Abstract
Data from earlier studies showed that minor structural changes at the surface of cytochrome c oxidase, near one of the proton-input pathways (the D pathway), result in dramatically decreased activity and a lower proton-pumping stoichiometry. To further investigate how changes around the D pathway orifice influence functionality of the enzyme, here we modified the nearby C-terminal loop of subunit I of the Rhodobacter sphaeroides cytochrome c oxidase. Removal of 16 residues form this flexible surface loop resulted in a decrease in the proton-pumping stoichiometry to <50 % of that of the wild-type enzyme. Replacement of the protonatable residue Glu552, part of the same loop, by an Ala, resulted in a similar decrease in the proton-pumping stoichiometry without loss of the O2-reduction activity or changes in the proton-uptake kinetics. The data show that minor structural changes at the orifice of the D pathway, at a distance of ~40 Å from the proton gate of cytochrome c oxidase, may alter the proton-pumping stoichiometry of the enzyme.
Introduction
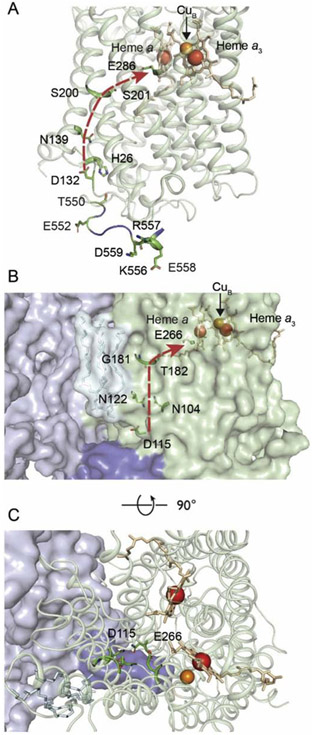
Cytochrome c oxidase (CytcO) is the terminal electron acceptor in the respiratory chain of mitochondria and aerobic bacteria where it receives electrons from cytochrome c and transfers these electrons to O2. The primary electron acceptor in CytcO is CuA, which donates electrons consecutively to an intermediate electron acceptor heme a and the O2-binding catalytic site, which is composed of heme a3 and CuB. The electron transfer is linked to proton pumping from the negative (n) to the positive (p) side of the membrane with a stoichiometry of ~1 H+ per electron transferred to O2 (for reviews on the structure and function of CytcO, see [1-3]). In the canonical type-A1 CytcOs (e.g. from mitochondria, Rhodobacter sphaeroides) [4, 5] protons used in the reduction of O2 to H2O are transferred from the n-side of the membrane to the catalytic site via two pathways called K and D, respectively. The latter pathway is also used for uptake of protons that are pumped across the membrane. It starts at Asp132 (R. sphaeroides numbering), at the n-side surface, and is composed of ~10 water molecules that span the distance from Asp132 to Glu286, both highly conserved and functionally essential residues of the D pathway [6, 7]. The Asp132 residue is located in the inner part of a cone-shaped cleft (Figure 1A), which is decorated by protonatable residues that form a protoncollecting antenna (Figure 1A) [8, 9].
Figure 1. The R. sphaeroides (A) and M. smegmatis (B, C) A1-type CytcOs.
(A) Membrane view of subunit I of the R. sphaeroides CytcO. Five protonatable residues of the C-terminus (E552-D559) are highlighted. Due to the flexibility of the C-terminus, its last six residues, including the protonatable residues E561, R562 and H566, are missing in the structure. The negatively charged E552 is situated about 10 Å from H26. The D pathway, which starts at D132 and ends at E286, is displayed by the red arrow. (B) Membrane view of subunit CtaD (corresponding to subunit I in R. sphaeroides CytcO) and subunit QcrB in M. smegmatis. A loop (blue), extending from subunit QcrB (light purple) of the cyt. bcc complex forms a "lid" that covers the entry point (D115) of the D pathway. The D pathway (D115 to E266) is shown by the red arrow. (C) Same as in B, but a top view. The figures were prepared using the program PyMOL from the PDB files 1M56 (R. sphaeroides) and 6HWH (M. smegmatis).
Results from earlier studies with e.g. the R. sphaeroides CytcO showed that replacement of Asp132 by a non-protonatable analog results in loss of activity and proton pumping [9, 10], but the effect of the mutation could be reversed by altering residues further "up" the D pathway [11], which suggests plasticity in the structural design of the D pathway and that the proton-transfer rates could be modulated by minor structural changes. Consequently, the question arises whether also structural changes outside of the D pathway, at the n-side surface, may alter the turnover activity or proton-pumping stoichiometry of the CytcO. Because the D pathway architecture is conserved among the A1 CytcOs [4, 5] and the functional properties of the CytcOs investigated to date are very similar, it is likely that the results obtained with any of the A1 CytcOs can be generalized.
In the R. sphaeroides CytcO residues near the orifice of the D pathway are found at the C terminus of subunit I. The last 16 amino-acid residues of this C terminus (residues 551-566, Figure 1A) are not observed in the two-subunit crystal structure of the CytcO [12], indicating that this tail may be flexible in nature [13]. The C terminus contains a large fraction protonatable residues (4 Asp/Glu, 1 His, 3 Arg/Lys), which are likely to collectively contribute to facilitate proton uptake. The C-terminal loop of subunit I is bound to residues of subunit III, but the last six residues of this loop are flexible, hence are not resolved in the four-subunit structure (residues 561-566, Figure 1A).
