Abstract
Purpose:
To feasibly analyze associations of Helicobacter pylori (H. pylori) with disease in large cohort studies, assays are needed to assess H. pylori prevalence in existing biospecimens. However, serology has traditionally been unable to distinguish active from past infection. We sought to determine the sensitivity of sero-positivity to H. pylori proteins to detect active infection.
Methods:
We measured antibody responses to 13 H. pylori proteins using multiplex serology in serum samples of a training (n=78) and validation set (n=49) collected concurrently from patients undergoing urea breath test (UBT). To determine sensitivity of sero-positivity to H. pylori proteins for active infection, a cut-off was applied to achieve 90% specificity. Antibody levels were retested in a subset of participants (n=16) 6 months after baseline.
Results:
With a specificity of 91%, sero-positivity to H. pylori proteins VacA, GroEl, HcpC, and HP1564 ascertained active infection from 100% to 75% sensitivity. Positivity to a combination of these proteins (≥2 out of the 4) resulted in specificity of 90% and sensitivity of 100%. The validation set replicated results from the training set. Among those participants with successful H. pylori eradication after baseline, antibody levels decreased significantly for VacA, HcpC, and HP1564 when assessed 6 months later.
Conclusion:
Utilizing the cut-offs for sero-positivity established through comparison with UBT, sero-positivity to ≥2 of the H. pylori proteins VacA, GroEl, HcpC, and HP1564 determines active H. pylori infection at high specificity and sensitivity and may approximate the prevalence of active H. pylori infection in large cohorts.
INTRODUCTION
Helicobacter pylori (H. pylori) is the main carcinogen for the development of gastric cancer worldwide.1 About half of the world’s population is infected with this bacterium,2 and within the US prevalence varies greatly by race/ethnicity, with African Americans twice as likely to be infected with H. pylori as compared to non-Hispanic whites.3 Beyond gastric cancer, research in the past years has assessed the association of H. pylori with other cancers, for example in the oesophagus or the colorectum, but also with non-cancerous diseases like Parkinson’s and diabetes.4,5
Gold standard assays to detect an active H. pylori infection include tests on biopsy material obtained during endoscopy, or, less invasively, performance of stool antigen test or Urea Breath test (UBT).6 However, these methodologies are not feasible in large-scale epidemiological studies, especially in already established cohort studies that have completed recruitment and biospecimen collection. For the majority of these current population health cohorts, stored biospecimens include blood samples and, thus, serology would be advantageous over clinically applied and/or invasive methodologies to detect H. pylori infection. Indeed, various studies have applied H. pylori serology, either whole cell ELISA, virulence factor Cytotoxin-associated antigen A (CagA)-specific ELISA, or multiplex serology to study associations with disease.7 Utilizing multiplex serology that assesses multiple H. pylori antigens in one reaction, we previously identified sero-positivity to H. pylori hypothetical proteins HP0305 and HP1564 to be strongly associated with the risk of developing gastric cancer in a consortium of East Asian prospective cohorts.8 Moreover, we have reported that sero-positivity to proteins Vacuolating cytotoxin A (VacA), Chaperonin GroEl, and hypothetical proteins HcpC, and HP1564, were significantly associated with colorectal cancer risk in diverse population across the United States, with the strongest associations observed among African Americans.9 The question remains, though, how the observed associations can be interpreted temporally, since serology is considered an indirect systemic measure that is not able to distinguish between current or past infection.
In the present study, we sought to assess the performance of H. pylori multiplex serology in comparison to a gold standard of active H. pylori infection as determined by UBT, as well as to explore potential changes in antibody response to H. pylori antigens after successful eradication treatment. The studies we analyzed to address these aims include a training and validation set with study participants of primarily African American descent, a US population, as described above, that presents with a high H. pylori prevalence, and is therefore of specific importance to study for potential H. pylori-related health outcomes.
METHODS
H. pylori multiplex serology
We measured antibody responses to H. pylori antigens using multiplex serology as described previously.9,10 Serological data was obtained for 16 recombinantly expressed GST-tagged fusion proteins originating from 13 H. pylori open reading frames (strains 26695 or G27) (Table 1), and for 2 recombinantly expressed GST-tagged fusion proteins (capsid proteins VP1) from Polyomavirus JC and BK as control antigens. Bacterially expressed fusion proteins were affinity-purified on polystyrene beads, filled with an internal fluorescent color distinct for each of the different proteins (SeroMap, Luminex Corp., Austin, TX, USA). Protein-coupled beads were mixed and incubated with serum, diluted 1:1000. Antigen-bound serum antibodies were labelled with a biotinylated goat anti-human IgG/IgA/IgM secondary antibody and subsequent incubation with a Streptavidin-coupled reporter fluorescent dye (Phycoerythrin). This secondary antibody was used to allow for detection of Ig subclasses of long-term (IgG/IgA) exposure as well as recently acquired infection (IgM). However, we only expect few if any recent exposures in this adult population and thus the measured antibody response mainly being driven by IgG and, to a lesser extent, IgA responses. A Luminex 200 analyzer (Luminex Corp., Austin, TX, USA) identified the bead type by its internal fluorescence and consequently the bound antigen, as well as quantified the amount of bound serum antibody by the reporter fluorescence. The output is the median fluorescence intensity (MFI) measured for at least 100 beads per type per serum. Final MFI values were generated by subtraction of GST-tag and individual bead background values, and addition of MFI values to N- and C-terminal protein fragments in the cases of CagA, VacA, and HyuA.
