Abstract
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) can be differentiated into many different cell types of the central nervous system. One challenge when using pluripotent stem cells is to develop robust and efficient differentiation protocols that result in homogenous cultures of the desired cell type. Here, we have utilized the SMAD-inhibitors SB431542 and Noggin in a fully defined monolayer culture model to differentiate human pluripotent cells into homogenous forebrain neural progenitors. Temporal fate analysis revealed that this protocol results in forebrain-patterned neural progenitor cells that start to express early neuronal markers after two weeks of differentiation, allowing for the analysis of gene expression changes during neurogenesis. Using this system, we were able to identify many previously uncharacterized long intergenic non-coding RNAs that display dynamic expression during human forebrain neurogenesis.
Keywords: Cell biology, Genetics, Neuroscience, Developmental genetics, Cellular neuroscience, lincRNAs, Forebrain development, Induced pluripotent stem cells, Neural progenitor cells, Differentiation
Cell biology; Genetics; Neuroscience; Developmental genetics; Cellular neuroscience; lincRNAs; Forebrain development; Induced pluripotent stem cells; Neural progenitor cells; Differentiation
1. Introduction
The cerebral cortex is a large and complex brain structure that is responsible for many functions considered unique to humans, such as advanced cognitive abilities and language, and is also involved in many neurodevelopmental and psychiatric disorders. During human brain development, the anterior part of the neural tube gives rise to forebrain neural progenitor cells (fbNPCs), which ultimately form the neurons and glial cells of the cerebral cortex. To date, much of our knowledge and understanding of brain development is acquired from studies using model organisms that are evolutionarily distant to humans. Although many conserved gene regulatory pathways are shared, there are also marked differences that distinguish human forebrain development from other species (Kyrousi and Cappello, 2019). Due to these differences, as well as the practical and ethical issues that limit the availability of tissue from the developing human brain, the ability to generate human fbNPCs in culture for research purposes is invaluable for studies of human brain development as well as for the investigation of neurodevelopmental disorders.
The use of cell culture techniques where human pluripotent stem cells (hPSCs), such as human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs), can be maintained and differentiated in culture has the potential to generate vast numbers of fbNPCs in vitro. Several protocols for generating fbNPCs from hPSCs have been developed over the past few decades. Early methods for neural induction included the formation of embryoid bodies, which results in heterogenic cultures and long differentiation times (Muratore et al., 2014; Watanabe et al., 2005). Subsequently, a monolayer differentiation protocol was developed using dual SMAD-inhibition (Chambers et al., 2009) that has been widely used for neuronal differentiation of hPSCs. This protocol relies on the action of Noggin and SB431542, which provides a synergistic inhibition of SMAD-signaling, resulting in a highly efficient neuronal differentiation of hPSCs.
In this study, we have developed and optimized a fully defined protocol to generate human fbNPCs from iPSCs and ESCs utilizing dual SMAD-inhibition on adherent monolayer cultures kept on laminin-111. Based on RNA-seq, qRT-PCR and immunocytochemistry we found that these cultures efficiently and robustly obtain a homogenous fbNPC identity after two weeks of differentiation. By using transcriptome analysis, we demonstrated that this model system can be used to monitor gene expression changes during neural differentiation of fbNPCs. We therefore used this system to study the expression of long intergenic non-coding RNA (lincRNAs) during the formation of human fbNPCs and identified several lincRNAs with dynamic expression during neurogenesis.
