Abstract
Various fusion tags are commonly employed to increase the heterologous expression and solubility of aggregation-prone proteins within Escherichia coli. Herein, we present a protocol for efficient recombinant expression and purification of the human RNA demethylases Alkbh5 and FTO. Our method incorporates a novel fusion tag (the N-terminal domain of bacterial enzyme I, EIN) that dramatically increases the solubility of its fusion partner and is promptly removed upon digestion with a protease. The presented protocol allows for the production of mg amounts of Alkbh5 and FTO in 1L of both rich and minimal media. We developed a liquid chromatography-mass spectrometry (LC-MS)-based assay to confirm that both proteins are enzymatically active. Furthermore, the LC-MS method developed here is applicable to other members of the AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases. The superior protein yield, afforded by our expression and purification method, will facilitate biochemical investigations into the biological function of the human RNA demethylases and endorse employment of EIN as a broadly applicable fusion tag for recombinant expression projects.
Keywords: Enzyme I, gene fusion, solubility tag, recombinant expression, N6-methyladenosine
Introduction
Recombinant expression of proteins in bacterial hosts has dramatically increased our ability to interrogate protein function at the molecular level. Indeed, such technology has allowed for expression and purification of a myriad of proteins in mg amounts. Furthermore, bacterial expression systems have facilitated sophisticated labeling schemes—both biomimetic and artificial—such as isotope labeling, site-specific modifications, and non-natural amino acid incorporation. However, several challenges, including low solubility of the target protein in the bacterial host, misfolding, and host cell toxicity, have hampered the use of the recombinant DNA technology in the production of several “hard to express” or unstable proteins.
The use of fusion tags is a common practice for improving the expression and solubility of challenging systems.1–16 Fusion tags are proteins or peptides that are fused in-frame and expressed with the target protein, either for improving expression and solubility, or to aid in the purification of the target proteins. Common fusion proteins known to enhance solubility and expression include Maltose Binding Protein (MBP)1–2, Small Ubiquitin-related Modifier (SUMO)3–4, Glutathione S-transferase (GST)5, N-utilization substance A (NusA),6 and B1 domain of Streptococcal protein G (GB1)7. Nevertheless, the discovery of new fusion tags is valuable as no single tag is capable of solving low expression and solubility issues for all target proteins.
Herein, we show that the N-terminal domain of Escherichia coli Enzyme I (EIN) functions as novel fusion tag that substantially enhances recombinant expression and solubility of the human RNA demethylases Alkbh5 and FTO, two oncoproteins whose overexpression has been linked to the development of leukemia, breast, and brain cancer (note that crystal structures of the human Alkbh5 and FTO are reported as Supporting Information, Figure S1).17–19 In addition, we developed a new Liquid Chromatography-Mass Spectrometry (LC-MS) assay to confirm that the purified proteins are enzymatically active. Therefore, our results will facilitate future biochemical and structural investigations on the molecular mechanisms connecting the human RNA demethylases to the development of cancer. Interestingly, we also show that the EIN fusion tag presented here has a similar impact on recombinant expression of Alkbh5 and FTO as MBP but is cleaved off the target protein more efficiently than MBP. Based on this data, we anticipate that EIN can be used to improve the yield of other “hard to express” genes and can be added to the portfolio of the fusion tags routinely employed in protein expression and purification applications.
Materials and Methods
Fusion plasmid construction and Recombinant expression in M9 minimal medium –
All plasmids used in this work were constructed using pET21a (Novagen) as the backbone vector. Gibson assembly20 was used to incorporate the fusion genes and TEV cleavage site, which are described in Results. Plasmids were transformed into Escherichia coli BL21 Star (DE3) (Invitrogen), and transformants were selected on LB-agar containing ampicillin (100 μg/ml). Cells were grown at 37 °C in LB or M9 minimal medium. The composition of the LB medium (Pre-buffered LB capsule from Fisher Bioreagents) was as follows: 5.0 g/L Casein peptone, 5.0 g/L Yeast extract, 5 g/L NaCl, and 1.5 g/L Tris-HCl (pH 7.2). The composition of the M9 minimal medium was as follows: 6.5 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl, 4 g/L Glucose, 1g/L NH4Cl, 100 mg/L ampicillin, 120 mg/L MgSO4, 11 mg/L CaCl2, and 10 mL/L of 100x MEM Vitamin mix (Gibco). At OD600nm ~ 0.6 the temperature was reduced to 16 °C, and expression was induced via addition of 1 mM isopropyl-D-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation after 16 h of induction.
