Abstract
gutMDisorder (http://bio-annotation.cn/gutMDisorder), a manually curated database, aims at providing a comprehensive resource of dysbiosis of the gut microbiota in disorders and interventions. Alterations in the composition of the gut microbial community play crucial roles in the development of chronic disorders. And the beneficial effects of drugs, foods and other intervention measures on disorders could be microbially mediated. The current version of gutMDisorder documents 2263 curated associations between 579 gut microbes and 123 disorders or 77 intervention measures in Human, and 930 curated associations between 273 gut microbes and 33 disorders or 151 intervention measures in Mouse. Each entry in the gutMDisorder contains detailed information on an association, including an intestinal microbe, a disorder name, intervention measures, experimental technology and platform, characteristic of samples, web sites for downloading the sequencing data, a brief description of the association, a literature reference, and so on. gutMDisorder provides a user-friendly interface to browse, retrieve each entry using gut microbes, disorders, and intervention measures. It also offers pages for downloading all the entries and submitting new experimentally validated associations.
INTRODUCTION
Human gut microbiota is the microbe population involving bacteria, archaea and eukarya inhabiting in our intestine, and has co-evolved with the host to live together (1,2). According to the latest estimation, the number of bacteria cells in colon reaches 4 × 1013, which is approximately equal to the number of Human cells (3). Evidence shows that gut microbiota plays a vital role in pathogen resistance, host immunity and metabolism (4–6).
With the advances of sequencing technologies and the importance of gut microbiota for health (7), it leads to a significant increase in the number of assembled genomes. Current resources mainly focus on restoring and managing microbial genomes, and thus to conduct comparative analysis. To provide gold-standard genomes, RefSeq microbial genomes database (8) manually collects the primary sequence records of the International Nucleotide Sequence Database public archives (INSDC). To the consistency of the organism, NCBI Taxonomy database (9) provides a standard nomenclature and taxonomic classification repository for INSDC. In addition, MBGD, IMG and MicrobesOnline (10–12) are frequently used tools and platforms using comparative analysis of microbial genome for exploring genome diversity in microbiota.
Over the past several years, accumulating evidence indicates that alterations in the composition of the gut microbial community, known as dysbiosis, could be closely related to the development of chronic disorders. In 2011, Koren et al. used 16S rRNA sequencing technology to investigate the diversity of gut microbiota in patients with atherosclerosis (13). As a result, they identified five bacterial taxa (Firmicutes, Proteobacteria, Lachnospiraceae, Erysipelotrichaceae and Pseudomonas luteola) in the gut that are correlated with atherosclerosis, which could be potential disease markers. Subsequently, hundreds of disorders that are associated with dysbiosis of gut microbiota have been experimentally validated, such as type 2 diabetes mellitus (T2DM), primary sclerosing cholangitis, acne and so on (14–16). Moreover, current discoveries suggest that the gut microbiota in disorders could be altered by drugs, diets and other intervention measures, which would lead to the beneficial effects on disorders. In 2017, the microbially mediated mechanism of metformin on glucose metabolism was revealed. To explore the therapeutic potential in patients by modulating the gut microbiota, experiments were designed on Mouse with intervention measures (17,18).
Although several online repositories have been developed for storing microbiota genome, few document the function of microbiota, e.g. associations between microbiota and disease (32), and none focus on the function of gut microbiota. Up to now, detailed information on dysbiosis of the gut microbiota in disorders and interventions is still scattered in papers. Thus, we developed a manually curated database entitled ‘gutMDisorder’ for collecting experimentally validated associations between gut microbiota and diseases or intervention measures from papers. The database could be freely available at: http://bio-annotation.cn/gutMDisorder.
