Abstract
DNA cytosine modifications are key epigenetic regulators of cellular processes in mammalian cells, with their misregulation leading to varied disease states. In the human malaria parasite Plasmodium falciparum, a unicellular eukaryotic pathogen, little is known about the predominant cytosine modifications, cytosine methylation (5mC) and hydroxymethylation (5hmC). Here, we report the first identification of a hydroxymethylcytosine-like (5hmC-like) modification in P. falciparum asexual blood stages using a suite of biochemical methods. In contrast to mammalian cells, we report 5hmC-like levels in the P. falciparum genome of 0.2–0.4%, which are significantly higher than the methylated cytosine (mC) levels of 0.01–0.05%. Immunoprecipitation of hydroxymethylated DNA followed by next generation sequencing (hmeDIP-seq) revealed that 5hmC-like modifications are enriched in gene bodies with minimal dynamic changes during asexual development. Moreover, levels of the 5hmC-like base in gene bodies positively correlated to transcript levels, with more than 2000 genes stably marked with this modification throughout asexual development. Our work highlights the existence of a new predominant cytosine DNA modification pathway in P. falciparum and opens up exciting avenues for gene regulation research and the development of antimalarials.
INTRODUCTION
In eukaryotic cells, gene regulation at the epigenetic level is integral to most, if not all, physiological processes. The main epigenetic factors include nuclear architecture, non-coding RNAs, the dynamic deposition and removal of histone modifications, and DNA methylation (1). Of these, DNA cytosine methylation is the most stable epigenetic mark and can be inherited over tens to hundreds of replication cycles and generations (2,3). It occurs at position 5 of the pyrimidine ring of cytosine (5mC) mainly in a CpG dinucleotide context—sequences that are located in CpG islands (occurring at 60% of all gene promoters), in repetitive sequences, or in CpG island shores (4)—and is catalysed by de novo C5-DNA methyltransferases or DNMTs (5). Methylation of promoter CpG islands in a symmetric manner results in transcriptional repression of the corresponding gene due to poor recognition by transcription factors, and the recruitment of proteins involved in chromatin remodelling such as methyl DNA-binding proteins (MBPs), which create a repressive chromatin environment (5). Moreover, deregulation of DNA methylation and establishment of new DNA methylation patterns are associated with the under- or over-expression of select genes, ultimately leading to inflammation, cancer and other diseases (1,3).
In 2009, 5-hydroxymethylcytosine (5hmC), a demethylation intermediate in the active [5mC to C] conversion pathway, garnered interest as an important epigenetic regulator (6,7). 5hmC is generated by the Fe2+- and 2-oxoglutarate-dependent oxidation of 5mC by ten-eleven translocation (TET) dioxygenase enzymes, with depletion of TET proteins resulting in global reduction of 5hmC levels (7,8). Although 5hmC levels are significantly lower than 5mC levels in most cellular systems studied to date, the first clue for its function comes from embyronic stem cells (ESCs) and cells from the central nervous system (CNS), where 5hmC constitutes up to 0.7% of all modified cytosines (6). In ESCs, 5hmC is enriched at gene bodies of actively transcribed genes as well as within extended promoter regions of Polycomb-repressed developmental regulators (9), whereas in differentiated neurons, its presence within gene bodies positively correlates to transcript levels (10–12): this indicates that 5hmC’s role in transcriptional regulation is cell type-, gene- and development stage-specific. Finally, misregulation of 5hmC is observed in neuronal disorders such as Alzheimer's disease, Huntington's disease, schizophrenia, etc. and cancers such as melanoma, pancreatic, haematopoietic malignancies, etc. (3,12–15) Taken together, 5hmC is now considered to be the sixth base in eukaryotic DNA and not just an intermediate in the 5mC demethylation pathway.
In the case of the lethal human malaria parasite, Plasmodium falciparum, epigenetic gene regulation, via specialized nuclear architecture, histone modifications, histone variants and chromatin-associated non-coding RNAs governs stage-specific biology, as the parasite develops within the human host and mosquito vector (16). For instance, during P. falciparum asexual blood stage development in humans, key processes such as clonally variant expression of surface-exposed virulence factors and commitment to transmission stages (gametocytes) are epigenetically regulated by all of the above mechanisms, and associated to disease pathogenesis (16,17). Furthermore, small molecules that specifically target P. falciparum histone methylation and acetylation have been shown to interfere with parasite growth and survival in vitro and in vivo (18–21). However, to date, the contribution of well-established cytosine modifications to parasite gene regulation remains poorly understood: one of the main deterrents has been the AT-rich nature (∼80% A+T content) of the P. falciparum genome (22). In fact, after decades of debate over the existence of 5mC in P. falciparum (23–28), in 2013, Le Roch and colleagues identified 5mC in genomic DNA prepared from P. falciparum asexual blood stages using mass spectrometry and bisulfite conversion followed by sequencing (BS-seq) (28). However, because BS-seq does not distinguish between 5mC and 5hmC (29), it still remains unclear as to what the levels of cytosine methylation and hydroxymethylation are in P. falciparum, and if indeed they play a role in regulating parasite gene expression.
To conclusively address this, we utilized a combination of biochemical and cell biological techniques to evaluate the presence of cytosine hydroxymethylation in the P. falciparum genome. We took particular care to work with parasite genomic DNA preparations devoid of contaminating DNA from human white blood cells, which are normally present during P. falciparum in vitro culture. We then used a validated antibody-dependent immunoprecipitation approach to delineate genome-wide hydroxymethyl-cytosine distribution at different stages of intra-erythrocytic development. Our work challenges the current view of DNA cytosine methylation in P. falciparum (28): we show that 5mC is present at very low levels in the parasite genome (0.05% from mass spectrometric analysis) and that a 5hmC signal cannot be detected by mass spectrometry. Instead, a signal more hydrophobic than 5hmC was consistently detected, and we refer to this modification as ‘5hmC-like’ throughout this report. We demonstrate that this modified form is the predominant cytosine modification in P. falciparum (0.2–0.4% from a variety of biochemical techniques) and that its levels in gene bodies positively correlate with gene expression. We speculate that the high oxygen level in infected erythrocytes may favor the deposition of unusual cytosine modifications, some of which may be 5mC derivatives, over 5mC in asexual stages of the malaria parasites.
MATERIALS AND METHODS
In vitro culture of Plasmodium falciparum
Blood stages of the P. falciparum laboratory strain 3D7, clone G7 and NF54varsporo were grown according to Trager and Jensen (30) with a few changes. Briefly, a mixed stage culture of P. falciparum was grown in white blood cell (WBC)-free O+ human erythrocytes (prepared from whole blood by treatment with leukocyte-specific filters) at a hematocrit of 4% in Roswell Park Memorial Institute (RPMI) 1640 medium containing L-glutamine (Invitrogen) supplemented with 1% v/v Albumax II (Invitrogen) and 200 μM hypoxanthine (C.C.Pro). The cultures were grown in a gas environment of 5% CO2, 1% O2 and 94% N2 to a parasitemia of 3–8% before harvesting for downstream analysis. For synchronization, knob-positive parasites were selected by gelatin flotation using Plasmion (Fresenius Kabi) (31) and after re-invasion, treated twice with 5% sorbitol (Sigma) (32) to obtain parasites that were synchronized within a window of approximately 4 h, as evaluated by Giemsa staining.
Genomic DNA and RNA isolation
Infected human erythrocyte cultures at different parasitemia and synchronized at the ring (8–12 h post-invasion (hpi)), trophozoite (28–32 hpi) or schizont (40–44 hpi) parasite stages were harvested and free parasites obtained by saponin lysis (33). Subsequently, the parasite pellet was split into two portions: one for genomic DNA extraction, the other for total RNA extraction. Genomic DNA was prepared using the DNeasy Blood and Tissue kit (Qiagen) or the Nucleospin Tissue kit (Machery-Nagel) as per manufacturer's instructions and treated with RNAse to removing any contaminating RNA molecules. Additionally, to remove heme and its derivatives from the genomic DNA, it was purified using magnetic bead-based clean up with Ampure XP beads (Beackman Coulter) at 1:1 ratio, according to manufacturer's instructions. Total RNA was prepared using the miRNeasy mini kit (Qiagen), as per manufacturer's instructions, along with on-column DNase digestion. DNA and RNA concentrations were measured using the NanoDrop Spectrophotometer (Thermo Fisher Scientific) and Qubit Colorimetric Assay (Thermo Fisher Scientific).
South-western blotting assays
South-western blotting assays of P. falciparum genomic DNA and DNA standards (Methylated DNA standard kit from ActiveMotif) were performed as previously described (34), but without using a DNA-binding protein. DNA prepared as described above from matched volumes of WBC-free uninfected blood served as a control. Primary antibodies used include anti-5mC (rabbit monoclonal; Abnova), anti-5mC (mouse monoclonal; Eurogentec), anti-5hmC (mouse monoclonal; ActiveMotif) and anti-5hmC (mouse monoclonal; Millipore) antibodies. No cross-reactivity was observed between 5hmC and 5mC, 5fc, 5cac and 5C for the anti-5hmC antibodies (Source: Active motif). HRP-conjugated secondary antibodies include anti-rabbit IgG and anti-mouse IgG from GE LifeSciences that were compatible with ECL Western Blotting Detection Reagents (Thermo Fisher Scientific).
ELISA-based 5hmC quantification in P. falciparum genomic DNA
Quantification of 5hmC/5hmc-like levels in P. falciparum genomic DNA was performed using the Global 5hmC quantification kit (ActiveMotif) with a few modifications to the manufacturer's protocol. Briefly, a 50 ml solution of genomic DNA (@ 50 ng/ml) was sonicated for 10 min (20 s ON; 20 s OFF) using a pre-chilled Bioruptor (Diagenode). This solution was then diluted to 2 ng/ml in sample dilution buffer provided with the kit, heat denatured at 98°C for 10 min and stored on ice. Simultaneously, 5hmC DNA standards (included in the kit and used to generate a standard curve) as well as unmethylated and hydroxymethylated DNA (Methylated DNA standard kit, ActiveMotif), included as negative and positive controls, respectively, were heat denatured at 98°C for 10 min and stored on ice. Next, assay wells were coated with a DNA binding solution and incubated for 1 h at room temperature (RT). Once coating was complete, 50 ml of genomic DNA samples, standards or controls, was added to an assay well and incubated for 2 h at 37°C. The subsequent steps, i.e. washing of wells, incubation with primary and secondary antibody solutions and developing of each reaction, were carried out as per manufacturer's instructions. Finally, the OD of the colorimetric reaction was measured at 450 nm within 5 min of development using a Synergy 2 Multi-Detection Microplate Reader (Biotek). The percentage 5hmC/5hmC-like levels in each sample were calculated after plotting a standard curve in Microsoft Excel. Note that the primary antibody used was a mouse anti-5hmC monoclonal antibody (ActiveMotif) at 1/1000 dilution and the secondary antibody used was anti-mouse-HRP antibody (GE Lifesciences) at 1/2000 dilution.