To further investigate the effect of changes at the protein surface on proton uptake by the CytcO, here, we prepared structural variants of CytcO from R. sphaeroides in which the 16-residue C-terminal loop of subunit I was altered. The study was motivated by analysis of a recently determined structure of a supercomplex composed of cyt. bcc and cytochrome c oxidase from Mycobacterium smegmatis [14, 15]. This structure showed that a C-terminal loop of the QcrB subunit of the cyt. bcc complex forms a "lid" that covers the orifice of the D pathway [14], which suggests a mechanism to regulate the proton-pumping stoichiometry and activity of this supercomplex by structural changes at the D pathway orifice. In the present study the C-terminal loop at the R. sphaeroides CytcO was either shortened or removed (CiΔ6, CiΔ16), or the first protonatable residue of the loop (i.e. the 16th residue from the C terminus), Glu552, located at the protein surface ~10 Å "below" Asp132, was replaced by a non-protonatable Ala (Glu552Ala). We crystallized the CiΔ16 as well as the CiΔ6 CytcO variants under conditions where the two-subunit CytcO forms crystals [12] and determined their structures. To investigate the effects of these alterations we measured the activity, proton-pumping stoichiometry as well as the electron and proton-uptake kinetics. The data show that the Glu552Ala replacement resulted in slowing proton uptake during O2 reduction and lowering the proton pumping stoichiometry to <50 % of that of the wild-type CytcO.
Results and Discussion
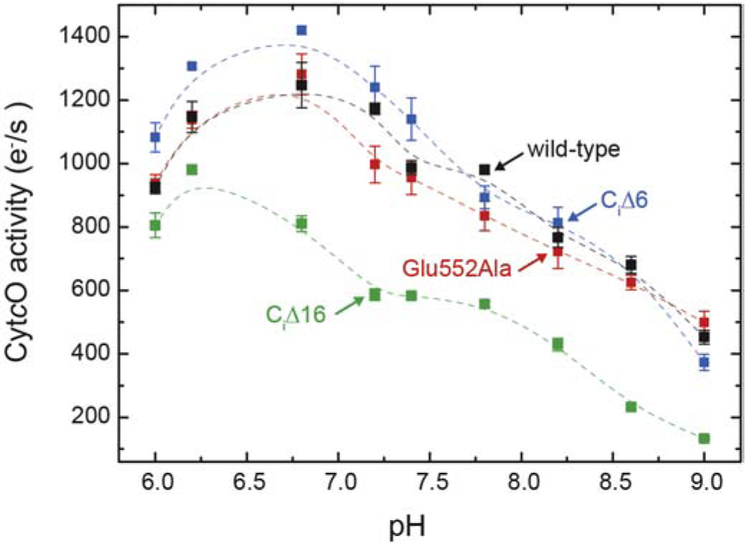
We first removed the entire 16-residue C terminus of the R. sphaeroides CytcO subunit I (CiΔ16). The activity was measured by following in time O2 reduction using a Clark electrode. As seen in Figure 2 the CiΔ16 CytcO displayed a lower activity as compared to the wild-type enzyme, but this lower activity was most likely a consequence of the smaller fraction of bound subunit III (see Figure S1) [16]. We also prepared a variant in which six residues from the C terminus of subunit I (CiΔ6) were removed (see description of the structure below). This construct yielded fully active CytcO without loss of subunit III. We also prepared a structural variant in which the first acidic residue of the 16-residue terminus, Glu552 (16 residues from the C terminus), was replaced by Ala. Here, the charge properties of the loop were altered as close to Asp132 as possible with retained subunit III. As seen in Figure S1, even though the subunit III band for the Glu552Ala variant was more narrow than that of the wild-type CytcO, it was stronger than that of CiΔ16 CytcO, which suggests that a larger fraction of the CytcO was intact with the single mutant CytcO. Furthermore, as seen in Figure 2, the steady-state activity of the purified Glu552Ala CytcO was the same as that of the wild-type CytcO in the entire measured pH range, which suggests that subunit III remained bound in the mutant. This conclusion is drawn because results from earlier studies (e.g. [16-18]) showed that the O2-reduction activity of CytcO without subunit III dropped to about 10 % of that of the wild-type CytcO above pH 8 (~100 s−1 at pH 8 and <20 s−1 at pH 9-10).
Figure 2. Steady-state O2 reduction activity.
The activities (in electrons per second per CytcO molecule) for the wild-type CytcO as well as the CiΔ6, CiΔ16, Glu552Ala variants (as indicated) were measured using an O2-electrode. See the Materials and Methods section for experimental conditions. Errors are SD based on three measurements. The lines are guides for the eye.
Proton pumping
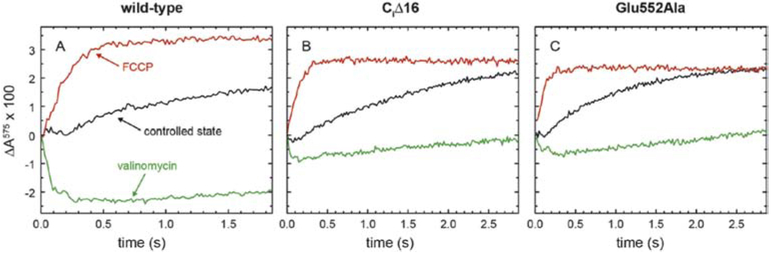
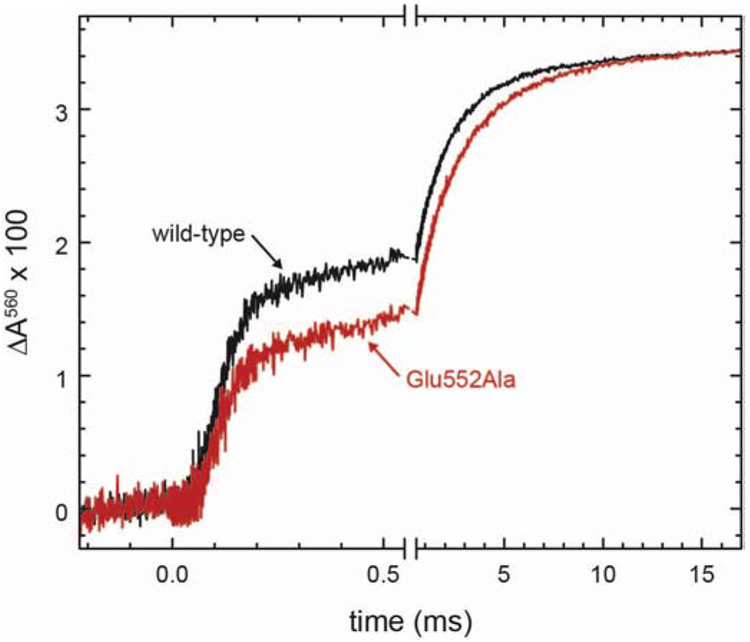
To determine the proton-pumping stoichiometry of the CytcO we reconstituted the enzyme into lipid vesicles and monitored absorbance changes of the pH dye phenol red, outside of the liposomes, upon addition of cyt. c to initiate CytcO turnover in a stopped-flow apparatus. The initial lag phase or slight decrease in absorbance is indicative of an initial proton extrusion to the outside of the liposomes (black traces, Figure 3). The appearance of this initial lag is dependent, e.g. on the relative rates of proton pumping and proton leaks in the controlled state. The slower increase in absorbance is due to a transmembrane proton leak across the membrane. Because this increase in absorbance was slower than that seen with the proton and K+ ionophores (red traces in Figure 3, see below), the data show that the liposomes were sufficiently tight to protons to allow measurements of proton pumping.