Purification of Alkbh5 and FTO –
Harvested cells were resuspended in 20 mL of 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl. The suspension was lysed using an EmulsiFlex-C3 microfluidizer (Avestin) and subsequently centrifuged at 40,000 ×g for 40 min. The supernatant traversed a 0.45 μM filter and was loaded onto a HisTrap HP column (5 mL; GE Healthcare). The protein was eluted in 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl using a 100 mL gradient from 0 mM to 375 mM Imidazole. The fractions containing the target protein were confirmed by SDS-polyacrylamide gel electrophoresis. The eluted protein was buffer exchanged into 50 mM Tris-HCl (pH 8.0) and 500 mM NaCl before being concentrated to 15 mL using an Amicon Ultra centrifugal filter (Millipore). The EIN or MBP tag was cleaved by incubation at room temperature with 0.25 mg of His-tagged Tobacco Etch Virus (TEV) protease. The TEV protease was expressed and purified in our lab. The same batch of protease was used throughout the entire work presented here. The length of incubation was optimized experimentally (see Results). After cleavage of the fusion tag, the protease and the liberated fusion tag was removed by a subtractive passage through the HisTrap HP column. The flow-through was concentrated and further purified on a Superdex-75 column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, 2 mM dithiothreitol (DTT), and 1 mM ethylenediaminetetraacetic acid (EDTA). Fractions containing the target protein were confirmed by SDS-polyacrylamide gel.
Enzymatic assay –
Enzymatic assays were performed in a final reaction volume of 300 μL. The reaction mixture consisted of 0.1 μM Alkbh5 or 2 μM FTO and varying substrate concentrations (0.5, 1,2, 3, 5, and 10 μM for Alkbh5 or 4, 8, 12, 20, and 40 μM for FTO) in the following reaction buffer: 2 mM L-ascorbic acid, 150 μM Fe (II), and 300 μM μKG in 50 mM Tris-HCl (pH 7.4). The 5-mer substrate, 5’-GG(m6A)CT-3’, was chosen based on previous reports indicating that both Alkbh5 and FTO are active against this oligonucleotide.21 Reactions were incubated at 37 °C. 50 μL aliquots were taken at regular time intervals (5, 10, 20, 30, 45, and 60 min for Alkbh5; 15, 30, 60, 120, and 240 min for FTO) and quenched with a 1:1 ratio of 20% (v/v) formic acid. The quenched reactions were passed through a 0.2 μm, 0.4 mL Ultrafree-MC Centrifugal Filter column (Millipore) by centrifugation (16,000 g for 5 min) to separate the enzyme from the nucleic acid. The pH of the flow-through, containing a mixture of the methylated and demethylated substrate, was neutralized by addition of HPLC grade NH4OH and analyzed by liquid chromatography – mass spectrometry (LC-MS). The LC-MS analysis was conducted using an Agilent Technologies 1290 Infinity Binary Pump UHPLC system coupled to an Agilent Technologies 6540 UHD Accurate-Mass Q-TOF mass spectrometer set to negative mode. The system was operated in high-resolution mode (4Gz) while scanning m/z 100-1700. The oligomers were separated on a ZORBAX Rapid Resolution HT Extended C18 column (80 Å, 1.8 μm, 2.1 x 50 mm). UHPLC separations were performed using a column temperature of 40°C with a flow rate of 0.3 mL/min. The solvent gradient began at 100% solvent A (5 mM NH4OH in H2O) progressing with a gradient to 25% solvent B (5 mM NH4OH in 70% acetonitrile) for 7 min, increased to 100% solvent B at 10 min, and returned to starting conditions at 12 min. A 3 min post-run with initial conditions was applied after each acquisition. The Q-TOF was used to determine the composition and purity of the peaks observed in the UHPLC dimension by mass-to-charge (m/z). The z = −2 species for both the methylated (m/z=757.533) and the demethylated (m/z=750.515) oligomers were predominately observed (see Results). The percent demethylation was calculated by the ratio of the integrated peak areas associated with each oligomer.
