Citrus polymethoxyflavones prevent metabolic syndrome by modulating gut dysbiosis and altering branched-chain amino acids.
Abstract
Metabolic syndrome (MetS) is intricately linked to dysregulation of gut microbiota and host metabolomes. Here, we first find that a purified citrus polymethoxyflavone-rich extract (PMFE) potently ameliorates high-fat diet (HFD)–induced MetS, alleviates gut dysbiosis, and regulates branched-chain amino acid (BCAA) metabolism using 16S rDNA amplicon sequencing and metabolomic profiling. The metabolic protective effects of PMFE are gut microbiota dependent, as demonstrated by antibiotic treatment and fecal microbiome transplantation (FMT). The modulation of gut microbiota altered BCAA levels in the host serum and feces, which were significantly associated with metabolic features and actively responsive to therapeutic interventions with PMFE. Notably, PMFE greatly enriched the commensal bacterium Bacteroides ovatus, and gavage with B. ovatus reduced BCAA concentrations and alleviated MetS in HFD mice. PMFE may be used as a prebiotic agent to attenuate MetS, and target-specific microbial species may have unique therapeutic promise for metabolic diseases.
INTRODUCTION
Metabolic syndrome (MetS) is a constellation of interrelated metabolic risk factors that include obesity, hyperlipidemia, insulin resistance, and hepatic steatosis. MetS affects at least 30% of adults in the Western world and has been proven to be associated with an increased incidence of type 2 diabetes, atherosclerosis, and coronary heart disease (1). Recently, mounting evidence suggests a causal relationship between the gut microbiota and MetS development with transferability of phenotypes through fecal microbiome transplantation (FMT) (2, 3). These effects may be mediated partly through the metabolome, especially the branched-chain amino acids (BCAAs) (4), because many bacterial species are capable of regulating biosynthesis, transport, and metabolism of BCAAs (5). Pedersen et al. (4) found that Prevotella copri and Bacteroides vulgatus are the main species driving the association between biosynthesis of BCAAs and insulin resistance (4). Gavage with Bacteroides thetaiotaomicron reduced serum BCAA concentrations and alleviated diet-induced body weight gain and adiposity in mice (6). However, there has been little discussion about whether “drugging the microbiome” is effective for the treatment of MetS and whether BCAA levels may correlate with the efficacy of drug interventions.
Epidemiological studies have demonstrated a positive relationship between high intake of flavonoid-rich foods and the reduction of MetS (7). The polymethoxyflavones (PMFs) extracted from citrus peels are of great interest due to their anti-obesity potential, favorable pharmacokinetic profiles, and no discernible toxicity in rodent models (8, 9). The physiological effects of flavonoids are in notable contrast to their poor bioavailability, and the critical mechanism for metabolic protective effects of PMFs remains elusive. Because products with low bioavailability may become potential substrates for gut microbiota, we hypothesize that PMFs protect against MetS through remodeling of the gut microbiota.
In the present study, we show that a PMF-rich extract (PMFE) isolated from citrus peels potently reduces body weight and accumulation of fat tissues in mice fed a high-fat diet (HFD). Using 16S ribosomal DNA (rDNA)–based microbiota analysis and metabolomics profiling, we demonstrate that PMFE treatment alleviates HFD-induced gut dysbiosis and dysregulation of BCAAs in a dose-dependent manner. Antibiotic treatment and horizontal-FMT experiments further demonstrate that remodeling of the gut microbiota is required for the high bioactivity of PMFE. We also shows that Bacteroides ovatus is sufficient to attenuate MetS as a target of PMFE. These findings thus demonstrate that the PMFE represents a potential prebiotic agent for the treatment of MetS and its complications.
RESULTS
Citrus PMFE shows potential metabolic protective effects via inhibition of mTOR/P70S6K/SREBP pathway
Here, we optimized the extraction and purification process of PMFE (see Supplementary Materials and Methods). The PMFs in the novel enriched citrus PMFE accounted for the largest proportion, with sinensetin, nobiletin, 3,5,6,7,8,3′,4′-heptamethoxyflavone, and tangeretin making up the major components and had increased purity from 1.27 to 60.85% (w/w; Fig. 1, A and B). Compared with the unenriched extracts of citrus peels (ECPs), PMFE showed no discernible toxicity in human liver HL-7702 cells (fig. S1A), even at 80 μg/ml (fig. S1B). Sterol regulatory element–binding proteins (SREBPs) play important roles in regulating lipid homeostasis and have been considered targets for the treatment of metabolic diseases (10). Therefore, we used a luciferase-based system to detect the bioactivity of PMFE as shown by the inhibition of SRE-containing promoter in HL-7702 cells. Notably, PMFE significantly decreased the SRE-luciferase activity in a dose- and time-dependent manner (Fig. 1C). We examined the expression of SREBP-1/2 in the cells treated with PMFE by Western blot. Both mature SREBP-1/2 levels were reduced by PMFE in a dose-dependent manner (Fig. 1D). We also observed significantly reduced expression of SREBP target genes, such as cholesterol metabolism genes DHCR24, HMGCR, and LDLR, as well as fatty acid metabolism genes SREBP-1c, FASN, ACC-1, and SCD-1 after PMFE treatment (Fig. 1E). Moreover, nile red staining indicated that PMFE markedly reduced the lipid accumulation in HL-7702 cells (Fig. 1F). Because SREBP processing and lipogenesis are regulated by mammalian target of rapamycin (mTOR) activity (10), the effect of PMFE on activity of mTOR and its downstream kinase P70S6K was further investigated (10, 11). Treatment with PMFE decreased the phosphorylation of mTOR and P70S6K (Fig. 1G). Together, the enriched PMFE down-regulating the mTOR/P70S6K/SREBP pathway represents potential lipid-lowering agents.
Fig. 1. Citrus PMFE shows potential metabolic protective effects via inhibition of the mTOR/P70S6K/SREBP pathway.
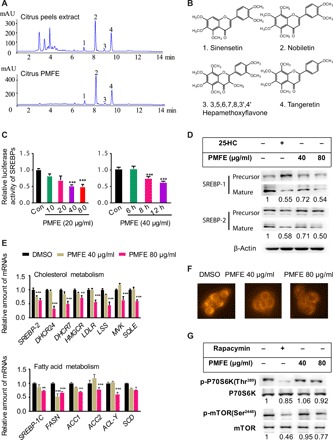
(A) Typical high-performance liquid chromatography (HPLC) chromatograms of citrus peel extract (ECP) and citrus PMFE. mAU, milli-absorbance unit. (B) Chemical structures of the four major PMFs. (C) PMFE inhibits SREBP luciferase activity in HL-7702/SRE-Luc reporter cells in a dose- and time-dependent manner. (D) PMFE reduces mature SREBP-1/2 levels in a dose-dependent manner. Representative Western blotting analysis of SREBP-1/2 is shown. Expression of SREBP (mature) protein was normalized to β-actin as the loading control, and the relative ratio to the vehicle group was labeled below (estimated using the ImageJ software). (E) PMFE inhibits de novo synthesis of cholesterol and fatty acid by decreasing SREBP target genes. HL-7702 cells were treated with dimethyl sulfoxide (DMSO) or indicated concentrations of PMFE for 18 hours. (F) HL-7702 cells were stained with nile red to assess lipids content. (G) PMFE inhibits the activity of mTOR and its downstream kinase P70S6K. Relative intensities of the bands were taken as a ratio of the phosphoprotein over total protein (normalized to internal controls). All experiments were repeated three times. Error bars represent SD. Significant differences compared with DMSO group are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001 (assessed by Student’s t test).
