Abstract
Genome-wide association studies (GWAS) have focused primarily on populations of European descent, but it is essential that diverse populations become better represented. Increasing diversity among study participants will advance our understanding of genetic architecture in all populations and ensure that genetic research is broadly applicable. To facilitate and promote research in multi-ancestry and admixed cohorts, we outline key methodological considerations and highlight opportunities, challenges, solutions, and areas in need of development. Despite the perception that analyzing genetic data from diverse populations is difficult, it is scientifically and ethically imperative, and there is an expanding analytical toolbox to do it well.
Keywords: GWAS, ancestry, diversity, cross-ancestry, trans-ancestry, trans-ethnic, population genetics, admixed populations, psychiatry, complex disease
A disproportionate majority (>78%) of participants in published genome-wide association studies (GWAS) are of European descent (Popejoy and Fullerton, 2016; Sirugo et al., 2019), with 71.8% of these individuals having been recruited from just three countries: the United States, the United Kingdom, and Iceland (Mills and Rahal, 2019). Studies of major psychiatric disorders are no exception, having focused largely on populations of European ancestry (Figure 1). Conducting GWAS in individuals of European ancestry was a practical starting point given the availability of samples and limited funding, genotyping technologies, and analytic methods. However, there is now widespread acknowledgement of the need for more diverse samples and for improved analytic methods. Broadening diversity of studied populations will improve the effectiveness of genomic medicine by expanding the scope of known human genomic variation and bolstering our understanding of disease etiology. Consensus in the field points to many benefits of increased representation of more diverse populations for locus discovery, fine-mapping, polygenic risk scores, and addressing existing health disparities (Duncan et al., 2018; Hindorff et al., 2018a; Lam et al., 2018; Martin et al., 2019; Walters et al., 2018).
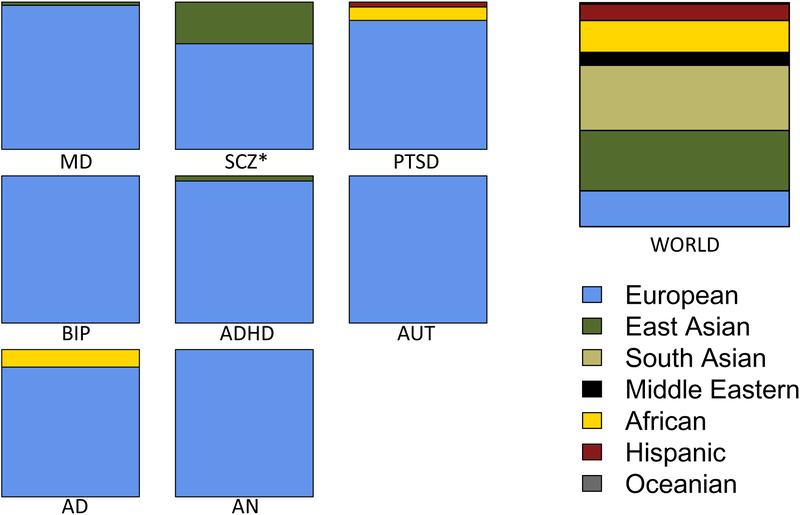
Figure 1. Diversity in GWAS of psychiatric disorders compared to global diversity.
Participant numbers were extracted from the largest consortium publication(s) for each psychiatric disorder and are shown as fractions of the total sample size for each disorder. Note: Sample sizes are given in parentheses. Numbers reflect cases and controls combined. MD=major depression (490,999), SCZ=schizophrenia (205,661), PTSD=post-traumatic stress disorder (188,932), BIP=bipolar disorder (51,710), ADHD=attention deficit hyperactivity disorder (55,230), AUT=autism (46,350), AD=alcohol dependence (52,848), AN=anorexia (14,477). *For schizophrenia, the African American samples from an earlier publication (2009, International Schizophrenia Consortium) were not included in the most recent PGC schizophrenia publication (2014). Ancestry information for each participant was based on principal components analysis of genetic data. See Supplemental Table S1 for consortium studies and references.
With increasing representation of global populations in GWAS, there is an opportunity for advanced methods development and a need for consensus “best practices” for analyzing the emerging complex datasets. Here, we provide background on the scientific and ethical importance of including underrepresented groups in genetics research and offer guidance for whole-genome analysis of ancestrally diverse study cohorts. We summarize currently available resources and make recommendations for avoiding practices that could lead to false-positives, loss of statistical power, or misinterpretation of results. Because this primer represents a collaborative product of the Cross-Population Special Interest Group of the Psychiatric Genomics Consortium (PGC) (https://www.med.unc.edu/pgc/cross-population/), we have framed our discussion within the context of psychiatric genetics. Nevertheless, the points and recommendations outlined herein are applicable to any complex biomedical phenotype.
Genetic ancestry is estimated from DNA and provides information about shared demographic history at the population level. Individuals with similar ancestral origins have shared genomic signatures due to migration of common ancestors, mutations and recombination, genetic drift, and natural selection. These processes yield differences in allele frequencies and linkage disequilibrium (LD) patterns across populations (Barrett and Cardon, 2006; International HapMap Consortium, 2005) that must be properly addressed to avoid false positive genetic findings. In addition to ancestral diversity, the current lack of racial and ethnic diversity, which are related but distinct from ancestry (see Box 1), hinder the development of more complete etiological models (Banda et al., 2015; Medina-Gomez et al., 2015; Race, Ethnicity, and Genetics Working Group, 2005). In complex disease research, race and ethnicity can provide information about social, cultural, and environmental factors that affect risk for disease, including having a lived experience of social injustice. Given that these socio-cultural measures are often inappropriately used as a proxy for genetic ancestry, researchers and clinicians should be careful to distinguish among them in order to tease apart specific biological, environmental, and social determinants of health.
Box 1: Race, Ethnicity, and Ancestry: Interpretation and Relevance for Genetic Diversity.
| ‘Race’, ‘ethnicity’ and ‘ancestry’ are often used interchangeably, yet they have no universal definitions. We provide brief descriptions of our usage below. For extensive discussion in the context of genomics, including recommendations from professional organizations see: (Banda et al., 2015; Mersha and Abebe, 2015; Race, Ethnicity, and Genetics Working Group, 2005). | |
|---|---|
| Race | A culturally and politically charged term, for which definitions and meaning are context-specific. Race is related to individual and/or group identity, and is often linked to stereotypes of visible physical attributes such as skin and hair pigmentation. The concept of ‘race’ is tightly linked to social power dynamics and has historically been used to justify hierarchies of power, discrimination, and oppression in an unequal society. Social and cultural conditions may differ among racial groups, on average, and these differences may lead to environmental effects such as chronic stress and unequal access to goods and services including healthcare and nutrition. These inequities can affect environmental risk for complex diseases and/or potentially interact with genetics to affect risk. |
| Ethnicity | Describes people as belonging to cultural groups, usually on the basis of shared language, traditions, foods, etc. Ethnicity has often been used interchangeably with ‘race,’ and is similarly ambiguous. To the extent that traits are affected by social and environmental differences, ‘ethnicity’ has previously served as a proxy for health and disease risk at the population level as a result of social, cultural, and community effects described above. There is no universal agreement on a system of ‘ethnic’ groupings worldwide. Some ‘ethnic’ groups may share genetic factors due to similar ancestral origins, other groups may be more social and cultural in nature. |
| Ancestry | Meaning varies by context. Here we use the term to denote genetic ancestry, a description of the population(s) from which an individual’s recent biological ancestors originated, as reflected in the DNA inherited from those ancestors. Genetic ancestry can be estimated via comparison of participants’ genotypes to global reference populations, so incomplete availability of these references can create biased estimates. We note that different methods of calculating genetic ancestry can yield different results. Thus, discrete labelling of ancestral populations over-simplifies the complexity of human genetic variation and demography. Nevertheless, accounting for systematic differences in allele frequencies and LD is necessary for genetic analyses. In this paper, diversity in genomics is described primarily in terms of ‘ancestry’. |
Inclusion of diverse study participants in genomics research has yielded important scientific insights for a range of human traits and diseases. The resolution of fine-mapping improves through cross-ancestry analysis (Wojcik et al., 2019). Estimates of effect-sizes derived from cohorts of diverse ancestries tend to be more accurate than from those of a single ancestry (Li and Keating, 2014). Genetic risk prediction attenuates with increasing divergence between the discovery and target populations, indicating that polygenic risk scores (PRS) based on Eurocentric GWAS are not equally predictive when applied to non-European populations (Duncan et al., 2018; Martin et al., 2019). Conversely, constructing individual-level scores from cross-ancestry meta-analysis results improves overall prediction (Grinde et al., 2019; Márquez-Luna et al., 2017).