Results and Discussion
Overexpression of soluble Alkbh5 and FTO –
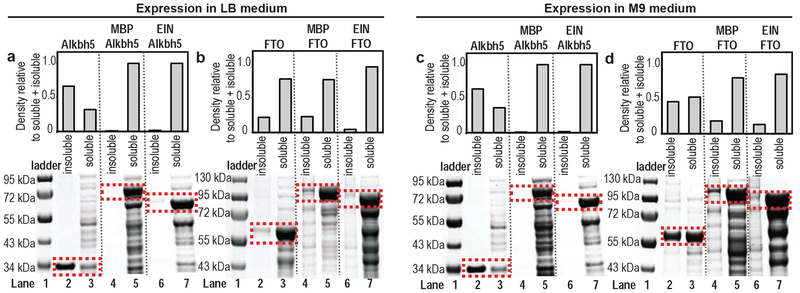
Several of the structural and biochemical characterizations of Alkbh5 and FTO reported in the literature were conducted on truncated enzymes (residues 66-292 and residues 32-505 for Alkbh5 and FTO, respectively).22–29 These truncated constructs were selected to facilitate recombinant expression and crystallization of the RNA demethylases and were shown to retain full enzymatic activity.22–29 Here, the truncated constructs of Alkbh5 and FTO were cloned into a pET21a, transformed into E. coli BL21 Star (DE3) competent cells, and expressed in Luria-Bertani (LB) or M9 minimal medium as described in Materials and Methods. Results reported in Figure 1 indicate that the two proteins are highly overexpressed but poorly soluble in E. coli. Indeed, overexpressed protein largely accumulates within inclusion bodies, despite the slow, low-temperature (16 °C), expression conditions (see Materials and Methods). The low solubility of the RNA demethylases obstructs analysis by high resolution techniques that require preparation of mg amounts of protein.
Figure 1.
Expression of Alkbh5 in (a) LB and (c) M9 medium, respectively. Expression of FTO in (b) LB and (d) M9 medium, respectively. Lane 1 is the molecular weight marker; lanes 2 and 3 correspond to the insoluble and soluble fractions of the target protein, respectively; lanes 4 and 5 correspond to the insoluble and soluble fractions of the MBP-target protein fusion construct, respectively; lanes 6 and 7 correspond to the insoluble and soluble fractions of the EIN-target protein fusion construct, respectively. Inclusion bodies were harvested by centrifugation of 20 mL of cell lysate and resuspended in 20 mL of 8 M urea. 10 μL of centrifuged cell lysate (soluble fraction) and 10 μL of resuspended inclusion bodies (insoluble fraction) were mixed with 10 μL of 2X SDS loading buffer, heated at 90 °C for 10 min and analyzed on 4-12% Bis-Tris SDS-PAGE gels. The bands on the SDS-PAGE gel relative to the relevant proteins are highlighted using red rectangles and were quantified using the densitometry functions of ImageJ software46. Measured intensities are reported in the top panel.
To increase expression of soluble Alkbh5 and FTO, we explored the use of an N-terminal solubility tag. Two different solubility tags were tested: the E. coli MBP (molecular weight: 40.7 kDa), and the E. coli EIN (molecular weight: 27 kDa). While MBP is one of the frequently implemented solubility tags for recombinant expression in E. coli30–32, EIN has not been employed in gene fusion applications yet. We opted to test EIN as a solubility tag because we have worked with this protein for several years33–42 and have noticed that (i) EIN is a well-folded protein with high thermodynamic stability (melting temperature ~ 55 °C), (ii) EIN is highly expressed (> 100 mg per L of culture) as a soluble protein in E. coli, and (iii) EIN is sufficient to recover overexpression and solubility of mutations of the EI C-terminus (EIC) that destabilize the EIC fold.
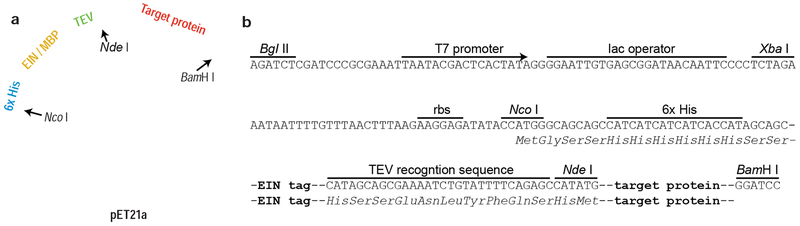
To test the capacity of an EIN-fusion to promote solubility, two constructs were designed in which the target protein (Alkbh5 or FTO) is fused to an N-terminal MBP or EIN tag. To facilitate purification of the target protein, the two constructs also included an N-terminal His-tag, and the TEV protease consensus sequence (ENLYFQ/S, where “/” signifies the cleaved peptide bond) was positioned between the solubility tag and the target protein. Further, the Nde I recognition sequence was used as a spacer between the TEV protease cleavage site and the target gene to facilitate TEV access to the cleavage site and expedite future cloning efforts. The synthesized constructs were cloned into a pET21a vector between the Xba I and BamH I restriction sites, and the resulting plasmid map is shown in Figure 2. The data reported in Figure 1 indicates that both the MBP and EIN tags have a similar impact on the expression of Alkbh5 and FTO, which significantly enhances the solubility of both RNA demethylases.