2. Results
2.1. Differentiation and characterization of fbNPCs
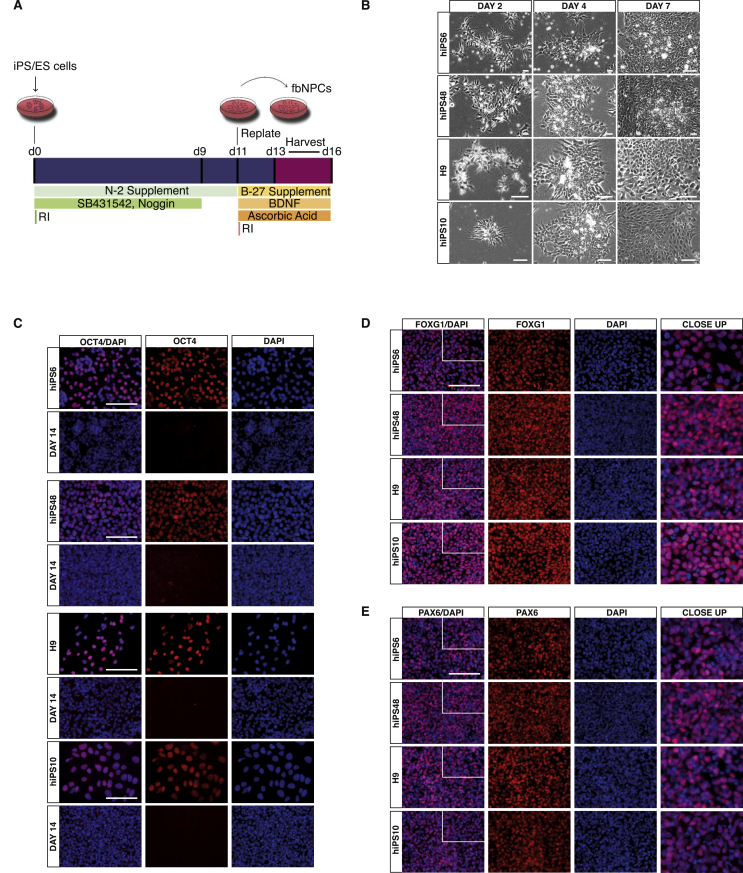
To efficiently differentiate hESCs and iPSCs to fbNPCs we optimized a fully defined 2D-protocol based on dual SMAD-inhibition (Noggin and SB431542) (Chambers et al., 2009; Nolbrant et al., 2017). Briefly, the hESCs and iPSCs were plated at low density (10,000 cells/cm2) on laminin-111-coated plastic in N2 supplemented media with Noggin and SB431542. The two inhibitors were present in the media up until day 9. The cells were dissociated and replated at high density (800,000 cells/cm2) on day 11 and kept in B27 supplemented media with BDNF and ascorbic acid to improve differentiation yields up to day 16 of differentiation (Figure 1A).
Figure 1.
Differentiation overview and characterization of fbNPCs at day 14 by immunocytochemistry. (A) Overview of the differentiation procedure. (B) Brightfield images of the four cell lines used in this study show a similar morphology at day 2, 4, and 7 of differentiation. (C) Immunocytochemical labeling of OCT4 in undifferentiated cells and at day 14 of differentiation. (D) Immunocytochemistry of the forebrain marker FOXG1 at day 14 of differentiation. (E) Immunocytochemistry of the forebrain marker PAX6 at day 14 of differentiation. Nuclei are counterstained with DAPI, shown in blue. Scale bar represents 100 μm.
We used this protocol to differentiate four human lines, one hESC line (H9, (Thomson et al., 1998) and three iPSC lines (RBRC-HPS0328 606A1, RBRC-HPS0360 648A1, and RBRC-HPS0331 610B1 from RIKEN; herein referred to as hiPS6, hiPS48, and hiPS10, respectively (Okita et al., 2013). Brightfield images of all four lines demonstrated a similar morphology during the differentiation at day 2, 4, and 7 (Figure 1B, Fig S1A). The resulting fbNPCs were characterized by immunocytochemistry at day 14, while qRT-PCR and RNA-seq were performed at four different time points (day 13–16). Immunocytochemistry of the cells at the pluripotent stage (i.e. prior to the start of differentiation) and at day 14 of differentiation, demonstrated that the expression of the pluripotency marker OCT4 was completely lost at day 14 (Figure 1C, Fig S1B), while the forebrain markers FOXG1 and PAX6 were expressed at a homogenous level in the vast majority of differentiated cells (Figure 1D-E, Fig S1C-D). Noteworthy, we found very similar results using all four hPSC lines.