Besides the strong scientific justifications for broader inclusion, there are important ethical, legal, and public health reasons for bolstering diversity in genomics (Hindorff et al., 2018b). Understanding how genetic risk and social inequities interact to influence disparities in disease risk and outcomes will be critical to improving public health.
Moreover, while integration of genomics into healthcare has the potential to improve disease prediction and optimize treatments, a lack of diversity will limit the utility of precision medicine efforts: individuals of non-European descent are more likely to receive ambiguous test results from genetic screening (e.g., variants of unknown or uncertain significance) (Petrovski and Goldstein, 2016) and false positive diagnoses (Manrai et al., 2016). There is also a higher chance of false negative diagnoses in individuals from ancestral backgrounds that are not well represented in clinical databases, due to missing information about additional disease-causing variants currently not on testing panels (Minster et al., 2016; Moltke et al., 2014; Wheeler et al., 2017). Similarly, the potential benefits of pharmacogenetics cannot be fully realized until there is equitable representation across ancestries, as some therapeutics may be more effective and/or safer in certain populations because of differences in allele frequency, effect size, and penetrance of variants associated with drug metabolism (Roden et al., 2011). Here, we provide an accessible framework for analyzing these data, while acknowledging that there are several important methodological areas in need of further development. Key terminology is bolded and defined in Box 2.
Box 2: General Terminology.
| Term | Definition/Comment |
|---|---|
| Admixed Population | A population of individuals with ancestors from two or more populations. Admixed can also be used to refer to individuals. |
| Fine-mapping | Analytical procedures designed to refine GWAS loci to a smaller set of likely causal variant candidates to facilitate interpretation and follow-up studies. |
| Genetic Correlation | The correlation of genome-wide genetic effects between two phenotypes, which is often estimated for a subset of genomic variants (e.g. SNPs in a GWAS). |
| Genotype Imputation | Estimation of genotypes at genetic sites that have not been directly measured, using data from a reference panel to infer genotypes based on LD and haplotype structures. Accuracy depends on availability of suitable reference panels. |
| GWAS | Genome-wide association study. Analysis of common genetic variants across the whole genome for association with a phenotype. |
| GxE | Gene by environment interaction refers to genetic effects on a phenotype that vary based on environment, or vice-versa. |
| Haplotype | A group of alleles that are correlated with one another because they are inherited together on a chromosome. |
| HWE | Hardy-Weinberg equilibrium, the expected balance of genotypes within a population assuming random mating, infinite population size, and no mutation, migration, or selection. Tests of deviations from HWE are used in quality control to detect technical issues with genotyping. Note that there are also non-technical reasons for deviation from HWE (e.g., selection, population structure, admixture, nonrandom mating). |
| LD | Linkage disequilibrium. Alleles in LD are physically linked on a chromosome, which leads to non-random coinheritance such that their frequencies in a population are correlated. |
| Major Population | A group of individuals with shared genetic ancestry. A heuristic simplification of the complexity of human demography, but useful for describing groups that are likely to have relatively similar allele frequencies and LD patterns due to shared ancestry. Common examples used in practice include continental ancestry groups or “super populations” as defined by the 1000 Genomes Project (e.g., African, Admixed American, East Asian). |
| PCA | Principal component analysis. PCA of genotype data is commonly used to examine population structure in a cohort by determining the average genome-wide genetic similarities of individual samples. Derived PCs can be used to group individuals with shared genetic ancestry, identify outliers, and as covariates to reduce false positives due to population stratification. |
| Population Stratification | Underlying population structure within a sample that is correlated with a phenotype, which can confound genetic association tests. |
| PRS | Polygenic risk score. A value computed from an individual’s genotype data that quantifies genetic influences on a particular phenotype; also known as polygenic score (PGS), genetic risk score (GRS), or risk profile score (RPS). |
| Reference Panel | A set of genetic variants from a population. Reference panels are used to design arrays, impute genotypes, catalogue genetic variants, and identify regions that are similar and different between populations. |
| SNP Heritability | Proportion of phenotypic variance that is explained by additive genetic effects of a set of SNPs. |
Methodological Considerations
In the analysis of multi-ancestry datasets, a significant concern is false positive genetic signals due to inflated test statistics from population stratification, which occurs when disease prevalence and allelic frequency differences are correlated within or between study cohorts (Marchini et al., 2004). Two typical strategies exist for addressing this challenge while analyzing samples from multiple major/admixed populations: (1) Empirically assign samples to major continental and/or admixed populations using genome-wide data, analyze each population separately, and conduct cross-ancestry meta-analysis (stratified meta-analysis approach), and (2) analyze samples from multiple populations together, most commonly with a mixed model (joint mixed model approach). The choice between these approaches is perhaps the most broadly impactful decision currently facing analysts of genome-wide data from multiple populations since it impacts methodological considerations in all analysis steps from quality control, to reference alignment in imputation, to association model, to the suitability of results for secondary analyses. We highlight elements of GWAS where the choice between the stratified meta-analysis and joint mixed model approaches is particularly salient. Figure 2 shows a general workflow for each approach.
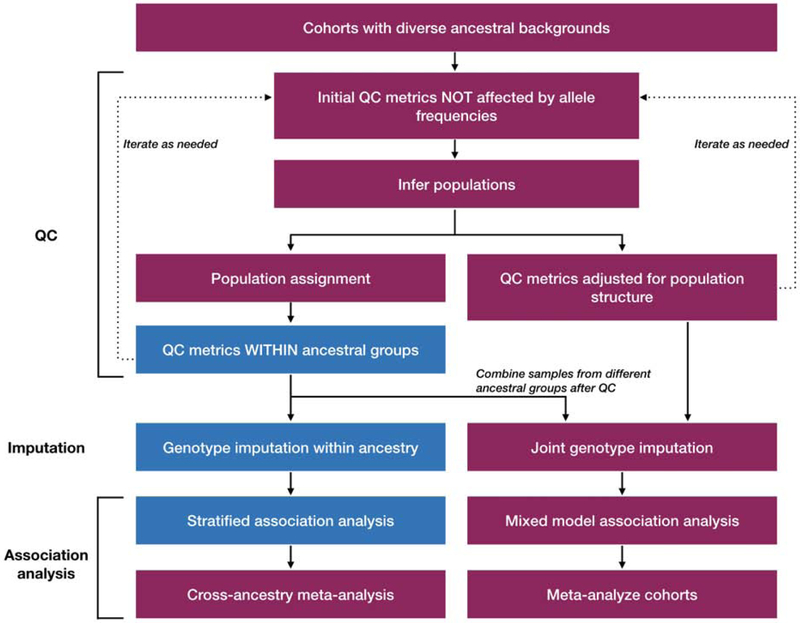
Figure 2: Flow chart for quality control, imputation, and association analysis in diverse population samples.
This flowchart depicts the general analysis framework for genome-wide association studies of participants with diverse ancestral backgrounds. Note: boxes with red headers indicate analyses done in samples with diverse ancestral backgrounds and blue denotes analysis done within samples in major population groups. The left path shows a strategy for the stratified meta-analysis approach and the right path shows steps for the joint mixed model approach [see Supplemental Table 2 for more detailed quality control (QC) considerations].
