Summary
Francisella bacteria rely on a phylogenetically distinct Type VI Secretion System (T6SS) to escape host phagosomes and cause the fatal disease tularemia, but the structural and molecular mechanisms involved are unknown. Here, we report the atomic structure of the Francisella T6SS central spike complex, obtained by cryo-electron microscopy. Our structural and functional studies demonstrate that, unlike the single-protein spike composition of other T6SS subtypes, Francisella T6SS’s central spike is formed by two proteins, PdpA and VgrG, akin to T4-bacteriophage gp27 and gp5, respectively, and that PdpA has unique characteristics, including a putative cargo within its cavity and an N-terminal helical lid. Structure-guided mutagenesis demonstrates that the PdpA N-terminal lid and C-terminal spike are essential to Francisella T6SS function. PdpA is thus both an adaptor, connecting VgrG to the tube, and a likely carrier of secreted cargo. These findings are important to understanding Francisella pathogenicity and designing therapeutics to combat tularemia.
Keywords: Type 6 Secretion System, central spike, cryo electron microscopy, Francisella, VgrG, PdpA, intracellular pathogen, contractile injection system
Graphical Abstract
eTOC Blurb
Type 6 secretion systems, critical to virulence of many bacteria, including Francisella, drive a central spike into target cells. Xue et al. used cryo-EM to determine the Francisella central spike atomic structure, revealing features absent from canonical T6SS, including an α-helical lid and putative cargo within its head cavity.
Introduction
Francisella tularensis is a highly infectious Gram-negative intracellular pathogen that causes the potentially fatal zoonotic disease tularemia in humans and animals (Ellis et al., 2002). The pathogenicity of F. tularensis relates to its ability to evade host cell defense systems, including preventing fusion of its phagosome with lysosomes, escaping from its phagosome, and ultimately replicating in the host cell cytoplasm (Chong and Celli, 2010; Clemens and Horwitz, 2007; Clemens et al., 2004), but the molecular mechanisms underlying these capabilities are not fully understood. It has been established that the gene cluster called the Francisella Pathogenicity Island (FPI), which encodes a non-canonical Type VI secretion system (T6SS), is critical for phagosomal escape and intracellular replication (Broms et al., 2010; Nano et al., 2004).
The FPI-encoded T6SS has been classified as a phylogenetically distinct T6SS subtype (T6SSii) for two reasons. First, many proteins of other T6SSs have no homologs identified in F. tularensis. Second, the level of sequence homology between known homologs of the F. tularensis T6SS and other T6SSs is extremely low (Russell et al., 2014). Several components of the FPI-encoded apparatus whose structures have been solved to date show strong structural homology to components of other T6SSs (Clemens et al., 2015; de Bruin et al., 2011; Robb et al., 2012). Nevertheless, the structure and function of the FPI-encoded T6SS is understood even less than that of other bacteria (Clemens et al., 2018).
F. tularensis VgrG has homology with the “gp27-like” and “gp5-like” T4 phage tail-spike proteins, but it is much smaller (17.5 kDa) than the VgrGs of other T6SSs (typically ~120 kDa). In addition, no protein with sequence homology to the PAAR-repeat motif that typically binds to VgrG in other systems has been identified in F. tularensis, although FPI proteins IglG and IglF have been proposed as a PAAR-like orthologue and associated effector protein, respectively (Rigard et al., 2016). While the tail-spike motif of VgrG is presumed to have membrane puncturing activity, it is unclear if this activity is sufficient to lyse the phagosome and mediate bacterial escape. Pathogenicity determinant protein A (PdpA), encoded by the pdpA gene, is the first cistron in the FPI and has been shown to be essential for the bacterium to escape the phagosome, replicate intracellularly, and cause disease in animals (Broms et al., 2010). Negative staining electron microscopy of purified PdpA and VgrG has revealed a “racket like” appearance structurally similar to the multi-domain VgrG proteins of other T6SSs, suggesting that PdpA serves as a gp27-like protein to cap VgrG (Eshraghi et al., 2016). However, an atomic structure is essential to reveal the interactions between VgrG and PdpA and between these proteins and the rest of the Francisella T6SS and to gain insights into how the apparatus enables the bacteria to lyse and escape from the phagosome.
F. novicida has considerable homology with F. tularensis, but it has only a single copy of the FPI. F. novicida has low virulence in humans and thus serves as a more practical subspecies for study. In this study, using cryo-electron microscopy (cryoEM), we present two structures of the Francisella central spike at up to 3.98 Å resolution for the complex of PdpA and VgrG, two virulence components of the FPI-encoded T6SS. Surprisingly, the structure of the complex shows a unique N-helical-lid domain of PdpA on the top of a T4-phage-like central spike complex structure and reveals the presence of a cargo molecule within the PdpA cavity.
Results
Isolation and overall structure of PdpA-VgrG complex
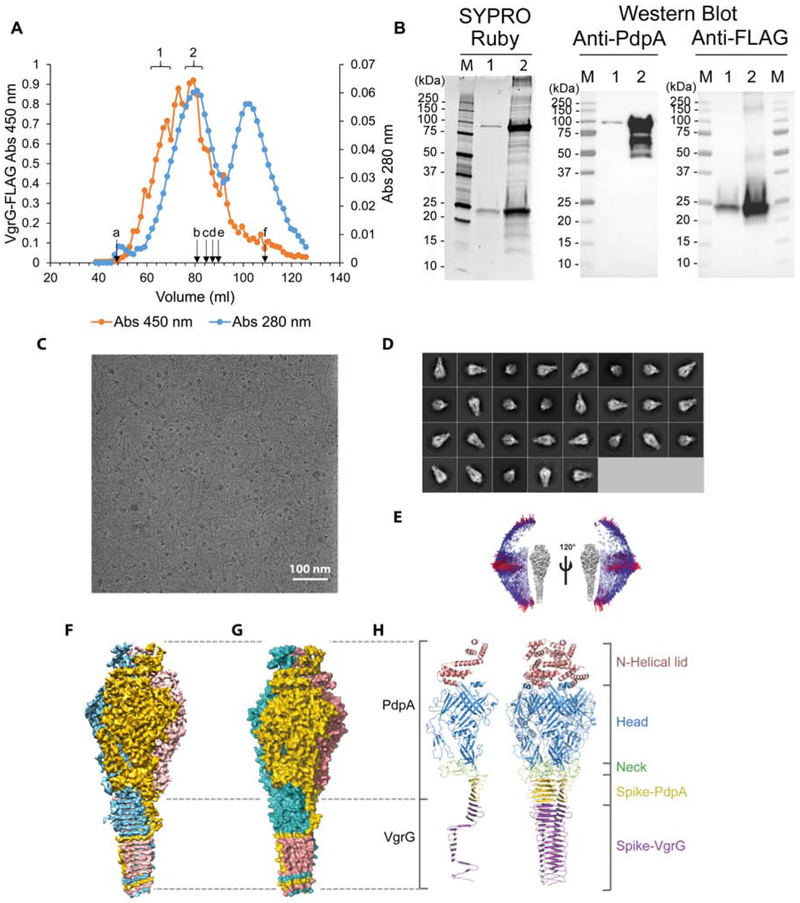
We cultured FLAG epitope-tagged VgrG-expressing strains of F. novicida in Trypticase Soy Broth, containing 5% KCl, to induce T6SS formation (Clemens et al., 2015), then purified VgrG by performing affinity chromatography using anti-FLAG agarose and gel filtration chromatography. In gel filtration chromatography, the FLAG-immunoreactive material correlated well with the peak of UV-absorbance but showed a broad elution profile, indicating size heterogeneity (Figure 1A). SDS-PAGE analysis of the purified material showed protein staining in two dominant bands at 95 kDa and 25 kDa, which, based on Western immunoblot staining, respectively correspond to PdpA and FLAG-VgrG (Figure 1B). The anti-PdpA monoclonal antibody used in our Western immunoblot binds to the C-terminus (Schmerk et al., 2009) and reveals some N-terminal degradation. However, the more quantitative SYPRO Ruby protein stain indicates that the extent of PdpA cleavage is minor (Figure 1B). Mass spectrometry-based proteomics analysis revealed that the dominant proteins in the sample were PdpA and VgrG (Table S1), consistent with our SDS-PAGE protein staining and Western immunoblotting results. To our surprise, we also identified in the sample the hypothetical protein FTN_0038 (Table S1), a VgrG-like protein encoded in the Francisella novicida island (FNI) (Rigard et al., 2016). With only 94 amino acid residues, this protein (FNI VgrG) is even smaller than the FPI VgrG (164 amino acid residues). While the structure of FTN_0038 (FNI-VgrG) has not been determined, its amino acid sequence shows 35.7% identify with the FPI-VgrG (E value 5 × 10−4). JPred4 (Drozdetskiy et al., 2015) and I-Tasser (Yang et al., 2015) predict it to have a secondary structure composed of repeating 5 – 6 residue β-strands separated by 2 – 3 residue coils or β-turns, consistent with the β-helical structure of VgrG.
