Endonuclease Q (EndoQ) is a lesion-specific DNA repair enzyme present in certain bacteria and archaea. To date, it remains unclear how EndoQ recognizes damaged bases. Understanding the mechanism of substrate recognition by EndoQ is important to grasp genome maintenance systems in organisms with EndoQ. Here, we find that EndoQ from the euryarchaeon Pyrococcus furiosus recognizes the imide structure in nucleobases by base flipping, and the cleavage activity is enhanced by the base pair instability of the target base, along with the discovery of its cleavage activity toward 5,6-dihydrouracil, 5-hydroxyuracil, 5-hydroxycytosine, and uridine in DNA. Furthermore, a potential role of EndoQ in Bacillus subtilis as an antiviral enzyme by digesting viral genome is demonstrated.
KEYWORDS: DNA repair, endonuclease Q, archaea, Pyrococcus furiosus, Bacillus subtilis, endonuclease
ABSTRACT
Endonuclease Q (EndoQ), a DNA repair endonuclease, was originally identified in the hyperthermophilic euryarchaeon Pyrococcus furiosus in 2015. EndoQ initiates DNA repair by generating a nick on DNA strands containing deaminated bases and an abasic site. Although EndoQ is thought to be important for maintaining genome integrity in certain bacteria and archaea, the underlying mechanism catalyzed by EndoQ remains unclear. Here, we provide insights into the molecular basis of substrate recognition by EndoQ from P. furiosus (PfuEndoQ) using biochemical approaches. Our results of the substrate specificity range and the kinetic properties of PfuEndoQ demonstrate that PfuEndoQ prefers the imide structure in nucleobases along with the discovery of its cleavage activity toward 5,6-dihydrouracil, 5-hydroxyuracil, 5-hydroxycytosine, and uridine in DNA. The combined results for EndoQ substrate binding and cleavage activity analyses indicated that PfuEndoQ flips the target base from the DNA duplex, and the cleavage activity is highly dependent on spontaneous base flipping of the target base. Furthermore, we find that PfuEndoQ has a relatively relaxed substrate specificity; therefore, the role of EndoQ in restriction modification systems was explored. The activity of the EndoQ homolog from Bacillus subtilis was found not to be inhibited by the uracil glycosylase inhibitor from B. subtilis bacteriophage PBS1, whose genome is completely replaced by uracil instead of thymine. Our findings suggest that EndoQ not only has additional functions in DNA repair but also could act as an antiviral enzyme in organisms with EndoQ.
IMPORTANCE Endonuclease Q (EndoQ) is a lesion-specific DNA repair enzyme present in certain bacteria and archaea. To date, it remains unclear how EndoQ recognizes damaged bases. Understanding the mechanism of substrate recognition by EndoQ is important to grasp genome maintenance systems in organisms with EndoQ. Here, we find that EndoQ from the euryarchaeon Pyrococcus furiosus recognizes the imide structure in nucleobases by base flipping, and the cleavage activity is enhanced by the base pair instability of the target base, along with the discovery of its cleavage activity toward 5,6-dihydrouracil, 5-hydroxyuracil, 5-hydroxycytosine, and uridine in DNA. Furthermore, a potential role of EndoQ in Bacillus subtilis as an antiviral enzyme by digesting viral genome is demonstrated.
INTRODUCTION
Chemical alternations of nucleobases are observed in all domains of life, Bacteria, Archaea, Eukarya, and their viruses. Modifications mediated by enzymes are known to play important roles in epigenetic regulations and in the context of the arms race between hosts and viruses (1–3). Variations also can be introduced into genomes by endogenous and exogenous damage (4) or the misincorporation of noncanonical deoxynucleotide monophosphates (dNMPs) in the nascent strand by DNA polymerase during DNA synthesis (1, 5).
Most of the noncanonical bases found in genomes have been reported to be derived from pyrimidine bases (6). The deamination of cytosine generates uracil via a hydrolytic reaction, which can be catalyzed by cytidine deaminases or caused by spontaneous decay (4, 7). Uracil is also known to act as a precursor of thymine (5-methyluracil). The methylation of cytosine is carried out by DNA methyltransferases. The methylation of the cytosine C-5 (5mC) is a prime epigenetic mark in eukaryotic genomes (8), and N4-cytosine methylation is observed more commonly in prokaryotic genomes (2, 9). The oxidation of cytosine with reactive oxygen species such as ·OH followed by the hydrolytic deamination can form 5-hydroxyuracil (5hU), 5-hydroxycytosine (5hC), and 5,6-dihydroxy-5,6-dihydrouracil as stable compounds (10) and 5hU and 5,6-dihydrouracil (DHU) as major products under anoxic conditions (11). One of the major products of the thymine oxidation is 5,6-dihydroxy-5,6-dihydrothymine (thymine glycol [Tg]) (12). Purine bases are also chemically modified because of damage or enzymatic reactions. The spontaneous deamination of adenine and guanine yields hypoxanthine (Hx) and xanthine (X), respectively (13, 14). The methylation of adenine as N6-adenine methylation (6mA) is commonly observed both in archaeal and bacterial domains and enables organisms to distinguish between self and nonself DNA (2, 9, 15).
Among all of these modified bases generated because of damage, many are known to cause genome instability. For example, uracil and its derivatives, which are generated from cytosine, are very mutagenic because uracil tends to pair with adenine instead of guanine, leading to a G·C to A·T transition in the next round of replication (4). Indeed, genomes from Bacteria and Eukarya have mutational biases that decrease the GC content, potentially due to cytosine deamination (16). The removal of uracil from DNA occurs via the base excision repair (BER) pathway (17). In this pathway, uracil-DNA glycosylase (UDG) is responsible for detecting uracil in DNA and hydrolyzing the DNA-N-glycosidic bond to release the base from the DNA strand. This system is widely distributed across all domains of life, which underscores its crucial contribution toward uracil removal from the very early period of life (18). To date, many such lesion-specific DNA glycosylases or endonucleases have been identified (17, 19).