Figure 3. Proton pumping.
CytcO reconstituted in liposomes was mixed with ascorbate and cyt. c in the presence of O2. Absorbance changes of the pH dye phenol red, associated with changes in the proton concentration, were measured as a function of time (black traces). The green traces were obtained after addition of the K+ ionophore valinomycin (2 μM), while the red traces were obtained after addition of both valinomycin and the proton ionophore FCCP (5 μM). Experimental conditions: 4.2 μM cyt. c, 0.04 μM CytcO in liposomes, which contained 50 mM HEPES-KOH, pH 7.4, 43 mM KCl.
When the same experiment was done in the presence of the K+ ionophore valinomycin we observed a decrease in absorbance associated with proton release to the outside of the liposomes (green traces, Figure 3). Valinomycin short-circuits the electrical component of the electrochemical gradient thereby relieving the membrane potential, allowing CytcO turnover and protons to be pumped. In the presence of the proton ionophore FCCP as well as valinomycin, the membrane is fully permeable to both H+ and K+(red traces in Figure 3). As a result, a net proton uptake is observed associated with O2 reduction to H2O. Because the stoichiometry of this proton uptake is well defined, 1 H+/e−, the signal could be used for normalization. Comparison of the amplitudes of the red and green traces yields the proton pumping stoichiometry, which was ~30 % and ~50 % of that of the wild-type CytcO for the CiΔ16 and Glu552Ala CytcOs, respectively. Hence, structural changes at the n-side surface of the CytcO resulted in lowering the proton-pumping stoichiometry of the CytcO. The lower stoichiometry obtained with the CiΔ16 than with the Glu552Ala CytcO is likely to be caused by the fractional loss of subunit III in the former (Figure S1) [16, 17, 19, 20], which is discussed in detail below.
Single-turnover electron transfer and proton uptake
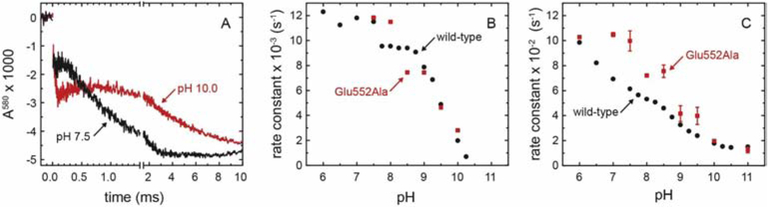
The purified Glu552Ala CytcO was reduced by four electrons per enzyme molecule and incubated under an atmosphere of CO, which results in formation of the reduced CytcO-CO complex. The CytcO-CO complex was mixed with an O2-saturated solution, followed in time by flash-photolysis of the CytcO-CO complex. The dissociation of the CO ligand allows O2 to bind to heme a3 to initiate the reaction, which was monitored using time-resolved absorption spectroscopy at wavelengths that are specific to redox changes at the cofactors of the CytcO [21]. At 580 nm (Figure 4A) the initial, unresolved decrease in absorbance is associated with dissociation of CO from heme a3 forming the reduced state of the CytcO, called R. Next, O2 binds to heme a3 with a time constant of ~10 μs at 1 mM O2, but this kinetic component is not resolved in Figure 4A. The first observed kinetic component (decrease in absorbance with a time constant of ~50 μs) is associated with formation of the so-called peroxy state, PR. The amplitude of this component was much smaller at pH 7.5 than at pH 10 because at the lower pH it is followed in time by an increase in absorbance, associated with formation of the ferryl state, F, with a similar time constant of 100 μs. This increase in absorbance was slower at pH 10 (τ ≅ 250 μs), which explains why the amplitude of the 50-μs absorbance decrease was larger at the higher pH. The slowest decrease in absorbance, with time constants of 1 ms and 5 ms at pH 7.5 and pH 10, respectively, is associated with the final oxidation step of the CytcO, which yields state O. A similar behavior was observed earlier for the wild-type CytcO [22].
Figure 4. Electron-transfer kinetics.
The reduced CytcO was incubated under an atmosphere of CO and mixed in a stopped-flow apparatus with an O2-saturated solution. (A) Absorbance changes at 580 nm. The reaction was initiated at t = 0 by photolysis of the CytcO-CO complex (unresolved decrease in absorbance). The following small decrease in absorbance is associated with formation of the PR state. The increase in absorbance with a time constant of ~100 μs or ~400 μs at pH 7.5 and pH 10, respectively, is associated with the PR → F reaction. The final decrease in absorbance with time constants of 1 ms and 5 ms, respectively, is associated with formation of the oxidized (O) CytcO. The traces are scaled to 1 μM reacting enzyme. A laser artifact at t = 0 has been truncated for clarity. (B,C) pH dependence of the PR → F (B) and F → O (C) rate constants (red) compared to the equivalent rates obtained with the wild-type CytcO (black trace, [23, 43]).