Figure 2.
(a) Map of the plasmid used for expression of Alkbh5 and FTO with an N-terminal fusion tag. A gene encoding a ribosome binding site (RBS), a N-terminal His-tag, the EIN or MBP solubility tag, a TEV recognition site, an Nde I restriction site, and the target protein was cloned between the Xba I and BamH I sites of a pET21a vector. The size of the vectors incorporating the MBP and EIN fusion partners are 6,588 bp and 6,222 bp, respectively. The Alkbh5 and FTO nucleotide sequences are 681 and 1,422 bp, respectively. Details about the cloning/expression region are shown in (b). The nucleotide sequence of the EIN-fusion construct is reported as Supporting Information.
Purification of enzymatically active Alkbh5 and FTO –
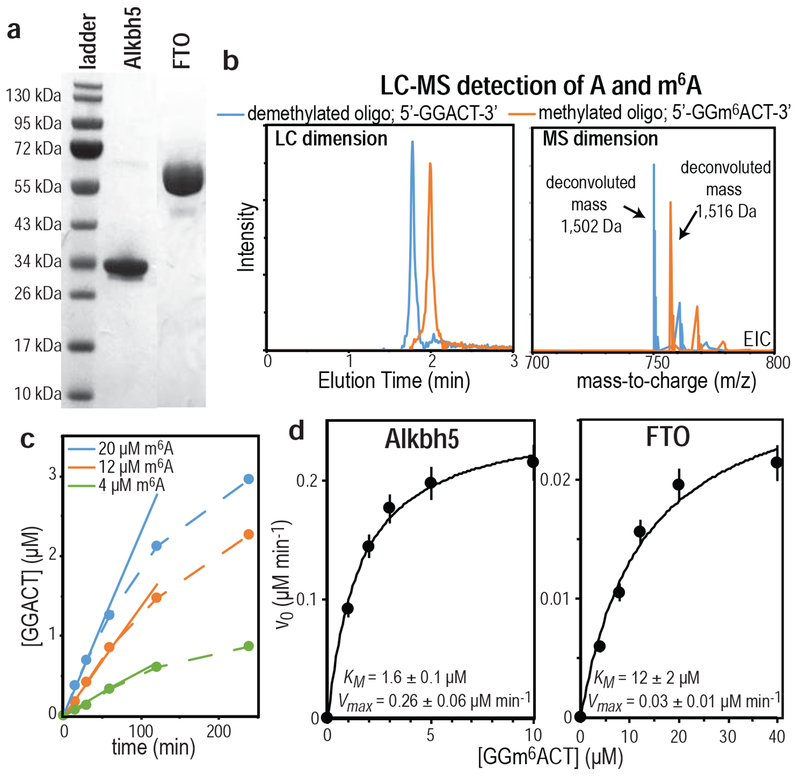
Purification of the target protein from the cell lysate was carried out using a four-step protocol (for details, see “Materials and Methods”). Briefly, a first His-tag purification step was used to isolate the target protein from most protein and nucleic acid contaminants (step 1). The fused construct was then incubated with 15 μg/mL of TEV protease to cleave the solubility tag away from the RNA demethylases (step 2). A second His-tag purification step was performed to separate the solubility tag and the protease (which incorporate an N-terminal His tag) from the target enzyme (step 3), and a final gel filtration step was added to eliminate any residual contamination (step 4). The final enzyme yields for both Alkbh5 and FTO were > 90% pure, as determined by SDS-PAGE (Figure 3a). It is worth noticing that the two RNA demethylases show a higher apparent molecular weight by SDS-PAGE than expected based on the amino acid sequence (26,375 Da and 54,810 Da for Alkbh5 and FTO, respectively). Higher apparent molecular weight in SDS-PAGE gels are commonly observed for proteins with significant acidic residue content (Asp and Glu), which affects binding of SDS to the protein.43–44 Both Alkbh5 and FTO have a relatively high number of acidic residues (~15% and ~16%, respectively). Complementary analysis of our purified samples by liquid chromatography with tandem mass spectrometry (LC-MS/MS) returns masses of 26,374 Da and 54,809 Da for Alkbh5 and FTO, respectively, confirming that the purified proteins are the truncated constructs of the two RNA demethylases. Favorably, the purified Alkbh5 and FTO remain soluble at room temperature and relatively high concentration (~0.5 mM) for several weeks under various buffer conditions (tested at pH 6.5, 7.4, and 8.0, NaCl concentrations of 0 mM, 100 mM, and 200 mM, and MnCl2 concentrations of 0 mM, 1 mM, and 2 mM).