Genotyping Technologies
Most genome-wide DNA microarrays were designed for individuals of European ancestry. The differences in LD structure and allele frequency among populations can lead to significantly worse coverage for other ancestry groups. For example, at imputation accuracy r2>0.8, the Affymetrix UK Biobank array covers 84% of the variants that have minor allele frequencies (MAF) > 1% in samples of European ancestry but only 46% of those for samples of African ancestry (Nelson et al., 2017). The large genetic diversity in African populations means that a larger number of variants are needed on arrays in order to provide similar coverage as in other populations (Barrett and Cardon, 2006). To address this issue, some groups, such as China Kadoorie Biobank (Chen et al., 2011), have designed population-specific arrays. Multi-ancestry arrays, such as the Multi-Ethnic Global Array (MEGA), Global Screening Array (GSA), and the H3Africa array (Mulder et al., 2018) were designed based on panels with more diverse ancestries, and are therefore recommended. An alternative strategy is to sequence whole genomes; low-depth sequencing has received recent attention for application in diverse samples due to cost-effectiveness and higher coverage with acceptable error rates ((Gilly et al., 2018; Peterson et al., 2017a); see Rare Variants).
Quality Control
Quality control (QC) of GWAS data aims to remove low quality data and technical artifacts in order to reduce the risk of false positive associations. In diverse ancestry cohorts, the main issue is that many common QC criteria assume the sample comes from a homogeneous population. Applying standard QC procedures without adjustment for population structure leads to the erroneous removal of too many variants and samples from minority subgroups and admixed samples, reducing statistical power.
QC criteria that are dependent on population allele frequencies can generally be adapted for application in diverse cohorts by either stratifying the cohort into major populations prior to filtering (the stratified meta-analysis approach) or by adjusting the QC measure to allow for varying allele frequencies (the joint mixed model approach; see Figure 2). For example, individuals are often removed based on excess autosomal heterozygosity, as a potential indication of sample contamination, but the standard heterozygosity statistic assumes each variant’s expected allele frequency is constant across individuals. In diverse cohorts, regressing this heterozygosity statistic on principal components prior to identifying outliers can avoid excessive exclusions of individuals from subgroups in the cohort. Step-by-step considerations for common QC criteria, including sample QC workflows for the stratified meta-analysis and joint mixed model approaches, are given in Supplemental Methods I (see also Supplemental Table S2, Supplemental Figure 1). In addition to these pre-imputation QC steps, post-imputation QC steps should also consider ancestry (see Imputation).
Inferring Population Structure
Estimating the genetic population structure of a cohort typically serves two primary goals in GWAS: 1) to characterize the ancestral diversity of the cohort as a descriptive measure and 2) to provide a quantitative estimate of population structure that can be used in QC and in GWAS association models to reduce the risk of false positives. We focus here on use for description and QC, and later discuss methods for controlling for population structure (see Genome-wide Association).
For cohorts with diverse ancestral backgrounds, we can estimate population structure based on genome-wide data. Currently the most common tool for estimating continuous population structure is principal component analysis (PCA); a listing of other approaches is included in Supplemental Methods II. PCA is a statistical method for reducing the complexity of high-dimensional data (e.g., thousands of measured variants across the genome) into orthogonal axes (principal components, PCs) that explain the largest fraction of variability in the data. The spread of data across these axes provides a visual guide to sub-structure among samples; when data points are estimated from each individual’s genetic markers, the PCs illustrate population structure. These PCs can be computed within the cohort, or can be estimated from an external reference (e.g., The 1000 Genomes Project (1KGP); (Sudmant et al., 2015)) and the GWAS sample can be projected onto the PC axes to allow comparison with the ancestries of known reference populations (Peterson et al., 2017b). However, the latter approach can be limited by the number and diversity of populations represented on the reference panel, highlighting the need for many additional diverse population references to be generated. PCs may also be used to control for ancestry structure in other QC metrics (see Quality Control and Supplemental Table S2).
This sample-wide estimation and visualization of genetic ancestry can be used to empirically assign genetically similar samples into more homogenous groups. This assignment is necessary for the stratified meta-analysis approach to GWAS of diverse cohorts, and is intended to reduce the risk of false positive genetic signals due to inflated test statistics from population stratification. Assigning samples to more homogeneous groups for analysis reduces stratification by limiting the degree of population structure remaining in the sample. Samples with a specific admixture can be assigned into their own major ancestral group, instead of being excluded from the analysis or forced into other ancestry groups, provided there are adequate numbers of individuals in the sample with comparable admixed backgrounds. However, it is often the case that genomic outliers (which tend to be from under-represented or admixed backgrounds) might need to be excluded if there is an insufficient number of other individuals who fall into a similar cluster. These assignment methods will not provide - and are not intended to provide - detailed ancestral background information for each individual. Rather, they provide a working solution to reduce false positives due to population stratification (Hellwege et al., 2017). We stress that sample group assignment and identifying appropriate reference population panels can be difficult, particularly for admixed ancestry, thus requiring careful inspection of data and methods (Medina-Gomez et al., 2015).
Imputation and Population Reference Panels
GWAS arrays genotype a portion of common variation. Genotype imputation is a cost-effective computational approach for inferring genotypes or genotype probabilities at variants that have not been directly genotyped on GWAS arrays, based on comparisons to genetic data from external reference samples. Imputation increases the number of markers available for association testing and can harmonize cohorts genotyped on different arrays for meta-analysis.
Imputation accuracy relies on having an appropriate reference panel that includes haplotypes from the population studied. Matching alleles and allele frequencies in the study cohort with reference panels as part of pre-imputation QC also relies on using reference data from a matched ancestral background. Reference panels with better coverage of haplotypes from the population of the genotyped cohort will yield a greater number of well-imputed variants for GWAS, especially among lower frequency variants (Ahmad et al., 2017; Howie et al., 2012). Table 1 lists major imputation panels that are currently publicly available. We note that although many ongoing projects are aiming at more diverse populations (Supplemental Table S3), additional efforts in more populations are needed to expand the diversity of imputation reference panels (Kelleher et al., 2018).
Current imputation methods are summarized in Supplemental Methods III. Joint imputation using the largest applicable reference panel is expected to perform at least as well as subsetting that reference panel to match the target population (Ahmad et al., 2017; Howie et al., 2012), possibly due to maintaining a larger sample size for phasing. Use of the same reference panel for all cohorts also avoids potential confounding with varying imputation quality. However, it may be necessary to consider imputation quality separately within subsets of individuals even if the samples are jointly imputed since imputation accuracy for a variant may vary widely across individuals of different ancestries.
Genome-wide Association
The core of GWAS analysis is testing the association between each variant and a target phenotype. As noted, a primary consideration for association testing in diverse cohorts is whether to stratify samples into major population groups or to analyze the full cohort jointly (assuming imputation was also done jointly). In either case, the major concern is proper control of population stratification to ensure that observed associations reflect genetic effects of each locus rather than correlations with ancestry.
Joint analysis using a mixed model approach is attractive because all participants are included irrespective of ancestry. Ideally, mixed model approaches control for population stratification by modelling distant relatedness between individuals due to ancestry (Sul et al., 2018; Wojcik et al., 2019). Several implementations exist and some are listed in Supplementary Methods Section IV and Supplementary Table S4. Mixed models may yield greater statistical power, both through increased sample size and by controlling for the variance explained by the genetic relatedness between individuals (i.e., a random effects component; (Loh et al., 2018)). However, there is evidence that basic mixed models may not fully control for population structure in diverse cohorts, especially if there is an environmental component to phenotypic associations with ancestry beyond the modelled genetic relatedness (Conomos et al., 2018; Heckerman et al., 2016; Zhang and Pan, 2015). Non-genetic factors such as environmental exposures may be correlated with ancestry due to a shared local environment (familial or community effects) or due to the relationship between ancestry and socio-cultural factors such as race and ethnicity. More methodological development is needed before mixed models or other strategies for joint GWAS of a diverse cohort can be confidently recommended as robust.