The rates in (B) were obtained from measurements at 580 nm while those in (C) were obtained from measurements at 445 nm, 580 nm and 605 nm. The error bars are SD based on 2-4 measurements. Experimental conditions: the mixing ratio was 1:5. Syringe 1 contained 5 μM CytcO, 5 mM Bis-trispropane pH 8.0, 100 μM EDTA, 0.05 % DDM. Syringe 2 contained 33 mM of Bis-tris-propane, CAPS, and CHES supplemented with 100 μM EDTA, 0.05 % DDM.
Panels B, C in Figure 4 show the pH dependence of the two reactions that are linked to proton uptake and pumping, i.e. PR → F and F → O, respectively. The pH dependence of the former reaction is determined by the protonation state of Glu286 [23], which has a pKa of ~9.4. As seen in the figure, this pKa in the pH dependence of the PR → F reaction was unaltered in the Glu552Ala variant indicating the pKa of Glu286 was unaltered. Furthermore, the data indicate that the kinetics of proton transfer through the D pathway was unaltered by the Glu552Ala replacement.
The pH dependence of the F → O rate titrates in two regimes with one pKa of ~8.5, presumably also determined by the protonation state of Glu286, and a pKa <6.5 [10], which has previously been attributed to reflect the protonation state of a heme propionate [24].
As already discussed above, we argue that subunit III is lost in the CiΔ16 variant, but intact in the major fraction of Glu552Ala population. We note that results from earlier studies showed that loss of subunit III results in a shift in the pH dependence of the F → O reaction to a lower pH range, yielding a decrease in the pKa by ~1.8 units [18]. The data from the present study show qualitatively a shift in the pH dependence in the opposite direction, i.e. the F → O rate with the Glu552Ala CytcO titrated at higher pH values than the wild-type CytcO. Hence, these data suggest that the effect of the mutation on the F → O rate is not caused by loss of subunit III.
It has been suggested that surface groups around the D pathway orifice may be involved in controlling the pH dependence of proton uptake [25], This scenario may explain the absence of the lower pKa in the pH titration of the F → O rate with the Glu552Ala variant.
Next, we measured proton uptake with detergent-solubilized CytcO by following in time absorbance changes of the pH dye phenol red added to the solution in the absence of buffer (Figure 5). With the wild-type CytcO, two components are observed [26], overlapping in time with the PR → F and F → O reactions, respectively, and with approximately equal amplitudes (~0.8 H+ in each reaction) [21]. The same two components were observed with the Glu552Ala variant, however, the amplitude of the faster, PR → F, component was 30 ± 7 % of the total absorbance change, i.e. equivalent to 0.5 ± 0.1 H+/CytcO while proton uptake during F → O amounted to 1.1 ± 0.1 H+/CytcO (SD of 11 and 6 measurements on each of two samples). The numbers were obtained from a fit of the data with a sum of two exponential functions. The data report the net proton uptake because the reaction was studied in detergent solution. Consequently, the signal amplitudes are expected to be the same for the wild-type and Glu552Ala CytcOs because the net reaction that is observed is reduction of O2 to H2O by the reduced CytcO. However, the relative amplitudes of the two components are modulated by the uptake and release of protons that are pumped. Even though proton uptake and release typically display the same time constants with e.g. the mitochondrial or R. sphaeroides wild-type CytcO [27, 28], a fraction of protons that are taken up during the PR → F reaction may be released in the F → O reaction, as has been observed with the wild-type ba3 oxidase from T. thermophilus [29]. The smaller amplitude with the Glu552Ala variant would then explain the increase of the absorbance-change amplitude of the F → O component on the expense of that of the PR → F component.
Figure 5. Proton-uptake kinetics.
The reduced CytcO was incubated under an atmosphere of CO in a solution containing the pH dye phenol red, in the absence of buffer. The experiment was carried out as described in Figure 4. An increase in absorbance is associated with proton uptake. For comparison of the relative amplitudes of the two kinetic components, the Glu552Ala (red) and wild-type (black) traces have been scaled to yield the same total amplitude. To remove absorbance changes that are not associated with changes in protonation, the experiments were repeated with buffer at pH 7.8 (syringe 2 contained 100 mM HEPES pH 7.8 instead of KCl) and the absorbance changes were subtracted from those obtained in the absence of buffer. Experimental conditions before mixing (1:1): syringe 1 contained: 100 mM KCl (pH 7.8), 100 μM EDTA, 40 μM phenol red, 0.05 % DDM; 2 mM ascorbate; 0.2 μM hexaammineruthenium(11) chloride (HexaRu(11)), 10-20 μM CytcO. Syringe 2 contained an oxygen-saturated solution (~1.2 mM) with the same composition as that in syringe 1, except that no ascorbate/HexaRu(11) were added.
Structures
Results from earlier studies have shown that the removal of the flexible portions of proteins can improve crystal quality. As already discussed above, the 16 amino acid residues at the C terminus of subunit I of the R. sphaeroides CytcO subunit I is presumably flexible and not observed in the two-subunit structural model of the CytcO. The last six residues were not seen in the four-subunit model. Here, we crystallized both the CiΔ16 and CiΔ6 CytcO variants and under conditions where the two-subunit CytcO forms crystals [12] and observed X-ray diffraction at 2.1 Å and 2.5 Å, respectively (Table S1). Although the crystal of CiΔ16 has a similar space group (P212121) to that seen with the wild-type CytcO, the cell dimensions of the crystals were slightly different from those of wild-type crystals (Table S1). This difference might be caused by the C-terminal deletion, which influences the crystal packing. Consequently, molecular replacement, instead of rigid body refinement, was performed to obtain the correct phase information of this crystal. The data show that the overall structure of this variant is essentially the same as that of the wild-type CytcO with structural changes only around the site of mutation (T550 region, Figure 6). The two-subunit crystals of CiΔ6 CytcO were essentially identical to those of the wild-type CytcO having the same space group (P212121) similar unit cell dimensions and crystal contacts. Even though we did not obtain the crystal structure of the Glu552Ala variant, the structural data from the more major alterations suggest that this single residue replacement would result in changes only around the orifice of the D pathway.
