table 2.
Top Single Nucleotide Polymorphisms and Genes in Acute Respiratory Distress Syndrome Literature—Genome-Wide Association Study Risk
| Gene | Predictive SNPs | Population | Study |
|---|---|---|---|
| GWAS risk SNPs | |||
| ABCC1 | Rs3887893 (p = 0.0001, meta) | European descent (MGH, Boston, MA) | 765 Stage II ARDS trauma population, 838 stage II ARDS sepsis population; direct vs. indirect ARDS association and meta-analysis (75) |
| ARSD | Rs78142040 (3.64e-47) | ARDSNet | 213 ARDS patients, 440 (379 EUR and 61 ASW) controls; Exome-seq case/control association (161) |
| FAAH | Rs324420 (p = 0.0131) | European descent (MGH, Boston, MA) | 765 Stage II ARDS trauma population, 838 stage II ARDS sepsis population; direct vs. indirect ARDS association and meta-analysis (75) |
| HEATR1 | Rs2115740(p = 6.53 × 10−5, unadjusted) | African American descent (Seattle, WA and Chicago, IL) | 232 ARDS cases, 162 ICU controls (53) |
| IL1RN | Rs315952 (p = 0.0023), rs380092 (p = 0.026) | European descent (Philadelphia, PA) | Association stage II (n = 606) ARDS and stage III (n = 561) ARDS (74) |
| PDE4B | Rs12080701 (p = 0.0005, meta), Rs17419964 (p = 0.0002, meta) | European descent (MGH, Boston, MA) | 765 Stage II ARDS trauma population, 838 stage II ARDS sepsis population; direct vs. indirect ARDS association and meta-analysis (75) |
| POPDC3 | rs1190286 (p = 0.0094) | European descent (MGH, Boston, MA) | 765 Stage II ARDS trauma population, 838 stage II ARDS sepsis population; direct vs. indirect ARDS association and meta-analysis (75) |
| PPFIA1 | Rs471931(p = 0.0021) | European descent (Philadelphia, PA) | Two-stage GWAS; phase 1 compared 600 ARDS trauma-associated ALI, 2266 population-based controls; phase 2 compared 212 ALI cases and 238 at-risk controls (71) |
| SELPLG | Rs109017898(p = 1.5 × e-04, p = 0.005, discovery, meta) | African American descent (Seattle, WA and Chicago, IL) | 232 ARDS cases, 162 ICU controls (53) |
| TACR2 | Rs61732394(p = 6.24 × 10−4) | African American descent (Seattle, WA and Chicago, IL) | 232 ARDS cases, 162 ICU controls (53) |
| TNFRSF11A | Rs9960450 (p = 5.3 × 10−3, meta), Rs17069902 (p = 0.0001, meta) | European descent (MGH, Boston, MA) | 765 Stage II ARDS trauma population, 838 stage II ARDS sepsis population; direct vs. indirect ARDS association and meta-analysis (75) |
| XKR3 | Rs9605146 (1.68 × 10−59) | ARDSNet | 213 ARDS patients, 440 (379 EUR and 61 ASW) controls; Exome-seq case/control association (161) |
| ZNF335 | Rs3848719 (p = 2.86e-04) | ARDSNet | 213 ARDS patients, 440 (379 EUR and 61 ASW) controls; Exome-seq case/control association (161) |
| GWAS survival-specific risk SNPs | |||
| ADIPOQ | Rs2082940 (p = 0.0039) | ICU (MGH and BIDMC, Boston) | 2067 ICU patients; 567 ARDS patients; prospective risk and mortality study (169) |
| FER | Rs4957796 (p = 0.0144) | European descent (Gottingen, Germany) | 441 Total ARDS patients, 274 ARDS patients with pneumonia; 90-day survival; prospective case–control (166) |
ALI, acute lung injury; ARDS, acute respiratory distress syndrome; ASW, African ancestry in the Southwest USA; EUR, European; GWAS, genome-wide association study; ICU, intensive care unit; SNP, single nucleotide polymorphism; XRK3, XK-related 3.
Among European ARDS GWAS studies, nine ARDS genes, including XK-related 3 (XKR3), arylsulfatase D (ARSD), and Zinc-Finger/Leucine-Zipper Co-Transducer NIF1 (ZNF335), were identified (Table 2) by case–control whole exome sequencing of Asian American and European American populations (162). Another study in multiple ARDS populations (trauma- and sepsis-induced ARDS) of European descent identified several genes (POPDC3, FAAH, PDE4B, ABCC1, TNFRSF11A) to reach population-wide significance in a meta-analysis (173). Two other independent ARDS studies in European descent patients (Philadelphia, PA) had two genes (IL1RN, PPFIA1) reaching population-wide significance in their respective case–control studies (47, 123). IL1RN is linked to the development of both ARDS and sepsis (123, 140); IL1RNA levels were shown to be significantly higher in ARDS patients compared with controls, and predicted mortality (140).
Another immunity-related gene, TNFRSF11A, is a member of the tumor necrosis factor receptor (TNFR) family, mechanistically important to developing ARDS (78, 145). TNFR1 mediates cell death, inflammation, which, in turn, leads to vascular leak and neutrophil infiltration of the alveolar space (145). TNFRSF11A encodes the receptor activator of NF-kB (RANK), which is the receptor for receptor activator of NF-kB ligand (RANKL), key to altered NF-kB signaling (78, 165). Although TNFRSF11A SNPs have been significantly associated with the severity of Paget's disease, the reported ARDS risk SNPs are unique (78, 123) and may influence TNFRSF11A alternative splicing, a largely unexplored mechanism in ARDS (165, 181), and NF-kB signaling (78, 165).
GWAS studies of ARDS mortality
In the ARDS genetic literature, GWAS studies are less common than candidate gene studies due to their expense as well as that ARDS GWAS studies require larger patient populations to overcome the limitations of multiple association testing (Bonferroni correction) (149). Only two GWAS studies evaluated ARDS as an end-point with approximation of severity (11, 89, 117, 195), and both were conducted in European (89) or European descent populations (11, 116, 117). Each study had a single gene reach population significance for mortality association (ADIPOQ, FER, ACE) with at least one SNP, although other SNPs in linkage disequilibrium were reported (Table 2).
ADIPOQ encodes adiponectin with an SNP associated with the ARDS mortality study (4), and it has been linked in several meta-analyses to type 2 diabetes or obesity (114, 178) in Caucasians, in a South Eastern Asian population (114) and with type 2 diabetes in Chinese and Taiwanese populations (4, 48, 178, 195). However, after adjusting for body mass index, a measure of obesity, and for diabetes status, rs2082940 remained significantly associated with ARDS mortality (4).
Unlike ADIPOQ, ACE has a history of mechanistic studies that implicate ACE as an important gene in ARDS pathology (77, 198, 201). ACE is the enzyme that degrades angiotensin I (Ang I) to angiotensin II (Ang II), which is the peptide that is primarily responsible for maintaining blood pressure homeostasis and fluid/salt balance in kidney filtration (198). In animal models, ACE is strongly associated with elevations in IL-6 and leukocyte counts (201) and it is elevated in human ARDS BALF (198, 201).
Ang I has also been implicated as having an insertion/deletion polymorphism that is associated with mortality in an ICU ARDS cohort (116). FER is a member of the FPS/FES non-transmembrane receptor tyrosine kinase family, and it is significantly associated with ARDS (Table 2) and survival in septic patients (148). In a multi-cohort study, rs4957796 was significantly associated with survival in sepsis patients (148) and with increased survival in ARDS (89). Many ARDS patients have sepsis as a comorbidity, and rs4957796 being an SNP associated with both mortality in ARDS and sepsis makes it a strong candidate gene to study in ARDS, and potentially useful in developing an ARDS risk SNP panel (89, 148).
ARDS genetic variants/genes identified by candidate gene studies
Candidate gene studies require an a priori hypothesis or prior knowledge of a gene's function, which make candidate gene studies popular for studying genes with known functional roles in ARDS (5). A limitation of candidate gene studies is a potential failure to account for genetic drift and population demographics on natural selection (5). In ARDS, this is a particularly important limitation because of observed health disparities between African, Hispanic, and European populations (64). The largest category of ARDS genetic studies are ARDS risk-association candidate gene studies (2, 12, 17, 24, 80, 93, 111, 154, 158, 174, 188). Candidate gene studies are the original genetic approaches in the ARDS literature and are often associated with a gene or protein's hypothesized role in lung injury (149).
The first reported candidate risk genes were SFTPB (SP-B variant), ACE, and IL-6 (68, 80, 111, 116, 117). The advantage of candidate gene studies is that analyses can be performed in smaller population case sizes to achieve significance when compared with GWAS approaches (111, 116, 117). Although candidate gene studies are numerous, replication of specific SNPs has proven difficult. However larger study populations have produced more robust results for specific risk genes (NFE2L2, NAMPT/PBEF, IL-4, IL-13, SP-B, AGER, PI3, MAP3K1, IL-6, MYLK) and specific SNPs of interest (Table 2). Among candidate gene risk studies, 18 of 23 studies (∼78%) have been performed exclusively in European populations or populations of European descent (11, 12, 24, 80, 93, 111, 154, 158, 174, 188).
The early research into candidate genes provided viable genotype–phenotype links for biomarker studies, and one of the most successful has been IL-6 (117). A haplotype of IL-6, -174G/C, has been identified and validated as a risk SNP for several ARDS case–control studies (68, 118). Increased levels of IL-6 cause a rise in ROS via the TLR4-TRIF-TRAF6 pathway (92). IL-6 is elevated in patients with ARDS and has been shown to have a significant role in the permeability of the lung endothelium in multiple ARDS mouse models (79). IL-6 has several risk SNPs and promoter haplotypes associated with sepsis-related ALI (66, 79). IL-6 levels are determined by many genetic factors, and the SNPs associated with ALI in sepsis patients were discovered in a Hispanic population (66).
Most importantly, two of the SNPs (−597G/−174G) are associated with a risk haplotype (118). A strong phenotype–genotype relationship with IL-6, genotypes, clinical outcomes, and ARDS severity or mortality has been documented, and the case for IL-6 genetics playing a role in ARDS severity risk and mortality is strong (66, 117, 118).
Candidate gene studies of ARDS mortality
Although there are more candidate gene studies that focus on ARDS risk genes, multiple candidate gene studies have evaluated ARDS mortality and associated risk genes (4, 12, 17, 61). When ARDS patients are stratified by the Berlin definition (mild, moderate, and severe), the more severe ARDS patients are significantly more likely to die from ARDS than patients with mild or moderate ARDS (16). Thus, patients who die from ARDS can be argued to have severe ARDS (16).