Endonuclease Q (EndoQ) was originally identified in the hyperthermophilic euryarchaeon Pyrococcus furiosus as a novel lesion-specific endonuclease (20). This enzyme recognizes three mutagenic deaminated bases (uracil, hypoxanthine, and xanthine) and an apurinic/apyrimidinic (AP) site, and directly cleaves the DNA strand at the 5′ sides of the lesions, leaving 3′-hydroxyl and 5′-phosphate groups (20). Previous work showed that the putative EndoQ homologs are present in certain prokaryotes from the following phyla: Firmicutes, Actinobacteria, Proteobacteria, and Spirochaetes in Bacteria and Euryarchaeota in Archaea (21). Although EndoQ appears to be conserved in a limited number of species, the substrate specificities of the homologs seem to be highly conserved among the homologs. The archaeal orthologs from Thermococcus kodakarensis and the methanogen Methanosarcina acetivorans and the bacterial ortholog from the Gram-positive bacterium Bacillus pumilus have all been shown to generate a nick on DNA strands containing uracil, hypoxanthine, and an AP site (20–22). However, the type of substrates EndoQ preferentially acts upon and the factors contributing to its preferences for various DNA substrates remain unknown. Notably, EndoQ is very unique with regard to its substrate specificity because it does not discriminate between pyrimidines and purines (20).
Here, we examined the substrate specificity range of EndoQ from P. furiosus (PfuEndoQ). Our results reveal its cleavage activities toward specific uracil derivatives (DHU, 5hU, and 5hC), all of which are mutagenic bases. Further, we determined the kinetic parameters in order to identify the preferential bases and DNA structures for PfuEndoQ. Moreover, to explore the role of EndoQ, we examined the activity of EndoQ from Bacillus subtilis in the presence of the uracil-glycosylase inhibitor (UGI) from B. subtilis bacteriophage PBS1, in whose genome uracil is fully substituted for thymine (23).
RESULTS
PfuEndoQ cleaves 5hU-, 5hC-, and DHU-containing DNA.
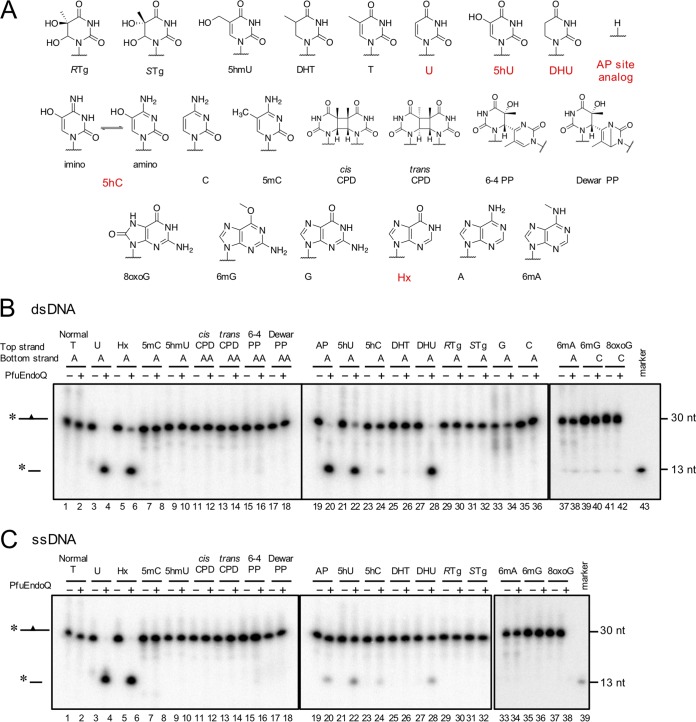
A previous study showed that EndoQ can recognize specific DNA lesions, including U, Hx, X, and an AP site in DNA, but cannot recognize a canonical base lesion (G·T mismatch) or dimerized bases (cyclobutane pyrimidine dimers [CPD]) (20). To understand the recognition mechanism of EndoQ, we investigated the substrate specificity range of EndoQ using damaged and modified bases. As shown in Fig. 1, the endonuclease activity of PfuEndoQ was tested against 21 different types of DNA substrates, and PfuEndoQ was found to exhibit cleavage activities toward 5hU, 5hC, and DHU in addition to the previously reported bases, including U, Hx, and an AP site. The activity toward 5hC seems to be much lower than that toward 5hU, DHU, U, Hx, and the AP site. Additionally, no cleavage activity was detected with 5mC, 5-hydroxymethyluracil (5hmU), UV photoproducts (cis/trans-CPD, [6-4]/Dewar photoproducts), 5,6-dihydrothymine (DHT), Tg, G·A or C·A mismatches, 6mA, 8-methylguanine, or 8-oxoguanine. When comparing the results of Fig. 1B and C, the activity range was conserved between single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA); however, the cleavage efficiency seemed to be suppressed with ssDNA. Overall, these results indicate that the activity of PfuEndoQ was limited to some mutagenic bases. Notably, PfuEndoQ can act on U and its derivatives (5hU and DHU) but not on thymine and its derivatives (5hmU [or 5-hydroxythymine], DHT, and Tg); this indicates that the methylation at the C-5 position of uracil might cause steric hinderance in the recognition pocket.
FIG 1.
Substrate screening for PfuEndoQ. Various modified or damaged bases containing DNA (A) (see Table S1 in the supplemental material) were incubated in a reaction mixture with or without PfuEndoQ at 60°C for 10 min. DNA products were separated by 8 M urea-12% PAGE, and 32P-labeled DNA strands were detected by autoradiography. (A) Cleaved substrates by PfuEndoQ are highlighted in red. (B) dsDNA. (C) ssDNA. −, no enzyme; +, 100 nM PfuEndoQ; marker, 13 nt.
PfuEndoQ cuts the DNA backbone at the 5′ side of 5hU, 5hC, and DHU in DNA.
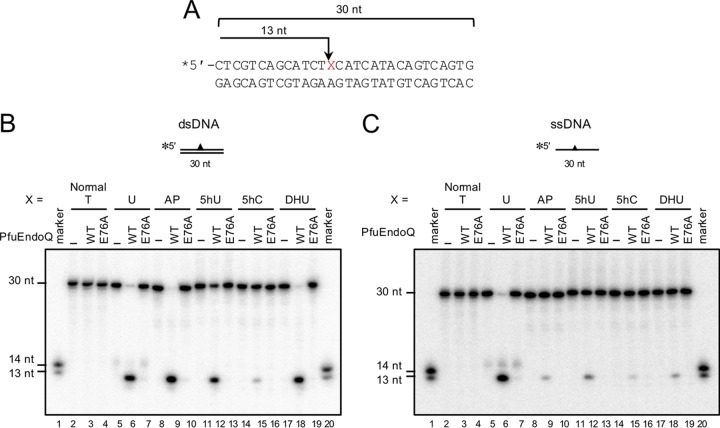
To determine the nicking sites of PfuEndoQ in 5hU-, 5hC-, and DHU-containing DNA, the cleavage products were analyzed on sequencing gels. As shown in Fig. 2, all of the cleavage products were 13 nucleotides (nt) long; this indicates that the nicks were generated at the 5′ side of the damaged site (Fig. 2B and C, lanes 6, 9, 12, 15, and 18). This property was conserved in both dsDNA and ssDNA (Fig. 2B and C), and it also corresponded to the activity observed with U, Hx, X, or the AP site. To ensure that this activity was attributed to PfuEndoQ, we prepared the inactive mutant, PfuEndoQE76A, in addition to the wild type (WT) (see Fig. S1 in the supplemental material). Previous mutational and structural analyses showed that E76 is essential for EndoQ cleavage activity (24). When PfuEndoQE76A was added to the reactions, no cleavage product was detected, which demonstrated that these cleavage activities were attributed to PfuEndoQ and not due to contaminating activities (Fig. 2B and C, lanes 7, 10, 13, 16, and 19).