ELISA-based 5mC quantification in P. falciparum genomic DNA
Quantification of 5mC levels in P. falciparum gDNA was performed using the methylated DNA quantification kit (Abcam) according the manufacturer's instructions. Briefly, 100 ng of DNA samples, positive and negative controls (provided in the kit), were bound to assay wells for 90 min at 37°C. Wells were washed three times and primary anti-5mC antibody was added to all assay wells for 1 h at RT. Wells were then washed and secondary antibody was added to all assay wells for 30 min at RT. The subsequent steps of fluorescence developpement were carried out according to manufacturer's instrucations, and RFU (Relative fluorescence units) was measured using a Synergy 2 Multi-Detection Microplate Reader (Biotek) at Excitation/Emission = 530/590nm. 5mC percentage in each sample was calculated after plotting a standard curve in Microsoft Excel.
Immunofluorescence assays
Anti-5hmC and anti-5mC immunofluorescent staining was performed as previously described (35) with a few modifications. Free P. falciparum parasites were obtained after saponin lysis (33), washed with phosphate-bufffered saline (PBS), and fixed with 0.0075% Glutaraldehyde / 4% Paraformaldehyde for 30 min at RT. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 15 min at RT and DNA denatured by adding 4N HCl for 20 min at RT. HCl was washed away and 100 mM Tris pH 8.0 was added for 10 min at RT for neutralization. Cells were then washed with PBS + 0.05% Tween-20 (PBST) and free aldehydes were quenched using 50 mM NH4Cl in PBS before blocking with PBS + 3% Bovine Serum Albumin. After blocking, primary anti-5hmC (Mouse monoclonal, Activemotif) or anti-5mC antibodies (Mouse polyclonal, Eurogentek) were added at 1/250 dilution for 1 h at RT. Anti-Histone3 (Rabbit; Abcam) primary antibodies were also added as a positive control to stain the nucleus. Cells were then washed three times with PBST and the following secondary antibodies were added at 1:2000 dilution for 30 min at RT: Goat anti-Mouse conjugated to Alexafluor-488 and Goat anti-Rabbit conjugated to Alexafluor-647. Finally, after washing with PBS, 5 ml of cells were mounted in Vectashield fluorescent mounting medium and images were acquired using a Deltavision Elite imaging system (GE Healthcare). Images were processed using Fiji/ImageJ.
Preparation of cytoplasmic and nuclear extracts
Plasmodium falciparum cytoplasmic and nuclear extracts were prepared from 3 ml of infected RBCs synchronized at the R, T or S stages, harvested at a parasitemia of 6 to 8% as previously described (36). The only change was that for nuclear fraction extraction, the extraction buffer was changed to 20 mM HEPES pH 7.9, 600 mM NaCl and 1 mM DTT. The resulting cytoplasmic and nuclear fractions were aliquoted, snap-frozen and stored at −80°C until further use. Protein concentration was measured using a Bradford assay and the Qubit Colorimetric Protein Assay.
TET activity assay using P. falciparum nuclear extracts
TET-like activity of P. falciparum nuclear extracts was measured using the TET hydroxylase activity quantification kit (fluorometric) from Abcam as per manufacturer's directions. A total of 5 μg of nuclear extracts was used per reaction. As a positive control, human recombinant TET1 protein (Abcam) was included in the assay. Extracts prepared as described above from matched volumes of WBC-free uninfected blood served as an additional control. The resulting fluorescence was measured at 530 nm excitation/590 nm emission using a Synergy 2 Multi-Detection Microplate Reader. TET hydroxylase activity is represented as nanograms of DNA converted per minute per milligram of protein. This was calculated based on a standard curve generated using Microsoft Excel.
Bisulfite (BS) and oxidative bisulfite (oxBS) sequencing
Treatment of genomic DNA prepared from P. falciparum asexual stage parasites with sodium bisulfite alone (BS) or with an oxidizing agent followed by sodium bisulfite (oxBS) was initially performed for pilot experiments using the TrueMethyl® Whole Genome Bisulfite Sequencing kit (Cambridge Epigenetix) according to manufacturer's instructions. Spike-in DNA controls to estimate bisulfite conversion and oxidation efficiencies were provided with the kit and added prior to the chemical treatments. The treated DNA was further processed with the same kit to generate DNA libraries that were compatible with Illumina sequencing-by-synthesis. For the data presented in this report, BS-seq and oxBS-seq libraries were prepared using the Ovation® Ultralow Methyl-Seq with TrueMethyl® oxBS module kit (NuGen) according to manufacturer's instructions; spike-in controls were obtained from Cambridge Epigenetix as above. All libraries were multiplexed and run on an Illumina NextSeq 500 as a paired-end run of 150 nt. The resulting fastq file was demultiplexed using bcl2fastq2 Conversion Software v2.17 (Illumina) and further analyzed as described below.
Analysis of BS-seq and oxBS-seq data
Quality control of fastq files was performed using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) after which Cutadapt v1.14 (37) was used to cut adapter sequences and trim low quality bases (Q < 20) from the 3′-end of reads. The resulting high-quality reads were aligned to the P. falciparum 3D7 reference genome (http://genedb.org; v3, PlasmoDB v13.0) using Bismark v.0.14.5 (38). Duplicated reads were removed from the raw alignments using SAMtools v0.1.19 (39) and further filtered for quality (Q < 30). Properties of the BS- and oxBS-seq datasets generated in this study and related statistics are provided in Supplementary Table S1.
Spike-in analysis
We estimated the methylation conversion rate of the BS- and oxBS-seq libaries using the six spike-in controls provided by the TrueMethyl kit. These spike-in controls contain an expected set of methylated (both 5mC and 5hmC) and unmethylated (truth set) loci. The fastq reads were aligned to the spike-in control sequences using the cegx-bsExpress algorithm (https://bitbucket.org/cegx-bfx/cegx_bsexpress_docker), and the number of true positives and false positives were calculated at varying read depths based on the conversion summary provided by cegx-bsExpress. For BS-seq libraries, sensitivity for 5mCs was defined as the proportion of 5mCs from the truth set that were called as methylated by our pipeline at a given depth cutoff. Similarly, sensitivity for 5hmCs was defined as the proportion of 5hmCs from the truth set that were called as methylated by our pipeline at a given depth cutoff. False positive rate (FPR) was calculated as the proportion of unmethylated cytosines from the truth set that were called as methylated by our pipeline at a given depth cutoff. For oxBS-seq libraries, sensitivity of 5mCs was calculated as described above, and FPR was calculated as the proportion of unmethylated cytosines and 5hmCs from the truth set that were called as methylated by our pipeline at a given depth cutoff. At depth ≥5× and % of methylated reads ≥10% (i.e. ≥1 read), we observed a sensitivity of ≥96% and a FPR of ≥2.5% (Supplementary Table S2). These cutoffs were used for all subsequent analyses.
Methylkit-based determination of 5hmC loci
De-duplicated and filtered alignments were used as inputs to methylKit v0.9.2 (40) to estimate methylation percentages at CpG, CHG and CHH contexts. Loci with a minimum read coverage of 5× with at least 1 methylation-containing read were considered for down-stream analysis based on our spike-in results. The resulting calls from the BS-seq and oxBS-seq samples were compared to identify 5mC and 5hmC/5hmC-like loci. Specifically, loci that were found to be methylated in both BS-seq and oxBS-seq were classified as 5mCs whilst those that were methylated in BS-seq and unmethylated in oxBS-seq were classified as 5hmCs or 5hmC-like bases. Alternately, we processed the de-duplicated alignments using Methpipe (41) and identified between 34–70% of the 5hmC/5hmC-like loci identified by Methylkit. We decided to use the Methylkit lists for downstream analysis.
Data were visualized using the Integrative Genomics Viewer (IGV) browser v.2.3.90 (42). For representation of 5hmC/5hmC-like genome-wide distribution, the coverage of each mark was calculated as average reads per million over bins of 2000 nt using bedtools v2.19.0 (43) and correlations between the genome-wide distributions were calculated using deeptools2 v3.1.1 (44).
hmeDIP-seq and RNA-seq library preparation and sequencing
Hydroxymethylated DNA Immunoprecipitation (hmeDIP) was performed using the hmeDIP kit from ActiveMotif according to manufaturer's instructions. The starting material was genomic DNA from the Ring stage sheared using a pre-chilled Bioruptor (Diagenode) for 10 min, 30 s ON and 30 s OFF to a size of 200–400 bp. The immunoprecipitated DNA was processed using the MicroPlex Library Preparation Kit v2 (Diagenode) with the KAPA Hifi polymerase for library amplification. All libraries were multiplexed and run on an Illumina NextSeq 500 as a paired-end run of 150 nt. The resulting fastq file was demultiplexed using bcl2fastq2 Conversion Software v2.17.
For transcriptomic analysis, 4 μg of total RNA from Ring or Schizont stage parasites was poly(A)-enriched using the Dynabeads mRNA purification kit (Thermo Fisher Scientific), and used for strand-specific RNA-seq library preparation with the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina) according to manufacturer's instructions. The only change was that library amplification was performed using KAPA Hifi polymerase (KAPA Biosystems). All libraries were multiplexed and run on an Illumina NextSeq 500 as a paired-end run of 150 nt. The resulting fastq file was demultiplexed using bcl2fastq2 Conversion Software v2.17.