Of the four candidate gene studies that reported on mortality, three genes (75%) were obtained in European descent populations, and one population is from a pediatric, Brazilian population (Table 3). In the European populations, NAMPT/PBEF, IL-1β, and PHD2 were identified as each having at least one SNP (NAMPT/PBEF has four) that is associated with ARDS mortality (4, 17, 61). In the Brazilian, pediatric ARDS population, TNF had two SNPS that were associated with death in septic and ARDS populations (4).
Table 3.
Top Single Nucleotide Polymorphisms and Genes in Acute Respiratory Distress Syndrome Literature—Candidate Gene Risk
| Gene | Predictive SNPs | Population | Study |
|---|---|---|---|
| Candidate gene risk SNPs | |||
| ACE | D/D (p = 0.00004, healthy population, p = 0.0008 CABG patients) | European descent (University of College London Hospitals) | 88 Respiratory failure patients, 174 CABG controls (192) |
| AGER | Rs2070600 (A/A, Ser/Ser, p < 0.0001) | European descent (Clermont-Ferrand, France) | 59 ARDS, 405 controls; log rank test with case–control (178) |
| AGT | Rs699 (0.028, dom) | European descent (Moscow, Russia) | 68 NP ARDS cases, 198 NP controls (179) |
| AhR | Rs2066853 (0.0012, dom) | European descent (Moscow, Russia) | 68 NP ARDS cases, 198 NP controls (179) |
| CYP1A1 | Rs2606345 (0.0027, dom) | European descent (Moscow, Russia) | 68 NP ARDS cases, 198 NP controls (179) |
| DIO2 | Rs12885300 (p = 0.039) | African and European descent (American-European Consensus Criteria) | 327 European Americans: 139 sever sepsis, 78 severe sepsis + ARDS/ALI, 188 controls; 261 African Americans: 78 severe sepsis, 41 severe sepsis + ARDS/ALI, 187 controls (187) |
| Rs225014 (p = 0.009) | |||
| EGF | Rs4444903 (p = 0.005, males), rs2298991 (p = 0.019, males), Rs7692976 (p = 0.005, males), Rs6533485 (p = 0.025, males) | European descent (MGH, Boston, MA) | 416 ARDS cases (246 survivors, 170 died), 1052 ICU controls; 887 males and 581 females (180) |
| GADD45a | Rs581000 (p = 0.009) | African Americans Chicago Study and Spanish Study (Chicago, IL) | African American Chicago cohort: 71 severe sepsis, 40 sepsis + ALI, 182 controls; Spanishcohort: 80 severe sepsis, 66 sepsis + ALI, 95 controls (61) |
| IL-13 | 1 SNP (431 A>Gr, p = 0.008) | European descent (Moscow, Russia) | 347 Controls, 74 ARDS cases; logistic regression adjusted case/control (98) |
| IL-4 | 1 SNP (−589 C>T, p = 0.01) | European descent (Moscow, Russia) | 345 Controls, 72 ARDS cases; logistic regression adjusted case/control (98) |
| IL-6 | −174G/C allele (p = 0.03) | European descent | Twin study on lung function, 427 twins (232 women, 195 men) (36, 41) |
| MAP3K1 | Rs832582 (p = 0.01, rec) | FACTT and ARDSNet (Seattle WA) | 241 ARDS patients, 346 healthy, locally matched controls (193) |
| MIF | 2 SNPs (rs755622, p = 0.03; Rs2070767, p = 0.04) | European descent, African descent | 288 European: 113 severe sepsis, 90 sepsis-associate ALI, 85 healthy controls; 218 African: 69 severe sepsis, 61 sepsis-associated ALI, 88 healthy controls (50) |
| MYLK | Rs820336 (p = 0.002) | European and African descent (John Hopkins University and Medical College of Wisconsin) | European: 92 ALI, 99 sepsis, 85 healthy controls; African: 43 ALI, 51 sepsis, 61 healthy control (91) |
| Rs936170 (p = 0.009) | |||
| Rs936170 (p = 0.025) | |||
| NAMPT/PBEF | 2 SNPs (−948, p = 0.015–2422, p = 0.03) | European descent | 374 ARDS patients and 787 at-risk controls; nested case–control (88) |
| NFE2L2 | 7 SNPS (p-values: 0.0069–0.0089) | Spanish Network | 321 Severe sepsis and ARDS; 871 population-based controls; case–control (76) |
| PI3 | Rs1983649 (p = 0.034, add), Rs2664581 (p = 0.004, 0.023, add, dom) | European descent (MGH, Boston, MA) | 449 ARDS patients, 1031 at-risk controls; case–control (181, 182) |
| S1PR3 | 2 SNPS (rs7022797, p = 0.017; Rs11137480, p = 0.042) | European and African descent (Chicago, IL) | 71 European ARDS and 24 African ARDS; 186 European controls, 185 African controls (66) |
| SP-B | 1 SNP (606-bp variant allele, 1580 C/T) | MGH (Boston, MA), European descent (German) | 72 ARDS cases, 117 controls; nested case–control (177) 52 ARDS patients, 46 healthy controls (44) |
| TLR1 | 1 SNP (rs5743551, p = 0.002) | European descent (Seattle, WA) | 138 Severe sepsis ARDS, 107 ALI, 167 healthy controls (23) |
| Candidate gene survival-specific risk SNPs | |||
| IL-1B | 1 SNP (−511 G>A, p = 0.0019) | European descent (Moscow, Russia) | 321 Controls, 91 mortality cases; adjusted logistic regression for case–control mortality (150) |
| NAMPT/PBEF | −1001G (p = 0.001) | European descent | 374 ARDS patients and 787 at-risk controls; nested case–control (101) |
| −1543T (p = 0.03) | |||
| PHD2 | 1 SNP (rs516651, p = 0.002) | European descent (Duisburg-Essen, Germany) | 264 ARDS (70 died); case–control (184) |
| TNF | TNF-308 (p = 0.0006) | Brazilian septic and ARDS pediatric patients | 490 Septic and ARDS patients; 610 controls (119) |
| TNF-863 (p = 0.01) | |||
CABG, coronary artery bypass graft; D/D, deletion/deletion; DIO2, iodothyronine deiodinase 2; GADD45a, growth arrest and DNA damage-inducible gene; NP, nosocomial pneumonia.
Two of these candidate genes (TNF and IL-1β) were chosen because they are molecules with a long history as ARDS biomarkers (11, 31, 120). IL-1β transcription is caused by stress and endotoxin triggers and is secreted by macrophages, thrombocytes, and injured endothelial cells (31, 55). The promoter for IL-1β includes NF-kB sites and activating protein-1 sites (55). In an attempt to link biomarkers to genotype and establish a genotype–phenotype relationship in IL-1β, a significant SNP was found in the IL-1β promoter region (-511 upstream from the TATA box and transcription start site) (17, 55). The site found to be related to ARDS and sepsis mortality was previously reported to be an important site for the secretion of IL-1β (143).
ARDS risk SNPs in African Americans
Racial and ethnic disparities in ARDS mortality and disease susceptibility have been reported (64); however, genetic studies in ARDS have focused on larger, European cohorts. Although this has provided a strong foundation for the understanding of ARDS genetics, population diversity among ARDS should not be discounted in the sub-phenotyping, diagnosis, and treatment of ARDS. Understanding genetic population diversity in ARDS is critical because there is a significant difference in mortality rates between European and African descent populations (49, 64).
Across multiple age populations (until the age of 65), African Americans have significantly higher rates of both sepsis and ARDS than their matched European American cohorts (16, 23, 49, 51, 129), greater duration on mechanical ventilation than European Americans (64), and a higher risk of ARDS mortality (16, 23, 129) when compared with age-matched European American counterparts. Hispanic Americans also have significantly higher mortality rates (5).
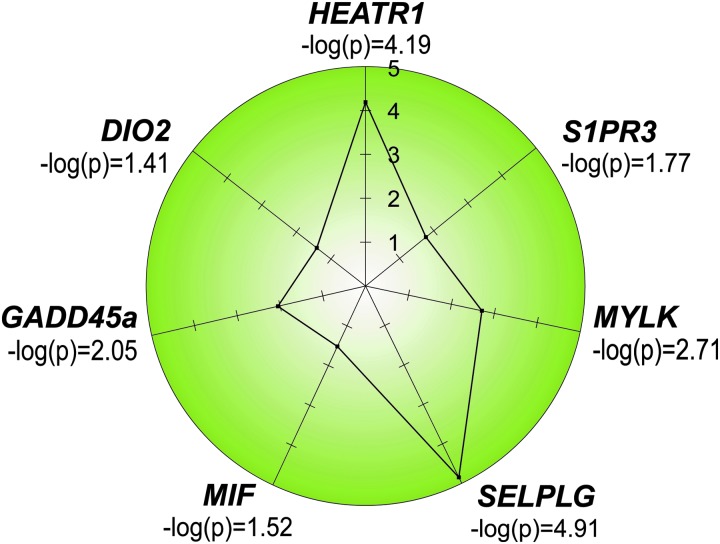
The health disparity borne by African Americans in ARDS and severe sepsis warrants additional research and attention to the role of genetics in identifying unique biomarkers and genetic markers for African Americans at risk for ARDS. Several unique candidate genes MYLK, HEATR1, MIF, GADD45a, DIO2, SELPLG, and S1PR3 are promising genetic markers for increased risk of ARDS and ARDS mortality among African Americans (Fig. 10) (24, 46, 71–73, 115, 155, 169). In the case of MYLK, the gene encoding myosin light chain kinase, a risk haplotype was identified consisting of coding SNPs with one of these SNPs verified in other inflammatory disorders, including sickle cell disease and severe asthma in African Americans (67).
FIG. 10.
Risk genes in African descent and Hispanic ethnicity. Depicted are the seven top risk single nucleotide polymorphisms/genes that are unique to ARDS risk in African descent individuals and to ARDS survival. The genes were identified via interrogation of GWAS results (HEATR1, SELPLG) or via candidate gene approaches and sequencing (S1PR3, MYLK, DIO2, GADD45a). DIO2, iodothyronine deiodinase 2; GADD45a, growth arrest and DNA damage-inducible gene; MYLK, myosin light chain kinase; S1Ps, sphingosine 1-phosphate receptors.
The functionality of the SNP has been shown to cause a delay in restoration of the vascular barrier in inflammatory models as well as to cause secondary mRNA structure alterations that promote excessive expression of this major cytoskeletal regulatory protein (67, 72, 73, 187, 204).