FIG 2.
PfuEndoQ generates a nick at the 5′ sides of 5hU, 5hC, and DHU. (A) The nicking sites of PfuEndoQ in 5hU-, 5hC-, and DHU-containing DNA. (B and C) DNA substrates were incubated with or without PfuEndoQ WT or E76A (10 nM) at 60°C for 10 min in a reaction mixture. DNA products were separated by 8 M urea-12% PAGE (sequencing gel), and 32P-labeled DNA strands were detected by autoradiography. (B) dsDNA. (C) ssDNA. −, no enzyme; marker, 13 and 14 nt.
Kinetics of PfuEndoQ activity.
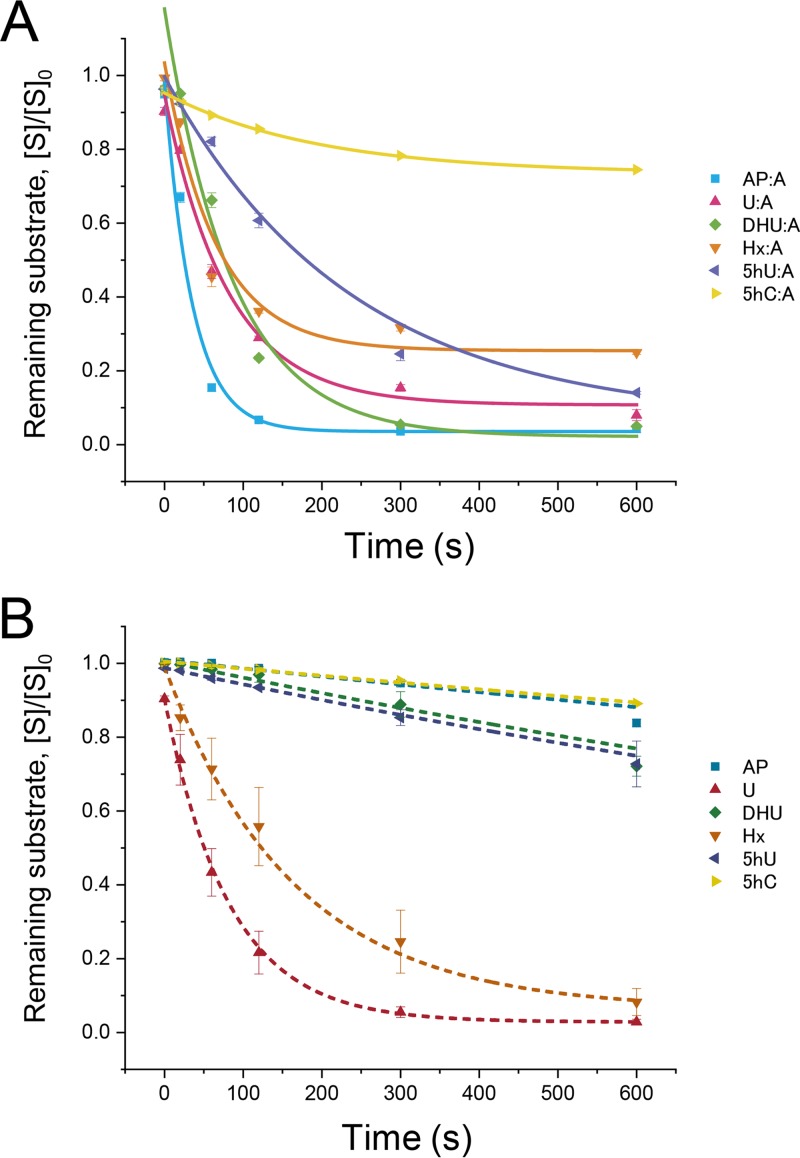
Although we have understood that PfuEndoQ can act on various damaged bases, the substrate structure type preferred by this enzyme remains unknown. Interestingly, EndoQ can recognize either dsDNA or ssDNA as well as pyrimidine and purine bases. To determine its primary target in cells, we estimated the kinetic parameters of PfuEndoQ. By monitoring the cleavage activities of PfuEndoQ over time, we found that the cleavage efficiency of AP·A > U·A > DHU·A > Hx·A > 5hU·A > 5hC·A for dsDNA, and U > Hx > 5hU > DHU > 5hC > AP for ssDNA (Table 1 and Fig. 3). Overall, EndoQ exhibited a strong preference for dsDNA over ssDNA. In particular, in the case of the AP site, PfuEndoQ significantly increased the activity by approximately 250-fold for dsDNA. In contrast, PfuEndoQ exhibited similar activity for U and Hx in both ssDNA and dsDNA. In comparison to 5hU and U, the 5-hydroxyl group appears to inhibit the activity. A large part, i.e., approximately 80%, of the 5hC substrate remained intact at 600 s, which showed that PfuEndoQ exhibits almost no cleavage activity toward 5hC, suggesting that 5hC may not be the natural target for PfuEndoQ because of the weak base detection.
TABLE 1.
Rate constants of cleavage reactions by PfuEndoQ
| Substrate | k ± SEM (s−1) |
|---|---|
| AP·A | (2.5 ± 0.48) × 10−2 |
| AP | (1.0 ± 0.37) × 10−4 |
| U·A | (9.8 ± 0.59) × 10−3 |
| U | (1.2 ± 0.053) × 10−2 |
| DHU·A | (1.1 ± 0.37) × 10−2 |
| DHU | (2.7 ± 0.27) × 10−4 |
| Hx·A | (8.5 ± 0.60) × 10−3 |
| Hx | (6.0 ± 0.75) × 10−3 |
| 5hU·A | (3.3 ± 0.59) × 10−3 |
| 5hU | (4.5 ± 0.16) × 10−4 |
| 5hC·A | (9.3 ± 1.3) × 10−4 |
| 5hC | (1.9 ± 0.23) × 10−4 |
FIG 3.