Citrus PMFE exhibits robust metabolic protective effects in HFD-fed mice
To address whether citrus PMFE displays a metabolic protective effect, we used HFD-fed mice as a MetS model (fig. S2A) (12). In HFD-fed mice, 8-week treatment of PMFE significantly decreased body weight gain compared to the HFD group (Fig. 2A and fig. S2B). No significant difference in food intake was observed among HFD-fed groups, suggesting that the effects of PMFE were not due to reduced food consumption (Fig. 2B). PMFE administration also reduced adipose deposition, lipid accumulation in liver tissues, and the cell size of both white and brown adipocyte tissue (Fig. 2, C to G, and fig. S2, C and D). Meanwhile, glucose tolerance and insulin resistance were markedly ameliorated in PMFE-treated HFD-fed mice (Fig. 2, F to H). The serum total cholesterol (TC), triglyceride (TG), and low-density lipoprotein cholesterol (LDL-C) were largely reduced, whereas the significant increase in high-density lipoprotein cholesterol (HDL-C) was not observed in PMFE-treated mice (Fig. 2I). PMFE also exhibited protective effects against the HFD-induced liver damage, supported by significantly lower plasma ALT (Alanine Aminotransferase) and AST (Aspartate Aminotransferase) level (fig. S2E). We further assessed the impact of PMFE on inflammatory cytokines and monocyte chemoattractant protein-1 (MCP-1). PMFE significantly reduced the serum levels of secreted tumor necrosis factor–α (TNF-α), interleukin-1β (IL-1β), IL-6, and their mRNA expression in hepatic tissues. Macrophage levels and MCP-1 mRNA expression decreased in the PMFE-treated mice (fig. S2, F to H). Consistent with in vitro results, immunohistochemistry analysis indicated that PMFE inhibited the phosphorylation of mTOR and P70S6K and decreased the expression of SREBP-1/2 compared with HFD-fed group (fig. S2I). We also observed that PMFE could potently ameliorate diet-induced obesity, hepatic steatosis, dyslipidemia, and insulin resistance in mice in a dose-dependent manner after long-term high-calorie challenge (fig. S3). The metabolic protective effect of a high dose of PMFE was almost equal or even better than that of lovastatin, while PMFE did not produce any apparent effects in chow-fed mice (fig. S4). These results imply that PMFE exhibits robust efficacy against MetS in HFD-fed mice.
Fig. 2. Citrus PMFE exhibits robust metabolic protection in HFD-fed mice.
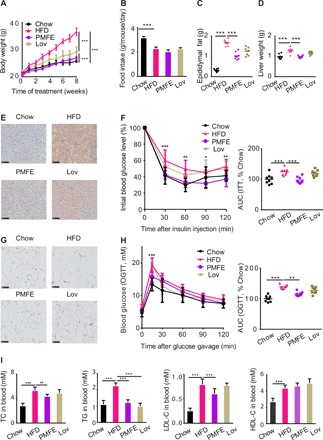
Mice were randomly divided into four groups (n = 8). Chow-fed mice were treated daily with 0.5% CMCNa suspension (Chow). HFD-fed mice were orally administrated 0.5% CMCNa suspension (HFD), PMFE (PMFE; 120 mg/kg per day), or lovastatin (Lov; 30 mg/kg per day). (A) Body weight of the chow- and HFD-fed mice treated daily with solvent (0.5% CMCNa), PMFE, or lovastatin for 8 weeks. (B) Average daily food intake for the above four groups of mice. (C) Epididymal fat. (D) Liver weight. (E) Liver lipid content was assessed using oil red O staining (scale bar, 100 μm). (F) Effect of PMFE on percentage of initial blood glucose level during insulin tolerance test (ITT). Right: Area under the curve (AUC). (G) Representative pictures of hematoxylin and eosin (H&E)–stained white adipose tissue (scale bar, 100 μm). (H) Effect of PMFE on glucose tolerance measured by oral glucose tolerance test (OGTT). Right: AUC. The PMFE-gavaged mice had significantly lower serum glucose levels compared to HFD mice [two-way analysis of variance (ANOVA)]. (I) Total TC, TG, LDL, and HDL levels in blood. Error bars are expressed as means ± SD. Statistical significance was determined by one-way or two-way ANOVA with Tukey tests for multiple-group comparisons. *P < 0.05, **P < 0.01, and ***P < 0.001.
Citrus PMFE alleviates HFD-induced gut dysbiosis
Emerging explorations of the interplay of phytochemicals and gut microbiota have revolutionized our understanding of their mechanisms (13). Because gut microbiota plays a pathogenic role in the development of MetS and modulation of the microbiome is a potential therapeutic approach for prevention of MetS (14, 15), we examined the effects of PMFE on gut microbiota composition by performing a pyrosequencing-based analysis of bacterial 16S rDNA in feces. We observed a distinct clustering of microbiota composition for chow, HFD, and PMFE treatment group using UniFrac-based principal coordinates analysis (PCoA) (Fig. 3A). To assess the overall composition of the bacterial community in different groups, we analyzed the degree of bacterial taxonomic similarity at the phylum level. Compared to chow-fed mice, HFD-fed mice displayed a significant increase in relative abundance of Firmicutes and higher Firmicutes-to-Bacteroidetes ratio, while PMFE treatment protected against this effect to a large extent (Fig. 3, B and C). Meanwhile, HFD mice exhibited epithelial disruption and the presence of abscesses, as well as increased edema and cellular infiltrate into the submucosal layer, which were significantly ameliorated after PMFE treatment (Fig. 3D). HFD may also affect epithelial integrity and hence lead to impaired gut permeability as well as the release of lipopolysaccharide into the circulation. PMFE treatment increased expression of the tight junction proteins zonula occludens-1 (ZO-1) and occludin, supporting a potential role in the regulation of intestinal permeability (fig. S5, A and B). Together, PMFE administration had a substantial effect on remodeling the gut microbiome in response to HFD.
Fig. 3. Citrus PMFE alleviates HFD-induced gut dysbiosis.
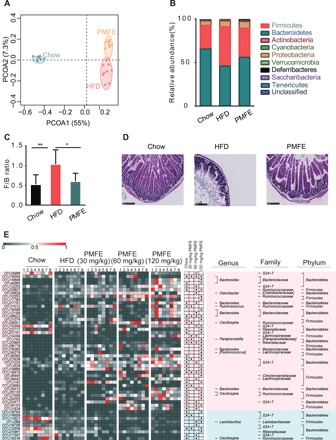
Microbiota composition of chow-fed mice and HFD mice treated with PMFE (30, 60, and 120 mg/kg per day) was analyzed by 16S rDNA pyrosequencing (n = 8 for each group). (A) Weighted UniFrac PCoA analysis of gut microbiota based on the OTU data of chow, HFD, and PMFE groups. (B) Bacterial taxonomic profiling at the phylum level of intestinal bacteria from different mouse groups. (C) Firmicutes-to-Bacteroidetes ratio in the indicated groups. (D) Representative H&E pictures of intestine (scale bars, 250 μm). (E) Heatmap of the 50 OTUs in Chow group altered by HFD responding to PMFE treatment. The color of the spots in the left panel represents the relative abundance of the OTU in each group. In the middle panel, white circles represent less abundant OTUs in Chow and PMFE compared with HFD; black diamonds represent more abundant OTUs in Chow and PMFE compared with HFD. The phylum, family, and genus names of the OTUs are shown on the right panel. The analyses were conducted using R software version 3.3.1.