Our group recently conducted a GWAS study of African American ARDS patients and ICU controls that was underpowered but after innovative pathway prioritization, it discovered three novel genes that achieved genome-wide significance (24). Unlike in the European American ARDS studies, the population size in this study was relatively small (n = 232) (Tables 2 and 3; Fig. 10) (24). Two unique risk SNPs for ARDS (rs2115740, HEATR1; rs109017898, SELPLG) were found in this African American GWAS study in two genes that had not previously been identified as ARDS risk genes. Further, higher-powered GWAS studies in non-European populations may potentially provide more novel genes and SNPS that are risk factors for ARDS in other under-studied populations.
Future Directions
ARDS is a severe, high-mortality complex and heterogeneous critical illness influenced by environmental and genetic factors. In this review, we have collated the available preclinical and human ARDS literature and identified 201 pooled ARDS candidate genes (Supplementary Table S2) in a multi-database approach. Although we highlighted risk SNPs from both candidate gene studies and GWAS, pathway analysis allowed genes without known SNPs but reported mRNA and protein fold change to be included in our pathway analysis (25, 167, 196).
Our pathway analysis strategies revealed results that were consistent with the concept that evolutionarily conserved inflammatory and ROS networks and vascular gene dysregulation are potent contributors to ARDS pathobiology (82, 83). A broader “omics” approach to ARDS allows for the focus on biologically relevant pathways and genotype–phenotype connections between established ARDS biomarkers and differentially expressed ARDS risk genes.
We have also chosen to evaluate ARDS studies that use mortality as an end-point as this captures the most severe outcome for ARDS patients and summarized the evidence from genetic studies in diverse populations that have the potential to uncover novel biomarkers for ARDS risk and mortality and potential therapeutic targets in ARDS. We highlighted information relevant to the role of genetic factors in ARDS susceptibility and mortality (23) that address the well-known health disparities that exist in susceptibility to and mortality from ARDS (23, 38, 75). Improved strategies for sub-phenotyping of diverse ARDS patients via molecular signatures or SNP panels will facilitate the potential for successful clinical trials in ARDS and yield a better fundamental understanding of ARDS pathobiology (105).
Supplementary Material
Abbreviations Used
- ACE
angiotensin-converting enzyme
- ALI
acute lung injury
- ARDS
acute respiratory distress syndrome
- ARE
antioxidant response elements
- BALF
bronchoalveolar lavage fluid
- CPDB
consensus pathway database
- DEG
differentially expressed gene
- DIO2
iodothyronine deiodinase 2
- eNAMPT
extracellular NAMPT
- GWAS
genome-wide association study
- HMGB1
high mobility group box 1
- ICU
intensive care unit
- IL
interleukin
- KEAP1
Kelch-like ECH associated protein 1
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KL-6
Krebs von den Lugen-6
- MAPK
mitogen-activated protein kinase
- MIF
macrophage migration inhibitory factor
- MMP
matrix metallopeptidase
- MYLK
myosin light chain kinase
- NADPH
nicotinamide adenine dinucleotide
- NAMPT
nicotinamide phosphoribosyl transferase
- NOX
NADPH oxidase
- PBMC
peripheral blood mononuclear cell
- PRXs
peroxiredoxin gene family
- RAGE
receptor of advanced glycation end products
- ROS
reactive oxygen species
- S1Ps
sphingosine 1-phosphate receptors
- SNP
single nucleotide polymorphism
- SOD
superoxide dismutase
- STAT
signal transducer and activator of transcription
- TLR4
toll-like receptor 4
- TNFR
tumor necrosis factor receptor
- VEGF
vascular endothelial growth factor
- VILI
ventilator-induced lung injury
- XKR3
XK-related 3
Supplementary Material
References
- 1. This reference has been deleted.
- 2. Acosta-Herrera M, Pino-Yanes M, Blanco J, Ballesteros JC, Ambrós A, Corrales A, Gandía F, Subirá C, Domínguez D, Baluja A, Añón JM, Adalia R, Pérez-Méndez L, Flores C, Villar J, and for the GRECIA and GE N-SEP Networks. Common variants of NFE2L2 gene predisposes to acute respiratory distress syndrome in patients with severe sepsis. Crit Care 19: 256, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adyshev DM, Elangovan VR, Moldobaeva N, Mapes B, Sun X, and Garcia JGN. Mechanical stress induces pre-B-cell colony-enhancing factor/NAMPT expression via epigenetic regulation by miR-374a and miR-568 in human lung endothelium. Am J Respir Cell Mol Biol 50: 409–418, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahasic AM, Zhao Y, Su L, Sheu C-C, Thompson BT, and Christiani DC. Adiponectin gene polymorphisms and acute respiratory distress syndrome susceptibility and mortality. PLoS One 9: e89170, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res 19: 711–722, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Albaiceta GM, Gutierrez-Fernández A, García-Prieto E, Puente XS, Parra D, Astudillo A, Campestre C, Cabrera S, Gonzalez-Lopez A, Fueyo A, Taboada F, and López-Otin C. Absence or inhibition of matrix metalloproteinase-8 decreases ventilator-induced lung injury. Am J Respir Cell Mol Biol 43: 555–563, 2010 [DOI] [PubMed] [Google Scholar]
- 7. Albaiceta GM, Gutiérrez-Fernández A, Parra D, Astudillo A, García-Prieto E, Taboada F, and Fueyo A. Lack of matrix metalloproteinase-9 worsens ventilator-induced lung injury. Am J Physiol Lung Cell Mol Physiol 294: L535–L543, 2008 [DOI] [PubMed] [Google Scholar]
- 8. ARDSNet. Ventilation with lower tidal volumes as compared with traditional tidal volumes for acute lung injury and the acute respiratory distress syndrome. The Acute Respiratory Distress Syndrome Network. N Engl J Med 342: 1301–1308, 2000 [DOI] [PubMed] [Google Scholar]
- 9. Armstrong L, Thickett DR, Christie SJ, Kendall H, and Millar AB. Increased expression of functionally active membrane-associated tumor necrosis factor in acute respiratory distress syndrome. Am J Respir Cell Mol Biol 22: 68–74, 2000 [DOI] [PubMed] [Google Scholar]
- 10. Ashbaugh D, Boyd Bigelow D, Petty T, and Levine B. ACUTE respiratory distress in adults. Lancet 290: 319–323, 1967 [DOI] [PubMed] [Google Scholar]
- 11. Azevedo ZM, Moore DB, Lima FC, Cardoso CC, Bougleux R, Matos GI, Luz RA, Xavier-Elsas P, Sampaio EP, Gaspar-Elsas MI, and Moraes MO. Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) single nucleotide polymorphisms: importance in ARDS in septic pediatric critically ill patients. Hum Immunol 73: 661–667, 2012 [DOI] [PubMed] [Google Scholar]
- 12. Bajwa EK, Yu C-L, Gong MN, Thompson BT, and Christiani DC. Pre-B-cell colony-enhancing factor gene polymorphisms and risk of acute respiratory distress syndrome. Crit Care Med 35: 1290–1295, 2007 [DOI] [PubMed] [Google Scholar]
- 13. Barkan D, Kleinman H, Simmons JL, Asmussen H, Kamaraju AK, Hoenorhoff MJ, Liu Z-Y, Costes SV, Cho EH, Lockett S, Khanna C, Chambers AF, and Green JE. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res 68: 6241–6250, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bauer TT, Montón C, Torres A, Cabello H, Fillela X, Maldonado A, Nicolás J-M, and Zavala E. Comparison of systemic cytokine levels in patients with acute respiratory distress syndrome, severe pneumonia, and controls. Thorax 55: 46, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Becker KG, Hosack DA, Dennis G, Jr., Lempicki RA, Bright TJ, Cheadle C, and Engel J. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics 4: 61, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bellani G, Laffey JG, Pham T, Fan E, Brochard L, Esteban A, Gattinoni L, van Haren F, Larsson A, McAuley DF, Ranieri M, Rubenfeld G, Thompson BT, Wrigge H, Slutsky AS, Pesenti A; LUNG SAFE Investigators; ESICM Trials Group. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in 50 countries. JAMA 315: 788–800, 2016 [DOI] [PubMed] [Google Scholar]
- 17. Belopolskaya OB, Smelaya TV, Moroz VV, Golubev AM, and Salnikova LE. Clinical associations of host genetic variations in the genes of cytokines in critically ill patients. Clin Exp Immunol 180: 531–541, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benveniste EN, Liu Y, McFarland BC, and Qin H. Involvement of the janus kinase/signal transducer and activator of transcription signaling pathway in multiple sclerosis and the animal model of experimental autoimmune encephalomyelitis. J Interferon Cytokine Res 34: 577–588, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bernard K, Hecker L, Luckhardt TR, Cheng G, and Thannickal VJ. NADPH oxidases in lung health and disease. Antioxid Redox Signal 20: 2838–2853, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bernhagen J, Calandra T, Mitchell RA, Martin SB, Tracey KJ, Voelter W, Manogue KR, Cerami A, and Bucala R. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature 365: 756, 1993 [DOI] [PubMed] [Google Scholar]
- 21. Bhargava M, Becker TL, Viken KJ, Jagtap PD, Dey S, Steinbach MS, Wu B, Kumar V, Bitterman PB, Ingbar DH, and Wendt CH. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PLoS One 9: e109713, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhargava M, Viken K, Wang Q, Jagtap P, Bitterman P, Ingbar D, and Wendt C. Bronchoalveolar lavage fluid protein expression in acute respiratory distress syndrome provides insights into pathways activated in subjects with different outcomes. Sci Rep 7: 7464, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bime C, Poongkunran C, Borgstrom M, Natt B, Desai H, Parthasarathy S, and Garcia JGN. Racial differences in mortality from severe acute respiratory failure in the United States, 2008–2012. Ann Am Thorac Soc 13: 2184–2189, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bime C, Pouladi N, Sammani S, Batai K, Casanova N, Zhou T, Kempf CL, Sun X, Camp SM, Wang T, Kittles RA, Lussier YA, Jones TK, Reilly JP, Meyer NJ, Christie JD, Karnes J, Gonzalez-Garay M, Christiani DC, Yates CR, Wurfel MM, Meduri GU, and Garcia JGN. Genome wide association study in african americans with acute respiratory distress syndrome identifies the selectin p ligand gene as a risk factor. Am J Respir Crit Care Med 197: 1421–1432, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bime C, Zhou T, Wang T, Slepian MJ, Garcia JGN, and Hecker L. Reactive oxygen species-associated molecular signature predicts survival in patients with sepsis. Pulm Cir 6: 196–201, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Birukov KG, Jacobson JR, Flores AA, Ye SQ, Birukova AA, Verin AD, and Garcia JGN. Magnitude-dependent regulation of pulmonary endothelial cell barrier function by cyclic stretch. Am J Physiol Lung Cell Mol Physiol 285: L785, 2003 [DOI] [PubMed] [Google Scholar]