Time course of cleavage reactions by PfuEndoQ. DNA substrates were incubated with PfuEndoQWT (100 nM) at 60°C for the indicated time (20, 60, 120, 300, and 600 s). Remaining substrates ([intact substrate]/[initial substrate], [S]/[S]0) were measured by 8 M urea-12% PAGE and autoradiography (mean ± standard error of the mean [SEM] for n = 3). The curves were fitted using a one-phase exponential decay equation using Origin software (LightStone). (A) dsDNA; (B) ssDNA.
Base pair stability affects EndoQ activity.
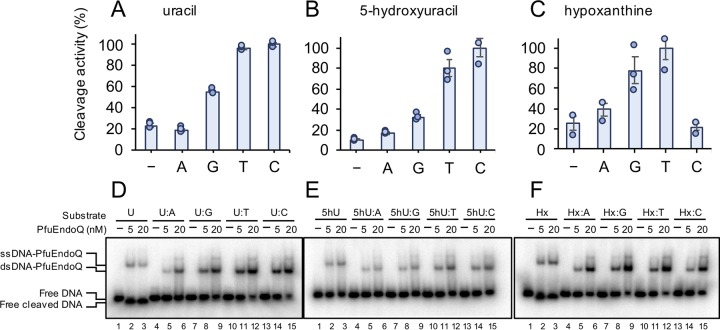
To investigate the mechanisms underlying the significant difference in the cleavage efficiencies depending on ss/dsDNA or lesions, base-pairing effects were investigated. The cleavage specificities of PfuEndoQ were measured with U, 5hU, and Hx opposite either A, G, T, or C (Fig. 4A to C) in dsDNA or in ssDNA. The rates of cleavage activity were observed to be in the following orders: U·C > U·T > U·G > U > U·A, 5hU·C > 5hU·T > 5hU·G > 5hU·A > 5hU and Hx·T > Hx·G > Hx·A > Hx > Hx·C. These findings indicated that the cleavage efficiency of PfuEndoQ was strongly affected by the stability of base pairs of the damaged bases. Uracil forms a stable base pair with adenine (U·A > U·G), and the pairing of 5hU occurs in the following order: 5hU·G > 5hU·A > 5hU·T ∼ 5hU·C (10). Hx forms pairs in the following order: Hx·C > Hx·A > Hx·G ∼ Hx·T (25). Therefore, it appears that there is a negative correlation between base pair stability and the EndoQ activity. This is in agreement with the following specific known facts: (i) U and 5hU exhibit similar preferences with regard to base pairing, which may be due to the fact that the 5-hydroxyl group is not engaged in base pairing, and (ii) the difference in the preferences of Hx and U/5hU showed that their preferences are not attributable to the presence of opposite bases alone or in the context of the DNA sequence. Further, to examine the binding activity along with the cleavage activity, an electrophoretic mobility shift assay (EMSA) was conducted using the same sets of substrates. As shown in Fig. 4D to F, the binding activity was found to be positively correlated with the nuclease activity. These results indicated that the stable binding of PfuEndoQ to DNA, which is promoted by the base pair instability, enhances its cleavage activity.
FIG 4.
Binding and cleavage specificities of PfuEndoQ. (A to C) Cleavage activity toward U-, 5hU-, and Hx-containing DNA. Damaged bases were paired with either A, G, T, or C in dsDNA or remained unpaired (−, ssDNA). DNA substrates (5 nM) were incubated with PfuEndoQ (U, 50 pM [A]; 5hU, 1 nM [B]; Hx, 50 pM [C]) at 60°C for 10 min. DNA products were separated by 8 M urea-12% PAGE, and 32P-labeled DNA strands were detected by autoradiography. (D to F) Binding activity toward U-containing (D), 5hU-containing (E), and Hx-containing (F) DNA. DNA substrates were incubated with or without PfuEndoQ (0 [−], 5, or 20 nM) at 40°C for 5 min. The products were separated by 4% PAGE, and 32P-labeled DNA strands were detected by autoradiography. Bars, mean ± SEM for n = 3 with the individual data (dots).
PfuEndoQ exhibits cleavage activity toward uridine-containing DNA.
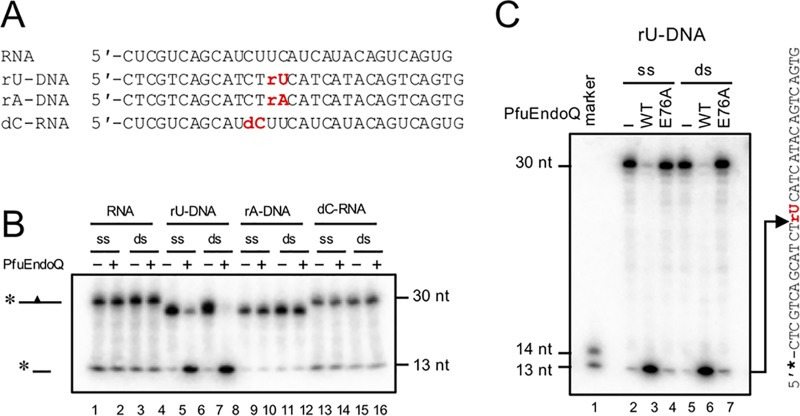
Previous work showed that PfuEndoQ does not exhibit cleavage activity toward RNA strands (20); however, the underlying mechanism remains uncertain. As shown in Fig. 5, cleavage activity of PfuEndoQ for ribose-containing DNA was investigated. The data indicated that PfuEndoQ exhibits the endonuclease activity at the 5′ side of uridine (rU)-containing DNA (Fig. 5B and C). Conversely, the cleavage activity was not observed for adenosine (rA)-containing DNA or deoxycytidine (dC)-containing RNA (Fig. 5B); this indicates that the specific base structure and its adjacent deoxyribose (more than one deoxyribose nucleotide) are critical factors for the cleavage activity.
FIG 5.
PfuEndoQ cuts uridine (rU)-containing DNA. (A) DNA/RNA substrates (see Table S1 in the supplemental material). (B) RNA substrate screening for PfuEndoQ. DNA/RNA substrates were incubated with (100 nM) or without (−) PfuEndoQWT at 60°C for 10 min in a reaction mixture. (C) rU-DNA substrates were incubated with or without PfuEndoQ WT or E76A (10 nM) at 60°C for 10 min in a reaction mixture. −, no enzyme. DNA products were separated by 8 M urea-12% PAGE, and 32P-labeled RNA/DNA strands were detected by autoradiography.
Comparison of substrate specificities of archaeal UDG and EndoQ.