Variance analysis was further used to identify the specific bacterial phylotypes that were altered by PMFE treatment. In HFD-fed mice, supplementation with PMFE significantly increased 87 operational taxonomic units (OTUs) and decreased 50 OTUs in a dose-dependent manner (fig. S5C). Family analysis revealed that most of these altered OTUs belong to Bacteroidetes S24-7 (34 OTU), Firmicutes Ruminococcaceae (26 OTU), Firmicutes Lachnospiraceae (11 OTU), and Bacteroidetes Bacteroidaceae (10 OTU). Among these 137 OTUs, PMFE treatment regulated 50 OTUs, which were altered upon HFD feeding compared with chow-fed mice (Fig. 3E). Notably, genus-level analysis showed that eight OTUs were from Bacteroides. Moreover, PMFE could reduce opportunistic pathogen species, such as Firmicutes Paraprevotella and Firmicutes Streptococcus. These results indicate that PMFE modulates the gut microbiota of HFD-fed mice, resulting in a microbiota composition similar to that of chow-fed mice.
Citrus PMFE alters MetS-associated BCAA levels in HFD mice
The profound influence of gut microbiota on the host is strongly associated with complex interactions comprising a series of host-microbe metabolic axes (6, 16). To assess metabolic alternations in response to the gut microbiota remodeled by PMFE, untargeted metabolome profiles were generated on serum and feces samples by ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF/MS). After peak alignment and removal of missing values, a total of 1769 positive ions and 663 negative ions were detected in feces. Distinct clustering of metabolites was apparent among chow-fed, HFD-fed, and PMFE-treated HFD groups by principal components analysis model (Fig. 4A). The orthogonal partial least squares–discrimination analysis (OPLS-DA) also showed distinct separation of these groups (fig. S6). HFD feeding was sufficient to trigger widespread changes of metabolites, with 68 and 23 metabolites significantly induced and repressed, respectively, compared with chow-fed mice (table S2). Notably, PMFE treatment partially regulated the metabolites altered upon HFD feeding, abrogating 24 of the HFD-induced metabolite changes (9 up-regulated and 15 down-regulated; Fig. 4B). The topology map generated by MetaboAnalyst (www.metaboanalyst.ca) described the impact of PMFE on these responsive metabolites and revealed the most significant pathway influenced by valine, leucine, and isoleucine degradation, phenylalanine tyrosine, and tryptophan biosynthesis (Fig. 4C). In serum, we also found that PMFE intervention resulted in a similar variation tendency (fig. S7). By cross-comparisons of different groups, a total of 73 (Chow versus HFD) and 48 (HFD versus PMFE) differential metabolites were identified (table S2). Treatment with PMFE regulated 13 differential metabolites (7 up-regulated and 6 down-regulated). The altered metabolic pathways showed an overlap for valine, leucine, and isoleucine synthesis, phenylalanine tyrosine, and tryptophan biosynthesis. Next, we performed targeted metabolomics profiling of the amino acid level in feces and serum samples from different groups by gas chromatography–MS (GC-MS). The levels of valine, leucine, isoleucine, and phenylalanine were significantly elevated in feces and serum of HFD groups compared to chow-fed mice, while PMFE treatment altered their level in the same direction in chow-fed mice (Fig. 4D and fig. S7). Spearman’s correlation analysis indicated that the levels of these amino acids were positively correlated with metabolic features, including body weight gain, blood lipids, and insulin resistance (Fig. 4E and fig. S7).
Fig. 4. Citrus PMFE alters MetS-associated BCAA levels in HFD mice.
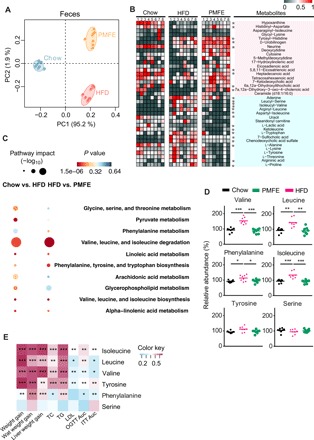
Chow-fed mice were treated daily with solvent (0.5% CMCNa) (Chow). HFD-fed mice were orally administered solvent (0.5% CMCNa) (HFD) or PMFE (120 mg/kg per day). (A) Principal components analysis (PCA) score plots for discriminating the fecal metabolome from Chow, HFD, and PMFE groups. (B) Heatmaps of the differential metabolites that were altered by HFD feeding compared with PMFE-fed mice. Asterisks represent metabolites whose abundance in chow-fed mice was altered by HFD and then regulated by PMFE. The differences of abundance distributions among metabolites between two groups were measured by the Mann-Whitney U test with Benjamini-Hochberg false discovery rate correction. Adjusted P values less than 0.05 were considered statistically significant. (C) Disturbed metabolic pathways in the Chow versus HFD and HFD versus PMFE groups. (D) Comparison of circulating levels of valine, leucine, isoleucine, serine, and phenylalanine in feces by GC-MS in the indicated groups. (E) Heatmap analysis of the Pearson correlation of fecal amino acids and metabolic syndrome–related indexes. Red represents positive correlation, and blue indicates negative correlation. Error bars are expressed as means ± SD. Statistical significance was determined by one-way ANOVA with Tukey tests for multiple-group comparisons. *P < 0.05, **P < 0.01, and ***P < 0.001.
Citrus PMFE attenuates MetS in HFD mice in a gut microbiota–dependent manner
To investigate whether the metabolic protective effects of PMFE are dependent on the presence of gut microbiota, we treated PMFE-fed HFD mice with a cocktail of antibiotics, which included vancomycin, neomycin sulfate, metronidazole, and ampicillin. Consistent with the above effects on metabolic disorder, PMFE greatly reduced body weight gains, adipose depositions, and dyslipidemia. However, when the gut microbiota was suppressed by antibiotics cocktail, the metabolic protective effects of PMFE were abolished. No significant difference was detected in the levels of body weight gain; total TC, TG, LDL, and HDL levels; and insulin sensitivity between the antibiotics group and antibiotics + PMFE group (fig. S8, A to E), indicating that PMFE attenuated diet-induced MetS in a gut microbiota–dependent manner. Consistently, targeted metabolomics profiling of serum and fecal amino acids revealed that the decrease in the concentrations of amino acids by PMFE treatment was blocked by antibiotic intervention (fig. S8F). These findings indicate that PMFE protects mice against diet-induced MetS in a gut microbiota–dependent manner and further affects circulating amino acid concentration.
The metabolic protection of citrus PMFE are transferable by fecal transplantation
To further illustrate the beneficial effects of PMFE mediated by the gut microbiota and its effects on the host circulating amino acid, we transferred the microbiota from PMFE-treated mice (PMFE, HFD fed) to conventional recipient mice fed with HFD, followed by examination of MetS-related traits (Fig. 5A). After 8 weeks of colonization, PMFE receivers (PMFE→HFD) already showed a significant decrease in body weight compared with HFD receivers (HFD→HFD) (Fig. 5B and fig. S9A). Horizontal fecal transfer from PMFE mice (PMFE→HFD) demonstrated similar metabolic protective effects as observed in PMFE-treated group. These recipients showed improved metabolic features of hepatic steatosis, diabetic symptoms, body weight, and epididymal fat; however, insulin sensitivity was not greatly ameliorated (Fig. 5, C to F, and fig. S9, B to H). Conversely, HFD receivers (HFD→HFD) failed to ameliorate MetS-related traits. We also detected decreased abundance of fecal amino acids including BCAAs, phenylalanine, and serine in the PMFE receivers, which were similar with the results shown in PMFE-treated mice (Fig. 5G and fig. S9I). To confirm that fecal transplantation modulates the gut microbiota, we conducted 16S rDNA sequencing to examine the composition of intestinal bacteria. PCoA revealed that the microbial communities exhibited higher similarity between the PMFE and PMFE→HFD treatment groups (a similar trend was observed in the HFD and HFD→HFD groups) (fig. S9J). Fecal transfer from the PMFE-treated group displayed a significant decrease in relative abundance of Firmicutes-to-Bacteroidetes ratio (fig. S9K). A heatmap revealed a large overlap between PMFE and PMFE→HFD group. Fecal transfer from PMFE mice altered OTUs mostly in the same direction as PMFE, especially for decreased taxonomic units (fig. S9L). Together, these results demonstrate that recipient mice recapitulated microbial and metabolic phenotypes as observed in their respective donor mice and gut microbiota mediates the metabolic protection of PMFE.
