Abstract
It's widely appreciated that liquid–liquid phase separation (LLPS) underlies the formation of membraneless organelles, which function to concentrate proteins and nucleic acids. In the past few decades, major efforts have been devoted to identify the phase separation associated proteins and elucidate their functions. To better utilize the knowledge dispersed in published literature, we developed PhaSepDB (http://db.phasep.pro/), a manually curated database of phase separation associated proteins. Currently, PhaSepDB includes 2914 non-redundant proteins localized in different organelles curated from published literature and database. PhaSepDB provides protein summary, publication reference and sequence features of phase separation associated proteins. The sequence features which reflect the LLPS behavior are also available for other human protein candidates. The online database provides a convenient interface for the research community to easily browse, search and download phase separation associated proteins. As a centralized resource, we believe PhaSepDB will facilitate the future study of phase separation.
INTRODUCTION
The densely packed eukaryotic cellular interior is comprised of numerous compartments or organelles. These compartments enable spatiotemporal control over biological materials, metabolic reactions and signaling pathways. Classic compartments bounded by surrounding lipid bilayer membranes include the endoplasmic reticulum, Golgi apparatus, mitochondrion etc. However, many compartments lacking membranes are widely distributed in essentially all eukaryotic cells. They are micron-scale supramolecular assemblies of proteins, nucleic acids and other molecular components. These include the nucleolus, Cajal body, PML bodies in the nucleus, and stress granules (SGs), processing bodies (P-bodies) in the cytoplasm (1). Recent studies show that these membraneless compartments exhibit similar liquid-like characteristics despite their distinct composition, location and functions (2–5). It's widely appreciated that membraneless compartments form through the liquid–liquid phase separation (LLPS) process, a phenomenon in which a supersaturated solution of components separates into stably coexisting dense phase and dilute phase (6).
Membraneless compartments are often enriched with multivalent molecules that can form networks of interactions to promote phase separation (7). Such molecules include proteins with linear repeats of modular interaction domains. For example, in the T cell receptor signal transduction system, pLAT2-Grb2 forms condensates via multiple SH3-PRM interactions (8). Proteins containing a large fraction of intrinsically disordered regions (IDRs), which lack defined 3D structure but often harbor repetitive sequences and low-complexity sequence regions (LCRs), represent another way to mediate multivalent interactions. Many IDR-containing proteins can undergo phase separation in vitro and localize in specific membraneless compartments in vivo, such as pgl-1, pgl-3 and laf-1 in P granules (4) and Nsp1 in nuclear pore complex (9). Especially, known RNA-binding proteins like FUS, hnRNPA1, hnRNPA2, TDP-43 contain prion-like domains (PrLDs), one of the most common types of LCRs, are frequently involved in phase separation (10–12). The combinations amongst these multivalent interactions between IDRs, modular domains and linear motifs regulate the formation and heterogeneity of assemblies, leading to structural and dynamic continuum from liquid to gel and solid state (6,13). Such heterogeneity could be a general principle underlying protein phase transitions. As a known example, various concentration of protein FUS can span the entire range of material state in vitro (14). In fact, some membraneless organelles harbor complex topologies with various states of subcompartments. For example, SGs was shown to contain non-liquid cores, suggesting a complex internal organization, which may be true for other membraneless organelles (15,16).
Membraneless compartments driven via LLPS and associated proteins are being identified rapidly, providing new insights into various cellular processes, as well as disease generation. For instance, HP1α mediates heterochromatin domain formation and maturation via phase separation (17,18). Sabari et al. suggest coactivators BRD4 and MED1 form phase separation droplets at super-enhancers and regulate gene expression (19). The aberrant liquid-solid phase transition of FUS in vivo reveals that aging diseases resulting from amyloidosis like amyotrophic lateral sclerosis (ALS) may arise from disability to maintain liquid phase homeostasis (14).