Analysis of hmeDIP-seq and RNA-seq data
Quality control of fastq files was performed using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Cutadapt. High quality reads were mapped to the P. falciparum 3D7 genome (http://genedb.org; v3, PlasmoDB v13.0) using the Burrows-Wheeler Alignment tool Bowtie 2 v.2.2.3 under default settings (45). SAMTools was used to remove duplicates for all hmeDIP-seq samples. Properties of the RNA-seq and hmeDIP-seq datasets generated in this study and related statistics are provided in Supplementary Table S1.
For hmeDIP-seq analysis, deduplicated bam files filtered for quality (Q > 30) using SAMtools served as a starting point to identify enriched peaks in immunoprecipitated DNA relative to the genomic DNA input. First, correlation of the bam files of the different stages was calculated using bedtools and deepTools2. Second, enrichment peaks were identified using the MACS2 software (46) after controlling for a false discovery rate (q-value) of 1%. Correlation of the different MACS2 peak files was determined using deepTools2 and bedGraphtobigwig as previously described (https://github.com/taoliu/MACS/wiki/Build-Signal-Track). Linear coverage plots were visualized using the IGV browser. Gene Ontology (GO) analysis was performed in PlasmoDB (https://plasmodb.org) and reductionist analysis of GO terms performed using Revigo (47).
For RNA-seq analysis, bam files were filtered down to either the gene body (exons and introns) or exonic regions alone using bedtools. The resulting alignments were used as inputs to the featureCounts (48) from Subread v1.5.2 to count the number of non-duplicate, uniquely aligned (mapping quality > 30) reads per gene. Custom R function was used to calculate FPKM (Fragments Per Kilobase of transcript per Million mapped reads) per gene from the raw counts.
Identification and quantification of 5hmdC and 5mdC in Plasmodium falciparum DNA by LC-MS/MS
Purified P. falciparum genomic DNA was hydrolyzed enzymatically as described (49–51) with a modified protocol. The digestion mix contained 10 mM Tris–HCl (pH 7.9), 1 mM MgCl2, 5 U Benzonase (99% Purity Merck 71206), 50 μM Desferroxamine (Sigma D9533), 0.1 μg/μl Pentostatin (Sigma SML0508), 100 μM Butylated hydroxytoluene (Sigma W218405), 0.5 μg/μl Tetrahydrouridine (Calbiochem 584222), 5 U Bacterial Alkaline Phosphatase (Thermo Fischer 18011015), 0.05 U Phosphodiesterase I (Affymetrix P3243). Hypersil GOLD aQ column [100 × 2.1 mm, 1.9 μm (Thermo Scientific 25305)] was used to resolve the digested deoxyribonucleosides in a two-buffer eluent system which was composed as follows: buffer A = 0.1% formic acid in water and buffer B = 0.1% formic acid in acetonitrile). Synthetic hm5dC and m5dC were purchased from Cayman Chemicals (Items 18 162 and 16 166, respectively). HPLC was performed at a flow rate of 300 μl/m at 25°C. The gradient of 0.1% formic acid in acetonitrile was as follows: 0–12 min, held at 0%; 12–15.3 min, 0%-1%; 15.3–18.7 min, 1%-6%; 18.7–20 min, held at 6%; 20–24 min, 6%-100%; 24–27.3 min, held at 100%; 27.3–28 min, 100%-0%; 28–41 min, 0%. The HPLC column was directly connected to an Agilent 6490 triple quadrupole mass spectrometer with ESI Jetstream ionization operated in positive ion mode. The voltages and source gas parameters were as follows: gas temperature: 200°C; gas flow: 15 l/min; nebulizer: 30 psi; sheath gas temperature: 300°C; sheath gas flow: 12 l/min; capillary voltage 1800 V and nozzle voltage, 0V. The molecular transition ions were quantified in multiple-reaction monitoring (MRM) mode.
Statistical analyses
For biochemical assays (5hmC quantification and TET activity assay), a minimum of three biological replicates were analyzed. Statistical analysis was performed in Microsoft Excel 2008. Results were plotted using Plotly (52).
For differences in mRNA expression between hydroxymethylated and unhydrooxymethylated gene sets, after a preliminary analysis in Microsoft Excel 2008 to determine sample variance, P-values were calculated using the non-parametric Mann–Whitney U-test. Box plots were generated using Plotly.
RESULTS
Identification of a 5hmC-like modification in the genome of the malaria parasite P. falciparum
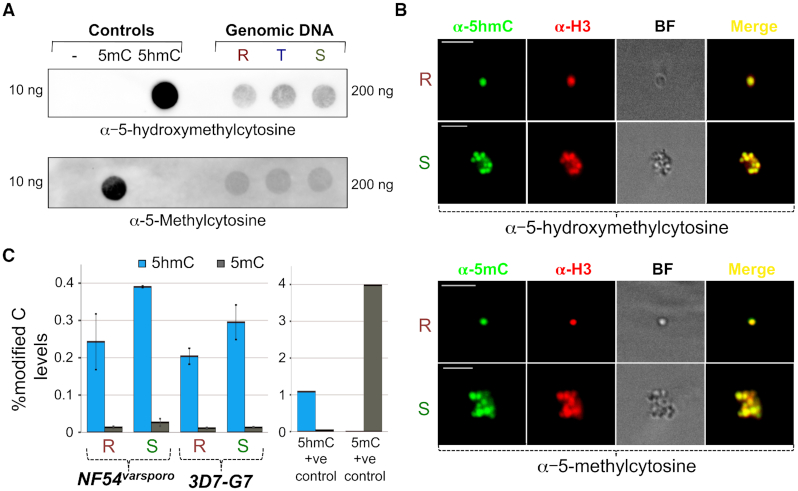
To examine the presence of unexplored cytosine modifications in P. falciparum, we denatured genomic DNA prepared from parasites of the 3D7-G7 strain (53) grown in WBC-free blood, spotted them onto nylon membranes and probed the membrane with either anti-5mC or anti-5hmC antibodies from different sources (see ‘Materials and Methods’ section). Genomic DNA prepared from all three stages (synchronized at the Ring (R; 8–14 h post-invasion (hpi)), Trophozoite (T; 24–28 hpi) and Schizont (S; 38–42 hpi)), reacted with the anti-5hmC antibodies, indicating the presence of a new DNA modification, 5hmC or 5hmC-like, in the parasite genome: Figure 1A and Supplementary Figure S1A show typical examples of our south-western blotting results, with DNA preparations from uninfected WBC-free blood showing zero reactivity to all antibodies tested (Supplementary Figure S1B). Additionally, we performed immunofluorescence assays on fixed 3D7-G7 parasites using the primary antibodies from Figure 1A and observed that the fluorescence signal was restricted to the nucleus, further supporting the existence of this novel DNA modification in vivo (Figure 1B). Finally, we used an ELISA-based assay to quantify the levels of this new modification in P. falciparum genomic DNA extracted from two strains, 3D7-G7 and NF54varsporo(54), and found that its levels varied between 0.19–0.38% during asexual development (Figure 1C). Interestingly, the ELISA-based quantification yielded very low numbers for 5mC levels in the P. falciparum genome, at 0.01–0.02%; this is in contrast to the report from Ponts et al. (28) that identified substantially higher 5mC levels (0.47–0.58%) in P. falciparum genomic DNA by mass spectrometry. The observed dominance of a 5hmC-like modification over 5mC in a eukaryotic genome is unprecedented (55–57) (Figure 1C and Supplementary Figure S1C) and indicates that the parasite might have evolved to use the 5hmC-like modification in a unique manner.
Figure 1.
A 5hmC-like modification is the predominant cytosine modification in the Plasmodium falciparum genome. (A) Southwestern blotting analysis of genomic DNA prepared from Ring (R), Trophozoite (T) or Schizont (S) stage P. falciparum 3D7-G7 parasites using antibodies against 5mC (Abnova) or 5hmC (Activemotif). Unmethylated, methylated or hydroxymethylated DNA standards (Activemotif) served as controls. The total amount of DNA used is indicated in ng. (B) Immunostaining of Ring (R) or Schizont (S) stage P. falciparum 3D7-G7 parasites with anti-5hmC (top) or anti-5mC (bottom) antibodies. Anti-histone H3 antibodies were used as a positive control for nuclear staining. BF = Brightfield. (C) ELISA-based quantification of 5hmC (blue) or 5mC (gray) levels in genomic DNA prepared from Ring (R) or Schizont (S) stage parasites of the NF54varsporo and 3D7-G7 strains. The positive controls were the methylated (5mC) and hydroxymethylated (5hmC) DNA standards used in part 1A. Data were calculated as a percentage and represent the means of three independent experiments + standard errors of means (S.E.M.).
We also queried if TET proteins, the writers of 5hmC, are expressed by P. falciparum asexual stage parasites. We first measured TET hydroxylase activity in nuclear extracts prepared from Ring and Schizont stages of P. falciparum using an ELISA-based colorimetric assay, with extracts from uninfected WBC-free blood serving as a negative control, and detected TET-like activity in all parasite samples, with highest activity in Ring stage nuclear extracts, indicating that a TET-like protein is indeed expressed by the parasite (Supplementary Figure S2). However, the latest annotation of the P. falciparum genome does not include a TET homologue or orthologue neither could we identify a TET-like protein in P. falciparum using hidden markov model (HMM)-based predictions. The identification of the gene(s) coding for the observed enzymatic activity, however, is beyond the scope of this report. Nonetheless, our data, for the first time, establish the presence of a 5hmC-like modification in the genome of a eukaryotic pathogen.