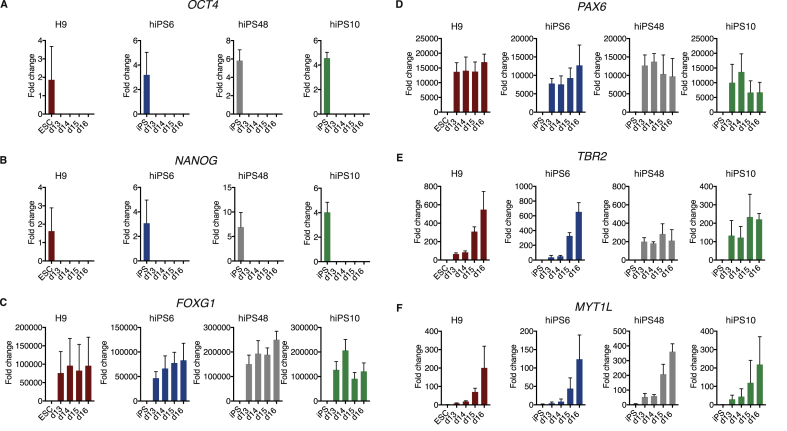
To determine if our protocol mimics the progress of neural development at the transcriptional level, we monitored gene expression between day 13 and 16. We performed qRT-PCR on three independent differentiation replicates from each day between day 13–16 for each of the cell lines as well as on three replicates of each line of undifferentiated cells. We found that expression of the pluripotency markers OCT4 and NANOG were consistently absent in all fbNPC samples (Figure 2A-B), while the forebrain markers FOXG1 and PAX6 were highly expressed in fbNPCs at these time points (Figure 2C-D). Interestingly, the neuronal marker MYT1L and the telencephalic marker TBR2 displayed a temporal increase in expression during the course of differentiation, suggesting that at these time points this protocol allows us to follow the transcriptional dynamics of the early neuronal differentiation of fbNPCs (Figure 2E-F).
Figure 2.
Characterization of fbNPCs by qRT-PCR at day 13 to 16 of differentiation. qRT-PCR data from undifferentiated cells and at day 13–16 of differentiation. The data represents the fold changes in relation to one of the H9 hESC samples for each gene. (A) OCT4. (B) NANOG. (C) FOXG1. (D) PAX6. (E) TBR2. (F) MYT1L. The bars and error bars represent mean with SD of three differentiation replicates.
Together these results demonstrate that this protocol results in the generation of homogenous cultures of human fbNPCs. Noteworthy, we found very similar results using independent differentiation rounds, indicating that the protocol is robust and reproducible. To confirm the potential of the fbNPCs to form mature human forebrain neurons, we also differentiated the cells for 45 days (see methods for details) and stained for the pan-neuronal markers NEUN and MAP2 as well as the forebrain specific neuronal marker TBR1. We found that these markers were all expressed in cells with a mature neuronal morphology at this timepoint (Fig S2, Fig S1E).
2.2. Temporal gene expression changes in differentiating fbNPCs
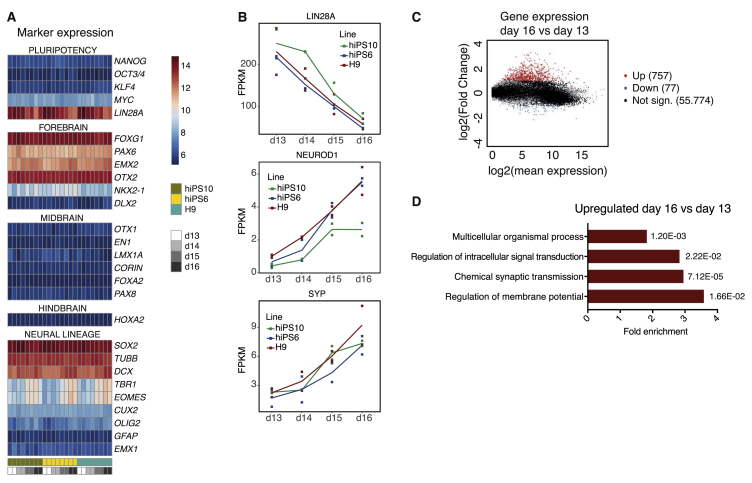
To perform a detailed analysis of the temporal gene expression patterns during differentiation, we performed RNA-seq analysis on two replicates of differentiating H9, hiPS6 and hiPS10 NPCs at d13, d14, d15, and d16 (Figure 3A). Similar to the qRT-PCR data, FOXG1 and PAX6 expression was evident in all samples, with no clear difference between the time points. Additionally, the forebrain marker EMX2 as well as the forebrain-midbrain marker OTX2 were expressed in all samples, whereas other markers of ventral forebrain, midbrain and hindbrain, were absent in our cells, confirming a dorsal forebrain identity of the fbNPCs. Dorsal and ventral fbNPCs correspond to the progenitor cells giving rise to the pallium and subpallium, respectively, in vivo (Campbell, 2003). We monitored in detail the expression of LIN28A, which is associated with undifferentiated cell states, and the neuronal lineage markers NEUROD1 and SYP over the course of day 13–16. This analysis demonstrated a temporal downregulation of LIN28A, whereas the neuronal lineage markers were upregulated (Figure 3B). Additionally, the replicates for each time point were highly consistent, demonstrating the robustness of the protocol.