Figure 1. Structure determination and overall structure of the PdpA-VgrG complex.
(A) Gel elution profile of FLAG-VgrG immunoreactive material on Sephacryl S400. Lettered arrows indicate elution positions of calibration markers: blue dextran (a, 2,000 kDa), thyroglobulin (b, 669 kDa), apoferritin (c, 443 kDa), beta-amylase (d, 200 kDa), alcohol dehydrogenase (e, 150 kDa), and acetone (f, 58 Da). Brackets 1 and 2 indicate fractions pooled for further analysis. Data shown are representative of two independent experiments.
(B) Fractions indicated by brackets 1 and 2 in (A) were pooled, concentrated with a 100 kDa MWCO filter, and evaluated by SYPRO Ruby protein stain and Western Immunoblotting using monoclonal antibodies directed against PdpA and the FLAG epitope. Data shown are representative of two independent experiments.
(C) A drift-corrected cryoEM micrograph of the PdpA-VgrG complex. Scale bar, 100 nm.
(D) Representative 2D class averages of the PdpA-VgrG complex obtained in Relion-2.1.
(E) Angular distribution of particles used for reconstruction of the lidded map of the PdpA-VgrG complex.
(F) Surface representation of the cryoEM density map of the PdpA-VgrG complex colored by individual protomers, each comprising a single PdpA-VgrG heterodimer.
(G) Atom surface representation of the atomic model of the PdpA-VgrG complex colored by individual protomers each comprising a single PdpA-VgrG heterodimer.
(H) Side view of a cartoon representation of the overall structure of a single PdpA-VgrG protomer (left) and a trimeric PdpA-VgrG complex (right). The N-Helical-lid, head domain, neck domain, spike of PdpA and spike of VgrG are colored pink, blue, green, yellow and purple, respectively. See also Figures S1, S2, S3, and Video S1.
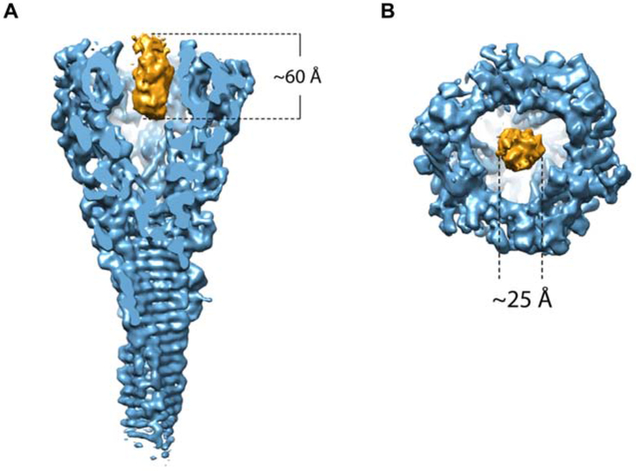
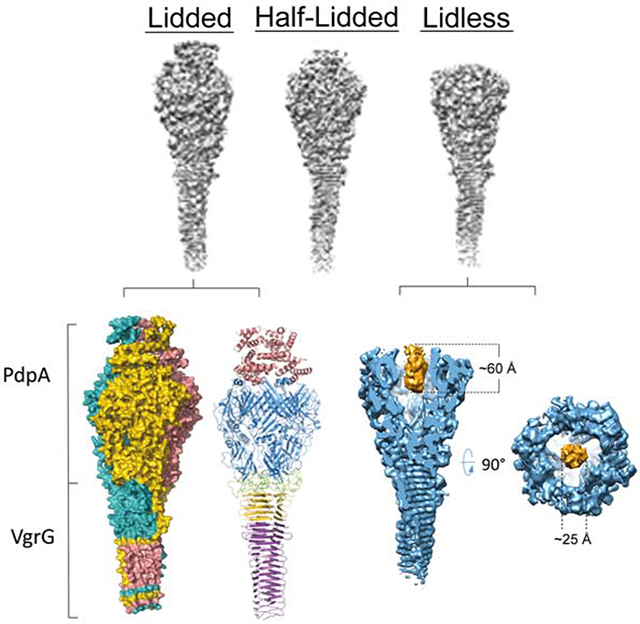
CryoEM revealed that our purified sample contains club-shaped particles with a globular head and a shaft of lengths varying from 600 to 3,000 Å (Figure 1C), similar to those of the VgrG-PdpA complex seen in negative stain transmission electron microscopy (TEM) images (Eshraghi et al., 2016). 2D classification of side views of the head reveals that the head also varies, with two main populations, one with a flat top and the other with an additional fuzzy density extending beyond the flat top (Figure 1D). Using electron-counting cryoEM with a Titan Krios and single-particle reconstruction with 3D classification, we obtained three different structures without imposing any symmetry (i.e., C1): the first with a lid (“lidded,” from 3,765 particles), the second with half of a lid (“half-lidded,” from 10,343 particles) and the third without lid (“lidless,” from 16,648 particles), at resolutions of 5.59 Å, 4.51 Å and 4.51 Å, respectively. Imposition of 3-fold symmetry (C3) further improved the average resolutions to 4.35 Å, 3.98 Å, and 4.21 Å, respectively (Figure S1A). The map was of sufficient quality for model building of major parts of the protein (Figure S2). In all three structures, the head is funnel-shaped with a central cavity. In the lidded structure, the lid covers this central cavity, but in the lidless (C1 symmetry) structure, the central cavity contains a cylindrical density plug with a height of 60 Å and a diameter of 25 Å (Figure 2A,B and Video S2), indicating that a “cargo” molecule might be loaded in the cavity of PdpA.
Figure 2. Surface representation of the cryoEM density map of the PdpA-VgrG central spike complex with a central plug within the PdpA cavity.
Side view (A) and top view (B) of the plug density of the PdpA-VgrG map. See also Video S2.
To establish the molecular identities in the complex, we first examined the half-lidded structure, since it had the highest resolution. It shows clear secondary structure elements with some side chain densities visible, providing landmarks for building atomic models. Matching the secondary structure elements and visible side chain densities with elements predicted from amino acid sequences for mass spectrometry-identified proteins (Table S1) indicated that this complex contains both PdpA and VgrG; hereafter, it will be designated as the PdpA-VgrG complex.
The structure of the PdpA-VgrG complex has a trimeric arrangement composed of 3 PdpA-VgrG heterodimers (Figure 1F,G, and Video S1). Each protomer of the trimeric complex consists of four major domains, with the lower part of the spike from VgrG (residues 3-135) and everything else from the PdpA monomer [N-helical lid (residues 1-169), head (residues 178-642, 691-714 and 734-750), neck (residues 643-690, 715-733 and 751-762) and upper part of the spike (residues 763-817)] (Figure 1H). Thus, our final atomic model of the PdpA-VgrG monomer includes 133 of 164 residues (residues 3-135) of VgrG and 717 of 820 residues of PdpA. As expected, the head, neck and spike domains bear great structural resemblance to the bacteriophage T4 tail spike, which comprises of gp27 and gp5. However, the lid in our PdpA-VgrG complex has no structural counterpart in the T4 phage tail spike complex (Taylor et al., 2016). Figure S3 shows that the F. novicida central spike complex closely resembles T4 phage and Pseudomonas aeruginosa central spikes, except for the presence of a taller head and the N-helical lid.
The N-helical lid is unique to Francisella T6SS
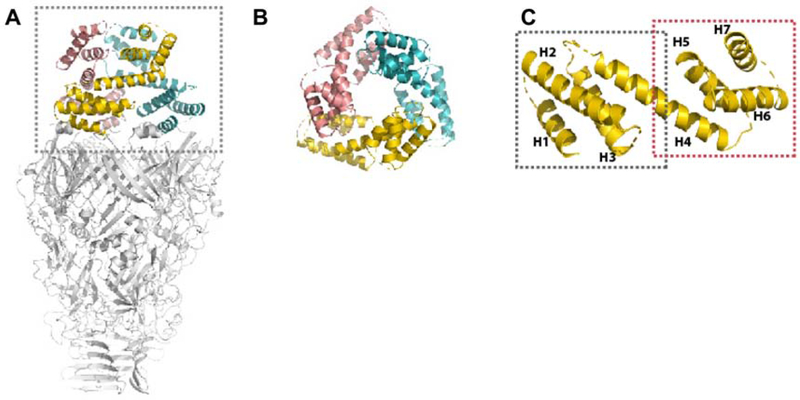
The N-helical-lid of the model (residues 1-169; Figure 3A,B) consists of 7 helices (Figure 3C); each monomer of the N-helical lid forms a dumbbell-like structure, with two triple-α-helical bundles connected by a long helix. Secondary structure prediction of the PdpA amino acid sequence, using the PSIPRED server (Buchan et al., 2013), indicates a high probability of helices in residues 1-169, further supporting this model. The result of 3D classification shows that only 10.5% of the particles have the whole N-terminal-lid, while 28.9% of particles have half of the N-helical-lid (Figure S1A). Because the density of the lid is not as well resolved as the densities in the head and neck domains, side-chains are not assigned in this region. SDS-PAGE of the sample showed a major band at 90 kDa corresponding to the expected molecular weight of PdpA (Figure 1B), indicating dynamic motion, but not degradation, of the N-helical lid. The N-helical lid is not present in other VgrG or gp27 homologous structures. A search through the Dali server (Holm and Rosenstrom, 2010) did not identify any proteins with strong homology corresponding to all 7 helices of residues 1-169. Numerous eukaryotic and some prokaryotic proteins show structural homology to the helices on either side of the PdpA lid “dumbbell” (e.g. residues 1 – 75 or residues 70 – 169), but the significance of this homology is unclear because of the relative simplicity of the structural fold, consisting of 4 helices. For example, the structure with the highest homology identified by the Dali server was yeast RNA helicase EIF4A (pdb 2vsx.E; Z-score 7.5, rmsd 4.2), which aligns with the central bar helix and the 3 dumbbell helices on one side. Intriguingly, the small Legionella pneumophila Type IV effector protein, Lem22 (pdb 5wd9), aligns well with part of the central bar and two of the three of the a-helices on one side of the dumbbell; its best structural alignment was with residues 70 – 169 (Z-score 5.3, rmsd 2.1), which also includes a 41-residue region with sequence similarity (24% identity, 51% similarity, E value 0.09). The function of the T4SS secreted effector protein Lem22 is unknown, but Kozlov et al. speculated that it might suppress host cell apoptosis, thereby allowing more rounds of L. pneumophila intracellular replication (Kozlov et al., 2018). The L. pneumophila Type IV and Francisella Type VI secretion systems are known to have homologous DotU and IcmF (PdpB) membrane complex proteins and, based on this evidence, could plausibly share an effector domain.