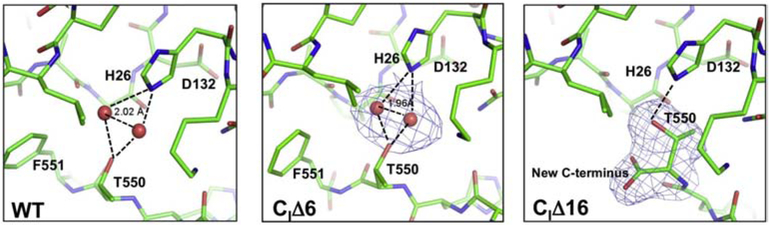
Figure 6. Structures of the C-terminus of wild-type, CiΔ6, and CiΔ16.
The major structural changes in the CiΔ16 strain (PDB ID: 6PW1) were found in the T550 region. In the wild-type [12] and CiΔ6 (PDB ID: 6PW0) CytcOs, there are electron densities attributed to two water molecules or a metal ion (e.g. Mg2+) and a water molecule bound between T550 and H26. In CiΔ16, the loss of the last 16 residues of subunit I make T550 the new C-terminus. Here the T550 side chain is folded closer to H26, forming a hydrogen bond. This region is very close to the entrance of the D-pathway at residue D132.
Conclusions
The data show that the proton-pumping stoichiometry of the Glu552Ala structural variant was <50 % of that of the wild-type CytcO. A similar effect was noted for the CiΔ16 CytcO in which Glu552 was also removed, but in this variant subunit III was lost in a fraction of the CytcOs. This observation complicates interpretation of the data because loss of subunit III also yields CytcO with a ~50 % proton pumping stoichiometry [17, 19, 20], presumably as a result of removal of subunit III residues that extend toward the orifice of the D pathway in subunit I [16]. As outlined above, we exclude the loss of subunit III in the Glu552Ala variant for the following reasons: (i) The subunit III band in the SDS-PAGE was stronger than with the CiΔ16 variant (although it was less broad than with the wild-type CytcO, as discussed above). (ii) The activity of CytcO without subunit III is lower than that of the wild-type CytcO at pH≥7 and approaches ~10 % at pH >8 [16-18], while the activity of the Glu552Ala variant was the same as that of the wild-type CytcO (Figure 2). (iii) The pH dependence of the F → O reaction with CytcO without subunit III was shifted to lower pH values [18], but with the Glu552Ala it was shifted to higher pH values (Figure 4C). (iv) Proton uptake during the PR → F reaction (τ ≅ 100 μs) was not observed without subunit III [18], but it was observed with the Glu552Ala variant.
The observation of a lower proton-pumping stoichiometry with the Glu552Ala variant is thus intriguing because the segment of the CytcO that controls proton pumping (often referred to as the proton-loading site, PLS) is presumably located in the membrane-spanning region of the CytcO, above the heme groups, i.e. at least 40 Å from Asp552 (see e.g. [1-3, 30, 31]). A lower proton-pumping stoichiometry was also observed earlier upon replacing specific residues along the D pathway (summarized in [32]). These variants could be divided into two categories, those in which the proton-uptake rate through the D pathway was slowed and those in which it was unaltered [9, 10]. The Glu552Ala variant studied here belongs to the latter category. Another example of a structural alteration that yields CytcO of this latter category is replacement of Asn139 by Asp, which resulted in completely impaired proton pumping while the proton-transfer rate through the D pathway was retained [22]. The residue is located at the lower part of the D pathway at a considerable distance from Glu286 and the PLS. Siegbahn and Blomberg presented data based on theoretical calculations, which showed that the lower pumping stoichiometry could be explained by an increased back-leak of protons from the PLS to the n-side, caused by changes in the energy level of the "resting state" for the proton near the orifice of the D pathway [33, 34]. They considered a scenario where proton transfer through the D pathway is initiated by protonation of His26 (Figure 1), which results in rotation of Asp132 placing the side chain close to Asn139 from where the proton is transferred through the pathway toward Glu286. According to these calculations, any changes in the pKa of His26 would alter the proton back-leak rate, which would lower the proton-pumping stoichiometry. Even though residue Glu552, investigated here, is located about 10 Å from His26, these calculations indicate that structural changes near the CytcO surface may influence the stoichiometry of proton pumping.
The data from the present study point to a possible mechanism by which structural changes around the D pathway orifice could modulate the proton-pumping activity of the CytcO. This mechanism may be discussed in the framework of a recently-determined structure of an obligate supercomplex from the gram-positive bacterium Mycobacterium smegmatis. It is composed of a cyt. bcc dimer flanked by two CytcO monomers [14, 15, 35]. The structure of this supercomplex shows that a loop in subunit QcrB (cyt. b of the cyt. bcc complex) consisting of ~100 amino-acid residues forms a "lid" that covers the Asp132 cleft [14] (Asp115 in M. smegmatis) (see Figure 1BC, blue). This arrangement would provide a mechanism to regulate the CytcO activity and pumping stoichiometry by changes in the cyt. bcc complex, as illustrated in the present study.
Materials and Methods
Genetic construction of CytcO variants from R. sphaeroides.
The expression strain for CiΔ16 was constructed by removing the last 16 amino acids of the subunit I C-terminus from the plasmid pJL-33. The expression strain for CiΔ6 was created by changing the codon of E561 to a stop codon, in the plasmid pJL-49, so that the last six residues of subunit I were not translated. The modified plasmid constructs were then conjugated into the R. sphaeroides D1D4 strain. To introduce the mutation Glu552Ala (subunit I) into the plasmid PJS3-SH the following PCR primers were used: CCGGAGCATACGTTCGCGCAGCTTCCCAAGCGG (upper primer), CCGCTTGGGAAGCTGCGCGAACGTATGCTCCGG (lower primer). After the mutation, a fragment containing subunit II-III operon genes was inserted into the Pst I site on PJS3-SH to form plasmid pJL-44. The DNA fragment containing subunit I with the Glu552Ala mutation and the subunit II-III operon was then ligated into pRK415-1 to form the expression plasmid pJL-45. pJL-45 was then conjugated into the R. sphaeroides strain Δ123. Histidine tags were added onto either the C-terminus of subunit I (done for Glu552Ala) or the C-terminus of subunit II (done for CiΔ6 and CiΔ16) as described in [36].