Despite growing interest in the area, there is a lack of centralized resources gathering related research from the dispersed literature. Catarina et al. (20) constructed a database of stress granule, but it doesn’t include other membraneless organelles. Hence, we have built PhaSepDB, a novel database that aims to provide a comprehensive map of LLPS associated proteins. A total of 2914 non-redundant LLPS related proteins located in different membraneless organelles were included in PhaSepDB after two rounds of manual curation from publications and public database. Also, sequence feature analysis of these proteins and all other human proteins is available. Our PhaSepDB database provides an convenient interface that helps users to browse, retrieve and download LLPS related proteins.
DATA COLLECTION AND PROCESSING
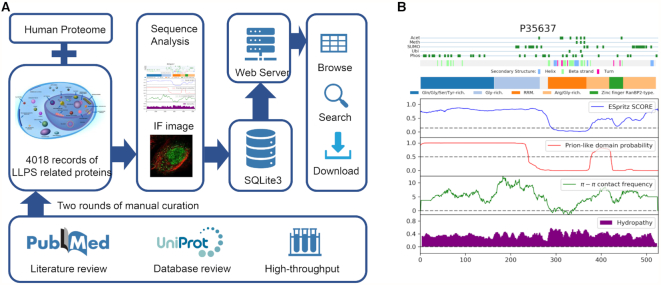
PhaSepDB is constructed based on the curated information derived from published literature after two rounds of manual verification (Figure 1A). The data sources can be classified into three groups: (i) Reviewed. Using keywords ‘((phase transition[Title/Abstract] OR phase separation[Title/Abstract] OR membraneless organelles[Title/Abstract] OR biomolecular condensates[Title/Abstract]) AND protein) AND cell’, 2763 publications from 1 January 2000 to 30 June 2019 were extracted from NCBI PubMed. We then manually rechecked the abstracts to obtain relevant publications that describe membraneless organelles and related proteins. By examining the full text of these publications, we extracted information of phase separation associated proteins that were verified by experiments, including reconstituting an LLPS assembly of purified components in vitro, fluorescence recovery after photobleaching (FRAP) assay in vitro or in cell, droplets formation examined by immunofluorescence with fluorescent marker in cell. (ii) UniProt reviewed. Some known membraneless organelles such as P-body have been investigated long before the emergence of the concept that LLPS underlies membraneless organelle formation. The UniProt protein knowledgebase (21) includes subcellular annotations of proteins localized in several membraneless compartments (a): PML body, nuclear speckle, Cajal body, GEM, nucleolus, nuclear pore complex, SGs, P-body and postsynaptic density. For these proteins, we collected the annotations with supporting literature and re-examine the original articles based on the criteria the same as the Reviewed part. Generally, the membraneless organelles localization of these proteins was mostly verified by in cell immunofluorescence experiments in which the protein showed colocalization with known markers of corresponding membraneless organelles or specific droplets patterns. It should be noted that the localization into specific membraneless organelle can be considered as LLPS related rather than direct LLPS behavior, which need more examination (7). (b) High-throughput. Phase separation associated compartments or proteins can be identified with a high-throughput method. We collected the research that utilized high throughput experiments during the literature search of Reviewed data. Then we further summarized different experiments into different types namely: (a) Organelle purification. The membraneless organelles can be isolated of purified and the components can be identified by coupled mass spectrometry (MS). For example, Hubstenberger et al. purified P-bodies using a fluorescence-activated particle sorting (FAPS) method and conduct MS. In this approach, they identified 125 proteins as significantly enriched in P-bodies (22); (b) Proximity labeling. In this approach, the proteins close to the marker of membranless organelles are labelled by chemical reactions and further identified. Markmiller et al. used ascorbate peroxidase (APEX) proximity labeling MS to identify SGs components (23); (c) Immunofluorescence image-based screen. The components of LLPS compartments display as droplets with higher fluorescence intensity compared to the background. LLPS related proteins can be identified via IF-image based method in a high throughput way. A systematic study targeted more than 1300 genes with siRNAs and imaged thousands of single cells to screen for regulatory genes that perturbed membraneless organelles (24); (d) Affinity purification. The proteins in membraneless organelles can be identified via the affinity with the known markers of the organelle. DDX6 as a unique marker of P-body, was utilized in tandem-affinity purification coupled to MS to identified DDX6-related P-body components (25).