Table 1:
H. pylori multiplex serology antigens
| Systematic name |
Name | Full name | Accession no. † | Selected amino acids |
Strain |
|---|---|---|---|---|---|
| HP0010 | GroEL | Chaperonin GroEL | AM_997163 | 1–547 | G27 |
| HP0073 | UreA | Urease alpha subunit | NP_206873 | 1–238 | 26695 |
| HP0231 | HP0231 | Hypothetical protein | NP_207029 | 1–265 | 26695 |
| HP0243 | NapA | Neutrophil-activating protein | NP_207041 | 1–144 | 26695 |
| HP0305 | HP0305 | Hypothetical protein | NP_207103 | 1–184 | 26695 |
| HP0410 | HpaA | Neuraminyllactose-binding hemagglutinin homolog | NP_207208 | 1–249 | 26695 |
| HP0547 | CagA N-Terminus | Cytotoxin-associated antigen A | NP_207343 | 1–650 | 26695 |
| CagA C-Terminus | 641–1186 | ||||
| HP0695 | HyuA N-Terminus | Hydantoin utilization protein A | NP_207208 | 1–220 | 26695 |
| HyuA C-Terminus | 210–713 | ||||
| HP0875 | Catalase | Catalase | NP_207669 | 1–505 | 26695 |
| HP0887 | VacA N-Terminus | Vacuolating cytotoxin A | NP_207680 | 34–363 | 26695 |
| VacA C-Terminus | 328–1008 | ||||
| HP1098 | HcpC | Conserved hypothetical secreted protein | NP_207889 | 1–290 | 26695 |
| HP1104 | Cad | Cinnamyl alcohol dehydrogenase ELI3–2 | NP_207895 | 1–348 | 26695 |
| HP1564 | HP1564 | Hypothetical protein | NP_208355 | 1–271 | 26695 |
National Center for Biotechnology Information Reference Sequence
Urea breath test (UBT)
The BreathTek kit (Otsuka America Pharmaceutical Inc., Rockville, MD, USA) was utilized to determine active H. pylori infection by UBT.11 The assay was performed according to the manufacturer’s instructions. Briefly, participants first provided a baseline breath sample by exhaling into a collection bag. Next, participants drank a solution containing a 13C-labelled urea substrate. After 15 minutes, participants exhaled into a second, post-dose collection bag. If the participant has an active H. pylori infection, the H. pylori enzyme urease decomposes the 13C-urea contained in the ingested solution and sets free 13CO2, which is then detected in the post-dose breath sample. The ratio of 13CO2 to 12CO2 in the post-dose collection bag is compared to that in the baseline collection bag using a 13CO2Urea Breath Analyzer (POCone, Otsuka America Pharmaceutical Inc., Rockville, MD, USA) either by Quest Diagnostics (Lenexa, KS, and Wood Dale, IL, USA) or at the Duke Cancer Institute (Durham, NC, USA). The manufacturer considers an increase in the ratio of at least 2.4‰ as being positive for H. pylori infection. To ensure a valid UBT result, participants had to fast for at least one hour prior to the performed test.
Study samples
Training set – Durham Initiative for Stomach Health (DISH)
The training set comprised serum samples and study data from the Durham Initiative for Stomach Health (DISH), a community-based H. pylori screening and engagement project to determine the H. pylori prevalence and feasibility of an H. pylori eradication trial in Durham County (NC, USA). The study was conducted in May 2018 in partnership with the River Church in Durham, NC, USA. A total number of 92 participants 40 years of age or older and with the ability to provide informed consent in English were enrolled onsite on one day in May 2018. At enrollment, participants completed a study questionnaire, donated a blood sample, and performed a UBT. Exclusion criteria for the study were prior gastric surgery and/or gastric cancer, and the use of antibiotics, Pepto Bismol or any proton pump inhibitor in the past two weeks prior to enrollment.
The study was approved by the Duke University Health System Institutional Review Board for Clinical Investigations.