- 27. Blondonnet R, Constantin J-M, Sapin V, and Jabaudon M. A pathophysiologic approach to biomarkers in acute respiratory distress syndrome. Dis Markers 2016: 3501373, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bouros D, Alexandrakis MG, Antoniou KM, Agouridakis P, Pneumatikos I, Anevlavis S, Pataka A, Patlakas G, Karkavitsas N, and Kyriakou D. The clinical significance of serum and bronchoalveolar lavage inflammatory cytokines in patients at risk for acute respiratory distress syndrome. BMC Pulm Med 4: 6, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Braun R. Systems analysis of high-throughput data. Adv Exp Med Biol 844: 153–187, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brown SM, Grissom CK, Rondina MT, Hoidal JR, Scholand MB, Wolff RK, Morris AH, Paine R, and Network NNA. Polymorphisms in key pulmonary inflammatory pathways and the development of acute respiratory distress syndrome. Exp Lung Res 41: 155–162, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a. Bryan AC. Conference on the scientific basis of respiratory therapy. Pulmonary physiotherapy in the pediatric age group: comments of a devil's advocate. Am Rev Respir Dis 110: 143–144, 1974 [DOI] [PubMed] [Google Scholar]
- 31. Butt Y, Kurdowska A, and Allen TC. Acute lung injury: a clinical and molecular review. Arch Pathol Lab Med 140: 345–350, 2016 [DOI] [PubMed] [Google Scholar]
- 32. Calfee CS, Delucchi K, Parsons PE, Thompson BT, Ware LB, Matthay MA, and Network NA. Subphenotypes in acute respiratory distress syndrome: latent class analysis of data from two randomised controlled trials. Lancet Respir Med 2: 611–620, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Calfee CS, Gallagher D, Abbott J, Thompson BT, Matthay MA, and the NAN. Plasma angiopoietin-2 in clinical acute lung injury: prognostic and pathogenetic significance. Crit Care Med 40: 1731–1737, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Calfee CS, Ware LB, Eisner MD, Parsons PE, Thompson BT, Wickersham N, Matthay MA, and Network NA. Plasma receptor for advanced glycation end products and clinical outcomes in acute lung injury. Thorax 63: 1083–1089, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camp SM, Ceco E, Evenoski CL, Danilov SM, Zhou T, Chiang ET, Moreno-Vinasco L, Mapes B, Zhao J, Gursoy G, Brown ME, Adyshev DM, Siddiqui SS, Quijada H, Sammani S, Letsiou E, Saadat L, Yousef M, Wang T, Liang J, and Garcia JGN. Unique toll-like receptor 4 activation by NAMPT/PBEF induces NFκB signaling and inflammatory lung injury. Sci Rep 5: 13135, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carnesecchi S, Dunand-Sauthier I, Zanetti F, Singovski G, Deffert C, Donati Y, Cagarelli T, Pache J-C, Krause K-H, Reith W, and Barazzone-Argiroffo C. NOX1 is responsible for cell death through STAT3 activation in hyperoxia and is associated with the pathogenesis of acute respiratory distress syndrome. Int J Clin Exp Pathol 7: 537–551, 2014 [PMC free article] [PubMed] [Google Scholar]
- 37. Cartin-Ceba R, Hubmayr RD, Qin R, Peters S, Determann RM, Schultz MJ, and Gajic O. Predictive value of plasma biomarkers for mortality and organ failure development in patients with acute respiratory distress syndrome. J Crit Care 30: 219.e211–219.e217, 2015 [DOI] [PubMed] [Google Scholar]
- 38. Celedón JC, Burchard EG, Schraufnagel D, Castillo-Salgado C, Schenker M, Balmes J, Neptune E, Cummings KJ, Holguin F, Riekert KA, Wisnivesky JP, Garcia JGN, Roman J, Kittles R, Ortega VE, Redline S, Mathias R, Thomas A, Samet J, Ford JG, and American Thoracic Society and the National Heart LaBI. An American Thoracic Society/National Heart, Lung, and Blood Institute workshop report: addressing respiratory health equality in the United States. Ann Am Thorac Soc 14: 814–826, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang DW, Hayashi S, Gharib SA, Vaisar T, King ST, Tsuchiya M, Ruzinski JT, Park DR, Matute-Bello G, Wurfel MM, Bumgarner R, Heinecke JW, and Martin TR. Proteomic and computational analysis of bronchoalveolar proteins during the course of the acute respiratory distress syndrome. Am J Respir Crit Care Med 178: 701–709, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Checkley W, Martin GS, Brown SM, Chang SY, Dabbagh O, Fremont RD, Girard TD, Rice TW, Howell MD, Johnson SB, O'Brien J, Park PK, Pastores SM, Patil NT, Pietropaoli AP, Putman M, Rotello L, Siner J, Sajid S, Murphy DJ, Sevransky JE, and United States Critical Illness and Injury Trials Group Critical Illness Outcomes Study I. Structure, process, and annual ICU mortality across 69 centers: United States Critical Illness and Injury Trials Group Critical Illness Outcomes Study. Crit Care Med 42: 344–356, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen J, Zhang Z, and Cai L. Diabetic cardiomyopathy and its prevention by nrf2: current status. Diab Metab J 38: 337–345, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chiu L-C, Tsai F-C, Hu H-C, Chang C-H, Hung C-Y, Lee C-S, Li S-H, Lin S-W, Li L-F, Huang C-C, Chen N-H, Yang C-T, Chen Y-C, and Kao K-C. Survival predictors in acute respiratory distress syndrome with extracorporeal membrane oxygenation. Ann Thorac Surg 99: 243–250, 2015 [DOI] [PubMed] [Google Scholar]
- 43. Cho H-Y. and Kleeberger SR. Nrf2 protects against airway disorders. Toxicol Appl Pharmacol 244: 43–56, 2010 [DOI] [PubMed] [Google Scholar]
- 44. Chow C-W, Abreu MTH, Suzuki T, and Downey GP. Oxidative stress and acute lung injury. Am J Respir Cell Mol Biol 29: 427–431, 2003 [DOI] [PubMed] [Google Scholar]
- 45. Chow C-W, Herrera Abreu MT, Suzuki T, and Downey GP. Oxidative stress and acute lung injury. Am J Respir Cell Mol Biol 29: 427–431, 2003 [DOI] [PubMed] [Google Scholar]
- 46. Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, and Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med 36: 2794–2800, 2008 [DOI] [PubMed] [Google Scholar]
- 47. Christie JD, Wurfel MM, Feng R, O'Keefe GE, Bradfield J, Ware LB, Christiani DC, Calfee CS, Cohen MJ, Matthay M, Meyer NJ, Kim C, Li M, Akey J, Barnes KC, Sevransky J, Lanken PN, May AK, Aplenc R, Maloney JP, Hakonarson H, and for the Trauma ALISNPCi. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS One 7: e28268, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chung H-F, Long KZ, Hsu C-C, Mamun AA, Chiu Y-F, Tu H-P, Chen P-S, Jhang H-R, Hwang S-J, and Huang M-C. Adiponectin gene (ADIPOQ) polymorphisms correlate with the progression of nephropathy in Taiwanese male patients with type 2 diabetes. Diab Res Clin Pract 105: 261–270, 2014 [DOI] [PubMed] [Google Scholar]
- 49. Cochi SE, Kempker JA, Annangi S, Kramer MR, and Martin GS. Mortality trends of acute respiratory distress syndrome in the United States from 1999 to 2013. Ann Am Thorac Soc 13: 1742–1751, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cohen MJ, Brohi K, Calfee CS, Rahn P, Chesebro BB, Christiaans SC, Christiaans, Carles M, Howard M, Pittet J-F. Early release of high mobility group box nuclear protein 1 after severe trauma in humans: role of injury severity and tissue hypoperfusion. Crit Care 13: R174, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cooke CR, Erickson SE, Eisner MD, and Martin GS. Trends in the incidence of noncardiogenic acute respiratory failure: the role of race. Crit Care Med 40: 1532–1538, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. D'Alessio FR, Craig JM, Singer BD, Files DC, Mock JR, Garibaldi BT, Fallica J, Tripathi A, Mandke P, Gans JH, Limjunyawong N, Sidhaye VK, Heller NM, Mitzner W, King LS, and Aggarwal NR. Enhanced resolution of experimental ARDS through IL-4-mediated lung macrophage reprogramming. Am J Physiol Lung Cell Mol Physiol 310: L733–L746, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Devaney J, Contreras M, and Laffey JG. Clinical review: gene-based therapies for ALI/ARDS: where are we now? Crit Care 15: 224, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Di A, Mehta D, and Malik AB. ROS-activated calcium signaling mechanisms regulating endothelial barrier function. Cell Calcium 60: 163–171, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dinarello CA. Biologic basis for interleukin-1 in disease. Blood 87: 2095, 1996 [PubMed] [Google Scholar]
- 56. Doyle IR, Hermans C, Bernard A, Nicholas TE, and Bersten AD. Clearance of clara cell secretory protein 16 (CC16) and surfactant proteins A and B from blood in acute respiratory failure. Am J Respir Crit Care Med 158: 1528–1535, 1998 [DOI] [PubMed] [Google Scholar]
- 57. Dreyfuss D. and Saumon G. Ventilator-induced lung injury. Am J Respir Crit Care Med 157: 294–323, 1998 [DOI] [PubMed] [Google Scholar]
- 58. Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, and Lin SM. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11: 587, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dudek SM. and Garcia JGN. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol 91: 1487–1500, 2001 [DOI] [PubMed] [Google Scholar]
- 60. Dustin ML, Rothlein R, Bhan AK, Dinarello CA, and Springer TA. Induction by IL 1 and interferon-gamma: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1). 137: 245–254, 1986 [PubMed] [Google Scholar]
- 61. Dötsch A, Eisele L, Rabeling M, Rump K, Walstein K, Bick A, Cox L, Engler A, Bachmann HS, Jöckel K-H, Adamzik M, Peters J, and Schäfer ST. Hypoxia inducible factor-2 alpha and prolinhydroxylase 2 polymorphisms in patients with acute respiratory distress syndrome (ARDS). Int J Mol Sci 18: 1266, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Eklund L. and Saharinen P. Angiopoietin signaling in the vasculature. Exp Cell Res 319: 1271–1280, 2013 [DOI] [PubMed] [Google Scholar]