UDG is the most prevalent uracil-acting protein in all domains of life. Except for some orders in Euryarchaeota, almost all archaea have family 4 UDGs (18, 22). Thus far, several family 4 UDGs from bacteria and archaea have been investigated (22, 26–31); however, their substrate specificity range has not been fully examined. We examined the substrate specificity of family 4 UDG from the closely related T. kodakarensis. As shown in Fig. S2 in the supplemental material, purified UDG from T. kodakarensis (TkoUDG) (see Fig. S1) exhibited glycosylase activity only toward U-containing DNA in both ss/dsDNA, which indicated that UDG exhibits a strict and more specific recognition of U compared to EndoQ. The data suggest that UDG is highly specialized with regard to U repair and that the substrate recognition mechanism of EndoQ seems to be more relaxed.
Reconstitution of a reconstitution-modification system using EndoQ from Bacillus subtilis.
To explore the biological benefits of the relaxed substrate recognition in EndoQ, we investigated whether EndoQ can act as an antiviral (bacteriophage) protein. Using purified recombinant proteins, WT, and its inactive mutant E90A (corresponding to E76 in PfuEndoQ), the activity of EndoQ from B. subtilis (BsuEndoQ) (Fig. S1) was examined in the presence or absence of the uracil DNA glycosylase inhibitor from B. subtilis bacteriophage PBS1 (UGI). Previous studies showed that the thymine bases in this phage genome were found to be completely replaced with uracil, and UGI functions as an inhibitor against host UDG through UDG-UGI direct interactions (32, 33). As shown in Fig. S3 in the supplemental material, UGI from B. subtilis bacteriophage PBS1 inhibited UDG from Escherichia coli (50 nM). Conversely, the cleavage activity of BsuEndoQ (50 nM) was not affected by the presence of UGI (up to 200 nM). Using the inactive mutant, it was confirmed that the U cleavage activity was attributed to BsuEndoQ. These findings suggest that EndoQ might counteract viral infection (such as Bacillus phage PBS1) in B. subtilis by digesting phage DNA.
DISCUSSION
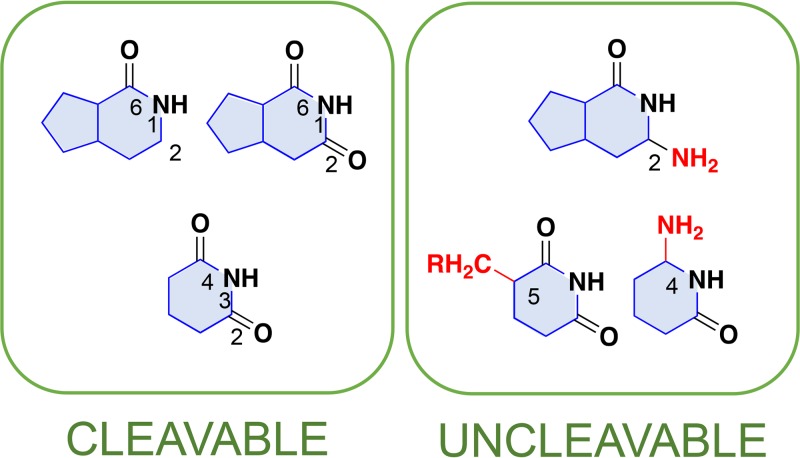
Our results revealed that EndoQ can incise DNA strands containing DHU, 5hU, and 5hC in addition to the previously reported damaged bases U, Hx, X, and an AP site. The cleavable and uncleavable nucleobases for PfuEndoQ are illustrated in Fig. 6. Given that the cleavage activity of PfuEndoQ is highly affected by the opposite base at the damaged site (Fig. 4), the affinity of the recognition pocket and the target base can be compared by the kinetic parameters for each damaged base in ssDNA, except for AP sites. The cleavage-favorable structures for PfuEndoQ, such as the imide structure [−(CO)NH(CO)−] in purine and pyrimidine bases, were identified. In contrast, the affinity between the base and the base recognition site is likely to be significantly reduced by some features, such as the C-5 methylation of pyrimidine or the amino groups at pyrimidine C-4 and purine C-2. Regarding the cleavage activity toward 5hC, we speculate that EndoQ recognizes the imino form of 5hC, as the 5-hydroxyl group significantly promotes the formation of the imino structure (34).
FIG 6.
Nucleobase structures that are cleavable/uncleavable by PfuEndoQ. The features in nucleobases, which are cleavable and uncleavable by PfuEndoQ, are summarized. The favorable structure for the EndoQ activity is the imide structure in C-2, N-1, and C-6 in purine bases (C-2: “=CO” or “≡CH”) and C-2, N-3, and C-4 in pyrimidine bases. Groups shown in red might cause the inhibition of the EndoQ activity as follows: C-4 amino group and C-5 methylation of pyrimidine, and C-2 amino group in purine.
Our findings regarding the negative correlation between base pair stability and cleavage by EndoQ support the idea obtained from the structural model described in a previous study of a PfuEndoQ-DNA complex, in which the target base is flipped out from DNA and trapped by PfuEndoQ (24). Because previous work showed that mismatched bases are more frequently flipped out than matched bases from dsDNA (35), it appears beneficial for EndoQ to use the intrinsic nature of damaged bases, which are mismatched with their original partner (e.g., the base flipping frequency is U·G > C·G). Although the underlying mechanisms have not been addressed, the same tendencies in base pair preferences have also been observed in other DNA repair enzymes (36, 37). Base flipping is a common strategy used for DNA repair or modification enzymes to gain access to the target base (38); however, it is still under debate whether the process of base flipping is catalyzed by enzymes or occurs spontaneously (39). Although it remains unclear whether PfuEndoQ promotes “base breathing” in general (as enzymatic base flipping), it is presumed that spontaneous base flipping is a key for the efficient detection of damaged bases because the cleavage activity toward the target base was highly affected by its opposite base (Fig. 4). Here, we propose a model for the recognition and cleavage processes catalyzed by EndoQ, as shown in Fig. S4 in the supplemental material. A spontaneously flipped out base is trapped in the recognition site, and if the affinity between the target base and the pocket is high enough, an amino acid residue in PfuEndoQ is inserted into the DNA duplex in place of the flipped-out base; this triggers the nicking activity. As an AP site can also be a substrate for PfuEndoQ, the process of recognition itself is not likely to be a trigger for the cleavage. Also, this model can explain that the most rapid reaction by PfuEndoQ was observed with an AP site containing DNA.