Validation set - Healthy Stomach Study (HSS)
Serum samples and study data from the Healthy Stomach Study (HSS) served as a validation set. The HSS is a pilot study within the Southern Community Cohort Study (SCCS), a cohort of 86,000 men and women, ages 40–79, recruited from community health clinics in 12 southeastern states between 2002 and 2009.12 The goal of the HSS was to assess patient demographic, lifestyle, and medical history characteristics associated with sero-prevalence of H. pylori.13 For the HSS, SCCS participants originally recruited from the Nashville (TN, USA) area were re-contacted to assess interest and eligibility for the HSS study, which took place from April to October 2016. HSS participants were required to have a current Tennessee address at the time of the study and the ability to provide informed consent in English. Exclusion criteria for the study comprised a diagnosis of HIV, a history of gastric cancer, prior gastric surgery, use of antibiotics or Pepto Bismol in the past month prior to participation, current use of proton pump inhibitors, and for women a current or planned pregnancy. Forty-nine Nashville-based SCCS participants consented to participate in the HSS and completed the study protocol, including a comprehensive questionnaire, collection of a blood sample, and performance of a UBT.
The study was approved by the Vanderbilt University Medical Center Institutional Review Board and de-identified data was moved to Duke University with the PI (M. Epplein) and then approved as an exempt study by the Duke University Health System Institutional Review Board for Clinical Investigations.
DISH follow-up assessment
All DISH study participants were provided with the UBT test results two weeks after the initial testing, and those with a positive test result were encouraged to seek H. pylori eradication treatment. A subset of the DISH study participants were then invited to attend a follow-up six months (T1) after the initial event (T0). Seven UBT-negative individuals at T0 and 9 UBT-positive participants at T0 were retested to assess active H. pylori infection at T1 using a second UBT. Follow-up participants also were asked to provide a second blood sample and answer a follow-up questionnaire. All UBT-negative individuals at T0 (Group I) as well as 8 out of the 9 UBT-positive participants at T0 (Group II) tested UBT-negative at T1. One baseline UBT-positive participant presented with a positive UBT at T1 and, thus, a persistent H. pylori infection.
Serum samples from DISH, DISH follow-up, and HSS were shipped on dry ice to the German Cancer Research Center (Heidelberg, Germany) for measurement by H. pylori multiplex serology. In total, the present analyses include all participants who provided both a valid UBT and were able to successfully donate blood – this includes 78 participants from DISH (13 of the other DISH participants were unable to provide a blood sample, 1 participant was unable to complete a valid UBT) and all 49 participants from the HSS.
Statistical analyses
Differences in study characteristics between the training and validation set were assessed using the Chi-Square test for categorical variables and the Wilcoxon rank-sum test for continuous variables. Differences in antibody response to H. pylori antigens between UBT-negative and UBT-positive participants within each, the training and validation set, were compared using the Wilcoxon rank-sum test.
We determined the sensitivity for sero-positivity to each individual antigen at a cut-off MFI value set to achieve at least 90% specificity in relation to UBT result in the training set (DISH). Due to technical limitations of the multiplex serology assay, the minimum cut-off applied was 50 MFI to avoid cut-offs within the background noise of detection. The newly determined cut-offs were then applied in the validation set (HSS). We further determined specificity, sensitivity, and the respective 95% CIs for sero-positivity to multiple proteins, either out of all 13 H. pylori proteins, or only among VacA, GroEl, HcpC, and HP1564, the antigens identified in this study to have highest sensitivity and specificity in comparison to UBT.
To compare antibody level [MFI] between baseline and 6 months follow-up in DISH (T0 and T1) within DISH participant Group I (UBT-negative at T0 and T1) and II (UBT-positive at T0 but negative at T1), we first normalized MFI level between T0 and T1 to exclude that observed effects may result from the two separate serum samplings. Polyomaviruses JC and BK cause life-long latent, usually asymptomatic infections, with about 80% of adults being infected with at least one of the two viruses and exhibiting stable antibody responses to capsid protein VP1 over many years.14–16 Thus, under the assumption that antibody responses to Polyomavirus JC and BK VP1 should remain stable over a six-month period of time, we compared the MFI level to JC and BK virus VP1 between the two time points and found indeed on average 1.8-fold higher MFI level at T0 compared to T1. The normalization between T0 and T1 for each participant was based on individual MFI level to JC virus VP1, or if this was found below the established antigen-specific cut-off of 250 MFI,17 based on MFI level to BK virus VP1. Factors for normalization of follow-up samples ranged between 0.6 and 4 (median: 1.6). MFI levels between T0 and T1 for H. pylori proteins were then compared using the Wilcoxon matched-pairs signed-rank test within both DISH participant groups, I and II. A sensitivity analysis with non-normalized MFI values showed, as expected, that the difference in antibody level to H.pylori proteins between T0 and T1 was more pronounced, specifically among group II, and that normalization was necessary to avoid over-interpretation of the results.
A two-sided p-value of <0.05 was considered statistically significant. All graphical presentations were prepared using GraphPad Prism 8 (GraphPad Software, San Diego, CA, USA), and all statistical analyses were carried out using SAS version 9.4 (SAS Institute, Cary, NC, USA).