Fig. 5. Fecal transplants of citrus PMFE exhibit metabolic protection in HFD mice.
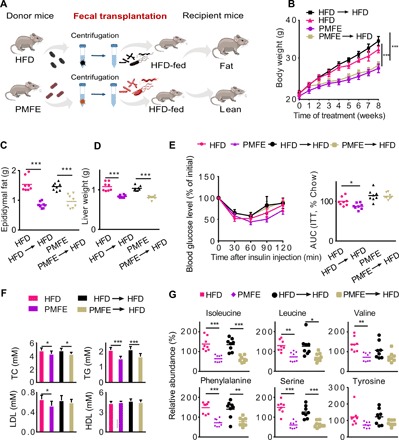
Mice were randomly divided into four groups (n = 8). HFD-fed mice were orally administered solvent (0.5% CMCNa) (HFD) or PMFE (120 mg/kg per day). Horizontal fecal transfer from solvent (0.5% CMCNa)–treated HFD mice is referred to as HFD receivers (HFD→HFD). Horizontal fecal transfer from PMFE-treated mice is referred to as PMFE receivers (PMFE→HFD). (A) Study design of fecal transplant experiment. (B) Body weight of the above four groups of mice. (C) Epididymal fat. (D) Liver weight. (E) Insulin tolerance measured by ITT. Right: AUC. (F) Total TC, TG, LDL, and HDL levels in blood. (G) Relative abundance of isoleucine, leucine, valine, phenylalanine, serine, and tyrosine in feces by GC-MS in the indicated groups. Error bars are expressed as means ± SD. Statistical significance was determined by one-way or two-way ANOVA with Tukey tests for multiple-group comparisons.
Citrus PMFE–mediated enrichment of B. ovatus attenuates MetS in HFD mice
The associations between gut microbiota, alterations of circulating amino acids, and the metabolic protection of PMFE have been investigated above. Bacteroides species were identified as key genus responding to PMFE treatment as shown in Fig. 3E. To directly assess the effects of PMFE on Bacteroides species, the abundance of 10 Bacteroides strains was measured after incubation with PMFE under anaerobic conditions by fecal batch culture fermentation in vitro (Fig. 6A). After 24-hour fermentation, we observed that PMFE could significantly enrich the abundance of B. ovatus, B. thetaiotaomicron, B. vulgatus, B. dorei, B. caccae, B. stercoris, and B. uniformis and decrease the abundance of B. fragilis, B. finegoldii, and B. coprophilus, as determined by quantitative polymerase chain reaction (qPCR) (Fig. 6B). The concentrations of amino acids were also monitored under anaerobic conditions at 0, 12, 24, and 48 hours throughout the fermentation by GC-MS metabolomics. The results reveal that PMFE could significantly decrease amino acid levels in batch culture in vitro (Fig. 6C).
Fig. 6. Citrus PMFE–mediated enrichment of B. ovatus attenuates MetS in HFD mice.
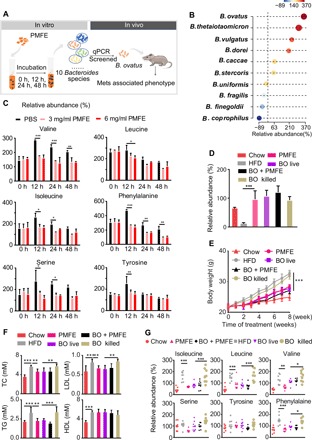
Chow-fed mice were treated daily with solvent (0.5% CMCNa) (Chow). HFD-fed mice were orally administered with high-dose PMFE (120 mg/kg per day, HFD and PMFE), B. ovatus (BO live), B. ovatus and high-dose PMFE (BO and PMFE), or killed B. ovatus (BO killed). (A) Study design of in vitro batch culture fermentation and in vivo verification experiment. (B) Relative abundance of Bacteroides species after incubation with PMFE (presented in percent initial levels). (C) Changes of the amino acid concentrations at 0, 12, 24, and 48 hours after incubation with different concentrations of PMFE. (D) Relative abundance of B. ovatus in the indicated groups measured by qPCR. (E) Body weight of the above four groups of mice. (F) Total TC, TG, LDL, and HDL levels in blood. (G) Relative abundance of isoleucine, leucine, valine, serine, tyrosine, and phenylalanine in feces by GC-MS in the indicated groups. Error bars are expressed as means ± SD. Statistical significance was determined by one-way or two-way ANOVA with Tukey tests for multiple-group comparisons.
Because the increase of B. ovatus exhibited the most notable change in Bacteroides species by PMFE treatment, we next examined the possible causal relationship between B. ovatus and MetS. HFD-fed mice were gavaged daily with commercial strains of B. ovatus for 8 weeks followed by examination of MetS-related traits. In fecal DNA, colonization was observed in HFD-fed mice treated with B. ovatus. Meanwhile, administration of PMFE also greatly enriched the abundance of B. ovatus (Fig. 6D). Gavage with live, but not heat-killed, B. ovatus significantly lowered body weight gain and fat accumulation of HFD mice (Fig. 6E and fig. S10, A to C). The administration of B. ovatus greatly reduced serum LDL-C and TG concentrations compared to HFD mice gavaged with heat-killed B. ovatus (Fig. 6F). Treatment with live B. ovatus also significantly ameliorated plasma ALT and AST (fig. S10D). We did not observe a considerable improvement in the glucose tolerance and insulin resistance after B. ovatus gavage (fig. S10E). Notably, we detected significantly lower abundance of fecal BCAAs in HFD mice gavaged with live B. ovatus compared to HFD mice gavaged with heat-killed B. ovatus (Fig. 6G). Similar results were also measured in the serum samples (fig. S10F). Together, these findings indicate that PMFE could enrich the commensal bacterium B. ovatus, which attenuates diet-induced obesity and metabolic disorders and modulates circulating amino acid concentration.
DISCUSSION
MetS is emerging as one of the most health-threatening diseases in the world (1). Hence, there remains an urgent need for the development of effective and safe therapeutic agents. Citrus PMFs are attractive candidates due to their potent protective effects against MetS as well as safety in rodent experimental models (17, 18). Here, we used an optimized and standardized method to purify PMFs from citrus peels based on macroporous resin chromatography. The citrus PMFEs enriched are qualitatively and quantitatively characterized, and the compounds contained are clearly elucidated (19). We found that PMFE is superior to ECP owing to no discernible toxicity even at high concentration in in vitro experiments. In addition, we also compared the effect of PMFE with a positive control statin, which is the preferred treatment of MetS in spite of potential adverse effects (1). Over 8 weeks of treatment, a high dose of PMFE exhibits equal or even better metabolic protective effect than that of lovastatin. We did not observe obvious side effects of high dose of PMFE in chow- and HFD-fed mice. These data indicate that the citrus PMFE might be a promising therapeutic agent for metabolic disorders in terms of their efficacy and safety.
Given that PMFs are groups of phenolic phytochemicals with poor absorption, we believe that the beneficial effects of PMFE are mostly due to remodeling of gut microbiota, which is pivotal in multiple phenotypes associated with MetS (2, 3). The evidence that strongly supports this idea is demonstrated by the diminishment of protective effects by suppression of gut microbiota using antibiotics and transferable metabolic protection by FMT. Our findings that PMFE could exert metabolic protection through gut microbiota provide novel mechanistic understanding of flavonoids with low bioavailability and could revolutionize therapeutic assessment of phytochemicals.