Hydroxymethylation is the predominant cytosine modification in P. falciparum asexual stages
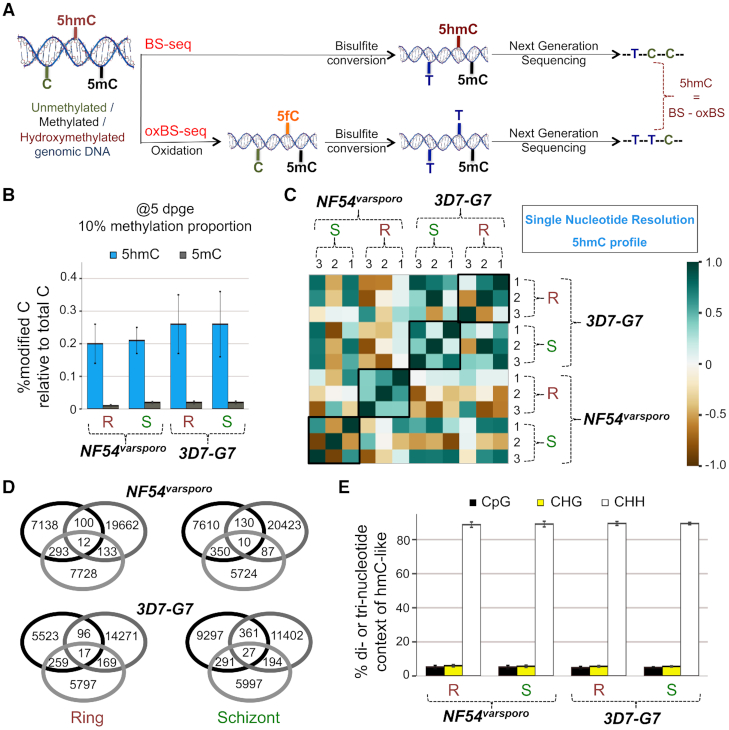
Thereafter, to determine the genome-wide distribution of the 5hmC-like modification at single nucleotide resolution, we performed bisulfite sequencing (BS-seq) and oxidative bisulfite sequencing (oxBS-seq) of genomic DNA derived from two developmental stages, Ring (R; 8–14 hpi) and Schizont (S; 38–42 hpi)—stages that have distinct transcriptomic profiles (58,59)—of two P. falciparum strains, 3D7-G7 and NF54varsporo, using the TrueMethyl technology (60,61) (Figure 2A). Briefly, we treated genomic DNA supplemented with tailed spike-in controls with sodium bisulfite alone (Figure 2A; top), or sequentially with the oxidizing agent potassium perruthenate and sodium bisulfite (Figure 2A; bottom) to yield BS (5mC + 5hmC (or 5hmC-like)) and oxBS (5mC) samples, respectively. These were then processed for multiplexed Illumina sequencing-by-synthesis and the resulting libraries sequenced using a paired-end run of 150 nt. After performing quality control analysis of fastq files (Supplementary Table S1), conversion efficiency of the spike-in controls was measured for each sample using cegx-BSexpress (Supplementary Figure S3A–H), and sensitivity and FPR values estimated at varying sequencing depths (≥5, ≥10, ≥20 and ≥30) and methylation proportion (i.e. % of reads supporting methylation at the given sequencing depth) with a P-value cutoff of 0.05 (Supplementary Table S2). Based on this, we chose a sequencing depth of ≥5 and a methylation proportion cutoff of 10% for downstream analysis.
Figure 2.
BS- and oxBS-seq profiling confirms that a majority of modified cytosines in Plasmodium falciparum are 5hmC-like in nature. (A) Schematic representation of the BS- and oxBS-seq methodology. Genomic DNA prepared from synchronized P. falciparum parasites was treated with sodium bisulfite alone (BS-seq; top panel) or with an oxidizing agent prior to sodium bisulfite treatment (oxBS-seq; bottom panel). The resulting DNA was processed for Next Generation Sequencing and a subtraction of oxBS-seq signals from BS-seq signals yielded 5hmC distribution at single nucleotide resolution. See ‘Materials and Methods’ section for additional details. (B) Measured 5hmC (blue) or 5mC (grey) levels (relative to total cytosines in the P. falciparum genome, at 5 dpge and 10% methylation proportion and represented as a percentage) in Ring (R) or Schizont (S) stage BS- and oxBS-seq samples for the NF54varsporo and 3D7-G7 strains. Data represent the means of three independent experiments + S.E.M. (C) Pearson correlation analysis of the 5hmC-like profiles (calculated for bigwig files normalized over 500 nt bins) for the different BS- and oxBS-seq replicates for genomic DNA extracted from Ring (R) or Schizont (S) stage parasites of the NF54varsporo and 3D7-G7 strains. The colour scale indicates the value of the Pearson Correlation Coefficient R from −1 to 1. (D) Overlap in the number of 5hmC-like loci identified in BS- and oxBS-seq replicates of the indicates samples of the NF54varsporo and 3D7-G7 strains. (E) The percentage of 5hmC-like loci found in the CpG (black), CHG (yellow) or CHH (white) context was calculated for the indicated parasite strain and growth stage: R = Ring and S = Schizont, from the BS- and oxBS-seq data. Data represent the means of three independent experiments + S.E.M.
Next, the fastq files were aligned to the P. falciparum genome (http://genedb.org; v3) using a bisulfite-aware algorithm, Bismark (38), and the numbers of methylated cytosines calculated for each sample (Supplementary Table S3); this preliminary examination indicated that ∼0.3–0.4% of the sequenced cytosines were modified in the three biological replicates of the R and S samples from both strains. The data were further processed using Methylkit (40) to identify 5hmC (or 5hmC-like) and 5mC loci at the parameters specified above, resulting in an average of 8897 5hmC-like loci for the NF54varsporo R stage or 0.20% of all genomic cytosines (Cs; numbering 4512612 in the P. falciparum genome), 9490 5hmC-like loci for the NF54varsporo S stage or 0.21% of all Cs, 11872 5hmC-like loci for the 3D7-G7 R stage or 0.26% of all Cs, and 11640 5hmC-like loci for the 3D7-G7 S stage or 0.26% of all Cs (Figure 2B; Supplementary Tables S4 and 5)—these values are comparable to those observed in Figure 1C. Moreover, we detected very low levels of 5mC in the parasite, with a total of 680–720 unique 5mC loci in the various samples accounting for 0.01–0.02% of genomic cytosines (Supplementary Figure S4A and Tables S6-7).
We next performed a Pearson Correlation analysis of the 5hmC-like profiles of NF54varsporo R & S and 3D7-G7 R & S replicates and found to our surprise that the correlation coefficient R ranged from −0.85 to 1 (Figure 2C), with replicates of the same strain and stage showing variable correlations—for example, the biological replicates of the NF54varsporo R and 3D7-G7 S stages were positively correlated, whilst those of the NF54varsporo S and 3D7-G7 R stages were not. This was emphasized by the low overlap in the exact nucleotide position of the 5hmC-like loci amongst biological replicates (Figure 2D). Technical replicates for a single genomic DNA sample further confirmed this observation (Supplementary Figure S5). Because our analysis of the spike-in DNA controls indicated that both oxidation and bisulfite conversion efficiencies of the BS- and oxBS-seq methods are close to 100% for non-parasite DNA (Supplementary Figure S3), we conclude that this method generates incomplete bisulfite modifications when applied to the high AT-rich P. falciparum genome (>80% AT content). We therefore decided to use another reference method to perform genome-wide 5hmC-like analysis of P. falciparum blood stage DNA.
hmeDIP-seq reproducibly identifies the genome-wide distribution of 5hmC-like modifications in P. falciparum
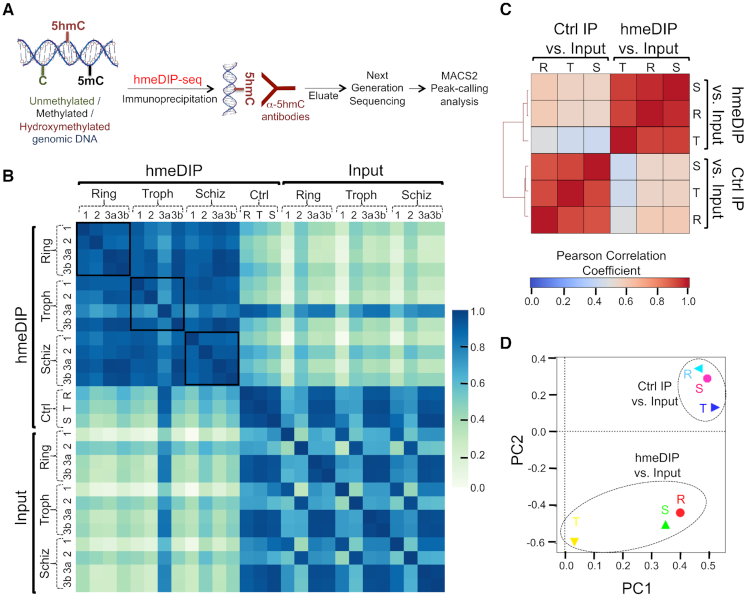
Hydroxymethylated DNA immunoprecipitation (hmeDIP) followed by next generation sequencing (-seq) is routinely used to characterize the distribution of 5hmC (62–64). For P. falciparum hmeDIP-seq, we isolated genomic DNA from three biological replicates of the 3D7-G7 strain synchronized at the Ring (R; 6–10 hpi), Trophozoite (T; 24–28 hpi) and Schizont (S; 38–42 hpi) stages, sheared the DNA to fragments that were 300–400 bp in size and immunoprecipitated 5hmC-enriched DNA using anti-5hmC antibodies (Figure 3A); rabbit IgG antibodies were used as a negative control for immunoprecipitation (IP). The immunoprecipitated DNA was processed for multiplexed Illumina sequencing-by-synthesis and the resulting libraries sequenced using a paired-end run of 150 nt. After performing quality control analysis of fastq files (Supplementary Table S1), the sequences were aligned against the P. falciparum genome (https://genedb.org; v3) to yield BAM files.
Figure 3.
hmeDIP-seq performs robustly for the AT-rich Plasmodium falciparum genome. (A) Schematic representation of the hmeDIP-seq methodology used to measure 5hmC (or 5hmC-like) distribution in P. falciparum asexual stages. Sheared P. falciparum genomic DNA was immunoprecipitated using anti-5hmC antibodies and the resulting DNA processed for Next Generation Sequencing (NGS). MACS2 was used for 5hmC-like peak-calling. See Materials and Methods for additional details. (B) Correlation between hmeDIP-seq biological replicates (calculated for bam alignment files derived from Illumina sequencing data) for the Ring (R), Trophozoite (Troph or T) and Schizont (Schiz or S) stages of 3D7-G7 was determined using a Pearson correlation analysis. The colour scale indicates values of the Pearson correlation coefficient R from 0 to 1. Ctrl = Control IP for biological replicate 1. A and B are technical replicates for the same genomic DNA sample. (C) Correlation of MACS2 fold enrichment (FE) profiles of hmeDIP-seq samples relative to Input DNA for the Ring (R), Trophozoite (T) and Schizont (S) stages of 3D7-G7 was compared to the IgG control IPs relative to Input DNA. For each stage, data were normalized across all replicates to generate a single FE profile (described in ‘Materials and Methods’ section). The colour scale indicates values of the Pearson correlation coefficient R from 0 to 1. Ctrl = Control IP for biological replicate 1. (D) Principal Component Analysis of the different MACS2-derived FE profiles of part C was performed using the plotPCA function of DeepTools on a multibigwigsummary file. The eigenvalues of the top two principal components PC1 (54.3% of variance explained) and PC2 (31.3% of variance explained) are shown and meaningful clustering of samples indicated.