Figure 3.
Temporal transcriptome changes from day 13 to day 16. RNA-seq data of H9, hiPS6 and hiPS10 fbNPCs at day 13, 14, 15, and 16 of differentiation. (A) Heatmap displaying the marker expression profile between day 13 and day 16, two differentiation replicates per time-point. (B) Expression of LIN28A, NEUROD1, and SYP transcripts plotted as fragments per kilobase of transcript per million mapped reads (FPKM), the line represents average values for each time-point and the squares represent each differentiation replicate. (C) MA plot displaying significantly upregulated (p-adj. < 0.0001 & log2(FC) > 1) genes in day 16 compared to day 13 plotted in red, significantly downregulated (p-adj. < 0.0001 & log2(FC) < -1) genes in blue and non-significant genes in black. (D) Gene ontology analysis of upregulated genes (as shown in C) showing the fold enrichment and p-values for each parent term.
We next set stringent criteria to identify genes that are up- or down-regulated upon differentiation (day 16 compared to day 13, p-adj. < 0.0001 & log2(fold change) > 1 or log2(fold change) < -1 for up- or down regulated genes, respectively). We found that 757 genes were significantly upregulated while 77 genes were downregulated between day 16 and 13 (Figure 3C, top 50 up- and down-regulated genes listed in Supplementary Tables 1 and 2, respectively). To investigate the functional roles of these genes we performed gene ontology analysis of biological processes. We found that genes involved in the regulation of intracellular signal transduction, synaptic transmission as well as regulation of membrane potential were more highly expressed at day 16 compared to day 13, confirming that transcriptional programs associated with neuronal maturation were activated during this period (Figure 3D, Supplementary Table 3). Together, these data demonstrate that this model system offers a possibility to identify transcripts that are dynamically regulated during human forebrain neurogenesis.
2.3. Identification of dynamically expressed lincRNAs upon neural differentiation
As mentioned above, the complex development of the human forebrain is thought to underlie many human-specific characteristics, but for many of these unique mechanisms the underlying genetic elements are unknown. However, it is known that the non-coding sequences, such as long non-coding RNAs (lncRNAs), are less conserved throughout evolution compared to the coding sequences and these are currently widely accepted to play important roles in a variety of biological processes (reviewed e.g. in (Aprea and Calegari, 2015). LncRNAs have the potential to affect gene expression in a variety of ways by regulating the transcription of genes in cis or in trans, as well as the translation efficiency of mRNAs. An interesting class of non-coding RNAs are the long intergenic non-coding RNAs (lincRNAs), which are a type of non-coding RNAs of at least 200 nucleotides in length that do not overlap with coding genes. However, so far only a handful of lincRNAs, such as MALAT1, have been implicated in neuronal differentiation (Guennewig and Cooper, 2014; Ng et al., 2012).