Figure 3.
(a) SDS-PAGE analysis of purified Alkbh5 (lane 2) and FTO (lane 3). The molecular weight marker is shown in lane 1. (b) Example of extracted LC-MS data measured for the m6A demethylation reaction catalyzed by FTO. The LC dimension (left panel) shows two well resolved signals (colored blue and orange, respectively). The MS dimension of the experiment (right panel) reveals that the blue and orange peaks correspond to the −2 ionized species of the demethylated and methylated nucleotide, respectively. The smaller peaks observed in the MS dimension correspond to Na+ adducts. (c) Plot of the concentration of demethylated nucleotide versus time measured for the demethylation reaction catalyzed by FTO at three different concentrations of substrate (4 μM, green; 12 μM, orange; 20 μM, blue). The slope of the linear portion of the plot (solid line) has been fit to determine the initial velocity of the reaction (v0). (d) Michaelis-Menten plot for the demethylation reaction catalyzed by 0.1 μM Alkbh5 (left panel) and 2.0 μM FTO (right panel).
To test whether the EIN tag promoted correct folding we have developped a Liquid Chromatography–Mass Spectrometry (LC-MS) method to monitor the demethylation of the 5-mer substrate, 5’-GG(m6A)CT-3’, catalyzed by Alkbh5 and FTO. Briefly, various concentrations of substrate were incubated with 0.1 μM Alkbh5 or 2 μM FTO at 37 °C. At regular intervals, 50 μL aliquots were mixed with a 1:1 ratio of 20% (v/v) formic acid to quench the reaction for subsequent analysis by LC-MS (see Materials and Methods). Results indicate the presence of two peaks in the LC dimension, which correspond to the methylated and demethylated nucleotides, as confirmed by the molecular masses deconvoluted from the MS dimension of the experiment (Figure 3b). Initial velocities (v0) at different substrate concentrations were obtained from analyzing plots of the concentration of demethylated nucleotide (calculated from the integrated areas of the LC peaks) versus time (Figure 3c) and used to fit the Michaelis-Menten parameters for the demethylation reactions catalyzed by Alkbh5 and FTO (Figure 3d). Km values of 1.6 μM and 12.6 μM, and kcat values of 2.6 min−1 and 0.02 min−1 were obtained for Alkbh5 and FTO, respectively, which are consistent with the slow reaction kinetics previously reported for these enzymes.21
Comparison between MBP and EIN solubility tags –
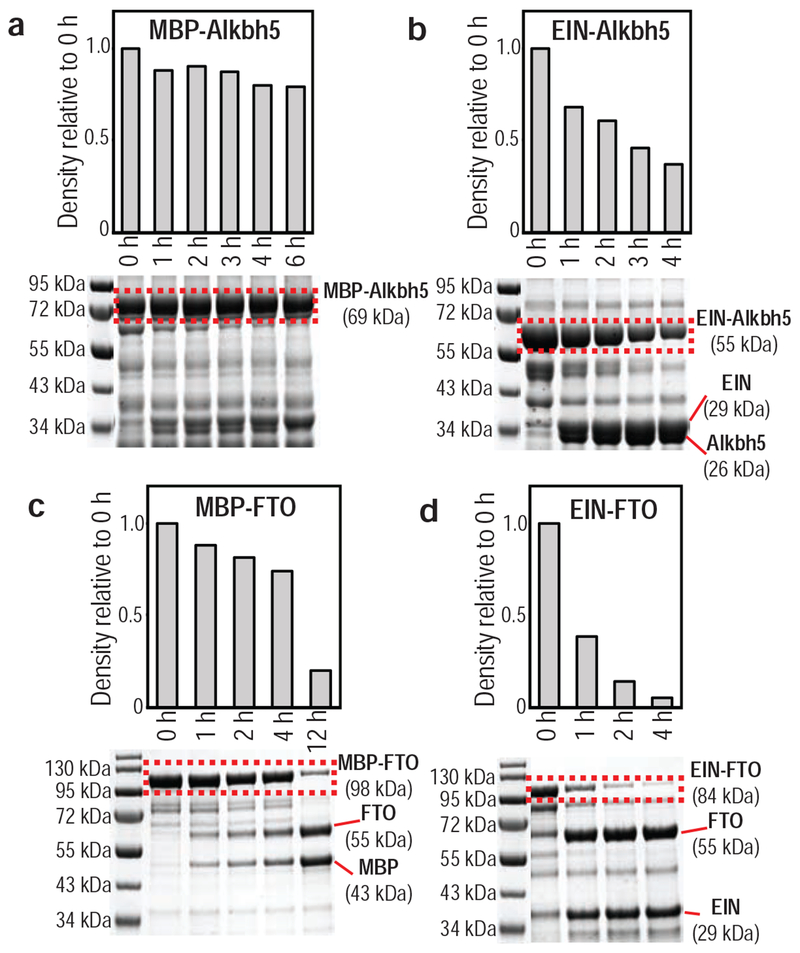