RESULTS
Demographic characteristics of training (DISH) and validation (HSS) set
Both studies, DISH and HSS, had a similar prevalence of active H. pylori infection, as determined by UBT (DISH: 26%, HSS: 20%), and consisted primarily of female participants (DISH: 77%, HSS: 88%). The training and validation set differed, however, in that DISH participants were younger (median 52 years versus HSS: 59 years), almost exclusively of African American descent (94% versus HSS: 55%), of higher education (81% >High school education versus HSS: 63%), and having a higher BMI (median 31 kg/m2 versus HSS: 28 kg/m2) (Table 2).
Table 2:
Characteristics of training set (DISH) and validation set (HSS) study participants.
| Training set (DISH) n=78 |
Validation set (HSS) n=49 |
|
|---|---|---|
| UBT, n (%) | ||
| Negative | 58 (74) | 39 (80) |
| Positive | 20 (26) | 10 (20) |
| Sex, n (%) | ||
| Female | 60 (77) | 43 (88) |
| Male | 18 (23) | 6 (12) |
| Age, median (IQR)† | 52 (47–60) | 59 (56–68) |
| Race, n (%)‡ | ||
| White | 2 (3) | 22 (45) |
| African American | 74 (94) | 27 (55) |
| Other | 2 (3) | 0 (0) |
| Education, n (%)‡ | ||
| ≤ High school | 15 (19) | 18 (37) |
| > High school | 63 (81) | 31 (63) |
| BMI, median (IQR)† | 31 (29–36) | 28 (23–35) |
Abbreviations: DISH, Durham Initiative for Stomach Health; HSS, Healthy Stomach Study; IQR, Interquartile range; BMI, Body mass index
p-value<0.05 with Wilcoxon signed-rank test
p-value<0.05 with Chi-Square test
Performance of H. pylori multiplex serology in comparison to UBT in the training (DISH) and validation (HSS) set
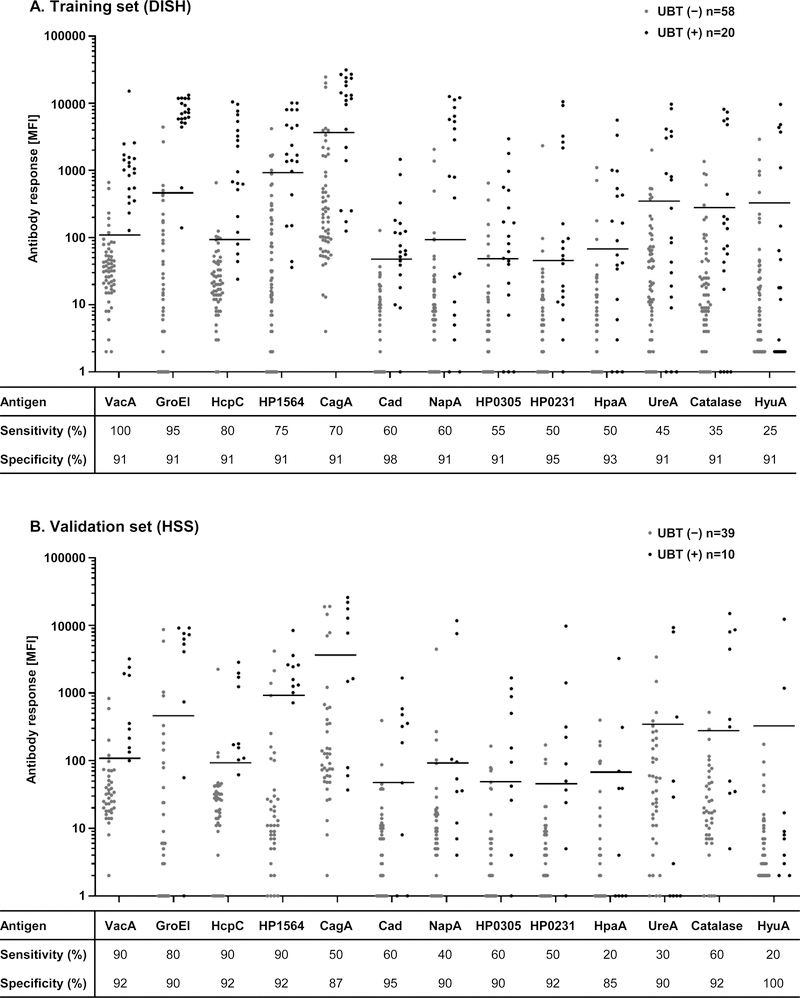
Antibody responses [MFI] to all H. pylori antigens, except HyuA in the training set and HpaA, UreA, and HyuA in the validation set, were statistically significantly higher in UBT-positive than in UBT-negative study participants (Figure 1).
Figure 1: Antibody responses and sero-positivity to H. pylori proteins in relation to active H. pylori infection determined by UBT, in A. DISH (training set), and B. HSS (validation set).