When stratifying by population backgrounds, covariates such as PCs should still be used to correct for population stratification. Conventional linear or logistic regression with these covariates can be used for association testing as long as QC included exclusion of related individuals; mixed models or other alternatives with PC covariates may be applied in family-based samples stratified by ancestry (Walters et al., 2018). Computing these PCs separately within each ancestry subset instead of the full study ensures better control for residual structure specific to that subset (e.g., fine structure, genotyping/technical artifacts), but at the cost of potentially reduced control for stratification related to population structure shared across subsets (Patterson et al. 2006). For analyses of admixed or multi-ancestry cohorts, PCs may still be included in the regression but additional covariates may be required to control for stratification that is not linear in PCA space (Conomos et al., 2018; Heckerman et al., 2016; Zhang and Pan, 2015). For example, race and ethnicity are often correlated with socio-economic status and other environmental risk factors for disease. Self-reported ethnicity or other variables that capture trait heterogeneity on the basis of socio-cultural factors may also be appropriate to consider as covariates in those instances (Banda et al., 2015; Medina-Gomez et al., 2015). Directly controlling for local ancestry tracts in variant-level association analyses may further improve power and reduce false positives in admixed samples (Li & Keating 2014).
The meta-analysis approach, combining separate analyses of samples stratified by similar genetic background, currently has several pragmatic advantages. First, computational pipelines developed for single-ancestry analyses can be used for each cohort. Separate analysis also naturally provides ancestry-specific results, which may be valuable for secondary analyses including PRS (Bulik-Sullivan et al., 2015; Lam et al., 2018). Reduced environmental variability within a subset may also improve power. On the other hand, splitting each cohort may be challenging due to continuous gradients of admixture or small sample sizes within an ancestry group. This loss of information from excluding individuals from diverse genomic backgrounds is a missed opportunity for discovery and validation of GWAS findings, and thus additional approaches need to be developed and leveraged.
Meta-analysis of GWAS Summary Statistics
Traditional meta-analytic approaches for GWAS rely on fixed-effects models that assume a given variant has the same true marginal effect size across all studies. This assumption is likely to be violated in meta-analyses across diverse cohorts. Even when the causal genetic effect of a variant is constant across populations, as seems common in cross-ancestry GWAS to date (Huang et al., 2017; Lam et al., 2018), marginal effect sizes may show heterogeneity when LD structures are different. Further heterogeneity across cohorts from different populations may arise due to differences in genetic background (e.g., gene × gene interactions) and/or environmental context (e.g., gene × environment interactions), as well as differences in study design (e.g., imputation artifacts, phenotyping). As a result, it is generally appropriate to model this cross-cohort heterogeneity in meta-analysis by using a random effects or trans-ancestral meta-analysis model (Supplementary Methods Section 5, Supplementary Table S4).
Fine-mapping
A trait-associated locus from GWAS typically implicates a large genomic region with many variants of similar significance. This set may contain a few causal variants, while the association of other variants is driven by their LD with the causal one(s). Fine-mapping refines GWAS loci to a smaller set of likely causal variants to facilitate interpretation and follow-up studies (Schaid et al., 2018). Fine-mapping studies in samples of European ancestry have made important advances, with some loci resolved even to single-variant resolution (Huang et al., 2017; Mahajan et al., 2018). Because fine-mapping assumes the causal variant(s) have been observed, non-European populations face a unique challenge due to the lack of representation of many variants as a result of incomplete sampling from these populations, suboptimal chip design, and limited imputation performance.
Combining samples across ancestries has an advantage for fine-mapping: the LD patterns that differ across populations can improve the resolution, assuming that many causal variants are shared across populations, which has been shown true for some traits, including schizophrenia (Lam et al., 2018; Marigorta and Navarro, 2013; Wojcik et al., 2019). Non-causal variants tagging the causal variants have marginal different effects across populations if LD is different, thus allowing the causal variant to be distinguished from non-causal variants. Furthermore, in certain populations (e.g., African), LD blocks are generally smaller, so fewer non-causal variants will tag the causal variants, improving the resolution of fine-mapping (International HapMap Consortium, 2005; Schaid et al., 2018).
Most fine-mapping algorithms (Huang et al., 2017; Schaid et al., 2018) can be applied to samples from multiple ancestries combined through meta-analysis. However, this strategy does not take full advantage of genomic diversity across populations. An alternate Bayesian fine-mapping strategy (Lam et al., 2018) more precisely mapped the schizophrenia genetic associations through explicitly modeling diversity in LD between East Asian and European samples. This approach works on a presumption that the causal variants and their effect sizes are identical across populations, which is not always true. PAINTOR (Kichaev and Pasaniuc, 2015) relaxes this presumption by allowing the effect size to vary across populations, although the causal variant still needs to be the same. Fine-mapping methods will benefit from continued development that appropriately models LD and relies on fewer assumptions.
Polygenic Risk Scores in Diverse Populations
PRS are individual-level estimates of the relative genetic contribution to a phenotype, computed for each genotyped individual in a target sample based on GWAS results from a discovery sample. PRS are useful for validating GWAS results in external cohorts and have the potential to provide individualized risk prediction from genetic data (Khera et al., 2018; Martin et al., 2019). The predictive value of PRS profiling depends both on the statistical power of the discovery (training) dataset— specifically, enrichment in the genome-wide distribution of association test statistics that is attributable to aggregate, additive genetic effects — and the relevant characteristics of the target (testing) dataset.
In particular, PRS accuracy is also a function of recent human demographic history, such that a greater proportion of phenotypic variance is explainable in target populations that are genetically more similar to the population studied in the discovery GWAS. Stated another way, with increasing genetic “distance” between the discovery and target datasets, there is often attenuation of polygenic predictive value. Furthermore, because most participants in large GWAS have been broadly European (Figure 1), most PRS currently perform best in target samples of European ancestries, with markedly worse performance in other populations, especially in individuals of African descent (Duncan et al., 2018; Martin et al., 2019).
A practical question is how to construct polygenic scores for recently admixed individuals or individuals who are genetically distant from those in the largest existing GWAS. Use of trans-ancestry meta-analytic results to weight alleles can increase prediction accuracy (Grinde et al., 2019), and MultiPred is an approach that combines PRS based on European training data with PRS based on training data from the target population (Márquez-Luna et al., 2017). Current methods development is focused on improving handling of allele frequency differences and LD within and across populations. Given current limitations in understanding similarities and differences in polygenic risk across populations, caution is advised in interpreting differences in PRS across ancestries (Novembre and Barton, 2018).
Heritability and Genetic Correlation
GWAS can provide insights into the genetic architecture of human traits, including SNP heritability and genetic correlation. Several methods have been proposed for estimating these parameters from genotype data (Supplemental Table S4; Supplemental Methods Section V), but estimation and interpretation of these quantities is more challenging in diverse populations. Heritability estimates may differ between populations due to variation in both environmental factors and population genetic forces. Cross-population differences in phenotype measurement (Section XI) may further complicate interpretation. In evaluating shared genetic variance across populations, genetic correlation between groups can be defined either as the correlation of allelic effect sizes (genetic-effect correlation) or the correlation of the relative contribution to total phenotypic variance (genetic-impact correlation), and for all variants or for common variants present in a study. Each value is potentially informative, but divergence in allele frequencies and LD patterns between populations will lead to differences between these parameters (Galinsky et al., 2019).
As detailed in the supplement, most common methods for estimating SNP heritability and genetic correlation either require modification or may not be suitable for use in multi-ancestry studies. Methods relying on relatedness estimation (e.g., genomic relatedness matrix restricted maximum likelihood; GREML) require estimation methods robust to population structure (Conomos et al., 2018; Thornton et al., 2012), and methods modelling LD (e.g., LD Score regression; LDSC) require either ancestry-matched reference panels or individual level data for LD calculations (Luo et al., 2018). Ancestry-matched reference panels, along with the large GWAS sample sizes required for robust estimation using these methods, may be especially challenging to acquire for studies in underrepresented or admixed groups.
Beyond these most common methods, local ancestry tracts in admixed population samples can be leveraged to estimate heritability (Zaitlen et al., 2014) and both genetic-effect and genetic-impact correlations of observed variants can be estimated using Popcorn (Brown et al., 2016) if LD information is available and the two populations are relatively homogeneous. Recent studies estimating cross-ancestry genetic effect correlations have found moderate to high correlations for most phenotypes (Bigdeli et al., 2017; Brick et al., 2019; Lam et al., 2018). The extent to which these cross-ancestry genetic correlations reflect consistent effects at any particular locus remains a question for fine mapping analyses.