- 63. Elangovan VR, Camp SM, Kelly GT, Desai AA, Adyshev D, Sun X, Black SM, Wang T, and Garcia JGN. Endotoxin- and mechanical stress-induced epigenetic changes in the regulation of the nicotinamide phosphoribosyltransferase promoter. Pulm Circ 6: 539–544, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Erickson SE, Shlipak MG, Martin GS, Wheeler AP, Ancukiewicz M, Matthay MA, and Eisner MD. Racial and ethnic disparities in mortality from acute lung injury. Crit Care Med 37: 1–6, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Famous KR, Delucchi K, Ware LB, Kangelaris KN, Liu KD, Thompson BT, and Calfee CS. Acute respiratory distress syndrome subphenotypes respond differently to randomized fluid management strategy. Am J Respir Crit Care Med 195: 331–338, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Flores C, del Mar Pino-Yanes M, and Villar J. A quality assessment of genetic association studies supporting susceptibility and outcome in acute lung injury. Crit Care 12: R130, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Flores C, Ma S, Maresso K, Ober C, and Garcia J. A variant of the myosin light chain kinase gene is associated with severe asthma in African Americans. Genet Epidemiol 41: 4, 2007 [DOI] [PubMed] [Google Scholar]
- 68. Flores C, Ma S-F, Maresso K, Wade MS, Villar J, and Garcia JGN. IL6 gene-wide haplotype is associated with susceptibility to acute lung injury. Transl Res 152: 11–17, 2008 [DOI] [PubMed] [Google Scholar]
- 69. Flores C, Pérez-Méndez L, Maca-Meyer N, Muriel A, Espinosa E, Blanco J, Sangüesa R, Muros M, Garcia JGN, and Villar J. A common haplotype of the LBP gene predisposes to severe sepsis. Crit Care Med 37: 2759–2766, 2009 [DOI] [PubMed] [Google Scholar]
- 70. Frank AJ. and Thompson BT. Pharmacological treatments for acute respiratory distress syndrome. Curr Opin Crit Care 16: 62–68, 2010 [DOI] [PubMed] [Google Scholar]
- 71. Gao L, Flores C, Fan-Ma S, Miller EJ, Moitra J, Moreno L, Wadgaonkar R, Simon B, Brower R, Sevransky J, Tuder RM, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Ye SQ, Barnes KC, and Garcia JGN. Macrophage migration inhibitory factor in acute lung injury: expression, biomarker, and associations. Transl Res 150: 18–29, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gao L, Grant A, Halder I, Brower R, Sevransky J, Maloney JP, Moss M, Shanholtz C, Yates CR, Meduri GU, Shriver MD, Ingersoll R, Scott AF, Beaty TH, Moitra J, Ma SF, Ye SQ, Barnes KC, and Garcia JGN. Novel polymorphisms in the myosin light chain kinase gene confer risk for acute lung injury. Am J Respir Cell Mol Biol 34: 487–495, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, Chi P, Muñoz M, Watson H, Dunston G, Togias A, Hansel N, Sevransky J, Maloney JP, Moss M, Shanholtz C, Brower R, Garcia JGN, Grigoryev DN, Cheadle C, Beaty TH, Mathias RA, and Barnes KC. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol 119: 1111–1118, 2007 [DOI] [PubMed] [Google Scholar]
- 74. Gao L, Tsai YJ, Grigoryev DN, and Barnes KC. Host defense genes in asthma and sepsis and the role of the environment. Curr Opin Allergy Clin Immunol 7: 459–467, 2007 [DOI] [PubMed] [Google Scholar]
- 75. Garcia JGN. and Sznajder JI. Healthcare disparities in patients with acute respiratory distress syndrome. toward equity. Am J Respir Crit Care Med 188: 631–632, 2013 [DOI] [PubMed] [Google Scholar]
- 76. Garcia JGN, Verin AD, and Schaphorst KL. Regulation of thrombin-mediated endothelial cell contraction and permeability. Semin Thromb Hemost 22: 309–315, 1996 [DOI] [PubMed] [Google Scholar]
- 77. Ghosh CC, David S, Zhang R, Berghelli A, Milam K, Higgins SJ, Hunter J, Mukherjee A, Wei Y, Tran M, Suber F, Kobzik L, Kain KC, Lu S, Santel A, Yano K, Guha P, Dumont DJ, Christiani DC, and Parikh SM. Gene control of tyrosine kinase TIE2 and vascular manifestations of infections. Proc Natl Acad Sci U S A 113: 2472, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gianfrancesco F, Rendina D, Di Stefano M, Mingione A, Esposito T, Merlotti D, Gallone S, Magliocca S, Goode A, Formicola D, Morello G, Layfield R, Frattini A, De Filippo G, Nuti R, Searle M, Strazzullo P, Isaia G, Mossetti G, and Gennari L. A nonsynonymous TNFRSF11A variation increases NFκB activity and the severity of Paget's disease. J Bone Miner Res 27: 443–452, 2011 [DOI] [PubMed] [Google Scholar]
- 79. Goldman JL, Sammani S, Kempf C, Saadat L, Letsiou E, Wang T, Moreno-Vinasco L, Rizzo AN, Fortman JD, and Garcia JGN. Pleiotropic effects of interleukin-6 in a “two-hit” murine model of acute respiratory distress syndrome. Pulm Circ 4: 280–288, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gong MN, Wei Z, Xu L-L, Miller DP, Thompson BT, and Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest 125: 203–211, 2004 [DOI] [PubMed] [Google Scholar]
- 81. Griffith B, Pendyala S, Hecker L, Lee PJ, Natarajan V, and Thannickal VJ. NOX enzymes and pulmonary disease. Antioxid Redox Signal 11: 2505–2516, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grigoryev DN, Finigan JH, Hassoun P, and Garcia JGN. Science review: searching for gene candidates in acute lung injury. Crit Care 8: 440–447, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Grigoryev DN, Ma S-F, Irizarry RA, Ye SQ, Quackenbush J, and Garcia JGN. Orthologous gene-expression profiling in multi-species models: search for candidate genes. Genome Biol 5: R34, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Grommes J. and Soehnlein O. Contribution of neutrophils to acute lung injury. Mol Med 17: 293–307, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Headley AS, Meduri GU, and Tolley E. Infections and the inflammatory response in acute respiratory distress syndrome. Chest 111: 1306–1321, 1997 [DOI] [PubMed] [Google Scholar]
- 86. Hecker L, Vittal R, Jones T, Jagirdar R, Luckhardt TR, Horowitz JC, Pennathur S, Martinez FJ, and Thannickal VJ. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med 15: 1077–1081, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hernandez-Pacheco N, Guillen-Guio B, Acosta-Herrera M, Pino-Yanes M, Corrales A, Ambros A, Nogales L, Muriel A, Gonzalez-Higueras E, Diaz-Dominguez FJ, Zavala E, Belda J, Ma SF, Villar J, and Flores C. A vascular endothelial growth factor receptor gene variant is associated with susceptibility to acute respiratory distress syndrome. Intens Care Med Exp 6: 16, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Herwig R, Hardt C, Lienhard M, and Kamburov A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc 11: 1889, 2016 [DOI] [PubMed] [Google Scholar]
- 89. Hinz J, Büttner B, Kriesel F, Steinau M, Frederik Popov A, Ghadimi M, Beissbarth T, Tzvetkov M, Bergmann I, and Mansur A. The FER rs4957796 TT genotype is associated with unfavorable 90-day survival in Caucasian patients with severe ARDS due to pneumonia. Sci Rep 7: 9887, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Hong S-B, Huang Y, Moreno-Vinasco L, Sammani S, Moitra J, Barnard JW, Ma S-F, Mirzapoiazova T, Evenoski C, Reeves RR, Chiang ET, Lang GD, Husain AN, Dudek SM, Jacobson JR, Ye SQ, Lussier YA, and Garcia JGN. Essential role of pre-B-cell colony enhancing factor in ventilator-induced lung injury. Am J Respir Crit Care Med 178: 605–617, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hästbacka J, Linko R, Tervahartiala T, Varpula T, Hovilehto S, Parviainen I, Vaara ST, Sorsa T, and Pettilä V. Serum MMP-8 and TIMP-1 in critically ill patients with acute respiratory failure: TIMP-1 is associated with increased 90-day mortality. Anesth Analg 118: 790–798, 2014 [DOI] [PubMed] [Google Scholar]
- 92. Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YHC, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JSM, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, and Penninger JM. Identification of oxidative stress and toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell 133: 235–249, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Jabaudon M, Berthelin P, Pranal T, Roszyk L, Godet T, Faure J-S, Chabanne R, Eisenmann N, Lautrette A, Belville C, Blondonnet R, Cayot S, Gillart T, Pascal J, Skrzypczak Y, Souweine B, Blanchon L, Sapin V, Pereira B, and Constantin J-M. Receptor for advanced glycation end-products and ARDS prediction: a multicentre observational study. Sci Rep 8: 2603, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jabaudon M, Blondonnet R, Roszyk L, Bouvier D, Audard J, Clairefond G, Fournier M, Marceau G, Déchelotte P, Pereira B, Sapin V, and Constantin J-M. Soluble receptor for advanced glycation end-products predicts impaired alveolar fluid clearance in acute respiratory distress syndrome. Am J Respir Crit Care Med 192: 191–199, 2015 [DOI] [PubMed] [Google Scholar]
- 95. Jabaudon M, Blondonnet R, Roszyk L, Pereira B, Guérin R, Perbet S, Cayot S, Bouvier D, Blanchon L, Sapin V, and Constantin J-M. soluble forms and ligands of the receptor for advanced glycation end-products in patients with acute respiratory distress syndrome: an observational prospective study. PLoS One 10: e0135857, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jabaudon M, Futier E, Roszyk L, Chalus E, Guerin R, Petit A, Mrozek S, Perbet S, Cayot-Constantin S, Chartier C, Sapin V, Bazin J-E, and Constantin J-M. Soluble form of the receptor for advanced glycation end products is a marker of acute lung injury but not of severe sepsis in critically ill patients. Crit Care Med 39: 480–488, 2011 [DOI] [PubMed] [Google Scholar]
- 97. Jabaudon M, Futier E, Roszyk L, Sapin V, Pereira B, and Constantin J-M. Association between intraoperative ventilator settings and plasma levels of soluble receptor for advanced glycation end-products in patients without pre-existing lung injury. Respirology 20: 1131–1138, 2015 [DOI] [PubMed] [Google Scholar]
- 98. Janz DR. and Ware LB. Biomarkers of ALI/ARDS: pathogenesis, discovery, and relevance to clinical trials. Semin Respir Crit Care Med 34: 537–548, 2013 [DOI] [PubMed] [Google Scholar]