Purification of CytcO enzyme.
The R. sphaeroides strains were grown as described in [36]. The proteins of PJL33, pJL-45 and PJL49 were purified with a Ni-NTA FPLC column and a further purification step with a DEAE-5pw column was also performed to facilitate the reconstitution of CytcO into liposomes (see below) [36]. For crystallization, the enzyme was purified via a single step Ni-NTA FPLC purification method [12].
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Approximately 10 μg of protein in sample buffer containing 25 mM Tris, pH 6.5, 40 % glycerol, 8 % SDS and 0.08 % bromophenol blue was loaded onto an 8 % acrylamide stacking gel at pH 6.8 on top of an 18 % acrylamide separating gel at pH 8.8. The gel was electrophoresed at 100 V at room temperature [37]. The gel was then stained with Coomassie blue and destained with 7.5% glacial acetic acid for three times [38]. Polypeptide sizes were estimated from low-molecular weight range markers from New England Biolabs.
CytcO activity assay.
The CytcO activity was measured using a Clark-type oxygen electrode (Gilson model 5/6H). The assay was performed at 25°C in a 1.75 mL reaction cell containing 50 mM KH2PO4, pH 6.5, 0.05 % dodecyl maltoside, 2.8 mM ascorbate, 0.55 mM TMPD and 30 μM of horse heart cytochrome c. The CytcO concentrations were in the range of 5 nM to 50 nM. Soybean phospholipids, asolectin (1 mg/mL) was also added to stimulate the enzyme for full activity. The activity of CytcO is calculated as described in [39, 40].
Reconstitution of CytcO into vesicles.
To investigate proton pumping ability purified CytcO was reconstituted into small unilamellar vesicles made of soybean polar lipid extract (asolectin) obtained from Avanti Polar Lipids (product number 541602) [38]. A cholate dialysis method [38] was used for the reconstitution. Before usage, the asolectin was precipitated with acetone and then extracted with diethyl ether. The cholate was removed by dialysis as previously described [39].
Proton Pumping assay.
For the measurement of alkalization inside the vesicle, 4.2 μM of horse heart cytochrome c was mixed with 0.04 μM of oxidase (in the proteoliposomes; inside buffer was 50 mM HEPES-KOH, pH 7.4, 43 mM KCl). Data scans were collected (1000 scans/s) using a stopped-flow apparatus (Applied Photophysics).
Flow-flash kinetics.
Flow-flash experiments were performed to study the single-turnover oxidation kinetics of CytcO as described in [41, 42]. Briefly, the purified CytcO was transferred to a Thunberg cuvette and air was exchanged for N2. The sample was fully reduced by addition of 2 mM ascorbate and 1 mM phenazine methosulfate (PMS). The N2 atmosphere was then exchanged for CO. Absorbance spectra were acquired on a Cary 4000 spectrophotometer (Agilent) to monitor the redox state of CytcO and the binding of CO to heme a3. The kinetic measurements were done using a flow-flash instrument (Applied Photophysics). The delay time between mixing and photo-dissociation of CO was 200 ms and the cuvette path length was 1.00 cm (for other conditions, see the Figure legend of Figure 4). The kinetic data were analyzed using the Pro-K software (Applied Photophysics).
Proton uptake measurements.
A PD-10 column (GE Healthcare) was used to exchange the sample buffer to 100 mM KCl (pH 7.8), 100 μM EDTA, 0.05 % DDM. The pH dye phenol red (40 μM) was added to the sample and the pH was set to 7.8. The sample was then prepared as for the flow-flash experiments (see above) except that 0.2 μM hexaammineruthenium(II) chloride was used as an electron mediator (see also the legend of Figure 5). The exhaust solution from the stopped-flow apparatus (unbuffered solution) was collected and titrated with HCl for calibration purposes.
Crystallization.
Purified PJL33 CytcO was crystallized with two-subunit crystallization conditions [12]: 6 μl of enzyme solution containing 10 mM Tris, pH 8.0, 50 mM NaCl, 0.20% decyl maltoside and 120 μM CytcO was mixed with 3 μL of reservoir solution containing 100 mM MES, pH 6.3, 27 % PEG-400 and 3 μl of crystallization additive solution containing 5 % heptanetriol, 32 mM MgCl2, 1.3 mM CdCl2 and 0.026 % Dodecyl maltoside. Two subunit crystals were observed after three days growing under 4 °C incubation. After two weeks, the crystals of PJL33 were collected and examined at Argonne National Lab. The best resolution of the crystals was 2.1 Å (Table S1). Two-subunit crystals of PJL49 CytcO were also obtained under the sitting drop condition with slightly different conditions: well solutions containing 100 mM MES, pH 6.3, 27-29% PEG-400. The drop also contained 1.3 % heptanetriol, 8 mM MgCl2, 0.4 mM CdCl2 and 0.06% Dodecyl maltoside. The best resolution of the crystal of PJL49 was 2.5 Å (Table S1).
Supplementary Material
Highlights.
Importance of the C-terminal of Cox1 (subunit I) for cytochrome c oxidase activity.
Changes in protonatable surface residues modulate the proton-pumping stoichiometry.
Possible implications for function of the M. smegmatis supercomplex.