DATA COLLECTION AND DATABASE CONTENT
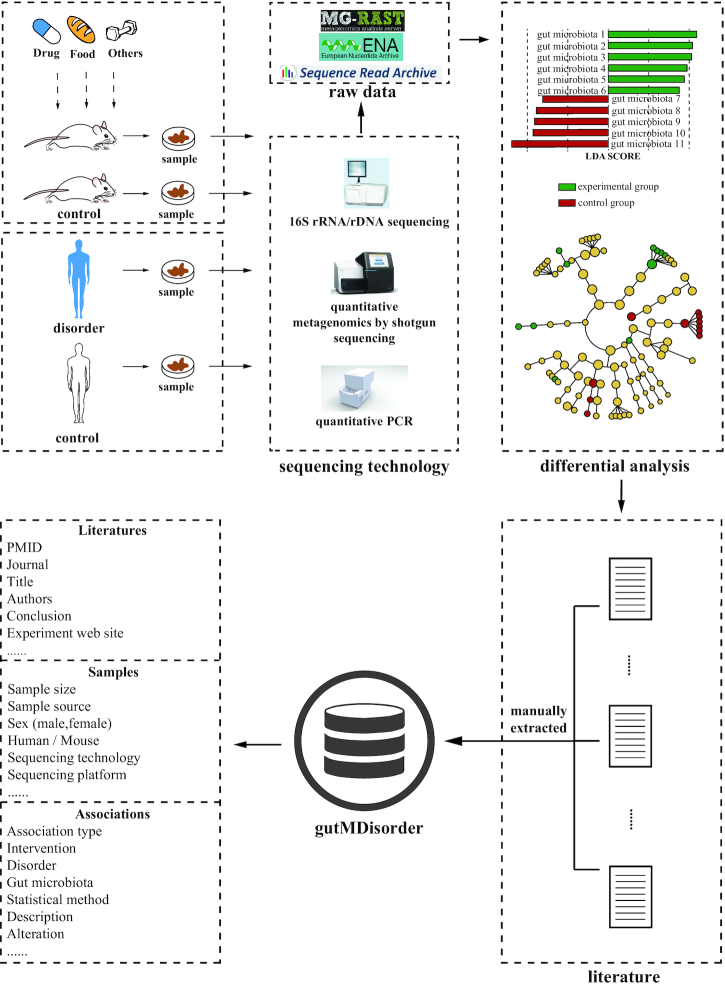
To collect high-quality data, all the associations between gut microbiota and disorders or intervention measures were manually extracted from publications as previous studies (19–22). Here we searched PubMed database with a list of keywords to acquire potentially relevant papers, such as ‘gut’, ‘intestinal’, ‘microbiota’, ‘microbiome’, ‘mice’, ‘rat’, ‘16S’, etc., and selected >1900 papers. Then we checked which papers documented experimentally validatedassociations. Subsequently, differential gut microbiota in disorders and intervention measures were extracted and double-checked from 443 papers (upper part of Figure 1). As a result, 2263 experimentally validated associations between 579 gut microbes and 123 disorders or 77 interventions in Human and 930 associations between 273 gut microbes and 33 disorders or 151 interventions in Mouse were collected in the current version of gutMDisorder (Table 1).
Figure 1.
The process of data collection.
Table 1.
The number of intestinal microbes, disorders, interventions and their associations in Human and Mouse
| Species | No. of intestinal microbes | No. of disorders | No. of intervention measures (drugs, foods, others) | No. of associations |
|---|---|---|---|---|
| Human | 579 | 123 | 77 (46, 15, 16) | 2263 |
| Mouse | 273 | 33 | 151 (66, 52, 33) | 930 |
Each entry in the gutMDisorder contains three sections for documenting an association (lower part of Figure 1). ‘Association’ section documents an association type, an intestinal microbe, a disorder, intervention measures (drug, food, or others), a brief description of the alteration in the intestinal microbe under the disorder or intervention. The gutMDisorder mainly documents two types of associations, ‘gut microbiota associated with disorder’ and ‘interventions change the composition of gut microbiota’. The gut microbe, disorder and drug terminologies were organized based on NCBI taxonomy database (9), Disease Ontology (DO) (23) and DrugBank (24), respectively. Such an organization provides not only a substantial advantage in terms of search but also further analysis as previously described (22). ‘Sample’ section documents species (Human or Mouse), sample size and source, sex, age, BMI, sequencing technology and platform and so on. The sequencing technology includes 16S rRNA/rDNA, quantitative metagenomics by shotgun sequencing, qPCR, RT-qPCR, and so on. ‘Literature’ section documents detailed description of the literature reference. Especially, it contains web sites of sequencing data, which can help researchers to obtain the original sequence data for further analysis.