In the archaeal domain, the following specific protein families have been reported to be capable of acting on uracil in DNA: EndoQ, UDG, and exonuclease III (ExoIII also known as Xth). ExoIII has been known to exhibit robust levels of AP endonuclease activity, but some ExoIII proteins (likely from Archaea to Eukarya) have been shown to directly incise the 5′ side of uracil in DNA with a relatively low efficiency (22, 40, 41). Our study showed that family 4 UDG has a strict specificity for uracil alone, while EndoQ has a more relaxed recognition mechanism. Other damaged bases that are not repaired by EndoQ can be repaired by other enzymes, such as 3-methyladenine DNA glycosylase and endonuclease V for Hx (42, 43) and EndoIII for 5hC, 5hU, DHU, and DHT (11, 44, 45). The substrate specificity of EndoQ overlaps with that of these enzymes; however, it should be noted that EndoQ activity is distinct, as it directly incises the DNA backbone. We speculate that this property of EndoQ may be important for the functioning of an efficient repair pathway, as it enables the generation of highly cytotoxic AP sites, which easily induce DNA strand breaks that are to be avoided. Furthermore, our results have shown that PfuEndoQ can cleave rU-containing DNA, suggesting its possible involvement in the removal of rU from genome as the euryarchaeal replicative DNA polymerases have been shown to incorporate ribonucleoside monophosphates (rNMPs) into nascent DNA at a significant rate (46, 47).
Our findings have shown that EndoQ from B. subtilis can cleave U-containing DNA, and its activity is not inhibited in the presence of UGI from Bacillus phage PBS1 (see Fig. S3 in the supplemental material). Notably, the genome of the Rhizobium phage RL38JI exhibited some tolerance against DNase because all cytosine bases were replaced with 5hC (48). The EndoQ homolog is present in certain Alphaproteobacteria, including in the phyletic order Rhizobiales (21) (as was shown by our BLAST search result, for example, for GenBank accession number OJU35833 for Rhizobiales bacterium 68-8). Further study is needed to investigate whether EndoQ proteins play a role in the degradation of viral genome, which contains noncanonical modified bases in certain organisms (see Fig. S5 in the supplemental material).
In conclusion, we have elucidated the substrate specificity range and favorable structure for PfuEndoQ and discovered the novel properties of this enzyme (the cleavage activities at the 5′ sides of 5hU, 5hC, DHU, and rU). In particular with the B. subtilis cells, EndoQ could act as a potential component of a restriction enzyme as well as a DNA repair enzyme.
MATERIALS AND METHODS
Oligonucleotides.
The oligonucleotides used in this study are shown in Table S1 in the supplemental material. The 30-nt oligonucleotides T, RTg, STg, CPD, 6-4PP, DewarPP, 6mG, and 8oxoG were chemically synthesized using an ABI 3400 DNA synthesizer (Applied Biosystems), followed by high-pressure liquid chromatography (HPLC) purification. The building blocks of (5R)- and (5S)-Tg, cis-syn CPD, trans-syn CPD, pyrimidine-pyrimidone (6-4) photoproduct, Dewar photoproduct, O6-methylguanine, and 8-oxoguanine were synthesized as described previously (49–54), while the other phosphoramidite building blocks were obtained from Glen Research. The other oligonucleotides were obtained from Tsukuba Oligo Service Co., Ltd. (Ibaraki, Japan), Integrated DNA Technologies, Inc. (IA), and Fasmac Co., Ltd. (Kanagawa, Japan). The 5′ termini of the oligonucleotides were labeled with T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (PerkinElmer), followed by purification using Illustra MicroSpin G-25 columns (GE Healthcare). The annealing of oligonucleotides was conducted by gradually decreasing the temperature.
Preparation of recombinant PfuEndoQ proteins.
The expression vectors encoding PfuEndoQWT (GenBank accession number AAL81675) and PfuEndoQE76A (E76A mutation) (pET21d-PfuEndoQWT and pET21d-PfuEndoQE76A) were gifted by Y. Ishino (Kyushu University, Fukuoka, Japan). Recombinant PfuEndoQ proteins were purified as described previously (20). The protein concentrations were determined by measuring the absorbance at 280 nm. The theoretical molar extinction coefficients of PfuEndoQWT (47,120 M−1 cm−1) and PfuEndoQE76A (47,120 M−1 cm−1) were estimated from the amino acid sequences using the ProtParam tool (55).
Endonuclease activity assay.
The cleavage reactions for PfuEndoQ were performed at 60°C for various time periods in a 20-μl reaction mixture (50 mM Tris-HCl [pH 8.0], 1 mM dithiothreitol [DTT], 1 mM MgCl2, 0.01% Tween 20, 50 mM NaCl, 5 nM 32P-labeled DNA) with PfuEndoQ as described in the figure legends. The reactions were terminated by adding 10 μl of stop solution (98% deionized formamide, 10 mM EDTA, 0.1% bromophenol blue [BPB]). After incubation at 95°C for 5 min, the samples were immediately placed on ice. The DNA samples were separated by 8 M urea-12% PAGE in Tris-borate-EDTA (TBE) buffer (89 mM Tris-borate and 2.5 mM EDTA). The gel was dried, exposed to a phosphor imaging plate, and analyzed with an FLA 7000 phosphorimager (Fujifilm). The rate constant (k) was obtained from the linear fitting, ln (S) = −kt, using Origin software (LightStone), in which S is the remaining substrate and t is the reaction time from 0 s to 120 s.
Electrophoretic mobility shift assay.
PfuEndoQWT (0, 5, and 20 nM) was incubated with 32P-labeled DNA (5 nM) in a 20-μl reaction mixture (50 mM Tris-HCl [pH 8.0], 1 mM DTT, 1 mM MgCl2, and 0.01% Tween 20) at 40°C for 5 min, followed by the addition of 5 μl loading buffer (20% glycerol, 250 mM Tris-HCl [pH 8.0], and 0.1% BPB). Samples were analyzed by 4% PAGE in TBE buffer. The gel was visualized as described above.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yoshizumi Ishino, Sonoko Ishino, and Naruto Makita for the generous gifts of PfuEndoQ expression vectors, B. subtilis genomic DNA, and for providing advice regarding the BsuEndoQ purification. We also thank Junpei Yamamoto for the help in DNA synthesis.
This work was supported by a research grant from the Agricultural Chemical Research Foundation, Japan (M.S.), and was partly supported by the Inamori Research Grant (2019) (M.S.).
We declare no conflicts of interest.