Acknowledgements
We would like to thank Dr. Jacob Schäfer for help with preparation of the figures. This work was supported by grants from the Knut and Alice Wallenberg Foundation (to PB), the Swedish Research Council (to PB), NIH R01 GM26916 (to S.F.-M.), the MSU Foundation Strategic Partnership Grant “Mitochondrial Science & Medicine” (S.F.-M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hosler JP, Ferguson-Miller S, Mills DA, Energy transduction: Proton transfer through the respiratory complexes, Annual Review of Biochemistry, 75 (2006) 165–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Belevich I, Verkhovsky MI, Molecular mechanism of proton translocation by cytochrome c oxidase, Antioxid Redox Signal, 10 (2008) 1–29. [DOI] [PubMed] [Google Scholar]
- [3].Kaila VRI, Verkhovsky MI, Wikström M, Proton-coupled electron transfer in cytochrome oxidase, Chem. Rev, 110 (2010) 7062–7081. [DOI] [PubMed] [Google Scholar]
- [4].Pereira MM, Santana M, Teixeira M, A novel scenario for the evolution of haem-copper oxygen reductases, Biochim. Biophys. Acta-Bioenerg, 1505 (2001) 185–208. [DOI] [PubMed] [Google Scholar]
- [5].Hemp J, Gennis RB, Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling, Results Probl Cell Differ, 45 (2008) 1–31. [DOI] [PubMed] [Google Scholar]
- [6].Brzezinski P, Gennis RB, Cytochrome c oxidase: exciting progress and remaining mysteries, J. Bioenerg. Biomembr, 40 (2008) 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee HJ, Reimann J, Huang Y, Ädelroth P, Functional proton transfer pathways in the heme-copper oxidase superfamily, Biochimica et Biophysica Acta - Bioenergetics, 1817 (2012) 537–544. [DOI] [PubMed] [Google Scholar]
- [8].Smirnova IA, Ädelroth P, Gennis RB, Brzezinski P, Aspartate-132 in cytochrome c oxidase from Rhodobacter sphaeroides is involved in a two-step proton transfer during oxo-ferryl formation, Biochemistry, 38 (1999) 6826–6833. [DOI] [PubMed] [Google Scholar]
- [9].Fetter JR, Qian J, Shapleigh J, Thomas JW, García-Horsman A, Schmidt E, Hosler J, Babcock GT, Gennis RB, Ferguson-Miller S, Possible proton relay pathways in cytochrome c oxidase, Proc. Natl. Acad. Sci. USA, 92 (1995) 1604–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Namslauer A, Brzezinski P, Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase, FEBS Lett, 567 (2004) 103–110. [DOI] [PubMed] [Google Scholar]
- [11].Johansson AL, Högbom M, Carlsson J, Gennis RB, Brzezinski P, Role of aspartate 132 at the orifice of a proton pathway in cytochrome c oxidase, Proc. Natl. Acad. Sci. U. S. A, 110 (2013) 8912–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S, Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase, Proc Natl Acad Sci USA, 103 (2006) 16117–16122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S, The X-ray Crystal Structures of Wild-Type and EQ(I-286) Mutant Cytochrome c Oxidases from Rhodobacter sphaeroides, J. Mol. Biol, 321 (2002) 329–339. [DOI] [PubMed] [Google Scholar]
- [14].Wiseman B, Nitharwal RG, Fedotovskaya O, Schäfer J, Guo H, Kuang Q, Benlekbir S, Sjöstrand D, Ädelroth P, Rubinstein JL, Brzezinski P, Högbom M, Structure of a functional obligate complex III2IV2 respiratory supercomplex from Mycobacterium smegmatis, Nature Structural and Molecular Biology, 25 (2018) 1128–1136. [DOI] [PubMed] [Google Scholar]
- [15].Gong H, Li J, Xu A, Tang Y, Ji W, Gao R, Wang S, Yu L, Tian C, Li J, Yen HY, Lam SM, Shui G, Yang X, Sun Y, Li X, Jia M, Yang C, Jiang B, Lou Z, Robinson CV, Wong LL, Guddat LW, Sun F, Wang Q, Rao Z, An electron transfer path connects subunits of a mycobacterial respiratory supercomplex, Science, 362 (2018). [DOI] [PubMed] [Google Scholar]
- [16].Varanasi L, Hosler JP, Subunit III-depleted cytochrome c oxidase provides insight into the process of proton uptake by proteins, Biochimica et Biophysica Acta - Bioenergetics, 1817 (2012) 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mills DA, Tan Z, Ferguson-Miller S, Hosler J, A role for subunit III in proton uptake into the D pathway and a possible proton exit pathway in Rhodobacter sphaeroides cytochrome c oxidase, Biochemistry, 42 (2003) 7410–7417. [DOI] [PubMed] [Google Scholar]
- [18].Gilderson G, Salomonsson L, Aagaard A, Gray J, Brzezinski P, Hosler J, Subunit III of cytochrome c oxidase of Rhodobacter sphaeroides is required to maintain rapid proton uptake through the D pathway at physiologic pH, Biochemistry, 42 (2003) 7400–7409. [DOI] [PubMed] [Google Scholar]
- [19].Thompson DA, Gregory L, Ferguson-Miller S, Cytochrome c oxidase depleted of subunit III: proton-pumping, respiratory control, and pH dependence of the midpoint potential of cytochrome a, J Inorg Biochem, 23 (1985) 357–364. [DOI] [PubMed] [Google Scholar]
- [20].Wilson KS, Prochaska LJ, Phospholipid vesicles containing bovine heart mitochondrial cytochrome c oxidase and subunit III-deficient enzyme: Analysis of respiratory control and proton translocating activities, Arch. Biochem. Biophys, 282 (1990) 413–420. [DOI] [PubMed] [Google Scholar]
- [21].Ädelroth P, Ek M, Brzezinski P, Factors Determining Electron-Transfer Rates in Cytochrome c Oxidase: Investigation of the Oxygen Reaction in the R. sphaeroides and Bovine Enzymes, Biochim. Biophys. Acta, 1367 (1998) 107–117. [DOI] [PubMed] [Google Scholar]