Rare Variant Association Analysis
Rare SNPs and structural variants have been implicated in complex disease (Bomba et al., 2017). Due to their more recent origin, rare variants tend to be more geographically clustered and can be population specific. They can also be particularly important from both clinical and biological perspectives because some confer a large increase in disease risk. However, there is severely limited power to identify trait associations of individual rare variants. Therefore, aggregation methods such as burden tests, variance-component tests, and hybrid tests have been developed to test the combined effect of several variants. Using this approach, variants can be combined within genes or regulatory genetic elements (Gilly et al., 2018; Kuchenbaecker and Appel, 2018). Ancestry groups may carry different driving variants at the same locus, as demonstrated by the association of different functional variants in ADH1B with alcohol use disorder in African Americans compared with European and Asian Americans (Edenberg and McClintick, 2018). Therefore, aggregate testing can be particularly suitable to projects involving different ancestral groups because they focus on functional units rather than individual variants and it is not necessary to observe the same variants or frequencies across cases. Meta-analysis methods have been developed that are able to encompass heterogeneous genetic effects across studies and are applicable to cross-ancestry meta-analysis (Lee et al., 2013; Tang and Lin, 2015).
Association testing for rare variants is particularly sensitive to population stratification, and adjusting for fine-scale patterns of population stratification can be difficult with traditional methods (Zhang et al., 2013). In simulation studies, adjusting for PCs failed to fully control inflation for collapsing and variance-component methods (Persyn et al., 2018). Mixed effects models that have been developed for related samples might improve on this (Jiang and McPeek, 2014). However, this area requires further methods development.
Non-Genetic Contributors to Trait Variability
Diversity in social, cultural, and environmental factors also affect disease risk, and can contribute to confounding in genetic studies. In the case of complex traits with strong environmental influences, such as psychiatric conditions, the need to account for non-genetic contributors to disease is important. Unfortunately, measurement of environmental factors can be difficult, so proxy measures such as zip code or insurance status can be used to model non-genetic risk factors such as air quality or accessibility to quality health care. PCs calculated from genotypes can control for population structure due to genetic relatedness, but this approach alone may not capture the social and environmental factors that are encompassed in self-reported “race” and “ethnicity”, even though these measures can be correlated with genetic ancestry. Self-reported measures of diversity can help in the modeling of societal determinants of health, such as increased stress due to the experience of racism and inequality and related variability in environmental factors (e.g., socio-economic status) that affect disease risk. However, the reliance on race and ethnicity as proxy variables for environmental effects or in order to control for population structure may be inappropriate. Better understanding and measurement of causal environmental risk factors is critical in order to advance discovery methods beyond these over-simplified and potentially harmful constructs of non-genetic contributors to trait variability.
Investigating complex traits in diverse populations, especially when samples are pooled from different research sites or cultural contexts, requires consistency and equivalence in the underlying construct and assessment measures across groups. Differences and variability in phenotypic measurement between study sites and populations may affect both gene discovery and the transferability of genetic findings between populations. Most psychiatric classification systems and diagnostic measures have been developed and validated in individuals from industrialized, Western societies (Henrich et al., 2010). This presents a substantial challenge for global and cross-cultural collaborations. Investigations into cross-cultural differences in the prevalence of major depression, for instance, have suggested that although there is a shared underlying disorder construct across groups (Kendler et al., 2015; Simon et al., 2002), individuals may differ culturally in terms of the level of symptomatology reached prior to seeking help (Simon et al., 2002). The inclusion and consideration of diverse populations in the development, validation, and deployment of diagnostic measures used in genetic studies is therefore critical for ensuring an unbiased picture of disease etiology (Supplemental Methods VII).
Despite known large effects of environmental exposures on complex disease risk, there have been limited efforts to incorporate these factors into large-scale genetic studies. Appropriate modeling of the environment is especially critical when a phenotype or trait of interest is influenced by gene-by-environment interactions (GxE). That is, genetic risk factors not only alter average risk but also influence sensitivity to the effects of environmental adversities. However, the majority of GxE studies have been underpowered and conducted using samples of primarily European descent, which limits the assessment of GxE and thereby the identification of modifiable targets for intervention and prevention among understudied groups (Duncan et al., 2014). We note that the statistical definition of GxE depends on the choice of modelling on an additive or multiplicative scale (Kendler and Gardner, 2010). Greater representation of diverse individuals is critically needed in order to increase our understanding of how the interrelated contributions of genes and environment vary across social and cultural groups, and how these factors may interact.
Perspectives and Recommendations
The lack of diversity in genetic studies is problematic for a variety of ethical and scientific reasons. Continued reliance on samples that only represent a fraction of genomic, socio-cultural, and environmental diversity limits our understanding of disease biology and may ultimately contribute to widening global health disparities. Greater ancestral diversity in study samples has the potential to accelerate the discovery of causal risk variants and is critical for a greater understanding of the biological causes of disease, including gene-by-environment interactions. In this Primer, we have highlighted the challenges and benefits of working with diverse populations, recommended practices based on current methods, and have noted specific areas that are in need of further methodological development (Box 3). In summarizing progress, remaining challenges, and requisite next steps, we consider three main domains: 1) researcher participation, 2) data resources, and 3) analytic methods.
Box 3: Common pitfalls, recommendations, and methods in need of development.
| Method | Pitfall | Recommendation | Needs |
|---|---|---|---|
| Genotyping | Many genotyping platforms do not cover non-European variation well. | Use or design population-specific array or multi-ancestry array; high array density can improve coverage in groups with high diversity | Continue improving coverage of diverse ancestries on genotyping arrays |
| Consider low-depth whole-genome sequencing | Encourage ongoing development and sharing of pipelines for analysis of low-depth sequencing data | ||
| QC | Unnecessary loss of data and/or incorrect inferences by using a one-size-fits-all approach | See Figure 2 for specific recommendations for each QC step and Supplemental Table 2 | Improve availability and convenience of implementing proposed QC methods robust to population structure |
| Imputation | Inaccurate imputation due to poor matching of reference panel to sample | Consider matching the ancestry of the reference panel as closely as possible to the sample ancestry if using a single ancestry sample. Consider the largest reference panel possible for imputation of multiple or admixed samples | Continue expanding diversity of imputation panels,through collection of whole-genome sequencing data, creation of imputation panels from that data, and promoting public sharing/accessibility of those panels |
| GWAS | Poor control of population stratification | Consider standard linear/logistic regression methods for analysis of single ancestry groups followed by meta-analysis. Consider mixed model approaches for admixed or multi-ancestry analyses | Continue investigating causes of - and solutions to - current incomplete control of population stratification from principal components and mixed models |
| Include PCs as covariates even when single ancestry groups analyzed. PCs should be computed individually for each major population group within a multi-ancestry cohort and included as covariates in the regression model. Additional covariates should be considered for the multi-ancestry analysis | |||
| Meta-analysis | False negative and false positive findings, effect heterogeneity | Use a random-effects (with possible bias towards the null), or modified random-effects meta-analysis model | Continue to investigate and find solutions to improve power for the detection of heterogeneous effects |
| Fine-mapping | LD improperly handled when all samples are meta-analyzed across populations | Use fine-mapping methods that explicitly model population- specific LD | Continue to develop fine-mapping methods that rely on fewer assumptions, and thoroughly evaluate their performance |
| Uneven genome coverage across populations because of the genotyping array and the imputation reference panel | See recommendations for Genotyping and Imputation above | ||
| Polygenic risk scores | Loss of accuracy in target population with increasing genetic distance from discovery cohort | Extrapolation of PRS from one ancestry to another is problematic with current approaches and data | Large discovery cohorts for all populations are needed. Develop methods for computing PRS that are not biased when applied across populations, potentially incorporating LD information and/or local ancestry information among diverse populations |
| Rare variants | Population stratification; low power to detect associations | Aggregate tests can improve power and handle separate causal variants in different populations | Approaches with better control of population stratification; more data on diverse populations needed |
| Heritability estimates | Differences in MAF and LD structures | For GREML, use admixture-aware relatedness estimation for admixed samples | Currently no method based entirely on summary statistics can handle admixed/diverse samples. Evaluate options for developing estimation methods with reduced requirements for access to genotype data or ancestry-matched LD reference panels |