M.S. conceived the project, conducted all of the experiments, and drafted the first version of the manuscript. S.I. synthesized some of the damaged DNA substrates and provided constructive advice. Both authors reviewed and approved of the final version of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Weigele P, Raleigh EA. 2016. Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 116:12655–12687. doi: 10.1021/acs.chemrev.6b00114. [DOI] [PubMed] [Google Scholar]
- 2.Sánchez-Romero MA, Cota I, Casadesús J. 2015. DNA methylation in bacteria: from the methyl group to the methylome. Curr Opin Microbiol 25:9–16. doi: 10.1016/j.mib.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Carell T, Kurz MQ, Müller M, Rossa M, Spada F. 2018. Non-canonical bases in the genome: the regulatory information layer in DNA. Angew Chem Int Ed Engl 57:4296–4312. doi: 10.1002/anie.201708228. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Olinski R, Jurgowiak M, Zaremba T. 2010. Uracil in DNA—its biological significance. Mutat Res 705:239–245. doi: 10.1016/j.mrrev.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Nabel CS, Manning SA, Kohli RM. 2012. The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem Biol 7:20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramiro AR, Barreto VM. 2015. Activation-induced cytidine deaminase and active cytidine demethylation. Trends Biochem Sci 40:172–181. doi: 10.1016/j.tibs.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Kumar S, Chinnusamy V, Mohapatra T. 2018. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front Genet 9:640. doi: 10.3389/fgene.2018.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couturier M, Lindås AC. 2018. The DNA methylome of the hyperthermoacidophilic crenarchaeon Sulfolobus acidocaldarius. Front Microbiol 9:137. doi: 10.3389/fmicb.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiviyanathan V, Somasunderam A, Volk DE, Gorenstein DG. 2005. 5-Hydroxyuracil can form stable base pairs with all four bases in a DNA duplex. Chem Commun (Camb) 2005:400–402. doi: 10.1039/b414474k. [DOI] [PubMed] [Google Scholar]
- 11.Dizdaroglu M, Laval J, Boiteux S. 1993. Substrate specificity of the Escherichia coli endonuclease III: excision of thymine- and cytosine-derived lesions in DNA produced by radiation-generated free radicals. Biochemistry 32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 12.Breimer LH, Lindahl T. 1985. Thymine lesions produced by ionizing radiation in double-stranded DNA. Biochemistry 24:4018–4022. doi: 10.1021/bi00336a032. [DOI] [PubMed] [Google Scholar]
- 13.Karran P, Lindahl T. 1980. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry 19:6005–6011. doi: 10.1021/bi00567a010. [DOI] [PubMed] [Google Scholar]
- 14.Caulfield JL, Wishnok JS, Tannenbaum SR. 1998. Nucleic acids, protein synthesis, and molecular genetics: nitric oxide-induced deamination of cytosine and guanine in deoxynucleoside sand oligonucleotides. J Biol Chem 273:12689–12695. doi: 10.1074/jbc.273.21.12689. [DOI] [PubMed] [Google Scholar]
- 15.Ouellette M, Jackson L, Chimileski S, Papke RT. 2015. Genome-wide DNA methylation analysis of Haloferax volcanii H26 and identification of DNA methyltransferase related PD-(D/E)XK nuclease family protein HVO_A0006. Front Microbiol 6:251. doi: 10.3389/fmicb.2015.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershberg R, Petrov DA. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet 6:e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krokan HE, Bjørås M. 2013. Base excision repair. Cold Spring Harb Perspect Biol 5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind L, Koonin EV. 2000. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol 1:research0007.1. doi: 10.1186/gb-2000-1-4-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yasui A. 2013. Alternative excision repair pathways. Cold Spring Harb Perspect Biol 5:a012617. doi: 10.1101/cshperspect.a012617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiraishi M, Ishino S, Yamagami T, Egashira Y, Kiyonari S, Ishino Y. 2015. A novel endonuclease that may be responsible for damaged DNA base repair in Pyrococcus furiosus. Nucleic Acids Res 43:2853–2863. doi: 10.1093/nar/gkv121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiraishi M, Ishino S, Cann I, Ishino Y. 2017. A functional endonuclease Q exists in the bacterial domain: identification and characterization of endonuclease Q from Bacillus pumilus. Biosci Biotechnol Biochem 81:931–937. doi: 10.1080/09168451.2016.1277946. [DOI] [PubMed] [Google Scholar]
- 22.Shiraishi M, Ishino S, Heffernan M, Cann I, Ishino Y. 2018. The mesophilic archaeon Methanosarcina acetivorans counteracts uracil in DNA with multiple enzymes: EndoQ, ExoIII, and UDG. Sci Rep 8:15791. doi: 10.1038/s41598-018-34000-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi I, Marmur J. 1963. Replacement of thymidylic acid by deoxyuridylic acid in the deoxyribonucleic acid of a transducing phage for Bacillus subtilis. Nature 197:794–795. doi: 10.1038/197794a0. [DOI] [PubMed] [Google Scholar]
- 24.Miyazono K, Ishino S, Makita N, Ito T, Ishino Y, Tanokura M. 2018. Crystal structure of the novel lesion-specific endonuclease PfuEndoQ from Pyrococcus furiosus. Nucleic Acids Res 46:4807–4818. doi: 10.1093/nar/gky261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Case-Green SC, Southern EM. 1994. Studies on the base pairing properties of deoxyinosine by solid phase hybridisation to oligonucleotides. Nucleic Acids Res 22:131–136. doi: 10.1093/nar/22.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sartori AA, Jiricny J. 2003. Enzymology of base excision repair in the hyperthermophilic archaeon Pyrobaculum aerophilum. J Biol Chem 278:24563–24576. doi: 10.1074/jbc.M302397200. [DOI] [PubMed] [Google Scholar]
- 27.Moen MN, Knævelsrud I, Haugland GT, Grøsvik K, Birkeland NK, Klungland A, Bjelland S. 2011. Uracil-DNA glycosylase of Thermoplasma acidophilum directs long-patch base excision repair, which is promoted by deoxynucleoside triphosphates and ATP/ADP, into short-patch repair. J Bacteriol 193:4495–4508. doi: 10.1128/JB.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knævelsrud I, Moen MN, Grøsvik K, Haugland GT, Birkeland NK, Klungland A, Leiros I, Bjelland S. 2010. The hyperthermophilic euryarchaeon Archaeoglobus fulgidus repairs uracil by single-nucleotide replacement. J Bacteriol 192:5755–5766. doi: 10.1128/JB.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyonari S, Uchimura M, Shirai T, Ishino Y. 2008. Physical and functional interactions between uracil-DNA glycosylase and proliferating cell nuclear antigen from the euryarchaeon Pyrococcus furiosus. J Biol Chem 283:24185–24193. doi: 10.1074/jbc.M802837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandigursky M, Franklin WA. 1999. Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr Biol 9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]