Figure 3. Structure of the N-Helical-lid.
(A) Side view of a PdpA trimer, with its N-helical lid colored and enclosed within the dotted box.
(B) Top view of the trimeric N-helical lid.
(C) Ribbon diagram of a monomeric unit of the trimeric N-helical-lid. The regions of residues 1-75 and 70-169 are marked by a grey and red dashed box, respectively. The 7 helices present in the N-helical-lid are labeled as H1 to H7.
The trimeric head domain has a helix on top and a positively charged cavity.
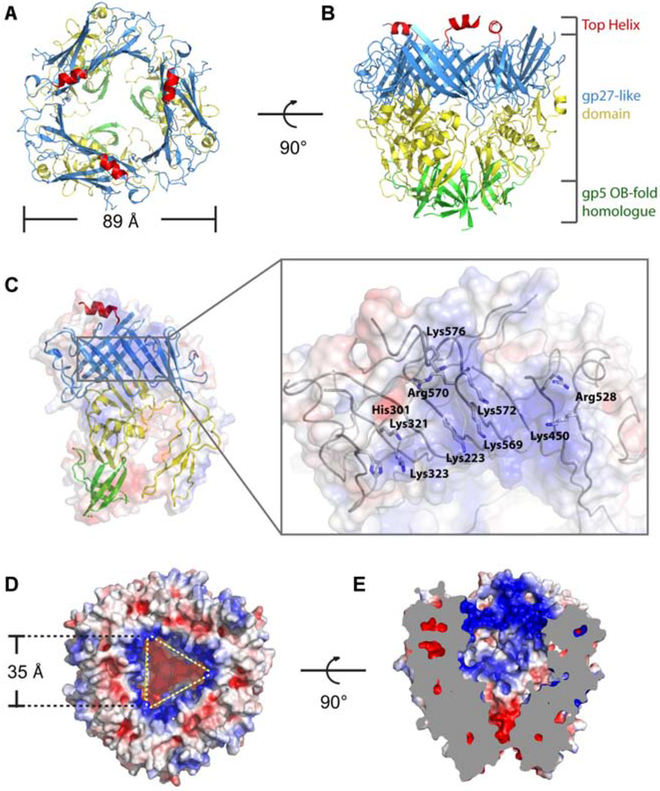
The head domain forms a funnel-shaped structure, with an external diameter of 89 Å and three 2-turn helical appendages at the top (Figure 4A,B and Video S3). The funnel is comprised of 3 stacked rings. The upper and middle rings comprise the gp27-like domain, while the lower ring is a homologue of the OB-fold region of the gp5 domain. The most significant difference from other gp27-like domains is the helix (residues 570-581) atop each gp27-like domain.
Figure 4. Structure of the head.
(A, B) Top (A) and side (B) views of the head. The top helix, upper ring, middle ring and lower ring are colored red, blue, yellow and green, respectively. (C) Electrostatic surface representation of the monomeric head domain showing the inner surface of the funnel. The electrostatic surface is shown semi-transparently, superposed with the ribbon diagram of the atomic model. Inset: atomic model of the positive charged “belt” region is shown as grey wires with key residues labeled. (D, E) Top (D) and side cut-away (E) views of electrostatic surface potential distribution of the head showing the cavity. See also Video S3.
The upper ring consists of 11 β-strands and 4 α-helices from each monomer of the head domain. The triangular-shaped β-barrel inner layer is formed of 24 β-strands; the outer layer is comprised of a 3-stranded antiparallel sheet and 2 short α-helices from each monomer, with an α-helix lying on the top of the inner layer of each monomer. The middle ring consists of 4 α-helices and 3 antiparallel sheets from each monomer. The lower ring is formed by a 5-stranded antiparallel sheet from each monomer. The helix on the top protrudes from each monomer, and may provide a docking site for the N-helical-lid.
We calculated the electric potential distribution of the lining of the cavity and found a positively charged “belt” at the entrance of the cavity (Figure 4C). This “belt” consists of 10 positively charged residues: Lys223, His301, Lys321, Lys323, Lys450, Arg528, Lys569, Arg570, Lys572, and Lys576. These residues form a positively charged ring inside the cavity (Figure 4D,E), the surface of which may be important for binding and carrying the aforementioned “cargo" molecule (Figure 2A,B, and Video S2).
Because of the limited resolution (4.5 Å) of the plug structure inside the PdpA cavity, we have been unable to determine the identity of this molecule. Based on its dimensions (60 Å × 25 Å), we estimate its size to be approximately 10 kDa, which is smaller than any previously identified Francisella T6SS effector (Eshraghi et al., 2016) and therefore potentially suggests a Francisella T6SS effector different from any of the previously reported effectors. Our mass spectrometry data did not identify any protein of appropriate size and abundance that would correspond to the plug molecule. While monomeric FNI VgrG (FTN_0038) is of the correct size and relative abundance in our samples (10 kDa and 1/3rd as abundant as FPI VgrG), FNI VgrG is expected to form a trimeric β-helical structure, which would be too large and would have C3 symmetry, which is inconsistent with the C1 symmetry that we observe for the plug molecule. Since we observe the plug only in the lidless structure, we cannot rule out the possibility that it is formed from the N-terminus of one PdpA monomer, rather than from a separate protein. In this scenario, while the N-terminus of one PdpA monomer forms the plug, the N-termini of the other two would be disordered. We also cannot rule out the possibility that the plug is a non-peptide molecule. While it has been recognized that cargo molecules or effectors may be packaged between the central spike and baseplate cavity (Nazarov et al., 2018) and within the Hcp tube (Silverman et al., 2013; Whitney et al., 2014), to our knowledge, this is the first direct observation of a putative T6SS cargo molecule packaged within the cavity of the gp27-like domain of the central spike.
The spike domain is comprised of VgrG and the C-terminus of PdpA
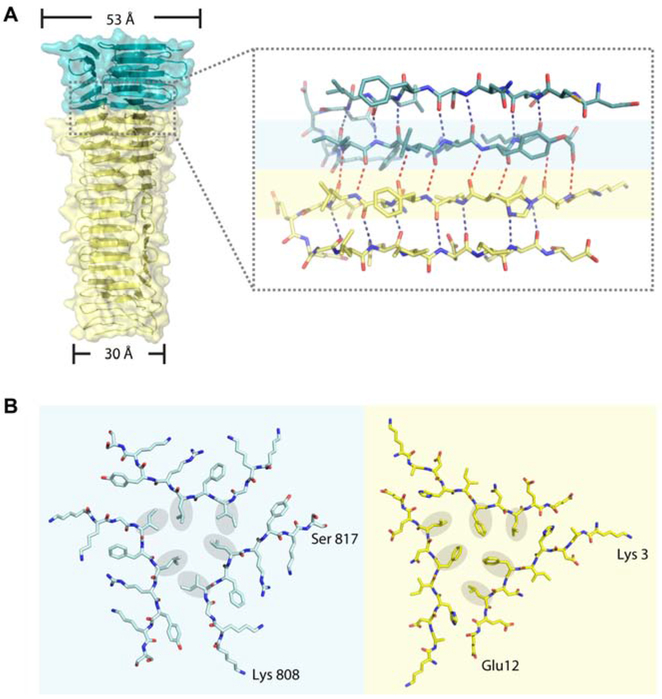
In our model, the spike domain is an integrated triple-stranded β-helix with a twisted triangular prism shape, comprised of 21 β-strands from each heterodimer of PdpA-VgrG. The width of the helix tapers gradually, from 53 Å at the top to 30 Å at the bottom (Figure 5A). The β-helix from PdpA consists of trimeric 5-stranded antiparallel sheets. Each of the three faces of the VgrG prism is composed of a long β-sheet with 16 strands, 12 antiparallel and 4 parallel, and each β-sheet comprises 6 strands from one of the three monomers, 2 strands from the second, and 8 strands from the third.