| Different environments | For LDSC, consider using cov- LDSC if in-sample genotype data is available Caution when comparing estimates between groups | ||
| Cross-ancestral genetic correlation | Requires large sample sizes and dense array; estimates influenced by genetic distance between groups | Use Popcorn or GREML with admixture-aware estimation of genetic relatedness | Improve robustness and user- friendliness of software for summary statistics; increase diversity of LD reference panels |
| Phenotypic measurement | Lack of consideration of potential measurement differences across groups | Consider and test for equivalence across populations. Be cautious when meta-analyzing or comparing across groups in which culturally sensitive measurement has not been demonstrated. | Interdisciplinary collaborations with local researchers across populations to continue developing and validating phenotypic measures |
| GxE | Lack of consideration of environmental factors that are relevant | Consider environmental factors that may be of particular relevance to different socio-cultural groups (e.g., “racial/ethnic” discrimination). | Large samples of diverse individuals and assessment of a broad range of environmental exposures and socio-cultural experiences |
| Consider running analyses separately for each group to gain understanding of GxE processes within populations, and be cautious when making comparisons across populations |
Researcher Participation
It is essential that cross-population research is carried out with careful consideration of its ethical, legal, and social implications (ELSI). This includes an ethos of trust-building, transparency, bi-directional knowledge sharing, and community engagement. This is especially true in low and middle income (LMIC) settings and in work with minority groups – contexts in which mistrust of researchers is warranted given historical mistreatment and ethical violations. As there is no single overarching legislative framework that covers this area, we draw attention to literature that (i) articulates key issues (e.g., consent-taking, data-sharing, sample governance, equal partnership, capacity building, community engagement, participants’ advisory boards (Akinhanmi et al., 2018; Claw et al., 2018; Parker and Kwiatkowski, 2016) and (ii) proposes effective working solutions to them (Beaton et al., 2017; Campbell et al., 2017; de Vries et al., 2015). Additionally, there is a need to overcome traditional barriers to research empowerment for under-represented groups. H3ABioNet (https://www.h3abionet.org/), GINGER (https://ginger.sph.harvard.edu/), AMARI (https://amari-africa.org/), MIND (https://minds-uf.org/), and BRAIN (https://advance.washington.edu/brains) are examples of initiatives that embed the targeted delivery of skills and training within broader programs of research. Additional funding mechanisms that support such an approach would be particularly beneficial.
Data Resources
There is a critical need for extensive collaborative efforts to generate large-scale discovery cohorts of diverse ancestry. Limited diversity in genetics research is a major factor limiting our ability to address important scientific questions. The 1KGP (Sudmant et al., 2015) serves as one of the most widely-used resources in genetics research, but expanding those reference panels is a priority. Here, we provide a selected catalogue of extant and emerging sources of whole-genome sequence data (Table 1 and Supplemental Table S3), to facilitate improved matching of diverse study cohorts to appropriate reference panels. Notably, some sources of non-European data are under-utilized, such as minority groups within the UK Biobank. Although diverse ancestry groups only account for about 5% of this data, that fraction amounts to over 35,000 samples of non-European and admixed ancestry (Bycroft et al., 2018) and yet only 7.3% of publications since 2008 that used this data included any of these diverse samples. Thus, there are opportunities to make better use of these and other existing resources.
Additionally, substantial efforts are needed for efficient and ethical international sample and data sharing. This is an issue under active debate, as countries have different approaches to weighing concerns about the privacy of individuals against the collective benefits of science, and the regulatory landscape of individual-level genotype data has been uneven. For example, while the UK allows open access of individual-level genotype data with a valid scientific proposal, other countries, such as Denmark, Iceland, and China, tightly regulate the sharing of such data. Some GWAS consortia, including the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) and Social Science Genetic Association Consortium (SSGAC), overcame these regulatory challenges using essentially a “federated sharing model” (Fiume et al., 2019). Without sharing individual-level genotype data, a study in these consortia follows the prespecified analytic protocol and contributes its summary statistics to the meta-analysis, allowing the participation of studies that do not have permission to share individual-level data. Researchers should be aware of such options and restrictions, and we recommend regular review of policies as scientific advances may change the ground on which they are based. The practice of sharing summary statistics is increasingly important, and facilitates meta-analyses and other secondary analyses like polygenic risk scoring and estimation of cross-trait genetic correlations. Journals and funding agencies should require sharing of summary statistics whenever it is ethically and legally possible.
Future directions for improving analytic methods
Many of the analytic challenges involved in genetic studies of diverse populations (Box 3) can be addressed by recent advances in methodologies. We reflect on two key issues that remain unresolved and are likely to be beneficial directions for methodological development: 1) the division of individuals into major population groups for analysis and 2) the extension of common secondary analyses of GWAS results to accommodate results from cross-population studies.
A primary question currently faced in genetic analyses of diverse cohorts is whether to follow a ‘combining’ approach (analyzing all individuals together, regardless of ancestry) or a ‘stratifying’ approach (dividing the cohort into major population groups for separate analysis, followed by cross-ancestry meta-analysis; Figure 2). Concerns regarding joint analysis methods (e.g., mixed models) include inadequate control for confounding population stratification and the limited options for secondary analyses such as polygenic risk scoring and genetic correlation estimates. To the extent that stratifying individuals into major population groups remains a feature of cross-population analyses, future methods and theoretical work may continue to refine standards for how best to assign individuals to more homogenous groups. The best solution currently available combines a priori analysis plans, exploratory examination of the data, and involving collaborators with expertise in analyzing globally representative datasets. Future work will benefit from increasing diversity in reference panels, formalizing how major populations should be defined for the purposes of genetic analyses, and evaluating the performance of such methods. Continued methodological work should help resolve the tension between these approaches, clarifying if and when stratifying samples is necessary and providing improved methods for joint analysis of diverse cohorts that addresses population stratification.
Many post-GWAS statistical methods have limited portability to association results from diverse and admixed populations, due to complexities with LD patterns. Caution should be taken in the downstream analysis of cross-population GWAS meta-analyses, as many common approaches such as gene-based testing (e.g., MAGMA (de Leeuw et al., 2015)), heritability and genetic correlation estimation (e.g., LD Score regression (Bulik-Sullivan et al., 2015)), and predicted gene expression (e.g., S-PrediXcan (Barbeira et al., 2018)) rely on external reference panels that may not be compatible with the ‘combining’ approach. Even methodologies such as Popcorn (Brown et al., 2016) that are specifically designed for cross-population analyses typically assume single-population summary statistics as input. Furthermore, it is unclear whether annotations of GWAS results based on observed associations in external studies (e.g., gene expression, Hi-C contacts, methylation) may also need to evaluate population specificity or include diverse samples to improve generalizability across populations. For example, 85% of GTEx eQTL annotations are from individuals of European ancestry (GTEx Consortium, 2013) and other functional genomics resources may be similarly limited.
The above-described methods of cross-population aggregation and comparison rely on an assumption that complex diseases are phenotypically similar across global populations and that measurement of such disorders is culturally unbiased. Given that we know these assumptions are not always accurate, the best practical steps are to be aware of potential phenotypic and environmental differences across populations and involve multi-disciplinary teams with expertise in global societal determinants of health and cultural competency. Suitable methods – such as those that account for cultural context of phenotype ascertainment and GxE – should then be developed and implemented to more precisely measure and treat disorders across cultures.
Conclusion
There is a growing need for investment in policies and practices to support the inclusion of diverse research participants and thus maximize the global potential of genetics research and precision medicine. Broadening participation of both study populations and researchers from many regions of the globe and LMIC in particular will likely be tremendously beneficial. Within the arenas of available data and analytic methods, short-term goals include improved sharing and openness of data. Longer-term goals include identifying ways in which the complex practical, cultural, social, legal and ethical issues inhibiting sample collection from under-represented populations are best resolved. Early, often, and meaningful engagement of stakeholders from diverse patient groups and communities, multi-disciplinary investigators including those with expertise in community-based participatory research, research institutions, scientific editors and reviewers, and funding agencies will all be critical to the success of these short- and long-term objectives towards fostering an environment of inclusive research. Knowing that the lack of representation of diverse populations in genetics research will hinder our understanding of disease etiology, it is clear that this is both an important ethical and scientific growth area for genomics research.