Given that hmeDIP-seq has been standardized for genomes with higher G+C content as compared to P. falciparum such as humans and mice, and given the poor performance of BS- and oXBS-seq for the P. falciparum genome (Figure 2), we first evaluated the reproducibility of our hmeDIP-seq experiments by measuring the Pearson correlation coefficient R values between the various BAM alignment files. As shown in Figure 3B, we observed a segregation of the hmeDIP-seq samples into a cluster that is distinct from the IgG control IPs and the Input DNA samples, which were also processed for Illumina sequencing-by-synthesis as described above. In fact, the control reactions were better correlated to the Input DNA samples as compared to the hmeDIP-seq samples. Interestingly, the values of R ranged from 0.8 to 1.0 between the Ring, Trophozoite and Schizont stage hmeDIP-seq samples suggesting that the 5hmC-like profiles of the P. falciparum genome at different growth stages might be similar.
Next, we used the peak-calling software ‘Model-based analysis of ChIP-seq’ or MACS2 (46) to delineate hmeDIP-enriched regions of the P. falciparum genome relative to the corresponding Input DNA. A similar analysis was performed for the IgG control IPs. When we evaluated the Pearson correlation coefficient R values between the various MACS2 fold enrichment (FE) profiles, we found that the ‘hmeDIP versus Input’ FE profiles of various growth stages clustered independently from the ‘control IP versus Input’ FE profiles (Figure 3C). A similar separation was observed using a Principal Component Analysis (PCA) analysis, along the second principal component axis (Figure 3D). Of note, in the PCA analysis, the Trophozoite stage ‘hmeDIP versus Input’ FE profile segregated distinctly from the Ring and Schizont stage FE profiles along the first principal component axis. Taken together, we conclude that hmeDIP-seq performs reproducibly and robustly to study the distribution of 5hmC-like marks in the P. falciparum genome.
The 5hmC-like modification is predominantly found in gene bodies and appears to be stably maintained during intra-erythrocytic development
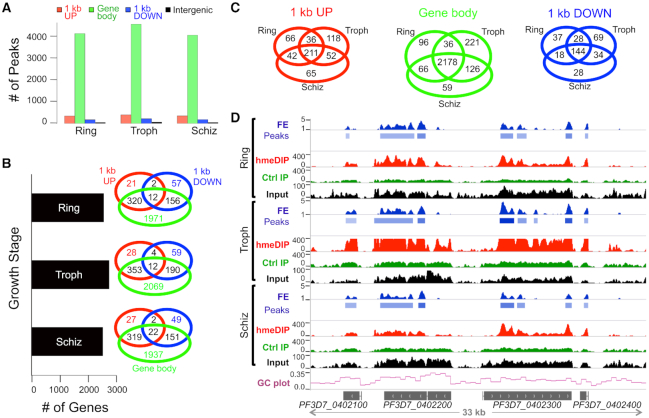
We calculated the number of MACS2-derived, hmeDIP-enriched peaks in the Ring, Trophozoite and Schizont samples at a log2(fold change) ≥ 1 and false discovery rate < 0.05 and identified 4148, 4603 and 4080 peaks, respectively (Supplementary Table S8). Of these, 98–98.5% of the peaks mapped to genic regions, described here as the region covering 1 kb upstream of START codon, gene body (introns and exons) and 1 kb downstream of STOP codon (Figure 4A). The remaining 1–1.5% of the so-called intergenic peaks mapped to chromosome ends, primarily to telomeric sequences (Supplementary Figure S6). This observation is not surprising given the abundance of cytosines in gene bodies and telomeric ends relative to the rest of the P. falciparum genome (22). Furthermore, the cytosines in these regions are predominantly in the CHH context, suggesting that a majority of the modified cytosines in the parasite genome may be in an asymmetric context, as was revealed by the BS- and oxBS-seq analysis in Figure 2E.
Figure 4.
5hmC-like bases occur predominantly in genic regions of the Plasmodium falciparum genome and are stably maintained during intra-erythrocytic development. (A) The number of hmeDIP-enriched peaks found in genic (red bars; 1 kb upstream, green bars; gene body and blue bars; 1 kb downstream) and intergenic (black bars) regions was calculated for the indicated growth stage from the hmeDIP-seq data: Troph = Trophozoite and S = Schizont. (B) Genes containing 5hmC-like loci either in the gene body or within their 1 kb upstream or downstream flanks were identified for the Ring, Trophozoite (Troph) and Schizont (Schiz) stages of the 3D7-G7 strain. A Venn diagram was used to represent the overlap in 5hmC-like-associated genes for the different sub-domains of a genic region. (C) A Venn diagram was used to represent the overlap in 5hmC-like-associated genes for the indicated sub-region across different P. falciparum growth stages: Troph = Trophozoite and S = Schizont. (D) The hmeDIP-seq and MACS2-derived fold enrichment (FE) pofiles for a 35 kb region of chromosome 4 (dark blue) are shown for the Ring, Trophozoite (Troph) and Schizont (Schiz) stages. MACS2-derived peaks are shown underneath the FE profiles. The coverage of bam files for hmeDIP-enriched DNA (red), DNA enriched in the control IP (green), or Input DNA (black) is shown and was calculated as reads per million over bins of 1000 nt. The GC plot (light blue) represents the G+C content of the P, falciparum genome normalized across bins of 1000 nt.
We then identified the genes that are associated with 5hmC-like loci by intersecting the hmeDIP-enriched peaks with annotated features of the P. falciparum genome (Supplementary Table S9). As shown in Figure 4B, 2539, 2735 and 2507 genes have this cytosine modification in the Ring, Trophozoite and Schizont stages, respectively, and a majority of these genes carry the modification within the gene body relative to the 1 kb upstream and downstream regions. However, there are some instances wherein a gene has 5hmC-like modifications in its gene body as well as in the 1 kb upstream and/or downstream regions, indicating that the MACS2-derived peak is spread across multiple areas of the same genic region (Figure 4B). We also analyzed the overlap in the 5hmC-like-associated genes between the different growth stages and found that ∼2200 genes, i. e. >80%, are stably marked with 5hmC-like modifications in the Ring, Trophozoite and Schizont stages (Figure 4C). An example of a stably modified region of chromosome 4 is shown in Figure 4D. Nonetheless, there are some genes that appear to be dynamically marked during the Ring-Trophozoite-Schizont-Ring transitions (Supplementary Figure S7) and point to potentially specific roles of the 5hmC-like mark in gene regulation and parasite development.
5hmC-like levels within gene bodies positively correlate with P. falciparum gene expression
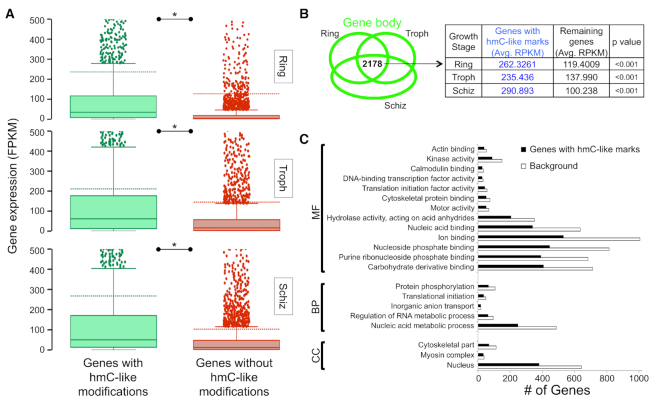
Given that a majority of hmeDIP-enriched peaks were distributed within genic regions, more specifically within gene bodies (Figures 4A and B), we asked if gene body 5hmC-like levels correlated with mRNA expression. We performed RNA-seq-based transcriptomic analysis of total RNA isolated from the parasite cultures that were used for genomic DNA preparation and extracted reads that mapped to ∼5670 P. falciparum genes (Supplementary Table S10; fragments per kilobase of transcript per million reads or FPKM). Next, for each stage and each replicate, we compared the average expression of genes that were hydroxymethylated in the gene body to unhydroxymethylated genes and found that hydroxymethylated genes had significantly higher levels of mRNA expression in all stages analysed in this study (Figure 5A; Supplementary Figure S8 and Table S11A). A similar correlation has been observed in neuronal cells, where 5hmC levels in gene bodies positively correlates with transcriptional activity (10–12). Interestingly, we found that hydroxymethylation in the 1 kb upstream and 1 kb downstream regions of genes also correlated with higher gene expression (Supplementary Tables S11B and C), with a stronger effect of 5hmC-like peaks in the 1 kb downstream regions on gene expression. However, because a majority of these peaks are contiguous with gene body peaks, we cannot attribute a regulatory role to them at this stage.
Figure 5.
5hmC-like distribution within gene bodies is positively correlated to mRNA levels. (A) The expression of genes with (green) or without (red) 5hmC-like peaks in the gene body is displayed as a box and whisker plot for one replicate of the Ring (Rep 1), Trophozoite (Troph; Rep 1) and Schizont (Schiz; Rep 1) stages of the 3D7-G7 strain. For each box and whisker plot, the mean is indicated as a dashed line and the median as a solid line within the box. Y-axis: FPKM (fragments per kilobase of transcript per million mapped reads) measured using strand-specific RNA-seq. For each comparison, P-values were calculated using the non-parametric Mann–Whitney U-test. * indicates a P-value < 0.001. Refer to Supplementary Figure S5 and Table SS5A for the other replicates (B) Left panel: The intersection of 5hmC-like-associated genes (peaks within gene body) identified in the Ring, Trophozoite (Troph) and Schizont (Schiz) stages of the 3D7-G7 strain are represented using a Venn diagram. Right panel: The table summarizes the expression (Average RPKM) of 2178 overlapping genes relative to the remaining genes in replicate 1 of the indicated growth stage. The P-value was calculated using the non-parametric Mann–Whitney U-test. Refer to Supplementary Table S5D for the remaining replicates. (C) Gene Ontology (GO) analysis of genes that contain 5hmC-like loci within the gene body in all three stages of the 3D7-G7 strain was performed using https://plasmodborg. Background refers to the number of genes annotated to the indicated GO term in the entire background set, which in this case is comprised of all annotated P. falciparum genes. MF = Molecular Function; BP = Biological Process; CC = Cellular Component. Refer to Supplementary Table S6 for additional details.