Antibody responses [MFI] to 13 H. pylori proteins were measured using multiplex serology and compared to UBT results. Antibody responses to all antigens except HyuA discriminated significantly (p<0.05, Wilcoxon rank-sum test) UBT (−) from UBT (+) in the training set (DISH, panel A), and except to HpaA, UreA, and HyuA in the validation set (HSS, panel B). Horizontal lines indicate cut-offs derived to achieve at least 90% specificity in the training set (panel A). Tables below the respective dot plots show specificity and sensitivity of sero-positivity in % with the applied cut-offs in the training (panel A) and validation set (panel B).
Application of a cut-off for sero-positivity defined at a specificity of at least 90% compared to UBT results in the training set (DISH) resulted in sensitivities ranging from 25% to 100%. The highest sensitivity in the training set in comparison to UBT results was achieved with sero-positivity to VacA (sensitivity: 100%; 95% CI: 84–100%), followed by GroEl (sensitivity: 95%; 95% CI: 76–100%), HcpC (sensitivity: 80%; 95% CI: 58–92%), and HP1564 (sensitivity: 75%; 95% CI: 53–89%) (Figure 1A).
The sensitivities obtained in the validation set (HSS) with application of the same cut-off values for sero-positivity as in the training set ranged from 20% to 90%, while maintaining a high specificity of over 85%. Furthermore, the four antigens with the highest sensitivities in the training set were reproduced as the top 4 antigen hits in the validation set with a sensitivity of 90% (95% CI: 71–100%) for VacA, HcpC, and HP1564, as well as a sensitivity of 80% (95% CI: 55–100%) for GroEl (Figure 1B).
We further determined specificity and sensitivity for sero-positivity to multiple proteins in comparison to the UBT result, either among all 13 H. pylori proteins, or only among the top four hits, VacA, GroEl, HcpC, and HP1564 (Table 3). With increasing number of positive proteins among all 13 proteins as criterion for sero-positivity, specificity increased (training set: 59% for ≥1 protein sero-positive to 100% for ≥8 proteins; validation set: 59% to 95%), while sensitivity decreased (training set: 100% to 65%; validation set: 100% to 40%). For two categorizations (≥3 and ≥4 positive proteins out of all 13 H. pylori proteins), 100% sensitivity and >85% specificity was reached in both the training and validation sets.
Table 3:
Sero-positivity to H. pylori proteins in relation to active H. pylori infection determined by UBT, DISH (training set) and HSS (validation set).
| N positive | Training set (DISH) n=58 UBT (−), n=20 UBT (+) |
Validation set (HSS) n=39 UBT (−), n=10 UBT (+) |
||||||
|---|---|---|---|---|---|---|---|---|
| proteins | Specificity | 95% CI | Sensitivity | 95% CI | Specificity | 95% CI | Sensitivity | 95% CI |
| Among all 13 proteins | ||||||||
| ≥ 1 | 59 | 46–71 | 100 | 100–100 | 59 | 44–74 | 100 | 100–100 |
| ≥ 2 | 78 | 67–88 | 100 | 100–100 | 74 | 61–88 | 100 | 100–100 |
| ≥ 3 | 86 | 77–95 | 100 | 100–100 | 90 | 80–99 | 100 | 100–100 |
| ≥ 4 | 86 | 77–95 | 100 | 100–100 | 92 | 84–100 | 100 | 100–100 |
| ≥ 5 | 95 | 89–100 | 90 | 77–100 | 92 | 84–100 | 100 | 100–100 |
| ≥ 6 | 98 | 95–100 | 80 | 62–98 | 92 | 84–100 | 70 | 42–98 |
| ≥ 7 | 98 | 95–100 | 70 | 50–90 | 95 | 88–100 | 50 | 19–81 |
| ≥ 8 | 100 | 100–100 | 65 | 44–86 | 95 | 88–100 | 40 | 10–70 |
| Among VacA, GroEl, HcpC, HP1564 | ||||||||
| ≥ 1 | 76 | 65–87 | 100 | 100–100 | 85 | 73–96 | 100 | 100–100 |
| ≥ 2 | 90 | 82–97 | 100 | 100–100 | 92 | 84–100 | 100 | 100–100 |
| ≥ 3 | 100 | 100–100 | 85 | 69–100 | 92 | 84–100 | 100 | 100–100 |
| = 4 | 100 | 100–100 | 65 | 44–86 | 97 | 92–100 | 50 | 19–81 |
Abbreviations: (−), Negative; (+), Positive; CI: confidence interval; DISH: Durham Initiative for Stomach Health; HSS: Healthy Stomach Study; UBT, Urea breath test
When considering VacA, GroEl, HcpC, and HP1564 only, specificity increased from 76% to 100% in the training set (validation set 85% to 97%) and sensitivity decreased from 100% to 65% (validation set: 100% to 50%) from being positive to ≥1 protein to being positive to all four (Table 3). Specifically, being positive to at least two proteins of VacA, GroEl, HcpC, and HP1564 resulted in only six false-sero-positive UBT-negative individuals in the training set (specificity: 90%; 95% CI: 82–97%), while detecting all UBT-positive participants as sero-positive (sensitivity: 100%; 95% CI: 100–100%). These results were reproduced in the validation set (specificity: 92%; 95% CI: 84–100%; sensitivity: 100%; 95% CI: 100–100%).