Supplementary Material
Table 1:
Listing of currently available imputation reference panels.
| Reference Panels | Haplotypes | Ancestries | Sites | Availability |
|---|---|---|---|---|
| TOPMed | 125,568 | African 32%, Asian 10%, European 40%, Hispanic 16% | 463,000,000 | forthcoming |
| Haplotype Reference Consortium (HRC; Version 1.1 2016) | 64,940 | predominantly European | 39,635,008 | *, ** |
| African Genome Resources | 9,912 | African populations + 1000 Genomes Project | 93,421,145 | ** |
| UK10K | 7,562 | British population | 24,128,798 | ** |
| 1000 Genomes Project Phase 3 (version 5) | 5,008 | African 26%, Admixed American 14%, East Asian 20%, European 20%, South Asian 20% | 85,167,453 | *, **, mathgen.stats.ox.ac.uk/impute |
| Consortium on Asthma among African-ancestry Populations in the Americas (CAAPA) | 1766 | Admixed African populations | 31,163,897 (autosomes only) | * |
| Genome of the Netherlands (GoNL) | 998 | Dutch population | ~20,000,000 | nlgenome.nl |
Note:
available via Michigan imputation server (https://imputationserver.sph.umich.edu).
available via Sanger imputation server (https://imputation.sanger.ac.uk). A listing of ongoing projects for imputation panels can be found in Supplemental Table S3.
Acknowledgements:
The authors acknowledge the support and helpful discussions with many members in the Psychiatric Genomics Consortium (PGC), which is supported by the National Institutes of Health (NIH) grants U01 MH109528, MH109539, MH109539, MH109536, MH109501, MH109514, MH109499, MH109532. REP is supported by NIH K01 grant MH113848. KK is supported by Wellcome Trust grant 212360/Z/18/Z. RKW is supported by NIH U01 MH094432. ABP is supported by a Postdoctoral Fellowship from the Stanford Center for Computational, Evolutionary, and Human Genomics (CEHG). RJS is supported by a UKRI Innovation- HDR- UK Fellowship (MR/S003061/1). ARM is supported by NIH grant K99MH117229. MLP is supported in part by grant CONICYT FONDECYT 1181365. HH is supported by NIH K01DK114379, R21AI139012, and the Stanley Center for Psychiatric Research. LD was supported by UL1 TR001085 and Stanford Department of Psychiatry and Behavioral Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Material:
Includes Supplemental Methods I-VII and Supplemental Tables S1–S4.
References
- Ahmad M, Sinha A, Ghosh S, Kumar V, Davila S, Yajnik CS, and Chandak GR (2017). Inclusion of Population-specific Reference Panel from India to the 1000 Genomes Phase 3 Panel Improves Imputation Accuracy. Sci. Rep 7, 6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinhanmi MO, Biernacka JM, Strakowski SM, McElroy SL, Balls Berry JE, Merikangas KR, Assari S, McInnis MG, Schulze TG, LeBoyer M, et al. (2018). Racial disparities in bipolar disorder treatment and research: a call to action. Bipolar Disord. 20, 506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H, Sabatti C, Croen LA, Dispensa BP, Henderson M, et al. (2015). Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics 200, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, et al. (2018). Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun 9, 1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, and Cardon LR (2006). Evaluating coverage of genome-wide association studies. Nat. Genet 38, 659–662. [DOI] [PubMed] [Google Scholar]
- Beaton A, Hudson M, Milne M, Port RV, Russell K, Smith B, Toki V, Uerata L, Wilcox P, Bartholomew K, et al. (2017). Engaging Māori in biobanking and genomic research: a model for biobanks to guide culturally informed governance, operational, and community engagement activities. Genet. Med 19, 345–351. [DOI] [PubMed] [Google Scholar]
- Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu S-A, Abdellaoui A, Andlauer TFM, Beekman ATF, Berger K, Blackwood DHR, et al. (2017). Genetic effects influencing risk for major depressive disorder in China and Europe. Transl. Psychiatry 7, e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomba L, Walter K, and Soranzo N (2017). The impact of rare and low-frequency genetic variants in common disease. Genome Biol. 18, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick LA, Keller MC, Knopik VS, McGeary JE, and Palmer RHC (2019). Shared additive genetic variation for alcohol dependence among subjects of African and European ancestry. Addict. Biol 24, 132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BC, Asian Genetic Epidemiology Network Type 2 Diabetes Consortium, Ye CJ, Price AL, and Zaitlen N (2016). Transethnic Genetic-Correlation Estimates from Summary Statistics. Am. J. Hum. Genet 99, 76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium, Patterson N, Daly MJ, Price AL, and Neale BM (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MM, Susser E, Mall S, Mqulwana SG, Mndini MM, Ntola OA, Nagdee M, Zingela Z, Van Wyk S, and Stein DJ (2017). Using iterative learning to improve understanding during the informed consent process in a South African psychiatric genomics study. PLoS One 12, e0188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Chen J, Collins R, Guo Y, Peto R, Wu F, Li L, and China Kadoorie Biobank (CKB) collaborative group (2011). China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int. J. Epidemiol 40, 1652–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claw KG, Anderson MZ, Begay RL, Tsosie KS, Fox K, Garrison NA, and Summer internship for INdigenous peoples in Genomics (SING) Consortium (2018). A framework for enhancing ethical genomic research with Indigenous communities. Nat. Commun 9, 2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conomos MP, Reiner AP, McPeek MS, and Thornton TA (2018). Genome-Wide Control of Population Structure and Relatedness in Genetic Association Studies via Linear Mixed Models with Orthogonally Partitioned Structure (bioRxiv).