Figure 5. Binding interface analysis of PdpA and VgrG.
(A) Side view of the spike showing detailed interactions between PdpA (teal) and VgrG (yellow). In the inset, oxygen and nitrogen atoms are colored red and blue, respectively; intra- and inter-molecular hydrogen bonds are shown by blue and red dotted lines, respectively.
(B) Stick representation of the bottom strands of the PdpA trimer (left, corresponding to residues on blue background in Figure 4A) and the top strands of the VgrG trimer (right, corresponding to residues on yellow background in Figure 4A) at their boundary. The hydrophobic residues are marked with grey oval background to show the “hydrophobic triangles”. The interfaces among both the PdpA strands and the VgrG strands are characterized by a mixture of hydrophobic interactions and hydrogen bonds.
The C-terminal face of the trimeric β-helix from PdpA (residues 763-820) is connected to the N-terminal face of the trimeric β-helix from VgrG. Each monomer forms eight backbone hydrogen bonds between PdpA and VgrG (Figure 5A). Residues 808-817 of the PdpA trimer (on blue background in Figure 5A,B) and residues 3-12 of the VgrG trimer (on yellow background in Figure 5A,B) form flat interfaces (Figure 5B). Six residues with hydrophobic sidechains from PdpA (the Ile 811 and Leu 812 from each PdpA monomer) form a hydrophobic triangle in the flat interface of PdpA, and six residues with hydrophobic sidechains from VgrG (Phe8 and Leu10 from each VgrG monomer) form another hydrophobic triangle in the flat interface of VgrG. The interactions between the two hydrophobic triangles (residues on grey background in Figure 5B) further stabilize the PdpA-VgrG interface. The buried surface area, calculated by AREAIMOL (Winn et al., 2011), is 1360.45 Å2 for the trimeric PdpA-VgrG interface. The interactions between PdpA and VgrG were analyzed using PDBePISA Assembly (Krissinel and Henrick, 2007). The Complex Formation Significance Score (CSS) for the interaction between the PdpA monomer and the VgrG monomer is 0.055, whereas the CSS value for the interaction between the PdpA trimer and the VgrG trimer is 1, indicating that PdpA and VgrG must form trimers in order to undergo complex assembly.
We expressed and purified FLAG-tagged VgrG G164R, which forms trimers but not long needles (Eshraghi et al., 2016), and PdpA in E. coli (Figure S4A,B), then examined the interaction between PdpA and VgrG by ELISA. As shown in Figure S4C, we were able to demonstrate binding of FLAG-tagged VgrG G164R to PdpA-coated ELISA wells.
The cryoEM density map corresponding to the C-terminal region (residues 136-164) of VgrG is of lower resolution than other parts of the model and therefore does not provide structural information to explain the length heterogeneity of the long filament forms in our sample (i.e., the heterogeneity of filament lengths in Figure 1C). The width of the β-helix in the N-terminal region of VgrG is greater than that in the C-terminal region; the different sizes of the VgrG β-helix’s two ends would make it difficult to form a long filament by a “head-to-tail” interaction, but “head-to-head” and “tail-to-tail” interactions would be possible. Meanwhile, the mass spectrometry data of the sample shows that, while the two most abundant proteins are PdpA and VgrG, the 3rd most abundant protein is a short (94 amino acid) VgrG-like protein (FNI-VgrG) from the Francisella novicida island (Table S1). Normalized Spectral Abundance Factor (NSAF) analysis (Florens et al., 2006) indicates that the relative abundance of FNI-VgrG:FPI-VgrG:PdpA is 1:3.3:2.2 (Table S1). Thus, the protein complex is most likely composed mainly of PdpA-VgrG, but some additional VgrG trimers interact with PdpA-VgrG, making the needles longer. Some of the additional length of our needles may also come from PdpA-VgrG’s interacting with the trimeric FNI-VgrG. While the FNI-VgrG may form complexes with PdpA-VgrG, it is unlikely to be of any functional importance in mammalian infections, since Rigard et al. have demonstrated that deletion of the FNI does not impair virulence either in vitro in macrophage cell culture or in vivo in a mouse infection model (Rigard et al., 2016).
PAAR proteins bind to the distal (C-terminal) end of the T6SS central spike complexes of Vibrio cholerae and P. aeruginosa and to gp5 of T4 phage. The PAAR proteins form conical extensions, sharpening the spike and serving as adaptors for binding of additional effectors to the spike (Shneider et al., 2013). No PAAR protein has been identified in the Francisella genome, but homology modeling predicts that IglG is a PAAR-like protein and may fulfill this role in the Francisella T6SS (Rigard et al., 2016). The distal surface of the T4 phage gp5 and V. cholerae and P. aeruginosa VgrG proteins are β-strand triangles with a hydrophilic exterior and a central hydrophobic patch that interact strongly with complementary surfaces of their respective PAAR proteins (Shneider et al., 2013). The distal surface of F. novicida VgrG also consists of a β-strand triangle, but the hydrophobic residues are deep and not surface exposed (Figure S3C). The β-strand triangle would be capable of interacting with a complementary β-strand of a PAAR-like protein such as IglG. The absence of a central hydrophobic patch may lead to a weaker interaction between F. novicida VgrG and its PAAR-like protein. The interaction between Francisella VgrG and its PAAR-like protein is probably stabilized by interaction with additional proteins within the baseplate, such that Francisella VgrG and IglG dissociate after secretion or during purification.
Structure guided mutagenesis of PdpA
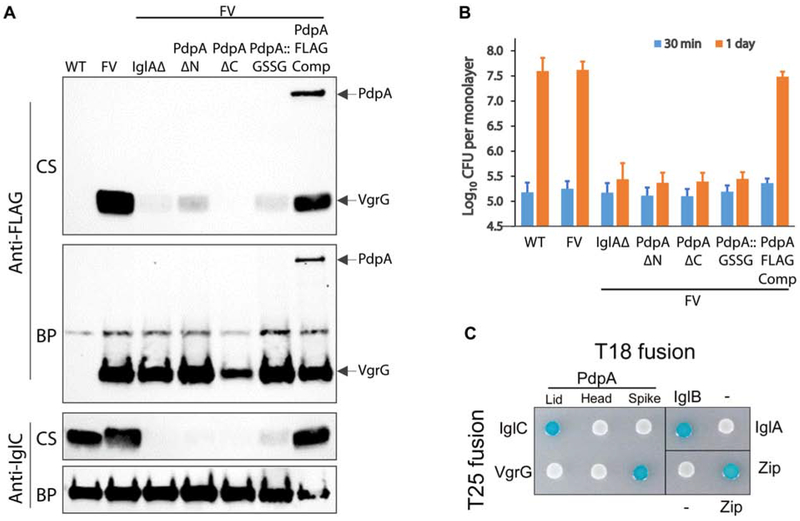
To evaluate whether the N-terminal lid and the C-terminus of PdpA are important to biological function, we prepared FLAG-VgrG expressing strains of F. novicida with N-terminal (residues 2-175) and C-terminal (residues 762-820) truncations in PdpA. We observed that deletion of either the N-terminus or the C-terminus eliminates secretion of FLAG-VgrG and IglC (Figure 6A) and also severely impedes intracellular replication within macrophages (Figure 6B). We observed that the PdpA sequence included a possible calmodulin binding site at residues 68 – 75 (GLISKLDY). To explore the functional importance of this sequence, we replaced these 8 amino acids with 2xGSSG, which is predicted by JPRED (Drozdetskiy et al., 2015) to have no impact on the alpha-helical structure. We expressed and purified the recombinant His-tagged PdpA::GSSG protein from E. coli and confirmed that it retained its capacity to bind VgrG in our ELISA assay. However, we found that neither wildtype PdpA nor the PdpA::GSSG protein were bound by a calmodulin-agarose column (data not shown). While this substitution did not adversely impact the binding of PdpA to VgrG in our ELISA assay (Figure S4C), we observed that the PdpA::GSSG substitution mutation negatively impacts both secretion of FLAG-VgrG and IglC and intracellular growth in macrophages (Figure 6A,B), indicating that relatively subtle changes in the helical lid region of PdpA severely disrupt biological function.
Figure 6. Structure-guided mutagenesis of lid, head, and spike of PdpA.