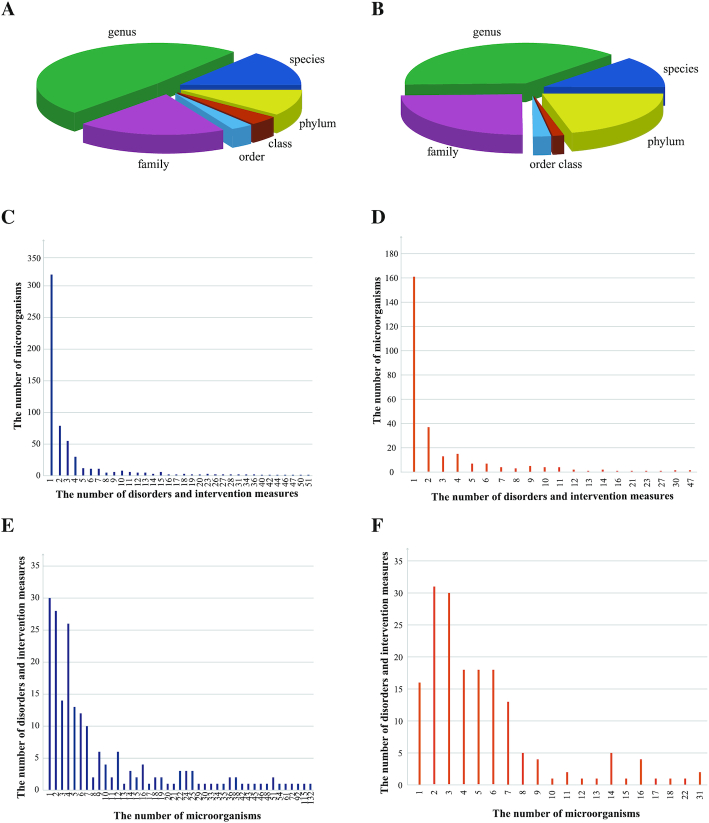
Figure 2A shows the pie chart of the distribution of gut microbiota in Human among different classifications of NCBI Taxonomy database (9). Current version of gutMDisorder documents microbes across phylum, class, order, family, genus and species levels. 279 (48.19%) microbes are at genus level, which make up almost half of our database. As in Human, a great many of the microbes (40.29%, 110/273) in Mouse are at genus level, which is demonstrated in Figure 2B.
Figure 2.
The distribution of gut microbes, disorders, and intervention measures in gutMDisorder. (A) The distribution of gut microbes in Human among different taxa. (B) The distribution of gut microbes in Mouse among different taxa. (C) Histogram of the number of microbes in Human associated with individual disease or intervention measure. (D) Histogram of the number of microbes in Mouse associated with individual disease or intervention measure. (E) Histogram of the number of diseases and intervention measures associated with individual microbe in Human. (F) Histogram of the number of diseases and intervention measures associated with individual microbe in Mouse.
Figure 2C and D shows the histogram of the number of diseases and intervention measures associated with individual microbe in Human and Mouse, respectively. The majority of Human (54.75%, 317/579) and Mouse (58.97%, 161/273) microbes are associated with only one disease and intervention measure, which could be a potential marker. The richness of bifidobacterium (at genus level) is the most likely to be altered in Human, since it is associated with the most number (51) of diseases and interventions.
Figure 2E and F demonstrates the histogram of the number of microbes associated with individual disease or intervention measure in Human and Mouse, respectively. Thirty diseases and intervention measures are associated with only one microbe in Human. While most of diseases and intervention measures (143) are associated with three or more microbes. The dysbiosis of gut microbiota could be very important in the development of these diseases.