We next evaluated whether the 2178 genes that maintain gene body hydroxymethylation in the Ring, Trophozoite and Schizont stages (Figure 4C) have higher expression throughout the parasite lifecycle. We measured average transcriptional activity of these genes for each stage (and each replicate) and compared them to the remaining genes. As shown in the inset in Figure 5B and Supplementary Table S11D, the steady state mRNA levels of the stably hydroxymethylated genes were significantly higher than the remaining P. falciparum genes in all three stages. Gene ontology (GO) analysis revealed that these genes are enriched for GO terms such as nucleus, myosin complex, cytoskeleton, regulation of RNA metabolic process, inorganic anion transport, translational initiation, protein phosphorylation, carbohydrate derivative binding, purine ribonucleoside triphosphate binding, ion binding, nucleic acid binding, hydrolase activity (acting on acid anhydrides), motor activity, DNA-binding transcription factor activity, calmodulin binding and actin binding (Figure 5C and Supplementary Table S12). Hence, it appears that housekeeping pathways that are associated with the regulation of gene expression, cellular signalling and cytoskeletal organization may be fine-tuned via this newly discovered 5hmC-like modification.
Mass spectrometry analysis posits the presence of a cytosine DNA modification in P. falciparum genomic DNA that is distinct from 5hmdC and 5mdC
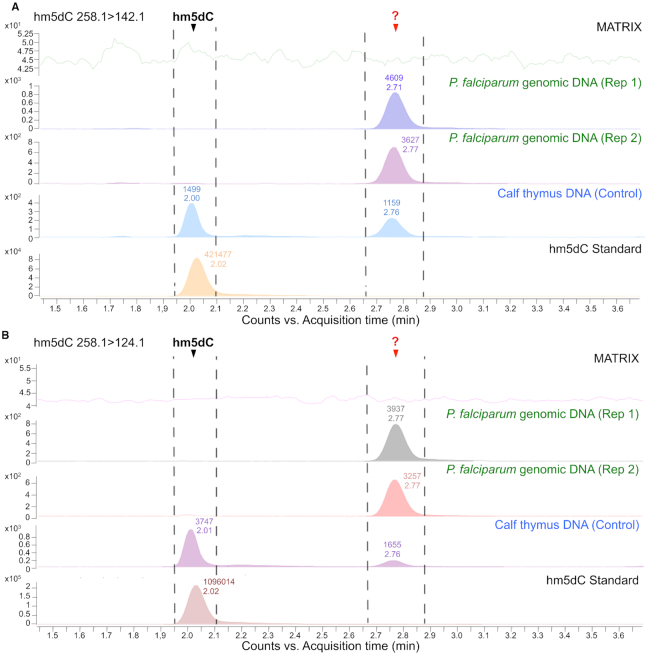
To verify the presence of cytosine modifications in P. falciparum, we surveyed the P. falciparum genome using mass spectrometry (MS). For this, we digested the genomic DNA (gDNA) extracted from asexual blood stage parasites in the presence of anti-oxidants and deaminase inhibitors. Collision-induced dissociation (CID) causes the loss of the 2′-deoxyribose moiety (m/z 116.1) from both 5-hydroxymethyl-2′-deoxycytidine (5hmdC) and 5-methyl-2′-deoxycytidine (5mdC), leaving a protonated pyrimidine ring for both bases with m/z ratios of 142.1 and 126.1, respectively, which was monitored using an Agilent 6490 QqQ mass spectrometer (Supplementary Table S13). In order to make our detection more robust, we used a second qualifier transition of 258.1>124.1 and 258.1>109.1, which corresponds to a further loss of a water and ammonia molecule from the pyrimidine base for 5hmdC and 5mdC, respectively (Figure 6 and Supplementary Figure S9). An analysis of the LC-MS chromatogram shows distinctly the elution of two species at 2.00 min and 3.71 min corresponding to the commercial 5hmdC and 5mdC standards respectively for both of the monitored transitions. These peaks are also visible in the calf thymus DNA, which was used as a positive control. Two biological replicates of hydrolysed P. falciparum gDNA provided a very small signal for 5mdC (∼0.05% of total dC content), confirming the results of the biochemical methods that we used (Figures 1 and 2), and failed to provide a signal for 5hmdC altogether. We, however, noticed the elution of a cryptic peak at 2.75 min and 5.72 min for the transitions that match 5hmdC and 5mdC respectively in P. falciparum gDNA (Figure 6 and Supplementary Figure S9). Spiking in commercial 5hmdC standard in hydrolysed gDNA showed a concomitant increase in the signal at 2.00 min and no change at 2.75 min indicating that the digestion matrix did not contribute to a shift in the retention time and could not account for the cryptic peak (Supplementary Figure S10). Taken together, the MS data indicate the presence of a cytosine modification in P. falciparum genomic DNA – corresponding to the unknown peak that eluted at ∼2.75 min in Figure 6—that is distinct from 5hmdC and 5mdC, and which remains to be characterized.
Figure 6.
hm5dC is not detectable in Plasmodium falciparum DNA by mass spectrometry but there is evidence for another type of modified nucleoside. LC-MS/MS spectra captured using Multiple Reaction Monitoring (MRM) mode are shown for two transitions, (A) 258.1>142.1 and (B) 258.1>124.1, for two replicates (Rep 1 and Rep 2) of P. falciparum genomic DNA. Calf Thymus DNA and a synthetic standard purchased from Cayman Chemicals served as positive controls, and showed hm5dC (i.e. 5hmC) elution at 2.00 min at room temperature. ? signifies an unknown peak that elutes at approximately 2.75 min for both transitions. The no DNA ‘Matrix’ control included enzymes, inhibitors and other buffers. Note that the y-axis scales of the various samples are not linked.
DISCUSSION
In the past 10 years, cytosine hydroxymethylation has emerged as an integral component of the epigenetic landscape in higher eukaryotes. Nevertheless, its regulatory role in model organisms and lower eukaryotes is relatively unexplored, partly due to the very low levels of 5mC and/or 5hmC in these organisms (0.03–0.06%) (65–67). Importantly, cytosine hydroxymethylation or similar derivatives have never been reported for a eukaryotic pathogen, making our study the first such report. Indeed, our findings that high levels of cytosine hydroxymethylation (0.2–0.4%) is correlated to general gene expression in the malaria parasite emphasize, once again, the need to study epigenetic processes in non-model systems to gain insights into organism-specific, novel modes of gene regulation.
Plasmodium falciparum epigenetic regulation governs pathogenesis in humans (16,17) and therefore, for several decades, there has been interest in studying the role of DNA modifications in P. falciparum gene regulation. A number of reports provided experimental evidence that the parasite genome does not contain the so-called fifth DNA base 5mC (23,24,26) and this was further supported by the absence of DNMT1 and DNMT3 homologues in the malaria parasite (22). One exception was the indirect detection of 5mC within the genomic locus of the dihydrofolate reductase-thymidine synthase (DHFR-TS) gene by restriction mapping (27). In 2013, Ponts et al. reported 5mC in genomic DNA prepared from an asynchronous P. falciparum blood stage culture using MS (0.36% to 1.31%) and BS-seq (28). In the light of our data - we detect very low levels of 5mC by MS in genomic DNA prepared from parasites grown in blood free of white blood cells (Supplementary Figure S9) - the reported values for cytosine methylation by Ponts et al. could be attributed to several experimental shortcomings. First, their methodology for genomic DNA preparation did not eliminate human white blood cells, which are present during standard P. falciparum in vitro culture (relevant to their MS observations); second, it is now well-established that 5mC and 5hmC are not distinguished by BS-seq (29); and third, the authors do not provide any information about the reproducibility of their BS-seq experiments. Given that biological and technical replicates of BS- and oxBS-seq were poorly correlated in our study, our data challenge the results of Ponts et al. Moreover, our subsequent MS analyses demonstrate that a new cytosine modification(s) 5hmC-like exists in the parasite genome, which may be less reactive than 5hmC with the oxidizing agent of oxBS-seq, potassium perruthenate, and more reactive than 5mC and 5hmC with sodium bisulfite. Overall, we caution that BS- and oxBS-seq has technical shortcomings that preclude this method to study cytosine modifications in P. falciparum.
Our study also indicates that the new cytosine form, 5hmC-like, is consistently detected by a reference anti-5hmC monoclonal antibody in asexual stages of P. falciparum. Because this monoclonal antibody has been widely used in several model systems to detect 5hmC (9,55,62–64), and because our biochemical studies are reproducible, we infer that the anti-5hmC antibody is cross-reacting with 5hmC-like in parasite genomic DNA. A similar cross-reactivity has been observed for RNA modifications such as N6-methyladenosine and N6,2′-O-dimethyladenosine (m6Am) (68), and other DNA and RNA modifications (69). The molecular basis of cross-reactivity awaits the chemical chracterization of 5hmC-like. Nonetheless, the application of anti-5hmC antibodies in hmeDIP-seq to study a new cytosine mark and its genome-wide distribution is an important outcome of our study. hmeDIP-seq also overcomes any concerns that arise from incomplete chemical conversion in BS- and oxBS-seq (70–72), especially in light of the AT-rich nature of the P. falciparum genome, which lacks CpG islands and contains Cs primarily in a CHH context (22).