We also compared the overall performance of the MBP and EIN solubility tags for recombinant expression of Alkbh5 and FTO in M9 minimal medium. The use of the MBP tag resulted in production of ~5 mg/L of purified Alkbh5 and ~11 mg/L of purified FTO, while the use of the EIN tag resulted in production of ~12 mg/L of purified Alkbh5 and ~20 mg/L of purified FTO, indicating that EIN is better suited for recombinant expression of the human RNA demethylases in E. coli. Interestingly, EIN is cleaved off the target protein more efficiently than MBP upon digestion with TEV protease. Indeed, near-complete removal of EIN from the fusion partner is obtained in less than 4 hours, while MBP requires overnight incubation with the protease to reach a sufficient level of cleavage (Figure 4). The more efficient cleavage of EIN from the target protein results in faster purification protocols, therefore reducing the risk of misfolding and sample degradation via proteolysis.45
Figure 4.
Time dependence for the cleavage of the fusion constructs catalyzed by 15 μg/mL TEV at 25 °C. (a) MBP-Alkbh5, (b) EIN-Alkbh5, (c) MBP-FTO, and (d) EIN-FTO. The bands highlighted by the red boxes were quantified using the densitometry functions of ImageJ software 46. Measured intensities are reported in the top panels.
Conclusion
In this contribution, we have presented protocols for recombinant expression and purification of the human RNA demethylases Alkbh5 and FTO. We have used a fusion partner to increase the solubility of the target enzymes and maximize the overall yield, required for structural and biochemical studies of these proteins. Our data indicate that (i) the N-terminal domain of bacterial enzyme I (EIN) is slightly more efficient than MBP at facilitating expression of soluble Alkbh5 and FTO in E. coli (see Comparison between MBP and EIN solubility tags in the Results section), and (ii) EIN is cleaved off the target enzyme considerably faster than MBP (Figure 4), therefore minimizing both the purification time and the risk for proteolytic cleavage from endogenous proteases. Furthemore, while the EIN tag was essential to afford high protein solubility in E. coli, the purified proteins retained high solubility after cleavage, indicating that the EIN does not stabilize Alkbh5 and FTO folds via tight interactions. We have shown that the purified proteins are enzymatically active, which permits detailed structural and functional studies on the interactions between the RNA demethylases and their physiological substrates. To the best of our knowledge, the use of EIN as an efficient solubility tag has not been previously reported yet. Our work demonstrates that EIN can be added to the catalog of the available fusion partners for protein production and purification in bacterial hosts. The plasmid used here to express Alkbh5 fused to the N-terminal EIN-tag (Figure 2) has been deposited on Addgene (plasmid # 125732).
Supplementary Material
Highlights.