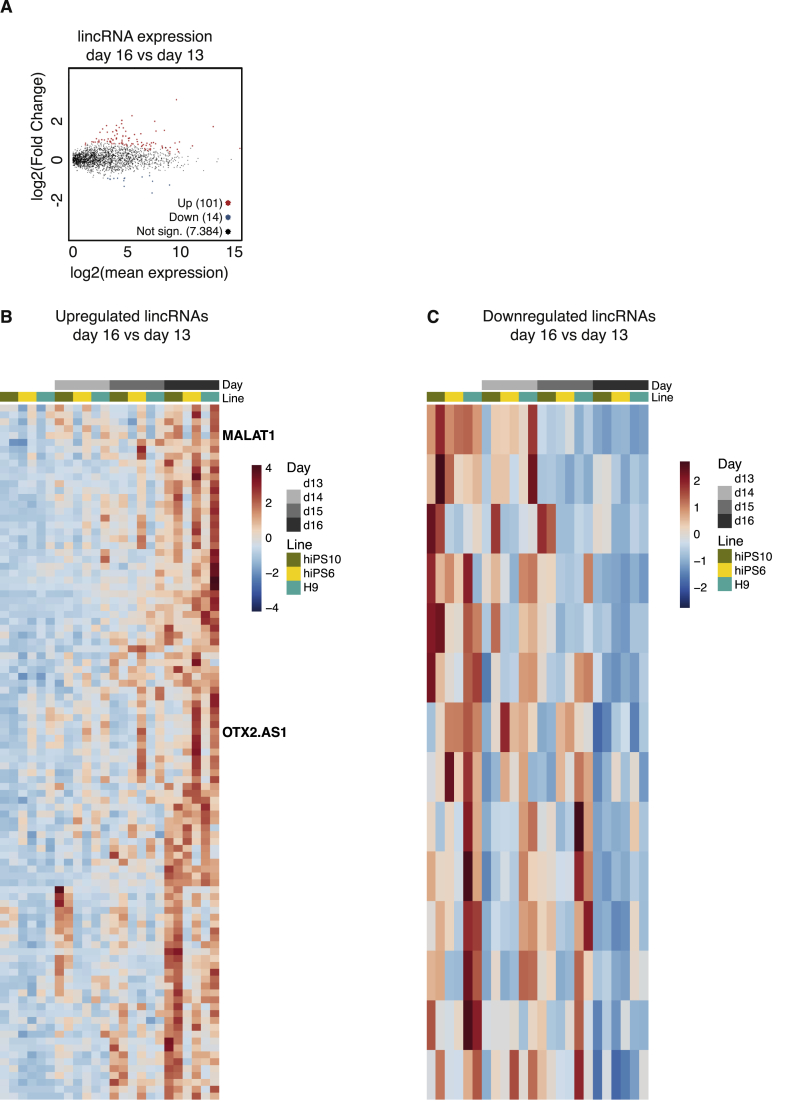
To identify novel lincRNAs that display dynamic expression during human forebrain development, we analyzed and compared the expression of lincRNAs using our fbNPC RNA-seq data-set. By selecting significantly changed lincRNAs (p-adj < 0.001), we found 101 lincRNAs that were more highly expressed at day 16 compared to day 13, and 14 lincRNAs that were down-regulated (Figure 4A-C). We found upregulation of expression during differentiation of lincRNAs that have previously been implicated in neurodevelopment, such as the previously mentioned MALAT1 as well as an antisense transcript of the transcription factor OTX2 (OTX2-AS1). However, the majority of the lincRNAs that we identified have previously not been implicated in human brain development (Supplementary Tables 4–5).
Figure 4.
Expression of lincRNAs changes upon differentiation. (A) MA plot illustrating the log2 fold changes of lincRNAs between day 16 and day 13 fbNPCs, upregulated lincRNAs shown in red and downregulated in blue (p-adj < 0.001). (B) Heatmap of upregulated lincRNAs (p-adj < 0.001). (C) Heatmap of downregulated lincRNAs (p-adj < 0.001).
3. Discussion
In this study, we describe an optimized protocol for 2D differentiation of hPSCs into fbNPCs that is robust and highly efficient. The differentiation protocol is highly reproducible between differentiation replicates and cell lines, and independent on the origin of the hPSCs (iPS- or hESC-line). The protocol is adapted from Nolbrant et al. (2017), in which ventral midbrain progenitors were generated using a fully defined protocol, where the cells are grown on laminin-111. Our current protocol uses a similar approach, but without the addition of the ventralizing and caudalizing factors (SHH–C24II, CHIR99021, FGF8b) in order to generate dorsal fbNPCs. Nolbrant et al. described their progenitors at day 16 as late caudal ventral midbrain progenitors which is the basis for the selection of time points of analysis in our study. Our protocol is slightly different from previously described protocols to generate fbNPCs, which include multiple different strategies for neural differentiation, e.g. through the formation of embryoid bodies (Zhang et al., 2018), single SMAD inhibition with Noggin (Espuny-Camacho et al., 2013), or by using a cocktail of small molecules and growth factors in addition to the dual-SMAD inhibitors (Maroof et al., 2013). Furthermore, the timing of dual SMAD-inhibition varies between previous studies. (Chambers et al., 2009) added dual SMAD-inhibitors for different time lengths, e.g. 11 days, whereas (Hu et al., 2010) used noggin throughout the first 15 days of differentiation and SB only on day 0–5. In our protocol Noggin and SB431542 were both kept in the media up until day 9.