As our previous resources (20,25,26), all the data of gutMDisorder were stored and managed in a cloud server called ucloud (27). It can be freely available at http://bio-annotation.cn/gutMDisorder.
USER INTERFACE
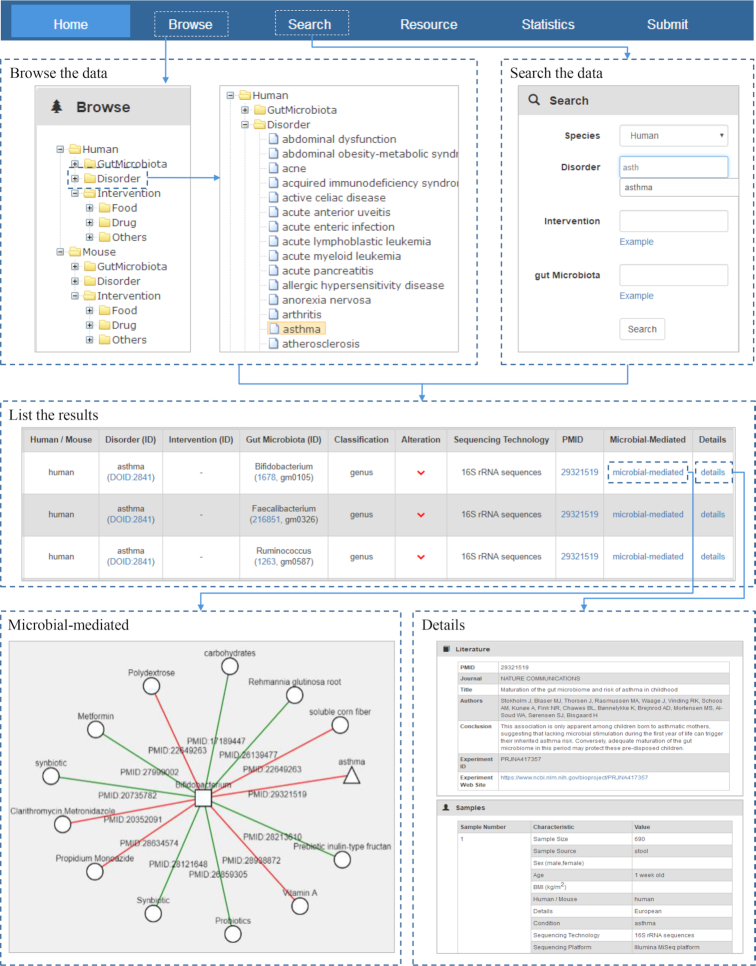
gutMDisorder provides a tree browser and a search engine to query detailed information about associations between gut microbiota and diseases or interventions. Figure 3 shows the schematic workflow.
Figure 3.
Schematic workflow of gutMDisorder.
The tree browser organizes the data according to species and uses ‘Human’ and ‘Mouse’ as root categories. Each of the species involves three sub-categories named ‘GutMicrobiota’, ‘Disorder’ and ‘Intervention’. By clicking ‘GutMicrobiota’ or ‘Disorder’ category, all the names of intestinal microbes or disorders belonged to the corresponding species would be listed as leaf nodes. The ‘Intervention’ category contains ‘Drug’, ‘Food’ and ‘Others’ sub-categories, which could be expanded to specific intervention measures. Figure 3 shows a partial list of Human disorders. After selecting a disorder ‘asthma’, all the associations between asthma and gut microbes would be retrieved and shown in a table, where an association with a brief introduction was represented into one row. The identifier of microbe, disorder, drug and PMID could be linked to NCBI taxonomy database, DO, DrugBank, and PubMed for detailed description of these entities. In the result table, clicking the ‘detail’ link of a row would lead to the detailed information about an association. For an association between a disease and a microbe in a row, clicking the ‘microbial-mediated’ link, the disease, and the microbe incorporated with the interventions that could alter it would be shown in a network.