Overexpression of Alkbh5 and/or FTO links to leukemia, breast, and brain cancer
The EIN domain of Enzyme I is an efficient solubility tag for recombinant expression
EIN is cleaved off the target protein more efficiently than MBP
EIN allows recombinant expression of mg amounts of Alkbh5 and FTO in LB and M9 media
Acknowledgments
We thank the Protein Facility of the Iowa State University Office of Biotechnology and the W. M. Keck Metabolomics Research Laboratory for acquisition of all reported mass spectrometry data. This work was supported by NIGMS R35GM133488 (V.V.) and by funds from the Roy J. Carver Charitable Trust and Iowa State University (V.V.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.di Guana C; Lib P; Riggsa PD; Inouyeb H, Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 1988, 67 (1), 21–30. [DOI] [PubMed] [Google Scholar]
- 2.Reuten R; Nikodemus D; Oliveira MB; Patel TR; Brachvogel B; Breloy I; Stetefeld J; Koch M; Riggs PD, Maltose-Binding Protein (MBP), a Secretion-Enhancing Tag for Mammalian Protein Expression Systems. PLoS One 2016, 11 (3), e0152386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butt TR; Edavettal SC; Hall JP; Mattern MR, SUMO fusion technology for difficult-to-express proteins. Protein Expression Purif. 2005, 43 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malakhov MP; Mattern MR; Malakhova OA; Drinker M; Weeks SD; Butt TR, SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 2004, 5 (1-2), 75–86. [DOI] [PubMed] [Google Scholar]
- 5.Smith DB; Johnson KS, Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67 (1), 31–40. [DOI] [PubMed] [Google Scholar]
- 6.Davis GD; Elisee C; Newham DM; Harrison RG, New fusion protein systems designed to give soluble expression inEscherichia coli. Biotechnol. Bioeng 1999, 65 (4), 382–388. [PubMed] [Google Scholar]
- 7.Cheng Y; Patel DJ, An efficient system for small protein expression and refolding. Biochem. Biophys. Res. Commun 2004, 317 (2), 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marblestone JG, Comparison of SUMO fusion technology with traditional gene fusion systems: Enhanced expression and solubility with SUMO. Protein Sci. 2006, 15 (1), 182–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer LAM; Meier MA; Dames SA, A fast and simple method for probing the interaction of peptides and proteins with lipids and membrane-mimetics using GB1 fusion proteins and NMR spectroscopy. Protein Sci. 2012, 21 (10), 1566–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waugh DS, Crystal structures of MBP fusion proteins. Protein Sci. 2016, 25 (3), 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee DK; Esposito D, Enhanced soluble protein expression using two new fusion tags. Protein Expression Purif. 2006, 46 (1), 122–129. [DOI] [PubMed] [Google Scholar]
- 12.Dattananda C, Fusion tags for protein expression and purification: Fusion tags can improve the yield and solubility of many recombinant proteins. Of course, no single tag or cleavage method will answer every need. BioPharm Int. 2008, (June, Suppl.). [Google Scholar]
- 13.McCoy J, Expression and purification of thioredoxin fusion proteins. Curr Protoc Protein Sci 2001, Chapter 6. [DOI] [PubMed] [Google Scholar]
- 14.Yadav DK; Yadav N; Yadav S; Haque S; Tuteja N, An insight into fusion technology aiding efficient recombinant protein production for functional proteomics. Arch. Biochem. Biophys 2016, 612, 57–77. [DOI] [PubMed] [Google Scholar]
- 15.Bernier SC; Cantin L; Salesse C, Systematic analysis of the expression, solubility and purification of a passenger protein in fusion with different tags. Protein Expression Purif. 2018, 152, 92–106. [DOI] [PubMed] [Google Scholar]
- 16.Niiranen L; Espelid S; Karlsen CR; Mustonen M; Paulsen SM; Heikinheimo P; Willassen NP, Comparative expression study to increase the solubility of cold adapted Vibrio proteins in Escherichia coli. Protein Expression Purif. 2007, 52 (1), 210–218. [DOI] [PubMed] [Google Scholar]
- 17.Deng X; Su R; Feng X; Wei M; Chen J, Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev 2018, 48, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X; Su R; Weng H; Huang H; Li Z; Chen J, RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018, 28 (5), 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esteller M; Pandolfi PP, The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov 2017, 7 (4), 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson DG, Chapter fifteen - Enzymatic Assembly of Overlapping DNA Fragments In Methods in Enzymology, Voigt C, Ed. Academic Press: 2011; Vol. 498, pp 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou S; Toh JD; Wong KH; Gao YG; Hong W; Woon EC, N(6)-Methyladenosine: a conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep 2016, 6, 25677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T; Hong T; Huang Y; Su H; Wu F; Chen Y; Wei L; Huang W; Hua X; Xia Y; Xu J; Gan J; Yuan B; Feng Y; Zhang X; Yang CG; Zhou X, Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein. Journal of the American Chemical Society 2015, 137 (43), 13736–9. [DOI] [PubMed] [Google Scholar]