Intriguingly, our biochemical data reveal a low methylation/hydroxymethylation ratio in malaria parasites, which contrasts to the situation described in a number of mammalian cell types, where 5mC is detected at higher levels compared to 5hmC (55–57). For example, in HEK293 cells, 5mC accounts for 4% of total Cs whilst 5hmC accounts for 0.12% (35); these numbers are even lower in human pancreatic cells (73) (Supplementary Figure S1B). The exceptions include ESCs and different brain tissues, where 5hmC has been detected at elevated levels within gene bodies and has been associated to a role in gene activation (10–12). In fact, the highest levels of 5hmC have been observed in brain cells from the frontal cortex of adult mice, with 5hmC at 0.87% of total Cs accounting for 17.2% of modified Cs (10) (Supplementary Figure S1B). However, the molecular basis of the dominance of 5hmC-like marks over 5mC in P. falciparum is currently unclear. One possibility is that the high oxidative environment within the infected erythrocyte may enhance the cytosine transition from methylation to hydroxymethylation or other similar derivatives such as 5hmC-like and determine their stability.
The genome-wide distribution pattern of 5hmC-like loci in P. falciparum revealed that this mark is predominantly genic in nature, with more than >95% of 5hmC-like peaks mapping to gene bodies. This has been observed in other cell types of higher eukaryotes where 5hmC was primarily detected in enhancers and transcribed sequences and was depleted from intergenic regions (9–11,62,64,74,75). Consequently, 5hmC-like levels in the gene bodies of several P. falciparum genes positively correlated to mRNA levels, which is in agreement with studies in mouse ESCs (11). Given that a majority of 5hmC-marked genes maintain the mark during asexual blood development, our data suggest that this mark may add a layer of epigenetic control that favors higher transcriptional rates by introducing subtle changes in the chromatin, by influencing nucleosome positioning and/or by recruiting RNA polymerase II to these genes. Nonetheless, genetic validation of the role of 5hmC-like in P. falciparum gene regulation awaits the identification of protein(s) involved in writing and reading this cytosine mark. Our inability to identify an ortholog to mammalian TET proteins by bioinformatic approaches is not surprising: for Plasmodium spp., orthologs of conserved pathways, structures or organelles are frequently difficult to identify by sequence homology alone—for example, only a very small fraction of proteins constituting the nuclear pore complex (NPC) can be identified in P. falciparum using bioinformatics tools (22) although the NPC can be visualized using several imaging approaches (76). We cannot exclude that this pathogen has evolved other pathways for modified cytosine synthesis as has been suggested for plants (77). Alternately, P. falciparum may co-opt the SOS response-associated peptidase (SRAP) domain-containing protein PfSrap1 (PF3D7_1202100) for cytosine dehydroxymethylation as has been observed in mouse embryos (78).
Lastly, we applied mass spectrometry analysis of P. falciparum genomic DNA (free from leukocyte contamination) to revisit the methylation of cytosine. For one, the Plasmodium sample peaks for 5mdC are extremely low (∼0.05% of total dC content) and undetectable for the second qualifier transition (242.1 > 109.1), corroborating our results from the ELISA-based 5mC quantification assay. This is possibly a resonant of the fact that CID fragmentation favours the glycosidic transition 1 (242.1n > 126.1), and the low overall abundance of the modification. Intriguingly, we were unable to detect 5hmdC in the P. falciparum DNA up to a limit of detection of 6 modifications per 107 nucleotides (Supplementary Figure S7), in spite of detecting ‘5hmC’ with various biochemical techniques. Nonetheless, the additional peak at 2.75 min is suggestive of a DNA modification that is similar to a hydroxymethyl-2′-deoxycytidine based on the nominal intact mass and the corresponding predictive fragments. Our method, which relies upon C18 chromatography, would suggest that this modification is more hydrophobic in nature than 5hmdC and could be a positional isomer, for example at position 4 of the nucleotide ring instead of position 5. The presence of a similar cryptic peak in the transitions for 5mdC at 5.72 min lends credence to the suggestion that this might be the precursor to the 5hmC-like modification. These observations based on the LC-MS chromatogram are, however, speculative, must be interpreted with caution and need further validation, which is beyond the scope of this report.
The study of DNA modifications in malaria parasites has also identified N6-Methyladenosine (m6A) in the genome of P. falciparum using a restriction enzyme-based approach (79) and based on the kinetics of the DNA polymerase during single molecule real-time sequencing and de novo genome assembly (80), predicted the genome-wide distribution of m6A and m4C; however, these marks need to be validated by MS studies. African trypanosomes have evolved a unique DNA modification called β-D-Glucopyranosyloxymethyluracil or Base J that acts as transcription terminator in certain kinetoplastid protozoa (81). Base J is generated by hydroxylation and a subsequent glycosylation of thymidine by an as yet unknown glycosyltransferase (82). This illustrates that different eukaryotic pathogens use diversified DNA modification mechanisms to enable parasitism. Additionally, in the green alga Chlamydomonas reinhardtii, a recent report by Xue et al. identified a new 5mC derivative that is generated by the conjugation of a glyceryl moiety from Vitamin C to the methyl group of 5mC by a Tet homologue (83); this strongly emphasises that our knowledge of DNA cytosine modifications is far from complete. Therefore, besides exploring the exact nature of the 5hmC-like base, it also remains to be seen if oxidative derivatives of 5mC, including 5-carboxylcytosine (5caC), 5-formylcytosine (5fC), 5-hydroxymethyluracil (5hmU) and glyceryl-5mC, all of which are predicted to be epigenetic in nature (8,12,84), exist in P. falciparum.
In conclusion, our study changes the current dogma on DNA cytosine methylation in malaria parasites. The identification of a novel predominant type of cytosine modification points to a new epigenetic layer that contributes to mRNA expression. Our work opens up avenues to explore the pathway that leads to the synthesis of the 5hmC-like DNA modification and the reader protein of this mark. Given that cytidine analogs and DNMT inhibitors block parasite growth ((28) and our unpublished data), our work also raises interest to exploit cytosine modifications for new intervention strategies against malaria parasites.
DATA AVAILABILITY OF DATA AND MATERIALS
All data generated or analysed during this study are included in this published article (and its supplementary information files).
All NGS files generated in this study are available for download from the Europeran Bioinformatics Institute's European Nucleotide Archive (ENA) under project accession number PRJEB32602 (http://www.ebi.ac.uk/ena/data/view/PRJEB32602).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Cameron Macpherson for troubleshooting assistance during bioinformatic analysis and the Plasmodium Genome Database (PlasmoDB) consortium for their continued efforts in maintaining https://plasmodb.org.
Author contributions: Conceptualization S.S.V. and A.S; Methodology S.S.V., E.H. and C.B.; Investigation S.S.V., E.H., Am.S., and C.B.; Analysis S.S.V., E.H., Am.S., and G.A.; Writing – S.S.V., E. H., G.A., P.C.D., and A.S.; Funding Acquisition A.S.
Notes
Present address: Shruthi S. Vembar, Institute of Bioinformatics and Applied Biotechnology, Bengaluru 560100, India; Guruprasad Ananda, Dana-Farber Cancer Institute, Boston 02215, MA, USA
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council Advanced Grant [PlasmoSilencing 670301 to A.S.]; French Parasitology consortium ParaFrap [ANR-11-LABX0024 to A.S.]; Institut Carnot-Pasteur Maladies Infectieuses Fellowship (to S.S.V.); National Research Foundation, Singapore, under its Singapore‐MIT Alliance for Research and Technology (SMART) Centre, Anti-Microbial Resistance IRG (to P.R.P.); Singapore-MIT Alliance (SMA) Graduate Fellowship Program (to Am.S); Funding for open access charge: PlasmoSilencing 670301.
Conflict of interest statement. None declared.