Gut microbiota, with Firmicutes and Bacteroidetes dominating, has been linked to obesity-related MetS (20). A shift toward an increase in Firmicutes-to-Bacteroidetes ratio was identified in obese mice (20, 21) and humans (6, 22), while weight loss intervention was accompanied by a higher abundance of Bacteroidetes. Comparison of gut microbial compositions between HFD and PMFE-treated HFD mice by 16S rDNA sequencing revealed a similar trend toward a decreased Firmicutes-to-Bacteroidetes ratio, which would contribute to the metabolic protective function of PMFE. At the family level, PMFE treatment enriched the abundance of Bacteroidetes S 24-7, which is actively involved in the degradation of particular carbohydrates (plant glycan, host glycan, and α-glucan), and the increased abundance has been described in mice fed a low-fat diet (23). PMFE also increased the abundance of Ruminococcus, which was reported to be negatively correlated with metabolic disorders and diabetes (24). Bacteroides is the key genus responding to PMFE treatment. By assessment of the fecal batch culture fermentation in vitro, we found that PMFE could significantly induce the enrichment of six species including B. ovatus, B. uniformis, and B. thetaiotaomicron. The glutamate-fermenting commensal B. thetaiotaomicron has been reported to protect mice against adiposity and is associated with weight loss in a human weight loss intervention (6). B. uniformis has also been known to alleviate HFD-induced obesity (25). PMFE incubation also led to a reduction of the level of three species, especially with a large decrease of B. fragilis, which was reported to result in higher body weight gain, impaired glucose tolerance, and lower insulin sensitivity (26). Given that B. fragilis decreased the levels of TβMCA and TUDCA, resulting in activated intestinal farnesoid X receptor (FXR) signaling in mice, PMFE may improve metabolic dysfunction by partly modulating the B. fragilis–GUDCA (glycoursodeoxycholic acid)–intestinal FXR axis (26). Also, we observed that PMFE could improve the impaired barrier function caused by epithelial damage and by dysregulation of tight junction proteins. Together, PMFE treatment can modulate the gut microbiome toward a healthier profile.
Drugging the microbiome has been emerging as a very attractive therapy, and an interesting question is whether the alteration of microbiome is correlative or causative to disease. In the present study, we demonstrate that the PMFE-altered microbiome has a causal role in protection of host metabolism by antibiotic treatment and FMT. In addition, we identified robust microbiome drug targets. In fecal batch culture fermentation, B. ovatus exhibited a notable increase after PMFE treatment. Because there is no direct evidence of the role of B. ovatus in host metabolism, here, we established a causal relationship between B. ovatus and metabolic homeostasis for the first time. Our data demonstrated that intervention in the gut ecosystem with B. ovatus could significantly lower body weight gain and reduced serum LDL-C and TG concentrations. These results further confirm that PMFE improves metabolic dysfunction involving the enrichment of B. ovatus, a potentially beneficial intestinal bacterium.
A growing body of evidence makes it clear that the composition of gut microbiota affects systemic metabolism through alterations in the host metabolome (27). Manipulating the gut microbiome could reverse dysregulation of microbial host co-metabolism associated with a pathological state (28). Here, we found the metabolic pathways involving valine, leucine, isoleucine, phenylalanine, tyrosine, and trypophan biosynthesis are strongly associated with dyslipidemia and actively responsive to therapeutic interventions of PMFE. These findings are consistent with the results of a cross-sectional study of sedentary MetS subjects (29). Targeted metabolomics profiles uncovered a prominent change in the levels of BCAAs in PMFE-treated mouse feces and serum. Nevertheless, there is less agreement on the association with obesity, abundance of Bacteroides species, and the interplay with host amino acid metabolism (30). We further demonstrated that B. ovatus, which was significantly enriched by the intervention of PMFE, had a positive association between weight loss and lower circulating levels of BCAAs. These data indicate a potentially causal role of B. ovatus in mediating the biosynthesis of BCAAs in metabolic disorders. Previous studies identified P. copri and B. vulgatus as main drivers for increased levels of circulating BCAAs. It seems controversial with our finding that PMFE treatment enriched the abundance of B. vulgatus as well as reduced BCAAs. The role of B. vulgatus in metabolic disorders seems complex, as shown by protection against the development of MetS in the low-fat diet mice model (31, 32) in spite of a positive correlation with MetS in human (4). The context-specific role of B. vulgatus after PMFE treatment in regulation of BCAA levels and metabolic disorders needs further exploration. Also, further studies are needed to explore the role of other Bacteroides species in host amino acid metabolism.
Presently, there is little mechanistic insight into how amino acid–derived metabolites contribute to disease. A recent study identified that imidazole propionate, a microbially produced amino acid–derived metabolite, impairs insulin signaling through activation of mTOR complex 1 (mTORC1) (33). Insulin resistance induced by BCAAs was accompanied by chronic activation of mTOR and P70S6K (34). Our results demonstrated that PMFE could decrease the levels of BCAAs in feces and serum. We further showed that PMFE suppressed the phosphorylation of mTOR and P70S6K, as well as decreased the expression of SREBPs in cells and HFD-fed mice. Therefore, we speculate that the decreased BCAAs by PMFE treatment contribute to the improvement of MetS through inhibition of the mTOR/P70S6K/SREBP pathway. The detailed mechanism of action needs to be investigated further.
In conclusion, our results demonstrate that the citrus PMFE could attenuate MetS by regulating amino acid metabolism via modulation of gut microbiota. PMFE improves metabolic dysfunction involving the enrichment of B. ovatus, a potentially beneficial intestinal bacterium. Our findings should be of value in considering drugging the microbiome as a novel mechanistic understanding of phytochemicals.
MATERIALS AND METHODS
Preparation of PMFE
The effects of extraction and purification on the yield of PMFs were investigated and optimized for a standardized production process of citrus PMFE. All the PMFE used throughout the trial was prepared by the same standardized process of macroporous resin chromatography (Supplementary Materials and Methods). The pale yellow PMFE powder was then qualitatively and quantitatively analyzed by high-performance liquid chromatography (HPLC)–QTOF/MS and HPLC-DAD (diode array detector). A total of 56 PMFs were chemically characterized from the PMFE. Sinensetin, nobiletin, 3,5,6,7,8,3′,4′-heptamethoxyflavone, and tangeretin were the main constituents in PMFE. After the extraction and purification of PMFE, the total contents of the four PMFs were increased to 60.85% [including 38.51% (w/w) nobiletin, 15.62% tangeretin, 3.43% sinensetin, and 3.29% 3,5,6,7,8,3′,4′-heptamethoxyflavone] in PMFE determined by HPLC-DAD.
Cell culture
HL-7702 cells were purchased from KeyGen Biotech (Nanjing China) and grown in Dulbecco’s modified eagle’s medium supplemented with penicillin (100 U/ml), streptomycin (100 μg/ml), and 10% (v/v) fetal bovine serum (FBS) under 37°C, 5% CO2.
Luciferase assays and Western blot analysis
HL-7702/SRE-Luc cells were plated in 96-well plates at a density of 2.5 × 105 cells per well and cultured for 18 hours. The cells were then switched to medium containing 5% LPDS, 10 μM compactin, and 10 μM mevalonate for 18 hours in the absence or presence of PMFE, and luciferase activity was measured as previously described (10). Normalized luciferase values were determined by dividing luciferase activity by the protein content in cell extracts quantified using the BCA (bicinchonininc acid) protein assay (Beyotime). Western blot was measured as previously described (35).
Real-time qPCR
Total RNA was extracted using TRIzol reagent (Life Technologies, USA) according to the manufacturer’s instructions. RNA concentrations were equalized and converted to cDNA using a kit (HiScript II reverse transcriptase, Vazyme, Nanjing, China). Gene expression was measured by qPCR (Roche, Basel, Switzerland) using SYBR Green (Roche, Basel, Switzerland). Expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or 18S ribosomal RNA (rRNA) control. Primer sets were designed from primer bank (36).