- 23.Feng C; Liu Y; Wang G; Deng Z; Zhang Q; Wu W; Tong Y; Cheng C; Chen Z, Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem 2014, 289 (17), 11571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu C; Liu K; Tempel W; Demetriades M; Aik W; Schofield CJ; Min J, Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6-methyladenosine RNA demethylation. J Biol Chem 2014, 289 (25), 17299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aik W; Demetriades M; Hamdan MK; Bagg EA; Yeoh KK; Lejeune C; Zhang Z; McDonough MA; Schofield CJ, Structural basis for inhibition of the fat mass and obesity associated protein (FTO). J Med Chem 2013, 56 (9), 3680–8. [DOI] [PubMed] [Google Scholar]
- 26.Han Z; Niu T; Chang J; Lei X; Zhao M; Wang Q; Cheng W; Wang J; Feng Y; Chai J, Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature 2010, 464 (7292), 1205–9. [DOI] [PubMed] [Google Scholar]
- 27.Aik W; Scotti JS; Choi H; Gong L; Demetriades M; Schofield CJ; McDonough MA, Structure of human RNA N(6)-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res 2014, 42 (7), 4741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y; Yan J; Li Q; Li J; Gong S; Zhou H; Gan J; Jiang H; Jia GF; Luo C; Yang CG, Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res 2015, 43 (1), 373–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X; Wei LH; Wang Y; Xiao Y; Liu J; Zhang W; Yan N; Amu G; Tang X; Zhang L; Jia G, Structural insights into FTO’s catalytic mechanism forthe demethylation of multiple RNA substrates. Proc Natl Acad Sci U S A 2019, 116 (8), 2919–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa S; Almeida A; Castro A; Domingues L, Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: the novel Fh8 system. Front Microbiol 2014, 5, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.di Guan C; Li P; Riggs PD; Inouye H, Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene 1988, 67 (1), 21–30. [DOI] [PubMed] [Google Scholar]
- 32.Pryor KD; Leiting B, High-level expression of soluble protein in Escherichia coli using a His6-tag and maltose-binding-protein double-affinity fusion system. Protein Expr Purif 1997, 10 (3), 309–19. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen TT; Ghirlando R; Venditti V, The oligomerization state of bacterial enzyme I (EI) determines EI’s allosteric stimulation or competitive inhibition by alpha-ketoglutarate. J Biol Chem 2018, 293, 2631–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venditti V; Fawzi NL; Clore GM, Automated sequence- and stereo-specific assignment of methyl-labeled proteins by paramagnetic relaxation and methyl-methyl nuclear Overhauser enhancement spectroscopy. J Biomol NMR 2011, 51 (3), 319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Venditti V; Schwieters CD; Grishaev A; Clore GM, Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering. Proc Natl Acad Sci U S A 2015, 112 (37), 11565–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venditti V; Tugarinov V; Schwieters CD; Grishaev A; Clore GM, Large interdomain rearrangement triggered by suppression of micro- to millisecond dynamics in bacterial Enzyme I. Nat Commun 2015, 6, 5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venditti V; Fawzi NL; Clore GM, An efficient protocol for incorporation of an unnatural amino acid in perdeuterated recombinant proteins using glucose-based media. J Biomol NMR 2012, 52 (3), 191–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dotas RR; Venditti V, (1)H, (15)N, (13)C backbone resonance assignment of the C-terminal domain of enzyme I from Thermoanaerobacter tengcongensis. Biomol NMR Assign 2017. [DOI] [PubMed] [Google Scholar]
- 39.Dotas RR; Venditti V, Resonance assignment of the 128 kDa enzyme I dimer from Thermoanaerobacter tengcongensis. Biomol NMR Assign 2019. [DOI] [PubMed] [Google Scholar]
- 40.Evangelidis T; Nerli S; Novacek J; Brereton AE; Karplus PA; Dotas RR; Venditti V; Sgourakis NG; Tripsianes K, Automated NMR resonance assignments and structure determination using a minimal set of 4D spectra. Nat Commun 2018, 9 (1), 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venditti V; Clore GM, Conformational selection and substrate binding regulate the monomer/dimer equilibrium of the C-terminal domain of Escherichia coli enzyme I. J Biol Chem 2012, 287 (32), 26989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venditti V; Ghirlando R; Clore GM, Structural basis for enzyme I inhibition by alpha-ketoglutarate. ACS Chem Biol 2013, 8 (6), 1232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graceffa P; Jancsó A; Mabuchi K, Modification of acidic residues normalizes sodium dodecyl sulfate-polyacrylamide gel electrophoresis of caldesmon and other proteins that migrate anomalously. Archives of Biochemistry and Biophysics 1992, 297 (1), 46–51. [DOI] [PubMed] [Google Scholar]
- 44.Alves VS; Pimenta DC; Sattlegger E; Castilho BA, Biophysical characterization of Gir2, a highly acidic protein of Saccharomyces cerevisiae with anomalous electrophoretic behavior. Biochemical and Biophysical Research Communications 2004, 314 (1), 229–234. [DOI] [PubMed] [Google Scholar]
- 45.Jenny RJ; Mann KG; Lundblad RL, A critical review of the methods for cleavage of fusion proteins with thrombin and factor Xa. Protein Expr Purif 2003, 31 (1), 1–11. [DOI] [PubMed] [Google Scholar]
- 46.Schneider CA; Rasband WS; Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012, 9 (7), 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.