Figure 1.
Overview of phase associated proteins collection, annotation and PhaSepDB features. (A) The workflow of the construction of PhaSepDB database (B) An example of easily interpreted per-residue plot.
During the curation process, we sought to collect as much useful information as possible, that is, original supporting sentences, organelle location, mutation/disease, verifying experiments, regulatory role, protein material state and corresponding cell lines or tissues. For example, Dao et al. show that UBQLN2, an ALS associated RNA-binding protein, colocalizes with SGs under stress conditions in vivo (in U2OS cells), and undergoes LLPS in vitro, forming liquid droplets. Using biophysical techniques including NMR, they demonstrate that the STI1-II, Pxx and UBA domains are obligatory to its LLPS behavior (26). Thus, organelle location, disease, liquid material state and cell line information were recorded in our data. It should be noted that the material state of the proteins can be dynamic according the different experiment conditions.
The molecular properties of proteins are essential to understand their potential phase behavior. The patterning of IDRs, interaction domains and sequences connecting structured domains fine-tune the protein's phase behavior. Disorder prediction tools such as PONDR series (27–30) and ESpritz (31) are often used to analyze disorder regions. Tools like PLAAC (32) were developed for the identification of proteins with prion-like amino acid composition. In addition, in the studies of natively unfolded or disorder proteins (33,34), the fraction and distribution of hydrophobic and charged residues were proposed as a unique feature. They are typical determinants of physicochemical and conformational properties, which in turn affect electrostatic interactions (7). Hence, for each protein, we integrated results of IDR prediction by ESpritz, prion-like sequence prediction by PLAAC, electrostatic interaction prediction Pi–Pi (35) as well as charged/hydrophobic residue distribution analysis by CIDER(36) into an easily interpreted per-residue plot. The plot also contains four types of post-translational modifications (PTMs) (37), secondary structure annotations, domain and compositional bias annotations (Figure 1B). The detail program parameters and annotation resources used were listed in Supplementary Table S1. Furthermore, the components of LLPS compartments display as droplets with higher fluorescence intensity compared to the background, which allows distinguishing phase-separated proteins. We therefore attach IF images available in the Human Protein Atlas (38).
However, despite the rapid growth of phase separation associated proteins, there are still a large amount of potential LLPS proteins waiting to be discovered. Therefore, for all human proteins (UniProt proteome UP000005640 release 2019_06) (21), we also provide the researchers with molecular analysis mentioned above to facilitate potential LLPS protein identification.
DATABASE CONTENT AND THE WEB INTERFACE
Database content
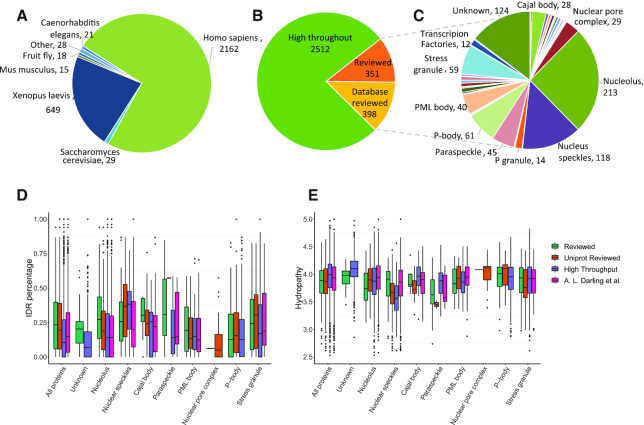
As of June 2019, PhaSepDB includes 4020 curated records from 635 publications, in total 2914 non-redundant LLPS related proteins of several organisms (Figure 2A). The reviewed and UniProt reviewed data accounts for approximately one fourth of the collection, including more than 30 different membraneless organelles, represented by nucleolus, nucleus speckles, P-bodies etc. (Figure 2B and C).