- 31.Kawai A, Higuchi S, Tsunoda M, Nakamura KT, Yamagata Y, Miyamoto S. 2015. Crystal structure of family 4 uracil-DNA glycosylase from Sulfolobus tokodaii and a function of tyrosine 170 in DNA binding. FEBS Lett 589:2675–2682. doi: 10.1016/j.febslet.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 32.Savva R, Pearl LH. 1995. Cloning and expression of the uracil-DNA glycosylase inhibitor (UGI) from bacteriophage PBS-1 and crystallization of a uracil-DNA glycosylase-UGI complex. Proteins 22:287–289. doi: 10.1002/prot.340220310. [DOI] [PubMed] [Google Scholar]
- 33.Katz GE, Price AR, Pomerantz MJ. 1976. Bacteriophage PBS2-induced inhibition of uracil-containing DNA degradation. J Virol 20:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suen W, Spiro TG, Sowers LC, Fresco JR. 1999. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc Natl Acad Sci U S A 96:4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin Y, Yang L, Zheng G, Gu C, Yi C, He C, Gao YQ, Zhao XS. 2014. Dynamics of spontaneous flipping of a mismatched base in DNA duplex. Proc Natl Acad Sci U S A 111:8043–8048. doi: 10.1073/pnas.1400667111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB. 2002. Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase. J Biol Chem 277:31673–31678. doi: 10.1074/jbc.M204475200. [DOI] [PubMed] [Google Scholar]
- 37.Waters TR, Swann PF. 1998. Kinetics of the action of thymine DNA glycosylase. J Biol Chem 273:20007–20014. doi: 10.1074/jbc.273.32.20007. [DOI] [PubMed] [Google Scholar]
- 38.Hong S, Cheng X. 2016. DNA base flipping: a general mechanism for writing, reading, and erasing DNA modifications. Adv Exp Med Biol 945:321–341. doi: 10.1007/978-3-319-43624-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker JB, Bianchet MA, Krosky DJ, Friedman JI, Amzel LM, Stivers JT. 2007. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 449:433–437. doi: 10.1038/nature06131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georg J, Schomacher L, Chong JPJ, Majerník AI, Raabe M, Urlaub H, Müller S, Ciirdaeva E, Kramer W, Fritz HJ. 2006. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res 34:5325–5336. doi: 10.1093/nar/gkl604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prorok P, Alili D, Saint-Pierre C, Gasparutto D, Zharkov DO, Ishchenko AA, Tudek B, Saparbaev MK. 2013. Uracil in duplex DNA is a substrate for the nucleotide incision repair pathway in human cells. Proc Natl Acad Sci U S A 110:E3695–E3703. doi: 10.1073/pnas.1305624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansfield C, Kerins SM, McCarthy TV. 2003. Characterisation of Archaeglobus fulgidus AlkA hypoxanthine DNA glycosylase activity. FEBS Lett 540:171–175. doi: 10.1016/s0014-5793(03)00257-6. [DOI] [PubMed] [Google Scholar]
- 43.Kiyonari S, Egashira Y, Ishino S, Ishino Y. 2014. Biochemical characterization of endonuclease v from the hyperthermophilic archaeon, Pyrococcus furiosus. J Biochem 155:325–333. doi: 10.1093/jb/mvu010. [DOI] [PubMed] [Google Scholar]
- 44.Asagoshi K, Odawara H, Nakano H, Miyano T, Terato H, Ohyama Y, Seki S, Ide H. 2000. Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry 39:11389–11398. doi: 10.1021/bi000422l. [DOI] [PubMed] [Google Scholar]
- 45.Shekhtman A, McNaughton L, Cunningham RP, Baxter SM. 1999. Identification of the Archaeoglobus fulgidus endonuclease III DNA interaction surface using heteronuclear NMR methods. Structure 7:919–930. doi: 10.1016/s0969-2126(99)80119-1. [DOI] [PubMed] [Google Scholar]
- 46.Lemor M, Kong Z, Henry E, Brizard R, Laurent S, Bossé A, Henneke G. 2018. Differential activities of DNA polymerases in processing ribonucleotides during DNA synthesis in archaea. J Mol Biol 430:4908–4924. doi: 10.1016/j.jmb.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Heider MR, Burkhart BW, Santangelo TJ, Gardner AF. 2017. Defining the RNaseH2 enzyme-initiated ribonucleotide excision repair pathway in Archaea. J Biol Chem 292:8835–8845. doi: 10.1074/jbc.M117.783472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swinton D, Hattman S, Benzinger R, Buchanan-Wollaston V, Beringer J. 1985. Replacement of the deoxycytidine residues in Rhizobium bacteriophage RL38JI DNA. FEBS Lett 184:294–298. doi: 10.1016/0014-5793(85)80625-6. [DOI] [PubMed] [Google Scholar]
- 49.Iwai S, Shimizu M, Kamiya H, Ohtsuka E. 1996. Synthesis of a phosphoramidite coupling unit of the pyrimidine (6-4) pyrimidone photoproduct and its incorporation into oligodeoxynucleotides. J Am Chem Soc 118:7642–7643. doi: 10.1021/ja9603158. [DOI] [Google Scholar]
- 50.Yamamoto J, Hitomi K, Todo T, Iwai S. 2006. Chemical synthesis of oligodeoxyribonucleotides containing the Dewar valence isomer of the (6-4) photoproduct and their use in (6-4) photolyase studies. Nucleic Acids Res 34:4406–4415. doi: 10.1093/nar/gkl572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takasawa K, Masutani C, Hanaoka F, Iwai S. 2004. Chemical synthesis and translesion replication of a cis-syn cyclobutane thymine-uracil dimer. Nucleic Acids Res 32:1738–1745. doi: 10.1093/nar/gkh342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor JS, Brockie IR. 1988. Synthesis of a trans-syn thymine dimer building block. Solid phase synthesis of CGTAT[t,s]TATGC. Nucleic Acids Res 16:5123–5136. doi: 10.1093/nar/16.11.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimizu T, Manabe K, Yoshikawa S, Kawasaki Y, Iwai S. 2006. Preferential formation of (5S,6R)-thymine glycol for oligodeoxyribonucleotide synthesis and analysis of drug binding to thymine glycol-containing DNA. Nucleic Acids Res 34:313–321. doi: 10.1093/nar/gkj443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwai S. 2000. Synthesis of thymine glycol containing oligonucleotides from a building block with the oxidized base. Angew Chem Int Ed 39:3874–3876. doi:. [DOI] [PubMed] [Google Scholar]
- 55.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.