By using our optimized protocol, we obtained homogeneous cultures of FOXG1-expressing fbNPCs and transcriptome analysis confirmed that they express appropriate neuronal and forebrain markers, while genes related to other brain regions or other tissues were not detected. Interestingly, when we analyzed the fbNPCs during day 13 to day 16 of differentiation we found that they corresponded to a differentiation state resembling early neuronal commitment, as demonstrated by the gradual increase in neuronal markers such as MYT1L, NEUROD1 and SYP. This suggest that our model system can be used to identify novel genes or transcripts that are activated or silenced during human forebrain neurogenesis.
This allowed us to expand our analysis to non-coding transcripts, such as lincRNAs, that have the ability to affect the translational efficiency of mRNAs and which therefore are important players in post-translational gene regulation. We searched for lincRNAs that were differentially expressed between day 13 and 16 of differentiation, since they may then play an important role during neurogenesis. Interestingly, we were able to identify many novel lincRNAs that have not been previously implicated in human neurogenesis and this data set should be a useful resource for future studies, for example investigations of complex neurodevelopmental disorders, such as autism spectrum disorder (ASD) and schizophrenia (Supplementary Table 4–5). In fact, previous studies have linked dysregulation of lncRNAs to ASD (Roberts et al., 2014; Ziats and Rennert, 2013). Furthermore, one study used an expression quantitative trait loci analysis to investigate the genetic variants of lincRNAs associated with clinical phenotypes, such as schizophrenia (Branco et al., 2018). These studies indicated that lincRNAs could play important roles in complex neural disorders, but to date the connection between lincRNAs and these disorders is not well understood. Our study provides a resource of lincRNAs associated with human neural differentiation, which could be of interest in the context of neurodevelopmental diseases.
In summary, we have generated a fully defined protocol for differentiation of human pluripotent stem cells into fbNPCs. Our protocol is highly efficient and robust, allowing for reliable differentiation into a vast number of fbNPCs that can be used for further studies of neurodevelopment, evolution, and drug screening.
4. Methods
4.1. hPSC culture
hESCs and iPSCs were maintained in iPS brew medium (StemMACS iPS-Brew XF and 0.5% penicillin/streptomycin (Gibco)) on Nunc Δ multidishes coated with LN521 (0.7 μg/cm2; Biolamina). Cells were passaged approximately 1:2–1:10 every 2–5 days, starting with one rinse with DPBS (Gibco) followed by dissociation using 0.5 mM EDTA (75 μl/cm2; Gibco) at 37 °C for 7 min. Following incubation, EDTA was carefully removed from the well and the cells were washed off and collected in 10 ml wash medium (9.5 ml DMEM/F-12 (31330–038; Gibco) with 0.5 ml knockout serum replacement (Gibco)). The cells were then centrifuged at 400 × g for 5 min, supernatant aspirated, and the cells were resuspended in iPS brew medium supplemented with 10 μM Y27632 (Rock inhibitor; Miltenyi) and plated for expansion. The media was changed daily to fresh iPS brew medium.
4.2. Differentiation into forebrain neural progenitors
hESCs and iPSCs were dissociated as previously described for passaging. After centrifugation, the cells were resuspended in N2 medium consisting of 1:1 DMEM/F-12 (21331–020; Gibco) and Neurobasal (21103–049; Gibco) supplemented with 1% N2 (Gibco), 2 mM L-glutamine (Gibco), and 0.2% penicillin/streptomycin. The cells were counted twice and plated at a density of 10,000 cells/cm2 in 250 μl medium/cm2 on LN111-coated Nunc Δ multidishes (1.1–1.7 μg/cm2; Biolamina). The culture medium was supplemented with the dual SMAD-inhibitors 10 μM SB431542 (Axon) and 100 ng/ml noggin (Miltenyi), as well as 10 μM Y27632 for increased survival. The medium was changed every 2–3 days (N2 medium supplemented with SB431542 and Noggin) up until day 9 of differentiation, when N2 medium without SB431542 and Noggin was used. On day 11, the cells were dissociated by washing twice with DPBS followed by incubation with StemPro accutase (75 μl/cm2; Gibco) for 10–20 min at 37 °C. The cells were washed off and collected in 10 ml wash medium, centrifuged for 5 min at 400 × g and resuspended in B27 medium consisting of Neurobasal supplemented with 1% B27 without vitamin A (Gibco), 2 mM L-glutamine and 0.2% penicillin/streptomycin. The cells were counted twice and replated at a high density of 800,000 cells/cm2 on LN111-coated plastic (1.1–1.7 μg/cm2) in B27 medium (600 μl medium/cm2) supplemented with Y27632 (10 μM), BDNF (20 ng/ml; R&D), and L-ascorbic acid (0.2 mM; Sigma-Aldrich). The cells were kept in the same medium until day 14, after which the medium was replaced with fresh B27 medium supplemented with BDNF and L-ascorbic acid.