- 99. Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, and Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest 113: 1318–1327, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jin L, Zuo X-Y, Su W-Y, Zhao X-L, Yuan M-Q, Han L-Z, Zhao X, Chen Y-D, and Rao S-Q. Pathway-based analysis tools for complex diseases: a review. Genomics Proteomics Bioinformatics 12: 210–220, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kallet RH, Zhuo H, Ho K, Lipnick MS, Gomez A, and Matthay MA. Lung injury etiology and other factors influencing the relationship between dead-space fraction and mortality in ARDS. Respir Care 62: 1241, 2017 [DOI] [PubMed] [Google Scholar]
- 102. Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, and Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res 39: D712–D717, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kamburov A, Stelzl U, Lehrach H, and Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res 41: D793–D800, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kangelaris KN, Calfee CS, May AK, Zhuo H, Matthay MA, and Ware LB. Is there still a role for the lung injury score in the era of the Berlin definition ARDS? Ann Intens Care 4: 4, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Kangelaris KN, Prakash A, Liu KD, Aouizerat B, Woodruff PG, Erle DJ, Rogers A, Seeley EJ, Chu J, Liu T, Osterberg-Deiss T, Zhuo H, Matthay MA, and Calfee CS. Increased expression of neutrophil-related genes in patients with early sepsis-induced ARDS. Am J Physiol Lung Cell Mol Physiol 308: L1102–L1113, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kovach MA, Stringer KA, Bunting R, Wu X, San Mateo L, Newstead MW, Paine R, and Standiford TJ. Microarray analysis identifies IL-1 receptor type 2 as a novel candidate biomarker in patients with acute respiratory distress syndrome. Respir Res 16: 29, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Laffey JG. and Kavanagh BP. Fifty years of research in ARDS. Insight into acute respiratory distress syndrome. from models to patients. Am J Respir Crit Care Med 196: 18–28, 2017 [DOI] [PubMed] [Google Scholar]
- 108. Lee K, Huh JW, Lim C-M, Koh Y, and Hong S-B. Clinical role of serum pre-B cell colony-enhancing factor in ventilated patients with sepsis and acute respiratory distress syndrome. Scand J Infect Dis 45: 760–765, 2013 [DOI] [PubMed] [Google Scholar]
- 109. Lee Y-L, Chen W, Chen L-Y, Chen C-H, Lin Y-C, Liang S-J, and Shih C-M. Systemic and bronchoalveolar cytokines as predictors of in-hospital mortality in severe community-acquired pneumonia. J Cri Care 25: 176.e7–176.e13, 2010 [DOI] [PubMed] [Google Scholar]
- 110. Li G, Malinchoc M, Cartin-Ceba R, Venkata CV, Kor DJ, Peters SG, Hubmayr RD, and Gajic O. Eight-year trend of acute respiratory distress syndrome. Am J Respir Crit Care Med 183: 59–66, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison U, and Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet 58: 181–191, 2001 [DOI] [PubMed] [Google Scholar]
- 112. Liu H, Zhang D, Zhao B, and Zhao J. Superoxide anion, the main species of ROS in the development of ARDS induced by oleic acid. Free Radic Res 38: 1281–1287, 2004 [DOI] [PubMed] [Google Scholar]
- 113. Liu P, Rojo de la Vega M, Sammani S, Mascarenhas J, Kerins M, Dodson M, Sun X, Wang T, Ooi A, Garcia J, and Zhang D. MYLK transcriptional repression by NRF2-ARE and RPA1 enhances vascular integrity. Proc Natl Acad Sci U S A 115:E10352–E10361, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Lu J-F, Zhou Y, Huang G-H, Jiang H-X, Hu B-L, and Qin S-Y. Association of ADIPOQ polymorphisms with obesity risk: a meta-analysis. Hum Immunol 75: 1062–1068, 2014 [DOI] [PubMed] [Google Scholar]
- 115. Ma S-F, Xie L, Pino-Yanes M, Sammani S, Wade MS, Letsiou E, Siegler J, Wang T, Infusino G, Kittles RA, Flores C, Zhou T, Prabhakar BS, Moreno-Vinasco L, Villar J, Jacobson JR, Dudek SM, and Garcia JGN. Type 2 deiodinase and host responses of sepsis and acute lung injury. Am J Respir Cell Mol Biol 45: 1203–1211, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Marshall RP, Webb S, Bellingan GJ, Montgomery HE, Chaudhari B, McAnulty RJ, Humphries SE, Hill MR, and Laurent GJ. Angiotensin converting enzyme insertion/deletion polymorphism is associated with susceptibility and outcome in acute respiratory distress syndrome. Am J Respir Crit Care Med 166: 646–650, 2002 [DOI] [PubMed] [Google Scholar]
- 117. Marshall RP, Webb S, Hill MR, Humphries SE, and Laurent GJ. Genetic polymorphisms associated with susceptibility and outcome in ARDS. Chest 121: 68S–69S, 2002 [DOI] [PubMed] [Google Scholar]
- 118. Martín-Loeches I, Solé-Violán J, Rodríguez de Castro F, García-Laorden MI, Borderías L, Blanquer J, Rajas O, Briones ML, Aspa J, Herrera-Ramos E, Marcos-Ramos JA, Sologuren I, González-Quevedo N, Ferrer-Agüero JM, Noda J, and Rodríguez-Gallego C. Variants at the promoter of the interleukin-6 gene are associated with severity and outcome of pneumococcal community-acquired pneumonia. Intens Care Med 38: 256–262, 2012 [DOI] [PubMed] [Google Scholar]
- 119. Mascarenhas JB, Tchourbanov AY, Fan H, Danilov SM, Wang T, and Garcia JGN. Mechanical stress and single nucleotide variants regulate alternative splicing of the MYLK gene. Am J Respir Cell Mol Biol 56: 29–37, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Meduri GU, Headley S, Kohler G, Stentz F, Tolley E, Umberger R, and Leeper K. Persistent elevation of inflammatory cytokines predicts a poor outcome in ARDS: plasma IL-1β and IL-6 levels are consistent and efficient predictors of outcome over time. Chest 107: 1062–1073, 1995 [DOI] [PubMed] [Google Scholar]
- 121. Mehta NN, Heffron SP, Patel PN, Ferguson J, Shah RD, Hinkle CC, Krishnamoorthy P, Shah R, Tabita-Martinez J, Terembula K, Master SR, Rickels MR, and Reilly MP. A human model of inflammatory cardio-metabolic dysfunction; a double blind placebo-controlled crossover trial. J Transl Med 10: 124, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Meyer NJ. Beyond SNPs—genetics, genomics and other ‘omic approaches to ARDS. Clin Chest Med 35: 673–684, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Meyer NJ, Feng R, Li M, Zhao Y, Sheu C-C, Tejera P, Gallop R, Bellamy S, Rushefski M, Lanken PN, Aplenc R, O'Keefe GE, Wurfel MM, Christiani DC, and Christie JD. IL1RN coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL-1 receptor antagonist. Am J Respir Crit Care Med 187: 950–959, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Meyer NJ, Huang Y, Singleton PA, Sammani S, Moitra J, Evenoski CL, Husain AN, Mitra S, Moreno-Vinasco L, Jacobson JR, Lussier YA, and Garcia JGN. GADD45a is a novel candidate gene in inflammatory lung injury via influences on Akt signaling. FASEB J 23: 1325–1337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Mirzapoiazova T, Moitra J, Moreno-Vinasco L, Sammani S, Turner JR, Chiang ET, Evenoski C, Wang T, Singleton PA, Huang Y, Lussier YA, Watterson DM, Dudek SM, and Garcia JGN. Muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol 44: 40–52, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Mitra S, Wade MS, Sun X, Moldobaeva N, Flores C, Ma S-F, Zhang W, Garcia JGN, and Jacobson JR. GADD45a promoter regulation by a functional genetic variant associated with acute lung injury. PLoS One 9: e100169, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Moreno-Vinasco L, Quijada H, Sammani S, Siegler J, Letsiou E, Deaton R, Saadat L, Zaidi RS, Messana J, Gann PH, Machado RF, Ma W, Camp SM, Wang T, and Garcia JGN. nicotinamide phosphoribosyltransferase inhibitor is a novel therapeutic candidate in murine models of inflammatory lung injury. Am J Respir Cell Mol Biol 51: 223–228, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Morrell ED, O'Mahony DS, Glavan BJ, Harju-Baker S, Nguyen C, Gunderson S, Abrahamson A, Radella F, Rona G, Black RA, and Wurfel MM. Genetic variation in MAP3K1 associates with ventilator-free days in acute respiratory distress syndrome. Am J Respir Cell Mol Biol 58: 117–125, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Moss M. and Mannino DM. Race and gender differences in acute respiratory distress syndrome deaths in the United States: an analysis of multiple-cause mortality data (1979–1996). Crit Care Med 30: 1679–1685, 2002 [DOI] [PubMed] [Google Scholar]
- 130. Murray JF, Matthay MA, Luce JM, and Flick MR. An expanded definition of the adult respiratory distress syndrome. Am Rev Respir Dis 138: 720–723, 1988 [DOI] [PubMed] [Google Scholar]
- 131. Nakamura T, Sato E, Fujiwara N, Kawagoe Y, Maeda S, and Yamagishi S-I. Increased levels of soluble receptor for advanced glycation end products (sRAGE) and high mobility group box 1 (HMGB1) are associated with death in patients with acute respiratory distress syndrome. Clin Biochem 44: 601–604, 2011 [DOI] [PubMed] [Google Scholar]
- 132. Natarajan V, Dudek SM, Jacobson JR, Moreno-Vinasco L, Huang LS, Abassi T, Mathew B, Zhao Y, Wang L, Bittman R, Weichselbaum R, Berdyshev E, and Garcia JGN. Sphingosine-1-phosphate, FTY720, and sphingosine-1-phosphate receptors in the pathobiology of acute lung injury. Am J Respir Cell Mol Biol 49: 6–17, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, and Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem 267: 14998–15004, 1992 [PubMed] [Google Scholar]
- 134. Nick JA, Caceres SM, Kret JE, Poch KR, Strand M, Faino AV, Nichols DP, Saavedra MT, Taylor-Cousar JL, Geraci MW, Burnham EL, Fessler MB, Suratt BT, Abraham E, Moss M, and Malcolm KC. Extremes of interferon-stimulated gene expression associate with worse outcomes in the acute respiratory distress syndrome. PLoS One 11: e0162490, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. O'Mahony DS, Glavan BJ, Holden TD, Fong C, Black RA, Rona G, Tejera P, Christiani DC, and Wurfel MM. Inflammation and immune-related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLoS One 7: e51104, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135a. O'Shea JJ. and Plenge R. JAKs and STATs in immunoregulation and immune-mediated disease. Immunity 36: 542–550, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. This reference has been deleted.