(A) Secreted products of F. novicida growing in broth with high KCl. Culture supernates (CS) and bacterial pellets (BP) were evaluated by Western immunoblotting. While the F. novicida wildtype (WT) and the strain expressing FLAG-VgrG from the chromosome (FV) secrete IglC and VgrG in response to high KCl, the capability to secrete IglC and VgrG is lost in FV strains lacking IglA (IglAΔ), with N-terminal truncation of PdpA (PdpA ΔN), with C-terminal truncation of PdpA (PdpA ΔC), or with the PdpA GSSG substitution mutation of residues 68 - 75 (PdpA::GSSG). Both VgrG and PdpA are secreted by FV strain in which the truncated PdpA is complemented with a FLAG-tagged PdpA (PdpA FLAG Comp).
(B) Strains defective in Type VI secretion are unable to replicate intracellularly in human THP-1 like macrophages. Strains are designated using the same nomenclature as in (A). Data shown are the means and standard deviations of three independent experiments each with biological triplicates.
(C) Bacterial-two-hybrid analysis shows interaction of the PdpA lid domain with IglC and of the PdpA spike domain with VgrG. Interactions between the head domain and IglC or VgrG were not observed. Positive controls: IglA-IglB and Zip-Zip constructs. Negative controls: IglA-null and Zip-null constructs.
See also Figure S4.
Our atomic model predicts that the N-terminal portion of PdpA corresponds to gp27, serving as a tube initiator, and that the C-terminal portion interacts with VgrG. Therefore, we examined the interaction of PdpA with the Francisella tube protein, IglC, by using a bacterial two-hybrid (BACTH) system in which protein-protein interaction leads to β-galactosidase production and formation of blue colored reporter bacteria in presence of X-gal. We fused amino acids of PdpA corresponding to its N-terminal lid, head domain or spike to the T18 domain and Francisella IglC or VgrG to the T25 domain of adenylate cyclase and interrogated their interactions using the BACTH system. Results from the study confirmed our atomic structure that PdpA spike interacts with VgrG, as evident by the blue spot of reporter bacteria harboring VgrG and PdpA spike. Moreover, we observed that the N-terminal helical lid, but not the head or spike domains of PdpA, interacts with IglC, consistent with the N-terminal helices playing a role in IglC tube initiation (Figure 6C).
Discussion
The present study describes the cryoEM structures of the PdpA-VgrG complex. We show that the Francisella T6SS central spike comprises two proteins, PdpA and VgrG, rather than the single VgrG protein as in the canonical T6SS. Bacteriophage T4 and R-pyocins also have two proteins (e.g., gp27 and gp5 in the case of T4 phage) that correspond to PdpA and VgrG, respectively; however, PdpA includes part of the spike domain, which in contractile phage and R-pyocins is encompassed entirely in the gp5-like protein. The division of the Francisella T6SS central spike into two proteins is most likely the result of a gene insertion event, i.e., insertion of the pdpB and iglE genes (tssM and tssJ orthologues, respectively) between the pdpA and vgrG genes.
Our results also demonstrate the following three remarkable features of the Francisella T6SS central spike: First, the N-helical lid, which contains a dumbbell-like structure with 7-α helices, is unique among all other known gp27-like structures. Second, according to the 3D classification results and SDS-PAGE, there are different structural states of the PdpA-VgrG complex with regard to the N-helical-lid—lidded, half-lidded, and lidless. The status of the N-helical-lid may control the binding of the PdpA-VgrG complex to other components of the T6SS in Francisella. Third, the lidless (C1 symmetry) structure contains a cylindrical density plug in the cavity.
The N-helical lid of PdpA is unique among all known gp27-like structures. In most cases, the gp27-like domain serves as an adaptor protein, with its small funnel-shaped trimer at the bottom connecting with the needle tip and its pseudo-6-fold wider funnel top interacting with the Hcp hexamer and triggering assembly of the phage tail-like structure. However, in our PdpA-VgrG structure, the N-helical lid would prevent interaction with the Hcp homologue when the lid sits on the top. We speculate that PdpA operates in two different modes: “lid on” and “lid off’. The cargo molecule can load into the cavity in the “lid off’ status; the loaded cargo keeps the lid open and allows Hcp homologue molecule (IglC) binding and efficient assembly. As our SDS-PAGE results indicate that PdpA is mostly uncleaved, the invisibility of the lid domain in the “lidless” structure suggests the existence of a flexible lid when cargo is loaded. As such, the N-terminus of the “lidless” structure assumes a conformation that interacts with the Hcp homologue. Our B2H analysis is consistent with interaction between IglC and the PdpA lid and with the IglC-lid interaction stabilizing the structure of the N-terminal helices in the assembled pre-contraction structure. Whether the alpha-helical lid structure has any effector function will require additional studies.
Intriguingly, we observed that a second form of our affinity-purified central spike complex preparation contains a cylindrical density plug. However, the limited resolution (4.5 Å) for this “plug” density molecule has precluded us from determining its identity. We also compared the map of the second and the third subset from the 3D classification result and found some clashes between the 5th helix and the density of the cargo molecule. These clashes suggest that the loaded cargo may keep the N-helical-lid open, or require the lid to assume a different configuration. Should we be able to confirm experimentally the possibility of the F. novicida central spike complex packing an effector protein as cargo within the PdpA cavity, it would open an exciting venue to investigate how this effector functions in F. novicida pathogenicity.
In conclusion, we have determined the atomic structure of the virulence associated PdpA-VgrG complex from the FPI-encoded T6SS in Francisella. Our atomic structure of the F. novicida T6SS and structure-based biological assays reveal the critical importance of the PdpA-VgrG complex for phagosomal escape, and reveal that PdpA not only acts as a structural adaptor in the T6SS complex, but also, that it could serve as a carrier and effector. This atomic model will facilitate the design and testing of therapeutics targeting F. tularensis.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcus A. Horwitz (MHorwitz@mednet.ucla.edu). Materials generated in this study will be made available on request, but payment for shipping and a completed Materials Transfer Agreement may be required.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
All F. novicida strains are derived from F. novicida Utah 112 obtained from Karl Klose, University of Texas, San Antonio. Strains were prepared as glycerol stocks and stored frozen at −80°C until used in experiments.
Human THP-1 monocytic cells
Prior to use in infection assays, the human monocytic THP-1 cell line (ATCC TIB-202) was grown in suspension in RPMI-1640 supplemented with 2 mM glutamine, 10% heat-inactivated fetal bovine serum, penicillin (100 IU) and streptomycin (100 μg/ml) at 37°C in a high humidity atmosphere containing 5% CO2-95% air.
METHOD DETAILS
Expression and purification of FLAG-VgrG
F. novicida expressing FLAG-VgrG, prepared as described (Clemens et al., 2015), was grown in trypticase soy broth supplemented with 0.2% L-cysteine (TSBC) at 37°C to an OD of 1.0 and used to inoculate 4 liters of TSBC containing 5% KCl at an OD of 0.05. The culture was grown at 37°C, rotating at 180 rpm, to an OD of 1.2 – 1.5, and pelleted by centrifugation at 4000 g for 1 hour. The supernatant was discarded, and the pellet was resuspended in 50 mM Tris HCl, pH 8, 1 mM EDTA, 1% Tween, and Calbiochem Protease inhibitors (1:100) 1 mM N-ethylmaleimide, 1mM phenylmethylsulfonyl fluoride, and benzonase (1:1000). Bacterial lysis was promoted by sonication with a probe tip sonicator (Cell Disruptor model W-375, Heat Systems Ultrasonics, Plainview, NY) while stirring in an ice bath. 1 M NaCl was added slowly while stirring to achieve a final concentration of 100 mM NaCl. The sample was centrifuged at 15,000 g for 30 min at 4°C. The pellet was discarded and the supernatant fluid was centrifuged at 44,000 g for 20 min 4°C. The pellet was discarded and the supernatant fluid was applied to a column containing 1 ml of mouse monoclonal (clone M2, Sigma Chemical Company) anti-FLAG agarose resin. The resin was washed sequentially with (a) 200 ml of 50 mM Tris, 0.3 M NaCl containing 1% Tween, (b) 10 ml of Tris-buffered saline (TBS) with 10% glycerol, 10 mM MgCl2 and 10 mM ATP (to remove heat shock proteins), and (c) 50 ml of 50 mM Tris HCl, pH 7.5, containing 0.3 M NaCl. The resin was eluted with 10 ml of 3X-FLAG peptide (0.1 mg/ml) in TBS. The eluate was concentrated to 1 ml with a 100,000 MW cut-off spin concentrator (Millipore) and applied to a Sephacryl S400HR gel filtration column that was pre-equilibrated with TBS. UV absorbance at 280 nm of the eluate from the column was monitored with a UV-monitor (2238 LKB Uvicord SII). FLAG-immunoreactivity of the eluate was evaluated by diluting aliquots from the column 100-fold with 0.05 M NaHCO3, pH 9.6, and adding 0.1 ml/well of high bind, polystyrene, 96-well ELISA plates (Costar, Corning Inc.). After 90 minutes at room temperature, the wells were blocked with 1% BSA in TBS, washed three times with TBS, and incubated for 60 min at room temperature with horseradish peroxidase (HRP)-conjugated mouse anti-FLAG monoclonal antibody (clone M2, Sigma-Aldrich) diluted 1:2000 with TBS-1% BSA. The wells were washed three times with TBS, and peroxidase activity was developed with 0.1 ml per well of tetramethylbenzidine (TMB) substrate (Thermo Scientific) according to the manufacturer’s directions. The reaction was stopped by adding 0.1 ml/well of 2 M H2SO4, and the absorbance at 450 nm was measured with a microplate reader (iMark, BioRad). The peak fractions containing FLAG-immunoreactive material were pooled and concentrated to 1 ml with a 100,000 MW cut-off spin concentrator (Millipore).