Decline in antibody response to H. pylori proteins after successful eradication treatment
A sub-set of DISH study participants attended a follow-up 6 months after baseline assessment. Two groups were assessed for changes in antibody response to the top four H. pylori antigen hits (VacA, GroEl, HcpC, and HP1564) over time (Figure 2, results for other antigens in Supplementary Figure S1): those that were UBT-negative at T0 and T1 (Group I, n=7); and those that were UBT-positive at T0 but UBT-negative at T1 due to successful eradication treatment (Group II, n=8). Additionally, we measured antibody responses at T0 and T1 in one study participant who presented with a persistent H. pylori infection at follow-up despite antibiotic treatment (Supplementary figure S2).
Figure 2: Antibody responses to H. pylori proteins VacA, GroEl, HcpC, and HP1564 at baseline (T0) and 6 months follow-up (T1) in DISH.
Antibody responses [MFI] between T0 and T1 were compared for DISH study participants that were UBT negative at T0, and received no H. pylori eradication treatment (grey, group I), as well as for UBT positive study participants at T0 and complete successful eradication treatment at T1 (black, group II). P-values to compare level of antibody response between T0 and T1 in the respective groups was determined using Wilcoxon matched-pairs signed-rank test. A p-value< 0.05 was considered statistically significant.
Among the participants in Group I, median antibody responses at T0 were similar to those at T1 (79 and 120 for VacA, 358 and 252 for GroEl, 83 and 76 for HcpC, and 321 and 389 for HP1564, respectively). In a pairwise comparison, we did not observe any statistical differences in MFI values of these H. pylori proteins. Among the participants in Group II, median antibody responses decreased for the top four antigen hits from T0 to T1 (997 to 294 for VacA, 10,868 to 6,968 for GroEl, 1,961 to 803 for HcpC, and 3,238 to 1,228 for HP1564) (Figure 2). This resulted in statistically significant differences by pairwise comparison for antigens VacA, HcpC, and HP1564 (all p=0.008), but not for GroEl (p=0.055). We even observed sero-reversion in a subset of Group II participants at T1 compared to T0, including sero-reversion to VacA for one participant and to HP1564 for three participants. When we define sero-positivity as ≥2 among VacA, GroEl, HcpC, and HP1564, we also find sero-reversion for one participant.
The one participant with persistent H. pylori infection was initially sero-positive to VacA and GroEl (and sero-negative to HcpC and HP1564) and antibody responses to these antigens did not change substantially from T0 to T1 (Supplementary figure S2).
DISCUSSION
In the present study, including both a training and validation set, we found that level of antibody responses to H. pylori proteins strongly associate with active H. pylori infection as determined by UBT. More specifically, utilizing cut-offs of MFI values for sero-positivity established to achieve at least 90% specificity in comparison to UBT, we reported 75% to 100% sensitivity for the use of the individual antigens VacA, GroEL, HcpC, and HP1564 in determining active H. pylori infection. Additionally, utilizing a criterion of ≥2 sero-positive results to any of these four antigens resulted in a specificity of at least 90% and a sensitivity of 100% in both the training and validation sets. Antibody levels to three (VacA, HcpC, and HP1564) of these four H. pylori proteins also showed a significant decline after a follow-up time of only 6 months among study participants with successful H. pylori eradication. To note, study participants in both the training and validation sets were primarily African American, a population presenting with high prevalence of H. pylori and concomitant health disparities in the US.3
H. pylori multiplex serology, a fluorescent bead-based suspension array including a set of 13 H. pylori antigens, has previously only been validated against a commercially available ELISA based on H. pylori whole cell protein.10 Those analyses found that sero-positivity to four or more of the included H. pylori proteins (15 during that study) resulted in a sensitivity of 89% and a specificity of 82% compared to the ELISA. Individually, antibody responses to VacA, GroEl, HcpC, and HP1564 yielded sensitivities varying between 63% to 88% and specificities between 71% and 92%. The results of this comparison, though, are dependent on the composition of the protein lysate in the ELISA, i.e. whether the specific proteins are represented in the ELISA or not. Thus, with the study presented here, we add important information to the validity of the H. pylori multiplex assay in reference to a non-serological determination, and more importantly, of an active H. pylori infection status.
Formichella et al. developed a similar serological assay including specific H. pylori proteins, however, instead of a bead-based suspension array, this assay is based on a line blot immunoassay (recomLine).18 Proteins CagA, VacA, GroEl, gGT, HcpC, and UreA are recombinantly expressed and immobilized on a nitrocellulose membrane to detect antibody responses in sera. In an initial validation study the authors compared the serological response on the recomLine assay with histologically ascertained H. pylori infection in stomach biopsies from a cohort in Germany and reported a sensitivity of 98% and specificity of 96%,18 similar to the sensitivity of 100% at a specificity of 90% reported in our study. The major advantages of multiplex serology, however, are its flexibility in including additional antigens of interest for the respective study question, as well as its applicability in a high-throughput manner. Although it should be acknowledged that this assay is not commercially available and thus not easily applicable outside a few specific laboratories.