Animals and drug administration
Animal experiments were conducted in accordance with the Guidelines for Animal Experimentation of China Pharmaceutical University (Nanjing, China), and the protocols were approved by the Animal Ethics Committee of this institution. Eight-week-old C57BL/6J male mice (18 to 20 g) were obtained from Sino-British Sippr/BK Lab Animals Ltd. (Shanghai, China). Before the experiment, they were acclimatized to the environment with free access to food and water for at least 1 week. Mice were randomly distributed and fed with either a standard chow diet (13.5% of energy from fat, normal diet, Nanjing Qinglong Mountain Laboratory Animal Co. Ltd.) or an HFD (60% of energy from fat; TP 23300, TROPHIC Animal Feed High-tech Co. Ltd.) plus daily administration of either 0.5% carboxymethylcellulose sodium (CMCNa) or PMFE at 30, 60, or 120 mg/kg suspended in 0.5% CMCNa by intragastric gavage.
Antibiotic treatment and fecal microbiota transplantation
Eight-week-old male C57BL/6 J mice fed an HFD were supplemented with either 0.5% CMCNa (solvent) or PMFE at 120 mg/kg in the absence or presence of antibiotics [vancomycin (0.5 g/liter), neomycin sulfate (1 g/liter), metronidazole (1 g/liter), and ampicillin (1 g/liter)]. Fecal transplant was performed on the basis of an established protocol (37). Briefly, 8-week-old male donor mice were fed with HFD or HFD + PMFE (120 mg/kg). The recipient mice were fed HFD and treated daily with fresh transplant material from either PMFE-treated mice or HFD mice. Stool was collected daily from donor mice and pooled. Donor stool (100 mg) was diluted with saline and homogenized for 1 min using a vortex to achieve a liquid slurry, and then centrifuged at 500g for 3 min to remove particulate matter to facilitate administration. Fresh transplant material was prepared within 10 min before oral gavage to prevent changes in bacterial composition. There was no pretreatment of HFD and HFD + PMFE before the fecal transplantation. Oral gavage with drug or fecal transplant material was conducted daily through the 8-week experiment.
Mouse intervention study with B. ovatus
B. ovatus (ATCC 8483) was purchased from the American Type Culture Collection (ATCC) and cultivated in sterilized thioglycollate medium containing trypticase peptone (15 g/liter), yeast extract (5 g/liter), glucose (5 g/liter), sodium thioglycollate (0.5 g/liter), l-cysteine (0.5 g/liter), sodium chloride (2.5 g/liter), resazurin (0.001 g/liter), and agar (0.75 g/liter) (HB 5190-5, QingDao Hopebio-Technology Co. Ltd.) anaerobically. B. ovatus was stored in 10% glycerol at −80°C until use. Cultures were collected in log phase and diluted with sterile phosphate-buffered saline (PBS) to 5 × 106 colony-forming units/ml for gavage. For the sham-controlled trials, the B. ovatus was heat-killed by high-pressure steam sterilization at 121°C for 15 min. Mice were gavaged daily with either live bacteria or sham (0.2 ml/10 g).
Metabolomics profiling of serum and fecal samples
Serum samples were separated from the blood by centrifugation and stored at −80°C until required for metabolomic analyses. Serum (50 μl) was precipitated with 150 μl of ice-cold methanol [containing chloro-d-phenylalanine (100 ng/ml) and ketoprofen (10 ng/ml), internal standards]. To ensure data quality for metabolic profiling, pooled quality control samples were prepared by mixing equal (10 μl) amounts of each serum sample. All samples were subsequently centrifuged at 13,000g for 10 min at 4°C. The supernatants were subjected to metabolomics profiling by UHPLC-QTOF/MS.
For fecal samples, 50 mg of lyophilized feces was homogenized with 600 μl of ultrapure water. The extracts were centrifuged at 13,000 rpm and 4°C for 10 min, and the supernatant was immediately transferred. The residue was further extracted with 600 μl of ice-cold methanol. Each 200 μl of supernatant was combined, and a volume of 80 μl was precipitated with 80 μl of methanol [containing chloro-d-phenylalanine (100 ng/ml) and ketoprofen (10 ng/ml)]. After the homogenization and centrifugation, 100 μl of supernatant was transferred to a sample vial for analysis.
Liquid chromatographic separation for processed serum was achieved on a Zorbax Eclipse Plus C18 column (100 mm × 2.1 mm, 1.8 μm) using a 1290 Infinity system, and MS was performed on a 6545 Quadrupole Time-of-Flight system (all devices from Agilent Technologies, Santa Clara, CA, USA). The mobile phase was composed of 0.1% formic acid–water (v/v; A) and 0.1% formic acid–acetonitrile (v/v; B) in positive ion modes, and 5 mM ammonium acetate in water (A) and 5 mM ammonium acetate in water, acetonitrile (10: 90, B) in negative ion mode. The flow rate was set at 0.4 ml min−1 with the following optimal gradient elution condition: 0 to 7 min, 5 to 80% B; 7 to 10 min, 80 to 100%; 10 to 13 min, 100 to 100% (serum); 0 to 3 min, 5 to 30% B; 3 to 9 min, 30 to 90%; 9 to 12 min, 90 to 100% (serum); 12 to 16 min, 100 to 100% B (feces).
The operation parameters of the mass spectrometer were set as follows: drying gas temperature, 350°C; nebulizing gas (N2) flow rate, 10 liters min−1; nebulizer, 35 psi; capillary, 3500 V; fragmentor voltage, 120 V. All operations, acquisition, and analysis of date were monitored by Agilent LC-QTOF/MS MassHunter Acquisition Software version A.01.05 (Agilent Technologies, Santa Clara, CA, USA) and operated under MassHunter Acquisition Software version B.06.00 (Agilent Technologies, Santa Clara, CA, USA).
The HPLC grade acetonitrile, methanol, and formic acid were purchased from Merck (Darmstadt, Germany). Ultrapure water was purified by a Milli-Q academic water purification system (Millipore, Bedford, MA, USA). Other reagents used were of at least analytical grade.
Targeted amino acid quantification
Six amino acids (isoleucine, leucine, valine, phenylalanine, tyrosine, and serine) were quantified by Agilent 7890B/5977A GC-MS equipped with HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm, Agilent J&W Scientific, USA). Aliquots of 50-μl serum were precipitated with 200 μl of methanol followed by vortex mixing for 5 min before centrifuged at 13,000g for 10 min at 4°C. Supernatant (200 μl) was transferred to a clean Eppendorf tube and dried under a gentle stream of nitrogen gas. The residue was derivatized by addition of 40 μl of methoxyamine hydrochloride (15 mg/ml in pyridine, 1 hour at 60°C) followed by trimethylsilyl derivatization using MSTFA [N-methyl-N-(trimethylsilyl)trifluoroacetamide 60 μl, 1 hour at 70°C]. After cooling down to room temperature, the mixture was centrifuged at 13,000 rpm for 10 min before GC-MS analysis. A 1-μl aliquot of derivatized sample was injected into an Agilent 7890B/5977A gas using helium as carrier gas (1 ml/min). Initially, the oven temperature was 80°C for 2 min, elevated to 250°C at a rate of 10°C/min, and maintained for 3 min. The temperature of the injector, transfer line, and ion source were set to 250°, 290°, and 230°C, respectively. Using a single-quadrupole MS detector in SIM mode, chromatographic system, ion source, and MS detector parameters were optimized to obtain desired sensitivity. The solvent delay time was set to 5 min. The results of method validation are shown in the Supplementary Materials.