Mass spectrometry based proteomics analysis
The purified sample was acetone precipitated by adding 0.4 ml of acetone (−20°C) to 0.1 ml of sample. The sample was maintained at −20°C for 14 hours and pelleted by centrifugation at 10,000 g for 10 min at 4°C. The sample was resuspended in 90% acetone/water (−20°C) and centrifuged as before. The supernatant was discarded, and the pellet was air dried for 10 min at room temperature and stored at −20°C. Further processing was conducted by the UCLA Proteome Research Center. The pellet was resuspended in 8 M urea, 100 mM Tris-HCl, pH 8.5; reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP); alkylated with 10 mM iodoacetamide; and digested with sequencing-grade trypsin. The peptide mixture was desalted using Pierce C18 Tips (Thermo Fisher Scientific), dried and resuspended in 5% formic acid, and fractionated on-line using a 19 cm long, 100 μm inner diameter fused silica capillary packed in-house with bulk C18 reverse phased resin (1.9 μm, 100 Å pores, Dr. Maisch GmbH). A water-acetonitrile gradient was delivered over 140 minutes to a maximum of 80% buffer B using an Easy nLC-1000 UHPLC system (Thermo Fisher Scientific) at a flow rate of 300 nL/min (Buffer A: water with 3% DMSO and 0.1% formic acid; Buffer B: acetonitrile with 3% DMSO and 0.1% formic acid). MS/MS spectra were generated by a Data Dependent acquisition strategy on a Q-Exactive tandem mass spectrometer (Thermo Fisher). Data acquisition consisted of cycles of one full MS spectrum at a resolution of 70,000 followed by MS/MS of precursor ions from the full MS scan using a resolution of 17,500.
Data analysis was performed using the Integrated Proteomics Pipeline 2 (Integrated Proteomics Applications, San Diego, CA). MS/MS spectra were searched against the F. novicida U112 fasta protein database (taxid: 401614) using the ProLuCID algorithm, and peptide-to-spectrum matches (PSM) were calculated by DTASelect and filtered using a decoy-database estimated false discovery rate of less than 0.01.
Cloning and mutagenesis
E. coli strains NEB 5-alpha and 5-alpha F′ Iq were used for general cloning purposes. For recombinant expression of the full length (residues 2-820) of PdpA (FTN 1309), the corresponding nucleotide sequences were amplified with PCR from the genomic DNA of F. novicida strain U112 and cloned into pET-28 derivatives for expression as His18-PdpA.
For analysis of protein-protein interaction by the bacterial adenylate cyclase two-hybrid system (BACTH, Euromedex), nucleotide sequences corresponding to IglC (FTN_1322), VgrG (FTN_1312), and PdpA residues 2-175 (PdpA N-helical lid domain), 172-621 (PdpA head domain), and 762-820 (PdpA spike) were cloned into pKT25 or pKNT25 for expression as a fusion partner to the T25 fragment and into pUT18 or pUT18C as a fusion partner to the T18 fragment.
Mutant strains of F. novicida expressing N-terminal (residues 2-175) or C-terminal (residues 762-820) truncation and GSSG substitution (residues 68-75) of PdpA were generated through PCR amplification of chromosomal DNA sequences ≥ 1 kb upstream and downstream of the mutation site; the resulting PCR fragments were subsequently joined together through overlapping PCR extension and cloned into the suicide plasmid pMP590 between BamHI and NotI sites. The resulting plasmid constructs were confirmed by nucleotide sequencing and were introduced into the F. novicida FLAG-VgrG strain (Clemens et al., 2015) through chemical transformation. The transformation mixture was selected on GCII chocolate agar plates containing 20 μg/ml kanamycin. F. novicida grown on the kanamycin containing agar plates were counter-selected on antibiotic-free agar plates containing 7% sucrose and screened by colony PCR for evidence of allelic replacement via homologous recombination.
Expression and purification of recombinant proteins
Overnight cultures of E. coli strain Rosetta 2(DE3) carrying either pET28-His18-PdpA, pET28-His18-PdpA with residues 68 – 75 (GLISKLDY) replaced with GSSGGSSG, or His6-3xFLAG-VgrG G164R were inoculated into 2xYT medium containing kanamycin (30 μg/ml) and chloramphenicol (34 μg/ml) and grown at 37°C, 250 rpm to OD600 of 0.5. Expression of the recombinant protein was induced by addition of 1 mM IPTG to the culture and incubation overnight at 18°C, 250 rpm. E. coli was harvested from the culture by centrifugation and was lysed by sonication in sodium phosphate buffer, pH 7.4 containing protease inhibitors. After addition of NaCl and imidazole to a final concentration of 0.3 M and 5 mM, respectively, the insoluble materials were removed from the lysate by centrifugation. Soluble histidine-tagged recombinant proteins were purified by chromatography on Ni-NTA affinity resin (Qiagen).
Assay of protein-protein interactions by BACTH Analysis
Genes of interest cloned in compatible vectors as T25 fusion or T18 fusion genes were co-transformed into E. coli reporter strain BTH101 by electroporation. Single colonies were used to inoculate Luria broth containing carbenicillin (100 μg/ml) kanamycin (30 μg/ml), and IPTG (0.5 mM). After cultivation at 30°C, 250 rpm overnight, 2 μl of the culture were spotted onto Luria broth agar containing the antibiotics, IPTG, and X-gal (40 μg/ml). Positive protein-protein interactions were evident after overnight incubation at 30°C as indicated by the blue color of the E. coli spots.
Assay of T6SS-mediated secretion
F. novicida strains were inoculated into 10 ml TSBC containing 5% KCl at an initial OD540 of 0.05 and cultivated for 10-12 hours to an OD540 of 1.2 to 1.6. The bacteria were pelleted by centrifugation, and the culture supernates were passed through 0.2/0.8 μm syringe filters and concentrated using Amicon Ultracel 10k centrifugal filter units. Samples of pelleted bacteria and concentrated filtrate were analyzed by Western immunoblotting, probing with anti-IglC polyclonal antibody or HRP-conjugated anti-FLAG mouse monoclonal antibody (clone M2, Sigma Aldrich).
Assay of F. novicida intracellular growth in THP-1 cells
The intracellular growth assay was performed using a modification of our previously published methods (Clemens et al., 2015). Human monocytic THP-1 cells, in RPMI-1640 supplemented with 2 mM glutamine, 10% heat-inactivated fetal bovine serum and 100 nM phorbol 12-myristate 13-acetate (PMA) without antibiotics, were seeded in poly-L-lysine coated 96-well glass bottom microplates (Matrical) at a density of 1 × 105 cells/200 μl/well for 3 days at 37°C in a high humidity atmosphere containing 5% CO2-95% air. F. novicida strains were grown overnight in TSBC to O.D. 540 nm of 1 – 1.5 and added to the monolayers of THP-1 cells at an multiplicity of infection of 2:1 (bacterium:cell) in DMEM containing 10% AB serum. After incubating the monolayers 2 h at 37°C, we replaced the culture medium with DMEM containing 10% FBS and 10 μg/ml gentamicin and incubated for 30 min at 37°C to kill extracellular bacteria. We washed the monolayers twice with Hank’s Balanced Salt Solution and added fresh DMEM containing 10% FBS and 0.1 μg/ml gentamicin to restrict extracellular bacterial growth. At the indicated time points, we determined the numbers of colony forming units (CFU) in each monolayer by lysing the monolayers with 1% saponin in PBS, serially diluting the lysate with PBS, and spotting aliquots of the diluted lysates on GCII agar plates.
CryoEM
An aliquot of 3.5 microliters of PdpA-VgrG sample was applied onto a glow discharged holey carbon film grid (300 mesh, R2/1, Quantifoil). The grid was blotted and flash-frozen in liquid ethane with FEI Vitrobot Mark IV. The grid was loaded onto an FEI Titan Krios electron microscope with a K2 Summit direct electron counting detector (Gatan). Movies were acquired with Leginon (Carragher et al., 2000) by electron counting in super-resolution mode at a pixel size of 0.535 Å/pixel. A total number of 60 frames were acquired in 12 seconds for each movie, giving a total dose of 46.65 e−/A2/movie. A total of 3690 movies were acquired from 2 grids in an imaging session of 72 h.