Figure 2.
PhaSepDB content. (A–C) Data statistics of PhaSepDB. (A) Organism distribution; (B) data source distribution and (C) membraneless organelles localization distribution in reviewed data. (D and E) Molecular properties of proteins in eight membraneless organelle. ‘Unknown’ refers to proteins that were not associated with any known organelles. ‘All proteins’ refers to all proteins analyzed in the plot. (D) IDR percentage was extracted from MobiDB database (40) and hydropathy (E) were calculated using CIDER (36).
As we mentioned above, the phase behavior is determined by the molecular properties to some extent. To see if proteins localized in different membraneless organelle harbor different molecular properties, we analysed human proteins in eight membraneless organelles that contain more than 20 proteins (in reviewed and UniProt reviewed data) as well as proteins that were not associated with any known organelles (Figure 2D and E). In addition, a systematic review discussed the prevalence of intrinsic disorder in proteins associated with membrane-less organelles and its functional role, using a list of more than 4000 human proteins mostly retrieved from QuickGO tool (https://www.ebi.ac.uk/QuickGO) as well as literature search (39). We also compared the list with the proteins curated in PhaSepDB (Supplementary Figure S1). The results suggested that proteins in reviewed data contained more IDRs content than those curated from other sources. There were more proteins that verified to be able to form LLPS droplets in vitro in the reviewed data, suggesting their potential to possess more IDRs (Figure 2D). Another possible reason is that in individual LLPS study, proteins with more IDRs are preferably chosen by researchers to carry on further study, causing the observed bias. Furthermore, compared with proteins with unknown organelle localization and average level, proteins in nucleus speckles contain a relative high proportion of IDRs, which was in consistent with previous analysis (39). In contrast, proteins in P-body and PML body contain a relative low proportion of IDRs. In terms of hydropathy, small difference between data sources and membraneless organelles were observed (Figure 2E).
Further work
The principle future aim is to include more LLPS related proteins as the field is developing rapidly. Another aim is to add more useful annotations that may help biological study. For example, some specific residue mutations were responsible for the protein's material state change, such as the disease mutation of FUS accelerated its liquid-to-solid transition (14). We noted such information in the curated data for some proteins. More annotations will be added and disease related datasets can be integrated in the future.
Web interface
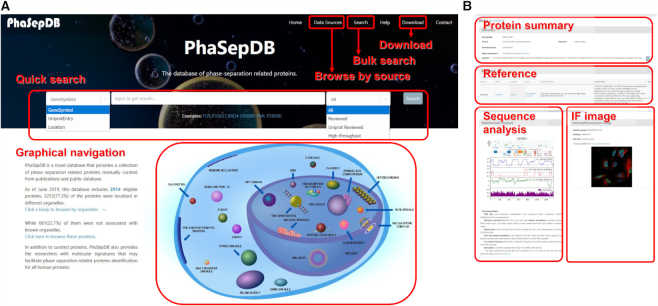
A freely available and fully functional website has been developed to access the collection (http://db.phasep.pro/). The website interface comprises six sections: ‘Home’, ‘Data Sources’, ‘Search’, ‘Help’, ‘Download’, ‘Contact’. From the ‘Home’ page, advanced search by gene symbol, UniProt ID or subcellular location is provided, together with data source drag-down options. A user-friendly graphical navigation is provided to browse through LLPS proteins based on specific membraneless body location upon click (Figure 3A). The data sources are described in detail in the ‘Data sources’ page, in which users can click to browse the proteins from the specific data sources. The query results are presented as a responsible table, which contains the basic information of proteins, including UniProt ID, gene symbol, supporting publication, IDR percentage, subcellular/membraneless body location and curation source. The ID or symbol can be clicked to navigate to the protein detail page, which includes a protein summary, references and protein properties (Figure 3B). The publication from which the protein is extracted is shown together with, when available, related original sentences, the research's experiment and used cell lines. The sequence analysis results and protein IF image are presented below the reference section. PhaSepDB provides protein summary and properties sections for not only curated proteins but also all other human proteins without supporting evidence. A detailed tutorial for the usage of the database can be found on the ‘Help’ page.