- 137. Ozolina A, Sarkele M, Sabelnikovs O, Skesters A, Jaunalksne I, Serova J, Ievins T, Bjertnaes LJ, and Vanags I. Activation of coagulation and fibrinolysis in acute respiratory distress syndrome: a prospective pilot study. Front Med 3: 64, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. This reference has been deleted.
- 139. Park J-B, Nagar H, Choi S, Jung S-B, Kim H-W, Kang SK, Lee JW, Lee JH, Park J-W, Irani K, Jeon BH, Song H-J, and Kim C-S. IDH2 deficiency impairs mitochondrial function in endothelial cells and endothelium-dependent vasomotor function. Free Radic Biol Med 94: 36–46, 2016 [DOI] [PubMed] [Google Scholar]
- 140. Parsons PE, Moss M, Vannice JL, Moore EE, Moore FA, and Repine JE. Circulating IL-1ra and IL-10 levels are increased but do not predict the development of acute respiratory distress syndrome in at-risk patients. Am J Respir Crit Care Med 155: 1469–1473, 1997 [DOI] [PubMed] [Google Scholar]
- 141. Pendyala S, Moitra J, Kalari S, Kleeberger SR, Zhao Y, Reddy SP, Garcia JGN, and Natarajan V. NRF2 Regulates hyperoxia-induced NOX4 expression in human lung endothelium: identification of functional antioxidant response elements on NOX4 promoter. Free Radic Biol Med 50: 1749–1759, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Pino-Yanes M, Ma S-F, Sun X, Tejera P, Corrales A, Blanco J, Pérez-Méndez L, Espinosa E, Muriel A, Blanch L, Garcia JGN, Villar J, and Flores C. Interleukin-1 receptor-associated kinase 3 gene associates with susceptibility to acute lung injury. Am J Respir Cell Mol Biol 45: 740–745, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Pociot F, MØLvig J, Wogensen L, Worsaae H, and Nerup J. A Taql polymorphism in the human interleukin-1β (IL-1β) gene correlates with IL-1β secretion in vitro. Eur J Clin Invest 22: 396–402, 1992 [DOI] [PubMed] [Google Scholar]
- 144. Prabhakaran P, Ware LB, White KE, Cross MT, Matthay MA, and Olman MA. Elevated levels of plasminogen activator inhibitor-1 in pulmonary edema fluid are associated with mortality in acute lung injury. Am J Physiol Lung Cell Mol Physiol 285: L20–L28, 2003 [DOI] [PubMed] [Google Scholar]
- 145. Proudfoot A, Bayliffe A, O'Kane CM, Wright T, Serone A, Bareille PJ, Brown V, Hamid UI, Chen Y, Wilson R, Cordy J, Morley P, de Wildt R, Elborn S, Hind M, Chilvers ER, Griffiths M, Summers C, and McAuley DF. Novel anti-tumour necrosis factor receptor-1 (TNFR1) domain antibody prevents pulmonary inflammation in experimental acute lung injury. Thorax 73: 723, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Qayyum R, Snively BM, Ziv E, Nalls MA, Liu Y, Tang W, Yanek LR, Lange L, Evans MK, Ganesh S, Austin MA, Lettre G, Becker DM, Zonderman AB, Singleton AB, Harris TB, Mohler ER, Logsdon BA, Kooperberg C, Folsom AR, Wilson JG, Becker LC, and Reiner AP. A meta-analysis and genome-wide association study of platelet count and mean platelet volume in African Americans. PLOS Genet 8: e1002491, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Qi D, Wang D, Zhang C, Tang X, He J, Zhao Y, Deng W, and Deng X. Vaspin protects against LPS-induced ARDS by inhibiting inflammation, apoptosis and reactive oxygen species generation in pulmonary endothelial cells via the Akt/GSK-3β pathway. Int J Mol Med 40: 1803–1817, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Rautanen A, Mills TC, Gordon AC, Hutton P, Steffens M, Nuamah R, Chiche J-D, Parks T, Chapman SJ, Davenport EE, Elliott KS, Bion J, Lichtner P, Meitinger T, Wienker TF, Caulfield MJ, Mein C, Bloos F, Bobek I, Cotogni P, Sramek V, Sarapuu S, Kobilay M, Ranieri VM, Rello J, Sirgo G, Weiss YG, Russwurm S, Schneider EM, Reinhart K, Holloway PAH, Knight JC, Garrard CS, Russell JA, Walley KR, Stüber F, Hill AVS, Hinds CJ, and for the EEGI. Genome-wide association study of survival from sepsis due to pneumonia: an observational cohort study. Lancet Respir Med 3: 53–60, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Reilly JP, Christie JD, and Meyer NJ. Fifty years of research in ARDS. Genomic contributions and opportunities. Am J Respir Crit Care Med 196: 1113–1121, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Rojo de la Vega M, Dodson M, Gross C, Manzour H, Lantz RC, Chapman E, Wang T, Black SM, Garcia JGN, and Zhang DD. Role of Nrf2 and autophagy in acute lung injury. Curr Pharmacol Rep 2: 91–101, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, Stern EJ, and Hudson LD. Incidence and outcomes of acute lung injury. N Engl J Med 353: 1685–1693, 2005 [DOI] [PubMed] [Google Scholar]
- 152. Ruhl AP, Huang M, Colantuoni E, Karmarkar T, Dinglas VD, Hopkins RO, Needham DM, and with the National Institutes of Health, National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome Network. Healthcare utilization and costs in ARDS survivors: a 1-year longitudinal national US multicenter study. Intensive Care Med 43: 980–991, 2017 [DOI] [PubMed] [Google Scholar]
- 153. Ryu H, Lee J, Zaman K, Kubilis J, Ferrante RJ, Ross BD, Neve R, and Ratan RR. Sp1 and Sp3 are oxidative stress-inducible, antideath transcription factors in cortical neurons. J Neurosci 23: 3597, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Salnikova LE, Smelaya TV, Vesnina IN, Golubev AM, and Moroz VV. Genetic susceptibility to nosocomial pneumonia, acute respiratory distress syndrome and poor outcome in patients at risk of critical illness. Inflammation 37: 295–305, 2014 [DOI] [PubMed] [Google Scholar]
- 155. Sammani S, Moreno-Vinasco L, Mirzapoiazova T, Singleton PA, Chiang ET, Evenoski CL, Wang T, Mathew B, Husain A, Moitra J, Sun X, Nunez L, Jacobson JR, Dudek SM, Natarajan V, and Garcia JGN. Differential effects of sphingosine 1-phosphate receptors on airway and vascular barrier function in the murine lung. Am J Respir Cell Mol Biol 43: 394–402, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Shaver CM. and Bastarache JA. Clinical and biological heterogeneity in ARDS: direct versus indirect lung injury. Clin Chest Med 35: 639–653, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Shen K, Ramirez B, Mapes B, Shen GR, Gokhale V, Brown ME, Santarsiero B, Ishii Y, Dudek SM, Wang T, and Garcia JGN. Structure–function analysis of the non-muscle myosin light chain kinase (nmMLCK) isoform by NMR spectroscopy and molecular modeling: influence of MYLK variants. PLoS One 10: e0130515, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Sheu CC, Zhai R, Su L, Tejera P, Gong MN, Thompson BT, Chen F, and Christiani DC. Sex-specific association of epidermal growth factor gene polymorphisms with ARDS. Eur Respir J 33: 543–550, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Shibuya M. Vascular endothelial growth factor and its receptor system: physiological functions in angiogenesis and pathological roles in various diseases. J Biochem 153: 13–19, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Shimizu Y, Camp SM, Sun X, Zhou T, Wang T, and Garcia JGN. Sp1-mediated nonmuscle myosin light chain kinase expression and enhanced activity in vascular endothelial growth factor-induced vascular permeability. Pulm Circ 5: 707–715, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Shirasawa M, Fujiwara N, Hirabayashi S, Ohno H, Iida J, Makita K, and Hata Y. Receptor for advanced glycation end-products is a marker of type I lung alveolar cells. Genes Cells 9: 165–174, 2004 [DOI] [PubMed] [Google Scholar]
- 162. Shortt K, Chaudhary S, Grigoryev D, Heruth DP, Venkitachalam L, Zhang LQ, and Ye SQ. Identification of novel single nucleotide polymorphisms associated with acute respiratory distress syndrome by exome-seq. PLoS One 9: e111953, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Simon BA, Easley RB, Grigoryev DN, Ma S-F, Ye SQ, Lavoie T, Tuder RM, and Garcia JGN. Microarray analysis of regional cellular responses to local mechanical stress in acute lung injury. Am J Physiol Lung Cell Mol Physiol 291: L851–L861, 2006 [DOI] [PubMed] [Google Scholar]
- 164. This reference has been deleted.