The search engine offers a way to query associations by inputting a term of microbe, disorder, and intervention measure. For ease of use, these inputting terms could be auto-completed by selecting a species. After submitting the input items, entries in the database that match with these items exactly will be returned and shown in a table as above.
gutMDisorder provides a ‘Submit’ page for researchers to submit a traceable introduction about important associations that are not documented in the database. Once approved by the reviewer committee, the associations with detailed information will be included in the update version. In addition, a ‘Resource’ page was also offered for downloading all the data.
FUTURE DEVELOPMENT
To make the collection process more systematic and data content more complete, we plan to adopt following three strategies. First, text-mining tools would be used to prescreen papers in MEDLINE Titles and Abstracts. Second, new associations submitted in web page would be included after reviewing. Third, gutMDisorder will be updated quarterly for adding the latest discoveries. With the development of high-throughput technologies, more characterize of gut microbiota would be revealed. This could lead to the rapid increase of microbial resources. Hence, it would be very valuable to give a convenient way to incorporate gutMDisorder to other potential related resources for annotating the function of gut microbiota. To this end, more links through gut microbiota, disorders, and intervention measures to other potential related resources would be provided.
CONCLUSION
gutMDisorder is a comprehensive resource for documenting dysbiosis of the gut microbiota in disorders and interventions, which provides an easy way to search, browse, and download all the experiment-based associations in Human and Mouse. Current version contains 2263 associations between 579 gut microbes and 123 disorders or 77 interventions in Human, and 930 associations between 273 gut microbes and 33 disorders or 151 interventions in Mouse. Since it is the first manually curated resource for annotating the function of gut microbiota, gutMDisorder provides a choice using previous methods and tools about predicting roles of molecules (28–31) in exploring new function of microbiota.
FUNDING
The Tou-Yan Innovation Team Program of the Heilongjiang Province (2019-15); National Natural Science Foundation of China [61871160]; China Postdoctoral Science Foundation [2018T110315, 2016M590291]; Heilongjiang Province Postdoctoral Fund [LBH-TZ20, LBH-Z15179]. Funding for open access charge: The Tou-Yan Innovation Team Program of the Heilongjiang Province (2019-15); National Natural Science Foundation of China [61871160]; China Postdoctoral Science Foundation [2018T110315, 2016M590291]; Heilongjiang Province Postdoctoral Fund [LBH-TZ20, LBH-Z15179].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gregory D., Chaudet H., Lagier J.C., Raoult D.. How mass spectrometric approaches applied to bacterial identification have revolutionized the study of human gut microbiota. Expert Review of Proteomics. 2018; 15:217–229. [DOI] [PubMed] [Google Scholar]
- 2. Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I.. Host-bacterial mutualism in the human intestine. Science. 2005; 307:1915–1920. [DOI] [PubMed] [Google Scholar]
- 3. Sender R., Fuchs S., Milo R.. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016; 14:e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S.. How colonization by microbiota in early life shapes the immune system. Science. 2016; 352:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R.. Diversity, stability and resilience of the human gut microbiota. Nature. 2012; 489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mithieux G. Gut microbiota and host metabolism: what relationship. Neuroendocrinology. 2018; 106:352–356. [DOI] [PubMed] [Google Scholar]
- 7. Roy S., Trinchieri G.. Microbiota: a key orchestrator of cancer therapy. Nature Reviews. Cancer. 2017; 17:271–285. [DOI] [PubMed] [Google Scholar]