4.3. Long term differentiation
At day 16 of differentiation, the fbNPCs were washed twice with DPBS and detached using StemPro accutase (approximately 10 min at 37 °C). Following dissociation, the cells were transferred to a tube with 10 ml wash medium and the cell number was quantified. The cell amount needed for replating (155,000 cells/cm2) was transferred to a new vial and the cells were spun at 400 × g for 5 min, resuspended and replated in B27 medium supplemented with 10 μM Y27632, 20 ng/ml BDNF, 0.2 mM Ascorbic Acid, 10 ng/ml GDNF (R&D), 500 μM cAMP (Merck), and 1μM DAPT (Tocris Bioscience) at a density of 500,000 cells/ml medium on plates coated with DPBS + Ca2-/+Mg2 (Thermo Fisher Scientific) with 1:200 laminin30 (Fisher Scientific), 1:100 poly-L-ornithine (Sigma-Aldrich), and 1:100 fibronectin (Thermo Fisher Scientific). 75% of the medium was exchanged to fresh medium with the before mentioned supplements, with the exception of Y27632, every two to three days up until day 45 of differentiation, at which point the cells were fixed for immunocytochemistry.
4.4. Immunocytochemistry
The cells were washed with DPBS and fixed at room temperature for 15 min with 4% paraformaldehyde (Merck Millipore), and washed with DPBS. The fixed cells were blocked for a minimum of 30 min in a blocking solution consisting of KPBS with 0.25% triton-X100 (Fisher Scientific) and 5% donkey serum. The primary antibody (rabbit anti-FOXG1, 1:50 dilution, Abcam, RRID: AB_732415; mouse anti-OCT3/4, 1:500, Santa Cruz Biotechnology, RRID: AB_628051; mouse anti-MAP2, 1:1000, Abcam, RRID: AB_2138153; rabbit anti-PAX6, 1:1000, Biolegend, RRID: AB_2565003; mouse anti-NEUN, 1:1000 dilution, Millipore, RRID: AB_2298772; rabbit anti-TBR1, 1:500, Abcam, RRID: AB_2200219) was added and incubated at room temperature overnight. On the following day, the cells were washed with KPBS and blocked for at least 10 min in donkey serum blocking solution. The secondary antibody (donkey anti-rabbit Cy3, donkey anti-rabbit Cy2, donkey anti-mouse Cy2; 1:200; Jackson Lab) was added with the nuclear stain DAPI (1:1000; Sigma-Aldrich) and incubated at room temperature for approximately 1 h, followed by 2–3 rinses with KPBS. The immunocytochemically labelled cells were then visualized with a Leica microscope (model DMI6000 B), and images were cropped and adjusted in Adobe Photoshop CC 2015.
4.5. Bulk RNA sequencing
On the day of harvest, the cells were washed once with DPBS and lysed with 350 μl RLT buffer with 1% β-mercaptoethanol (Thermo Fisher). The RNA was isolated using the RNeasy mini kit (Qiagen) according to manufacturer's protocol. The quality and concentration of the RNA samples was assessed using 2100 Bioanalyzer (RNA nano; Agilent) and Qubit (RNA HS assay kit). Libraries for sequencing were prepared with the TruSeq RNA Library Prep kit v2 (Illumina) and the quality of the libraries was assessed using the Bioanalyzer (high-sensitivity DNA assay) and Qubit (dsDNA HS assay kit). Finally, the libraries were sequenced 150x paired-end reads (300 cycles) with Illumina NextSeq 500.
The reads were mapped to the human reference genome (GRCh38) using STAR aligner v2.5.0a (Dobin et al., 2013), allowing a ratio of mismatches per mapped length less than 0.03 and multimapping at a maximum of 10 loci. Gencode v27 (Harrow et al., 2012) gene models were used for splice junction annotation. Gene counts were quantified using the Subread package FeatureCounts (Liao et al., 2014), using the Gencode (v27) gene annotations. Normalization and differential expression analysis was performed with the R package DESeq2 (Love et al., 2014), including all genes. The R package limma was used to correct batch effects for the analysis shown in Figure 3A (Ritchie et al., 2015). In-house scripts used for analysis can be found on https://github.com/perllb.