The advantages of the multiplex serology assay become specifically apparent in epidemiological studies of disease associations. For example, the same top four antigen hits in the present study were recently described to be associated with colorectal cancer risk in a large consortium of cohorts in the United States including over 4,000 prospectively ascertained colorectal cancer cases and the same number of control samples, with the association being strongest in African American study participants.9 Moreover, we found that 93% of H. pylori sero-positive individuals were sero-positive to GroEl, independent of race, suggesting this antigen represents a general H. pylori infection marker. The sero-prevalence to well-known H. pylori virulence factors CagA and VacA, however, varied strongly by race (CagA: 60% among H. pylori sero-positive whites versus 86% among H. pylori sero-positive African Americans; VacA: 86% versus 92%, respectively) providing information beyond just a general marker for active H. pylori infection. These findings furthermore highlight the importance of assessing multiple antigens in the application of the assay to both account for heterogeneity in different populations and to explore differences therein. To note, we did not have matching tissue samples in these studies, and thus could not determine concurrent information on the infecting bacterial strain – i.e. whether antibodies detected to CagA indeed correlate with presence of CagA-carrying H. pylori.
Formichella et al. also examined the antibody response detected in the recomLine assay after H. pylori eradication therapy.19 The authors reported a decline in antibody response to all antigens included (except CagA) to below the cut-off over the follow-up period of 17 years, with the strongest decline in level of antibody responses within the first 5 years after eradication. In our study, H. pylori UBT-positive DISH participants were informed by mail about their infection status along with a letter for the primary care physician two weeks after baseline assessment. Within the 6 months follow-up time participants then received treatment from their primary care physician although the exact time point of treatment relative to baseline was unknown to us, which may have caused inter-individual variation in antibody decline. Remarkably, however, we observed a significant decline in antibody response to the majority of antigens, including CagA after this short period of follow-up time (Figure 2 and Supplementary Figure S1), although sero-reversion according to the here defined classification of sero-positivity (≥2 sero-positive of VacA, GroEl, HcpC, and HP1564) was observed in only one of the eight participants that underwent successful eradication therapy. A longer follow-up would have been necessary to assess effects as observed in Formichella et al., however, this was not part of our current study protocol. Thus, we cannot conclude a time-point of sero-reversion after eradication treatment from our data. However, published data suggests low test-and-treat rates for H. pylori in the US, especially in African Americans20, suggesting that serology indeed may be applicable in these populations to study associations with active H. pylori infection.
A limitation of our study is that the analyses were performed within one US geographic region only. As discussed above, it will be important to validate the developed cut-offs and algorithms for sero-positivity in different populations that most likely develop immune response of differing strengths to the infecting agent.21 Moreover, as mentioned above, different populations might carry distinct H. pylori strains potentially contributing to differential immune responses. In this respect it will also be important to test different serum dilutions in addition to the 1:1000 dilution applied here. We specifically chose this serum dilution since a previous study in African Americans and Whites has shown that this dilution is appropriate to study disease associations – i.e. signal-to-noise ratios in the lower end of antibody level where low and high titer antigens were not in saturation allowing quantitative analyses.9 For some of the antigens like Cad, this might have resulted in lowered sensitivity in the data set presented here. However, we sought to allow for a high-throughput methodology, which would be more difficult to achieve and more costly when adding the analysis of an additional serum dilution. It should be noted, too, that the lower sensitivity of certain antigens may not only be dependent on the population under study but also loss in immunogenicity due to conformational changes as a result of the recombinant expression as GST-fusion proteins in E. coli. As an example, HyuA was recombinantly expressed in two parts due to its size, and we do not have information whether this might have affected the immunogenicity of the protein even further. Thus, future studies are needed to validate the present reported findings in different study populations to assess the generalizability of the reported results. In the direction to achieve this, the major strength of our study is its design with a training and validation set and although the validation set differed significantly in factors like race, socioeconomic status, age and BMI, we were still able to reproduce our main findings from the training set in the validation set.
In conclusion, we found that sero-positivity to at least two of the H. pylori proteins VacA, GroEl, HcpC, and HP1564 indicates active H. pylori infection at high specificity and sensitivity utilizing cut-offs for sero-positivity established through comparison with UBT. However, a longer follow-up would be necessary to determine the time-point of sero-reversion after successful eradication therapy and consequently the impact of eradication on the interpretation of multiplex serology results. Thus, we conclude that in populations with generally low rates of H. pylori eradication therapy like the US H. pylori multiplex serology may be applicable to approximate the prevalence of active H. pylori infection in large epidemiological cohorts.