Image processing and 3D reconstruction by single-particle analysis
The recorded movies were processed by MotionCor2 (Zheng et al., 2017) for a 5 × 5 patches drift correction with dose weighting and binned 2-fold, resulting in a pixel size of 1.07 Å/pixel. The non-dose-weighted images were used to estimate defocus, astigmatism, and astigmatism angle for the contrast transfer function (CTF) by CTFFIND 4.18 (Rohou and Grigorieff, 2015). The dose-weighted images were used for particle picking. 726,724 particles were semi-automatically picked by Gautomatch (https://www.mrc-lmb.cam.ac.uk/kzhang/) and extracted by Relion-2.1 (Fernandez-Leiro and Scheres, 2017) in a box size of 250 pixels. Three rounds of 2D classification were performed in Relion-2.1 to remove contaminating matter, ice, and bad particles, yielding 35,747 good particles. The selected particles were then used to generate an initial model through the ab initio method in CryoSparc (Punjani et al., 2017). Auto refinement of these particles by CryoSparc yielded a map with an average resolution of 4.2 Å.
The previously refined 4.2 Å model was low pass-filtered to 30 Å resolution as an initial model for 3D auto refine in Relion-2.1. The 3D classification was performed without particle rotation and translation alignment using the 3D-auto-refined particles. The particles were divided into 3 classes by 3D classification. 10,343 particles in 1 class, corresponding to the “half-lidded” map, were selected for further refinement. The selected particles were re-centered and re-extracted in a box size of 350 pixels. Further auto-refinement of the re-extracted particles yielded a map with an average resolution of 3.98 Å for the C3 symmetry, which was selected as the final map for model building. 16,889 particles in another class, corresponding to the “lidless map”, were also re-centered and re-extracted, yielded a map with a 4.21 Å resolution after 3D auto-refinement. To build the whole model of the “N-helical-lid,” we applied two additional rounds of 3D classification, focusing on the region of “N-helical-lid”; 3,765 particles were selected and re-extracted, yielding a map with an average resolution of 4.35 Å for the C3 symmetry. The maps were sharpened with B-factors of −150 Å2. The stated resolutions were evaluated using “gold-standard” FSC=0.143 criterion. Data collection and reconstruction statistics are presented in Table S2.
Atomic model building and refinement
Model building of the head region of PdpA took advantage of the VgrG1 structure from P. aeruginosa (PDB: 4MTK). Residues 5 to 350 of chain A from the model of VgrG1 (Pa) were fitted into the “half-lidded” map of PdpA-VgrG in Chimera (Pettersen et al., 2004), and the pre-fitted model was refined by PHENIX (Adams et al., 2010) in real space with secondary structure and geometry restraints. The residue assignment and sidechain orientation were further refined in COOT software (Emsley et al., 2010) according to the density map. The VgrG and the remaining parts of PdpA were built manually using COOT; sequence assignment was mainly guided by secondary structure prediction results from PSIPRED (McGuffin et al., 2000) and visible densities of residues with bulky side chains (Trp, Phe, Tyr and Arg). The model was refined by PHENIX real_space_refinement with secondary structure and geometry restraints. The model of the “lidless” and “lidded” structure were built and modified from the refined “half-lidded” model based on the “lidless” and “lidded” map, respectively. The quality of the models was assessed with MolProbity (Chen et al., 2010). The Molprobity scores of the final atomic models for the lidded, half-lidded, and lidless models were 2.05, 1.91 and 1.96, respectively. Refinement statistics of PdpA-VgrG complex and validation statistics for the lidded, half-lidded, and lidless models are shown in Table S2.
Evaluation of PdpA interaction with VgrG by ELISA
Wells of a 96-well high-binding, polystyrene ELISA plate were coated with His-epitope tagged PdpA or PdpA::GSSG (0 – 2 μg/ml) in 0.05 M NaHCO3, pH 8.3, for 1 hour at room temperature. Wells were blocked by incubation with 2% BSA, 5% gelatin in TBS; washed three times with TBS; and incubated for 60 min with His-FLAG-VgrG (0.1 μg/ml) in TBS containing 2% BSA, 5% gelatin, and 1% Tween-20. The wells were washed three times with TBS; incubated with HRP-conjugated mouse anti-FLAG M2 monoclonal antibody diluted 1:2000 in TBS containing 1% BSA and 0.2% Tween; washed with TBS; and peroxidase was developed with TMB substrate and read with a microplate reader as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
All ELISA data shown represent the means and standard errors of the mean of independent duplicate wells. All ELISA experiments were conducted at least twice on separate days. Bacterial growth (colony forming unit) data shown are the means and standard deviations of three independent experiments each with biological triplicates.
DATA AND SOFTWARE AVAILABILITY
The cryo-EM density maps and atomic models for the lidded, half-lidded, and lidless forms of the F. novicida central spike have been deposited in the Electron Microscopy Data Bank (EMD-20696, EMD-20698, EMD-20695) and the Protein Data Bank (PDB ID 6U9F, 6U9G, 6U9E), respectively.
Supplementary Material
Video S1. Related to Figure 1. Shaded surface view of the cryoEM density map of the PdpA-VgrG complex. Individual PdpA-VgrG protomers are colored differently.
Video S2. Related to Figure 2. Surface representation of the cryoEM density map of the PdpA-VgrG central spike complex with a central plug within the PdpA cavity.
Video S3. Related to Figure 4. Same as Video S1 except that the lid is cut away to reveal the cavity.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-FLAG Peroxidase (clone M2) | MilliporeSigma | Cat# A8592; RRID:AB_439702 |
| Mouse monoclonal antibody to PdpA | BEI Resources | Cat# NR-3197 |
| Goat anti-mouse IgG peroxidase conjugate | Pierce-Thermo | Cat# 32230 |
| Goat anti-rabbit IgG peroxidase conjugate | Bio-Rad | Cat# 170-6515 |
| Rabbit polyclonal antibody to IglC | Clemens et al., 2015 | N/A |
| Bacterial and Virus Strains | ||
| E. coli NEB 5-alpha | New England BioLabs | Cat# C2987H |
| E. coli NEB 5-alpha F’ Iq | New England BioLabs | Cat# C2992H |
| E. coli Rosetta 2(DE3) | MilliporeSigma | Cat# 71400 |
| E. coli BTH101 | Euromedex | Cat# EUB001 |
| F. novicida IglAΔ | Clemens et al., 2015 | Fn IglAΔ |
| F. novicida PdpAΔN | This Study | Fn PdpAΔN |
| F. novicida PdpAΔC | This Study | Fn PdpAΔC |
| F. novicida PdpA::GSSG | This Study | Fn PdpA::GSSG |
| F. novicida PdpA-FLAG | This Study | Fn PdpA-FLAG |
| F. novicida FLAG-VgrG | Clemens et al., 2015 | FV, Fn-FLAG-VgrG |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Adenosine triphosphate | Sigma-Aldrich | Cat# A-6419 |
| 3X FLAG peptide | Sigma-Aldrich | Cat# F4799-25MG |
| N-ethylmaleimide | Sigma-Aldrich | Cat# E3876 |
| Phenylmethylsulfonyl fluoride | Sigma-Aldrich | Cat# P-7626 |
| Benzonase Nuclease | VWR | Cat# EM71206-3 |
| Tween 20 | Fisher Chemical | Cat# BP337-500 |
| Critical Commercial Assays | ||
| Tetramethyl benzidine substrate kit | Thermo Fisher Scientific | Cat# 34021 |
| Deposited Data | ||
| P. aeruginosa PA0091 VgrG1 X-ray structure | PDB | PDB 4MTK |
| T4 phage (gp27-gp5)3 X-ray structure | PDB (Koshyiama et. al, 2008) | PDB 2Z6B |
| P. aeruginosa VgrG1 X-ray structure | PDB (Quentin et al., 2018) | PDB 6H3L |
| F. novicida central spike complex, lidded atomic model | This Study | PDB 6U9F |
| F. novicida central spike complex, half-lidded atomic model | This Study | PDB 6U9G |
| F. novicida central spike complex, lidless atomic model | This Study | PDB 6U9E |
| F. novicida central spike complex, lidded cryo-EM density map | This Study | EMD-20696 |
| F. novicida central spike complex, half-lidded cryo-EM density map | This Study | EMD-20698 |
| F. novicida central spike complex, lidless cryo-EM density map | This Study | EMD-20695 |
| Experimental Models: Cell Lines | ||
| THP-1 Cells | ATCC | NIH-ARP Cat# 9942-142, RRID:CVCL_0006 |
| Experimental Models: Organisms/Strains | ||
| Francisella novicida Utah 112 | Karl Klose, University of Texas San Antonio | WT |
| Recombinant DNA | ||
| pET28a | MilliporeSigma | Cat# 69864 |
| pET28-His18-PdpA | This Study | N/A |
| pET28-His18-PdpA::GSSG | This Study | N/A |
| pET28-3xFLAG-VgrG G164R | This Study | N/A |
| pKNT25 | Euromedex | Cat# EUP-25N |
| pKT25 | Euromedex | Cat# EUP-25C |
| pKT25-zip | Euromedex | Cat# EUP-25Z |
| pUT18 | Euromedex | Cat# EUP-18N |
| pUT18C | Euromedex | Cat# EUP-18C |
| pUT18C-zip | Euromedex | Cat# EUP-18Z |
| pKNT25-IglA | This Study | N/A |
| pKNT25-VgrG | This Study | N/A |
| pKNT25-IglC | This Study | N/A |
| pKT25-IglC | This Study | N/A |
| pKT25-VgrG | This Study | N/A |
| pUT18-IglB | This Study | N/A |
| pUT18-PdpA2-175 | This Study | N/A |
| pUT18-PdpA172-621 | This Study | N/A |
| pUT18-PdpA762-820 | This Study | N/A |
| pUT18C-PdpA2-175 | This Study | N/A |