- 8. Tatusova T., Ciufo S., Fedorov B., O’Neill K., Tolstoy I.. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014; 42:D553–D559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen I.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., Huntemann M., Varghese N., White J.R., Seshadri R. et al.. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019; 47:D666–D677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dehal P.S., Joachimiak M.P., Price M.N., Bates J.T., Baumohl J.K., Chivian D., Friedland G.D., Huang K.H., Keller K., Novichkov P.S. et al.. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010; 38:D396–D400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uchiyama I., Mihara M., Nishide H., Chiba H.. MBGD update 2013: the microbial genome database for exploring the diversity of microbial world. Nucleic Acids Res. 2013; 41:D631–D635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koren O., Spor A., Felin J., Fak F., Stombaugh J., Tremaroli V., Behre C.J., Knight R., Fagerberg B., Ley R.E. et al.. Human oral, gut, and plaque microbiota in patients with atherosclerosis. PNAS. 2011; 108(Suppl. 1):4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackson M.A., Verdi S., Maxan M.E., Shin C.M., Zierer J., Bowyer R.C.E., Martin T., Williams F.M.K., Menni C., Bell J.T. et al.. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nature Communications. 2018; 9:2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sabino J., Vieira-Silva S., Machiels K., Joossens M., Falony G., Ballet V., Ferrante M., Van Assche G., Van der Merwe S., Vermeire S. et al.. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016; 65:1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sedighi M., Razavi S., Navab-Moghadam F., Khamseh M.E., Alaei-Shahmiri F., Mehrtash A., Amirmozafari N.. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb. Pathog. 2017; 111:362–369. [DOI] [PubMed] [Google Scholar]
- 17. Anhe F.F., Nachbar R.T., Varin T.V., Trottier J., Dudonne S., Le Barz M., Feutry P., Pilon G., Barbier O., Desjardins Y. et al.. Treatment with camu camu (Myrciaria dubia) prevents obesity by altering the gut microbiota and increasing energy expenditure in diet-induced obese mice. Gut. 2018; 68:453–464. [DOI] [PubMed] [Google Scholar]
- 18. Ponziani F.R., Nicoletti A., Gasbarrini A., Pompili M.. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Therapeutic Advances in Medical Oncology. 2019; 11:1758835919848184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D.. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019; 47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., Zhou W., Liu G., Jiang H., Jiang Q.. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019; 47:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang Z., Shi J., Gao Y., Cui C., Zhang S., Li J., Zhou Y., Cui Q.. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019; 47:D1013–D1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Q., Wang Y., Hao Y., Juan L., Teng M., Zhang X., Li M., Wang G., Liu Y.. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009; 37:D98–D104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kibbe W.A., Arze C., Felix V., Mitraka E., Bolton E., Fu G., Mungall C.J., Binder J.X., Malone J., Vasant D. et al.. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015; 43:D1071–D1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng L., Sun J., Xu W., Dong L., Hu Y., Zhou M.. OAHG: an integrated resource for annotating human genes with multi-level ontologies. Sci. Rep. 2016; 6:34820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng L., Yang H., Zhao H., Pei X., Shi H., Sun J., Zhang Y., Wang Z., Zhou M.. MetSigDis: a manually curated resource for the metabolic signatures of diseases. Brief. Bioinform. 2019; 20:203–209. [DOI] [PubMed] [Google Scholar]
- 27. Sqalli M.H., Alsaeedi M., Binbeshr F., Siddiqui M.. UCloud: A simulated Hybrid Cloud for a university environment. IEEE International Conference on Cloud Networking. 2012; 170–172. [Google Scholar]
- 28. Wang D., Wang J., Lu M., Song F., Cui Q.. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010; 26:1644–1650. [DOI] [PubMed] [Google Scholar]
- 29. Chen X., Wang L.Y., Huang L.. NDAMDA: Network distance analysis for MiRNA-disease association prediction. J. Cell Mol. Med. 2018; 22:2884–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng L., Jiang Y., Ju H., Sun J., Peng J., Zhou M., Hu Y.. InfAcrOnt: calculating cross-ontology term similarities using information flow by a random walk. BMC Genomics. 2018; 19:919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zou Q., Li J., Hong Q., Lin Z., Wu Y., Shi H., Ju Y.. Prediction of MicroRNA-Disease Associations Based on Social Network Analysis Methods. BioMed Research International. 2015; 2015:810514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Janssens Y., Nielandt J., Bronselaer A., Debunne N., Verbeke F., Wynendaele E., Van Immerseel F., Vandewynckel Y.P., De Tré G., De Spiegeleer B.. Disbiome database: linking the microbiome to disease. BMC Microbiol. 2018; 18:50. [DOI] [PMC free article] [PubMed] [Google Scholar]