Figure 3.
An illustration of PhaSepDB. (A) Screenshot of the main page. A user can query the database by gene symbol, UniProt entry or cellular location via the quick search panel. The graphical navigation provides a membraneless organelle view of the data. A user can mouseover the specific body to see the number of records and click to browse. (B) The protein detail page. The detail page contains protein summary, reference, sequence analysis and IF image.
Database implementation
The data is stored in a SQLite v3.24.0 (https://www.sqlite.org/) database engine. The web framework is constructed based on Django v2.1.3 (https://www.djangoproject.com/) running on CentOS Linux operating system. We have tested it in Google Chrome, Mozilla Firefox, Microsoft Edge and Apple Safari browsers. The PhaSepDB database is freely available to the research community online at http://db.phasep.pro/.
CONCLUSION
The emergence of the concept that LLPS underlies cellular compartmentalization formation has expanded our understanding of cell biology and motivated tremendous work to delineate the function of these membraneless organelles. Systemic identification and study of phase separation related proteins and membraneless organelles are urgently needed. Innovative bioinformatic approaches will speed up the process to thoroughly understand phase separation. The PhaSepDB database is the first comprehensive database of phase separation related proteins. Based on manual curation from >600 publications, it integrates more than 2900 phase separation associated proteins. Moreover, it provides a useful sequence analysis of each LLPS protein as well as candidates. The database and website will be periodically updated according to new research on phase separation. We believe that as a data resource, it will benefit not only the traditional molecular biology research but also the progression of promising computational approaches.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kostas Sidiropoulos (EMBL-EBI) for providing suggestions on the website construction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Program of China [2018YFA0507504, 2017YFA0505503]; National Natural Science Foundation of China [61773025, 31871343]; European Bioinformatics Institute core funding. Funding for open access charge: National Key Research and Development Program of China [2018YFA0507504]; National Natural Science Foundation of China [61773025].
Conflict of interest statement. None declared.
REFERENCES
- 1. Banani S.F., Lee H.O., Hyman A.A., Rosen M.K.. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017; 18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deryusheva S., Gall J.G.. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:4810–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li L., Roy K., Katyal S., Sun X., Bléoo S., Godbout R.. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol. Biol. Cell. 2006; 17:1126–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Julicher F., Hyman A.A., Jülicher F., Hyman A.A.. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009; 324:1729–1732. [DOI] [PubMed] [Google Scholar]
- 5. Brangwynne C.P., Mitchison T.J., Hyman A.A.. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:4334–4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L. et al.. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018; 28:420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alberti S., Gladfelter A., Mittag T.. Considerations and challenges in studying liquid–liquid phase separation and biomolecular condensates. Cell. 2019; 176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D.. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016; 352:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frey S., Richter R.P., Görlich D.. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science. 2006; 314:815–817. [DOI] [PubMed] [Google Scholar]
- 10. Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. et al.. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012; 149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin Y., Protter D.S.W., Rosen M.K., Parker R.. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryan V.H., Dignon G.L., Zerze G.H., Chabata C.V., Silva R., Conicella A.E., Amaya J., Burke K.A., Mittal J., Fawzi N.L.. Mechanistic view of hnRNPA2 low-complexity domain structure, interactions, and phase separation altered by mutation and arginine methylation. Mol. Cell. 2018; 69:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu H., Fuxreiter M.. The structure and dynamics of higher-order assemblies: amyloids, signalosomes, and granules. Cell. 2016; 165:1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M. et al.. A Liquid-to-Solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015; 162:1066–1077. [DOI] [PubMed] [Google Scholar]