| pUT18C-PdpA172-621 | This Study | N/A |
| pUT18C-PdpA762-820 | This Study | N/A |
| pMP590 | Martin Pavelka, University of Rochester | N/A |
| pMP590-pdpAΔ2-175-updn | This Study | N/A |
| pMP590-pdpAΔ762-820-updn | This Study | N/A |
| pMP590-pdpA::GSSG-updn | This Study | N/A |
| pMP590-pdpA-FLAG-updn | This Study | N/A |
| Software and Algorithms | ||
| Leginon | Carragher et al., 2000 | http://nramm.nysbc.org/downloads |
| MotionCor2 | Zheng et al., 2017 | http://emcore.ucsf.edu/ucsf-motioncor2 |
| CTFFIND 4.18 | Rohou and Grigorieff, 2015 | http://grigoriefflab.janelia.org/ctf |
| Gautomatch | Dr. Zhang, MRC Laboratory of Molecular Biology | http://www.mrc-lmb.cam.ac.uk/kzhang |
| Relion-2.1 | Fernandez-Leiro and Scheres, 2017 | http://www3.mrc-lmb.cam.ac.uk/relion |
| CryoSparc | Punjani et al., 2017 | http://cryosparc.com/ |
| PHENIX | Adams et al., 2010 | http://www.phenixonline.org/ |
| Dali Server | Holm and Rosenstrom, 2010 | http://ekhidna2.biocenter.helsinki.fi/dali/ |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/. |
| PDBePISA Assembly | Krissinel and Henrick, 2007 | http://www.ebi.ac.uk/pdbe/pisa/ |
| PsiPred server | McGuffin et al., 2000 | http://bioinf.cs.ucl.ac.uk/psipred/ |
| I-Tasser server | Yang et al., 2015 | http://zhanglab.ccmb.med.umich.edu/I-TASSER/ |
| JPred4 | Drozdetskiy et al., 2015 | http://www.compbio.dundee.ac.uk/jpred4/ |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu/ |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera/download.html |
| Other | ||
| SYPRO Ruby | Thermo Fisher Scientific | Cat# S11791 |
| Anti-FLAG M2 affinity gel | MilliporeSigma | Cat# A2220; RRID: AB_10063035 |
| Ni-NTA Agarose affinity gel | Qiagen | Cat# 30210 |
| Trypticase soy broth | BD Difco | Cat# 211768 |
| Protease Inhibitor Cocktail Set III, EDTA-Free | MilliporeSigma | Cat# 539134 |
| Sephacryl S400HR gel filtration resin | Pharmacia | Cat# 17-0609-01 |
Highlights.
The atomic structure of the Francisella T6SS central spike complex is presented
The structure, comprised of PdpA and VgrG, has a unique α-helical lid
The central spike assumes three conformations: lidded, half-lidded, and lidless
The lidless form has a density within the PdpA cavity suggesting a cargo molecule
Acknowledgements:
We thank James Wolfschegel and the UCLA Proteomics Core Facility for mass spectrometry analysis and Peng Ge for helpful discussion throughout the project. This work was supported by National Institute of Health (NIH) grant AI125497 and in part by NIH grant GM071940. The authors acknowledge the use of instruments at the Electron Imaging Center for NanoMachines supported by NIH (1S10RR23057, 1S10OD018111, and 1U24GM116792), NSF (DBI-1338135 and DMR-1548924) and CNSI at UCLA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms JE, Sjostedt A, and Lavander M (2010). The role of the Francisella tularensis pathogenicity island in Type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol 1, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, and Jones DT (2013). Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 41, W349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, Pulokas J, and Reilein A (2000). Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol 132, 33–45. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A, and Celli J (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol 1, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Ge P, Lee BY, Horwitz MA, and Zhou ZH (2015). Atomic structure of T6SS reveals interlaced array essential to function. Cell 160, 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, and Horwitz MA (2007). Uptake and intracellular fate of Francisella tularensis in human macrophages. Ann. N. Y Acad. Sci 1105, 160–186. [DOI] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, and Horwitz MA (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun 72, 3204–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens DL, Lee BY, and Horwitz MA (2018). The Francisella Type VI Secretion System. Front. Cell Infect. Microbiol. 8, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin OM, Duplantis BN, Ludu JS, Hare RF, Nix EB, Schmerk CL, Robb CS, Boraston AB, Hueffer K, and Nano FE (2011). The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion. Microbiology 157, 3483–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, and Barton GJ (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Oyston PC, Green M, and Titball RW (2002). Tularemia. Clin. Microbiol. Rev 15, 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi A, Kim J, Walls AC, Ledvina HE, Miller CN, Ramsey KM, Whitney JC, Radey MC, Peterson SB, Ruhland BR, et al. (2016). Secreted Effectors Encoded within and outside of the Francisella Pathogenicity Island Promote Intramacrophage Growth. Cell Host Microbe 20, 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Leiro R, and Scheres SHW (2017). A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D Struct. Biol 73, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, and Washburn MP (2006). Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, and Rosenstrom P (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiyama T , Yokoi N , Ueno T , Kanamaru S , Nagano S , Shiro Y , Arisaka F and Watanabe Y (2008). Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor. Small 4, 50–54. [DOI] [PubMed] [Google Scholar]
- Kozlov G, Wong K, and Gehring K (2018). Crystal structure of the Legionella effector Lem22. Proteins 86, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 372, 774–797. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, and Jones DT (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- Nano FE, Zhang N, Cowley SC, Klose KE, Cheung KK, Roberts MJ, Ludu JS, Letendre GW, Meierovics AI, Stephens G, et al. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol 186, 6430–6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov S, Schneider JP, Brackmann M, Goldie KN, Stahlberg H, and Basler M (2018). Cryo-EM reconstruction of Type VI secretion system baseplate and sheath distal end. EMBO J. 37, e97103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. [DOI] [PubMed] [Google Scholar]
- Quentin D, Ahmad S, Shanthamoorthy P, Mougous JD, Whitney JC , Raunser S (2018). Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol 3, 1142–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigard M, Broms JE, Mosnier A, Hologne M, Martin A, Lindgren L, Punginelli C, Lays C, Walker O, Charbit A, et al. (2016). Francisella tularensis IglG belongs to a novel family of PAAR-like T6SS proteins and harbors a unique N-terminal extension required for virulence. PLoS Pathog. 12, e1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb CS, Nano FE, and Boraston AB (2012). The structure of the conserved type six secretion protein TssL (DotU) from Francisella novicida. J. Mol. Biol 419, 277–283. [DOI] [PubMed] [Google Scholar]
- Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol 192, 216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, et al. (2014). A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerk CL, Duplantis BN, Wang D, Burke RD, Chou AY, Elkins KL, Ludu JS, and Nano FE (2009). Characterization of the pathogenicity island protein PdpA and its role in the virulence of Francisella novicida. Microbiology 155, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, and Leiman PG (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman Julie M., Agnello Danielle M., Zheng H, Andrews Benjamin T., Li M, Catalano Carlos E., Gonen T, and Mougous Joseph D. (2013). Haemolysin Coregulated Protein Is an Exported Receptor and Chaperone of Type VI Secretion Substrates. Mol. Cell 51, 584–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NM, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, and Leiman PG (2016). Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533, 346–352. [DOI] [PubMed] [Google Scholar]
- Whitney JC, Beck CM, Goo YA, Russell AB, Harding BN, De Leon JA, Cunningham DA, Tran BQ, Low DA, Goodlett DR, et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol 92, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, and Zhang Y (2015). The I-TASSER Suite: protein structure and function prediction. Nat. Methods 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Related to Figure 1. Shaded surface view of the cryoEM density map of the PdpA-VgrG complex. Individual PdpA-VgrG protomers are colored differently.
Video S2. Related to Figure 2. Surface representation of the cryoEM density map of the PdpA-VgrG central spike complex with a central plug within the PdpA cavity.
Video S3. Related to Figure 4. Same as Video S1 except that the lid is cut away to reveal the cavity.
Data Availability Statement
The cryo-EM density maps and atomic models for the lidded, half-lidded, and lidless forms of the F. novicida central spike have been deposited in the Electron Microscopy Data Bank (EMD-20696, EMD-20698, EMD-20695) and the Protein Data Bank (PDB ID 6U9F, 6U9G, 6U9E), respectively.