Gene ontology enrichment analysis was performed using the Panther Overrepresentation test (Released, 20190711) with PANTHER v14.1, released 2019-03-12 (Mi et al., 2013). To evaluate significance, all genes with a mean expression of >2 in all samples were used as the reference background list, to avoid bias from only analyzing a subset of all possible genes. Fisher's exact test with Bonferroni correction for multiple testing was used to test for overrepresentation with GO-slim biological process.
4.6. qRT-PCR
250 ng RNA of each sample was used to synthesize cDNA using Maxima cDNA synthesis kit (Thermo Scientific). qRT-PCR was performed with the LightCycler® 480 instrument (Roche) using the primers listed in supplementary table 6 and SYBR green master mix (Roche). Three technical replicates were performed for each sample and the cycle values were normalized to GAPDH and ß-actin.
Declarations
Author contribution statement
Daniela A Grassi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Per Ludvik Brattås: Analyzed and interpreted the data.
Marie E Jönsson, Diahann Atacho, Ofelia Karlsson: Performed the experiments.
Sara Nolbrant, Malin Parmar: Contributed reagents, materials, analysis tools or data.
Johan Jakobsson: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
The work was supported by grants from the Swedish Research Council, the Swedish Foundation for Strategic Research, the Swedish Brain Foundation, the Swedish excellence project Basal Ganglia Disorders Linnaeus Consortium (Bagadilico), and the Swedish Government Initiative for Strategic Research Areas (MultiPark & StemTherapy).
Competing interest statement
The authors declare the following conflict of interests: Malin Parmar; owner of Parmar Cells AB and co-inventor of the U.S. patent application 15/093,927 owned by Biolamina AB and EP17181588 owned by Miltenyi Biotec.
Additional information
No additional information is available for this paper.
The accession number for the RNA-seq data presented in this paper is NCBI GEO: GSE131697.
Acknowledgements
We are grateful to all members of the Jakobsson lab, we would especially like to thank J. Johansson, M. Persson Vejgården, U. Jarl, and A. Hammarberg for technical assistance.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Aprea J., Calegari F. Long non-coding RNAs in corticogenesis: deciphering the non-coding code of the brain. EMBO J. 2015;34:2865–2884. doi: 10.15252/embj.201592655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco P.R., de Araujo G.S., Barrera J., Suarez-Kurtz G., de Souza S.J. Uncovering association networks through an eQTL analysis involving human miRNAs and lincRNAs. Sci. Rep. 2018;8:15050. doi: 10.1038/s41598-018-33420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr. Opin. Neurobiol. 2003;13:50–56. doi: 10.1016/s0959-4388(03)00009-6. [DOI] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I., Michelsen K.A., Gall D., Linaro D., Hasche A., Bonnefont J., Bali C., Orduz D., Bilheu A., Herpoel A. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Guennewig B., Cooper A.A. The central role of noncoding RNA in the brain. Int. Rev. Neurobiol. 2014;116:153–194. doi: 10.1016/B978-0-12-801105-8.00007-2. [DOI] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrousi C., Cappello S. Wiley Interdiscip Rev Dev Biol; 2019. Using brain organoids to study human neurodevelopment, evolution and disease; p. e347. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof A.M., Keros S., Tyson J.A., Ying S.W., Ganat Y.M., Merkle F.T., Liu B., Goulburn A., Stanley E.G., Elefanty A.G. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore C.R., Srikanth P., Callahan D.G., Young-Pearse T.L. Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.Y., Johnson R., Stanton L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolbrant S., Heuer A., Parmar M., Kirkeby A. Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 2017;12:1962–1979. doi: 10.1038/nprot.2017.078. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.C., Morris K.V., Wood M.J. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Kamiya D., Nishiyama A., Katayama T., Nozaki S., Kawasaki H., Watanabe Y., Mizuseki K., Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Zhang M., Ngo J., Pirozzi F., Sun Y.P., Wynshaw-Boris A. Highly efficient methods to obtain homogeneous dorsal neural progenitor cells from human and mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res. Ther. 2018;9:67. doi: 10.1186/s13287-018-0812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats M.N., Rennert O.M. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013;49:589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.