16S rDNA amplicon sequencing
Genomic DNA of fecal samples was extracted using a DNA kit (TIANGEN Biotech Co. Ltd., Beijing, China) and quantified using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA). DNA (30 to 50 ng) was used to generate amplicons using a MetaVx Library Preparation kit. V3, V4, and V5 hypervariable regions of prokaryotic 16S rDNA were selected for generating amplicons and following taxonomy analysis. DNA libraries were validated by Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified by Qubit 2.0 Fluorometer. DNA libraries were multiplexed and loaded on an Illumina MiSeq instrument according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). Sequencing was performed using paired-end configuration; image analysis and base-calling were conducted by the MiSeq Control Software (MCS) embedded in the MiSeq instrument.
Fecal batch-culture fermentation in vitro
Fresh fecal samples from HFD mice were collected and processed within 30 min. The 20% (w/v) fecal slurry was prepared by diluting the homogenized fecal sample in sterile PBS medium. The total volume of each culture system was 5 ml. PMFE treatment cultures consisted of 3.5 ml of culture medium (thioglycollate medium), 0.5 ml of fecal slurry, and 1 ml of PMFE of different concentrations (3 and 6 mg/ml). Negative control cultures consisted of 3.5 ml of culture medium and 0.5 ml of fecal slurry with 1 ml of PBS. Batch cultures were incubated in an anaerobic chamber (H2:CO2:N2, 10:10:80) at 37°C without stirring, and samples were dynamically collected at 0, 12, 24, and 48 hours for DNA extraction and amino acid quantification. Fermentations were conducted in triplicate (38).
Fecal DNA extraction and quantification of the abundance of Bacteroides species
The stool sample of each mouse was collected at the end of the experiment (between weeks 7 and 8) and frozen (−80°C) until analysis. Fecal DNA was extracted using the TIANamp Stool DNA Kit [TIANGEN Biotech (Beijing) Co. Ltd., DP 328] according to the manufacturer’s instructions, and the concentration was measured by a Nano-100 micro-spectrophotometer (Hangzhou AllSheng Instruments Co. Ltd.). Real-time qPCR (LightCycler 96, Roche Diagnostics) and AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co. Ltd.) were used to determine the amounts of B. ovatus. A set of specific primers used to amplify the bacterial is described in table S1. Standard curves were constructed using the diluted DNA of cultured B. ovatus. Standard and quantified samples were performed in triplicate (39).
Statistical analysis
The results of biological assay are presented as means ± SD. The differences between two groups were analyzed by Student’s t test. Datasets that involved more than two groups were assessed by one-way or two-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison’s test. Statistical analyses were performed using GraphPad Prism (USA).
All results were considered statistically significant at P < 0.05. The differences of abundance distributions among metabolites between two groups were measured by the Mann-Whitney U test with Benjamini-Hochberg false discovery rate correction. Adjusted P values less than 0.05 were considered statistically significant. OPLS-DA was conducted to identify the discrimination of variables. Differential metabolites were defined as those with variable importance in the projection (VIP) >1.0 obtained from OPLS-DA and adjusted P values less than 0.05.
Supplementary Material
Acknowledgments
We thank C. Zeng for contribution to the discussions. We thank T.-Q. Huang for help in the molecular biology experiments and J. Folz for critical reading of this manuscript. Funding: We greatly appreciate financial support from the National Natural Science Foundation of China (81922072, 81673569, 81973443, and 81421005), National Key Research and Development Program of China (2017YFC1701105), “Double First-Class” University project (CPU2018PZQ16 and CPU2018GF04), National Modern Agricultural Industrial Park of China (no. njf [2017] 110), and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions. Author contributions: E.-H.L., J.L., P.L., and S.-L.Z. designed the study protocol and supervised all parts of the project. S.-L.Z. and S.-Z.L. conducted animal experiments. P.-T.X. performed the molecular biology experiments. Y.-Y.C. did the bioinformatics analyses in close collaboration with S.-L.Z. S.-L.Z. drafted the first versions. E.-H.L., J.L., P.L., B.-Z.C., and C.C. contributed to text revision and discussion. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaax6208/DC1
Supplementary Materials and Methods
Table S1. PCR primers for detection of 10 Bacteroides species.
Table S2. (Excel file) The differential metabolites identified by cross-comparisons of different groups (Chow versus HFD and HFD versus PMFE) in feces and serum.
Table S3. (Excel file) The method validation of the GC-MS for the quantification of six amino acids.
Fig. S1. Citrus PMFE shows negligible cytotoxicity in HL-7702 cells.
Fig. S2. Oral treatment of citrus PMFE prevents HFD-induced lipid deposition and inflammation in mice.
Fig. S3. Robust dose-dependent metabolic protection of citrus PMFE in HFD mice.
Fig. S4. Citrus PMFE does not produce any apparent effects in chow-fed mice.
Fig. S5. Citrus PMFE increases intestinal tight junction in HFD mice.
Fig. S6. Citrus PMFE regulates host fecal and serum metabolome.
Fig. S7. Citrus PMFE alters MetS-associated BCAAs in HFD mice.
Fig. S8. Citrus PMFE attenuates MetS in HFD mice in a gut microbiota–dependent manner.
Fig. S9. Fecal transplantation of citrus PMFE exhibits metabolic protection in HFD mice.
Fig. S10. Citrus PMFE–mediated enrichment of B. ovatus prevents metabolic syndrome in HFD mice.
REFERENCES AND NOTES
- 1.Ford E. S., Li C., Zhao G., Prevalence and correlates of metabolic syndrome based on a harmonious definition among adults in the US. J. Diabetes 2, 180–193 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Tang W. H. W., Kitai T., Hazen S. L., Gut microbiota in cardiovascular health and disease. Circ. Res. 120, 1183–1196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ussar S., Griffin N. W., Bezy O., Fujisaka S., Vienberg S., Softic S., Deng L., Bry L., Gordon J. I., Kahn C. R., Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 22, 516–530 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen H. K., Gudmundsdottir V., Nielsen H. B., Hyotylainen T., Nielsen T., Jensen B. A. H., Forslund K., Hildebrand F., Prifti E., Falony G., Chatelier E. L., Levenez F., Doré J., Mattila I., Plichta D. R., Pöhö P., Hellgren L. I., Arumugam M., Sunagawa S., Vieira-Silva S., Jørgensen T., Holm J. B., Trošt K.; MetaHIT Consortium, Kristiansen K., Brix S., Raes J., Wang J., Hansen T., Bork P., Brunak S., Oresic M., Ehrlich S. D., Pedersen O., Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Yoon M.-S., The emerging role of branched-chain amino acids in insulin resistance and metabolism. Nutrients 8, 405 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu R., Hong J., Xu X., Feng Q., Zhang D., Gu Y., Shi J., Zhao S., Liu W., Wang X., Xia H., Liu Z., Cui B., Liang P., Xi L., Jin J., Ying X., Wang X., Zhao X., Li W., Jia H., Lan Z., Li F., Wang R., Sun Y., Yang M., Shen Y., Jie Z., Li J., Chen X., Zhong H., Xie H., Zhang Y., Gu W., Deng X., Shen B., Xu X., Yang H., Xu G., Bi Y., Lai S., Wang J., Qi L., Madsen L., Wang J., Ning G., Kristiansen K., Wang W., Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Del Rio D., Rodriguez-Mateos A., Spencer J. P., Tognolini M., Borges G., Crozier A., Dietary (poly) phenolics in human health: Structures, bioavailability, and evidence of protective effects against chronic diseases. Antioxid. Redox Signal. 18, 1818–1892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saigusa D., Shibuya M., Jinno D., Yamakoshi H., Iwabuchi Y., Yokosuka A., Mimaki Y., Naganuma A., Ohizumi Y., Tomioka Y., Yamakuni T., High-performance liquid chromatography with photodiode array detection for determination of nobiletin content in the brain and serum of mice administrated the natural compound. Anal. Bioanal. Chem. 400, 3635–3641 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Guo J., Tao H., Cao Y., Ho C.