REFERENCES
- 1. Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007; 447:433–440. [DOI] [PubMed] [Google Scholar]
- 2. Law J.A., Jacobsen S.E.. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010; 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mikeska T., Craig J.M.. DNA methylation biomarkers: cancer and beyond. Genes (Basel). 2014; 5:821–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeltsch A., Jurkowska R.Z.. New concepts in DNA methylation. Trends Biochem. Sci. 2014; 39:310–318. [DOI] [PubMed] [Google Scholar]
- 5. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007; 447:425–432. [DOI] [PubMed] [Google Scholar]
- 6. Kriaucionis S., Heintz N.. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009; 324:929–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al.. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pastor W.A., Aravind L., Rao A.. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013; 14:341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu H., D’Alessio A.C., Ito S., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y.. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011; 25:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D. et al.. Global epigenomic reconfiguration during mammalian brain development. Science. 2013; 341:626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mellén M., Ayata P., Dewell S., Kriaucionis S., Heintz N.. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012; 151:1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu X., Zhang Y.. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat. Rev. Genet. 2017; 18:517–534. [DOI] [PubMed] [Google Scholar]
- 13. Al-Mahdawi S., Virmouni S.A., Pook M.A.. The emerging role of 5-hydroxymethylcytosine in neurodegenerative diseases. Front. Neurosci. 2014; 8:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madrid A., Papale L.A., Alisch R.S.. New hope: the emerging role of 5-hydroxymethylcytosine in mental health and disease. Epigenomics. 2016; 8:981–991. [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Tang J., Lai M., Zhang H.. 5-Hydroxymethylcytosine and disease. Mutat. Res. Rev. Mutat. Res. 2014; 762:167–175. [DOI] [PubMed] [Google Scholar]
- 16. Cortés A., Deitsch K.W.. Malaria Epigenetics. Cold Spring Harb. Perspect. Med. 2017; 7:a025528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duraisingh M.T., Horn D.. Epigenetic regulation of virulence gene expression in parasitic protozoa. Cell Host Microbe. 2016; 19:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hailu G.S., Robaa D., Forgione M., Sippl W., Rotili D., Mai A.. Lysine deacetylase inhibitors in parasites: past, present, and future perspectives. J. Med. Chem. 2017; 60:4780–4804. [DOI] [PubMed] [Google Scholar]
- 19. Malmquist N.A., Moss T.A., Mecheri S., Scherf A., Fuchter M.J.. Small-molecule histone methyltransferase inhibitors display rapid antimalarial activity against all blood stage forms in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16708–16713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malmquist N.A., Sundriyal S., Caron J., Chen P., Witkowski B., Menard D., Suwanarusk R., Renia L., Nosten F., Jiménez-Díaz M.B. et al.. Histone methyltransferase inhibitors are orally bioavailable, fast-acting molecules with activity against different species causing malaria in humans. Antimicrob. Agents Chemother. 2015; 59:950–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trenholme K., Marek L., Duffy S., Pradel G., Fisher G., Hansen F.K., Skinner-Adams T.S., Butterworth A., Ngwa C.J., Moecking J. et al.. Lysine acetylation in sexual stage malaria parasites is a target for antimalarial small molecules. Antimicrob. Agents Chemother. 2014; 58:3666–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S. et al.. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002; 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Choi S.W., Keyes M.K., Horrocks P.. LC/ESI-MS demonstrates the absence of 5-methyl-2′-deoxycytosine in Plasmodium falciparum genomic DNA. Mol. Biochem. Parasitol. 2006; 150:350–352. [DOI] [PubMed] [Google Scholar]
- 24. Gissot M., Choi S.W., Thompson R.F., Greally J.M., Kim K.. Toxoplasma gondii and Cryptosporidium parvum lack detectable DNA cytosine methylation. Eukaryot. Cell. 2008; 7:537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Govindaraju G., Jabeena C.A., Sethumadhavan D.V., Rajaram N., Rajavelu A.. DNA methyltransferase homologue TRDMT1 in Plasmodium falciparum specifically methylates endogenous aspartic acid tRNA. Biochim. Biophys. Acta. 2017; 1860:1047–1057. [DOI] [PubMed] [Google Scholar]
- 26. Pollack Y., Katzen A.L., Spira D.T., Golenser J.. The genome of Plasmodium falciparum. I: DNA base composition. Nucleic Acids Res. 1982; 10:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pollack Y., Kogan N., Golenser J.. Plasmodium falciparum: evidence for a DNA methylation pattern. Exp. Parasitol. 1991; 72:339–344. [DOI] [PubMed] [Google Scholar]
- 28. Ponts N., Fu L., Harris E.Y., Zhang J., Chung D.W., Cervantes M.C., Prudhomme J., Atanasova-Penichon V., Zehraoui E., Bunnik E.M. et al.. Genome-wide mapping of DNA methylation in the human malaria parasite Plasmodium falciparum. Cell Host Microbe. 2013; 14:696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., Rao A.. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010; 5:e8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trager W., Jensen J.B.. Human malaria parasites in continuous culture. Science. 1976; 193:673–675. [DOI] [PubMed] [Google Scholar]
- 31. Lelièvre J., Berry A., Benoit-Vical F.. An alternative method for Plasmodium culture synchronization. Exp. Parasitol. 2005; 109:195–197. [DOI] [PubMed] [Google Scholar]
- 32. Lambros C., Vanderberg J.P.. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979; 65:418–420. [PubMed] [Google Scholar]
- 33. Moll K, Kaneko A, Scherf A, Wahlgren M. Methods in malaria research. 2013; Sixth Edition.
- 34. Bowen B., Steinberg J., Laemmli U.K., Weintraub H.. The detection of DNA-binding proteins by protein blotting. Nucleic Acids Res. 1980; 8:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y.. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011; 333:1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chene A., Vembar S.S., Riviere L., Lopez-Rubio J.J., Claes A., Siegel T.N., Sakamoto H., Scheidig-Benatar C., Hernandez-Rivas R., Scherf A.. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 2012; 40:3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011; 17:doi:10.14806/ej.17.1.200. [Google Scholar]
- 38. Krueger F., Andrews S.R.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000, G.P.D.P.S. . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012; 13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song Q., Decato B., Hong E.E., Zhou M., Fang F., Qu J., Garvin T., Kessler M., Zhou J., Smith A.D.. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One. 2013; 8:e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Supek F., Bošnjak M., Škunca N., Šmuc T.. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 49. Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O. et al.. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014; 10:574–581. [DOI] [PubMed] [Google Scholar]
- 50. Ng C.S., Sinha A., Aniweh Y., Nah Q., Babu I.R., Gu C., Chionh Y.H., Dedon P.C., Preiser P.R.. tRNA epitranscriptomics and biased codon are linked to proteome expression in Plasmodium falciparum. Mol. Syst. Biol. 2018; 14:e8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cao H., Wang Y.. Collisionally activated dissociation of protonated 2′-deoxycytidine, 2′-deoxyuridine, and their oxidatively damaged derivatives. J. Am. Soc. Mass Spectrom. 2006; 17:1335–1341. [DOI] [PubMed] [Google Scholar]
- 52. Inc.b, P.T. Collaborative Data Science. 2015; Montreal, QC: Plotly Technologies Inc. [Google Scholar]
- 53. Zhang Q., Huang Y., Zhang Y., Fang X., Claes A., Duchateau M., Namane A., Lopez-Rubio J.J., Pan W., Scherf A.. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011; 10:451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zanghì G., Vembar S.S., Baumgarten S., Ding S., Guizetti J., Bryant J.M., Mattei D., Jensen A.T.R., Rénia L., Goh Y.S. et al.. A Specific PfEMP1 Is Expressed in P. falciparum Sporozoites and Plays a Role in Hepatocyte Infection. Cell Rep. 2018; 22:2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li W., Liu M.. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids. 2011; 2011:870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nestor C.E., Ottaviano R., Reddington J., Sproul D., Reinhardt D., Dunican D., Katz E., Dixon J.M., Harrison D.J., Meehan R.R.. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012; 22:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ruzov A., Tsenkina Y., Serio A., Dudnakova T., Fletcher J., Bai Y., Chebotareva T., Pells S., Hannoun Z., Sullivan G. et al.. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011; 21:1332–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foth B.J., Zhang N., Chaal B.K., Sze S.K., Preiser P.R., Bozdech Z.. Quantitative time-course profiling of parasite and host cell proteins in the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics. 2011; 10:M110.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bozdech Z., Llinás M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L.. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003; 1:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Booth M.J., Ost T.W., Beraldi D., Bell N.M., Branco M.R., Reik W., Balasubramanian S.. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat. Protoc. 2013; 8:1841–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Booth M.J., Branco M.R., Ficz G., Oxley D., Krueger F., Reik W., Balasubramanian S.. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012; 336:934–937. [DOI] [PubMed] [Google Scholar]
- 62. Stroud H., Feng S., Morey Kinney S., Pradhan S., Jacobsen S.E.. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011; 12:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K.. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011; 473:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S. et al.. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011; 42:451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Capuano F., Mülleder M., Kok R., Blom H.J., Ralser M.. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal. Chem. 2014; 86:3697–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Erdmann R.M., Souza A.L., Clish C.B., Gehring M.. 5-hydroxymethylcytosine is not present in appreciable quantities in Arabidopsis DNA. G3 (Bethesda). 2014; 5:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tang Y., Gao X.D., Wang Y., Yuan B.F., Feng Y.Q.. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal. Chem. 2012; 84:7249–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R.. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015; 12:767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Feederle R., Schepers A.. Antibodies specific for nucleic acid modifications. RNA Biol. 2017; 14:1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McInroy G.R., Beraldi D., Raiber E.A., Modrzynska K., van Delft P., Billker O., Balasubramanian S.. Enhanced methylation analysis by recovery of unsequenceable fragments. PLoS One. 2016; 11:e0152322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Warnecke P.M., Stirzaker C., Song J., Grunau C., Melki J.R., Clark S.J.. Identification and resolution of artifacts in bisulfite sequencing. Methods. 2002; 27:101–107. [DOI] [PubMed] [Google Scholar]
- 72. Genereux D.P., Johnson W.C., Burden A.F., Stöger R., Laird C.D.. Errors in the bisulfite conversion of DNA: modulating inappropriate- and failed-conversion frequencies. Nucleic Acids Res. 2008; 36:e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bhattacharyya S., Pradhan K., Campbell N., Mazdo J., Vasantkumar A., Maqbool S., Bhagat T.D., Gupta S., Suzuki M., Yu Y. et al.. Altered hydroxymethylation is seen at regulatory regions in pancreatic cancer and regulates oncogenic pathways. Genome Res. 2017; 27:1830–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X. et al.. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011; 29:68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mellén M., Ayata P., Heintz N.. 5-hydroxymethylcytosine accumulation in postmitotic neurons results in functional demethylation of expressed genes. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E7812–E7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weiner A., Dahan-Pasternak N., Shimoni E., Shinder V., von Huth P., Elbaum M., Dzikowski R.. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell. Microbiol. 2011; 13:967–977. [DOI] [PubMed] [Google Scholar]
- 77. Shi D.Q., Ali I., Tang J., Yang W.C.. New insights into 5hmC DNA modification: generation, distribution and function. Front. Genet. 2017; 8:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kweon S.M., Zhu B., Chen Y., Aravind L., Xu S.Y., Feldman D.E.. Erasure of Tet-Oxidized 5-Methylcytosine by a SRAP Nuclease. Cell Rep. 2017; 21:482–494. [DOI] [PubMed] [Google Scholar]
- 79. Luo G.Z., Wang F., Weng X., Chen K., Hao Z., Yu M., Deng X., Liu J., He C.. Characterization of eukaryotic DNA N(6)-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat. Commun. 2016; 7:11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Vembar S.S., Seetin M., Lambert C., Nattestad M., Schatz M.C., Baybayan P., Scherf A., Smith M.L.. Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (>11 kb), single molecule, real-time sequencing. DNA Res. 2016; 23:339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. van Luenen H.G., Farris C., Jan S., Genest P.A., Tripathi P., Velds A., Kerkhoven R.M., Nieuwland M., Haydock A., Ramasamy G. et al.. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012; 150:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Borst P., Sabatini R.. Base J: discovery, biosynthesis, and possible functions. Annu. Rev. Microbiol. 2008; 62:235–251. [DOI] [PubMed] [Google Scholar]
- 83. Xue J.H., Chen G.D., Hao F., Chen H., Fang Z., Chen F.F., Pang B., Yang Q.L., Wei X., Fan Q.Q. et al.. A vitamin-C-derived DNA modification catalysed by an algal TET homologue. Nature. 2019; 569:581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu H., Zhang Y.. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011; 25:2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files).
All NGS files generated in this study are available for download from the Europeran Bioinformatics Institute's European Nucleotide Archive (ENA) under project accession number PRJEB32602 (http://www.ebi.ac.uk/ena/data/view/PRJEB32602).