- Duncan LE, Pollastri AR, and Smoller JW (2014). Mind the gap: why many geneticists and psychological scientists have discrepant views about gene-environment interaction (G×E) research. Am. Psychol 69, 249–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Shen H, Gelaye B, Ressler KJ, Feldman MW, Peterson RE, and Domingue BW (2018). Analysis of Polygenic Score Usage and Performance in Diverse Human Populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, and McClintick JN (2018). Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol. Clin. Exp. Res 42, 2281–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume M, Cupak M, Keenan S, Rambla J, de la Torre S, Dyke SOM, Brookes AJ, Carey K, Lloyd D, Goodhand P, et al. (2019). Federated discovery and sharing of genomic data using Beacons. Nat. Biotechnol 37, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinsky KJ, Reshef YA, Finucane HK, Loh P-R, Zaitlen N, Patterson NJ, Brown BC, and Price AL (2019). Estimating cross-population genetic correlations of causal effect sizes. Genet. Epidemiol 43, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilly A, Suveges D, Kuchenbaecker K, Pollard M, Southam L, Hatzikotoulas K, Farmaki A-E, Bjornland T, Waples R, Appel EVR, et al. (2018). Cohort-wide deep whole genome sequencing and the allelic architecture of complex traits. Nat. Commun 9, 4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinde KE, Qi Q, Thornton TA, Liu S, Shadyab AH, Chan KHK, Reiner AP, and Sofer T (2019). Generalizing polygenic risk scores from Europeans to Hispanics/Latinos. Genet. Epidemiol 43, 50–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckerman D, Gurdasani D, Kadie C, Pomilla C, Carstensen T, Martin H, Ekoru K, Nsubuga RN, Ssenyomo G, Kamali A, et al. (2016). Linear mixed model for heritability estimation that explicitly addresses environmental variation. Proc. Natl. Acad. Sci. U. S. A 113, 7377–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellwege JN, Keaton JM, Giri A, Gao X, Velez Edwards DR, and Edwards TL (2017). Population Stratification in Genetic Association Studies. Curr. Protoc. Hum. Genet 95, 1.22.1–1.22.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich J, Heine SJ, and Norenzayan A (2010). The weirdest people in the world? Behav. Brain Sci 33, 61–83; discussion 83–135. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, Manolio TA, and Green ED (2018a). Prioritizing diversity in human genomics research. Nat. Rev. Genet 19, 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Bonham VL, and Ohno-Machado L (2018b). Enhancing diversity to reduce health information disparities and build an evidence base for genomic medicine. Per. Med 15, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, and Abecasis GR (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet 44, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Fang M, Jostins L, Umićević Mirkov M, Boucher G, Anderson CA, Andersen V, Cleynen I, Cortes A, Crins F, et al. (2017). Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium (2005). A haplotype map of the human genome. Nature 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, and McPeek MS (2014). Robust rare variant association testing for quantitative traits in samples with related individuals. Genet. Epidemiol 38, 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher J, Wong Y, Albers P, Wohns AW, and McVean G (2018). Inferring the ancestry of everyone. [Google Scholar]
- Kendler KS, and Gardner CO (2010). Interpretation of interactions: guide for the perplexed. Br. J. Psychiatry 197, 170–171. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Li Y, Lewis CM, Breen G, Boomsma DI, Bot M, Penninx BWJH, and Flint J (2015). The similarity of the structure of DSM-IV criteria for major depression in depressed women from China, the United States and Europe. Psychol. Med 45, 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. (2018). Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet 50, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichaev G, and Pasaniuc B (2015). Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. Am. J. Hum. Genet 97, 260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker K, and Appel EVR (2018). Assessing Rare Variation in Complex Traits. Methods Mol. Biol 1793, 51–71. [DOI] [PubMed] [Google Scholar]
- Lam M, Chen C-Y, Li Z, Martin A, Bryois J, Ma X, Gaspar H, Ikeda M, Benyamin B, Brown B, et al. (2018). Comparative genetic architectures of schizophrenia in East Asian and European populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Teslovich TM, Boehnke M, and Lin X (2013). General framework for meta-analysis of rare variants in sequencing association studies. Am. J. Hum. Genet 93, 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, and Posthuma D (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, and Keating BJ (2014). Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P-R, Kichaev G, Gazal S, Schoech AP, and Price AL (2018). Mixed-model association for biobank-scale datasets. Nat. Genet 50, 906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Li X, Wang X, Gazal S, Mercader JM, Neale B, Florez JC, Auton A, Price A, Finucane HK, et al. (2018). Estimating heritability of complex traits in admixed populations with summary statistics. [Google Scholar]
- Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. (2018). Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet 50, 1505–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, and Kohane IS (2016). Genetic Misdiagnoses and the Potential for Health Disparities. N. Engl. J. Med 375, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, and Donnelly P (2004). The effects of human population structure on large genetic association studies. Nat. Genet 36, 512–517. [DOI] [PubMed] [Google Scholar]
- Marigorta UM, and Navarro A (2013). High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 9, e1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Luna C, Loh P-R, South Asian Type 2 Diabetes (SAT2D) Consortium, SIGMA Type 2 Diabetes Consortium, and Price AL (2017). Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet. Epidemiol 41, 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, and Daly MJ (2019). Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51, 584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Gomez C, Felix JF, Estrada K, Peters MJ, Herrera L, Kruithof CJ, Duijts L, Hofman A, van Duijn CM, Uitterlinden AG, et al. (2015). Challenges in conducting genome-wide association studies in highly admixed multi-ethnic populations: the Generation R Study. Eur. J. Epidemiol 30, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersha TB, and Abebe T (2015). Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum. Genomics 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills MC, and Rahal C (2019). A scientometric review of genome-wide association studies. Commun Biol 2, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minster RL, Hawley NL, Su C-T, Sun G, Kershaw EE, Cheng H, Buhule OD, Lin J, Reupena MS, Viali S ‘itea, et al. (2016). A thrifty variant in CREBRF strongly influences body mass index in Samoans. Nat. Genet 48, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltke I, Grarup N, Jørgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT, et al. (2014). A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512, 190–193. [DOI] [PubMed] [Google Scholar]
- Mulder N, Abimiku A ‘le, Adebamowo SN, de Vries J, Matimba A, Olowoyo P, Ramsay M, Skelton M, and Stein DJ (2018). H3Africa: current perspectives. Pharmgenomics. Pers. Med 11, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SC, Romm JM, Doheny KF, Pugh EW, and Laurie CC (2017). Imputation-Based Genomic Coverage Assessments of Current Genotyping Arrays: Illumina HumanCore, OmniExpress, Multi-Ethnic global array and sub-arrays, Global Screening Array, Omni2.5M, Omni5M, and Affymetrix UK Biobank
- Novembre J, and Barton NH (2018). Tread Lightly Interpreting Polygenic Tests of Selection. Genetics 208, 1351–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M, and Kwiatkowski DP (2016). The ethics of sustainable genomic research in Africa. Genome Biol. 17, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persyn E, Redon R, Bellanger L, and Dina C (2018). The impact of a fine-scale population stratification on rare variant association test results. PLoS One 13, e0207677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Cai N, Bigdeli TB, Li Y, Reimers M, Nikulova A, Webb BT, Bacanu S-A, Riley BP, Flint J, et al. (2017a). The Genetic Architecture of Major Depressive Disorder in Han Chinese Women. JAMA Psychiatry 74, 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Edwards AC, Bacanu S-A, Dick DM, Kendler KS, and Webb BT (2017b). The utility of empirically assigning ancestry groups in cross-population genetic studies of addiction. Am. J. Addict 26, 494–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, and Goldstein DB (2016). Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine. Genome Biol. 17, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popejoy AB, and Fullerton SM (2016). Genomics is failing on diversity. Nature 538, 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race Ethnicity, and Genetics Working Group (2005). The use of racial, ethnic, and ancestral categories in human genetics research. Am. J. Hum. Genet 77, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden DM, Wilke RA, Kroemer HK, and Stein CM (2011). Pharmacogenomics: the genetics of variable drug responses. Circulation 123, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Chen W, and Larson NB (2018). From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat. Rev. Genet 19, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GE, Goldberg DP, Von Korff M, and Ustün TB (2002). Understanding cross-national differences in depression prevalence. Psychol. Med 32, 585–594. [DOI] [PubMed] [Google Scholar]
- Sirugo G, Williams SM, and Tishkoff SA (2019). The Missing Diversity in Human Genetic Studies. Cell 177, 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH-Y, et al. (2015). An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sul JH, Martin LS, and Eskin E (2018). Population structure in genetic studies: Confounding factors and mixed models. PLoS Genet. 14, e1007309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z-Z, and Lin D-Y (2015). Meta-analysis for Discovering Rare-Variant Associations: Statistical Methods and Software Programs. Am. J. Hum. Genet 97, 35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, Tang H, Hoffmann TJ, Ochs-Balcom HM, Caan BJ, and Risch N (2012). Estimating kinship in admixed populations. Am. J. Hum. Genet 91, 122–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries J, Tindana P, Littler K, Ramsay M, Rotimi C, Abayomi A, Mulder N, and Mayosi BM (2015). The H3Africa policy framework: negotiating fairness in genomics. Trends Genet. 31, 117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, et al. (2018). Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci 21, 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler E, Leong A, Liu C-T, Hivert M-F, Strawbridge RJ, Podmore C, Li M, Yao J, Sim X, Hong J, et al. (2017). Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 14, e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik GL, Graff M, Nishimura KK, Tao R, and Haessler J (2019). Genetic analyses of diverse populations improves discovery for complex traits. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitlen N, Pasaniuc B, Sankararaman S, Bhatia G, Zhang J, Gusev A, Young T, Tandon A, Pollack S, Vilhjálmsson BJ, et al. (2014). Leveraging population admixture to characterize the heritability of complex traits. Nat. Genet 46, 1356–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, and Pan W (2015). Principal component regression and linear mixed model in association analysis of structured samples: competitors or complements? Genet. Epidemiol 39, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shen X, and Pan W (2013). Adjusting for population stratification in a fine scale with principal components and sequencing data. Genet. Epidemiol 37, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.