Supplementary Material
Acknowledgments
Funding: The Durham Initiative for Stomach Health (DISH) was funded in part by the Duke Cancer Institute. The Healthy Stomach Study (HSS) was funded in part by the Vanderbilt Institute for Clinical and Translational Research (VICTR) and the National Cancer Institute: Southern Community Cohort Study (U01 CA202979, PI: Blot); HSS enrollment, data collection, sample storage and processing for this study were conducted by the Survey and Biospecimen Shared Resource, which is supported in part by P30 CA068485.
Footnotes
Conflict of interest: None declared
REFERENCES
- 1.Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 2.Hooi JKY, Lai WY, Ng WK, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153(2):420–429. [DOI] [PubMed] [Google Scholar]
- 3.Grad YH, Lipsitch M, Aiello AE. Secular Trends in Helicobacter pylori Seroprevalence in Adults in the United States: Evidence for Sustained Race/Ethnic Disparities. Am J Epidemiol. 2012;175(1):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschi F, Gasbarrini A, Polyzos SA, Kountouras J. Extragastric Diseases and Helicobacter pylori. Helicobacter. 2015;20 Suppl 1:40–46. [DOI] [PubMed] [Google Scholar]
- 5.Venerito M, Vasapolli R, Rokkas T, Delchier JC, Malfertheiner P. Helicobacter pylori, gastric cancer and other gastrointestinal malignancies. Helicobacter. 2017;22 Suppl 1. [DOI] [PubMed] [Google Scholar]
- 6.Skrebinska S, Megraud F, Bessede E. Diagnosis of Helicobacter pylori infection. Helicobacter. 2018;23 Suppl 1:e12515. [DOI] [PubMed] [Google Scholar]
- 7.Megraud F, Floch P, Labenz J, Lehours P. Diagnostic of Helicobacter pylori infection. Helicobacter. 2016;21 Suppl 1:8–13. [DOI] [PubMed] [Google Scholar]
- 8.Cai H, Ye F, Michel A, et al. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol. 2016;45(3):774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butt J, Varga MG, Blot WJ, et al. Serologic Response to Helicobacter pylori Proteins Associated With Risk of Colorectal Cancer Among Diverse Populations in the United States. Gastroenterology. 2019;156(1):175–186 e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michel A, Waterboer T, Kist M, Pawlita M. Helicobacter pylori multiplex serology. Helicobacter. 2009;14(6):525–535. [DOI] [PubMed] [Google Scholar]
- 11.Graham DY, Miftahussurur M. Helicobacter pylori urease for diagnosis of Helicobacter pylori infection: A mini review. J Adv Res. 2018;13:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Signorello LB, Hargreaves MK, Steinwandel MD, et al. Southern community cohort study: establishing a cohort to investigate health disparities. J Natl Med Assoc. 2005;97(7):972–979. [PMC free article] [PubMed] [Google Scholar]
- 13.Epplein M, Signorello LB, Zheng W, et al. Race, African ancestry, and Helicobacter pylori infection in a low-income United States population. Cancer Epidemiol Biomarkers Prev. 2011;20(5):826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonsson A, Green AC, Mallitt KA, et al. Prevalence and stability of antibodies to the BK and JC polyomaviruses: a long-term longitudinal study of Australians. J Gen Virol. 2010;91(Pt 7):1849–1853. [DOI] [PubMed] [Google Scholar]
- 15.Pinto M, Dobson S. BK and JC virus: a review. J Infect. 2014;68 Suppl 1:S2–8. [DOI] [PubMed] [Google Scholar]
- 16.Syrjanen S, Waterboer T, Sarkola M, et al. Dynamics of human papillomavirus serology in women followed up for 36 months after pregnancy. J Gen Virol. 2009;90(Pt 6):1515–1526. [DOI] [PubMed] [Google Scholar]
- 17.Teras LR, Rollison DE, Pawlita M, et al. Prediagnostic circulating polyomavirus antibody levels and risk of non-Hodgkin lymphoma. Cancer Epidemiol Biomarkers Prev. 2015;24(2):477–480. [DOI] [PubMed] [Google Scholar]
- 18.Formichella L, Romberg L, Bolz C, et al. A novel line immunoassay based on recombinant virulence factors enables highly specific and sensitive serologic diagnosis of Helicobacter pylori infection. Clin Vaccine Immunol. 2013;20(11):1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Formichella L, Romberg L, Meyer H, et al. Validation of a Novel Immunoline Assay for Patient Stratification according to Virulence of the Infecting Helicobacter pylori Strain and Eradication Status. J Immunol Res. 2017;2017:8394593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Florea A, Brown HE, Harris RB, Oren E. Ethnic Disparities in Gastric Cancer Presentation and Screening Practice in the United States: Analysis of 1997–2010 Surveillance, Epidemiology, and End Results-Medicare Data. Cancer Epidemiol Biomarkers Prev. 2019;28(4):659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedelec Y, Sanz J, Baharian G, et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167(3):657–669 e621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.