- [22].Namslauer A, Pawate AS, Gennis RB, Brzezinski P, Redox-coupled proton translocation in biological systems: Proton shuttling in cytochrome c oxidase, Proc. Natl. Acad. Sci. U. S. A, 100 (2003) 15543–15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Namslauer A, Aagaard A, Katsonouri A, Brzezinski P, Intramolecular proton-transfer reactions in a membrane-bound proton pump: the effect of pH on the peroxy to ferryl transition in cytochrome c oxidase, Biochemsitry, 42 (2003) 1488–1498. [DOI] [PubMed] [Google Scholar]
- [24].Brändén G, Brändén M, Schmidt B, Mills DA, Ferguson-Miller S, Brzezinski P, The protonation state of a heme propionate controls electron transfer in cytochrome c oxidase, Biochemistry, 44 (2005) 10466–10474. [DOI] [PubMed] [Google Scholar]
- [25].Wikström M, Verkhovsky MI, The D-channel of cytochrome oxidase: An alternative view, Biochimica et Biophysica Acta - Bioenergetics, 1807 (2011) 1273–1278. [DOI] [PubMed] [Google Scholar]
- [26].Salomonsson L, Faxén K, Ädelroth P, Brzezinski P, The timing of proton migration in membrane-reconstituted cytochrome c oxidase, Proc. Natl. Acad. Sci. U S A, 102 (2005) 17624–17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Verkhovsky MI, Morgan JE, Verkhovskaya ML, Wikström M, Translocation of electrical charge during a single turnover of cytochrome-c oxidase, Biochim. Biophys. Acta, 1318 (1997) 6–10. [Google Scholar]
- [28].Faxén K, Gilderson G, Ädelroth P, Brzezinski P, A mechanistic principle for proton pumping by cytochrome c oxidase, Nature, 437 (2005) 286–289. [DOI] [PubMed] [Google Scholar]
- [29].Von Ballmoos C, Lachmann P, Gennis RB, Ädelroth P, Brzezinski P, Timing of electron and proton transfer in the ba 3 cytochrome c oxidase from Thermus thermophilus, Biochemistry, 51 (2012) 4507–4517. [DOI] [PubMed] [Google Scholar]
- [30].Mills DA, Florens L, Hiser C, Qian J, Ferguson-Miller S, Where is 'outside' in cytochrome c oxidase and how and when do protons get there?, Biochim. Biophys. Acta, 1458 (2000) 180–187. [DOI] [PubMed] [Google Scholar]
- [31].Popović DM, Stuchebrukhov AA, Proton pumping mechanism and catalytic cycle of cytochrome c oxidase: Coulomb pump model with kinetic gating, FEBS Lett., 566 (2004) 126–130. [DOI] [PubMed] [Google Scholar]
- [32].Brzezinski P, Johansson AL, Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase, Biochimica et Biophysica Acta - Bioenergetics, 1797 (2010) 710–723. [DOI] [PubMed] [Google Scholar]
- [33].Blomberg MRA, Siegbahn PEM, The mechanism for proton pumping in cytochrome c oxidase from an electrostatic and quantum chemical perspective, Biochimica et Biophysica Acta - Bioenergetics, 1817 (2012) 495–505. [DOI] [PubMed] [Google Scholar]
- [34].Siegbahn PEM, Blomberg MRA, Mutations in the D-channel of cytochrome c oxidase causes leakage of the proton pump, FEBS Lett., 588 (2014) 545–548. [DOI] [PubMed] [Google Scholar]
- [35].Kao W-C, Kleinschroth T, Nitschke W, Baymann F, Neehaul Y, Hellwig P, Richers S, Vonck J, Bott M, Hunte C, The obligate respiratory supercomplex from Actinobacteria, Biochimica et Biophysica Acta (BBA) - Bioenergetics. [DOI] [PubMed] [Google Scholar]
- [36].Zhen Y, Qian J, Follmann K, Hosler J, Hayward T, Nilsson T, Dahn M, Hilmi Y, Hamer A, Hosler J, Ferguson-Miller S, Overexpression and purification of cytochrome coxidase from Rhodobacter sphaeroides. Protein Expression and Purification, Protein Expr. Purif., 13 (1998) 326–336. [DOI] [PubMed] [Google Scholar]
- [37].Peiffer WE, Ingle RT, Ferguson-Miller S, Structurally unique plant cytochrome c oxidase isolated from wheat germ, a rich source of plant mitochondrial enzymes, Biochemistry, 29 (1990) 8696–8701. [DOI] [PubMed] [Google Scholar]
- [38].Hiser C, Mills DA, Schall M, Ferguson-Miller S, C-terminal truncation and histidinetagging of cytochrome c oxidase subunit II reveals the native processing site, shows involvement of the C-terminus in cytochrome c binding, and improves the assay for proton pumping, Biochemistry, 40 (2001) 1606–1615. [DOI] [PubMed] [Google Scholar]
- [39].Hosler JP, Fetter J, Tecklenburg MM, Espe M, Lerma C, Ferguson-Miller S, Cytochrome aa3 of Rhodobacter sphaeroides as a model for mitochondrial cytochrome c oxidase. Purification, kinetics, proton pumping, and spectral analysis, J Biol Chem, 267 (1992) 24264–24272. [PubMed] [Google Scholar]
- [40].Thompson DA, Ferguson-Miller S, Lipid and subunit III depleted cytochrome c oxidase purified by horse cytochrome c affinity chromatography in lauryl maltoside, Biochemistry, 22 (1983) 3178–3187. [DOI] [PubMed] [Google Scholar]
- [41].Brändén M, Sigurdson H, Namslauer A, Gennis RB, Ädelroth P, Brzezinski P, On the role of the K-proton transfer pathway in cytochrome c oxidase, Proc. Natl. Acad. Sci. U. S. A, 98 (2001) 5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rydström Lundin C, Von Ballmoos C, Ott M, Ädelroth P, Brzezinski P, Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A, 113 (2016) E4476–E4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Namslauer A, Lepp H, Brändén M, Jasaitis A, Verkhovsky MI, Brzezinski P, Plasticity of proton pathway structure and water coordination in cytochrome c oxidase, J Biol Chem, 282 (2007) 15148–15158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.