-T., Jin S., Huang Q., Prevention of obesity and type 2 diabetes with aged citrus peel (Chenpi) extract. J. Agric. Food Chem. 64, 2053–2061 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Soyal S. M., Nofziger C., Dossena S., Paulmichl M., Patsch W., Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol. Sci. 36, 406–416 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Owen J. L., Zhang Y., Bae S.-H., Farooqi M. S., Liang G., Hammer R. E., Goldstein J. L., Brown M. S., Insulin stimulation of SREBP-1c processing in transgenic rat hepatocytes requires p70 S6-kinase. Proc. Natl. Acad. Sci. U.S.A. 109, 16184–16189 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He B., Nohara K., Park N., Park Y.-S., Guillory B., Zhao Z., Garcia J. M., Koike N., Lee C. C., Takahashi J. S., Yoo S.-H., The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab. 23, 610–621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen F., Wen Q., Jiang J., Li H.-L., Tan Y.-F., Li Y.-H., Zeng N.-K., Could the gut microbiota reconcile the oral bioavailability conundrum of traditional herbs? J. Ethnopharmacol. 179, 253–264 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Chen M.-l., Yi L., Zhang Y., Zhou X., Ran L., Yang J., Zhu J.-d., Zhang Q.-y., Mi M.-t., Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio 7, e02210–e02215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Roberts A. B., Buffa J. A., Levison B. S., Zhu W., Org E., Gu X., Huang Y., Zamanian-Daryoush M., Culley M. K., Di Donato A. J., Fu X., Hazen J. E., Krajcik D., Di Donato J. A., Lusis A. J., Hazen S. L., Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X., Xie G., Zhao A., Zhao L., Yao C., Chiu N. H. L., Zhou Z., Bao Y., Jia W., Nicholson J. K., The footprints of gut microbial–mammalian co-metabolism. J. Proteome Res. 10, 5512–5522 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Tung Y.-C., Chang W.-T., Li S., Wu J.-C., Badmeav V., Ho C.-T., Pan M.-H., Citrus peel extracts attenuated obesity and modulated gut microbiota in mice with high-fat diet-induced obesity. Food Funct. 9, 3363–3373 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Malkanthi E., Judy W. V., Wilson D., Rumberger J. A., Guthrie N., Randomized, double-blind, placebo-controlled, clinical study on the effect of Diabetinol® on glycemic control of subjects with impaired fasting glucose. Diabetes, Metab. Syndr. Obes. 8, 275–286 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng S.-L., Duan L., Chen B.-Z., Li P., Liu E.-H., Chemicalome and metabolome profiling of polymethoxylated flavonoids in Citri Reticulatae Pericarpium based on an integrated strategy combining background subtraction and modified mass defect filter in a Microsoft Excel Platform. J. Chromatogr. A 1508, 106–120 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh P. J., Bäckhed F., Fulton L., Gordon J. I., Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome. Cell Host Microbe 3, 213–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I., Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley R. E., Turnbaugh P. J., Samuel K., Gordon J. I., Microbial ecology: Human gut microbes associated with obesity. Nature 444, 1022–1023 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Ormerod K. L., Wood D. L. A., Lachner N., Gellatly S. L., Daly J. N., Parsons J. D., Dal’Molin C. G. O., Palfreyman R. W., Nielsen L. K., Cooper M. A., Morrison M., Hansbro P. M., Hugenholtz P., Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 4, 36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., Kurilshikov A., Bonder M. J., Valles-Colomer M., Vandeputte D., Tito R. Y., Chaffron S., Rymenans L., Verspecht C., De Sutter L., Lima-Mendez G., D’hoe K., Jonckheere K., Homola D., Garcia R., Tigchelaar E. F., Eeckhaudt L., Fu J., Henckaerts L., Zhernakova A., Wijmenga C., Raes J., Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gauffin C. P., Santacruz A., Moya Á., Sanz Y., Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLOS ONE 7, e41079 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delzenne N. M., Bindels L. B., Microbiome metabolomics reveals new drivers of human liver steatosis. Nat. Med. 24, 906–907 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S., Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Chang C.-J., Lin C.-S., Lu C.-C., Martel J., Ko Y.-F., Ojcius D. M., Tseng S.-F., Wu T.-R., Chen Y.-Y., Young J. D., Lai H.-C., Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat. Commun. 6, 7489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tai E. S., Tan M. L. S., Stevens R. D., Low Y. L., Muehlbauer M. J., Goh D. L. M., Ilkayeva O. R., Wenner B. R., Bain J. R., Lee J. J. M., Lim S. C., Khoo C. M., Shah S. H., Newgard C. B., Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia 53, 757–767 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wexler A. G., Goodman A. L., An insider's perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch C. J., Adams S. H., Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 10, 723–736 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridaura V. K., Faith J. J., Rey F. E., Jiye C., Duncan A. E., Kau A. L., Griffin N. W., Lombard V., Henrissat B., Bain J. R., Muehlbauer M. J., Ilkayeva O., Semenkovich C. F., Funai K., Hayashi D. K., Lyle B. J., Martini M. C., Ursell L. K., Clemente J. C., Van Treuren W., Walters W. A., Knight R., Newgard C. B., Heath A. C., Gordon J. I., Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh A., Molinaro A., Ståhlman M., Khan M. T., Schmidt C., Mannerås-Holm L., Wu H., Carreras A., Jeong H., Olofsson L. E., Bergh P.-O., Gerdes V., Hartstra A., de Brauw M., Perkins R., Nieuwdorp M., Bergström G., Bäckhed F., Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Newgard C. B., Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 15, 606–614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu D., Wang Z., Zhang Y., Jiang W., Pan Y., Song B.-L., Chen Y., PAQR3 modulates cholesterol homeostasis by anchoring Scap/SREBP complex to the Golgi apparatus. Nat. Commun. 6, 8100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spandidos A., Wang X., Wang H., Seed B., PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 38, D792–D799 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borody T. J., Paramsothy S., Agrawal G., Fecal microbiota transplantation: Indications, methods, evidence, and future directions. Curr. Gastroenterol. Rep. 15, 337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koutsos A., Lima M., Conterno L., Gasperotti M., Bianchi M., Fava F., Vrhovsek U., Lovegrove J. A., Tuohy K. M., Effects of commercial apple varieties on human gut microbiota composition and metabolic output using an in vitro colonic model. Nutrients 9, 533 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R.-F., Cao W.-W., Cerniglia C. E., PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62, 1242–1247 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaax6208/DC1
Supplementary Materials and Methods
Table S1. PCR primers for detection of 10 Bacteroides species.
Table S2. (Excel file) The differential metabolites identified by cross-comparisons of different groups (Chow versus HFD and HFD versus PMFE) in feces and serum.
Table S3. (Excel file) The method validation of the GC-MS for the quantification of six amino acids.
Fig. S1. Citrus PMFE shows negligible cytotoxicity in HL-7702 cells.
Fig. S2. Oral treatment of citrus PMFE prevents HFD-induced lipid deposition and inflammation in mice.
Fig. S3. Robust dose-dependent metabolic protection of citrus PMFE in HFD mice.
Fig. S4. Citrus PMFE does not produce any apparent effects in chow-fed mice.
Fig. S5. Citrus PMFE increases intestinal tight junction in HFD mice.
Fig. S6. Citrus PMFE regulates host fecal and serum metabolome.
Fig. S7. Citrus PMFE alters MetS-associated BCAAs in HFD mice.
Fig. S8. Citrus PMFE attenuates MetS in HFD mice in a gut microbiota–dependent manner.
Fig. S9. Fecal transplantation of citrus PMFE exhibits metabolic protection in HFD mice.
Fig. S10. Citrus PMFE–mediated enrichment of B. ovatus prevents metabolic syndrome in HFD mice.