- 165. Sirinian C, Papanastasiou AD, Zarkadis IK, and Kalofonos HP. Alternative splicing generates a truncated isoform of human TNFRSF11A (RANK) with an altered capacity to activate NF-κB. Gene 525: 124–129, 2013 [DOI] [PubMed] [Google Scholar]
- 166. Soubrier F, Chung WK, Machado R, Grünig E, Aldred M, Geraci M, Loyd JE, Elliott CG, Trembath RC, Newman JH, and Humbert M. Genetics and genomics of pulmonary arterial hypertension. J Am Coll Cardiol 62: D13–D21, 2013 [DOI] [PubMed] [Google Scholar]
- 167. Steinberg K, Hudson L, Goodman R, Hough C, Lanken P, Hyzy R, Thompson B, Ancukiewicz M, and National Heart, Lung, and Blood Institute Acute Respiratory Distress Syndrome (ARDS) Clinical Trials Network. Efficacy and safety of corticosteroids for persistent acute respiratory distress syndrome. N Engl J Med 354: 1671–1684, 2006 [DOI] [PubMed] [Google Scholar]
- 168. Sun X, Elangovan VR, Mapes B, Camp SM, Sammani S, Saadat L, Ceco E, Ma S-F, Flores C, MacDougall MS, Quijada H, Liu B, Kempf CL, Wang T, Chiang ET, and Garcia JGN. The NAMPT promoter is regulated by mechanical stress, signal transducer and activator of transcription 5, and acute respiratory distress syndrome-associated genetic variants. Am J Respir Cell Mol Biol 51: 660–667, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Sun X, Ma S-F, Wade MS, Acosta-Herrera M, Villar J, Pino-Yanes M, Zhou T, Liu B, Belvitch P, Moitra J, Han Y-J, Machado R, Noth I, Natarajan V, Dudek SM, Jacobson JR, Flores C, and Garcia JGN. Functional promoter variants in sphingosine 1-phosphate receptor 3 associate with susceptibility to sepsis-associated acute respiratory distress syndrome. Am J Physiol Lung Cell Mol Physiol 305: L467–L477, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sun X, Singleton PA, Letsiou E, Zhao J, Belvitch P, Sammani S, Chiang ET, Moreno-Vinasco L, Wade MS, Zhou T, Liu B, Parastatidis I, Thomson L, Ischiropoulos H, Natarajan V, Jacobson JR, Machado RF, Dudek SM, and Garcia JGN. Sphingosine-1-phosphate receptor-3 is a novel biomarker in acute lung injury. Am J Respir Cell Mol Biol 47: 628–636, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Suter PM, Fairley HB, and Isenberg MD. Optimum end-expiratory airway pressure in patients with acute pulmonary failure. N Engl J Med 292: 284–289, 1975 [DOI] [PubMed] [Google Scholar]
- 172. Tao S, Rojo de la Vega M, Quijada H, Wondrak GT, Wang T, Garcia JGN, and Zhang DD. Bixin protects mice against ventilation-induced lung injury in an NRF2-dependent manner. Sci Rep 6: 18760, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Tejera P, Meyer N, Chen F, Feng R, Zhao Y, O'Mahony DS, Li L, Sheu C-C, Zhai R, Wang Z, Su L, Bajwa E, Ahasic AM, Clardy P, Gong MN, Frank AJ, Lanken PN, Thompson BT, Christie JD, Wurfel M, O'Keefe G, and Christiani DC. Distinct and replicable genetic risk factors for acute respiratory distress syndrome of pulmonary or extrapulmonary origin. J Med Genet 49: 671–680, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Tejera P, Wang Z, Zhai R, Su L, Sheu C-C, Taylor DM, Chen F, Gong MN, Thompson BT, and Christiani DC. Genetic polymorphisms of peptidase inhibitor 3 (Elafin) are associated with acute respiratory distress syndrome. Am J Respir Cell Mol Biol 41: 696–704, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Terpstra ML, Aman J, van Nieuw Amerongen GP, and Groeneveld ABJ. Plasma biomarkers for acute respiratory distress syndrome: a systematic review and meta-analysis. Crit Care Med 42: 691–700, 2014 [DOI] [PubMed] [Google Scholar]
- 176. The ADTF. Acute respiratory distress syndrome: the berlin definition. JAMA 307: 2526–2533, 2012 [DOI] [PubMed] [Google Scholar]
- 177. Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, Cambien F, and Soubrier F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet 51: 197–205, 1992 [PMC free article] [PubMed] [Google Scholar]
- 178. Tsai M-K, Wang H-MD, Shiang J-C, Chen IH, Wang C-C, Shiao Y-F, Liu W-S, Lin T-J, Chen T-M, and Chen Y-H. Sequence variants of ADIPOQ and association with type 2 diabetes mellitus in Taiwan Chinese Han population. Sci World J 2014: 650393, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Uchida T, Shirasawa M, Ware LB, Kojima K, Hata Y, Makita K, Mednick G, Matthay ZA, and Matthay MA. Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am J Respir Crit Care Med 173: 1008–1015, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. van den Borst B, Souren NY, Gielen M, Loos RJF, Paulussen ADC, Derom C, Schols AMWJ, and Zeegers MPA. Association between the IL6-174G/C SNP and maximally attained lung function. Thorax 66: 179, 2011 [DOI] [PubMed] [Google Scholar]
- 181. Varley I, Hughes DC, Greeves JP, Stellingwerff T, Ranson C, Fraser WD, and Sale C. RANK/RANKL/OPG pathway: genetic associations with stress fracture period prevalence in elite athletes. Bone 71: 131–136, 2015 [DOI] [PubMed] [Google Scholar]
- 182. Villarino AV, Kanno Y, and O'Shea JJ. Mechanisms of Jak/STAT signaling in immunity and disease. J Immunol 194: 21–27, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Wacharasint P, Nakada TA, Boyd John H, Russell James A, and Walley Keith R. AA genotype of IL-8–251A/T is associated with low PaO2/FiO2 in critically ill patients and with increased IL-8 expression. Respirology 17: 1253–1260, 2012 [DOI] [PubMed] [Google Scholar]
- 184. Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, Manogue KR, Faist E, Abraham E, Andersson J, Andersson U, Molina PE, Abumrad NN, Sama A, and Tracey KJ. HMG-1 as a late mediator of endotoxin lethality in mice. Science 285: 248, 1999 [DOI] [PubMed] [Google Scholar]
- 185. Wang H-W, Yang W, Lu J-Y, Li F, Sun J-Z, Zhang W, Guo N-N, Gao L, and Kang J-R. N-acetylcysteine administration is associated with reduced activation of NF-kB and preserves lung dendritic cells function in a zymosan-induced generalized inflammation model. J Clin Immunol 33: 649–660, 2013 [DOI] [PubMed] [Google Scholar]
- 186. Wang M, Yan J, He X, Zhong Q, Zhan C, and Li S. Candidate genes and pathogenesis investigation for sepsis-related acute respiratory distress syndrome based on gene expression profile. Biol Res 49: 25, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Wang T, Brown ME, Kelly GT, Camp SM, Mascarenhas JB, Sun X, Dudek SM, and Garcia JGN. Myosin light chain kinase (MYLK) coding polymorphisms modulate human lung endothelial cell barrier responses via altered tyrosine phosphorylation, spatial localization, and lamellipodial protrusions. Pulm Circ 8: 2045894018764171, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wang Z, Beach D, Su L, Zhai R, and Christiani DC. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am J Respir Cell Mol Biol 38: 724–732, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Ware LB. Prognostic determinants of acute respiratory distress syndrome in adults: impact on clinical trial design. Crit Care Med 33: S217–S222, 2005 [DOI] [PubMed] [Google Scholar]
- 190. Ware LB, Koyama T, Billheimer DD, Wu W, Bernard GR, Thompson BT, Brower RG, Standiford TJ, Martin TR, Matthay MA, and Network NACT. Prognostic and pathogenetic value of combining clinical and biochemical indices in patients with acute lung injury. Chest 137: 288–296, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Ware LB, Koyama T, Zhao Z, Janz DR, Wickersham N, Bernard GR, May AK, Calfee CS, and Matthay MA. Biomarkers of lung epithelial injury and inflammation distinguish severe sepsis patients with acute respiratory distress syndrome. Crit Care 17: R253, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Wei Y, Tejera P, Wang Z, Zhang R, Chen F, Su L, Lin X, Bajwa EK, Thompson BT, and Christiani DC. A missense genetic variant in LRRC16A/CARMIL1 improves acute respiratory distress syndrome survival by attenuating platelet count decline. Am J Respir Crit Care Med 195: 1353–1361, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Wei Y, Wang Z, Su L, Chen F, Tejera P, Bajwa EK, Wurfel MM, Lin X, and Christiani DC. Platelet count mediates the contribution of a genetic variant in LRRC 16A to ARDS risk. Chest 147: 607–617, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Wolfson RK, Mapes B, and Garcia JGN. Excessive mechanical stress increases HMGB1 expression in human lung microvascular endothelial cells via STAT3. Microvasc Res 92: 50–55, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Wu J, Liu Z, Meng K, and Zhang L. Association of adiponectin gene (ADIPOQ) rs2241766 polymorphism with obesity in adults: a meta-analysis. PLoS One 9: e95270, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Wurfel MM. Microarray-based analysis of ventilator-induced lung injury. Proc Am Thorac Soc 4: 77–84, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Wurfel MM, Gordon AC, Holden TD, Radella F, Strout J, Kajikawa O, Ruzinski JT, Rona G, Black RA, Stratton S, Jarvik GP, Hajjar AM, Nickerson DA, Rieder M, Sevransky J, Maloney JP, Moss M, Martin G, Shanholtz C, Garcia JGN, Gao L, Brower R, Barnes KC, Walley KR, Russell JA, and Martin TR. Toll-like receptor 1 polymorphisms affect innate immune responses and outcomes in sepsis. Am J Respir Crit Care Med 178: 710–720, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Wösten-van Asperen RM, Lutter R, Specht PA, Moll GN, van Woensel JB, van der Loos CM, van Goor H, Kamilic J, Florquin S, and Bos AP. Acute respiratory distress syndrome leads to reduced ratio of ACE/ACE2 activities and is prevented by angiotensin-(1–7) or an angiotensin II receptor antagonist. J Pathol 225: 618–627, 2011 [DOI] [PubMed] [Google Scholar]
- 199. Ye SQ, Simon BA, Maloney JP, Zambelli-Weiner A, Gao L, Grant A, Easley RB, McVerry BJ, Tuder RM, Standiford T, Brower RG, Barnes KC, and Garcia JGN. Pre-B-cell colony-enhancing factor as a potential novel biomarker in acute lung injury. Am J Respir Crit Care Med 171: 361–370, 2005 [DOI] [PubMed] [Google Scholar]
- 200. Ye SQ, Zhang LQ, Adyshev D, Usatyuk PV, Garcia AN, Lavoie TL, Verin AD, Natarajan V, and Garcia JGN. Pre-B-cell-colony-enhancing factor is critically involved in thrombin-induced lung endothelial cell barrier dysregulation. Microvasc Res 70: 142–151, 2005 [DOI] [PubMed] [Google Scholar]
- 201. Yilin Z, Yandong N, and Faguang J. Role of angiotensin-converting enzyme (ACE) and ACE2 in a rat model of smoke inhalation induced acute respiratory distress syndrome. Burns 41: 1468–1477, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Zhang LQ, Adyshev DM, Singleton P, Li H, Cepeda J, Huang S-Y, Zou X, Verin AD, Tu J, Garcia JGN, and Ye SQ. Interactions between PBEF and oxidative stress proteins-a potential new mechanism underlying PBEF in the pathogenesis of acute lung injury. FEBS Lett 582: 1802–1808, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Zhao J, Yu H, Liu Y, Gibson SA, Yan Z, Xu X, Gaggar A, Li P-K, Li C, Wei S, Benveniste EN, and Qin H. Protective effect of suppressing STAT3 activity in LPS-induced acute lung injury. Am J Physiol Lung Cell Mol Physiol 311: L868–L880, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Zhou T, Wang T, and Garcia JGN. A nonmuscle myosin light chain kinase-dependent gene signature in peripheral blood mononuclear cells is linked to human asthma severity and exacerbation status. Pulm Circ 5: 335–338, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Zhou T, Wang T, and Garcia JGN. Genes influenced by the non-muscle isoform of myosin light chain kinase impact human cancer prognosis. PLoS One 9: e94325, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Zimlichman E, Henderson D, Tamir O, Franz C, Song P, Yamin CK, Keohane C, Denham CR, and Bates DW. Health care-associated infections: a meta-analysis of costs and financial impact on the us health care system. JAMA Intern Med 173: 2039–2046, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.