- 15. Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., Parker R.. ATPase-Modulated stress granules contain a diverse proteome and substructure. Cell. 2016; 164:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheeler J.R., Matheny T., Jain S., Abrisch R., Parker R.. Distinct stages in stress granule assembly and disassembly. Elife. 2016; 5:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larson A.G., Elnatan D., Keenen M.M., Trnka M.J., Johnston J.B., Burlingame A.L., Agard D.A., Redding S., Narlikar G.J.. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017; 547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strom A.R., Emelyanov A.V., Mir M., Fyodorov D.V., Darzacq X., Karpen G.H.. Phase separation drives heterochromatin domain formation. Nature. 2017; 547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C. et al.. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018; 361:eaar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nunes C., Mestre I., Marcelo A., Koppenol R., Matos C.A., Nóbrega C.. MSGP: the first database of the protein components of the mammalian stress granules. Database. 2019; 2019:doi:10.1093/database/baz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bateman A., Martin M.J., O’Donovan C., Magrane M., Alpi E., Antunes R., Bely B., Bingley M., Bonilla C., Britto R. et al.. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hubstenberger A., Courel M., Bénard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M. et al.. P-body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017; 68:144–157. [DOI] [PubMed] [Google Scholar]
- 23. Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.C., Krach F., Yang D., Sen A. et al.. Context-Dependent and Disease-Specific diversity in protein interactions within stress granules. Cell. 2018; 172:590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berchtold D., Battich N., Pelkmans L.. A Systems-Level study reveals regulators of Membrane-less organelles in human cells. Mol. Cell. 2018; 72:1035–1049. [DOI] [PubMed] [Google Scholar]
- 25. Ayache J., Bénard M., Ernoult-Lange M., Minshall N., Standart N., Kress M., Weil D.. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell. 2015; 26:2579–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dao T.P., Kolaitis R.-M., Kim H.J., O’Donovan K., Martyniak B., Colicino E., Hehnly H., Taylor J.P., Castañeda C.A., O’Donovan K. et al.. Ubiquitin modulates liquid–liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell. 2018; 69:965–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Romero P., Obradovic Z., Li X., Garner E.C., Brown C.J., Dunker A.K.. Sequence complexity of disordered protein. Proteins. 2001; 42:38–48. [DOI] [PubMed] [Google Scholar]
- 28. Peng K., Vucetic S., Radivojac P., Brown C.J., Dunker A.K., Obradovic Z.. Optimizing long intrinsic disorder predictors with protein evolutionary information. J. Bioinform. Comput. Biol. 2005; 3:35–60. [DOI] [PubMed] [Google Scholar]
- 29. Peng K., Radivojac P., Vucetic S., Dunker A.K., Obradovic Z.. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006; 7:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N.. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010; 1804:996–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walsh I., Martin A.J.M., Di Domenico T., Tosatto S.C.E.. ESpritz: accurate and fast prediction of protein disorder. Bioinformatics. 2012; 28:503–509. [DOI] [PubMed] [Google Scholar]
- 32. Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D.. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014; 30:2501–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uversky V.N., Gillespie J.R., Fink A.L.. Why are ‘natively unfolded’ proteins unstructured under physiologic conditions. Proteins. 2000; 41:415–427. [DOI] [PubMed] [Google Scholar]
- 34. Oldfield C.J., Cheng Y., Cortese M.S., Brown C.J., Uversky V.N., Dunker A.K.. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005; 44:1989–2000. [DOI] [PubMed] [Google Scholar]
- 35. Vernon R.M., Chong P.A., Bah A., Lin H., Farber P., Tsang B., Kim T.H., Forman-Kay J.D.. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. Elife. 2018; 7:1–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Holehouse A.S., Das R.K., Ahad J.N., Richardson M.O.G., Pappu R.V.. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys. J. 2017; 112:16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hornbeck P.V, Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E.. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015; 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M. et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 39. Darling A.L., Liu Y., Oldfield C.J., Uversky V.N.. Intrinsically disordered proteome of human membrane-less organelles. Proteomics. 2018; 18:1–12. [DOI] [PubMed] [Google Scholar]
- 40. Piovesan D., Tabaro F., Paladin L., Necci M., Micetic I., Camilloni C., Davey N., Dosztányi Z., Mészáros B., Monzon A.M. et al.. MobiDB 3.0: more annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res. 2018; 46:D471–D476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.